Abstract
The intestinal microbiota is critical for the development of gut-associated lymphoid tissues, including Peyer’s patches and mesenteric lymph nodes, and is instrumental in educating the local as well as systemic immune system. In addition, it also impacts the development and function of peripheral organs, such as liver, lung, and the brain, in health and disease. However, whether and how the intestinal microbiota has an impact on T cell ontogeny in the hymus remains largely unclear. Recently, the impact of molecules and metabolites derived from the intestinal microbiota on T cell ontogeny in the thymus has been investigated in more detail. In this review, we will discuss the recent findings in the emerging field of the gut-thymus axis and we will highlight the current questions and challenges in the field.
Keywords: Positive selection, Negative selection, Clonal deletion, Clonal diversion, Thymic regulatory T cells, Microbial metabolites, Antigen-presenting cells, Germ-free mice
Introduction
The mammalian gastrointestinal tract is a particularly unique environment due to the presence of extraordinary numbers of mostly commensal microbes that can include bacteria, fungi, viruses, and parasites. These microbes form our intestinal microbiota, and the collective genomes are referred to as the microbiome, which has a tremendous impact on the development, maturation, and regulation of many, if not all, of our organs, body systems, and physiology, particularly our immune system [1, 2]. The host–microbiota relationship is mostly mutualistic because the host provides a nutrient-rich niche to the microbiota, while the microbiota assists the host in digesting complex food molecules which provides important metabolites, such as short-chain fatty acids (SCFA), as well as essential vitamins B and K for example. Many studies in germ-free, gnotobiotic (defined microbiota), or antibiotic-treated mouse models have demonstrated a pivotal role of a healthy microbiome in the optimal functioning of the immune system, and this is also observed in human studies [3–5]. At homeostasis, the immune system and microbiota co-exist peacefully where innate and adaptive immune compartments are well regulated and respond to the microbiota appropriately without causing any harm to the host. However, this coalition is affected under various circumstances, such as the use of antibiotics, the consumption of processed food, and host genetics, leading to the dysregulated host immune responses toward microbiota. This can result in the development of severe immune-mediated chronic diseases, such as autoimmune diseases and inflammatory bowel diseases [2]. One important avenue by which the microbiome impacts not only the local mucosal but also the systemic immune system is through the production of microbial metabolites [6].
Whether microbial exposure impacts the incidence of immune-mediated disorders in humans is epidemiologically best described by the “hygiene hypothesis” which was first proposed by Strachan in 1989 after he observed a positive correlation between hay fever and families with higher cleanliness practice [7]. Later, this hypothesis was extended from the field of allergy to autoimmune diseases, such as type 1 diabetes and multiple sclerosis [8, 9]. Several epidemiological observations in humans and experiments in various mouse models embraced this hypothesis where the studies found that the reduced exposure to microbes leads to a dysregulated immune response [10]. For instance, children exposed to farming and high levels of endotoxins in early life showed a reduced incidence of allergies and autoimmunity [11, 12]. These observations were recapitulated in the germ-free mice that display a dysregulated immune phenotype reflected by very high serum Immunoglobulin (Ig) E levels for example [13, 14]. These findings suggested that the microorganisms around and colonizing us can protect against a spectrum of immune-mediated disorders.
The uterine environment and the amniotic fluid are generally considered sterile with no evidence of actively colonizing microbes present under healthy conditions. There is some controversy about this in the field [15, 16]. However, it has been shown that microbial metabolites derived from the mother’s microbiome can reach the fetus and influence the fetus’ immune system prenatally as well as postnatally and not only in adult life [17–19]. For example, Gomez de Aguero et al. [19] have shown that transient colonization during pregnancy increased the number of type 3 innate lymphoid cells (ILC3) and F4/80+CD11c+ mononuclear cells in the intestines of the offspring compared to pups that were born from control germ-free mother’s that were never exposed to intestinal microbes during pregnancy. Similarly, maternal microbiota-derived metabolites, such as SCFA, particularly acetate, have been shown to modulate the immune system of the lung during the prenatal stage to promote regulatory T cell (Treg) generation later in adulthood [20, 21]. Therefore, prenatal and postnatal exposure to microbiota-derived metabolites is critical to the development of a balanced immune system [22], and induction of appropriate immune regulation early in life has a long-term impact on the health of the offspring.
A prominent local effect of the gut microbiota is the influence on the development of gut-associated lymphoid tissues (GALT), including Peyer’s patches (PP) and crypto patches. Further, the influence of the microbiota on various organs, such as the liver, lung, and brain, in health and disease has been documented as well [23–25]. Because small amounts of microbial molecules, such as polysaccharide A (PSA), bacterial cell wall peptidoglycans (PGN), and metabolites, such as SCFA or retinoic acid (RA), get absorbed from the gastrointestinal tract into the bloodstream and transported into different peripheral organs, they can influence local as well as systemic immune responses [26, 27]. In support of this, broad-spectrum antibiotic treatment in mice before challenging systemically with lymphocytic choriomeningitis virus (LCMV) or intranasally by influenza virus, for example, impaired both innate and adaptive antiviral immunity [23, 28].
The impact of the intestinal microbiota on T cell development in the thymus is suggested in germ-free mice, which have reduced thymus size and cellularity [29]. The existing dogma of central tolerance and T cell development in the thymus suggests presentation of self-antigens by thymic antigen-presenting cells (APCs) is key in T cell selection; however, the recent findings suggest that microbial molecules and metabolites derived from the intestinal microbiota might also play a role in this process either directly (by the presentation of microbial epitopes) or indirectly (by modulating the selection process). Commensal metabolites could traffic from the mucosal surfaces to the thymus where they could modulate the development and maturation of T cells. In this review, we will discuss the recent findings on how the gut microbiota potentially influences thymus functions (the gut-thymus axis), and how bacterial molecules and metabolites derived from the intestine can determine the development of both conventional and unconventional T cells.
Overview on T cell development in the thymus
Conventional T cell development
Although thymic organogenesis is largely conserved between mice and humans, the thymic output significantly differs between mice and humans. In mice, the first wave of T cells that emerge from the thymus on embryonic day 15 consists of γδTCR T cells [30, 31]. Mice are born with a smaller αβTCR T cell population in the periphery and depend on T cell output from the thymus throughout life [32]. In contrast, thymic output in humans starts, relatively speaking, earlier than in mice at around 12–14 weeks of gestation and the first of wave of T cells consists of γδTCR and αβTCR T cells (including regulatory T cells) [33, 34] and the majority of T cell development in humans occurs in utero. Therefore, in humans, cardiac surgery-associated neonatal thymectomy does not lead to susceptibility to infections and autoimmune diseases due to the expansion of already existing naïve T cells in the periphery [32, 35].
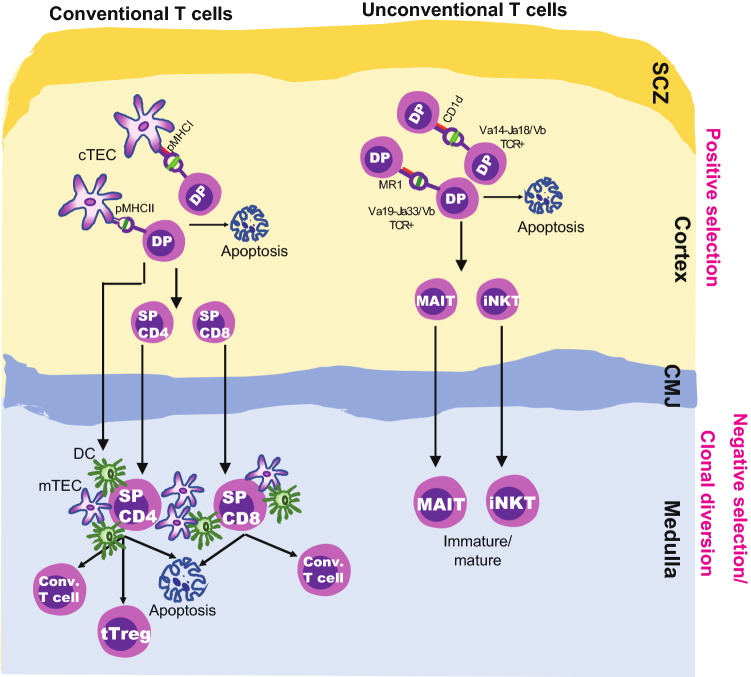
The thymus is a bi-lobular primary lymphoid organ consisting of two regions, the cortex and the medulla, and is composed of thymic epithelial cells (TECs), mesenchymal cells, endothelial cells, and dendritic cells (DCs). It provides a milieu that dictates the selection and maturation of T cells from lymphoid progenitors that have developed from hematopoietic stem cells in the bone marrow [36, 37]. The development of T cells starts with the seeding of common lymphoid progenitors (CLPs) in the parenchyma of the thymus close to the corticomedullary junction (CMJ). CLPs undergo a series of developmental stages: first, they differentiate into early thymic progenitors (ETP), also known as CD4– CD8– double-negative (DN) thymocytes; and later, DN thymocytes start to express the adhesion molecule, CD44, and cytokine receptor Interleukin (IL)-2Rα (CD25) and rearrange the genes for the δ, γ, or β T cell receptor (TCR) chains in the sub-capsular zone (SCZ) [38, 39]. The expression of recombination-activating gene1 (Rag1) and Rag2 at this stage is crucial for TCR assembly. A thymocyte with a successfully rearranged β chain pairs the β chain with the surrogate α chain to form the pre-TCR. Expression of the pre-TCR on DN cells leads to proliferation and development of double-positive (DP) thymocyte by expressing both CD4 and CD8 co-receptors. At this stage, Rag1 and Rag2 re-express for TCRα chain rearrangement. DP cells with successful αβTCR rearrangement undergo three potential fates, (a) death by neglect, (b) process of positive (in the thymic cortex), and (c) negative selection (in the thymic cortex and medulla) (Fig. 1). The event of death by neglect occurs when TCR fail to receive stimulation by APC. Positive selection in the thymic cortex aids the survival and differentiation of thymocytes that are able to recognize self-major histocompatibility complex (MHC) class I or II molecules on cortical thymic epithelial cells (cTEC) into single-positive (SP) CD8+ or CD4+ T cells, respectively. In addition, positive selection triggers the expression of CCR7 that aids in traveling of newly developed thymocytes into the medulla. In the cortex and medulla, the additional mechanisms of negative selection by clonal deletion and generation of thymic (or natural) factor forkhead box protein3 (FoxP3)+ Tregs by clonal diversion ensure that the positively selected T cells are tolerant to self-antigens. APCs (medullary TECs and DCs) provide a specialized microenvironment by expressing both ubiquitous and tissue-specific autoantigens (TSA), in an autoimmune regulator (AIRE)-independent and -dependent manner, respectively [40, 41]. Both mTECs and DCs are key regulators of negative selection and thymic Foxp3+ Treg development. Single-positive CD8+ or CD4+ thymocytes that recognize self-pMHCs with high affinity undergo negative selection or clonal deletion; this process is key in eliminating some, but not all, self-reactive T cells (Fig. 1). A combination of complex factors controls whether the fraction of CD4+ SP thymocytes that recognize self-pMHC undergoes clonal deletion or differentiates into thymic (or natural) Foxp3+ Tregs [42], which provides numerous potential targets for microbial-derived metabolites to regulate this process. Tregs generated in the thymus are known as natural or thymic Tregs (nTregs or tTregs, in this paper we use tTregs), which are effective in limiting many autoreactive immune responses that rise by T cells that escaped negative selection [43, 44]. tTregs express CD25 and transcription FoxP3 and Helios [44–47]. There are two distinct subsets of tTregs precursors (CD25+FoxP− and CD25−FoxP3+ SP CD4+ thymocytes) that have been identified in mouse thymus and these precursors lead to the development of CD25+FoxP3+ Tregs [48, 49].
Fig. 1.
Positive and negative selection of T cells in thymus. In thymus, positive and negative selection take place in the cortex and medulla, respectively. In conventional T cell compartment, DP (CD4+CD8+) thymocytes undergo positive selection after binding to self-pMHC I or II complexes on cTECs, then differentiate into SPCD8 or SPCD4 T cells, respectively. DP thymocytes which do not recognize or bind to pMHC with undergo apoptosis. SPCD8 or SPCD4 T cells undergo negative selection or clonal deletion if they recognize the pMHC with high-affinity on APCs such as mTECs and DCs; whereas the low and intermediate-affinity interactions lead to the development of conventional T cells and thymic Treg cells (tTreg; clonal diversion), respectively. In unconventional T cell compartment, MAIT and iNKT cells develop by positive selection of DP thymocytes by recognizing antigens on MR1 or CD1d molecules, respectively, on another DP thymocytes. Majority of MAIT and iNKT mature in the thymus but few leave thymus as immature cells but undergo maturation in peripheral tissues. Arrows in the figure indicates the sequence of the developmental stage or an event. DC dendritic cell, double negative, DP double positive, SP single positive, cTEC cortical thymic epithelial cells, mTEC medullary thymic epithelial cells, pMHC peptide major histocompatibility complex, MR1 non-classical MHC-related Ib molecule restricted to MAIT cells, CD1d non-classical MHC I-like molecule restricted to iNKT cells, MAIT mucosal-associated invariant T cell, iNKT invariant natural killer T cell, Treg regulatory T cell, CMJ corticomedullary junction, SCZ sub-capsular zone
Unconventional T cell development
Certain T cells possess functional traits of both innate and adaptive immune cells and are denoted as unconventional T cells. These cells are of limited TCR diversity and recognize cognate (non-peptide) antigens that are presented on non-classical MHC molecules, such as CD1d and MR1. Unconventional T cells mainly reside in the gut mucosa, liver, and skin and have the capacity to rapidly release cytokines in response to stimulation and play a central role in maintaining tissue homeostasis and in responding to stress conditions. Defects and deficiencies in unconventional T cells can lead to autoimmunity and chronic inflammatory conditions [50–52]. The key subsets of unconventional T cells are Mucosal-Associated Invariant T (MAIT) cells, invariant Natural Killer T (iNKT) cells, and γδT cells. Unconventional T cells develop in the thymus either from DN (γδT cells and iNKT cells) or DP thymocytes (iNKT and MAIT cells) [53–55].
MAIT cells express an invariant TCRα chain (mice, Vα19Jα33; human, Vα7.2Jα33) and have a biased TCRβ chain repertoire to recognize vitamin B2 precursor derivatives [5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU)] of bacteria and yeast or self-antigen presented on non-classical MHC-related Ib molecule MR1 within the thymus [56, 57]. They have been implicated in both intestinal inflammation and homeostasis [58, 59]. Their development depends on the transcription factor promyelocytic leukemia zinc finger (PLZF) and it occurs in three consecutive stages (S1, S2, and S3, indicated based on the CD24 and CD44 expression) [60, 61]. The selection of MAIT cells depends on the type of exogenous antigens presented on DP thymocytes [53, 62, 63]; however, recent work showed the role of TECs in their selection [64]. Unlike conventional CD4+ and CD8+ T cells, which develop into naïve T cells and acquire effector functions only after antigen encounter in secondary lymphoid organs, MAIT cells develop and mature into T-bet+ MAIT1 or RORγt+ MAIT17 effectors already in the thymus (Fig. 2B) [60, 61, 64]. Unlike for iNKT cells, the negative selection has not been studied for MAIT cells.
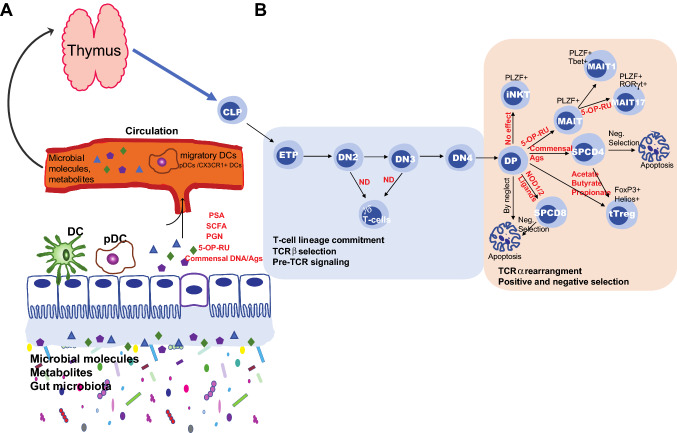
Fig. 2.
The effect of microbes, microbial molecules, and metabolites on the development of T cells in thymus. A The microbial molecules and metabolites such as PGN, PSA, DNA, antigens and SCFA produced in the intestine translocate into the thymus via circulation, possibly by migratory DCs such as pDCs and CX3CR1+ DCs, where they could influence the development of both conventional and unconventional T cells. B Overview on the developmental stages of T cells and the effect of microbial molecules or metabolites on T cell development in the thymus. The development of unconventional and conventional T cells starts by seeding CLP that is derived from the hematopoietic stem cell (HSC) of bone marrow into the thymus. The CLP could differentiate into ETP. T cell lineage-committed ETP develops though four DN stages (DN1 through DN4); at DN2 and DN3 stages thymocytes could progress into either TCRγδ or TCRαβ rearrangement (light blue box). TCRαβ committed CD4+CD8+ DP thymocytes undergo the process of positive and negative selection. DP thymocytes could develop into either iNKT, MAIT, SPCD4, SPCD8, or tTreg. Thymocytes that do not recognize self-pMHC complex or bind to self-pMHCs with high-affinity undergo death by neglect or by apoptosis, respectively (light orange box). The effect of different microbial molecules or metabolites on the development of unconventional and conventional T cells are denoted (red fonts) in the figure. Arrows in the light orange or green box indicates the sequence of the developmental stage or an event. DC dendritic cell, TCR T cell receptor, PGN peptidoglycan, PSA polysaccharide A, SCFA small-chain fatty acid, NOD1/2 nucleotide-binding oligomerization domain protein 1 or 2, 5-OP-RU 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil, CLP, ETP early thymic progenitors, DN double negative, DP double positive, SP single positive, MAIT mucosal-associated invariant T cell, iNKT invariant natural killer T cell, tTreg thymic regulatory T cell, PLZF promyelocytic leukemia zinc finger, FoxP3 Forkhead box protein3, ND not determined
iNKT cells express semi-invariant TCRs (mice, Vα14-Jα18/Vβ8.2/Vβ7/Vβ2; humans, Vα24-Jα18/Vβ11) and recognize lipid-based antigens presented on class I-like MHC molecule CD1d [65, 66]. Neonatally thymectomized and athymic mice are devoid of iNKT cells, suggesting their development takes place in the thymus after birth [67, 68]. Like MAIT cells, iNKT developmental selection is dependent on DP thymocytes presenting lipid antigens on CD1d molecules [69–71]. iNKT cell selection is also known as agonist selection because they require strong TCR signals during their development, and it results in the upregulation of two key transcription factors, Egr2 and PLZF [72–74]. Following DP thymocyte-dependent positive selection, iNKT cells are likely to undergo negative selection because it is evident in mice where the injection of α-galactosylceramide (α-GalCer, a sphingolipid that acts as iNKT cell ligand), into neonatal mice, led to the loss of developing iNKT cells. The negative selection was mainly mediated by CD1d-expressing DCs. Importantly, the selection of iNKT cells in the thymus is mainly driven by the recognition of endogenous antigens. The majority of iNKT cells complete their development in the thymus and then migrate into different peripheral tissues or organs, albeit few egress thymus and complete their maturation in the peripheral tissues [68, 75].
In the end, conventional and unconventional T cells that survive the thymic selection process egress from the thymus into the periphery.
The effect of microbiota on conventional T cell development
Mounting evidence in mice suggests a functional impact of microbiota-derived metabolites and molecules on T cell ontogeny in the thymus. Germ-free mice display general lymphopenia and also reduced thymus size and cellularity compared to colonized specific pathogen-free (SPF) mice. This supports the notion that the microbiota can influence thymic events as well. Mature T cells recirculate into the thymus from the periphery, albeit in much lower numbers as compared to T cells that mature in the thymus [76, 77]. Therefore, reduced thymic size and cellularity found in germ-free mice were apparently related to the absence of microbiota [29, 78, 79]. In support of this, the depletion of gut microbiota by oral antibiotics led to reduced thymic cellularity, particularly reduced number of CD4+ and Treg cells in SPF mice, but not in germ-free mice, suggesting the critical role of microbiota on T cell development in the thymus [80].
Nakajima et al. addressed the role of microbial molecules on the development of αβTCR T cells in mice [81]. They found a significant difference in the expression of the transcription factor AIRE, which is important for TSA expression in the thymus. AIRE was reduced at the mRNA and protein level in TECs of germ-free compared to SPF mice [81]. However, this study has not found significant differences in thymocytes numbers and their subsets (DN, DP, SPCD4, and SPCD8). SPF mice used in the studies were generated by fostering the newborn germ-free pups by SPF mothers. Nucleotide-binding oligomerization domain protein 1 (NOD1) is a pattern recognizing receptor sensing PGN-derived molecules and NOD1 is expressed in myeloid and epithelial cells. Nakajima et al. found that mean fluorescence intensity (MFI) and mRNA expression of AIRE were significantly reduced in mTECs that were derived from the germ-free mice as compared to SPF. This suggests that commensals are important for augmenting AIRE expression in mTECs. In a separate experiment, the authors found that SPF NOD1-deficient mice displayed a reduced AIRE expression in TECs as compared to NOD1-expressing WT mice. These results suggest, though this experiment was not performed in germ-free conditions, the recognition of microbial components by TECs is important for AIRE expression [81]. The Receptor Activation of Nuclear factor Kappa B Ligand (RANKL) produced by thymocytes binds to the RANK receptor on mTECs, and this binding enhances the expression of AIRE [82–84]. Nakajima et al. also showed AIRE expression in TECs was mediated by upregulation of RANKL in the thymocytes in response to NOD-1 ligand Staphylococcal enterotoxin B [81]. Both NOD1 and 2 are involved in promoting the positive selection of SP CD8 thymocytes. NOD1 and 2-deficient mice showed a reduction in SP CD8 cells in the thymus as compared to wild-type mice and this reduction was due to the intrinsic requirement of NOD ligands recognition by NOD1 or 2 for efficient TCR-mediated extracellular signal-regulated kinase (ERK) phosphorylation, which is a key element for T cell differentiation and proliferation [85].
The effect of the human commensal microbe Lactobacillus reuteri on the thymus has also been studied in mice [86, 87]. L. reuteri, originally isolated from human breast milk, has been used in probiotics. L. reuteri might have a beneficial effect in infectious and non-infectious gastrointestinal tract disorders in mice and humans [88, 89]. The use of L. reuteri in probiotics increased thymus size and cellularity mainly by upregulating transcription factor forkhead box protein N1 (FoxN1) in the TECs of C57BL/6 ApcMIN (APNMin/+) mutant mice that are predisposed to develop cancer-associated cachexia (wasting syndrome) due to higher levels of IL-6 [86]. FoxN1 induces the differentiation of TECs by regulating the expression of numerous transcription factors that are essential for thymus organogenesis [90–93]. However, this study has not addressed whether L. reuteri has any effect on enhancing the generation and survival of DP thymocytes, Tregs development, or increasing T cell repertoires in C57BL/6 ApcMIN (APNMin/+) or in wild-type C57BL/6 mice. Also, this study has not used any other probiotic microbes/bacteria as a control to rule out the observed effect is specific to L. reuteri.
The microbiota composition impacts the development of immune cells and their functions. In addition, maternal microbiota not only impacts the fetal and postnatal immune development [19, 94, 95] but also modulates fetal thymic and Treg development [79, 96]. Increased intake of dietary fibers during pregnancy and lactation in mice increased SCFA in the serum, which in turn favored tTreg differentiation in the offspring mainly via G protein-coupled receptor 41-mediated upregulation of AIRE in the mTEC. High percentages of FoxP3+ tTregs were found in the spleen and thymus of offspring derived from high-fiber diet (HFD)-fed SPF mice as compared to low-fiber diet-fed mice [79]. However, it is unclear whether the high frequencies of tTregs in offspring are due to fetal exposure to the maternal SCFA or the direct exposure of offspring to the mother’s HFD-altered microbiota after the birth as the study did not use any auxotrophic commensals in pregnant mothers or germ-free controls.
The influence of maternal microbial products, particularly SCFA, on the organogenesis of thymus and generation of tTregs has also been recently studied under preeclampsia condition in humans. Preeclampsia is a pregnancy-associated immune disorder that affects maternal–fetal tolerance. Consequently, offspring are predisposed to atopic and cardiovascular diseases [97–99]. Reduced thymus diameter in the fetus is considered as one of the parameters to predict preeclampsia and its adverse effect on the fetal or postnatal immune system in humans [100, 101]. An important study by Hu et al. has shown the link between low maternal serum acetate and reduced fetus thymic size, cellularity, and architecture as well as tTreg development in preeclampsia. Importantly, longitudinal monitoring from infancy to early childhood of the children born to preeclamptic mothers indicated a persistently reduced naïve Tregs during the first four years of early life, suggesting the importance of maternal commensal metabolites on tTregs development [96]. This study also investigated the influence of maternal acetate on the development of fetal T cell development by feeding germ-free pregnant mice with acetate. The supplementation of acetate in the drinking water during pregnancy increased total CD4+ T cells and promoted stable expression of AIRE in mTECs of offspring [96].
A recent study in murine models showed the role of microbiota in the development of microbiota-specific T cells in the thymus [80]. The colonization of Segmented Filamentous Bacteria (SFB)-negative SPF mice (young vs adult) with SFB or Escherichia coli (E. coli) led to the trafficking of microbial-specific antigens from the intestine to thymus by CX3CR1+ DCs. Antigen presentation by these DCs induced the expansion of microbiota-specific T cells in the thymus as measured by SFB-3340 protein-specific tetramer. The expansion of antigen-specific T cells in the thymus was predominantly found in young mice that were colonized with SFB than in adult mice. Interestingly, SFB colonization into TCR transgenic mice (expressing SFB-specific TCR) also increased the total number of thymic CD4+ T cells. The expanded antigen-specific T cells were not regulatory in nature (did not express FoxP3 and CD25) but expressed variable levels of CD44 and CD69, which is indicative of their positive selection and antigen experience. The study has used mice expressing green fluorescent protein (GFP) under the control of the Rag2 gene promoter, which identifies recent thymic emigrants (RTE) in the periphery [102]. Importantly, in Rag2-GFP mice, this study found that the expanded thymic SFB-specific CD4+ T cells were GFP+CD73–, indicating that these are likely not recirculating mature T cells but rather RTEs [80].
Together, from the above discussion, it is clear that the intestinal microbiota and its molecules and metabolites influence T cell and Treg development mainly in two ways, (a) enhancing the expression of two transcription factors, AIRE and FoxN1, in TECs and (b) inducing the proliferation of antigen-specific T cells. The effect of different microbial metabolites or molecules on the development of unconventional and conventional T cells found in different studies is listed in Table 1.
Table 1.
The effect of microbial metabolites or molecules on the development of unconventional and conventional T cells in the thymus
| Type of T cell | MHC restriction | TCR repertoire | Positive selection | Selecting cells | Commensal, molecules, or metabolite involved | Mode of transport to thymus | Mechanism involved in selection | References |
|---|---|---|---|---|---|---|---|---|
| MAIT cells | MR1 | Vα19Jα33/ Vβ8, Vβ6 | Yes | DP thymocytes and TECs | 5-OP-RU, a vitamin B2 precursor | By circulation | Presentation of 5-OP-RU by DP thymocytes and TECs for the positive selection | [78] |
| iNKT cells | CD1d | Vα14-Jα18/Vβ8.2, Vβ7, Vβ2 | Yes | DP thymocytes | Development independent of microbiota | – | – | [78, 105] |
| γδ T cells | ND | Vγ1.1Vδ6.3 | No | ND | ND | – | – | |
| T cells /Tregs | MHC I or MHC II | Diverse | Yes | TECs and APCs | SCFAs acetate, butyrate, and propionate | By circulation | G protein-coupled receptor 41-mediated upregulation of AIRE in mTECs | [79] |
| Lactobacillus reuteri | Unknown | Upregulation of FoxN1 in TECs | [86] | |||||
| Acetate | By circulation | Upregulation of AIRE expression in mTECs | [96] | |||||
| Peptidoglycan (PGN) | By circulation | Upregulation of AIRE expression in TECs | [81] | |||||
|
Segmented Filamentous Bacteria (SFB) E. coli |
By circulation/CX3CR1 + DCs | Presentation of microbiota-derived Ags to expansion cognate thymic T cells | [80] |
MAIT mucosal-associated invariant T cells, iNKT invariant natural killer T cells, TCR T cell receptor, MR1 non-classical MHC-related Ib molecule restricted to MAIT cells, CD1d non-classical MHC I-like molecule restricted to iNKT cells, Tregs regulatory T cells, MHC I or II major histocompatibility complexes I or II, TECs thymic epithelial cells, mTECs medullary thymic epithelial cells, APCs antigen-presenting cells, DP double positive, pDCs plasmacytoid dendritic cells, PGN peptidoglycan, PSA polysaccharide A, SCFAs small-chain fatty acids, 5-OP-RU 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil, AIRE autoimmune regulator, FoxN1 Forkhead box protein N1, ND not determined
The effect of microbiota on unconventional T cell development
MAIT cells are present in lower numbers in the thymus, spleen, lung, and colon of germ-free mice as compared to SPF mice, suggesting the involvement of foreign antigens in their selection and development [56, 60, 78]. Colonization of germ-free mice with a human or SPF microbiota was able to restore the MAIT cells to SPF levels but not to the higher levels as found in humans [103, 104]. A recent study reported that vitamin B2 precursors derived from the intestinal commensals get transferred to the thymus and presented by DP thymocytes to drive the positive selection of MAIT cells [78]. Besides, in germ-free mice, heat-stable antigen (HSA)-expressing immature MAIT cells (HSA+CD44–) were very low, and only a few cells reached a mature stage (HSA– CD44+) while expressing optimal levels of PLZF, suggesting a very few matured MAIT cells could be developed against self-antigens in germ-free mice. Further, these mice contain low levels of RORγt+ MAIT17. Most importantly, cohousing of germ-free mice with SPF mice resulted in increased frequencies of MAIT cells, particularly MAIT17, first in the thymus, and later in the spleen and lungs, suggesting the importance of commensals for the development of MAIT17 cells in the thymus [78]. Thus, this work demonstrates that commensal products, such as the derivatives of vitamin B2, influence the gut barrier homeostasis by guiding the maturation of MAIT cells in the thymus.
The role of microbiota on the ontogeny of iNKT cells was studied almost 20 years ago by Park, S.H. et al.in mice. The study found that iNKT cells do not alter in their frequency, phenotype, and functions in germ-free C57BL/6 mice as compared to SPF mice [105]. The analysis of iNKT cells was done in the study before the advent of CD1d tetramer and based on the expression of the NK1.1, CD4, and αβTCR, which do not reflect the true frequencies of iNKT cells. However, recent studies also reiterated the same findings in germ-free mice [61, 78, 106]. The development of iNKT cells in the thymus was unaffected in the absence of the microbiota in germ-free mice, but their frequencies were increased in the small intestine and colon with less mature and hypo-responsive phenotype, suggesting the role of commensal-derived antigens in iNKT functions. Although MAIT and iNKT cells share similarities in thymic differentiation, their dependency on gut microbiota-derived antigens for their development differs in that MAIT cells rely on microbial metabolite but not iNKT cells [78, 105]. This suggests that their development could be mainly dependent on CD1d-mediated self-antigens presentation by DP thymocytes.
Unlike unconventional T cells (iNKT, MAIT, and γδ T cells), ILCs do not express unique antigen-specific receptors or undergo clonal selection, but they do transcriptionally and functionally mirror αβTCR T cell subsets. ILCs react rapidly to the stimuli of infected or injured tissues by releasing an array of cytokines, which exert immune responses that are directed against infection or injury. ILCs are divided into four subsets: the conventional lineage consists of T-bet- and Eomes-expressing cytotoxic NK cell subsets and the ILC-1, ILC-2, and ILC-3 subsets, which express T-bet, GATA-3, and RoRγt and secrete IFNγ, type 2 cytokines (IL-4, IL-5 and IL-13), and IL-17, respectively [107–109]. A common multilineage progenitor that differentiates into ILC subsets expresses the signature transcription factor, PLZF, which is also expressed by iNKT and MAIT cells [110–112]. Intestinal microbes influence the function of the innate immune system in early life [19, 113]. In the periphery, the influence of the gut microbiota on the frequencies of ILC subsets is widely studied in germ-free mice as well as in mice treated with antibiotics [114–116]. In the thymus, a recent study revealed a pivotal role of intestinal microbes, particularly PSA-expressing Bacteroides fragilis, in the development of ILC precursors in the thymus [29]. In this study, germ-free mice were monocolonized with PSA-expressing and PSA-deficient B. fragilis. PSA-expressing B. fragilis monocolonization restored thymic and splenic cellularity to the levels of SPF mice, but PSA-deficient B. fragilis did not. Interestingly, this study found a higher percentage of PLZF+ thymocytes in PSA-expressing B. fragilis colonized mice compared to germ-free mice or mice colonized with PSA-deficient B. fragilis. The deletion of plasmacytoid dendritic cells (pDCs) in BDCA2-DTR mice reduced the levels of PLZF+ innate-like cells in the thymus compared to wild-type mice. Therefore, migratory pDCs were considered to be the major entero-thymic communicators, though the precise mechanism involved in the transportation of intestinal microbial components was not described [29].
The transcription factor PLZF is expressed by several innate T cell lineages, including iNKT, ILC precursors, MAIT, and Vγ1.1/Vδ6.3+ NKT cells [73, 110–112, 117]. Its expression provides IFNγ and IL-4 secretion ability to these cells and their migration from the thymus to non-lymphoid tissues. Interleukine-2 (IL-2) inducible T cell kinase (Itk) is a member tyrosine kinases family, expressed in both naïve and activated T cells, and is participated in TCR signaling. Therefore, Itk-gene-deficient (Itk−/−) mice present decreased T cell numbers and proliferation in the periphery due to impaired TCR-induced Ca2+ mobilization in the absence of Itk [118]. Interestingly, these mice harbor increased numbers of PLZF-expressing innate CD4+PLZF+ T cells (αβTCR unconventional T cells) in the thymus and their development in the thymus was independent of CD1d or MR1. The treatment of Itk−/− mice with antibiotics decreased the frequencies of innate CD4+PLZF+ T cells in the thymus, suggesting their expansion is dependent on the gut microbiota [111].
During T cell ontogeny, γδTCR T cells are the first T cells to appear in the thymus in mice and humans with the ability to secrete copious amount IFNγ [30, 31]. These cells primarily participate in immune protection and immune regulation of neonates because of their tropism to epithelial tissues. Unlike αβTCR T cells, γδTCR T cells do not completely dependent on MHC-I or II restricted antigen recognition. γδTCR T cells respond to a variety of stimuli, including toll-like receptors (TLRs). The protective role and frequencies of γδTCR T cells in the intestinal surface are greatly impacted by microbiota [119]. However, until now, the role of the microbiota and microbiota-derived molecules or metabolites in the ontogeny of γδTCR T cells in the thymus is not well understood.
Potential routes for the transportation of microbial components to the thymus
The absorbed molecules and metabolites from the gut microbiota circulate and migrate into various distant lymphoid organs and tissues, including the thymus. Almost 40 years ago, seminal work by Raviola and Karnovsky, using different molecular sizes of electron opaque tracers, has demonstrated that blood-borne antigens can reach the thymic corticomedullary space by venules but not the thymic cortex region as it is surrounded by impermeable capillaries [120]. Recently, a study by Uchimura et al. used 13C radio-labeled E. coli HA107 to assess the penetration of microbial metabolites in host tissues. Interestingly, it was found that (at both 2 h and 18 h after gavage) a wide range of 13C-labeled bacterial metabolites had penetrated almost all fluids and organs, including the thymus [121].
The blood–thymus barrier permits low molecular weight proteins or particles, but not high molecular proteins, to enter the thymus [122, 123]. Consequently, blood-borne antigens get trapped in the perivascular system of the thymic medulla where they get presented by thymic stromal cells, mainly TECs. In addition, the thymic medulla conduit system—the structure of collagen fibers surrounded by basement membrane is enwrapped by thymic stromal cells—creates a specific milieu for the development of thymocytes where small blood-borne molecules get captured by TECs [124]. Passage of blood-borne antigens to thymus is well defined in recent studies where peptides and proteins were used for intravenous administrations to follow their migration to the thymus; intravenously injected ovalbumin peptides and hen egg-white lysozyme entered the thymus and induced clonal deletion of thymocytes in TCR transgenic models [125–127]. Thus, commensal antigens, metabolites, or products can potentially enter the thymic environment via the blood–thymus barrier or by the conduit system where APCs, such as epithelial cells and DCs, could present the antigens into thymocytes, thereby influencing the ontogeny of T cells.
Another potential mode of transportation of microbial molecules from the gut to the thymus is by DCs, particularly by pDCs and CX3CR1+ DCs. pDCs are known to produce type 1 interferon in response to viral infections and are defined as CD11c+, CD4+, MHC-II+, and BTLA+ (mouse, B220+, BST2+, SiglecH+, Ly6C+; human, CD303+ CD304+) cells. Although they play an important role in viral immunity in the periphery, pDCs induce tolerance by deleting autoreactive thymocytes and favoring the generation of Tregs in the thymus [128–131]. CCR9 is a chemokine receptor involved in the homing of progenitor T cells into the thymus and also T cells into the small intestine [132, 133]. Tolerogenic (immature) pDCs present in the lymphoid tissues express CCR9, demonstrating their potential to migrate into the thymus [133, 134]. Consistent with this, pDCs carry soluble blood-borne antigens from the periphery into the thymus, and their antigen presentation contributes to the deletion of antigen-specific thymocytes [129, 135, 136]. In the thymus, mTEC cells express many but not all TSA in an AIRE-dependent manner. pDCs are known for their increased endocytic activity, and hence, they could present many peripheral endocytosed antigens, including TSA and innocuous antigens derived from microbiota [137]. Studies have found that pDCs play an important role in exposing thymocytes to commensal antigens or metabolites, and impact the development of thymocytes [29, 129]. From these observations, it is possible, yet to be tested, that pDCs carrying commensal peptide antigens could have a role in inducing central tolerance either by eliminating commensal-reactive thymocytes or by inducing commensal-specific Tregs in the thymus.
The CX3CR1+ DCs can be found in the intestine and continuously patrol the surrounding environment and capture luminal antigens through dendritic extensions [138, 139]. A recent study by Zegarra-Ruiz et al. found the role of CX3CR1+ DCs in the transportation of microbial DNA and antigens from the intestine into the thymus. The study detected the microbial DNA related to a broad range of microbial phyla (for example, SFB or E. coli) in the thymus and other organs (liver and heart) of SPF mice, using 16s rDNA sequencing. The 16s rDNA found in the thymus overlapped with fecal microbial phyla. The oral antibiotics treatment in mice reduced the bacterial DNA load in different organs, including the thymus [80]. The depletion of CX3CR1+ DCs and also ablation of MHCII antigen presentation on CX3CR1+ DCs in mice reduced the level of bacterial 16s rDNA and reduced microbiota-specific T cell expansion in the thymus, respectively, suggesting the role of CX3CR1+ DCs in trafficking gut-microbial molecules into the thymus [80]. This finding on the role of CX3CR1+ DCs is novel but in contrast to well-recognized functions of thymic APCs (TEC and DCs) where they are involved in clonal deletion and induction of tTregs instead of inducing proliferation of T cells in an antigen-specific manner.
TLRs are expressed on various immune cells, including TECs, resident or circulating DCs, and thymocytes. Recently, Voboril et al. [140] showed that CD11c+ DC, mTEChi (TLR2, 3, 4, 5, and 9), and mTEClow (TLR2, 3, and 9) express different TLRs at various levels. For example, TLR9 is highly expressed in mTEChi. Notably, this study revealed that the recognition of TLR9 ligands by mTECs (TLR/MyD88 signaling activation) is important for the generation of tTregs. This effect was mediated by TLR-induced chemokine production by mTECs that led to the recruitment of CD14+ monocytes (CD14+moDC) from the periphery. CD14+moDC differentiated into CD14+ Signal regulatory proteinα (Sirpα)+ conventional type 2 DC (cDC2: CD14+ Sirpα+cDC2). Furthermore, MyD88-deficient TECs failed to exhibit the recruitment of CD14+moDCs, resulting in reduced tTreg output and functionality. These findings illustrate the importance of sensing TLR ligands by mTECs in the thymus.
Open questions in the field and concluding remarks
Early-life exposure to microbiota is important for a balanced regulation and education of the innate and adaptive immune systems [141, 142]. This is also evident for the thymus with recent emerging work in the field. It will be important to better understand to what extent the influence of the microbiota on T cells ontogeny in the thymus has an impact on health and diseases, such as autoimmune diseases or inflammatory bowel diseases for example. TCR repertoire analyses of colonic T cells indicated that colonic T cells utilize a different TCR repertoire than T cells from other secondary lymphoid organs at steady state and this repertoire is affected by broad-spectrum antibiotics, suggesting a role of intestinal bacteria in shaping the TCR repertoire [143, 144]. Based on these findings one could hypothesize that there are differences in the peptide repertoire presented in the thymi of SPF and germ-free mice, resulting in different peripheral TCR repertoires, which might be more diverse in SPF mice.
In addition, it is also important to better understand the role of epitope/peptide mimicry between a commensal-derived and self-antigen on T cell ontogeny. The commensal microbiota is the source of an enormous amount of peptide antigens. A handful of studies demonstrated the presence of peptide mimicry between gut microbiota and autoantigens that are targeted in autoimmune diseases [145–147]. For instance, E. coli express a peptide mimic to pyruvate dehydrogenase complex-E2 (PDC-E2) of humans that activates CD4+ and CD8+ T cell response and promote primary biliary cholangitis (PBC) [148, 149]. Similarly, Bacteroides thetaiotaomicron expresses a peptide mimic to myosin heavy chain 6 (MYH6) a motor protein in humans that activates CD4+ T cell response and promotes cardiomyopathy [147]. Recent studies also demonstrated that pDCs and CX3CR1+ DCs are involved in the transportation of commensal-related molecules/proteins from the intestine to the thymus [29, 80]. Therefore, it would be interesting to understand the consequences of the presentation of commensal peptide mimics to autoantigens on T cell ontogeny. Such presentation would lead to either (a) the proliferation of antigen-specific T cells as found in Ref. [80], (b) negative selection of T cells, or (c) tTreg cells development. These consequences would be dependent on numerous factors, such as the amount of peptide/protein transported into the thymus, the affinity of the peptide presented on APCs, the type of APCs involved in the antigen presentation, and the genetics of the host. Notably, this new paradigm would challenge the existing dogma of central tolerance and T cell development in the thymus where the presentation of self-antigens by thymic APCs is considered to be paramount in T cell ontogeny.
It is also important to consider the role of the microbiota at other body sites, such as the skin or airways. A recent study found the transportation of commensal-derived molecules from the skin to the thymus [78]. Application of the vitamin B2 precursor derivative 5-OP-RU onto intact skin of mice resulted in activated thymic and splenic MAIT cells. Therefore, one can also speculate that peptides or commensal molecules present on the skin and in the airways can reach the thymus and affect the development of T cells. A detailed understanding of the cellular and molecular mechanisms involved in this putative phenomenon would be required to design novel therapies to treat various autoimmune diseases or inflammatory conditions.
A number of diverse natural ligands, such as riboflavin-based and folic acid-based ligands and synthetic analogs, such as uracil analogs, that bind to MR1 have been identified [150, 151]. These ligands can induce both weak and potent activation of MAIT cells. Because vitamin B2 precursors derived from intestinal commensal bacteria have been shown to be involved in positive selection of MAIT cells in the thymus [78], it would be interesting to understand the role of other natural MR1 ligands, for instance, folic acid-based ligands. Likewise, a variety of soluble mediators, such as TLR ligands, retinoic acid, and lactate, are also produced by intestinal commensal bacteria. There are receptors expressed to sense these microbial molecules or mediators on different cells in the thymus, including thymic resident DCs, mTECs, cTECs, as well as thymocytes (both conventional and unconventional T cell precursors) [140, 152]. A detailed understanding of various microbial molecules on the various thymic cell types would further improve our knowledge of how our intestinal microbiota shapes host immunity.
Furthermore, T cells complete their maturation in the periphery in secondary lymphoid organs after egressing from the thymus as recent thymic emigrants (RTEs). Maturation into mature naïve (Mn)T cells in the periphery takes about 2–3 weeks. RTEs are hypo-functional versions of mature T cells because they possess the diminished proliferative capacity and produce lower levels of cytokines, such as IL-2, IFNγ, TNFα, IL-4, and IL-17 under Th1 and Th17 polarizing conditions. There are many cell-intrinsic and -extrinsic factors, such as DCs and intact lymphoid structure, that play important role in the transition of RTEs into MnT cells [153–156]. However, there are currently no studies that have addressed the role of intestinal commensals bacteria on the maturation of RTEs in secondary lymphoid organs.
In conclusion, recent discoveries in the field of the gut-thymus axis have uncovered the role of commensals in the development of both conventional and unconventional T cells. This opens up many new questions, such as whether there is any effect on the development and maturation of thymic APCs particularly on resident DCs and cortical and medullary TECs by commensal-derived metabolites reaching the thymus for example. Germ-free and gnotobiotic infrastructure in combination with the ability to genetically manipulate not only the host (mouse) but also commensal bacteria will allow us to experimentally address these questions.
Author contributions
MBG and RHN contributed to conceptualization:. RHN, CSU, and MBG were involved in writing and editing.
Funding
This work was supported by the CIHR grant (PJT-156073) to M.B. Geuking.
Data availability
Not applicable.
Declarations
Conflict of interest
The authors have no relevant financial and non-financial interests to disclose.
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4(137):13rv77. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshmukh HS, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20(5):524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F, et al. The microbiota maintain homeostasis of liver-resident gammadeltaT-17 cells in a lipid antigen/CD1d-dependent manner. Nat Commun. 2017;7:13839. doi: 10.1038/ncomms13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Perez G, et al. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J Immunol. 2016;196(9):3768–3779. doi: 10.4049/jimmunol.1502322. [DOI] [PubMed] [Google Scholar]
- 6.Geuking MB, Burkhard R. Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol. 2020;13(6):855–866. doi: 10.1038/s41385-020-00335-w. [DOI] [PubMed] [Google Scholar]
- 7.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 9.Azad MB, et al. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9(1):15. doi: 10.1186/1710-1492-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deckers J, et al. Protection against allergies: microbes, immunity, and the farming effect. Eur J Immunol. 2021;51(10):2387–2398. doi: 10.1002/eji.202048938. [DOI] [PubMed] [Google Scholar]
- 11.Riedler J, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358(9288):1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 12.Braun-Fahrlander C, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 13.Cahenzli J, et al. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14(5):559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy KD, et al. Natural IgE production in the absence of MHC Class II cognate help. Immunity. 2006;24(3):329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Stinson LF, et al. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. 2019;10:1124. doi: 10.3389/fmicb.2019.01124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milani C et al (2017) The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81(4) [DOI] [PMC free article] [PubMed]
- 17.Romano-Keeler J, Weitkamp JH. Maternal influences on fetal microbial colonization and immune development. Pediatr Res. 2015;77(1–2):189–195. doi: 10.1038/pr.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuriel-Ohayon M, et al. Microbial changes during pregnancy, birth, and infancy. Front Microbiol. 2016;7:1031. doi: 10.3389/fmicb.2016.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351(6279):1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 20.Thorburn AN, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 21.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Stiemsma LT, Turvey SE. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin Immunol. 2017;13:3. doi: 10.1186/s13223-016-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry RJ, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buffington SA, et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165(7):1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ennamorati M, et al. Intestinal microbes influence development of thymic lymphocytes in early life. Proc Natl Acad Sci USA. 2020;117(5):2570–2578. doi: 10.1073/pnas.1915047117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikuta K, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62(5):863–874. doi: 10.1016/0092-8674(90)90262-D. [DOI] [PubMed] [Google Scholar]
- 31.Havran WL, Allison JP. Developmentally ordered appearance of thymocytes expressing different T-cell antigen receptors. Nature. 1988;335(6189):443–445. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- 32.den Braber I, et al. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity. 2012;36(2):288–297. doi: 10.1016/j.immuni.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Hartvigsson O, et al. Associations of maternal and infant metabolomes with immune maturation and allergy development at 12 months in the Swedish NICE-cohort. Sci Rep. 2021;11(1):12706. doi: 10.1038/s41598-021-92239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, et al. T cell tolerance in early life. Front Immunol. 2020;11:576261. doi: 10.3389/fimmu.2020.576261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prelog M, et al. Thymectomy in early childhood: significant alterations of the CD4(+)CD45RA(+)CD62L(+) T cell compartment in later life. Clin Immunol. 2009;130(2):123–132. doi: 10.1016/j.clim.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 36.Dzhagalov I, Phee H. How to find your way through the thymus: a practical guide for aspiring T cells. Cell Mol Life Sci. 2012;69(5):663–682. doi: 10.1007/s00018-011-0791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aghaallaei N, Bajoghli B. Making thymus visible: understanding T-cell development from a new perspective. Front Immunol. 2018;9:375. doi: 10.3389/fimmu.2018.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 39.Koch U, Radtke F. Mechanisms of T cell development and transformation. Annu Rev Cell Dev Biol. 2011;27:539–562. doi: 10.1146/annurev-cellbio-092910-154008. [DOI] [PubMed] [Google Scholar]
- 40.Yu W, et al. Clonal deletion prunes but does not eliminate self-specific alphabeta CD8(+) T lymphocytes. Immunity. 2015;42(5):929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry JSA, et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 2014;41(3):414–426. doi: 10.1016/j.immuni.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savage PA, et al. Regulatory T cell development. Annu Rev Immunol. 2020;38:421–453. doi: 10.1146/annurev-immunol-100219-020937. [DOI] [PubMed] [Google Scholar]
- 43.Kim JM, et al. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 44.Sakaguchi S, et al. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155(3):1151–1164. [PubMed] [Google Scholar]
- 45.Fontenot JD, et al. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 46.Hori S, et al. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 47.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28(1):100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tai X, et al. Foxp3 transcription factor is proapoptotic and lethal to developing regulatory T cells unless counterbalanced by cytokine survival signals. Immunity. 2013;38(6):1116–1128. doi: 10.1016/j.immuni.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiba A, et al. Mucosal-associated invariant T cells in autoimmune diseases. Front Immunol. 2018;9:1333. doi: 10.3389/fimmu.2018.01333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Kaer L, Wu L. Therapeutic potential of invariant natural killer T cells in autoimmunity. Front Immunol. 2018;9:519. doi: 10.3389/fimmu.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiromizu CM, Jancic CC. gammadelta T lymphocytes: an effector cell in autoimmunity and infection. Front Immunol. 2018;9:2389. doi: 10.3389/fimmu.2018.02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seach N, et al. Double-positive thymocytes select mucosal-associated invariant T cells. J Immunol. 2013;191(12):6002–6009. doi: 10.4049/jimmunol.1301212. [DOI] [PubMed] [Google Scholar]
- 54.Gold MC, et al. Human thymic MR1-restricted MAIT cells are innate pathogen-reactive effectors that adapt following thymic egress. Mucosal Immunol. 2013;6(1):35–44. doi: 10.1038/mi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Z et al (2019) NKT cells in mice originate from cytoplasmic CD3-positive, CD4(-)CD8(-) double-negative thymocytes that express CD44 and IL-7Ralpha. Sci Rep 9(1):1874 [DOI] [PMC free article] [PubMed]
- 56.Treiner E, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422(6928):164–169. doi: 10.1038/nature01433. [DOI] [PubMed] [Google Scholar]
- 57.Franciszkiewicz K, et al. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol Rev. 2016;272(1):120–138. doi: 10.1111/imr.12423. [DOI] [PubMed] [Google Scholar]
- 58.Magalhaes I, et al. Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J Clin Invest. 2015;125(4):1752–1762. doi: 10.1172/JCI78941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rouxel O, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. 2017;18(12):1321–1331. doi: 10.1038/ni.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koay HF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. 2016;17(11):1300–1311. doi: 10.1038/ni.3565. [DOI] [PubMed] [Google Scholar]
- 61.Salou M, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019;216(1):133–151. doi: 10.1084/jem.20181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, et al. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat Immunol. 2019;20(4):447–457. doi: 10.1038/s41590-019-0334-0. [DOI] [PubMed] [Google Scholar]
- 63.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27(5):751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Legoux F, et al. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat Immunol. 2019;20(9):1244–1255. doi: 10.1038/s41590-019-0465-3. [DOI] [PubMed] [Google Scholar]
- 65.Bendelac A, et al. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 66.Godfrey DI, et al. Raising the NKT cell family. Nat Immunol. 2010;11(3):197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 67.Hammond K, et al. Three day neonatal thymectomy selectively depletes NK1.1+ T cells. Int Immunol. 1998;10(10):1491–1499. doi: 10.1093/intimm/10.10.1491. [DOI] [PubMed] [Google Scholar]
- 68.Pellicci DG, et al. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1(−)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195(7):835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 71.Benlagha K, et al. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 72.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10(3):306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H, Hogquist KA. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. Elife. 2018;7:e34793. doi: 10.7554/eLife.34793.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thiault N, et al. Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors. Nat Immunol. 2015;16(6):628–634. doi: 10.1038/ni.3150. [DOI] [PubMed] [Google Scholar]
- 77.Cowan JE, et al. CCR7 controls thymus recirculation, but not production and emigration, of Foxp3(+) T Cells. Cell Rep. 2016;14(5):1041–1048. doi: 10.1016/j.celrep.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Legoux F, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019;366(6464):494–499. doi: 10.1126/science.aaw2719. [DOI] [PubMed] [Google Scholar]
- 79.Nakajima A, et al. Maternal high fiber diet during pregnancy and lactation influences regulatory T cell differentiation in offspring in mice. J Immunol. 2017;199(10):3516–3524. doi: 10.4049/jimmunol.1700248. [DOI] [PubMed] [Google Scholar]
- 80.Zegarra-Ruiz DF, et al. Thymic development of gut-microbiota-specific T cells. Nature. 2021;594(7863):413–417. doi: 10.1038/s41586-021-03531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakajima A, et al. Commensal bacteria regulate thymic Aire expression. PLoS ONE. 2014;9(8):e105904. doi: 10.1371/journal.pone.0105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hikosaka Y, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29(3):438–450. doi: 10.1016/j.immuni.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 83.White AJ, et al. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol. 2014;192(6):2659–2666. doi: 10.4049/jimmunol.1303057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roberts NA, et al. Rank signaling links the development of invariant gammadelta T cell progenitors and Aire(+) medullary epithelium. Immunity. 2012;36(3):427–437. doi: 10.1016/j.immuni.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinic MM, et al. The bacterial peptidoglycan-sensing molecules NOD1 and NOD2 promote CD8(+) thymocyte selection. J Immunol. 2017;198(7):2649–2660. doi: 10.4049/jimmunol.1601462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Varian BJ, et al. Beneficial bacteria inhibit cachexia. Oncotarget. 2016;7(11):11803–11816. doi: 10.18632/oncotarget.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Varian BJ, et al. Microbial lysate upregulates host oxytocin. Brain Behav Immun. 2017;61:36–49. doi: 10.1016/j.bbi.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ojetti V, et al. The effect of Lactobacillus reuteri supplementation in adults with chronic functional constipation: a randomized, double-blind, placebo-controlled trial. J Gastrointest Liver Dis. 2014;23(4):387–391. doi: 10.15403/jgld.2014.1121.234.elr. [DOI] [PubMed] [Google Scholar]
- 89.Hou C, et al. Study and use of the probiotic Lactobacillus reuteri in pigs: a review. J Anim Sci Biotechnol. 2015;6(1):14. doi: 10.1186/s40104-015-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amorosi S, et al. FOXN1 homozygous mutation associated with anencephaly and severe neural tube defect in human athymic Nude/SCID fetus. Clin Genet. 2008;73(4):380–384. doi: 10.1111/j.1399-0004.2008.00977.x. [DOI] [PubMed] [Google Scholar]
- 91.Markert ML, et al. First use of thymus transplantation therapy for FOXN1 deficiency (nude/SCID): a report of 2 cases. Blood. 2011;117(2):688–696. doi: 10.1182/blood-2010-06-292490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zuklys S, et al. Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nat Immunol. 2016;17(10):1206–1215. doi: 10.1038/ni.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nowell CS, et al. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 2011;7(11):e1002348. doi: 10.1371/journal.pgen.1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ege MJ, et al. Prenatal exposure to a farm environment modifies atopic sensitization at birth. J Allergy Clin Immunol. 2008;122(2):407–412. doi: 10.1016/j.jaci.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 95.Conrad ML, et al. Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med. 2009;206(13):2869–2877. doi: 10.1084/jem.20090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu M, et al. Decreased maternal serum acetate and impaired fetal thymic and regulatory T cell development in preeclampsia. Nat Commun. 2019;10(1):3031. doi: 10.1038/s41467-019-10703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santner-Nanan B, et al. Fetal-maternal alignment of regulatory T cells correlates with IL-10 and Bcl-2 upregulation in pregnancy. J Immunol. 2013;191(1):145–153. doi: 10.4049/jimmunol.1203165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stokholm J, et al. Preeclampsia associates with asthma, allergy, and eczema in childhood. Am J Respir Crit Care Med. 2017;195(5):614–621. doi: 10.1164/rccm.201604-0806OC. [DOI] [PubMed] [Google Scholar]
- 99.Byberg KK, et al. Birth after preeclamptic pregnancies: association with allergic sensitization and allergic rhinoconjunctivitis in late childhood; a historically matched cohort study. BMC Pediatr. 2014;14:101. doi: 10.1186/1471-2431-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eviston DP, et al. Impaired fetal thymic growth precedes clinical preeclampsia: a case-control study. J Reprod Immunol. 2012;94(2):183–189. doi: 10.1016/j.jri.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 101.Eviston DP, et al. Altered fetal head growth in preeclampsia: a retrospective cohort proof-of-concept study. Front Pediatr. 2015;3:83. doi: 10.3389/fped.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Owen DL, et al. Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol. 2019;20(2):195–205. doi: 10.1038/s41590-018-0289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui Y, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. 2015;125(11):4171–4185. doi: 10.1172/JCI82424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Le Bourhis L, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. 2010;11(8):701–708. doi: 10.1038/ni.1890. [DOI] [PubMed] [Google Scholar]
- 105.Park SH, et al. Unaltered phenotype, tissue distribution and function of Valpha14(+) NKT cells in germ-free mice. Eur J Immunol. 2000;30(2):620–625. doi: 10.1002/1521-4141(200002)30:2<620::AID-IMMU620>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 106.Wingender G, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143(2):418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang Q, Bhandoola A. The development of adult innate lymphoid cells. Curr Opin Immunol. 2016;39:114–120. doi: 10.1016/j.coi.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517(7534):293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 109.Eberl G, et al. The brave new world of innate lymphoid cells. Nat Immunol. 2015;16(1):1–5. doi: 10.1038/ni.3059. [DOI] [PubMed] [Google Scholar]
- 110.Constantinides MG, et al. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prince AL, et al. Innate PLZF+CD4+ alphabeta T cells develop and expand in the absence of Itk. J Immunol. 2014;193(2):673–687. doi: 10.4049/jimmunol.1302058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu W, et al. An Id2(RFP)-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity. 2019;50(4):1054–1068 e3. doi: 10.1016/j.immuni.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pronovost GN, Hsiao EY. Perinatal interactions between the microbiome, immunity, and neurodevelopment. Immunity. 2019;50(1):18–36. doi: 10.1016/j.immuni.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gury-BenAri M, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166(5):1231–1246 e3. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 115.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10(1):83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29(6):958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 117.Yin CC, et al. The Tec kinase ITK regulates thymic expansion, emigration, and maturation of gammadelta NKT cells. J Immunol. 2013;190(6):2659–2669. doi: 10.4049/jimmunol.1202531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liao XC, Littman DR. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3(6):757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 119.Yang Y, et al. gammadelta T cells: crosstalk between microbiota, chronic inflammation, and colorectal cancer. Front Immunol. 2018;9:1483. doi: 10.3389/fimmu.2018.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Raviola E, Karnovsky MJ. Evidence for a blood-thymus barrier using electron-opaque tracers. J Exp Med. 1972;136(3):466–498. doi: 10.1084/jem.136.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Uchimura Y, et al. Antibodies set boundaries limiting microbial metabolite penetration and the resultant mammalian host response. Immunity. 2018;49(3):545–559 e5. doi: 10.1016/j.immuni.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kyewski BA, et al. Intrathymic presentation of circulating non-major histocompatibility complex antigens. Nature. 1984;308(5955):196–199. doi: 10.1038/308196a0. [DOI] [PubMed] [Google Scholar]
- 123.Atibalentja DF, et al. Thymus-blood protein interactions are highly effective in negative selection and regulatory T cell induction. J Immunol. 2009;183(12):7909–7918. doi: 10.4049/jimmunol.0902632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drumea-Mirancea M, et al. Characterization of a conduit system containing laminin-5 in the human thymus: a potential transport system for small molecules. J Cell Sci. 2006;119(Pt 7):1396–1405. doi: 10.1242/jcs.02840. [DOI] [PubMed] [Google Scholar]
- 125.Murphy KM, et al. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250(4988):1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 126.Volkmann A, et al. Antigen-presenting cells in the thymus that can negatively select MHC class II-restricted T cells recognizing a circulating self antigen. J Immunol. 1997;158(2):693–706. [PubMed] [Google Scholar]
- 127.Liblau RS, et al. Intravenous injection of soluble antigen induces thymic and peripheral T-cells apoptosis. Proc Natl Acad Sci USA. 1996;93(7):3031–3036. doi: 10.1073/pnas.93.7.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang H, et al. A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc Natl Acad Sci USA. 2017;114(8):1988–1993. doi: 10.1073/pnas.1610630114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hadeiba H, et al. Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity. 2012;36(3):438–450. doi: 10.1016/j.immuni.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ito T, et al. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood. 2006;107(6):2423–2431. doi: 10.1182/blood-2005-07-2709. [DOI] [PubMed] [Google Scholar]
- 131.Barchet W, et al. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J Exp Med. 2002;195(4):507–516. doi: 10.1084/jem.20011666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wurbel MA, et al. Mice lacking the CCR9 CC-chemokine receptor show a mild impairment of early T- and B-cell development and a reduction in T-cell receptor gammadelta(+) gut intraepithelial lymphocytes. Blood. 2001;98(9):2626–2632. doi: 10.1182/blood.V98.9.2626. [DOI] [PubMed] [Google Scholar]
- 133.Uehara S, et al. A role for CCR9 in T lymphocyte development and migration. J Immunol. 2002;168(6):2811–2819. doi: 10.4049/jimmunol.168.6.2811. [DOI] [PubMed] [Google Scholar]
- 134.Hadeiba H, et al. CCR9 expression defines tolerogenic plasmacytoid dendritic cells able to suppress acute graft-versus-host disease. Nat Immunol. 2008;9(11):1253–1260. doi: 10.1038/ni.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hanabuchi S, et al. Thymic stromal lymphopoietin-activated plasmacytoid dendritic cells induce the generation of FOXP3+ regulatory T cells in human thymus. J Immunol. 2010;184(6):2999–3007. doi: 10.4049/jimmunol.0804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Proietto AI, et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc Natl Acad Sci U S A. 2008;105(50):19869–19874. doi: 10.1073/pnas.0810268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Proietto AI, et al. The impact of circulating dendritic cells on the development and differentiation of thymocytes. Immunol Cell Biol. 2009;87(1):39–45. doi: 10.1038/icb.2008.86. [DOI] [PubMed] [Google Scholar]
- 138.Hapfelmeier S, et al. Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med. 2008;205(2):437–450. doi: 10.1084/jem.20070633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Niess JH, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307(5707):254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 140.Voboril M, et al. Toll-like receptor signaling in thymic epithelium controls monocyte-derived dendritic cell recruitment and Treg generation. Nat Commun. 2020;11(1):2361. doi: 10.1038/s41467-020-16081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brown EM, et al. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013;14(7):660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 142.Hooper LV, et al. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cebula A, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497(7448):258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Greiling TM et al (2018) Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med 10(434) [DOI] [PMC free article] [PubMed]
- 146.Hebbandi Nanjundappa R, et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell. 2017;171(3):655–667 e17. doi: 10.1016/j.cell.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 147.Gil-Cruz C, et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science. 2019;366(6467):881–886. doi: 10.1126/science.aav3487. [DOI] [PubMed] [Google Scholar]
- 148.Kita H, et al. Analysis of TCR antagonism and molecular mimicry of an HLA-A0201-restricted CTL epitope in primary biliary cirrhosis. Hepatology. 2002;36(4 Pt 1):918–926. doi: 10.1053/jhep.2002.35616. [DOI] [PubMed] [Google Scholar]
- 149.Bogdanos DP, et al. Microbial mimics are major targets of crossreactivity with human pyruvate dehydrogenase in primary biliary cirrhosis. J Hepatol. 2004;40(1):31–39. doi: 10.1016/S0168-8278(03)00501-4. [DOI] [PubMed] [Google Scholar]
- 150.Ioannidis M, et al. The immune modulating properties of mucosal-associated invariant T cells. Front Immunol. 2020;11:1556. doi: 10.3389/fimmu.2020.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McWilliam HE, Villadangos JA. MR1 antigen presentation to MAIT cells: new ligands, diverse pathways? Curr Opin Immunol. 2018;52:108–113. doi: 10.1016/j.coi.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 152.Bernasconi P, et al. Increased toll-like receptor 4 expression in thymus of myasthenic patients with thymitis and thymic involution. Am J Pathol. 2005;167(1):129–139. doi: 10.1016/S0002-9440(10)62960-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Boursalian TE, et al. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5(4):418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 154.Hendricks DW, Fink PJ. Recent thymic emigrants are biased against the T-helper type 1 and toward the T-helper type 2 effector lineage. Blood. 2011;117(4):1239–1249. doi: 10.1182/blood-2010-07-299263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Makaroff LE, et al. Postthymic maturation influences the CD8 T cell response to antigen. Proc Natl Acad Sci USA. 2009;106(12):4799–4804. doi: 10.1073/pnas.0812354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cunningham CA, et al. Cutting edge: defective aerobic glycolysis defines the distinct effector function in antigen-activated CD8(+) recent thymic emigrants. J Immunol. 2017;198(12):4575–4580. doi: 10.4049/jimmunol.1700465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.