Abstract
Male moths utilize their pheromone communication systems to distinguish potential mates from other sympatric species, which contributes to maintaining reproductive isolation and even drives speciation. The molecular mechanisms underlying the evolution of pheromone communication systems are usually studied between closely-related moth species for their similar but divergent traits associated with pheromone production, detection, and/or processing. In this study, we first identified the functional differentiation in two orthologous pheromone receptors, OR14b, and OR16, in four Helicoverpa species, Helicoverpa armigera, H. assulta, H. zea, and H. gelotopoeon. To understand the substrate response specificity of these two PRs, we performed all-atom molecular dynamics simulations of OR14b and OR16 based on AlphaFold2 structural prediction, and molecular docking, allowing us to predict a few key amino acids involved in substrate binding. These candidate residues were further tested and validated by site-directed mutagenesis and functional analysis. These results together identified two hydrophobic amino acids at positions 164 and 232 are the determinants of the response specificity of HarmOR14b and HzeaOR14b to Z9-14:Ald and Z9-16:Ald by directly interacting with the substrates. Interestingly, in OR16 orthologs, we found that position 66 alone determines the specific binding of Z11-16:OH, likely via allosteric interactions. Overall, we have developed an effective integrated method to identify the critical residues for substrate selectivity of ORs and elucidated the molecular mechanism of the diversification of pheromone recognition systems.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-04837-1.
Keywords: Helicoverpa, Pheromone receptor, AlphaFold2, Molecular docking, Molecular dynamics simulations, Site-directed mutagenesis
Introduction
A crucial event in sexual reproduction is to locate or recruit a mate, and in many insect groups, intraspecific communication between sexes is primarily mediated by sex pheromones [1]. Typically, in Lepidoptera, female moths act as signal senders and males are signal receivers, constituting a species-specific communication pathway that serves as a behavioral prezygotic barrier to mating, which is sufficient to drive reproductive isolation and can lead to speciation [2–4]. Pheromone signals are under strong stabilizing selection because subtle shifts in composition can disfavor the upwind flight behavior of males of one species and turn on the response of males of a different species [5–7]. One longstanding but unsolved question in the study of pheromone communication is how the species-specific pheromone sensing systems can adapt to novel sex-pheromone-related molecules or changes in ratios of the existing molecular blends because strong sexual selection between signal sender and receiver appears to allow little possibility for change. According to the asymmetric tracking hypothesis, male preference varies more frequently than female pheromones. Thus, mutations that produced divergent sex pheromones could still be tracked by male receivers that are pre-adapted to a broader range of signals [4, 8, 9]. However, it is still elusive how the novel pheromone communication channel was ultimately fixed. Illustrating the genetic basis and evolutionary mechanisms of pheromone communication systems requires us to comprehensively dissect divergences in the compositions and recognition traits of pheromones in closely related taxa.
In moths, sex pheromones usually consist of a bouquet of structurally similar blends [10, 11], with species-specificity due to the use of a few novel components within a critical range of ratios [5, 12, 13]. Differences in pheromone compositions (qualitative or/and quantitative differences) are usually associated with genetic variation in biosynthesis pathways [6, 14–16]. Additionally, the pheromone perception systems of different moths also exhibit high diversity corresponding to their species-specific sex pheromones. In the peripheral olfactory system of moths, pheromone signals are detected and processed by several proteins, including pheromone binding proteins (PBPs) [17, 18], sensory neuron membrane proteins (SNMPs) [19], pheromone degrading enzymes (PDEs) [20, 21], and pheromone receptors (PRs) [22–24]. Of these, PRs determine the selectivity and specificity of the sex pheromone reception. The functional differentiation of essential PRs results in behavioral preference divergence in closely-related moth species [25, 26]. So far, numerous studies have been conducted on the identification [27–30] and functional deorphanization [24, 31–34] of PRs in moths, which together open an avenue for the discovery of genetic architectures and evolution in pheromone preferences on the molecular level. However, lacking information on PR function in closely-related moth species greatly limits our understanding of the relationships between PR evolution and speciation.
Helicoverpa is an important genus in Lepidoptera, containing 18 species, some of which cause massive economic impacts on a global scale annually [35, 36]. Helicoverpa gelotopoeon (Hgel), H. assulta (Hass), H. armigera (Harm) and H. zea (Hzea) are four closely-related moths, which have diverse geographic distributions. While some of the species have overlapping habitats, such as Harm and Hass, which have the same distributions in Africa, Asia, and Oceania (including Australia), and Hgel and Hzea are distributed in the Americas, they show reproductive isolation from each other [37]. Therefore, they are excellent candidates for studying the evolution of pheromone communication systems concerning divergent traits associated with pheromone production, detection, and processing [7].
The components of the pheromones of the four species have been identified and functionally specified through sex gland composition, behavior, electrophysiology, and field trap attraction investigation [23, 38–48]. The four species use a different chemical as their major pheromone component: Hass uses Z9-16:Ald, and Hgel uses 16:Ald and Z9-16:Ald [42, 46], while Harm and Hzea both use Z11-16:Ald [38–40, 47]. In addition to minor pheromone components, there is evidence indicating that the behavioral effects also exhibit significant divergence between the four species [38–48]. For example, Z11-16:OH, which was identified in the sex glands of female Harm, is antagonistic to Harm and Hzea males but does not affect the behavior of Hass or Hgel [23, 45]. Trace amounts of Z9-14:Ald (~ 0.3%) identified in the sex glands of female Harm were shown to increase attraction to male Harm, but to be behaviorally antagonistic to male Hass, when added to the optimal ratio of their two-component blend respectively [43, 47, 48].
These examples suggest that functional differentiation may have occurred in the PRs of the four species. This hypothesis has been partially verified in the following PR function study on Harm and Hass. In Harm and Hass, seven and six PR genes, respectively, have been identified by analyzing their antennal transcriptome data [29, 49], and their functions have been characterized in a Xenopus oocytes expression system [26, 33, 50]. Functional differentiation was found in two PRs, OR14b and OR16. HarmOR14b and HassOR14b were both narrowly tuned to Z9-14:Ald and Z9-16:Ald but presented different tuning specificities [26]. For OR16, HarmOR16 responded to Z9-14:Ald and Z11-16:OH [33], while HassOR16 was only tuned to Z9-14:Ald [50]. However, the sequences and functional information of PRs are still lacking in other Helicoverpa species, which greatly limits our understanding of the molecular evolution of these ecologically relevant receptors and their contributions to speciation.
In our study, we identified all candidate PRs in Hgel and Hzea and deciphered their functions using the heterologous Xenopus oocytes expression system. After a functional comparison of orthologous genes in the four species, we determined functional differentiation occurred in two PR clades, OR14b and OR16. We then endeavored to determine which residue(s) resulted in the functional shifts of the two genes by AlphaFold2 structural prediction, molecular docking, and molecular dynamics simulations of each PR, followed by site-directed mutagenesis and functional characterization. We found two hydrophobic residues at positions 164 and 232 are responsible for the difference in response specificity towards Z9-14:Ald and Z9-16:Ald in HarmOR14b and HzeaOR14b, and a single hydrophobic residue at position 66 contributes to the functional differentiation of OR16 across the four species. Our results provide critical evidence of PR evolution in Helicoverpa species, which expands our understanding of mechanisms driving the diversification of pheromone recognition.
Materials and methods
OR identification and gene clone
The sequences of 1 odorant receptor co-receptor (Orco) and 7 PR genes from Hzea including HzeaOR6, HzeaOR11, HzeaOR13, HzeaOR14, HzeaOR14b, HzeaOR15, HzeaOR16, and 1 Orco, 5 PR genes from Hgel including HgelOR6, HgelOR11, HgelOR13, HgelOR14b, and HgelOR16 were identified by analyzing the antennal transcriptome data of each species.
The antennae RNA of Hzea and Hgel were provided by Fred Gould’s Lab at North Carolina State University. 1 μg of RNA from male and female antennae of each species was used to synthesize cDNA with a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) following the manufacturer’s instructions.
The mixed cDNA of both sexes was used as a template to amplify the full-length sequences of each gene with gene-specific primers (listed in Table S1) designed with Primer Premier 5.0 software. PrimeSTAR HS DNA polymerase (Takara, Dalian, China) was used to amplify target genes in PCR under the following conditions: 98 °C for 3 min, followed by 35 cycles of 98 °C for 10 s, 57 °C for 5 s and 72 °C for 80 s, with final extension 72 °C for 10 min. PCR products were purified and then subcloned into a cloning vector pEASY-Blunt (TransGen Biotech, Beijing, China) and transformed into Trans-T1 Escherichia coli competent cells (TransGen Biotech, Beijing, China). Successful transformation of colonies was confirmed by colony PCR and sent for sequencing. The verified sequence of each OR was then subcloned into an expression vector pT7Ts with primers containing a restriction site and a Kozak sequence to the 5′ end as well as a different restriction site to the 3′ end. Primers utilized for vector construction were listed in Table S1. Sequences of Orco and PRs from the two species were submitted to GenBank databases under accession numbers MN399784-MN399797.
The PRs and Orco sequences of Harm and Hass used in the experiments were obtained from previous studies [33, 50] and were newly submitted to GenBank under accession number MN399769-MN399783.
Sequence and phylogenetic analysis of OR genes in the four species
The sequences of 29 ORs (including 4 Orco and 25 PRs) in the four species were aligned using ClustalW. A phylogenetic tree was constructed using MEGA-X software. The maximum likelihood method was used to construct the phylogenetic tree with Jones–Taylor–Thornton (JTT) amino acid substitution model. Node support was assessed using a bootstrap procedure based on 1000 replicates. The phylogenetic tree was rooted in a subfamily of the conserved odorant receptor coreceptor, Orco. The selective pressures acting on each OR clade (Orco, OR6, OR11, OR13, OR14, OR14b, OR15, and OR16) were calculated using the CodeML program implemented in the PAML 4.9 package that estimates ratios of the normalized nonsynonymous (dN) to synonymous (dS) substitution rate (ω) via the maximum likelihood method. For each lineage, the codon sequences were aligned using the ClustalW procedure, and a maximum likelihood phylogenetic tree was reconstructed with MEGA-X software. The substitution rate (ω) of each lineage was calculated using the CodeML procedure with Site Model 0: one-ratio.
Sequence alignment of OR14b and OR16 were conducted using DNAMAN 8 software (Version 8, Lynnon Biosoft, Quebec, Canada), and the transmembrane domains were predicted by AlphaFold2.
Pheromone components
Hexadecenal (16:Ald), (Z)-11-hexadecenal (Z11-16:Ald), (Z)-9-hexadecenal (Z9-16:Ald), (Z)-9-tetradecenal (Z9-14:Ald), (Z)-11-hexadecen-1-ol (Z11-16:OH), (Z)-9-hexadecen-1-ol (Z9-16:OH), and (Z)-11-hexadecenyl acetate (Z11-16:OAc) (all over 95% purity) were purchased from Nimrod Inc. (Changzhou, China).
Stock solutions (1 M) used for the two-electrode voltage clamp experiments were prepared in dimethyl sulfoxide (DMSO) and stored at − 20 °C. Before experiments, the stock solution was diluted in 1 × Ringer’s buffer (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 0.8 mM CaCl2, and 5 mM HEPES, pH 7.6) to a working concentration of 100 μM. 1 × Ringer’s buffer containing 0.1% DMSO was used as a negative control.
Preparation of site-directed mutants
The OR mutants were generated by PCR with the methods described by Zhu [51]. To introduce specific mutations into genes encoding OR14b and OR16, a primer bearing the modified nucleotide(s) and another primer complementary either to another region of the gene sequence were used to amplify a short DNA fragment (250–500 bp long) with PrimeSTAR HS DNA polymerase under the following conditions: 98 °C for 1 min, then 35 cycles of 98 °C for 10 s, 58 °C for 5 s, 72 °C for 30 s, and final elongation for 10 min. The target band was excised from the gel, and after extraction, the product was used as a primer in the second round PCR with Phusion DNA Polymerase (Thermo Scientific, Waltham, MA, USA) under the following conditions: 9 cycles of 95 °C for 30 s, 68 °C for 6 min, followed by 68 °C for 16 min. The PCR product was then treated with DpnI (New England Biolabs, Beverly, MA) at 37 °C for 2 h to digest the plasmid templates. The digested product was used for transformation into E. coli competent cells (TransGen Biotech, China) as described above. Five individual colonies were sent to sequence to check for the right mutations. All the primers used to prepare mutants were listed in Table S1.
Functional characterization of PRs and their mutants
The two-electrode voltage-clamp (TEVC) recording in combination with the Xenopus laevis oocytes heterologous expression system has been widely used to deorphanize the ligands of insect odorant receptors (ORs) [24–26]. The caped RNA (cRNA), encoding the candidate OR and Orco were co-injected into Xenopus oocytes. cRNA was synthesized with linearized vector templates of each OR and mutants using an mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions.
Mature oocytes were surgically removed from female X. laevis frogs. Follicle cells were removed by treating them with collagenase I for 1 h at room temperature. 27.6 ng of cRNA encoding each PR subunit together with 27.6 ng of cRNA encoding the Orco subunit were microinjected into healthy oocytes. The injected oocytes were incubated at 18 °C in 1 × Ringer’s buffer supplemented with 5% dialyzed horse serum, 50 μg/mL tetracycline, 100 μg/mL streptomycin and 550 μg/mL sodium pyruvate for 3–5 days prior to electrophysiological recording. Pheromone-inducing currents were recorded using a two-electrode voltage clamp with an OC-725C oocyte clamp (Warner Instruments, Hamden, CT, USA) from the PR-expressed oocytes. Oocytes injected with Orco only were used as a negative control. During the recording, each pheromone was applied for 15 s at a flow rate of 5 mL/min, with extensive washing with 1 × Ringer solution until the response curve back to the baseline after each stimulus. The holding potential was − 80 mV. Data acquisition and analysis were conducted using Digidata 1440A and Pclamp10.0 software (Axon Instruments Inc., Union City, CA, USA).
Structural prediction and molecular docking
We utilized the updated version of AlphaFold2, AF2complex [52], installed on our local computer cluster with default pipeline to predict the heteromeric structures of HarmOrco and HarmOR14b/HarmOR16 in a stoichiometry of two ORs and two Orcos. In principle, these two ORs could be assembled in an adjacent or diagonal position, resulting in two different arrangements. To check which stoichiometry is more favorable, we predicted ten models for the HarmOR14b-HarmOrco complex. The results suggested that the diagonal form has a higher probability (70%) than the adjacent form. The AF2 structures of HarmOR14b and HarmOR16 were subsequently used to dock with the corresponding pheromone compounds (Z9-14:Ald and Z9-16:Ald for HarmOR14b, and Z9-14:Ald and Z11-16:OH for HarmOR16, respectively) using Autodock Vina [53]. The structures of pheromone molecules were obtained from the PubChem database (compound ID 5364643, 5364471, and 5283305 for Z9-16:Ald, Z9-14:Ald, and Z11-16:OH, respectively). The pheromone binding pocket of HarmOR14b and Harm16 is highly similar to the substrate binding site in the cryoEM structures of MhOR5. In our models, the binding modes in two OR pockets of the homotetrameric OR-Orco complex are identical, considering the symmetry.
MD simulations and analysis
The docked structures with lower binding energy and hydrophilic head groups oriented in the direction of the solvent served as the starting models of the pheromone-bound systems, including HarmOR14b with Z9-14:Ald and Z9-16:Ald, and HarmOR16 and HarmOR16-A66L with Z11-16:OH. These models were embedded into a lipid bilayer consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and solvated in a cubic water box containing 0.15 M NaCl to mimic physiological conditions. The systems were organized using the CHARMM-GUI web server [54] and underwent an energy minimization step using the steepest descent algorithm followed by a six-step equilibration during which position constraints were gradually removed. The CHARMM36m force field [55] was used for the protein alongside the POPC lipids. The TIP3P water model was used for the explicit solvent. The force field parameters for pheromone compounds were generated by the CHARMM General Force Field (CGenFF) [56]. In the MD simulations, the temperature was kept constant at 310 K using a Nose–Hoover thermostat with a 1 ps coupling constant, and the pressure at 1.0 bar using the Parrinello–Rahman barostat with a 5 ps time coupling constant. A cut-off of 1.2 nm was applied for the van der Waals interactions using a switch function beginning at 1.0 nm. The cut-off for the short-range electrostatic interactions was also at 1.2 nm and the long-range electrostatic interactions were calculated using the particle mesh Ewald decomposition algorithm with a mesh spacing of 0.12 nm. A reciprocal grid of 144 × 144 × 108 cells was used with fourth-order B-spline interpolation. All simulations were performed using a GPU-accelerated version of Gromacs 2021.5 [57]. For each system, we performed two repeats of 500 ns or 1 μs simulations, as summarized in Table S2.
We used Gromacs gmx tools for data processing. Protein–ligand interactions were analyzed using GetContacts (https://getcontacts.github.io/). The residue distances were calculated with VMD tcl scripts [58]. The molecular structures were visualized using Pymol.
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). The response value among different groups (in Fig. 1, 2, 5, and 6) was analyzed with a general linear model (PROC-GLM) followed by Duncan’s multiple range test with SAS 9.2 for Windows. The response value between Z9-14:Ald and Z9-16:Ald in HarmOR14b, HzeaOR14b, and each mutant (in Fig. 5) was analyzed with paired t-test. Statistical significance was determined at α = 0.05 level.
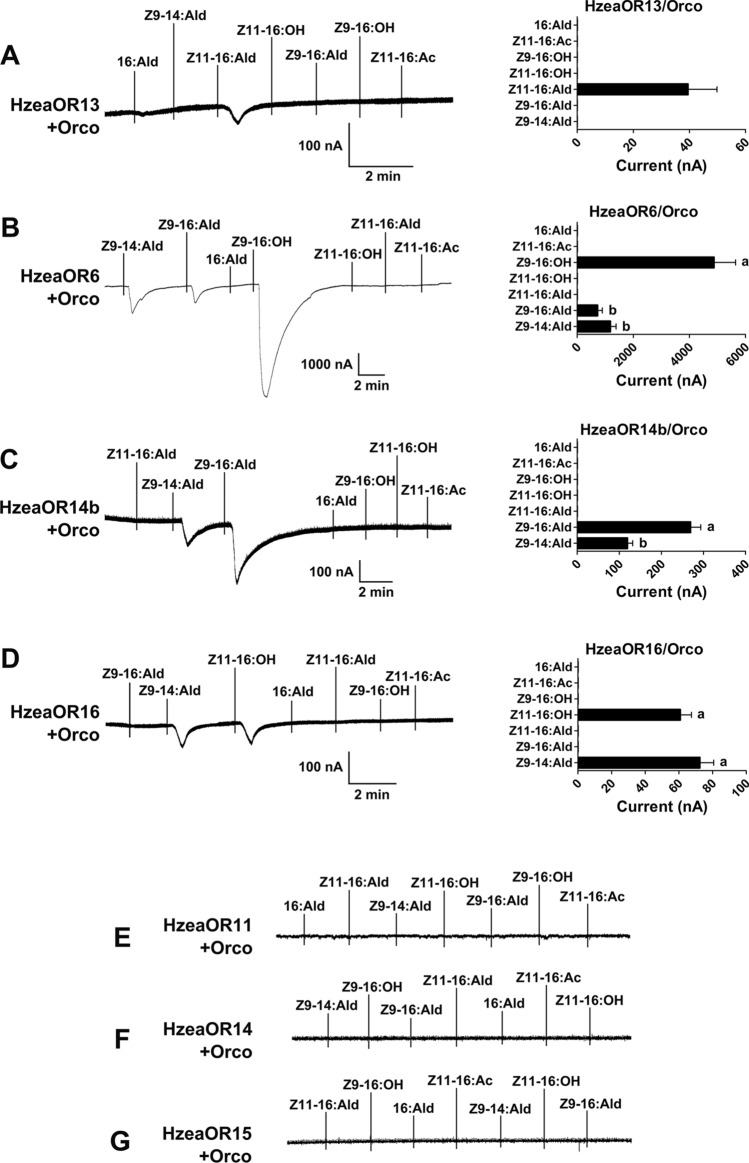
Fig. 1.
Inward current responses (left) and response profiles (right) of co-expressed HzeaPR/HzeaOrco to 10−4 M of each pheromone solution. A HzeaOR13/HzeaOrco (n = 7). B HzeaOR6/HzeaOrco (n = 8, F = 23.34, P < 0.0001). C HzeaOR14b/HzeaOrco (n = 27, F = 30.35, P < 0.0001). D HzeaOR16/HzeaOrco (n = 9, F = 1.21, P = 0.2874). E HzeaOR11/HzeaOrco. F HzeaOR14/HzeaOrco. G HzeaOR15/HzeaOrco. Error bars indicate SEM, one-way ANOVA, and Duncan test, each column with different letters indicates a significant difference, α = 0.05
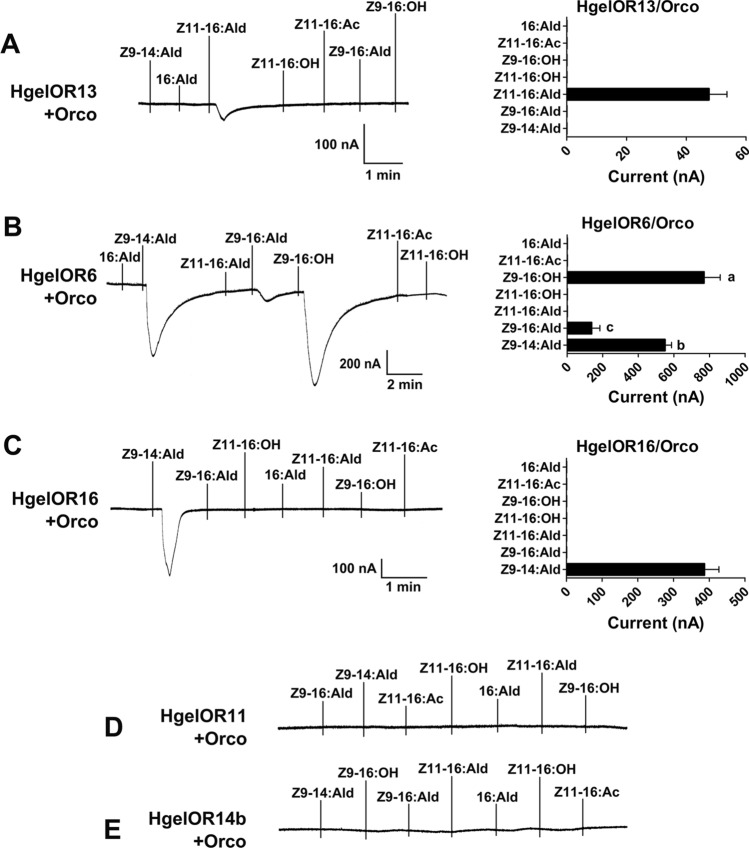
Fig. 2.
Inward current responses (left) and response profiles (right) of co-expressed HgelPR/HgelOrco to 10−4 M of each pheromone solution. A HgelOR13/HgelOrco (n = 6). B HgelOR6/HgelOrco (n = 7, F = 26.06, P < 0.0001). C HgelOR16/HgelOrco (n = 23). D HgelOR11/HgelOrco. E HgelOR14b/HgelOrco. Error bars indicate SEM, one-way ANOVA, and Duncan test, each column with different letters indicates a significant difference, α = 0.05
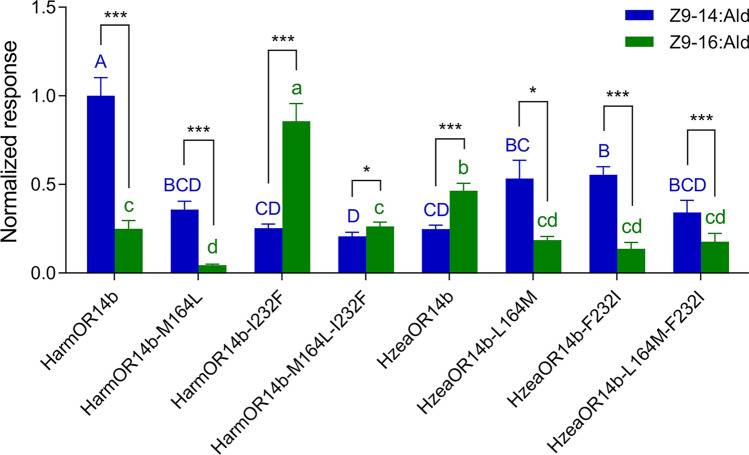
Fig. 5.
Comparison of the response of HarmOR14b, HzeaOR14b, and their mutants at position 232 to Z9-14:Ald and Z9-16:Ald. The response of HarmOR14b, HzeaOR14b, and their mutants to Z9-14:Ald and Z9-16:Ald at a concentration of 10−4 M. Response values were normalized by defining the mean response of HarmOR14b to Z9-14:Ald as 1. The response to Z9-14:Ald and Z9-16:Ald of the two receptors and mutants are compared individually, with each column with a different letter indicating a significant difference. Error bars indicate SEM (n = 8–13), one-way ANOVA, Duncan test; paired t-test
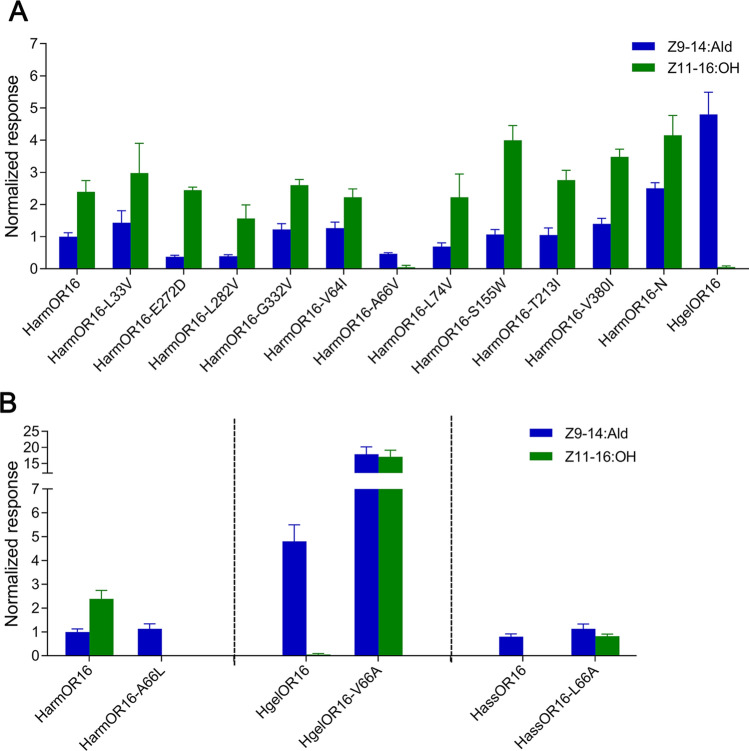
Fig. 6.
Comparison of the response of OR16 and its mutants to Z9-14:Ald and Z9-16:Ald. Response values were normalized by defining the mean response value of HarmOR16 to Z9-14:Ald as 1. A The response of HarmOR16 and its mutants to Z9-14:Ald and Z11-16:OH at a concentration of 10−4 M. Error bars indicate SEM (n = 3–20). B Left: The response of HarmOR16 and its mutants to Z9-14:Ald and Z11-16:OH at a concentration of 10−4 M. (Middle) The response of HgelOR16 and its mutants to Z9-14:Ald and Z11-16:OH at a concentration of 10−4 M. Right: The response of HassOR16 and its mutants to Z9-14:Ald and Z11-16:OH at a concentration of 10−4 M. Error bars indicate SEM (n = 6–20). HarmOR16-N represents the mutant mutating all the nine candidate residues except A66 in HarmOR16
Results
Sequence analysis of candidate ORs in the four Helicoverpa species
In this study, seven candidate PR genes, namely, HzeaOR6, HzeaOR11, HzeaOR13, HzeaOR14, HzeaOR14b, HzeaOR15, and HzeaOR16, were identified in Hzea from antennal transcriptome data. In the transcriptome of Hgel, only five candidate PRs genes (HgelOR6, HgelOR11, HgelOR13, HgelOR14b, and HgelOR16) were identified. HgelOR14 and HgelOR15 were found to be pseudogenes due to the presence of premature termination codes in their open reading frames (ORFs). The full-length sequences of the seven candidate PR genes in Hzea and the five candidate PR genes in Hgel were cloned using PCR. The coding length of these PRs ranged from 422 to 440 amino acid and were all predicted to possess seven transmembrane domains (TMDs). The sequence identities of each orthologous PRs—OR6, OR11, OR13, OR14, OR14b, OR15, and OR16—in the four species were 96.94%, 99.25%, 97.76%, 97.95%, 95.34%, 96.36%, and 96.74%, respectively.
Functional study of candidate PRs in Hzea and Hgel
The seven candidate PRs in Hzea were functionally investigated by co-expression with HzeaOrco in Xenopus oocytes, followed by testing using a panel of pheromone components from the four selected species. The oocytes expressing HzeaOR13 specifically responded to its major pheromone component Z11-16:Ald (Fig. 1A). HzeaOR6 broadly responded to Z9-14:Ald, Z9-16:Ald, and Z9-16:OH, with the largest response to Z9-16:OH (F = 23.34, P < 0.0001) (Fig. 1B). HzeaOR14b could be activated by both Z9-14:Ald and Z9-16:Ald, with a greater response to Z9-16:Ald (F = 30.35, P < 0.0001) (Fig. 1C). HzeaOR16 showed similar responses to Z9-14:Ald and Z11-16:OH (F = 1.21, P = 0.2874) (Fig. 1D). The other three candidate PRs, HzeaOR11, HzeaOR14, and HzeaOR15, failed to respond to any tested pheromone components (Fig. 1E–G).
Similarly, the five candidate PRs in Hgel were co-expressed with HgelOrco in Xenopus oocytes and functionally deorphanized as described for Hzea. HgelOR13-expressing oocytes specifically responded to Z11-16:Ald (Fig. 2A), which is not included in the pheromone components of Hgel. Oocytes expressing HgelOR6 could be activated by three pheromone compounds, Z9-14:Ald, Z9-16:Ald, and Z9-16:OH, with the most significant response to Z9-16:OH, and the most negligible response to Z9-16:Ald (F = 26.06, P < 0.0001) (Fig. 2B). HgelOR16 was specifically and strongly tuned to Z9-14:Ald, but had no response to other test chemicals (Fig. 2C). HgelOR11 and HgelOR14b could not be activated by the tested pheromone compounds (Fig. 2D, E).
Phylogenetic analysis and functional comparison of candidate ORs of the four Helicoverpa species
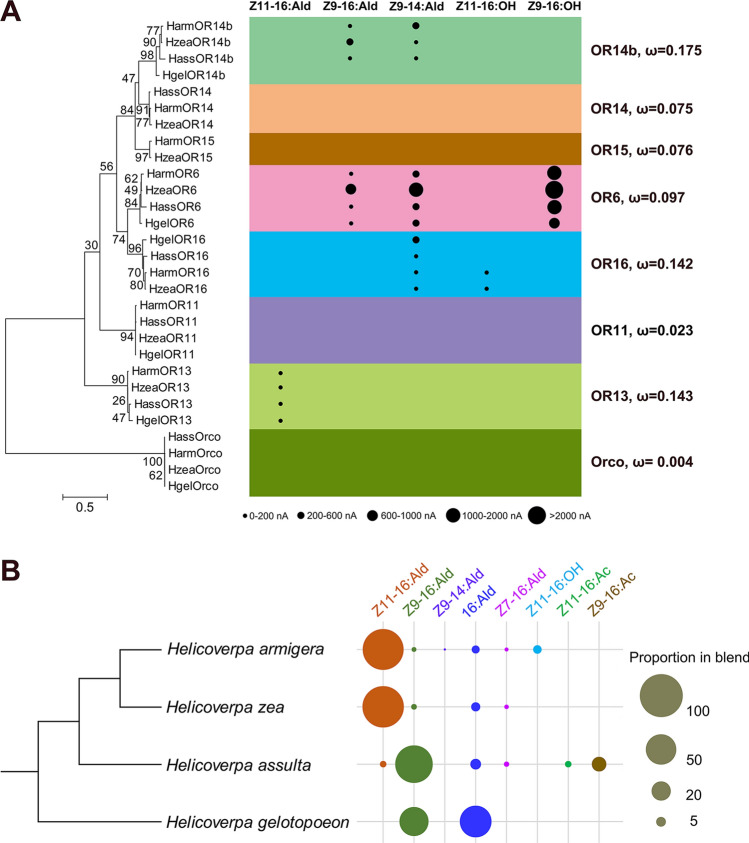
4 Orco genes and 25 PR genes (NCBI accession number MN399769-MN399797) from four Helicoverpa moths were used to construct a maximum likelihood phylogenetic tree (Fig. 3A). These 29 OR sequences clustered into 8 orthologous lineages, one conserved Orco lineage, and seven traditional moth PR lineages. The normalized nonsynonymous (dN) to synonymous (dS) substitution rate (ω) of each clade ranged from 0.004 to 0.175 (Fig. 3A).
Fig. 3.
Comparison of sex pheromone components and PR functions in the four Helicoverpa species. A Phylogenetic analysis and functional comparison of candidate pheromone receptors. The phylogenetic tree was constructed using MEGA-X software with the maximum likelihood method, and bootstrap values are based on 1000 replicates. The size of each dot represents the magnitude of the response of each pheromone receptor towards its agonists. Each PR clade is marked in a different color, and the nonsynonymous (dN) to synonymous (dS) substitution ratio (ω) is labeled alongside. B Composition of female sex pheromones in the four species. The size of each dot is proportional to the ratio of an individual pheromone component. The evolutionary relationships of the four specified Helicoverpa species were estimated as previously described [70]. The pheromone compositions of each species were arranged according to data from previous studies [38, 39, 42, 46, 47]
To effectively showcase and contrast the roles of equivalent PRs in each group, their response values were categorized into five sections and depicted by dots of varying sizes in ascending order (Fig. 3A). In the four species, the function of OR13 orthologs was conserved and specifically responded to Z11-16:Ald (Fig. 3A), although Z11-16:Ald is not the major pheromone component of Hgel or Hass (Fig. 3B). OR6 responded to three of the same chemicals in all four species, Z9-16:OH, Z9-16:Ald, and Z9-14:Ald. Although each species-specific ligand had a different response value, they all had the highest response to Z9-16:OH and the lowest response to Z9-16:Ald. Functional differentiation was found in the OR14b and OR16 clades. HgelOR14b showed no response to any tested pheromones, but the remaining three OR14b all responded to Z9-16:Ald and Z9-14:Ald with different specificities. HzeaOR14b and HassOR14b showed greater responses to Z9-16:Ald (in Fig. 3A, the response values of HassOR14b to the two aldehydes were represented by the same dot because the responses to them were both less than 200 nA, but the response values, while small, were quite different; see Ref. [24]), the major pheromone component of Hass (Fig. 3B), while HarmOR14b exhibited a greater response to Z9-14:Ald, a pheromone component only identified in female Harm, albeit in trace amounts (Fig. 3B). In particular, the functional acquisition was detected in the OR16 orthologs of these four species. HassOR16 and HgelOR16 were specifically tuned to Z9-14:Ald, while HarmOR16 and HzeaOR16 could be activated by either Z9-14:Ald or Z11-16:OH (Fig. 3A). Z11-16:OH is a minor pheromone of Harm (Fig. 3B), and acts as a behavioral antagonist in Harm and Hzea [40, 45].
Two residues at positions 164 and 232 in the substrate binding site determine the pheromone selectivity of HarmOR14b and HzeaOR14b
Three of the four OR14b orthologs, HarmOR14b, HzeaOR14b, and HassOR14b, are tuned to both Z9-14:Ald and Z9-16:Ald, with different specificities. HzeaOR14b and HassOR14b [26] exhibited higher responses to Z9-16:Ald, while HarmOR14b showed a higher response to Z9-14:Ald. It has been demonstrated that altering the amino acids residues at positions 232 and 355 can affect the ligand selectivity of HarmOR14b and HassOR14b [26]. Therefore, our goal was to pinpoint the specific residues that result in the functional differentiation of HarmOR14b and HzeaOR14b which share 97.05% sequence identity with 13 divergent residues (13 out of 440; Fig. S1). This is a very difficult task and requires substantial work, considering the exponentially possible combinations of these candidate residues.
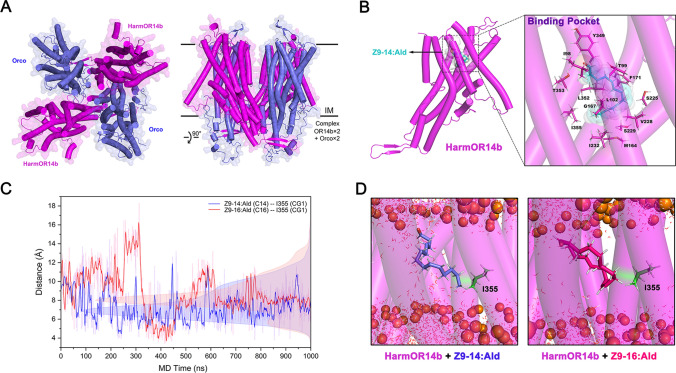
To gain insight into the specific residues that play a role in functional differentiation, we further resorted to the computational structural biology method, since the experimentally determined structure of pheromone receptors from Helicoverpa species is not available so far. A recently developed machine learning-based approach, AlphaFold2 (AF2) [59], is able to yield highly accurate structure prediction of proteins, including membrane proteins. We used its updated version AF2complex [52] to predict the heteromeric structures of HarmOR14b in complex with HarmOrco. Based on previous studies, a hypothetical stoichiometry of two ORs and two Orcos was proposed [60], while the precise stoichiometry of OR and Orco is still unknown in the heteromeric complex. In principle, these two ORs/Orcos could assemble in an adjacent or diagonal position, resulting in two different sub-stoichiometries. We generated ten AF2 models for the (HarmOR14b)2–(HarmOrco)2 complex of which 70% were predicted to be in the diagonal form than in the adjacent form. Therefore, based on the AF2 structures of heteromeric complexes in a diagonal assembly form (Fig. 4A), we predicted the substrate-bound structures of HarmOR14b-HarmOrco heterotetramer by symmetrically docking pheromone compounds (Z9-14:Ald and Z9-16:Ald, respectively) into the substrate pockets which are similar to the substrate-bound pockets captured in the cryoEM structure of MhOR5 in complex with eugenol [61] (Fig. 4A, B).
Fig. 4.
Molecular dynamics simulations of substrate-bound HarmOR14b. A Representative structure of the heteromeric structures of HarmOR14b in complex with HarmOrco in a stoichiometry of two ORs and two Orcos predicted by Alphafold2. B Close-up view of the docking structure of HarmOR14b in complex with Z9-14:Ald (represented by cyan sticks). The transparent surface and the sticks around Z9-14:Ald show the surrounding residues in the binding pocket. C shows the MD trajectories of the distance between I355 and the endmost carbon of Z9-14:Ald (blue line) and Z9-16:Ald (red line). D Structural snapshots to show the interactions between HarmOR14b and Z9-14:Ald (left), and Z9-16:Ald (right), respectively, observed in 1 μs MD simulations. The phosphate head groups of POPC are shown as orange spheres. The transmembrane helices of HarmOR14b are represented by transparent cylinders
To further identify the key residues and quantify their contributions to the pheromone interactions, we performed explicit solvent all-atom MD simulations of HarmOR14b complexed with Z9-14:Ald and Z9-16:Ald based on these docked structures (Fig. 4B). The simulations suggested that Z9-16:Ald was less stable than Z9-14:Ald in the binding site as evident from the more significant fluctuations and the larger average distance of Z9-16:Ald from the pocket (Fig. 4C), consistent with the observation that HarmOR14b exhibited a greater response to Z9-14:Ald than Z9-16:Ald. This may be due to the long tail of Z9-16:Ald leading to a less proper fit with the pocket than Z9-14:Ald (Fig. 4D). Moreover, the MD simulations allowed us to identify a few critical residues contributing to substrate binding as ranked by their binding frequencies (Fig. S2). Among these residues, only two at positions 164 and 232 differ between HarmOR14b and HzeaOR14b, as indicated by the sequence alignment (Fig. S1).
To verify that these two sites are the molecular determinant for the functional differentiation between OR14b of these two species, we altered the residues at positions 164 and 232 in HarmOR14b and HzeaOR14b and measured the impacts on substrate response. The results suggested that a single mutation at position 164 (M164L) of HarmOR14b still had a stronger response to Z9-14:Ald (t = 7.14, P < 0.001) but with lower electrophysiological responses to both chemicals, having the same specificity as the wide type of HarmOR14b (Fig. 5). However, the L164M mutant reverses the substrate response of HzeaOR14b, resulting in a similar response to HarmOR14b, with a stronger response to Z9-14:Ald (Fig. 5, t = 3.76, P = 0.013). Remarkably, the mutants at position 232 (HarmOR14b-I232F and HzeaOR14b-F232I) resulted in the alteration of substrate response in comparison to their wide types, and the HarmOR14b-1232F mutant even has a stronger response to Z9-16:Ald than HzeaOR14b (Fig. 5), indicating the essential role of this site in the functional differentiation of OR14b in Helicoverpa species.
To further uncover the synergistic effects of both positions, we mutated these two sites simultaneously in HzeaOR14b and HarmOR14b. The results suggested that the double mutation in HarmOR14b resulted in a significantly lower response to Z9-14:Ald and an unaltered response to Z9-16:Ald. This was coupled with a slightly stronger response to Z9-16:Ald than to Z9-14:Ald (t = − 3.04, P = 0.016) but a weaker response to Z9-16:Ald in comparison with the wide type HzeaOR14b (Fig. 5). Moreover, the double mutant of HzeaOR14b (L164M-F232I) had a weaker but similar response pattern to HarmOR14b with a stronger response to Z9-14:Ald than Z9-16:Ald (t = 6.88, P < 0.001).
One single position determines the binding selectivity of OR16 to Z11-16:OH via indirect interactions
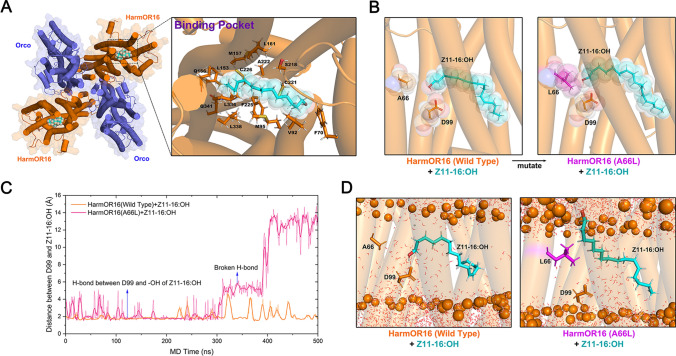
Functional differentiation was detected in OR16 orthologs. HassOR16 and HgelOR16 specifically respond to Z9-14:Ald, whereas HzeaOR16 and HarmOR16 respond to both Z9-14:Ald and Z11-16:OH [33]. To uncover the critical residues contributing to the function acquisition in HzeaOR16 and HarmOR16, we separated the four OR16 orthologs into two groups according to their response profiles. The sequence alignment indicated a total of ten different sites between group 1 (HarmOR16 and HzeaOR16) and group 2 (HassOR16 and HgelOR16) (Fig. S3). We individually mutated all ten residues from HarmOR16 to HgelOR16, and the results indicated that only the mutant HarmOR16-A66V lost the response to Z11-16:OH (Fig. 6A). To verify the importance of A66, we mutated all other nine residues together except A66 in HarmOR16 to the corresponding sites in Hgel, named HarmOR16-N. Those results showed that the mutant HarmOR16-N still could be activated by Z11-16:OH (Fig. 6A), suggesting that A66 is essential for HarmOR16 to bind Z11-16:OH. To further confirm the residue at position 66 was the key site triggering the functional differentiation of OR16 orthologs, we mutated it in HgelOR16 (V66) and HassOR16 (L66) to the corresponding residue in HarmOR16 (A66) and, mutated it in HarmOR16 (A66) to HassOR16 (L66). The results demonstrated that the HgelOR16-V66A and HassOR16-L66A mutants gained the response to Z11-16:OH, and as expected, the mutant HarmOR16-A66L lost its response to Z11-16:OH (Fig. 6B).
To provide molecular insights into the specificity, we again resorted to a similar computational structural biology protocol as was done for HarmOR14b. The MD simulations of HarmOR16 in complex with Z11-16:OH (Fig. 7A) suggested that A66 in HarmOR16 was distant from the substrates and did not participate in the direct interaction with pheromones, while the side chain of D99 formed a stable hydrogen bonding interaction with the hydroxide radical of Z11-16:OH. To understand how the distant A66 could impact pheromone recognition, we also performed MD simulations of the HarmOR16-A66L mutant based on the same docked structure (Fig. 7B). The simulations revealed that the hydrogen bonding interaction with the Z11-16:OH substrate was unstable and broken after a few hundred nanoseconds (Fig. 7C, D). Considering Leu is bulkier and has a higher hydrophobicity than Ala, it seems that a larger and stronger hydrophobic residue at position 66 is unfavorable to the binding of Z11-16:OH. We, therefore, propose that in wild-type HarmOR16, the side chain of A66 is small enough to give sufficient space for accommodating Z11-16:OH. In contrast, in the A66L mutant, the bulkier side chain may stretch into the neighboring hydrophilic domain and weaken the hydrogen bonding interactions between Z11-16:OH and the hydrophilic D99, resulting in the loss of the response to Z11-16:OH.
Fig. 7.
Molecular dynamics simulations of substrate-bound HarmOR16. A Structure of the HarmOR16-Z11-16:OH complex after MD relaxation. HarmOR16 and Orco subunits are colored blue and orange, respectively. Z11-16:OH is represented by spheres. The binding pocket is zoomed-in to show the residues surrounding Z11-16:OH. B The A66L mutation of HarmOR16 would cause steric clashes with the bound Z11-16:OH. C MD trajectories of the distance between the sidechain oxygen atom of Asp 99 and the hydroxyl radical of Z11-16:OH (wild-type in red, A66L in blue). D Representative structures to show typical substrate binding modes of the wide-type HarmOR16 and its A66L mutant observed in MD simulations. The phosphate headgroups of POPC are shown as orange spheres and the transmembrane helices of HarmOR16 are represented by transparent cylinders. The hydrogen bonding interaction with Z11-16:OH was quickly broken in the mutant
Discussion
In moths, reproductive isolation between closely related species is often operationalized by species-specific sex pheromones emitted by females [2, 4, 12], which function to minimize male and female copulation errors and to optimize conspecific mate finding and the reproductive fitness of both sexes. Likewise, the species-specific sex pheromone detection system of male moths is also essential for reproductive isolation. The pheromone preferences of male moths can be associated with odor recognition specificity of the peripheral nervous system and/or with odor processing and discrimination in the central nervous system. However, PRs are a key element of the peripheral nervous system, directly binding to pheromone molecules and initiating signal transduction, making them essential targets for studying the pheromone detection evolution in closely related species.
In this study, to investigate the evolution of pheromone recognition systems as well as a process that may have given rise to four Helicoverpa species, we made a detailed and prolific comparison of the functions of their PRs to identify the changes most likely to be responsible for PR functional shifts. Of the seven PR clades, OR6, OR13, OR14b, and OR16, can be activated by pheromones or analogs (Fig. 3A). In all four species, OR13 responds specifically to Z11-16:Ald, although it accounts for different ratios and acts in divergent roles in the four species (Fig. 3B). Z11-16:Ald is not a sex pheromone component in Hgel but acts as a pheromone antagonist in the species [46]. HgelOR13 is the receptor for Z11-16:Ald and enables Hgel to recognize this component, but the opposite behavioral response to Z11-16:Ald in Hgel may be due to a change in the projection position of the HgelOR13 neurons in the brain. In Harm, Hzea, and Hass, the proportion of olfactory receptor neurons sensing Z11-16:Ald corresponds to the proportion of this component in their pheromone blends [50, 62] (Fig. 3B). In the four species, OR6 responds to three of the same chemicals with different magnitudes across each case. However, they all show a higher response to Z9-16:OH than to Z9-16:Ald, even though Z9-16:OH is not a pheromone component in any of the four species (Fig. 3B). Its behavioral effect has not yet been verified. We found functional differentiation in both the OR14b and OR16 orthologs. HgelOR14b could not be activated by any tested chemicals, but the other three OR14b orthologs all responded to Z9-16:Ald and Z9-14:Ald with different degrees of specificity (Fig. 3A). In the case of OR16, HarmOR16, and HzeaOR16 are tuned to Z9-14:Ald and Z11-16:OH, while HassOR16 and HgelOR16 both responded to Z9-14:Ald only, which is consistent with the previous results by expressing OR16 in Drosophila T1 neurons [63]. These findings indicate that the pheromone communication system of the four Helicoverpa species has the ability to recognize a wider range of pheromone components than those emitted by their conspecific females. It aligns well with the asymmetric tracking hypothesis [8, 9].
Structural biology should provide valuable guidance on the key sites, but unfortunately, to date, only the structures of the homotetrameric Orco from the parasitic fig wasp (Apocrypta bakeri, AbakOrco) and the homotetrameric OR from a promiscuous OR from the evolutionarily primitive jumping bristletail (Machilis harabi, MhOR5) have been solved by cryogenic electron microscopy (cryoEM) [61, 64]. To further investigate the key residues that are responsible for the functional differentiation of the two PR clades respectively, AlphaFold2 structure prediction, followed by molecular docking and MD simulations, were utilized. The odorant receptor-phospholipid bilayer system was prepared using a well-established protocol [65, 66] and thoroughly relaxed via a sufficiently long equilibrium simulation. Extensive site-directed mutagenesis of candidate sites in the two PR groups revealed that two residues at positions 164 and 232 contributed to the response specificity of HarmOR14b and HzeaOR14b (Fig. 5), and a single residue at position 66 determined the selectivity of OR16 (Fig. 6). However, we noticed that the identified key residues have divergent roles in the functional shifts of the two PRs. In OR14b, the two residues located at positions 164 and 232 are directly involved in the binding to the ligands Z9-14:Ald and Z9-16:Ald (Fig. 4A), while in OR16, the residue at position 66 does not directly interact with Z11-16:Ald but may reduce the binding of Z11-16:OH by weakening the hydrogen bonding interacting between the hydroxide radical of Z11-16:OH and the residue at position 99. In prior studies, a few residues in several insect ORs have been demonstrated to be essential for their ligand binding, especially those residues in the TMDs or the extracellular loops (ECLs), which were thought to form the ligand-binding pocket [25, 26, 67–69]. However, no evidence has demonstrated that these residues are involved in the direct binding to the ligands. Our results give new insight into the relationship between the structure and function of insect ORs.
Only OR14b in Hgel did not respond to the tested pheromone components in the four Helicoverpa species, this is because HgelOR14b duplicated more recently than OR14b of the other Helicoverpa species. Likely, HgelOR14b has yet to gain this specific function. Interestingly, HgelOR16 and HassOR16 have the same ligand, which differs from the other two OR16 orthologs found in Harm and Hzea. It is consistent with the evolutionary relationships of the four species, with Hass and Hgel being basal to Harm and Hzea [70]. Furthermore, no additional compounds are required to prevent the cross-attraction of Hgel with other Helicoverpa species. Although we did not find a receptor in Hgel that could be activated by 16:Ald, it is one of its major pheromone components, implying that there must be one another receptor that is responsible for sensing this component, nonetheless. It is easier to understand the functional differences between the Hgel PRs and those of the other three species if we consider that of the Helicoverpa species we studied, Hgel was first to diverge and also possesses divergent pheromone components compared with them.
The divergence of Hass is in turn basal to that of Harm and Hzea, having some different characteristics with the latter two species, such as their major pheromone components and a lacking OR15 transcript in antennal transcriptome data. However, functional differentiation only occurred in OR16, but not OR14b, when comparing Hass with Harm/Hzea. OR16 has acquired the ability to recognize Z11-16:OH in Harm and Hzea during evolution, as evidenced by Z11-16:OH being a pheromone antagonist in both Harm and Hzea [23, 44, 45]. There is still no evidence that Z11-16:OH impacts the behaviors of Hass or Hgel. A single residue located at the first transmembrane domain could change the binding activities of OR16 concerning Z11-16:OH in both directions in the two groups, HarmOR16/HzeaOR16 and HassOR16/HgelOR16. It is likely that the residue has been recently evolved during the separation of the two groups.
Of the reported species, Harm and Hzea are the most closely related [70], and remain similar in reproductive aspects. It has been demonstrated that they can mate and produce fertile offspring in reciprocal crosses inbred for two generations or in lines backcrossed for four generations in the laboratory [71]. In recent studies, unequivocal evidence of hybrids between the two species was found in South America, raising great concern that novel hybrid ecotypes may form and spread in the region [72]. Among their seven orthologous PRs, only OR14b showed different binding profiles to their ligands, which may partly explain why they could mate in the lab or even in nature, but still present partial reproductive isolation given the small amount of morphological divergence in their genitalia [73]. Hzea was reported to have formed after Harm entered the Americas ~ 1.5 Mya [74, 75], and the slight difference in PR function between them could be explained by genetic drift resulting from the long period of geographic isolation. There is no evidence that their PRs are under selective pressure to differentiate from each other.
Our findings not only assist us understand the structure–function relationship of moth PRs, but also provide new insights into the relationship between the evolution of PRs and the speciation of the four Helicoverpa species.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Fred Gould (North Carolina State University, Raleigh, NC, USA) for providing the experimental materials and comments. We also thank Dr. Emmanuelle Jacquin-Joly (INRAE, Sorbonne Universite, CNRS, IRD, UPEC, Universite de Paris, Institute of Ecology and Environmental Sciences of Paris) for helpful discussions. Y.W acknowledges the access to computational resources from the Information Technology Center and State Key Lab of CAD&CG, Zhejiang University.
Abbreviations
- OR
Odorant receptor
- Orco
Odorant receptor co-receptor
- PBP
Pheromone binding protein
- SNMP
Sensory neuron membrane protein
- PDE
Pheromone degrading enzyme
- PR
Pheromone receptor
- cRNA
Caped RNA
- TMD
Transmembrane domain
- ECL
Extracellular loop
- cryoEM
Cryogenic electron microscopy
- CGenFF
CHARMM general force field
Author contributions
Conceptualization, YL, GW and YW; conducted experiments, SC, YL, CS and BW; data analysis and visualization, SC and CS; writing—original draft, SC, YL and CS; writing—review and editing, SC, CS, BW, PX, YW, YL and GW; funding acquisition, SC, PX, YW, YL and GW.
Funding
This work was funded by National Natural Science Foundation of China (32130089 to G.W., 32272540, 32072509 to Y.L., and 11932017 to P.X.), the National Key R&D Program of China (No. 2021YFF1200404 to Y.W.), the Fundamental Research Funds for the Central Universities of China (No. K20220228 to Y.W.), China Postdoctoral Science Foundation (2020M680785 to S.C.), and the Shenzhen Science and Technology Program (Grant No. KQTD20180411143628272 to G.W.), Central Public-interest Scientific Institution Basal Research Fund (CAAS-ZDRW202108), Projects subsidized by Special Funds for Science Technology Innovation and Industrial Development of Shenzhen Dapeng New District (PT202101-02), and the Agricultural Science and Technology Innovation Program (ASTIP). The funder had no role in study design, data collection and interpretation or manuscript preparation.
Data availability
All data generated or analyzed during this study are included in this article (and its Supplementary files).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Song Cao and Chen Shi contributed equally.
Contributor Information
Yong Wang, Email: yongwang_isb@zju.edu.cn.
Yang Liu, Email: yangliu@ippcaas.cn.
Guirong Wang, Email: wangguirong@caas.cn.
References
- 1.Cardé RT, Baker TC. Sexual communication with pheromones. In: Bell WJ, Cardé RT, editors. Chemical ecology of insects. Boston: Springer; 1984. pp. 355–383. [Google Scholar]
- 2.Baker TC. Mechanism for saltational shifts in pheromone communication systems. Proc Natl Acad Sci USA. 2002;99:13368–13370. doi: 10.1073/pnas.222539799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groot AT, Horovitz JL, Hamilton J, Santangelo RG, Schal C, Gould F. Experimental evidence for interspecific directional selection on moth pheromone communication. Proc Natl Acad Sci USA. 2006;103:5858–5863. doi: 10.1073/pnas.0508609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smadja CM, Butlin RK. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity. 2009;102:77–97. doi: 10.1038/hdy.2008.55. [DOI] [PubMed] [Google Scholar]
- 5.Löfstedt C. Moth pheromone genetics and evolution. Philos Trans R Soc Lond B Biol Sci. 1993;340:167–177. doi: 10.1098/rstb.1993.0055. [DOI] [Google Scholar]
- 6.Roelofs WL, Liu W, Hao G, Jiao H, Rooney AP, Jr, Linn CE. Evolution of moth sex pheromones via ancestral genes. Proc Natl Acad Sci USA. 2002;99:13621–13626. doi: 10.1073/pnas.152445399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillier NK, Baker TC. Pheromones of Heliothine moths. In: Allison JD, Cardé RT, editors. Pheromone communication in moths: evolution, behavior, and application. Berkeley: University of California Press; 2016. pp. 301–333. [Google Scholar]
- 8.Phelan PL. Evolution of sex pheromones and the role of asymmetric tracking. In: Roitberg BD, Isman MB, editors. Insect chemical ecology: an evolutionary approach. New York: Chapman and Hall; 1992. pp. 265–314. [Google Scholar]
- 9.Phelan PL. Genetic and phylogenetics in the evolution of sex pheromones. In: Cardé TR, Minks KA, editors. Insect pheromone research. New York: Chapman and Hall; 1997. pp. 563–579. [Google Scholar]
- 10.Jurenka RA, Subchev M, Abad J-L, Choi M-Y, Fabriàs G. Sex pheromone biosynthetic pathway for disparlure in the gypsy moth, Lymantria dispar. Proc Natl Acad Sci USA. 2003;100:809–814. doi: 10.1073/pnas.0236060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yew JY, Chung H. Insect pheromones: an overview of function, form, and discovery. Prog Lipid Res. 2015;59:88–105. doi: 10.1016/j.plipres.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Löfstedt C, Herrebout WM, Menken SBJ. Sex pheromones and their potential role in the evolution of reproductive isolation in small ermine moths (Yponomeutidae) Chemoecology. 1991;2:20–28. doi: 10.1007/BF01240662. [DOI] [Google Scholar]
- 13.Groot AT, Dekker T, Heckel DG. The genetic basis of pheromone evolution in moths. Annu Rev Entomol. 2016;61:99–117. doi: 10.1146/annurev-ento-010715-023638. [DOI] [PubMed] [Google Scholar]
- 14.Lassance J-M, Groot AT, Liénard MA, Antony B, Borgwardt C, Andersson F, Hedenström E, Heckel DG, Löfstedt C. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature. 2010;466:486–489. doi: 10.1038/nature09058. [DOI] [PubMed] [Google Scholar]
- 15.Lassance J-M, Liénard MA, Antony B, Qian S, Fujii T, Tabata J, Ishikawa Y, Löfstedt C. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc Natl Acad Sci USA. 2013;110:3967–3972. doi: 10.1073/pnas.1208706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buček A, Matoušková P, Vogel H, Šebesta P, Jahn U, Weiβflog J, Svatoš A, Pichová I. Evolution of moth sex pheromone composition by a single amino acid substitution in a fatty acid desaturase. Proc Natl Acad Sci USA. 2015;112:12586–12591. doi: 10.1073/pnas.1514566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosse-Wilde E, Svatoš A, Krieger J. A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem Senses. 2006;31:547–555. doi: 10.1093/chemse/bjj059. [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Xu J, Cui Z, Dong X, Ye Z, Niu D, Huang Y, Dong S. Functional characterization of SlitPBP3 in Spodoptera litura by CRISPR/Cas9 mediated genome editing. Insect Biochem Mol. 2016;75:1–9. doi: 10.1016/j.ibmb.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Pregitzer P, Greschista M, Breer H, Krieger J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol Biol. 2014;23:733–742. doi: 10.1111/imb.12119. [DOI] [PubMed] [Google Scholar]
- 20.Ishida Y, Leal WS. Rapid inactivation of a moth pheromone. Proc Natl Acad Sci USA. 2005;102:14075–14079. doi: 10.1073/pnas.0505340102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand N, Carot-Sans G, Bozzolan F, Rosell G, Siaussat D, Debernard S, Chertemps T, Maïbèche-Coisne M. Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS One. 2011;6:e29147. doi: 10.1371/journal.pone.0029147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakurai T, Mitsuno H, Haupt SS, Uchino K, Yokohari F, Nishioka T, Kobayashi I, Sezutsu H, Tamura T, Kanzaki R. A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genet. 2011;7:e1002115. doi: 10.1371/journal.pgen.1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang H, Liu Y, Ai D, Jiang X, Dong S, Wang G. A pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr Biol. 2017;27:1610–1615. doi: 10.1016/j.cub.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Bastin-Héline L, de Fouchier A, Cao S, Koutroumpa F, Caballero-Vidal G, Robakiewicz S, Monsempes C, François M-C, Ribeyre T, Maria A, et al. A novel lineage of candidate pheromone receptors for sex communication in moths. Elife. 2019;8:e49826. doi: 10.7554/eLife.49826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leary GP, Allen JE, Bunger PL, Luginbill JB, Jr, Linn CE, Macallister IE, Kavanaugh MP, Wanner KW. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang K, Huang L, Ning C, Wang C. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. Elife. 2017;6:e29100. doi: 10.7554/eLife.29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krieger J, Grosse-Wilde E, Gohl T, Dewer Y, Raming K, Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens) Proc Natl Acad Sci USA. 2004;101:11845–11850. doi: 10.1073/pnas.0403052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta. Proc Natl Acad Sci USA. 2011;108:7449–7454. doi: 10.1073/pnas.1017963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Gu S, Zhang Y, Guo Y, Wang G. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One. 2012;7:e48260. doi: 10.1371/journal.pone.0048260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuvaraj JK, Andersson MN, Zhang D, Löfstedt C. Antennal transcriptome analysis of the chemosensory gene families from trichoptera and basal Lepidoptera. Front Physiol. 2018;9:1365. doi: 10.3389/fphys.2018.01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai T, Nakagawa T, Mitsuno H, Mori H, Endo Y, Tanoue S, Yasukochi Y, Touhara K, Nishioka T. Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc Natl Acad Sci USA. 2004;101:16653–16658. doi: 10.1073/pnas.0407596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638–1642. doi: 10.1126/science.1106267. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Liu C, Lin K, Wang G. Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One. 2013;8:e62094. doi: 10.1371/journal.pone.0062094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuvaraj JK, Andersson MN, Corcoran JA, Anderbrant O, Löfstedt C. Functional characterization of odorant receptors from Lampronia capitella suggests a non-ditrysian origin of the lepidopteran pheromone receptor clade. Insect Biochem Mol. 2018;100:39–47. doi: 10.1016/j.ibmb.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Fitt GP. The ecology of Heliothis species in relation to agroecosystems. Annu Rev Entomol. 1989;34:17–53. doi: 10.1146/annurev.en.34.010189.000313. [DOI] [Google Scholar]
- 36.Behere G, Tay WT, Russell DA, Batterham P. Molecular markers to discriminate among four pest species of Helicoverpa (Lepidoptera: Noctuidae) Bull Entomol Res. 2008;98:599–603. doi: 10.1017/S0007485308005956. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, Dong J. Interspecific hybridization of Helicoverpa armigera and H. assulta (Lepidoptera: Noctuidae) Chin Sci Bull. 2001;46:489–491. doi: 10.1007/BF03187264. [DOI] [Google Scholar]
- 38.Klun JA, Plimmer JR, Bierl-Leonhardt BA, Sparks AN, Primiani M, Chapman OL, Lee GH, Lepone G. Sex pheromone chemistry of female corn earworm moth, Heliothis zea. J Chem Ecol. 1980;6:165–175. doi: 10.1007/BF00987535. [DOI] [Google Scholar]
- 39.Pope MM, Gaston LK, Baker TC. Composition, quantification, and periodicity of sex pheromone volatiles from individual Heliothis zea females. J Insect Physiol. 1984;30:943–945. doi: 10.1016/0022-1910(84)90072-6. [DOI] [Google Scholar]
- 40.Kehat M, Dunkelblum E. Behavioral responses of male Heliothis armigera (Lepidoptera: Noctuidae) moths in a flight tunnel to combinations of components identified from female sex pheromone glands. J Insect Behav. 1990;3:75–83. doi: 10.1007/BF01049196. [DOI] [Google Scholar]
- 41.Vickers NJ, Christensen TA, Mustaparta H, Baker TC. Chemical communication in heliothine moths III. Flight behavior of male Helicoverpa zea and Heliothis virescens in ratios of intra- and interspecific sex pheromone components. J Comp Physiol A. 1991;169:275–280. [Google Scholar]
- 42.Cork A, Boo KS, Dunkelblum E, Hall DR, Jee-Rajunga K, Kehat M, Jie EK, Park KC, Tepgidagarn P, Xun L. Female sex pheromone of oriental tobacco budworm, Helicoverpa assulta (Guenee) (Lepidoptera: Noctuidae): identification and field testing. J Chem Ecol. 1992;18:403–418. doi: 10.1007/BF00994240. [DOI] [PubMed] [Google Scholar]
- 43.Boo KS, Park KC, Hall DR, Cork A, Berg BG, Mustaparta H. (Z)-9-tetradecenal: a potent inhibitor of pheromone-mediated communication in the oriental tobacco budworm moth, Helicoverpa assulta. J Comp Physiol A. 1995;177:695–699. doi: 10.1007/BF00187628. [DOI] [Google Scholar]
- 44.Fadamiro HY, Baker TC. Helicoverpa zea males (Lepidoptera: Noctuidae) respond to the intermittent fine structure of their sex pheromone plume and an antagonist in a flight tunnel. Physiol Entomol. 1997;22:316–324. doi: 10.1111/j.1365-3032.1997.tb01175.x. [DOI] [Google Scholar]
- 45.Quero C, Baker TC. Antagonistic effect of (Z)-11-Hexadecen-1-ol on the pheromone-mediated flight of Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) J Insect Behav. 1999;12:701–710. doi: 10.1023/A:1020935905506. [DOI] [Google Scholar]
- 46.Cork A, Lobos EA. Female sex pheromone components of Helicoverpa gelotopoeon: first heliothine pheromone without (Z)-11-hexadecenal. Entomol Exp Appl. 2003;107:201–206. doi: 10.1046/j.1570-7458.2003.00052.x. [DOI] [Google Scholar]
- 47.Zhang J, Salcedo C, Fang Y, Zhang R, Zhang Z. An overlooked component: (Z)-9-tetradecenal as a sex pheromone in Helicoverpa armigera. J Insect Physiol. 2012;58:1209–1216. doi: 10.1016/j.jinsphys.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 48.Wu H, Xu M, Hou C, Huang L, Dong J, Wang C. Specific olfactory neurons and glomeruli are associated to differences in behavioral responses to pheromone components between two Helicoverpa species. Front Behav Neurosci. 2015;9:206. doi: 10.3389/fnbeh.2015.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Wang B, Dong S, Cao D, Dong J, Walker WB, Liu Y, Wang G. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One. 2015;10:e0117054. doi: 10.1371/journal.pone.0117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang H, Guo M, Wang B, Liu Y, Dong S, Wang G. Sensillar expression and responses of olfactory receptors reveal different peripheral coding in two Helicoverpa species using the same pheromone components. Sci Rep. 2016;6:18742. doi: 10.1038/srep18742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, Ban L, Song L, Liu Y, Pelosi P, Wang G. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem Mol. 2016;72:10–19. doi: 10.1016/j.ibmb.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Gao M, Nakajima An D, Parks JM, Skolnick J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat Commun. 2022;13:1744. doi: 10.1038/s41467-022-29394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61:3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmüller H, Jr, MacKerell AD. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 58.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 59.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan H, Jafari S, Pask G, Zhou X, Reinberg D, Desplan C. Evolution, developmental expression and function of odorant receptors in insects. J Exp Biol. 2020;223:jeb208215. doi: 10.1242/jeb.208215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.del Mármol J, Yedlin MA, Ruta V. The structural basis of odorant recognition in insect olfactory receptors. Nature. 2021;597:126–131. doi: 10.1038/s41586-021-03794-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cossé AA, Todd JL, Baker TC. Neurons discovered in male Helicoverpa zea antennae that correlate with pheromone-mediated attraction and interspecific antagonism. J Comp Physiol A. 1998;182:585–594. doi: 10.1007/s003590050205. [DOI] [Google Scholar]
- 63.Guo H, Huang L, Gong X, Wang C. Comparison of functions of pheromone receptor repertoires in Helicoverpa armigera and Helicoverpa assulta using a Drosophila expression system. Insect Biochem Mol. 2022;141:103702. doi: 10.1016/j.ibmb.2021.103702. [DOI] [PubMed] [Google Scholar]
- 64.Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V. Cryo-EM structure of the insect olfactory receptor Orco. Nature. 2018;560:447–452. doi: 10.1038/s41586-018-0420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stansfeld PJ, Sansom MS. Molecular simulation approaches to membrane proteins. Structure. 2011;19:1562–1572. doi: 10.1016/j.str.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, Bugge K, Kragelund BB, Lindorff-Larsen K. Role of protein dynamics in transmembrane receptor signalling. Curr Opin Struct Biol. 2018;48:74–82. doi: 10.1016/j.sbi.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 67.Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature. 2011;478:511–514. doi: 10.1038/nature10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hughes DT, Wang G, Zwiebel LJ, Luetje CW. A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem Senses. 2014;39:761–769. doi: 10.1093/chemse/bju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao S, Liu Y, Wang B, Wang G. A single point mutation causes one-way alteration of pheromone receptor function in two Heliothis species. iScience. 2021;24:102981. doi: 10.1016/j.isci.2021.102981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho S, Mitchell A, Mitter C, Regier J, Matthews M, Robertson R. Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Syst Entomol. 2008;33:581–594. doi: 10.1111/j.1365-3113.2008.00427.x. [DOI] [Google Scholar]
- 71.Laster ML, Sheng CF. Search for hybrid sterility for Helicoverpa zea in crosses between the north American H. zea and H. armigera (Lepidoptera: Noctuidae) from China. J Econ Entomol. 1995;88:1288–1291. doi: 10.1093/jee/88.5.1288. [DOI] [Google Scholar]
- 72.Anderson CJ, Oakeshott JG, Tay WT, Gordon KHJ, Zwick A, Walsh TK. Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proc Natl Acad Sci USA. 2018;115:5034–5039. doi: 10.1073/pnas.1718831115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pogue MG. A new synonym of Helicoverpa zea (Boddie) and differentiation of adult males of H. zea and H. armigera (Hübner) (Lepidoptera: Noctuidae: Heliothinae) Ann Entomol Soc Am. 2004;97:1222–1226. doi: 10.1603/0013-8746(2004)097[1222:ANSOHZ]2.0.CO;2. [DOI] [Google Scholar]
- 74.Mallet J, Korman A, Heckel DG, King P. Biochemical genetics of Heliothis and Helicoverpa (Lepidoptera: Noctuidae) and evidence for a founder event in Helicoverpa zea. Ann Entomol Soc Am. 1993;86:189–197. doi: 10.1093/aesa/86.2.189. [DOI] [Google Scholar]
- 75.Pearce SL, Clarke DF, East PD, Elfekih S, Gordon KHJ, Jermiin LS, McGaughran A, Oakeshott JG, Papanikolaou A, Perera OP, et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017;15:63. doi: 10.1186/s12915-017-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its Supplementary files).