Abstract
Two modes of motility have been reported for bi-directional kinesin-5 motors: (a) context-dependent directionality reversal, a mode in which motors undergo persistent minus-end directed motility at the single-molecule level and switch to plus-end directed motility in different assays or under different conditions, such as during MT gliding or antiparallel sliding or as a function of motor clustering; and (b) bi-directional motility, defined as movement in two directions in the same assay, without persistent unidirectional motility. Here, we examine how modulation of motor–microtubule (MT) interactions affects these two modes of motility for the bi-directional kinesin-5, Cin8. We report that the large insert in loop 8 (L8) within the motor domain of Cin8 increases the MT affinity of Cin8 in vivo and in vitro and is required for Cin8 intracellular functions. We consistently found that recombinant purified L8 directly binds MTs and L8 induces single Cin8 motors to behave according to context-dependent directionality reversal and bi-directional motility modes at intermediate ionic strength and according to a bi-directional motility mode in an MT surface-gliding assay under low motor density conditions. We propose that the largely unstructured L8 facilitates flexible anchoring of Cin8 to the MTs. This flexible anchoring enables the direct observation of bi-directional motility in motility assays. Remarkably, although L8-deleted Cin8 variants exhibit a strong minus-end directed bias at the single-molecule level, they also exhibit plus-end directed motility in an MT-gliding assay. Thus, L8-induced flexible MT anchoring is required for bi-directional motility of single Cin8 molecules but is not necessary for context-dependent directionality reversal of Cin8 in an MT-gliding assay.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03891-x.
Keywords: Kinesin-5, Cin8, Motor–microtubule interaction, Context-dependent directionality reversal, Bi-directional motility
Introduction
Kinesin-5 motors perform essential mitotic functions in spindle assembly, maintenance and elongation (reviewed in [1–3]). These motors are unique in that they function as bipolar homotetramers, with dimeric motor domains positioned at opposite sides of the active tetrameric complex [4, 5]. This architecture enables kinesin-5 motors to crosslink and slide apart two antiparallel spindle microtubules (MTs) by simultaneous plus-end directed stepping on the two crosslinked MTs [6, 7]. It is believed that by this mechanism kinesin-5 motors are able to perform their mitotic functions in both spindle assembly and maintenance of the bipolar spindle structure [8–12] and in anaphase B spindle elongation [13–19].
Kinesin-5 motor molecules include a conserved N-terminal motor domain, a characteristic that they share with all plus-end directed kinesins. Interestingly, it has been shown that three fungal kinesin-5 motors—Saccharomyces cerevisiae Cin8 and Kip1 and Schizosaccharomyces pombe Cut7—are minus-end directed at the single-molecule level but switch directionality toward MT plus-ends under different experimental conditions [20–27]. For fungal kinesin-5 motors, such bi-directional motility was suggested to play an important role in the physiological functions of spindle assembly [20, 28]. Bi-directionality has also been demonstrated in other motors; for example, studies have shown that two kinesin-14 motors are bi-directional [29, 30] and that cytoplasmic dynein changes the directionality of force generation upon an increase of the motor number from a single molecule to the multi-motor level [31]. These studies thus indicate that switchable directionality may be more common among the motor proteins than was previously appreciated.
The different phenomena observed in the directional switching of kinesin-5 motors can be divided into two categories or modes. The first mode, referred to here as “context-dependent directionality reversal,” may be defined as a change from fast and processive minus-end directed motility, usually at the single-molecule level under high ionic strength, to plus-end directed motility in different assays, such as in MT-gliding assays. Here, motor density is locally high due to surface-concentrated motors interacting with the same MT, as has been demonstrated for all three bi-directional kinesin-5 motors [23, 25–27]. Context-dependent directionality reversal has also been observed in antiparallel MT sliding assays for Cin8 [20, 26, 27] and in parallel MT sliding for Cut7 [23]. Finally, it has been shown that Cin8 motors exhibit context-dependent directionality reversal due to motor clustering while they move along MTs [20, 32]. The second mode, referred to here as “bi-directional motility,” may be defined as successive movement in two opposite directions that can be detected in the same assay. For example, a dimeric truncated variant of S. cerevisiae Cin8 was shown to exhibit bi-directional motility at the single-molecule level [33]. Similarly, full-length Cin8 and Kip1 were shown to exhibit bi-directional motility in single-molecule motility assays when the ionic strength was decreased [21, 27]. However, it was not clear whether this effect was caused by motor clustering [15], which is expected to increase under lower ionic strength conditions. Finally, it has also been shown that Cin8 induces MT motility in both directions in MT-gliding assays, at low motor density, when several or individual surface-bound motors interact with the same MT [26, 34].
The above body of knowledge notwithstanding, how kinesin-5 motors can switch directionality is poorly understood. One of the reasons is the lack of systematic characterization of the different motility modes of the same kinesin motor. It has been shown experimentally that S. cerevisiae Cin8 shows both modes of directional switching, and this motor can thus be studied as a model to address the above-described longstanding gap in the understanding of bi-directional motility. Here, we examined how the two directionality modes of Cin8 are modulated by interactions of its motor domain with MTs. We found that the large sequence insert in loop 8 (L8), within the motor domain of Cin8, directly binds MTs, increases the affinity of motors for MTs in vivo and in vitro, and plays an essential role in the intracellular functions of Cin8. In vitro, L8 induces both context-dependent directionality reversal and bi-directional motility of single Cin8 motors, most likely through their direct MT binding. In MT-gliding assays, L8 facilitates MT attachment to surface-bound Cin8 high ionic strength and low motor density—conditions under which the bi-directional motility of MTs can be observed. Remarkably, however, L8 is not required for the context-dependent directionality reversal that takes place in MT-gliding assays under high motor density conditions. Since this large L8 of Cin8 is intrinsically disordered, we propose that it induces flexible anchoring of Cin8 motors to the MTs, thereby increasing Cin8–MT affinity and facilitating bi-directional motility in a given motility assay.
Materials and methods
The strains and plasmids used in this study are listed in Tables S3 and S4, respectively, in the Supporting Information section.
In silico analysis
Multiple sequence alignment was calculated by the MUSCLE algorithm via Unipro UGENE program [35]. Organism abbreviations used are: Sc—Saccharomyces cerevisiae; Spo—Schizosaccharomyces pombe; Kl—Kluyveromyces lactis; Ag—Ashbya gossypii; Cg—Candida glabrata; Dm—Drosophila melanogaster; Xl—Xenopus laevis; Hs—Homo sapiens; Ce—Caenorhabditis elegans. Residue-wise intrinsic disorder regions for the Cin8 motor domain (aa 1–530) were predicted by DISOPRED3 and ProDOS algorithms [36, 37], with a prediction false-positive rate of 5% and a disorder confidence score above 0.5 taken as the threshold for predicting disorder (Fig. S2B).
Yeast viability assay
A viability assay of cells expressing HA-tagged Cin8 variants as the sole source of kinesin-5 was performed as previously described [15, 38]. Briefly, a S. cerevisiae strain with chromosomal deletions of CIN8 and KIP1 and with an endogenic recessive cycloheximide-resistance gene (cin8Δkip1Δcyhr) was used. The double-deletion cin8Δkip1Δ was covered by a shuffle-out plasmid (pMA1208) containing a WT cycloheximide sensitivity gene and encoding for WT Cin8. Following transformation with centromeric plasmids expressing WT Cin8 or L8-deleted variants, the pMA1208 plasmid was shuffled out using 7.5 μg/ml cycloheximide in the YPD growth medium for 3–4 days at 26 °C and 37 °C.
Live-cell imaging
Live-cell imaging was performed as previously described [15, 38] using S. cerevisiae strains expressing 3GFP-tagged Cin8 variants, listed in Table S3 and S4. Cells were grown overnight in a medium lacking tryptophan, and 2 h prior to imaging the cultures were diluted tenfold. A sample of cells was placed on a low fluorescence agarose gel on a slide. Images were acquired at room temperature using a Zeiss Axiovert 200 M-based microscope, as described below for the single-molecule motility assay. Images of Z stacks of 15 planes were obtained in the bright field, and green and red fluorescence channels, with 0.5-μm separation between planes. Simultaneously, images of spindle localization of Cin8 variants were obtained by Z-projection of fluorescence images of the Z-planes using ImageJ-Fiji software.
Overexpression and purification of Cin8 variants for the single-molecule motility assay
6xHis-labeled full-length GFP-tagged Cin8 variants were used for the single-molecule motility assay. Cin8 variants were overexpressed and purified from a protease-deficient S. cerevisiae strain, as previously described [20, 27]. Briefly, S. cerevisiae cultures were grown with 2% raffinose, and overexpression of the motors was induced by 2% galactose. The harvested cells were ground in liquid nitrogen and centrifuged. The supernatant was loaded onto a nickel-nitrilotriacetic acid (Ni2+-NTA) column pre-equilibrated with equilibration buffer (50 mM Tris–HCl, 30 mM PIPES, 500 mM KCl, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, 0.05% Triton X-100, pH 8). After washing with equilibration buffer supplemented with 25 mM imidazole, Cin8 motors were eluted with elution buffer (50 mM Tris–HCl, 30 mM Pipes, 500 mM KCl, 250 mM imidazole, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, 0.05% Triton X-100, pH 7.2). The fractions were analyzed by SDS-PAGE and western blotting for the detection of Cin8-GFP. The fractions containing Cin8-GFP were pooled and subsequently purified by size-exclusion chromatography (SEC) on a Superose-6 10/300GL column, pre-equilibrated with SEC buffer (50 mM Tris–HCl, 30 mM Pipes, 500 mM KCl, 10% glycerol, 2 mM DTT, 1 mM MgCl2, 0.1 mM ATP, 0.05% Triton X-100, pH 7.2) on an ÄKTA FPLC system (GE Healthcare). The selected fractions were aliquoted, snap frozen, and stored at − 80 °C until use.
Cloning of bacterially expressed Cin8, L8-deleted variants, L8 and chimeric constructs
The plasmids for different Cin8 constructs were prepared using standard cloning methods. The Cin8 motor domain, aa 39–530 (WT Cin8 39–530, Cin8 39–530 ΔL8, or Cin8 39–530 ΔL8-D254A), L8 and extended L8 were amplified from synthesized primers (Sigma) and then cloned into a pET26b(+) vector (Novagen) with a C-terminal 6xHis tag. For the L8-deleted constructs, Cin8 39–530 ΔL8 and Cin8 39–530 ΔL8-D254A, the 99-aa insert in L8 of Cin8 was replaced with the 7-aa acid sequence of Kip1 L8 [Cin8: aa S255–G353 were replaced by Kip1 aa N234–S240 (see Fig. 1A)]. All the motor domain constructs of Cin8 used in this study also included the 18-aa neck-linker of Cin8 (aa K513–L530).
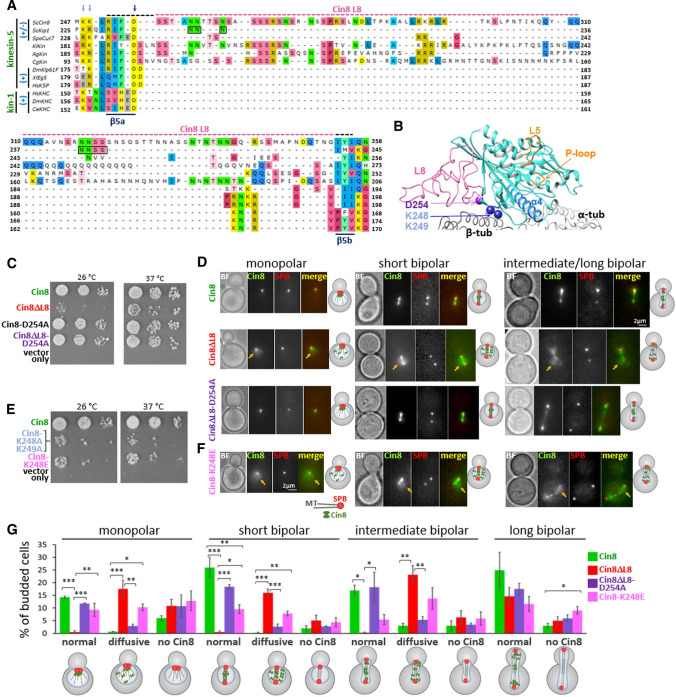
Fig. 1.
Intracellular functions of Cin8 variants mutated in the MT–motor interface. A Amino acid (aa) sequence alignment of the L8 region of kinesin homologs listed in Materials and methods. Kinesin family (green) and directionality (blue) of the kinesin motors are indicated on the left [(+) plus-end directed; (±) bi-directional]. Conserved aa D254, K248 and K249 are indicated by purple and blue arrows, respectively. The L8 of Cin8 (aa 255–353) is highlighted with a pink dashed line; the black extension of this pink dashed line represents the extended L8 variant. Residues of Kip1 L8 (aa 234–240) that replace L8 of Cin8 in Cin8ΔL8 are indicated by black boxes. β5-strands are indicated at the bottom of the panel. B Model of Cin8 motor domain bound to an αβ-tubulin-dimer, based on the cryo-EM structure of S. pombe Cut7 (PDB: 5M5I). This model is presented because a high-resolution atomic structure of WT Cin8 containing L8 is not available. L5 (gold), L8 (pink), P-loop (orange) and α4 (light blue) are shown. D254 (purple sphere) and K248 and K249 (blue spheres) are conserved in bi-directional kinesin-5 motors. C and E Yeast viability assay of Cin8 variants at two temperatures [15, 38]. D and F Localization of Cin8-3GFP variants in S. cerevisiae cells expressing tdTomato-tagged SPB component Spc42. Bright field (BF) Cin8, SPB and red and green merged images are shown. Arrows show diffusive localization of Cin8 in the nucleus. Schematic representation of budded cells with spindles of different lengths and morphologies are shown on the right of each panel. Spindle morphology and length (L) were categorized as follows: monopolar spindles—the red signals of the two SPBs are indistinguishable; short bipolar (pre-anaphase) spindles—L < 1.8 µm; intermediate bipolar (early anaphase) spindles—1.8 µm < L < 4 µm; long bipolar (late anaphase) spindles—4 µm < L. (G) Distribution of Cin8 localization phenotypes, presented in D and F. In each experiment, 200 cells were categorized. Columns and bars represent averages (± SEM) of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, calculated by one-way ANOVA, followed by Tukey’s test
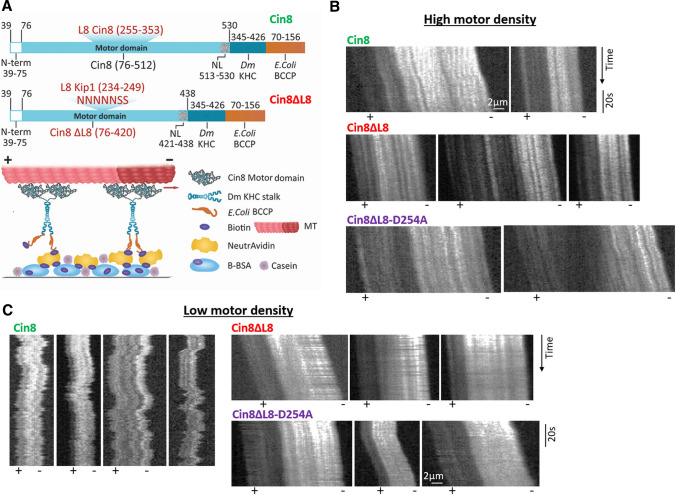
For the in vitro MT-gliding assay, chimeric constructs were prepared (Fig. 4A), in which the motor domain of Cin8, starting from 39, was cloned upstream to Drosophila melanogaster (Dm) kinesin-1-KHC (dimerization domain, aa 345–426, accession no.: P17210 (Gene ID:36,810). In addition, the 87 amino acids of the biotin COOH-terminal acceptor domain (aa 70–156) of Escherichia coli biotin carboxyl carrier protein (BCCP) (accession no.: P0ABD8; Gene ID: 947,758)[39–41] was cloned downstream to the KHC domain with a C-terminal 6xHis tag, into the pET26b(+) vector. The motor domain of Dm kinesin-1 from residue 1 to residue 401 was similarly cloned upstream to BCCP for use as the control in the MT-gliding assay. The DNA sequencing of all the cloned constructs confirmed the homogeneity of the desired sequences.
Fig. 4.
L8 of Cin8 induces bi-directional motility in a multi-motor gliding assay. A Schematic representation of the multi-motor MT-gliding assay. Top panel: bacterially expressed dimeric chimera proteins, containing the biotin-binding BCCP subunit. Bottom panel: motors adsorbed onto a glass surface via avidin–biotin interaction, followed by addition of polarity-marked MTs and ATP (see Materials and Methods). B and C Representative kymographs of MT gliding driven by the different Cin8 variants at high (B) and low (C) motor densities. Bright seed indicates the minus-end of MTs. When visible, MT polarities are indicated at the bottom
Purification of bacterially expressed Cin8 motor domain variants
Cells of E. coli strain BL21 codon plus (New England Biolabs) were transformed with plasmids encoding for the motor domains of the Cin8 variants, Cin8 39–530, Cin8 39-530ΔL8, and Cin8 39–530 ΔL8-D254A. Similarly, E. coli strain NiCo21 (DE3) cells were transformed with Cin8 L8, extended L8, a Cin8-Kin1-BCCP chimeric construct, or a kinesin-1-BCCP clone construct and subsequently grown in LB medium. Overexpression was induced with 0.2 mM isopropyl-1-thio-d-galactopyranoside (IPTG), and the cells were incubated at 18 °C for 6 h. For the Cin8-Kin1-BCCP construct and the Dm-kinesin-1-BCCP construct, 24 mg/l of biotin were added together with the IPTG.
The cells harvested from these cultures were resuspended in lysis buffer (50 mM HEPES, 500 mM KCl, 10 mM MgCl2, 0.01% Triton X-100, 0.5 mM TCEP and 10% glycerol, pH 7.5) and disrupted with a probe-type ultrasonicator, followed by centrifugation. For the kinesin-1-BCCP construct, 300 mM NaCl was used in the lysis buffer. The 6xHis tagged Cin8 constructs were subsequently purified using a Ni2+-NTA–agarose affinity column (Invitrogen) equilibrated with lysis buffer. After washing with 20 mM and 30 mM imidazole-supplemented lysis buffers, the proteins were eluted using 400 mM imidazole in the lysis buffer. The recombinant Cin8 variants were then purified by SEC using a Superdex 200 HR 16/60 column (GE Healthcare) in an ÄKTA FPLC system (GE Healthcare) equilibrated with 50 mM HEPES, 250 mM KCl, 1 mM MgCl2, 0.002% Triton X-100, 0.5 mM TCEP and 6% glycerol (pH 7.5). The eluted proteins were fractionated on SDS-PAGE, and the selected fractions were aliquoted, snap frozen, and stored at − 80 °C until use.
SEC-MALS of recombinant L8 proteins
The purified L8 and extended L8 proteins were separated on a Superdex 75 HR 10/300 column using an ÄKTA FPLC system. The proteins were concentrated to 1.5 mg/ml (in 50 mM HEPES pH 7.0, 25 mM KCl, 0.1 mM MgCl2, 1% glycerol and 0.5 mM TCEP). The SEC-MALS analysis was performed using theoretical molecular weights of 11.76 kDa and 12.68 kDa and extinction coefficients of 1490 M−1 cm−1 and 2980 M−1 cm−1 for L8 and extended L8, respectively. Peak alignment and band broadening correction between the UV, MALS, and RI detectors were conducted using Astra software algorithms [42].
Circular dichroism of recombinant L8 proteins
Far-UV circular dichroism (CD) measurements were obtained on a JASCO J-815 spectropolarimeter calibrated with ammonium (+)-10-camphorsulfonate, with a 1-mm path length cell at 25 °C. The values obtained were normalized by subtracting the baseline recorded for the buffer under similar conditions. A protein concentration of 10 μM was used for both L8 and the extended L8 protein. The protein was incubated for 3 h in a potassium phosphate buffer (pH 7.0) with different concentrations of KCl before taking the readings. The secondary structure and fold deconvolution were obtained from the CD spectra using CDNN software, and the CAPITO and BESTSEL online servers.
Single-molecule motor motility assay
In vitro single-molecule motility assays were performed as previously described [20]. Briefly, GMPCPP-stabilized, fluorescently labeled, biotinylated MTs were polymerized with 1 mg/ml tubulin, 0.08 mg/ml biotinylated tubulin and 0.08 mg/ml rhodamine-labeled tubulin, in the presence of 1 mM GMPCPP for 1 h at 37 °C in tubulin buffer (80 mM Pipes, 0.5 mM EGTA, 2 mM MgCl2, pH 6.9). For plus-end labeling of MTs, an additional 0.08 mg/ml of rhodamine-labeled tubulin was added after 1 h, and the mixture was incubated at 37 °C for another 45 min. Commercially available tubulin (Cytoskeleton Inc., USA) was used.
Coverslips, cleaned by ultrasonication in piranha solution and subsequently treated with 0.1% dimethyldichlorosilane in trichloroethylene, were assembled into flow cells of ⁓10 µl volume using double-sided tape. A silanized coverslip was coated with biotinylated bovine serum albumin (Sigma-Aldrich) and then incubated with NeutrAvidin (Life Technologies). Following the attachment of the biotinylated MTs, 20 µl of WT Cin8-GFP variants in motility buffer (50 mM Tris–HCl, 30 mM PIPES, 165/110 mM KCl, 2 mM MgCl2, 1 mM EGTA, 50 µg/ml casein, 1 mM DTT, 1 mM ATP, 10 mM glucose, 100 µg/ml glucose oxidase, 80 µg/ml catalase, 10 mM phosphocreatine, 50 µg/ml creatine kinase, pH 7.2) were added to the flow cell. The final concentration of all Cin8 variants in the single-molecule motility assay was ⁓10 pM, except for WT Cin8 at 110 mM KCl for which the concentration was ⁓7 pM, unless otherwise indicated. For the MT attachment analysis, the final concentration of all Cin8 variants was ⁓6 pM (Figs. 2A–C and S4). MTs and Cin8-GFP were imaged using a wide-field illumination Zeiss Axiovert 200 M inverted microscope equipped with an sCMOS camera (Neo, Andor) and a Plan-Apochromat DIC 100x/1.4NA objective (pixel size: 124 nm). Two filter sets were used: EGFP #49,002 and TRITC #49,004 (Chroma). Motility data were acquired using MicroManager controller software [43]. For Cin8-GFP motility and photobleaching experiments, MT images were acquired first, and thereafter 90–110 images were captured with the EGFP filter set, at 1 frame/s with an exposure time of 800 ms. All measurements were acquired in at least three independent experiments using two different protein preparations.
Fig. 2.
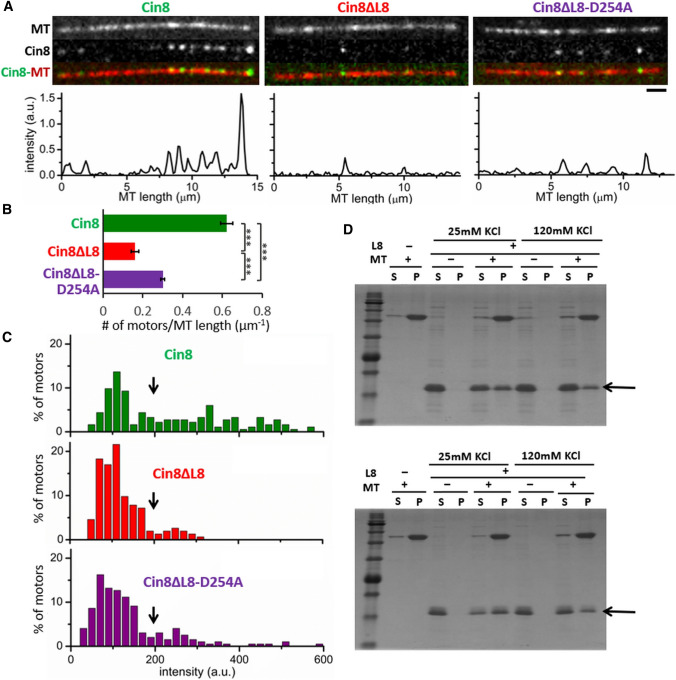
L8 induces MT attachment and clustering of Cin8 via direct binding to MTs. A Binding of full-length Cin8 variants to surface-bound MTs, in the presence of 1 mM ATP and 110 mM KCl. Top: Representative images of MT-bound Cin8 variants; bottom: corresponding intensity profile of the GFP signal. Bar: 2 µm. B Average number of MT-bound motors per MT length, calculated on 16 fields of 3844 µm2 (± SEM) at 110 mM KCl. ***P < 0.001, calculated by one-way ANOVA, followed by Tukey's test. C Intensity distribution histograms of MT-bound Cin8 variants in the assay described in (A). Maximal possible intensity of a single Cin8 monomer is indicated by black arrows (Fig. S3). D Representative SDS-PAGE profile of co-pelleting of recombinant L8 variants with MTs; L8 (aa 255–353) top panel and extended L8 (aa 251–356) bottom panel. SDS-PAGE stained with CBB. Supernatant (S) and pellet (P) of each reaction mixture is indicated on the top of each lane; Mw marker is shown on the left; Arrows indicate the sizes of the recombinant L8. Panels are representative of three experiments
Analysis of single-molecule motility data
Polarity of MTs was assigned on the basis of their bright plus-end labeling and/or the direction of fast-moving minus-end-directed Cin8 molecules [44]. ImageJ-Fiji software was used for image analysis and building kymographs [45]. Mean displacement (MD) analysis was performed as previously described [21, 33]. The coordinates of motile GFP-labeled motors were determined with the TrackMate plugin of the ImageJ-Fiji software or by manual tracking of the intensity center over time. Only those Cin8-GFP motors that moved more than 3 pixels were considered motile, and only those with motility events of at least 4 s were tracked. The MD values were obtained by averaging the displacements, calculated for all motility recordings of Cin8-GFP on the MTs. Mean velocities (V) were derived by fitting the MD functions to the linear equation MD = V·t. Each motor was categorized as MT-plus-end directed if its net displacement was in the plus-end direction and if it remained continuously plus-end directed for at least three quarters of the length of its overall run. All other motile motors were classified as minus-end directed.
Determination of the cluster size of Cin8
To evaluate the size of Cin8 clusters, photobleaching experiments of Cin8-GFP were performed to determine the contribution of single GFP molecules to the total intensity of these clusters, as previously described [32]. The fluorescence intensity of fluorescent Cin8 molecules was followed as a function of time within a circle of radius 4 pixels, using the TrackMate plugin of the ImageJ-Fiji software [46]. After correction for uneven illumination and background subtraction, we found that the intensity of the fluorescent Cin8 molecules was reduced in steps of ⁓50 a.u., with each step representing the photobleaching of 1 GFP (Fig. S3A). Since Cin8 is a homotetrameric motor and one GFP is attached to each subunit chain, the Cin8 tetramer (referred to as a single Cin8 molecule in this article) contains four GFPs. Hence, all Cin8 motors having an intensity ≤ 200 a.u. were likely to be single tetrameric Cin8 molecules. In addition, the following measures were taken to minimize the effect of GFP photobleaching on the determination of the Cin8 cluster size. We first determined the lifetime of a GFP molecule before photobleaching under our experimental conditions, which was found to be 26 ± 4 (± SEM) s (n = 23). Consequently, based on this estimation, all our motility measurements were performed only on those Cin8 motors that started moving within the first 30 s of each measurement. Finally, for motile Cin8 molecules and clusters, we measured the fluorescence intensity only in the first frame of their appearance, thereby significantly reducing the likelihood of estimating the cluster size of a photobleached motor. Using this method, we assigned intensity ranges of Cin8 motor fluorescence as < 200 a.u. and > 200 a.u. for single Cin8 molecules and Cin8 clusters, respectively.
Quantitative analysis of bi-directional motility
To determine the extent of bi-directional motility of single Cin8 molecules, we plotted the instantaneous 3-s displacement () over a moving window of 1 s. The extent of bi-directional motility was determined based on the average crossover frequency (υ, s−1) of crossing of the () vs. time plot of the horizontal axis ( = 0) (Fig. 3D). Statistical significance was calculated by one-way ANOVA followed by Tukey’s test.
Fig. 3.
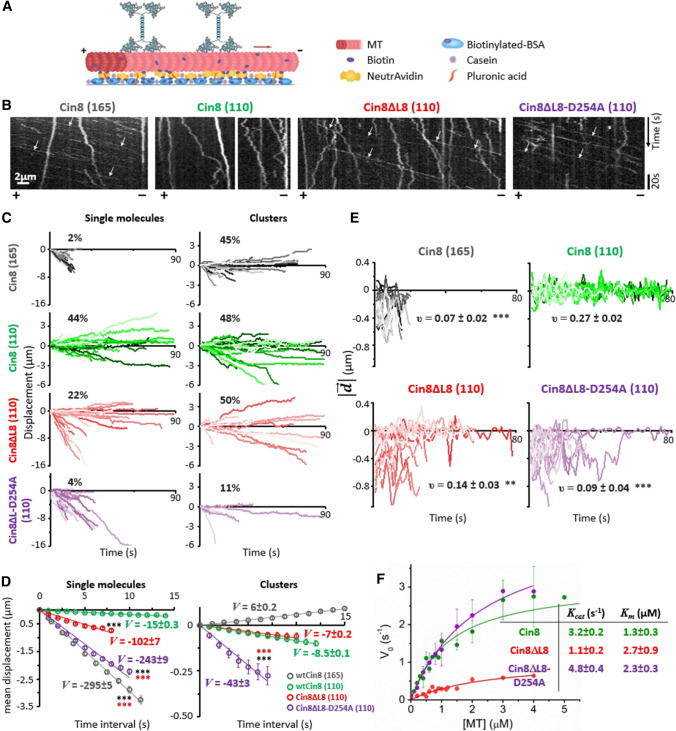
L8 induces bi-directional motility of single Cin8 molecules. A Schematic representation of the single-molecule motility assay. Red arrow represents the direction of movement of single Cin8 molecules under high ionic strength conditions. B Representative kymographs of motility of three variants of Cin8 on surface-bound MTs in the presence of 1 mM ATP. Polarity of the MTs is indicated at the bottom of the kymographs. Arrows indicate selected fast-moving and minus-end directed motility trajectories. C Displacement trajectories of Cin8 variants of single Cin8 molecules and clusters in the presence of 1 mM ATP. The percentage of plus-end directed trajectories is indicated in each plot. D Mean displacement (MD) analysis of trajectories of single Cin8 molecules and clusters shown in C. At t = 7 s, ***P < 0.001, compared to WT Cin8 (black asterisks) and to Cin8ΔL8 (red asterisks), calculated using two way ANOVA, followed by Tukey's test. The solid lines represent linear (MD = V·t) fits. Velocity values (V, nm/s ± SD) are indicated. E Instantaneous displacement () over a 3-s moving window, with 1-s interval, as a function of time. The variants of Cin8 and salt conditions (mM KCl) are indicated on top of each panel. υ indicates the average frequency (s−1 ± SEM) of directionality changes (n = 20). **P < 0.01; ***P < 0.001, compared to WT Cin8 at 110 mM KCl, calculated using one way ANOVA, followed by Tukey's test. B–E KCl concentrations, 165 or 110 mM, are indicated in parentheses. F ATPase activity of recombinant motor domain variants (Fig. S5) in the presence of 1 mM ATP. Kcat and Km values are indicated
MT-gliding assay
NeutrAvidin-coated coverslips were prepared, and flow cells were assembled as described for the single-molecule motility assay. Biotinylated BCCP-tagged motor proteins (Fig. 4) were incubated in motility buffer for 5 min at room temperature and washed. Two different buffers were used: for kinesin-1: 80 mM Pipes, pH 6.9, 4 mM MgCl2, 1 mM EGTA, 12 mM DTT, and 50 µg/ml casein, and for Cin8 variants: 50 mM Tris, 30 mM Pipes, pH 7.2, 1 mM EGTA, 10 µM Taxol, 2 mM MgCl2, 10 mM DTT, and 50 µg/ml casein. Taxol-stabilized minus-end labeled MTs were prepared as previously described [27] and added to the flow cell in the relevant motility buffer. According to the specific experiment performed, 1 mM ATP or AMP-PNP was added. In the case of ATP, an ATP regeneration system was also added; it contained 10 mM glucose, 100 µg/ml glucose oxidase, 80 µg/ml catalase 40, 10 mM phosphocreatine, and 50 µg/ml creatine phosphokinase. MT motility or attachment was monitored with the microscopy setup described for the single-molecule motility assay, using the TRITC filter set (#49,004, Chroma), with a 1-s time interval. Data were analyzed using the ImageJ-Fiji software. The following motor concentrations were used for high motor density experiments: WT Cin8—62 µg/ml; Cin8ΔL8—47 µg/ml; and Cin8ΔL8-D254A—43 µg/ml. In these experiments, the following KCl concentrations were used: Low KCl: WT Cin8—110 mM; Cin8ΔL8—45 mM; and Cin8ΔL8-D254A—60 mM; high KCl: WT Cin8—165 mM; Cin8ΔL8—60 mM; and Cin8ΔL8-D254A—110 mM (see Table S2 and Fig. S7 of the Supporting Information section). For low motor density experiments, the following motor and KCl concentrations were used: WT Cin8—5.2 µg/ml; 165 mM KCl; Cin8ΔL8—15.7 µg/ml; 60 mM KCl; and Cin8ΔL8-D254A—7.1 µg/ml; 110 mM KCl (see Table S2 and Fig. S7 of the Supporting Information section). The velocity of MTs was calculated as follows: a kymograph along the MT path was made using the Fiji software. The velocity of the MT movement was determined by measuring the slopes of motility trajectories in the kymograph. Only MTs with a displacement > 0.38 µm (3 pixels) and a motility duration longer than 10 s were included in the average velocity and directionality analysis. Velocities were estimated for each motility episode within a single run. Stops in motility were excluded from the velocity averaging. Only polarity-marked MTs were considered for the determination of directionality. Bi-directional motility of MTs was defined as motility with the appearance of at least two incidences of movement in two directions (i.e., back and forth) of 0.25–0.38 µm (2–3 pixels) over at least 1 s.
ATPase assay
The ATPase activity of Cin8 motor domain 39–530, Cin8 39–530-ΔL8, and Cin8 39–530-ΔL8-D254A were determined in ATPase buffer by measuring phosphate production in the presence of a minimum of 2.5-fold molar excess of Taxol-stabilized-MTs [47], using a commercially available kit (EnzChek, Molecular Probes) [48, 49]. ATPase activity was measured according to the manufacturer's instructions, and all the reagents were provided in the kit, except ATP and the PIPES buffer (pH 7.0). Protein concentrations from 80 to 200 nM were used. The ATPase assay was monitored with a Jasco V-570 UV/VIS/NIR Spectrophotometer.
MT co-sedimentation assay
The binding of the purified L8 and extended L8 proteins to MTs was monitored as previously described [50]. To this end, 750 ng of recombinant L8 or extended L8 were incubated with 250 ng of Taxol-stabilized MTs for 30 min at 28 °C, with no added nucleotide. MT-L8 complexes were pelleted by centrifugation at 21,000×g for 45 min at 28 °C. The pellets were resuspended in buffer of volume equal to that of the supernatant. The pellet and supernatant samples were fractionated on 15% SDS-PAGE and stained with Coomassie Brilliant Blue (CBB).
Results
Mutations in the motor–MT interface affect the in vivo function and spindle localization of Cin8
The Cin8 sequence contains a large insert in loop 8 (L8) in its N-terminal motor domain (Fig. 1A, B). Previous reports have indicated that this insert may interact directly with MTs [51]. Thus, we replaced this loop (aa 255–353) with the short L8 of the homologous Kip1 loop (aa 234–240), referred to herein as Cin8ΔL8 (Fig. 1A). We examined how this replacement modulates motor–MT interactions and how it affects the intracellular functions and motor activity of Cin8. First, we investigated the ability of Cin8ΔL8 to support yeast viability as the sole source of kinesin-5. It is known that at least one of the two S. cerevisiae kinesin-5 motors, Cin8 and Kip1, is essential for cell viability [9, 52], and we, therefore, examined the ability of Cin8 variants to cover the double chromosomal deletion of CIN8 and KIP1 (Fig. 1C) [38]. We found that expression of Cin8ΔL8 as the sole source of kinesin-5 reduced the viability of cin8Δkip1Δ cells (Fig. 1C), indicating that L8 deletion impairs important Cin8 functions that overlap with those of Kip1. Since Cin8 and Kip1 are believed to perform their mitotic roles by binding to and moving along MTs, our data suggest that deletion of L8 impairs these functions in Cin8 (see below).
To determine the cause of the reduced viability of cells expressing Cin8ΔL8, we followed the localization of 3GFP-tagged Cin8 variants in cells expressing the tdTomato-tagged SPB component Spc42 at different stages of the cell cycle (Fig. 1D). Consistent with previous reports, we found that, prior to spindle assembly, when the two SPBs are not yet separated, WT Cin8 was localized near the SPBs at the minus-ends of the nuclear MTs (Fig. 1D, left). This localization was found to be dependent on the minus-end directed motility of single Cin8 molecules in vitro [20]. We also found that WT Cin8 was bound along spindle MTs in pre-anaphase and early anaphase spindles but exhibited diffusive localization in the divided nuclei of late-anaphase cells [15, 16]. In contrast, Cin8ΔL8 exhibited diffusive nuclear localization (Fig. 1D) in monopolar, pre-anaphase, and mid-anaphase spindles (Fig. 1D, G). This finding indicates a reduction in the affinity of Cin8ΔL8 for spindle MTs compared to its WT counterpart. It should be noted that some cells did not produce a detectable Cin8 signal (Fig. S1 of the Supporting Information section), but there was no substantial difference between the different variants in the percentage of such cells (Fig. 1G).
We then examined the effect of mutating the conserved aspartic acid D254 to alanine (Cin8-D254A) on cell viability, since the sequence of Cin8 in the original study reporting on the mitotic functions of Cin8 contained this mutation [8]. In the absence of Kip1, cells expressing WT Cin8 or the mutant Cin8-D254A exhibited similar viability (Fig. 1C). This finding indicates that although D254 is located at the motor–MT interface (Fig. 1B), the D254A mutation had no effect on the function of WT Cin8 containing the native large L8 insert. This finding also explains the lack of observed phenotype for Cin8 carrying this mutation, reported in the original study [8]. Remarkably, however, the viability of cells expressing the combination of D254A mutation with the L8 deletion (referred to as Cin8ΔL8-D254A herein) was significantly improved, compared to cells expressing L8-deleted Cin8 (Cin8ΔL8) (Fig. 1C). The D254A mutation also consistently improved spindle localization of Cin8ΔL8, inducing SPB localization in monopolar spindles (similar to WT Cin8) and spindle attachment prior to and during anaphase B (Fig. 1D, G). These findings indicate that in the L8-deleted Cin8, a single replacement of aspartic acid with alanine at the motor–MT interface rescued the defects in intracellular functions and spindle attachment caused by deletion of the large L8 of Cin8. Since the surface of the MT lattice is negatively charged due to the outwardly exposed αβ-tubulin C-termini (including at and near the motor–MT interface [53, 54]), the replacement in Cin8 of a negatively charged aspartic acid with a neutral alanine at the motor–MT interface appeared to strengthen motor–MT interactions, thus improving spindle localization and the intracellular functionality of Cin8. When the D254A mutation was present and the large L8 of Cin8 was intact, this mutation had only a minor effect on Cin8 functionality (Fig. 1C). However, the effect of the D254A mutation became more apparent when the large L8 of Cin8 was deleted and the motor–MT interactions were consequently weakened (see below). In support of this notion, we found that mutations to either alanine or glutamate of the lysine residues located in the motor–MT interface (K248 and K249), which are conserved in bi-directional kinesin-5 motors (Fig. 1A, B), led to a reduction in cell viability, when the mutants were expressed as the sole source of kinesin-5 motors, and to diffusive localization of the mutants in cells (Fig. 1E–G). Replacing the positively charged lysines of Cin8 with a neutral alanine or a negatively charged glutamate at the motor–MT interface probably weakens motor–MT interactions. Thus, this result supports the notion that L8 deletion or K248 and K249 mutations interfere with the motor–MT interface, reduce motor affinity for the spindle MTs, and impair kinesin-5 functions in cells.
The native L8 insert in Cin8 induces MT attachment and clustering via direct binding to MTs
To validate the notion that the native L8 insert in Cin8 induces MT attachment and clustering via direct binding to MTs, we first examined how L8 deletion affects the binding of full-length GFP-labeled Cin8 variants, overexpressed and purified from S. cerevisiae cells, to surface-attached MTs in vitro (Fig. 2A [20]). We found that, compared to WT Cin8, considerably lower numbers of Cin8ΔL8 motors are bound to MTs (Fig. 2A, B). This finding indicates that—in keeping with its reduced affinity to spindle MTs (Fig. 1D)—Cin8ΔL8 exhibits reduced affinity to MTs in vitro. Moreover, the fluorescence intensity of MT-bound Cin8 motors was considerably higher for WT Cin8 than for the Cin8ΔL8 variant (Fig. 2A,C), indicating that the large L8 induces cluster accumulation of MT-bound Cin8 motors. These results are consistent with a previous report indicating that L8 of Cin8 mediates the super-stoichiometric MT binding of the bacterially expressed motor domain of Cin8 [51].
The D254A mutation alone did not substantially affect the activity of Cin8 motors in vitro, compared to WT Cin8 [Fig. S4 and 3C, Cin8 (110)]. However, combining the D254A mutation with deletion of L8 increased the number of MT-bound motors, compared to L8 deletion alone (Cin8ΔL8 vs. Cin8ΔL8-D254A, Fig. 2B), indicating that in the L8-deleted variant the D254A mutation increased the affinity of Cin8 for MTs. This result is consistent with the improved cell viability and spindle attachment observed in vivo (Fig. 1C, D, G). However, both L8-deleted variants exhibited reduced numbers of MT-bound motors, suggesting that their affinity for MTs is reduced compared to that of WT Cin8 (Fig. 2B). Cin8 motors carrying a mutation in the motor–MT interface (Cin8-K248E) that reduces cell viability and binding of Cin8 to spindle MTs (Fig. 1E–G) also consistently exhibited reduced attachments to MTs in vitro (Fig. S4B). Finally, the fluorescence intensity distributions of MT-bound Cin8 motors are similar for both Cin8ΔL8 and Cin8ΔL8-D254A variants, indicating that the D254A mutation does not affect the tendency of MT-bound Cin8 to accumulate in clusters.
We have shown that deletion of L8 reduces the binding of Cin8 to MTs in vivo (Fig. 1D) and in vitro (Fig. 2A,B). These findings, taken together with a previous demonstration that L8 promotes non-canonical MT binding of the bacterially expressed Cin8 motor domain [51], led us to hypothesize that the large L8 insert in Cin8 can directly bind MTs, in the absence of most of its motor domain and of the canonical MT-binding interface of kinesin motors. To examine this hypothesis, we generated two recombinant L8 proteins, expressed and purified from E. coli: a “native” version containing the insert in L8 of Cin8 (aa 255–353) and an extended version containing sequences of beta strand 5 (β5), on both sides of insert in L8 (aa 251–356) (Fig. 1A). Secondary structure analysis, using CD spectroscopy, of the recombinant L8 constructs revealed that both L8 versions assumes a random-coil conformation in solution, and that increasing the salt concentration does not dramatically alter the folding of L8 (Fig. S2A in the Supporting Information section) [55–57]. Our bioinformatics analysis also predicted that L8 is intrinsically disordered (Fig. S2B), in keeping with a previous analysis of L8 [51]. Based on the SEC-MALS data, both L8 versions are likely to be monomeric in solution (Table S1 and Fig. S2) [58].
To test whether the recombinant L8 interacts directly with MTs, the two L8 variants were subjected to co-pelleting assays with MTs at two salt concentrations (see Materials and Methods). L8 was found in the pellet only when MTs were present in the reaction mixture (Fig. 2D), directly demonstrating that Cin8 contains a MT-binding site in its large L8. In the absence of the motor domain and its canonical kinesin–MT interface, L8 of Cin8 can directly and solely bind MTs.
L8 of Cin8 promotes bi-directional motility of single Cin8 motors
To examine how MT binding via L8 affects the directionality of Cin8, we examined the motility of full-length Cin8 variants on plus-end labeled MTs (Fig. 3A) [20]. Experiments were performed at a saturating ATP concentration and at different ionic strengths. In keeping with previous reports [21], we observed that at high KCl concentrations (165 mM KCl) the majority of the motility trajectories of WT Cin8 were minus-end directed (Fig. 3B, and movie M1). However, at lower KCl concentrations (110 mM KCl), WT Cin8 exhibited slower and bi-directional motility (see below; also Figs. 3B, S4, and movie M2). In contrast to WT Cin8, the two L8-deleted variants did not bind to MTs at high ionic strength (not shown), and at lower ionic strength both L8-deleted variants exhibited faster and more minus-end directed motility, compared to WT Cin8 (Fig. 3B,C and movies M3 and M4).
We previously reported that one of the factors affecting the directionality and velocity of Cin8 is its accumulation in clusters on MTs [20]. Since we found here that L8 affects the accumulation of full-length Cin8 in MT-bound clusters (Fig. 2C), we sought to determine whether L8 deletion alters the motility of single Cin8 motors directly or indirectly by inducing their accumulation into clusters. Thus, to quantitatively characterize the effect of L8 on the motility of Cin8, we used a procedure that allowed us to distinguish between single molecules and clusters of Cin8 by following the fluorescence intensity of GFP-tagged Cin8 over time and determining size of Cin8 clusters based in their fluorescence [32] (see Materials and methods and Fig. S3).
We used the above method to examine the effect of L8 deletion on the motile properties of single molecules and clusters of Cin8. At intermediate KCl concentrations, we observed that ~ 40% of the trajectories of single molecules and clusters of WT Cin8 exhibited a net displacement toward the plus-end of the MTs and were thus defined as plus-end directed (Fig. 3C, see Materials and methods). Interestingly, although L8-induced clustering of Cin8 (Fig. 2C), deletion of L8 affected the motility of single Cin8 molecules by increasing their velocity and reducing the number of plus-end directed trajectories by a factor of two, from ~ 40 to ~ 20% (Fig. 3C). A similar effect on single-molecule motility was also observed for the Cin8-K248E mutant, whose affinity for MTs is also reduced in vivo and in vitro (Figs. 1F,G and S4), supporting the notion that increased minus-end directed bias and faster velocities are the result of weaker interactions between single motors and MTs. In contrast, expression of the D254A mutation, with the L8 present, had only a minor effect on the motility of single motors and clusters of Cin8 in vitro (Figs. S4, Cin8-D254A). This result reinforces the finding that that when the large L8 of Cin8 is present, the D254A mutation does not significantly affect motor–MT interactions and motor motility. MD analysis revealed that the average minus-end directed velocity of single Cin8ΔL8 molecules was considerably higher than that of WT Cin8 (Fig. 3D and Table 1) and similar to that of the Cin8-K248E mutant (Fig. S4). We also found that the average minus-end directed velocities of the single molecules and clusters of Cin8ΔL8-D254A were considerably higher than those of WT Cin8 and Cin8ΔL8 (Fig. 3D and Table 1). Finally, in contrast to WT Cin8 and Cin8ΔL8, there were almost no plus-end directed motility trajectories of single molecules and clusters of the Cin8ΔL8-D254A variant (Fig. 3C). These results indicate that L8 directly modulates the directionality of single molecules of Cin8 and promotes context-dependent directionality reversal toward the MT plus-end, when the conditions of the assay change from high to intermediate ionic strength.
Table 1.
Motile properties of WT and L8-deleted variants of Cin8
| Variant | Single-molecule motility assay a,b | Multi-motor MT-gliding assay | |||||||
|---|---|---|---|---|---|---|---|---|---|
| High-motor density d | Low-motor density h | ||||||||
| V (nm/s) c | Low KCl e | High KCl e | |||||||
| Single molecules | Clusters | V a (nm/s) | Net displacement (%)f | V a (nm/s) | Net displacement (%)f | V a (nm/s) | Net displacement (%)f | Bi-directional motility (%) | |
| WT CiIn8 | − 15.0 ± 0.3 (41) | − 8.5 ± 0.1 (47) | 30 ± 3 (94) g | 29 (177) | 30 ± 2 (114) g | 50 (389) | n.d. i | 18 (159) | 45 |
| Cin8ΔL8 | − 102 ± 7 (37) | − 7.0 ± 0.2 (12) | 24 ± 2 (130) g | 81 (210) | 24 ± 2 (126) g | 56 (384) | 26 ± 4 (13) g | 32 (59) | 0 |
| Cin8ΔL8-D254A | − 243 ± 9 (53) | − 43 ± 3 (9) | 38 ± 2*** (132) g | 75 (284) | 36 ± 2*** (122) g | 87 (219) | 38 ± 5 (38) g | 56 (91) | 0 |
aAverages ± SEM are presented. Values in parentheses represent the number of trajectories in the single-molecule assay, or number of MTs that exhibited a net displacement ≥ 0.37 µm
bConditions: 110 mM KCl, 1 mM ATP
cVelocities evaluated from MD analysis; negative values represent minus-end directed motility
dSee Materials and methods for motor concentrations
eSee Materials and methods for KCl concentrations
f% of MTs that exhibited a net displacement of ≥ 0.37 µm. Numbers in parentheses represent the total number of MTs in a field of 2460 µm2
g100% of the movements were plus-end directed
hSee Materials and methods for motor and KCl concentrations
iNot determined due to low number (only ten) of polarity-labeled MTs with displacement ≥ 0.37 µm, out of which seven were plus-end directed
***P < 0.001 compared to Cin8ΔL8, calculated using ANOVA, followed by Tukey's test
We then examined whether modulating motor–MT interactions by the deletion of L8 affects the bi-directional motility of Cin8 at the single-molecule level. To quantify the switches in directionality within continuous runs of Cin8, we plotted the instantaneous 3-s (frames) displacement of single motors () with a 1-s moving window, as a function of time (Fig. 3E). This analysis is sensitive to instantaneous changes in directionality and can thus quantitatively assess the bi-directional motility of the motors. In this presentation, switches in directionality are depicted by crossing the =0 axis by the 3 s displacement vs. time plot (Fig. 3E). As can be seen, single WT Cin8 motors exhibit higher frequency of directionality switching under intermediate, compared to high, ionic strength conditions (Fig. 3E). Thus, at the intermediate KCl concentration, WT Cin8 exhibited bi-directional motility within the same processive run. In addition, under intermediate ionic strength conditions, the average frequency (υ) of directionality switches was significantly higher for WT Cin8 than for Cin8ΔL8 or Cin8ΔL8-D254A (Fig. 3E). This finding indicates that under these conditions, the motility of WT Cin8 is significantly more bi-directional, while the motility of the L8-deleted variants is more minus-end directed. This finding thus shows that, while Cin8 moves on single MTs, the large L8 insert of Cin8 provides the functional context that directly influences the motility of single molecules of Cin8 by inducing both bi-directional motility and context-dependent directionality reversal.
The differences in single-molecule motility of WT and L8-deleted Cin8 variants can partially be attributed to differences in the steady-state ATPase activity of their motor domains (Fig. 3F) [47, 48]. For example, deletion of L8 significantly decreased Cin8 affinity for MTs (increasing Km) (Fig. 3F), consistent with the decreased MT affinity of full-length variants in single-molecule assay (Fig. 2B) and in cells (Fig. 1D). We can hypothesize that during the ATPase cycle, detachment from the MTs occurred faster for the L8-deleted motors than for WT Cin8 due to the loss of a second MT-binding site, resulting in decreased overall MT affinity. In addition, consistent with the faster velocity of full-length single molecules (Fig. 3D and Table 1), the ATPase rate of Cin8ΔL8-D254A was considerably higher than that of Cin8ΔL8 (Fig. 3F), indicating that modulations of the motor–MT interface directly affect the rate of ATP hydrolysis by Cin8. It was also found that Cin8ΔL8 exhibited faster minus-end directed motility, which could be attributed, at least in part, to its decreased affinity for MTs (Fig. 2A,B), as was suggested previously [21]. However, reduced affinity to MTs is probably not the sole cause of fast and minus-end directed motility, since the Cin8ΔL8-D254A variant is considerably faster and more minus-end directed than the Cin8ΔL8 variant (Fig. 3C,D) but exhibits higher affinity for MTs in vitro (Fig. 2A,B) and in vivo (Fig. 1C). Additional experiments are needed to understand the effect of the D254A mutation on the motility of L8-deleted variants of Cin8.
L8 of Cin8 promotes bi-directional motility in an MT-gliding assay
It has previously been demonstrated that Cin8, Kip1, and Cut7 motors reverse directionality from minus-end directed motility in a single-molecule assay to plus-end directed motility in MT-gliding assays [23–27, 34]. Since the L8-deleted variants of Cin8 exhibit a strong minus-end directed bias at the single-molecule level (Fig. 3B–E), we questioned how this bias would affect the motor directionality in MT-gliding assays. For this purpose, we generated dimeric chimera motor proteins containing the motor domain of WT Cin8, Cin8ΔL8 or Cin8ΔL8-D254A fused to the kinesin-1 dimerization domain [33] and containing a part of the E. coli BCCP, previously shown to undergo biotinylation in bacterial cells [39–41] (Fig. 4A). We previously demonstrated that similar single dimeric chimera Cin8 motors, lacking BCCP, can move in two directions on single MTs [33], indicating that bi-directional motility is an intrinsic property of Cin8 motors. As a control, we used a similar construct containing the kinesin-1 motor domain and confirmed that the kinesin-1 protein induced fast plus-end directed gliding of MTs following its attachment to an avidin surface (Fig. S6 and movie M5). Control experiments also indicated that incubation of the avidin surface with biotin prior to the attachment of kinesin-1 and Cin8 variant motors abolished MT binding to the surface (not shown), indicating that this MT attachment is mediated by the specific binding of biotinylated motors to the surface via avidin–biotin interactions (Fig. 4A).
At the same motor concentrations, the different dimeric Cin8 variants require different KCl concentrations for minimal MT attachment to the surface in the presence of a saturating ATP concentration (Table S2). WT Cin8 mediates the attachment of MTs at a concentration of 165 mM KCl. In contrast, L8-deleted Cin8ΔL8 fails to induce MT binding at KCl concentrations above 60 mM KCl, consistent with the notion that L8 deletion lowers the affinity of Cin8 for MTs in the single-molecule assay (Fig. 2A,B). The maximal concentration at which Cin8ΔL8-D254A induced MT binding was 110 mM KCl, which was higher than that for Cin8ΔL8. This finding indicates that, similar to full-length variants (Fig. 2A,B), a combination of the D254A mutation with the L8 deletion increased the affinity of the variant for MTs. Finally, both Cin8ΔL8 and Cin8ΔL8-D254A variants exhibited a lower affinity for MTs compared to WT Cin8 (Table S2), consistent with full-length motor binding to MTs (Fig. 2B) and kinetic ATPase data (Fig. 3F).
Based on the salt concentration requirement for MT attachment (Table S2), we examined the MT-gliding activity for each variant at two KCl concentrations, including the highest concentration at which MT attachment was observed (Tabl S2). Interestingly, we found that both L8-deleted variants exhibited fast minus-end directed motility at the single-molecule level (Fig. 3B–D), but at a high motor density both these variants induced plus-end directed MT gliding, similarly to WT Cin8 (Fig. 4B and movies M6-M8). Remarkably, the average MT plus-end directed gliding velocity of Cin8ΔL8 was slower than that of Cin8ΔL8-D254A (Table 1), in agreement with the velocities of single molecules toward the MT minus-end (Fig. 3E and Table 1) and the observed rate of ATPase activity (Fig. 3F).
At low protein concentrations, full-length Cin8 induced MT gliding in both plus-end and minus-end directions [34]. Thus, to examine whether L8 affects the directionality in the MT-gliding assay, we reduced the concentrations of the Cin8 variants to those at which minimal binding of MTs to the protein surface was achieved in the presence of ATP (Fig. S7). We found that at low protein concentrations, WT Cin8 induced bi-directional movement of MTs, with the MTs being observed as moving back and forth (Fig. 4C, Table 1, and movie M9). In most cases, no net displacement of the MTs was observed (Fig. 4B and movie M9). Remarkably, at the lowest motor density at which MT attachment could be observed in the presence of ATP, both L8-deleted variants induced only robust and processive plus-end directed MT motility, with no bi-directional episodes (Fig. 4B, Table 1, and movies M10 and M11). These results indicate that, similar to the single-molecule scenario (Fig. 3C), L8 also enables bi-directional motility in the MT-gliding assay. In contrast, the L8-deleted variants exhibited mainly unidirectional motility, either in the minus-end direction as single molecules or in the plus-end direction when inducing surface gliding of MTs.
In summary, our data indicate that by binding to MTs, the large L8 insert directly affects the motility of single Cin8 molecules. Our ability to separate single molecules from clusters enabled us to show that trajectories of single Cin8 molecules are bi-directional (Fig. 3C,E), indicating that single Cin8 molecules can move in both directions (Fig. 5Ai and Biii). In the multi-motor MT-gliding assay, under high motor density conditions (Fig. 5ii), all the examined variants exhibited context-dependent directionality reversal from minus-end directed motility of single molecules to plus-end directed motility (Figs. 4B and 5A). However, in that assay (Figs. 4C and 5Biv), bi-directional motility could be observed only under low motor density conditions when L8 was present, with L8 probably enabling the interaction of very few surface-bound motors with the MTs.
Fig. 5.
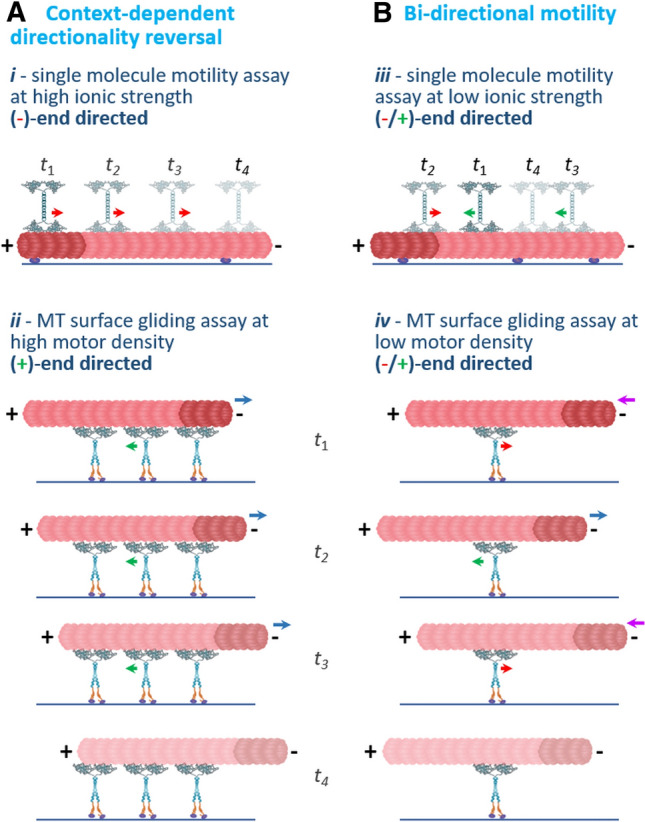
Schematic representation of the two modes of switchable directionality of Cin8 discussed in this study: context-dependent directionality reversal and bi-directional motility. t1–t4 represent four consecutive time points. A An example of context-dependent directionality reversal, where the same motors exhibit different directionalities in different assays or under different conditions. (i) In a single-molecule motility assay under high ionic strength condition, the motility of single Cin8 motors is minus-end directed (Fig. 3B, E). Red arrow indicates minus-end directionality of the motors. (ii) In contrast, in a multi-motor MT-gliding assay, at high motor density, when several motors interact with the same MT, motility is plus-end directed (Fig. 4B). Green arrows indicate plus-end directionality of the motors; blue arrows indicate the direction of MT gliding, with the minus-end leading. B Examples of bi-directional motility, where the motors move in two directions in the same assay and under the same conditions. (iii) In a single-molecule motility assay, under intermediate ionic strength conditions, single Cin8 motors move in both plus- and minus-end directions of the MTs, indicated by green and red arrows, respectively. This bi-directional motility is enhanced by the presence of the large L8 (Fig. 3B, C, E). (iv) In an MT-gliding assay, under low motor density conditions, the motor directionality is plus- and minus-end directed, indicated by green and red arrows, respectively. This bi-directional motility is manifested by bi-directional movement of the MT with the plus- and minus-ends leading at different time points (purple and blue arrows). This bi-directional motility is also dependent on the large L8 of Cin8 (Fig. 4C)
Discussion
Previous experimental and theoretical studies have indicated that, in cells dividing via closed mitosis in which the nuclear envelope remains intact, the switchable directionality of kinesin-5 motors is important for their physiological functions [20, 28]. According to a recently suggested mechanism, prior to spindle assembly, kinesin-5 motors first localize near the spindle poles of these cells via their minus-end directed motility on nuclear MTs. At this location, they then crosslink MTs emanating from the neighboring poles, reverse directionality, and mediate the initial separation of the spindle poles by sliding apart antiparallel MTs through plus-end directed motility on the two crosslinked MTs [20]. Thus, minus-end directed motility and context-dependent directionality reversal of fungal kinesin-5 motors are important for their physiological function, and understanding this mechanism is crucial for elucidating how bi-directional kinesin-5 motors perform essential mitotic functions. To date, context-dependent directionality reversal has been observed for the three bi-directional kinesin-5 motors [20, 23, 25–27] characterized by minus-end directed motility on the single-molecule level and plus-end directed motility in MT-gliding or sliding assays and as a function of motor clustering. In addition, bi-directional motility has been reported for Cin8 and Kip1 in the single-molecule assay [25, 33] and for Cin8 in MT-gliding assays [34] (Fig. 4). It should be noted that for Kip1, it has not been determined whether the bi-directional motility is an intrinsic property of single Kip1 motors or is a result of motor clustering, as was demonstrated for Cin8 [20, 32]. Since both modes of motility were previously reported for Cin8, we used Cin8 in the current study to examine the factors considered important for switchable directionality.
An accumulating body of evidence suggests that the two modes of the switchable directionality originate from the ability of bi-directional kinesin-5 motors to step in two directions on MTs. First, we have previously demonstrated that single dimeric Cin8 motors move in two directions on single MTs [33], indicating that bi-directional motility is an intrinsic property of Cin8 motors. Second, force measurements of Cin8 ensembles have revealed that the single-motor stall force is similar in the plus- and minus-end directions, indicating a fundamentally similar mechanism of stepping in the two directions [34]. Finally, a recent study indicated that increasing MT crowding induces a switch from minus- to plus-end directed motility of S. pombe Cut7, suggesting that stepping in two directions is affected differently by the presence of other MT-bound proteins [23]. The findings presented here also support this notion. By separately examining the motility of single molecules and clusters of Cin8, we show that at an intermediate KCl concentration single molecules of Cin8 move in two directions, even within the same run (Fig. 3E), consistent with bi-directional stepping. Finally, bi-directional movement of MTs was observed in the MT-gliding assay (Fig. 4C) when the attachment between surface-bound motors and MTs was minimal, which is also consistent with stepping in two directions.
Stepping in two directions most probably involves an intermediate stage in which the two motor heads of a dimer interact simultaneously with the same MT [59]. Such two-headed motor binding is inhibited in exclusively plus-end directed kinesin-1 motors to prevent back-stepping [60]. The two-headed strong bound state and stepping in two directions can be achieved by reduced inter-head tension of kinesin dimers, as was demonstrated by extending the length of the neck-linker, allowing back-stepping of the kinesin-1 dimers [59]. An additional mechanism for stepping in two directions can be provided by weaker and more flexible interactions between the motor domains and the MTs. The differences in the ionic strength requirements in in vitro motility assays of the different kinesin motors support this idea. Multiple studies indicate that the bi-directional kinesin-5 motors require a significantly higher ionic strength for single-molecule motility [24–27, 44] compared to the exclusively plus-end directed kinesin-5 motors [6, 61]. While single molecules of bi-directional kinesin motors interact with the MT lattice at high ionic strength, the plus-end directed kinesin-5 motors detach from the MTs under the same conditions [6]. Since a higher ionic strength appears to screen the net charges at the motor–MT interface [21], it is likely to decrease the strength of the non-covalent interactions between motors and MTs. Thus, under physiological conditions, the attachment between the MTs and the bi-directional kinesin motors is expected to be weaker and more flexible compared to that of the exclusively plus-end directed kinesins.
We recently proposed a model for context-dependent directionality reversal of bi-directional kinesin motors, based on the characterization of the motility of single motors, pairs, and clusters of Cin8 [32]. According to this model, context-dependent directionality reversal is achieved due to the asymmetric response to drag of stepping in the two directions. The predictions of this model show excellent agreement with experimental data and explain seemingly unrelated observations reported previously [32]. Based on this model and on the results presented in the current study, we propose that the large L8 insert of Cin8, which interacts directly with the MTs (Fig. 2), is one of the factors providing such drag, leading to both context-dependent directionality reversal and bi-directional motility at the single-molecule level under intermediate ionic strength conditions (Fig. 3). We also propose that the large and flexible L8 insert in Cin8, which is also present in other fungi kinesin-5 motors [38], is one of the structural elements that can facilitate flexible MT anchoring by mediating simultaneous binding of two motor heads to MTs and allowing stepping in two directions. We show that this additional MT anchoring, provided by the flexible L8 of Cin8, is required for the direct observation of the bi-directional motility in the MT surface-gliding assay. MT attachment can then be induced under conditions in which motor–MT interactions are minimal, such as high ionic strength and/or low surface motor density (Fig. 4), where bi-directional motility can be observed. For the L8-deleted variants, minimal MT attachment was achieved under lower ionic strength conditions and with higher motor density compared to the conditions for WT Cin8 (Fig. S7 and Table S2). Under these conditions, in the presence of L8-deleted variants, bi-directional motility of surface-bound MTs was not observed.
We show here that the L8 of Cin8 is required for bi-directional motility of Cin8 at the single-molecule level (Fig. 3E) and in the MT-gliding assay (Fig. 4C). Surprisingly, a recent study reported that in an MT-gliding assay of monomeric Cin8, no bi-directional motility was observed, independently of the presence of the large L8 [62]. These findings may indicate that for Cin8 to exhibit bi-directional motility, the two motor domains of an active dimer should interact with the same MT, similarly to the dimeric construct of Cin8 reported here (Fig. 4).
Although the L8 of Cin8 is required for bi-directional motility, it is not required for context-dependent directionality reversal in a multi-motor gliding assay (Fig. 4). The two L8-deleted variants, which exhibited strong minus-end directional bias at the single-molecule level (Fig. 3C, E), reversed directionality to robust plus-end directed motility in our MT-gliding assay (Fig. 4). Based on our recently proposed model [32], this finding may indicate that, under the conditions where surface-bound L8-deleted variants mediate MT attachment to the surface, the number of motors interacting with the same MT provide sufficient drag to induce directionality reversal. For these variants, minimal interaction of MTs (which enables bi-directional MT motility) is probably prevented by the lack of L8-induced flexible anchoring and the resulting lower MT affinity of these variants, compared to WT Cin8 (Fig. 1B and 2A, B). Thus, in keeping with a previous report [32], we propose that context-dependent directionality reversal can be achieved by increased drag in the motility assay, accomplished either by increasing motor–MT interactions in the single-molecule assay or by interaction of several motors with the same MT in the gliding assay.
The L8-deleted Cin8 variants and other bi-directional kinesins, such as Cut7 [23] and Kip1 [25], that lack the large L8 can reverse directionality, indicating that these motors can step in the two directions in the absence of the large L8. Based on our arguments above, the MT attachment of these kinesins is likely to be more flexible than that of the exclusively plus-end directed kinesins. Thus, it is likely that additional structural elements within the bi-directional kinesins facilitate flexible MT attachment, similarly to the K-loop within loop 12 specific to the Kif1A kinesin motors [63–65]. The bi-directional kinesin-5 motors share a common extended ~ 70 aa long N-terminal non-motor region [2, 22]. It is possible that this region also serves as a flexible anchor to MTs. Reports that the N-terminal region of the bi-directional Cut7 and of Aspergillus nidulans kinesin-5 BimC contain a non-canonical MT-binding site [23, 66] support this notion. These findings further indicate that flexible attachment to MTs may be achieved via different structural elements within the motor proteins and may be a common trait of bi-directional kinesins.
To summarize, we have shown here how modulation of motor–MT interactions affects the two modes of switchable directionality—context-dependent directionality reversal and bi-directional motility—of the kinesin-5, Cin8. We have demonstrated that single Cin8 molecules can reverse directionality and move in a bi-directional manner on the same MT. Both modes of motility of the single molecules of Cin8 are dependent on flexible anchoring to MTs mediated by an additional MT-binding site located in its large L8. The L8-induced flexible MT anchoring is also required for the bi-directional motility to be experimentally observed in MT surface-gliding assays. Since all bi-directional kinesin-5 motors exhibiting context-dependent directionality reversal execute stepping in two directions, we propose that the MT attachment of these motors allows higher flexibility than that of the exclusively plus-end directed kinesins.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Alina Goldstein-Levitin from the Gheber lab for critical reading of this manuscript and Tatiana Zvagelsky from the Gheber laboratory for providing images of yeast cells expressing GFP-tagged WT Cin8.
Author contributions
HP and SKS planned and performed experiments, analyzed the data and wrote the manuscript; MS, MP, MS, GD and SI performed experiments and analyzed the data; JAB, RZ, SSR analyzed the data and wrote the manuscript; LG conceived and supervised the study, planned the experiments, analyzed the data and wrote the manuscript.
Funding
This research was supported in part by the Israel Science Foundation grant (ISF-386/18) awarded to L.G.; the National Science Foundation (NSF-1615991) and United States—Israel Binational Science Foundation grant (BSF-2015851), awarded to L.G. and J.A.B.; the United States—Israel Binational Science Foundation grant (BSF-2019008), awarded to L.G. and S.R.; and the National Institutes of Health (grant NIH-R01-GM11283), awarded to J.A.B.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no competing interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable. All the authors agreed to publish this manuscript under Author Contributions.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Himanshu Pandey and Sudhir Kumar Singh have equal contribution.
Change history
11/4/2021
ESM of the article which was missed has been updated.
References
- 1.Mann BJ, Wadsworth P. Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 2019;29(1):66–79. doi: 10.1016/j.tcb.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK, Pandey H, Al-Bassam J, Gheber L (2018) Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell Mol Life Sci (75(10)):1757–1771.10.1007/s00018-018-2754-7 [DOI] [PMC free article] [PubMed]
- 3.Goulet A, Moores C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int Rev Cell Mol Biol. 2013;304:419–466. doi: 10.1016/b978-0-12-407696-9.00008-7. [DOI] [PubMed] [Google Scholar]
- 4.Scholey JE, Nithianantham S, Scholey JM, Al-Bassam J. Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers. Elife. 2014;3:e02217. doi: 10.7554/eLife.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379(6562):270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ. Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol. 2008;182(3):421–428. doi: 10.1083/jcb.200801145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435(7038):114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 8.Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118(1):109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70(3):451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- 10.Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83(7):1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 11.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286(5441):971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 12.Leary A, Sim S, Nazarova E, Shulist K, Genthial R, Yang SK, Bui KH, Francois P, Vogel J (2019) Successive Kinesin-5 Microtubule Crosslinking and Sliding Promote Fast, Irreversible Formation of a Stereotyped Bipolar Spindle. Curr Biol 29 (22):3825–3837 e3823 [DOI] [PubMed]
- 13.Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144(1):125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143(3):687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein A, Siegler N, Goldman D, Judah H, Valk E, Koivomagi M, Loog M, Gheber L. Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor localization and activity during anaphase. Cell Mol Life Sci. 2017;74(18):3395–3412. doi: 10.1007/s00018-017-2523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124(Pt 6):873–878. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerson-Gurwitz A, Movshovich N, Avunie R, Fridman V, Moyal K, Katz B, Hoyt MA, Gheber L (2009) Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell Mol Life Sci 66 (2):301–313 [DOI] [PMC free article] [PubMed]
- 18.Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10(4):387–393. doi: 10.1038/embor.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Movshovich N, Fridman V, Gerson-Gurwitz A, Shumacher I, Gertsberg I, Fich A, Hoyt MA, Katz B, Gheber L. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J Cell Sci. 2008;121(15):2529–2539. doi: 10.1242/jcs.022996. [DOI] [PubMed] [Google Scholar]
- 20.Shapira O, Goldstein A, Al-Bassam J, Gheber L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J Cell Sci. 2017;130(4):725–734. doi: 10.1242/jcs.195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shapira O, Gheber L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci Rep. 2016;6(25597):25597. doi: 10.1038/srep25597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edamatsu M. Molecular properties of the N-terminal extension of the fission yeast kinesin-5, Cut7. Genet Mol Res. 2016;15(1):15017799. doi: 10.4238/gmr.15017799. [DOI] [PubMed] [Google Scholar]
- 23.Britto M, Goulet A, Rizvi S, von Loeffelholz O, Moores CA, Cross RA. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proc Natl Acad Sci U S A. 2016;113(47):E7483–E7489. doi: 10.1073/pnas.1611581113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edamatsu M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochem Biophys Res Commun. 2014;446(1):231–234. doi: 10.1016/j.bbrc.2014.02.106. [DOI] [PubMed] [Google Scholar]
- 25.Fridman V, Gerson-Gurwitz A, Shapira O, Movshovich N, Lakamper S, Schmidt CF, Gheber L. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013;126(Pt 18):4147–4159. doi: 10.1242/jcs.125153. [DOI] [PubMed] [Google Scholar]
- 26.Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the Kinesin cin8 through motor coupling. Science. 2011;332(6025):94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 27.Gerson-Gurwitz A, Thiede C, Movshovich N, Fridman V, Podolskaya M, Danieli T, Lakamper S, Klopfenstein DR, Schmidt CF, Gheber L. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 2011;30(24):4942–4954. doi: 10.1038/emboj.2011.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwell R, Edelmaier C, Sweezy-Schindler O, Lamson A, Gergely ZR, O'Toole E, Crapo A, Hough LE, McIntosh JR, Glaser MA, Betterton MD (2017) Physical determinants of bipolar mitotic spindle assembly and stability in fission yeast. Sci Adv 3 (1):e1601603 [DOI] [PMC free article] [PubMed]
- 29.Popchock AR, Tseng KF, Wang P, Karplus PA, Xiang X, Qiu W. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat Commun. 2017;8(13999):13999. doi: 10.1038/ncomms13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molodtsov MI, Mieck C, Dobbelaere J, Dammermann A, Westermann S, Vaziri A. A Force-Induced Directional Switch of a Molecular Motor Enables Parallel Microtubule Bundle Formation. Cell. 2016;167(2):539–552.e514. doi: 10.1016/j.cell.2016.09.029. [DOI] [PubMed] [Google Scholar]
- 31.Nicholas MP, Höök P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun. 2015;6(6206):6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey H, Reithmann E, Goldstein-Levitin A, Al-Bassam J, Frey E, Gheber L (2021) Drag-induced directionality switching of kinesin-5 Cin8 revealed by cluster-motility analysis. Sci Adv 7 (6) [DOI] [PMC free article] [PubMed]
- 33.Duselder A, Fridman V, Thiede C, Wiesbaum A, Goldstein A, Klopfenstein DR, Zaitseva O, Janson ME, Gheber L, Schmidt CF (2015) Deletion of the Tail Domain of the Kinesin-5 Cin8 affects its directionality. J Biol Chem 19: 620–799 [DOI] [PMC free article] [PubMed]
- 34.Fallesen T, Roostalu J, Duellberg C, Pruessner G, Surrey T. Ensembles of Bidirectional Kinesin Cin8 Produce Additive Forces in Both Directions of Movement. Biophys J. 2017;113(9):2055–2067. doi: 10.1016/j.bpj.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5(113):113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31(6):857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishida T, Kinoshita K (2007) PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res 35 (Web Server issue):W460–464 [DOI] [PMC free article] [PubMed]
- 38.Goldstein A, Goldman D, Valk E, Loog M, Holt LJ, Gheber L. Synthetic-evolution reveals narrow paths to regulation of the saccharomyces cerevisiae mitotic kinesin-5 Cin8. Int J Biol Sci. 2019;15(6):1125–1138. doi: 10.7150/ijbs.30543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nenortas E, Beckett D. Purification and characterization of intact and truncated forms of the Escherichia coli biotin carboxyl carrier subunit of acetyl-CoA carboxylase. J Biol Chem. 1996;271(13):7559–7567. doi: 10.1074/jbc.271.13.7559. [DOI] [PubMed] [Google Scholar]
- 40.Li SJ, Cronan JE., Jr The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267(2):855–863. [PubMed] [Google Scholar]
- 41.Berliner E, Mahtani HK, Karki S, Chu LF, Cronan JE, Jr, Gelles J. Microtubule movement by a biotinated kinesin bound to streptavidin-coated surface. J Biol Chem. 1994;269(11):8610–8615. [PubMed] [Google Scholar]
- 42.Some D, Amartely H, Tsadok A, Lebendiker M. Characterization of proteins by size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) J Vis Exp. 2019;20(148):59615. doi: 10.3791/59615. [DOI] [PubMed] [Google Scholar]
- 43.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1(2):36. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapira O, Goldstein A, Al-Bassam J, Gheber L. Regulation and Possible Physiological Role of BI-Directional Motility of the Kinesin-5 Cin8. Biophys J. 2016;110(3):460a. doi: 10.1016/j.bpj.2015.11.2462. [DOI] [Google Scholar]
- 45.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Bodrug T, Wilson-Kubalek EM, Nithianantham S, Thompson AF, Alfieri A, Gaska I, Major J, Debs G, Inagaki S, Gutierrez P, Gheber L, McKenney RJ, Sindelar CV, Milligan R, Stumpff J, Rosenfeld SS, Forth ST, Al-Bassam J. The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. Elife. 2020;9(9):51131. doi: 10.7554/eLife.51131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muretta JM, Jun Y, Gross SP, Major J, Thomas DD, Rosenfeld SS. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc Natl Acad Sci USA. 2015;112(48):E6606–6613. doi: 10.1073/pnas.1512305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89(11):4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cochran JC. Kinesin motor enzymology: chemistry, structure, and physics of nanoscale molecular machines. Biophys Rev. 2015;7(3):269–299. doi: 10.1007/s12551-014-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell KM, Cha HK, Sindelar CV, Cochran JC (2017) The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner. J Biol Chem 12 (797662):797662 [DOI] [PMC free article] [PubMed]
- 52.Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper JR, Wordeman L. The diffusive interaction of microtubule binding proteins. Curr Opin Cell Biol. 2009;21(1):68–73. doi: 10.1016/j.ceb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang D, Asenjo AB, Greenbaum M, Xie L, Sharp DJ, Sosa H (2013) A second tubulin binding site on the kinesin-13 motor head domain is important during mitosis. PLoS One 8 (8):e73075 [DOI] [PMC free article] [PubMed]
- 55.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1(6):2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiedemann C, Bellstedt P, Görlach M. CAPITO–a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics. 2013;29(14):1750–1757. doi: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
- 57.Micsonai A, Wien F, Bulyáki É, Kun J, Moussong É, Lee YH, Goto Y, Réfrégiers M, Kardos J. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46(W1):W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Folta-Stogniew E (2006) Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. In: New and Emerging Proteomic Techniques. Springer, pp 97–112 [DOI] [PubMed]
- 59.Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18(9):1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Block SM. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J. 2007;92(9):2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, Hancock WO. Kinesin-5 is a microtubule polymerase. Nat Commun. 2015;6(8160):8160. doi: 10.1038/ncomms9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamagishi M, Maruyama Y, Sugawa M, Yajima J. Characterization of the motility of monomeric kinesin-5/Cin8. Biochem Biophys Res Commun. 2021;555:115–120. doi: 10.1016/j.bbrc.2021.03.134. [DOI] [PubMed] [Google Scholar]
- 63.Okada Y, Hirokawa N. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 1999;283(5405):1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 64.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA. 2000;97(2):640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lessard DV, Zinder OJ, Hotta T, Verhey KJ, Ohi R, Berger CL. Polyglutamylation of tubulin's C-terminal tail controls pausing and motility of kinesin-3 family member KIF1A. J Biol Chem. 2019;294(16):6353–6363. doi: 10.1074/jbc.RA118.005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stock MF, Chu J, Hackney DD. The kinesin family member BimC contains a second microtubule binding region attached to the N terminus of the motor domain. J Biol Chem. 2003;278(52):52315–52322. doi: 10.1074/jbc.M309419200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.