Abstract
Osteoarthritis (OA) is mainly characterized by articular cartilage degeneration, synovial fibrosis, and inflammation. LncRNA CRNDE (colorectal neoplasia differentially expressed) has been reported to be down-regulated in age-related OA, but its role in injury-induced OA needs to be further explored. In this study, an OA rat model was established using anterior cruciate ligament transection, and the adenovirus-mediated CRNDE overexpression (Ad-CRNDE) or DACT1 (dapper antagonist of catenin-1) interference (sh-DACT1) vectors were administered by intraarticular injection. Moreover, chondrocyte‑like ATDC5 cells were treated with IL-1β (10 ng/mL) to simulate OA conditions in vitro. We found that overexpression of CRNDE alleviated cartilage damage and synovitis in OA rats, and suppressed IL-1β-induced apoptosis, inflammation, and extracellular matrix (ECM) degradation in chondrocyte‑like ATDC5 cells, while silencing DACT1 effectively antagonized the protective effect of CRNDE both in vivo and in vitro. Mechanism studies revealed that DACT1 could act as a downstream target of CRNDE. By recruiting p300, CRNDE promoted the enrichment of H3K27ac in the DACT1 promoter, thus promoting DACT1 transcription. In addition, CRNDE hindered the activation of the Wnt/β-catenin pathway in IL-1β-stimulated cells by inducing DACT1 expression. In conclusion, CRNDE promoted DACT1 expression through epigenetic modification and restrained the activation of Wnt/β-catenin signaling to impede the progression of OA.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04427-7.
Keywords: lncRNA CRNDE, DACT1, H3K27 acetylation, Osteoarthritis, Chondrocyte‑like ATDC5 cells
Introduction
Osteoarthritis (OA) is a degenerative disease involving joints, and usually characterized by articular cartilage degeneration, synovial fibrosis and inflammation, subchondral bone sclerosis, and osteophyte formation [1]. In the world, among people older than 60, approximately 10% of men and 18% of women suffer from this disease [2]. The incidence of OA is high, and currently there are no effective drugs to prevent or reverse the disease progression [3]. Joint replacement is effective in the treatment of end-stage disease, but its prognosis is poor [4]. This prompted us to conduct a more in-depth study on the pathogenesis of OA, so as to provide new ideas for the early prevention of OA.
With the continuous advancement of high-throughput technologies, such as transcriptome and whole-genome sequencing, people have gradually realized that non-coding RNAs account for the vast majority (98%) of the human genome, and protein-coding RNAs account for only a small portion (2%) [5]. Long non-coding RNAs (lncRNAs) are RNA molecules with limited protein-coding ability due to the lack of open reading frames, and their length is greater than 200 nucleotides [6]. Recently, lncRNAs as novel non-coding RNAs have received widespread attention. Growing evidence indicates that some lncRNAs are involved in many biological and pathological regulation processes through epigenetic regulation (including acetylation, methylation and ubiquitination) [7–9].
LncRNA CRNDE (colorectal neoplasia differentially expressed) has been reported to play a carcinogenic effect in various human cancers, including colorectal cancer [10], liver cancer [11], osteosarcoma [12], and cervical cancer [13]. The mechanisms by which CRNDE works are multiple, and a previous research demonstrated that CRNDE can promote the progression of colorectal cancer through epigenetic silencing gene expression [14]. In addition, CRNDE has been identified as a key gene in age-related OA, with decreased expression in OA [15]. However, the role of CRNDE in injury-related OA and the mechanisms involved need to be further studied.
It has been reported that the classic Wnt/β-catenin signaling pathway is abnormally activated in OA, and effective inhibitors of the Wnt pathway can alleviate the severity of OA and reduce cartilage degeneration and synovitis in vivo [16]. The Dapper Antagonist of Catenin-1 (DACT1) gene is located at 14q22.3 and has been reported as an effective inhibitor of the Wnt signaling, playing an important role in the progression of many human cancers [17–19]. Moreover, SIRT1 regulates DACT1 expression through deacetylation [20]. In addition, a whole genome chip sequencing data showed that DACT1 was down-regulated in human OA knee tissues [21], suggesting its possible involvement in the occurrence of OA.
In this study, we demonstrated that CRNDE was downregulated in the OA rat model, and its high expression impeded the development of OA. CRNDE bound to DACT1, and knockdown of DACT1 functionally antagonized the protective effect of CRNDE on cartilage injury. In addition, we found that CRNDE might regulate DACT1 transcription through epigenetic modification. Our data may provide evidence for CRNDE as a potential therapeutic target for OA.
Materials and methods
Model establishment
Healthy specific pathogen-free 6-week-old Sprague Dawley (SD) rats (weighing 200 ± 20 g) were purchased from Vital River Laboratory Animal Technology (Beijing, China). Rats were fed in a clean animal room (21 ± 3 ℃; relative humidity, 50 ± 10%) with free access to food and drink under a 12 h dark/light cycle. Rats were randomly assigned into six main groups: (1) OA group (n = 8): rats underwent anterior cruciate ligament transection surgery as previously described [22] to destabilize the joints and induce OA; (2) Sham group (n = 8): a similar incision was made in the right joint capsule of the rat without cutting the ligament; (3) OA + Vector group (n = 8): On day 2 after OA induction, control vector (30 μL) was injected into the joint; (4) OA + Vector + sh-NC group (n = 8): On day 2 after OA induction, control vector (20 μL) + sh-NC (20 μL) were injected into the joint; (5) OA + Ad-CRNDE group (n = 8): On day 2 after OA induction, adenovirus-mediated CRNDE overexpression vector (30 μL) was injected into the joint; (6) OA + Ad-CRNDE + sh-DACT1 group (n = 8): On day 2 after OA induction, Ad-CRNDE (20 μL) + adenovirus-mediated DACT1 interference vector (Ad-sh-DACT1, 20 μL) were injected into the joint. All intra-articular injections were performed within a 1-cm2 area around the ligament transection every 2 weeks for 6 weeks. The dosage of adenovirus solution was 2 × 1010 pfu/mL. The rats were euthanized 6 weeks after surgery and the knee joint tissues were separated for subsequent experiments. All animal experiments were approved by the Second Affiliated Hospital of Xi’an Jiaotong University Animal Ethics Committee and were performed in accordance with the Committee’s guidelines.
Cell differentiation and culture
ATDC5 cell line was purchased from Riken BioResource Center (Tsukuba, Japan). Undifferentiated ATDC5 cells were cultured in DMEM/F12 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% fetal bovine serum (Gibco, Rockville, MD) and 1% P/S (penicillin/streptomycinat) at 37 °C in 5% CO2. When the cell confluence reached about 75%, 1% insulin–transferrin–selenite (ITS) was added to the medium, and then the medium was changed every two days to induce the cells to differentiate into chondrocyte-like cells. Cells were used for the subsequent experiments after 2 weeks of differentiation in culture. And cells were kept in monolayer culture in this study.
Isolation and culture of human primary chondrocyte
Normal articular cartilage tissues from 3 patients (aged 55–65 years) undergoing joint replacement surgery were collected for primary chondrocyte extraction. In brief, cartilage tissues were cut into small pieces, and treated respectively with 0.2% trypsin for 0.5 h and 0.2% collagenase II for 2 h. The harvested cells were placed in DMEM/F12 medium (Thermo) containing 10% fetal bovine serum (Gibco) and 1% penicillin–streptomycin and incubated at 37 °C in 5% CO2. In this study, primary cells were kept in monolayer culture. The second passage cells were seeded in 6-well plates for cell transfection or treatment.
Cell transfection
Adenovirus-mediated CRNDE, DACT1 and p300 overexpression vectors were constructed using full-length sequences of CRNDE, DACT1 and p300. The adenovirus vectors containing small hairpin RNA (shRNA) sequence targeting CRNDE (sh-CRNDE), DACT1 (sh-DACT1), p300 (sh-p300) or negative control vector (sh-NC) were amplified and cloned by GeneChem (Shanghai, China). All transfection was performed using Lipofectamine® 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Cell proliferation
Cells were seeded into 96-well plates and incubated at 37 ℃ under 5% CO2 for 24 h. After treatment, 10 μL of MTT reagent was added into each well, and then cells were incubated at 37℃ for 4–6 h. After that, the supernatant was discarded and 100 μL of DMSO was added into each well. The absorbance of each well was measured at a wavelength of 450 nm after the crystals were sufficiently dissolved.
Cell apoptosis
Apoptosis analysis was carried out using Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (Multisciences, Shanghai, China). In brief, the treated cells were washed with pre-cooled PBS and cell suspension was prepared. Next, 100 μL of cell suspension was incubated with 5 μL of Annexin V-FITC and 5 μL of PI in the dark for 20 min. Fluorescence activated cell sorting (FACS) was used to detect cell apoptosis.
ELISA
The concentrations of IL-6, IL-8, and TNF-α in cell supernatant were measured using ELISA kits (R&D Systems, USA) according to the manufacturer’s instructions. The absorbance of each well at the wavelength of 450 nm was measured. The experiment was repeated three times to obtain the mean value.
Western blotting
Total protein extracted from cells and rat knee cartilage tissues was lysed with RIPA lysis buffer. CelLytic™ NuCLEAR™ Extraction Kit (Sigma, St. Louis, MO, USA) was used to extract nuclear proteins. Protein concentrations were determined using the BCA Assay Kit (Beyotime, Shanghai, China). Equal amounts of protein were separated on 10% SDS/PAGE gels and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking in 5% skim milk at room temperature for 2 h, membranes were incubated at 4 ℃ overnight with primary antibodies including anti-ADAMTS-5 (ab41037, 1:250, Abcam, Cambridge, UK), anti-MMP13 (ab51072, 1:1000, Abcam), anti-Collagen 2 (ab188570, 1:1000, Abcam), anti-Aggrecan (ab3778, 1:1000, Abcam), anti-β-catenin (ab68183, 1:500, Abcam), anti-DACT1 (ab42547, 1:1000, Abcam), anti-c-Myc (ab32072, 1:1000, Abcam) and anti-Cyclin D1 (ab16663, 1:200, Abcam). Then, membranes were incubated with HRP-conjugated specific secondary antibodies for 2 h at room temperature. Protein band signals were measured using enhanced chemiluminescence reagents (Beyotime). Image J software was used to analyze the gray level of bands.
RT-qPCR
Total RNA was extracted from cells and rat knee cartilage and synovial tissues using TRIzol kit (Invitrogen, Carlsbad, CA, USA). 1 μg of RNA was reverse-transcribed to cDNA using a Reverse Transcription Kit (Takara, Dalian, China). Synthesized cDNAs were used for RT-qPCR using SYBR Premix Ex Taq Kit (Takara). The thermal cycling conditions were as follows: 3 min at 95 °C, and 35 cycles for 15 s at 95 °C, 30 s at 60 °C, and 15 s at 72 °C. The relative mRNA levels were normalized to GAPDH and calculated using 2−ΔΔCT method. DACT1: 5ʹ-GGA AGA GGA CAG GCT TGG AAA C-3ʹ (S) and 5ʹ-GTC CCA TTG TTC AGA GAA GGT ATC-3ʹ (R). CRNDE: 5ʹ-TGA AGG AAG GAA GTG GTG CA-3ʹ (S) and 5ʹ-TCC AGT GGC ATC CTA CAA GA-3ʹ (R).
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was carried out using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Burlington, MA) in accordance with the manufacturer’s protocol. In brief, cells were incubated with formaldehyde for 10 min to produce DNA–protein crosslinks. Next, the cell lysates were sonicated into 200–1000 bp chromatin fragments and immuno-precipitated with anti-IgG (#3900, Cell Signaling Technology, Boston, MA, USA), anti-H3K27ac (ab4729, Abcam) and anti-p300 (ab2832, Abcam).
RNA immunoprecipitation (RIP) assay
RIP assay was performed using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). In brief, the treated cells were washed by ice-cold PBS and lysed in a radioimmu-noprecipitation buffer for 30 min at 4 °C. Next, the whole cell lysate was incubated with A/G sepharose beads coupled with anti-DACT1 (Abcam) or normal human IgG (Millipore) antibodies. The precipitated RNA was analyzed by qPCR.
RNA pull-down assay
The transcribed RNA was biotin-labeled with Biotin RNA Labeling Mix (Ambio Life), treated with RNase-free DNase I (Roche, Indianapolis, IN, USA), and purified with an RNeasy Mini Kit (Qiagen, Valencia, CA). 1 mg of whole-cell lysates were incubated with 3 μg of purified biotinylated transcripts at room temperature for 1 h. And then the complex was isolated with streptavidin agarose beads (Invitrogen, San Diego, CA, USA). The eluted proteins were measured by Western blot analysis.
Fluorescence in situ hybridization analysis (FISH)
RNA FISH probe was synthesized by Rochen Pharma (Shanghai, China). Briefly, ATDC5 cells were fixed in 4% formaldehyde for 15 min, washed with PBS, treated with pepsin, and dehydrated through graded ethanol. After the cells were air-dried, they were incubated with 40 nM FISH probe in hybridization buffer for 2 min at 80 °C, followed by hybridization at 55 °C for 2 h. Subsequently, the slides were washed with 0.1 × SSC and dehydrated with graded ethanol. Then, the slides were mounted with Prolong Gold Antifade Reagent with DAPI, and a fluorescence microscope (DMI4000B, Leica) was used to observe the images.
Hematoxylin and eosin (H&E) and safranin O-fast green staining
The knee joints were fixed with 10% formalin for 12 h at room temperature, decalcified with 0.5 M EDTA at pH 8.0 for 4 weeks, and dehydrated and embedded in paraffin. The tissue sections were cut into 4–6 μm thickness along the sagittal direction, and the intervals of serial sections were 200 μm. Next, H&E and Safranin O-Fast Green staining was performed. At least five sections for each rat were selected to assess the degree of cartilage degeneration using the Osteoarthritis Research Society International (OARSI) scoring system on a scale of 0–5 [23]. The sum of the scores in the medial tibia and medial femur quadrants was averaged as the final score. Moreover, synovial membrane inflammation was evaluated using a synovial scoring system on a scale of 0–4 [23].
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay
Paraffin sections of knee joint cartilage tissues were stained using TNUEL (Roche, Basel, Switzerland) method in accordance with the manufacturer's protocol. After staining, normal cells showed purple-blue, and apoptotic cells showed yellow–brown. At least 5 sections were selected from each rat to evaluate the percentage of TUNEL-positive cells. The apoptotic cells and total cells were counted using Image J software, and apoptosis rate = 100% × (apoptotic cells / total cells).
Statistical analysis
All data in this study were analyzed using SPSS 22.0 software (IBM Corp., Armonk, NY). Data were presented as mean ± standard error of mean (mean ± SEM). The Shapiro–Wilk test was used to verify the normal distribution of data. Levene’s test was used for variance homogeneity verification. The statistical significance of groups was detected using Student’s t test and analysis of variance (ANOVA). The level of significance was set at *P < 0.05.
Results
Overexpression of CRNDE alleviated cartilage injury in an OA rat model
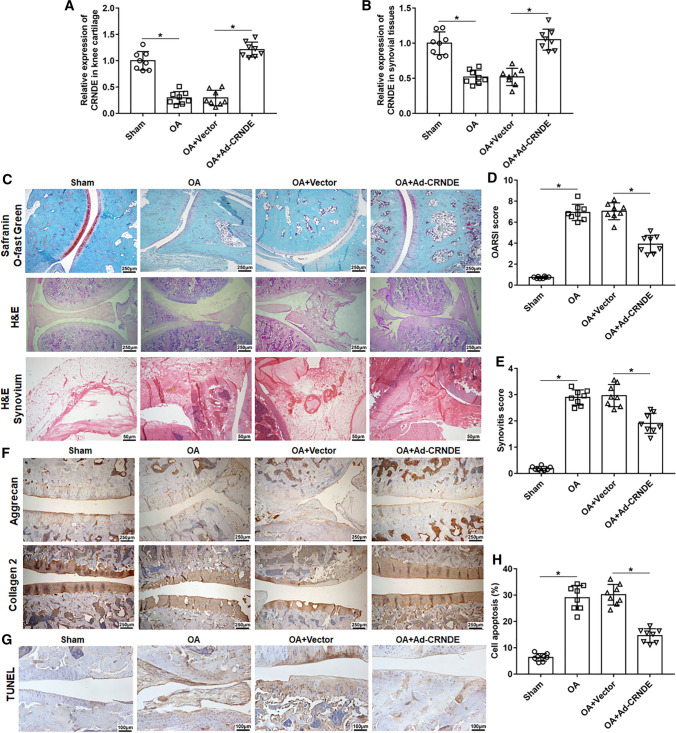
To explore the role of CRNDE in OA, we used anterior cruciate ligament transection to establish an OA rat model, and Ad-CRNDE was administered by intraarticular injection. Our results showed that CRNDE expression in knee cartilage and synovial tissues was significantly reduced in OA rats compared with that in the sham group, and the injection of Ad-CRNDE observably increased the expression of CRNDE (Fig. 1A, B). Next, optical microscopy was used to observe the pathological changes of knee cartilage in each group. As shown in Fig. 1C–E, in the sham group, the cartilage surface was smooth, and chondrocytes were orderly arranged with no cartilage hyperplasia, cartilage sclerosis and synovial hyperplasia, and no significant inflammatory cells and blood vessels were observed in the synovial tissues. Compared with the sham group, the OA group had higher OARSI scores and synovial scores, and significant proteoglycan loss, cartilage wear, synovial hyperplasia, and inflammatory cell infiltration were observed in the OA group (Fig. 1C–E). And overexpression of CRNDE markedly improved proteoglycan loss, cartilage damage, and synovial inflammation (Fig. 1C–E). Moreover, immunostaining analysis indicated that CRNDE overexpression notably increased the expression of Collagen 2 and Aggrecan in OA rats (Fig. 1F). In addition, we found that high expression of CRNDE rescued OA-induced apoptosis of chondrocytes in the knee joint (Fig. 1G, H).
Fig. 1.
Overexpression of CRNDE hinders the progression of post-traumatic osteoarthritis in rats. An OA rat model was established using anterior cruciate ligament transectectomy. Ad-CRNDE (30 μL) or adenovirus empty vector (30 μL) were injected into the knee of OA rat. Six weeks after the operation. A, B The expression of CRNDE in knee cartilage tissues and synovial tissues was detected using qPCR. C H&E staining (Bar = 1 cm) and Safranin-O-fast green staining (Bar = 1 cm) were performed to analyze the pathological changes of the medial tibial and medial femur plateau joints in rats. And H&E staining (Bar = 1 cm) was used to assess inflammation of synovial tissues at the margin of articular cartilage. D, E Osteoarthritis Research Society International (OARSI) scores and synovitis scores. F The expression of Collagen 2 and Aggrecan in knee articular cartilage was detected with immunohistochemistry (Bar = 1 cm). G, H TUNEL staining (Bar = 1 cm) of the medial tibial and medial femur plateau joints was conducted to assess cell apoptosis, and the apoptotic cells and total cells were counted using Image J software. The sample size was n = 8, and each test was independently repeated at least three times. One-way ANOVA was used to detect differences among groups. *P < 0.05, **P < 0.01
Overexpression of CRNDE promoted cell proliferation and suppressed apoptosis, inflammation and ECM degradation in IL-1β-induced chondrocyte‑like ATDC5 cells
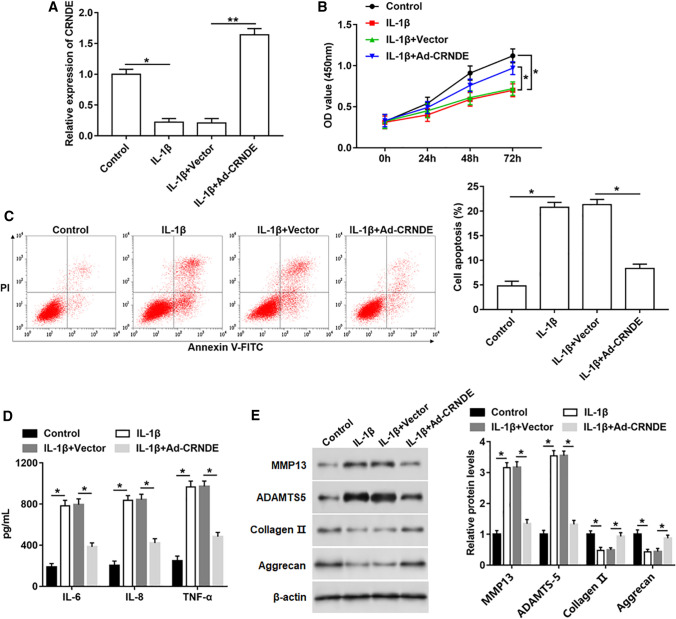
To further explore the role of CRNDE in OA, IL-1β was used to stimulate chondrocyte‑like ATDC5 cells to simulate OA condition. Meanwhile, CRNDE was overexpressed in IL-1β-induced cells. Firstly, the CRNDE expression was measured, and it was downregulated after IL-1β treatment, but increased after transfection with Ad-CRNDE (Fig. 2A). Besides, IL-1β induction led to reduced cell proliferation, and increased apoptosis and inflammatory factor (including IL-6, IL-8 and TNF-α) secretion, which was partially abolished by CRNDE overexpression (Fig. 2B–D). Moreover, the expression of ECM degradation-related biomarkers was evaluated, and higher expression of MMP13 and ADAMTS-5, and lower protein levels of Collagen 2 and Aggrecan were observed in IL-1β treatment group than in control group (Fig. 2E). However, overexpression of CRNDE reversed the promotion effect of IL-1β on ECM degradation (Fig. 2E).
Fig. 2.
Protective effects of CRNDE on IL-1β-induced apoptosis, inflammation and ECM degradation in chondrocyte‑like ATDC5 cells. Ad-CRNDE was used to infect chondrocyte‑like ATDC5 cells for 24 h, and then IL-1β (10 ng/mL) was used to treat cells for 24 h. A CRNDE expression was measured with qPCR. B Cell proliferation was detected using MTT assay. C Fluorescence activated cell sorting (FACS) was performed to analyze cell apoptosis. D The concentrations of IL-6, IL-8 and TNF-α in cell supernatant were determined by ELISA. E The protein levels of MMP13, ADAMTS-5, Collagen 2 and Aggrecan were measured using Western blotting. The sample size was n = 5, and each test was independently repeated at least three times. One-way ANOVA was used in A and C–E, and two-way ANOVA was used in B. *P < 0.05, **P < 0.01
CRNDE targeted DACT1 and modulated its expression
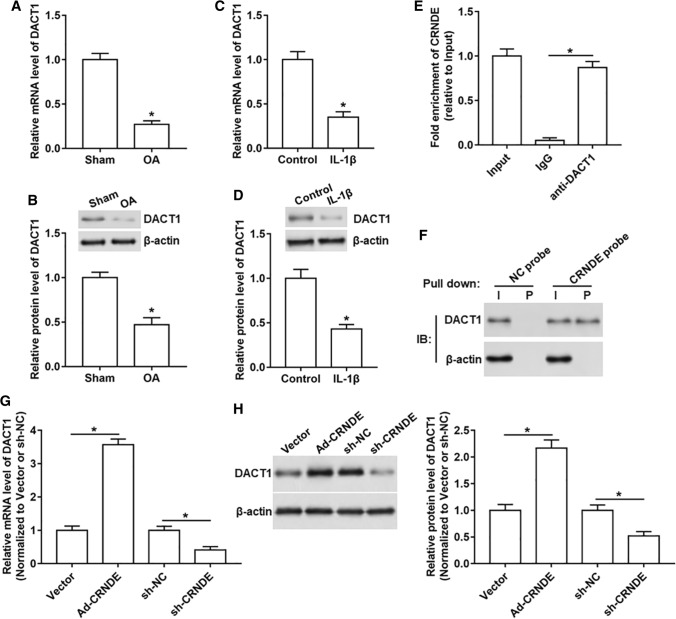
It is reported that DACT1 is down-regulated in OA knee tissues, and its low expression may be related to the progression of OA [21]. So we determined DACT1 expression in OA in vivo and in vitro models, and the data displayed that the mRNA and protein levels of DACT1 were decreased both in OA rat tissues and IL-1β-treated chondrocyte‑like ATDC5 cells compared with that in normal cartilaginous tissues and un-treated cells (Fig. 3A–D). Next, to explore whether DACT1 was a downstream target of CRNDE, RIP and RNA pull-down assays were performed. The results in RIP assay showed a significant enrichment of CRNDE using DACT1 antibody compared with IgG antibody (Fig. 3E). And RNA pull-down assay showed a prominent enrichment of DACT1 protein in CRNDE-protein complexes (Fig. 3F). Furthermore, we also found that the mRNA and protein levels of DACT1 were upregulated after overexpressing CRNDE, while downregulated after silencing CRNDE (Fig. 3G, H).
Fig. 3.
DACT1 is a downstream target of CRNDE. A, B The mRNA and protein levels of DACT1 in the OA rat model were assessed. C, D The mRNA and protein levels of DACT1 in IL-1β-stimulated chondrocyte‑like ATDC5 cells were detected. E RIP assay was performed in chondrocyte‑like ATDC5 cells to confirm the binding of CRNDE and DACT1. F RNA pull-down was carried out in chondrocyte‑like ATDC5 cells to verify the binding of CRNDE and DACT1 using CRNDE probe. G, H Ad-CRNDE, sh-CRNDE or their respective controls were transfected into chondrocyte‑like ATDC5 cells, and the mRNA and protein levels of DACT1 were analyzed. Ad-CRNDE was normalized to Vector and sh-CRNDE was normalized to sh-NC. The sample size was n = 5, and each test was independently repeated at least three times. Student’s t test was used to detect differences between two groups. *P < 0.05
DACT1 inhibition abolished the protective effects of CRNDE on IL-1β-stimulated chondrocyte‑like ATDC5 cells
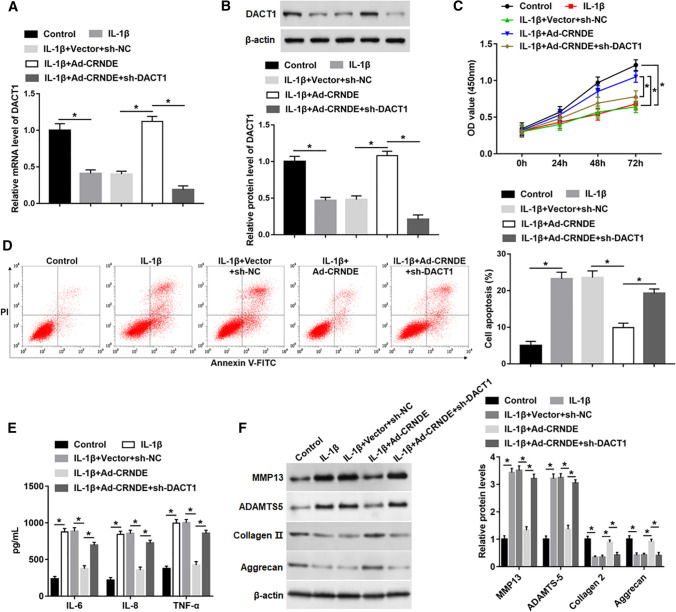
To study whether CRNDE modulated cell functions by regulating DACT1 expression, Ad-CRNDE was transfected into IL-1β-stimulated cells alone, or together with sh-DACT1. The results indicated that CRNDE overexpression upregulated the mRNA and protein levels of DACT1 in IL-1β-treated cells, which was observably reduced by transfection with sh-DACT1 (Fig. 4A, B). Moreover, silencing DACT1 markedly hindered the reversal effect of CRNDE overexpression on decreased cell proliferation, and increased apoptosis and inflammatory mediator (including IL-6, IL-8 and TNF-α) release caused by IL-1β (Fig. 4C–E). In addition, the knockdown of DACT1 also abolished the inhibitory action of CRNDE on IL-1β-induced ECM degradation by upregulating MMP13 and ADAMTS-5 expression and downregulating the protein levels of Collagen 2 and Aggrecan (Fig. 4F).
Fig. 4.
Silencing DACT1 abolished the protective effect of CRNDE on IL-1β-induced cell damage. Ad-CRNDE was transfected into chondrocyte‑like ATDC5 cells alone or together with sh-DACT1 for 24 h, and then IL-1β (10 ng/mL) was used to treat cells for 24 h. A, B The mRNA and protein expression of DACT1 was detected with qPCR and Western blot analysis. C Cell proliferation was detected using MTT assay. D FACS was performed to analyze cell apoptosis. E The concentrations of IL-6, IL-8 and TNF-α in cell supernatant were determined by ELISA. F The protein levels of MMP13, ADAMTS-5, Collagen 2 and Aggrecan were measured using Western blot analysis. The sample size was n = 5, and each test was independently repeated at least three times. One-way ANOVA was used in A, B and D, F, and two-way ANOVA was used in C. *P < 0.05
Additionally, we also explored the roles of CRNDE and DACT1 in human primary chondrocytes. Our results showed that the expression of CRNDE and DACT1 was downregulated in chondrocytes after treatment with IL-1β compared with the control, which was reversed by overexpression of CRNDE. And treatment with DACT1 shRNA effectively abolished the increase of DACT1 caused by CRNDE overexpression (Fig. S1A, B). Moreover, IL-1β stimulation significantly promoted cell apoptosis and inflammatory factor secretion, which was suppressed by CRNDE overexpression. And compared with Ad-CRNDE infection group, silencing of DACT1 observably increased cell apoptosis and the secretion of IL-6, IL-8 and TNF-α (Fig. S1C, D). Furthermore, treatment with IL-1β promoted ECM degradation by upregulating MMP13 and ADAMTS-5 expression and reducing Collagen 2 and Aggrecan expression. Highly expressed CRNDE inhibited ECM degradation compared with the IL-1β group. And the knockdown of DACT1 facilitated ECM degradation in primary chondrocytes compared with the CRNDE overexpression group (Fig. S1E). These results suggested that the roles of CRNDE and DACT1 in human primary chondrocytes were similar to their roles in chondrocyte‑like ATDC5 cells.
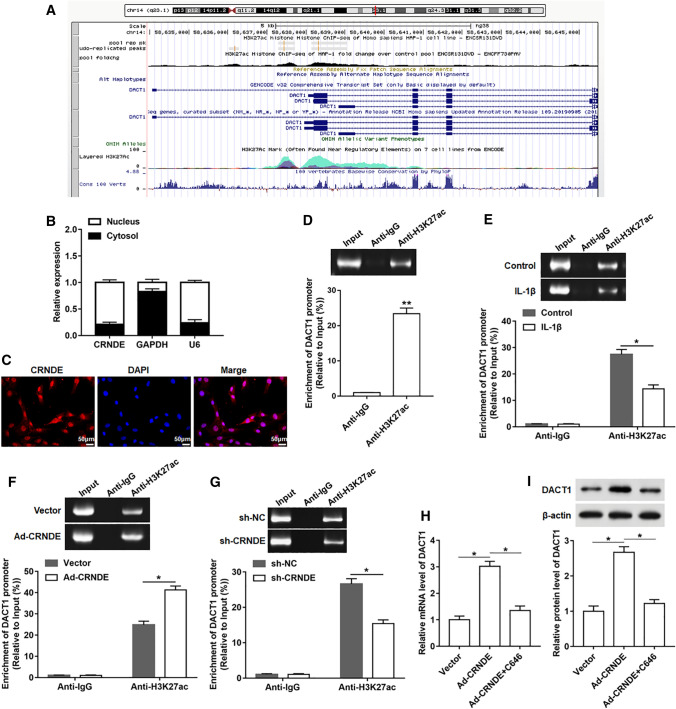
Overexpression of CRNDE promoted DACT1 expression by regulating H3K27 acetylation
To deeply investigate the regulatory relationship between CRNDE and DACT1, we explored the possible regulatory mechanisms using online databases of UCSC and ENCODE (Fig. 5A). And we found that H3K27ac was significantly enriched in the promoter region of DACT1. Next, nuclear and cytoplasmic RNA isolation assay was used to analyze the cellular localization of CRNDE. The results showed that CRNDE was mainly located in nucleus (Fig. 5B). Consistently, FISH assay showed that CRNDE was mainly enriched in the nucleus (Fig. 5C). Therefore, we speculated that CRNDE might regulate the DACT1 expression through histone modification at the transcriptional level. To test our hypothesis, ChIP assay was performed in chondrocyte‑like ATDC5 cells. The data showed that DACT1 promoter had a significant enrichment in the H3K27ac antibody-precipitated complex compared to the IgG (Fig. 5D). Additionally, a notably reduced enrichment of H3K27ac was observed in IL-1β-treated cells compared to normal cells (Fig. 5E). We also evaluated the effect of CRNDE on the enrichment of H3K27ac in the DACT1 promoter region, and the results displayed that overexpression of CRNDE promoted the enrichment of H3K27ac, while silencing CRNDE hindered the H3K27ac enrichment (Fig. 5F, G). In addition, a histone acetyltransferase inhibitor (C646) was used to treat chondrocyte‑like ATDC5 cells, and the data indicated that treatment with C646 reversed the increase in DACT1 mRNA and protein levels induced by CRNDE overexpression (Fig. 5H, I).
Fig. 5.
CRNDE promoted DACT1 expression by increasing H3K27 acetylation in chondrocyte‑like ATDC5 cells. A Using UCSC and ENCODE online databases, H3K27ac was found to be highly enriched in the DACT1 promoter region. B Cytoplasmic & Nuclear RNA Purification Kit was used to isolate cytoplasmic and nuclear RNA. And the expression of CRNDE in the nucleus and cytoplasm was evaluated using qPCR. U6 and GAPDH were used as nuclear and cytoplasmic markers, respectively. C FISH analysis (Bar = 2 cm) of the enrichment level of CRNDE in nucleus of chondrocyte‑like ATDC5 cells. D ChIP assay performed in chondrocyte‑like ATDC5 cells showed the enrichment of H3K27ac in the DACT1 promoter region. E Chondrocyte‑like ATDC5 cells were treated by IL-1β for 24 h, and then the the enrichment of H3K27ac in the DACT1 promoter was analyzed using ChIP assay. F, G After overexpression or interference with CRNDE in chondrocyte‑like ATDC5 cells, the enrichment of H3K27ac in DACT1 promoter was detected using ChIP assay. H, I Chondrocyte‑like ATDC5 cells were infected with Ad-CRNDE for 48 h, and then a histone acetyltransferase inhibitor (C646, 25 μM) was used to treat cells for 1 h. The mRNA and protein levels of DACT1 were determined. The sample size was n = 5, and each test was independently repeated at least three times. Student’s t test was used to detect differences between two groups. *P < 0.05, **P < 0.01
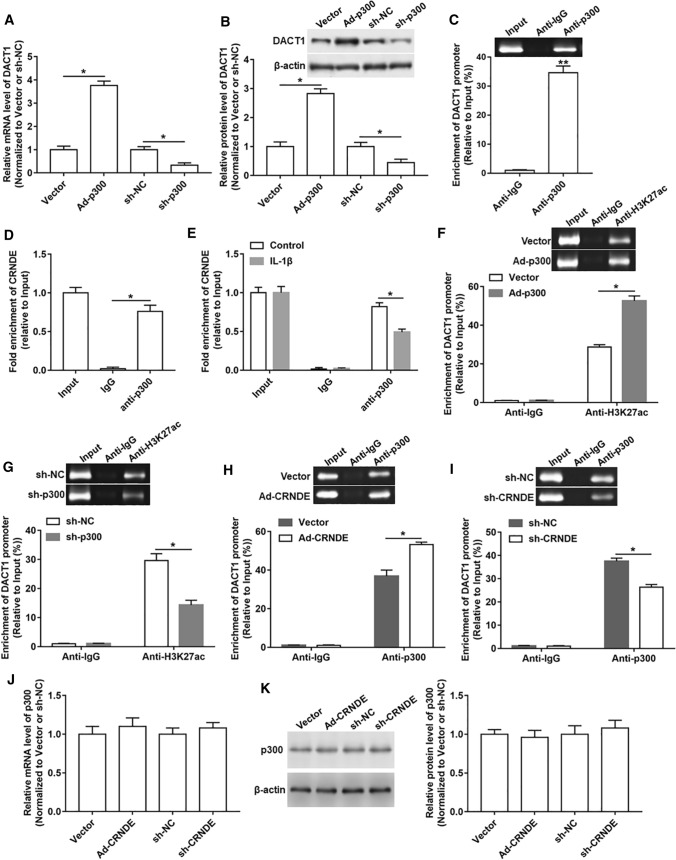
CRNDE regulated H3K27 acetylation at the promoter of DACT1 by binding to p300
Since C646 is an effective inhibitor of p300, the role of p300 in the regulatory effect of CRNDE on DACT1 expression was investigated. We first detected whether the expression of DACT1 responded to changes in p300 expression levels, and the results revealed that the mRNA and protein levels of DACT1 was prominently upregulated after overexpressing p300 and downregulated after the knockdown of p300 (Fig. 6A, B). The ChIP assay showed a prominent enrichment of DACT1 promoter in the p300 antibody-precipitated complex compared to the IgG (Fig. 6C). Moreover, RIP assay was carried out to measure the interaction between CRNDE and p300. As shown in Fig. 6D and E, a significant amount of CRNDE was detected using p300 antibody compared to IgG antibody, and treatment with IL-1β attenuated this interaction between them (Fig. 6D, E). Additionally, we found that p300 could also regulate the enrichment of H3K27ac in the DACT1 promoter region. Because overexpression of p300 increased the enrichment of H3K27ac in the DACT1 promoter region, while silencing p300 had an opposite effect (Fig. 6F, G). In addition, a markedly increased enrichment of p300 in the DACT1 promoter region was observed in response to overexpression of CRNDE, while silencing of CRNDE showed an opposite role (Fig. 6H, I). However, the mRNA and protein levels of p300 were not altered by either high or low expression of CRNDE (Fig. 6J, K). Our results suggested that CRNDE might function by recruiting p300 to target downstream genes.
Fig. 6.
CRNDE mediated H3K27 acetylation at DACT1 promoter region by binding to p300. A, B Ad-p300, sh-p300 or their respective controls were transfected into chondrocyte‑like ATDC5 cells, and the mRNA and protein levels of DACT1 were detected. Ad-p300 was normalized to Vector and sh-p300 was normalized to sh-NC. C ChIP assay showed the enrichment of p300 in the DACT1 promoter region. D RIP assay was performed to determine the binding between CRNDE and p300. E After IL-1β treatment, RIP assay was used to measure the binding of CRNDE and p300 in chondrocyte‑like ATDC5 cells. F, G After overexpression or interference with p300 in chondrocyte‑like ATDC5 cells, the enrichment of H3K27ac in DACT1 promoter was analyzed using ChIP assay. H, I After overexpression or interference with CRNDE in chondrocyte‑like ATDC5 cells, the enrichment of p300 in DACT1 promoter was assessed. J, K The mRNA and protein levels of p300 were determined after overexpressing or inhibiting CRNDE in chondrocyte‑like ATDC5 cells. Ad-CRNDE was normalized to Vector and sh-CRNDE was normalized to sh-NC. The sample size was n = 5, and each test was independently repeated at least three times. Student’s t test was used to detect differences between two groups. *P < 0.05, **P < 0.01
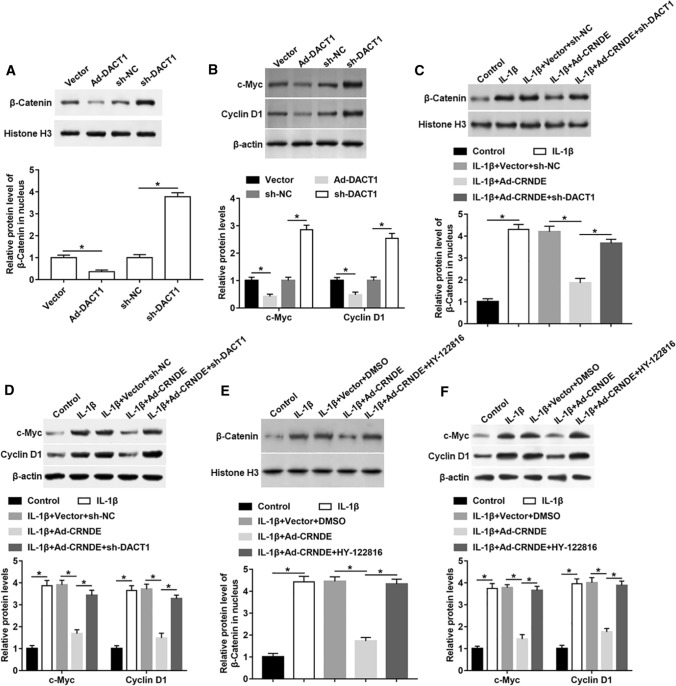
CRNDE restrained IL-Iβ-induced activation of Wnt/β-catenin signaling by regulating DACT1 in chondrocyte‑like ATDC5 cells
DACT1 is reported to be an effective inhibitor of the Wnt signaling pathway [24]. We overexpressed and interfered DACT1 in chondrocyte‑like ATDC5 cells, and measured Wnt signaling activity and its downstream gene expression. The protein levels of β-catenin in nucleus, and c-Myc and Cyclin D1 in the whole cells were downregulated in DACT1 overexpression group, but increased in DACT1 silencing group (Fig. 7A, B). To explore whether CRNDE regulated Wnt signaling pathway activity through regulating DACT1, Ad-CRNDE was transfected into IL-1β-stimulated cells alone, or together with sh-DACT1. IL-1β treatment resulted in a significant increase in the expression of β-catenin, c-Myc and Cyclin D1, which was rescued by overexpression of CRNDE (Fig. 7C, D). Additionally, silencing of DACT1 abrogated the effect of CRNDE on Wnt/β-catenin pathway activity (Fig. 7C, D). Next, we replaced the sh-DACT1 with HY-122816 (a specific activator of the Wnt/β-catenin pathway). Similar to the knockdown of DACT1, HY-122816 treatment could also hinder the decrease of β-catenin, c-Myc and Cyclin D1 expression caused by CRNDE overexpression in IL-1β-induced cells (Fig. 7E, F).
Fig. 7.
CRNDE restrained Wnt/β-catenin pathway activation by up-regulating DACT1 expression in IL-Iβ-treated chondrocyte‑like ATDC5 cells. A, B Ad-DACT1, sh-DACT1 or their respective controls were transfected into chondrocyte‑like ATDC5 cells, and the expression of β-catenin in nuclear and the expression of c-Myc and Cyclin D1 in the whole cell lysate were measured with Western blot analysis. Ad-DACT1 was normalized to Vector and sh-DACT1 was normalized to sh-NC. C, D Ad-CRNDE was transfected into cells alone or together with sh-DACT1 for 24 h, and then IL-Iβ was used to stimulate cells for 24 h. The expression of β-catenin in nuclear and the expression of c-Myc and Cyclin D1 in the whole cell lysate were evaluated using Western blot analysis. E, F Chondrocyte‑like ATDC5 cells were treated by IL-Iβ alone or together with Ad-CRNDE, and then cells were treated with HY-122816. The expression of β-catenin in nuclear and the expression of c-Myc and Cyclin D1 in the whole cell lysate were analyzed with Western blot analysis. The sample size was n = 5, and each test was independently repeated at least three times. Student’s t test was used in A, B, and one-way ANOVA was used in C–F. *P < 0.05
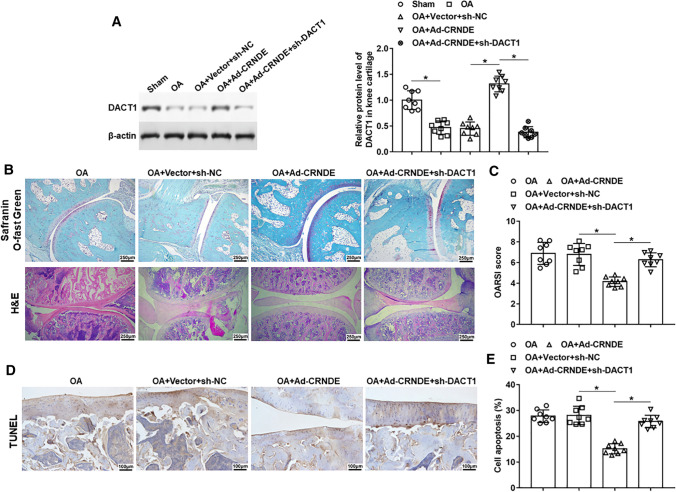
Overexpression of CRNDE mitigated cartilage injury in an OA rat model by modulating DACT1
Ad-CRNDE was administration alone or together with sh-DACT1 in the OA rat model to study the effect of DACT1 in OA progress. The data showed that DACT1 expression was downregulated in knee cartilage of OA rats. And sh-DACT1 injection reversed CRNDE-induced increase in DACT1 expression (Fig. 8A). As shown in Fig. 8B and C, HE staining and Safranin O staining displayed that overexpression of CRNDE alleviated OA-induced cartilage wear and loss of Safranin O, and increased OARSI score. Silencing of DACT1 hindered the protective effect of CRNDE on knee joint damage (Fig. 8B, C). In addition, silencing DACT1 could also reverse the inhibitory effect of CRNDE overexpression on OA-induced apoptosis of chondrocytes in the knee joint (Fig. 8D, E).
Fig. 8.
CRNDE slowed the progression of post-traumatic OA in rats by modulating DACT1. An OA rat model was established using anterior cruciate ligament transectectomy. Ad-CRNDE was injected into the knee of OA rats alone or together with sh-DACT1. Six weeks after the operation, A the protein level of DACT1 in knee cartilage tissues was detected using Western blot analysis. B HE staining (Bar = 1 cm) and Safranin-O-fast green staining (Bar = 1 cm) were used to assess the pathological changes of the medial tibial and medial femur plateau joints in rats. C Osteoarthritis Research Society International (OARSI) scores. D, E TUNEL staining (Bar = 1 cm) of the medial tibial plateau joint was performed to analyze cell apoptosis, and the apoptotic cells and total cells were counted using Image J software. The sample size was n = 8, and each test was independently repeated at least three times. One-way ANOVA was used to detect differences among groups. *P < 0.05
Discussion
As shown in Fig. 9, CRNDE promotes DACT1 transcription by promoting the enrichment of H3K27ac in the DACT1 promoter. Next, the DACT1 protein hinders the activity of the Wnt signaling pathway by blocking the entry of β-catenin into the nucleus, thereby suppressing the transcription of the Wnt downstream target genes c-Myc and Cyclin D1, thus inhibiting OA-induced chondrocyte damage.
Fig. 9.
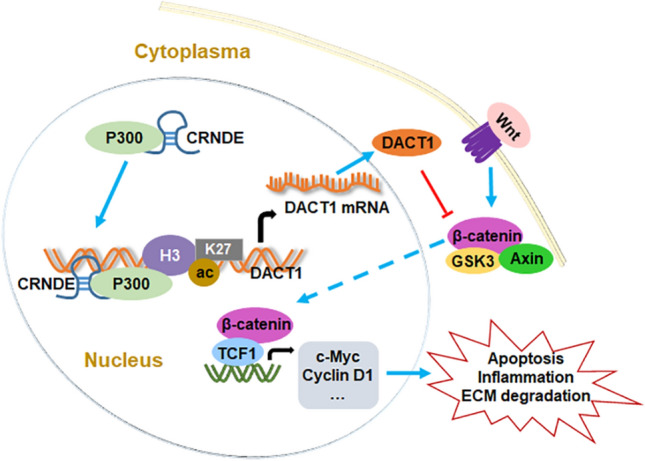
A schematic diagram for the effect of CRNDE on OA progression. By recruiting p300, CRNDE promotes the enrichment of H3K27ac in the DACT1 promoter, thereby promoting DACT1 transcription and translation. DACT1 impedes the nuclear transfer of β-catenin, and then inhibiting the activity of Wnt signaling pathway and the transcription of downstream target genes c-Myc and Cyclin D1, thereby suppressing OA-induced chondrocyte apoptosis, inflammation and ECM degradation
OA is induced by many factors, including obesity, mechanical injury, aging, heredity, etc. It usually occurs in the knee, spine and hip, and patients show swelling and pain in the joints and decreased ability to walk [25]. Healthy cartilage consists mainly of chondrocytes [26]. Chondrocytes can produce cartilage extracellular matrix rich in collagen (mainly type II collagen) and proteoglycans. Under pathological conditions, chondrocytes can also activate mechanisms that lead to the production of metalloproteinases to degrade cartilage matrix [27]. In addition, after the occurrence of OA, chondrocyte functions including proliferation and secretion have undergone many changes [28]. Interleukin-1β (IL-1β) has been widely reported to be associated with several pathological features of OA [29, 30], and it is commonly used in the in vitro model of OA to simulate the environment driving cartilage degradation in vivo [31]. Our results showed that treatment with IL-1β significantly promoted cell apoptosis, inflammation and extracellular matrix degradation in chondrocyte‑like ATDC5 cells. Stav et al. indicated that IL-1β treatment for 24 h can induce ATDC5 cells to exit G1 phase and enter S phase, and facilitate cell proliferation [32], which is inconsistent with our results. Our results showed that the proliferation of ATDC5 cells was not significantly changed at 24 h after treatment with IL-1β, but was significantly inhibited at 48 or 72 h after IL-1β treatment. The reason for this difference may be that Stav et al. only observes changes in cell proliferation within 24 h, and the results are not comprehensive and contingent.
It is reported that lncRNAs regulate various biological processes, including proliferation, metastasis, apoptosis, cell cycle transformation and cell structural integrity, through chromatin remodeling, adsorption of miRNAs and epigenetic modification [33, 34]. The potential importance of lncRNAs in OA has been revealed by Fu et al. [35]. CRNDE is up-regulated in many solid and hematopoietic cancers, and its high expression contributes to malignant progression of cancers [36–38]. But in osteoblasts, CRNDE appeared to be protective, and it promoted proliferation and differentiation of osteoblasts and could be used as a novel regulator of bone metabolism [39]. In addition, in age-induced OA, CRNDE is low-expressed and may serve as a key regulator for OA [15]. Our findings indicated that local overexpression of CRNDE by intra-articular injection of Ad-CRNDE attenuated cartilage damage and synovial inflammation in OA rats. Moreover, the adenoviral vector does not integrate into the host cell genome and we intervened the OA individuals (animals) with Ad-CRNDE strictly locally, so that it is improbable that CRNDE is overexpressed in other tissues of the animals to play a carcinogenic role. Therefore, injection of CRNDE overexpression vector in knee tissues may be used as a potential treatment for OA. In vitro, the high expression of CRNDE hindered IL-1β-induced apoptosis, inflammation and ECM degradation both in chondrocyte‑like ATDC5 cells and human primary chondrocytes. Our data indicated the protective role of CRNDE in OA.
It is generally believed that the Wnt/β-catenin signaling pathway is a unique signaling pathway that regulates the occurrence and development of OA. Numerous studies have shown that Wnt/β-catenin signaling plays an important role in joint pathology by acting directly on bone, cartilage and synovial tissues [40, 41]. Consistent with previous reports, our results showed that the Wnt signaling was activated in IL-1β-stimulated chondrocyte‑like ATDC5 cells, and the expression of its downstream genes c-Myc and Cyclin D1 was also up-regulated. As we all know, DACT1 is an inhibitor of the Wnt pathway and can also regulate the expression of Wnt downstream genes [24]. We found that overexpression of DACT1 suppressed the activity of Wnt signaling in chondrocyte‑like ATDC5 cells. Both silencing DACT1 and treatment with a Wnt-specific activator (HY-122816) effectively reversed the inactivation of the Wnt pathway caused by CRNDE overexpression. DACT1 plays a dual role in cancers. It can not only act as a tumor suppressor in breast and liver cancers [24, 42], but also promotes the malignant progression of colon and ovarian cancers [17, 43]. In addition, the sequencing data of Chou et al. indicated that DACT1 was lower expressed in OA knee tissues than in normal knee tissues [21]. In in vivo and in vitro OA models, DACT1 expression was significantly reduced. And silencing DACT1 partially abrogated the protective effects of CRNDE on IL-1β-induced apoptosis, inflammation, and ECM degradation in chondrocyte‑like ATDC5 cells, demonstrating that DACT1 acted as a downstream target gene of CRNDE function.
Increasing evidence shows that lncRNAs can activate or inhibit target gene expression through epigenetic modifications (including acetylation, methylation, ubiquitination, phosphorylation) [44]. The mechanism of lncRNAs is closely related to its cellular localization, and lncRNAs located in the nucleus are likely to regulate gene expression through transcriptional regulation and chromatin modification [45]. The results in this study displayed that CRNDE was mainly expressed in the nucleus, suggesting that it might affect gene expression through epigenetic modifications. CRNDE has been reported to promote EZH2-mediated methylation modification of target genes by binding to EZH2, thereby silencing gene expression [14]. We found that CRNDE could bind to DACT1 and might modulate its expression by mediating histone acetylation. Acetylation is the most common histone acyl modification and a large number of studies have demonstrated the relationship between increased histone acetylation levels and transcriptional activation [46]. Histone acetylation modification regulated by lncRNA has been widely reported. IL-7-AS-V1 regulates H3K9ac and H3K14ac at several inflammatory gene promoters, and promotes gene expression by recruiting histone acetyltransferase p300 [47]. LncRNA AGAP2-AS1 increases MyD88 expression by promoting H3K27ac in the MyD88 promoter [48]. Moreover, it is reported that SIRT1 regulates DACT1 expression through deacetylation [20]. The bioinformatics analysis showed that H3K27ac enriched significantly in the promoter region of DACT1, and CRNDE could promote the enrichment of H3K27ac in the promoter region of DACT1. In addition, IL-1β stimulation restrained the enrichment of H3K27ac in DACT1 promoter in chondrocyte‑like ATDC5 cells.
According to reports, many lncRNAs regulate gene expression by binding to various histone-modifying enzymes to affect cell functions [14]. For example, lncRNA Khps1 upregulates gene expression through recruitment of histone acetyltransferase p300 / CBP [49]. P300 is a global co-activator of various transcription factors with histone acetyltransferase (HAT) activity [50]. Besides, it is reported that RNA can directly bind to a unique region in the HAT domain of p300 to stimulate the HAT activity of p300 [51]. Our data revealed that CRNDE modulated H3K27ac enrichment in the DACT1 promoter by recruiting p300 to the DACT1 promoter and binding to p300 to form a complex, which ultimately affected the expression of DACT1.
In conclusion, our study provided evidence that highly expressed CRNDE might hinder the progression of OA. And the mitigation effect of CRNDE on knee articular cartilage injury in vivo, and chondrocyte damage in vitro might be achieved by epigenetic promotion of DACT1 transcription by binding to p300. CRNDE might be used as a promising target for OA diagnosis and treatment.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
ZQZ: research design, manuscript preparation and data analysis; PY: experimental material preparation and rat feeding; CHW: completion of molecular biology experiments; RT: histology analysis.
Funding
Not applicable.
Data availability
All data used during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
The study was approved by the Ethics Committee of the Second Affiliated Hospital of Xi’an Jiaotong University. All methods were performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Millerand M, Berenbaum F, Jacques C. Danger signals and inflammaging in osteoarthritis. Clin Exp Rheumatol. 2019;37(Suppl 120):48–56. [PubMed] [Google Scholar]
- 2.Glyn-Jones S, Palmer AJR, Agricola R, Price AJ, Vincent TL, Weinans H, Carr AJ. Osteoarthritis. Lancet. 2015;386(9991):376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z, Leong DJ, Xu L, He Z, Wang A, Navati M, Kim SJ, Hirsh DM, Hardin JA, Cobelli NJ, Friedman JM, Sun HB. Curcumin slows osteoarthritis progression and relieves osteoarthritis-associated pain symptoms in a post-traumatic osteoarthritis mouse model. Arthritis Res Ther. 2016;18(1):128. doi: 10.1186/s13075-016-1025-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 4.Carr AJ, Robertsson O, Graves S, Price AJ, Arden NK, Judge A, Beard DJ. Knee replacement. Lancet. 2012;379(9823):1331–1340. doi: 10.1016/S0140-6736(11)60752-6. [DOI] [PubMed] [Google Scholar]
- 5.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, Sauvageau M, Tazon-Vega B, Kelley DR, Hendrickson DG. Long noncoding RNAs regulate adipogenesis. P Nnal Acad Sci USA. 2013;110(9):3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 7.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, Xie J, Giang K, Chung H, Sun X. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6(1):1–13. doi: 10.1038/ncomms10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, Chen K, Chen L, Ding Y. A novel lncRNA uc. 134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10(1):91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6(10):2299. [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Tang Y, Xing W, Dong W, Wang Z. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp Mol Pathol. 2018;104(1):19–25. doi: 10.1016/j.yexmp.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Bai X, Wang W, Zhao P, Wen J, Guo X, Shen T, Shen J, Yang X. LncRNA CRNDE acts as an oncogene in cervical cancer through sponging miR-183 to regulate CCNB1 expression. Carcinogenesis. 2020;41(1):111–121. doi: 10.1093/carcin/bgz166. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Li J, Wang H, Tian Y, Xie M, He X, Ji H, Ma Z, Hui B, Wang K, Ji G. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P, Sun F, Ran J, Wu L. Identify CRNDE and LINC00152 as the key lncRNAs in age-related degeneration of articular cartilage through comprehensive and integrative analysis. PeerJ. 2019;7:e7024. doi: 10.7717/peerj.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lietman C, Wu B, Lechner S, Shinar A, Sehgal M, Rossomacha E, Datta P, Sharma A, Gandhi R, Kapoor M, Young PP. Inhibition of Wnt/beta-catenin signaling ameliorates osteoarthritis in a murine model of experimental osteoarthritis. JCI Insight. 2018 doi: 10.1172/jci.insight.96308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li RN, Liu B, Li XM, Hou LS, Mu XL, Wang H, Linghu H. DACT1 Overexpression in type I ovarian cancer inhibits malignant expansion and cis-platinum resistance by modulating canonical Wnt signalling and autophagy. Sci Rep. 2017;7(1):9285. doi: 10.1038/s41598-017-08249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang JH, Lin LK, Zhang S. Effects of DACT1 methylation status on invasion and metastasis of nasopharyngeal carcinoma. Biol Res. 2019;52(1):31. doi: 10.1186/s40659-019-0238-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu K, Jiang B, Yang Y, Hu R, Liu Z. DACT1 overexpression inhibits proliferation, enhances apoptosis, and increases daunorubicin chemosensitivity in KG-1alpha cells. Tumour Biol. 2017;39(10):1010428317711089. doi: 10.1177/1010428317711089. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Song T, Peng J, Zhou Z, Wei H, Zhou R, Jiang S, Peng J. SIRT1 suppresses adipogenesis by activating Wnt/β-catenin signaling in vivo and in vitro. Oncotarget. 2016;7(47):77707. doi: 10.18632/oncotarget.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou CH, Lee MT, Song IW, Lu LS, Shen HC, Lee CH, Wu JY, Chen YT, Kraus VB, Wu CC. Insights into osteoarthritis progression revealed by analyses of both knee tibiofemoral compartments. Osteoarthr Cartil. 2015;23(4):571–580. doi: 10.1016/j.joca.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoop R, Buma P, van der Kraan PM, Hollander AP, Billinghurst RC, Meijers TH, Poole AR, van den Berg WB. Type II collagen degradation in articular cartilage fibrillation after anterior cruciate ligament transection in rats. Osteoarthr Cartil. 2001;9(4):308–315. doi: 10.1053/joca.2000.0390. [DOI] [PubMed] [Google Scholar]
- 23.Gerwin N, Bendele AM, Glasson S, Carlson CS. The OARSI histopathology initiative – recommendations for histological assessments of osteoarthritis in the rat. Osteoarthr Cartil. 2010;18:S24–S34. doi: 10.1016/j.joca.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 24.Yin X, Xiang T, Li L, Su X, Shu X, Luo X, Huang J, Yuan Y, Peng W, Oberst M. DACT1, an antagonist to Wnt/β-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 2013;15(2):1–12. doi: 10.1186/bcr3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Z, Guo XH, Tang C, Yue ST, Shi L, Qiang B. Effects of artesunate on the expressions of insulin-like growth factor-1, osteopontin and C-Telopeptides of type II collagen in a rat model of osteoarthritis. Pharmacology. 2018;101(1–2):1–8. doi: 10.1159/000479160. [DOI] [PubMed] [Google Scholar]
- 26.Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!) Osteoarthr Cartil. 2013;21(1):16–21. doi: 10.1016/j.joca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Yao Z, Zhang Y, Yang Y, Liu J, Shi Y, Zhang C. Metformin mitigates cartilage degradation by activating AMPK/SIRT1-mediated autophagy in a mouse osteoarthritis model. Front Pharmacol. 2020;11:1114. doi: 10.3389/fphar.2020.01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlier E, Deroyer C, Ciregia F, Malaise O, Neuville S, Plener Z, Malaise M, de Seny D. Chondrocyte dedifferentiation and osteoarthritis (OA) Biochem Pharmacol. 2019;165:49–65. doi: 10.1016/j.bcp.2019.02.036. [DOI] [PubMed] [Google Scholar]
- 29.Daheshia M, Yao JQ. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. J Rheumatol. 2008;35(12):2306–2312. doi: 10.3899/jrheum.080346. [DOI] [PubMed] [Google Scholar]
- 30.Pascarelli NA, Collodel G, Moretti E, Cheleschi S, Fioravanti A. Changes in ultrastructure and cytoskeletal aspects of human normal and osteoarthritic chondrocytes exposed to interleukin-1beta and cyclical hydrostatic pressure. Int J Mol Sci. 2015;16(11):26019–26034. doi: 10.3390/ijms161125936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheleschi S, Fioravanti A, De Palma A, Corallo C, Franci D, Volpi N, Bedogni G, Giannotti S, Giordano N. Methylsulfonylmethane and mobilee prevent negative effect of IL-1beta in human chondrocyte cultures via NF-kappaB signaling pathway. Int Immunopharmacol. 2018;65:129–139. doi: 10.1016/j.intimp.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Simsa-Maziel S, Monsonego-Ornan E. Interleukin-1beta promotes proliferation and inhibits differentiation of chondrocytes through a mechanism involving down-regulation of FGFR-3 and p21. Endocrinology. 2012;153(5):2296–2310. doi: 10.1210/en.2011-1756. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Ge Z, Zhang E, Zuo Q, Huang S, Yang N, Wu D, Zhang Y, Chen Y, Xu H, Huang H, Jiang Z, Sun L. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8(10):e3104. doi: 10.1038/cddis.2017.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang SD, Lu J, Deng ZH, Li YS, Lei GH. Long noncoding RNAs in osteoarthritis. Joint Bone Spine. 2017;84(5):553–556. doi: 10.1016/j.jbspin.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Fu M, Huang G, Zhang Z, Liu J, Huang Z, Yu B, Meng F. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthr Cartil. 2015;23(3):423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Ma X, Zhang W, Zhao M, Li S, Jin W, Wang K. Oncogenic role of lncRNA CRNDE in acute promyelocytic leukemia and NPM1-mutant acute myeloid leukemia. Cell Death Discov. 2020;6(1):1–13. doi: 10.1038/s41420-020-00359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, He L, Ma L, Lu T, Wei J, Xie K, Wang X. LncRNA CRNDE promotes cell proliferation, invasion and migration by competitively binding miR-384 in papillary thyroid cancer. Oncotarget. 2017;8(66):110552. doi: 10.18632/oncotarget.22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie S-C, Zhang J-Q, Jiang X-L, Hua Y-Y, Xie S-W, Qin Y-A, Yang Y-J. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020;11(8):1–17. doi: 10.1038/s41419-020-02853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulati M, Kobayashi Y, Takahashi A, Numata H, Saito M, Hiraoka Y, Ochi H, Sato S, Ezura Y, Yuasa M, Hirai T, Yoshii T, Okawa A, Inose H. The long noncoding RNA Crnde regulates osteoblast proliferation through the Wnt/beta-catenin signaling pathway in mice. Bone. 2020;130:115076. doi: 10.1016/j.bone.2019.115076. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Fan X, Xing L, Tian F. Wnt signaling: a promising target for osteoarthritis therapy. Cell Commun Signal. 2019;17(1):97. doi: 10.1186/s12964-019-0411-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Wang T, Hamilton JL, Chen D. Wnt/beta-catenin signaling in osteoarthritis and in other forms of arthritis. Curr Rheumatol Rep. 2017;19(9):53. doi: 10.1007/s11926-017-0679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuo H, Wang Y, Wang L, Yao B, Li Q, Wang C, Liu Z, Han S, Yin G, Tu K. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/β-catenin pathway in hepatocellular carcinoma. Oncotarget. 2017;8(39):65687. doi: 10.18632/oncotarget.20058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan G, Wang C, Ma C, Chen N, Tian Q, Zhang T, Fu W. Oncogenic function of DACT1 in colon cancer through the regulation of beta-catenin. PLoS ONE. 2012;7(3):e34004. doi: 10.1371/journal.pone.0034004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 45.Ma H, Chang H, Yang W, Lu Y, Hu J, Jin S. A novel IFNalpha-induced long noncoding RNA negatively regulates immunosuppression by interrupting H3K27 acetylation in head and neck squamous cell carcinoma. Mol Cancer. 2020;19(1):4. doi: 10.1186/s12943-019-1123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes CE, English DM, Cowley SM. Acetylation & Co: an expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem. 2019;63(1):97–107. doi: 10.1042/EBC20180061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Lu Y, Zhu J, Liu M, Xie M, Ye M, Li M, Wang S, Ming Z, Tong Q, Liu F, Zhou R. A long noncoding RNA, Antisense IL-7, promotes inflammatory gene transcription through facilitating histone acetylation and switch/sucrose nonfermentable chromatin remodeling. J Immunol. 2019;203(6):1548–1559. doi: 10.4049/jimmunol.1900256. [DOI] [PubMed] [Google Scholar]
- 48.Dong H, Wang W, Mo S, Chen R, Zou K, Han J, Zhang F, Hu J. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202. doi: 10.1186/s13046-018-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell. 2015;60(4):626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Khilji S, Hamed M, Chen J, Li Q. Loci-specific histone acetylation profiles associated with transcriptional coactivator p300 during early myoblast differentiation. Epigenetics. 2018;13(6):642–654. doi: 10.1080/15592294.2018.1489659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017;168(1–2):135–149. doi: 10.1016/j.cell.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used during the current study are available from the corresponding author on reasonable request.