Abstract
Velvet ants (Hymenoptera: Mutillidae) are a family of solitary parasitoid wasps that are renowned for their painful stings. We explored the chemistry underlying the stings of mutillid wasps of the genus Dasymutilla Ashmead. Detailed analyses of the venom composition of five species revealed that they are composed primarily of peptides. We found that two kinds of mutillid venom peptide appear to be primarily responsible for the painful effects of envenomation. These same peptides also have defensive utility against invertebrates, since they were able to incapacitate and kill honeybees. Both act directly on cell membranes where they directly increase ion conductivity. The defensive venom peptides of Dasymutilla bear a striking similarity, in structure and mode of action, to those of the ant Myrmecia gulosa (Fabricius), suggesting either retention of ancestral toxins, or convergence driven by similar life histories and defensive selection pressures. Finally, we propose that other highly expressed Dasymutilla venom peptides may play a role in parasitisation, possible in delay or arrest of host development. This study represents the first detailed account of the composition and function of the venoms of the Mutillidae.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03847-1.
Keywords: Dasymutilla, Cow killer, Wasp, Pain, Parasitoid
Introduction
Mutillid wasps (Hymenoptera: Mutillidae) are a large family of approximately 4300 described species of solitary parasitoid wasps [1]. They are commonly known as “velvet ants”, in reference to the dense “velvet-like” pilosity covering the exoskeleton of most species and the fact that females are wingless and, therefore, resemble large ants (Hymenoptera: Formicidae).
Mutillid wasps are probably best known for their characteristic defensive adaptations: females have a very strong, rounded and smooth exoskeleton [2]; almost all diurnal species display striking aposematic coloration; and when under duress, they release alarm pheromones and use a stridulatory organ to produce a characteristic squeaking sound [2]. While the strong exoskeleton provides an initial protection against predatory strikes [3], the visual, auditory and olfactory cues serve to warn or remind potential predators that these wasps harbour an exceptionally powerful sting.
Stings from mutillid wasps are notoriously painful. Those of the larger species are considered to be among the most painful of the Hymenoptera [4], which has earned them the common name of “cow-killers”. While evidence indicates that stings are unlikely to be directly lethal to female Bos taurus [5], the nickname does effectively convey the human perception of the sting as being “intense enough to kill a cow”. Mutillid wasp stings cause nociception in a diverse array of potential predators including reptiles, amphibians, birds and mammals [2, 6]. The effectiveness of the mutillid sting has resulted in one of the largest Müllerian mimicry complexes so far reported [7] as well as several diverse examples of Batesian mimicry [8–12].
The chemistry underlying stings of the Mutillidae remains unknown. To date, only one study has examined the venom bioactivity of a single species, Dasymutilla lepeletierii (Fox) (now synonymized under Dasymutilla bioculata (Cresson) [13]), which was reported to have esterase activity and low levels of phospholipase and hyaluronidase activity [14]. Here, we present detailed blueprints of the venom compositions of five species of the mutillid wasp genus Dasymutilla Ashmead. We report the pain-causing agents and their mode of action, and we examine the activity of other venom components in the context of parasitisation.
Results
The venom composition of Dasymutilla klugii (Gray)
We used a combined transcriptomic and mass spectrometry (MS)-based proteomic strategy to generate a full profile of the polypeptidic venom composition of D. klugii (Fig. 1a). We collected venom by inciting individual D. klugii to sting a thin layer of parafilm. Unless otherwise stated, the venom samples analysed in subsequent experiments was “stung” venom droplets pooled from three individuals. We dissected and pooled the venom apparatus (venom gland filaments, venom reservoir and venom duct; Fig. 1b) from the three individuals from which venom had been collected, and from these, we extracted mRNA, which was used to generate a venom apparatus transcriptome. We obtained 10,034,381 demultiplexed paired-end reads from Illumina NextSeq RNA sequencing, and following adaptor trimming, quality trimming and filtering and error correction, they were assembled de novo using Trinity to yield a total of 20,749 contigs.
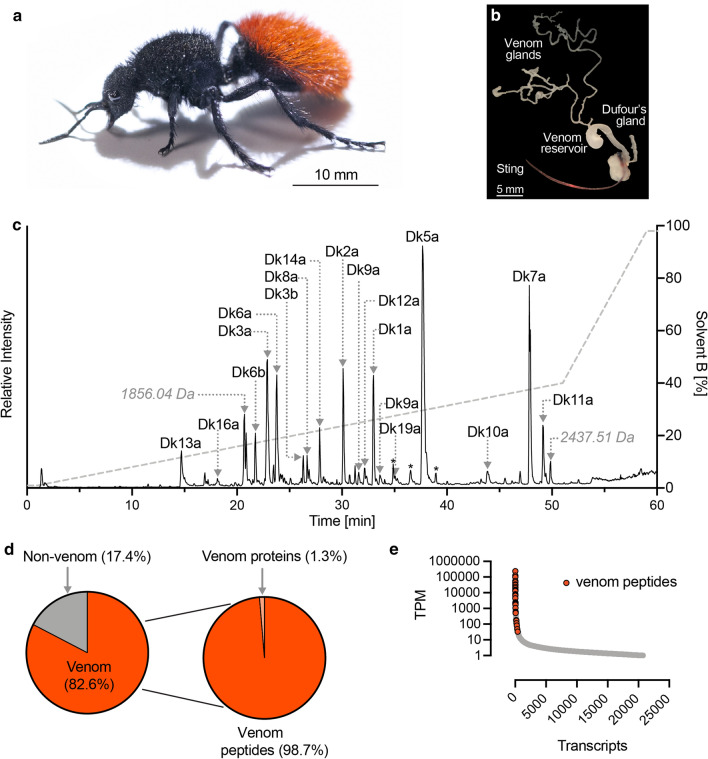
Fig. 1.
The venom composition of Dasymutilla klugii. a Adult female D. klugii. b Venom apparatus of D. klugii. c Total ion chromatogram of reduced and alkylated D. klugii stung venom. Venom (10 μg) was fractionated using C18 ultrahigh-performance liquid chromatography (uHPLC), using a gradient of 1–40% solvent B [90% acetonitrile (ACN) and 0.1% FA] over 50 min and analysed using MS/MS. Peaks corresponding to identified venom peptides are labelled, peaks identified as corresponding to fragments of venom peptides are labelled with an asterisk, and peaks for which no corresponding venom peptide was identified are labelled with their deconvoluted monoisotopic mass. d Venom component-encoding transcripts (that is, those encoding peptides detected in the venom itself) comprised 82.6% of total venom apparatus transcript expression. Of these, transcripts encoding aculeatoxin peptides and venom proteins comprised 98.7 and 1.3%, respectively, of venom component expression. e Transcript abundance expressed as transcripts per million (TPM). Venom peptide-encoding transcripts (highlighted in red) are found exclusively in the highly expressed portion of the venom apparatus transcriptome, where they constitute most of the most highly expressed transcripts
Liquid chromatography-tandem MS (LC–MS/MS) data from three venom samples (native; reduced and alkylated; reduced, alkylated, and trypsin-digested) were searched against a database comprising the translated venom apparatus transcriptome. We confirmed (by top-down sequencing of reduced and alkylated venom) the complete mature peptide sequence of 18 unique peptides (Fig. 1c). An additional seven peptides that were not detected in our proteomic data were included as probable venom components based on high sequence similarity and similarly high-expression level estimates to identified venom peptides. Transcripts encoding venom components accounted for 82.6% of all venom apparatus–derived sequencing reads (Fig. 1d). Of these, 98.7% encoded peptides (as opposed to proteins). Venom peptide-encoding transcripts were confined to the high-expression portion of the venom apparatus transcriptome, where they constituted almost all of the most highly expressed transcripts (Fig. 1e). The high proportion of venom apparatus-derived reads encoding venom peptides, the high-expression level of each venom peptide-encoding transcript, and the assignment of all major peaks in the total ion chromatogram of venom strongly suggest that these 25 peptides represent the major components in D. klugii venom. Of the 25 total peptides (Table 1), 13 lacked cysteine residues, while the remaining 12 peptides each had 2 cysteine residues. Analysis of the native venom revealed that the cysteine-containing peptides were present as monomers, each with a single intrachain disulfide bond. No evidence of homo- or hetero-dimerisation was detected.
Table 1.
Polypeptides found in D. klugii venom
| TPM | Sequence | |
|---|---|---|
| Dk1a | 251710b | LSPAVIASLAa |
| Dk2a | 137436b | VFVLPDICNERPRIPQCRa |
| Dk3a | 99830 | VFDVPEICNKKPHILRCRa |
| Dk3bc | 46905 | VFDIPEICKKRPYILRCRa |
| Dk4ac | 56683 | SALRICRRKPSSPLCWa |
| Dk4bc | 33563 | SALRICRRNPRSPLCWa |
| Dk5a | 32233 | RFGGILKILKKVLPKAIKVAAEMAPPQNE |
| Dk6a | 31244 | SKYAICKIKPRLPWCRa |
| Dk6b | 23878 | SIYNICKIKPRLPRCRa |
| Dk7a | 19527 | HLGDVITDLVNKALNSLa |
| Dk8a | 14890 | WKHPVCGLFPKAPMCSVRAE |
| Dk9a | 14403 | KRKKRKGSKFGKDILTSFGKGAAEAAGEATVNAAVDQILEKQREGEYLY |
| Dk9bc | 2439 | KRKKRKGSKFGKNILTSFGKGAAEAAGEATVNAAVDQILEAQGEGEYLY |
| Dk10ac | 12959 | KRGRSKGDGKKGKKPKDKIASKIGDIIKNSLNKFGVVTGETIVEKTVEAVKDALQSGEASAEEEAPSE |
| Dk11a | 9871b | LVGAVLGAVELGELIHHLIKEa |
| Dk12ac | 7130b | VSPVVLASLTa |
| Dk13a | 7036 | KRKWKKKLKKLVKKALKHGAGALLSa |
| Dk14ac | 4486 | SKPSLCKLVPHIPLCRa |
| Dk15ac | 2379b | FRSKPRICRKKPNIPLCRS |
| Dk15bc | 550 | FRSKPRICRKKPNIPLCREYPFIPYHGVDPPPN |
| Dk16ac | 2068b | RRAPPLPY |
| Dk17a | 1912 | EGDPPQQRPL |
| Dk17b | 166 | ERDPPQQRPL |
| Dk18ac | 1186b | RRRGFGNENLDKIYRGVINPQSPGVDVGKLPDDIFMQQARREFAEALAREKAERARIIYKQAQREKAERARa |
| Dk19a | 512 | FKHSLCAVFPFLPHCGVSYGNST |
| Kunitz-domain proteinsc | 3373 | |
| M13-metalloproteasesc | 3616 | |
| Phospholipase-A2c | 483 | |
| Hyaluronidasec | 166 |
TPM transcripts per million
aC-terminal amidation
bThe TPM value is the sum of more than one assembled transcript encoding the same mature peptide
cComplete primary structure not confirmed by MS/MS; Equivalent tables for D. gloriosa, D. bioculata, D. sicheliana and D. occidentalis are included in the supplementary material as Tables S1–4, respectively
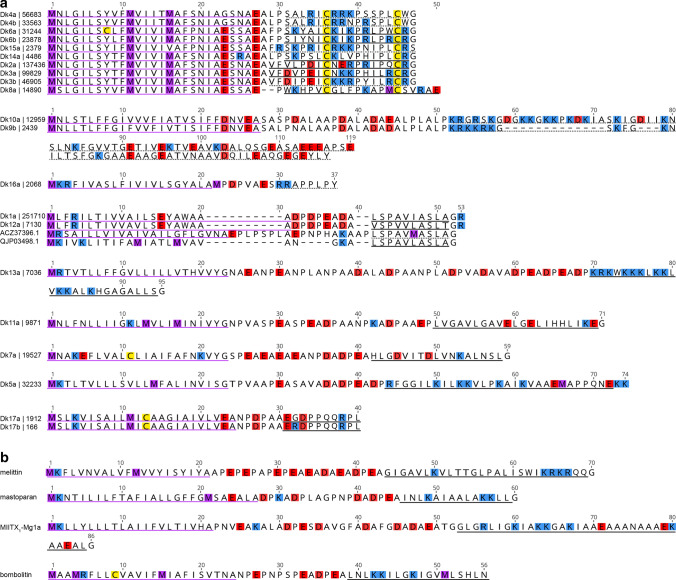
In an effort to predict function of the D. klugii venom peptides, we used the BLASTp algorithm to search the UniProt/GenBank protein database for related sequences. Significant alignments were detected for the two closely related peptides, Dk1a and Dk12a, both to aculeate venom peptides; VP4a protein precursor (ACZ37396.1) from the potter wasp Eumenes pomiformis (Fabricius) [15] and U12-MYRTX-Tb1a precursor (QJP03498.1) from the ant Tetramorium bicarinatum (Nylander) [16] (Fig. 2a). While the complete prepropeptide sequences share limited similarity, the mature peptides are very similar, with mature VP4a and U12-MYRTX-Tb1a both differing from Dk1a by only a single conservative substitution. No biological activity has been reported for these peptides, although given the similarity in primary structure it is reasonable to predict they have similar activity as Dk1a and Dk12a. The precursor sequences of the venom peptides of D. klugii are generally similar to those of most venom peptides identified in other hymenopteran venoms—they contain a signal peptide, followed by a highly repetitive and net anionic propeptide, followed by a single copy of a largely hydrophobic and often cationic mature peptide (Fig. 2b). While not detected in our BLAST search, some of the encoded mature venom peptides of Dasymutilla, namely Dk5a and Dk13a, resemble other previously described hymenopteran venom peptides (an observation elaborated in the Discussion section).
Fig. 2.
Venom peptides of D. klugii are related to those of other Hymenoptera. a Precursor sequences of D. klugii venom peptides. For clarity, prepropeptide sequences have been aligned according to subgroup. Estimated expression level (in transcripts per million) is indicated alongside each peptide name. Signal peptides, experimentally confirmed mature peptides and partially confirmed/predicted mature peptides are underlined in purple, black and black dashes, respectively. Methionine, lysine/arginine, aspartate/glutamate, and cysteine residues are highlighted in purple, blue, red, and yellow, respectively. ACZ37396.1, VP4a protein precursor (Eumenes pomiformis) [15]; QJP03498.1, U12-MYRTX-Tb1a precursor (Tetramorium bicarinatum) [16]. b Precursor sequence of selected aculeatoxins showing similar precursor structures and overall biophysical properties: Melittin (P01501), Apis mellifera (Apidae); Mastoparan-M (S4S3F3), Vespa mandarinia (Vespidae); MIITX1-Mg1a (P0DSJ4), Myrmecia gulosa (Formicidae); Bombolitin (D0VDZ4), Bombus ignitus (Apidae)
Proteins made up the remaining 1.3% of venom component expression and those detected (by bottom-up sequencing of reduced, alkylated, and trypsin-digested venom) included M13-metalloproteases (e.g. neprilysins), phospholipase A2 (PLA2), hyaluronidase, and Kunitz-domain proteins. Metalloproteases were detected as multiple isoforms and were the most highly expressed protein class in the venom, consistent with a significant functional role. The major isoform (shown in Fig. S1) retains the active site residues of endogenous insect neprilysins, but the cytoplasmic and transmembrane domains have been replaced with a secretory signal peptide, consistent with “weaponisation” of an intracellular enzyme as a venom toxin in D. klugii. PLA2 and hyaluronidase were detected at lower expression levels and as a single isoform per species, consistent with the previous report of low but measurable phospholipase and hyaluronidase activity in Dasymutilla venom [14]. One to three isoforms of Kunitz-domain proteins were detected per species.
The venom composition of other Dasymutilla species
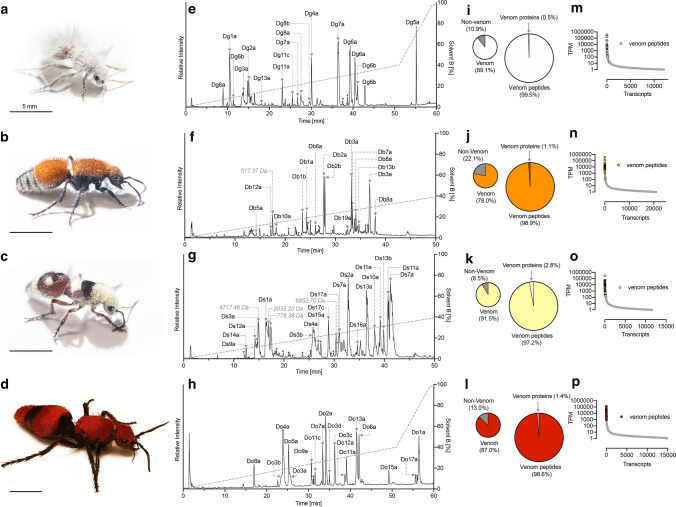
We repeated the aforementioned combined transcriptomic and MS-based proteomic strategy to investigate the stung venom composition of four additional species: Dasymutilla gloriosa (Saussure), Dasymutilla bioculata, Dasymutilla sicheliana (Saussure) and Dasymutilla occidentalis (Lin.) (Fig. 3). These species have a similar venom composition, comprising 23–29 aculeatoxin peptides (Tables S1–4) encompassing homologues of each of the venom peptides identified in D. klugii. The percentage of transcripts encoding venom components ranged from 78.0 to 91.5% of all venom apparatus-derived reads (Fig. 3i–l). Of these, between 97.2 and 99.5% encoded peptides. Venom peptide-encoding transcripts were invariably confined to the high-expression portion of each venom apparatus transcriptome, where, again, they constituted almost all of the most highly expressed transcripts (Fig. 3m–p). Venom proteins detected in all species were M13-metalloproteases (e.g. neprilysins), PLA2, hyaluronidase and Kunitz-domain proteins at similar expression levels to those observed for D. klugii. Thus, all the Dasymutilla species examined appear to share a generally similar venom composition, marked only by mostly minor differences in individual peptide sequences.
Fig. 3.
The venom compositions of D. gloriosa, D. bioculata, D. sicheliana and D. occidentalis. a–d Adult female D. gloriosa, D. bioculata, D. sicheliana and D. occidentalis, respectively. Scale bars = 5 mm. e–h Total ion chromatogram of reduced and alkylated D. gloriosa, D. bioculata, D. sicheliana and D. occidentalis venoms. Venoms (10 μg) were separated by C18 uHPLC, using a gradient of 1–40% solvent B (90% ACN and 0.1% FA) over 50 min and analyzed using MS/MS. Peaks are labelled as in Fig. 1c. i–l Proportion of venom component-encoding transcripts and proportion of transcripts encoding aculeatoxin peptides and venom proteins. m–p Transcript abundance expressed as TPM. For each species, venom peptide-encoding transcripts (coloured) are found exclusively in the highly expressed portion of each venom apparatus transcriptome, where they consistently constitute most of the most highly expressed transcripts
Identification of pain-causing agents in Dasymutilla venoms
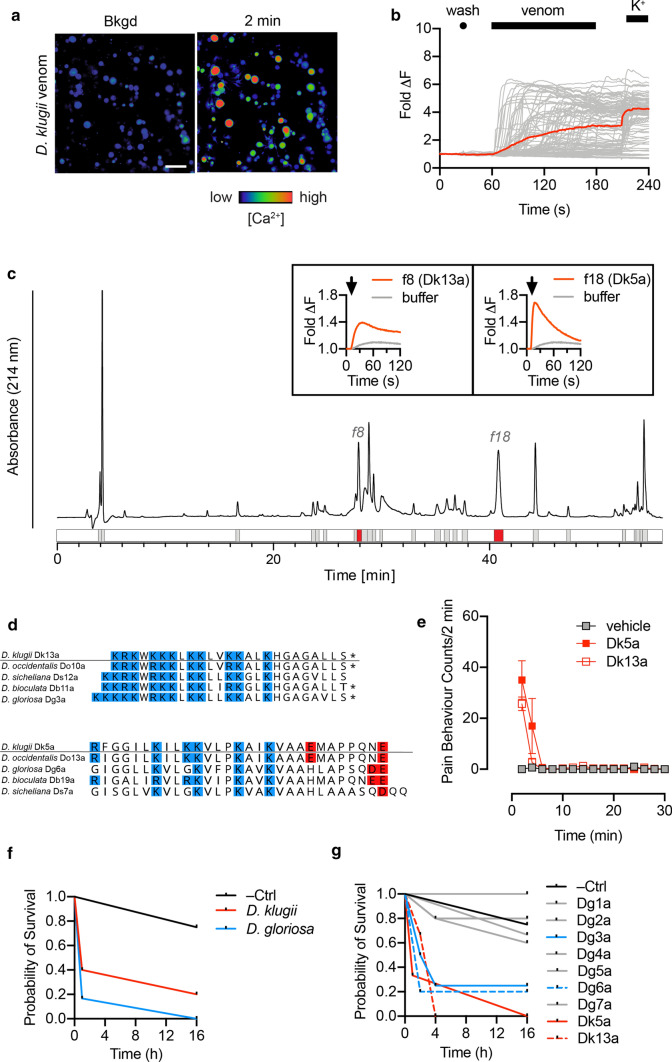
Dasymutilla are renowned for their painful stings. To confirm that our venom samples retained algogenic activity, we tested their capacity to directly activate cultured mouse dorsal root ganglion (DRG) neurons, which include the neurons responsible for detecting painful stimuli. Application of D. klugii venom (100 μg/mL) to mouse DRG cultures caused an immediate, rapid and sustained increase in intracellular Ca2+ concentration in all cells (neuronal and non-neuronal) (Fig. 4a, b).
Fig. 4.
Identification of defensive peptides in Dasymutilla venoms. a Application of D. klugii venom (100 μg/mL) to DRG cells produced a rapid, non-cell-specific increase in [Ca2+]i. Micrographs show recordings before and 2 min after venom application. Scale bar = 100 μm. b Each trace represents the fluorescence of a single cell in the field of view, with the average of all traces displayed in color. c D. klugii venom (250 μg) was fractionated using reversed-phase HPLC and each fraction was screened for activity on F11 cells, which yielded two active fractions (f8 and f18). Traces corresponding to the change in [Ca2+]i for f8 and f18, and representing the mean of two experiments, are displayed in the insets. d Active fractions were identified as Dk13a (D. klugii f8) and Dk5a (D. klugii f18). Alignment of Dk5a and Dk13a with homologous peptides from the venom of other species. Lysine/arginine and aspartate/glutamate are colored in blue and red, respectively; *, C-terminal amidation. e Intraplantar injection of synthetic Dk13a and Dk5a (600 pmol) induced spontaneous nocifensive behavior in mice. Data are expressed as mean ± SEM (n = 3). i Intrathoracic injection of crude D. klugii or D. gloriosa venom (15 μg/g) reduced survival probability in Apis mellifera (n = 4–6 per condition). (j) Intrathoracic injection of Dg3a, Dg6a, Dk5a and Dk13a, but not Dg1a, Dg2a, Dg4a, Dg5a and Dg7a (15 nmol/g) reduced survival probability in Apis mellifera (n = 4–6 per condition)
To identify the active algogenic agents, we fractionated D. klugii venom by reversed-phase HPLC and screened fractions for direct activity on mammalian sensory neurons using the mouse neuroblastoma × rat DRG F11 cell line. Two active fractions were identified (Fig. 4c) (all other fractions were inactive). The active component in each fraction determined using MALDI-MS, corresponded to peptides Dk13a (fraction 8) and Dk5a (fraction 18) (Fig. 4d). Dk13a is a cysteine-free peptide of 25 residues, with a net cationic charge of + 11, while Dk5a is cysteine-free peptide of 29 residues with net charge of + 4. Homologous peptides of both Dk5a and Dk13a were present in venom of each of the other four species of Dasymutilla examined in this study (Fig. 4d).
We tested the capacity of synthetic Dk5a and Dk13a to induce spontaneous pain behaviour in mice following intraplantar injection in the hindpaw. Both peptides (600 pmol) caused immediate and short-lasting spontaneous pain behaviour (Fig. 4e). The intense but brief time-course (2–4 min) of pain behaviour is consistent with stings of Dasymutilla species (author observations, J.O.S., S.D.R.) and these data are, therefore, consistent with these peptides being the primary vertebrate-active algogenic agents in Dasymutilla venoms. Dasymutilla stings can also cause pronounced inflammatory effects including oedema at the sting site (author observation, J.O.S., S.D.R.). Neither of the peptides tested here caused oedema of the injected paw, suggesting that other venom components may be responsible for those effects.
To test whether this trend was shared by other Dasymutilla species, we synthesised an additional seven venom peptides (Dg1a, Dg2a, Dg3a, Dg4a, Dg5a, Dg6a, Dg7a; Table S1), which represent most of the structural diversity present in the venom of D. gloriosa, and tested their capacity to activate F11 cells and DRG neurons. Dg3a and Dg6a, which are homologues of Dk13a and Dk5a, respectively, were active in both assays, causing effects similar to those of their D. klugii counterparts (Fig. S2), whereas the other peptides were inactive (up to 100 μM). We also investigated whether the peptides that did not have an independent effect in these assays could potentiate the effects of the direct pain-causing agents (Dg3a and Dg6a), but found that none of these peptides had any effect on the potency or efficacy of Dg3a or Dg6a. We also found that neither PLA2 nor the venom metalloproteases could potentiate the effects of Dk5a and Dk13a or contributed to the algogenic activity of D. klugii venom.
Activity of velvet ant venoms and venom peptides against insects
Several reports suggest that mutillid wasps use their sting against other invertebrates, including in defence against potential predators such as arachnids and insects (author observation, J.O.S.) and in combat against host adults defending their nests [17–19]. We explored this by testing the effects of crude venoms and a broad range of venom peptides on bees. When whole venoms of D. klugii and D. gloriosa were injected intrathoracically (15 μg/g) into worker honeybees, Apis mellifera Linnaeus, they both caused a gradual immobilisation of injected bees, and, in most cases, eventual death (Fig. 4f).
We then tested synthetic Dg1a, Dg2a, Dg3a, Dg4a, Dg5a, Dg6a, Dg7a, Dk5a, and Dk13a (at a dose of 15 nmol/g), by intrathoracic injection in A. mellifera. Both Dk5a and Dk13a caused similar effects to the whole venoms (i.e. gradual immobilisation and eventual death), while two of the seven D. gloriosa venom peptides produced similar results (Fig. 4g). Notably, the two active peptides from D. gloriosa venom were Dg3a and Dg6a, homologues of Dk13a and Dk5a, respectively. Thus, Dasymutilla venoms are active against insects, and the insecticidal activity appears to be due to the same peptides that confer algogenic activity against vertebrates.
Activity of velvet ant venoms and venom peptides against insect larvae
Many parasitoid wasps use their sting on host larvae/pupae to modify their physiology to the advantage of the parasitoid larva [20]—in some cases, this is by a direct paralytic action on the host, in others, it is by interference with host development or immune function. The literature on Mutillidae in this context is inconsistent: Hoffer reported that the mutillid wasp Mutilla europaea Linnaeus paralyses its host with its sting [21], but this was later suggested not to be the case [22]. However, two more recent studies have reported use of the sting by mutillid wasps on the host prior to oviposition, as well as changes in host development following parasitisation [19, 23].
We injected whole venoms of D. gloriosa and D. klugii (30 μg/g) and each of the nine synthetic venom peptides (3 nmol/g) intra-abdominally into black soldier fly (Hermetia illucens (Linnaeus)) larvae. Neither venom, nor any of the venom peptides, produced any effect that could be distinguished from negative control injections with water (i.e. the larvae continued to move freely following injection). When checked at 24 h post-injection, larval behaviour was unaltered. While it is possible that the mutillid venoms and venom peptides we tested are active only against specific hosts and not H. illucens, these data are consistent with a lack of acute paralytic action of Dasymutilla venoms on insect larvae, but do not rule out other effects, e.g. arrest of development.
Mode of action of Dasymutilla venom peptides Dk5a and Dk13a
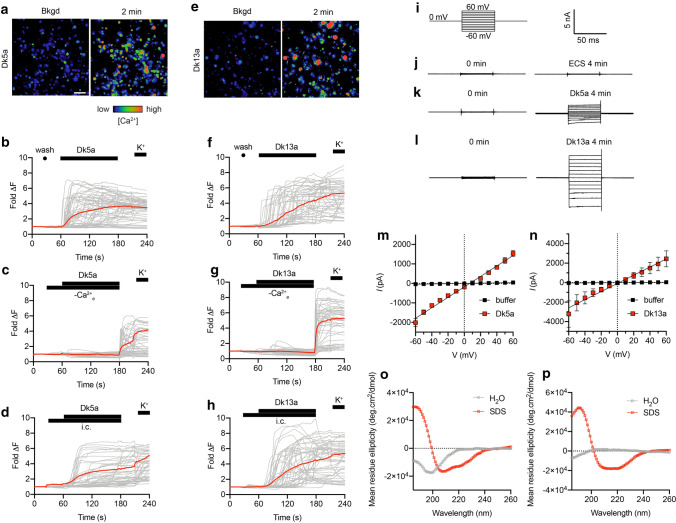
We used a range of assays to investigate the mode of action of Dk5a and Dk13a. In cultures of mouse DRG neurons, Dk5a caused an immediate and simultaneous increase in [Ca2+]i in all cells (Fig. 5b). Dk13a also caused a sustained increase in [Ca2+]i in all cells, but which was variable in onset (i.e. not simultaneous) (Fig. 5f). For both peptides, the effects were observed in all cells (neuronal and non-neuronal) in the heterogeneous primary culture. For each peptide, the increase in [Ca2+]i was prevented by the absence of extracellular Ca2+, but largely unaffected by the presence of a cocktail of ion channel inhibitors, including 1-μM tetrodotoxin (TTX; a blocker of voltage-gated sodium channels); 100-μM Cd2+ (a blocker of voltage-sensitive calcium channels); 20-μM ruthenium red (a non-selective antagonist of transient receptor potential cation (TRP) channels and ryanodine receptors); 20-μM amiloride (a non-selective inhibitor of acid-sensing ion channels (ASICs)). Together these data strongly suggest that the increase in [Ca2+]i caused by these peptides is due to influx of Ca2+ ions from the extracellular medium, and it is likely not mediated by a specific ion channel. Such effects are diagnostic of a membrane-mediated mode of action.
Fig. 5.
Mode of action of Dasymutilla venom peptides. a–b Application of Dk5a (10 μM) to DRG cells produced an immediate and sustained, non-cell-specific increase in [Ca2+]i. c–d The increase in [Ca2+]i caused by Dk5a was prevented by removal of extracellular Ca2+, but unaffected by the presence of a cocktail of ion channel inhibitors [TTX (1 µM), Cd2+ (100 µM), amiloride (20 µM) and RR (20 µM)]. e–f Application of Dk13a (10 μM) to DRG cells produced a rapid and sustained, but variable onset, non-cell-specific increase in [Ca2+]i. g–h The increase in [Ca2+]i caused by Dk13a was prevented by removal of extracellular Ca2+, but unaffected by the presence of a cocktail of ion channel inhibitors. i Voltage protocol used to investigate leak current induced by Dk5a and Dk13a in HEK293 cells. j Scale bar for panels (j–l). j–l Representative current traces at voltages of − 60 to + 60 mV (10 mV steps) acquired before application and 4 min after application of (j) extracellular solution (ECS), k 10-μM Dk5a, or (l) 10-μM Dk13a. m) Current–voltage (I–V) relationship 4 min after addition of ECS (black) or Dk5a (10 μM; red). n I–V relationship 4 min after addition of ECS (black) or Dk13a (10 μM; red). Data are expressed as mean ± SEM (n = 5 cells) and fitted to a simple linear regression. o Far-UV CD spectra (260–185 nm) of Dk5a and Dk13a (25 µM) in 100% H2O and 20-mM SDS. Dk5a is unordered in water but adopts helical secondary structure in 20-mM SDS, as indicated by the ellipticity minima at 208 and 222 nm. p Dk13a is unordered in water but adopts helical secondary structure in 20-mM SDS
With this in mind, we tested, by whole-cell patch-clamp electrophysiology, the capacity of each peptide to directly induce leak currents in cell membranes. For these experiments, we used HEK293 cells, which lack appreciable expression of the ion channels found in neurons. At 4 min after application of Dk5a (10 μM) or Dk13a (10 μM), we recorded leak currents (at − 60 mV) of − 2.0 ± 0.2 nA (n = 5 cells) and − 3.2 ± 1.3 nA (n = 4 cells), respectively, compared with − 0.04 ± 0.02 nA (n = 5 cells) for time-matched negative controls [application of extracellular solution (ECS)] (Fig. 5a–f). These data are consistent with a membrane-mediated mode of action for the peptides. Other hymenopteran venom peptides, including melittin, the major pain-causing agent in the venom of A. mellifera, cause leak currents in cell membranes via the formation of ion-permeable “pores” [24, 25]. While Dk5a and Dk13a are structurally distinct from melittin, their similarities in biophysical properties such as charge and hydrophobicity appear to impart a related mode of action.
Melittin and some related hymenopteran venom peptides are structurally disordered in solution but adopt helical secondary structure when interacting with membranes or membrane-mimicking solvents, an effect which is important in their action of membrane permeation [26–28]. To investigate whether Dk5a and Dk13a shared similar properties, we used circular dichroism (CD) spectropolarimetry to observe the secondary structure of each peptide in water and in the membrane-mimicking detergent sodium dodecyl sulfate (SDS). Far-UV (260–185 nm) CD data of Dk5a (25 µM) and Dk13a (25 µM) were consistent with a mostly disordered conformation in 100% H2O indicated by minima < 200 nm, which shifted to helical conformation in SDS (20 mM), as indicated by minima ~ 208 and 220 nm (Fig. 5g–h). These data indicate that, like melittin, Dk5a and Dk13a have the capacity to adopt helical secondary structure when presented with a membrane-mimicking environment.
On the basis of these data, we propose a model whereby Dk5a and Dk13a interact with cell membranes where they increase ion conductivity, possibly via pore formation, which in excitable cells such as mammalian sensory neurons leads to depolarisation, action potential generation, and pain signalling.
Discussion
In this study, we determined the venom composition of five species of mutillid wasps of the genus Dasymutilla. Each venom was rich in peptides. From the overall biophysical properties of the peptide toxins and their precursor architecture, it appears that most, if not all, are derived from the aculeatoxin gene superfamily which encompasses the majority of the venom peptides reported in the aculeate Hymenoptera [29]. However, only two peptides from D. klugii showed detectable BLAST homology to previously described aculeatoxins. Thus, with few exceptions (described later in the Discussion), the “cocktail” of aculeatoxin peptides in Dasymutilla venoms appear to be, at present, largely unique to this lineage. It will be of considerable interest to investigate the venom composition of other Mutillidae and that of closely related aculeate Hymenoptera.
We observed little difference between the venom compositions of the different Dasymutilla species. At first, this may seem surprising given that stings from larger species, such as D. klugii, are substantially more painful than those of smaller species, such as D. gloriosa [4] (author observations J.O.S. and S.D.R.). Differences between the algogenic venom peptides of each species (see Fig. 4d) could explain this difference, however, the structural differences are minimal (and conservative), making this explanation unlikely. One obvious difference between species, which does correlate with sting pain, was the volume of venom delivered during a sting. While collecting “stung” venom, the larger species consistently delivered a larger volume than their smaller counterparts. This was also mirrored in the scale of their venom apparatus (i.e. venom glands, venom reservoir, venom duct and sting). Thus, we suspect that the difference in sting pain between species may be primarily due to a dosage effect—larger species such as D. klugii can, and do, deliver more venom than smaller species such as D. sicheliana.
Our data suggest that the pain experienced from stings of D. klugii, and each of the other Dasymutilla investigated in this study, is primarily due to the action of two kinds of membrane-targeting peptide represented by Dk5a and Dk13a and their orthologues. The effects of whole D. klugii venom on sensory neurons appeared to be very similar to the combined effects of Dk5a and Dk13a, a result which suggested that these constitute the dominant mode of action in regards to algogenic activity. We found no evidence for the hypothesis that some of the “silent” venom components serve as cofactors that potentiate the effects of these direct pain-causing agents, as occurs in spitting cobra venoms, where the algogenic effects of cytotoxins are potentiated by venom PLA2 [30].
An obvious question that arises is to the function of the remaining venom peptides, most of which are also very highly expressed and abundant components of the “stung” venom. We tested two additional hypotheses regarding the roles of these peptides, namely: (a) they serve as defensive agents against invertebrates, or (b) they play a functional role in parasitisation. When we tested the ability of a range of peptides to immobilise or kill honeybees, we found that only Dk5a and Dk13a, and the two homologous peptides in D. gloriosa venom (Dg6a and Dg3a, respectively), were able to immobilise or kill honeybees. These results indicate that these two classes of peptide have a dual function, able to serve as defensive weapons against both vertebrates and invertebrates, but we are still left with the question of the functional role of the remaining venom peptides.
Another possibility that warrants further investigation is that the non-defensive venom components play a functional role in parasitisation. Hosts of mutillid wasps are the pupae or post-fed prepupae of other Hymenoptera (wasps or bees) and occasionally Diptera, Lepidoptera or Hemiptera [19, 22], and are, without exception, in a life stage where they are not actively feeding and are enclosed in some sort of package (cell, cocoon, puparium) [19, 31]. Paralysis of an immobile host may, therefore, seem redundant, although it has been proposed that paralysis of host pupa may still serve to minimise damage to the parasitoid egg or larva [19]. We found that neither whole venoms nor any of the individual peptides we tested affected the mobility of prepupal soldierfly larvae, consistent with a lack of paralytic action, although we note H. illucens is not the natural host of Dasymutilla. Regardless of whether host immobilisation is a function of Dasymutilla venom, delay or arrest of host development seems likely, based on reports of arrested development of Lasioglossum zephyrum (Smith) pupae parasitised by the mutilid wasp Pseudomethoca frigida (Smith) [19]. We anticipate that such a function may explain the highly expressed non-defensive components present in the venoms of Dasymutilla and the venom peptides reported in this study could form the foundation of future work exploring this hypothesis.
The two defensive agents in D. klugii venom, Dk5a and Dk13a, bear some resemblance in primary structure to two peptides previously reported in venom of the giant red bull ant Myrmecia gulosa (Fabricius) (Hymenoptera: Formicidae), [29] (Fig. S3). Dk5a and MIITX1-Mg1a are of a similar length and share a similar charge distribution. Dk13a is much shorter than MIITX1-Mg7a, but it shares the same repetitive lysine-rich domain. When biological function and mode of action are considered, the similarity between these peptides becomes more apparent: like their mutillid venom counterparts, MIITX1-Mg1a and MIITX1-Mg7a interact with cell membranes increasing ion conductivity [29] (Manuscript in preparation, S.D.R.). Activation of sensory neurons by MIITX1-Mg1a is immediate and simultaneous (like Dk5a), while activation of sensory neurons by MIITX1-Mg7a is variable in onset (like Dk13a). Both MIITX1-Mg1a and MIITX1-Mg7a cause nocifensive behaviour in mice and are active against invertebrates [29] (Manuscript in preparation, S.D.R.). In short, the two defensive peptides of D. klugii, Dk5a and Dk13a, mirror, in terms of both structure and function, two defensive peptides of the ant M. gulosa.
M. gulosa are large ants (up to 30 mm in length) and defend themselves against potential predators with a notoriously painful sting that is qualitatively similar to that of the some Dasymutilla species [4] (author observations, J.O.S., S.D.R.). While they are eusocial, workers are solitary hunters, and from the viewpoint of potential predators there may be considerable overlap between the ‘ant-like’ female Dasymutilla and the solitary foraging workers of the true ant M. gulosa. This overlap may have resulted in similar selection pressures on the defensive venom components of each lineage, resulting in the structurally and functionally similar pair of defensive toxins, we now see for each. The Mutillidae and Formicidae are separated by > 150 million years of evolution [32] and the similarity in the defensive venom repertoire of these two distantly related lineages could represent either (i) retention of ancestral toxins or (ii) convergence, albeit from the same ancestral aculeatoxin building blocks. Investigations of the venom composition and function of additional lineages of the aculeate Hymenoptera will resolve this question.
Methods
Collection of venom
Female individuals of D. bioculata, D. klugii and D. sicheliana were collected in Santa Cruz County Arizona. D. gloriosa were collected in Cochise County, Arizona and a single individual of D. occidentalis was collected in Tallahassee, Florida. Species identifications were based on keys by Mickel [33] and Manley and Pitts [34] and comparison with specimens in the University of Arizona insect collection.
To obtain stung venom, individuals were held with forceps and incited to repeatedly sting a thin layer of parafilm. Stung venom droplets were collected in 10 μL of ultrapure water. For some individuals, additional venom was collected from venom ducts and reservoirs during dissection (dissected tissue was placed in 10-μL H2O, then venom was squeezed out and the tissue discarded). Venoms of the same species and collection method were pooled and dried by vacuum centrifugation prior to transportation and storage.
RNA extraction, transcriptome sequencing and assembly
The venom duct, venom reservoir, and venom glands (together referred to as the venom apparatus) were dissected out from individuals from which stung venom was collected, and placed in RNALater. Total RNA was extracted from each venom apparatus using TRIzol reagent (Thermo Fisher Scientific Inc.), according to the manufacturer’s instructions. cDNA library preparation and sequencing was performed by the Institute for Molecular Bioscience Sequencing Facility, The University of Queensland. A dual-indexed library was constructed with the TruSeq-3 Stranded mRNA Sample Prep Kit (Illumina) with oligo (dT) selection and an average insert size of 180 base pairs. 150-cycle paired-end sequencing was performed on an Illumina NextSeq 500 instrument. Adapter trimming of demultiplexed raw reads was performed using fqtrim (Pertea, G. 2015. fqtrim: v0.9.4 release), followed by quality trimming and filtering using prinseq-lite [35] and error correction using BBnorm tadpole, which is a part of the BBtools package. Trimmed and error-corrected reads were assembled using Trinity (version 2.4.0) [36] with a k-mer length of 31 and a minimum k-mer coverage of 2. Assembled transcripts were annotated using a BLASTX search [37] with E value setting of 1e−3 against a UniRef90 database. Estimates of transcript abundance was made using the RSEM [38] plug-in of Trinity (align_and_estimate_abundance). Using TransDecoder, transcripts were translated and filtered to open-reading frames (> 50 amino acids), which was used as a search database for ProteinPilot.
Venom peptide-encoding transcripts confirmed by MS/MS of venom were combined into a single database which was used to search the remaining transcriptome for homologous sequences (BLASTX; e value of 1e−6). All venom peptide-encoding transcripts were manually examined using the Map-to-Reference tool of Geneious Prime version 10.2.6 (Biomatters, Ltd.) [39]. Transcript abundance was calculated in TPM using the Trinity plug-in RSEM [38].
Mass spectrometry
Dried venom samples were reconstituted in 20-μL ultrapure water and total protein concentration determined by absorbance at 280 nm using a NanoDrop spectrophotometer. When not used immediately, samples were stored at − 20 °C. A combination of top-down analysis of native and reduced and alkylated stung venom, and bottom-up proteomics of reduced, alkylated and trypsin-digested stung venom was used to determine the venom composition of each species. Two aliquots of stung venom from each species (10 μg each) were dried by vacuum centrifugation. Gas phase reduction and alkylation was performed as described [40]. 100 μL of reduction/alkylation reagent (50% (v/v) ammonium carbonate, 48.75% ACN, 1% 2-iodoethanol, 0.25% triethylphosphine) was added to the lid of each 1.5-mL tube containing dried venom, which was then inverted, closed and incubated at 37 °C for 90 min. One aliquot of reduced and alkylated venom was then digested by incubating with trypsin (20 ng/μL) overnight at 37 °C, according to the manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO).
Three samples—native venom, reduced and alkylated venom, and reduced, alkylated and trypsin-digested venom—were analysed using LC–MS/MS. Samples were fractionated on a Shimadzu (Japan) Nexera uHPLC with an Agilent Zorbax stable-bond C18 column (2.1 × 100 mm; particle size, 1.8 μm; pore size, 300 Å), using a flow rate of 180 μL/min and a gradient of 1–40% solvent B (90% ACN, 0.1% FA) in 0.1% FA over 25 min, 40–80% solvent B over 4 min, and analyzed on an AB Sciex 5600 TripleTOF mass spectrometer equipped with a Turbo-V source heated to 550 °C. MS survey scans were acquired at 300–1800 mass/charge ratio (m/z) over 250 ms, and the 20 most intense ions with a charge of + 2 to + 5 and an intensity of at least 120 counts were selected for MS/MS. The unit mass precursor ion inclusion window mass within 0.7 Da and isotopes within 2 Da were excluded from MS/MS, with scans acquired at 80–1400 m/z over 100 ms and optimised for high resolution. Using ProteinPilot version 5.0 (ABSciex), MS/MS spectra were searched against each corresponding translated venom apparatus transcriptome (described above).
For each species, transcripts identified as encoding venom components in the first ProteinPilot search were used to generate a BLAST database, which, in an effort to identify all venom component homologues in the corresponding transcriptome, were aligned using BLASTN (e value 1e−6) back to the complete transcriptome. Transcripts encoding venom components and identified homologues were then manually examined using the Map-to-Reference tool of Geneious version 10.2.6 [39], where numerous “masked” homologues were extricated from assembled transcripts and erroneous transcripts discarded. These were then reincorporated back into the complete transcriptome, estimation of transcript abundance repeated, and a second, final ProteinPilot search performed. Peptides identified by ProteinPilot were validated by comparison of experimentally derived MS/MS peaks against a theoretical peak list generated using MS-Product in ProteinProspector version 5.22.1 (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct).
Activity-guided fractionation of Dasymutilla venoms
Approximately 250 µg of each of D. klugii and D. gloriosa venom was separated on a Phenomenex Gemini NX-C18 column (250 × 4.6 mm; particle size, 3 µm; pore size, 110 Å) using a gradient of 5–80% solvent B (90% ACN, 0.05% TFA) over 60 min at a flow rate of 1 mL min−1. Fractions were collected on the basis of absorbance at 214 nm and MALDI-MS was used to confirm the identity of the peptide/s present in each fraction. Fractions were dried by vacuum concentration and resuspended in 25 µL of pure water from which 1 µL-aliquots were used (in a final volume of 30 µL) for calcium imaging experiments.
F11 (mouse neuroblastoma × DRG neuron hybrid) were cultured as previously described [41]. Cells were maintained on Ham’s F12 media supplemented with 10% FBS, 100-µM hypoxanthine, 0.4-µM aminopterin, and 16-µM thymidine (Hybri-Max, Sigma Aldrich). 384-well imaging plates (Corning, Lowell, MA, USA) were seeded 24 h prior to calcium imaging, resulting in ~ 90% confluence at the time of imaging. Cells were loaded for 30 min at 37 °C with Calcium 4 assay component A in physiological salt solution (PSS; 140-mM NaCl, 11.5-mM D-glucose, 5.9-mM KCl, 1.4-mM MgCl2, 1.2-mM NaH2PO4, 5-mM NaHCO3, 1.8-mM CaCl2, 10-mM HEPES) according to the manufacturer’s instructions (Molecular Devices, Sunnyvale, CA). Ca2+ responses were measured using a FLIPRTETRA fluorescent plate reader equipped with a CCD camera (Ex: 470–490 nm, Em: 515–575 nM) (Molecular Devices, Sunnyvale, CA). Signals were read every second for 10 s before, and 300 s after, the addition of fractions (in PSS supplemented with 0.1% BSA) in duplicate.
Peptide synthesis
Peptides were produced at 0.1 mmol scale by Fmoc solid-phase peptide synthesis. Protecting groups used were Lys/Trp/His(Boc), Ser/Thr/Tyr(tBu), Asp/Glu(OtBu), Asn/Gln/Cys/His(Trt), and Arg(Pbf). Dg1a, Dg2a, Dg4a, Dg5a and Dg7a were assembled on Polystyrene AM RAM (Rapp Polymere GmbH), Dg3a and Dk13a on Rink-amide ProTide resin (CEM, Matthews, NC), and Dg6a and Dk5a on preloaded Wang-polystyrene resins (ChemImpex, Wood Dale, IL). Peptides were assembled on a CEM Liberty Prime HT24 microwave synthesiser (CEM Corp, Matthews, NC, USA) using N,N'-diisopropylcarbodiimide (DIC)/oxyma and Fmoc groups were removed with 20% pyrrolidine, as per manufacturer protocols.
Peptides were cleaved from resin by treatment with 92.5% TFA/2.5% water/2.5% triisopropylsilane/2.5% 2,2ʹ-(ethylenedioxy)diethanethiol on a CEM Razor Rapid Peptide Cleavage System (CEM, Matthews, NC) for 30 min at 40ºC. Peptides were precipitated with 15 mL ice-cold ether, extracted in A/B 50/50 (A: 0.05% TFA; B: 90% ACN, 0.05% TFA) and lyophilised prior to purification. Peptides were purified on a LC-20AP HPLC system (Shimadzu Corp.) using a Agilent Zorbax 300SB-C18 column (150 × 21.2 mm; particle size, 5 μm) at 16 mL/min. Gradients used were 10–50% B over 40 min (Dg1a, Dg2a, Dg3a, Dg4a, Dg6a, Dg7a, Dk5a and Dk13a) and 20–70% B over 50 min (Dg5a). Fractions were lyophilised and purity assessed using LC–MS. Pure fractions of each peptide were lyophilised, pooled and stored at room temperature until use. Purified Dg1a and Dg2a were oxidised in a 0.1 M ammonium bicarbonate (0.5 mg/mL; pH 8.2; overnight), repurified and analysed as above.
Calcium imaging assay of vertebrate sensory neurons
DRG cells were isolated from 4 to 6-week-old male C57BL/6 mice that were purchased from the Animal Resources Centre (WA, Australia). DRGs were dissociated, then cells were plated in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, MD, USA) containing 10% fetal bovine serum (FBS) (Assaymatrix, VIC, Australia) and penicillin/streptomycin (Gibco) on a 96-well poly-D-lysine-coated culture plate (Corning, ME, USA) and maintained overnight. Cells were loaded with Fluo-4 AM calcium indicator, according to the manufacturer’s instructions (ThermoFisher Scientific, MA, USA). After loading (1 h), the dye-containing solution was replaced with assay solution (1 × Hanks’ balanced salt solution, 20 mM HEPES). Images were acquired at 10 × objective at 1 frame/s (excitation 485 nm, emission 521 nm). Fluorescence corresponding to [Ca2+]i of ~ 200 cells per experiment was monitored in parallel using a Nikon Ti-E Deconvolution inverted microscope, equipped with a Lumencor Spectra LED Lightsource. Baseline fluorescence was monitored for 30 s. At 30 s, assay solution was replaced with either assay solution, Ca2+-free assay solution with BAPTA (1 mM), or assay solution containing a cocktail of ion channel inhibitors [TTX (1 µM), Cd2+ (100 µM), amiloride (20 µM) and RR (20 µM)], then at 1 min with test peptide (10 µM in assay solution ± Ca2+-free assay solution or ion channel inhibitor cocktail) and monitored for 2 min before being replaced with assay solution and then KCl (30 mM; positive control). Experiments involving the use of mouse tissue were approved by the University of Queensland Animal Ethics Committee (UQ AEC; approval number TRI/IMB/093/17).
Pain behaviour experiments
Male adult (4–6 weeks old) C57BL/6 J mice used for behavioural experiments were purchased from the Animal Resources Centre (WA, Australia). They were housed in groups of up to four per cage, maintained on a 12/12 h light–dark cycle, and fed standard rodent chow and water ad libitum. Each peptide (600 pmol) diluted in saline containing 0.1% BSA was administered in a volume of 20 µL into the hindpaw by shallow intraplantar injection. Negative control animals were injected with saline containing 0.1% BSA. Following injection, spontaneous pain behaviour events were counted from video recordings, by a blinded observer. Experiments involving animals were approved by UQ AEC (approval number PHARM/526/18).
Insecticidal activity
A. mellifera worker bees were injected intrathoracically with 2-µL water (negative control) or 2 µL of whole venom (15 µg/g) or venom peptide (15 nmol/g). The injected bees were monitored for 10 min and then assessed at 1, 2, 4 and 16 h post-injection.
Black soldier fly (Hermetia illucens) larvae were purchased from Biosupplies (Yagoona, NSW, Australia). Late instar larvae, still actively foraging, were selected for assay and injected intra-abdominally with 2-µL water (negative control) or 2 µL of whole venom (30 µg/g) or venom peptide (3 nmol/g). The injected larvae were monitored for 30 min and then assessed at 24 h post-injection.
Data are expressed as mean ± SEM and are representative of three experiments. Data were fitted with a nonlinear regression with variable slope (four parameters) in GraphPad Prism 8.
Whole-cell voltage-clamp electrophysiology
HEK293 cells were cultured as previously described [41]. Cells were maintained on DMEM supplemented with 10% heat-inactivated FBS, 2-mM L-glutamine, pyridoxine and 110-mg/mL sodium pyruvate. Whole-cell patch-clamp experiments were performed using a QPatch 16X automated electrophysiology platform (Sophion Bioscience). The extracellular solution contained the following: 70-mM NaCl, 70-mM choline chloride, 4-mM KCl, 2-mM CaCl2, 1-mM MgCl2, 10-mM Hepes, and 10-mM glucose (pH 7.4; 305 mosmol). The intracellular solution contained the following: 140-mM CsF, 1 mM:5 mM EGTA/CsOH, 10-mM Hepes, and 10-mM NaCl (pH 7.3 with CsOH; 320 mosmol). From a holding potential of 0 mV, each recorded cell was subjected to a series of 50-ms voltage pulses that ranged from − 60 to + 60 mV in 10-mV increments. Recordings were made prior to and 4 min after the addition of either ECS (negative control) or peptide (10 µM). Data are mean ± SEM of five experiments and fitted to a simple linear regression.
Circular dichroism spectroscopy
Far-UV CD spectra were recorded for Dk5a and Dk13a (25 µM) in 100% H2O and in 20-mM SDS, using a Jasco J-810 CD spectropolarimeter (Easton, MD). Spectra were acquired from 260 to 185 nm, using a 1-mm path-length quartz cuvette, 1-nm step size, and averaging of five spectra.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Australian Research Council (Discovery Project DP190103787 to G.F.K., S.D.R, I.V.) and a National Geographic Society early career grant (EC-58468R-19 to S.D.R.). GFK is supported by Principal Research Fellowship APP1136889 from the Australian National Health & Medical Research Council. Henrik Y. O’Brien assisted with specimen collection. Apis mellifera were provided by Peter Ryan.
Author contributions
Conceptualisation, JOS, SDR; methodology, IV, JOS and SDR; investigation, TJ, AA, SHN, AHJ, JRD, JOS and SDR; writing—original draft, TJ, SDR; writing—review and editing, all authors; funding acquisition, IV, GFK and SDR; resources, IV, GFK, JOS and SDR; supervision, IV, GFK and SDR.
Funding
This work was funded by the Australian Research Council (Discovery Project DP190103787 to G.F.K., S.D.R, I.V.) and a National Geographic Society early career grant (EC-58468R-19 to S.D.R.). GFK is supported by Principal Research Fellowship APP1136889 from the Australian National Health & Medical Research Council.
Availability of data and materials
Precursor sequences of venom peptides have been deposited with GenBank under accessions MW323130–MW323158 (D. klugii), MW323159–MW323187 (D. bioculata), MW323188–MW323215 (D. gloriosa), MW323216–MW323243 (D. occidentalis), and MW323244–MW323300 (D. sicheliana). Raw sequencing data have been deposited in the NCBI sequence read archive under accessions SRR13038346 (D. klugii), SRR13038345 (D. gloriosa), SRR13038344 (D. bioculata), SRR13038343 (D. sicheliana), and SRR13038342 (D. occidentalis).
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Experiments involving animals or animal tissue were approved by The University of Queensland animal ethics committee (UQ AEC).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brothers DJ, Lelej AS. Phylogeny and higher classification of Mutillidae (Hymenoptera) based on morphological reanalyses. J Hymenopt Res. 2017;60:1–97. doi: 10.3897/jhr.60.20091. [DOI] [Google Scholar]
- 2.Schmidt JO, Blum MS. Adaptations and responses of Dasymutilla occidentalis (Hymenoptera: Mutillidae) to predators. Entomol Exp Appl. 1977;21(2):99–111. doi: 10.1111/j.1570-7458.1977.tb02663.x. [DOI] [Google Scholar]
- 3.Vitt LJ, Cooper WE. Feeding responses of skinks (Eumeces laticeps) to velvet ants (Dasymutilla occidentalis) J Herpetol. 1988;22(4):485–488. doi: 10.2307/1564347. [DOI] [Google Scholar]
- 4.Schmidt JO. The sting of the wild. Baltimore: Johns Hopkins University Press; 2016. [Google Scholar]
- 5.Schmidt JO, Blum MS, Overal WL. Hemolytic activities of stinging insect venoms. Arch Insect Biochem Physiol. 1983;1(2):155–160. doi: 10.1002/arch.940010205. [DOI] [Google Scholar]
- 6.Gall BG, Spivey KL, Chapman TL, Delph RJ, Brodie ED, Jr, Wilson JS. The indestructible insect: velvet ants from across the United States avoid predation by representatives from all major tetrapod clades. Ecol Evol. 2018;8(11):5852–5862. doi: 10.1002/ece3.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JS, Jahner JP, Forister ML, Sheehan ES, Williams KA, Pitts JP. North American velvet ants form one of the world’s largest known Mullerian mimicry complexes. Curr Biol. 2015;25(16):R704–706. doi: 10.1016/j.cub.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 8.John HA. Mimetic tiger beetles and the puzzle of cicindelid coloration (Coleoptera: Cicindelidae) Coleopt Bull. 1988;42(1):28–33. [Google Scholar]
- 9.Jonathan RM. Mimicry in Cleridae (Coleoptera) Coleopt Bull. 1994;48(2):115–125. [Google Scholar]
- 10.Analia AL, Río MGD. Taxonomy of the monotypic genus Trichaptus Pascoe (Coleoptera: Curculionidae: Entiminae), a potential weevil mimic of Mutillidae. Coleopt Bull. 2005;59(1):47–54. doi: 10.1649/683. [DOI] [Google Scholar]
- 11.Edwards G. Mimicry of velvet ants (Hymenoptera: Mutillidae) by jumping spiders (Araneae: Salticidae) Peckhamia. 1984;2(4):46–49. [Google Scholar]
- 12.Nentwig W. A mimicry complex between multillid wasps (Hymenoptera: Mutillidae) and spiders (Araneae) Studies on neotropical fauna and environment. 1985;20(2):113–116. doi: 10.1080/01650528509360679. [DOI] [Google Scholar]
- 13.Williams KA, Manley DG, Pilgrim EM, Von Dohlen CD, Pitts JP. Multifaceted assessment of species validity in the Dasymutilla bioculata species group (Hymenoptera: Mutillidae) Syst Entomol. 2011;36(1):180–191. doi: 10.1111/j.1365-3113.2010.00555.x. [DOI] [Google Scholar]
- 14.Schmidt JO, Blum MS, Overal WL. Comparative enzymology of venoms from stinging Hymenoptera. Toxicon. 1986;24(9):907–921. doi: 10.1016/0041-0101(86)90091-7. [DOI] [PubMed] [Google Scholar]
- 15.Baek JH, Lee SH. Differential gene expression profiles in the venom gland/sac of Eumenes pomiformis (Hymenoptera: Eumenidae) Toxicon. 2010;55(6):1147–1156. doi: 10.1016/j.toxicon.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Touchard A, Tene N, Song PCT, Lefranc B, Leprince J, Treilhou M, Bonnafe E. Deciphering the molecular diversity of an ant venom peptidome through a venomics approach. J Proteome Res. 2018;17(10):3503–3516. doi: 10.1021/acs.jproteome.8b00452. [DOI] [PubMed] [Google Scholar]
- 17.Lin N. Increased parasitic pressure as a major factor in the evolution of social behavior in halictine bees. Insectes Soc. 1964;11(2):187–192. doi: 10.1007/BF02222936. [DOI] [Google Scholar]
- 18.Jordan R. Die spinnenameise, Mutilla europaea, ein Bienenschadling! Deutsche Imker. 1935;48:421–427. [Google Scholar]
- 19.Brothers D. Biology and immature stages of Pseudomethoca f. frigida, with notes on other species (Hymenoptera: Mutillidae) Univ Kansas Sci Bull. 1972;50:1–38. [Google Scholar]
- 20.Piek T. Venoms of the hymenoptera: biochemical, pharmacological, and behavioural aspects. Orlando: Academic Press, London; 1986. [Google Scholar]
- 21.Hoffer E. Zur biologie der Mutilla europaea L. Zoologische Jahrbücher. 1886;1:679–686. [Google Scholar]
- 22.Mickel CE. Biological and taxonomic investigations on the mutillid wasps. Bull US Natl Mus. 1928;143:1–351. [Google Scholar]
- 23.Katayama E. Oviposition behavior of Mutilla mikado Cameron (Hymenoptera, Mutillidae), an ectoparasitoid of bumblebees (Hymenoptera, Apidae) Japanese Journal of Entomology. 2008;11(2):57–68. [Google Scholar]
- 24.Tosteson MT, Tosteson DC. The sting. Melittin forms channels in lipid bilayers. Biophys J. 1981;36(1):109–116. doi: 10.1016/S0006-3495(81)84719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okumura K, Inui K-I, Hirai Y, Nakajima T. The effect of mastoparan on ion movement in black lipid membrane. Biomed Res. 1981;2(4):450–452. doi: 10.2220/biomedres.2.450. [DOI] [Google Scholar]
- 26.Bazzo R, Tappin MJ, Pastore A, Harvey TS, Carver JA, Campbell ID. The structure of melittin: a 1H-NMR study in methanol. Eur J Biochem. 1988;173(1):139–146. doi: 10.1111/j.1432-1033.1988.tb13977.x. [DOI] [PubMed] [Google Scholar]
- 27.Inagaki F, Shimada I, Kawaguchi K, Hirano M, Terasawa I, Ikura T, Gō N. Structure of melittin bound to perdeuterated dodecylphosphocholine micelles as studied by two-dimensional NMR and distance geometry calculations. Biochemistry. 1989;28(14):5985–5991. doi: 10.1021/bi00440a040. [DOI] [Google Scholar]
- 28.Higashijima T, Wakamatsu K, Takemitsu M, Fujino M, Nakajima T, Miyazawa T. Conformational change of mastoparan from wasp venom on binding with phospholipid membrane. FEBS Lett. 1983;152(2):227–230. doi: 10.1016/0014-5793(83)80385-8. [DOI] [PubMed] [Google Scholar]
- 29.Robinson SD, Mueller A, Clayton D, Starobova H, Hamilton BR, Payne RJ, Vetter I, King GF, Undheim EAB. A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci Adv. 2018;4(9):eaau4640. doi: 10.1126/sciadv.aau4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazandjian TD, Petras D, Robinson SD, van Thiel J, Greene HW, Arbuckle K, Barlow A, Carter DA, Wouters RM, Whiteley G, Wagstaff SC, Arias AS, Albulescu LO, Plettenberg Laing A, Hall C, Heap A, Penrhyn-Lowe S, McCabe CV, Ainsworth S, da Silva RR, Dorrestein PC, Richardson MK, Gutiérrez JM, Calvete JJ, Harrison RA, Vetter I, Undheim EAB, Wüster W, Casewell NR. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science. 2021;371(6527):386. doi: 10.1126/science.abb9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brothers DJ. Alternative life-history styles of mutillid wasps (Insecta, Hymenoptera) In: Bruton MN, editor. Alternative life-history styles of animals. Netherlands, Dordrecht: Springer; 1989. pp. 279–291. [Google Scholar]
- 32.Peters RS, Krogmann L, Mayer C, Donath A, Gunkel S, Meusemann K, Kozlov A, Podsiadlowski L, Petersen M, Lanfear R, Diez PA, Heraty J, Kjer KM, Klopfstein S, Meier R, Polidori C, Schmitt T, Liu S, Zhou X, Wappler T, Rust J, Misof B, Niehuis O. Evolutionary history of the Hymenoptera. Curr Biol. 2017;27(7):1013–1018. doi: 10.1016/j.cub.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 33.Mickel CE. New species and records of nearctic mutillid wasps of the genus Dasymutilla (Hymenoptera) Ann Entomol Soc Am. 1936;29(1):29–60. doi: 10.1093/aesa/29.1.29. [DOI] [Google Scholar]
- 34.Manley DG, Pitts JP. Tropical and subtropical velvet ants of the genus Dasymutilla Ashmead (Hymenoptera: Mutillidae) with descriptions of 45 new species. Zootaxa. 2007;1487(1):1–128. doi: 10.11646/zootaxa.1487.1.1. [DOI] [Google Scholar]
- 35.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):1–16. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hale JE, Butler JP, Gelfanova V, You JS, Knierman MD. A simplified procedure for the reduction and alkylation of cysteine residues in proteins prior to proteolytic digestion and mass spectral analysis. Anal Biochem. 2004;333(1):174–181. doi: 10.1016/j.ab.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Vetter I, Lewis RJ. Characterization of endogenous calcium responses in neuronal cell lines. Biochem Pharmacol. 2010;79(6):908–920. doi: 10.1016/j.bcp.2009.10.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Precursor sequences of venom peptides have been deposited with GenBank under accessions MW323130–MW323158 (D. klugii), MW323159–MW323187 (D. bioculata), MW323188–MW323215 (D. gloriosa), MW323216–MW323243 (D. occidentalis), and MW323244–MW323300 (D. sicheliana). Raw sequencing data have been deposited in the NCBI sequence read archive under accessions SRR13038346 (D. klugii), SRR13038345 (D. gloriosa), SRR13038344 (D. bioculata), SRR13038343 (D. sicheliana), and SRR13038342 (D. occidentalis).