Abstract
Misaligned feeding may lead to pancreatic insufficiency, however, whether and how it affects circadian clock in the exocrine pancreas is not known. We exposed rats to a reversed restricted feeding regimen (rRF) for 10 or 20 days and analyzed locomotor activity, daily profiles of hormone levels (insulin, glucagon, and corticosterone) in plasma, and clock gene expression in the liver and endocrine and exocrine pancreas. In addition, we monitored responses of the exocrine pancreatic clock in organotypic explants of mPer2Luc mice in real time to acetylcholine, insulin, and glucocorticoids. rRF phase-reversed the clock in the endocrine pancreas, similar to the clock in the liver, but completely abolished clock gene rhythmicity and significantly downregulated the expression of Cpb1 and Cel in the exocrine pancreas. rRF desynchronized the rhythms of plasma insulin and corticosterone. Daily profiles of their receptor expression differed in the two parts of the pancreas and responded differently to rRF. Additionally, the pancreatic exocrine clock responded differently to treatments with insulin and the glucocorticoid analog dexamethasone in vitro. Mathematical simulation confirmed that the long-term misalignment between these two hormonal signals, as occurred under rRF, may lead to dampening of the exocrine pancreatic clock. In summary, our data suggest that misaligned meals impair the clock in the exocrine part of the pancreas by uncoupling insulin and corticosterone rhythms. These findings suggest a new mechanism by which adverse dietary habits, often associated with shift work in humans, may impair the clock in the exocrine pancreas and potentially contribute to exocrine pancreatic insufficiency.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04354-7.
Keywords: Circadian clock, Dexamethasone, Insulin, mPer2Luc mouse, Misaligned feeding, Pancreas
Introduction
Food intake is controlled by both homeostatic and temporal (circadian) mechanisms to ensure that sufficient nutrient acquisition is coordinated with active foraging and food availability. The circadian system orchestrates daily behavior and physiology in preparation for regular feeding times and nutrient uptake. Misalignment of food intake with the natural sleep/wake schedules (i.e. feeding during the sleep phase) is a critical factor contributing to reduced metabolic health, including the development of obesity [1, 2] and disturbed glucose homeostasis leading to type 2 diabetes mellitus [3].
The circadian mechanism is based on cellular clocks generating rhythmic signals via transcription-translation feedback loops which drive daily cycles in the expression of so-called clock genes, namely Clock/Npas2, Bmal1, Per1/Per2, Cry1/Cry2, and Nr1d1 (Rev-Erbα) [4]. The mammalian circadian system is hierarchical, with the master clock residing in the suprachiasmatic nuclei (SCN) in hypothalamus, which receive and internalize information about external light/dark conditions and synchronize so-called peripheral clocks located in cells of neuronal and nonneuronal origin elsewhere in the body via multiple signaling pathways (reviewed in [5, 6]).
The daily regime in food intake is a potent cue that rapidly adjusts the phases of clocks in various tissues involved in food acquisition and metabolisms (liver, kidney, colon, etc.). When entrained appropriately, these clocks align tissue function with patterns of food availability, thereby enabling food anticipation [7–9] irrespective of the signals from the light entrained SCN clock. As a result of misaligned food timing, the SCN and food-sensitive peripheral clocks become uncoupled [7, 8]. Notably, some peripheral oscillators do not synchronize with the feeding regimen [10], which leads to a further compromise of synchrony among the individual tissue clocks and metabolic disturbances [11]. Additionally, the dynamics of shifting peripheral clocks according to misaligned food timing are slowed down by glucocorticoids [12]
In the pancreas, secretion of hormones (endocrine part) and enzymes (exocrine part) is under dual control. Circadian control operates via clocks in pancreatic β-cells and acinar cells, which secrete insulin [13, 14] and digestive enzymes [15–17], respectively, in anticipation of food intake, and homeostatic control adjusts their production according to actual demand. To date, studies have mostly focused on a circadian clock in the endocrine pancreas (reviewed in [18]) which drives the circadian rhythm in insulin secretion in both fed and fasted animals [19, 20]. Disruption of the pancreatic clock impairs glucose homeostasis [14] and insulin secretion leading to hypo-insulinemia and a reduction in insulin sensitivity [21, 22]. Circadian disruption is sufficient to accelerate the development of diabetes in diabetes-prone rats [23] and has been implicated in the development of type 2 diabetes mellitus in night-shift workers [24]. Although the effect of a misaligned feeding regime on the pancreatic clock has been previously studied [22, 25], there is a significant gap in our knowledge of its impact on the clocks operating selectively in the exocrine part of the pancreas. In this study, we tested the hypothesis that misaligned feeding imposes different demands on the endo- and exocrine pancreas which will produce a difference in their circadian responses to the presentation of food. To test this hypothesis, we exposed animals to a protocol of reversed restricted feeding (rRF), in which food was presented only for a limited time interval during the light phase of the light/dark cycle, when they normally rest or sleep, and attempted to identify the mechanism underlying the resulting impact on the clock in the exocrine pancreas.
Materials and methods
Experimental animals and procedures
Adult male Wistar rats (Institute of Physiology, Czech Academy of Sciences) were housed individually in a temperature-controlled (21 ± 2 °C) animal facility under a light/dark cycle with 12 h of light and 12 h of darkness (LD12:12), with lights on between 06:00 and 18:00, assigned as Zeitgeber time (ZT) 0 and ZT12, respectively. The light intensity was 150–300 lx, depending on the cage position. Rats were randomly divided into 3 groups. The control group was fed ad libitum throughout the experiment (designated as “ad libitum” or “ad lib”), and two groups had restricted access to food; the group designated as “rRF10” had food available between ZT3 and ZT9 for 10 days, and the group designated as “rRF20” had food available between ZT3 and ZT10 for 20 days; food availability on rRF20 was thus longer by 1 h compared to rRF10 to prevent undernutrition of animals due to the prolonged restricted feeding schedule.
Male and female mPer2Luc mice (strain B6.129S6-Per2tm1Jt/J, JAX, USA) aged 3 to 6 months housed in a separate animal facility under LD12:12 and fed ad libitum were used to prepare organotypic explants of pancreatic tissue.
All experiments were approved by the Animal Care and Use Committee of the Institute of Physiology and were in agreement with the Animal Protection Law of the Czech Republic, as well as the European Community Council directives 86/609/EEC. All efforts were made to lessen the suffering of animals.
Behavioral activity monitoring
Locomotor activity of all rats was monitored throughout the entire experiment as previously described [26]. The resulting data were analyzed using the ClockLab toolbox (Actimetrics, Illinois, USA).
Collection of samples, RNA isolation and real-time RT-qPCR
Wistar rats were sacrificed under deep anesthesia with 150 mg/kg ketamine (Vétoquinol, s.r.o., Czech Republic) and 15 mg/kg xylazine (Bioveta a.s., Czech Republic) every 4 h during a 24-h cycle, starting at ZT0 (n = 5 per time point). Liver and pancreatic tissues were processed as described elsewhere [27]. Samples of exocrine (approx. 10 mm2) and endocrine (approx. 2 mm2) pancreas were dissected from 25 µm-thick frozen slices of pancreatic tissue using a laser microdissector (LMD6000, Leica), collected in a microfuge tube, and stored in RLT buffer from the RNeasy Micro kit (Qiagen, Valencia, CA, USA) until RNA isolation was performed according to the manufacturer’s instructions.
After the reverse transcription (High-Capacity cDNA Reverse Transcription Kit; Applied Biosystems, Foster City, CA), the cDNA samples were analyzed by real-time qPCR using PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA) on a ViiA7 Real-Time PCR System (Life Technologies, Carlsbad, USA). For specific primers, see Supplementary Table S1. The ΔΔCt method was used for the quantification of relative cDNA concentration. Relative expression was achieved by normalization to β2-microglobulin (B2m) for the liver and TATA-binding protein (Tbp) and ribosomal protein S18 (Rps18) for the pancreas and pancreatic dissections. For each tissue, samples from the ad libitum, RF10 and RF20 groups were assayed in the same real-time RT-qPCR run.
ELISA
Blood was collected by cardiac puncture under anesthesia before collection of tissue samples (n = 3–5 per time point). Plasma samples were assayed by ELISA to detect daily profiles of insulin (Mouse Insulin High Sensitivity ELISA kit; BioVendor, Brno, CZ), glucagon (Rat Glucagon ELISA Kit; Crystal Chem, Elk Grove Village, USA) and corticosterone (Corticosterone rat/mouse ELISA kit; LDN, Nordhorn, Germany) concentrations according to respective the manufacturer’s instructions.
Bioluminescence monitoring of organotypic explants and in vitro treatment
Explants of the exocrine pancreas were prepared from mPer2Luc mice as previously described [27]. To abolish β-cells, the explants were treated overnight with 1.5 mM streptozocin (STZ) in the recording medium. The efficiency of β-cell destruction in the explanted tissue was confirmed by detection of Pdx1, Ins1 and Ins2 mRNA by real-time RT-qPCR; expression was normalized to the housekeeping genes ribosomal protein S18 (Rps18) and hypoxanthine–guanine phosphoribosyltransferase (Hprt).
Bioluminescence was monitored in a LumiCycle apparatus (Actimetrics, Wilmette, IL, USA) and the raw bioluminescence traces were analyzed using LumiCycle Analysis software (Actimetrics). After 3 days of culturing, the explants were treated with acetylcholine (ACh; 10 µM, Sigma-Aldrich), bovine insulin (INS; 500 nM, Sigma-Aldrich) and/or dexamethasone (DEX; 100 nM, Sigma-Aldrich), and with the corresponding vehicles VEHACh—ddH2O, VEHINS—7 µM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; Sigma-Aldrich) in ddH2O, and VEHDEX—0.00001% ethanol in ddH2O. The phase shifts were quantified by fitting a sine curve to at least two full circadian cycles of a 24-h running average baseline-subtracted rhythm, extrapolating it beyond the time of treatment, and then measuring the difference between the time of the first peak after the treatment and the corresponding peak of the extrapolated rhythm. The calculated phase shift was designated as a phase advance ( +) or a phase delay (−). The phase response curves (PRCs) were constructed by plotting the calculated phase shift as a function of treatment time normalized to the endogenous period in vitro and expressed relative to the trough (time 0) or peak (time 12) of the rhythm.
Separate set of explants was monitored in a motorized Luminoview LV200 luminescence microscope (Olympus, Tokyo, Japan) with LUCPLFLN20X or UPLSAPO10X2 objectives (Olympus) and an ImageEM X2 EMCCD camera (Hamamatsu, Hamamatsu City, Japan) water cooled by a Minichiller 280 (Huber, Offenburg, Germany) with an exposure time of 10 min. Single cells were outlined and tracked using the Trackmate [28] plugin in ImageJ/Fiji.
Mathematical simulation
Data on phase shifts and relative amplitude changes acquired during in vitro treatments of INS and DEX were interpolated by linear regression with polynomial features using Scikit-learn [29]. The degree of polynomial function was determined by the evaluation of the mean squared error. For the circadian oscillation model, a cosine curve defined by the equation Y = mesor + (amplitude*cos(2*π*(X-acrophase)/wavelength)) was used as the default function. Wavelength (24 h) and mesor (0) were set constant and the rise of amplitude was restricted. Two model conditions were established: (1) simulated “control” ad libitum conditions, when induced changes in rhythm phase and amplitude (i.e., simulated “treatment”) by the combination of DEX and INS occurred every 24 h, and (2) simulated rRF conditions when “treatment” by DEX and INS alternately occurred every 12 h. At each “treatment” time, the cosine curve was replotted with shifted phase and modified amplitude according to parameters obtained by polynomial regression curves. The time of the initial treatment was established to be 2.3 h after the trough, as it was at this “treatment” time when the model curve became stabilized. All code was written in Python 3.8.3.
Statistical analysis
Parameters of locomotor activity were assessed by one-way ANOVA followed by the post hoc analysis with Sidak's multiple comparison method. The daily profiles of gene expression and insulin, glucagon and corticosterone blood plasma concentrations were assessed using two-way ANOVA (effect of the group) followed by the post hoc analyses with Sidak’s multiple comparison method. Changes in body weight were compared using paired t-tests, and differences in levels of hormones during light/dark period, functional and marker gene expression and periods of PER2-driven bioluminescence rhythms in pancreatic explants were compared using unpaired t-tests. The daily profiles of gene expression and hormonal levels were analyzed by cosinor analysis for detection of the presence/absence of a circadian rhythm as previously described [26]. Briefly, two alternative regression models to differentiate between rhythmic and non-rhythmic expression were used: either a horizontal straight line (null hypothesis) or a single cosine curve (alternative hypothesis), defined by the equation Y = mesor + (amplitude*cos(2*π* (X-acrophase)/wavelength)) with a constant wavelength of 24 h. The extra sum-of-squares F test was used for comparison, and the cosine curve parameters were calculated unless the P value exceeded 0.05.
All statistics were performed using Prism 7 software (GraphPad, CA, USA). P < 0.05 was required for significance; in case of multiple testing P was adjusted accordingly.
Results
rRF re-organizes daily locomotor activity patterns and has no major effect on body weight gain
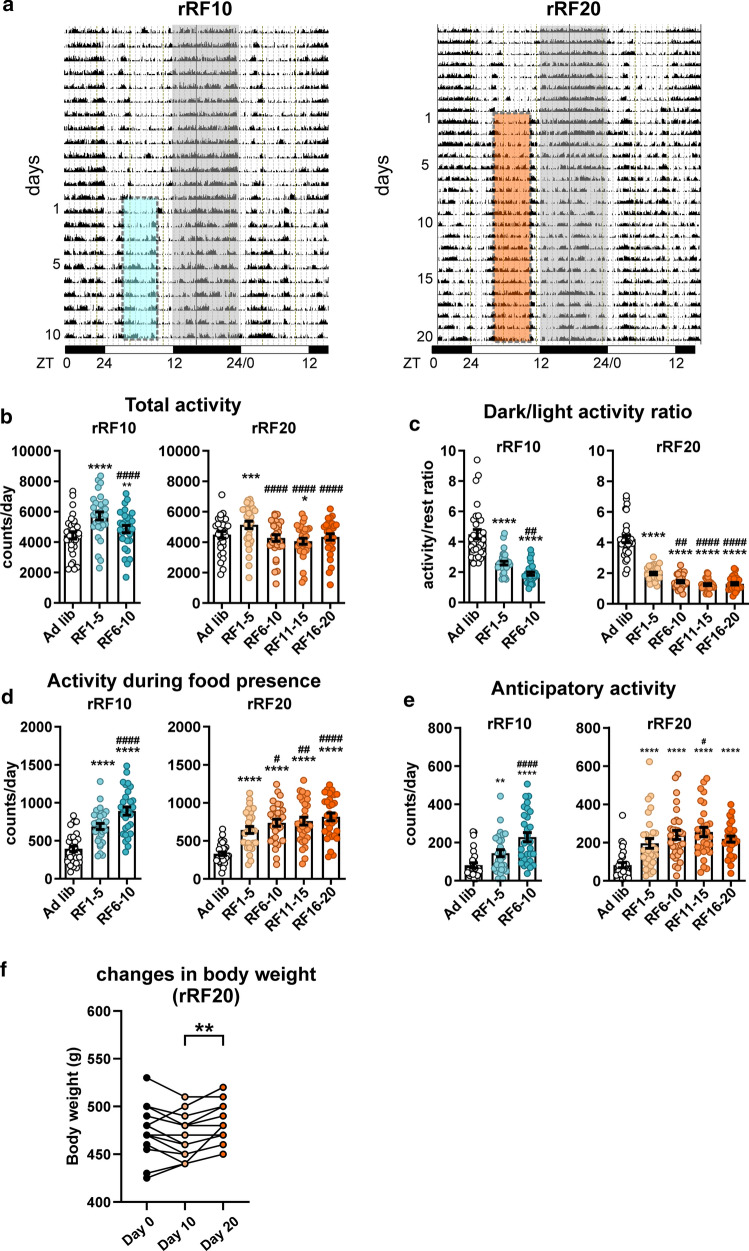
Locomotor activity of Wistar rats was monitored throughout the experiment (Fig. 1a, for statistical comparisons, see Supplementary Table S2). Total activity (Fig. 1b) initially increased during the first 5 days of rRF and then dropped back significantly to the ad libitum level. However, over time rats became more active during light phase, i.e., the ratios of dark:light activity decreased (Fig. 1c). It was mainly due to increased activity during food presence (Fig. 1d) as well as during its anticipation measured 2 h before food availability (Fig. 1e).
Fig. 1.
Analysis of locomotor activity of animals maintained under two experimental protocols of reversed restricted feeding (rRF). Rats maintained in 12 h light:12 h dark cycle were fed ad libitum and, thereafter, they were exposed to reversed restricted feeding protocol lasting for either 10 days (rRF10; n = 35) or 20 days (rRF20; n = 35). For more details, see “Materials and methods” section a Representative activity records (double-plotted actograms) of one rat from each experimental group are depicted. The dark phase of the LD cycle is shaded and the interval when food was provided under each of the rRF regimes is depicted by colored dashed rectangles. The activity records were analyzed under ad libitum conditions and in 5-day intervals throughout rRF protocols. b Total activity. c Dark/light activity ratio (ratio between activity during the dark and light phases of the LD cycle). d Activity during presence of food. e Anticipatory activity measured during 2 h before the food presence. Data are expressed as mean ± SEM. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001 vs. ad libitum; #P < 0.05; ##P < 0.01; ####P < 0.0001 vs. RF1-5. f Changes in body weight throughout rRF20 protocol. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001
The amount of food the rats consumed per day on RF20 (24.8 ± 0.5 g; n = 34; mean ± SEM) was only slightly lower than under ad libitum feeding conditions (27.2 ± 0.3 g; n = 29; mean ± SEM). Importantly, the rRF20 protocol had no effect on body weight (Fig. 1f); it caused only a mild loss in body weight during the first 10 days (day0 vs. day10: P = 0.0728), but then, most animals gained their weight back or weighed even more by the end of the rRF20 protocol (day 10 vs. day 20: P = 0.0011).
rRF produces tissue-specific adjustment of daily clock gene expression profiles
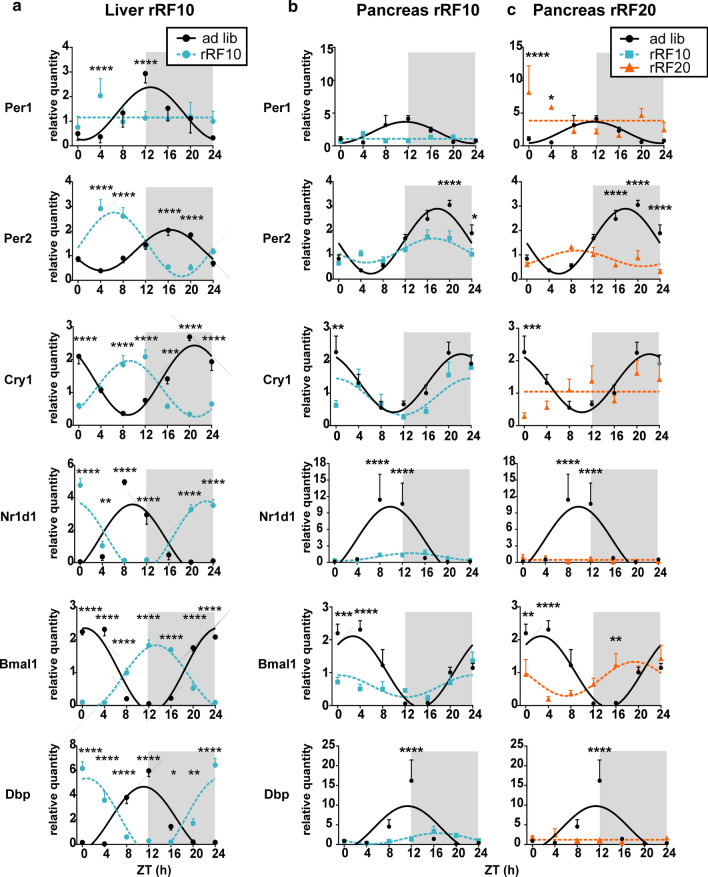
We assessed the effect of rRF on the clocks in tissue homogenates prepared from rats fed ad libitum and those exposed to one of each of the restricted feeding regimens. The mRNA levels and cosinor fits of the profiles are shown in Fig. 2; for better clarity, the rRF10 and rRF20 comparisons with ad libitum fed groups are presented separately. The acrophases and amplitudes of the rhythms are provided in Supplementary Table S3 and Supplementary Table S4 for the liver and pancreas, respectively.
Fig. 2.
Clock gene expression in the liver and pancreas homogenates. Comparison of daily profiles of relative expression of Per1, Per2, Cry1, Nr1d1, Bmal1 and Dbp detected by RT-qPCR in animals fed ad libitum (ad lib) and exposed to reversed restricted feeding protocols (rRF10 and rRF20). Data were fit with cosine curves (for details, see “Materials and methods” section). a Liver; ad libitum circles and full line, rRF10 circles and dashed line, significant differences between groups at individual time points are depicted as indicated by two-way ANOVA post hoc analysis *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001. b Pancreas; ad libitum circles and full line, rRF10 squares and dashed line. c Pancreas; ad libitum circles and full line, rRF20 triangles and dashed line. Data are expressed as mean ± SEM. Data were fitted with a horizontal straight line (null hypothesis) or a cosine curve (alternative hypothesis), compared by the extra sum-of-squares F test and plotted accordingly. Significant differences between groups at individual time points are depicted as indicated by two-way ANOVA post hoc analysis, due to multiple comparisons, alpha was adjusted *P < 0.025, **P < 0.005; ***P < 0.0005; ****P < 0.00005
In agreement with previous data [8, 30], exposure of animals to the rRF10 protocol used in this study completely phase reversed expression profiles in the liver for Per2, Cry1, Nr1d1, Bmal1, Dbp (Fig. 2a), with the exception of Per1, which lost rhythmicity. Surprisingly, the pancreatic tissue from the same animals exposed to rRF10 responded quite differently (Fig. 2b). After 10 days on rRF, the expression rhythm was either not shifted (Per2), or the shift was only very small (Cry1, Bmal1, Nr1d1, Dbp). In the group exposed to rRF for 20 days (Fig. 2c) pancreatic rhythms of Per2 and Bmal1 expression phase advanced (almost to antiphase) compared to the ad libitum. Surprisingly, all other gene expression profiles in this study became completely arrhythmic. In contrast to the liver (Fig. 2a), the amplitudes of the pancreatic profiles were significantly dampened on both rRF10 and RF20 protocols compared with ad libitum feeding (Fig. 2b, c).
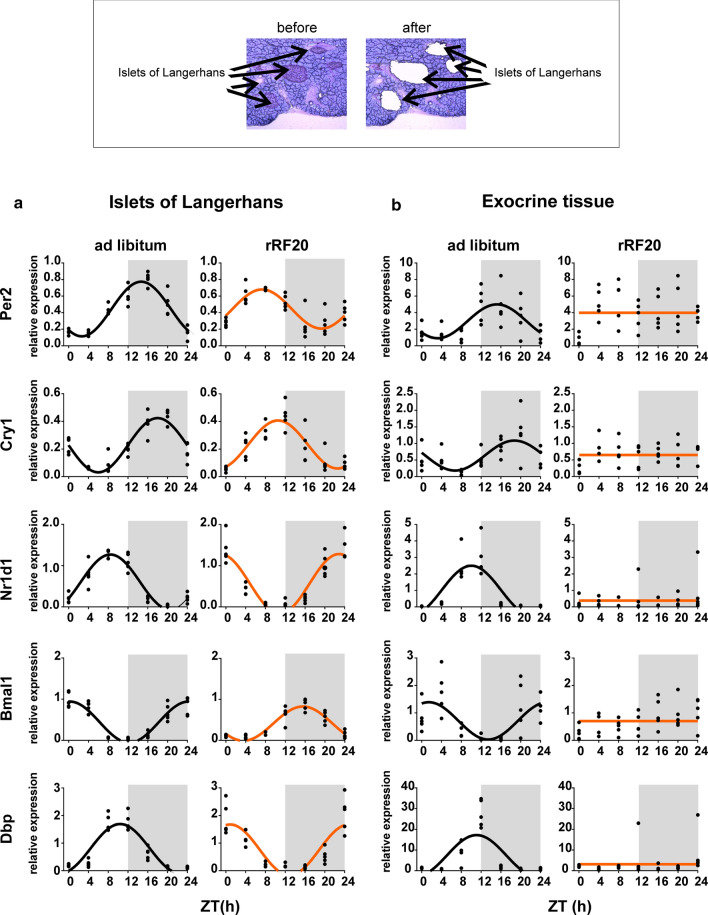
Clocks in the exocrine and endocrine pancreas respond differently to rRF
The findings described above prompted us to investigate the effect of rRF on clocks located separately in the exocrine and endocrine parts of the pancreas. Each part was precisely dissected from frozen pancreatic tissue sections using a laser-dissection technique (shown in Fig. 3; upper part). The samples from the ad libitum-fed group and the rRF20 group (which showed the most significant impact on the profiles detected at the whole-tissue level, as shown in Fig. 2c) were assessed within the same RT-qPCR assay for each of the two parts of the pancreas (Fig. 3; for the results of cosinor analysis, see Supplementary Table S5). Whereas in the ad libitum group, the clocks in both pancreatic parts exhibited robust and synchronized circadian rhythms, their responses to rRF20 differed. In the islets (Fig. 3a), the expression profiles of all studied genes remained rhythmic and shifted to antiphase compared to those in the ad libitum group similar to the clock in the liver. In contrast, in the exocrine part (Fig. 3b), the profiles of all studied genes lost rhythmicity, and, additionally, the expression of Nr1d1 and Dbp was significantly downregulated.
Fig. 3.
Gene expression in the separated endocrine and exocrine pancreatic tissue. Representative sections of the pancreas to demonstrate separation of the endocrine pancreas (islets of Langerhans) and the exocrine pancreas (acinar cells) using laser microdissection (upper part). Daily profiles of relative expression of Per2, Cry1, Nr1d1, Bmal1 and Dbp in animals fed ad libitum compared to animals on rRF20 protocol were detected using real-time RT-qPCR. Data were fitted with a horizontal straight line (null hypothesis) or a cosine curve (alternative hypothesis), compared by the extra sum-of-squares F test and plotted accordingly. a Endocrine pancreas (islets of Langerhans). b Exocrine (acinar cells) pancreatic tissue. Individual values at each time point are shown
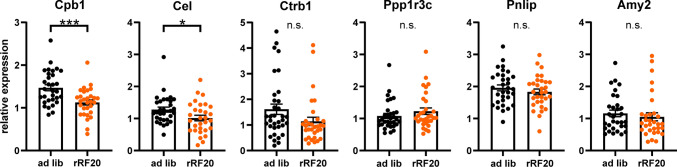
To assess the impact of rRF20 on the function of the exocrine pancreas, we compared the expression of several genes coding important digestive enzymes produced in the acinar cells, namely carboxyl ester lipase (Cel), carboxypeptidase (Cpb1), chymotrypsinogen B1 (Ctrb1), pancreatic triacylglycerol lipase (Pnlip), protein phosphatase 1 regulatory subunit 3C (Ppp1r3c), and pancreatic alpha-amylase (Amy2), in animals fed ad libitum (n = 35) and those exposed to rRF20 (n = 35) (Fig. 4). rRF20 significantly downregulated two of the genes (Cpb1: P = 0.0005 and Cel: P = 0.0305), there was only a non-significant trend for lower expression for Ctrb1 (P = 0.0757) and expressions of Ppp1r3c, Pnlip and Amy2 were not affected (P = 0.2473, 0.3377 and 0.5211, respectively). The data suggest that the production of some enzymes in the acinar pancreatic cells may be impaired due to exposure to the long-term rRF.
Fig. 4.
Gene expression of exocrine pancreas-specific genes. Expression of exocrine pancreas-specific genes (Cel, Cpb1, Ctrb1, Pnlip, Ppp1r3c and Amy2) in pancreatic homogenates from animals fed ad libitum (ad lib; n = 35) and exposed to rRF20 (n = 35). The data from samples collected around the clock (data shown in Fig. 3c) were pooled to compare the expression levels between both groups because none of the gene expression profile exhibited circadian variation (data not shown). Data are expressed as individual values and mean ± SEM, *P < 0.05; ***P < 0.001
Daily profiles of insulin and corticosterone levels in plasma are uncoupled due to the rRF schedules
To understand the implication of the rRF regimen for pancreatic function, we measured plasma levels of insulin (Fig. 5a) and corticosterone (Fig. 5b), which are reflecting feeding and activity patterns, respectively, in animals fed ad libitum and those exposed to rRF10 and rRF20. Plasma levels of glucagon are shown in Supplementary Fig. S1.
Fig. 5.
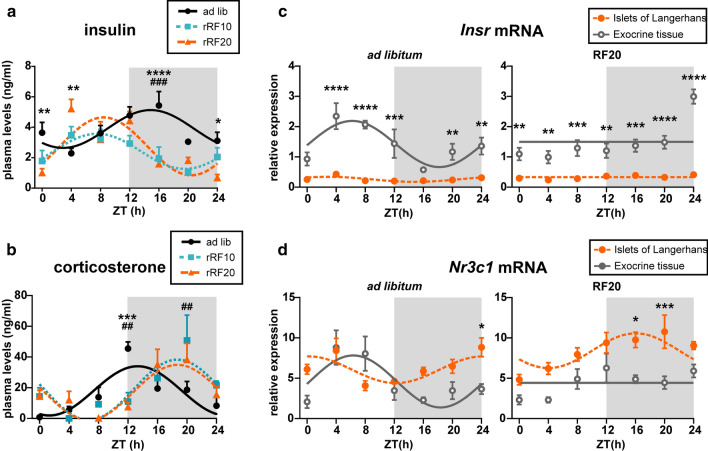
Levels of insulin and corticosterone in blood plasma and their receptors in the endocrine and exocrine pancreas. Daily profiles of a insulin and b corticosterone levels in the plasma of animals which were fed ad libitum (circles, full line), or exposed to 10 days (squares, dotted line) and 20 days (triangles, dashed line) of restricted feeding protocols (rRF10 and rRF20). Daily profiles of c Insr and d Nr3c1 mRNA in laser-dissected endocrine (islets of Langerhans; full circles) and exocrine pancreas (open circles) of animals fed ad libitum and exposed to rRF20. Data are depicted as mean ± SEM and fitted cosine curves. Significant differences between groups at individual time points identified by two-way ANOVA post hoc analysis are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 ad libitum vs. RF20; ##P < 0.01, ###P < 0.001 ad libitum vs. RF10
Insulin levels (Fig. 5a) in the ad libitum fed group exhibited circadian variation (cosinor: P = 0.0033) with elevated levels during the activity (dark) phase (acrophase mean ± SEM: ZT14.9 ± 1.1), when rats consume most of the food. After 10 days of exposure to rRF, the plasma insulin levels remained rhythmic (cosinor: P = 0.0066) but the peak of the rhythm shifted according to mealtime into the inactive part of the light/dark cycle (acrophase mean ± SEM: ZT7.6 ± 1.0). After 20 days, the phase-reversed insulin profile was still rhythmic (cosinor: P = 0.0002) with approximately the same phase (acrophase mean ± SEM: ZT8.4 ± 0.7) as on rRF10. The significant effect of rRF on the insulin profiles was also confirmed by 2-way ANOVA (group: P = 0.0001; time: P < 0.0001; interaction: P = 0.0001).
For corticosterone levels (Fig. 5b), cosinor analysis detected significant circadian rhythmicity in all three groups (ad libitum: P = 0.0006, rRF10: P = 0.0019, rRF20: P = 0.0009). In animals fed ad libitum, corticosterone levels peaked around the beginning of the nocturnal activity (acrophase mean ± SEM: ZT13.3 ± 0.9), however, exposure to rRF shifted the peak to later nighttime hours (acrophases mean ± SEM: rRF: ZT18.7 ± 0.8 and rRF20: ZT18.7 ± 0.7). Therefore, misaligned feeding affected the daily profile of corticosterone plasma levels in an opposite way compared to insulin. As a result, synchronously elevated insulin and corticosterone levels in ad libitum fed animals attained opposite phase-relationship after exposure to rRF.
Expression levels of insulin and corticosterone receptors differ in endocrine and exocrine pancreas and in their responses to the rRF20
We detected daily expression profiles of genes for the insulin receptor Insr (Fig. 5c) and the corticosterone receptor Nr3c1 (Fig. 5d) in the endocrine (islets of Langerhans) and exocrine pancreas under ad libitum feeding and rRF20 conditions. We compared the profiles between both parts of the pancreas for each feeding condition by 2-way ANOVA (significant differences at individual time points are shown in the graphs). Results of cosinor analysis are shown in Supplementary Table S6.
Expression of Insr in the endocrine pancreas was very low throughout the 24 h and significantly downregulated compared with the exocrine part under both ad libitum and rRF20 conditions (Fig. 5c). In the exocrine pancreas, Insr expression was higher than in the endocrine part and exhibited a circadian rhythm under ad libitum feeding that was completely abolished under rRF20. The rhythm of Insr expression in the exocrine pancreas detected under ad libitum feeding was roughly in the opposite phase to the rhythm of plasma insulin levels (Fig. 5a), probably to compensate for the hormone daily variations. The results suggest that cells in the exocrine pancreas are more sensitive to insulin signaling than cells in the endocrine part.
Expression of Nr3c1 in the endocrine pancreas exhibited rhythms under both ad libitum and rRF20 (Fig. 5d). Under ad libitum feeding, Nr3c1 expression in the endocrine part peaked at the end of the subjective night, which was approximately the opposite phase to the plasma corticosterone rhythm (Fig. 5b), probably to compensate for the hormone daily variations. However, under rRF20, Nr3c1 rhythm in the endocrine part shifted to earlier nighttime hours. In the exocrine pancreas, similar to Insr expression (Fig. 5c), Nr3c1 expression was also rhythmic under ad libitum feeding, but rhythmicity was lost under rRF20 (Fig. 5d). Intriguingly, in control conditions Nr3c1 expression rhythms were not synchronous in the exocrine and endocrine parts, suggesting a different sensitivity of these pancreatic tissues to glucocorticoids. Most importantly, Nr3c1 expression under rRF20 was significantly lower in the exocrine part than in the endocrine part (Fig. 5d), which was quite opposite to Insr expression (Fig. 5c). Differences in the receptor mRNA expression levels and patterns between the two tissues suggest that the responses of the endocrine and exocrine pancreas to each of the hormones under ad libitum feeding and rRF20 conditions may differ. The abolition of the Insr and Nr3c1 expression rhythms in the exocrine pancreas under rRF20 correlated with the impact of rRF20 on the circadian clock in the exocrine pancreas (Fig. 3b). The data suggest that the clock in the exocrine pancreas is dependent on the synchrony of insulin and corticosterone rhythms.
Clock in the exocrine pancreas dampens under rRF due to conflicting signaling from insulin and glucocorticoids
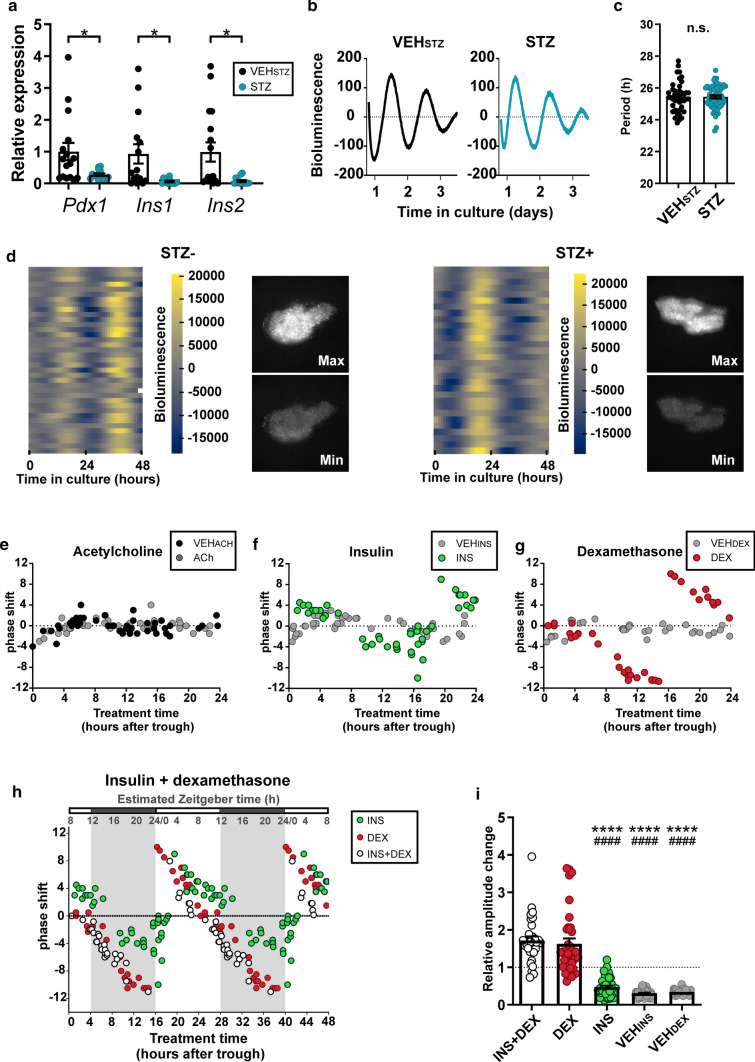
To study mechanisms potentially involved in entrainment of the exocrine pancreatic clock, we prepared organotypic explants from mPer2Luc mice which were devoid of viable β-cells as confirmed by a significant reduction in Pdx1 (P = 0.0194), Ins1 (P = 0.0175) and Ins2 (P = 0.0136) mRNA levels (Fig. 6a). The explanted tissue maintained viability, period of the circadian bioluminescence rhythms at the tissue level (Fig. 6b, c) and the synchrony among cellular oscillators (Fig. 6d). The explants were treated with either acetylcholine (ACh), dexamethasone (DEX) or insulin (INS) and corresponding vehicles (VEHACh/DEX/INS) at various times relative to the peak (time 12) in PER2-driven bioluminescence rhythms and the resulting phase shifts of the rhythms were plotted as PRCs (for more details, see “Materials and methods” section). Treatments with ACh (Fig. 6e) did not induce phase shifts that were significantly different from those induced by VEHACh treatments at any time of the cycle. In contrast, treatments with INS (Fig. 6f) and DEX (Fig. 6g) (but not with VEHINS and VEHDEX) produced phase advances or phase delays, the magnitudes of which were highly dependent on the timing of their application. The DEX-induced shifts were larger than the ones caused by INS. The same pattern of phase shift magnitudes and directions was obtained when DEX was cotreated with INS, as clearly visible from double-plotted PRCs for the effects of INS, DEX and INS + DEX (Fig. 6h). Accordingly, whereas INS alone had no effect on the amplitude of the bioluminescence rhythms, both DEX and INS + DEX increased the ratio of amplitudes before and after treatment significantly above the ratios for corresponding vehicles (for statistical significance, see Fig. 6i). To determine sensitivity windows for DEX, INS and their combination in external (Zeitgeber) time, we transposed the in vitro time into the in vivo time based on the daily Per2 expression profile in the exocrine pancreas we had detected in ad libitum fed animals (Fig. 3b). The peak in Per2 mRNA levels was identified at ZT15.22 ± 0.76 (see Supplementary Table S5), and assuming the PER2 protein levels likely lag the transcription by approximately 4 h [31], we can expect that PER2 protein levels peak in vivo at approximately ZT20. Therefore, we assigned the in vitro time 12, which determines the PER2::LUC peak, as ZT20 (see estimated Zeitgeber time in Fig. 6h). This extrapolation revealed that the DEX-induced shifts were the smallest during the transition from the day to the night and the INS-induced shifts were the smallest during the first part of the night (Fig. 6h).
Fig. 6.
Bioluminescence recording in exocrine pancreatic explants from mPer2Luc mice. a Relative expression of Pdx1, Ins1 and Ins2 (β-cell markers) in STZ treated organotypic explants. b Representative examples of bioluminescence recording of the pancreatic organotypic explants. c STZ treatment did not affect the period of the clock in the pancreatic explant. d Bioluminescence recording of cellular oscillators in the pancreatic explants without (left side) and with STZ (right side) treatments. Normalized heatmaps, in which each line represents the bioluminescence of a tracked single cell. Snapshots of the same exocrine pancreatic explant depict the maximum and minimum of PER2-driven bioluminescence in the whole sample. e–g Phase response curves (PRC) constructed for the treatments of the exocrine pancreatic explants with e acetylcholine (ACh), f insulin (INS) and g dexamethasone (DEX) and corresponding vehicles (VEHACh/INS/DEX). h PRCs for treatment with DEX and INS were double plotted for better comparison with PRC for treatment with a combination of insulin and dexamethasone (INS + DEX). The external time (Zeitgeber time; ZT) is depicted in the upper scale (shaded area denotes dark period of the light/dark cycle). ZT was extrapolated by correlating the peaks of PER2-driven bioluminescence in vitro and Per2 mRNA levels detected in the exocrine pancreatic tissue in vivo (for more details, see “Materials and methods” section). i Relative amplitude changes of bioluminescence rhythms after the treatments of exocrine pancreatic explants with DEX, INS, DEX + INS and corresponding vehicles (VEHDEX and VEHINS). Data are expressed as mean ± SEM. *P < 0.05; ****P < 0.0001 vs. INS + DEX; ####P < 0.0001 vs. DEX
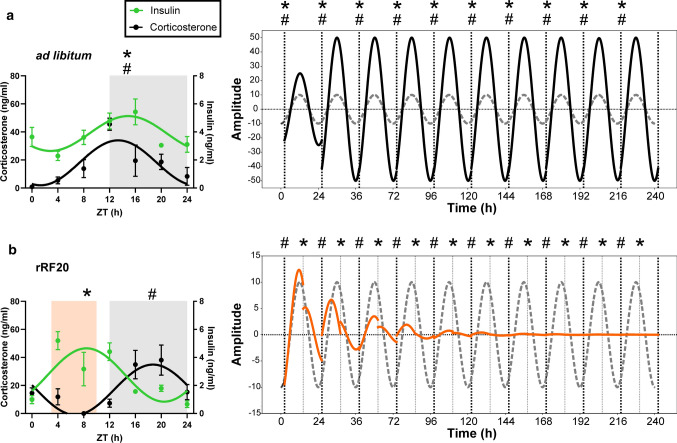
Finally, we used our biological data for an in silico approach to model how misaligned insulin and glucocorticoid signals during the long-term rRF impact the clock in the exocrine pancreas (Fig. 7). We obtained phase and amplitude responses to INS and DEX across the whole PER2-bioluminescence cycle in vitro (Fig. 6h, see “Materials and methods” section) and used them to infer the long-term response of an undamped sine curve representing the rhythmic PER2 expression in vivo. We simulated either ad libitum feeding conditions as concurrent daily stimulation with INS + DEX or rRF conditions as misaligned stimulation with INS and DEX separately. While concurrent stimulation produced a stable high amplitude rhythm (Fig. 7a), misaligned stimulation resulted in a rapid decrease in amplitude leading to a loss of oscillation within several circadian cycles (Fig. 7b), accurately reflecting the observed in vivo data (Fig. 3).
Fig. 7.
Mathematical simulation of the effect of misaligned insulin and glucocorticoid signaling on the clock in the exocrine pancreas. The effect of a concurrent (# *; ad libitum conditions) or b alternating (rRF conditions) timing of insulin (*) and corticosterone (#) signals (as observed in daily profiles of hormone levels) on phase and amplitude of sine curve representing responses of PER2-driven bioluminescence rhythm in the exocrine pancreas. Resulting curves are accompanied by the baseline curve for reference (dashed line). For more details, see “Materials and methods” section
Discussion
Both rRF schedules used in this study represent a significant metabolic challenge forcing the animals to reorganize their activity, in spite of their SCN clock still remaining entrained with the light/dark cycle (as evidenced by previously published daily clock gene expression profiles in the SCN of the same rat strain exposed to an identical rRF procedure [8]). Notably, the clock gene expression profiles in homogenates of the hepatic and pancreatic tissues of the same animals responded differently to exposure of rRF10. Whereas in the liver, the clock gene Per2, Cry1, Nr1d1, and Bmal1 (but not Per1), and a clock-controlled gene, Dbp, rhythms persisted and were completely reversed compared to controls, in the pancreas, they became either severely dampened and/or only negligibly shifted. The response in the liver was in accordance with our previous data [8, 30] as well as data from others [7, 32, 33] and served us here as proof of concept confirming that 10 days of the misaligned feeding schedule was sufficient to affect metabolic functions and peripheral clocks. After 20 days of rRF, the expression of only two of the studied clock genes (Per2 and Bmal1) remained rhythmic in the pancreas. The downregulation of Dbp due to rRF is in discordance with a previous study [7], in which rhythmic Dbp expression profile shifted after 12-h reversal of the feeding schedule for 6 days. The discrepancy is likely due to the different experimental protocols of these two studies, which differed in animal strain (rats versus mice), interval of food availability during the day, as well as in the duration of the rRF exposure. The slower adjustment of the clock in pancreatic homogenates compared to liver was previously noticed [7, 25]; however, its mechanism had not been elucidated [22]. Analyzes of separate clock gene expression profiles in the islets and in the exocrine pancreatic tissue revealed that exposure to rRF induced dyssynchrony among clocks in these functionally distinct cellular subpopulations; it reversed rhythmic clock gene expression profiles in the islets (in accordance with insulin levels), but completely abolished the rhythmicity in the exocrine part. Importantly, the expression of Nr1d1 and Dbp, which are genes involved in the metabolic input to and output from the clock, was not only arrhythmic but also severely downregulated. Therefore, the clock in the islets was able to adjust to changes in the feeding regime similarly to the clock in the liver, but the most abundant exocrine pancreatic tissue lost rhythmicity, which may explain the dampened expression profiles in the whole pancreas homogenates.
We hypothesized that the loss of rhythmicity in the exocrine pancreas was caused by rRF-induced impairment of clock-entraining signals. We tested entraining capability of two hormones, insulin as a hormone tracking the feeding regime [34] and corticosterone as a hormone tracking the activity state [35]. We demonstrate that under ad libitum feeding, daily rhythms of insulin and corticosterone are synchronous and their levels peak approximately at the beginning of the nocturnal activity. However, under rRF conditions these two rhythms rapidly uncouple as the insulin peak levels shift to the daytime and the corticosterone peak levels shift later into the nighttime. The shift in insulin peak tracked the meal timing and corresponded with the shift in the circadian clock in the islets. The shift in corticosterone peak tracked the delay in nocturnal activity onset into later hours (Supplementary Figure S2). It has been shown previously that the corticosterone peak responds to rRF with different patterns that depend strongly on species, biological sex, and timing and duration of food availability [10, 36–40]. In our experimental design, the corticosterone peak shifted to late subjective night. This was likely due to the fact that the relatively long food availability (8 h) in the rRF resulted in a bimodal activity state, i.e., relatively long food-driven daytime activity bout and delayed nighttime activity-related bout. The resulting corticosterone peak could represent a consolidated state between these two conflicting activity states. The clock in beta cells reportedly responds to both DEX [41] and insulin [42], although PRCs for their effects have not been shown. Interestingly, we found that pancreatic islets exhibit very low Insr mRNA levels (significantly downregulated compared to the exocrine pancreas). In the exocrine pancreas, we found that both Insr and Nr3c1 were highly expressed and their expression profiles were synchronously rhythmic under ad libitum feeding. Intriguingly, their rhythmicity was completely abolished under rRF20, similar to the rhythmicity of clock gene expression. Therefore, we speculate that, compared to the clock in the endocrine part, the clock in the exocrine pancreas is more sensitive to both hormones and is, therefore, more dependent on their synchrony.
To test the entrainment of the exocrine clock formally, we compared the potency of three plausible signals, namely, (1) acetylcholine, representing the tonus of the parasympathetic nervous system connecting the pancreas with the SCN [43], (2) DEX, representing the SCN-driven elevation of glucocorticoid levels during the activity state and (3) INS, representing a food-responsive signal, which was administered either without or with DEX. For these tests, we used organotypic explants of mPer2Luc mice treated with β-cell-specific toxin STZ in which persistence of cellular oscillators in the exocrine part was confirmed. The explants still contained α-cells, which, however, represent only a mere minor cell type (approximately 20%) of the sparsely distributed islets in the tissue, and thus they could not interfere with the rhythmicity of most abundant acinar cells in the explanted tissue. Although both β-cells [44] and acinar cells [45] respond to cholinergic signaling, our data excluded this pathway from participation in the entrainment of the clock in the exocrine pancreas. In contrast, INS and DEX both shifted the clock with magnitudes and directions which were distinct and highly dependent on the time of their application. The transition of the INS and DEX treatment time into the external time for the clock in the exocrine pancreas revealed that the clock does not respond to the treatments at the times that closely correspond to each of the hormone’s respective peak under ad libitum feeding conditions. However, responses to INS and DEX are maximal and opposite after treatments are performed at the times corresponding to the rRF-shifted peaks for each of the hormone. This time-dependent sensitivity of the clock to these two hormones reflects the fact that the clock does not change its phase much when their levels peak approximately at expected time during the day, as occurs under ad libitum conditions in the laboratory. However, the clock does respond with the largest shifts when the hormonal levels are unexpectedly elevated at an incorrect time of day in order to achieve the fastest synchronization with new conditions. Based on the data, we suggest that the rRF-induced discordance between the insulin and glucocorticoid peak levels provides the clock in the exocrine pancreas with contradictory signaling forcing the clock to opposite phase shifts, which consequently may lead to impairment of rhythmicity. Mathematical modeling used to simulate such a situation that occurs under the long-term exposure to rRF regime fully confirmed the hypothesis.
The importance of our study is strengthened by the fact that impairment of the exocrine pancreatic clock induced by misaligned mealtime correlated with significant downregulation of two enzyme-coding genes, Cel and Cpb1, involved in the acinar cells’ function. Importantly, the implication of Cel in various pancreatic diseases has been well recognized (summarized in [46]) and Cpb1 has recently been associated with pancreatic cancer susceptibility [47]. Therefore, aberrant regulation of those genes due to a misaligned feeding schedule may implicate serious health consequences.
Conclusions
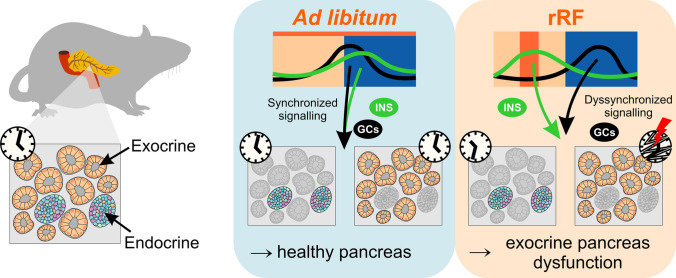
Misaligned meals become a frequent lifestyle transgression related to current societal pressure imposed on human behavior. Here we provide the first evidence that whereas the clock in the endocrine pancreas shifts according to mealtime, the clock in the exocrine pancreas is severely impaired, causing discordance among these normally synchronous clocks of the two functionally distinct parts of the pancreas. Our data suggest that this effect might be mediated via food presence-induced divergent timing in hormonal signaling (Fig. 8). From a general perspective, these results represent the discovery of an additional level of internal dyssynchrony in the body caused by a misaligned feeding schedule; it uncouples not only (1) central and peripheral clocks [7] and (2) clocks of individual peripheral organs [7, 10] but also (3) clocks in functionally distinct cellular subpopulations within the same organ (this study). We speculate that the distinct responses of the endocrine and exocrine pancreas have a physiologically relevant basis. When food resources are available at an inconvenient (sleep) time, the clock in the endocrine pancreas responds with a phase shift to match the anticipated time of food availability in synchrony with the clock in the liver. In contrast, the exocrine part of the pancreas responds by deactivating the clock regulation, allowing it to respond on demand basis so that the digestive system can continue to function at any time when food is available. Such an effect may help to survive a short interval before food resources and activity/rest cycles resynchronize, but their long-term misalignment can have negative effects. In our study, the long-term misaligned feeding-induced clock impairment was accompanied by a decrease in the capability of acinar cells to express selective functional genes previously associated with human disorders. In humans, misaligned meal schedules resembling long-term rRF (which disrupted the exocrine pancreatic clock) occur in shift workers who frequently develop type 2 diabetes mellitus [48]. In diabetic patients, exocrine pancreas insufficiency has been reported [49]. Therefore, the mechanism underlying the clinical symptoms associated with the exocrine pancreatic insufficiency may relate to the disruption of the clock in the exocrine part. Our findings pave a new avenue for future investigation of the relationship between long-term circadian misalignment and a higher risk of development of metabolic disorders.
Fig. 8.
Schematic summary of the results. Feeding aligned with activity status (as occurs in rats under ad libitum conditions) is associated with a coupling between the peaks in insulin and corticosterone levels and the phases of the clocks in the endocrine and exocrine parts of the pancreas. Misaligned feeding uncouples the hormone peaks, causing a discordance between these normally synchronous clocks of these two functionally distinct parts of the pancreas and severely compromising the clock and function of the exocrine pancreas
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Mgr. Karolina Liska for help with IPGTT and Eva Tlusta for technical assistance.
Author contributions
PeH data acquisition, investigation, methodology, writing a draft, manuscript reviewing, statistical analysis; ZN data acquisition, methodology; PaH data acquisition, investigation, methodology; MS methodology, manuscript reviewing; AS conceptualization, study supervision, formal analysis, methodology, writing draft, manuscript reviewing.
Funding
The study was supported by the OPPK BrainView CZ.2.16/3.1.00/21544, MEYS (LM2015062 Czech-BioImaging), and the Research Project RV0: 67985823.
Data availability
All data are presented.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
All experiments were approved by the Animal Care and Use Committee of the Institute of Physiology and were in agreement with the Animal Protection Law of the Czech Republic, as well as the European Community Council directives 86/609/EEC.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Salgado-Delgado R, Angeles-Castellanos M, Saderi N, et al. Food intake during the normal activity phase prevents obesity and circadian desynchrony in a rat model of night work. Endocrinology. 2010;151:1019–1029. doi: 10.1210/en.2009-0864. [DOI] [PubMed] [Google Scholar]
- 2.Arble DM, Bass J, Laposky AD, et al. Circadian timing of food intake contributes to weight gain. Obesity. 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenvers DJ, Scheer FAJL, Schrauwen P, et al. Circadian clocks and insulin resistance. Nat Rev Endocrinol. 2019;15:75–89. doi: 10.1038/s41574-018-0122-1. [DOI] [PubMed] [Google Scholar]
- 4.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102. doi: 10.1152/physrev.00009.2009. [DOI] [PubMed] [Google Scholar]
- 6.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 7.Damiola F, Le Minli N, Preitner N, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polidarová L, Sládek M, Nováková M, et al. Increased sensitivity of the circadian system to temporal changes in the feeding regime of spontaneously hypertensive rats—a potential role for Bmal2 in the liver. PLoS ONE. 2013;8:e75690. doi: 10.1371/journal.pone.0075690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sladek M, Rybova M, Jindrakova Z, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133:1240–1249. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 10.Oishi K, Yasumoto Y, Higo-Yamamoto S, et al. Feeding cycle-dependent circulating insulin fluctuation is not a dominant Zeitgeber for mouse peripheral clocks except in the liver: differences between endogenous and exogenous insulin effects. Biochem Biophys Res Commun. 2017;483:165–170. doi: 10.1016/j.bbrc.2016.12.173. [DOI] [PubMed] [Google Scholar]
- 11.Bray MS, Ratcliffe WF, Grenett MH, et al. Quantitative analysis of light-phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes. 2013;37:843–852. doi: 10.1038/ijo.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Minh N, Damiola F, Tronche F, et al. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perelis M, Marcheva B, Moynihan Ramsey K, et al. Pancreatic cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science (80-) 2015;350:aac4250–aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadacca LA, Lamia KA, DeLemos AS, et al. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maouyo D, Sarfati P, Guan D, et al. Circadian rhythm of exocrine pancreatic secretion in rats: Major and minor cycles. Am J Physiol - Gastrointest Liver Physiol. 1993;264:G792–G800. doi: 10.1152/ajpgi.1993.264.4.g792. [DOI] [PubMed] [Google Scholar]
- 16.Thaela M-J, Pierzynowski SG, Jensen MS, et al. The pattern of the circadian rhythm of pancreatic secretion in fed pigs. J Anim Sci. 1995;73:3402–3408. doi: 10.2527/1995.73113402x. [DOI] [PubMed] [Google Scholar]
- 17.Keller J, Gröger G, Cherian L, et al. Circadian coupling between pancreatic secretion and intestinal motility in humans. Am J Physiol Liver Physiol. 2001;280:G273–G278. doi: 10.1152/ajpgi.2001.280.2.G273. [DOI] [PubMed] [Google Scholar]
- 18.Seshadri N, Doucette CA. Circadian regulation of the pancreatic beta cell. Endocrinology. 2021 doi: 10.1210/endocr/bqab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellinger LL, Mendel VE, Moberg GP. Circadian insulin, GH, prolactin, corticosterone and glucose rhythms in fed and fasted rats. Horm Metab Res. 1975;7:132–135. doi: 10.1055/s-0028-1093763. [DOI] [Google Scholar]
- 20.Kalsbeek A, Strubbe JH. Circadian control of insulin secretion is independent of the temporal distribution of feeding. Physiol Behav. 1998;63:553–560. doi: 10.1016/S0031-9384(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 21.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woller A, Gonze D. Modeling clock-related metabolic syndrome due to conflicting light and food cues. Sci Rep. 2018;8:13641. doi: 10.1038/s41598-018-31804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gale JE, Cox HI, Qian J, et al. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms. 2011;26:423–433. doi: 10.1177/0748730411416341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reutrakul S, Knutson KL. Consequences of circadian disruption on cardiometabolic health. Sleep Med Clin. 2015;10:455–468. doi: 10.1016/j.jsmc.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherji A, Kobiita A, Damara M, et al. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci. 2015;112:E6691–E6698. doi: 10.1073/pnas.1519807112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sládek M, Polidarová L, Nováková M, et al. Early chronotype and tissue-specific alterations of circadian clock function in spontaneously hypertensive rats. PLoS ONE. 2012;7:e46951. doi: 10.1371/journal.pone.0046951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novosadová Z, Polidarová L, Sládek M, Sumová A. Alteration in glucose homeostasis and persistence of the pancreatic clock in aged mPer2Luc mice. Sci Rep. 2018;8:11668. doi: 10.1038/s41598-018-30225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tinevez J-Y, Perry N, Schindelin J, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 29.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 30.Sládek M, Houdek P, Sumová A. Circadian profiling reveals distinct regulation of endocannabinoid system in the rat plasma, liver and adrenal glands by light-dark and feeding cycles. Biochim Biophys Acta - Mol Cell Biol Lipids. 2019;1864:158533. doi: 10.1016/j.bbalip.2019.158533. [DOI] [PubMed] [Google Scholar]
- 31.Harbour VL, Weigl Y, Robinson B, Amir S. Phase differences in expression of circadian clock genes in the central nucleus of the amygdala, dentate gyrus, and suprachiasmatic nucleus in the rat. PLoS ONE. 2014;9:e103309. doi: 10.1371/journal.pone.0103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara R, Wan K, Wakamatsu H, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 33.García-Gaytán AC, Miranda-Anaya M, Turrubiate I, et al. Synchronization of the circadian clock by time-restricted feeding with progressive increasing calorie intake. Resemblances and differences regarding a sustained hypocaloric restriction. Sci Rep. 2020;10:10036. doi: 10.1038/s41598-020-66538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crosby P, Hamnett R, Putker M, et al. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell. 2019;177:896–909.e20. doi: 10.1016/j.cell.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oster H, Damerow S, Kiessling S, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Mendoza M, Rivera-Zavala JB, Díaz-Muñoz M. Daytime restricted feeding modifies the daily variations of liver gluconeogenesis: adaptations in biochemical and endocrine regulators. Chronobiol Int. 2014;31:815–828. doi: 10.3109/07420528.2014.908898. [DOI] [PubMed] [Google Scholar]
- 37.Luna-Moreno D, Aguilar-Roblero R, Díaz-Muñoz M. Restricted feeding entrains rhythms of inflammation-related factors without promoting an acute-phase response. Chronobiol Int. 2009;26:1409–1429. doi: 10.3109/07420520903417003. [DOI] [PubMed] [Google Scholar]
- 38.Kalsbeek A, van Heerikhuize JJ, Wortel J, Buijs RM. Restricted daytime feeding modifies suprachiasmatic nucleus vasopressin release in rats. J Biol Rhythms. 1998;13:18–29. doi: 10.1177/074873098128999880. [DOI] [PubMed] [Google Scholar]
- 39.Selmaoui B, Bah TM, Brazzini-Poisson V, Godbout R. Daily changes of plasma corticosterone by an 8-h daytime feeding occur without body weight loss or severe food restriction in the rat. Biol Rhythm Res. 2003;34:423–434. doi: 10.1076/brhm.34.5.423.27856. [DOI] [Google Scholar]
- 40.Honma KI, Honma S, Hiroshige T. Critical role of food amount for prefeeding corticosterone peak in rats. Am J Physiol Integr Comp Physiol. 1983;245:R339–R344. doi: 10.1152/ajpregu.1983.245.3.R339. [DOI] [PubMed] [Google Scholar]
- 41.Pulimeno P, Mannic T, Sage D, et al. Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia. 2013;56:497–507. doi: 10.1007/s00125-012-2779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrenko V, Saini C, Giovannoni L, et al. Pancreatic α- and β-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev. 2017;31:383–398. doi: 10.1101/gad.290379.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buijs RM, Chun SJ, Niijima A, et al. Parasympathetic and sympathetic control of the pancreas: a role for the suprachiasmatic nucleus and other hypothalamic centers that are involved in the regulation of food intake. J Comp Neurol. 2001;431:405–423. doi: 10.1002/1096-9861(20010319)431:4<405::AID-CNE1079>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 44.Adablah JE, Vinson R, Roper MG, Bertram R. Synchronization of pancreatic islets by periodic or non-periodic muscarinic agonist pulse trains. PLoS ONE. 2019;14:e0211832. doi: 10.1371/journal.pone.0211832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura K, Hamada K, Terauchi A, et al. Distinct roles of M1 and M3 muscarinic acetylcholine receptors controlling oscillatory and non-oscillatory [Ca2+]i increase. Cell Calcium. 2013;54:111–119. doi: 10.1016/j.ceca.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Johansson BB, Fjeld K, El Jellas K, et al. The role of the carboxyl ester lipase (CEL) gene in pancreatic disease. Pancreatology. 2018;18:12–19. doi: 10.1016/j.pan.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Yu J, Hata T, et al. Mutations in the pancreatic secretory enzymes CPA1 and CPB1 are associated with pancreatic cancer. Proc Natl Acad Sci. 2018;115:4767–4772. doi: 10.1073/pnas.1720588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antunes LC, Levandovski R, Dantas G, et al. Obesity and shift work: chronobiological aspects. Nutr Res Rev. 2010;23:155–168. doi: 10.1017/S0954422410000016. [DOI] [PubMed] [Google Scholar]
- 49.Nunes ACR, Pontes JM, Rosa A, et al. Screening for pancreatic exocrine insufficiency in patients with diabetes mellitus. Am J Gastroenterol. 2003;98:2672–2675. doi: 10.1111/j.1572-0241.2003.08730.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented.