Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) is an important regulator of cellular metabolism that is commonly hyperactivated in cancer. Recent cancer genome screens have identified multiple mutations in Ras-homolog enriched in brain (Rheb), the primary activator of mTORC1 that might act as driver oncogenes by causing hyperactivation of mTORC1. Here, we show that a number of recurrently occurring Rheb mutants drive hyperactive mTORC1 signalling through differing levels of insensitivity to the primary inactivator of Rheb, tuberous sclerosis complex. We show that two activated mutants, Rheb-T23M and E40K, strongly drive increased cell growth, proliferation and anchorage-independent growth resulting in enhanced tumour growth in vivo. Proteomic analysis of cells expressing the mutations revealed, surprisingly, that these two mutants promote distinct oncogenic pathways with Rheb-T23M driving an increased rate of anaerobic glycolysis, while Rheb-E40K regulates the translation factor eEF2 and autophagy, likely through differential interactions with 5′ AMP-activated protein kinase (AMPK) which modulate its activity. Our findings suggest that unique, personalized, combination therapies may be utilised to treat cancers according to which Rheb mutant they harbour.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03825-7.
Keywords: Rheb, mTOR, TSC, eEF2, PKM, AMPK
Introduction
The mechanistic target of rapamycin complex 1 (mTORC1) is a serine/threonine kinase that is activated by diverse signals, in particular amino acid availability and growth factors, to drive anabolic processes and inhibit some catabolic ones, notably autophagy [1]. This is achieved through phosphorylation of a range of proteins, the best-characterized of which include the ribosomal protein S6 (rpS6) kinases (S6Ks) [2] and eukaryotic initiation factor 4E-binding proteins (4E-BPs) [3]. S6Ks phosphorylate rpS6 [4], a component of the 40S ribosomal subunit, and positively regulate cell size, while the 4E-BPs control the initiation of mRNA translation and cell proliferation [4, 5]. 4E-BPs bind to eIF4E in competition with eIF4G to prevent formation of the eIF4F complex thus inhibiting cap-dependent translation [6]. mTORC1-catalysed phosphorylation of the 4E-BPs decreases their affinity for eIF4E allowing eIF4E to form translation initiation complexes and promote translation initiation [7].
Rheb (Ras homolog enriched in brain) is a small (21 kDa) GTPase that, in its GTP-bound form, activates mTORC1. Hydrolysis of Rheb-GTP is promoted by the GTPase-activating protein (GAP) tuberous sclerosis complex (TSC), comprised of TSC1, TSC2 and TBC1D7, yielding inactive, GDP-bound Rheb [8–10]. The ability of TSC1/2 to impair Rheb function is inhibited by signalling events which are activated by hormones, mitogenic stimuli and growth factors involving the PI3K and MAPK signalling pathways [1, 9, 11]. This involves the phosphorylation of TSC2, the subunit responsible for GAP activity [12], downstream of oncogenic pathways such as phosphoinositide 3-kinase (PI3K)/protein kinase B (PKB) and rat sarcoma (Ras)/extracellular signal regulated kinase (ERK) [37]. As the pathways upstream of TSC are often aberrantly active in cancer, mTORC1 is also activated in many cancers.
Several mutations in the RHEB gene have also been identified in a range of human tumours [13]. Several of these Rheb mutants have been shown to be constitutively active [14–17]. Some mutants are insensitive to TSC’s GAP activity and thus confer constitutive activity on Rheb and mTORC1 [16, 18]. In contrast, it has recently been suggested that the constitutively active Rheb mutant Rheb-Y35N drives continuous mTORC1 activation by activating the MAPK pathway via direct inhibition of AMPKα or heterodimerisation with BRAF [19, 20]. Given that mTORC1 signalling is hyperactive in many cancers and some Rheb mutants can promote oncogenic transformation [15, 19], it is important to understand the underlying mechanisms and whether or not all Rheb mutants exert the same effects.
Here, we show that previously uncharacterised Rheb mutants which occur in human tumours drive constitutive mTORC1 signalling and drive oncogenic transformation and tumour growth in vivo. Importantly, in addition to activating mTORC1, some mutants affect other processes, including glycolysis and the translational machinery, effects which differ between mutants. These unexpected findings may require or offer additional therapeutic approaches for treating cancers bearing these mutations.
Materials and methods
Reagents
All reagents were from Merck (NSW, Australia) unless otherwise specified. Bradford assay reagent was from Bio-Rad (NSW, Australia). AZD8055 was purchased from Jomar Life Research (VIC, Australia).
Cell culture, site-directed mutagenesis and transfection
NIH3T3, HEK293 and HeLa cells were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% foetal bovine serum and 1% penicillin/streptomycin at 37 °C with 5% (v/v) CO2 and regularly tested for mycoplasma contamination. pRK7-FLAG-Rheb vector [10] was purchased from Addgene and was used to generate Rheb mutants via site-directed mutagenesis. Briefly, mutagenesis primers were used for PCR amplification of pRK7-FLAG-Rheb using pfu Turbo Polymerase (Promega, NSW, Australia) to induce specific mutations. Donor plasmids were digested with DpnI. Transfection was performed using Lipofectamine 3000 (Thermo Fisher Scientific, SA, Australia) according to the manufacturer’s instructions.
Generation of NIH3T3 cell lines stably expressing Rheb variants
cDNAs encoding wildtype Rheb (Rheb-WT), or Rheb-T23M or E40K were cloned into a pEGFP-N1 vector using the EcoRI and BamHI restriction sites. Vectors were transfected into NIH3T3 cells which were cultured in the presence of 1 mg/ml G418 for 6 weeks to select stable transfectants. Surviving cells were transferred to 96-well plates at 1 cell/well and monitored for colonies. Rheb expression was determined via SDS-PAGE Western Blot; monoclonal colonies expressing low levels of exogenous Rheb were selected for use.
SDS-PAGE and immunoblot analysis
SDS-PAGE/Western blot analysis was carried out as previously described [21]. Briefly, cells were lysed in ice-cold lysis buffer containing 1% Triton X-100, 150 mM NaCl, 20 mM Tris–HCl pH 7.5, 2.5 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM sodium orthovanadate, 1 mM dithiothreitol (DTT) and 1 mM β-glycerophosphate supplemented with protease inhibitor cocktail. Tumour xenograft tissues were homogenised in RIPA lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 50 mM β-glycerophosphate, 1 mM EDTA, 0.5 mM NaVO3, 1 mM dithiothreitol and protease inhibitor cocktail) using a tissue grinder (Thermo Fisher Scientific). Lysates were spun at 16,000×g for 10 min. Protein concentrations were determined [22] and normalized. Equal aliquots of protein (40 μg) were denatured in Laemmli loading buffer, heated at 95 °C for 3 min and separated by SDS-PAGE using gels containing 7–13% acrylamide and 0.1–0.36% bis-acrylamide. Proteins were transferred to nitrocellulose membranes, which were blocked and incubated with primary antibody as indicated (Supplementary Table S1). Fluorescently conjugated secondary antibody was applied and signals were imaged using a LiCor Odyssey® CLx imager (Millennium Science, VIC, Australia).
Immunoprecipitation
HEK293 cells were lysed in CHAPS lysis buffer [40 mM HEPES pH 7.5, 120 mM NaCl, 1 mM EDTA pH 8, 10 mM Na2H2P2O7, 10 mM β-glycerophosphate, 50 mM NaF, 0.5 mM Na3VO4, 0.3% (m/v) CHAPS and protease inhibitor cocktail] and protein complexes associated with Rheb were immunoprecipitated with protein A beads. Briefly, protein lysates were centrifuged for 10 min at 16,000×g. The supernatants were incubated with anti-Rheb antibody (Genesearch, Arundel, QLD, Australia, Cat No. 13879) for 16 h at 4 °C with constant rotation, followed by the incubation with protein A Sepharose beads for a further period of 2 h at 4 °C with rotation. Beads were washed twice with CHAPS lysis buffer and then resuspended in 2× Laemmli sample buffer before SDS-PAGE analysis.
Purification of recombinant GST-Rheb
pGEX-4T2-Rheb vectors were a kind gift from Dr. John Blenis (now at Weill Cornell Medicine, New York, NY, USA). Site-directed mutagenesis was performed using pfu Turbo Polymerase. Recombinant GST-Rheb proteins were expressed in and purified from E. coli BL21 cells after incubation with 25 µM IPTG (6 h, 30 °C). Bacterial lysates were prepared by sonication in ‘Rheb lysis buffer’ (0.2% Triton X-100, 50 mM HEPES–KOH pH 7.4, 140 mM NaCl, 1 mM EDTA pH 8, 1 mM DTT, plus protease inhibitor cocktail). Lysates were incubated with 0.03 U DNase I, 12 mM MgCl2 and 0.25 mg/ml lysozyme for 30 min at 4 °C, before spinning down at 13,000×g for 10 min at 4 °C. Supernatants were incubated with glutathione-agarose (Thermo Fisher Scientific) for 2 h at 4 °C. GST-Rheb was eluted in 30 mM reduced l-glutathione, 50 mM HEPES–KOH pH7.4, 140 mM NaCl, 2.7 mM KCl, 0.1 mg/ml bovine serum albumin (BSA) plus protease inhibitor cocktail. Eluted GST-Rheb (approximately 1 µg/µl) was then analysed and quantitated by SDS-PAGE and Coomassie staining of resolved proteins and BSA standards.
GAP assay
Rheb-GAP assays were performed on complexes immunoprecipitated from HEK293 cells transfected with WT FLAG-TSC1 and WT FLAG-TSC2. Cells were lysed in 1 ml NP-40 lysis buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P-40, 10% glycerol, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, and protease inhibitor cocktail). Flag-tagged TSC1 and TSC2 were then immunoprecipitated with FLAG antibody coupled to protein-G beads for 2 h at 4 °C. Immune complexes on beads were washed thrice in IP wash buffer (20 mM HEPES–KOH pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 mM DTT, 50 mM β-glycerophosphate, 50 mM NaF, and protease inhibitor cocktail), and once in 1 ml Rheb exchange buffer (50 mM HEPES–KOH pH 7.4, 1 mM MgCl2, 100 mM KCl, 0.1 mg/ml BSA, 1 mM DTT, and protease inhibitor cocktail). GST-Rheb (10 μg) was loaded with 100 μCi [α-32P]GTP by incubation for 5 min at 37 °C in 100 μl GTP-loading buffer (50 mM HEPES–KOH pH 7.5, 5 mM EDTA, 5 mg/ml BSA, and protease inhibitor cocktail). After 5 min, 2.5 μl 1 M MgCl2, 100 μl ice-cold 50 mM HEPES–KOH pH 7.4, and 20 μl 10 mM GDP were added to the [α-32P]GTP-loaded Rheb. GAP assays were initiated by adding 20 μl GTP-loaded Rheb mixture (1 μg GST-Rheb) to each aliquot of FLAG-TSC1/2-protein-G agarose immune complexes. Assays were performed at room temperature with constant agitation for 60 min. Reactions were stopped by adding 300 μl Rheb wash buffer containing 1 mg/ml BSA. Immune complexes were removed by brief centrifugation, and nucleotide-bound GST-Rheb was purified from supernatants by incubating with glutathione beads for 2 h at 4 °C. After three washes with Rheb wash-buffer (50 mM HEPES–KOH pH 7.5, 0.5 M NaCl, 0.1% Triton X-100, 5 mM MgCl2, 0.005% SDS plus protease inhibitor cocktail), radiolabelled GTP and GDP were eluted from Rheb with 20 µl elution buffer (0.5 mM GDP, 0.5 mM GTP, 5 mM DTT, 5 mM EDTA, and 0.2% SDS) at 68 °C for 20 min. One microliter (μl) of each eluted reaction was resolved by thin-layer chromatography on PEI cellulose with 0.75 M KH2PO4 pH 3.4 as solvent. Relative levels of [α-32P]-labelled GTP and GDP were detected and quantitated with Typhoon phosphor-imager (GE Healthcare, NSW, Australia).
In silico modelling
In silico modelling was carried out using ICM Pro (Molsoft LLC, La Jolla, CA, USA). PDB accession code 6BCU was used as a starting template. Mutants were created in ICM Pro and subjected to 20 rounds of energy minimization and annealing. All molecular visualizations were obtained using the PyMOL graphic tool (the PyMOL molecular graphics system, Version 2.2.3. Schrödinger, LLC).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolim bromide (MTT) assay
For MTT assay, 3000 HEK293 cells expressing Rheb mutants were seeded into 96-well plates. Five mg/ml of MTT was added to each well and plates were incubated at 37 °C for 4 h. Medium was carefully aspirated, and crystals were dissolved in 50 µl DMSO. Absorbance was read at 540 nm using a Glomax Discover Multimode Microplate Reader (Promega); cell number was calculated against a standard curve generated by 1:2 serial dilutions of HEK293 cells.
Bromodeoxyuridine (BrdU) assay
BrdU Cell Proliferation Assay Kit (Cell Signaling Technology, Danvers, MA #6813) was performed as per the manufacturer’s instructions. Cells were plated at 10,000 cells per well in a 96-well plate 2 h prior to addition of 10 µl of 10× BrdU solution. 2 h after adding BrdU, cells were fixed/denatured for 30 min. BrdU detection antibody diluted 1:100 and added to cells for 1 h. Cells were washed 3× in wash buffer before addition of HRP-linked secondary antibody for 30 min. Cells were washed 3× before addition of 3,3′,5,5′-tetramethylbenzidine. After 30 min, ‘STOP solution’ was added and absorbance determined at 450 nm.
Colony formation assay
To perform colony formation assays, 1 ml base layer containing 0.5% agarose in DMEM was plated in six-well plates and overlaid with 1 ml of 0.3% agarose in DMEM containing 3000 NIH3T3 cells stably expressing Rheb-WT, T23M or E40K. Cells were fed every 2–3 days with 1 ml of DMEM containing DMSO, AZD8055 or JAN-384 as indicated. After 6 weeks, cells were stained with 0.05% crystal violet in phosphate-buffered saline containing 2% ethanol. Colonies larger than 100 nm were counted by hand and images captured on a dissecting microscope.
In vivo tumour model
All animal work was conducted in accordance with the National Health and Medical Research Council’s Care and use of Animals for Scientific Purposes guidelines and with approval (No. SAM339) from the South Australian Health and Medical Research Institute’s animal ethics committee. Tumours were generated by subcutaneous injection of 1 × 106 NIH3T3 cells stably expressing Rheb-WT, T23M or E40K into both flanks of 8–10-week-old, male NOD SCID gamma (NSG) mice (The Jackson Laboratory, Stock number 005557) (five mice per group generating ten tumours). Tumours were measured daily with Vernier callipers and volumes were calculated using the standard formula (volume = length × width2 × 0.5). When tumours reached a volume of 60 mm3, mice were randomly assigned into groups to be injected daily with 20 mg/kg AZD2014 or vehicle (DMSO in PBS containing 5% Tween-80 and 5% PEG) via intraperitoneal injection. After 7 days, or when total tumour bearing load exceeded 2000 mm3 or individual tumours exceeded 1000 mm3 (in strict accordance with our approved animal ethics application), animals were immediately culled by CO2 immersion and tumours were removed.
Immunohistochemistry
Tumours were embedded in cryo-embedding medium (OCT) and 8 µm sections cut using a Shandon Cryotome E Cryostat at − 20 °C. Sections were allowed to dry for 30 min before fixation in 10% neutral buffered formaldehyde. Endogenous peroxidase activity was blocked with 3% peroxide and further blocked in 5% BSA to prevent non-specific binding. Indicated primary antibodies were diluted 1:100 in blocking buffer and added to sections at 4 °C overnight. Sections were incubated with SignalStain® Boost IHC-Detection Reagent (HRP, Rabbit; Cell Signalling Technologies #8114) at room temperature for 30 min and developed using SignalStain® DAB Substrate Kit (Cell Signalling Technologies #8059). Sections were counterstained with hematoxylin for 60 s and observed under a light microscope. DAB staining was quantified using ImageJ.
Mass spectrometry analysis
Polyclonal NIH3T3 cells stably expressing Rheb-WT, T23M or E40K were grown in DMEM either containing or lacking FBS for 72 h. Cells were washed 5× in ice cold PBS, lysed in lysis buffer containing 10% sodium deoxycholate, 0.1 M Tris–HCl pH 8.5. Protein concentration was determined using the BCA method and samples normalized. Protein was digested with trypsin (Sigma, T6567) and Lys-C (Wako Chemicals, 129-02541) at a 1:100 ratio of enzyme-to-protein. Peptides were desalted and concentrated using SDB-RPS StageTips as described [23] with minor modifications. Briefly, peptides were diluted 1:10 in H2O, then 100 µl was mixed 1:1 with loading buffer (99% ethyl acetate, 1% TFA), vortexed vigorously, and then loaded directly onto StageTips packed with three layers of SDB-RPS material. Samples were spun through to dryness using a custom 3D printed 96-well StageTip adapter at 1000×g [24]. StageTips were washed with a further 100 µl loading buffer, followed by 100 µl wash buffer 1 (99% isopropanol, 1% TFA), then 100 µl wash buffer 2 (5% acetonitrile, 0.2% TFA). Samples were subsequently eluted directly into clean PCR strip tubes with 60 µl 60% acetonitrile/5% ammonium hydroxide (25% ammonia solution). Samples were then dried in a SpeedVac vacuum concentrator, resuspended in MS loading buffer (2% acetonitrile/0.3% TFA), and peptides were normalized by A280 absorbance. A Dionex Ultimate 3000 (Thermo Fisher Scientific) UHPLC was connected to a Q Exactive HF-X Orbitrap mass spectrometer, and 1 µg peptides were loaded directly onto a 75 μm I.D., 60 cm column packed with 1.9 μm C18 material (Dr. Maisch ReproSil Pur AQ) and separated over a gradient of 3 to 24% acetonitrile in 0.1% formic acid, over 2 h. Column temperature was maintained at 60 °C. The mass spectrometer was operated in data-dependent mode, with one full scan of 350–1400 m/z performed with resolution 60,000 at a target of 3e6 ions, followed by 20 data-dependent HCD MS/MS scans with a target of 1e5 ions, max IT 28 ms, isolation window 1.4 m/z, normalized collision energy 27, minimum AGC target 1e4, and resolution 15,000. Dynamic exclusion was switched on (30 s). RAW data were processed using MaxQuant version 1.6.6.0, searched against the Mouse UniProt sequence database (June 2019 release) with default settings, with the addition of “Match between runs” (match time window 0.7 min), and “MaxLFQ” enabled. Bioinformatic analysis was performed using Perseus version 1.6.10.43. Raw data are available in Supplementary Table S2.
Transwell migration and invasion assays
This was performed as previously described [25]. For migration assays, transwells (8 µm pore size, BD Biosciences, NSW, Australia) were pre-coated with 1% gelatin in serum-free medium. Assays were then performed with 1.5 × 104 cells plus 0.5 μg/μl mitomycin C for 24 h using 10 μg/ml collagen and 20% FBS as chemo-attractant. For invasion assays, transwells were pre-coated with Matrigel (1:3 in DMEM, BD Biosciences) and assays performed with 1.5 × 104 cells plus 0.5 μg/μl mitomycin C for 72 h using 10 μg/ml collagen and 20% FBS as chemo-attractants. Transwells were stained with DAPI (4′,6-diamidino-2-phenylindole, 1:20,000) and visualized with a Nikon Eclipse Ni microscope (× 10 objective lens). DAPI-stained cell numbers were quantified using the Fiji (Java 8) software.
Fluorescence associated cell sorting (FACS)
FACS analysis was performed as previously described [26]. Briefly, tf-LC3 HeLa cells were transfected with the indicated Rheb mutant (Calcium phosphate method) and grown in DMEM containing 10% FBS for 24 h. Cells were collected, strained into a FACS tube and analysed by flow cytometry on a BD LSR Fortessa X20 Analyser (BD Bioscience). Cells were gated on SSC-H and FSC-H. Analysis of red to green fluorescence ratios (mRFP1-H:EGFP-H) was performed using FlowJo v10.6.1 software package.
Seahorse glycolysis stress test
Agilent Seahorse XF Glycolysis Stress Test (Agilent Technologies, Santa Clara, CA) was performed as per the manufacturer’s instructions. HEK293 cells were transfected with vectors encoding Rheb and allowed to grow for 24 h. 1 × 104 cells were seeded in Seahorse XF microplate and kept in fully supplemented medium for 6 h to allow attachment. Growth medium was replaced for DMEM lacking FBS overnight. Meanwhile, a 96-well sensor cartridge was hydrated with Seahorse XF calibrant in a non-CO2 incubator overnight at 37 °C. The following day, sensor cartridge was loaded with 10 mM glucose in port A, 1 µM oligomycin in port B and 50 mM 2-deoxyglucose in port C. Cells were prepared for the assay by replacing growth medium with Seahorse XF Base Medium supplemented with 2 mM glutamine, and adjusted to pH 7.4 for 1 h. Glycolysis stress test was then conducted in a Seahorse XFe96 Analyser using the Wave software package. Extracellular acidification rate (ECAR) was normalized to Hoechst stain and graphed using GraphPad Prism 8.
Results
Rheb mutants commonly found in cancer promote constitutively active mTORC1 signalling independently of MAPK
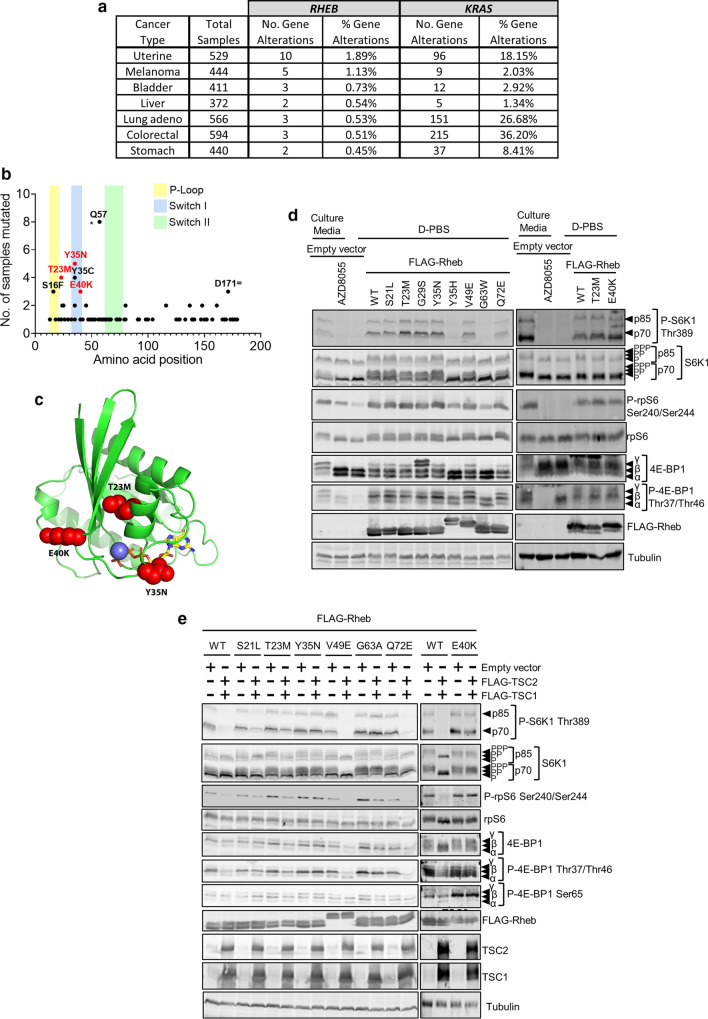
To assess the extent of RHEB mutations in cancer, we evaluated data from The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) and COSMIC [13]. We also showed KRAS alterations, a frequently mutated gene in cancer, as a comparison. We found that, similarly to KRAS mutations, RHEB mutations occur in a wide variety of cancers (Fig. 1a). We identified several mutations that were present in multiple samples with the six most frequent being S16F, T23M, Y35N, Y35C, E40K and Q57* (Fig. 1b; Supplementary Table S3). To gain further insights into the molecular basis of these mutations, we performed in silico modelling for Rheb-T23M, -Y35N and -E40K. A loop covers the active site (Fig. 1c), implying it is important in binding substrate (GTP) due to its mobility and direct contacts with substrate. Additionally, there is an α-helix which flanks the substrate, GTP, making direct contact with the bound GTP. The T23M side chain lies between the afore-mentioned α-helix and the β-sheet (Fig. 1c). The larger Met here likely forces the α-helix closer to the substrate, potentially altering binding and affecting catalysis. E40K is at the base of the loop that covers the active site (Fig. 1c); this mutation to a large positively charged residue would likely perturb the loop’s conformation, altering its mobility and substrate binding, hydrolysis and/or release. Thus, we hypothesised that the mutations promote constitutive mTORC1 activation through decreased sensitivity to TSC-mediated GAP activity.
Fig. 1.
Rheb mutants promote hyperactive mTORC1 signalling independently of the MAPK pathway. a Data from the TCGA showing the number and rate of identified RHEB and KRAS mutations across various cancer types. b The number of times each RHEB mutation appears in the TCGA and COSMIC databases. Key domains of the protein are shaded and hotspot mutations subjected to further investigation are shown in red. c In silico analysis of the 3D structure of Rheb (green ribbons). GTP is represented as sticks (coloured by element) while the mutations T23M, Y35N and E40K are represented as spheres (red). d Immunoblot analysis of lysates from HEK293 cells that had been transfected with either a pcDNA3.1 empty vector or a vector encoding the indicated FLAG-tagged Rheb mutant. After 24 h, the medium was replaced with D-PBS for 1 h. As a control to inhibit mTORC1 signalling, one plate of cells was treated with 1 µM AZD8055 for 30 min. e Immunoblots of lysates from HEK293 cells previously transfected with either the indicated Rheb mutant and a pcDNA3.1 empty vector or the indicated Rheb mutant and vectors encoding FLAG-TSC1 and FLAG-TSC2. After 24 h, the medium was replaced with fresh DMEM without FBS for 16 h followed by D-PBS for 1 h. Immunoblots were probed with antibodies for the indicated proteins or phosphorylation sites
To determine whether these mutations drive hyperactive mTORC1 signalling, we employed site-directed mutagenesis to create selected mutations in a vector encoding FLAG-tagged Rheb. We then transfected HEK293 cells with these vectors, one encoding Rheb-WT or an empty vector, as negative control. To assess the effect of the Rheb mutants on mTORC1 activity, transfected cells were cultured for 16 h in DMEM lacking FBS, conditions under which mTORC1 is generally inactivated, while active Rheb can restore mTORC1 signalling [27]. Cell lysates were then analysed by immunoblot for downstream effectors of mTORC1, including its direct substrates 4E-BP1 and S6K1, and the S6K1 substrate rpS6.
As expected, in fully supplemented growth medium, conditions where the TSC complex is not active against Rheb-GTP, WT Rheb and mutants did not promote levels of mTORC1 signalling above those seen in cells transfected with empty vector (Supplementary Fig. S1a). In every case, the mTOR kinase inhibitor AZD8055 inhibited the phosphorylation both of S6K1 at Thr389 and of rpS6 at Ser240/Ser244 (Supplementary Fig. S1a) in fully supplemented medium showing the effect of every Rheb variant tested is mediated through mTOR. In contrast, when in cells that were subsequently transferred to D-PBS (conditions that cause a profound inhibition of mTORC1 due to the lack of amino acids, key activators of mTORC1), WT Rheb supported the sustained phosphorylation of S6K1, rpS6 and 4E-BP1, while several mutants (T23M, G29S, Y35N and E40K) did so more strongly (Fig. 1d). Thus, under these conditions, it appears that some Rheb mutants are more potent activators of mTORC1 signalling than WT Rheb (Fig. 1d). We noted that the Rheb mutants Rheb-Y35H, -V49E, and -E40K showed a lower mobility (compared to WT Rheb) on polyacrylamide gel electrophoresis. To determine the cause, we performed mass spectrometry to compare possible post-translational modifications of Rheb-WT and E40K; however, we were unable to detect any differences that might explain its altered mobility (data not shown).
It has previously been reported [19, 20] that Rheb-Y35N drives hyperactive mTORC1 signalling through constitutive activation of the MAPK pathway. While we did observe increased P-ERK1/2 levels in cells expressing Rheb-Y35N and -E40K (Supplementary Fig. S1b–d), this was not statistically significant (Supplementary Fig. S1d). This observation has previously been reported and thoroughly examined [19, 20], so we have not followed it up again in detail here, although we did note that P-ERK-1/2 was lost in cells expressing Rheb mutants when cells were deprived of serum whereas mTORC1 signalling was not, which suggests that Rheb mutants are not promoting mTORC1 signalling via the ERK pathway under the conditions tested here (Supplementary Fig. S1c). Furthermore, treatment with the MEK inhibitor AZD6244 did not decrease mTORC1 signalling in cells expressing Rheb mutants (Supplementary Fig. S1b). Taken together, these data suggest that these Rheb mutants promote mTORC1 signalling independently of the MAPK/ERK pathway.
Rheb are insensitive to inhibition by TSC
It was possible that the ability of some ectopically expressed Rheb mutants to promote mTORC1 signalling under serum-starved conditions might arise because their levels (likely well above endogenous Rheb) exceeded those of TSC1/2, rather than due to intrinsic constitutive activity. To test this, we co-expressed Rheb mutants alongside vectors encoding FLAG-TSC1/2, to increase the available levels of this complex (Fig. 1e; Supplementary Fig. S1e; quantified in Supplementary Fig. S1f; TBC1D7 is not essential for function of the TSC1/2 complex). Strikingly, TSC1/2 decreased the phosphorylation of S6K1 at Thr389 and rpS6 at Ser240/244 in cells expressing wild-type Rheb but not in cells expressing Rheb-G63A (Supplementary Fig. S1e; quantified in Supplementary Fig. S1f), a mutant designed, based on the structure of Rheb, to be catalytically dead [16]. Likewise, phosphorylation of these proteins was less sensitive to TSC1/2 in cells expressing Rheb-T23M, -Y35N or -E40K than in cells expressing Rheb-WT. Cells expressing Rheb-S21L, -G29S or -Q64L still displayed increased P-S6K1 levels compared to cells expressing Rheb-WT when co-expressed with TSC1/TSC2; however, P-S6K1 was more sensitive to TSC1/2 in cells expressing Rheb-S21L, G29S or Q64L than in those expressing Rheb-T23M, Y35N, E40K or G63A (Fig. 1e; Supplementary Fig. S1e; quantified in Supplementary Fig. S1f). These data suggest that certain Rheb mutants promote hyperactivation of mTORC1 because they are insensitive to TSC1/2.
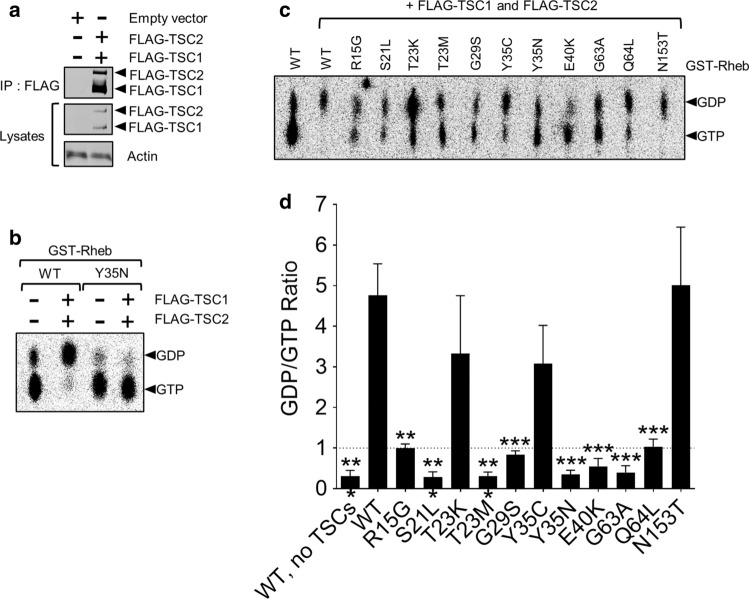
To study this further, we assessed whether specific Rheb mutations affected the ability of TSC1/TSC2 to promote the hydrolysis of Rheb-bound GTP, we performed GAP assays using recombinant GST-tagged Rheb purified from E. coli BL21 cells and FLAG-tagged TSC1/TSC2 co-expressed in, and immunoprecipitated from, HEK293 cells (Fig. 2a). To validate the assay, Rheb-WT and Rheb-Y35N were ‘preloaded’ with [α-32P]GTP and then incubated with or without TSC1/TSC2. TSC1/TSC2 efficiently promoted hydrolysis of [α-32P]GTP bound to Rheb-WT, but not GTP bound to Rheb-Y35N (Fig. 2b), which has been previously shown to be resistant to TSC1/2’s GAP function [19, 28]. This validated the Rheb-GAP assay, allowing us to test further mutants. TSC1/TSC2 promoted hydrolysis of [α-32P]GTP bound to Rheb-T23K, Y35C and N153T similarly to Rheb-WT (GDP/GTP ratio > 1); in contrast, hydrolysis of GTP bound to Rheb-R15G and Q64L was partially resistant to stimulation by TSC1/2 (GDP/GTP ratio ≈ 1). Furthermore, TSC1/TSC2 complexes were completely unable to promote hydrolysis of [α-32P]GTP bound to Rheb-S21L, G29S, T23M, Y35N, E40K or G63A (GDP/GTP ratio < 1). This striking difference is notable as this property would allow these Rheb mutants to escape inhibition by the TSC1/TSC2 tumour suppressor complex and thus display constitutive activity (Fig. 2c; quantified in Fig. 2d). To determine whether mutations are insensitive to TSC inhibition through insensitivity to its GAP function and not to altered binding to the TSC1/2 heterodimer, we performed immunoprecipitation from lysates of HEK293 cells over expressing Rheb mutants and TSC1/2. We found that Rheb-WT, T23M, Y35N and E40K all bind TSC1/TSC2 to a similar extent suggesting TSC binding is not grossly affected (Supplementary Fig. S2a). Likewise, binding to mTORC1 was also not affected (Supplementary Fig. S2b). These data again suggest that the Rheb mutants S21L, T23M, G29S, Y35N and E40K promote hyperactive mTORC1 signalling (under serum-starved conditions, Fig. 1d, e; Supplementary Fig. S1e) because they are insensitive to the GAP function of TSC.
Fig. 2.
Rheb mutants are resistant to TSC GAP activity in vitro. a Immunoblot analysis of FLAG-tagged TSC1 and TSC2 immunoprecipitated (using anti-FLAG antibody) from lysates of HEK293 cells which had been transfected 24 h earlier with vectors encoding FLAG-TSC1 and FLAG-TSC2. b GTPase assay with GST-purified Rheb WT or Y35N, prebound to [α-32P]GTP and then mixed with or without FLAG-immunoprecipitated TSC1 and TSC2 as indicated. Reaction products were resolved by thin layer chromatography and detected using a phosphorimager. Positions of GDP and GTP are indicated. c As B but using the indicated mutants of Rheb. Rheb-WT without TSC1/2 acts as a control. GDP:GTP ratios are quantified in d for n = 3 (means ± S.D.). The data were analysed using Student’s t-test, ratios of GDP:GDP being compared to those for Rheb-WT. ***P < 0.001
Rheb T23M and E40K drive increased cell proliferation and anchorage independent growth
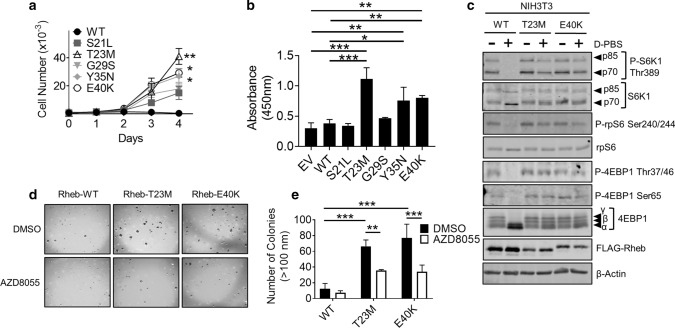
To determine whether specific Rheb mutants can drive cell proliferation, HEK293 cells were transfected with vectors encoding the Rheb mutants Rheb-S21L, T23M, G29S, Y35N and E40K and subjected to MTT and BrdU incorporation assays with/without FBS. MTT assays, which estimate cell number (and thus, over time, cell number and hence cell proliferation) by measuring mitochondrial activity [29], were performed every 24 h for 4 days to generate growth curves. While there was no difference in MTT activity between cells expressing WT Rheb or its mutants in fully supplemented medium (Supplementary Fig. S3a), we saw striking differences when cells were placed in medium lacking FBS (Fig. 3a). While there was no increase in MTT activity over 4 days in cells expressing Rheb-WT, each of the Rheb mutants promoted increased cell number (MTT activity; Fig. 3a). In particular, cells expressing Rheb-T23M did so to a greater extent than any other Rheb mutants, while Rheb-Y35N and E40K showed mutually similar effects on cell number, which were greater than those of Rheb-S21L and G29S (or Rheb-WT).
Fig. 3.
Rheb mutants drive increased cell proliferation and growth in soft agar. a MTT assay data for HEK293 cells transiently expressing the indicated Rheb mutants in medium lacking FBS. Cell number is calculated against a standard curve generated by 1:2 serial dilution of HEK293 cells. b BrdU incorporation assay for HEK293 cells transiently expressing the indicated Rheb mutants. Growth medium was replaced with medium lacking FBS for 24 h prior to analysis. BrdU was allowed to be incorporated for 2 h. c Western blot analysis of monoclonal NIH3T3 cells stably over-expressing Rheb-WT, T23M or E40K. Cell medium was replaced with medium either containing or lacking FBS 16 h and then replaced with D-PBS as indicated for 1 h prior to harvest. d Colony formation assay for NIH3T3 cells stably over expressing Rheb-WT, T23M or E40K. Liquid medium was replaced twice per week with medium containing 1 µM AZD8055 or DMSO as indicated. Number of colonies larger than 100 nM were counted by hand and are quantified in (e). For panels a, b and e, n = 3 (means ± S.D.). Statistical significance was determined by Student’s t-test where *0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001
However, as the potential of a cell to reduce MTT depends on its metabolic rate, it is an imprecise measure of cell number and proliferation. It was therefore important to measure cell proliferation by other means. To do this, we performed BrdU incorporation assays, which measure DNA replication. Cells expressing Rheb-T23M, Y35N and E40K cells showed significantly greater BrdU incorporation than those transfected with Rheb-WT or the empty vector, either in the absence or presence of FBS in the growth medium (Fig. 3b; Supplementary Fig. S3b). Rheb-S21L and G29S did not increase BrdU incorporation either in the presence or absence of FBS (Fig. 3b; Supplementary Fig. S3b). Taken together, these data strongly suggest that Rheb-T23M, Y35N and E40K each increase cell proliferation under serum-starved conditions. Given that Rheb-Y35N has previously been characterised [19, 20, 28, 30], Rheb-S21L and G29S show only a minor effect on cell proliferation, we subsequently focused only on Rheb T23M and E40K. These findings are tabulated in Supplementary Fig. S3c.
As hyperactive mTORC1 signalling occurs in multiple cancers [31], we hypothesised that Rheb mutants that drive hyperactive mTORC1 signalling may be oncogenic and drive tumour growth. To test this hypothesis, we stably expressed Rheb-WT, T23M or E40K in mouse NIH3T3 cells (Fig. 3c; Supplementary Fig. S4a). Given that Rheb-Y35N has previously been shown to be oncogenic, i.e., to drive tumour growth [19], we chose not to include Rheb-Y35N in our analysis. To determine whether Rheb mutants cause oncogenic transformation of NIH3T3 cells, we performed assays for anchorage-independent growth in soft agar. Cells expressing Rheb-T23M and E40K grew robustly in soft agar, forming large colonies, while cells expressing Rheb-WT did not (Fig. 3d; quantified Fig. 3e). Treatment with AZD8055 prevented this, indicating that Rheb-T23M and E40K each promote oncogenic transformation in a manner that requires mTOR signalling, a pathway that is accepted as being tightly associated with cancer development in vivo [32].
Rheb-T23M and E40K are oncogenic and drive increased tumour growth in vivo
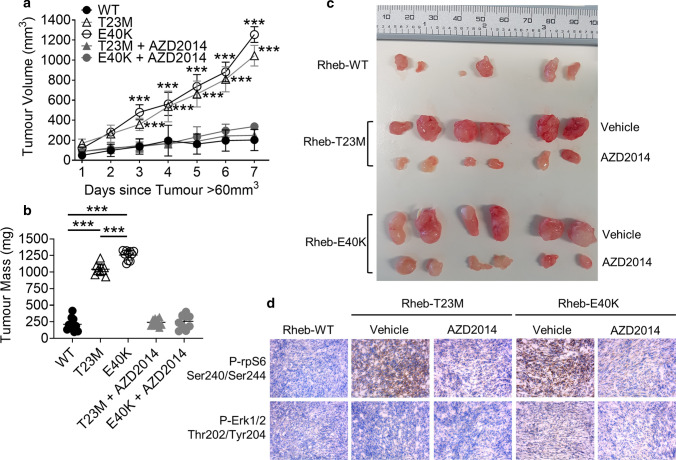
To test whether Rheb mutants drive tumour growth in vivo, NIH3T3 cells stably expressing Rheb-WT, T23M or E40K were injected subcutaneously into the flanks of 8–10-week old, male, immunodeficient NSG mice. When tumour volumes exceeded 60 mm3, some animals were treated with 20 mg/kg AZD2014 (an mTOR inhibitor suitable for use in vivo [33]) or vehicle via intraperitoneal injection daily for 7 days. Tumours from cells expressing Rheb-T23M and E40K grew significantly faster than those expressing Rheb-WT (Fig. 4a) so that, over the time of the experiment, Rheb-T23M and E40K tumours also reached a greater volume (up to 1000 mm3; within the permitted end-point of two tumours/mouse, each < 1000 mm3; Fig. 4a, b) and mass (approximately 1200 mg; Fig. 4c; full data for both tumours in every mouse are provided in Supplementary Table S4) compared to Rheb-WT tumours (approximately 300 mm3 and 250 mg). Treatment of mice with AZD2014 greatly impaired tumour growth indicating that Rheb-driven tumour growth depends on mTOR (Fig. 4a–c). Interestingly, while rates of growth and tumour mass were similar between Rheb-T23M and E40K-generated tumours, the times for tumour growth to become evident differed, with Rheb-E40K-driven tumours appearing on average 5 days earlier than Rheb-T23M or Rheb-WT tumours (Supplementary Fig. S4b). Immunohistochemical staining with P-rpS6 Ser240/244 and P-ERK Thr202/Tyr204 antibodies was performed on 8 µm sections to assess the mTORC1 and MAPK pathways within the tumours. As expected, tumours expressing Rheb-T23M and E40K showed increased P-rpS6 compared Rheb-WT tumours (Fig. 4d; quantified Supplementary Fig. S4c) while P-rpS6 was absent from tumours from animals treated with AZD2014. There was no difference in P-ERK between any of the tumours showing that MAPK was not activated (Fig. 4d; Supplementary Fig. S4d).
Fig. 4.
Rheb-T23M and Rheb-E40K drive mTOR-dependent tumour growth in vivo independently of MAPK. a Tumour size was measured every day for 7 days after the tumour volume had exceeded 60 mm3. Each animal produced two tumours. Each point represents means ± S.D.; n = 10 tumours. b Masses of tumours from panel a after 7 days of measured growth. c Images of tumours taken from three representative animals per group. d Immunohistochemistry of frozen sections taken from tumours probing for P-rpS6 Ser240/244 and P-ERK Thr202/Tyr204. Figure is representative of three replicates of randomly chosen tumours. Statistical significance was assessed by Student’s t-test where ***P < 0.001
Rheb-T23M and E40K promote cancer through distinct changes to the proteome
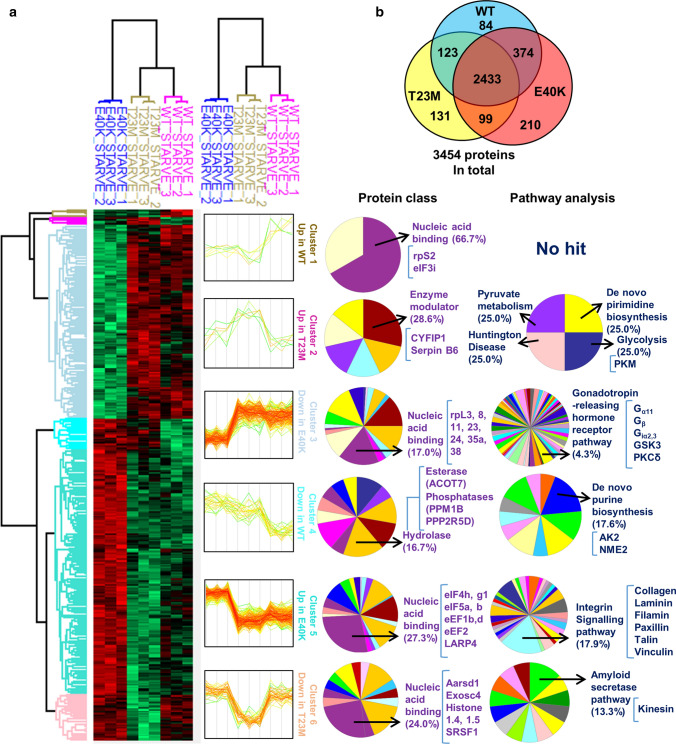
Given that we observe hyperactive mTORC1 activation driven by four previously unknown Rheb mutants, while two of them (Rheb-T23M and E40K) appear to cause a greater effect on cell proliferation than the others (Rheb-S21L and G29S; Fig. 3a), we hypothesised that Rheb-T23M and E40K may exert additional, perhaps mTORC1-independent, functions that aid tumour growth. To test this, we starved NIH3T3 cells stably expressing Rheb-WT, T23M and E40K of serum for 72 h before performing HPLC-mass spectrometry (MS) to evaluate changes in the global proteome (to assess the effects of the differences in sensitivity to TSC-GAP function; Fig. 5a; Supplementary Fig. S5a–c). Given that monoclonal cell lines can display distinct clonal differences that may be impossible to differentiate from the effect of Rheb mutation itself, we performed MS and all follow-up experiments using stably transfected, polyclonal NIH3T3 cell lines. Unexpectedly, we found that Rheb-T23M and E40K induce distinct changes in the proteome (Fig. 5b) with Rheb-T23M appearing more similar to Rheb-WT than Rheb-E40K. In cells expressing Rheb-T23M, we observed increases in the key glycolytic enzyme pyruvate kinase (PKM). In contrast, we found that Rheb-E40K upregulated proteins involved in protein synthesis or its control, such as La Ribonucleoprotein Domain 4 (LARP4). We also found that integrin signalling proteins were increased or decreased consistent with their role in either activating or inhibiting cell migration by both Rheb-T23M and E40K (Fig. 5a).
Fig. 5.
MS analysis of Rheb-WT, T23M and E40K expressing cells show distinct changes to the proteome. a NIH3T3 cells stably over-expressing Rheb-WT, T23M or E40K were starved of serum for 72 h before being subjected to MS analysis. Heat-map illustrates six distinct clusters as indicated. Proteins identified from each cluster were classified using the online bioinformatics source portal gene ontology (geneontology.org) according to their class and involvement in signalling pathways or other processes. Top hits from each protein class or signalling pathway group are shown. b Venn diagram illustrates the number of identified proteins whose abundances differ significantly from other cell groups
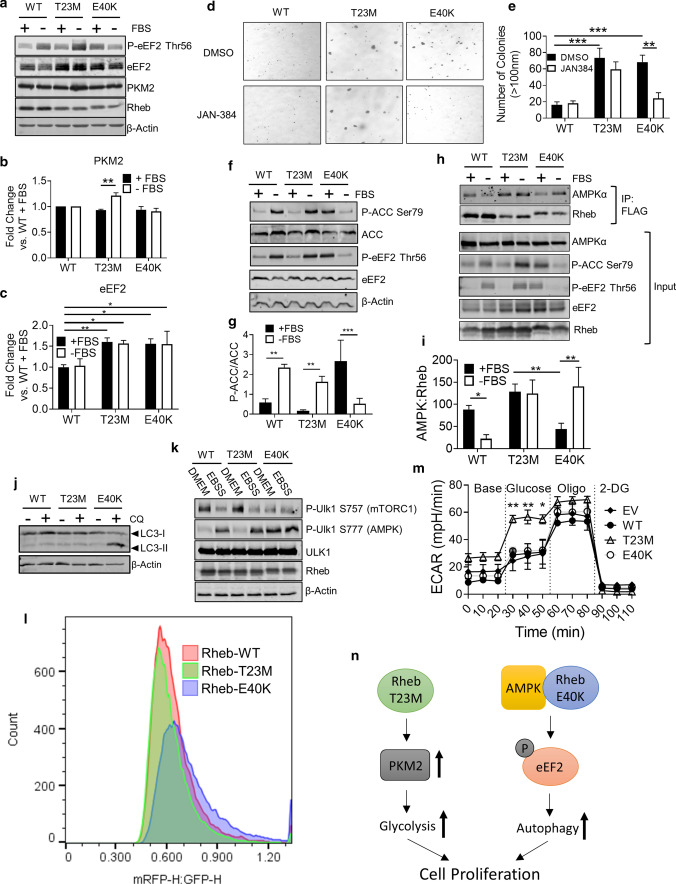
To validate our MS data, we performed western blot analysis for selected proteins, namely PKM2, eEF2, collagen and paxillin. This confirmed that PKM2 was increased only in Rheb-T23M cells (Fig. 6a; quantified in Fig. 6b) and that eEF2 is higher in both Rheb-T23M and E40K cells compared to those expressing Rheb-WT (Fig. 6a; quantified in Fig. 6c). To assess whether changes in protein levels reflected altered mRNA levels, we performed RT-qPCR analysis for the mRNAs encoding two proteins of particular interest, PKM2 and eEF2. RT-qPCR analysis showed no significant change in mRNA abundance of either PKM2 or EEF2 under any of the conditions tested and thus the changes in these proteins are not the result of altered mRNA levels (Supplementary Fig. S6a, b). Levels of collagen 2α1, an extracellular matrix protein involved in cell adhesion and mobility, were also increased in Rheb-T23M and E40K cells consistent with the MS data (Supplementary Fig. S6c, d). To determine if the MS data were relevant in vivo, we performed western blot analysis on samples from tumours (as generated in Fig. 4). eEF2 and PKM2 were increased in cells expressing Rheb T23M or E40K with eEF2 increased under all conditions. Interestingly, PKM2 was decreased in Rheb T23M when cells were pre-treated with the mTOR inhibitor AZD2014 (Supplementary Fig. S6e; quantified Supplementary Fig. S6f). How long-term mTOR inhibition causes decreases PKM2 levels warrants further investigation, which lies beyond the scope of this study.
Fig. 6.
Rheb-T23M and E40K drive cancer through distinct mechanisms. a Immunoblot analysis of lysates harvested from NIH3T3 cells stably expressing indicated Rheb mutant. Immunoblots were probed with antibodies for the indicated proteins or phosphorylation sites. Cells were grown in DMEM lacking FBS for 72 h prior to harvest. b Quantification of a for ratio of PKM2: β-actin. c Quantification of a for ratio of eEF2: β-actin. d Colony formation assay for 1 × 105 NIH3T3 cells stably expressing the indicated Rheb mutant. Liquid medium was changed twice per week for fresh medium containing either DMSO or 1 µM JAN-384. The number of colonies larger than 100 nM was counted by hand and is quantified in e. f Immunoblot analysis using the indicated antibodies of NIH3T3 cells stably expressing the indicated Rheb mutant. Cells were grown in either fully supplemented media or starved of serum for 24 h prior to harvest. g Quantification of f for the ratio of P-ACC:ACC normalized to β-actin. h Immunoprecipitation with anti-FLAG antibody of lysates harvested from NIH3T3 cells stably expressing the indicated Rheb mutant, followed by immunoblot analysis for the indicated proteins or phosphoproteins. i Quantification of h for co-immunoprecipitated AMPK normalized to immunoprecipitated FLAG-Rheb. j Western blot analysis for LC3 of NIH3T3 cells stably expressing the indicated Rheb mutant (positions of LC3-I and -II are shown). Cells were treated with either DMSO or 10 µM CQ, as indicated, for 2 h. k NIH3T3 cells stably expressing the indicated Rheb mutant we grown in fully supplemented media followed by treatments with EBSS as indicated for 2 h. Lysates were collected and western blot analysis performed with the indicated antibodies. l Flow cytometric analysis of tf-LC3 HeLa cells transiently expressing Rheb-T23M or E40K. Cells were gated to include only singlets and to exclude dead cells. Flow was stopped after 30,000 cells had been detected. Figure represents the ratio of mRFP1-H:EGFP-H. m Seahorse assay of extracellular acidification rate of HEK293 cells transfected with empty vector control or indicated Rheb mutant. For all figures error bars indicate SD for three independent experiments. n Diagram depicting how Rheb T23M and E40K drive cell proliferation via different pathways. For panels b, c, e, g, i and m, n = 3 (means ± S.D.). Statistical significance was determined by Student’s t-test where **0.001 ≤ P < 0.01; ***P < 0.001
Given that integrin signalling is important for tumour metastasis [34], we tested whether mutant Rheb proteins affected cells’ ability to migrate or invade using 3D ‘transwell’ assays. Cells expressing Rheb-T23M, Y35N or E40K all showed strikingly higher rates of migration than those transfected with empty vector or expressing Rheb-WT (Supplementary Fig. S7a; quantified Supplementary Fig. S7b). To measure invasion, cells were placed in Transwell chambers in which the bottom had been coated with matrigel. Cells expressing the Rheb mutants T23M, Y35N or E40K showed a significantly greater invasive ability than cells expressing Rheb-WT (Supplementary Fig. S7a–c).
Rheb T23M and E40K differentially regulate AMPK
Interestingly, phosphorylation of eEF2 at Thr56 differed between the mutants with P-eEF2 Thr56 increasing under serum starvation in cells expressing Rheb-WT or T23M but being lower under the same conditions in cells expressing Rheb-E40K (Fig. 6a; quantified Supplementary Fig. S6g). Under fully supplemented conditions, P-eEF2 was higher in Rheb-E40K cells compared to those expressing Rheb-WT or T23M (Fig. 6a; quantified Supplementary Fig. S6g). Thr56 in eEF2 is only phosphorylated by eEF2K [35], an atypical protein kinase which has been shown to play important roles in cancer cells [36]. Given the elevated levels of p-eEF2 in cells expressing Rheb-E40K and extensive previous work [36], we hypothesised that eEF2K may be required for growth of cells expressing Rheb-E40K. As expected from the data presented above (Fig. 3d), in soft agar assays cells expressing Rheb-T23M or E40K formed large colonies, while treatment with the eEF2K inhibitor JAN-384 prevented colony growth for cells expressing Rheb-E40K but not those expressing Rheb-WT or Rheb-T23M (Fig. 6d; quantified in Fig. 6e). This indicates that phosphorylation of eEF2 is required for the growth, under fully supplemented conditions, of cells expressing Rheb-E40K but not cells expressing Rheb-T23M. This striking difference warranted further study.
AMPK is an important upstream activator of eEF2K [37] whose input overrides the inhibitory effect on eEF2K activity of mTORC1 signalling (the latter pathway being activated by both Rheb mutants, so cannot explain their differing effects on p-eEF2). Given that Rheb-Y35N has previously been shown to regulate AMPK [19], we hypothesised that AMPK signalling might actually be differentially regulated by Rheb-T23M and -E40K. To test this, we starved NIH3T3 cells of serum for 24 h, a condition that activates AMPK to a similar extent as treatment with 2-deoxyglucose (2-DG) (Supplementary Fig. S7d) as judged by the phosphorylation of acetyl-CoA-carboxylase (ACC; at Ser79). This is a well-characterised substrate of AMPK and thus an indicator of AMPK activity. P-ACC was strongly increased above its low basal levels under serum-starved conditions in cells expressing Rheb-T23M (Fig. 6f; quantified in Fig. 6g). In contrast, P-ACC actually fell when cells expressing Rheb-E40K were starved of serum, indicating that AMPK is inhibited under this condition in cells expressing this mutant. When cells were treated with the mTOR inhibitor AZD8055 under fully supplemented conditions, P-ACC and P-eEF2 rose in cells expressing Rheb-WT and T23M consistent with the known effects of mTOR inhibition (Supplementary Fig. S7e). Cells expressing Rheb-E40K maintained elevated levels of P-ACC and P-eEF2, but phosphorylation was not increased by mTOR inhibition (Supplementary Fig. S7e). Total eEF2 and PKM2 protein levels were not affected (Supplementary Fig. S7e) by mTOR inhibition suggesting the decrease in PKM2 protein observed in vivo (Supplementary Fig. S6e) likely requires longer duration mTOR inhibition.
To study the interaction between Rheb and AMPK, Rheb (WT or mutant) was immunoprecipitated from cell lysates (using anti-FLAG) and samples were analysed, by immunoblot for FLAG and AMPK (Fig. 6h). In samples from cells in fully supplemented medium, AMPK bound less to Rheb-E40K than to Rheb-WT or T23M, whereas under serum-starved conditions binding of Rheb-E40K to AMPK actually increased (Fig. 6h; quantified in Fig. 6i). These data suggest that while Rheb-T23M and E40K can each bind to AMPK, their binding responds differently to serum starvation.
To assess further whether increased binding of Rheb-E40K to AMPK under serum-starvation conditions inhibits AMPK activity, we investigated the effects of Rheb and its mutants on another process which is well known to be regulated by AMPK, autophagy [38]. Like eEF2K, autophagy is positively regulated by AMPK and negatively regulated by mTORC1. We hypothesised that if expression of Rheb-E40K allowed for activation of AMPK in fully supplemented medium, it might drive autophagy even in the face of activated mTORC1 signalling. To test this, we treated NIH3T3 cells stably expressing Rheb-WT, T23M or E40K with a widely employed inhibitor of autophagy, chloroquine (CQ), for 2 h. Cells were then lysed, and western blot analysis performed for LC3. LC3-II is a 17-kDa protein that is recruited to the autophagosomal membrane and is degraded upon fusion with the lysosome. It therefore acts as an indicator of autophagic flux since inhibition of autophagy results in accumulation of LC3-II to an extent that depends on the prior rate of autophagy. LC3-II levels were greatly increased in serum-supplemented cells expressing Rheb-E40K that were treated with CQ compared to cells expressing Rheb-WT or T23M. The observed accumulation of LC3-II is indicative of increased autophagic flux (Fig. 6j), but is not diagnostic [39]. Given that mTORC1 inhibits autophagy via phosphorylation of ULK1 at Ser757 [38], it was important to determine if Rheb-T23M or E40K affected the mTORC1-mediated phosphorylation of ULK1. As expected, P-ULK1 Ser757 was decreased upon nutrient starvation in NIH3T3 cells stably expressing Rheb-WT or T23M (Fig. 6k); however, cells expressing Rheb E40K showed reduced basal phosphorylation of ULK1 at Ser757 that was insensitive to starvation. In contrast, upon nutrient deprivation, AMPK-mediated phosphorylation of ULK1 at Ser777 was increased in cells expressing Rheb WT or T23M but was already elevated in fully supplemented conditions in cells expressing Rheb E40K (Fig. 6k), consistent with the data for AMPK. Treatment with the mTOR inhibitor AZD8055 caused a loss of P-ULK1 Ser757 in cells stably expressing Rheb WT or T23M but not in those expressing Rheb E40K (Supplementary Fig. S7f).
To gain further insight into the effects on autophagy, we utilised a previously described [26] tandem-fluorescent LC3 (tf-LC3) HeLa cell line. As GFP’s fluorescence is quenched at the low intralysosomal pH, an increase in the mRFP1:EGFP fluorescence ratio indicates LC3 has entered lysosomes pointing to increased autophagic flux. Under fully supplemented growth conditions, Rheb-E40K caused a modest increase in the mRFP1:EGFP ratio compared to Rheb-WT or T23M (Fig. 6l) indicating that more tagged LC3 is present within the lysosomes of these cells and thus that the rate of autophagy is increased. Thus, unlike Rheb-WT or -T23M, Rheb-E40K actually promotes autophagy, despite also activating mTORC1; its effect on autophagy (presumably via AMPK) overrides its ability to switch on mTORC1.
Together, these data indicate that Rheb-T23M and Rheb-E40K differentially regulate AMPK through distinct features of their binding to AMPK under specific conditions, with Rheb-T23M binding to, and likely inhibiting, AMPK under fully supplemented conditions but not under serum-starvation and Rheb-E40K binding to AMPK and inhibiting it under serum-starvation but not in fully supplemented conditions. These Rheb mutants thus differentially affect autophagy and eEF2K; future studies are required to fully establish the mechanisms by which they differentially bind to and regulate AMPK. Since both are resistant to the GAP activity of TSC1/2 (which is enhanced under serum-starved conditions), their differing behaviour towards eEF2K is presumably not a consequence of their guanine nucleotide binding status.
Altered metabolism in cells expressing Rheb-T23M
Given that inhibition of eEF2K did not affect the growth of Rheb-T23M-expressing cells in soft agar, we suspected that another pathway(s) might be required for Rheb-T23M to drive cancer. We identified PKM2 levels as being slightly higher in cells expressing Rheb-T23M (Fig. 6a; quantified in Fig. 6b). If PKM2 is indeed elevated in cells expressing Rheb-T23M, it is likely that the rate anaerobic glycolysis may be increased. This effect has long been known to aid cancer cell growth, proliferation, and survival [40]. To test this, we performed a metabolic activity (‘Seahorse®’) assay to measure extracellular acidification rate (ECAR, i.e., due to production of lactate by anaerobic glycolysis).
Cells expressing Rheb-T23M showed much higher basal rates of anaerobic glycolysis upon addition of the substrate glucose than cells expressing Rheb-WT, E40K or empty-vector (Fig. 6m). As expected, adding oligomycin, which inhibits oxidative phosphorylation by blocking ATP synthase and thus ‘forces’ cells to increase flux through anaerobic glycolysis, markedly enhanced ECAR in Rheb-WT, E40K and empty-vector control cells. In contrast, cells expressing Rheb-T23M showed only a modest further increase in their already elevated ECAR. Maximal rates of ECAR (‘glycolytic capacity’) were rather similar in all cases. These data indicate that Rheb-T23M drives basal glycolytic rates much closer to the cells’ glycolytic capacity than is normally the case; thus, expression of Rheb-T23M appear to favour anaerobic glycolysis over oxidative mitochondrial metabolism, which would be consistent with increased levels of PKM2. This likely also contributes to their enhanced oncogenic potential.
Discussion
It is now clear that signalling through mTORC1 is deregulated in many cancers, due to mutation or loss of components of oncogenic upstream signalling pathways that impinge on TSC and thus Rheb [41], such as proteins involved in the classical MAP kinase (ERK) and PI 3-kinase/PKB (Akt) pathways including Ras or PI 3-kinase, which are mutated in many cancers. Here we show that multiple mutations in the Rheb-GTPase, a proximal regulator of mTORC1 [8, 9, 42–44], that have been identified in genomic studies of patient-derived tumours but previously considered to be background mutations or genetic noise [19], also lead to constitutive Rheb activity and thus promote signalling through mTORC1. Our results show that, in addition to the previously reported Rheb-Y35N mutant [19, 20, 28, 30], several further mutants identified in multiple cancer genome studies on a variety of different cancers are resistant to negative control by TSC1/2 and thus promote aberrantly active mTORC1 signalling.
We found that the Rheb mutants S21L, T23M, G29S, Y35N and E40K, are partially or wholly resistant to the GAP activity of TSC2 both when TSC1/2 is overexpressed in cells as well an in an in vitro GTPase assay. These data therefore suggest that these Rheb mutants do not drive constitutive mTORC1 signalling through Rheb-mediated activation of MAPK [19, 20], but rather through reduced sensitivity to the GAP activity of TSC2. Recent studies have also reported that Rheb-Y35N causes constitutive activation of MAPK leading to dysregulated mTORC1 signalling through continual inhibition of TSC2 [19, 20]. We show that Rheb-Y35N and E40K each increase P-ERK1/2 in fully supplemented medium (Supplementary Fig. S1b, c); however, our data suggest that neither pharmacological inhibition nor upstream inhibition of MAPK prevents the stimulation of mTORC1 signalling by Rheb mutants.
Combined data from three cancer genomics databases (COSMIC, The Cancer Genome Atlas and the Broad Institute Cancer Cell Line Encyclopedia) revealed that Rheb mutations occur in some cancers with the most common ones being, in descending order, Q57*, Y35N, T23M, Y35C, S16F, D171* and E40K. The frequency of Rheb-Q57* seems perhaps surprising as this truncation removes several important structural and functional domains, most notably the Switch II region (residues 63–79) which is crucial for Rheb function. It is therefore unlikely that Rheb-Q57* is constitutively active or even functional. Given that NIH3T3 cells stably expressing Rheb-T23M and E40K showed faster proliferation and robust growth in soft agar as well as increased growth in an in vivo syngeneic tumour graft, the Rheb-T23M and E40K variants are not simply background (‘passenger’) mutations, but instead likely act as strong driver oncogenes.
To assess the global protein expression changes regulated by Rheb-driven, constitutively active mTORC1 signalling, we performed MS analysis on NIH3T3 cells stably expressing Rheb-WT, T23M or E40K grown in DMEM lacking FBS. As our data imply that both mutants drive constitutive mTORC1 signalling through the same mechanism, we were surprised to observe other quite distinct changes in the proteome suggesting that Rheb, or at least these Rheb mutants, may exert functions additional to activation of mTORC1 signalling. Given that E40 is located within the Switch II region of Rheb which has been shown to be important for mTORC1 binding [45], it is possible this mutation alters the interaction between Rheb and other binding partners. In contrast, since T23 is located adjacent to, but not within, the P-loop, the T23M mutation may only affect GTP hydrolysis and would thus be expected only to maintain Rheb activity without significantly altering protein–protein interactions. This notion of differential effects of the two mutations is supported by our observation that Rheb-T23M and E40K interact differently with AMPK, a known binding partner of Rheb [19, 46]. The crystal structures of Rheb-T23M and E40K would need to be resolved to determine whether these mutations do indeed differentially alter Rheb-partner protein interactions.
We show that, as well as promoting constitutive mTORC1 activity, Rheb-T23M and E40K each modulate additional pathways that may help to drive cancer (Fig. 6n, i.e., PKM2 and AMPK, see elsewhere in text for more details). We show that, in NIH3T3 cells expressing Rheb-E40K, eEF2K activity is required for anchorage-independent growth, suggesting that constitutive mTORC1 activation alone is insufficient to drive cancer in this context and eEF2K activity is also required. Why eEF2K should be necessary to drive cancer growth in cells expressing Rheb-E40K but not Rheb-T23M is puzzling, although several earlier studies have implicated eEF2K in tumour growth (reviewed [35]). It has previously been shown that TSC1/2 null cells, which mimic the phenotype of constitutively active Rheb mutants, expend more ATP and become reliant on glucose for survival due to the increase in protein synthesis that accompanies mTORC1 activation [47]. We (and others) have also previously shown that AMPK phosphorylates eEF2K in response to poor intracellular energy status [37]. It is possible that the pro-survival effects of AMPK complement the pro-growth effects of mTORC1, thus producing the ideal conditions for sustained cancer growth. This would further be aided by the AMPK-mediated activation of autophagy (which in cells expressing Rheb-E40K appears to over-ride the inhibitory effects of mTORC1 on autophagy), as autophagy helps to provide the necessary macronutrients (precursors) to sustain biosynthetic processes and cell growth, such as amino acids. The ability to drive both mTORC1 signalling and autophagy would be a significant advantage to cancer cells especially in poor nutritional conditions where autophagy can provide precursors for the macromolecular synthesis pathways that are driven by mTORC1. Rheb-T23M cells instead show increased rates of anaerobic glycolysis, which may be related to changes in the levels of PKM, especially the PKM2 isoform. This effect is a well-known feature of tumour cells and allows for the rapid production of ATP, while conserving carbon atoms for use in biosynthetic processes (they would other be lost if they entered the Krebs cycle). In this context, this may act as a compensatory mechanism to provide cells with the ATP required for sustained growth without the activation of a pro-survival pathway such as AMPK and its downstream target, eEF2K. Future studies are required to test this hypothesis and to understand both the biological relevance of this observed difference between Rheb-T23M and E40K as well as the mechanisms responsible.
In summary, we show here that several tumour-associated mutations in Rheb are constitutively active and can act as driver oncogenes. We show that they drive constitutive mTORC1 signalling through insensitivity to TSC2’s GAP activity and not through constitutive activation of MAPK as previously suggested for another Rheb mutant. Finally, we show that Rheb-T23M and E40K regulate distinct, cancer-associated pathways. These findings also suggest that unique, personalized, combination therapies might be utilised to treat cancers harbouring different Rheb mutants.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (TIF 1892 KB) Supplementary Figure S1 Activation by Rheb mutants of signalling downstream of mTOR is blocked by AZD8055 but not AZD6244. Related to Fig. 1. (a) Immunoblot analysis of HEK293 cells transfected with the indicated Rheb mutants. Cells were treated with 1 µM AZD8055 1 h prior to harvest. (b) Immunoblot of lysates from HEK293 cells that had been transfected with a pcDNA3.1 empty vector (EV) or vector encoding wild-type Rheb or the indicated mutant. After 24 h, cells were treated with either DMSO or 5 µM AZD6244 for 2 h. All figures are representative images of three independent replicate experiments. (c) Immunoblot of lysates from HEK293 cells that had been transfected with a pcDNA3.1 empty vector or vector encoding wild-type Rheb or the indicated mutant. After 24 h, the medium was replaced with fresh medium either with or without FBS. After 16 h, cells were lysed. Immunoblots were probed with antibodies for the indicated proteins or phosphorylation sites. (d) Quantification of Supplementary Fig. S1c. P-ERK and total ERK were normalised to β-actin and the ratio of P-ERK:ERK graphed). Data are means ± S.D.; n = 3. (e) Immunoblots of lysates from HEK293 cells previously transfected with either the indicated Rheb mutant and a pcDNA3.1 empty vector or the indicated Rheb mutant and vectors encoding FLAG-TSC1 and FLAG-TSC2. After 24 h, the medium was replaced with fresh DMEM without FBS for 16 h followed by D-PBS for 1 h. (f) Quantification of Supplementary Fig. S1e. P-S6K1 Thr389 (top) and P-4EBP1 Ser65 (bottom) normalized to Tubulin loading control for means ± S.D.; n = 3. Data were analysed using Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Supplementary file2 (TIF 787 KB) Supplementary Figure S2: Rheb Mutants co-immunoprecipitate with TSC1/2 and mTORC1. Related to Fig. 2. (a) HEK293 cells were transfected with indicated vectors for FLAG-Rheb for 48 h, for both (a) and (b) Rheb and associated proteins were pulled down from cell lysates and both immunoprecipitates and input lysates were subjected to immunoblotting analysis for the indicated proteins.
Supplementary file3 (TIF 696 KB) Supplementary Figure S3: Rheb mutants do not drive faster proliferation in fully supplemented media. Related to Fig. 3. (a) MTT assay data for HEK293 cells transiently expressing the indicated Rheb mutants in fully supplemented medium. Data are means ± S.D.; n = 3. (b) BrdU incorporation data for HEK293 cells transiently expressing the indicated Rheb mutants in fully supplemented medium. (c) Table summarizing the phenotypes of various Rheb mutants as determined in Figs. 1d, e, 2c, d, 3a and b.
Supplementary file4 (TIF 1142 KB) Supplementary Figure S4: Characterisation of NIH3T3 cells stably over-expressing Rheb mutants. Related to Figs. 3 and 4. (a) Sanger sequencing results of monoclonal cell lines selected for use in the in vivo study. Cells were grown for 8 weeks before DNA was extracted and sequenced using primers specific to the inserted plasmid. (b) Survival curve showing the number of days post-injection until tumour volume reached 60 mm3. Statistics were calculated using the Mantel-Cox logrank test where @: P < 0.001. (c) Quantification of DAB stain in Fig. 4d for P-rpS6:rpS6 ratio. (d) As in (c) for P-ERK1/2:ERK1/2 ratio. Error bars indicate SD for three independent experiments. For panels c and d, data are means ± S.D.; n = 3. Statistical significance was determined by Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01.
Supplementary file5 (TIF 2923 KB) Supplementary Figure S5: MS analysis of proteome changes between fully supplemented and serum starved NIH3T3 cells. Related to Fig. 5. NIH3T3 cells stably over-expressing Rheb-WT, T23M or E40K were either cultured in full growth medium (with 10% FBS) or starved of serum for 72 h before being subjected to MS analysis. (a) Heat-map illustrates six proteins whose abundances differ among the cell groups cultured in fully supplemented growth medium. (b) Cluster analysis of proteins that are either upregulated when cultured in full growth medium or when serum starved. (c) Heat-map illustrates four different protein clusters among the treatment groups.
Supplementary file6 (TIF 852 KB) Supplementary Figure S6: Validation of MS data. Related to Fig. 5 a) RT-qPCR analysis of cDNA generated from whole cell RNA extracts from NIH3T3 cells stably expressing the indicated Rheb vector grown in fully supplemented media (+FBS) or starved of serum for 72 h (-FBS) using primers specific for PKM2. b) As in a) with primers for EEF2. c) Immunoblot analysis of NIH3T3 cells stably expressing the indicated Rheb mutant. Cells were grown in fully supplemented media or starved of serum for 72 h, as indicated. d) Quantification of c) normalized to β-actin. e) Proteins from tumour tissue (from Fig. 4) lysates were separated on SDS-PAGE gels, indicated proteins were immunoblotted with their corresponding antibodies. f) The indicated protein ratios in e) were quantified. g) Quantification of Fig. 6a for ratio of P-eEF2:eEF2. For panels XXX, data are means ± S.D.; n = 3. Statistical significance was determined by 2-way ANOVA with multiple comparisons to WT + FBS where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Supplementary file7 (TIF 1161 KB) Supplementary Figure S7: CA-Rheb mutants drive an increase in migration and invasion in trans-well assays. Related to Fig. 6. a) Transwell migration and invasion assays of 1.5 × 104 HEK293 cells transfected with vectors encoding the indicated Rheb mutant. For invasion assays, transwells were pre-coated with Matrigel. Cells were treated with 0.5 µg/µl for the duration of the experiment. Cells were allowed to migrate or invade for 72 h. b) Quantification of a) for migration. c) Quantification of a) for invasion. d) Immunoblot analysis of NIH3T3 cells. Cells were either grown in fully supplemented media or serum starved for 2h prior to harvest as indicated. As a positive control for AMPK activation, one well had media preplaced for media lacking glucose for 1 h prior to the addition of 2-deoxyglucose (2-DG) for 2 h. e) NIH3T3 cells stably expressing the indicated Rheb mutants were treated with AZD8055 or DMSO for 1h as indicated. f) NIH3T3 cells stably expressing the indicated Rheb mutants were treated with either 1 µM AZD8055 or DMSO, as indicated, for 1 h. Lysates were collected, and western blot analysis performed with the indicated antibodies. For panels b and c, data are means ± S.D.; n = 3. Statistical significance was determined by Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Acknowledgements
We would like to thank Professor Andrew Tee (Cardiff University, Cardiff, UK) and Dr. Brendan Manning (Harvard Medical School, Boston, MA, USA) for their valuable suggestions in regard to the GAP assay.
Financial support
This work was initiated with funding from the UK Biotechnology and Biological Sciences Research Council (to CGP) and then supported by SAHMRI. SDP acknowledges support by a Scholarship from the Australian Government Research Training Program (RTP). JX acknowledges a SAHMRI early/mid-career seed funding grant. LAS is supported by a Principal Cancer Research Fellowship awarded by Cancer Council’s Beat Cancer project on behalf of its donors, the state Government through the Department of Health, and the Australian Government through the Medical Research Future Fund.
Data availability
All RAW MS-based proteomics data have been deposited to ProteomeXchange via the PRIDE partner repository with the identifier PXD017006 and can be accessed at https://www.ebi.ac.uk/pride/archive/login.
Declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethics approval
All animal work was conducted in accordance with the National Health and Medical Research Council’s Care and use of Animals for Scientific Purposes guidelines and with approval (No. SAM339) from the South Australian Health and Medical Research Institute’s animal ethics committee.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jianling Xie and Stuart P. De Poi contributed equally to this work.
References
- 1.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-Q. [DOI] [PubMed] [Google Scholar]
- 3.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. doi: 10.1002/j.1460-2075.1996.tb00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeno P, Ballou LM, Novak-Hofer I, Thomas G. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA. 1988;85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 8.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 10.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 11.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Demetriades C, Plescher M, Teleman AA. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun. 2016;7:10662. doi: 10.1038/ncomms10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–d783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan L, Findlay GM, Jones R, Procter J, Cao Y, Lamb RF. Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J Biol Chem. 2006;281:19793–19797. doi: 10.1074/jbc.C600028200. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Vogt PK. Constitutively active Rheb induces oncogenic transformation. Oncogene. 2008;27:5729–5740. doi: 10.1038/onc.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazhab-Jafari MT, Marshall CB, Ho J, Ishiyama N, Stambolic V, Ikura M. Structure-guided mutation of the conserved G3-box glycine in Rheb generates a constitutively activated regulator of mammalian target of rapamycin (mTOR) J Biol Chem. 2014;289:12195–12201. doi: 10.1074/jbc.C113.543736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armijo ME, Campos T, Fuentes-Villalobos F, Palma ME, Pincheira R, Castro AF. Rheb signaling and tumorigenesis: mTORC1 and new horizons. Int J Cancer. 2016;138:1815–1823. doi: 10.1002/ijc.29707. [DOI] [PubMed] [Google Scholar]
- 18.Kim SR, Kareva T, Yarygina O, Kholodilov N, Burke RE. AAV transduction of dopamine neurons with constitutively active Rheb protects from neurodegeneration and mediates axon regrowth. Mol Ther. 2012;20:275–286. doi: 10.1038/mt.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Hong X, Wang J, Yin Y, Zhang Y, Zhou Y, Piao HL, Liang Z, Zhang L, Li G, Xu G, Kwiatkowski DJ, Liu Y. Inhibition of MAPK pathway is essential for suppressing Rheb-Y35N driven tumor growth. Oncogene. 2017;36:756–765. doi: 10.1038/onc.2016.246. [DOI] [PubMed] [Google Scholar]
- 20.Heard JJ, Phung I, Potes MI, Tamanoi F. An oncogenic mutant of RHEB, RHEB Y35N, exhibits an altered interaction with BRAF resulting in cancer transformation. BMC Cancer. 2018;18:69. doi: 10.1186/s12885-017-3938-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie J, de Souza AV, von der Haar T, O'Keefe L, Lenchine RV, Jensen KB, Liu R, Coldwell MJ, Wang X, Proud CG. Regulation of the elongation phase of protein synthesis enhances translation accuracy and modulates lifespan. Curr Biol. 2019;29:737–749.e735. doi: 10.1016/j.cub.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;77:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Humphrey SJ, Karayel O, James DE, Mann M. High-throughput and high-sensitivity phosphoproteomics with the EasyPhos platform. Nat Protoc. 2018;13:1897–1916. doi: 10.1038/s41596-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 24.Harney DJ, Hutchison AT, Hatchwell L, Humphrey SJ, James DE, Hocking S, Heilbronn LK, Larance M. Proteomic analysis of human plasma during Intermittent fasting. J Proteome Res. 2019;18:2228–2240. doi: 10.1021/acs.jproteome.9b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J, Shen K, Lenchine RV, Gethings LA, Trim PJ, Snel MF, Zhou Y, Kenney JW, Kamei M, Kochetkova M, Wang X, Proud CG. Eukaryotic elongation factor 2 kinase upregulates the expression of proteins implicated in cell migration and cancer cell metastasis. Int J Cancer. 2018;142:1865–1877. doi: 10.1002/ijc.31210. [DOI] [PubMed] [Google Scholar]
- 26.Hein LK, Apaja PM, Hattersley K, Grose RH, Xie J, Proud CG, Sargeant TJ. A novel fluorescent probe reveals starvation controls the commitment of amyloid precursor protein to the lysosome. Biochim Biophys Acta Mol Cell Res. 2017;1864:1554–1565. doi: 10.1016/j.bbamcr.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh AP, Marshall CB, Coric T, Shim EH, Kirkman R, Ballestas ME, Ikura M, Bjornsti MA, Sudarshan S. Point mutations of the mTOR-RHEB pathway in renal cell carcinoma. Oncotarget. 2015;6:17895–17910. doi: 10.18632/oncotarget.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 30.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.cd-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J, Wang X, Proud CG. mTOR inhibitors in cancer therapy. F1000Res. 2016 doi: 10.12688/f1000research.9207.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer. 2018;18:744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 33.Pike KG, Malagu K, Hummersone MG, Menear KA, Duggan HM, Gomez S, Martin NM, Ruston L, Pass SL, Pass M. Optimization of potent and selective dual mTORC1 and mTORC2 inhibitors: the discovery of AZD8055 and AZD2014. Bioorg Med Chem Lett. 2013;23:1212–1216. doi: 10.1016/j.bmcl.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 34.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore CE, Mikolajek H, Regufe da Mota S, Wang X, Kenney JW, Werner JM, Proud CG. Elongation factor 2 kinase is regulated by proline hydroxylation and protects cells during hypoxia. Mol Cell Biol. 2015;35:1788–1804. doi: 10.1128/mcb.01457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Proud CG. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol Sin. 2016;37:285–294. doi: 10.1038/aps.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 38.Kim J, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klionsky DJ, et al. Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Racker E. Bioenergetics and the problem of tumor growth. Am Sci. 1972;60:56–63. [PubMed] [Google Scholar]
- 41.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/mTOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 43.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 45.Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. doi: 10.1038/nature25023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacher MD, Pincheira R, Zhu Z, Camoretti-Mercado B, Matli M, Warren RS, Castro AF. Rheb activates AMPK and reduces p27Kip1 levels in Tsc2-null cells via mTORC1-independent mechanisms: implications for cell proliferation and tumorigenesis. Oncogene. 2010;29:6543–6556. doi: 10.1038/onc.2010.393. [DOI] [PubMed] [Google Scholar]
- 47.Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (TIF 1892 KB) Supplementary Figure S1 Activation by Rheb mutants of signalling downstream of mTOR is blocked by AZD8055 but not AZD6244. Related to Fig. 1. (a) Immunoblot analysis of HEK293 cells transfected with the indicated Rheb mutants. Cells were treated with 1 µM AZD8055 1 h prior to harvest. (b) Immunoblot of lysates from HEK293 cells that had been transfected with a pcDNA3.1 empty vector (EV) or vector encoding wild-type Rheb or the indicated mutant. After 24 h, cells were treated with either DMSO or 5 µM AZD6244 for 2 h. All figures are representative images of three independent replicate experiments. (c) Immunoblot of lysates from HEK293 cells that had been transfected with a pcDNA3.1 empty vector or vector encoding wild-type Rheb or the indicated mutant. After 24 h, the medium was replaced with fresh medium either with or without FBS. After 16 h, cells were lysed. Immunoblots were probed with antibodies for the indicated proteins or phosphorylation sites. (d) Quantification of Supplementary Fig. S1c. P-ERK and total ERK were normalised to β-actin and the ratio of P-ERK:ERK graphed). Data are means ± S.D.; n = 3. (e) Immunoblots of lysates from HEK293 cells previously transfected with either the indicated Rheb mutant and a pcDNA3.1 empty vector or the indicated Rheb mutant and vectors encoding FLAG-TSC1 and FLAG-TSC2. After 24 h, the medium was replaced with fresh DMEM without FBS for 16 h followed by D-PBS for 1 h. (f) Quantification of Supplementary Fig. S1e. P-S6K1 Thr389 (top) and P-4EBP1 Ser65 (bottom) normalized to Tubulin loading control for means ± S.D.; n = 3. Data were analysed using Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Supplementary file2 (TIF 787 KB) Supplementary Figure S2: Rheb Mutants co-immunoprecipitate with TSC1/2 and mTORC1. Related to Fig. 2. (a) HEK293 cells were transfected with indicated vectors for FLAG-Rheb for 48 h, for both (a) and (b) Rheb and associated proteins were pulled down from cell lysates and both immunoprecipitates and input lysates were subjected to immunoblotting analysis for the indicated proteins.
Supplementary file3 (TIF 696 KB) Supplementary Figure S3: Rheb mutants do not drive faster proliferation in fully supplemented media. Related to Fig. 3. (a) MTT assay data for HEK293 cells transiently expressing the indicated Rheb mutants in fully supplemented medium. Data are means ± S.D.; n = 3. (b) BrdU incorporation data for HEK293 cells transiently expressing the indicated Rheb mutants in fully supplemented medium. (c) Table summarizing the phenotypes of various Rheb mutants as determined in Figs. 1d, e, 2c, d, 3a and b.
Supplementary file4 (TIF 1142 KB) Supplementary Figure S4: Characterisation of NIH3T3 cells stably over-expressing Rheb mutants. Related to Figs. 3 and 4. (a) Sanger sequencing results of monoclonal cell lines selected for use in the in vivo study. Cells were grown for 8 weeks before DNA was extracted and sequenced using primers specific to the inserted plasmid. (b) Survival curve showing the number of days post-injection until tumour volume reached 60 mm3. Statistics were calculated using the Mantel-Cox logrank test where @: P < 0.001. (c) Quantification of DAB stain in Fig. 4d for P-rpS6:rpS6 ratio. (d) As in (c) for P-ERK1/2:ERK1/2 ratio. Error bars indicate SD for three independent experiments. For panels c and d, data are means ± S.D.; n = 3. Statistical significance was determined by Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01.
Supplementary file5 (TIF 2923 KB) Supplementary Figure S5: MS analysis of proteome changes between fully supplemented and serum starved NIH3T3 cells. Related to Fig. 5. NIH3T3 cells stably over-expressing Rheb-WT, T23M or E40K were either cultured in full growth medium (with 10% FBS) or starved of serum for 72 h before being subjected to MS analysis. (a) Heat-map illustrates six proteins whose abundances differ among the cell groups cultured in fully supplemented growth medium. (b) Cluster analysis of proteins that are either upregulated when cultured in full growth medium or when serum starved. (c) Heat-map illustrates four different protein clusters among the treatment groups.
Supplementary file6 (TIF 852 KB) Supplementary Figure S6: Validation of MS data. Related to Fig. 5 a) RT-qPCR analysis of cDNA generated from whole cell RNA extracts from NIH3T3 cells stably expressing the indicated Rheb vector grown in fully supplemented media (+FBS) or starved of serum for 72 h (-FBS) using primers specific for PKM2. b) As in a) with primers for EEF2. c) Immunoblot analysis of NIH3T3 cells stably expressing the indicated Rheb mutant. Cells were grown in fully supplemented media or starved of serum for 72 h, as indicated. d) Quantification of c) normalized to β-actin. e) Proteins from tumour tissue (from Fig. 4) lysates were separated on SDS-PAGE gels, indicated proteins were immunoblotted with their corresponding antibodies. f) The indicated protein ratios in e) were quantified. g) Quantification of Fig. 6a for ratio of P-eEF2:eEF2. For panels XXX, data are means ± S.D.; n = 3. Statistical significance was determined by 2-way ANOVA with multiple comparisons to WT + FBS where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Supplementary file7 (TIF 1161 KB) Supplementary Figure S7: CA-Rheb mutants drive an increase in migration and invasion in trans-well assays. Related to Fig. 6. a) Transwell migration and invasion assays of 1.5 × 104 HEK293 cells transfected with vectors encoding the indicated Rheb mutant. For invasion assays, transwells were pre-coated with Matrigel. Cells were treated with 0.5 µg/µl for the duration of the experiment. Cells were allowed to migrate or invade for 72 h. b) Quantification of a) for migration. c) Quantification of a) for invasion. d) Immunoblot analysis of NIH3T3 cells. Cells were either grown in fully supplemented media or serum starved for 2h prior to harvest as indicated. As a positive control for AMPK activation, one well had media preplaced for media lacking glucose for 1 h prior to the addition of 2-deoxyglucose (2-DG) for 2 h. e) NIH3T3 cells stably expressing the indicated Rheb mutants were treated with AZD8055 or DMSO for 1h as indicated. f) NIH3T3 cells stably expressing the indicated Rheb mutants were treated with either 1 µM AZD8055 or DMSO, as indicated, for 1 h. Lysates were collected, and western blot analysis performed with the indicated antibodies. For panels b and c, data are means ± S.D.; n = 3. Statistical significance was determined by Student’s t-test where *: 0.01 ≤ P < 0.05; **: 0.001 ≤ P < 0.01; ***: P < 0.001.
Data Availability Statement
All RAW MS-based proteomics data have been deposited to ProteomeXchange via the PRIDE partner repository with the identifier PXD017006 and can be accessed at https://www.ebi.ac.uk/pride/archive/login.