Abstract
Breast cancer stem cells (BCSCs) are positively correlated with the metastasis, chemoresistance, and recurrence of breast cancer. However, there are still no drugs targeting BCSCs in clinical using for breast cancer treatment. Here, we tried to screen out small-molecule compounds targeting BCSCs from the phenazine library established by us before. We focused on the compounds without affecting cell viability and screened out three potential compounds (CPUL119, CPUL129, CPUL149) that can significantly attenuate the stemness of breast cancer cells, as evident by the decrease of stemness marker expression, CD44+/CD24– subpopulation, mammary spheroid-formation ability, and tumor-initiating capacity. Additionally, these compounds suppressed the metastatic ability of breast cancer cells in vitro and in vivo. Combined with the transcriptome sequencing analysis, ferroptosis was shown on the top of the most upregulated pathways by CPUL119, CPUL129, and CPUL149, respectively. Mechanistically, we found that these three compounds could trigger ferroptosis by accumulating and sequestering iron in lysosomes through interacting with iron, and by regulating the expression of proteins (IRP2, TfR1, ferritin) engaged in iron transport and storage. Furthermore, inhibition of ferroptosis rescued the suppression of these three compounds on breast cancer cell stemness. This study suggests that CPUL119, CPUL129, and CPUL149 can specifically inhibit the stemness of breast cancer cells through triggering ferroptosis and may be the potential compounds for breast cancer treatment.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04384-1.
Keywords: Breast cancer stem cell, Phenazine compounds, Stemness, Ferroptosis, Iron, Lysosomes
Background
Cancer stem cells (CSCs) have been considered as a root of tumor progression [1–3]. Breast cancer displays characteristics of recurrence, metastasis, and chemoresistance due to its heterogeneity, which could be contributed by BCSCs [4]. Targeting BCSCs has been considered as a potential way for breast cancer treatment. However, there are still no drugs targeting BCSCs approved for clinical using in breast cancer treatment.
Many signaling pathways, such as Wnt, Hedgehog and Hippo, have been confirmed to be critically involved in BCSC progression [5]. Small molecular compounds and biologics have been developed for targeting CSCs, such as Glasdegib targeting leukemia stem cells (LSCs) [6], and ELZONRIS targeting CD123 that is highly expressed in LSCs has already been approved by the USA Food and Drug Administration (FDA) [7]. However, there are still no drugs targeting BCSCs approved for clinical application in breast cancer treatment. Thus, it is extremely important to find or construct drug repositioning to explore compounds targeting BCSCs.
Recently, the iron metabolism has been found to be dysregulated in BCSCs [8]. The iron level is increased in BCSCs, which confers BCSCs to be more sensitive to iron chelation [9]. Consistently, Basuli et al. indicated that ovarian cancer tumor-initiating cells (TICs) exhibited an accumulation of excess intracellular iron and an augmented dependence on iron for proliferation [10]. Iron also can produce OH. by inducing Fenton reaction which is the strongest oxygen-free radical in the reactive oxygen species (ROS), and OH. can then attack lipid to increase lipid peroxidation, which is a critical contributor of ferroptosis [5]. Additionally, iron homeostasis is modulated by different iron regulatory proteins, such as iron-responsive element-binding protein 2 (IRP2), transferrin receptor 1 (TfR1), and ferritin. TfR1 and transferrin degradation in lysosomes have been shown to be increased in BCSCs, while ferroportin (FPN) is decreased [11]. Furthermore, the decrease of transferrin suppresses ROS production and induces ferroptosis [12]. Ferritin is a container to store iron and superoxide radicals can trigger the release of Fe2+ from ferritin [13]. Notably, a lysosome disruptor, siramesine, can increase iron level in breast cancer cells resulting in ROS production and ferroptosis [12]. Besides, the induction of ferroptosis is accompanied by the degradation of ferritin [14, 15].
Lysosomal activity has been shown to be engaged in lipid ROS-mediated ferroptosis through regulation of cellular iron equilibria and ROS generation [16]. In 2017, Salinomycin and its derivatives were the first ever molecules described for killing BCSCs by inducing lysosomal iron sequestration and thus leading to ferroptosis [17]; this novel mechanism for killing BCSCs was further confirmed by numerous studies, for example, it was found that the decrease of iron concentration in lysosomes could maintain the stemness of cancer cells [18]; Xu et al. demonstrated that Itraconazole attenuates the stemness of nasopharyngeal carcinoma cells via triggering ferroptosis through this same mechanism [19]. Similarly, Shi et al. phenocopied this mechanism by which Dichloroacetate attenuates the stemness of colorectal cancer cells [20]. Notably, the decrease of iron output-related genes and the increase of iron input-related genes represent a better prognosis in breast cancer patients [21], and the CSC marker CD44 can mediate iron endocytosis in CSCs [22]. Furthermore, lysosome iron release has been shown to be essential for chemotherapy-induced stemness of small cell lung cancer cells [23]. Recently, iron nanoparticles that can target CSCs through ferroptosis and apoptosis were approved by the FDA, which bring up a new strategy to treat cancer by applying iron [24]. These results amplify that sequestering iron in lysosomes is a promising method in targeting BCSCs [25], and suggest that iron increase in BCSCs promotes the activity of iron-dependent proteins and avoids the damage caused by iron overload through reducing iron reserve in lysosomes, thus reaching an “adjusted iron homeostasis” in line with CSC metabolism level.
It has been reported that clofazimine, an imiphenazine drug, can inhibit the intercellular communication mediated by connexin Cx46 and induce the apoptosis of glioma CSC [26]. In consistent, clofazimine has recently been revealed to suppress leukemia stem cells [27]. Moreover, cloroprine can increase the sensitivity of leukemic cells to cisplatin and the sensitivity of three negative breast cancer cells to doxorubicin [28, 29]. These results suggest that phenazine compounds might inhibit BCSC. In the present study, we reported three phenazine derivatives (CPUL119, CPUL129, CPUL149) that can also significantly attenuate the stemness of breast cancer cells through sequestering iron in lysosomes by interacting with and sequestering iron in lysosomes and thus triggering ferroptosis. This study indicates that CPUL119, CPUL129, and CPUL149 may be the potential drugs for breast cancer treatment through targeting BCSCs.
Results
Identification of CPUL119, CPUL129, and CPUL149 as novel small-molecules attenuating breast cancer cell stemness
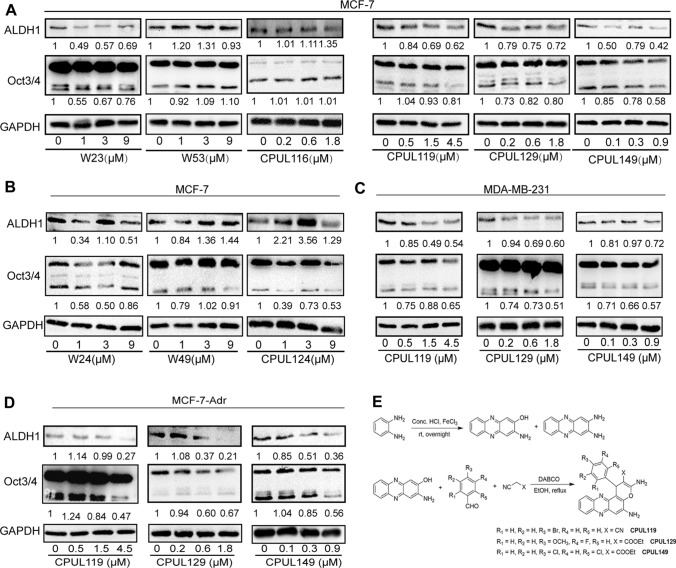
We tried to screen out potential compounds that can specifically attenuate the stemness of breast cancer cells without affecting cell viability from the phenazine compound library we established before [30–33]. It was found that nine compounds (W23, W24, W49, W53, CPUL116, CPUL124, CPUL119, CPUL129, CPUL149) had little effect on breast cancer cell viability (Table 1), which were chosen for the further studies. We initially performed qPCR analysis to evaluate these compounds on the mRNA expression levels of stemness markers. As shown in Supplementary Fig. S1A–F, the mRNA levels of at least two of three stemness markers (Oct4, Nanog, ALDH1A1) were decreased in MCF-7 cells treated with W23, W53, CPUL116, CPUL119, CPUL129, and CPUL149, and the inhibition displayed a concentration-dependent manner. Although the mRNA levels of Sox2, Oct4 and Nanog were reduced in MCF-7 cells treated with W24, the inhibition was not in a concentration-dependent manner (Supplementary Fig. S1G). And only the mRNA level of Sox2 or Nanog was reduced in MCF-7 cells treated with W49 or CPUL124 (Supplementary Fig. S1H and I). However, as shown in Fig. 1A and B, only CPUL119, CPUL129, and CPU149 decreased the protein expression of stemness markers in a concentration-dependent manner. To further confirm the effects of CPUL119, CPUL129, and CPUL149 on the stemness of breast cancer cells, we conducted western blot experiments in another two breast cancer cell lines (MDA-MB-231 and MCF-7-Adr) and obtained the consistent results (Fig. 1C and D). Therefore, these three compounds (CPUL119, CPUL129, CPUL149) were chosen for the further investigation. The synthetic routes and structures of these three compounds are shown in Fig. 1E. Furthermore, MTT analysis confirmed that CPUL119, CPUL129, and CPUL149 had no effect on the cell viability of breast cancer cells and mammary epithelial cells within these concentrations (Supplementary Fig. S2).
Table 1.
The information of compounds
| Number | Molecular weight | IC50 in MCF-7 (μM) |
|---|---|---|
| W23 | 605.23 | > 50 |
| W24 | 646.16 | 50 |
| W49 | 645.12 | 33.24 ± 2.29 |
| W53 | 605.23 | > 50 |
| CPUL116 | 446.89 | 11.02 ± 0.83 |
| CPUL119 | 444.28 | 21.23 ± 5.78 |
| CPUL124 | 455.47 | nd |
| CPUL129 | 460.46 | nd |
| CPUL149 | 481.33 | nd |
Fig. 1.
Identification of CPUL119, CPUL129 and CPUL149 as novel compounds targeting BCSCs. A and B Western blot analysis on the protein expression of stemness markers in MCF-7 cells treated with or without compounds (W23, W53, CPUL116, CPUL119, CPUL129 and CPUL149) for 48 h. C Western blot analysis on the protein expression of stemness markers in MDA-MB-231 cells treated with or without compounds (CPUL119, CPUL129, and CPUL149) for 48 h. D Western blot analysis on the protein expression of stemness markers in MCF-7-Adr cells treated with or without compounds (CPUL119, CPUL129, and CPUL149) for 48 h. E The synthetic route of CPUL119, CPUL129 and CPUL149
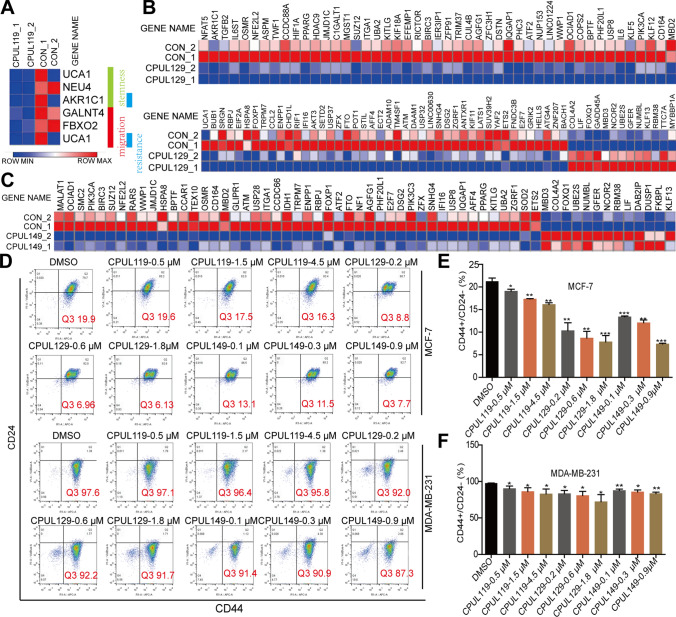
Additionally, we conducted the transcriptome sequencing in breast cancer cells with or without CPUL119, CPUL129, CPUL149 treatment, respectively. As shown in Fig. 2A–C, the transcription of stemness-related genes (such as UCA1 [34], MALAT1 [35]) was suppressed in cells treated with these three compounds, respectively. Furthermore, Gene Set Enrichment Analysis (GSEA) demonstrated that multiple signaling pathways related to stem cell differentiation were enriched in cells treated with these three compounds, respectively (Supplement Figs. S3 and S4). Moreover, the CD44+/CD24− subpopulation with stemness was decreased by these three compounds (Fig. 2D–F). And CPUL119, CPUL129, and CPUL149 significantly suppressed the mammary spheroid-formation ability which is positively correlated with cancer cell stemness, as characterized by the decrease of spheroid size and number (Supplement Fig. S5). What’s more, we preformed clonogenic assay, which is also positively correlated with tumor cell stemness, and found the colony-formation potential was suppressed after being treated with CPUL119, CPUL129, and CPUL149 (Supplementary Fig. S6). These results suggest that the compounds CPUL119, CPUL129, and CPUL149 can inhibit the stemness of breast cancer cells in vitro.
Fig. 2.
CPUL119, CPUL129 and CPUL149 inhibit the stemness of breast cancer cells. A Expression of stemness-, migration- and chemo-resistance-related genes was analyzed in cells treated with or without CPUL119 treatment based on RNA-seq analysis. B and C Expression of stemness-related genes was evaluated in cells treated with or without CPUL129 (B) and CPUL149 (C) based on RNA-seq analysis. D–F The subpopulation of CD44+/CD24− was analyzed by flow cytometry in breast cancer cells treated with or without CPUL119, CPUL129, and CPUL149, respectively (D), and quantified (E and F). Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 vs. control group
CPUL119, CPUL129, and CPUL149 inhibit the migration, invasion, and chemoresistance of breast cancer cells
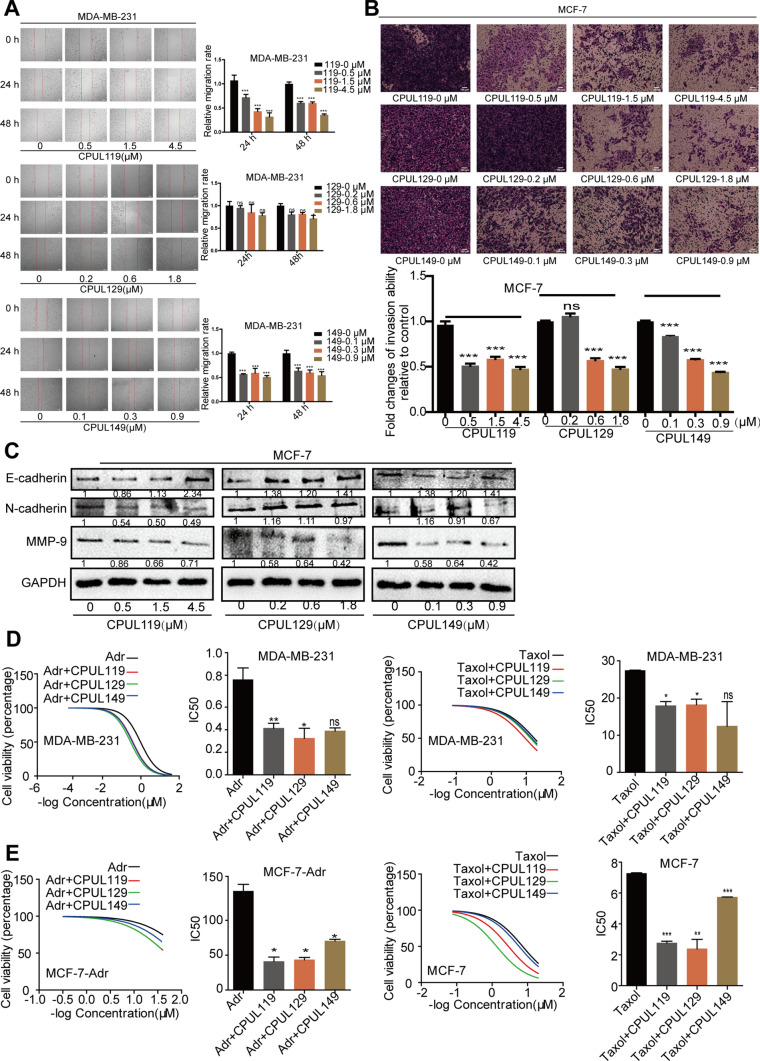
As CSCs have been regarded as one of a root of tumor metastasis and drug resistance, we continued to investigate the effects of CPUL119, CPUL129, and CPUL149 on the migration, invasion, epithelial–mesenchymal transition (EMT) process and drug sensitivity of breast cancer cells. First, it was found that these three compounds could inhibit the expression of metastasis- and drug resistance-related genes based on the transcriptome sequencing analysis (Fig. 2A and Supplementary Fig. S7). We then performed the wound-healing assay to determine whether these three compounds could influence the migration ability of breast cancer cells. As shown in the Fig. 3A and Supplementary Fig. S8A, cell migration ability was restrained after being treated with these three compounds for 24 h and 48 h. The invasion ability of breast cancer cells was also inhibited by these three compounds for 48 h (Fig. 3B and Supplementary Fig. S8B). In consistent, cell adhesion ability was significantly suppressed by these three compounds (Supplementary Fig. S8C). Additionally, it was found that the protein expression of epithelial marker (E-cadherin) was increased and mesenchymal marker (MMP-9) expression was decreased by these three compounds, while N-cadherin expression, another mesenchymal marker, was not decreased significantly (Fig. 3C). Moreover, the effects of these three compounds on drug resistance were further explored. Since paclitaxel (Taxol) and Adriamycin (Adr) are the first-line chemotherapeutic drugs for breast cancer treatment, we chose them as the research subjects. As shown in Fig. 3D and E, compared to the cells that only was treated with Taxol or Adr alone, a distinct decrease of IC50 values of Taxol and Adr was observed in different types of breast cancer cells treated with Taxol or Adr as well as CPUL119, CPUL129 or CPUL149, respectively, including Adr-sensitive (MCF-7 and MDA-MB-231) and Adr-resistant (MCF-7-Adr) cells. Thus, our results demonstrate that CPUL119, CPUL129 and CPUL149 can attenuate the metastasis and chemoresistance of breast cancer cells in vitro.
Fig. 3.
CPUL119, CPUL129 and CPUL149 inhibit the migration, invasion, EMT process, and drug resistance of breast cancer cells. A and B The migration and invasion ability were measured and quantified by wound-healing and transwell invasion analysis in cells treated with or without CPUL119, CPUL129, and CPUL149 for 24 h and 48 h, respectively. C The protein expression of EMT markers (E-cadherin, N-cadherin, MMP9) was measured in MCF-7 cells treated with or without CPUL119, CPUL129, and CPUL149 for 48 h. D and E Cell viability was determined in cells treated with CPUL119 (4.5 μM), CPUL129 (1.8 μM), and CPUL149 (0.9 μM) combined with Adr or Taxol for 48 h through MTT analysis. Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 vs. control group
CPUL119, CPUL129, and CPUL149 inhibit the stemness and metastasis of breast cancer cells in vivo
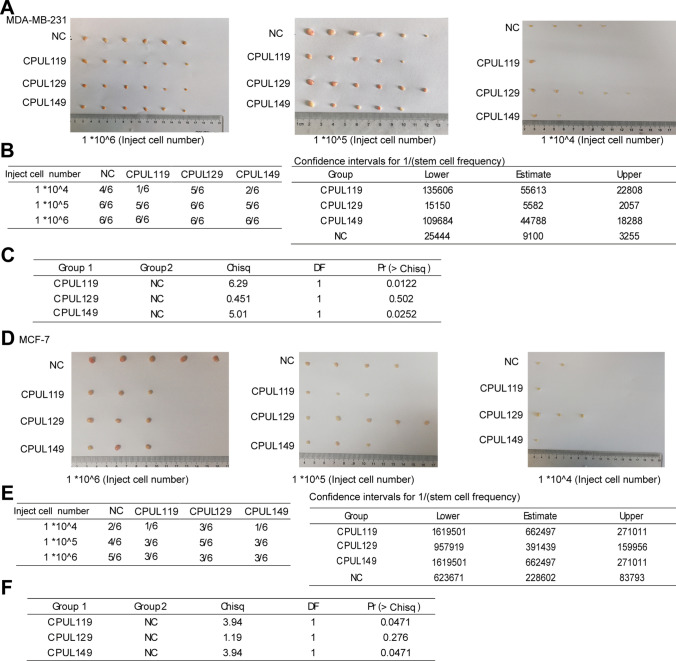
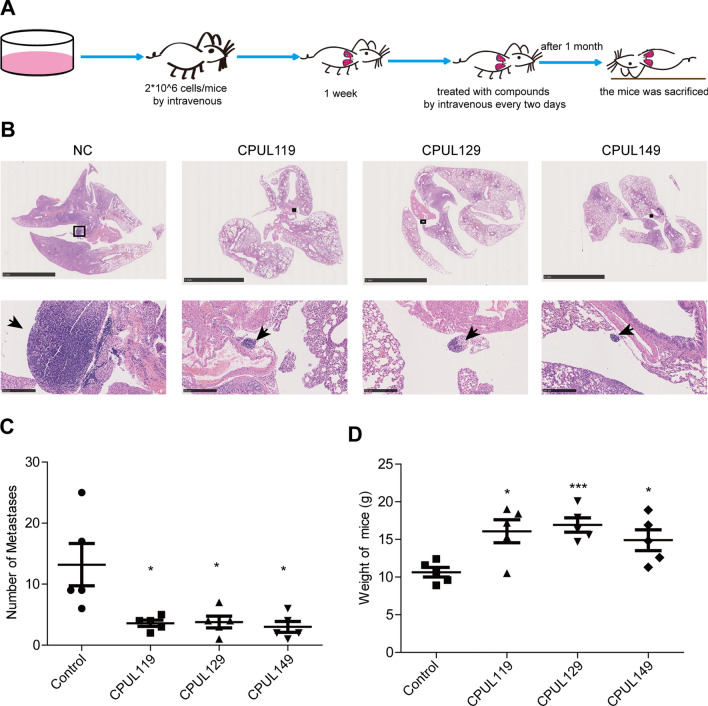
We then evaluated the tumor-initiating ability of MCF-7 and MDA-MB-231 cells pre-treated with or without CPUL119, CPUL129, or CPUL149, respectively. As shown in Fig. 4A–F, the tumor-initiating ability was decreased by the pre-treatment of CPUL119, CPUL129 or CPUL149, respectively, which was characterized by the decreased tumor formation rate and confidence intervals for 1/(stem cell frequency). Notably, CPUL129 just impaired the tumor-initiating ability of MCF-7 cells in the high concentration of injected cell number. This result suggested that CPUL129 may not inhibit the stemness of breast cancer cells in vivo, at least in the lower cell concentrations. Additionally, we constructed a lung metastatic model through injecting MDA-MB-231 cells by tail intravenous in nude mice. After one week, these nude mice were treated with CPUL119, CPUL129, and CPUL149 by tail intravenous every three days, respectively. One month later, these mice were sacrificed (Fig. 5A). As shown in Fig. 5B and C, through H&E staining analysis, mice treated with these three compounds displayed smaller and fewer lung metastatic nodules compared with control mice which were treated with solvent. Importantly, mice treated with these three compounds exhibited a healthier phenotype by detecting weight (Fig. 5D). Consequently, these results demonstrate that CPUL119, CPUL129, and CPUL149 can inhibit the stemness and metastasis of breast cancer cells in vivo.
Fig. 4.
CPUL119, CPUL129 and CPUL149 inhibit the stemness of breast cancer cells in vivo. A and B Images of tumors harvested when serially diluted MDA-MB-231 cells with or without CPUL119, CPUL129 or CPUL149 pre-treatment for 48 h, respectively (A) and tumor formation rate was evaluated (B). C Tumor Initiation Cells (TIC) frequencies (left), χ2 values, and associated probabilities (right) of cells described in (A). D and E Images of tumors harvested when serially diluted MCF-7 cells with or without CPUL119, CPUL129 or CPUL149 pre-treatment, respectively (D) and tumor formation rate was evaluated (E). F TIC (Tumor Initiation Cells) frequencies (left), χ2 values, and associated probabilities (right) of cells described in (D)
Fig. 5.
CPUL119, CPUL129 and CPUL149 inhibit the metastasis of breast cancer cells in vivo. A The diagram to construct the lung metastatic model in nude mice. B H&E staining of lungs derived from mice treated with or without CPUL119, CPUL129 or CPUL149, respectively. C The numbers of metastatic nodules were quantified. D The weight of nude mice was measured when sacrificed. Data are presented as the mean ± SD, n = 5, *p < 0.05, ***p < 0.001 vs. control group
CPUL119, CPUL129, and CPUL149 directly binds to iron (II), regulates iron homeostasis, and ferroptosis in breast cancer cells
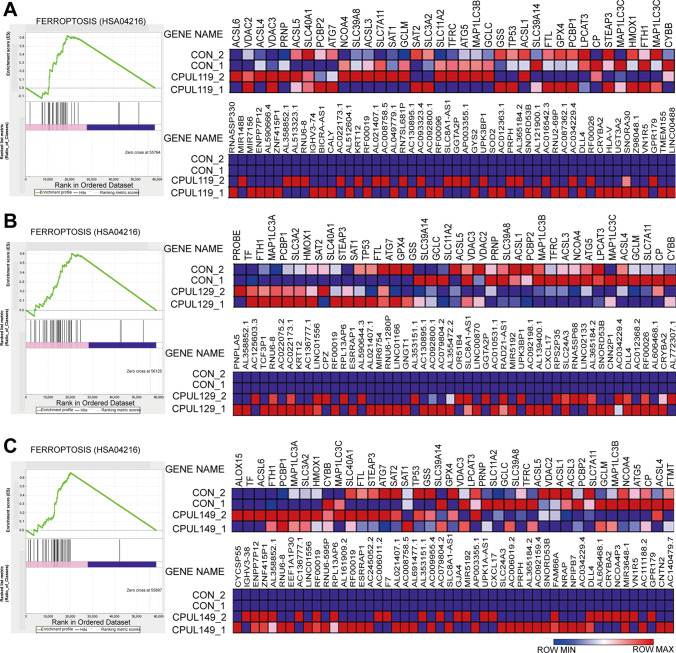
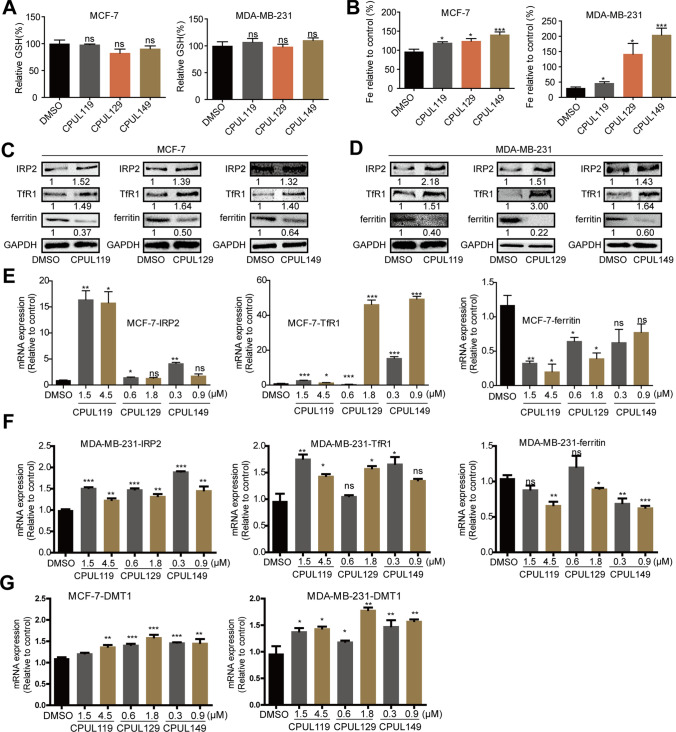
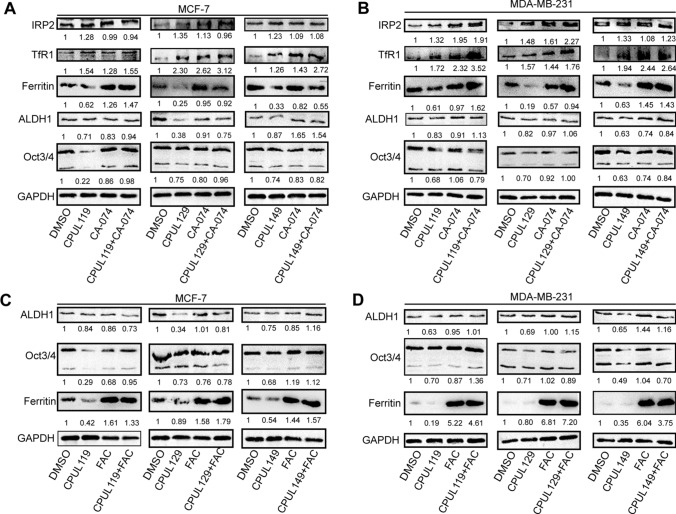
To explore the potential mechanisms underlying the effects of CPUL119, CPUL129, and CPUL149 on the stemness of breast cancer cells, we re-analyzed the transcriptome sequencing datasets and found that the ferroptosis (HSA04216) was significantly enriched in cells treated with these three compounds, respectively (Fig. 6A–C). Consistently, GO analysis revealed that some biological process-related lysosome function and iron transport were enriched, such as lysosomal lumen, iron–sulfur cluster assembly, protein maturation by iron–sulfur (Supplementary Figs. S9–11). KEGG pathway analysis showed that glutathione (GSH) metabolism, which is also involved in ferroptosis, was enriched too (Supplementary Figs. S9–11). As there are two factors regulating lipid peroxides which are the critical contributors for ferroptosis, including GSH and iron concentration [5], we first detected the effects of these three compounds on GSH level; however, it was found that GSH level was not changed by these three compounds (Fig. 7A). Then, we evaluated iron concentration in breast cancer cells (cells were serum-starved before treatment with compounds) with or without treatment of CPUL119, CPUL129, or CPUL149, respectively, and found that iron concentration was significantly increased by these three compounds (Fig. 7B). Furthermore, we examined the effect of these compounds on iron homeostasis. Treatment of CPUL119, CPUL129, or CPUL149 induced a cytoplasmic deletion of iron, which was characterized by the increase of TfR1, IRP2, and DMT1 (divalent-metal transporter-1) expression along with decreased ferritin expression (Fig. 7C–G). Besides, an increase of iron was observed in cells treated with these three compounds, respectively (Fig. 8A). What’s more, the compounds displayed an interaction with iron in MCF-7 cells based on the co-localization-coefficient Rr values (> 0.5) (Fig. 8B).
Fig. 6.
Transcriptome analysis on MD-MB-231 cells treated with or without CPUL119, CPUL129 and CPUL149. A–C GSEA enrichment analysis showed that the ferroptosis pathways were enriched in MDA-MB-231 cells treated with CPUL119, CPUL129 or CPUL149, respectively
Fig. 7.
CPUL119, CPUL129 and CPUL149 regulate the iron homeostasis and ferroptosis in breast cancer cells. A GSH level was detected in MCF-7 and MDA-MB-231 cells treated with or without CPUL119 (4.5 μM), CPUL129 (1.8 μM) and CPUL149 (0.9 μM) for 48 h. B Iron concentration was measured in MCF-7 and MDA-MB-231 cells treated with or without CPUL119 (4.5 μM), CPUL129 (1.8 μM) and CPUL149 (0.9 μM) for 48 h. C, D The protein expression of iron metabolism-related genes including IRP2, TfR1, and ferritin was analyzed by western blot in MCF-7 and MDA-MB-231 cells treated with or without CPUL119 (4.5 μM), CPUL129 (1.8 μM) and CPUL149 (0.9 μM) for 48 h. E–G The expression of iron metabolism-related genes including IRP2, TfR1, DMT1, and ferritin was analyzed by qPCR in MCF-7 and MDA-MB-231 cells treated with or without CPUL119, CPUL129 and CPUL149 for 48 h. Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 vs. DMSO group
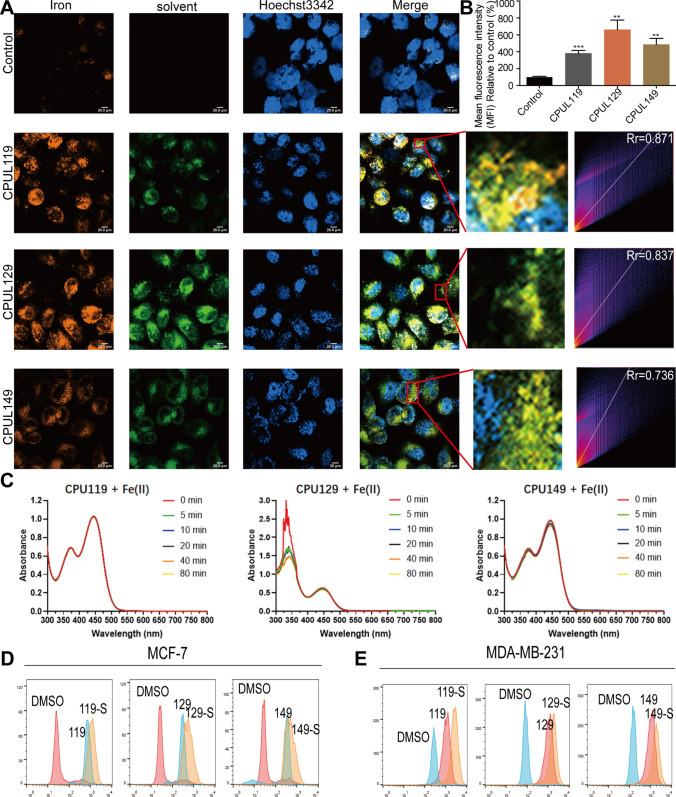
Fig. 8.
CPUL119, CPUL129 and CPUL149 regulate iron homeostasis and ferroptosis in breast cancer cells. A The localization of iron and CPUL119, CPUL129 and CPUL149 was detected using a laser confocal microscope and the co-localization-coefficient was analyzed. B Quantification of iron concentration in A. C UV–vis spectroscopy of CPU119, CPU129- and CPU149-binding iron (II) at different time points of 0, 5, 10, 20, 40 and 80 min. D and E ROS level was measured in MCF-7 and MDA-MB-231 cells treated with or without CPUL119, CPUL129 and CPUL149 for 48 h. Compound-S represents the cell stained with ROS probe. Data are presented as the mean ± SD, n = 3, **p < 0.01, ***p < 0.001 vs. control group
Then UV–vis spectrometry was performed to evaluate the binding of CPUL119, CPUL129, and CPUL149 to iron (II) and it was found that CPUL119 and CPUL149 displayed the absorbance peak around 374 nm and CPUL129 displayed the absorbance peak around 340 nm, as well as all compounds showed the absorbance peak around 445 nm (Fig. 8C). Therein, we observed little to no loss of absorbance at the λmax wavelength (445 nm) for CPUL119 and CPUL149 following addition of ammonium iron (II) sulfate hexahydrate. In contrast, addition of iron (II) to CPUL129 presents a distinct loss of absorbance at the λmax of 340 nm. It seemed apparent that CPUL119 and CPUL149 exhibited weak chelation to copper (II). Significantly, CPUL129 was more inclined to chelate copper (II) in a manner representative of expected 2:1 phenazine: iron(II) stoichiometry, which indicated a phenazine − iron (II) complex [in a 2:1 phenazine/iron(II) ratio] was apparently formed over the indicated time points along with yielding a loss in absorbance.
We then wondered whether the increased iron concentration could increase the production of lipid peroxides. As expected, ROS level was increased in cells treated with these three compounds, respectively (Fig. 8D and E). Consistently, GO analysis showed that gene associated with oxidation–reduction processes were enriched, such as oxidoreductase activity (Supplementary Fig. S10), and lipid peroxidation was increased by the treatment of these three compounds (Supplementary Figs. S12A–D).
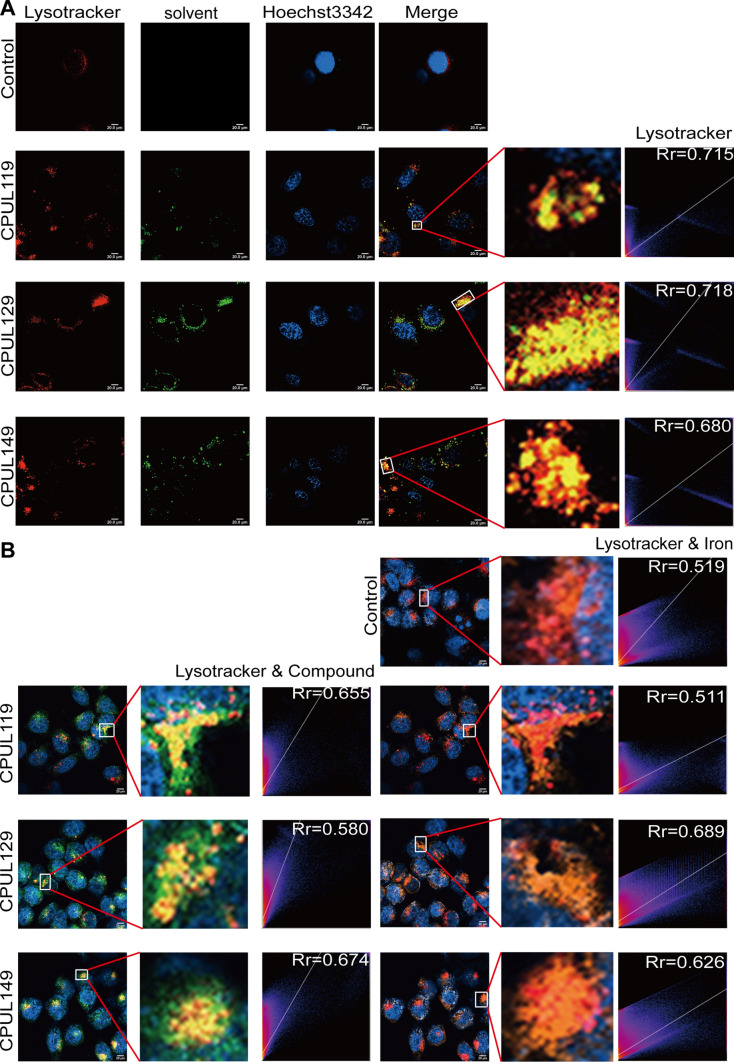
We then detected the localization of CPUL119, CPUL129, and CPUL149 in breast cancer cells. As shown in Fig. 9A, CPUL119, CPUL129, and CPUL149 were located in lysosomes, while a few of them were also located in mitochondria which plays a critical role in oxidative metabolism (Supplementary Fig. S13A) [36], demonstrating that these three compounds mostly accumulate in lysosomal compartment. Additionally, it was found that CPUL119, CPUL129, and CPUL149 interacted with iron and led to the iron aggregation in lysosomes, while iron was diffused in the cytosol of untreated-cells (Supplementary Fig. S13B and Fig. 9B). These data indicated that CPUL119, CPUL129, and CPUL149 can interact and induce iron aggregation in lysosomes.
Fig. 9.
CPUL119, CPUL129 and CPUL149 interact and sequester iron in lysosomes in breast cancer cells. A The localization of CPUL119, CPUL129, and CPUL149 was determined using lysosome probe (Lysotracker), and the co-localization-coefficient was analyzed. B The co-localization of CPUL119, CPUL129, or CPUL149 and iron was quantified with Image J. Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 vs. control group
CPUL119, CPUL129, and CPUL149 attenuate the stemness of breast cancer cells partially through triggering ferroptosis
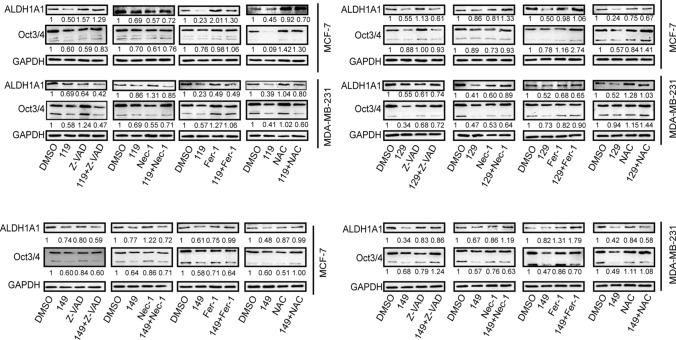
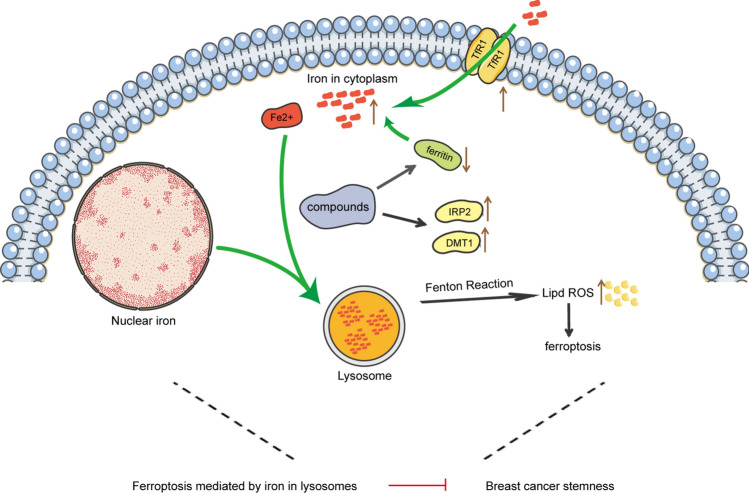
Finally, we investigated whether CPUL119, CPUL129, and CPUL149 attenuate the stemness of breast cancer cells through triggering ferroptosis. To determine whether other cell death pathways, including necrosis and apoptosis, are involved in these three compound-mediated inhibitions on breast cancer cell stemness, apoptosis inhibitor Z-VAD-FMK, necrosis inhibitor Nec-1, ferroptosis inhibitor Fer-1, or ROS scavenger NAC were added in cells with CPUL119, CPUL129, or CPUL149 treatment, respectively. As shown in Fig. 10, ferroptosis inhibitor or ROS scavenger NAC, but not Z-VAD-FMK or Nec-1, rescued the suppression of CPUL119, CPUL129, or CPUL149 on stemness marker expression. Besides, we explored whether the effects of CPUL119, CPUL129 or CPUL149 on the stemness of breast cancer cells were indeed dependent on lysosome function or iron. CA-074 (an inhibitor of the lysosomal protease cathepsin B) or iron-chelating agent deferoxamine (DFO) was added in cells treated with these three compounds, respectively. As shown in Supplementary Figure S12A-D, the increased lipid peroxides led by these three compounds were attenuated by CA-074, and enhanced by DFO. Additionally, the decreased subpopulation of CD44+/CD24− ratio led by CPUL119, CPUL129, or CPUL149 was partially abrogated by Fer-1 or NAC, respectively (Supplementary Fig. S14A–D). Furthermore, a consistent result was obtained when spheroid-formation ability was evaluated, including the increased spheroid number and size (Supplementary Fig. S14E and F). Consistently, CA-074 rescued the effects of CPUL119, CPUL129, and CPUL149 on the expression of ferritin and stemness markers (Fig. 11A and B). These above results link the lysosomal degradation of ferritin to the production of ROS in this organelle through the release of additional soluble redox-active iron, thus attenuating the stemness of breast cancer cells. Ferric ammonium citrate (FAC) was added into breast cancer cells to increase the iron concentration and the results indicated that FAC indeed promoted the expression of stemness markers and ferritin and even reversed the effects of CPUL119, CPUL129 and CPUL149 on the expression of ferritin and stemness markers (Fig. 11C and D). Overall, our results demonstrate that CPUL119, CPUL129 and CPUL149 suppress the stemness of breast cancer cells through sequestering iron in lysosomes and triggering ferroptosis (Fig. 12).
Fig. 10.
CPUL119, CPUL129 and CPUL149 attenuate the expression of stemness markers of breast cancer cells partially through triggering ferroptosis. The expression of stemness markers was detected in MCF-7 and MDA-MB-231 cells treated as indicated
Fig. 11.
CPUL119, CPUL129 and CPUL149 attenuate the stemness of breast cancer cells partially through triggering ferroptosis. A and B Western blot analysis on the iron regulatory genes and stemness markers in MCF-7 and MDA-MB-231 cells treated with CPUL119, CPUL129, CPUL149 as well as CA-074 or not. C and D Western blot analysis on the ferritin and stemness marker expression in MCF-7 and MDA-MB-231 cells treated with CPUL119, CPUL129, CPUL149 as well as FAC (60 μM) or not. Data are presented as the mean ± SD, n = 3, *p < 0.05, **p < 0.01, ***p < 0.001 vs. DMSO/CPUL119/CPUL129/CPUL149 group
Fig. 12.
The summary of compounds’ effects on the stemness of breast cancer cells. These three compounds could trigger ferroptosis by accumulating and sequestering iron in lysosomes through interacting with iron, and by regulating the expression of proteins (IRP2, TfR1, ferritin) engaged in iron transport and storage, thereby attenuating the stemness of breast cancer cells
Discussion
BCSCs have been considered to be the root of breast cancer progression; thus, targeting BCSCs may be a promising way to treat breast cancer thoroughly [4, 37–39]. In the present work, we aim to screen out small-molecule compounds that could inhibit the stemness of breast cancer cells from the phenazine library established by us before. We finally screened out CPUL119, CPUL129 and CPUL149 as promising compounds which can specifically attenuate the stemness of breast cancer cells without affecting cell viability, as evident by the ability to reduce the stemness, invasion and chemoresistance in vitro and in vivo, and CPUL149 showed a better activity among the three compounds. Although CPUL 149 at concentrations of 0.01 and 0.03 μM exhibited no difference in MCF-7 cell spheroid formation, and no difference in the size of MDA-MB-231 cell spheres, this might be due to the low drug concentrations and lower stemness of MCF-7 cells, leading to that the effects of compounds on MDA-MB-231 cells were more obvious than that in MCF-7 cells.
Targeting the abnormal signaling pathways, such as Wnt, Notch, Hedgehog and autophagy, has been shown to potentially killing CSC as we reviewed in a recent study [5]; however, most of small-molecule compounds are just in the preclinic stage. Targeting iron metabolism using the iron nanoparticles can be used to treat cancer through ferroptosis, which has been approved by FDA [40]. Besides, inducing ferroptosis through sequestering iron in lysosome has been confirmed to specifically killing CSCs including BCSCs [17, 19, 20, 41]. Similarly, Luca Laraia et al. found an autophagy inhibitor autoquin, which could also sequester Fe2+ in lysosomes, resulting in an increase of lysosomal ROS and ultimately cell death [42]. In this work, we found CPUL119, CPUL129 and CPUL149 could increase the content of iron in breast cancer cells accompanying with the change of iron regulatory protein, such as IRP2, TfR1 and ferritin. Specifically, these three compounds can interact, induce iron aggregation in lysosome and thus trigger ferroptosis. We noted that although iron in lysosomes could be further increased after the treatment of these compounds, iron also locates in lysosomes in the normal situation (Fig. 11C and D), this indicates an iron-rich environment in lysosomes [43]. Furthermore, based on the UV–vis spectrometry analysis, we think that these three molecules reversibly bind to iron and thus induce iron aggregation in lysosomes. Additionally, the decreased stemness resulted by these three compounds was attenuated or even reversed by ferroptosis inhibitor (fer-1), NAC and ferritin degradation inhibitor (CA-074), and the stemness was increased if we used the FAC to increase the iron. Notably, a small number of CPUL119, CPUL129 and CPUl149 were located in mitochondria which play a critical role in oxidative metabolism; this result hints that these three compounds might also affect iron homeostasis via modulating oxidative stress in mitochondria as oxidative stress has recently been shown to be linked with iron homeostasis [44]. Besides, apoptosis or necrosis inhibitor could also rescue the effects of these three compounds on the stemness of breast cancer cells although without significance. The GO analysis enriched necrosis correlated biological processes, such as tumor necrosis factor receptor and tumor necrosis factor receptor binding (Supplementary Fig. S4A). These results suggest that CPUL119, CPUL129 or CPUL149 might suppress the stemness of breast cancer cells through other signaling pathways, or these three compounds suppress the stemness of breast cancer cells through different signaling pathways as shown in the GO- or KEGG-enrichment analysis. Furthermore, since cancer cells can be transformed into CSCs under certain conditions [45, 46], the effects of apoptosis or necrosis inhibitor on the stemness of breast cancer cells might be led by the effects on cancer cells. Moreover, we determined the location of these compounds only using lysosome probe and mitochondrion probe, while other organelle probes, such as endoplasmic reticulum or Golgi apparatus, were not used. What’s more, it must be noted that although N-cadherin expression did not change by CPUL129 treatment (Fig. 3C), another two EMT markers (E-cadherin and MMP9) expression was altered, indicating the inhibition of CPUL129 on EMT process and that CPUL129 might not suppress all of the EMT markers. Therefore, it is still unclear whether CPUL119, CPUL129 and CPUL149 locate in other organelles. Finally, supplementing with FAC enhanced the stemness of breast cancer cells, this is consistent with the previous study indicating that glioblastoma stem-like cells prefer to iron trafficking [47]. The decreased stemness of breast cancer cells could inhibit the recurrence and resistance of breast cancer. In this article, we proved CPUL119, CPUL129 and CPUL149 could inhibit the stemness of breast cancer cells, and these three compounds through further structure modification may become a new adjuvant therapy in treating breast cancer. Besides, we also found that CPUL119, CPUL129 and CPUL149 could interact with Fe2+ to trigger ferroptosis, the structure of compounds binding with Fe2+ reminds us to explore more similar compounds that may have more effects to reduce the stemness of breast cancer cells.
Although the compounds inhibiting the stemness of breast cancer cells through ferroptosis have been shown to have better research trend, the risks should be considered. At first, the iron metabolism is very vital in hematologic, cardiomyopathy and liver [48–50]; thus, the iron overload may cause the unpredictable effects to this system; however, these three compounds may have no effect on hematopoietic system as a healthier phenotype was observed in mice treated with these three compounds by detecting mice weight (Fig. 7D). Besides, once the ferroptosis happens, abundant oxidative substance can be produced, such as OH·, these oxidative substances may attack the normal tissue. Therefore, more experiments are still needed to make these compounds in the clinic. Notably, in terms of the molecular structure of CPUL119, CPUL129 and CPUL149, we speculated these three compounds are likely to act as a kind of metal chelator, which means that the amino substituent in the phenazine skeleton and oxygen atom in the pyran ring could chelate iron ions in lysosomes to form complexes resulting in the decrease of intracellular concentrations of free iron ions and finally cause ferroptosis, this speculation should be confirmed by other experiments, such as Nuclear Magnetic Resonance (NMR), and the detailed binding sites should be further explored.
Conclusion
Nowadays, the chemotherapeutic drugs used for treating breast cancer usually appear resistance and accompany by tumor metastasis [51]. CSCs are considered as the root of tumor occurrence and recurrence, and breast cancer stemness is positively correlated with resistance and migration. Hence, the compounds targeting BCSCs maybe show better results in clinical. Currently, there are still no compounds targeting BCSCs approved by FDA and breast cancer treatment is mainly aimed at the most differentiated cells in tumor tissue, which can kill most tumor cells rather than BCSCs in a very short time, but enrich BCSCs to a certain extent [52]. Subsequently, the surviving BCSCs lead to the recurrence and metastasis. Therefore, our findings suggest that CPUL119, CPUL129, and CPUL149 may become a new adjuvant therapy or lead compounds to further enrich the library of molecules targeting BCSCs, and may be the potential compounds for breast cancer treatment.
Methods
Cell culture
Human breast cancer cell lines including MCF-7 cells, MDA-MB-231 cells and Adriamycin resistant MCF-7-Adr cells were stored in our laboratory. The MCF-7 and MDA-MB-231 cells were cultured in DMEM medium (KeyGEN, Cat# KGM12800-500) with 10% fetal bovine serum (FBS, BI, Cat# 04-001-1ACS). MCF-7-Adr cells were cultured in 1640 medium (KeyGEN, Cat# KGM31800-500) with 10% fetal bovine serum. All cells were cultured at 37 °C under a humidified atmosphere with 5% CO2.
Reagents
Z-VAD-FMK (APExBIO, Cat# A1902), Necrostatin-1 (APExBIO, Cat# A4213), Ferrostatin-1 (APExBIO, Cat# A4371), NAC (Beyotime, Cat# S0077), CA-074 methyl ester (MCE, Cat# HY-100350), Ammonium ferric citrate (Sigma, Cat# F5879), Deferoxamine mesylate salt (YuanyeBio, Cat# S61301).
Quantitative real-time PCR (qPCR)
The process of this experiment was referred to our previous study [53]. And others sequences of primers used for qPCR were listed in Table 2.
Table 2.
Sequences of primers used for qPCR
| Name | Sequence | |
|---|---|---|
| IRP2 | Forward (5′–3′) | ACCAGAGGTGGTTGGATGTGAGTT |
| Reverse (5′–3′) | ACTCCTACTTGCCTGAGGTGCTTT | |
| TfR1 | Forward (5′–3′) | AGTTGAACAAAGTGGCACGAGCAG |
| Reverse (5′–3′) | AGCAGTTGGCTGTTGTACCTCTCA | |
| Ferritin | Forward (5′–3′) | CATCAACCGCCAGATCAAC |
| Reverse (5′–3′) | GATGGC TTTCACCTGCTCAT | |
| DMT1 | Forward (5′–3′) | CCTGTGGCTAATGGTGGAGTTGG |
| Reverse (5′–3′) | GGAGATTGATGGCGATGGCTGAC | |
Western blot
The cells were collected by 1 × PBS (phosphate buffer saline) (diluted with double distilled water, Servicebio, Cat# G4207-500ML) and were lysed with RIPA Lysis Buffer (Beyotime, Cat# P0013B) with PMSF (1:100, Beyotime, Cat# ST506) on the ice. After SDS-PAGE (sodium dodecyl sulfate—polyacrylamide gel electrophoresis) analysis, the proteins were transferred on the PVDF (polyvinylidene fluoride) membranes (Millipore, Cat# IPVH00010), which were then incubated with the primary antibodies at 4℃ overnight. The membranes were incubated with the secondary antibodies (Goat Anti-Rabbit IgG (H + L), HRP conjugate; Goat Anti-Mouse, IgG (H + L), HRP conjugate, proteintech, Cat# SA00001-2, Cat# SA00001-1) and protein signals were detected with High-sig ECL Western Blotting Substrate (Tanon, Cat# 180–5001) on Tanon5200. The antibody used in this study included ALDH1A1 (Proteintech, 15910-1-AP), Oct3/4 (Wanleibio, Cat# WL01728), GAPDH (Proteintech, Cat# 60004-1-l g), E-cadherin (Proteintech, Cat# 20874-1-AP), N-cadherin (Proteintech, Cat# 22018-1-AP), MMP9 (Affinity, Cat# AF5228), IRP2 (Proteintech, Cat# 23829-1-AP), TfR1 (Santa, Cat# SC-32272), ferritin (Abcam, Cat# ab75973).
Flow cytometry analysis
MCF-7 and MDA-MB-231 cells were seeded into 6-well plates and treated with different compounds or inhibitors, after 48 h, cells were stained with Hu CD44 APC (BD Pharmingen, Cat# 559942) and Hu CD24 BV421 (BD Pharmingen, Cat# 562789) on ice in 30 min and then the subpopulation of CD44+/CD24− ratio was analyzed on a MACSQuant flow cytometer (Miltenyi Biotec).
Cell spheroid-formation assay
MCF-7 and MDA-MB-231 cells were seeded into low-adherent 24-well plates at a density of 3000–5000 cells/well and cultured with the MammoCult™ Human Medium Kit (stemcell, Cat# 05620) supplemented with compounds for 8 days. Then spheroid number was calculated and size was measured under a microscope (Leica).
MTT
Cell viability was measured by MTT (Solarbio, Cat# M8180-250 mg). MCF-7, MDA-MB-231 and MCF-7-Adr cells were seeded into 96-well plates at a density of 3000–5000 cells/well. After 24 h, different concentrations of W23, W24, W49, W53, CPUL116, CPUL119, CPUL124, CPUL129, and CPUL149 were added. 48 h later, 20 μl 5 mg/ml MTT was added into the medium for a 4 h culturing. Then the medium was removed and 150 μl DMSO was added into each well. After shocking for 10 min, the absorbance at 490 nm was analyzed on a Biorad iMark.
Detection of lipid peroxidation
MCF-7 and MDA-MB-231 cells were seeded into 6-well plates. After 24 h, cells were treated with compounds (4.5 μM CPUL119, 1.8 μM CPUL129 and 0.9 μM CPUL149), NAC (5 mM), and DFO (2 μM). 48 h later, cells were stained with 10 μM Lipid peroxidation sensor (ThermoFisher, Cat# B3932) for 30 min at 37℃. Then lipid peroxidation level was analyzed with a flow cytometer.
Detection of ROS level
MCF-7 and MDA-MB-231 cells were seeded into 6-well plates. After 24 h, cells were treated with compounds (4.5 μM CPUL119, 1.8 μM CPUL129 and 0.9 μM CPUL149). 48 h later, the production of ROS was analyzed by reactive oxygen species assay kit (Beyotime, Cat# S0033S) through a flow cytometer.
GSH determination
The analysis of GSH was performed by Micro Reduced Glutathione (GSH) Assay Kit (Solarbio, Cat# BC1175) following the manufacturer’s recommendation.
Analysis of iron content
The content of iron in the cells was measured by Iron Colorimetric Assay Kit (Applygen, Cat# E1042-100) according to the standard protocol. Briefly, MCF-7 and MDA-MB-231 cells were seeded into 24-well plates. After 24 h, cells were treated with compounds (4.5 μM CPUL119, 1.8 μM CPUL129 and 0.9 μM CPUL149). 48 h later, then iron concentration was analyzed by Iron Colorimetric Assay Kit.
UV − vis for HPs binding iron (II)
We tried to obtain the evidence that CPU119, CPU129 and CPU149 were able to directly bind to iron (II). The process of phenazine–iron (II) complex formation was independently evaluated by UV–vis spectrometry. Ammonium iron (II) sulfate hexahydrate (0.5 equiv) was added to a stirring solution of a phenazine derivative (5 mM, 5 ml) in dimethyl sulfoxide. Aliquots (20 μl) were then removed from the resulting mixture and added to dimethyl sulfoxide (980 μl) in a cuvette. Spectral scanning was performed from 300 to 800 nm in 2 nm increments [54, 55].
Wound-healing assay
MCF-7 and MDA-MB-231 cells were seeded into 6-well plates and cultured in complete medium until growing up to 90%. Then, a wound was produced on cell surface, and washed it with PBS twice to remove cell fragments. Cells were cultured in the medium with low serum concentration (≤ 2%) contained compounds or solvent. After 24 h and 48 h, wounds were observed and taken with a microscope. Migration rate = (Dt − D0) / D0, Dt represents the distance between the wounds at different times.
Invasion assay
Cell invasion ability was assayed by transwell invasion analysis. Finally, the Millicell Hanging cell culture inserts (Merck, Cat# MCEP24H48) coated with Matrigel matrix (Corning, Cat# 356234) were prepared in the 24-well plates. The 2 × 105 MCF-7 or MDA-MB-231 cells within 200 μl serum-free medium treated with different compounds or solvent were added into the upper chamber, and the lower chamber was filled with medium containing 20% serum. After 48 h, cells were fixed with 70% ethanol for 20 min at room temperature. Then, the invaded cells were stained with crystal violet staining solution (Beyotime, Cat# C0121-100 ml), and observed under a microscope. After then, the chambers were washed with 33% glacial acetic and the absorbance at 570 nm was analyzed on a Biorad iMark, which represents the invaded cell number.
Cell adhesion assay
Detailed procedure was referred to our previous study [56].
Fluorescence microscopy analysis
MCF-7 cells were seeded into glass-bottom culture dishes (NEST, Cat# 801001) to carry on this experiment. After 24 h, cells were treated with different compounds (4.5 μM CPUL119, 1.8 μM CPUL129 and 0.9 μM CPUL149). 48 h later, cells were stained with different probes as the following methods. 50 nM LysoTracker Deep Red (ThermoFisher, Cat# L12492) was incubated with cells for 90 min at 37 °C. 50 nM MitoTracker Deep Red (ThermoFisher, Cat# M22426) was incubated with cells for 30 min at 37 °C. 1 μM FerroOrange (DOJINDO, Cat# F374) was incubated with cells for 30 min at 37 °C. 2 μg/ml Hoechst 3342 was incubated with cells for 30 min at 37 °C. After stained with different probes, we used a LSM800 to analyze.
RNA sequencing and data analysis
RNA sequencing and data analysis were performed by Novogen (Beijing, China). The data are available in the Gene Expression Omnibus (GEO) database as GSE168188.
Tumorigenesis in vivo
The BALB/c-nude mice (Gempharmatech, Cat# D000521) were female, 3–4 weeks, and were cultured in standard pathogen-free conditions. Each group had six mice. All the experiments were obtained the approval of the Ethics Committee for Animal Experimentation of China Pharmaceutical University. In the tumor-limiting dilution assay, 1 × 106, 1 × 105 and 1 × 104 MCF-7 and MDA-MB-231 cells pre-treated with compounds (4.5 μM CPUL119, 1.8 μM CPUL129 and 0.9 μM CPUL149) for 48 h were implanted in the inguinal mammary gland of mice orthotopically. After 10 days, mice were sacrificed, and the amounts of tumors in each group were counted. And the stem cell frequencies were analyzed by ELDA (http://bioinf.wehi.edu.au/software/elda/).
Metastasis in vivo
The BALB/c-nude mice (Gempharmatech, Cat# D000521) were female, 3–4 weeks, and were cultured in standard pathogen-free conditions. Each group had five mice. All the experiments were obtained the approval of the Ethics Committee for Animal Experimentation of China Pharmaceutical University. For metastasis assay, each mouse was injected with 2 × 106 MDA-MB-231 cells by intravenous. One week later, mice were injected with different compounds (10 mg/kg 119, 10 mg/kg 129, 5 mg/kg 129) or solvent (23.7% ethanol, 17% PEG400, 59.3% saline) intravenous every 2 days. One month later, mice were sacrificed, and the lungs were subjected to H&E analysis by Servicebio (Wuhan, China).
Statistical analysis
The results were presented as mean ± SD. Student’s t test was used to perform the analysis using GraphPad Prism 5 software. P values less than 0.05 were considered to be statistically significant. (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- BCSCs
Breast cancer stem cells
- CSCs
Cancer stem cells
- LSCs
Leukemia stem cells
- FDA
Food and Drug Administration
- ROS
Reactive oxygen species
- IRP2
Iron-responsive element-binding protein 2
- TfR1
Transferrin receptor 1
- IRE
Iron regulatory element
- FPN
Ferroportin
- GSEA
Gene Set Enrichment Analysis
- EMT
Epithelial–mesenchymal transition
- Taxol
Paclitaxel
- Adr
Adriamycin
- GSH
Glutathione
- FAC
Ferric ammonium citrate
- NMR
Nuclear Magnetic Resonance
Author contributions
LZ, FJ and TX designed the research. YL and FJ approved the compounds. YY, CZ, ZX, JL, XC, TW, QG analyzed the data. YY, CZ performed the research. YY and LZwrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the Project Program of National Nature Science Foundation of China (Grant No. 82173842, 81872757), Nature Science Foundation of Jiangsu Province of China (Grant No. BK20201329), the Fundamental Research Funds for the Central Universities (grant No. 2632018ZD01, No. 2632020ZD10), Innovation and Entrepreneurship Training Program for Undergraduate (No. 201910316220, No. 202010316245), the Medical Science and Technology Research Project of Henan Province (no. SBGJ202003010), the Science and Technology Research Project of Henan Province (no. 202102310158) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Availability of data and materials
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
All the experiments were obtained the approval of the Ethics Committee for Animal Experimentation of China Pharmaceutical University.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yue Yang, Yuanyuan Lu, and Chunhua Zhang contributed equally to this work.
Contributor Information
Tao Xi, Email: xitao18@hotmail.com.
Feng Jiang, Email: jiangfeng@CPU.edu.cn.
Lufeng Zheng, Email: zhlf@CPU.edu.cn.
References
- 1.Chen W, Qin Y, Liu S. Cytokines, breast cancer stem cells (BCSCs) and chemoresistance. Clin Transl Med. 2018;7(1):27. doi: 10.1186/s40169-018-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiorentino S, Urueña C, Lasso P, Prieto K, Barreto A. Phyto-immunotherapy, a complementary therapeutic option to decrease metastasis and attack breast cancer stem cells. Front Oncol. 2020;10:1334. doi: 10.3389/fonc.2020.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Yang F, Xu J, Tang L, Guan X. Breast cancer stem cell: the roles and therapeutic implications. Cell Mol Life Sci. 2017;74(6):951–966. doi: 10.1007/s00018-016-2334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Li X, Wang T, Guo Q, Xi T, Zheng L. Emerging agents that target signaling pathways in cancer stem cells. J Hematol Oncol. 2020;13(1):60. doi: 10.1186/s13045-020-00901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norsworthy KJ, By K, Subramaniam S, Zhuang L, Del Valle PL, Przepiorka D, Shen YL, Sheth CM, Liu C, Leong R, et al. FDA approval summary: Glasdegib for newly diagnosed acute myeloid Leukemia. Clin Cancer Res. 2019;25(20):6021–6025. doi: 10.1158/1078-0432.CCR-19-0365. [DOI] [PubMed] [Google Scholar]
- 7.Beziat G, Ysebaert L. Tagraxofusp for the treatment of Blastic Plasmacytoid Dendritic Cell Neoplasm (BPDCN): a brief report on emerging data. Onco Targets Ther. 2020;13:5199–5205. doi: 10.2147/OTT.S228342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raggi C, Gammella E, Correnti M, Buratti P, Forti E, Andersen JB, Alpini G, Glaser S, Alvaro D, Invernizzi P, et al. Dysregulation of iron metabolism in Cholangiocarcinoma stem-like cells. Sci Rep. 2017;7(1):17667. doi: 10.1038/s41598-017-17804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozer U. The role of Iron on breast cancer stem-like cells. Cell Mol Biol (Noisy-le-grand) 2016;62(4):25–30. [PubMed] [Google Scholar]
- 10.Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, Xian W, McKeon F, Lynch M, Crum CP, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene. 2017;36(29):4089–4099. doi: 10.1038/onc.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajbouj K, Shafarin J, Hamad M. Estrogen-dependent disruption of intracellular iron metabolism augments the cytotoxic effects of doxorubicin in select breast and ovarian cancer cells. Cancer Manage Res. 2019;11:4655–4668. doi: 10.2147/CMAR.S204852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma S, Henson ES, Chen Y, Gibson SB. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7(7):e2307. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang VC, Cotterchio M, Khoo E. Iron intake, body iron status, and risk of breast cancer: a systematic review and meta-analysis. BMC Cancer. 2019;19(1):543. doi: 10.1186/s12885-019-5642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Dielschneider RF, Henson ES, Xiao W, Gibson SB. Ferroptosis and autophagy induced cell death occur independently after siramesine and lapatinib treatment in breast cancer cells. PLoS One. 2017;12(8):e0182921. doi: 10.1371/journal.pone.0182921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plays M, Müller S, Rodriguez R. Chemistry and biology of ferritin. Metallomics. 2021;13(5):mfab021. doi: 10.1093/mtomcs/mfab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torii S, Shintoku R, Kubota C, Yaegashi M, Torii R, Sasaki M, Suzuki T, Mori M, Yoshimoto Y, Takeuchi T, et al. An essential role for functional lysosomes in ferroptosis of cancer cells. Biochem J. 2016;473(6):769–777. doi: 10.1042/BJ20150658. [DOI] [PubMed] [Google Scholar]
- 17.Mai TT, Hamai A, Hienzsch A, Caneque T, Muller S, Wicinski J, Cabaud O, Leroy C, David A, Acevedo V, et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat Chem. 2017;9(10):1025–1033. doi: 10.1038/nchem.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corte-Rodríguez M, Blanco-González E, Bettmer J, Montes-Bayón M. Quantitative analysis of transferrin receptor 1 (TfR1) in individual breast cancer cells by means of labeled antibodies and elemental (ICP-MS) detection. Anal Chem. 2019;91(24):15532–15538. doi: 10.1021/acs.analchem.9b03438. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Wang Q, Li X, Chen Y, Xu G. Itraconazole attenuates the stemness of nasopharyngeal carcinoma cells via triggering ferroptosis. Environ Toxicol. 2021;36(2):257–266. doi: 10.1002/tox.23031. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Cheng X, Pan S, Wang L, Dou W, Liu J, Shi X. Dichloroacetate attenuates the stemness of colorectal cancer cells via trigerring ferroptosis through sequestering iron in lysosomes. Environ Toxicol. 2020;36(4):520–529. doi: 10.1002/tox.23057. [DOI] [PubMed] [Google Scholar]
- 21.Miller LD, Coffman LG, Chou JW, Black MA, Bergh J, D'Agostino R, Jr, Torti SV, Torti FM. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71(21):6728–6737. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller S, Sindikubwabo F, Cañeque T, Lafon A, Versini A, Lombard B, Loew D, Wu TD, Ginestier C, Charafe-Jauffret E, et al. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat Chem. 2020;12(10):929–938. doi: 10.1038/s41557-020-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Chen X, Zuyi W, Chen L, Fu W. Lysosome Fe(2+) release is responsible for etoposide- and cisplatin-induced stemness of small cell lung cancer cells. Environ Toxicol. 2021;36(8):1654–1663. doi: 10.1002/tox.23161. [DOI] [PubMed] [Google Scholar]
- 24.Zhao B, Li X, Wang Y, Shang P. Iron-dependent cell death as executioner of cancer stem cells. J Exp Clin Cancer Res. 2018;37(1):79. doi: 10.1186/s13046-018-0733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma S, Dielschneider RF, Henson ES, Xiao W, Choquette TR, Blankstein AR, Chen Y, Gibson SB. Ferroptosis and autophagy induced cell death occur independently after siramesine and lapatinib treatment in breast cancer cells. PLoS One. 2017;12(8):e0182921. doi: 10.1371/journal.pone.0182921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulkearns-Hubert EE, Torre-Healy LA, Silver DJ, Eurich JT, Bayik D, Serbinowski E, Hitomi M, Zhou J, Przychodzen B, Zhang R, et al. Development of a Cx46 targeting strategy for cancer stem cells. Cell Rep. 2019;27(4):1062–1072. doi: 10.1016/j.celrep.2019.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar H, Chattopadhyay S, Das N, Shree S, Patel D, Mohapatra J, Gurjar A, Kushwaha S, Singh AK, Dubey S, et al. Leprosy drug clofazimine activates peroxisome proliferator-activated receptor-γ and synergizes with imatinib to inhibit chronic myeloid leukemia cells. Haematologica. 2020;105(4):971–986. doi: 10.3324/haematol.2018.194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durusu İZ, Hüsnügil HH, Ataş H, Biber A, Gerekçi S, Güleç EA, Özen C. Anti-cancer effect of clofazimine as a single agent and in combination with cisplatin on U266 multiple myeloma cell line. Leuk Res. 2017;55:33–40. doi: 10.1016/j.leukres.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 29.Ahmed K, Koval A, Xu J, Bodmer A, Katanaev VL. Towards the first targeted therapy for triple-negative breast cancer: repositioning of clofazimine as a chemotherapy-compatible selective Wnt pathway inhibitor. Cancer Lett. 2019;449:45–55. doi: 10.1016/j.canlet.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 30.Lu Y, Wang L, Xiaobing X, Tao L, Jianmin Z. Design, combinatorial synthesis and biological evaluations of novel 3-amino-1′-((1-aryl-1H-1,2,3-triazol-5-yl)methyl)-2′-oxospiro[benzo[a] pyrano[2,3-c]phenazine-1,3′-indoline]-2-carbonitrile antitumor hybrid molecules. Eur J Med Chem. 2017;135:125–141. doi: 10.1016/j.ejmech.2017.04.040. [DOI] [PubMed] [Google Scholar]
- 31.Mei-Chen Z, Shu-Hui G, Guang-Pan L, Chen-Cheng L, Han-Mei X. Facile synthesis and cytotoxicity of phenazine-chromene hybrid molecules derived from phenazine natural product. Combin Chem High Throughput Screen. 2019;22(1):35–40. doi: 10.2174/1386207322666190307125015. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Yan Y, Wang L, Wang X, Gao J, Xi T, Wang Z, Jiang F. Design, facile synthesis and biological evaluations of novel pyrano[3,2-a]phenazine hybrid molecules as antitumor agents. Eur J Med Chem. 2016;127:928–943. doi: 10.1016/j.ejmech.2016.10.068. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Chen M, Tong X, Zhu H, Yan H, Liu D, Li W, Qi S, Xiao D, Wang Y. Synthesis, antitumor activity, and structure-activity relationship of some benzo[a]pyrano[2,3-c]phenazine derivatives. Combin Chem High Throughput Screen. 2015;18(10):960–974. doi: 10.2174/1386207318666150915113549. [DOI] [PubMed] [Google Scholar]
- 34.Ghafouri-Fard S, Taheri M. UCA1 long non-coding RNA: an update on its roles in malignant behavior of cancers. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120:109459. doi: 10.1016/j.biopha.2019.109459. [DOI] [PubMed] [Google Scholar]
- 35.Klinge CM. Non-coding RNAs in breast cancer: intracellular and intercellular communication. Non-coding RNA. 2018;4(4):40. doi: 10.3390/ncrna4040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M, Yi J, Zhu J, Minikes AM, Monian P, Thompson CB, Jiang X. Role of Mitochondria in Ferroptosis. Mol Cell. 2019;73(2):354–363.e353. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50(3):117–125. doi: 10.5483/BMBRep.2017.50.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Shigdar S, Gantier MP, Hou Y, Wang L, Li Y, Shamaileh HA, Yin W, Zhou SF, Zhao X, et al. Cancer stem cell targeted therapy: progress amid controversies. Oncotarget. 2015;6(42):44191–44206. doi: 10.18632/oncotarget.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Conor CJ, Chen T, González I, Cao D, Peng Y. Cancer stem cells in triple-negative breast cancer: a potential target and prognostic marker. Biomark Med. 2018;12(7):813–820. doi: 10.2217/bmm-2017-0398. [DOI] [PubMed] [Google Scholar]
- 40.Kim SE, Zhang L, Ma K, Riegman M, Chen F, Ingold I, Conrad M, Turker MZ, Gao M, Jiang X, et al. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat Nanotechnol. 2016;11(11):977–985. doi: 10.1038/nnano.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Versini A, Colombeau L, Hienzsch A, Gaillet C, Retailleau P, Debieu S, Muller S, Caneque T, Rodriguez R. Salinomycin derivatives kill breast cancer stem cells via Lysosomal iron targeting. Chemistry. 2020;26(33):7416–7424. doi: 10.1002/chem.202000335. [DOI] [PubMed] [Google Scholar]
- 42.Laraia L, Garivet G, Foley DJ, Kaiser N, Müller S, Zinken S, Pinkert T, Wilke J, Corkery D, Pahl A, et al. Image-based morphological profiling identifies a Lysosomotropic, iron-sequestering autophagy inhibitor. Angew Chem Int Ed Engl. 2020;59(14):5721–5729. doi: 10.1002/anie.201913712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurz T, Terman A, Gustafsson B, Brunk UT. Lysosomes in iron metabolism, ageing and apoptosis. Histochem Cell Biol. 2008;129(4):389–406. doi: 10.1007/s00418-008-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goncalves J, Moog S, Morin A, Gentric G, Müller S, Morrell AP, Kluckova K, Stewart TJ, Andoniadou CL, Lussey-Lepoutre C, et al. Loss of SDHB promotes dysregulated iron homeostasis, oxidative stress, and sensitivity to ascorbate. Cancer Res. 2021;81(13):3480–3494. doi: 10.1158/0008-5472.CAN-20-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rycaj K, Tang DG. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Res. 2015;75(19):4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, Wang M, Luo M, Xing Y, Chen F, et al. CD10(+)GPR77(+) cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172(4):841–856.e816. doi: 10.1016/j.cell.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Schonberg DL, Miller TE, Wu Q, Flavahan WA, Das NK, Hale JS, Hubert CG, Mack SC, Jarrar AM, Karl RT, et al. Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell. 2015;28(4):441–455. doi: 10.1016/j.ccell.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghupathy R, Manwani D, Little JA. Iron overload in sickle cell disease. Adv Hematol. 2010;2010:272940. doi: 10.1155/2010/272940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pietrangelo A. Iron and the liver. Liver Int. 2016;36(Suppl 1):116–123. doi: 10.1111/liv.13020. [DOI] [PubMed] [Google Scholar]
- 50.Kremastinos DT, Farmakis D. Iron overload cardiomyopathy in clinical practice. Circulation. 2011;124(20):2253–2263. doi: 10.1161/CIRCULATIONAHA.111.050773. [DOI] [PubMed] [Google Scholar]
- 51.Zhao W, Liu J, Li Y, Chen Z, Qi D, Zhang Z. Immune effect of active components of traditional Chinese medicine on triple-negative breast cancer. Front Pharmacol. 2021;12:731741. doi: 10.3389/fphar.2021.731741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tharmapalan P, Mahendralingam M, Berman HK, Khokha R. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;38(14):e100852. doi: 10.15252/embj.2018100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Q, Wang T, Yang Y, Gao L, Zhao Q, Zhang W, Xi T, Zheng L. Transcriptional Factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity. Mol Ther Nucleic acids. 2020;21:527–541. doi: 10.1016/j.omtn.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrison AT, Abouelhassan Y, Kallifidas D, Tan H, Kim YS, Jin S, Luesch H, Huigens RW., 3rd An efficient buchwald-hartwig/reductive cyclization for the scaffold diversification of halogenated phenazines: potent antibacterial targeting, biofilm eradication, and prodrug exploration. J Med Chem. 2018;61(9):3962–3983. doi: 10.1021/acs.jmedchem.7b01903. [DOI] [PubMed] [Google Scholar]
- 55.Yang H, Kundra S, Chojnacki M, Liu K, Fuse MA, Abouelhassan Y, Kallifidas D, Zhang P, Huang G, Jin S, et al. A modular synthetic route involving N-aryl-2-nitrosoaniline intermediates leads to a new series of 3-substituted halogenated phenazine antibacterial agents. J Med Chem. 2021;64(11):7275–7295. doi: 10.1021/acs.jmedchem.1c00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J, Li T, Gao C, Lv X, Liu K, Song H, Xing Y, Xi T. FOXO1 3'UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett. 2014;588(17):3218–3224. doi: 10.1016/j.febslet.2014.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.