Abstract
The PHD finger, a Cys4–His–Cys3 zinc finger, is found in many regulatory proteins from plants or animals which are frequently associated with chromatin-mediated transcriptional regulation. We show here that the PHD finger activates transcription in yeast, plant and animal cells. In plant homeodomain transcription factors the PHD finger is combined with an upstream leucine zipper. Both domains together form a highly conserved 180 amino acid region called the ZIP/PHDf motif and transcriptional activity of the PHD finger is masked when embedded in this motif. Our results indicate that the ZIP/PHDf domain is a potential regulatory domain of PHDf-HD proteins. The leucine zipper upstream of the PHD finger interacts with 14-3-3GF14µ from Arabidopsis thaliana and 14-3-3GF14-12 from maize via a leucine zipper conserved in helix 4 of various 14-3-3 proteins from plants and animals. PHD-type plant homeodomain proteins consequently may represent potential targets of 14-3-3 signalling.
INTRODUCTION
The PHD finger was originally identified by comparison of the maize homeodomain (HD) protein ZMHOX1a (1) to its Arabidopsis relative HAT3.1 (2) and named plant homeodomain (PHD) finger due to its association with the DNA-binding HD in both genes. Initially this zinc finger was implicated in DNA recognition (2). However, this motif often occurs in various regulatory genes, such as members of the trithorax (TRX-G) or polycomb (PC-G) groups (3) and leukaemia-associated proteins (LAP finger) (4). The established function of TRX-G and PC-G genes in chromatin modulation in Drosophila led to the suggestion that the PHD finger is involved in chromatin-mediated transcriptional control (3). Recent data provide evidence that PHD finger proteins are associated with chromatin remodelling complexes (5) or contribute to histone acetylation (6).
Based on the position of the unique His residue, the cysteine scaffold of the PHD finger (Cys4–His–Cys3) is clearly distinct from RING fingers (Cys3–His–Cys4) (7) and LIM domains (Cys2–His–Cys5) (8) and from DRIL domains (9), where two RING finger motifs are closely linked. Both the RING finger and the LIM domain mediate protein–protein interactions (10,11) and are involved in transcriptional control, either by directly affecting transcription (12) or recruiting co-activators or co-repressors (13,14). LIM domains also contribute to various signalling pathways. They may interact with protein kinases and anchor gene products to large protein complexes or to cellular compartments. Individual functions are often associated with specific subgroups of the LIM domain (11). In contrast to the accumulating knowledge about LIM domains, functional data concerning the PHD finger remain rare. Recent studies on the AIRE gene from humans have shed more light on the importance of this motif, since all clinically significant mutations in the AIRE gene coincide with alteration in two PHD fingers (15), resulting in the rare autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED).
Similar to the linkage of PHD fingers (PHDf) with HDs in plant PHDf-HD proteins, LIM domains were originally identified as motifs of animal HD gene products. LIM-HD transcription factors regulate expression of genes that pattern the body and generate cell specificity during development in vertebrates and invertebrates. LIM-HD proteins are themselves regulated by both intramolecular and intermolecular interactions mediated by the LIM domains, which negatively regulate LIM-HD activity by preventing HD association with DNA. Interactions of LIM domains with other proteins relieve this interference, permitting DNA binding and providing a mechanism for restoring LIM-HD activity (reviewed in 16). Similar intramolecular contacts between the PHD finger and the HD have not been discovered in plant PHDf-HD proteins so far. Beside the structural similarities between animal LIM-HD and plant PHDf-HD proteins, the expression patterns of four ZMHOX genes in maize are intriguing. Expression occurs early in embryogenesis and is restricted to meristems and initiating organ primordia (17). ZMHOX gene products have been shown to enter the plant cell nucleus (18) and bind to DNA (1,19), suggesting that these DNA-binding proteins regulate transcription. Unfortunately, loss of function mutations in plant PHDf-HD genes are neither available for the ZMHOX genes (ZMHOX1a/b and ZMHOX2a/b) (20) nor the Arabidopsis HAT3.1 (2) or PRHA gene or PRHP, its relative from Petroselinum crispum (21). The main function of PHDf-HD genes in plants therefore remains to be established.
GYMNOS, a recently described member of the SWI2/SNF2 protein family in plants (22), also contains a PHD finger. Here, the characteristic Zn finger is combined with a myb-type DNA-binding domain, instead of a HD. GYMNOS is involved in establishment of carpel polarity, indicating that PHD finger motifs take part in the control of plant development. Due to significant similarity to the Drosophila MI-2 gene, a hunchback interacting protein that functions in polycomb-mediated gene repression (23), GYMNOS has been implicated in chromatin modulation (22). While the PHD finger is an isolated motif in GYMNOS, the characteristic Cys4–His–Cys3 scaffold in PHDf-HD plant genes is embedded in a larger region, more than twice as long as the separate PHD finger motif. This 180 amino acid region shares 60% identical residues between seven genes of maize, parsley and Arabidopsis and is more highly conserved than the HD (40%). This conservation suggests that the PHD finger is part of a larger functional unit, which is analysed in detail here. Our experimental data suggest that the PHD finger activates transcription in plant, yeast and animal cells. Combined with a leucine zipper in the surrounding conserved 180 amino acid region in the PHDf-HD proteins, PHD finger activity is masked. This leucine zipper upstream of the PHD finger mediates a novel kind of interaction with helix 4 of plant 14-3-3 proteins, thus identifying PHDf-HD proteins as potential targets of 14-3-3 signalling pathways.
MATERIALS AND METHODS
Plasmid constructs
All fragments inserted into the pACT2 (24) or pAS2 (25) vectors (Clontech) were obtained by PCR from cDNA clones using primers that provide appropriate in-frame restriction sites and confirmed by DNA sequencing. In the following the numbers indicate the first and last amino acid residue relative to each accession number in the SwissProt protein database. ZMHOX1a (P46605): ZIP1a (124–200), PHDf1a (197–303) and ZIP/PHDf1a (124–303); ZMHOX1b (CAA63156): PHDf1b (197–303), Zmhox2a (CAA61909), PHDf2a (658–564) and ZIP/PHDf2a (385–564); ZMHOX2b (S65775): PHDf (461–567), HAT3.1 (Q04996), PHDf-HAT3.1 (187–289) and ZIP/PHDf-HAT3.1 (118–289); PRHA (P48785): PHDf-PRHA (175–287) and ZIP/PHDf-PRHA (105–278); PRHP (P47786): PHDf-PRHP (563–671) and ZIP/PHDf-PRHP (493–671) and the second PHD finger of the Drosophila dMI-2 (AF119716) gene PHD2-dMI-2 (435–497). All fragments were cloned directionally in the SfiI and BamHI sites of pACT2 (24) or the NcoI and BamHI sites of pAS2 (25).
Full-length cDNA and subclones of 14-3-3GF14µ (Q96299) from Arabidopsis thaliana and 14-3-3GF14-12 (JQ1680) from Zea mays were fused to the Gal4 transactivation domain, using the EcoRI and XhoI sites in pACT2. Subclones of 14-3-3GF14µ contained residues 1–45 (cc1), 76–121 (cc2), 134–167 (cc3) and 195–263 (cc4) or carried an N-terminal deletion lacking the first 76 residues. In 14-3-3GF14-12 cc2 spans residues 83–124. The coiled-coil predictions are based on programs at the ISREC Coils server (EMBn, Switzerland). The A.thaliana expression library was a kind gift of Dr K. Salchert (MPI, Cologne). The cDNA prepared from a cell suspension culture was cloned directionally into the EcoRI and XhoI sites of pACT2; the average insert length was 1.8 kb.
Most plant effector and reporter plasmids are derivatives of pRTΩNot/Asc (26), which was modified in the polylinker region by insertion of HindIII, BamHI and XhoI sites. The Gal4DB, Gal4DB-PHDf1a and Gal4DB-ZIP/PHDf1a coding sequences were amplified by PCR and inserted directionally behind the CaMV 35S promoter into the NotI and BamHI or NotI and XbaI sites. The Gal4-UAS pentameric sequences were inserted into a HindIII site preceding the CaMV 35S minimal promoter (–90) in pK373, a GUS reporter construct kindly provided by Dr R. Thompson (MPI, Cologne). The 35S::Gal4DB-VP16, 35S::luciferase (27) and 35S::GFP constructs were described previously (18). Chimeric effector constructs in the zebrafish transient gene expression system (28) were driven by the CMV (cytomegalovirus) promoter. The PHDf1a fragment was inserted into the SmaI site and the ZIP/PHDf1a fragment between the EcoRI and SmaI sites of pCMV GAL4DB (29). Glutathione S-transferase (GST) fusion proteins were constructed in pGEX-2T (Pharmacia Biotech) by in-frame cloning of the ZMHOX1a coding region and ZIP1a, PHDf1a and ZIP/PHDf1a motifs with GST into BamHI and EcoRI sites. Full-length 14-3-3GF14µ and 14-3-3GF14-12 were provided by Dr R. Ferl (University of Florida, Gainesville, FL) in the pET-15b vector (Novagen).
Protein interaction analysis in yeast
Use of the MATCHMAKER Gal4 two-hybrid system and the liquid ONPG β-galactosidase assay followed the Clontech protocol. Clones (7 × 106) of the A.thaliana library in pACT2 were screened with the ZMHOX1a leucine zipper in pAS2 as bait. The Saccharomyces cerevisiae strain Y190 was sequentially transformed with the bait and prey constructs by lithium acetate. Clones that specifically interact with the bait were rescued, sequenced and compared to database entries by BLAST search. The clones isolated in the yeast two-hybrid screen share extensive homology with the ESTs 303D8T7, 127M1T7, 96E13T7, 152J23T7, 113M5T7 and 147E13T7 (14-3-3GF14µ, Q96299). Yeast protein extracts were prepared from overnight cultures of single transformed Y190 cells using the urea/SDS method (30). Equal amounts of proteins from the transformed and untransformed yeast cells were resolved by 14% SDS–PAGE (31) and analysed by western blotting using an anti-Gal4BD monoclonal antibody (0.5 µg/ml; Clontech). Detection was performed with a horseradish peroxidase-conjugated polyclonal goat anti-mouse IgG (1:15000; Jackson ImmunoResearch Laboratories) and enhanced chemiluminescence (Amersham Pharmacia Biotech).
Transient gene expression assays in plant and animal cells
Arabidopsis protoplasts were isolated and transfected with plasmid DNA prepared from an Escherichia coli dam– strain (GM2163) as described (27). Reporter (UAS-core::GUS) and effector plasmids encoding either Gal4DB, Gal4DB-PHDf1a, Gal4DB-ZIP/PHDf1a or Gal4DB-VP16 constructs behind the CaMV 35S promoter were mixed in a 10:1 ratio of reporter to effector. Additionally, each transfection contained 5 µg of the 35S::luc reference plasmid. Generally eight independent co-transformations were analysed for each effector.
Microinjection of zebrafish embryos (28) was performed into transgenic UAS::myc-notch:intra progeny at the 1–2 cell stage by injecting ∼5 nl of effector plasmid (50 ng DNA/µl, 0.2% phenol red) into the cytoplasm. After incubation for 13 h at 29°C, embryos were fixed in 4% paraformaldehyde at 4°C overnight. Antibody staining was performed according to The Zebrafish Book (32) with slight modifications. Embryos were treated with a hybridoma supernatant containing monoclonal anti-myc antibody. Detection was performed with a horseradish peroxidase-coupled polyclonal goat anti-mouse antibody (1:1000; Dianova).
Co-precipitation analysis
The ZMHOX1a, ZIP1a and ZIP/PHDf1a GST fusions in pGEX-2T were expressed in E.coli BL21 cells. Expression was induced at an OD260 of 0.4–0.5 by addition of 0.1 mM isopropypl-1-thio-β-d-galactoside (IPTG). Cells were collected after 2 h at 37°C by centrifugation, resuspended in 10 ml of LSD (50 mM Tris, pH 8.0, 20% glycerol, 1 mM DTT, 0.1% Nonident P-40) supplemented with 500 mM KCl and 0.1 mM PMSF at 4°C. After sonification cell debris was collected by centrifugation and the supernatant either stored at –70°C or GFP fusion proteins were purified by glutathione–Sepharose 4B (Pharmacia) chromatography. In vitro transcription and translation of 14-3-3GF14µ and 14-3-3GF14-12 proteins was performed using the T7 promoter in pET-15b in the TNT coupled reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer’s protocol. For co-precipitation, glutathione–Sepharose beads loaded with GST fusion proteins were washed five times in LSD (3 × 500 mM KCl and 2 × 100 mM KCl) and incubated with in vitro translated radioactively labelled 14-3-3 protein for 2 h at 4°C. After washing in LSD supplemented with 150 mM KCl, proteins were eluted and analysed by SDS–PAGE. Radioactively labelled 14-3-3 protein bound to the ZMHOX1a peptides was visualised by fluorography.
RESULTS
Functional association of PHD fingers and leucine zippers in plant HD transcription factors
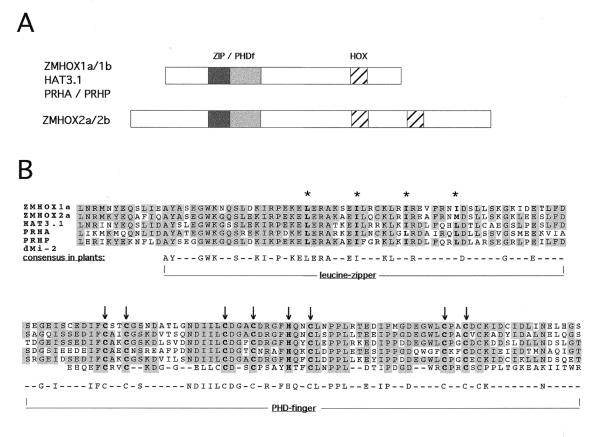
The PHD finger characterises one class of plant HD proteins (PHDf-HD), where it comprises the C-terminal part of a larger, highly conserved 180 amino acid residue region. Sequence comparison of this region between the seven published plant gene products revealed a leucine zipper preceding the PHD finger. The amino acid sequence of the 180 amino acid region of five plant proteins from maize, parsley and Arabidopsis is compared to the Cys4–His–Cys3 motif of the second PHD finger from Drosophila MI-2 protein in Figure 1. The ZMHOX1b and 2b sequences have been omitted in this comparison, since they are almost identical to the ZMHOX1a/2a sequences. The structural similarity between PHD motifs and RING fingers or LIM domains suggested that this highly conserved 180 amino acid region within the leucine zipper in PHDf-HD transcription factors comprises a close association of two potential protein–protein interaction surfaces. The different plant sequences were named ZIP/PHDf for the whole domain and ZIP and PHDf for the single leucine zipper and PHD finger motif, respectively. To identify potential interaction partners in the yeast two-hybrid system, different parts of the ZMHOX1a ZIP/PHDf domain were cloned into the pACT2 (prey) and pAS2 (bait) (24) vectors and expressed in translational fusion with the Gal4 DNA-binding domain (Gal4DB) or transactivation domain (Gal4AD).
Figure 1.
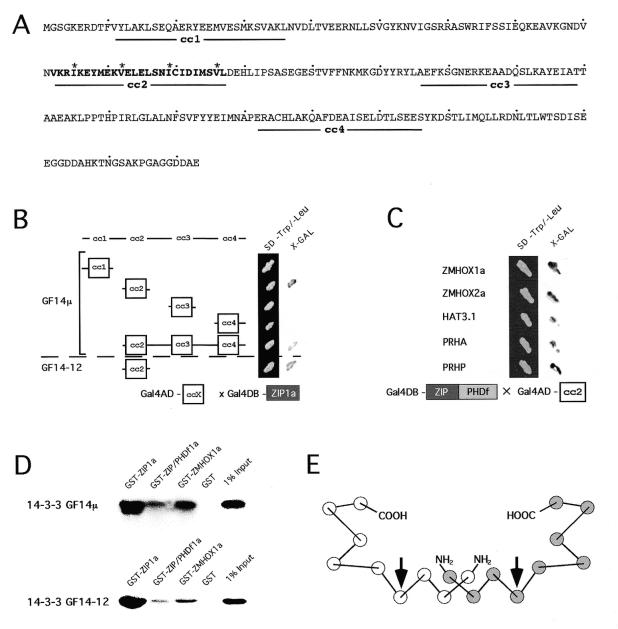
Position and conservation of the ZIP/PHDf domain in plant HD proteins. (A) Position of the conserved 180 amino acid region of the ZIP/PHDf domain in different PHD-HD proteins. (B) Alignment of five plant PHDf and the second PHD finger in Drosophila dMI-2 protein. Asterisks indicate leucine residues of the leucine zipper; arrows mark the cysteine residues in the PHD finger. Shading indicates amino acid conservation in at least three genes. The identity between the five plant sequences exceeds 60%.
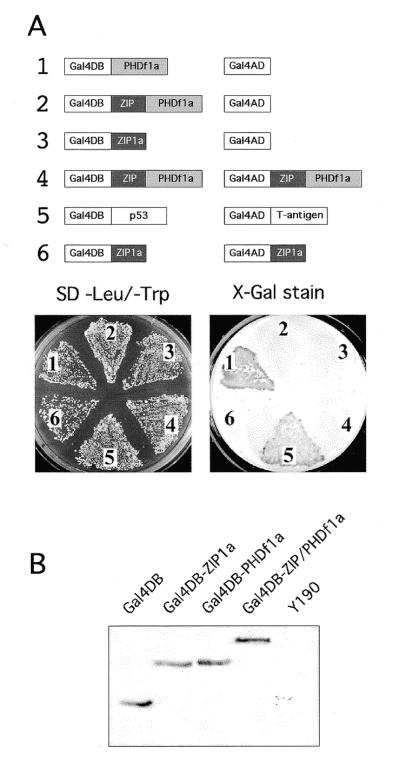
In a first experiment, individual bait and prey constructs or combinations were introduced into yeast strain Y190 and stained for β-galactosidase (lacZ) activity. As far as detectable in the yeast two-hybrid system, neither the ZMHOX1a leucine zipper nor the larger ZIP/PHDf domain mediate homodimerisation (Fig. 2A). This result is compatible with previous native protein gel analysis and co-precipitation experiments, which showed no evidence for the formation of ZMHOX homodimers (data not shown). Surprisingly, the PHD finger provides a transcriptional activation motif when introduced in combination with the empty prey vector. The kinetics and the intensity of lacZ staining are comparable to the positive control provided by the p53–T antigen interaction, reaching saturation within 3 h. However, no lacZ activity was detectable with the complete ZIP/PHDf domain, which carries the upstream leucine zipper and the PHD finger. This difference is not explained by protein instability. Western blot analysis of yeast protein extracts with a monoclonal antibody against the Gal4DB always confirmed Gal4DB-ZIP/PHDf steady-state levels higher than those observed with the individual motifs Gal4DB-PHD or Gal4DB-ZIP (see Fig. 2B). Consequently, the transcriptional activation provided by the isolated PHD finger appears to be silenced within the environment of the ZIP/PHDf domain.
Figure 2.
Dissectional analysis of the ZMHOX1a ZIP/PHDf domain in the yeast two-hybrid system. (A) Combinations of constructs 1–6 as indicated were used for co-transformation of Y190 yeast cells. Staining for lacZ activity was performed after 3 days at 30°C on minimal selection medium devoid of leucine and tryptophan. Galactosidase activity with the ZMHOX1a PHD finger (1), as for the positive control (5) comprising the p53–T antigen interaction, was detectable after 3 h. Note that the ZIP/PHDf domain (2) and the isolated leucine zipper (3) do not exert lacZ activity and show no homodimerisation (4 and 6). Identical results were observed with constructs 1–3 on single selection medium (leu–). See also Figure 3A. (B) Western blot analysis of protein extracts prepared from Y190 cells transformed with constructs as indicated above each lane. Equal amounts of protein extract were analysed with a monoclonal antibody against the Gal4DB domain and subsequent chemoluminiscent detection. Polypeptides exhibited the predicted sizes: Gal4DB, 22 kDa; Gal4DB-ZIP1a, 29 kDa; Gal4DB-PHDf1a, 31 kDa; Gal4DB-ZIP/PHDf1a, 40 kDa. No reacting polypeptide was detectable in untransformed Y190 cells.
Transcriptional activation mediated by the PHD finger in yeast, plant and animal cells
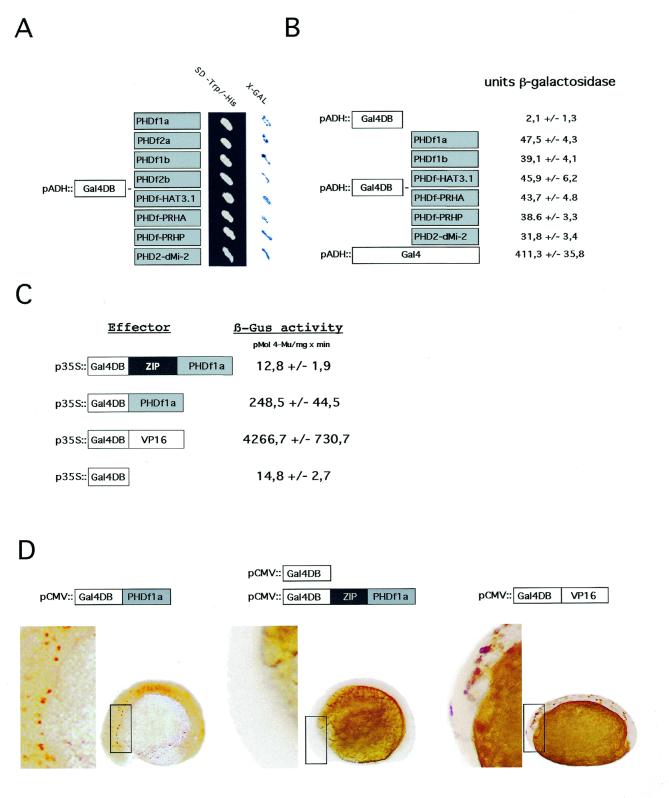
To exclude that transcriptional activation by the PHD finger is a peculiarity of the ZMHOX1a protein, we tested other PHD fingers in yeast (Fig. 3A). These are the PHD fingers from the ZMHOX1b, ZMHOX2a and ZMHOX2b genes of Z.mays, the A.thaliana HAT3.1 and PRHA genes and the P.crispum PRHP homologue. As a reference for animal counterparts, the second PHD finger of the dMI-2 protein from Drosophila melanogaster was included in the study. This Drosophila motif shares high sequence conservation to known plant PHD fingers and there is significant structural similarity between the dMI-2 and GYMNOS gene products. All PHD fingers were expressed in translational fusion with the Gal4DB and tested for transcriptional activation in yeast. The corresponding X-gal staining pattern is shown in Figure 3A. Quantitative liquid assays (Fig. 3B) confirm similar lacZ activities with every construct, although generally 10-fold lower than with the native Gal4 protein expressed behind the ADH promoter. Thus transcriptional activation is a common feature of all tested PHD fingers (eight in total), including the PHD finger from Drosophila dMI-2 protein. In the context of the ZIP/PHDf domains, however, any transcriptional activation provided by the isolated ZMHOX2a, HAT3.1, PRHA and PRHP PHD fingers is blocked in yeast (data not shown). These results are consistent with those obtained for PHD1a and ZIP/PHDf1a and suggest that the conserved upstream sequences in the ZIP/PHDf domain control activity of the PHD finger.
Figure 3.
Transcriptional activation is a common feature of PHD fingers in yeast, plant and animal cells. (A) Galactosidase staining obtained after transformation of Y190 yeast cells with various GAL4DB-PHDf fusions as schematically indicated to the left. All signals appeared within 2 h incubation at 37°C. (B) Quantification of lacZ activity [O-nitrophenyl-β-d-galactopyranoside (ONPG) test] in yeast protein extracts transformed with different PHD finger constructs. The values are calculated from three independent transformation experiments. (C) Relative GUS activities calculated from transient gene expression experiments performed in Arabidopsis protoplasts. Effector construct Gal4DB-PHDf1a, Gal4DB-ZIP/PHDf1a or Gal4DB-VP16, as a strong transcriptional activation domain, was combined with the UAS core::GUS reporter and a 35S::luciferase marker as internal reference. The relative GUS activities represent data from eight independent transfection experiments and were normalised to the luciferase standard. (D) Transcriptional activation by PHDf1a observed after injection of zebrafish embryos. The Gal4DB-PHDf1a, Gal4DB-ZIP/PHDf1a and Gal4DB effector proteins were expressed under the constitutive CMV promoter and monitored by the UAS::myc-notch:intra reporter. Antibody staining of a myc-tagged notch-intra marker monitors reporter gene activation. Note that staining is observed with Gal4DB-PHDf1a (left) and Gal4DB-VP16 (right) but not with Gal4DB-ZIP/PHDf1 (middle) or Gal4DB (data not shown)
To substantiate the results obtained in yeast cells the assay system was adapted and transferred to A.thaliana protoplasts. A reporter was constructed by inserting two Gal4-UAS (upstream activation sequence) pentamers in front of the CaMV 35S-core promoter (UAS-core) controlling the β-glucuronidase (GUS) marker. As effectors, the Gal4DB, Gal4DB-PHD1a and Gal4DB-ZIP/PHDf1a coding regions were inserted into vector pRTΩNot (33) and expressed under the strong CaMV 35S promoter. Gal4DB-VP16 (27) served as a positive control, containing a fusion of the strong VP16 activation domain to the Gal4DB. All effector constructs were driven by the CaMV 35S promoter and transformed into Arabidopsis protoplasts with the UAS-core::GUS reporter gene and a 35S::luciferase reference marker. Reporter GUS activity was estimated after 24 h and normalised to the luciferase standard. The results of eight independent experiments are combined in the block diagram in Figure 3C. They indicate that the PHD finger activates transcription in Arabidopsis cells as well as in yeast. In comparison to the background level provided by the Gal4DB alone, GUS activity derived from the UAS-core promoter is ∼17-fold higher when combined with the Gal4DB-PHD1a effector, although still 17-fold weaker than observed with the strong VP16 domain. The Gal4DB-ZIP/PHDf effector construct fails to activate transcription in Arabidopsis protoplasts, as observed in yeast cells. Therefore, transcriptional activity of the PHD finger is silenced in plant cells if included in the ZIP/PHDf domain.
Because the PHD fingers are conserved between plant and animal species, we tested for transcriptional activation in animal cells and chose the zebrafish embryo as a test system. Gal4DB-PHDf1a, Gal4DB-ZIP/PHDf1a and Gal4DB-VP16 were expressed under the constitutive CMV promoter. Plasmids providing the different constructs were microinjected at the 1–2 cell stage into transgenic zebrafish embryos carrying a UAS controlled myc-notch:intra reporter gene (28). After incubation for 13 h at 29°C, activation of the reporter gene was analysed by staining with a monoclonal anti-myc antibody. The results combined in Figure 3D confirm that Gal4DB-PHDf1a activates transcription in the zebrafish embryo, although the staining intensity is higher with Gal4DB-VP16. Transcriptional activation thus seems to be a general feature of the PHD finger motif in eukaryotic cells. These microinjection experiments into fish embryos again confirmed inactivity of the complete ZIP/PHDf1a domain. Thus masking of PHD finger activity by the upstream sequences within the plant-specific ZIP/PHDf motif is an inherent feature of the complete domain.
The leucine zipper of the ZIP/PHD domain mediates interactions with 14-3-3 proteins
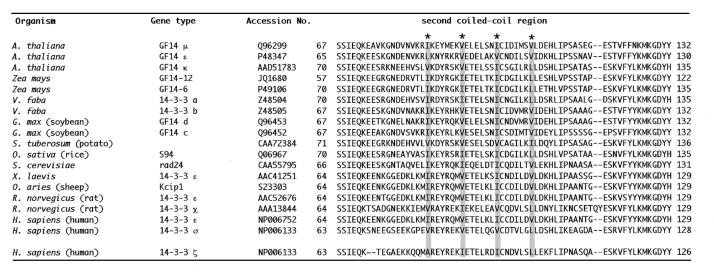
Since the ZMHOX1a leucine zipper failed to mediate homodimerisation (see Fig. 2), we tried to identify interacting protein partners by use of the yeast two-hybrid system. Gal4DB-ZIP1a was used as bait against an A.thaliana cDNA expression library fused to Gal4AD in the vector pACT2. Among 7 × 106 transformants six potential partners corresponding to known ESTs were identified. The individual EST numbers are included in Materials and Methods. Common to all these clones were potential coiled-coil motifs. EST clone 147E13T17 represented 14-3-3 GF14µ, an isoform of the Arabidopsis 14-3-3 family. The clone isolated in the yeast two-hybrid screen covered the C-terminal part of 14-3-3 GF14µ, starting with amino acid residue 80 in front of helix 4. Full-length cDNA clones encoding 14-3-3GF14µ and 14-3-3GF14-12, a close relative from maize, were kindly provided by Dr R. Ferl (University of Florida, Gainesville, FL). A conserved feature of 14-3-3 proteins are four coiled-coil (cc) motifs, either evident from the X-ray diffraction studies (34) or predicted by the ISREC Coils server (EMBn, Switzerland). The second cc motif is comprised of helix 4 and coincides with a potential leucine zipper (see Table 1; 35).
Table 1. Alignment of potential leucine zippers in helix 4 of various 14-3-3 proteins from plants, animals and yeast (with SwissProt accession nos).
Asterisks indicate the heptad repeat spacing of hydrophobic residues (shadowed). The sequence of crystallised h14-3-3ζ is compared at the bottom.
In a detailed analysis performed with the 14-3-3GF14µ clone we tried to address two major questions. First, we wanted to identify the interacting surface in the 14-3-3 protein and, second, confirm interactions between full-length ZMHOX1a PHDf-HD protein and 14-3-3 proteins. Since the cc structure is linked to leucine zipper function, we tested cc motifs of 14-3-3GF14µ as interaction domains. Different parts of the 14-3-3GF14µ reading frame and the four individual cc motifs were therefore cloned into the pACT2 prey vector and tested in the yeast two-hybrid system for interaction with Gal4DB-ZIP1a. Transformations with the complete 14-3-3 coding regions in pACT2 either failed (maize) or resulted in slow growth and instability (Arabidopsis). We were able to express a truncated version of 14-3-3 GF14µ (amino acids 76–263) that lacked residues 1–75. The results obtained with the four separate 14-3-3 cc domains in the yeast two-hybrid system are summarised in Figure 4B. Only the cc2 motif of 14-3-3GF14µ mediates interactions with Gal4DB-ZIP1a, whereas none of the residual three cc motifs (cc1, cc3 and cc4) exhibit any interaction in the yeast two-hybrid system. Similarly, the cc2 motif in 14-3-3GF14-12 from maize interacts with Gal4DB-ZIP1a. We have not isolated the individual leucine zippers preceding the PHD fingers in all the different plant HD proteins. However, the 14-3-3GF14µ cc2 motif interacts with all ZIP/PHDf domains (Fig. 4C). The leucine zippers preceding the PHD finger are therefore generally functional dimerisation motifs, although we have not observed any specificity.
Figure 4.
In vivo and in vitro analysis of the 14-3-3 protein interactions. (A) Coiled-coil predictions of 14-3-3 proteins. The highly conserved leucine zipper in cc2 is bold; asterisks mark its heptad repeat spacing. (B) Interactions between cc motifs of 14-3-3GF14µ and 14-3-3GF14-12 fused to the activation domain (Gal4AD-ccX) and the ZMHOX1a leucine zipper bait (Gal4DB-ZIP1a). The cc2 motif of 14-3-3GF14µ and 14-3-3GF14-12 is sufficient to mediate interactions in the yeast two-hybrid system. The N-terminally truncated peptide (Δ1–75) of 14-3-3GF14µ lacking region cc1 binds to the ZIP1a motif. Expression of full-length maize and Arabidopsis 14-3-3 proteins in yeast unfortunately failed. (C) Interactions between Gal4AD-cc2 of Arabidopsis 14-3-3GF14µ and different ZIP/PHDf domains as bait (Gal4DB-ZIP/PHDf). Individual ZIP/PHDf domains are indicated in the margin. (D) Co-precipitation of full-length 14-3-3GF14µ and 14-3-3 GF14-12 proteins by different ZMHOX1a peptides. The radioactively labelled 14-3-3GF14µ and 14-3-3GF14-12 proteins of maize and Arabidopsis bind to ZIP1a, ZIP/PHDf1a and the complete ZMHOX1a protein, but not to GST. (E) Helical structure of the 14-3-3 dimer. Circles indicate antiparallel α-helices, nine in each monomer. The two arrows mark α-helix 4 and the orientation of the leucine zipper interaction surface.
To verify interactions between the complete ZMHOX1a and 14-3-3 proteins from Arabidopsis and maize, co-precipitation assays were performed. ZIP1a, ZIP/PHDf1a and the complete ZMHOX1a coding regions were expressed in fusion with GST. The partner proteins 14-3-3GF14µ and 14-3-3GF14-12 were labelled with [35S]methionine by in vitro transcription/translation. The different GST fusion proteins comprising the ZIP1a and ZIP/PHDf1a domains and the full-length ZMHOX1a protein were incubated with equal amounts of labelled 14-3-3 proteins, immobilised on glutathione–Sepharose and washed intensively prior to elution and analysis of polypeptides by SDS–PAGE and fluorography. As shown in Figure 4D, the complete 14-3-3GF14µ and 14-3-3GF14-12 proteins complex with the GST–ZIP1a, GST–ZIP/PHDf1a and GST–ZMHOX1a fusion proteins but not with GST alone. In vitro co-precipitation thus confirms interactions between full-length ZMHOX1a and 14-3-3 proteins from maize and Arabidopsis. In conclusion, the leucine zippers in the ZIP/PHDf domain from ZMHOX1a and both 14-3-3 proteins are accessible to protein partners in the full-length polypeptides.
DISCUSSION
Due to its presence in the PC-G and TRX-G-like gene products, the PHD finger has been implicated in chromatin modulation (3). The data presented here provide evidence that PHD fingers, which are found in numerous regulatory eukaryotic genes, may directly stimulate transcription. This conclusion is based on results obtained with seven plant PHD fingers (ZMHOX1a/b, 2a/b, HAT3.1, PRHA and PRHP) and the Drosophila dMI-2 PHD finger, which all activated transcription in yeast cells. It remains to be elucidated whether the PHD finger directly interacts with a component of the transcription initiation complex or if its positive effect on transcription is mediated via auxiliary protein interactions. The latter possibility is compatible with data elaborated for human Mi-2β, where the PHD finger is required to recruit histone deacetylase into a large protein complex (36). Both assumptions, however, involve PHD finger-mediated protein–protein interactions. The activity of the ZMHOX1a PHD finger in yeast, plant and animal cells can be taken as evidence that interacting protein partners are widely present in eukaryotic cells.
In plant genes, PHD fingers are always found in combination with DNA-binding domains, either HDs in PHDf-HD genes or the myb domain in GYMNOS. Nuclear import of PHDf-HD proteins and DNA-binding have been demonstrated for the ZMHOX gene product (18). All plant PHD fingers tested in our studies are identical in spacing of the Cys/His residues and share a high degree of similarity between residual amino acid sequences. Also, the second PHD finger motif from dMI-2 is rather similar to these plant PHD fingers. Other members of the PHD finger family are more divergent in sequence and spacing of the cysteine scaffold. Transcriptional activation therefore may not necessarily be a common feature and individual subgroups of PHD fingers may perform different functions, as described for the LIM domain (11) and the RING finger (7). The presence of PHD fingers in genes up-regulated in leukaemia, associated with the autoimmune disease APECED or participating in euchromatin to heterochromatin modulation, like the TRX-G or PC-G genes, indicates that this motif may be involved in a variety of pathways. Similar functional differences are known for RING fingers. The RING finger motif of the Arabidopsis COP1 photomorphogenic repressor was used to isolate a novel RING H2-type protein partner in the yeast two-hybrid system (37), a screen impossible if this RING finger is a transcriptional activator. On the other hand, the RING finger of the SNURF transcriptional co-regulator activates transcription in monkey kidney CV-1 cells by directly contacting the TATA box-binding protein (TBP) (38).
Data presented here for yeast, plant and animal cells consistently indicate that complete ZIP/PHDf domains, in contrast to single PHD fingers, are inefficient in transcriptional activation. The strong conservation of surrounding sequences (60% identity in seven plant PHDf-HD proteins; 20) and the conserved functionality of the leucine zipper demonstrated here make it tempting to speculate as to whether and how ZIP/PHDf domains can act as regulatory units. Surrounding sequences may interfere sterically with accession of the PHD finger and its exposure could eventually depend on binding of a protein partner to the leucine zipper. In the full-length mouse LIM-HD protein ISL1 (39) intramolecular masking of the HD inhibits DNA-binding activity. Inhibition is released by protein interactions mediated by the LIM domain, presumably causing a conformational change of the ISL1 protein. Alternatively, the leucine zipper in ZIP/PHDf domains might recruit protein partners which mask PHD finger-mediated transcriptional activation. To explain the inefficiency in transcriptional activation observed with ZIP/PHDf domains in yeast, plant and animal cells by the latter hypothesis, such potential protein partners should exhibit a ubiquitous distribution. This prediction is realised for 14-3-3 proteins.
Leucine zipper-mediated interactions between PHDf-HD transcription factors and 14-3-3 proteins
Our experiments reveal that the leucine zipper adjacent to the PHD finger in ZIP/PHDf motifs binds to a conserved region of 14-3-3 proteins. The 14-3-3 family of multifunctional proteins is highly conserved between animals, plants and yeast. Loss of function mutations in yeast 14-3-3 genes cause lethality, but can be rescued by plant 14-3-3 proteins (40), indicating functional conservation. 14-3-3 proteins are involved in various signalling pathways that include, for example, Raf, BAD, Bcr/Bcr-Abl, KSR (kinase supressor of Ras), PKC, PI-3 kinase and cdc25C phosphatase (41). Others enter the nucleus and are associated with DNA-binding complexes (42). Recent data even indicate contacts to TBP, TFIIB and the human TBP-associated factor hTAF(II)32 (43). According to X-ray diffraction studies, 14-3-3 proteins form ω-shaped head-to-head dimers via N-terminal sequences. Each monomer provides an amphipathic groove for protein interactions, which is composed of nine helices in an antiparallel array (34). In many 14-3-3 proteins the second cc (cc2; see Table 1) evident in X-ray diffraction studies spans a leucine zipper with the characteristic heptad spacing. Our data show that this cc2 motif from 14-3-3GF14µ and 14-3-3GF14-12 of Arabidopsis and maize interacts with the leucine zipper motifs preceding the PHD finger in the ZMHOX1a, ZMHOX2a, HAT3.1, PRHA and PRHP HD proteins when tested in the yeast two-hybrid system. Comparable to the CLIM (cofactor of LIM) proteins, which interact with all LIM-HD proteins (44), the maize and Arabidopsis 14-3-3 leucine zippers do not discriminate between individual PHDf-HD proteins.
Due to the dimeric nature of 14-3-3 proteins and their capacity to form homo- and heterodimers (35), members of the 14-3-3 protein family function as scaffolds promoting association of protein complexes (45,46). The leucine zipper mediating the interaction with the ZIP/PHDf domain is located in helix 4 of 14-3-3 proteins. According to X-ray analysis of h14-3-3ζ (34) this helix is located on the convex side of the monomer, but the surface of the zipper is directed towards the concave amphipathic groove of the monomer (see Fig. 4E). In the ω-shaped dimer, helix 4 is partially covered by helix 3, crossing the N-terminal part of the leucine zipper motif. Although recent data elaborated for two other Arabidopsis 14-3-3 isoforms indicate that helix 4 contributes to 14-3-3 dimerisation (47), these authors also demonstrated that the equilibrium between monomers and dimers depends on the calcium level. At least in the monomer the leucine zipper might be free for heterotypic protein interactions. The co-precipitation experiments performed here provide evidence that full-length 14-3-3 and the ZMHOX1a gene product represent potential interaction partners within the nuclear compartment. Conservation of this leucine zipper motif in helix 4 of many 14-3-3 proteins makes it tempting to speculate about a general contribution to 14-3-3 signalling. Located in helix 4, this interaction surface is different to that in helix 7, which has been shown recently to interact with human TBP and TFIIB (43). However, both sets of experimental data support a contribution of 14-3-3 proteins to transcriptional control.
We presently do not know whether or how the 14-3-3 proteins are involved in control of PHD finger activity in the context of the ZIP/PHDf domain. Any answer to this question will presumably require the establishment of an in vitro test system, since 14-3-3 proteins are ubiquitous, highly conserved and essential in all eukaryotes. So far we will take the compatibility of leucine zippers in 14-3-3 and PHDf-HD proteins as an indication that plant HD transcription factors might be potential targets for 14-3-3 signalling. Linkage to the abscisic acid (ABA) response via transcription factor VP1 (48), fusicoccin perception at the plasma membrane (49) and gene regulation in plant pathogen responses (50) assign 14-3-3 proteins essential roles in signal transduction in plant cells. The identification of PHDf-HD proteins as potential interaction partners of 14-3-3 proteins in the plant cell nucleus therefore represents an interesting end-point of these signalling cascades. Concerning the established contributions of 14-3-3 proteins to mitogenic and apoptotic signalling pathways in animal cells, association with the ZMHOX gene transcripts, which are confined to meristems and descending proliferating cells of the maize plant, is significant.
In conclusion, the experiments described here have established the PHD finger as a general transcriptional activation domain in yeast, plant and animal cells. Embedded in the conserved ZIP/PHDf motif the transcriptional activity is repressed in plant transcription factors. This provides the potential for the leucine zipper to modulate PHDf-HD activity and suggests that the ZIP/PHDf motif represents a regulatory domain of these transcription factors. Consistent with this assumption, the leucine zipper upstream of the PHD finger in the ZIP/PHDf domain may mediate interactions with a corresponding motif that is conserved in helix 4 of two 14-3-3 proteins. This novel type of 14-3-3 protein–protein interaction may be of general importance and incorporate HD transcription factors into plant signalling pathways.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Rob Ferl (University of Florida, Gainsville) for providing the 14-3-3 cDNA clones, Dr Jürg Müller (MPI, Tübingen) for the dMI-2 cDNA, Dr Klaus Salchert (MPI, Köln) for the A.thaliana cDNA expression library, Dr Imre Somssich (MPI, Köln) for the PRHA and PRHP cDNA clones, Dr Richard Thompson (MPI, Köln) for the K373 plasmid and Dr Gerd Wohlfahrt (Institut für Biochemie, Köln) for help with the 14-3-3 3-dimensional structure. We gratefully acknowledge support by Dr Bernd Weisshaar (MPI, Köln) with the Arabidopsis transient gene expression system and Drs Rüdiger Simon and Richard Waites (Institut für Entwicklungsbiologie, Köln) for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through SFB 274.
REFERENCES
- 1.Bellmann R. and Werr,W. (1992) EMBO J., 11, 3367–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schindler U., Beckmann,H. and Cashmore,A.R. (1993) Plant J., 4, 137–150. [DOI] [PubMed] [Google Scholar]
- 3.Aasland R., Gibson,T.J. and Stewart,A.F. (1995) Trends Biochem. Sci., 20, 56–59. [DOI] [PubMed] [Google Scholar]
- 4.Saha V., Chaplin,T., Gregorini,A., Ayton,P. and Young,B.D. (1995) Proc. Natl Acad. Sci. USA, 92, 9737–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochar D.A., Savard,J., Wang,W., Lafleur,D.W., Moore,P., Cote,J. and Shiekhattar,R. (2000) Proc. Natl Acad. Sci. USA, 97, 1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loewith R., Meijer,M., Lees-Miller,S.P., Riabowol,K. and Young,D. (2000) Mol. Cell. Biol., 20, 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freemont P.S. (1993) Ann. NY Acad. Sci., 684, 174–192. [DOI] [PubMed] [Google Scholar]
- 8.Freyd G., Kim,S.K. and Horvitz,H.R. (1990) Nature, 344, 876–879. [DOI] [PubMed] [Google Scholar]
- 9.van der Reijden B.A., Erpelinck-Verschueren,C.A., Lowenberg,B. and Jansen,J.H. (1999) Protein Sci., 8, 1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smit L. and Borst,J. (1997) Crit. Rev. Oncogen., 8, 359–379. [DOI] [PubMed] [Google Scholar]
- 11.Dawid I.B., Breen,J.J. and Toyama,R. (1998) Trends Genet., 14, 156–162. [DOI] [PubMed] [Google Scholar]
- 12.Glenn D.J. and Maurer,R.A. (1999) J. Biol. Chem., 274, 36159–36167. [DOI] [PubMed] [Google Scholar]
- 13.Bach I., Rodriguez-Esteban,C., Carriere,C., Bhushan,A., Krones,A., Rose,D.W., Glass,C.K., Andersen,B., Izpisua Belmonte,J.C. and Rosenfeld,M.G. (1999) Nature Genet., 22, 394–399. [DOI] [PubMed] [Google Scholar]
- 14.Dobbelstein M., Wienzek,S., Konig,C. and Roth,J. (1999) Oncogene, 18, 2101–2106. [DOI] [PubMed] [Google Scholar]
- 15.Scott H.S., Heino,M., Peterson,P., Mittaz,L., Lalioti,M.D., Betterle,C., Cohen,A., Seri,M., Lerone,M., Romeo,G., Collin,P., Salo,M., Metcalfe,R., Weetman,A., Papasavvas,M.P., Rossier,C., Nagamine,K., Kudoh,J., Shimizu,N., Krohn,K.J. and Antonarakis,S.E. (1998) Mol. Endocrinol., 12, 1112–1119. [DOI] [PubMed] [Google Scholar]
- 16.Curtiss J. and Heilig,J.S. (1998) Bioessays, 20, 58–69. [DOI] [PubMed] [Google Scholar]
- 17.Klinge B. and Werr,W. (1995) Dev. Genet., 16, 349–357. [Google Scholar]
- 18.Comelli P., Konig,J. and Werr,W. (1999) Plant Mol. Biol., 41, 615–625. [DOI] [PubMed] [Google Scholar]
- 19.Kirch T., Bitter,S., Kisters-Woike,B. and Werr,W. (1998) Nucleic Acids Res., 26, 4714–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinge B., Überlacker,B., Korfhage,C. and Werr,W. (1996) Plant Mol. Biol., 30, 439–453. [DOI] [PubMed] [Google Scholar]
- 21.Korfhage U., Trezzini,G.F., Meier,I., Hahlbrock,K. and Somssich,I. (1994) Plant Cell, 6, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eshed Y., Baum,S.F. and Bowman,J.L. (1999) Cell, 99, 199–209. [DOI] [PubMed] [Google Scholar]
- 23.Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- 24.Bartel P., Chien,C.T., Sternglanz,R. and Fields,S. (1993) Biotechniques, 14, 920–924. [PubMed] [Google Scholar]
- 25.Harper J.W., Adami,G.R., Wei,N., Keyomarsi,K. and Elledge,S.J. (1993) Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 26.Überlacker B. and Werr,W. (1996) Mol. Breeding, 2, 293–295. [Google Scholar]
- 27.Sprenger-Haussels M. and Weisshaar,B. (2000) Plant J., 22, 1–8. [DOI] [PubMed] [Google Scholar]
- 28.Scheer N. and Campos-Ortega,J.A. (1999) Mech. Dev., 80, 153–158. [DOI] [PubMed] [Google Scholar]
- 29.Argenton F., Arava,Y., Aronheim,A. and Walker,M.D. (1996) Mol. Cell. Biol., 16, 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Printen J.A. and Sprague,G.F.Jr (1994) Genetics, 138, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U.K. (1970) Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 32.Westerfield M. (1993) A Guide to the Laboratory Use of the Zebrafish (Brachydanio rerio). The Zebrafish Book. Oregon University Press, OR.
- 33.Überlacker B., Klinge,B. and Werr,W. (1996) Plant Cell, 8, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., Bienkowska,J., Petosa,C., Collier,R.J., Fu,H. and Liddington,R. (1995) Nature, 376, 191–194. [DOI] [PubMed] [Google Scholar]
- 35.Wu K., Lu,G., Sehnke,P. and Ferl,R.J. (1997) Arch. Biochem. Biophys., 339, 2–8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]
- 37.Torii K.U., Stoop-Myer,C.D., Okamoto,H., Coleman,J.E., Matsui,M. and Deng,X.W. (1999) J. Biol. Chem., 274, 27674–27681. [DOI] [PubMed] [Google Scholar]
- 38.Moilanen A.M., Poukka,H., Karvonen,U., Hakli,M., Janne,O.A. and Palvimo,J.J. (1998) Mol. Cell. Biol., 18, 5128–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Garcia I., Osada,H., Forster,A. and Rabbitts,T.H. (1993) EMBO J., 12, 4243–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Heusden G.P., Griffiths,D.J., Ford,J.C., Chin,A.W.T.F., Schrader,P.A., Carr,A.M. and Steensma,H.Y. (1995) Eur. J. Biochem., 229, 45–53. [PubMed] [Google Scholar]
- 41.Lopaczynski W. (1999) Acta Biochim. Pol., 46, 51–60. [PubMed] [Google Scholar]
- 42.Bihn E.A., Paul,A.L., Wang,S.W., Erdos,G.W. and Ferl,R.J. (1997) Plant J., 12, 1439–1445. [DOI] [PubMed] [Google Scholar]
- 43.Pan S., Sehnke,P.C., Ferl,R.J. and Gurley,W.B. (1999) Plant Cell, 11, 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bach I., Carriere,C., Ostendorff,H.P., Andersen,B. and Rosenfeld,M.G. (1997) Genes Dev., 11, 1370–1380. [DOI] [PubMed] [Google Scholar]
- 45.Xing H., Kornfeld,K. and Muslin,A.J. (1997) Curr. Biol., 7, 294–300. [DOI] [PubMed] [Google Scholar]
- 46.Braselmann S. and McCormick,F. (1995) EMBO J., 14, 4839–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abarca D., Madueno,F., Martinez-Zapater,J.M. and Salinas,J. (1999) FEBS Lett., 462, 377–382. [DOI] [PubMed] [Google Scholar]
- 48.Schultz T.F., Medina,J., Hill,A. and Quatrano,R.S. (1998) Plant Cell, 10, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuglsang A.T., Visconti,S., Drumm,K., Jahn,T., Stensballe,A., Mattei,B., Jensen,O.N., Aducci,P. and Palmgren,M.G. (1999) J. Biol. Chem., 274, 36774–36780. [DOI] [PubMed] [Google Scholar]
- 50.Roberts M.R. and Bowles,D.J. (1999) Plant Physiol., 119, 1243–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]