Abstract
The development of the vertebrate central nervous system (CNS) is tightly regulated by many highly conserved cell signalling pathways. These pathways ensure that differentiation and migration events occur in a specific and spatiotemporally restricted manner. Two of these pathways, Notch and Hedgehog (Hh) signalling, have been shown to form a complex web of interaction throughout different stages of CNS development. Strikingly, some processes employ Notch signalling to regulate Hh response, while others utilise Hh signalling to modulate Notch response. Notch signalling functions upstream of Hh response through controlling the trafficking of integral pathway components as well as through modulating protein levels and transcription of downstream transcriptional factors. In contrast, Hh signalling regulates Notch response by either indirectly controlling expression of key Notch ligands and regulatory proteins or directly through transcriptional control of canonical Notch target genes. Here, we review these interactions and demonstrate the level of interconnectivity between the pathways, highlighting context-dependent modes of crosstalk. Since many other developmental signalling pathways are active in these tissues, it is likely that the interplay between Notch and Hh signalling is not only an example of signalling crosstalk but also functions as a component of a wider, multi-pathway signalling network.
Keywords: Notch signalling, Hedgehog signalling, Spinal cord, Central nervous system, Crosstalk
Introduction
The mature vertebrate central nervous system (CNS) consists of many distinct classes of neurons throughout the brain, spinal cord and retina. Each individual structure of the CNS is composed of a plethora of unique cell types organised in a specific spatiotemporal pattern that arises from a sheet of equivalent neural progenitor cells (NPCs), known as the neural plate, during development [1]. This pattern formation occurs in three-dimensional space, as highlighted by the layered structure of the spinal cord and cortex, demonstrating a requirement for local communication. In order to create functional neural networks, specific cell types must form within this three-dimensional space in distinct time windows. This occurs through selective temporal interactions between cells, providing a critical fourth dimension of control in the complex patterning of the CNS [2]. Understanding how these interactions are mediated is crucial in understanding how one of the most complex and important systems in the body is formed. Due to the delicate balance of control, when problems arise during CNS development, the resulting phenotypes are highly variable. Minor inaccuracies can cause anything from mild learning difficulties, to major defects in cognitive function, disrupting motor control and behaviour [3, 4]. As many of the conditions caused by incorrect CNS development are non-embryonic lethal but life-altering from an early stage, there has been a vast amount of research conducted across all aspects of the field. Despite this, whether different signalling cues that function to control the pattern formation of the CNS act in parallel or integrate into a multi-faceted signalling network remains poorly understood.
Cell signalling during CNS development
During early neurulation the embryonic neural plate forms the neural tube which, through tightly controlled spatiotemporal proliferation, then gives rise to the anterior anatomical structures of the brain and the more posterior spinal cord [5]. Continuously throughout this delineation, the NPCs activate specific transcriptional and morphological activity dependent on a variety of extracellular signalling events [6, 7]. Multiple conserved developmental pathways have been implicated in this process, and some are reused in several processes. Bone Morphogenic Protein (BMP) signalling, via Noggin inhibition, controls early neural induction while also playing a role in dorsal spinal cord patterning [8, 9], whereas Hedgehog (Hh) secreted from the notochord induces the floor plate, which organises the specification of cell fates in the ventral spinal cord [10–13]. Alongside this, Fibroblast Growth Factor (FGF) and Retinoic Acid (RA) signalling are required for correct anterior–posterior neural development along the hindbrain and spinal cord [14]. In the developing cortex, neurons receive multiple migratory cues controlling their differentiation programme and localisation [3]. Wnt signalling cascades regulate the migration and differentiation of neural progenitors in multiple regions of the developing CNS [15, 16]. Throughout all early patterning events, Notch signalling is continuously required for neural progenitor maintenance across the entire CNS [17–20].
A considerable amount of research has been conducted in the individual activity of these pathways and there are many corresponding reviews [12, 21–31], so this review will not focus on each pathway in isolation. Rather, this review will focus on the interaction of signalling pathways in the developing CNS. This area of research is scattered and understudied; however, there has been a recent emergence of studies focusing on the interaction of two of these pathways, the Notch and Hh signalling pathways. While these interactions are primarily in the spinal cord, there is evidence for crosstalk throughout the CNS. When we connect this research together, we begin to see trends emerging that could elucidate the complex, integrated network these two major pathways employ during the development of the CNS.
Hedgehog signalling
Hedgehog signalling is a conserved cell signalling pathway that is essential for pattern formation and cell fate specification in both invertebrates and vertebrates [12, 32]. For Hh signalling to function correctly in vertebrates, it requires the primary cilium, a microtubule-based organelle present on the surface of most cells [33]. When the Hh ligand is absent, the transmembrane receptor Patched (Ptc) is localised at the primary cilia. The presence of Ptc prevents the ciliary localisation of the transmembrane protein Smoothened (Smo), thereby repressing signal transduction by allowing for the proteolytic processing of the Gli proteins into a repressor form (Fig. 1a). In order for Smo inhibition to be released, a Hh ligand must bind to Ptc, which allows Smo to translocate to the primary cilia [34, 35]. This leads to the activation of the Gli family of transcription factors by preventing the proteolytic processing. This results in full-length Gli activators that drive the transcription of downstream target genes, such as ptc (Fig. 1b) [36, 37]. In mouse, Gli2 is the main activator and its expression is independent of Hh pathway activity, while Gli1 acts primarily as a signalling amplifier and its expression requires active Hh signalling [38, 39]. Gli3 generally acts as the main repressor [40]. Interestingly, Gli1 is the main activator in zebrafish. Although gli1 is a direct target of Hh signalling, low-level gli1 expression is maintained in the absence of Hh signalling via an unknown mechanism [41]. It is thought that Hh-independent gli expression allows cells to respond to Hh signals. Similar to the mouse model, Gli3 functions as the main repressor, though Gli2a and Gli2b have demonstrated repressor capabilities [42, 43]. The Hh pathway is critical in many developmental processes during CNS development. It is most well studied in the ventral spinal cord where it has emerged as one of the classical examples of graded morphogen signalling [44, 45]. In this system, it has been shown that both the level and duration of Hh signalling are critical to the correct formation of the discrete neural progenitor domains along the dorsoventral axis [10, 46].
Fig. 1.
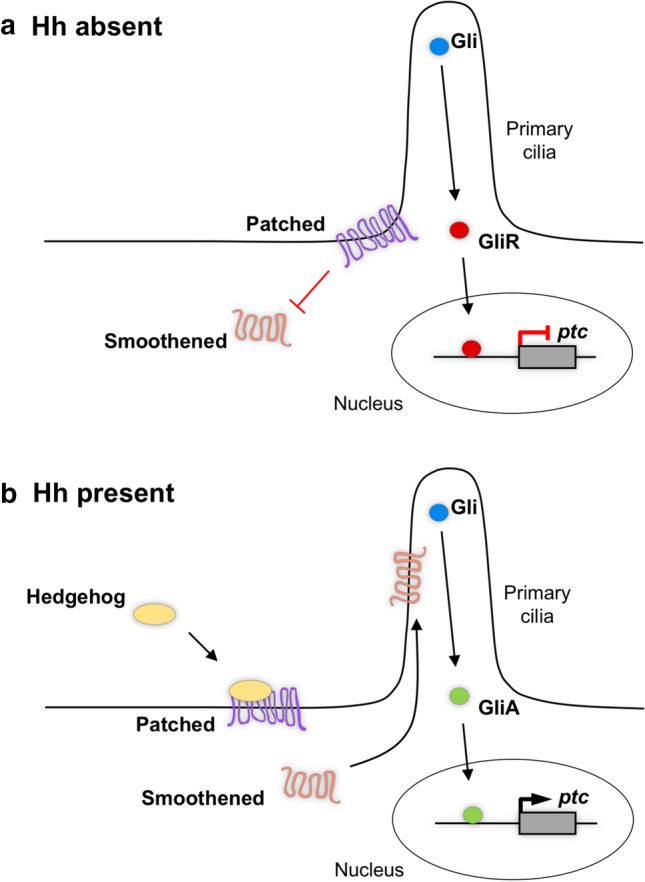
The Hedgehog signalling pathway. a In the absence of a Hh ligand, Patched (Ptc) localises to the primary cilium and excludes Smoothened (Smo). This leads to the processing of Gli transcription factors into a repressor form (GliR) which inhibits the transcription of downstream targets, such as ptc. b When a Hh ligand binds to Ptc, Smo can now localise to the primary cilium where it functions to prevent the processing of the Gli proteins. This results in full-length Gli proteins functioning as transcriptional activators (GliA), translocating to the nucleus to drive transcription of downstream targets
Notch signalling
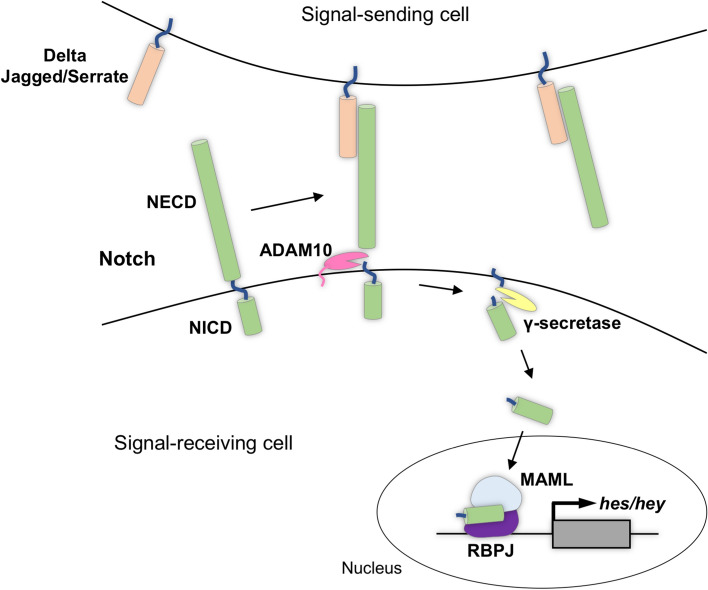
While the Hh pathway relies on secreted ligands to achieve long-range signalling, the Notch signalling pathway requires direct cell–cell interaction (Fig. 2). Unlike many other cell signalling pathways, both the receptor and ligand of the Notch pathway are membrane-bound proteins [47]. The Delta and Jagged/Serrate family of ligands are present at the membrane of the signal-sending cell, and their interaction with the extracellular domain of the Notch receptor (NECD), present at the membrane of the neighbouring signal-receiving cell, causes downstream pathway activation. Ligand binding results in two cleavage events of the Notch receptor. First, the NECD is cleaved by ADAM10, revealing a residual transmembrane fragment. This fragment is then targeted by a γ-secretase complex that releases the Notch intracellular domain (NICD). NICD then translocates to the nucleus where it forms a ternary transcription activation complex through interactions with the mastermind-like (MAML) coactivator and the DNA binding protein RBPJ. This activation complex drives the transcription of downstream targets, such as the Hes/Hey family of transcription factors (Fig. 2) [48, 49]. Two canonical roles of Notch signalling in neural development are to generate binary cell fate decisions through lateral inhibition and to maintain neural progenitor state [50, 51].
Fig. 2.
The Notch signalling pathway. When a ligand on the surface of the signal-sending cell interacts with the Notch receptor present at the membrane of the neighbouring signal-receiving cell, two cleavage events are activated: First, ADAM10 releases the NECD. Next, γ-secretase frees the NICD from the transmembrane domain. The NICD then translocates to the nucleus where it forms an activation complex and drives the transcription of downstream targets
Interactions of Hh and Notch signalling in the CNS
During the development of the CNS, many processes are restricted in their capabilities in a spatiotemporal manner. While at one time, a certain signal will cause cells to differentiate into a particular neuronal subtype, at a later time point that same signal drives a different fate. The Notch and Hh pathways behave in a similar mode in regard to their interactions, the level of interplay appears to be context dependent. In order to describe the complex, and often overlapping, crosstalk, we can create two categories of interaction: First, Hh signalling can control Notch response either directly or indirectly via downstream activity. Second, and more widely studied, Notch signalling can control Hh response. This broad categorisation allows us to visualise the complexity of the loops these two pathways create throughout neural development (Fig. 3).
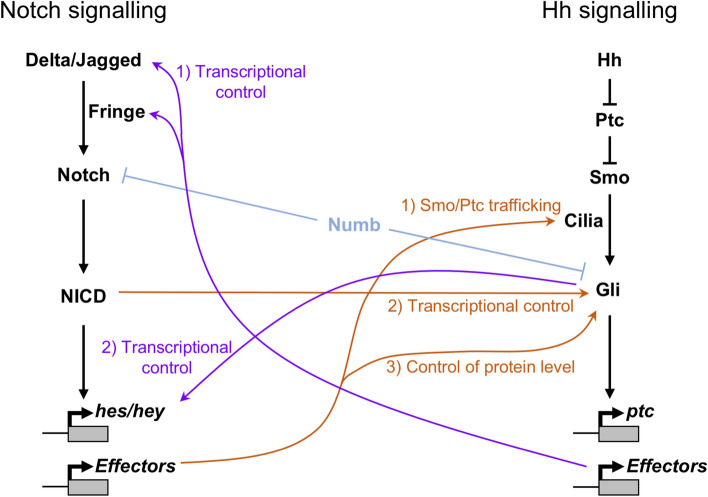
Fig. 3.
Complex crosstalk between Notch and Hh signalling pathways. The complex web of regulation between the Notch and Hh signalling pathways is shown. Hh control of Notch output is shown in purple: (1) Downstream effectors of the Hh pathway control transcription of Notch ligands and regulators, such as Fringe proteins. (2) The Gli proteins provide direct, Notch-independent transcriptional control of some Notch target genes. Notch signalling control of Hh output is shown in orange: (1) Downstream effectors of Notch signalling control the trafficking of Hh pathway components, such as Ptc and Smo, to the primary cilia. (2) NICD directly controls transcription of the Gli genes. (3) Downstream effectors of Notch signalling control Gli protein levels, independent of transcription. Both Notch and Hh signalling pathways are subject to Numb-regulated suppression (shown in light blue)
Hh signalling functions upstream of Notch response
The development of the spinal cord is highly conserved and tightly controlled. Multiple distinct classes of neurons are formed in a stereotypical dorsoventral pattern. This pattern is controlled through the highly specific expression of homeodomain (HD) transcription factors, alongside proneural basic helix-loop-helix proteins (bHLH), in discrete domains along the dorsoventral axis [13]. In order to achieve this precise pattern, the developing spinal cord employs anti-parallel signalling gradients of BMP from the dorsal roof plate and Hh from the ventral notochord and floor plate [14]. Cell fate is assigned through this dual signal interpretation, allowing for more refined positional information than sensing a single pathway in isolation [52]. Critically, the expression of HD and bHLH proteins in the ventral spinal cord requires a specific concentration and duration of Hh signalling [46]. In the ventral spinal cord, the combinatorial expression of seven homeodomain proteins defines the specific ventral progenitor domains. Once induced, these proteins display cross-repressive interactions that establish sharp boundaries, driving cell fate specification. For example, the transcription factor Nkx6.1 is expressed throughout the three ventral-most domains, the p2, pMN and p3 domains, while Dbx2 expression extends from the dorsal domains into the p0 and p1 domains of the intermediate spinal cord. The intersection of these expression domains forms the p1/p2 boundary, establishing spatial cell identity [53]. This transcription factor code exemplifies the high level of control Hh signalling exerts over spinal cord patterning.
First, Hh signalling can indirectly modulate Notch response by controlling the expression of Notch pathway components through downstream effectors (Fig. 3). The expression patterns of components of the Notch pathway are spatially restricted in the developing spinal cord [54, 55]. Alongside the previously mentioned Delta/Jagged ligands and Notch receptors, the Fringe family of glycosyltransferases also occupy specific dorsoventral (DV) domains [56, 57]. Fringe proteins modulate Notch activity through glycosylation of Notch receptors, mostly through regulating either Delta or Jagged specific interactions [58–60]. The expression of these Notch pathway components corresponds to specific DV progenitor domains, with Dll1 and Lfng both co-localising with the Nkx6.1 domain, while Jag1 is present in the Dbx2 domain. The localisation of Delta/Jagged or Fringe proteins in these domains is spatially controlled through the corresponding HD transcription factor: Nkx6.1 or Dbx2. When Nkx6.1 is knocked out, the Jag1 domain expands and the Dll1 domain is reduced. Conversely, forced expression of Nkx6.1 causes induced ectopic expression of Dll1 and Lfng at the expense of Jag1 [55]. This is likely because Dbx2 and Nkx6.1 have a cross-repressive interaction to generate a strict boundary [53], so the absence of Nkx6.1-mediated repression leads to an expansion of Dbx2 expression, while ectopic Nkx6.1 can overcome the repressive boundary, leading to a loss of Dbx2 expression. This shows that Nkx6.1 in the ventral spinal cord controls the expression profile of Notch ligands and Fringe proteins, which will have direct consequences on the levels of Notch signalling in this tissue. The spatial expression of Nkx6.1 requires Hh signalling [13, 53, 61], thus demonstrating an indirect mechanism by which Hh signalling regulates the expression of Notch pathway components through mediating the expression of downstream effectors. This interaction facilitates the ability for Notch signalling to control the neurogenesis of the specific neuronal populations in the developing mouse and chick spinal cord [55]. A similar mechanism has also been uncovered in zebrafish, where the Hh-dependent bHLH transcription factor olig2 is required for jagged2 expression [62]. When Olig2 activity is inhibited, via morpholino knockdown, there is a loss of jagged2 expression and an increase in differentiation in the neighbouring p3 domain [62]. This Jagged2-Notch signalling control over motor neuron and oligodendrocyte progenitor (OLP) differentiation is also present in the chick spinal cord, where absence of Hh-dependent Jagged2 results in the premature generation of OLPs and accelerated motor neuron differentiation [63]. Together, it highlights the indirect role Hh signalling plays in controlling Notch response, as the Hh-dependent transcription factor network has specific regulatory effects on the Notch signalling pathway.
Second, Hh signalling can also provide additional, direct control over the expression of canonical Notch target genes (Fig. 3). One of the most well-described roles of Notch signalling in the CNS is in binary cell fate decisions, repressing proneural gene expression in order to maintain progenitor populations. This is achieved through Notch-mediated transcription of certain members of the Hes/Hey family of transcription factors which then act as repressors of neural fate specifiers, such as Neurog2 and Ascl1 [19, 30, 31, 64–66]. These genes, such as Hes1, are classical Notch target genes and as such are often used as readouts for Notch signalling activity. Interestingly, there are examples of direct Hh-dependent control of Hes1 within the mouse retina and neocortex [67, 68]. Neuronal differentiation in the mammalian retina is a tightly controlled spatiotemporal process that generates all required cell types from a common pool of multipotent retinal progenitor cells (RPCs). This pool undergoes dramatic transcriptional remodelling during development in order to facilitate the differentiation of neuronal cell types in the right place and at the right time [69]. Notch signalling is integral in this process, with multiple tiers of control at different stages of the pathway; the deletion of Rbpj [70], Dll1 [71], Notch1 [72] or Hes1 [73, 74] results in subtly different phenotypes, suggesting a more complex regulation than purely canonical Notch signalling. Within this system, Hes1 has been shown to be activated by Hh signalling, independent from NICD activity. This activation occurs through direct Gli2 binding of the Hes1 promoter, exclusively in the presence of Hh signalling [67]. In the mouse neocortex Hes1 is similarly upregulated when Hh signalling is increased through genetic inactivation of Ptc1, although only mildly as the majority of Hes1 activity requires canonical Notch signalling [68]. A similar effect can be seen in the adult dentate gyrus, where increasing Hh signalling through genetic depletion of Ptc1 results in a mild upregulation of the Notch target Hey1 and, interestingly, Notch2 [75]. This Hh-dependent control of Hes1/Hey1 activity allows for Notch output to be spatial fine-tuned during specific time windows, adding another level of control over cell fate decisions.
Notch signalling functions upstream of Hh response
Many studies, primarily in the spinal cord, demonstrate a role for Notch signalling in the maintenance of Hh response. Intriguingly, this crosstalk is more complex than simply direct control and there are multiple mechanisms that form a diverse web of interplay to fine-tune Hh response during neural development (Fig. 3).
During spinal cord patterning, cells are maintained as progenitors which can respond to Hh signals in order to undergo cell fate determination. As these cells differentiate, they lose their competence to respond to incoming Hh signals [11, 76]. This loss of competence could either be an indirect consequence of differentiation or a precisely controlled process. Studies in the zebrafish lateral floor plate have shown that both Notch and Hh signalling pathways are required for cell fate induction. Temporal attenuation of both signalling pathways generates Kolmer-Agduhr” (KA”) interneurons, while continued signalling activity maintains the progenitor population. The progenitor cells with active Notch signalling remain Hh responsive, whereas the terminally differentiated KA” interneurons no longer have active Notch signalling and consequently lose their Hh response. This loss of Hh response occurs even when the Hh pathway is activated through overexpression a constitutively active form of Smo (rSmoM2) [77]. Sequential specification of oligodendrocyte lineages in the zebrafish spinal cord also utilises a similar mechanism. Oligodendrocyte precursor cells (OPCs) either differentiate into oligodendrocytes or are maintained as OPCs, dependent on the level of Hh signalling they receive. Inhibition of Notch signalling in this context results in an inability for OPCs to respond to high level Hh signalling, leading to a decrease in oligodendrocyte differentiation [78]. Taken together, this suggests a mechanism by which Notch signalling can maintain Hh response at the single cell level to permit cell fate specification events in multiple regions, and at different times, in the spinal cord.
One mode of action for this mechanism has been described in the developing mouse and chick spinal cord, where Notch signalling regulates the dynamic localisation of key pathway components at the primary cilia (Fig. 3) [79, 80]. Blockade of Notch signalling results in a dramatic decrease in the levels of the transcriptional Hh response reporter expressing luciferase under the control of Gli binding sites, highlighting a direct mechanism of interaction between Notch and Hh signalling [79]. For Hh signals to be interpreted and transduced, Ptc must be sequestered away from, while Smo must be trafficked to, the primary cilia [2, 33]. Manipulation of Notch signalling results in a profound effect on the levels of Ptc and Smo that are present at the primary cilia. The activation of Notch signalling leads to the accumulation of Smo at the primary cilia, resulting in elevated levels of Hh response. In contrast, Notch inhibition abrogates this accumulation and abolishes Hh response. Critically, this Notch-dependent trafficking of Smo to the primary cilia occurs without the presence of the Hh ligand, demonstrating a direct mechanism by which Notch signalling regulates the Hh pathway [80]. Interestingly, activated Notch signalling also leads to a significant increase in the length of primary cilia which provides a supplementary mechanism Notch signalling can implement to increase Hh responsiveness [79–82]. How does Notch signalling modulate Smo trafficking to the cilia? Inhibition of Notch signalling causes higher levels of Ptc to accumulate at the cilia, which would gate the ability of Smo to localise there. Remarkably, Notch inhibition does not reduce the levels of ciliary Smo in Ptc1-null cells, demonstrating that the key regulatory step centres on Ptc [79]. Overall, Notch signalling is required in order for Smo to localise to the primary cilia, demonstrating a necessary role for Notch signalling in activating Hh response.
Notch signalling also controls Hh signalling through maintaining transcription of the Gli genes (Fig. 3). In the zebrafish lateral floor plate, Hh signalling can still be terminated despite the loss of ptc1 and ptc2 [77], suggesting another level of interaction between Notch and Hh signalling. Indeed, further studies in the zebrafish spinal cord uncover a cilium-independent mechanism centring on the maintenance of the gli genes [83]. Inhibition of Notch signalling (Notchoff) results in a complete loss of Hh response throughout the developing spinal cord and abolishes the expression of the Hh-dependent transcription factor olig2. Stimulation of the Hh pathway in Notchoff spinal cords through ectopic expression of rSmoM2 is insufficient to restore Hh response; demonstrating regulation at the Ptc/Smo level is unlikely the only mechanism used to maintain Hh response in the zebrafish spinal cord [83]. In zebrafish, the absence of primary cilia through the mutant iguana leads to constitutive activation of endogenous Gli1 resulting in expanded, though lower level, Hh response [84–87]. Notchoff spinal cords in iguana mutants are unable to activate Hh response, indicating crosstalk further downstream of the primary cilia. This crosstalk indeed occurs at the Gli level, as ectopic expression of Gli1 is able to partially restore Hh response and the expression of all gli family genes requires Notch signalling [83]. This highlights a transcriptional mechanism of control, demonstrating that Notch signalling controls the transcription or mRNA stability of all members of the Gli family transcription factors. Consistent with this model, studies in mouse cortical neural stem cells have identified that Gli2 and Gli3 are direct targets of Notch signalling, as N1ICD/RBPJ binding regulates their expression [88].
Notch signalling also exerts further control of Hh response through maintaining Gli protein levels (Fig. 3). In the Müller glia of the mouse retina Notch signalling does not alter Gli2 transcript levels but rather controls Gli2 protein abundance. Notch activation increases levels of Gli2 protein, but is not capable of activating Hh-induced proliferation, suggesting the primary role of Notch signalling control of Gli2 is to prime retinal progenitors to respond to incoming Hh signals [89]. As ectopic Gli1 was only able to partially rescue Hh response during Notch inhibition in the zebrafish spinal cord [83], it is likely that this translation/protein stability mechanism functions alongside the Gli transcriptional control to facilitate full Hh responsiveness. Therefore, through ciliary trafficking of key components, direct transcriptional maintenance of Gli genes and translational/protein stability control of Gli transcription factors, Notch signalling has a complex, multi-faceted role upstream of Hh response (Fig. 3).
Conclusions
Overall, the level of crosstalk between Notch and Hh signalling is determined by various spatiotemporal cues and is not uniform throughout different developmental processes. Alongside the links discussed above, the two pathways also converge in methods of shared regulation. Asymmetric cell divisions during neural specification utilise a highly conserved protein segregation mechanism. In both Drosophila and mammalian nervous systems, the asymmetric segregation of Numb plays an integral role in binary fate decisions through antagonising Notch via E3-ligase-dependent ubiquitination [90–93]. Intriguingly, Numb also controls Hh output in a similar manner in the mouse cerebellum through targeting Gli1 for Itch-dependent ubiquitination [94]. This suggests a joint mechanism of control between Hh and Notch signalling, adding another, indirect level of interplay between these two pathways (Fig. 3). While this review has focused on interactions throughout development, we would be remiss not to recognise the likelihood of Notch and Hh signalling coordination in the adult. Both pathways are continually required in adult neurogenesis and appear to interact through the transcription factor Sox2. Expression of Sox2 in the postnatal cortex is Notch dependent, and it has been shown that Hh signalling is Sox2 dependent, suggesting an upstream regulatory role for Notch signalling in controlling Hh output [95–97]. Interestingly, the regulation of both Notch and Hh through Numb has been implicated in spinal plasticity in models of motoneuron disease, hinting at a further relationship in regeneration [98]. Another amyotrophic lateral sclerosis model demonstrates a temporal reduction of NICD in motor neurons that is accompanied by a decrease in Gli protein in spinal motor neurons [99], reminiscent of results seen in the developing spinal cord and retina. These conclusions rely on observational data as few mechanistic studies have been conducted in adult neurogenesis due to technical constraints. With the rapid advancements in technical capabilities, and the mechanistic conclusions from studies in embryos, this field will surely grow in the future.
Viewed together, we can begin to visualise the complex loops formed between these two signalling pathways (Fig. 3). Feedback loops can be found throughout development and it is plausible to suggest that Notch and Hh signalling control one another in this manner. For example, Hh signalling is required for early expression of Notch components in the oligodendrocyte precursor domain of the spinal cord, while Notch signalling is required for Hh signal sensing in later oligodendrocyte differentiation events [62, 63, 78]. Tightly controlled loops allow for fine-tuning of response and the maintenance of specific progenitor populations during waves of differentiation, preventing both runaway proliferation and incorrect patterning. In the case of Notch and Hh signalling, this is demonstrated during spinal cord, retinal and cortical development. The field of cell signalling crosstalk is rapidly expanding and it is likely that more of these multi-pathway signalling loops will be uncovered. All the systems described in this review require other signalling pathways, alongside Notch and Hh, to steer their development—one would expect they are all part of the same conversation.
Acknowledgements
The authors thank members of the Huang lab for discussions. This study was supported by grants to P.H. from the Natural Sciences and Engineering Research Council (NSERC) (RGPIN-2015-06343), Canada Foundation for Innovation John R. Evans Leaders Fund (Project 32920) and Startup Fund from the Alberta Children’s Hospital Research Institute (ACHRI). C.T.J. was supported by the ACHRI Graduate Scholarship and the University of Calgary Eyes High International Doctoral Scholarship.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/S1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briscoe J, Novitch BG. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philos Trans R Soc B Biol Sci. 2008;363:57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchsbaum IY, Cappello S. Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Development. 2019;146(1):dev163766. doi: 10.1242/dev.163766. [DOI] [PubMed] [Google Scholar]
- 4.Guerrini R, Dobyns WB. Malformations of cortical development: clinical features and genetic causes. Lancet Neurol. 2014;13:710–726. doi: 10.1016/S1474-4422(14)70040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern CD. Neural induction: old problem, new findings, yet more questions. Development. 2005;132:2007–2021. doi: 10.1242/dev.01794. [DOI] [PubMed] [Google Scholar]
- 6.Nikolopoulou E, Galea GL, Rolo A, et al. Neural tube closure: cellular, molecular and biomechanical mechanisms. Development. 2017;144:552–566. doi: 10.1242/dev.145904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilde JJ, Petersen JR, Niswander L. Genetic, epigenetic, and environmental contributions to neural tube closure. Annu Rev Genet. 2014;48:583–611. doi: 10.1146/annurev-genet-120213-092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Dréau G, Martí E. Dorsal-ventral patterning of the neural tube: a tale of three signals. Dev Neurobiol. 2012;72:1471–1481. doi: 10.1002/dneu.22015. [DOI] [PubMed] [Google Scholar]
- 9.Alaynick WA, Jessell TM, Pfaff SL. SnapShot: spinal cord development. Cell. 2011;146:178–178.e1. doi: 10.1016/j.cell.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dessaud E, Ribes V, Balaskas N, et al. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribes V, Balaskas N, Sasai N, et al. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010;24:1186–1200. doi: 10.1101/gad.559910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:418–431. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 13.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 14.Gouti M, Metzis V, Briscoe J. The route to spinal cord cell types: a tale of signals and switches. Trends Genet. 2015;31:282–289. doi: 10.1016/j.tig.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Inestrosa NC, Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015;359:215–223. doi: 10.1007/s00441-014-1996-4. [DOI] [PubMed] [Google Scholar]
- 16.Machon O, Backman M, Machonova O, et al. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev Biol. 2007;311:223–237. doi: 10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 17.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 18.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal transduction in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 20.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 21.Brazil DP, Church RH, Surae S, et al. BMP signalling: agony and antagony in the family. Trends Cell Biol. 2015;25:249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 23.Carter EP, Fearon AE, Grose RP. Careless talk costs lives: fibroblast growth factor receptor signalling and the consequences of pathway malfunction. Trends Cell Biol. 2015;25:221–233. doi: 10.1016/j.tcb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16:110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 26.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Gough NR. Focus issue: Wnt and β-catenin signaling in development and disease. Sci Signal. 2012;5:eg2–eg2. doi: 10.1126/scisignal.2002806. [DOI] [PubMed] [Google Scholar]
- 28.Ray LB. Dissecting wnt signaling. Sci Signal. 2012;5(254):ec319. doi: 10.1126/scisignal.2003850. [DOI] [PubMed] [Google Scholar]
- 29.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 31.Koch U, Lehal R, Radtke F. Stem cells living with a Notch. Development. 2013;140:689–704. doi: 10.1242/dev.080614. [DOI] [PubMed] [Google Scholar]
- 32.Lee RTH, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–372. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 33.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corbit KC, Aanstad P, Singla V, et al. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 35.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 36.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 37.Humke EW, Dorn KV, Milenkovic L, et al. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai CB, Auerbach W, Lee JS, et al. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- 39.Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- 40.Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO Rep. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlstrom RO, Tyurina OV, Kawakami A, et al. Genetic analysis of zebrafish Gli1 and Gli2 reveals divergent requirements for Gli genes in vertebrate development. Development. 2003;130:1549–1564. doi: 10.1242/dev.00364. [DOI] [PubMed] [Google Scholar]
- 42.Ke Z, Kondrichin I, Gong Z, Korzh V. Combined activity of the two Gli2 genes of zebrafish play a major role in Hedgehog signaling during zebrafish neurodevelopment. Mol Cell Neurosci. 2008;37:388–401. doi: 10.1016/j.mcn.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Tyurina OV, Guner B, Popova E, et al. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev Biol. 2005;277:537–556. doi: 10.1016/j.ydbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 45.Cohen M, Briscoe J, Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Curr Opin Genet Dev. 2013;23:423–428. doi: 10.1016/j.gde.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Dessaud E, Yang LL, Hill K, et al. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 47.Kopan R, Ilagan MXG. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90220-K. [DOI] [PubMed] [Google Scholar]
- 49.Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 50.Formosa-Jordan P, Ibañes M, Ares S, Frade J-M. Lateral inhibition and neurogenesis: novel aspects in motion. Int J Dev Biol. 2013;57:341–350. doi: 10.1387/ijdb.120259jf. [DOI] [PubMed] [Google Scholar]
- 51.Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat Neurosci. 2008;11:1247–1251. doi: 10.1038/nn.2208. [DOI] [PubMed] [Google Scholar]
- 52.Zagorski M, Tabata Y, Brandenberg N, et al. Decoding of position in the developing neural tube from antiparallel morphogen gradients. Science. 2017;356:1379–1383. doi: 10.1126/science.aam5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/S0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 54.Lindsell CE, Boulter J, DiSibio G, et al. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996 doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 55.Marklund U, Hansson EM, Sundstrom E, et al. Domain-specific control of neurogenesis achieved through patterned regulation of Notch ligand expression. Development. 2010;137:437–445. doi: 10.1242/dev.036806. [DOI] [PubMed] [Google Scholar]
- 56.Johnston SH, Rauskolb C, Wilson R, et al. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 57.Skaggs K, Martin DM, Novitch BG. Regulation of spinal interneuron development by the olig-related protein Bhlhb5 and notch signaling. Development. 2011;138:3199–3211. doi: 10.1242/dev.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hicks C, Johnston SH, DiSibio G, et al. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 59.Xu A, Haines N, Dlugosz M, et al. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by fringe. J Biol Chem. 2007;282:35153–35162. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 60.Kakuda S, Haltiwanger RS. Deciphering the fringe-mediated notch code: identification of activating and inhibiting sites allowing discrimination between ligands. Dev Cell. 2017;40:193–201. doi: 10.1016/j.devcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uygur A, Young J, Huycke TR, et al. Scaling pattern to variations in size during development of the vertebrate neural tube article scaling pattern to variations in size during development of the vertebrate neural tube. Dev Cell. 2016;37:127–135. doi: 10.1016/j.devcel.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeo S-Y, Chitnis AB. Jagged-mediated Notch signaling maintains proliferating neural progenitors and regulates cell diversity in the ventral spinal cord. Proc Natl Acad Sci U S A. 2007;104:5913–5918. doi: 10.1073/pnas.0607062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabadán MA, Cayuso J, Le Dréau G, et al. Jagged2 controls the generation of motor neuron and oligodendrocyte progenitors in the ventral spinal cord. Cell Death Differ. 2012;19:209–219. doi: 10.1038/cdd.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 65.Alcolea MP, Jones PH. Cell competition: winning out by losing notch. Cell Cycle. 2015;14:9–17. doi: 10.4161/15384101.2014.988027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boareto M, Iber D, Taylor V. Differential interactions between Notch and ID factors control neurogenesis by modulating Hes factor autoregulation. Development. 2017;144:3465–3474. doi: 10.1242/dev.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wall DS, Mears AJ, McNeill B, et al. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J Cell Biol. 2009;184:101–112. doi: 10.1083/jcb.200805155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dave RK, Ellis T, Toumpas MC, et al. Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PLoS ONE. 2011;6(2):e14680. doi: 10.1371/journal.pone.0014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clark BS, Stein-O’Brien GL, Shiau F, et al. Single-cell RNA-Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102:1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Riesenberg AN, Liu Z, Kopan R, Brown NL. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci. 2009;29:12865–12877. doi: 10.1523/JNEUROSCI.3382-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riesenberg AN, Brown NL. Cell autonomous and nonautonomous requirements for Delltalike1 during early mouse retinal neurogenesis. Dev Dyn. 2016;245:631–640. doi: 10.1002/dvdy.24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yaron O, Farhy C, Marquardt T, et al. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- 73.Imayoshi I, Isomura A, Harima Y, et al. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- 74.Bosze B, Moon MS, Kageyama R, Brown NL. Simultaneous requirements for Hes1 in retinal neurogenesis and optic cup-stalk boundary maintenance. J Neurosci. 2020;40:1501–1513. doi: 10.1523/JNEUROSCI.2327-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antonelli F, Casciati A, Tanori M, et al. Alterations in morphology and adult neurogenesis in the dentate Gyrus of Patched1 Heterozygous mice. Front Mol Neurosci. 2018;11:168. doi: 10.3389/fnmol.2018.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ericson J, Morton S, Kawakami A, et al. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996;87:661–673. doi: 10.1016/S0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 77.Huang P, Xiong F, Megason SG, Schier AF. Attenuation of Notch and Hedgehog signaling is required for fate specification in the spinal cord. PLoS Genet. 2012;8:e1002762. doi: 10.1371/journal.pgen.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ravanelli AM, Kearns CA, Powers RK, et al. Sequential specification of oligodendrocyte lineage cells by distinct levels of Hedgehog and Notch signaling. Dev Biol. 2018;444:93–106. doi: 10.1016/j.ydbio.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kong JH, Yang L, Dessaud E, et al. Notch activity modulates the responsiveness of neural progenitors to sonic Hedgehog signaling. Dev Cell. 2015;33:373–387. doi: 10.1016/j.devcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stasiulewicz M, Gray SD, Mastromina I, et al. A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development. 2015;142:2291–2303. doi: 10.1242/dev.125237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hudish LI, Galati DF, Ravanelli AM, et al. miR-219 regulates neural progenitors by dampening apical Par protein-dependent Hedgehog signaling. Development. 2016;143:2292–2304. doi: 10.1242/dev.137844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pusapati GV, Kong JH, Patel BB, et al. CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen Sonic Hedgehog in brief. Dev Cell. 2018;44:113–129.e8. doi: 10.1016/j.devcel.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jacobs CT, Huang P. Notch signalling maintains hedgehog responsiveness via a Gli-dependent mechanism during spinal cord patterning in zebrafish. Elife. 2019;8:1–24. doi: 10.7554/eLife.49252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glazer AM, Wilkinson AW, Backer CB, et al. The Zn Finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol. 2010;337:148–156. doi: 10.1016/j.ydbio.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang P, Schier AF. Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development. 2009;136:3089–3098. doi: 10.1242/dev.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolff C, Roy S, Lewis KE, et al. iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 2004;18:1565–1576. doi: 10.1101/gad.296004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sekimizu K, Nishioka N, Sasaki H, et al. The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development. 2004;131:2521–2532. doi: 10.1242/dev.01059dev.01059[pii]. [DOI] [PubMed] [Google Scholar]
- 88.Li Y, Hibbs MA, Gard AL, et al. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem Cells. 2012;30:741–752. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ringuette R, Atkins M, Lagali PS, et al. A Notch-Gli2 axis sustains Hedgehog responsiveness of neural progenitors and Müller glia. Dev Biol. 2016;411:85–100. doi: 10.1016/j.ydbio.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 90.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/S0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 91.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 92.Zhong W, Jiang MM, Weinmaster G, et al. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 93.Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat Neurosci. 2002;5:1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- 94.Di Marcotullio L, Ferretti E, Greco A, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 95.Choe Y, Pleasure SJ, Mira H. Control of adult neurogenesis by short-range morphogenic-signaling molecules. Cold Spring Harb Perspect Biol. 2016;8:a018887. doi: 10.1101/cshperspect.a018887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Favaro R, Valotta M, Ferri ALM, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 97.Ehm O, Göritz C, Covic M, et al. RBPJκ-dependent signaling is essential for long-term maintenance of neural stem cells in the adult hippocampus. J Neurosci. 2010;30:13794–13807. doi: 10.1523/JNEUROSCI.1567-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gulino R, Parenti R, Gulisano M. Novel mechanisms of spinal cord plasticity in a mouse model of motoneuron disease. Biomed Res Int. 2015;2015:654637. doi: 10.1155/2015/654637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma X, Drannik A, Jiang F, et al. Crosstalk between Notch and Sonic hedgehog signaling in a mouse model of amyotrophic lateral sclerosis. NeuroReport. 2017;28:141–148. doi: 10.1097/WNR.0000000000000725. [DOI] [PMC free article] [PubMed] [Google Scholar]