Abstract
Maintenance of oxygen homeostasis is an indispensable criterion for the existence of multicellular life-forms. Disruption of this homeostasis due to inadequate oxygenation of the respiring tissues leads to pathological hypoxia, which acts as a significant stressor in several pathophysiological conditions including cancer, cardiovascular defects, bacterial infections, and neurological disorders. Consequently, the hypoxic tissues develop necessary adaptations both at the tissue and cellular level. The cellular adaptations involve a dramatic alteration in gene expression, post-transcriptional and post-translational modification of gene products, bioenergetics, and metabolism. Among the key responses to oxygen-deprivation is the skewing of cellular alternative splicing program. Herein, we discuss the current concepts of oxygen tension-dependent alternative splicing relevant to various pathophysiological conditions. Following a brief description of cellular response to hypoxia and the pre-mRNA splicing mechanism, we outline the impressive number of hypoxia-elicited alternative splicing events associated with maladies like cancer, cardiovascular diseases, and neurological disorders. Furthermore, we discuss how manipulation of hypoxia-induced alternative splicing may pose promising strategies for novel translational diagnosis and therapeutic interventions.
Keywords: Hypoxia, Alternative splicing, Hallmarks of cancer, Cardiovascular disease, Neurodegenerative diseases, Splicing therapeutics
Introduction
Oxygen serves as an electron acceptor in a broad array of biochemical reactions and plays a critical role in the pursuance of legitimate cellular energetics and tissue homeostasis. A decline in the tissue oxygen concentration owing to respiratory failure, inadequate blood circulation to an organ, anemia, or specific chemical insults might give rise to hypoxic stress. The emergence of hypoxic niches in respiring tissues is manifested in many pathological conditions, including cancer, neurodegenerative diseases, and cardiovascular defects. To deal with inadequate tissue oxygenation, it is necessary to employ adaptive measures both at the histological and cellular front. The fundamental objectives of tissue-level adaptations in response to reduced oxygen availability are the attainment of increased perfusion, neovasculogenesis, and hemoglobin production. Concurrently, the reprogramming of vital cellular processes encompassing gene expression, post-transcriptional and post-translational modification of gene products, mitochondrial respiration, and energy metabolism serves as integral means in harmonizing cellular responses to oxygen-deprivation. It is interesting to note that the preferable expression of certain splice variants over others is one of the major strategies co-opted by cells while adapting to hypoxic stress [1], and an amended blueprint of alternative pre-mRNA splicing contributes substantially to the pathogenesis of various diseases. Therefore, the manipulation of these hypoxia-induced alternative splicing events may present novel targets for therapeutic approaches and biomarkers for disease progression [2]. This review provides a comprehensive analysis of currently available data about:
How cells sense and respond to hypoxic stress,
How pre-mRNAs splicing contributes to cellular adaptation under hypoxia,
Alternative splicing events driven by hypoxia in several human diseases, and
Probable therapeutic strategies targeting hypoxia-induced alternative splicing events in several diseases, especially in cancers.
Sensing and responding to hypoxia: the PHD-pVHL-HIF pathway and beyond
It was widely appreciated for a long time that oxygen deprivation results in the transcriptional upregulation of several genes, including some glycolytic enzymes and the hemopoietic growth factor, erythropoietin (EPO) [3, 4]. However, the molecular mechanism of this remained enigmatic until Gregg Semenza and colleagues [5, 6] identified that the biological responses to hypoxia are manifested by the stabilization of a group of transcription factors called hypoxia-inducible factors (HIFs). HIFs recognize the hypoxia response element (HRE) motif in the promoter and/or enhancer regions of target genes. HIFs are heterodimer complexes composed of an oxygen-regulated α subunit and a constitutively expressed β subunit (also known as the aryl hydrocarbon receptor nuclear translocator, ARNT). The HIF family members HIF-1 and HIF-2 are regulators of oxygen homeostasis [7], whereas the role of HIF-3 remains to be fully understood. Nevertheless, few reports suggested that HIF-3α (also known as inhibitory PAS domain protein, IPAS) forms an abortive complex with HIF-1α and renders it incapable of recognizing the HRE motif [8]. Under aerobic conditions, HIF-1α is hydroxylated at either (or both) of two conserved proline residues (Pro402 and Pro564 in HIF-1α and Pro405 and Pro531 in HIF-2α) located near the N-terminal transactivation domain (NTAD) by prolyl hydroxylase domain proteins (PHDs) in a Fe2+ and 2-oxoglutarate-dependent manner [9]. Hydroxylated prolyl residues bind with the Von Hippel–Lindau (pVHL) protein, a ubiquitin ligase complex component. Consequently, in well-oxygenated conditions, HIF-1α is polyubiquitylated and directed to proteasomal degradation. Furthermore, factor inhibiting HIF (FIH) is another Fe2 +—and 2-oxoglutarate-dependent dioxygenase that hydroxylates a conserved asparagine residue within the HIF-1α and HIF-2α (N803 and N851, respectively) C-terminal transactivation domains (CTADs), hindering the recruitment of the coactivators p300 and CBP [10]. Under hypoxic conditions, both PHDs and FIH fail to hydroxylate HIF-1α leading to HIF-1α accumulation and transcriptional activation of hundreds of genes related to angiogenesis, erythropoiesis, autophagy, energy metabolism, and many more [9, 11] (Fig. 1). Despite the majority of target genes of HIF-1 and HIF-2 being the same, there are specific roles assigned to different types of HIFs. While HIF-1α is ubiquitously expressed, the expression of HIF-2α is transient and tissue-specific [12–15]. Moreover, while HIF-1α is shown to be involved in short-term hypoxic stress, with a gradual decrease in expression over time, HIF-2α expression increases over time in hypoxic cancer cells [16]. Furthermore, FIH1 binding to HIF-2α seems to be weaker than that to HIF-1α, and HIF-2α is less prone to oxygen-driven degradation [17]. Besides the HIF pathway, hypoxia induces other signaling pathways, including PI3K/AKT/mTOR [18], ERK [19], TGF-β [20], Wnt/β-catenin [21], and NFκB [22, 23] pathways, which are known to regulate processes like cell proliferation, apoptosis, metabolism, migration, and inflammation. Remarkably, some of these pathways can also activate HIFs in a hypoxia-independent manner [24, 25].
Fig. 1.
Schematic for oxygen sensing by HIFs. In the presence of oxygen, the hydroxylation of HIF-α by prolyl hydroxylases (PHDs) leads to its proteasomal degradation. Hydroxylation of an Asn residue by FIH-1 prevents p300/CBP binding. Hypoxia inactivates both PHDs and FIH-1, leading to the formation of HIF-α and HIF-β-complex and induction of the HIF inducible genes
Alternative pre-mRNA splicing as an adaptation to the hypoxic microenvironment
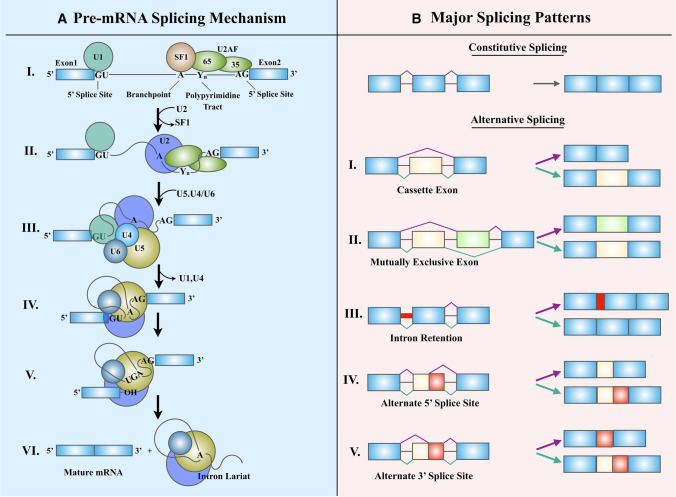
RNA splicing is a catalytic process that involves the excision of introns from pre-mRNAs and long noncoding RNAs followed by the precise joining of exons to form the mature RNA. The splicing process is catalyzed by complex macromolecular machinery known as a spliceosome. In metazoans, RNA splicing is carried out by either of two different spliceosomes, namely major and minor, depending on the intron type they recognize [26]. About 99.5% of the spliceosome machinery belong to the major category and are composed of five small nuclear ribonucleoproteins (snRNP) (U1, U2, U4, U5, and U6) complexes and more than 200 auxiliary proteins. Rest are minor type spliceosomes, which contains five snRNPs (U11, U12, U5, U4atac, and U6atac), most of which are distinct from its major counterpart [27, 28]. The spliceosome recognizes consensus splice sites located at the exon–intron junction, as well as the branch point. All splice sites are either constitutive or alternative, depending on whether they are always or only sometimes recognized by the spliceosome, respectively. The efficient recruitment of spliceosomal proteins to splice sites relies on the binding of additional regulatory splicing factors that either promote or repress splicing based on whether they bind to enhancer or silencer motifs, respectively [reviewed by Dvinge et al. [29] and Scotti & Swanson [30] (Fig. 2a). These trans-acting splicing factors belong to two major classes: heterogeneous nuclear ribonucleoproteins (hnRNPs) [31] and serine- and arginine-rich splicing factors (SR proteins) [32]. Apart from these two classes, RNA splicing is known to be regulated by many other RNA-binding proteins families that include CELF [33], MBNL [34], RBFOX proteins, RBM [35], STAR, NOVA [36] ESRPs, TIA1, TIAL1, etc. [37]. Alternative splicing (AS) happens to be one of the main engines that drive transcriptome and proteome diversity, and nearly 94% of all human genes are alternatively spliced [38, 39]. Apart from the constitutive alternative splicing, there are five major alternative splicing patterns (Fig. 2b) that operate through the recognition of cis-acting enhancer and silencer motifs by trans-acting splicing factors at the intron–exon junctions [40]. These patterns include exon skipping, intron retention, mutually exclusive exons, alternative 5′ splice site, and alternative 3′ splice site. A large body of research works has described the relevance of splice variants specific to cellular development stages and disease [41, 42].
Fig. 2.
Splicing machinery and the patterns of alternative splicing. a The mechanism of pre-mRNAs spicing by the spliceosome.I. E Complex formation: U1 snRNP attaches to the ‘CAGGURAGU' motif at the 5′ splice site. SF1, U2AF65, and U2AF35 recognize the branch-point, polypyrimidine tract, and AG dinucleotide at 3′ splice site, respectively. II. Complex A formation: The U2 snRNP displaces SF1 from the branchpoint. III. B complex formation: The U5.U4/U6 tri-snRNP adheres to the U1 snRNP. IV. B* complex formation: U1 and U4 are released, and the activated B* complex performs the first transesterification reaction at the 5′ splice site. V. C complex formation: This generates a lariat attached to 3′ exon and a free 5′ exon. VI. A second transesterification reaction at the 3′ splice site detaches the 3′ exon, leading to the removal of the lariat. The exons are stitched together, and the mature mRNA is channelized for translation. b The five major alternative splicing patterns. I. Exon skipping: Mature RNA is devoid of one or more exons. II. Intron retention: An intron is retained in the mature mRNA (marked in red). This could lead to the production of a non-functional protein. III. Mutually exclusive exons: Either of two exons is selected at a time for splicing. IV. Alternative 5′ splice site: An alternative donor site is recognized. V. Alternative 3′ splice site: An alternative acceptor splice site is used
Along with the reprogramming of alternative splicing patterns of genes, alterations in splicing factor expression and post-translational modification have been observed under certain pathological conditions, including hypoxia. With the mounting shreds of evidence, it is becoming increasingly evident that AS regulates many critical cellular processes while adapting to hypoxic stress [43–45]. Hence, the relevance of studying the link between AS and hypoxia is underscored by the possibility that some alternatively spliced isoforms could represent attractive therapeutic targets and unique splice variants could be used as biomarkers of hypoxia-elicited disease progression.
Hypoxia-driven AS in cancer
Cancer cells within rapidly growing solid tumors are often confronted with a severe reduction in oxygen availability (pO2 ≤ 2.5 mmHg). Primary pathogenetic mechanisms involved in the development of hypoxia in solid tumors are:
-
∙
Acute hypoxia (Perfusion-limited O2 delivery): Severe structural and functional abnormalities of tumor microvessels,
-
1
Chronic hypoxia (Diffusion-limited O2 delivery): Inefficient oxygen delivery to the cells distant (> 70 µm) from the blood vessels.
-
2
Anemic hypoxia: tumor-associated and therapy-induced anemia (hemoglobin levels < 12 g/dl), leading to a reduced O2 transport capacity of the blood.
There is abundant evidence for substantial heterogeneity in the tissue oxygenation status, predominantly due to the first two mechanisms mentioned above [46]. Nonetheless, tumor hypoxia is intensified when anemia is coupled with poor perfusion. The oxygen-deprivation exerts a selective pressure on tumor cells, which eventually elicits tumor aggressiveness by acquiring additional oncogenic properties, summarised as hallmarks of cancer by Hanahan and Weinberg [47, 48]. Hypoxic tumor cells achieve these cancer hallmarks by robustly customizing the cellular transcriptome and proteome diversity. Differential alternative splicing of pre-mRNA serves as a crucial adaptive gear to accomplish various hallmarks of cancer [1, 40, 47–49]. In this section, we provide a comprehensive outline of the studies related to hypoxia-elicited AS events in cancer.
Sustaining proliferative signaling
One of the most striking features of cancer cells is their ability to sustain autonomous neoplastic growth. Normal tissues maintain homeostasis of cell numbers and tissue architecture by closely controlling a complex network of signaling pathways that respond to growth factors. On the other hand, cancer cells acquire growth signal autonomy through a myriad of abnormal modifications of mitogenic signaling cascades. This particular trait of cancer cells is often achieved by hijacking the AS program and preferentially expressing isoforms that promote proliferation. Interestingly, many of these alternative splicing events are guided by the hypoxic tumor microenvironment. For instance, hypoxia promotes the production of a constitutively active type III variant of epithelial growth factor receptor (EGFR) that lacks exons 2–7 (EGFRvIII) (Fig. 3a), which is correlated with poor prognosis in several tumor types, including glioblastoma multiforme [50]. Similar to EGFR, hypoxia directs the AS of Neurotrophin tyrosine kinase receptor type 1 (TrkA) (Fig. 3a). TrkA is the preferred receptor for the nerve growth factor (NGF) that plays a critical role in nervous system development. Hypoxia-induced AS of TrkA produces constitutively active oncogenic isoform, TrkAIII that lacks exons 6, 7, and 9, and antagonizes antioncogenic NGF/TrkAI signaling in neuroblastoma [51]. KRAS is another proto-oncogene that participates in the Ras/MAPK pathway and undergoes alternative splicing of the last exon to generate two nearly identical isoforms, KRAS-4A and KRAS-4B, the latter being devoid of exon 4a. Although, when mutated, both the isoform perform oncogenic roles, including cell proliferation induction, they might behave differently in many cellular aspects. For instance, KRAS-4B is expressed ubiquitously in human tissues, whereas KRAS-4A is mainly restricted to tissues of endodermal origin [52]. While wild-type KRAS-4A classically promotes apoptosis, wild-type KRAS-4B is considered to be anti-apoptotic [53]. Moreover, when KRAS is mutated, both the isoforms differ in their ability to induce anchorage‐independent growth and cell migration [54, 55]. As to the choice of predominant isoform during hypoxia-driven AS of KRAS, there is some disparity in the data recorded in the literature. According to a study by Sena et al., the expression of KRAS-4B isoform is induced in hypoxic Hep3B cells [56], thereby reducing the KRAS-4A/4B ratio, which is often observed in human colorectal cancer [57]. On the other hand, KRAS-4A is shown to be induced by hypoxia in A549, SUIT2, and AsPC1 cells [58] (Fig. 3a). Along the same lines, Bowler et al. showed that in prostate cancer cells, hypoxia-induced AS of the tyrosine-protein phosphatase PTPN13 renders it non-catalytic and enhances tyrosine kinase-dependent signaling and proliferation. The same study also showed that hypoxia treatment induces the AS of TTC23 (involved in hedgehog signaling) and RAP1GDS1 (Stimulates GDP/GTP exchange reaction of a group of G proteins) and promotes proliferation and autonomous growth, respectively [59]. In colorectal cancer cells, hypoxia induces lncRNA LUCAT1 that interacts with polypyrimidine tract binding protein 1 (PTBP1), which skews the AS of a set of genes (APP, CD44, CLSTN1, MBNL1, and ZNF207) related to cell growth or DNA damage. An elevated level of LUCAT1 has been correlated with chemotherapy resistance and poor prognosis in patients. Therefore, interfering with the LUCAT1/PTBP1 axis represents a promising strategy to treat refractory hypoxic colorectal cancer [60]. The hypoxic milieu of late-stage pancreatic ductal adenocarcinoma stimulates the expression of a secreted splice variant of tissue factor, termed asTF (alternatively spliced tissue factor) (Fig. 3a), which then activates carbonic anhydrase IX (CA IX) that is implicated in hypoxia-driven cancer progression [61].
Fig. 3.
Schematic of hypoxia-driven splicing events in several hallmarks of cancer. The splicing patterns shown by solid blue lines represent hypoxia-specific splicing, whereas the grey lines represent normoxia-specific events. Hx above the blue arrow indicates the hypoxia-induced transcripts. a Examples of hypoxia-specific AS events contributing to sustaining proliferative signaling. b Examples of hypoxia-specific AS events contributing to metastasis. c Examples of hypoxia-specific AS events contributing to angiogenesis. d Examples of hypoxia-specific AS events contributing to metabolic changes. e Examples of hypoxia-specific AS events contributing to immune evasion. f Examples of hypoxia-specific AS events contributing to apoptosis avoidance. g Examples of hypoxia-specific AS events contributing to replicative immortality
Activating metastasis
More than 90% of cancer-mortality is attributable to metastases, not the primary tumors themselves [62, 63]. Metastasis is a multistep cell-biological process that involves disseminating cancer cells to distant organ sites, followed by adaptation to anatomically distinct tissue microenvironments. This incredible phenotypic plasticity is afforded by co-opting epithelial-mesenchymal transition (EMT) and the reverse, mesenchymal-epithelial transitions (MET), which are orchestrated by de-differentiation and re-differentiation of cancer cells. Several molecular players, including transcription factors, growth factors, cytokines, cell adhesion molecules, cytoskeleton remodelers, and extracellular matrix remodeling enzymes, are instrumental in EMT/MET cycles [64].
Classically, EMT has been thought to be regulated primarily at the transcription level by the players mentioned above. However, recent studies have begun to highlight additional layers of EMT control, which include epigenetic reprogramming, small noncoding RNAs, translational and post-translational regulations, and alternative splicing changes [65]. In particular, a wealth of studies indicates that splicing regulation alone can drive critical aspects of EMT acquisition [40]. Over the past few years, tumor hypoxia has been incriminated as a key driver of metastasis and has been shown to regulate several AS events related to invasion and metastasis. Our group recently reported that the hypoxic microenvironment promotes the exclusion of exon 11a of a master actin regulator, hMENA (also known as ENAH), generating a pro-metastatic and pro-invasive splice variant. Mechanistically, the repression of Epithelial splicing regulatory protein 1 (ESRP1) via the TGF-β-SLUG/RBFOX2 axis under hypoxia brings about the aberrant AS of hMENA [66] (Fig. 3b). Another attractive example of hypoxia-induced splicing change is the AS of Cluster of differentiation 44 (CD44), a transmembrane receptor that can bind essential components of the extracellular matrix, such as hyaluronic acid, collagen, fibronectin, laminin, and matrix metalloproteinases (MMPs), and promotes the migration and invasion processes involved in metastases. Out of 20 exons of CD44, 10 undergo alternative splicing, thus generating multiple CD44 isoforms (v1–v10) with different molecular weights and diverse extracellular domains [67]. The low-molecular-weight CD44 isoform, CD44s (90 kDa standard form), is expressed by several tissues, including hematopoietic and mesenchymal cells, whereas high-molecular-weight CD44v variants (140–230 kDa) are expressed in epithelial cells and abundant in several types of carcinoma, including breast and colorectal cancers [68, 69]. In particular, hypoxic regions of breast cancer specimens represent an elevated expression of CD44, and HIF-1α is shown to upregulate CD44 variants containing exons v6 and v7/8 [70] (Fig. 3b). Additional studies are necessary to identify the molecular events that regulate CD44 alternative splicing under hypoxia. The cysteine-rich angiogenic inducer 61 (CYR61) provides another attractive example of hypoxia-induced alternative splicing changes. CYR61 protein is involved in cellular events like cell proliferation, adhesion, and migration, thereby promoting critical pathophysiological processes during vascular development, angiogenesis, wound healing, and cancer progression [71, 72]. The role of CYR61 in tumor progression, however, is highly context-specific. It promotes cancer cell growth, migration, and invasion in the breast, gastric and ovarian cancers, gliomas, and pancreatic neuroendocrine tumors, whereas it appears to have a tumor-suppressive role in non-small-cell lung cancer [69, 70]. Notably, the CYR61 expression is induced by HIF-1α under hypoxia [71]. Two different alternatively spliced CYR61 isoforms are generated in cancer cells—one contains the correctly spliced mRNA stitching all four exons (intron spliced isoform). In contrast, the other isoform retains intron 3 (intron-retaining isoform) that is likely to undergo nonsense-mediated decay (NMD), since it harbors two stop codons within the intronic sequence. Hypoxia steers the alternative splicing of CYR61 toward the intron spliced isoform, having biological consequences in cancer cells [73] (Fig. 3b). However, further studies are required to decipher the molecular mechanisms that lead to this splicing change.
Inducing angiogenesis
The adequate supply of oxygen and nutrients is an absolute requirement to sustain the rapid proliferation and metastasis of tumor cells. Hence, new growth in the vascular network (angiogenesis) is essential for tumor growth and metastatic progression [74]. Vascular Endothelial Growth Factors (VEGFs), the key regulators of angiogenesis, are secreted by tumor cells and causes endothelial cells to detach and migrate into the neighboring stroma. The VEGF expression is primarily regulated by hypoxia, as VEGFs are a direct target of HIF-1α and HIF-2α transcription factors [75, 76]. The biochemical and biological properties of the VEGF family of ligands, as well as their receptors, are tightly fine-tuned by RNA processing [77, 78]. In humans, the VEGF family consists of five ligands (VEGF-A to D and placental growth factor) and three signaling receptors (VEGFR1, VEGFR2, and VEGFR3). The VEGF ligands and the receptors are known to undergo AS to form splice isoforms with different expression patterns and bioactivity. VEGF-A is composed of a total of eight exons and is studied extensively among all VEGF family members. Various splice variants of VEGF-A (121,145,165, 183, 189, and 206 amino acid long) with distinct C-terminal domains are generated by AS. Additional AS of exon 8 at the mutually exclusive proximal or distal splice site results in a pro-angiogenic (VEGF-Axxx) or an anti-angiogenic (VEGF-Axxxb) isoform, respectively, where xxx denotes the number of amino acids [79]. Tumor hypoxia is also known to augment the expression pro-angiogenic VEGF-Axxx isoforms, besides upregulating panVEGF-A [80] (Fig. 3c). On the other hand, the anti-angiogenic VEGF-Axxxb formation is promoted preferentially in normal tissues and under normoxia, while their expression diminished in cancerous tissues [81]. The cellular choice between proximal or distal 5′ splice site in exon 8 is determined by the activity of the serine/arginine-rich (SR) proteins SRSF1 and SRSF6. SRSF1 preferentially activates the proximal splice site, whereas SRSF6 mainly recognizes the distal splice site [82]. SR protein kinase 1 (SRPK1) specifically phosphorylates SR proteins and is upregulated under hypoxic stress [59, 83] and in various cancer types of cancer, including pancreatic, breast, and colon carcinoma [84]. Insulin-like Growth Factor 1 (IGF-1) increases the expression of VEGF165 isoforms through protein kinase C (PKC) and SRPK1 [77]. Hence, it is highly likely that tumor hypoxia acts as a determining factor to rewire the VEGF splicing to facilitate angiogenesis, thereby providing new insights into anti-angiogenic therapies.
Deregulating cellular energetics
Cancer cells need to rewire their metabolism to maintain upregulated cellular proliferation and survival under the metabolically compromised conditions posed by the tumor microenvironment. One of the significant molecular differences between cancer cells and noncancerous tissue is that cancer cells consume a large amount of glucose to produce pyruvate from glycolysis, which is then directed away from the mitochondria to produce a prodigious amount of lactate through the action of lactate dehydrogenase (LDH/LDHA)—a process otherwise reserved for the low oxygen state. This induction in glycolysis in the presence of oxygen is known as aerobic glycolysis or the Warburg effect and is exaggerated by hypoxia [85].
LDHA is a direct target of HIF1α transcription, and its overexpression is correlated with poor prognosis in multiple cancers. Among multiple LDHA isoforms annotated in Ensembl, LDHA-201 (ENST00000540430), that has an open reading frame starting from the first exon and encodes a 361 amino acid long protein, is significantly reduced during acute and chronic hypoxia with a corresponding increase in a novel isoform that retains the first intron [86] (Fig. 3d). AS of another metabolic enzyme, Pyruvate kinase (PK) that catalyzes the final step reaction of glycolysis, converting phosphoenolpyruvate (PEP) to pyruvate, is a critical determinant of how glucose is used in cancerous versus differentiated cells. Pyruvate kinase muscle isozyme PKM, expressed in all mammalian tissues except liver and erythrocytes [87], generates PKM1 and PKM2 splice variants using mutually exclusive exon 9 and exon 10, respectively (Fig. 3d). While PKM1 is expressed in most adult tissues and promotes oxidative phosphorylation, PKM2 is upregulated in cancers and promotes aerobic glycolysis. The universal switching of tumors to PKM2 is determined by splicing regulators hnRNPA1, hnRNPA2, hnRNPI, and SRSF3, etc. [88], most of which are known to be upregulated by hypoxia [59]. Of note, HIF-1α-directly stimulates PKM transcription, while PKM2 further enhances HIF-1α-mediated gene transcription forming a positive feedback loop [89, 90]. Another vital regulator of glycolysis 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase (PFKFB), is associated with many aspects of cancer, proliferation, angiogenesis, drug resistance, and tumor hypoxia. All four isozymes of PFKFB (PFKFB-1–4) are known to be upregulated by hypoxia [91]. Furthermore, hypoxia is shown to influence the AS pattern of PFKFB-3 and -4 [92, 93]. Thus, hypoxia-led changes in AS contributes substantially to maintaining the glycolytic metabotype in tumor cells, which fuels enhanced malignancy.
Avoiding immune destruction
Tumor cells have developed an incredible arsenal of strategies to escape elimination by the immune system. As investigated in a wealth of studies, tumor hypoxia is regarded as a driving force to diminish anticancer immunity by skewing the expression of tumor-associated immune checkpoint molecules (e.g., CD47, PDL1, and HLA-G), impairing immune effectors, and altered cellular metabolism [94–96]., Hypoxic stress is also known to influence the interaction of the Fas receptor (also known as CD95) with Fas-Ligand (FasL), an essential mechanism for the maintenance of immune homeostasis during tumor progression. The Fas receptor is a membrane-bound protein that, when bound to FasL expressed on cytotoxic T cells, triggers a Caspase cascade that eventually leads to apoptosis [97]. Interestingly, AS of the Fas protein exhibits a critical means by which tumor cells avoid immune destruction. Along with the full-length Fas protein, the AS of Fas pre-mRNA gives rise to several shorter isoforms, including FasΔEx6, which is the most abundant among the shorter isoforms and lacks the 63-nt long exon 6 [98]. FasΔEx6 translates into a soluble Fas protein (sFas), which is secreted in the extracellular milieu and capable of inhibiting Fas-mediated apoptosis, presumably by blocking the interaction of FasL with cell-surface Fas receptor [99, 100]. A recent study claimed that the AS of Fas is regulated by hypoxia, and it favors the production of sFas over the full-length isoform [101] (Fig. 3e). Of note, the total cellular expression of Fas receptors is reduced by hypoxia treatment in various cell lines, which further contributes to the avoidance of immune destruction [101]. Hypoxia is also found to modulate the alternative splicing of co-stimulatory receptor CD137 (4-1BB, TNSFR9), which is a member of the TNF-receptor (TNFR) superfamily, usually found on activated T lymphocytes and endothelial cells. Hypoxia induces the expression of a soluble variant of CD137 (CD137s) lacking the transmembrane domain, which interacts with CD137L to block the co-stimulatory signals to primed T lymphocytes (Fig. 3e). Thus, tumor hypoxia thwarts the CD137/CD137L co-stimulatory system, which would otherwise elicit cytotoxic immune responses against cancer cells [102].
By-passing apoptosis
Evasion of programmed cell death (Apoptosis) is one of the most studied hallmarks of cancer concerning AS. There are two routes to apoptosis: the extrinsic/death receptor pathway and the intrinsic/mitochondrial pathway. In the extrinsic pathway, a death-inducing signal complex (DISC) is formed in response to the interaction of cell surface receptors belonging to the Tumor-necrosis factor (TNF) receptor superfamily, with extracellular ligands, such as TNF ligand, TNF ligand superfamily member 10 (TNFSF10), Fas ligand and TNF-related apoptosis-inducing ligand (TRAIL). The intrinsic pathway of apoptosis is triggered by mitochondrial outer membrane permeabilization (MOMP), which is regulated by the B cell lymphoma 2 (BCL2) family of proteins [103]. Studies have shown that the AS regulation of BCL-x, a BCL2 family member, is crucial for the normal apoptotic response with aberrant control of the protein leading to tumorigenesis and therapy insensitivity. BCL-x is alternatively spliced to produce two isoforms with opposing function: either a long anti-apoptotic isoform, BCL-xL that contains three exons, or a short pro-apoptotic isoform, BCL-xS, in which the 3′ portion of exon 2 is spliced out [104]. High BCL-xL/BCL-xS ratio is a hallmark of a variety of cancer types, which is consistent with the crucial role of BCL-xL isoform in cancer cell survival [105–107]. A wealth of studies have demonstrated that hypoxia imparts resistance to cell death by favoring the expression of BCL-xL in a HIF-1α dependent manner [108–110] (Fig. 3f) while downregulating some pro-apoptotic BCL2 family members, including Bax, Bad, and Bid. On the contrary, in vivo studies have demonstrated that neonatal hypoxia–ischemia reciprocally alters BCL-x splicing toward its short form in the rat hippocampus and cortex [111]. Moreover, there are several RNA-binding proteins and signaling pathways that influence the Bcl-xL/Bcl-xS ratio (Reviewed by Stevens and Oltean [112]). Interestingly, many of these trans-acting factors are hypoxia sensitive. For example, hypoxia-induced lncRNA LUCAT1 complexes with PTPB1, a direct regulator of BCL-x AS, and promotes survival and therapy-resistance [60, 113].
Similar to BCL-x, BNIP3 is also a BCL2 family member responsible for provoking mitochondrial perturbations and cell death in response to various stresses. A hypoxia-induced AS of exon 3 of BNIP3 generates BNIP3ΔEx3 isoform lacking putative BH3-like domain (Fig. 3f) and carboxyl-terminal transmembrane domain critical for mitochondrial targeting and cell death while antagonizing the death induction by its full-length counterpart [114]. Thus, the hypoxic microenvironment seems to have an essential role in driving the AS events that preferably contribute to cell death resistance.
Furthermore, apoptosis is known to be affected by the altered splicing of a significant number of other genes, including Caspases, APAF1, MCL1, TP53, Survivin, and BIM, among others (Reviewed by Lin et al.[115]). However, whether and how hypoxia has any role in these events is yet to be elucidated.
Enabling replicative immortality
An essential property of cancer cells is to attain unlimited replicative potential circumventing the cellular impediments in the form of senescence and apoptosis. Accumulating shreds of evidence have attributed the unlimited proliferation of tumor cells to their ability to maintain telomere length typically by enhanced expression of telomerase [116, 117]. Telomerase reverse transcriptase (hTERT) is the catalytic subunit of the telomerase holoenzyme complex, and its aberrant expression is closely associated with various cancer phenotypes, including senescence evasion. A full-scale comprehension of the transcriptional and post-transcriptional regulation of hTERT expression is pivotal in understanding the pathogenesis and searching for therapeutic approaches. At the promoter level, a wide variety of mechanisms, including transcription factor (e.g., c-MYC, NF-κB, STAT3, and HIF-1α) binding, promoter mutation, promoter methylation, and histone acetylation fine-tune the hTERT expression (for review, refer [118]). At the post-transcriptional level, AS of the hTERT gene, which comprises 16 exons, plays a vital role in determining telomerase activity. The splicing reactions occurring in different combinations of selective splicing sites, which include three exclusion sites (α, β, and γ) and four intron retention sites, generate several alternative splice variants [119] (Fig. 3g). However, the telomerase activity is conserved to only the wild-type full-length hTERT isoform (FL hTERT) [120]. Hypoxia plays a substantial role in orchestrating the hTERT expression both at the transcription and splicing levels. Interestingly, hypoxia induces the production of FL hTERT isoform, which would otherwise be absent in differentiated cells, rather than merely elevating total hTERT expression in some cases [121]. Thus, hypoxic regulation of hTERT splicing may provide novel perspectives for translational diagnosis and therapeutic strategies targeting replicative immortality.
Hypoxia-driven AS in neurological diseases
Neurodegenerative diseases are a heterogeneous group of disorders that are characterized by the progressive and often irreversible damage of the central or peripheral nervous system. Aberrant alternative splicing is increasingly being recognized as a driving force in several neurological diseases, which is perhaps not surprising given the significance of splicing in the proper functioning of neurons. Many of these splicing events are governed by hypoxia–ischemia. Hypoxic brain injury is often the result of hypoperfusion due to either cardiac arrest, vascular injury, strangulation, smoke inhalation, poisoning from a drug overdose such as opiates, intoxication from carbon monoxide, or head trauma. Here, we outline a few examples of neurologic conditions related to anomalistic splicing changes in response to hypoxic stress.
Spinal muscular atrophy (SMA)
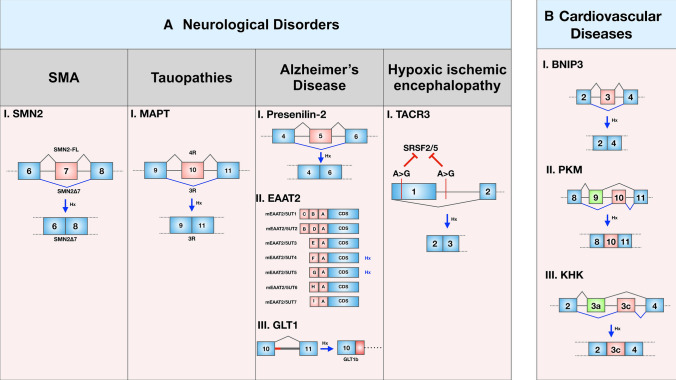
SMA is one of the most common lethal genetic diseases in infants characterized by wasting in muscles and progressive paralysis. These neuromuscular symptoms result from reduced levels of the survival motor neuron (SMN) protein. Humans have two nearly identical copies of the SMN gene: SMN1 and SMN2, which are basically two inverted repeats located on chromosome 5 [122]. While SMA mostly results from homozygous loss of the SMN1 gene, SMN2 often fails to compensate for the loss of SMN1 due to the predominant skipping of exon 7. Therefore, significant efforts have been made to develop small molecules and antisense oligonucleotides that can correct the splicing error of SMN2 exon 7 [123–125]. Remarkably, hypoxia treatment resulted in increased production of the exon 7-skipped isoform (SMN2Δ7) and reduced total SMN protein levels in cultured cells [126] (Fig. 4a).
Fig. 4.
Schematic of hypoxia-driven splicing events in neurological disorders and cardiovascular defects. The splicing patterns shown by blue lines represent hypoxia-specific splicing, whereas the grey lines represent normoxia-specific events. a Examples of hypoxia-specific AS events contributing to neurological disorders like Spinal muscular atrophy, Tauopathies, Alzheimer's Disease, and Hypoxic-ischemic encephalopathy. (Among EAAT2 splice variants, mEAAT2/5UT4 and mEAAT2/5UT5 are modulated by hypoxia). b Examples of hypoxia-specific AS events contributing to cardiovascular disease
Tauopathies
Tauopathies are a multifarious class of neurodegenerative conditions characterized by intracellular accumulation of misfolded tau proteins. Microtubule-associated protein tau (MAPT) plays an indispensable role in microtubule organization to maintain cytoskeletal structure in neurons. Out of six splice variants of MAPT genes, three‐repeat (3R) tau lacks exon 10 and is expressed during the embryonic stage, whereas the expression of exon 10 containing isoform, four‐repeat (4R) tau increases with maturation. In the adult human brain, both the isoforms are equally abundant, and any alteration in the 4R/3R isoform ratio affects microtubule binding and assembly. Proteasomal dysfunction caused by hypoxic‐ischemic injury skews the 4R/3R ratio by decreasing exon 10 inclusion (Fig. 4a). Of note, a skewed 4R/3R ratio is a signature of several tauopathies like Frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP‐17), Alzheimer's disease (AD), progressive supranuclear palsy (PSP), and Pick's disease (PiD).
Alzheimer's disease (AD)
Alzheimer's disease is a heterogeneous neurodegenerative disorder that is pathologically characterized by intraneural neurofibrillary tangles and extracellular deposition of β-amyloid peptide contributing to senile plaques. The preferential expression of aberrant Presenilin-2 isoform (termed as PS2V) lacking exon 5 is a hallmark of the sporadic AD brain, and is induced by hypoxic stress (Fig. 4a). Mechanistically, hypoxia upregulates the expression of high mobility group protein A1a protein (HMGA1a) that co-localizes with splicing factor SRSF2 in the nuclear speckles and binds to the exon 5 of Presenilin-2 pre-mRNA to splice it out [127]. Another study in a transgenic mouse model for Alzheimer's disease revealed that chemical hypoxia stimulates AS of excitatory amino acid transporter 2 (EAAT2), which is also referred to as GLT1 in rodent terminology (Fig. 4a). EAAT2 is a key player in glutamatergic synaptic signaling as it is the primary carrier of the neurotransmitter glutamate, and its dysregulation has been associated with AD. Several of the seven identified EAAT2 splice variants (mEAAT2/5UT1–7) are reported to be hypoxia-regulated and are expressed in a region-specific manner. For instance, mEAAT2/5UT4 is upregulated in the frontal cortex and cerebellum and downregulated in the hippocampus, whereas mEAAT2/5UT5 is upregulated in the frontal cortex and downregulated in the cerebellum after hypoxia induction by 3-nitropropionic acid treatment [128]. As far as splicing of GLT1 is concerned, GLT1b and 1c are the well-characterized functionals variants other than full-length GLT1 isoform. The expression of GLT1 is increased in the neonatal rat brain cortex and decreased in the striatum following hypoxic preconditioning [129]. On the other hand, while GLT1b is induced, GLT1 level is diminished in neurons of neonatal pigs that experienced severe hypoxic insults [130]. Another study in newborn piglets showed that GLT1 expression is diminished in the astroglial compartment but elevated in cells appearing as neurons when challenged with hypoxia–ischemia [131].
Hypoxic ischemic encephalopathy (HIE)
Neonatal hypoxic-ischemic encephalopathy (HIE) (also known as intrapartum asphyxia) is a type of brain damage caused by oxygen deprivation to the brain. In a recent study, Xue et al. (2020) showed that the number of SNP exhibiting genes is induced during HIE. These SNPs, in turn, play a pivotal role in AS of the associated genes. For example, Tachykinin receptor 3 (Tacr3) contributes significantly to the pathogenesis of HIE via SNP-induced AS of exon 1 (Fig. 4a). The SNPs (G > A) within and outside exon 1 disrupted the binding site of splicing factors SRSF2 and SRSF5, and thereby reduced their binding efficiency [132]. Therefore, it is evident that hypoxia–ischemia has earned remarkable attention in effectuating neurological disorders due to its role in driving AS to produce isoforms with pathological relevance. Nevertheless, it requires more in-depth studies to comprehend the role of hypoxia-driven splicing in neurologic diseases.
Hypoxia-driven AS in cardiovascular diseases
Ischemic heart disease is a significant cause of morbidity and mortality across the globe. Myocardial ischemic injury is characterized by a disparity between the supply and demand of oxygen in myocardial tissues, resulting from the inadequate coronary blood supply. The clinical manifestations of myocardial ischemia include cardiac dysfunction, arrhythmias, myocardial infarction, and sudden death [133]. The aberrant cardiomyocyte apoptosis is a fundamental underlying cause of the decline in cardiovascular performance following myocardial ischemia. Hypoxic milieu prevalent in ischemic cardiac tissues triggers the expression of BNIP3 (BCL2/adenovirus E1B 19 kDa protein-interacting protein 3), which is known to provoke apoptosis and autophagy in cardiomyocytes.
Interestingly, a recent study by Gang et al. showed that hypoxia not only induces BNIP3 expression but orchestrates the AS of BNIP3 to produce an abnormal isoform lacking exon 3 (BNIP3Δex3) in cardiomyocytes [114] (Fig. 4b). The exclusion of exon 3 offered a frame-shift mutation revealing a stop codon that terminates transcription up-stream of exon5 and exon6. This premature termination generates a truncated BNIP3 variant devoid of the putative BH3-like domain and carboxyl-terminal transmembrane domain crucial for mitochondrial localization. BNIP3Δex3 antagonistically interacted with the full-length BNIP3 protein and reduced the programmed cell death in hypoxic cardiomyocytes. Furthermore, a study about ischemic injury of myocardial infarction claimed that HIF-1α drives the AS of PKM to induce the glycolytic enzyme PKM2 isoform while reducing the expression of the predominant isoform PKM1 [134] (Fig. 4b). Another interesting study by Mirtschink et al. [135] showed that myocardial hypoxia drives the AS of the fructose metabolizing enzyme ketohexokinase (KHK) by activating the splicing factor SF3B1 in the heart of hypertrophic cardiomyopathy patients. Hypoxia-induced SF3B1 caused the shift from KHK-A isoform to KHK-C isoform that has a better affinity for sucrose (Fig. 4b). The elevated expression of KHK-C in the heart stimulated fructolysis that is otherwise restricted exclusively to the liver. Overall, AS events induced by hypoxia–ischemia are increasingly being recognized as an important pathogenetic factor of cardiovascular defects. However, it requires large-scale investigations to dissect the molecular mechanisms that account for AS events in cardiac diseases under hypoxia, which is an active area of investigation, nonetheless.
Diagnostic and therapeutic potentials of hypoxia-induced alternative splicing
Hypoxic milieu has been known to interfere with the efficacy of canonical therapeutic paradigms, for instance, the slow-growing cells in hypoxic regions by-pass apoptosis induced by chemotherapy that targets proliferating cells. Besides, the hypoxic tumor region's acidic nature interferes with immune cell recruitment and reduces the efficacy of immunotherapy. As opposed to the traditional approaches, targeting of splice isoforms serves as an outstanding therapeutic strategy in hypoxia-associated diseases. Moreover, it is not surprising that the development of diagnostic tests based on alternatively spliced biomarkers is gaining tremendous attention, given the crucial roles of hypoxia-driven AS in pathogenesis and disease progression. Here, we have outlined a few examples of the large number of therapeutic candidates that can potentially target hypoxia-induced AS events.
HIF inhibitors
HIFs are the determinative factors of molecular adaptations acquired by the cells under hypoxia. Even most of the hypoxia-elicited signaling pathways eventually converge to the HIFs. Given the fact that many of the hypoxia-induced splicing events require just the stable expression of HIFs, not hypoxia per se, HIF inhibition is believed to be an effective strategy against aberrant hypoxia-elicited AS. The HIF inhibitors may target one of the following steps: (1) HIF transcription and translation, (2) HIF dimerization, (3) DNA-binding activity, and (4) Proteasomal degradation of HIF (Table 1). The strategies to interfere with the HIF expression transcriptionally and translationally include the use of long noncoding RNA, antisense oligonucleotides, and topoisomerase I inhibitors [136–139]. Alternatively, there have been significant efforts to develop small molecules that inhibit HIF recruitment to HRE. Echinomycin, and Anthracyclines are examples of such inhibitors [140, 141]. In addition, inhibitors of HIF accumulation and transcriptional activity, include the Hsp90 inhibitors, Chetomin and Indenopyrazoles [142–145]. Furthermore, some drugs induce the proteasomal degradation of HIFs. For example, LW6 stimulates HIF degradation by inducing pVHL expression [146].
Table 1.
HIF inhibitors
| Inhibitory regimens | Examples | References |
|---|---|---|
| HIF transcription and translation inhibitors | lncRNA PIN1-v2 | [136] |
| HIF-1α antisense oligonucleotides: EZN- 2698, EZN-2208 | [137, 138] | |
| The topoisomerase I poison topotecan | [139] | |
| DNA-binding inhibitors | Echinomycin (NSC-13502), a small molecule inhibitor prevents HIF-1 binding to the HRE of VEGF promoter | [140] |
| Anthracyclines (particularly idarubicin), that inhibits HIF binding to HRE | [141] | |
| Inhibitors of HIF-driven transcription of target genes | Small molecule Hsp90 inhibitors EC154, 17-AAG etc | [142, 143] |
| Chetomin: reduces the expression of VEGF and CA IX | [144] | |
| Indenopyrazoles: Reduce the transcriptional activity of HIF1 | [145] | |
| Inducers of Proteasomal degradation of HIF | LW6: Induces pVHL expression | [146] |
Targeting the spliceosome
As we have discussed earlier, deregulated expression and even mutations of the spliceosomal components might play a key role in hypoxia-driven AS. Therefore, the emergence of small molecule inhibitors of spliceosome components as tools to inhibit hypoxia-induced AS is not surprising. Spliceosome inhibitors encompass a variety of small molecules that prevent different steps of the splicing cascade (Table 2). For example, many inhibitory regimens, such as Spliceostatin A, Meayamycin, and pladienolide B, target SF3B1, the core component of U2 snRNP, and destabilize its interactions with pre-mRNA [147]. On the other hand, small molecule inhibitor cp028 makes the spliceosomal complexes kinetically stalled at an intermediate stage of the spliceosome activation [148]. Isoginkgetin blocks the first step of splicing by preventing stable recruitment of U4/U6.U5 tri-snRNPs [149]. Another small-molecule inhibitor called Madrasin stalls spliceosome assembly at the A complex[150]. However, their clinical potential, safety, and usefulness is still a matter of great debate. Much work is required to understand the landscape of splicing changes brought about by these small molecules.
Table 2.
Inhibitors of spliceosomal components
| Compounds | Functions | References |
|---|---|---|
| Spliceostatin A, Meayamycin and pladienolide B | Target SF3B1 and alter the conformation of SF3B1 disrupting the U2 snRNP binding to pre-mRNA | [147] |
| cp028 | Inhibits splicing at an intermediate stage of the spliceosome activation | [148] |
| Isoginkgetin | Prevents the recruitment of U4, U5 and U6 tri-snRNP | [149] |
| Madrasin | Intervenes with the early stages of spliceosome assembly and stalls spliceosome assembly at the A complex | [150] |
Splice-switching oligonucleotides
Over 95% of all human genes are alternatively spliced to express splice variant proteins, and AS has implications in a multitude of diseases. Hence, splice-switching oligonucleotides (SSOs), which redirect AS by imparting steric hindrance to the splicing machinery, have promising therapeutic potential. The chemical nature of the SSOs falls under either of the following categories: (1) Peptide nucleic acids (PNAs), (2) Alternating locked nucleic acids (LNAs), (3) 2′-substituted oligonucleotides, and (4) Phosphorodiamidate morpholino oligomers-based oligomers [151]. Few examples of successful SSO mediated therapy are discussed below. A highly effective 2′-O-(2-methoxyethyl)-phosphorothioate targeting a silencer element in the last intron of SMN2 promoted exon 7 inclusion [152, 153]. In a severe genetic disorder called Duchenne muscular dystrophy (DMD), the deletions within the Dystrophin gene sequence shifts the reading frame, abrogating expression of the dystrophin protein. A similar disease called Becker muscular dystrophy (BMD) with milder symptoms arises when the deletions preserve the reading frame but produce a truncated protein. Intriguingly, the SSO-induced exclusion of the adjacent out-of-frame exon is shown to reinstate the reading frame converting severe DMD to a milder BMD [154, 155]. MDM4 splicing is another excellent target for cancer therapy. The exclusion of its exon 6 followed by NMD renders MDM4 protein undetectable in most normal adult tissues, whereas in cancer cells, exon 6 is often included to produce the full-length MDM4 that dampens the tumor-suppression effect of p53. In multiple melanoma cell lines and xenograft mouse models, SSO-mediated skipping of exon 6 promoted melanoma-regression [156]. Furthermore, a broad repertoire of splicing events can potentially be targeted by SSOs in different disease conditions, and some SSOs like Eteplirsen and Nusinersen have even received FDA approval for the treatment of Duchenne muscular dystrophy and spinal muscular atrophy, respectively [157]. However, the major drawbacks of this strategy are that the intracellular delivery and stability of SSOs are difficult to control. Additionally, SSOs are not cost-efficient for large scale studies. Nevertheless, the advantage of oligonucleotides is their reduced off-target effect and high specificity.
Targeting splicing factors
As the splicing factors are the central modulator of AS, therapeutic targeting of splicing factors offers an effective strategy to redirect the splicing mechanism. For example, targeting of nuclear-cytoplasmic shuttling of hnRNP K and A1 by KPT330 (selinexor) in acute myeloid leukemia cells showed encouraging anti-tumor activity [158]. The knockdown of HnRNP A2/ B1 induces apoptosis and reduces the invasion and chemoresistance of glioma cell lines [159]. Bortezomib (Velcal) was the first FDA-approved proteasome inhibitor to treat refractory multiple myeloma patients. Mechanistically, bortezomib treatment diminished the expression of high-molecular-weight hnRNP K (sumoylated) along with c-Myc while stimulating low-molecular-weight hnRNP K (desumoylated), and thus reduced the proliferation of Daudi cells [160].
Targeting splicing factor kinases
The activity of many splicing factors is phosphoregulated by the splicing factor kinases. Therefore, therapeutic targeting of splicing factor kinases may also skew the expression of hypoxia-induced splice isoforms. For instance, the phosphorylation levels of SR proteins are controlled majorly by three families of kinases: CDC2-like kinases (CLKs), dual-specificity tyrosine-regulated kinases (DYRKs), and SR-rich splicing factor protein kinases (SRPKs). One of the first potent CLK inhibitors was TG003 [161, 162], which has shown great promises in the treatment of Duchenne muscular dystrophy [163]. Another small molecule inhibitor of CLKs, CX-4945, reduced PI3K/Akt signaling and HIF1α transcription and is currently in a clinical trial for bile duct cholangiocarcinoma [164]. Furthermore, CLK inhibitor SM08502 is reported to inhibit cancer-progression by reducing Wnt pathway gene expression [165]. Many CLK inhibitors also show potent activity on DYRKs [166]. For example, INDY [167], a close relative of TG003, also shows potent activity on DYRK1A. In parallel, efforts to discover other inhibitors of SR protein phosphorylation resulted in the development of SRPK inhibitors, which include SRPIN340 [168] and trifluoroanilino-disubstituted furans [169]. Additionally, In screening for small molecules for modulating RNA splicing in hepatocellular carcinoma cell line Huh-7, amiloride was found to induce isoforms of BCL-x, HIPK3, and RON/MISTR1 transcripts with less oncogenic properties [170].
Although therapeutic targeting of splicing factor kinases poses a promising strategy to revert the pathogenic splicing events, the rising concern over potential off-target effects of splicing factor kinase inhibition. High throughput next-generation sequencing will allow for a more comprehensive understanding of changes to the transcriptome in response to kinase inhibition and provide insight into these potential off-target effects. In fact, an understanding of transcriptome changes in response to alterations in all splicing factor kinases would help identify which kinase, or combination of kinases represents the best therapeutic target, which kinases to avoid due to potential off-target effects, and the degree of redundancy between kinases which could be a potential mechanism of resistance for splicing kinase inhibition.
Artificial splicing factors
The latest advancement in the field of specific modulation of AS happens to be the use of artificial splicing factors (ASFs). It all started with the landmark study by Wang et al. [171] in 2009, wherein the authors engineered a ‘designer’ splicing factor by fusing a sequence-specific RNA-binding module (PUF domain of human Pumilio1) with functional domains of endogenous splicing factors. When they applied these ASFs to modulate the AS of Bcl-X, it augmented the Bcl-xS variant, thereby favoring apoptosis and fostering chemotherapeutic sensitivity. Further studies have achieved successful modulation of AS events by combining various other RNA-binding modules (e.g., MS2 coat protein (MCP), PP7 coat protein (PCP), and λN) with RNA regulatory proteins [172–174]. In a very recent study, Du et al. adopted a distinct strategy for splicing adjustments, which does not require extensive protein engineering. They devised CRISPR Artificial Splicing Factors (CASFx) by stitching RNA-targeting Cas proteins with splicing regulatory domains. The CASFx system was capable of modulating SMN2 splicing n spinal muscular atrophy (SMA) fibroblast cells to induce the inclusion of exon 7 [175]. Taken together, ASFs offer an excellent arsenal of tools to target virtually any AS event, including the ones induced by hypoxia.
Concluding remarks and future directions
Extensive studies have clearly established the tissue-hypoxia as a master regulator of alternative splicing, which often exhibits tremendous implications in pathological conditions. Hypoxia-elicited AS is more common in the context of various cancer hallmarks than other maladies, probably because hypoxia and AS are mostly studied in cancer. Nevertheless, other clinical conditions, including neurological disorders and cardiovascular disease, have been linked to a growing number of hypoxia-elicited AS events. Obviously, hypoxia-induced AS provides a remarkable set of biomarkers of prognostic potential and therapeutic targets. Targeted modulation of the hypoxia signaling with the intent of manipulating the hypoxia-driven AS is of great interest in several cancers. However, future studies will need to dissect the proper molecular mechanisms to determine useful treatment paradigms. Although many of the proposed molecular targets appear promising, it requires more stringent in-vivo studies to extrapolate to clinical applications.
Acknowledgements
This work is supported by DBT/Wellcome Trust India Alliance Fellowship Grant IA/I/16/2/502719 [to S.S.]. S.N. was supported by the Department of Science and Technology, Ministry of Science and Technology. C.A. was supported by Indian Institute of Science Education and Research Bhopal.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dales J-P, Beaufils N, Silvy M, Picard C, Pauly V, Pradel V, Formisano-Tréziny C, Bonnier P, Giusiano S, Charpin C. Hypoxia inducible factor 1α gene (HIF-1α) splice variants: potential prognostic biomarkers in breast cancer. BMC Med. 2010;8(1):44. doi: 10.1186/1741-7015-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fandrey J. Hypoxia-inducible gene expression. Respir Physiol. 1995;101(1):1–10. doi: 10.1016/0034-5687(95)00013-4. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988;242(4884):1412–1415. doi: 10.1126/science.2849206. [DOI] [PubMed] [Google Scholar]
- 5.Semenza G, Roth P, Fang H, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. Transcriptional regulation by hypoxia-inducible factor 1 molecular mechanisms of oxygen homeostasis. Trends Cardiovasc Med. 1996;6(5):151–157. doi: 10.1016/1050-1738(96)00039-4. [DOI] [PubMed] [Google Scholar]
- 7.Jiang B-H, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol-Cell Physiol. 1996;271(4):C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- 8.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414(6863):550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin W, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Kaelin WG., Jr Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 11.Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Nat Acad Sci. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian H. Hammer RE, Matsumoto AM, Russell DW, and McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F. Loss of HIF-2α and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8(7):702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- 14.Peng J, Zhang L, Drysdale L, Fong G-H. The transcription factor EPAS-1/hypoxia-inducible factor 2α plays an important role in vascular remodeling. Proc Natl Acad Sci. 2000;97(15):8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scortegagna M, Morris MA, Oktay Y, Bennett M, Garcia JA. The HIF family member EPAS1/HIF-2α is required for normal hematopoiesis in mice. Blood. 2003;102(5):1634–1640. doi: 10.1182/blood-2003-02-0448. [DOI] [PubMed] [Google Scholar]
- 16.Holmquist-Mengelbier L, Fredlund E, Löfstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg Å, Gradin K. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10(5):413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, Peet DJ. Cell-specific regulation of hypoxia-inducible factor (HIF)-1α and HIF-2α stabilization and transactivation in a graded oxygen environment. J Biol Chem. 2006;281(32):22575–22585. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Huang X, Luo Z, He J, Haider F, Song C, Peng L, Chen T, Wu B. Hypoxia-activated PI3K/Akt inhibits oxidative stress via the regulation of reactive oxygen species in human dental pulp cells. Oxid Med Cell Longev. 2019;2019:1. doi: 10.1155/2019/6595189. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Minet E, Arnould T, Michel G, Roland I, Mottet D, Raes M, Remacle J, Michiels C. ERK activation upon hypoxia: involvement in HIF-1 activation. FEBS Lett. 2000;468(1):53–58. doi: 10.1016/S0014-5793(00)01181-9. [DOI] [PubMed] [Google Scholar]
- 20.Mingyuan X, Qianqian P, Shengquan X, Chenyi Y, Rui L, Yichen S, Jinghong X. Hypoxia-inducible factor-1α activates transforming growth factor-β1/Smad signaling and increases collagen deposition in dermal fibroblasts. Oncotarget. 2018;9(3):3188. doi: 10.18632/oncotarget.23225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu W, Zhou W, Cheng M, Wang J, Liu Z, He S, Luo X, Huang W, Chen T, Yan W. Hypoxia activates Wnt/β-catenin signaling by regulating the expression of BCL9 in human hepatocellular carcinoma. Sci Rep. 2017;7(1):1–13. doi: 10.1038/srep40446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Ignazio L, Bandarra D, Rocha S. NF-κB and HIF crosstalk in immune responses. FEBS J. 2016;283(3):413–424. doi: 10.1111/febs.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koong AC, Chen EY, Giaccia AJ. Hypoxia causes the activation of nuclear factor κB through the phosphorylation of IκBα on tyrosine residues. Can Res. 1994;54(6):1425–1430. [PubMed] [Google Scholar]
- 24.Agani F, Jiang B-H. Oxygen-independent regulation of HIF-1: novel involvement of PI3K/AKT/mTOR pathway in cancer. Curr Cancer Drug Targets. 2013;13(3):245–251. doi: 10.2174/1568009611313030003. [DOI] [PubMed] [Google Scholar]
- 25.Du J, Xu R, Hu Z, Tian Y, Zhu Y, Gu L, Zhou L. PI3K and ERK-induced Rac1 activation mediates hypoxia-induced HIF-1α expression in MCF-7 breast cancer cells. PLoS ONE. 2011;6(9):e25213. doi: 10.1371/journal.pone.0025213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4(12):960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 27.Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. Amsterdam: Elsevier; 1994. [DOI] [PubMed] [Google Scholar]
- 28.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol Cell. 1998;2(6):773–785. doi: 10.1016/S1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 29.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;3(11):363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 32.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417(1):15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 33.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdisciplin Rev RNA. 2012;3(1):104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konieczny P, Stepniak-Konieczna E, Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42(17):10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94(1):5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 36.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444(7119):580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 37.Fu X-D, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15(10):689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan Q. Shai, o., lee, lJ, Frey, bJ & blencowe, bJ (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 39.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biamonti G, Bonomi S, Gallo S, Ghigna C. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT) Cell Mol Life Sci. 2012;69(15):2515–2526. doi: 10.1007/s00018-012-0931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sveen A, Kilpinen S, Ruusulehto A, Lothe R, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene. 2016;35(19):2413–2427. doi: 10.1038/onc.2015.318. [DOI] [PubMed] [Google Scholar]
- 42.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16(6):670. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 43.Kanopka A. Cell survival: Interplay between hypoxia and pre-mRNA splicing. Exp Cell Res. 2017;356(2):187–191. doi: 10.1016/j.yexcr.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama K, Kataoka N. Regulation of gene expression under hypoxic conditions. Int J Mol Sci. 2019;20(13):3278. doi: 10.3390/ijms20133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farina AR, Cappabianca L, Sebastiano M, Zelli V, Guadagni S, Mackay AR. Hypoxia-induced alternative splicing: the 11th Hallmark of Cancer. J Exp Clin Cancer Res. 2020;39(1):1–30. doi: 10.1186/s13046-020-01616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Meta Rev. 2007;26(2):225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9(8):556–570. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Han L, Dong Y, Tan Y, Li Y, Zhao M, Xie H, Ju H, Wang H, Zhao Y. EGFRvIII/integrin β3 interaction in hypoxic and vitronectinenriching microenvironment promote GBM progression and metastasis. Oncotarget. 2016;7(4):4680. doi: 10.18632/oncotarget.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mardy S, Miura Y, Endo F, Matsuda I, Sztriha L, Frossard P, Moosa A, Ismail EA, Macaya A, Andria G. Congenital insensitivity to pain with anhidrosis: novel mutations in the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor. AM J Hum Genet. 1999;64(6):1570–1579. doi: 10.1086/302422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riffo-Campos ÁL, Gimeno-Valiente F, Rodríguez FM, Cervantes A, López-Rodas G, Franco L, Castillo J. Role of epigenetic factors in the selection of the alternative splicing isoforms of human KRAS in colorectal cancer cell lines. Oncotarget. 2018;9(29):20578. doi: 10.18632/oncotarget.25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voice JK, Klemke RL, Le A, Jackson JH. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem. 1999;274(24):17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 54.Plowman SJ, Arends MJ, Brownstein DG, Luo F, Devenney PS, Rose L, Ritchie A-M, Berry RL, Harrison DJ, Hooper ML. The K-Ras 4A isoform promotes apoptosis but does not affect either lifespan or spontaneous tumor incidence in aging mice. Exp Cell Res. 2006;312(1):16–26. doi: 10.1016/j.yexcr.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Luo F, Ye H, Hamoudi R, Dong G, Zhang W, Patek CE, Poulogiannis G, Arends MJ. K-ras exon 4A has a tumour suppressor effect on carcinogen-induced murine colonic adenoma formation. J Pathol J Pathol Soc GB Irel. 2010;220(5):542–550. doi: 10.1002/path.2672. [DOI] [PubMed] [Google Scholar]
- 56.Sena JA, Wang L, Heasley LE, Hu C-J. Hypoxia regulates alternative splicing of HIF and non-HIF target genes. Mol Cancer Res. 2014;12(9):1233–1243. doi: 10.1158/1541-7786.MCR-14-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plowman S, Berry R, Bader S, Luo F, Arends M, Harrison D, Hooper M, Patek C. K-ras 4A and 4B are co-expressed widely in human tissues, and their ratio is altered in sporadic colorectal cancer. J Exp Clin Cancer Res. 2006;25(2):259–267. [PubMed] [Google Scholar]
- 58.Chen W-C, To MD, Westcott PM, Delrosario R, Kim I-J, Philips M, Tran Q, Bayani N, Balmain A (2019) Regulation of KRAS4A/B splicing in cancer stem cells by the RBM39 splicing complex. Preprint at bioRxiv. 10.1101/646125
- 59.Bowler E, Porazinski S, Uzor S, Thibault P, Durand M, Lapointe E, Rouschop KM, Hancock J, Wilson I, Ladomery M. Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC cancer. 2018;18(1):355. doi: 10.1186/s12885-018-4227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y, Liang L, He X. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol Cancer. 2020;19(1):1–17. doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramchandani D, Unruh D, Lewis CS, Bogdanov VY, Weber GF. Activation of carbonic anhydrase IX by alternatively spliced tissue factor under late-stage tumor conditions. Lab Invest. 2016;96(12):1234–1245. doi: 10.1038/labinvest.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 64.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Investig. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 66.Ahuja N, Ashok C, Natua S, Pant D, Cherian A, Pandkar MR, Yadav P, Narayanan SSV, Mishra J, Samaiya A, Shukla S. Hypoxia-induced TGF-β–RBFOX2–ESRP1 axis regulates human MENA alternative splicing and promotes EMT in breast cancer. NAR Cancer. 2020;2(3):1–17. doi: 10.1093/narcan/zcaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 68.Auvinen P, Tammi R, Tammi M, Johansson R, Kosma VM. Expression of CD44s, CD44v3 and CD44v6 in benign and malignant breast lesions: correlation and colocalization with hyaluronan. Histopathology. 2005;47(4):420–428. doi: 10.1111/j.1365-2559.2005.02220.x. [DOI] [PubMed] [Google Scholar]
- 69.Saito S, Okabe H, Watanabe M, Ishimoto T, Iwatsuki M, Baba Y, Tanaka Y, Kurashige J, Miyamoto Y, Baba H. CD44v6 expression is related to mesenchymal phenotype and poor prognosis in patients with colorectal cancer. Oncol Rep. 2013;29(4):1570–1578. doi: 10.3892/or.2013.2273. [DOI] [PubMed] [Google Scholar]
- 70.Krishnamachary B, Penet M-F, Nimmagadda S, Mironchik Y, Raman V, Solaiyappan M, Semenza GL, Pomper MG, Bhujwalla ZM. Hypoxia regulates CD44 and its variant isoforms through HIF-1α in triple negative breast cancer. PLoS ONE. 2012;7(8):e44078. doi: 10.1371/journal.pone.0044078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Babic A. Kiree a ML, Kolesniko a TV and Lau LF: CYR61, a product of a growth factor-inducible immediate early gene, promotes angiogenesis and tumor growth. Proc Natl Acad Sci USA. 1998;95:6355–6360. doi: 10.1073/pnas.95.11.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y-T, Lan Q, Lorusso G, Duffey N, Rüegg C. The matricellular protein CYR61 promotes breast cancer lung metastasis by facilitating tumor cell extravasation and suppressing anoikis. Oncotarget. 2017;8(6):9200. doi: 10.18632/oncotarget.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirschfeld M, zur Hausen A, Bettendorf H, Jäger M, Stickeler E, Alternative splicing of Cyr61 is regulated by hypoxia and significantly changed in breast cancer. Can Res. 2009;69(5):2082–2090. doi: 10.1158/0008-5472.CAN-08-1997. [DOI] [PubMed] [Google Scholar]
- 74.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1(1):27–30. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 75.McCarthy N. ALK takes the rap. Nat Rev Cancer. 2008;8(11):833–833. doi: 10.1038/nrc2529. [DOI] [Google Scholar]
- 76.Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1α versus HIF-2α in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Can Res. 2006;66(12):6264–6270. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- 77.Nowak D, Amin E, Rennel E, Hoareau-Aveilla C. Gam¬ mons M, Damodoran G, Hagiwara M. Harper SJ, Woolard J, Ladomery MR, Bates DO Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angio genesis. J Biol Chem. 2010;285:5532–5540. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Houck KA, Leung D, Rowland A, Winer J, Ferrara N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J Biol Chem. 1992;267(36):26031–26037. doi: 10.1016/S0021-9258(18)35712-0. [DOI] [PubMed] [Google Scholar]
- 79.Pritchard-Jones R, Dunn D, Qiu Y, Varey A, Orlando A, Rigby H, Harper S, Bates D. Expression of VEGF xxx b, the inhibitory isoforms of VEGF, in malignant melanoma. Br J Cancer. 2007;97(2):223–230. doi: 10.1038/sj.bjc.6603839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barratt SL, Blythe T, Ourradi K, Jarrett C, Welsh GI, Bates DO, Millar AB. Effects of hypoxia and hyperoxia on the differential expression of VEGF-A isoforms and receptors in Idiopathic Pulmonary Fibrosis (IPF) Respir Res. 2018;19(1):1–5. doi: 10.1186/s12931-017-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu Y, Hoareau-Aveilla C, Oltean S, Harper SJ, Bates DO (2009) The anti-angiogenic isoforms of VEGF in health and disease. Portland Press Ltd., [DOI] [PMC free article] [PubMed]
- 82.Nowak D, Woolard J, Amin EM, Konopatskaya O, Saleem MA, Churchill AJ, Ladomery MR, Harper SJ, Bates DO. Expression of proand anti-angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J Cell Sci. 2008;121:3487–4349. doi: 10.1242/jcs.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jakubauskiene E, Vilys L, Makino Y, Poellinger L, Kanopka A. Increased serine-arginine (SR) protein phosphorylation changes pre-mRNA splicing in hypoxia. J Biol Chem. 2015;290(29):18079–18089. doi: 10.1074/jbc.M115.639690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Can Res. 2007;67(5):2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- 85.Semenza GL, Jiang B-H, Leung SW, Passantino R, Concordet J-P, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 86.Han J, Li J, Ho JC, Chia GS, Kato H, Jha S, Yang H, Poellinger L, Lee KL. Hypoxia is a key driver of alternative splicing in human breast cancer cells. Sci Rep. 2017;7(1):1–17. doi: 10.1038/s41598-017-04333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin cancer biol. 2005;4:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Yang P. Li Z (2014) The multifaceted regulation and functions of PKM2 in tumor progression. Biochimica et Biophysica Acta (BBA) Rev Cancer. 1846;2:285–296. doi: 10.1016/j.bbcan.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 89.Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prigione A, Rohwer N, Hoffmann S, Mlody B, Drews K, Bukowiecki R, Blümlein K, Wanker EE, Ralser M, Cramer T, Adjaye J. HIF1α modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1–3 and PKM2. Stem Cells. 2014;32:364–376. doi: 10.1002/stem.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Minchenko O, Opentanova I, Caro J. Hypoxic regulation of the 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase gene family (PFKFB-1–4) expression in vivo. FEBS Lett. 2003;554(3):264–270. doi: 10.1016/S0014-5793(03)01179-7. [DOI] [PubMed] [Google Scholar]
- 92.Minchenko OH, Ogura T, Opentanova IL, Minchenko DO, Esumi H. Splice isoform of 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-4: expression and hypoxic regulation. Mol Cell Biochem. 2005;280(1–2):227–234. doi: 10.1007/s11010-005-8009-6. [DOI] [PubMed] [Google Scholar]
- 93.Mykhalchenko V, Minchenko D, Tsuchihara K, Moenner M, Komisarenko S, Bikfalvi A, Esumi H, Minchenko O. Expression of mouse 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3 mRNA alternative splice variants in hypoxia. Ukr Biokhim Zh. 2008;80(1):19. [PubMed] [Google Scholar]
- 94.Barsoum IB, Koti M, Siemens DR, Graham CH. Mechanisms of hypoxia-mediated immune escape in cancer. Can Res. 2014;74(24):7185–7190. doi: 10.1158/0008-5472.CAN-14-2598. [DOI] [PubMed] [Google Scholar]
- 95.Li Y, Patel SP, Roszik J, Qin Y. Hypoxia-driven immunosuppressive metabolites in the tumor microenvironment: new approaches for combinational immunotherapy. Front immunol. 2018;9:1591. doi: 10.3389/fimmu.2018.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Noman MZ, Hasmim M, Messai Y, Terry S, Kieda C, Janji B, Chouaib S. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am J Physiol-Cell Physiol. 2015;309(9):C569–C579. doi: 10.1152/ajpcell.00207.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang J-G, Belz GT. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461(7264):659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheng J, Zhou T, Liu C, Shapiro JP, Brauer MJ, Kiefer MC, Barr PJ, Mountz JD. Protection from Fas-mediated apoptosis by a soluble form of the Fas molecule. Science. 1994;263(5154):1759–1762. doi: 10.1126/science.7510905. [DOI] [PubMed] [Google Scholar]
- 99.Zheng Z. A complete checklist of species and subspecies of the Chinese birds. China: Science Press; 1994. [Google Scholar]
- 100.Hohlbaum AM, Moe S, Marshak-Rothstein A. Opposing effects of transmembrane and soluble Fas ligand expression on inflammation and tumor cell survival. J Exp Med. 2000;191(7):1209–1220. doi: 10.1084/jem.191.7.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peciuliene I, Vilys L, Jakubauskiene E, Zaliauskiene L, Kanopka A. Hypoxia alters splicing of the cancer associated Fas gene. Exp Cell Res. 2019;380(1):29–35. doi: 10.1016/j.yexcr.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 102.Labiano S, Palazón A, Bolanos E, Azpilikueta A, Sánchez-Paulete AR, Morales-Kastresana A, Quetglas JI, Perez-Gracia JL, Gúrpide A, Rodriguez-Ruiz M. Hypoxia-induced soluble CD137 in malignant cells blocks CD137L-costimulation as an immune escape mechanism. Oncoimmunology. 2016;5(1):e1062967. doi: 10.1080/2162402X.2015.1062967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nuñez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74(4):597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 105.Xerri L, Parc P, Brousset P, Schlaifer D, Hassoun J, Reed JC, Krajewski S, Birnbaum D. Predominant expression of the long isoform of Bcl-x (Bcl-xL) in human lymphomas. Br J Haematol. 1996;92(4):900–906. doi: 10.1046/j.1365-2141.1996.423958.x. [DOI] [PubMed] [Google Scholar]
- 106.Olopade OI, Adeyanju MO, Safa AR, Hagos F, Mick R, Thompson CB, Recant WM. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J Sci Am. 1997;3(4):230–237. [PubMed] [Google Scholar]
- 107.Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34(1):55–61. doi: 10.1053/jhep.2001.25387. [DOI] [PubMed] [Google Scholar]
- 108.Chen N, Chen X, Huang R, Zeng H, Gong J, Meng W, Lu Y, Zhao F, Wang L, Zhou Q. BCL-xL is a target gene regulated by hypoxia-inducible factor-1α. J Biol Chem. 2009;284(15):10004–10012. doi: 10.1074/jbc.M805997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sasabe E, Tatemoto Y, Li D, Yamamoto T, Osaki T. Mechanism of HIF-1α-dependent suppression of hypoxia-induced apoptosis in squamous cell carcinoma cells. Cancer Sci. 2005;96(7):394–402. doi: 10.1111/j.1349-7006.2005.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sermeus A, Genin M, Maincent A, Fransolet M, Notte A, Leclere L, Riquier H, Arnould T, Michiels C. Hypoxia-induced modulation of apoptosis and BCL-2 family proteins in different cancer cell types. PLoS ONE. 2012;7(11):e47519. doi: 10.1371/journal.pone.0047519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiao Q, Ford AL, Xu J, Yan P, Lee K-Y, Gonzales E, West T, Holtzman DM, Lee J-M. Bcl-x pre-mRNA splicing regulates brain injury after neonatal hypoxia-ischemia. J Neurosci. 2012;32(39):13587–13596. doi: 10.1523/JNEUROSCI.2617-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stevens M, Oltean S. Modulation of the apoptosis gene Bcl-x function through alternative splicing. Front Genet. 2019;10(804):1–9. doi: 10.3389/fgene.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bielli P, Bordi M, Biasio VD, Sette C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res. 2014;42(19):12070–12081. doi: 10.1093/nar/gku922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gang H, Hai Y, Dhingra R, Gordon JW, Yurkova N, Aviv Y, Li H, Aguilar F, Marshall A, Leygue E. A novel hypoxia-inducible spliced variant of mitochondrial death gene Bnip3 promotes survival of ventricular myocytes. Circ Res. 2011;108(9):1084–1092. doi: 10.1161/CIRCRESAHA.110.238709. [DOI] [PubMed] [Google Scholar]
- 115.Lin J-C, Tsao M-F, Lin Y-J. Differential impacts of alternative splicing networks on apoptosis. Int J Mol Sci. 2016;17(12):2097. doi: 10.3390/ijms17122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet. 2005;6(8):611–622. doi: 10.1038/nrg1656. [DOI] [PubMed] [Google Scholar]
- 117.Wright WE, Shay JW. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat Med. 2000;6(8):849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 118.Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21(2):220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 119.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer. 2001;91(5):644–649. doi: 10.1002/1097-0215(200002)9999:9999<::AID-IJC1103>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 120.Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. The major reverse transcriptase–incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Can Res. 2013;73(9):2817–2828. doi: 10.1158/0008-5472.CAN-12-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Anderson C, Hoare S, Ashcroft M, Bilsland A, Keith W. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene. 2006;25(1):61–69. doi: 10.1038/sj.onc.1209011. [DOI] [PubMed] [Google Scholar]
- 122.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 123.Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG. SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11(7):511. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 124.Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KK, Karp GM, Qi H, Woll MG. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345(6197):688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 125.Wang J, Schultz PG, Johnson KA. Mechanistic studies of a small-molecule modulator of SMN2 splicing. Proc Natl Acad Sci. 2018;115(20):E4604–E4612. doi: 10.1073/pnas.1800260115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bebee TW, Dominguez CE, Samadzadeh-Tarighat S, Akehurst KL, Chandler DS. Hypoxia is a modifier of SMN2 splicing and disease severity in a severe SMA mouse model. Hum Mol Genet. 2012;21(19):4301–4313. doi: 10.1093/hmg/dds263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Manabe T, Katayama T, Sato N, Gomi F, Hitomi J, Yanagita T, Kudo T, Honda A, Mori Y, Matsuzaki S. Induced HMGA1a expression causes aberrant splicing of Presenilin-2 pre-mRNA in sporadic Alzheimer's disease. Cell Death Differ. 2003;10(6):698–708. doi: 10.1038/sj.cdd.4401221. [DOI] [PubMed] [Google Scholar]
- 128.Münch C, Bg Z, Leven A, Stamm S, Einkörn H, Schwalenstöcker B, Ludolph AC, Riepe MW, Meyer T. Differential regulation of 5′ splice variants of the glutamate transporter EAAT2 in an in vivo model of chemical hypoxia induced by 3-nitropropionic acid. J Neurosci Res. 2003;71(6):819–825. doi: 10.1002/jnr.10536. [DOI] [PubMed] [Google Scholar]
- 129.Cimarosti H, Jones NM, O'Shea RD, Pow DV, Salbego C, Beart PM. Hypoxic preconditioning in neonatal rat brain involves regulation of excitatory amino acid transporter 2 and estrogen receptor alpha. Neurosci Lett. 2005;385(1):52–57. doi: 10.1016/j.neulet.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 130.Pow DV, Naidoo T, Lingwood BE, Healy GN, Williams SM, Sullivan RK, O'Driscoll S, Colditz PB. Loss of glial glutamate transporters and induction of neuronal expression of GLT-1B in the hypoxic neonatal pig brain. Dev Brain Res. 2004;153(1):1–11. doi: 10.1016/j.devbrainres.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 131.Martin LJ, Brambrink AM, Lehmann C, Portera-Cailliau C, Koehler R, Rothstein J, Traystman RJ. Hypoxia—ischemia causes abnormalities in glutamate transporters and death of astroglia and neurons in newborn striatum. Ann Neurol Off J Am Neurol Assoc Child Neurol Soc. 1997;42(3):335–348. doi: 10.1002/ana.410420310. [DOI] [PubMed] [Google Scholar]
- 132.Xue L-L, Wang F, Xiong L-L, Du R-L, Zhou H-L, Zou Y, Wu M-X, Yang M-A, Dai J, He M-X. A single-nucleotide polymorphism induced alternative splicing in Tacr3 involves in hypoxic-ischemic brain damage. Brain Res Bull. 2020;154:106–115. doi: 10.1016/j.brainresbull.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 133.Shimokawa H, Yasuda S. Myocardial ischemia: current concepts and future perspectives. J Cardiol. 2008;52(2):67–78. doi: 10.1016/j.jjcc.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 134.Williams AL, Khadka V, Tang M, Avelar A, Schunke KJ, Menor M, Shohet RV. Genomic and “Polyomic” Studies of Cardiovascular and Inflammatory Diseases: HIF1 mediates a switch in pyruvate kinase isoforms after myocardial infarction. Physiol Genomics. 2018;50(7):479. doi: 10.1152/physiolgenomics.00130.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mirtschink P, Krishnan J, Grimm F, Sarre A, Hörl M, Kayikci M, Fankhauser N, Christinat Y, Cortijo C, Feehan O. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 2015;522(7557):444–449. doi: 10.1038/nature14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choi Y-J, Kim I, Lee JE, Park J-W. PIN1 transcript variant 2 acts as a long non-coding RNA that controls the HIF-1-driven hypoxic response. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-47071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jeong W, Rapisarda A, Park SR, Kinders RJ, Chen A, Melillo G, Turkbey B, Steinberg SM, Choyke P, Doroshow JH. Pilot trial of EZN-2968, an antisense oligonucleotide inhibitor of hypoxia-inducible factor-1 alpha (HIF-1α), in patients with refractory solid tumors. Cancer Chemother Pharmacol. 2014;73(2):343–348. doi: 10.1007/s00280-013-2362-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coltella N, Valsecchi R, Ponente M, Ponzoni M, Bernardi R. Synergistic Leukemia Eradication by Combined Treatment with Retinoic Acid and HIF Inhibition by EZN-2208 (PEG-SN38) in Preclinical Models of PML-RARα and PLZF-RARα–Driven Leukemia. Clin Cancer Res. 2015;21(16):3685–3694. doi: 10.1158/1078-0432.CCR-14-3022. [DOI] [PubMed] [Google Scholar]
- 139.Rapisarda A, Uranchimeg B, Sordet O, Pommier Y, Shoemaker RH, Melillo G. Topoisomerase I-mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Can Res. 2004;64(4):1475–1482. doi: 10.1158/0008-5472.CAN-03-3139. [DOI] [PubMed] [Google Scholar]
- 140.Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, Fisher RJ, Shoemaker RH, Melillo G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Can Res. 2005;65(19):9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 141.Pang Y, Yang C, Schovanek J, Wang H, Bullova P, Caisova V, Gupta G, Wolf KI, Semenza GL, Zhuang Z. Anthracyclines suppress pheochromocytoma cell characteristics, including metastasis, through inhibition of the hypoxia signaling pathway. Oncotarget. 2017;8(14):22313. doi: 10.18632/oncotarget.16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lang SA, Moser C, Gaumann A, Klein D, Glockzin G, Popp FC, Dahlke MH, Piso P, Schlitt HJ, Geissler EK. Targeting heat shock protein 90 in pancreatic cancer impairs insulin-like growth factor-I receptor signaling, disrupts an interleukin-6/signal-transducer and activator of transcription 3/hypoxia-inducible factor-1α autocrine loop, and reduces orthotopic tumor growth. Clin Cancer Res. 2007;13(21):6459–6468. doi: 10.1158/1078-0432.CCR-07-1104. [DOI] [PubMed] [Google Scholar]
- 143.Bohonowych JE, Peng S, Gopal U, Hance MW, Wing SB, Argraves KM, Lundgren K, Isaacs JS. Comparative analysis of novel and conventional Hsp90 inhibitors on HIF activity and angiogenic potential in clear cell renal cell carcinoma: implications for clinical evaluation. BMC Cancer. 2011;11(1):520. doi: 10.1186/1471-2407-11-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Staab A, Loeffler J, Said HM, Diehlmann D, Katzer A, Beyer M, Fleischer M, Schwab F, Baier K, Einsele H. Effects of HIF-1 inhibition by chetomin on hypoxia-related transcription and radiosensitivity in HT 1080 human fibrosarcoma cells. BMC Cancer. 2007;7(1):1–7. doi: 10.1186/1471-2407-7-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minegishi H, Fukashiro S, Ban HS, Nakamura H. Discovery of indenopyrazoles as a new class of hypoxia inducible factor (HIF)-1 inhibitors. ACS Med Chem Lett. 2013;4(2):297–301. doi: 10.1021/ml3004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee K, Kang JE, Park S-K, Jin Y, Chung K-S, Kim H-M, Lee K, Kang MR, Lee MK, Song KB. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1α via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol. 2010;80(7):982–989. doi: 10.1016/j.bcp.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 147.Carvalho T, Martins S, Rino J, Marinho S, Carmo-Fonseca M. Pharmacological inhibition of the spliceosome subunit SF3b triggers exon junction complex-independent nonsense-mediated decay. J Cell Sci. 2017;130(9):1519–1531. doi: 10.1242/jcs.202200. [DOI] [PubMed] [Google Scholar]
- 148.Sidarovich A, Will CL, Anokhina MM, Ceballos J, Sievers S, Agafonov DE, Samatov T, Bao P, Kastner B, Urlaub H. Identification of a small molecule inhibitor that stalls splicing at an early step of spliceosome activation. Elife. 2017;6:e23533. doi: 10.7554/eLife.23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.O'Brien K, Matlin AJ, Lowell AM, Moore MJ. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem. 2008;283(48):33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pawellek A, McElroy S, Samatov T, Mitchell L, Woodland A, Ryder U, Gray D, Lührmann R, Lamond AI. Identification of small molecule inhibitors of pre-mRNA splicing. J Biol Chem. 2014;289(50):34683–34698. doi: 10.1074/jbc.M114.590976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bashaw ED, Huang S-M, Coté TR, Pariser AR, Garnett CE, Burckart G, Zhang L, Men AY, Le CD, Charlab R. Clinical pharmacology as a cornerstone of orphan drug development. Nat Rev Drug Discov. 2011;10(11):795–796. doi: 10.1038/nrd3595. [DOI] [PubMed] [Google Scholar]
- 152.Zhou HM, Slominski RM, Seymour LJ, Bell MC, Dave P, Atumonye J, Wright W, III, Dawes A, Griesenauer B, Paczesny S. Ex vivo culture of mouse skin activates an interleukin 1 alpha-dependent inflammatory response. Exp Dermatol. 2020;29(1):102–106. doi: 10.1111/exd.14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Matsuo M, Masumura T, Nishio H, Nakajima T, Kitoh Y, Takumi T, Koga J, Nakamura H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Investig. 1991;87(6):2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, Van Deutekom J, van Ommen GJ, Den Dunnen JT. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30(3):293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 156.Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A. Antisense oligonucleotide–mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Investig. 2016;126(1):68–84. doi: 10.1172/JCI82534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rinaldi C, Wood MJ. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 158.Cloe A, Chen L, Li Y, Liu H, Cheng JX. Identification of Specific Hnrnps As Novel Therapeutic Targets and Responsive Indicators of KPT330 (selinexor) in Leukemia. Washington, DC: American Society of Hematology; 2016. [Google Scholar]
- 159.Deng J, Chen S, Wang F, Zhao H, Xie Z, Xu Z, Zhang Q, Liang P, Zhai X, Cheng Y. Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol Neurobiol. 2016;53(2):1132–1144. doi: 10.1007/s12035-014-9080-3. [DOI] [PubMed] [Google Scholar]
- 160.Suk F-M, Lin S-Y, Lin R-J, Hsine Y-H, Liao Y-J, Fang S-U, Liang Y-C. Bortezomib inhibits Burkitt's lymphoma cell proliferation by downregulating sumoylated hnRNP K and c-Myc expression. Oncotarget. 2015;6(28):25988. doi: 10.18632/oncotarget.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J-i, Murray MV, Kimura H. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279(23):24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- 162.Nishida A, Kataoka N, Takeshima Y, Yagi M, Awano H, Ota M, Itoh K, Hagiwara M, Matsuo M. Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nature Commun. 2011;2(1):1–8. doi: 10.1038/ncomms1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sako Y, Ninomiya K, Okuno Y, Toyomoto M, Nishida A, Koike Y, Ohe K, Kii I, Yoshida S, Hashimoto N. Development of an orally available inhibitor of CLK1 for skipping a mutated dystrophin exon in Duchenne muscular dystrophy. Sci Rep. 2017;7:46126. doi: 10.1038/srep46126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lee JY, Yun J-S, Kim W-K, Chun H-S, Jin H, Cho S, Chang JH. Structural basis for the selective inhibition of Cdc2-like kinases by CX-4945. BioMed Res Int. 2019;2019:1. doi: 10.1155/2019/6125068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tam BY, Chiu K, Chung H, Bossard C, Nguyen JD, Creger E, Eastman BW, Mak CC, Ibanez M, Ghias A. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 2020;473:186–197. doi: 10.1016/j.canlet.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 166.Bates DO, Morris JC, Oltean S, Donaldson LF. Pharmacology of modulators of alternative splicing. Pharmacol Rev. 2017;69(1):63–79. doi: 10.1124/pr.115.011239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ogawa Y, Nonaka Y, Goto T, Ohnishi E, Hiramatsu T, Kii I, Yoshida M, Ikura T, Onogi H, Shibuya H. Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A. Nature Commun. 2010;1(1):1–9. doi: 10.1038/ncomms1090. [DOI] [PubMed] [Google Scholar]
- 168.Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci. 2006;103(30):11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gammons MV, Fedorov O, Ivison D, Du C, Clark T, Hopkins C, Hagiwara M, Dick AD, Cox R, Harper SJ. Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD. Invest Ophthalmol Vis Sci. 2013;54(9):6052–6062. doi: 10.1167/iovs.13-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chang J-G, Yang D-M, Chang W-H, Chow L-P, Chan W-L, Lin H-H, Huang H-D, Chang Y-S, Hung C-H, Yang W-K. Small molecule amiloride modulates oncogenic RNA alternative splicing to devitalize human cancer cells. PLoS ONE. 2011;6(6):e18643. doi: 10.1371/journal.pone.0018643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Y, Cheong C-G, Hall TMT, Wang Z. Engineering splicing factors with designed specificities. Nat Methods. 2009;6(11):825–830. doi: 10.1038/nmeth.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sun S, Zhang Z, Fregoso O, Krainer AR. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA. 2012;18(2):274–283. doi: 10.1261/rna.030486.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3(1):1–8. doi: 10.1038/ncomms2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G. N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Du M, Jillette N, Zhu JJ, Li S, Cheng AW. CRISPR artificial splicing factors. Nature. Communications. 2020;11(1):1–11. doi: 10.1038/s41467-020-16806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]