Abstract
This paper presents the first example of a complete gene sequence coding for and expressing a biologically functional human tRNA methyltransferase: the hTRM1 gene product tRNA(m22G)dimethyltransferase. We isolated a human cDNA (1980 bp) made from placental mRNA coding for the full-length (659 amino acids) human TRM1 polypeptide. The sequence was fairly similar to Saccharomyces cerevisiae Trm1p, to Caenorhabditis elegans TRM1p and to open reading frames (ORFs) found in mouse and a plant (Arabidopsis thaliana) DNA. The human TRM1 gene was expressed at low temperature in Escherichia coli as a functional recombinant protein, able to catalyze the formation of dimethylguanosine in E.coli tRNA in vivo. It targeted solely position G26 in T7 transcribed spliced and unspliced human tRNATyr in vitro and in yeast trm1 mutant tRNA. Thus, the human TRM1 protein is a tRNA(m22G26)dimethyltransferase. Compared with yeast Trm1p, hTRM1p has a C-terminal protrusion of ∼90 amino acids which shows similarities to a mouse protein related to RNA splicing. A deletion of these 90 C-terminal amino acids left the modification activity in vitro intact. Among point mutations in the hTRM1 gene, only those located in conserved regions of hTRM1p completely eliminated modification activity.
INTRODUCTION
An increasing number, but still very few, of the modified nucleosides in RNA have been linked to functional roles in the living cell, e.g. to the translational machinery (1). In tRNAs more than 80 modified nucleosides have been identified (2). Some of them occur only scarcely, while others, like 5-methyluracil, are present in almost every tRNA species (3). One fairly abundant modified nucleoside is N2,N2-dimethylguanosine (m22G), present in tRNAs from archaea and eukaryotes, but not in eubacterial tRNAs (3). In eukaryotic tRNAs m22G is only found at position 26 in the bend between the D stem and the anticodon stem, although two early reports claimed that it is also present at position 27 in tRNATyr from bovine liver (4) and human placenta (5). In some archaeal tRNAs m22G is found at position 10 as well as position 26 (6).
Transfer RNA modifications occur at the polynucleotide level. The frequency of modification is low in bacterial tRNAs, with about one base out of 10 modified, but the frequency increases to ∼25% of the bases in human tRNAs (3). This suggests that a multitude of eukaryotic genes which code for tRNA-modifying enzymes must exist. Very few such eukaryotic genes and their gene products have been identified and studied to date (7–9). Only one human gene for a tRNA-modifying enzyme (modifying adenosine in tRNA to inosine) has been cloned and characterized (9), while no gene coding for a human tRNA-methylating enzyme has been characterized and studied until now.
The tRNA-modifying enzymes are in essence site-specific enzymes (1,10,11). Those with the greatest and strictest specificity are active at only one single site, producing one specific product (12–15). Others have multisite specificity, i.e. one enzyme mediates the formation of one specific modification, but does so at more than one site (16–19). The yeast enzyme tRNA(m22G)dimethyltransferase, encoded by a single nuclear TRM1 gene (20,21) modifies G26 to m22G in both nuclear encoded and mitochondrial yeast tRNAs (21,22). TRM1 homologs from several different organisms can be identified as ORFs (23). Since there are examples in which the polypeptide sequences are quite similar but the proteins interact with very different types of substrates (13,19,24,25), it is necessary to identify and analyze the expression products to validate whether these ORFs code for tRNA(m22G)dimethyltransferases.
In this study we show that a human ORF with a previously unknown function is the human gene that corresponds to yeast TRM1. Placental mRNA was used as template for isolating the gene, which was functionally expressed as a tRNA(m22G)dimethyltransferase that modified only position G26. Via mutations in the cloned hTRM1 gene causing loss of enzyme activity, we localized regions in the expressed polypeptide that were vital for modification activity.
MATERIALS AND METHODS
Strains and plasmids
The Escherichia coli strains used were DH5α [supE44, ΔlacU169(Φ80lacZΔM15), hsdR17, recA1, endA1, gyrA96, thi-1, relA1] (26) for plasmid propagations and BL21(DE3) [F–, ompT, hsdSB(rB–mB–), gal, dcm(DE3)] and BL21(DE3)plysS (Novagen) for expression of recombinant human TRM1 proteins (hTRM1p). Bulk tRNAs were isolated from Saccharomyces cerevisiae strains D4 (His–, trm1) and YF+ (wild-type) (27). Plasmids pHtY1 and pHtY1Δ [referred to as pHtT1 and pHtT1Δ in van Tol et al. (5)] carrying human tDNA(Tyr) and its precursor were kind gifts from Prof. Hildburg Beier (Wurtzburg, Germany). Colon cell line HT29, HeLa cell lines and placental cells (gifts from nearby departments and from Umeå University Hospital, Umeå) were used to make cell extracts. Standard procedures for molecular and gene manipulations were essentially as in Sambrook et al. (26).
Cloning of the human TRM1 gene and its expression in E.coli
The reverse transcription reaction (10 µl) contained ∼1 µg human placental mRNA (Clontech) and was performed as described (Clontech). An aliquot of 1/50 of the cDNA obtained was used as template in a 50 µl PCR reaction (PCR Kit; Perkin-Elmer). All oligonucleotides used were synthesized at Cell and Molecular Biology, Umeå University, Sweden. The primers flanked the whole predicted TRM1 coding region in the human mRNA. An NdeI restriction site (underlined) was introduced at the 5′-ATG start codon (5′-gga att cca tAT GCA AGG ATC GTC TCT GTG-3′) and an XhoI restriction site (underlined) was inserted immediately downstream of the 3′-TGA stop codon (5′-gat ccg ctc gag TCA GTC TAT GCC TGG CCC AGC G-3′). The PCR amplification covered 30 cycles of 97°C for 45 s, 55°C for 45 s, 72°C for 6 min and a final extension step at 72°C for 10 min, resulting in a single fragment (∼2 kb size on a 1% agarose gel) which, after extraction from the gel (QiaexII Gel Extraction Kit; Qiagen, Germany) and end trimming with the restriction enzymes (New England Biolabs, USA) NdeI and XhoI, was inserted into pET15b downstream of the T7 lac promoter to give plasmid pET-hTRM1. The His-tagged N-terminus of the recombinant hTRM1p will then be MGSSHHHHHHSSGLVPRGSHMQG, where underlining refers to the hTRM1 enzyme.
Plasmids with correctly sized inserts were sequenced from two directions (ABI PRISM™ Dye Terminator Cycle Sequencing Ready Reaction Kit; Perkin-Elmer). Our determination of the hTRM1 cDNA is available in GenBank (accession no. AF196479). Escherichia coli strains BL21(DE3) and BL21(DE3)plysS, transformed with plasmid pET-hTRM1 and grown (28) to an OD590 of 0.6–0.7, were induced by IPTG (1 mM final concentration; Sigma) for 3–5 h at 22, 30 and 37°C. Harvested cells were resuspended in 1/10 of the culture volume of 20 mM Tris–HCl, pH 8.0, 2 mM EDTA and stored at –20°C until used.
Assay of tRNA(m22G26)methyltransferase activity in cell extracts in vitro
The method was essentially as in Kjellin-Stråby and Boman (29). An aliquot of 5 µl of crude extract in a total of 20 µl at pH 8.0 containing methyl-deficient tRNA from S.cerevisiae D4 (trm1) or from E.coli tRNA as substrate and [14C]AdoMet (53 mCi/mmol; Amersham, UK) as methyl donor was incubated at 30°C for 30 min. [14C]CH3 in tRNA was measured by liquid scintillation counting. No incorporation was obtained into yeast wild-type YF+ tRNA with any extracts from bacterial cells harboring cloned TRM1 genes.
HPLC analysis of modified nucleosides in tRNA formed in vivo
Total tRNA was isolated from E.coli cells with hTRM1 expression plasmids induced for 5 h at 22°C (30). tRNA was further purified on Nucleobond (A×500) columns as described by the manufacturer (Macherey-Nagel, Germany). About 100 µg of purified bulk tRNA was then digested with nuclease P1 (Boehringer-Mannheim, Germany) and bacterial alkaline phosphatase and the resulting nucleosides were analyzed by HPLC (31).
Construction of human tRNATyr- and pre-tRNATyr-carrying plasmids
The T7 promoter was introduced immediately upstream of the tRNATyr gene (from plasmid pHtY1Δ) and the pre-tRNATyr gene (intron-containing, from plasmid pHtY1) by PCR using the synthetic oligonucleotides JM56 (5′-GGA ATT CGC TGC AGT AAT ACG ACT CAC TAT AGC TTC GAT AGC TCA GCT GGT AG-3′) and JM57 (5′-CGG GAT CCT GGT GCT TCG AGC CGG AAT CGA ACC-3′), then cloned into pUC18 (Promega, UK), giving plasmids pUC-HtY1Δ and pUC-HtY1, respectively. The tDNA then coded for a CCA 3′-end and had a G1–C72 pair instead of T1–A72, to enhance T7 promoted transcription in vitro (32). The tDNA constructs were found to have the correct DNA sequence, especially at positions 26 and 27, which coded for guanosines.
Preparation of 32P-labeled tRNA transcripts in vitro
The T7 RNA polymerase run-off radiolabeled tRNA transcripts were prepared in the presence of 2 µg linearized (BstN1) plasmid template using the Riboprobe System-T7 (Promega) as described elsewhere (33). The tRNA transcripts were utilized in experiments within 1 week.
In vitro methylation of the 32P-labeled tRNA transcripts and 2-dimensional TLC analysis of modified nucleotides
Nearest neighbor analysis was used to identify the modified nucleotides formed (34). About 4 × 106 c.p.m. of [α-32P]GTP- or [α-32P]ATP-labeled tRNA transcripts were methylated in vitro with [14C]AdoMet as methyl donor. The enzyme source was either a crude extract from pET-hTRM1 plasmid-carrying E.coli cells induced by IPTG or nuclear extract from colon cell lines, placental cells, mammalian HeLa cells prepared as previously described (35) or purchased from Promega. In vitro methylated tRNA (incubated at 30 or 37°C for up to 180 min) was re-isolated by phenol extraction and ethanol precipitation and run on 10% PAGE–8 M urea gels. Full-length tRNA molecules were eluted, precipitated and then hydrolyzed completely by RNase T2 (3′-mononucleotides) (Sigma) or nuclease P1 (5′-mononucleotides) at 37°C overnight (33). The modified bases were identified by 2-dimensional TLC (Polygram Cel 300, 0.1 mm cellulose; Macherey-Nagel, Germany) with isobutyric acid:NH4OH:H2O (66:1:33 v/v/v) as the first dimension solvent system and 0.1 M Na2HPO4/NaH2PO4 buffer (pH 6.8):(NH4)2SO4:n-propanol (100:60:2 v/w/v) as the second dimension solvent (36). The modified nucleotides were identified by comparison with reference maps (37) and quantified (38) by use of a phosphorimager (Molecular Dynamics, supplied with ImageQuant software) in mol m22G/mol tRNA.
RESULTS
Identification and cloning of human TRM1 cDNA (hTRM1)
Dimethylguanosine (m22G) is present in many tRNAs from archaea and eucarya, but not in tRNAs from eubacteria (3). In a BLAST search (39) of the complete non-redundant GenBank database we identified one ORF, present on human chromosome 19, which might encode a human tRNA methyltransferase. It had similarities to the tRNA(m22G26)dimethyltransferase from yeast, Caenorhabditis elegans (7) and archaeal bacteria (40). The possible mode of m22G formation in human tRNAs where, besides G26, two exceptional cases of G27 might also be dimethylated (4,5), prompted us to clone the ORF and to experimentally define its specificity and biological function.
Using human placental mRNA as template, the ORF was cloned via RT–PCR into an E.coli plasmid, sequenced and shown to be 1980 bp long. This corresponds to 659 amino acids (GenBank accession no. AF196479). The cDNA sequence was identical to a genomic sequence in GenBank (AC005546) except that our clone encoded four additional amino acids. The corresponding bases in the DNA, C15305…C15406CAGCAAC15413, are not, as claimed (GenBank accession no. AA005546.1), part of an intron, since data from all our nine cDNA clones investigated contained information for the corresponding four additional amino acids R500CWE503. Sequence alignment of this region with other Trm1 polypeptides matched well. We conclude that the small region corresponding to R500CWE503 was in fact expressed and that this expression did not introduce any possibility for alternative splicing or any changed position for the stop codon used. Our hTRM1 sequence is identical to a recently published sequence for an ORF from colon mucosa cells (accession no. BAA91031).
Amino acid sequence alignment with the biologically functional Trm1 polypeptides from yeast (41) and nematode (7) demonstrated that the deduced human enzyme was 26% identical (42% similar) to the yeast Trm1 polypeptide and 31% identical (45% similar) to C.elegans TRM1p. The sequences have seven fairly well conserved regions, in essence as recently described by Stanford et al. (23). One of these regions contains VLEGLAASGLRSIRF, which, together with a conserved DA some 30 amino acids downstream, could be a potential AdoMet-binding domain (42,43). The first 55 N-terminal amino acids contain a potential mitochondrial import signal. Compared with the nematode and yeast sequences (7,41) the human sequence has a 90 amino acid protruding C-terminal end which harbors a zinc finger C-X8-C-X5-C-X3-H motif (606–624) and an arginine/proline-rich region (R+P 27.6%, 561–659). The predicted molecular weight is 72.2 kDa. The predicted pI is 9.2, which is the same as that postulated for yeast Trm1p.
Functional expression of the human TRM1 gene product in E.coli
Escherichia coli has no inherent tRNA(m22G)dimethyltransferase activity. Thus, if a C.elegans, archaeal, yeast or human methyltransferase gene could be expressed in E.coli, the resultant methyltransferase would then be the only G26-modifying enzyme in the bacterial cell (7,44,45). Human TRM1 protein expressed in E.coli cells, induced at 22, 30 and 37°C, was tested for methyltransferase activity in vitro using crude cell extracts with either E.coli bulk tRNA, in which all G26 positions are unmodified, or yeast D4 trm1 tRNA, which is specifically deficient in methylation at position G26, as substrate (29). Only expression at 22°C gave an active human methyltransferase (data not shown), while both the C.elegans and yeast TRM1 genes can be functionally expressed in bacteria at both 30 and 37°C (7,21,45). The E.coli strain BL21(DE3)plysS, which carries a plasmid that increases the tolerance of the host to plasmids with toxic inserts (46), gave about four times higher methyltransferase activity than BL21(DE3); it was thereafter used as the host strain. Even then expression was comparatively low. Although only induction at 22°C or lower gave an active enzyme, methylation in vitro still occurred at 30°C. This indicated that once a functional enzyme had been formed, it could retain activity at temperatures that did not allow synthesis of an active enzyme. The bacterial cell extract containing the recombinant protein was unable to modify any position in wild-type yeast tRNA, but could modify the specifically G26 methyl-deficient tRNA from yeast mutant D4. Thus, no site in yeast tRNA other than G26 functioned as a target for modification mediated by the recombinant hTRM1p. We therefore conclude that the only additional activity expressed in bacteria carrying the cDNA we had cloned was from a structural human gene coding for a human tRNA(G26)methyltransferase which was expressed in E.coli cells as a functional enzyme. Therefore, the gene should from now on be named hTRM1.
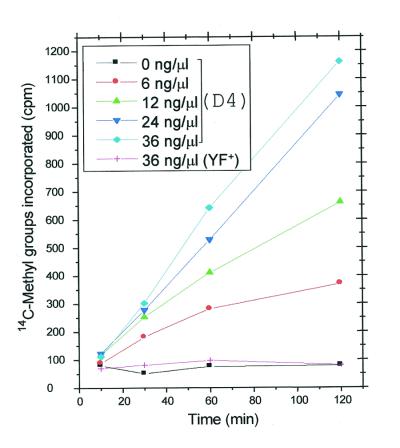
In a time course study of the in vitro interaction between recombinant human TRM1 protein and yeast tRNA (Fig. 1) no methyl groups were transferred to wild-type YF+ tRNA, whereas the incorporation of methyl groups into increasing amounts of G26 methyl-deficient D4 tRNA was RNA concentration dependent. However, in the truly homologous situation with yeast tRNA and the yeast enzyme comparable amounts of D4 tRNA became fully methylated in <60 min (47). In the present case the reaction rates also varied when tRNA was added in excess and plateau levels were only reached for low amounts of tRNA and after a long incubation time. This made more elaborate kinetic studies using this enzyme extract less relevant.
Figure 1.
In vitro incorporation of methyl groups into G26 methyl-deficient D4 tRNA catalyzed by the human TRM1 enzyme. The assay was performed at 30°C for different times in the presence of 5 µl crude enzyme extract from IPTG-induced E.coli BL21(DE3)plysS cells harboring the pET-hTRM1 plasmid, [14C]methyl-AdoMet, the methyl donor (53 mCi/mmol), and different amounts of tRNA substrates from S.cerevisiae strain D4 (trm1, filled symbols) or wild-type strain YF+ (TRM1, + symbol).
Modified nucleotides formed by heterologous methylation of E.coli tRNA with the human TRM1 enzyme in vivo
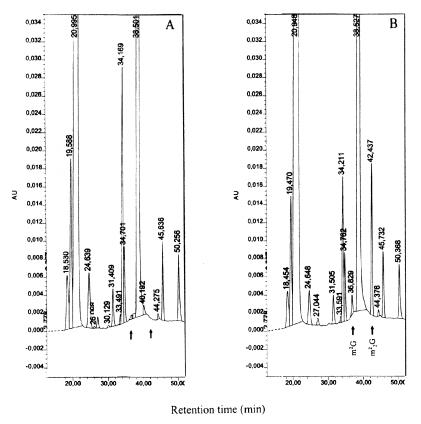
The in vitro modification we demonstrated in Figure 1 did not discriminate between the human TRM1 protein being a monomethyltransferase and/or a dimethyltransferase. Therefore, in vivo methylated bulk bacterial tRNA, isolated from IPTG-induced plasmid pET-hTRM1-containing E.coli cells, was hydrolyzed to mononucleosides and analyzed by HPLC (31). Figure 2 shows that, as compared with tRNA from E.coli control cells, the tRNA from the hTRM1-carrying bacterial cells had gained both m22G and m2G in their tRNA. Thus, the cloned hTRM1 gene encoded a functional tRNA(m22G)dimethyltransferase which recognizes E.coli tRNA as a substrate in vivo. The enzyme has both G mono- and dimethylating activity on E.coli tRNA and was efficient enough in vivo to produce the final product m22G (88%) with only 12% m2G, the transient precursor form (33).
Figure 2.
HPLC analyses of modified nucleosides in E.coli tRNA from wild-type cells and from E.coli carrying a plasmid with a human TRM1 gene. Isolated tRNA was digested with both nuclease P1 and bacterial alkaline phosphatase before HPLC chromatography of the nucleosides. (A) Escherichia coli BL21(DE3)plysS carrying the pET15b plasmid. (B) IPTG-induced E.coli BL21(DE3)plysS harboring the pET-hTRM1 plasmid. Nucleosides were identified by their UV spectra and their relative retention times (only times between 15 and 50 min are shown). The positions for m2G and m22G are indicated by arrows.
The homologous human case: modification of G26 in human tRNATyr catalyzed by the recombinant or native human TRM1 enzyme in vitro
The specificity of the recombinant hTRM1 protein towards the homologous human tRNA substrate was then tested in vitro using as substrate both spliced and intron-containing human tRNATyr with the relevant partial sequence …pC25–pG26–pG27–pA28…. The in vitro transcribed, completely unmodified, 32P-labeled human tRNATyr was modified in vitro using an extract from E.coli cells in which the hTRM1 protein had been expressed (E.coli has no inherent TRM1 enzyme) and full-length tRNA was isolated, degraded and analyzed by 2-dimensional TLC.
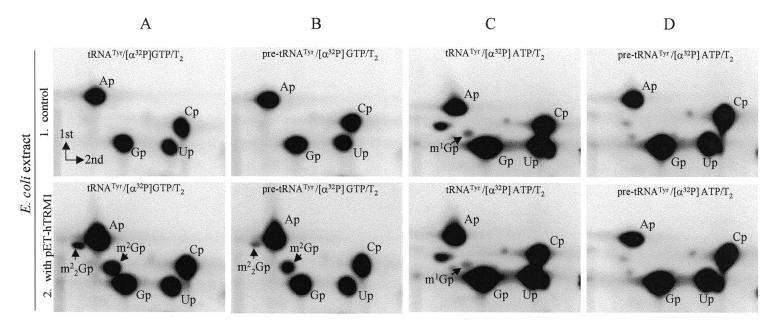
Transfer RNA transcripts, labeled with [α-32P]GTP and digested with RNase T2, will release G26 (but not G27) as a 32P-labeled nucleotide (…pC25–32P↓G26–32P↓G27–pA28…). Figure 3 shows that two additional spots appeared when tRNA incubated with extract from cells harboring the human TRM1 gene was degraded with RNase T2 (Fig. 3A2), as compared with incubation with the control extract from cells without hTRM1 (Fig. 3A1). These two spots were dimethylated guanosine (m22G, ∼0.1 mol/mol tRNA) and monomethylated guanosine (m2G, ∼0.6 mol/mol tRNA). The proportion of m22G over m2G increased with increasing incubation times. The presence of an intron in the human tRNATyr molecule did not significantly affect recognition of the substrate by the methylase (Fig. 3B2). Thus, homologous methylation mediated by the hTRM1 enzyme resulted in the final formation of m22G26, with m2G26 as a transient product, in human tRNA.
Figure 3.
Autoradiograms of 2-dimensional TLC showing the formation of modified nucleotides m22G and m2G in human tRNATyr and pre-tRNATyr molecules with recombinant hTRM1 enzyme. Unmodified [α-32P]GTP-labeled or [α-32P]ATP-labeled in vitro transcripts of human tRNATyr and pre-tRNATyr were incubated with methyl donor and cell extracts containing or lacking the hTRM1 gene product. The full-length tRNA was isolated, degraded with RNase T2, analyzed by 2-dimensional TLC and spots were identified (see Materials and Methods). The [α-32P] label in the tRNA transcripts and the nuclease used are indicated at the top of each panel. Spots corresponding to m2G and m22G are indicated. (A) [α-32P]GTP-labeled tRNATyr incubated with extract from E.coli pET15b without (A1) and with (A2) the hTRM1 gene. (B) [α-32P]GTP-labeled pre-tRNATyr incubated with extract from E.coli pET15b without (B1) and with (B2) the hTRM1 gene. (C) [α-32P]ATP-labeled tRNATyr incubated with extract from E.coli pET15b without (C1) and with (C2) the hTRM1 gene. (D) [α-32P]ATP-labeled pre-tRNATyr incubated with extract from E.coli pET15b without (D1) and with (D2) the hTRM1 gene. Note that the m1G spots in (C1) and (C2) originated from position G37 (label came from the neighboring A38) and were modified by an inherent E.coli enzyme. In unspliced tRNA G37 is adjacent to the intron and not to a labeled A and therefore no m1G was found in (D1) and (D2).
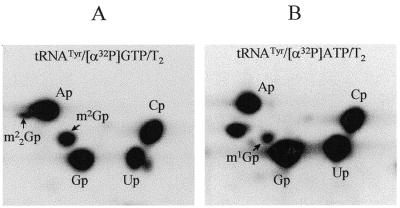
When human tRNATyr was modified by the recombinant hTRM1 enzyme in vitro the formation of m22G was less efficient (∼0.1 mol/mol tRNA) than if the enzyme was from S.cerevisiae (∼1.0 mol m22G/mol tRNATyr in 30 min at 30°C). Furthermore, in E.coli cells in vivo recombinant hTRM1 enzyme resulted in a m22G:m2G ratio of 88:12 in the bacterial tRNA (see above). Thus, when the recombinant hTRM1 was present in an intact cell it seemed to be more stable than in crude or partly purified cell extracts. We therefore compared these activities with the activity of the native TRM1 enzyme present in extracts of different human cells, e.g. placental and colon cancer cells and different HeLa cell lines. As expected, such nuclear extracts catalyzed homologous modification (m22G26) of completely unmodified human tRNATyr (Fig. 4A) at a somewhat higher efficiency (up to ∼0.3 mol m22G/mol tRNATyr) than the in vitro case with the recombinant hTRM1p, and in both cases modifying only at position G26. Thus, both the recombinant and native hTRM1 enzymes modified position 26 in tRNA to dimethylguanosine.
Figure 4.
Autoradiograms of 2-dimensional TLC showing the formation of modified nucleotides of m22G and m2G by human HeLa cell extracts. Unmodified [α-32P]GTP-labeled or [α-32P]ATP-labeled in vitro transcripts of human tRNATyr were incubated with HeLa cell extracts at 30°C for 60 min. The isolated tRNA was purified, degraded with nuclease T2, analyzed by 2-dimensional TLC and the spots identified (see Materials and Methods). The spots corresponding to m2G and m22G are indicated. (A) [α-32P]GTP-labeled tRNATyr. (B) [α-32P]ATP-labeled tRNATyr.
That the hTRM1 enzyme was specific only for the G26 position was demonstrated by using in vitro transcribed tRNATyr labeled with [α-32P]ATP (…pC25–pG26–p↓G27–32P↓A28…). In such tRNA, methylated in vitro with the hTRM1 enzyme and then re-isolated, the 32P-label from A28 would be transferred to the nearest neighbor (G27) when it is degraded with RNase T2. In no case could we identify a modified G at position 27 (Fig. 3C1, C2, D1 and D2). This was also valid when native methylating enzymes from human HeLa (Fig. 4B), placental or colon cell extracts (data not shown) were used. We conclude that the cloned hTRM1 enzyme is a tRNA(m22G)dimethyltransferase, unable to methylate position G27, and absolutely specific for position 26 in human, yeast and bacterial tRNAs.
Human TRM1 mutants that affect human TRM1 enzyme activity
Different hTRM1 alleles containing point mutations were expressed in E.coli and crude extracts were tested for tRNA G26 methyltransferase activity. Mutations having no effect on enzyme activity were all located outside the seven conserved regions, while all mutants causing a decrease in or total lack of enzyme activity were found in conserved regions. Thus, exchange of the conserved Cys348 for Arg, D319 for V, as well as mutations S237N and F320S, eliminated enzyme activity. The same was true for point mutations in yeast and C.elegans TRM1 genes (7,45). A specific characteristic of the human TRM1 polypeptides was a long protrusion at the C-terminus, ∼90 amino acids longer than yeast Trm1p. Deleting exactly these 90 C-terminal amino acids did not affect methylating activity.
DISCUSSION
Here we have reported the first successful cloning of a human tRNA methylating enzyme gene, hTRM1, and its expression into a biologically functional product, tRNA(m22G26)dimethyltransferase. No other ORF among the so far sequenced parts of the human genome can be a TRM1 allele or homolog. The human TRM1 gene seems to be ubiquitously transcribed (EST database) and thus this enzyme can be expected to be found in almost all human tissues.
Does the protruding C-terminus of the hTRM1 enzyme have its own function?
When aligned with all other known or postulated TRM1 gene products, including the truncated one from mouse (AI117630), that from the plant Arabidopsis (AA651222) and some archaeal TRM1 proteins, the deduced amino acid sequences of the human and mouse genes are unique in having a long C-terminal protrusion. When compared with yeast Trm1p the extension was ∼90 amino acids. Since this protrusion in the human TRM1 enzyme was shown not to be necessary for methyltransferase activity, it presumably has another function. The human TRM1 protein could well be a multifunctional protein, especially with reference to the expression products from the bacterial TrmA and TrmE genes, both of which simultaneously harbor tRNA methyltransferase activity and a second function (48,49). To identify the function of this C-terminal tail is a challenge. It shows rather high homology to a protein, the U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 2 (50), which is connected with an mRNA splicing system. It is arginine/proline-rich and has a Zn finger motif, pointing to a nucleic acid-binding capacity. Thus, the protruding C-terminus might be involved in other activities such as splicing and folding or as a handle to trigger an activity in some other protein- or DNA-complex. It might be rational if proteins, enzymes and factors involved in the tRNA maturation process were in fact physically co-localized somewhere within the nucleus, e.g. in complexes in or near the human nuclear pore or in the nucleolus, as exemplified by the yeast pseudouridine-modifying enzyme Pus1p, which is present in the yeast nuclear pore complex (51).
Substrate and site specificity of tRNA enzymes modifying guanosines
The recombinant human tRNA methyltransferase enzyme, expressed in the bacterium E.coli, was capable of dimethylating G to m22G in tRNA, either in vivo in E.coli cells or in vitro as a cell extract with tRNAs from yeast or human as substrate. Comparing the efficiencies of TRM1 gene expression in bacteria, the genes from C.elegans and yeast were fairly well expressed at 30 or 37°C, while the human gene gave an active enzyme only at lower temperatures. Even if the human enzyme, when in a crude extract, was also active at 30°C, it might have become non-optimally folded when synthesized in the bacterial cell. This assumption agrees with the idea that the enzyme is rather unstable during continued purification steps. Although the reaction in vitro was kinetically fairly slow, the product in all cases was dimethylation of G via transient formation of monomethylated G.
The yeast tRNA pseudouridine synthetase (Pus1p), the m5C-methyltransferase (Trm4p) working on tRNA and a dimethyltransferase catalyzing the formation of m26A1779m26A1780 in 18S rRNA (Dim1p) (52) are examples of enzymes that modify multiple sites in their respective RNA substrates. In contrast, the recombinant hTRM1 enzyme is specific for position G26 in all of the tRNA substrates we have tested and for dimethylation to m22G26 via m2G26. The recombinant hTRM1 enzyme turned out to be independent of the presence of an intron in the tRNA, in agreement with earlier findings where the yeast TRM1 enzyme recognized its target G26 site even if the tRNA substrate was unspliced (53), if it lacked the whole anticodon loop (47) or if the amino acceptor stem was deleted (47). The recognition elements for G26 dimethylation might, as in yeast (10), reside in the inner core of the tRNA molecule.
The hTRM1 enzyme did not catalyze formation of m2G at positions G6 and G10 nor did it mono- or dimethylate G27, a position reported to be dimethylated in human placental and bovine liver tRNATyr. Whether G27 is indeed methylated and, if so, which enzyme catalyzes such a reaction will be further investigated, although all our data so far suggest that the human cell systems we have studied in vitro and in vivo with the recombinant and native enzymes seem to lack G27-methylating activity. Most so far sequenced animal tRNATyr species have identical primary sequences. Expression of a Xenopus laevis tRNATyr gene in monkey cells did not lead to methylation of G27 (54). We will continue our efforts to clarify the question on possible modification of G27. This paper, however, has as its focus the identification, determination and characterization of the interaction between a human TRM1 dimethyltransferase and its substrate site at position G26 in human tRNAs leading to a dimethylated product.
Conserved domains turn out to be crucial for modification activity
All point mutations in TRM1 genes from yeast (45), C.elegans (7) and human cells that cause loss of enzyme activity are localized in conserved regions in the genes. Of the seven conserved regions (I–VII) we have identified in TRM1 genes, mutations were found in conserved regions II (166–184), III (226–261), IV (288–328) and VI (467–500) and conserved position C348. Region II is supposed to be the binding site for the methyl donor AdoMet, based on its homology to AdoMet-binding sites in other methyltransferases (43). Region III has a striking similarity to sequences in DNA methyltransferases important for methyl group transfer (55) and could have this function in tRNA(m22G26)dimethyltransferase. The detailed roles of regions IV and VI and position C348 will have to await the results of future studies. When all known sequences for TRM1 genes from yeast to human, also including archaean ones, were compared, the overall sequence similarity was quite low, but enzymatic function was well conserved. Comparisons across species borders and continued elucidation of motifs and conserved regions of importance for TRM1p enzymatic activity might lead to an understanding of the enzymatic and kinetic properties needed for this protein–RNA interaction and might even make this tRNA-modifying enzyme system a good object for evolutionary studies.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr H. Gross and Dr H. Beier for generous gifts of human tRNATyr encoding plasmids, Dr J. Edqvist for valuable technical advice, Dr R. Rosqvist, Dr S. Hammarström and Ms A. Tjernström (Umeå University) for HeLa and Colon HT29 cell donations, the Umeå University Hospital for placental cells, Mrs K. Jacobsson and Mrs S. Sääf for skillfully performing the HPLC and the DNA sequencing determinations and Mr S. Li for kind assistance. Dr G. Björk is thanked for critically reading the manuscript. This project was supported by The National Science Research Council, Sweden, project no. 2708 (K.B.S.) and the J.C. Kempe Foundation (J.L.).
DDBJ/EMBL/GenBank accession no. AF196479
REFERENCES
- 1.Björk G.R. (1995) In Söll,D. and Rajbhandary,U.L. (eds), tRNA: Structure, Biosynthesis, and Function. ASM Press, Washington, DC, pp. 165–205.
- 2.McCloskey J.A. and Crain,P.F. (1998) Nucleic Acids Res., 26, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sprinzl M., Horn,C., Brown,M., Loudovitch,A. and Steinberg,S. (1998) Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson G.D., Pirtle,I.L. and Pirtle,R.M. (1985) Arch. Biochem. Biophys., 236, 448–453. [DOI] [PubMed] [Google Scholar]
- 5.van Tol H., Stange,N., Gross,H.J. and Beier,H. (1987) EMBO J., 6, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosjean H., Sprinzl,M. and Steinberg,S. (1995) Biochimie, 77, 139–141. [DOI] [PubMed] [Google Scholar]
- 7.Liu J., Zhou,G.Q. and Sråby,K.B. (1999) Gene, 226, 73–81. [DOI] [PubMed] [Google Scholar]
- 8.Chen J. and Patton,J.R. (1999) RNA, 5, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maas S., Gerber,A.P. and Rich,A. (1999) Proc. Natl Acad. Sci. USA, 96, 8895–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grosjean H., Edqvist,J., Stråby,K.B. and Giege,R. (1996) J. Mol. Biol., 255, 67–85. [DOI] [PubMed] [Google Scholar]
- 11.Garcia G.A. and Goodenough-Lashua,D.M. (1998) In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp.135–168.
- 12.Becker H.F., Motorin,Y., Planta,R.J. and Grosjean,H. (1997) Nucleic Acids Res., 25, 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerber A.P., Grosjean,H., Melcher,T. and Keller,W. (1998) EMBO J., 17, 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber A.P. and Keller,W. (1999) Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 15.Cavaille J., Chetouani,F. and Bachellerie,J.P. (1999) RNA, 5, 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortese R., Kammen,H.O., Spengler,S.J. and Ames,B.N. (1974) J. Biol. Chem., 249, 1103–1108. [PubMed] [Google Scholar]
- 17.Motorin Y., Keith,G., Simon,C., Foiret,D., Simos,G., Hurt,E. and Grosjean,H. (1998) RNA, 4, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lecointe F., Simos,G., Sauer,A., Hurt,E.C., Motorin,Y. and Grosjean,H. (1998) J. Biol. Chem., 273, 1316–1323. [DOI] [PubMed] [Google Scholar]
- 19.Motorin Y. and Grosjean,H. (1999) RNA, 5, 1105–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips J.H. and Kjellin-Stråby,K. (1967) J. Mol. Biol., 26, 509–518. [DOI] [PubMed] [Google Scholar]
- 21.Ellis S.R., Morales,M.J., Li,J.M., Hopper,A.K. and Martin,N.C. (1986) J. Biol. Chem., 261, 9703–9709. [PubMed] [Google Scholar]
- 22.Hopper A.K., Furukawa,A.H., Pham,H.D. and Martin,N.C. (1982) Cell, 28, 543–550. [DOI] [PubMed] [Google Scholar]
- 23.Stanford D.R., Martin,N.C. and Hopper,A.K. (2000) Nucleic Acids Res., 28, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maas S., Melcher,T. and Seeburg,P.H. (1996) Curr. Opin. Cell Biol., 9, 343–349. [DOI] [PubMed] [Google Scholar]
- 25.Tscherne J.S., Nurse,K., Popienick,P., Michel,H., Sochacki,M. and Ofengand,J. (1999) Biochemistry, 38, 1884–1892. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Björk G.R. and Kjellin-Stråby,K. (1977) In Salvatore,F., Borek,E., Zappia,V., Williams-Ashman,H.G. and Schlenk,F. (eds), Biochemistry of Adenoyslmethionine. Columbia University Press, New York, NY, pp. 216–230.
- 28.Bertani G. (1951) J. Bacteriol., 62, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kjellin-Stråby K. and Boman,H.G. (1965) Proc. Natl Acad. Sci. USA, 53, 1346–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck M., Connick,M. and Ames,B.N. (1983) Anal. Biochem. 129, 1–13. [DOI] [PubMed] [Google Scholar]
- 31.Gehrke C.W. and Kuo,K.C. (1989) J. Chromatogr., 471, 3–36. [DOI] [PubMed] [Google Scholar]
- 32.Dunn J.J. and Studier,F.W. (1983) J. Mol. Biol., 166, 477–535. [DOI] [PubMed] [Google Scholar]
- 33.Edqvist J., Grosjean,H. and Stråby,K.B. (1992) Nucleic Acids Res., 20, 6575–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grosjean H., Motorin,Y. and Morin,A. (1998) In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 21–46.
- 35.Krämer A. and Keller,W. (1990) Methods Enzymol., 181, 3–19. [DOI] [PubMed] [Google Scholar]
- 36.Silberklang M., Gillum,A.M. and RajBhandary,U.L. (1979) Methods Enzymol., 59, 58–109. [DOI] [PubMed] [Google Scholar]
- 37.Keith G. (1995) Biochimie, 77, 142–144. [DOI] [PubMed] [Google Scholar]
- 38.Grosjean H., Droogmans,L., Giege,R. and Uhlenbeck,O.C. (1990) Biochim. Biophys. Acta, 1050, 267–273. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Constantinesco F., Benachenhou,N., Motorin,Y. and Grosjean,H. (1998) Nucleic Acids Res., 26, 3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis S.R., Hopper,A.K. and Martin,N.C. (1987) Proc. Natl Acad. Sci. USA, 84, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kagan R.M. and Clarke,S. (1994) Arch Biochem. Biophys., 310, 417–427. [DOI] [PubMed] [Google Scholar]
- 43.Koonin E.V., Tatusov,R.L. and Rudd,K.E. (1995) Proc. Natl Acad. Sci. USA, 92, 11921–11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Constantinesco F., Motorin,Y. and Grosjean,H. (1999) J. Mol. Biol., 291, 375–392. [DOI] [PubMed] [Google Scholar]
- 45.Liu J., Liu,J. and Stråby,K.B. (1998) Nucleic Acids Res., 26, 5102–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studier F.W. (1991) J. Mol. Biol., 219, 37–44. [DOI] [PubMed] [Google Scholar]
- 47.Edqvist J., Blomqvist,K. and Stråby,K.B. (1994) Biochemistry, 33, 9546–9551. [DOI] [PubMed] [Google Scholar]
- 48.Persson B.C., Gustafsson,C., Berg,D.E. and Björk,G.R. (1992) Proc. Natl Acad. Sci. USA, 89, 3995–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabedo H., Macian,F., Villarroya,M., Escudero,J.C., Martinez-Vicente,M., Knecht,E. and Armengod,M.E. (1999) EMBO J., 18, 7063–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaoka T., Hatada,I., Kitagawa,K., Wang,X. and Mukai,T. (1995) Genomics, 27, 337–340. [DOI] [PubMed] [Google Scholar]
- 51.Simos G., Tekotte,H., Grosjean,H., Segref,A., Sharma,K., Tollervey,D. and Hurt,E.C. (1996) EMBO J., 15, 2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 52.Lafontaine D., Delcour,J., Glasser,A.L., Desgres,J. and Vandenhaute,J. (1994) J. Mol. Biol., 241, 492–497. [DOI] [PubMed] [Google Scholar]
- 53.Kang H.S., Ogden,C., Knapp,G., Peebles,C.L. and Abelson,J. (1979) In Axel,R., Maniatis,T. and Fox,F. (eds), Eucaryotic Gene Regulations. Academic Press, New York, NY, pp. 69–84.
- 54.Laski F.A., Alzner-DeWeerd,B., RajBhandary,U.L. and Sharp,P.A. (1982) Nucleic Acids Res., 10, 4609–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willcock D.F., Dryden,D.T. and Murray,N.E. (1994) EMBO J., 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]