Abstract
The ability to rejoin broken chromosomes is fundamental to the maintenance of genetic integrity. Mammalian cells possess at least five DNA ligases, including three isoforms of DNA ligase III (Lig-3). Lig-3 proteins differ from other DNA ligases in the presence of an N-terminal zinc finger (Zn-f) motif that exhibits extensive homology with two zinc fingers in poly(ADP-ribose) polymerase (PARP). Here we report that the Zn-f confers upon Lig-3 the ability to bind DNA duplexes harbouring a variety of DNA secondary structures, including single-strand gaps and single-strand flaps. Moreover, the Zn-f stimulates intermolecular end joining of duplexes that harbour these structures up to 16-fold. The Zn-f also stimulates end joining between duplexes lacking secondary structure, but to a lesser extent (up to 4-fold). We conclude that the Zn-f may enable Lig-3 to rejoin chromosomal DNA strand breaks located at sites of clustered damage induced by ionising radiation or near to secondary structure intermediates of DNA metabolism.
INTRODUCTION
The Lig-3 gene encodes two nuclear polypeptides and a mitochondrial polypeptide (1–3). The two nuclear isoforms are denoted Lig-3α and Lig-3β and differ at their C-termini as a consequence of tissue- and cell type-specific splicing (4,5). Lig-3α is expressed in both somatic and germ cells and contains a 76 amino acid C-terminal BRCT domain that binds the single-strand break repair protein XRCC1 (4,5). The interaction with XRCC1 is required for intracellular stability of Lig-3α and consequently for efficient DNA ligation following the excision of damaged DNA bases (6–9). In contrast, Lig-3β lacks a C-terminal BRCT domain and does not interact with XRCC1, is selectively expressed in male meiotic cells and has been proposed to function in homologous recombination (4).
A feature of Lig-3 that is unique amongst DNA ligases is the presence of a zinc finger (Zn-f) motif at the N-terminus (3). The Zn-f exhibits extensive homology with two zinc fingers present in poly(ADP-ribose) polymerase (PARP). The Zn-f motifs in PARP are required for binding of this protein to DNA strand breaks (10,11). Here we report that the Zn-f in Lig-3 is required for a different activity, namely the ability to bind DNA containing secondary structures and to stimulate ligation of DNA strand breaks located near such structures. This novel activity may enable Lig-3 to rejoin DNA strand breaks at highly toxic sites of clustered damage induced by ionising radiation or near to secondary structure intermediates of DNA metabolism.
MATERIALS AND METHODS
Recombinant proteins
Histidine-tagged human Lig-3 proteins were expressed and affinity purified from Escherichia coli as previously described (12). Expression of Lig-3 in E.coli results in C-terminally truncated polypeptides in addition to full-length protein, particularly with the α-isoform. The truncated polypeptides co-purify with full-length Lig-3 during affinity chromatography, due to the presence of an N-terminal decahistidine tag, but are largely removed by subsequent gel filtration (>95%). Gel filtration was conducted using a BioCad Sprint Perfusion Chromatography System (Superdex 200; Amersham Pharmacia Biotech) in 10 mM Tris, pH 7.5, 500 mM NaCl, 1 mM DTT and 0.25% Tween-20. Fractions containing Lig-3 were dialysed against 10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM DTT and 10% glycerol. Human DNA polymerase β (Pol β) was expressed in E.coli from plasmid pLW-11 (kindly provided by S. Wilson) and purified as follows. Cells (0.125 l) were grown to an OD of ∼0.5 at 30°C and expression induced by continued incubation for 4 h at 42°C. Pelleted cells were resuspended and frozen in 50 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF and 1 µg/ml pepstatin. The soluble protein extract recovered from thawed cells lysed by sonication was diluted to a final NaCl concentration of 0.1 M and fractionated through 1 ml of HiTrap (Pharmacia) Q-Sepharose and SP-Sepharose columns in series using a Pharmacia Gradifrac. The columns were washed with 10 column vol of 50 mM Tris–HCl, pH 7.5, 1 mM EDTA and 0.1 M NaCl, the Q-Sepharose column removed and the proteins eluted from the SP-Sepharose column with a 0.1–1 M NaCl gradient over 10 column vol in 50 mM Tris–HCl, pH 7.5, and 1 mM EDTA. Fractions containing DNA Pol β (>95% homogeneity) were pooled, dialysed (50 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, 0.1 M NaCl, 1 mM DTT, 5% glycerol and protease inhibitors) and stored at –80°C.
DNA duplex substrates
Oligonucleotides were synthesised by AstraZeneca Pharmaceuticals and are depicted (5′→3′) below. All oligonucleotides were gel purified and phosphorylated, where appropriate, with T4 polynucleotide kinase (Boehringer Mannheim) in the presence of [γ-32P]ATP. Unincorporated nucleotides were removed using Nuc-Trap columns (Stratagene). 32P-labelled oligonucleotides were incubated with equimolar amounts of the appropriate unlabelled oligonucleotide(s) at 85°C for 10 min, prior to slow cooling to room temperature to allow annealing. Intact 70mer or 42mer duplexes were generated by annealing together oligonucleotides 1 and 2 or 3 and 4, respectively, and nicked 70mer by annealing together 2, 5 and 6. Gapped 70mer (1 bp gap) and gapped 42mer (5 bp gap) were created by annealing 2, 6 and 7 or 4, 8 and 9, respectively. 70mer duplex containing a 9 bp 3′ or 5′ single-strand flap was formed by annealing oligonucleotide 5 with duplex preformed with 2 and 10 (3′ flap) or by annealing oligonucleotide 10 with duplex preformed with 2 and 5 (5′ flap). All substrates migrated as a single band when fractionated by non-denaturing gel electrophoresis, indicating the presence of a single major labelled species. In particular, it was confirmed that the mobility of properly annealed 5′ and 3′ flap structures differed significantly from that of partially annealed structures, enabling us to ensure that the flapped structures had formed properly (data not shown). Holliday junctions were created and purified as described elsewhere (13). All oligonucleotide substrates were stored at –20°C in the presence of 50 mM NaCl. 1, 70mer, GCA TAT GGA AAT CGT CAG TTA GGA GCA CTC TAG CCT GAA GTG CAT GCC TAG CAC GTT CTA CCC AGT TCC G; 2, 70mer, CGG AAC TGG GTA GAA CGT GCT AGG CAT GCA CTT CAG GCT AGA GTG CTC CTA ACT GAC GAT TTC CAT ATG C; 3, 42mer, GAT TAA GAC TCC TTA TTC GGA AGT ATG TTA GCA AAC CTA GAA; 4, 42mer, TTC TAG GTT TGC TAA CAT ACT TCC GAA TAA GGA GTC TTA ATC; 5, 30mer, GCA TAT GGA AAT CGT CAG TTA GGA GCA CTC; 6, 40mer, TAG CCT GAA GTG CAT GCC TAG CAC GTT CTA CCC AGT TCC G; 7, 29mer, GCA TAT GGA AAT CGT CAG TTA GGA GCA CT; 8, 15mer, GAT TAA GAC TCC TTA; 9, 22mer, AAG TAT GTT AGC AAA CCT AGA A; 10, 49mer, GGA GCA CTC TAG CCT GAA GTG CAT GCC TAG CAC GTT CTA CCC AGT TCC G.
DNA ligase assays
Reactions contained radiolabelled substrate and recombinant Lig-3 protein in either ‘physiological’ buffer (14) (22 mM Na+, 130 mM K+, 1 mM Mg2+, <0.3 µM free Ca2+, 132 mM Cl–, 11 mM phosphate, 1 mM ATP and 1 mM DTT, pH 7.4) or in 60 mM Tris–HCl, pH 8.0, 10 mM MgCl, 50 µg/ml BSA, 5 mM DTT, 1 mM ATP and NaCl at the concentrations indicated. Reactions were incubated at 30°C for 30 min and terminated by the addition of 4 vol sample buffer (90% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol FF in 1× TBE), heated at 85°C for 10 min, cooled rapidly on ice and fractionated through 10% polyacrylamide–8 M urea gels. Dried gels were analysed by autoradiography or in a Fujix BAS2000 phosphorimager.
Electrophoretic mobility shift assay (EMSA)
32P-labelled substrates were incubated with excess (50-fold by weight) supercoiled competitor plasmid and the indicated amount of full-length or truncated Lig-3 on ice for 20 min in binding buffer (20 mM Tris–HCl, pH 7.5, 1 mM DTT and 0.1 mg/ml BSA) at the indicated salt concentration. Binding buffer for experiments employing Holliday junction intermediates was 50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 5 mM EDTA, 1 mM DTT and 0.1 mg/ml BSA. A one-fifth volume of loading buffer (30% glycerol and 0.25% bromophenol blue in 1× TBE) was added and samples fractionated through 5% non-denaturing polyacrylamide gels at 15 mA in pre-chilled 1× TBE (Bio-Rad Mini Protean II apparatus). Gels were fixed, dried and subjected to autoradiography.
Gap repair reactions
Repair reactions were conducted essentially as described with some modifications (15). Briefly, reactions (10 µl) were conducted in 40 mM HEPES, pH 7.8, 70 mM KCl, 7 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 2 mM ATP, 20 µM each dNTP, 500 µg/ml DNase-free BSA and 100 fmol 70mer 1 bp gapped duplex. DNA Pol β (1 pmol) and/or Lig-3 (1–135 fmol) were present where indicated. Reactions were incubated at 30°C for 30 min and terminated by addition of 4 vol sample buffer (90% formamide, 0.05% bromophenol blue and 0.05% xylene cyanol FF in 1× TBE). Samples were heated to 85°C for 10 min, cooled rapidly on ice and fractionated through 10% polyacrylamide–8 M urea gels. Dried gels were analysed by autoradiography and densitometry.
RESULTS
The Zn-f binds DNA harbouring secondary structure intermediates of DNA metabolism
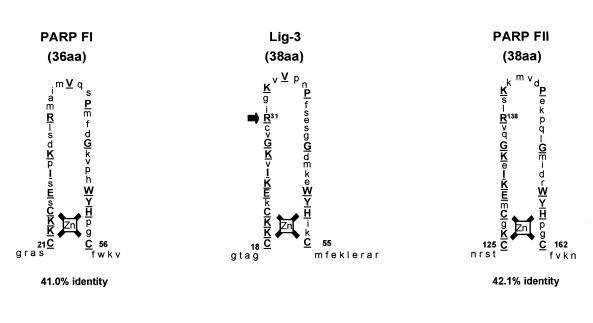
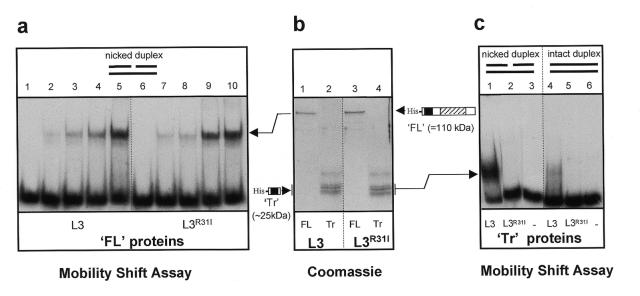
The experiments described in this study were conducted with both the α- and β-isoforms of Lig-3 with essentially identical results. Thus, for simplicity, we do not distinguish between the two isoforms in this report and refer to them collectively as Lig-3. The homologous zinc fingers of Lig-3 and PARP are shown (Fig. 1). The second zinc finger (FII) of PARP is required for binding of this enzyme to nicked DNA, since substitution of the critical Arg138 residue with Ile ablates this activity (10,11). To examine if this is also true for the Lig-3 Zn-f we created the analogous mutation in Lig-3 (Fig. 1, R31I) and compared binding of the wild-type and mutant proteins to a nicked 70mer duplex (Fig. 2a). In contrast to the PARP mutation, the analogous mutation in the Lig-3 Zn-f failed to ablate binding of this enzyme to nicked DNA (Fig. 2a, compare lanes 1–5 with 6–10). Neither wild-type nor mutant Lig-3 greatly bind intact duplex in control experiments conducted in parallel (data not shown and see later, Fig. 3c). It is unlikely that the apparent dispensability of the Zn-f for binding of Lig-3 to nicked DNA reflects ‘leakiness’ of the point mutation because we have observed similar results with deletion mutations that remove the putative zinc coordinating residues (12). Rather, this dispensability presumably reflects the additional presence in Lig-3 of a catalytic domain that is highly conserved in eukaryotic DNA ligases and which can also specifically bind nicked DNA (16–18). Consistent with this notion, binding to nicked DNA by small N-terminal fragments of Lig-3 that lack the catalytic domain, and which are thus dependent upon the Zn-f for binding, was ablated by the Zn-f mutation (Fig. 2b, lanes 2 and 4, and Fig. 2c, lanes 1 and 2).
Figure 1.
Homologous Zn-f motifs in Lig-3 and PARP. Schematic comparing the putative Zn-f motif of Lig-3 with the homologous motifs in PARP (FI and FII). The per cent identity between the Lig-3 Zn-f and those of PARP is given (comparing only the region between and including the first and last zinc-coordinating cysteines). Identical amino acids are in upper case and underlined. Arg31 of the Zn-f in Lig-3 (arrow) is substituted by Ile in Lig-3R31I. The position of the analagous mutation in PARP FII (R138I) is shown at Arg138.
Figure 2.
Zn-f-dependent and Zn-f-independent binding of Lig-3 to nicked duplex. (a) 32P-labelled nicked duplex (500 fmol) was incubated with 0, 0.25, 0.5, 1 or 2 pmol full-length (FL) Lig-3 (lanes 1–5, respectively) or Lig-3R31I (lanes 6–10, respectively). All protein–DNA complexes were formed in the presence of 175 mM NaCl and resolved by non-denaturing PAGE. (b) Histidine-tagged full-length (FL) or truncated (Tr) human Lig-3 or Lig-3R31I, fractionated by SDS–PAGE and stained with Coomassie blue. The schematic locates the histidine tag (His), the Zn-f (solid box) and the DNA-binding catalytic domain (hatched box). (c) 32P-labelled nicked or intact 70mer (250 fmol) was incubated in the absence (–) or presence of 6.25 pmol truncated (Tr) Lig-3 (L3) or Lig-3R31I (L3R31I).
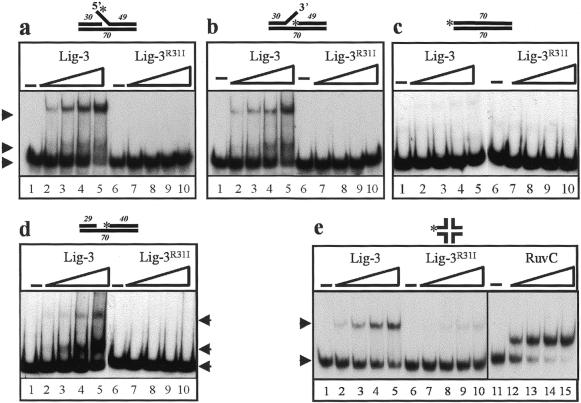
Figure 3.
The Zn-f confers the ability to bind DNA duplexes containing secondary structure on Lig-3. (a–d) 0, 1, 2, 4 or 8 pmol full-length Lig-3 (lanes 1–5, respectively) or Lig-3R31I (lanes 6–10, respectively) was incubated in the presence of 250 mM NaCl with 250 fmol 70mer duplex containing either (a) a 9 bp 5′-flap, (b) a 9 bp 3′-flap, (c) no secondary structure, i.e. intact duplex, (d) or a 1 bp gap. (e) Aliquots of 0, 30, 60, 120 or 240 pmol full-length Lig-3 (lanes 1–5) or Lig-3R31I (lanes 6–10) or 0, 3, 6, 12 or 18 pmol RuvC (lanes 11–15) was incubated with 125 fmol Holliday junction. The positions of unbound substrate (bottom arrows) or substrate bound by truncated Lig-3 (middle arrows) or full-length Lig-3 (top arrows) are indicated. The sizes of the oligonucleotides employed for each substrate and the positions of 32P-labelled 5′-termini (asterisks) are indicated.
Since the Zn-f is largely redundant for nick binding by full-length Lig-3 in vitro, it was considered possible that this motif serves to bind DNA structures that the catalytic domain cannot bind, thereby increasing the substrate specificity of Lig-3. Consistent with this notion, we previously reported that the R31I mutation abolished binding of full-length Lig-3 to a nicked RNA/DNA homopolymer and suggested that the Zn-f might recognise single-strand gaps or single-strand flaps formed in this substrate during its preparation (12). Similar secondary structures may arise in cellular DNA as intermediates of DNA replication, DNA repair and DNA recombination. We therefore examined whether Lig-3 was able to bind oligonucleotide duplexes possessing single-strand gaps or single-strand flaps and, if so, whether the Zn-f was required for binding. Wild-type Lig-3 polypeptide bound DNA duplexes containing a 9 bp single-strand flap in a concentration-dependent manner (Fig. 3a and b, lanes 1–5). Furthermore, binding was entirely Zn-f dependent because Lig-3R31I failed to form a detectable protein–DNA complex at any protein concentration examined (Fig. 3a and b, lanes 6–10). The binding observed with wild-type Lig-3 was dependent on the presence of the single-strand flap because binding to duplex lacking this structure was greatly reduced (Fig. 3c, lanes 1–5). Interestingly, the small amount of binding that was observed to duplex lacking the flap was also Zn-f dependent (Fig. 3c, compare lanes 1–5 with 6–10) and presumably reflected weak binding of the Zn-f to DNA ends, since it occurred in the presence of a 50-fold excess (by weight) of closed circular competitor. Similar results to those described above were observed with duplexes harbouring a 1 bp (Fig. 3d) or a 5 bp gap (data not shown), though the efficiency of binding by full-length Lig-3 was less than was observed for flapped substrates. Binding by Lig-3 was also observed to Holliday junction recombination intermediates and was again largely dependent upon an intact Zn-f (Fig. 3e, compare lanes 1–5 with 6–10). Confirmation that the substrate employed was a Holliday junction was provided by the observation that it was bound by the E.coli Holliday junction resolvase RuvC (Fig. 3e, lanes 11–15). The smaller protein–DNA complexes observed with gapped and flapped substrates (Fig. 3a, b and d, middle arrows) reflect complexes formed by very small amounts of contaminating truncated Lig-3 molecules that remain after gel filtration (see Fig. 2b, lanes 1 and 3).
In summary, the Zn-f confers upon Lig-3 the ability to bind a variety of DNA secondary structures including single-strand flaps, single-strand gaps and Holliday junction intermediates of homologous recombination.
The Lig-3 Zn-f does not enhance gap repair in vitro
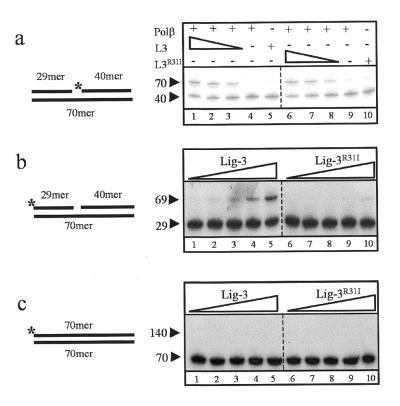
One base pair gaps are formed during short patch base excision repair by the concerted action of DNA glycosylases and AP endonuclease. Given the ability of the Lig-3 Zn-f to bind gapped DNA and the involvement of Lig-3α in their repair (9), we examined whether the Zn-f stimulated gap repair in vitro. The 1 bp gapped substrate employed above was incubated with recombinant human DNA Pol β and Lig-3 proteins and gap repair detected by conversion of the 32P-labelled 40mer to 70mer (see schematic in Fig. 4a). Gap repair required the presence of both polypeptides since 70mer product was not detected if the substrate was incubated with either Lig-3 or Pol β alone (Fig. 4a, lanes 4, 5, 9 and 10). When the substrate was incubated with both proteins efficient repair of the 1 bp gap was observed, with repair proficiency dependent upon Lig-3 concentration under the conditions employed (Fig. 4a, lanes 1–3). However, repair proficiency was not reduced if wild-type Lig-3 was replaced with Lig-3R31I (lanes 6–8), suggesting that the Zn-f does not stimulate gap repair in vitro.
Figure 4.
The Lig-3 Zn-f stimulates end joining but not gap filling in vitro. (a) An aliquot of 100 fmol 70mer harbouring a 1 bp gap was incubated with either 1 pmol Pol β (lanes 4 and 9) or 135 fmol Lig-3 or Lig-3R31I (lanes 5 and 10) or with 1 pmol Pol β plus 135 (lanes 1 and 6), 45 (lanes 2 and 7) or 15 fmol (lanes 3 and 8) Lig-3 or Lig-3R31I. (b and c) An aliquot of 330 fmol 70mer duplex harbouring a 1 bp gap (b) or intact duplex (c) was incubated with 15, 30, 60, 120 or 240 fmol Lig-3 (lanes 1–5) or Lig-3R31I (lanes 6–10). The observed products of intermolecular ligation are labelled 69mer (b) and 140mer (c). The positions of 32P-labelled 5′-termini are indicated (asterisks).
The Lig-3 zinc finger stimulates end joining in vitro
A possible role for Zn-f-mediated binding of Lig-3 to secondary structures might be to facilitate rejoining of DNA strand breaks located near such structures. To examine this possibility, we compared Lig-3 and Lig-3R31I for the ability to conduct intermolecular ligation between the ends of the 1 bp gapped 70mer DNA duplexes described above. To minimise the number of possible ligation products only one of the two 5′-termini of each duplex was phosphorylated (see schematic in Fig. 4b, [32P]phosphate indicated by an asterisk). The product of Lig-3-mediated ligation between duplexes aligned head-to-tail was detected as a 69 bp product resulting from ligation of the 29mer and 40mer (Fig. 4b, lanes 1–5). Strikingly, however, the reaction was ∼8-fold less efficient if wild-type Lig-3 was substituted with Lig-3R31I, suggesting that the Zn-f stimulated the end joining reaction (Fig. 4b, lanes 6–10). Furthermore, this stimulation was largely dependent on the presence of the 1 bp gap, since Lig-3 did not ligate duplexes lacking the gap under comparable conditions (Fig. 4c). The inability of Lig-3R31I to join gapped duplexes was not due to general instability of the mutant protein because Lig-3 and Lig-3R31I exhibited similar activity during gap repair (Fig. 4a) and on a simple nicked duplex (12).
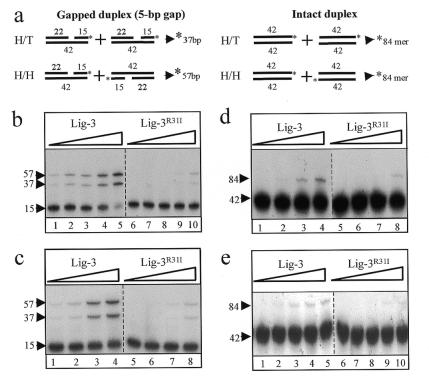
To further examine the role of the Zn-f we compared Lig-3 and Lig-3R31I for their ability to conduct intermolecular ligation between 42mer duplexes harbouring a 5 bp single-strand gap (Fig. 5a, schematic). Efficient ligation was observed at both low and ‘physiological’ salt concentrations, as indicated by appearance of the predicted 37 bp (head-to-tail end joining) and 57 bp (head-to-head end joining) 32P-labelled reaction products (Fig. 5b, lanes 1–5 and Fig. 5c, lanes 1–4). Moreover, ligation mediated by Lig-3R31I was up to 16-fold less efficient, suggesting that this reaction was largely Zn-f dependent (Fig. 5b, lanes 6–10 and Fig. 5c, lanes 5–8). To examine whether ligation stimulated by the Zn-f finger was dependent upon the presence of the 5 bp single-strand gap the activity of Lig-3 and Lig-3R31I was compared on intact duplex. Indeed, intermolecular ligation between intact duplexes was much less efficient overall and the Zn-f stimulated this reaction only up to 4-fold (Fig. 5d and e). This weak stimulation may reflect weak binding of the Zn-f to the ends of intact duplex (see Fig. 3c). Similar results to those described with the 5 bp gapped duplex were also observed when the activity of Lig-3 and Lig-3R31I was compared using duplex harbouring a single-strand flap (data not shown).
Figure 5.
The Lig-3 Zn-f stimulates end joining. (a) Structure of the DNA duplexes employed and the products of intermolecular ligation between duplexes aligned head-to-tail (H/T) and head-to-head (H/H). Asterisks denote 32P-labelled 5′-termini. (b) An aliquot of 550 fmol 5 bp gapped 42mer duplex was incubated with 15, 30, 60, 120 or 240 fmol Lig-3 (lanes 1–5) or Lig-3R31I (lanes 6–10) in the presence of 60 mM salt. (c) An aliquot of 550 fmol 5 bp gapped 42mer duplex was incubated with 5, 10, 20 or 40 fmol Lig-3 (lanes 1–4, respectively) or Lig-3R31I (lanes 4–8, respectively) in the presence of ‘physiological’ buffer (see Materials and Methods). (d) An aliquot of 550 fmol intact 42mer duplex was incubated with 15, 30, 60 or 120 fmol Lig-3 (lanes 1–4) or Lig-3R31I (lanes 5–8) in the presence of 60 mM salt. (e) An aliquot of 550 fmol intact 42mer duplex was incubated with 5, 10, 20, 40 or 80 fmol Lig-3 (lanes 1–5) or Lig-3R31I (lanes 6–10) in the presence of ‘physiological’ buffer.
DISCUSSION
The data described here indicate that the Lig-3 Zn-f promotes binding of this enzyme to DNA duplexes harbouring secondary structures and stimulates intermolecular ligation between the ends of such duplexes. The Zn-f thus expands the substrate specificity of Lig-3, by facilitating binding to DNA substrates to which the catalytic domain cannot bind. Given the diversity in structures ‘recognised’ by the Zn-f it is likely that this motif recognises a feature common to each rather than the specific structures themselves. Since Lig-3 does not bind or ligate single-stranded DNA (data not shown), one possibility is that the Zn-f recognises regions of transition from single-stranded to double-stranded DNA. What might the physiological role of the Lig-3 Zn-f be? Single-strand flaps and single-strand gaps are putative intermediates of various DNA repair processes, raising the possibility that the Zn-f can target Lig-3 to sites of DNA repair in preparation for DNA ligation. Of particular interest in this respect is the repair of single-strand gaps, since a role for Lig-3α in this process has been demonstrated in vitro (9). However, we failed to uncover any effect of mutating the Zn-f on this process, even under conditions in which the concentration of Lig-3 was limiting. In contrast, mutation of the Zn-f did reduce rejoining of DNA strand breaks located near single-strand gaps and single-strand flaps, suggesting that the Zn-f may function to promote ligation of strand breaks that arise near DNA secondary structures. Strand breaks located near single-strand gaps or other strand interruptions could arise at highly toxic sites of clustered damage induced by ionising radiation. Such sites appear to be more difficult to repair than single lesions and may require specialised activities. Perhaps the Zn-f confers one such activity, by mediating Lig-3 binding and activity at such sites. Consistent with this notion, a role for Lig-3α in the repair of DNA strand breaks induced by ionising radiation is suggested by its interaction with XRCC1 protein (7,8). Single-strand breaks also arise ‘spontaneously’ in cells from endogenous reactive oxygen species and base damage and single-strand flaps and single-strand gaps can arise at sites of replication and recombination. Thus, it is also possible that the Lig-3 Zn-f is required for repair of ‘spontaneous’ DNA strand breaks arising near secondary structure intermediates of DNA metabolism. It will now be of interest to determine what role is played by the Zn-f on Lig-3 activity and on the maintenance of chromosome integrity in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Steve West for the gift of RuvC protein. R.M.T. and C.J.W. are funded by Medical Research Council grants to K.W.C. (G9809326 and G9821041).
REFERENCES
- 1.Chen J., Tomkinson,A.E., Ramos,W., Mackey,Z.B., Danehower,S., Walter,C.A., Schultz,R.A., Besterman,J.M. and Husain,I. (1995) Mol. Cell. Biol., 15, 5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakshmipathy U. and Campbell,C. (1999) Mol. Cell. Biol., 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei Y.F., Robins,P., Carter,K., Caldecott,K., Pappin,D.J., Yu,G.L., Wang,R.P., Shell,B.K., Nash,R.A. and Schar,P. (1995) Mol. Cell. Biol., 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackey Z.B., Ramos,W., Levin,D.S., Walter,C.A., McCarrey,J.R. and Tomkinson,A.E. (1997) Mol. Cell. Biol., 17, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 6.Tebbs R.S., Flannery,M.L., Meneses,J.J., Hartmann,A., Tucker,J.D., Thompson,L.H., Cleaver,J.E. and Pedersen,R.A. (1999) Dev. Biol., 208, 513–529. [DOI] [PubMed] [Google Scholar]
- 7.Caldecott K.W., McKeown,C.K., Tucker,J.D., Ljungquist,S. and Thompson,L.H. (1994) Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldecott K.W., Tucker,J.D., Stanker,L.H. and Thompson,L.H. (1995) Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappelli E., Taylor,R., Cevasco,M., Abbondandolo,A., Caldecott,K. and Frosina,G. (1997) J. Biol. Chem., 272, 23970–23975. [DOI] [PubMed] [Google Scholar]
- 10.Gradwohl G., Menissier de Murcia,J.M., Molinete,M., Simonin,F., Koken,M., Hoeijmakers,J.H. and de Murcia,G. (1990) Proc. Natl Acad. Sci. USA, 87, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikejima M., Noguchi,S., Yamashita,R., Ogura,T., Sugimura,T., Gill,D.M. and Miwa,M. (1990) J. Biol. Chem., 265, 21907–21913. [PubMed] [Google Scholar]
- 12.Taylor R.M., Whitehouse,J., Cappelli,E., Frosina,G. and Caldecott,K.W. (1998) Nucleic Acids Res., 26, 4804–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons C.A. and West,S.C. (1993) J. Mol. Biol., 232, 397–405. [DOI] [PubMed] [Google Scholar]
- 14.Jackson D.A., Bartlett,J. and Cook,P.R. (1996) Nucleic Acids Res., 24, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty A.J. and Dafforn,T.R. (2000) J. Mol. Biol., 296, 43–56. [DOI] [PubMed] [Google Scholar]
- 17.Odell M. and Shuman,S. (1999) J. Biol. Chem., 274, 14032–14039. [DOI] [PubMed] [Google Scholar]
- 18.Subramanya H.S., Doherty,A.J., Ashford,S.R. and Wigley,D.B. (1996) Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]