Abstract
Traumatic spinal cord injury is an overwhelming condition that strongly and suddenly impacts the patient’s life and her/his entourage. There are currently no predictable treatments to repair the spinal cord, while many strategies are proposed and evaluated by researchers throughout the world. One of the most promising avenues is the transplantation of stem cells, although its therapeutic efficiency is limited by several factors, among which cell survival at the lesion site. In our previous study, we showed that the implantation of a human dental apical papilla, residence of stem cells of the apical papilla (SCAP), supported functional recovery in a rat model of spinal cord hemisection. In this study, we employed protein multiplex, immunohistochemistry, cytokine arrays, RT- qPCR, and RNAseq technology to decipher the mechanism by which the dental papilla promotes repair of the injured spinal cord. We found that the apical papilla reduced inflammation at the lesion site, had a neuroprotective effect on motoneurons, and increased the apoptosis of activated macrophages/ microglia. This therapeutic effect is likely driven by the secretome of the implanted papilla since it is known to secrete an entourage of immunomodulatory or pro-angiogenic factors. Therefore, we hypothesize that the secreted molecules were mainly produced by SCAP, and that by anchoring and protecting them, the human papilla provides a protective niche ensuring that SCAP could exert their therapeutic actions. Therapeutic abilities of the papilla were demonstrated in the scope of spinal cord injury but could very well be beneficial to other types of tissue.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04210-8.
Keywords: Stem cells, Apical papilla, SCAP, Spinal cord injury, Inflammation, Immunomodulation, Secretome, RNAseq, Cell therapy, Tissue repair
Introduction
Spinal cord injury (SCI) is an overwhelming traumatic injury that causes severe neurological disorders. Following the initial trauma (e.g., contusion, laceration or compression), mechanical disruption leads to a primary injury (cell death, edema, and ischemia). However, most of the neurological damages occur during the secondary phase of the injury, from minutes to years post-injury [1]. The injury leads to dramatic cell degeneration, releasing many deleterious molecules, such as free radicals, excitatory neurotransmitters, and inflammatory molecules that promote neuronal death and severe hamper any neuroregenerative potential [2]. In addition, over time, a chronic non-resolved inflammation state becomes established and prevents attempts at regeneration [1].
A multitude of strategies has been developed to try to repair the spinal cord and several clinical trials are ongoing [3]. One of the main approaches currently explored is cell-based therapy, mostly using neural and mesenchymal stem cells (MSCs) [4]. Although MSC derived from the bone marrow (BMSC) are currently the most investigated, alternatives have been sought, like adipose tissue-derived MSC or dental tissue-derived MSC. However, the SCI environment is cytotoxic for any potential cell transplantation, and triggers a degenerative cascade and scar formation.
Dental MSC can be obtained from extracted teeth (mostly wisdom teeth), making their isolation simpler and less invasive than aspiration of bone marrow [5]. They advantageously combine MSC-like properties, such as immune modulation with neural characteristics, due to their neural crest origin, such as the expression of neural markers and secretion of neurotrophic factors [5]. Among MSC present in different dental tissues, stem cells from the apical papilla (SCAP) [6] are easy to obtain, isolate, and expand [7]. In addition, SCAP has a high proliferation rate and express neural and glial markers [8, 9]. Thus, the apical papilla and its residing cells (SCAP) provide unique opportunities for its translational clinical use. In our previous work, we showed that SCAP can reduce TNFα expression and secretion in inflamed spinal cord tissues and can stimulate oligodendrocyte progenitor cell (OPC) differentiation via activin-A secretion [10]. However, implantation of the cells in a rat spinal cord hemisection model resulted in only modest recovery of locomotor activity, while the implantation of a whole papilla significantly improved rat gait [11].
The objective of this work was thus to decipher the mechanisms behind the apical papilla therapeutic action and more particularly, to understand how it could impact the SCI microenvironment after implantation and conduct to functional improvement. Influence of the apical papilla on inflammatory marker expression at the lesion site, as well as on microglial activation and neuroprotection, was evaluated by protein quantitative multiplex assays and immunofluorescence, respectively. The papilla secretome, and more particularly immunomodulatory molecules, was analyzed using a cytokine array, ELISA, and RT-qPCR. Finally, a RNA sequencing analysis (RNAseq) was conducted to study the global transcriptomic changes underlying the apical papilla-mediated repair in an acute SCI model.
Material and methods
Experimental settings
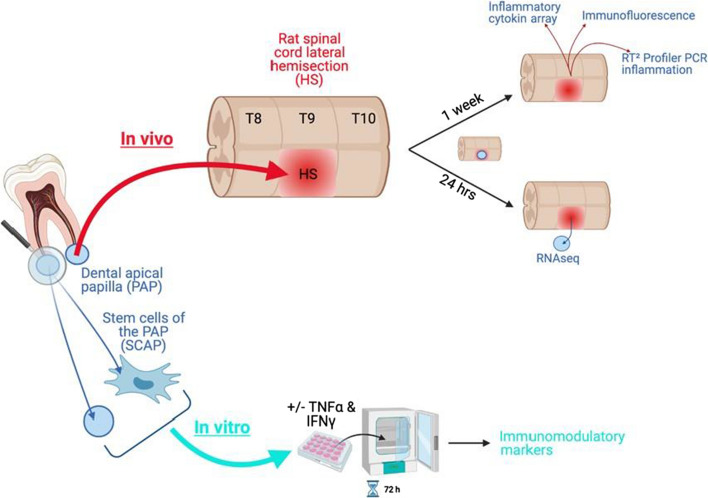
This work was performed in two main steps: (1) in vivo experiments aimed at studying the impact of the papilla on the spinal cord injury environment and vice versa, and (2) in vitro experiments looking into papilla secretome when subjected to a pro-inflammatory-like stimulus (Fig. 1). As a comparison, production of immunomodulatory molecules was also analyzed for SCAP.
Fig. 1.
Experimental settings. Created with Biorender.com
Apical papilla extraction
Wisdom teeth were collected from 10 healthy individuals aged between 16 and 18 years old. An informed consent was obtained from all donors (UCL/2012/14JUN/283). Apical papilla tissue was loosely attached and could be easily separated from the apex of the developing root with a pair of tweezers [12]. Papillae were then placed in 1% Penicillin-Streptomycin (PEST) in HBSS at 4 °C overnight before use.
Culture of dental stem cells of the apical papilla (SCAP)
Previously characterized human SCAP (RP89 cells [13]) between passages 3 and 10 was used. Mesenchymal stem cell markers (CD90, CD73, CD105) are homogeneously coexpressed by RP89 cells (97% of the population [13]), and RP89 cells are multipotent [14]. SCAP was grown in MEM (Sigma), supplemented with 10% Fetal Bovine Serum (FBS) (Sigma), 1% PEST, and 1% Glutamine (ThermoFisher) at 37 °C under 5% CO2.
Spinal cord hemisection and papilla implantation
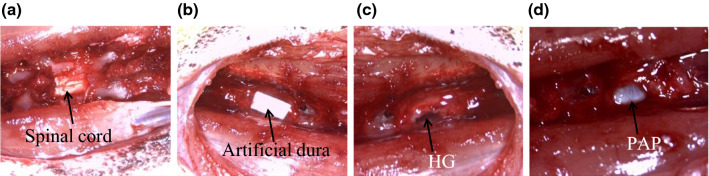
Animal experiments were approved by the ethical committee for animal care (2016/UCL/MD/011). Spinal cord lesions in rats (Long Evans, female, 8 weeks) were performed as described [11] (Fig. 2). A piece of artificial dura was placed above the hemisected spinal cord after administration of the treatment. The animals were given cyclosporine (15 mg/kg, subcutaneous) 1 day prior to the surgery and every day during the entire course of the experiment [15] to prevent immunologic responses due to cross-species transplantation. Rat bladder was emptied twice per day until bladder function recovery. Rat body weight was recorded for each animal over the duration of the experiment.
Fig. 2.
Papilla implantation in a rat thoracic spinal cord hemisection a Spinal cord T8-T10 segment after laminectomy. b Placement of artificial dura over the implants. c Hemisected spinal cord and injection of the fibrin hydrogel (HG). d Hemisected spinal cord and implantation of the papilla (PAP).
Animals were divided in 3 experimental groups (n = 7/group): (1) untreated group (UT), where hemisection was performed with no treatment, (2) hydrogel group (HG), where the lesion site was filled with 10 µL of fibrin hydrogel, and (3) papilla group (PAP), where a whole human papilla was implanted in the lesion site and sealed with 10 μL of fibrin hydrogel [11]. Rats returned to a normal nutritional habit after surgery and all groups of rats followed the same body weight curve as non-operated animals (data not shown). Areflexic detrusor resulting in urinary dysfunction was observed for all hemisected rats immediately after surgery. All rats that received apical papilla implantation recovered bladder function within 2 days while hemisected and fibrin treated animals recovered bladder function within 3 days (data not shown).
Impact of the papilla implantation on spinal cord tissue at the lesion site
After 1 week, the spinal cords were retrieved and four spinal cords out of 7 (per condition) were treated for gene and protein analysis, while three were processed for histological analysis.
Genetic and proteomic analysis of inflammatory markers on tissue extracts
The papilla was removed from the spinal cord and a 2 cm section of the spinal cord centered on the lesion (1 cm in each direction) was retrieved. Then, this segment was cut into four pieces centered on the lesion (Fig. 3). Each piece was weighted, snap frozen, and kept at − 80 °C until further analysis. One part was processed for gene expression analysis and another for protein analysis (n = 4).
Fig. 3.
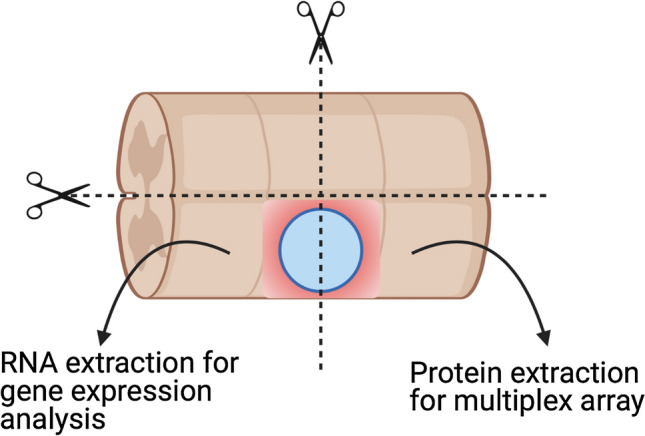
Experimental setting for the analysis of the spinal cord tissue around the lesion Spinal cords were retrieved, cut around the lesion (1 cm in each direction) and in 4 pieces. The two pieces rostral and caudal were processed for RNA and protein extraction, respectively. Created with Biorender.com
Gene expression
Spinal cords were homogenized in Trizol and RNA was extracted using the RNAeasy Mini kit (Qiagen, Valencia, CA). A reverse transcription kit (RT2 HT First Strand kit, Qiagen) was used to synthesize cDNA from 1 μg of total RNA. Real-time quantitative polymerase chain reaction (qPCR) was performed with a STEP one PLUS instrument and software (Applied Biosystems, Foster City, CA, USA) using a RT2 Profiler™ PCR Array Rat Inflammatory Cytokines and Receptors (#PARN-011Z) as per supplier instructions. Data analysis was performed using GeneGlobe (https://geneglobe.qiagen.com/us/). The CT cut-off was set to 38 and the normalization was done by GeneGlobe platform using Actin Beta (Actb), Beta-2-Microglobulin (B2m), and Ribosomal Protein Lateral Stalk Subunit P1 (Rplp1) setting the Fold Regulation Threshold at 1.5. The p values are calculated based on a Student’s t test of the replicate 2^ (- Delta CT) values for each gene in the control group and treatment groups.
Protein expression analysis
Harvested spinal cord samples were homogenized in RIPA lysis buffer, centrifuged, and the supernatants collected followed by protein concentration determination using BCA protein assay per supplier instruction. Samples were then shipped to Eve Technology (Calgary, CA) to be analyzed in duplicates using the Rat Cytokine Array / Chemokine Array 27 Plex.
Histological analysis of the spinal cord lesion and surrounding tissue Staining
Intact spinal cords samples (n = 3) were processed for immunofluorescence staining as described [16]. Three consecutive sections were stained for (1) CD68 and ionized calcium-binding adapter molecule 1 (Iba1), (2) glial fibrillary acidic protein (GFAP) and 5-hydroxytryptamine (5-HT), and (3) CD68, inducible nitric oxide synthase (iNOS) and Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL). In brief, spinal cord segments centered on the lesion site and including 1 cm of surrounding region (in each direction) were harvested and fixed in 4% formaldehyde in 0.9% sodium chloride for 24 h. Samples were then incubated in 30% sucrose and embedded in Tissue-Tek® O.C.T.™ compound (Sakura, AJ Alphen aan den Rijn, NL). Spinal cords were serially sliced at 10 μm with a cryostat (Leica), mounted on Superfrost Plus slides (Fisher Scientific), and stored at − 20 °C until use. Slides were allowed to dry at room temperature overnight before staining. Prior to the immunofluorescence staining, sections were stained for TUNEL using the DeadEnd™ Fluorometric TUNEL System kit (Promega, Leiden, NL) according to supplier instructions. Sectioned fixed tissues were then permeabilized, blocked, and incubated with primary antibodies (Supplementary Table 1) for 1 h at room temperature. Negative controls were obtained by omitting the primary antibodies. Samples were then washed and incubated with appropriate secondary antibodies for 1 h at room temperature. Cell nuclei were counterstained with DAPI (1 µM, Sigma). Samples were coverslipped with HIGHDEF® IHC fluoromount (Enzo life Sciences Inc., Belgium).
Quantification
Immunofluorescent staining was visualized and digitalized using a Pannoramic 250 FlashIII scanner (3DHistech) at × 20 magnification. Scanned slides were then analyzed using the image analysis tool Author version 2017.2 (Visiopharm, Hørsholm, DNK). On each slide, tissue sections were automatically delimited at low magnification. Lesions were then manually delineated and automatically surrounded by concentric regions distant up to 1 mm from the lesion borders (one concentric circle every 50 µm up to 200 µm followed by one concentric circle every 100 µm up to 1 mm away from the lesion delimitation).
To quantify CD68, Iba1, GFAP, and 5-HT staining, stained pixels in the lesion and concentric circles were detected (high resolution [× 20])) using a thresholding classification method and summed. Results are expressed as a percentage of stained area within analyzed region.
To quantify activated microglia, nuclei were detected (high resolution [× 20]) with a cell classification relying on the DAPI staining. Following segmentation, detected cells were identified by the algorithm according to their expression of both CD68 and Iba1 (CD68+Iba1+) and counted. Results are expressed as number of cells/mm2 of tissue within lesion and concentric circles. To evaluate apoptosis of microglia, nuclei were detected (high resolution [× 20]) with a cell classification relying on the DAPI staining. Following segmentation, post-processing steps were applied to separate successively cells stained for TUNEL, CD68, and iNOS according to fluorescence detection within (TUNEL) or around (CD68, iNOS) the detected nuclei. Detected cells (TUNEL+CD68+iNOS+ and TUNEL+CD68+iNOS−) were finally quantified. Results are expressed as number of cells/mm2 of tissue within lesion and concentric circles.
Thresholds were adjusted on representative stained versus not stained regions. The same parameters were kept constant for all slides. At least 6 serial sections per condition were analyzed.
Influence of a pro-inflammatory stimulus on papilla secretome
Papillae were placed in 48-well plates (1 per well) for 72 h in MEM supplemented with 1% PEST, 1% Glutamine, and 0.5% FBS. Papilla exposed to a pro-inflammatory stimulus (Activated) were incubated in the same medium supplemented with 20 ng/ml of tumor necrosis factor alpha (TNFγ) and 20 ng/ml of interferon gamma (IFNγ). At the end of the experiment, papillae were collected, weighted, snap frozen, and kept at − 80 °C. Supernatants were also stored at − 80 °C. For each independent experiment (N = 3), at least 3 papillae/patient/condition from 3 different patients were used (n = 9 per condition). In parallel, the same experiment was performed on SCAP (N = 3, n = 4).
Papilla secretome was analyzed using a Proteome Profiler Human Cytokine Array Kit following supplier instructions (ab133998, abcam, Cambridge, UK). Two membranes were used per condition and incubated with 2 pools of supernatants (samples from 3 different experiments where supernatants from 3 to 4 patients and 9 to 10 papillae were pooled).
Gene expression and concentration of tumor necrosis factor stimulated gene-6 (TSG-6) were measured by RT-qPCR [17] and ELISA (RAB1092-1KT, Sigma-Aldrich), respectively, while hepatic growth factor (HGF) concentration in supernatant was quantified by ELISA, according to the supplier instructions (BGK14210, Peprotech, respectively). Indoleamine 2, 3- dioxygenase (IDO) activity was measured as described [18]. Briefly, samples were incubated with trichloroacetic acid (30%) for 30 min at 50 °C. Then Ehrlich solution (2% 4- diméthylaminobenzaldehyde in acetic acid) was added (1:1 v/v) to the samples and incubated 10 min at RT. The optical density was measured at 490 nm and IDO activity was quantified using a standard curve obtained with different concentrations of L-Kynurenine (200 to 0 µM). Prostaglandin E2 (PGE2) was quantified based on a previously published procedure [19]. Briefly, methanol (500µL) and d4-PGE2 (as internal standard) were added to the samples (150µL). After incubation for 2 h at − 20 °C, samples were centrifuged and supernatant dried under a nitrogen flux. Samples were then reconstituted in methanol (20 µL) and analyzed using an Acquity UPLC class H system (Waters) coupled to a Xevo TQ-S mass spectrometer (Waters). For lipid separation, we used an Acquity UPLC BEH (150 × 2.1; 1.7 µm) from Waters maintained at 40 °C and a gradient between water–acetonitrile–acetic acid (94.9:5:0.1; v/v/v) and acetonitrile-acetic acid (99.9:0.1; v/v). Electrospray negative ionization mode was used. The transitions used were 351.10 > 315.18 (Q) and 351.10 > 271.15 (q) for PGE2 and 355.10 > 275.15 (Q) and 355.10 > 319.18 (q) for d4-PGE2. For data processing, the MassLynx software was used and the ratio between the area under the curve of the PGE2 signal and the area under the curve of the d4-PGE2 signal was used for data normalization. Calibration curves were obtained in the same experimental conditions to determine sample PGE2 amount.
Statistics
Statistical analysis was performed using PRISM (GraphPad Software, CA, USA) (p < 0.05). Error bars represent the standard error of the mean in all figures. The specific test used is specified in each figure legend.
Impact of papilla transplantation in hemisected rat spinal cord on its transcriptome
Surgery
The papilla implantation has been performed as described in 2.4 (n = 3 from 2 different patients). For each papilla implanted, another one from the same patient was weighted and snap frozen. Twenty-four hours after the implantation, papillae were retrieved, weighted, and snap frozen. All samples were kept at − 80 °C until analysis.
RNA sequencing
Total RNA was extracted from non-implanted and implanted papillae using RNeasy Micro kit (Qiagen) and samples were treated with DNase according to the manufacturer’s instructions. In total, RNA from 6 samples were quantified by Qubit RNA HS assay kit (Thermo Fisher Scientific, Q10211) on a Qubit 4 Fluorometer (Thermo Fisher Scientific). RNA integrity was evaluated on the Agilent 2100 Bioanalyzer using the RNA 6000 nano kit (Agilent, 5067–1511). All samples had RNA integrity number values between 7.7 and 8.3.
Libraries were prepared starting from 150 ng of total RNA using the KAPA RNA HyperPrep Kit with RiboErase (HMR) (KAPA Biosystems, KK8560) following the manufacturer’s recommendations (KR1351–v1.16). Libraries were equimolarly pooled and sequenced on a single lane on an Illumina NovaSeq 6000 platform. All libraries were paired-end (2 × 100 bp reads) sequenced and a minimum of 25 million paired-end reads were generated per sample.
RNAseq data analysis
Data were processed using a standard RNAseq pipeline, including FastQC v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) for the quality control of the raw data, and trimmomatic v0.38 [20] to remove low quality reads and trim the adapter from the sequences. Hisat2 v 2.1.0 [21] was used to align reads to the human genome (GRCh38). Gene expression levels were evaluated using featureCounts v2.0.0 [22] and Homo_sapiens.GRCh38.94.gtf. DESeq2 Bioconductor package v1.26.0 [23] was used for differential expression analyses. Genes with an adjusted p value < 0.05 and an absolute log2 fold change > 0.5 were considered differentially expressed. Gene Set Enrichment Analysis (GSEA) was done using clusterProfiler_3.14.3 Bioconductor package [24]. Raw data can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191140.
Results
Impact of the papilla implantation on inflammatory markers at the lesion site
The effect of papilla implantation (PAP) on the expression of inflammatory markers in the spinal cord tissue at the lesion site1 week after implantation was evaluated.
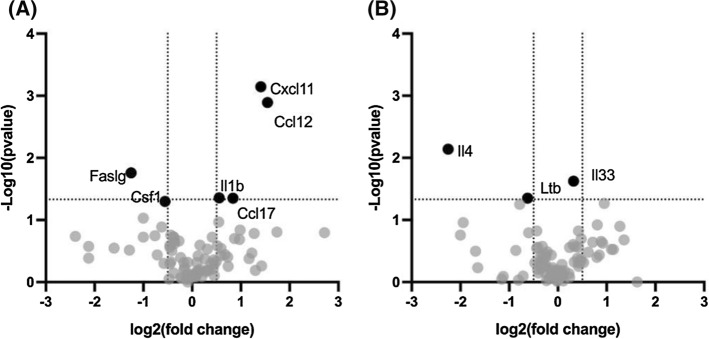
Spinal cord tissue was first analyzed for a panel of genes related to inflammation. A total of 6 genes were significantly regulated following PAP implantation (Fig 4): 4 were up-regulated (Interleukin-1β (IL1β), Chemokine (C-C motif) ligand 12 (Ccl12), Ccl17, and Chemokine (C-X motif) ligand 11 (Cxcl11)), while 2 were down-regulated (Fas ligand gene (Faslg) and colony-stimulating factor 1 (Csf1)). Injection of the hydrogel alone (HG) did not significantly impact gene expression. Only interleukin-4 (IL4) and lymphotoxin β (Ltb) were significantly down-regulated.
Fig. 4.
Impact of papilla implantation on gene expression of inflammatory cytokines and receptors at the lesion site Papillae were implanted in a hemi-section spinal cord lesions. Gene expression was analyzed using a RT2 Profiler™ PCR Array Rat Inflammatory Cytokines and Receptors. A PAP vs UT, B HG vs UT (n = 4). Only the cytokines identified by a black dot were significantly differentially expressed (p < 0.05)
Next, the concentration of certain cytokines was quantified in the spinal cord lesion. Pro- inflammatory cytokines IL6, IFNγ, IL18, and monocyte chemoattractant protein-1 (MCP-1 or CCL2) had the highest concentrations (40–600 pg/mg of tissue) in the hemisected spinal cord (UT), while the rest of the molecules analyzed ranged from 4 to 40 pg/mg, including macrophage inflammatory protein 2 (MIP-2), Leptin, IL17α, IL1β, IL1α, IL13, and IL2 (Fig. 5). Papilla implantation induced a decrease of most of the quantified cytokines to a higher extent that the hydrogel alone compared to the UT condition.
Fig. 5.
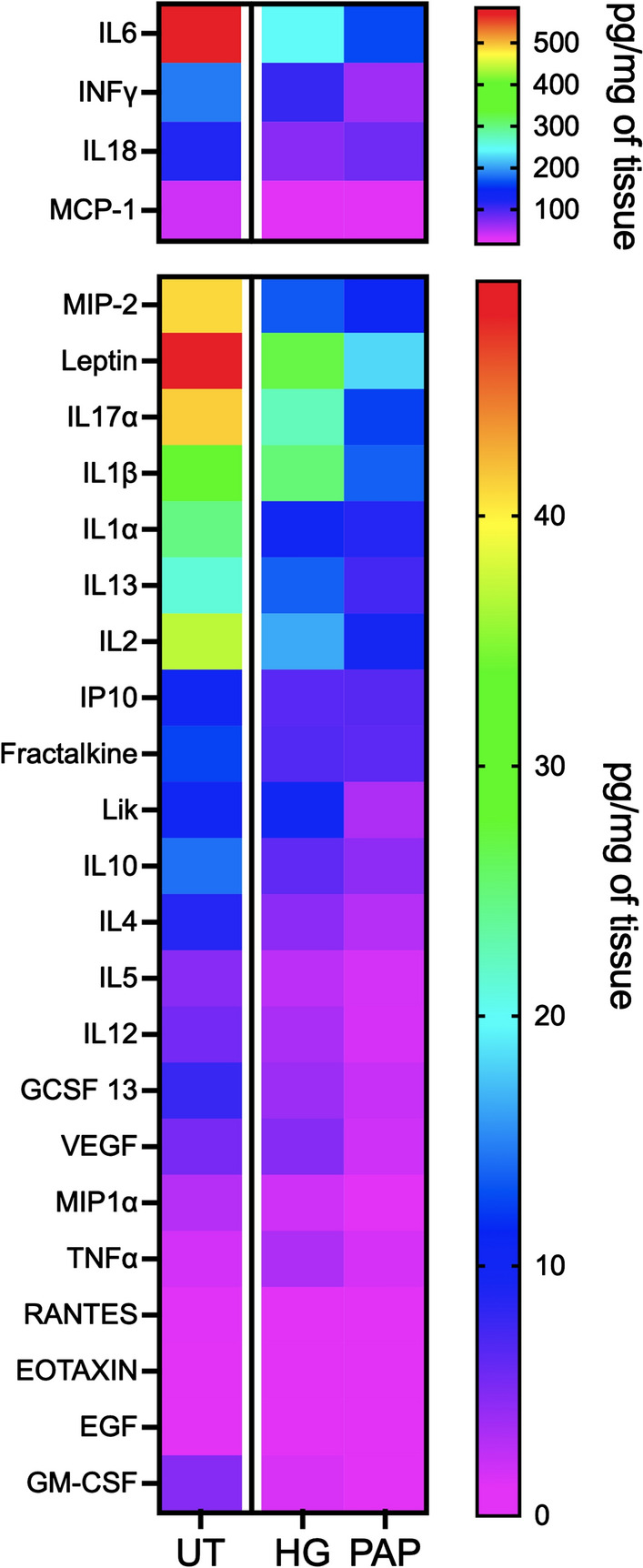
Papillae implantation significantly reduced the increase of cytokine expression observed after spinal cord injury Papillae were implanted in a hemi-section lesion, spinal cords were collected at the lesion site after 1 week and proteins were extracted. Cytokine concentrations were measured using a multiplex array (Rat Cytokine Array / Chemokine Array 27 Plex) (n = 4).
Influence of the papilla implantation on cell components at the spinal cord lesion site
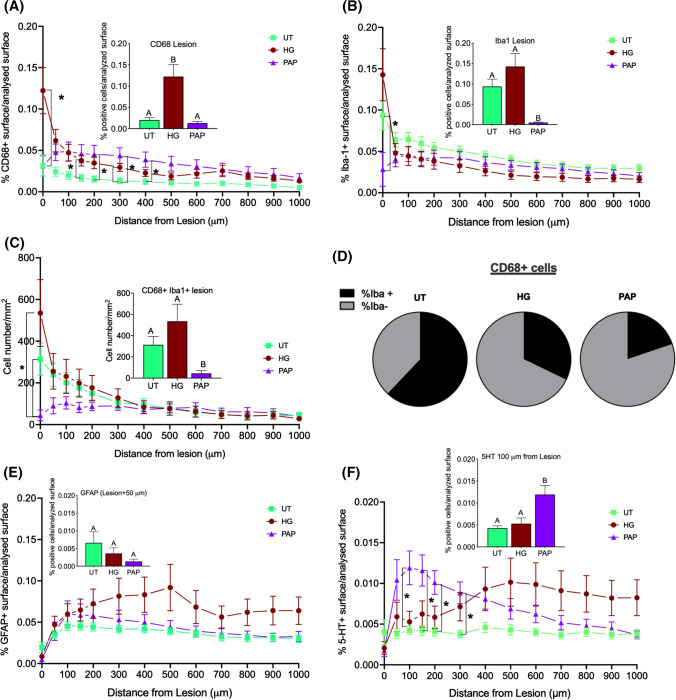
The objective was to study how the papilla would influence the surrounding spinal cord tissue with a focus on the activated glial cells (microglial cells with CD68 and Iba1, astrocytes with GFAP) and on motoneurons (5-HT) [25]. The analysis was performed within the lesion and in concentric circles around the lesion as described in 2.5.2.
Immunoreactive macrophages/microglia (CD68) were significantly increased in the lesion by the injection of the fibrin hydrogel (Fig. 6A). At a further distance from the lesion (50 to 400 um), the percentage of tissue positive for CD68 was significantly higher in the PAP condition and tended to be higher in the HG condition compared to UT. The percentage of tissue positive for activated microglia (Iba1) was significantly lower in the PAP condition compared to the controls (Fig. 6B) at the lesion site but was similar whatever the conditions away from the lesion. The number of Iba1/CD68 double positive cells per mm2 was significantly decreased by papilla implantation compared to controls at the lesion and up to 300 µm away from the lesion (Fig. 6C). Additionally, the percentage of CD68-positive cells that were also positive for Iba-1 was lower after papilla implantation compared to controls (Fig. 6D). Papilla implantation tended to decrease the percentage of GFAP staining but the differences were not significant (Fig. 6E). Finally, the percentage of tissue positive for 5-HT was significantly higher in PAP samples in the immediate proximity of the lesion (up to 300 µm from the lesion) than in the UT group (Fig. 6F).
Fig. 6.
Papilla implantation significantly reduce the number of activated macrophages and increase the percentage of 5-HT positive cells in the spinal cord lesion Papillae (PAP) were implanted in a rat spinal cord hemisection model (controls were fibrin hydrogel (HG) and untreated hemisection (UT)) for one week. Then, spinal cord tissue centered on the lesion (1 cm in each direction) was processed for immunofluorescence. Staining was quantified at the lesion (histograms inserted in A, B, C, D and F) and away from the lesion in concentric circles up to 1 mm (A, B, C, D and F) (n = 3). Stained area for activated microglia: CD68 A and Iba1 B, C Number of cells positive for both CD68 and Iba1. D Proportion of CD68 macrophages that are positive also for Iba1, Stained area for astrocytes: GFAP(E and for motoneurons: serotonin (5HT), F. Significant differences were analyzed using a mixed-effects model with geisser-Greenhouse correction. A Tukey test was used for simple effect within rows. Conditions not linked by the same letter are significantly different; *p < 0.05
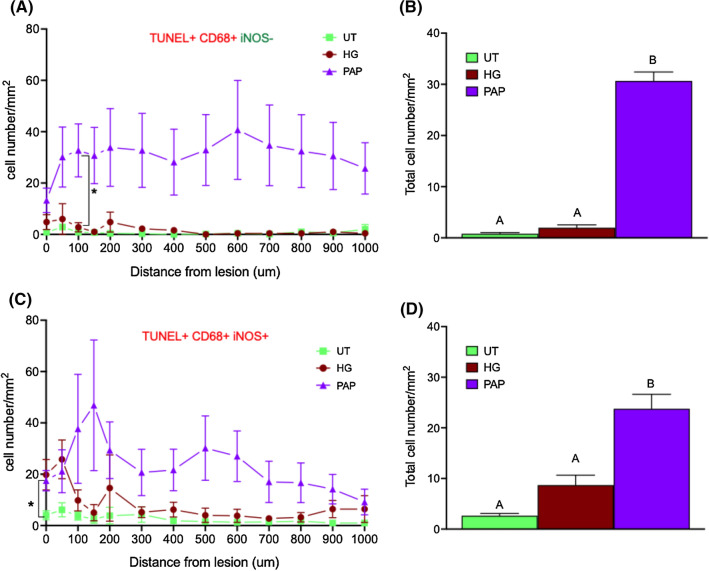
In order to determine whether the decreased activated microglia staining observed after papilla implantation could be related to the apoptosis of CD68 and iNOS (a common marker of activated glia cells) positive cells, further experiments were carried out. The number of apoptotic macrophages (CD68 + TUNEL +) expressing or not iNOS was thus quantified at the lesion site and away from the lesion up to 1 mm. Papilla implantation induced a significant increase of apoptotic macrophages (irrespective to their iNOS expression) compared to controls (Fig. 7).
Fig. 7.
Dental papilla implantation promoted the apoptosis of pro-inflammatory microglia apoptosis at the lesion site Quantification of cells positive for TUNEL and CD68 but negative for iNOS in the lesion and in each concentric circles around the lesion (A) and in the total surface analyzed (lesion + all concentric circles) (B) one week after implantation. Quantification of cells positive for TUNEL, and stained for CD68 and iNOS in concentric circles around the lesion (C) and in total surface analyzed (D). n = 3 Significant differences were analyzed using a mixed-effects model with geisser-Greenhouse correction. A Tukey test was used for simple effect within rows. Conditions not linked by the same letter are significantly different; *p < 0.05
Papilla secretome
The objective of this part was to analyze papilla secretome and how it could be influenced by pro-inflammatory conditions.
Inflammatory cytokines production by PAP
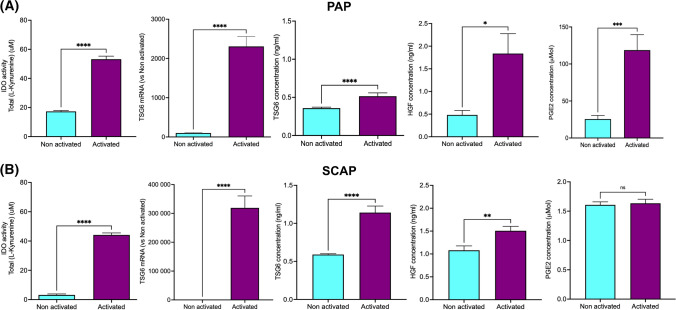
As far as we know, the secretome of human dental papillae has never been described. Here, the cytokines secreted by papillae incubated 72 h in culture medium were analyzed using a cytokine membrane array, either in steady-state or pro-inflammatory conditions. Fifty-nine cytokines were detected in papillae-conditioned medium regardless of the condition (Fig. 8A). Most of them were from the CC, CXC, and interleukin families but molecules stimulating neuroregeneration, like neurotrophin 3 (NT3), leukemia inhibitory factor (LIF), or glial-derived neurotrophic factor (GDNF) were also secreted. Pro-angiogenic molecules, like vascular endothelial growth factor (VEGF), and platelet-derived growth factor bb (PDGFbb) and pro-resolutive cytokines, like HGF, transforming growth factor β (TGF β), IL10, IL13, and IL4 were also detected. Only one was specific to the non-activated state (i.e., CCL26), while 20 were found only in papillae incubated with TNFα and IFNγ (Fig. 8B). They belonged mainly to the CC and interleukin families, but other cytokines, like neurotrophin 4 (NT4), granulocyte colony-stimulating factor (GCSF), or placental growth factor (PIGF), were also detected. When comparing the 59 cytokines common to the two conditions, 5 of them were significantly up-regulated by the incubation with TNFα and IFNγ (Fig. 8C): CCL5 (RANTES), CCL8 (monocyte chemoattractant protein-2 (MCP-2)), CCL7 (MCP-3), and osteopontin (OPN).
Fig. 8.
Impact of pro-inflammatory stimulus on papilla secretome A Venn diagram (https://bioinfogp.cnb.csic.es/tools/venny/) of all the cytokines detected in papilla supernatant. In blue, the cytokines detected in the supernatant of papilla non activated only, in yellow, the cytokines detected in the supernatant of papilla activated only and, in the middle, the cytokines detected in the supernatant of both. B List of the cytokines detected in the activated state only or in both conditions. C Volcano plot of all the cytokines comparing activated with non activated papillae. Multiple t tests with a False discovery approach: Two stage set up method of Benjamini, Krieger and Yekutieli (Q = 1%). n = 2, samples from different patients were pooled
Immunomodulatory molecules production by SCAP
The impact of a pro-inflammatory environment on the expression and secretion of molecules specifically known for their immunomodulatory properties was then studied. Papilla resident stem cells, SCAP, were studied as a comparison, as stem cells are considered strong producer of immunomodulatory molecules [26].
IDO activity also increased significantly both in papilla and SCAP-conditioned medium (3- and 14-folds, respectively) (Fig. 9). The expression of the TSG6 gene was strongly up-regulated by TNFα and IFNγ in papillae (23-folds), while the increase of TSG6 protein concentration in the medium was more modest (1.4-folds) in the presence of a pro-inflammatory stimulus (Fig. 9A). In comparison, TSG6 gene expression was robustly increased (3192-folds), while its secretion into the media had a twofold increase (Fig. 9B). The secretion of HGF secretion was also stimulated both in papillae and SCAP-conditioned medium but to a higher extent in papillae (3.8- and 1.4-folds, respectively). PGE2 was only affected in papillae with a 4.6-fold increase in the presence of TNFα and IFNγ. TGFβ1 was expressed by papillae and SCAP but was not impacted by the incubation with pro-inflammatory cytokines (data not shown).
Fig. 9.
Pro-inflammatory cytokines regulate human dental papilla and SCAP secretion of immunomodulatory molecules.22 Gene expression and concentrations of different immunomodulatory molecules have been analyzed for (A) papillae (PAP) and (B) SCAP in steady-state condition (non activated) or in presence of a pro-inflammatory stimulus (N = 3, n = 4). Significant differences were analyzed using a t test; *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001
Influence of papilla implantation in a spinal cord lesion on its gene expression Next, the transcriptomic changes within the dental papilla following implantation into spinal cord lesions were investigated using RNAseq
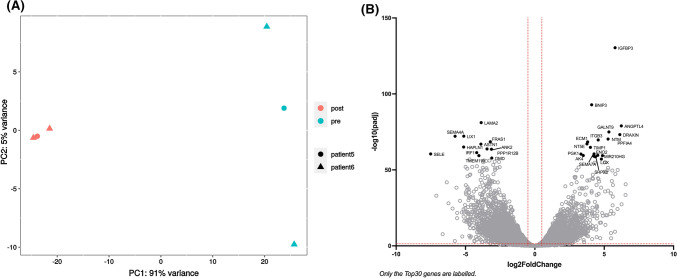
The sequencing identified 44 835 expressed genes, among which 7 308 genes had an adjusted p value (padj) smaller than 0.05 and a log fold change above 1 (log2FoldChange) between the implanted and the non-implanted papillae.
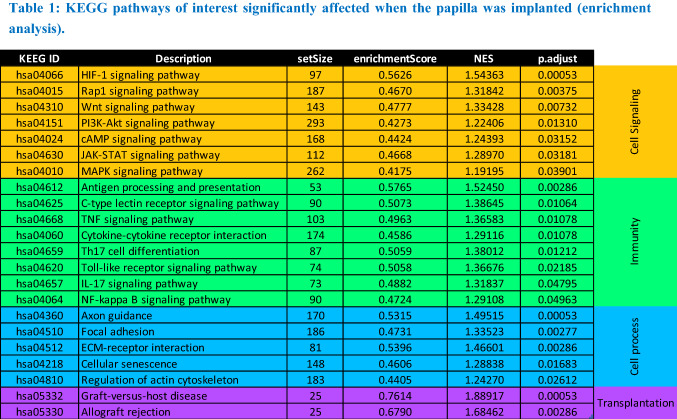
The Principal Component Analysis (PCA) highlighted that most of the differences observed between the samples can be attributed to the implantation rather than to the origin of the papilla. (Fig. 10A). As visible on the volcano plot, the papilla gene expression seemed equally up- and down-regulated upon implantation in the spinal cord lesion (Fig. 10B). A KEEG pathway enrichment analysis was performed and allowed the identification of 64 pathways significantly affected by the implantation (padj < 0.05), among which 22 relevant pathways have been selected (Table 1).
Fig. 10.
Papilla genes whose expression has been affected by acute implantation in a spinal cord hemisection. Papillae were implanted in a rat spinal cord hemisection (n = 3 papillae from 2 different patients) and retrieved 24 h later. RNA was extracted and papilla transcriptome was analyzed by RNAseq. A Principal Component Analysis (PCA), B Volcano plot (red dotted lines: padj < 0.05 and log2FoldChange < 2)
Table 1.
KEGG pathways of interest significantly affected when the papilla was implanted (enrichment analysis)
Implantation of the papilla in the spinal lesion strongly affected its transcriptome, involving mostly cell signaling pathways, like MAP kinase (MAPK), PI3-Akt or JAK-STAT pathways, immunity-related pathways, like nuclear factor-kappa B (NFκB), TNFβ and cytokine-cytokine receptor signaling pathways (Table 1). Importantly, PI3K-Akt, JAK-STAT and Toll-like receptor signaling pathways are also associated with macrophage polarization [27]. Pathways associated with cell processes, like focal adhesion, or cellular senescence, as well as transplantation were also enriched.
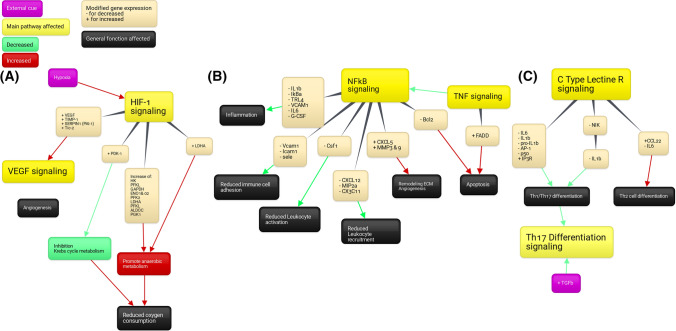
Genes belonging to the MAPK signaling pathways tended to be more down-regulated than up- regulated, while genes belonging to the PI3K-Akt pathway seemed equally up- and down- regulated (Supplementary data 1A&B). Certain genes associated with hypoxia-inducible factor-1 (HIF-1) signaling pathway tended to be up-regulated (Supplementary data 1C), among which TIMP metallopeptidase inhibitor 1 (TIMP1) and VEGF leading to the activation of the VEGF signaling pathways and phosphoinositide-dependent kinase-1 (PDK-1), phosphoglycerate kinase 1 (PGK1), enolase 1 (ENO1), and ENO2 involved in the promotion of the anaerobic metabolism and angiogenesis (Figure 11A).
Fig. 11.
Pathways impacted by papilla implantation in an acute SCI A HIF-1 pathway, B and C Pathways related to inflammation. Created with Bubbl.us.
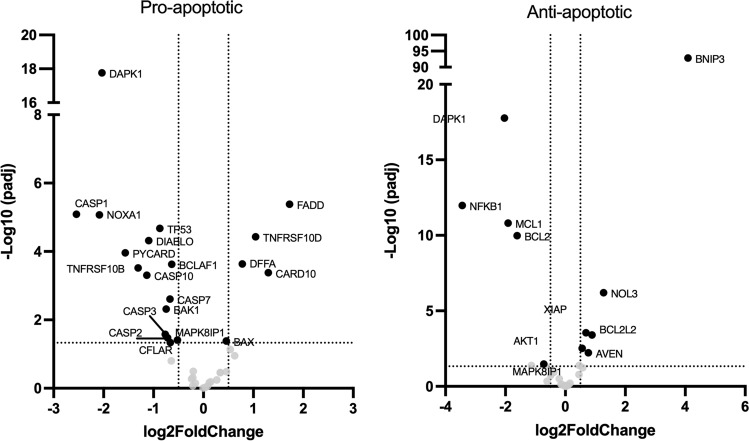
Genes included in pathways related to immunity were mostly down-regulated (TNF, NFKB, IL17, C-type lectin receptor, Th17, and toll-like receptor (TLR) signaling pathways). The gene expression of several markers of inflammation decreased, like TLR4, NFκB/IκBα, IL1β, IL6, and NOD-like receptor family pyrin domain containing 3 (NLRP3) (Figure 11 B and C), potentially leading to a reduced inflammation, immune cell activation and recruitment as well as decreased Th1/Th17 differentiation, while promoting ECM remodeling, angiogenesis, and Th2 cell differentiation. Two of the main anti-apoptotic genes, B-cell lymphoma 2 (Bcl-2) and induced myeloid leukemia cell differentiation (Mcl-1), decreased but overall, most of anti-apoptotic genes increased, while most pro-apoptotic gene expression decreased (Fig 12).
Fig. 12.
Influence of papilla implantation on genes related to apoptosis Genes whose expression is involved in apoptosis have been selected in the RNAseq data set, separated in function of their pro- or anti-angiogenic effect and their differential expression has been plotted in volcan plots (dotted lines: padj < 0.05 and log2FoldChange < 2).
Two KEGG pathways related to tissue transplantation were enriched: graft vs host disease and allograft rejection. Genes involved in both pathways were significantly down-regulated (Supplementary data 2).
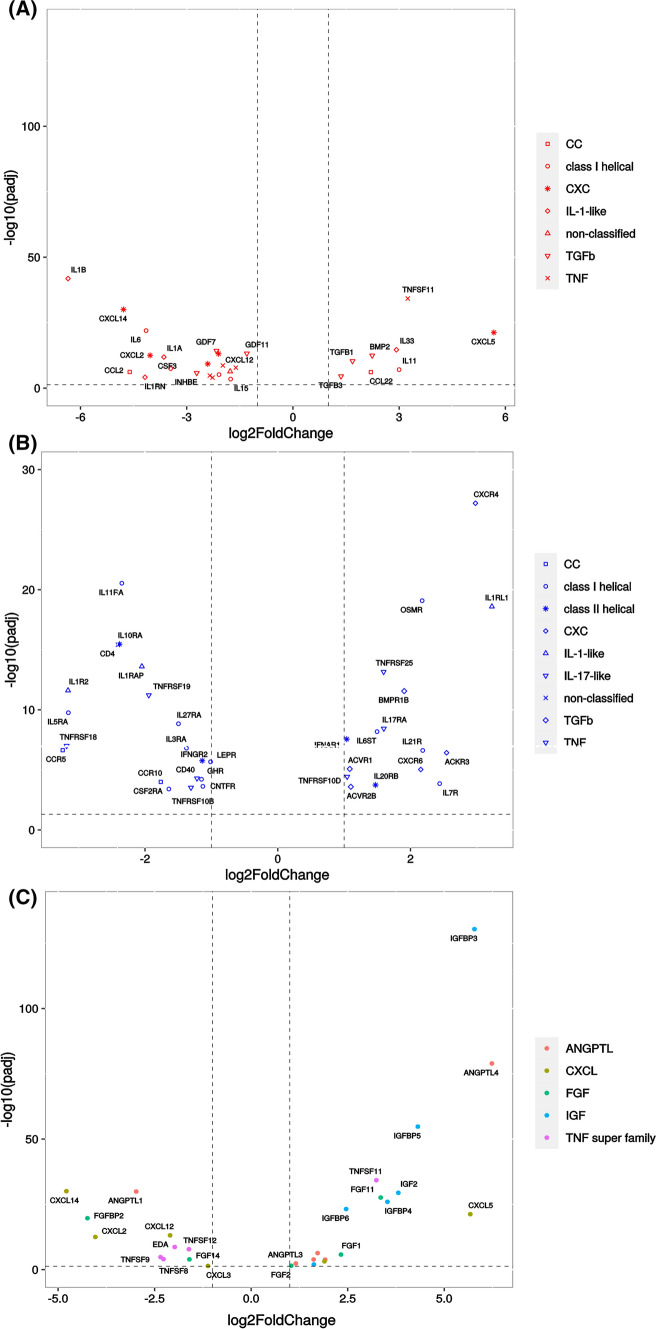
Genes related to cytokines, growth factors, and their receptors were analyzed specifically as they could influence the surrounding tissue (Fig. 13). The gene expression of 28 cytokines, belonging to 6 main families, has been affected by the papilla transplantation (Fig. 13A). Their expression was mostly down-regulated (8 up-regulated for 20 down-regulated). They are mainly related to the immune response, like IL1β andα, IL6, MCP1 (CCL2), or CXCL12 that are down-regulated or CXCL5, IL33, or TGFβ1 that are up-regulated. Regarding the gene expression of cytokine receptors in the papilla, 35, belonging to 8 main families, were differentially regulated by papilla implantation, equally down- and up-regulated (19 down and 16 up) (Fig. 13B). With the addition of class II helical and IL-17-like cytokines, the genes related to receptors belonged to the same families than the cytokine genes.
Fig. 13.
Influence of implantation on papilla gene expression of cytokines, growth factors and their receptors A Cytokines, B Cytokine receptors, C Growth factors from main families (only families with more than five members are plotted)
Gene expression of growth factors was also influenced by papilla implantation, as the gene expression of 63 growth factors was significantly changed, mostly toward up-regulation (38 vs 25 down-regulated) (Supplementary data 3). These genes belonged mainly (at least 5 genes per family) to the TNF super family, the angiopoietin (ANGPTL), Insulin Growth factor (IGF), Chemokine ligand (CXCL), and Fibroblast Growth Factor (FGF) families (Fig. 13C).
Discussion
The human dental apical papilla is the tissue giving rise to the tooth roots, but while SCAP and the apical papilla have been intensively studied recently, the papilla tissue itself has never been considered as a therapy before. In our previous work, we showed that implantation of the papilla in a rat spinal cord hemisection stimulated rat functional improvement and reduced microglia activation in the spinal cord lumbar section [11].
The objective of this work was then to gain a better understanding on how the papilla could exert its therapeutic actions. In this study, we conducted a thorough study where we demonstrated that the papilla implantation in the SCI decreased the pro-inflammatory markers in the spinal cord, dampened the microglial/macrophage activation, at least partially by apoptosis of activated cells, and increased the percentage of immunoreactive 5-HT-positive cells. We also provided for the first time information on the papilla secretome when subjected to a pro-inflammatory stimulus and showed that, like SCAP, the papilla responded to inflammation by secretion or expression and secretion of immunomodulatory molecules. Finally, an analysis of the papilla genomic expression post-implantation provided also, for the first time, insights on the pathways activated in the papilla implanted in the spinal cord that might explain its therapeutic action. We showed here that the papilla, most likely by protecting and anchoring its native stem cells, has a great healing potential mostly driven via its secretome, that could benefit any tissue needing repair. Analysis of gene expression and the protein secretion of markers associated with the inflammation at the lesion showed that one week after implantation, only a few genes were still regulated by the papilla implantation, while the concentration of most of the pro-inflammatory proteins was decreased. This difference might be due to the timing differences between gene expression modulation and protein secretion. Indeed, one week after implantation the gene expression might not be longer affected as much as the protein secretion. The two genes significantly activated by the papilla implantation (CXCL11 and CCL12) are related to the chemotaxis of immune cells. They are both produced by activated glial cells. CXCL11 attracts activated T cells and is induced by IFNγ. It is up-regulated after nerve injury and mediates inhibitory synaptic transmission but does not seem to contribute to neuropathic pain [28]. CCL12 is a potent monocyte attractant but does not seem involved in neuropathic pain either [29]. Of note, in our previous study, no neuropathic pain was detected after papilla implantation [11]. The only gene that was significantly down-regulated was the Fasl gene (Faslg). Fasl is a member of the TNF superfamily and the activation of Fas/Fasl plays an important role in apoptosis, inflammation, gliosis, and neurodegeneration in human SCI [30]. Zhang et al. showed that minocycline inhibition of mitochondrial-dependent cell death after SCI was linked to a reduced expression of Fasl [31]. As mentioned, papilla implantation significantly decreased the concentration of the main pro-inflammatory cytokines detected at the lesion site. This was associated with a strong reduction of activated microglia at the lesion site. Then, the papilla had a strong anti-inflammatory effect on the spinal cord tissue, either by reducing the number of activated macrophages and consequently the concentration of pro-inflammatory molecules in the spinal cord or by providing a lower pro-inflammatory environment that reduced the number of activated cells. Several studies report that dampening the inflammation is most usually associated with a better outcome [4], as activated macrophages produce cytotoxic mediators, high levels of oxidative metabolites and induce neural necrosis. They slow down recovery of patient functions and are consequently harmful, while pro-resolutive macrophages are considered beneficial [27]. As far as we know, no other studies used a whole tissue as a treatment for SCI, the comparison with published work is thus limited. As the papilla is the SCAP niche, a parallel could be then drawn between what was observed for the papilla and what was reported in the literature when isolated MSC was implanted at the lesion site. Numerous studies report the beneficial effect of MSC on spinal cord injury and most authors linked this therapeutic action to MSC secretome and anti-inflammatory/pro-resolutive action [32, 33].
As Lloyd et al. showed that activated microglial death was necessary for the regeneration of the central nervous system [34], although in a different model, we quantified the number of activated microglia that were apoptotic. We hypothesized that inducing the death of activated microglia was one of the ways the papilla could exert its therapeutic action. As clearly much more CD68-positive cells were apoptotic in the spinal cord tissue around the lesion for the PAP condition compared to the controls, this hypothesis might be indeed plausible.
We also showed that implantation of the papilla in the lesion resulted in a significant increase of 5-HT immuno-reactivity up to 300 µm away from the lesion. 5-HT plays a prominent role in locomotion and increased 5-HT neurotransmission often correlates with functional recovery after SCI [35]. Usually, 5-HT axons caudal to the lesion epicenter degenerate whereas there is an increase in 5-HT axon density rostral to the lesion [35]. Here we observed that most of the 5-HT staining visible in PAP spinal cord sections was caudal to the lesion (Supplementary data 4). As for cell transplantation, several studies report that injection of neural progenitors or MSC at the lesion site induced increased 5-HT axon sprouting caudal to the lesion and functional recovery [35]. We recently showed that SCAP seeded on brain derived neurotrophic factor (BDNF)-loaded microcarriers increased 5-HT level and Basso, Beattie and Bresnahan (BBB) score after injection in a rat spinal cord contusion immediately post-injury [16]. It can then be suggested here that the decrease of pro-inflammatory molecules and activated macrophages, combined with an increase of 5-HT caudally to the lesion, could explain why the papilla implantation supports functional recovery.
We analyzed papilla gene expression and protein secretion at a steady-state or under pro- inflammatory conditions. From our perspective, it was crucial to analyze how the papilla reacted to its environment to understand how its secretome could stimulate spinal cord repair. First, numerous cytokines were secreted by the papillae, most of them were chemo-attractants, proteins involved in the inflammatory response, and pro-regenerative and neurotrophic factors, like GDNF, NT3, NT4, and BDNF. MSCs secrete VEGF, HGF, IGF-1, stanniocalcin-1, TGFβ, and granulocyte–macrophage colony-stimulating factor (GM-CSF) that promote the survival of damaged neurons and oligodendrocytes [36]. They also stimulate angiogenesis via the secretion of PIGF, MCP-1, bFGF, and IL-6 [37] as well as proliferation and regeneration of the remaining neurons by the GDNF, BDNF, and NGF [38]. MSCs exert an immunomodulatory effect via secretion of IL-10, TGFβ, PGE2, galectin-1, IDO, and HGF [36, 39]. Yu et al. profiled the secretome of SCAP by mass spectrometry and found more than 2000 proteins in their conditioned medium, including chemokines and angiogenic, immunomodulatory, anti- apoptotic, and neuroprotective factors [40], but as far as we know, no study reported the same analysis for the papilla secretome. As most of these molecules have been detected in the papilla-conditioned medium in this work, we can hypothesize that the apical papilla supports tissue repair by a paracrine effect as described for MSC. Of note, most of the cells residing in the papilla were positive for immunofluorescence staining against mesenchymal markers [13]. This would explain the properties of the papilla and why a parallel can be observed with SCAP secretome. Thus, the papillae would act as a reservoir of MSC, presenting the advantage of keeping the cells in their native niche. This would ensure a better functionality and viability post-implantation than when the cells are injected as a suspension. In addition, the cells composing the papilla are mostly quiescent (staining against mitochondria cytochrome oxidase, Supplementary data 5) in a poorly vascularized niche [41]. This might be one of the reasons why cells from the apical papilla were able to survive and modulate the aggressive SCI environment significantly better than the implanted dissociated and cultured cells (SCAP) [11, 42].
Although the ability of certain MSC to secrete immunomodulatory factors in the presence of a pro- inflammatory stimulus is well known and well described [43], far less was about the human papilla. Here, in addition to the detection of IFNγ, IL4, IL10, IL13, and FGFs in the cytokine array, we demonstrated that like SCAP, papillae were able to respond to TNFα and IFNβ by an increased production of immunomodulators. These results further confirmed the paracrine action of the papilla and were consistent with an immunomodulatory effect on the SCI. Indeed, HGF, for instance, is a crucial factor for SCI as it promotes neuronal survival through pro-angiogenic, anti-inflammatory, and immune-modulatory mechanisms [44]. Song et al. demonstrated that the therapeutic effect of MSC-conditioned medium in a rat spinal cord contusion model was associated to HGF [39]. Li et al. showed that the beneficial effect of MSC in a mouse model of multiple sclerosis was directly linked with IDO [45], while Zhang et al. showed that the therapeutic efficacy of MSC was linked to the IDO-TSG6 axis, where kynurenine or kynurenic acid, controlled by IDO activity, augmented the expression of TSG-6 through activating their common receptor aryl hydrocarbon receptor [46]. Of note, an increasing body of evidences reports that molecules of the MSC secretome are transported by extracellular vesicles and demonstrates that MSC therapeutic effect could be recapitulated by their extracellular vesicles [47, 48].
Finally, when looking at the transgenic expression of the papilla after implantation, the first observation was that it was strongly impacted, with more than 7000 genes significantly regulated. The dental papilla was found to react to the implantation environment and promote immune modulation, despite its removal from its native site. Interestingly, the HIF-1α pathway was activated, probably due to the fact that the papilla was subjected to hypoxia after its implantation. It then induced an increase of the expression of genes related to angiogenesis and anaerobic metabolism to, at the same time, increase supplies and reduce oxygen consumption. Hypoxia combined with the toxic environment explain probably why some of the pro-apoptosis genes were up-regulated and some of the anti-apoptosis genes were down-regulated (i.e., Bcl2). However, few cells were positive for the TUNEL staining within the papillae (Supplementary data 6). Interestingly, pathways associated with inflammation, like the NFκB, TNF, or Th1/Th17 pathways were down-regulated in the papilla, while the Th2 pathway was activated. This was reflected by a decrease of the expression of pro-inflammatory cytokines, like IL1β, CXCL2, or IL6 and the increase of the expression of anti-inflammatory or pro-resolutive cytokines, like IL33, CCL22, or TGFβ1. The gene expression of pro-regenerative growth factors, like IGF2 (neuron survival), FGF1, or insulin-like growth factor binding protein-6 (IGFBP6) was also up-regulated.
Overall, this information corroborates the anti-inflammatory and pro-regenerative role of the papilla.
Conclusion
In our previous work and in this study, we have shown that the papilla is very reactive to its environment, has a strong healing potential, mainly driven by its secretome, and can resist to an unwelcoming environment better than isolated cells. As the papilla is mainly composed of MSC, it combines the advantages usually associated with stem cell therapy with the advantages of using a whole tissue. Indeed, transplanting the papilla as a healing patch removes the need for cell isolation, characterization, and expansion, preserves the cell initial niche, and protects and anchor them at the site to repair without the need to a complicated scaffold/cell construct. Here the human apical papilla has been studied for the repair of spinal cord tissue, but due to its robust immunomodulatory and pro-regenerative properties, it could potentially be used as a dressing for other tissues needing repair.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the Stockel Dental Practice for their support and for providing wisdom teeth as well as Prof. Julian Leprince (LDRI/UCLouvain) for fruitful discussions. The authors would also like to thank Prof. Catherine Levisage (Nantes University, RMeS) for constructive criticisms that contributed to improve the manuscript.
Author contributions
All the authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by PDB, KV, BU, VG, VP, and BB. CB: designed the algorithms and did the quantification of the immunofluorescence staining. AP and GGM: performed the PG2E quantification. A. Diogenes performed the genetic and proteomic analysis of spinal cord tissues. AL and LG: did the statistical analysis of the RNAseq data. The first draft of the manuscript was written by PDB: and all the authors commented on previous versions of the manuscript. All the authors read and approved the final manuscript.
Funding
Anne des Rieux is a F.R.S.-FNRS Senior Research Associate and is a recipient of a grant from the International Foundation of Research in Paraplegia [P155]. This work was supported by the F.R.S.-FNRS and a grant from the International Foundation of Research in Paraplegia [P155]. The authors have no relevant financial or non-financial interests to disclose.
Data availability
The dataset generated and analyzed during the current study is available in the GEO repository under the number GSE191140 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191140.
Declarations
Conflict of interest
The authors have not disclosed any competing interests.
Consent for publication
All the authors have consented for a publication in the CMLS.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vismara I, Papa S, Rossi F, Forloni G, Veglianese P. Current options for cell therapy in spinal cord injury. Trends Mol Med. 2017;23(9):831–849. doi: 10.1016/j.molmed.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Silva NA, Sousa N, Reis RL, Salgado AJ. From basics to clinical: a comprehensive review on spinal cord injury. Prog Neurobiol. 2014;114:25–57. doi: 10.1016/j.pneurobio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Flack JA, Sharma KD, Xie JY. Delving into the recent advancements of spinal cord injury treatment: a review of recent progress. Neural Regen Res. 2022;17(2):283–291. doi: 10.4103/1673-5374.317961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.des Rieux A. Stem cells and their extracellular vesicles as natural and bioinspired carriers for the treatment of neurological disorders. Curr Opin Colloid Interface Sci. 2021;54:101460. doi: 10.1016/j.cocis.2021.101460. [DOI] [Google Scholar]
- 5.Bonaventura G, Incontro S, Iemmolo R, La Cognata V, Barbagallo I, Costanzo E, Barcellona ML, Pellitteri R, Cavallaro S. Dental mesenchymal stem cells and neuro- regeneration: a focus on spinal cord injury. Cell Tissue Res. 2020;379(3):421–428. doi: 10.1007/s00441-019-03109-4. [DOI] [PubMed] [Google Scholar]
- 6.Bianco J, De Berdt P, Deumens R, des Rieux A. Taking a bite out of spinal cord injury: do dental stem cells have the teeth for it? Cell Mol Life Sci. 2016;73(7):1413–37. doi: 10.1007/s00018-015-2126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88(9):792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang S, Shi S, Huang GT. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008;34(2):166–171. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang GT, Sonoyama W, Liu Y, Liu H, Wang S, Shi S. The hidden treasure in apical papilla: the potential role in pulp/dentin regeneration and bioroot engineering. J Endod. 2008;34(6):645–651. doi: 10.1016/j.joen.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Berdt P, Bottemanne P, Bianco J, Alhouayek M, Diogenes A, Lloyd A, Llyod A, Gerardo-Nava J, Brook GA, Miron V, Muccioli GG, Rieux AD. Stem cells from human apical papilla decrease neuro-inflammation and stimulate oligodendrocyte progenitor differentiation via activin-A secretion. Cell Mol Life Sci. 2018;75(15):2843–2856. doi: 10.1007/s00018-018-2764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Berdt P, Vanacker J, Ucakar B, Elens L, Diogenes A, Leprince JG, Deumens R, des Rieux A. Dental apical papilla as therapy for spinal cord injury. J Dent Res. 2015;94(11):1575–81. doi: 10.1177/0022034515604612. [DOI] [PubMed] [Google Scholar]
- 12.Huang GT. A paradigm shift in endodontic management of immature teeth: conservation of stem cells for regeneration. J Dent. 2008;36(6):379–386. doi: 10.1016/j.jdent.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Ruparel NB, de Almeida JF, Henry MA, Diogenes A. Characterization of a stem cell of apical papilla cell line: effect of passage on cellular phenotype. J Endod. 2013;39(3):357–363. doi: 10.1016/j.joen.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 14.Vanacker J, Viswanath A, De Berdt P, Everard A, Cani PD, Bouzin C, Feron O, Diogenes A, Leprince JG, des Rieux A. Hypoxia modulates the differentiation potential of stem cells of the apical papilla. J Endod. 2014;40(9):1410–1418. doi: 10.1016/j.joen.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Schira J, Gasis M, Estrada V, Hendricks M, Schmitz C, Trapp T, Kruse F, Kogler G, Wernet P, Hartung HP, Muller HW. Significant clinical, neuropathological and behavioural recovery from acute spinal cord trauma by transplantation of a well-defined somatic stem cell from human umbilical cord blood. Brain. 2012;135(Pt 2):431–446. doi: 10.1093/brain/awr222. [DOI] [PubMed] [Google Scholar]
- 16.Kandalam S, De Berdt P, Ucakar B, Vanvarenberg K, Bouzin C, Gratpain V, Diogenes A, Montero-Menei CN, des Rieux A. Human dental stem cells of the apical papilla associated to BDNF-loaded pharmacologically active microcarriers (PAMs) enhance locomotor function after spinal cord injury. Int J Pharm. 2020;587:119685. doi: 10.1016/j.ijpharm.2020.119685. [DOI] [PubMed] [Google Scholar]
- 17.De Berdt P, Bottemanne P, Bianco J, Alhouayek M, Diogenes A, Llyod A, Gerardo-Nava J, Brook GA, Miron V, Muccioli GG, Rieux AD. Stem cells from human apical papilla decrease neuro-inflammation and stimulate oligodendrocyte progenitor differentiation via activin-A secretion. Cell Mol Life Sci. 2018;75(15):2843–2856. doi: 10.1007/s00018-018-2764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hached F, Vinatier C, Pinta PG, Hulin P, Le Visage C, Weiss P, Guicheux J, Billon- Chabaud A, Grimandi G. Polysaccharide hydrogels support the long-term viability of encapsulated human mesenchymal stem cells and their ability to secrete immunomodulatory factors. Stem Cells Int. 2017;2017:9303598. doi: 10.1155/2017/9303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buisseret B, Guillemot-Legris O, Ben Kouidar Y, Paquot A, Muccioli GG, Alhouayek M. Effects of R-flurbiprofen and the oxygenated metabolites of endocannabinoids in inflammatory pain mice models. FASEB J. 2021;35(4):e21411. doi: 10.1096/fj.202002468R. [DOI] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 23.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh M, Pearse DD. The role of the serotonergic system in locomotor recovery after spinal cord injury. Front Neural Circuits. 2014;8:151. doi: 10.3389/fncir.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarikaya A, Aydin G, Ozyuncu O, Sahin E, Uckan-Cetinkaya D, Aerts-Kaya F. Comparison of immune modulatory properties of human multipotent mesenchymal stromal cells derived from bone marrow and placenta. Biotech Histochem. 2021 doi: 10.1080/10520295.2021.1885739. [DOI] [PubMed] [Google Scholar]
- 27.An N, Yang J, Wang H, Sun S, Wu H, Li L, Li M. Mechanism of mesenchymal stem cells in spinal cord injury repair through macrophage polarization. Cell Biosci. 2021;11(1):41. doi: 10.1186/s13578-021-00554-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu XB, He LN, Jiang BC, Shi H, Bai XQ, Zhang WW, Gao YJ. Spinal CXCL9 and CXCL11 are not involved in neuropathic pain despite an upregulation in the spinal cord following spinal nerve injury. Mol Pain. 2018;14:1744806918777401. doi: 10.1177/1744806918777401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwiatkowski K, Popiolek-Barczyk K, Piotrowska A, Rojewska E, Ciapała K, Makuch W, Mika J. Chemokines CCL2 and CCL7, but not CCL12, play a significant role in the development of pain-related behavior and opioid-induced analgesia. Cytokine. 2019;119:202–213. doi: 10.1016/j.cyto.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Yu WR, Fehlings MG. Fas/FasL-mediated apoptosis and inflammation are key features of acute human spinal cord injury: implications for translational, clinical application. Acta Neuropathol. 2011;122(6):747–761. doi: 10.1007/s00401-011-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, Zha J, Liu J, Di J. Minocycline impedes mitochondrial-dependent cell death and stabilizes expression of hypoxia inducible factor-1alpha in spinal cord injury. Arch Med Sci. 2019;15(2):475–483. doi: 10.5114/aoms.2018.73520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakazaki M, Morita T, Lankford KL, Askenase PW, Kocsis JD. Small extracellular vesicles released by infused mesenchymal stromal cells target M2 macrophages and promote TGF-beta upregulation, microvascular stabilization and functional recovery in a rodent model of severe spinal cord injury. J Extracell Vesicles. 2021;10(11):e12137. doi: 10.1002/jev2.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sykova E, Cizkova D, Kubinova S. Mesenchymal stem cells in treatment of spinal cord injury and amyotrophic lateral sclerosis. Front Cell Dev Biol. 2021;9:695900. doi: 10.3389/fcell.2021.695900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd AF, Davies CL, Holloway RK, Labrak Y, Ireland G, Carradori D, Dillenburg A, Borger E, Soong D, Richardson JC, Kuhlmann T, Williams A, Pollard JW, des Rieux A, Priller J, Miron VE. Central nervous system regeneration is driven by microglia necroptosis and repopulation. Nat Neurosci. 2019;22(7):1046–1052. doi: 10.1038/s41593-019-0418-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrin FE, Noristani HN. Serotonergic mechanisms in spinal cord injury. Exp Neurol. 2019;318:174–191. doi: 10.1016/j.expneurol.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Liau LL, Looi QH, Chia WC, Subramaniam T, Ng MH, Law JX. Treatment of spinal cord injury with mesenchymal stem cells. Cell Biosci. 2020;10:112. doi: 10.1186/s13578-020-00475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorrell JM, Baber MA, Caplan AI. Influence of adult mesenchymal stem cells on in vitro vascular formation. Tissue Eng Part A. 2009;15(7):1751–1761. doi: 10.1089/ten.tea.2008.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofer HR, Tuan RS. Secreted trophic factors of mesenchymal stem cells support neurovascular and musculoskeletal therapies. Stem Cell Res Ther. 2016;7(1):131. doi: 10.1186/s13287-016-0394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song P, Han T, Xiang X, Wang Y, Fang H, Niu Y, Shen C. The role of hepatocyte growth factor in mesenchymal stem cell-induced recovery in spinal cord injured rats. Stem Cell Res Ther. 2020;11(1):178. doi: 10.1186/s13287-020-01691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu S, Zhao Y, Ma Y, Ge L. Profiling the secretome of human stem cells from dental apical papilla. Stem Cells Dev. 2016;25(6):499–508. doi: 10.1089/scd.2015.0298. [DOI] [PubMed] [Google Scholar]
- 41.Diogenes A, Henry MA, Teixeira FB, Hargreaves KM. An update on clinical regenerative endodontics. Endodontic Topics. 2013;28(1):2–23. doi: 10.1111/etp.12040. [DOI] [Google Scholar]
- 42.Alekseenko LL, Shilina MA, Lyublinskaya OG, Kornienko JS, Anatskaya OV, Vinogradov AE, Grinchuk TM, Fridlyanskaya II, Nikolsky NN. Quiescent human mesenchymal stem cells are more resistant to heat stress than cycling cells. Stem Cells Int. 2018;2018:3753547. doi: 10.1155/2018/3753547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song N, Scholtemeijer M, Shah K. Mesenchymal stem cell immunomodulation: mechanisms and therapeutic potential. Trends Pharmacol Sci. 2020;41(9):653–664. doi: 10.1016/j.tips.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desole C, Gallo S, Vitacolonna A, Montarolo F, Bertolotto A, Vivien D, Comoglio P, Crepaldi T. HGF and MET: from brain development to neurological disorders. Front Cell Dev Biol. 2021;9:683609. doi: 10.3389/fcell.2021.683609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Deng Y, Liang J, Huang F, Qiu W, Zhang M, Long Y, Hu X, Lu Z, Liu W, Zheng SG. Mesenchymal stromal cells attenuate multiple sclerosis via IDO-dependent increasing the suppressive proportion of CD5+ IL-10+ B cells. Am J Transl Res. 2019;11(9):5673–5688. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Fang J, Liu Z, Hou P, Cao L, Zhang Y, Liu R, Li Y, Shang Q, Chen Y, Feng C, Wang G, Melino G, Wang Y, Shao C, Shi Y. Inflammatory cytokines-stimulated human muscle stem cells ameliorate ulcerative colitis via the IDO-TSG6 axis. Stem Cell Res Ther. 2021;12(1):50. doi: 10.1186/s13287-020-02118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xin D, Li T, Chu X, Ke H, Yu Z, Cao L, Bai X, Liu D, Wang Z. Mesenchymal stromal cell-derived extracellular vesicles modulate microglia/macrophage polarization and protect the brain against hypoxia-ischemic injury in neonatal mice by targeting delivery of miR-21a-5p. Acta Biomater. 2020;113:597–613. doi: 10.1016/j.actbio.2020.06.037. [DOI] [PubMed] [Google Scholar]
- 48.Khare D, Or R, Resnick I, Barkatz C, Almogi-Hazan O, Avni B. Mesenchymal stromal cell-derived exosomes affect mRNA expression and function of B-Lymphocytes. Front Immunol. 2018;9:3053. doi: 10.3389/fimmu.2018.03053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated and analyzed during the current study is available in the GEO repository under the number GSE191140 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191140.