Abstract
The morphogen Sonic Hedgehog (SHH) plays an important role in coordinating embryonic development. Short- and long-range SHH signalling occurs through a variety of membrane-associated and membrane-free forms. However, the molecular mechanisms that govern the early events of the trafficking of neosynthesised SHH in mammalian cells are still poorly understood. Here, we employed the retention using selective hooks (RUSH) system to show that newly-synthesised SHH is trafficked through the classical biosynthetic secretory pathway, using TMED10 as an endoplasmic reticulum (ER) cargo receptor for efficient ER-to-Golgi transport and Rab6 vesicles for Golgi-to-cell surface trafficking. TMED10 and SHH colocalized at ER exit sites (ERES), and TMED10 depletion significantly delays SHH loading onto ERES and subsequent exit leading to significant SHH release defects. Finally, we utilised the Drosophila wing imaginal disc model to demonstrate that the homologue of TMED10, Baiser (Bai), participates in Hedgehog (Hh) secretion and signalling in vivo. In conclusion, our work highlights the role of TMED10 in cargo-specific egress from the ER and sheds light on novel important partners of neosynthesised SHH secretion with potential impact on embryonic development.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-04918-1.
Keywords: Biosynthetic pathway, p24 proteins, ERES, Membrane trafficking, Endoplasmic reticulum
Introduction
Intercellular communication plays an essential role in directing cell fate during embryonic development [1]. The Hedgehog signalling pathway illustrates very well this statement as the Hedgehog (Hh) family proteins have been shown to play pivotal roles such as acting as a morphogen, embryonic patterning during development, and regeneration of adult tissues [2–7]. Abnormal Hh expression and signalling has been shown to be responsible for cerebellar hypoplasia, and also for numerous cancers, including medulloblastoma and glioblastoma [8–10]. In metazoans, Hh has been shown to mediate cell differentiation and proliferation in the central nervous system [11–13]. In addition, Sonic Hedgehog (SHH), one of three Hh isoforms expressed in mammals, plays a pleiotropic and pivotal role in human physiopathology, such as holoprosencephaly and Pallister–Hall syndrome [14, 15]. However, the mechanisms governing the secretion of neosynthesized Hh family proteins remain incompletely understood.
The biosynthetic secretory pathway is a tightly regulated process to ensure precise coordinated trafficking of cargoes to their destined compartments. Newly synthesised proteins are transported into the endoplasmic reticulum (ER) through a protein translocon and undergo protein quality control through various molecular chaperones. Proteins are then sorted and concentrated into ER exit sites (ERES) by the coat protein complex II (COPII), and bud off as ER-derived vesicles [16, 17]. These vesicles fuse with other vesicular-tubular structures in the ER-Golgi intermediate compartment (ERGIC) for microtubule-dependent sorting to the Golgi complex [18, 19]. At the Golgi complex, cargoes are packaged and sorted to be delivered to various subcellular compartments, such as endosomes, lysosomes and the plasma membrane (PM), with the help of the recruitment of small Rab GTPases, including Rab6 and Rab7 [20–22].
SHH is first synthesised as a full-length 45 kDa precursor in the cytoplasm, with an N-terminal ER signal sequence. Upon entering into the ER, the signal peptide is cleaved, revealing a cysteine as the extreme N-terminal residue. The C-terminal domain of SHH contains an auto-processing catalytic domain, which mediates the self-cleavage of the precursor protein into two halves. The SHH-C protein (26 kDa) is degraded in the ER. In contrast, the SHH-N fragment possesses all signalling activity of SHH that is post-translationally modified with a cholesterol molecule covalently bound to the C-terminus of SHH-N and a palmitoyl-CoA on the N-terminal cysteine residue of SHH-N [23–25]. These modifications are critical for the signalling activity of the resulting dual-lipidated SHH-N (19 kDa) that represents the mature, biologically active molecule that exits the ER and is subsequently delivered via the secretory pathway to the cell surface [14, 26]. Given the importance of these post-translational modifications, it is particularly difficult to monitor the sequential steps governing the subcellular transport of neosynthesised SHH using conventional N- and/or C-terminal tagging strategies.
The further processing/delivery of mature SHH requires the activity of several small GTPases. In polarized cells, it is established, that Hh is first introduced to the plasma membrane, and then endocytosed in a clathrin dependent manner to Rab5 positive early endosomes [27–30]. Subsequently, SHH is sorted in a Rab8/Rab11 dependent way to its final site of release. Importantly however, these studies were performed in the context of apicobasal secretion, while the secretion of SHH in non-polarized cells remains to be determined. Moreover, the biosynthetic secretory pathway often represents a marginal fraction of the secretory pool at steady-state, convolved with the recycling and endo/exocytic trafficking routes. Hence, it is essential to develop tools to study the biosynthetic secretory pathway independently of the other SHH trafficking routes.
The Retention Using Selective Hook (RUSH) system allows for retention of a given protein in the ER and synchronously release upon biotin addition to follow its trafficking pathway in real time [31]. Here, we adapted the SHH-RUSH system that we previously developed [32] to study the secretion of SHH from its neosynthesis in the ER up to the plasma membrane (PM) and secretion into the extracellular media. We found that SHH-RUSH is functional, retaining signalling activity and is transported from the ER to the Golgi up to the plasma membrane within 45 min. We highlight that SHH exits the Golgi in vesicles harbouring Rab6, a small GTPase protein required for SHH to reach the PM. Furthermore, we report that TMED10, a member of the p24 family of transmembrane proteins known to interact with COPII machinery, is necessary for efficient ER-to-Golgi transport of SHH, and that silencing of TMED10 leads to decreased SHH secretion. This was confirmed in Drosophila, as depletion of Baiser (Bai; TMED10 homologue), leads to reduced extracellular Hh levels in producing cells, partial inhibition of Hh-dependent imaginal disc outgrowth and affects both short- and long-range Hh activity.
Results
Characterisation of the SHH-RUSH system
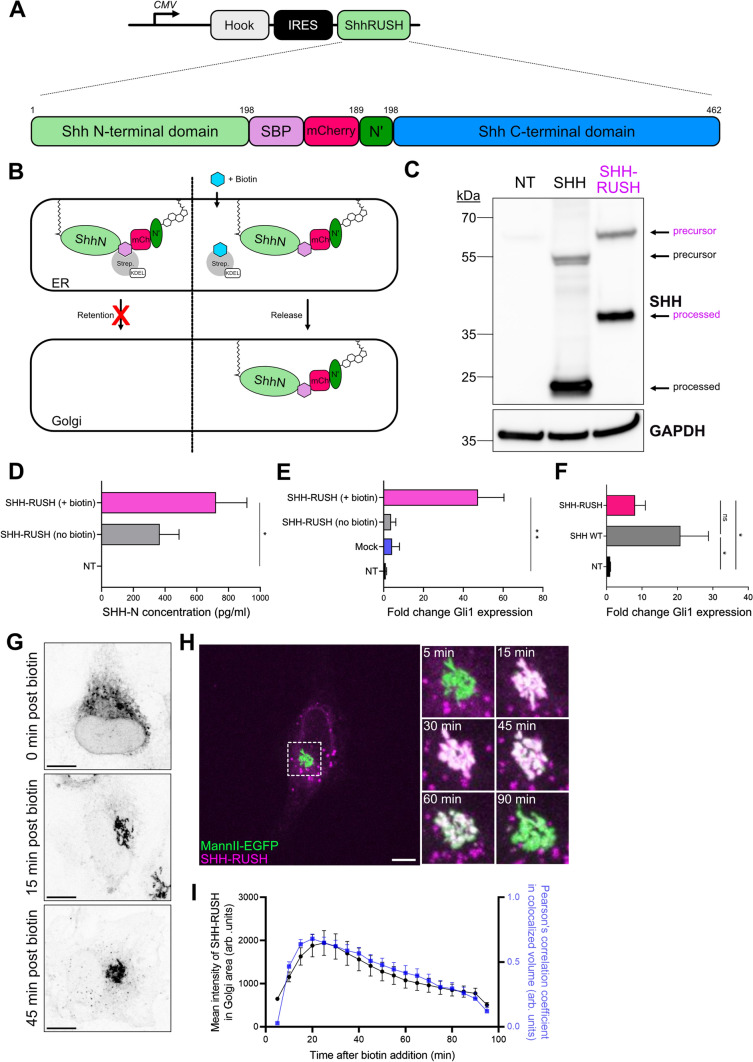
In order to investigate the SHH biosynthetic secretory pathway, we tagged murine SHH with a streptavidin-binding peptide (SBP) and an mCherry fluorescent protein to produce the SHH-RUSH reporter protein (Fig. 1A; Thauvin et al. [32]). The SBP and mCherry tags were introduced internally to preserve the N-terminal palmitate and C-terminal cholesterol modifications in order to track the biologically-active form of SHH [33]. This modification conditionally traps SHH in the ER in cells expressing Streptavidin-KDEL, a “hook” that retain SBP-expressing proteins in the lumen of the ER. This system allows to synchronise SHH-RUSH release and transport along the secretory pathway upon the addition of biotin (Fig. 1B).
Fig. 1.
Characterisation of SHH-RUSH system. A Schematic illustrating the SHH-RUSH reporter system. The Streptavidin-KDEL hook (grey) was encoded on the same plasmid as SHH-RUSH (green) with an internal ribosome entry site (IRES, black) to enable expression of both genes. B Cartoon illustrating the SHH-RUSH retention and release upon biotin addition. C Representative western blot of SVG-A cell lysates either non-transfected (NT), transfected with untagged SHH (SHH) or with SHH-RUSH probed with antibodies against SHH. GAPDH was used as a loading control. D SHH-ELISA of supernatant taken from cells either non-transfected (NT) or transfected with SHH-RUSH and cultured in the absence or presence of biotin for 24 h. Data is represented as mean ± SD from two individual experiments. Unpaired t-test p value < 0.05 (*). E RT-qPCR analysis of Gli1 mRNA expression (using primer set a) in RNA extracted from 3T3 fibroblasts treated with supernatants taken from cells either non-transfected (NT), mock-transfected (Mock), or transfected with SHH-RUSH and cultured in the absence or presence of biotin for 24 h. Data is represented as mean ± SD from two individual experiments. Unpaired t-test p value < 0.01 (**). F RT-qPCR analysis of Gli1 mRNA expression (using primer set b) from NIH 3T3 fibroblasts treated with supernatants taken from cells either non-transfected (NT), or transfected with untagged SHH (SHH WT) or SHH-RUSH. Data is represented as mean ± SD from two independent experiments. Unpaired t-test p value < 0.05 (*). ns non-significant. G Representative confocal microscopy images of SVG-A cells expressing SHH-RUSH and fixed at indicated timepoints after biotin addition. Scale bar = 10 µm. H Representative confocal images of SVG-A cells co-transfected with SHH-RUSH (purple) and a medial Golgi marker, Mannosidase-II-EGFP (Mann-II-EGFP; green). The left micrograph shows a snapshot at 5 min post biotin addition. Scale bar = 10 µm. The right panel highlights zoomed insets of the Golgi area at indicated timepoints post biotin addition from the white dotted square on the left image. I Quantification of the mean fluorescence intensity of SHH-RUSH in the Golgi area (black line) and colocalisation (blue line) of the two channels over time. Data is represented as mean ± SD from three individual experiments.
To verify that SHH-RUSH still undergoes the necessary processing for SHH secretion, we performed western blotting alongside lysates from the human astroglial cell line SVG-A either mock-transfected or transfected with a plasmid encoding untagged SHH or SHH-RUSH. Immunoblotting revealed little to no degradation of SHH proteins and demonstrated that the SHH-RUSH construct expressed the precursor form of SHH and underwent processing similar to untagged SHH, while wild-type cells did not express endogenous SHH (Fig. 1C). SHH protein secretion from cell culture supernatant by ELISA showed a strong increase upon incubating cells with biotin-rich media (Fig. 1D). Of note, SHH was also detected in the supernatant of cells that did not receive biotin treatment, which is likely due to some SHH-RUSH not being retained by the hook. This residual secreted SHH was poorly able to induce SHH-dependent Gli1 RNA expression, suggesting that the leaky forms of SHH might not be fully processed into an active form and/or loaded onto biologically active carriers, such as extracellular vesicles (EVs). Importantly, biotin addition induced secretion of SHH in an active state, able to induce Gli1 RNA expression in NIH 3T3 fibroblasts almost as efficiently as wild-type non-tagged SHH does (Fig. 1E, F). Together, this data indicates that the fully matured and secreted SHH-RUSH protein is biologically functional.
To visualise the trafficking pathway of SHH-RUSH, we performed high-resolution confocal imaging on SHH-RUSH cells fixed at different timepoints after biotin addition. Before the addition of biotin, SHH-RUSH displays an ER distribution (Fig. 1G, upper panel), at 15 min post-biotin addition, SHH-RUSH is enriched in the Golgi apparatus (Fig. 1G, middle panel), and at 45 min post-biotin addition, SHH-RUSH positive vesicles are trafficking towards the cell periphery and localise at the cell surface (Fig. 1G, lower panel). We confirmed that the cell population had relatively homogenous synchronization as most cells showed SHH-RUSH in the ER prior biotin addition, while expressing a Golgi-like signal 30 min post-biotin addition (Supplemental Fig. S1A). Further confocal analysis by co-staining with markers against the ER (Calnexin), cis-Golgi (GM130), trans-Golgi (TGN46) and the cell periphery (Actin) demonstrated that SHH-RUSH was trafficked through the canonical biosynthetic secretory pathway (Supplemental Fig. S1B).
To better understand the trafficking of SHH-RUSH between subcellular compartments, the construct was co-transfected with the medial Golgi marker, Mannosidase-II-EGFP. Approximately 5 min after biotin addition, SHH-RUSH was accumulating at dotted structures within the ER, which likely represent ER exit sites (ERES) (Fig. 1H + Video S1). An increase in SHH-RUSH intensity was observed at the Golgi at approximately 15–20 min post biotin addition (Fig. 1H-I). At 30 min post-biotin addition, a majority of SHH-RUSH had accumulated at the Golgi, and at 90 min post-biotin addition, SHH-RUSH had vacated the Golgi complex and was observed at the plasma membrane (Video S2).
Altogether, these results demonstrate that our SHH-RUSH reporter is a suitable system to study the secretion and signalling of neosynthesised active SHH.
SHH-RUSH follows the classical biosynthetic pathway
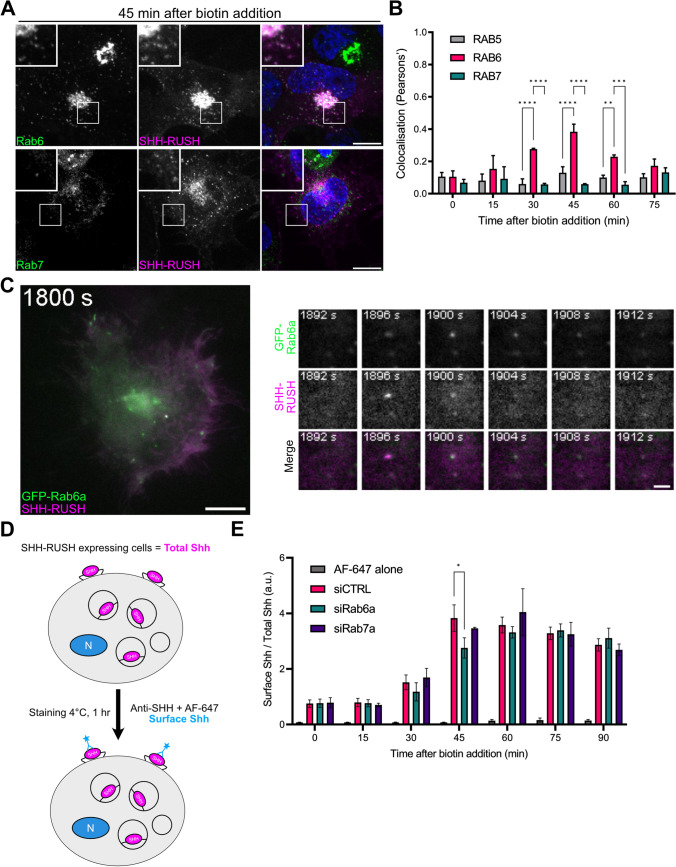
To further investigate the SHH secretory pathway, we looked at the role of Rab GTPases in SHH-RUSH trafficking to the cell surface, as they play important roles in the biosynthetic pathway. Rab6 is a well-described marker of anterograde and retrograde vesicular transport toward and from the Golgi complex [22]. Rab7, a key regulator of late endosome maturation, has recently been shown to be involved as an intermediate compartment of some post-Golgi cargoes [21]. By fixing cells transfected with SHH-RUSH at different timepoints after biotin addition, then performing immunostaining with different Rab GTPases (Supplemental Fig. S2A), we observed a significant subset of post-Golgi SHH-RUSH vesicles colocalising with Rab6-positive vesicles at 45 min post-biotin addition (Fig. 2A, B). By contrast, Rab5- and Rab7-positive vesicles did not colocalise with SHH-RUSH at this time-point, suggesting that post-Golgi SHH-RUSH is not trafficking through the Rab7 secretory pathway and is predominantly transported post-Golgi through the Rab6 secretory pathway en route to the plasma membrane (Supplemental Fig. S2A). Rab11 has previously been shown to be involved in secretion of endocytosed SHH at steady-state [30], but in our assay, investigating specifically neosynthesized SHH trafficking toward the plasma membrane, we could not detect significant enrichment of Rab11 in the SHH-containing vesicles at the investigated time points (Supplemental Fig. S2B). Of note, we were unable to investigate Rab8 localization as we lacked good anti-Rab8 antibodies.
Fig. 2.
SHH-RUSH follows the classical biosynthetic pathway. A Representative confocal images from a single plane in a z-stack of images from cells transfected with SHH-RUSH and stained with antibodies against Rab6 or Rab7, and fixed at 45 min post biotin addition. B Quantification of colocalisation between SHH-RUSH and endogenous Rab5, Rab6 or Rab7 from whole cells in which the Golgi was masked, at different timepoints post biotin addition. Data is represented as mean ± SD, n = 30 cells. Unpaired t-test p value < 0.0001 (****) or < 0.001 (***) or < 0.01 (**). C Representative TIRF images of cells co-transfected with SHH-RUSH and GFP-Rab6a under the weak L30 promoter associated to Video S3. The left panel shows a still image taken at 1898s post biotin addition. Scale bar = 10 µm. The right panel highlights zoomed insets showing a double-positive SHH-RUSH/Rab6a vesicle arriving into the focal plane then fusing with the plasma membrane. D Schematic illustrating the SHH-RUSH cell surface staining assay. E Quantification by flow cytometry of the ratio of surface staining (anti-SHH + AF-647) over the total mCherry signal mean fluorescence intensities is represented at different timepoints post biotin addition in cells treated with siRNA against Rab6a (siRab6a), or Rab7a (siRab7a), or a non-targeting control (siCTRL). Data was normalised to t = 0 and plotted as mean ± SD from four independent experiments. Unpaired t-test p value < 0.05 (*).
To define the fate of Rab6-containing post-Golgi SHH-RUSH vesicles, total internal reflection fluorescence (TIRF) microscopy was performed on cells co-transfected with GFP-Rab6a and SHH-RUSH (Fig. 2C (left panel) and Video S3). At approximately 30 min post biotin addition, we observed the arrival of SHH-RUSH in the evanescent TIRF field. We observed that GFP-Rab6a remained associated with SHH-RUSH vesicles until fusion with the plasma membrane (Fig. 2C (right panels) and Video S4).
To further support the evidence that SHH-RUSH is transported through Rab6-positive vesicles, we wanted to observe the effects of Rab6 depletion on the SHH secretory pathway. First, we used small interfering RNA (siRNA) against Rab6a or Rab7a, and validated their specificity at the mRNA and protein levels (Supplemental Fig. S2C, D). To quantitatively measure the kinetics of SHH-RUSH arriving at the cell surface, we performed a cell surface stain flow cytometry assay as previously described ([21], see Materials & Methods). At specific timepoints following the addition of biotin, cells were incubated on ice to arrest membrane trafficking and SHH-RUSH localised at the surface of non-permeabilised cells was labelled using an anti-SHH antibody (Fig. 2D). In cells treated with siCTRL, SHH surface labelling peaked at 45 min post-biotin addition. While the profile of Rab7a-depleted cells was similar to siCTRL, a significant decrease in surface SHH fluorescence was observed in siRab6a-treated cells (Fig. 2E). Our data indicates that, after exiting the Golgi, SHH is transported in Rab6-harbouring vesicles en route to the cell surface in mammalian cells, a role that was never reported previously to our knowledge.
TMED10 regulates SHH transport through the biosynthetic pathway
Although we demonstrated that Rab6 is an important player for post-Golgi transport of SHH to the plasma membrane, the molecular mechanism underlying ER-to-Golgi trafficking of SHH remains obscure. The transmembrane emp24 domain (TMED) family of proteins are key players in the regulation of protein trafficking and secretion, exhibiting evolutionary conservation across eukaryotic species [34]. Following heterodimer formation, TMED proteins oligomerize to form the p24 complex, which contains a single member from each of the four subfamilies (α, β, γ, δ) [35]. The p24 complex facilitates the interaction between soluble cargoes in the ER lumen and the cytosolic COPII machinery [36, 37].
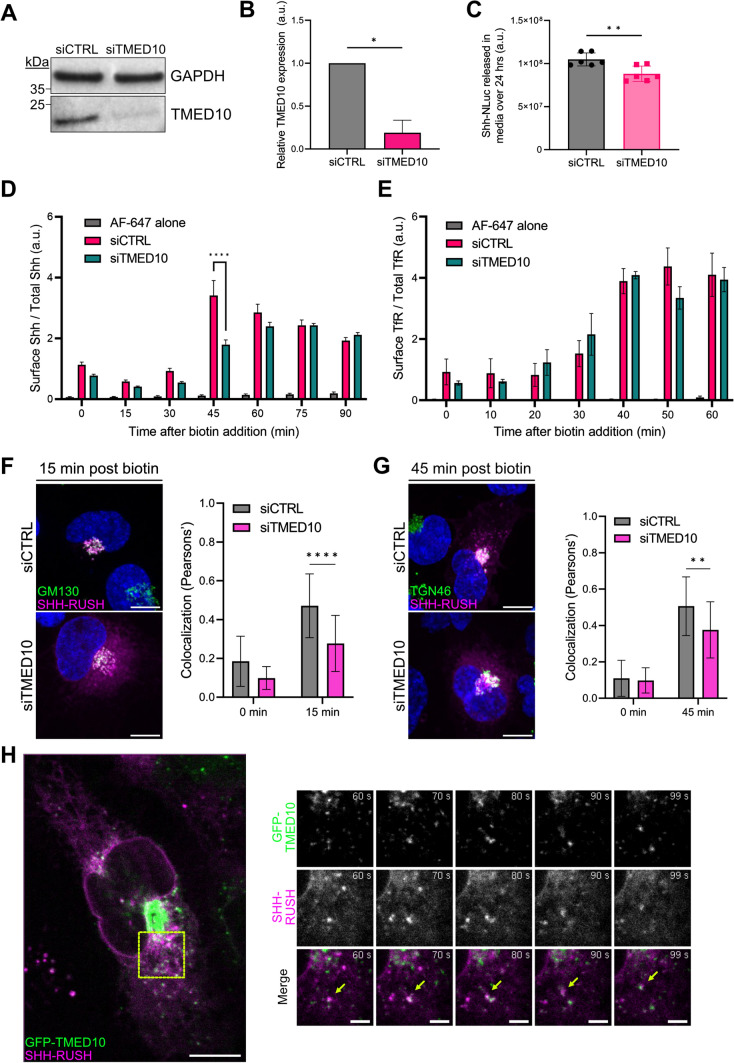
Due to its distribution and function at the ER-Golgi interface [38], we next investigated the potential role of TMED10, also known as Tmp21 or p24 delta (p24δ), in intracellular SHH trafficking. First, TMED10 protein expression was silenced in SVG-A cells, using siRNA (Fig. 3A, B). Using a SHH construct coupled to Nanoluciferase (SHH-NLuc; [39]), we observed that TMED10-depleted cells secreted significantly less SHH than control cells, although the decrease was modest at steady state (Fig. 3C). We observed that the decreased secretion was only observed in TMED10-depleted cells transfected with SHH-NLuc, while other NLuc constructs were not significantly impacted by the silencing of TMED10 (Supplemental Fig. S3A).
Fig. 3.
TMED10 regulates SHH transport through the biosynthetic pathway. A Representative western blot of lysates taken from SVG-A cells treated with siCTRL or siTMED10. Blots were then probed with antibodies against TMED10. GAPDH was used as a loading control. B Quantification of western blot in (A), data plotted as mean ± SD, n = 3 independent experiments, *p < 0.05. C Quantification of SHH-NLuc released into cell culture media from cells treated with siCTRL or siTMED10 over 24 h. Data plotted as mean ± SD, n = 6 independent experiments, **p < 0.01. D Quantification by flow cytometry of the ratio of surface staining (anti-SHH + AF-647) over the total mCherry signal mean fluorescence intensities is represented at different timepoints post biotin addition in cells treated with siRNA against TMED10 (siTMED10) or siCTRL. Data was normalised to t = 0 and plotted as mean ± SD from 3 independent experiments. Unpaired t-test p value < 0.0001 (****). E Flow cytometry analysis of TfR surface staining at different timepoints post biotin addition from SVG-A cells transfected with TfR-RUSH and treated with siTMED10 or siCTRL. Data was normalised to t = 0 and plotted as mean ± SD from 3 independent experiments. No significant differences were measured between the siTMED10 and siCTRL. F Colocalisation analysis of SHH-RUSH with cis-Golgi marker GM130. The left panel shows representative confocal images of cells treated with siCTRL or siTMED10 and transfected with SHH-RUSH (magenta), fixed at 15 min post biotin addition, permeabilized and stained for GM130 (green). Scale bar = 10 µm. The right panel shows the quantification of colocalisation between SHH-RUSH and GM130 at 0 min and 15 min post-biotin addition. Data is plotted as mean ± SD (n = 20 cells). Unpaired t-test p value < 0.0001 (****). G Colocalisation analysis of SHH-RUSH with trans-Golgi marker TGN46. The left panel shows representative confocal images of cells treated with siCTRL or siTMED10 and transfected with SHH-RUSH (purple), fixed at 45 min post biotin addition, permeabilized and stained for TGN46 (green). Scale bar = 10 µm. The right panel shows the quantification of colocalisation between SHH-RUSH and TGN46 at 0 min and 45 min post-biotin addition. Data is plotted as mean ± SD (n = 20 cells). Unpaired t-test p value < 0.05 (*). H TMED10 dynamics during SHH intracellular dynamics associated to Video S5. The left panel shows a still image at 0 s post biotin addition of SVG-A cells co-transfected with GFP-TMED10 and SHH-RUSH. Scale bar = 10 µm. The right panel highlights zoomed images of a double-positive TMED10:SHH-RUSH vesicle trafficking together towards a perinuclear region at indicated timepoints. Scale bar = 2 µm.
These experiments were performed at steady state, and the contribution of neosynthesised SHH (and thus TMED10) in this system may be masked by the large amount of SHH that would be already synthesised and recycled. To obtain more information on the role of TMED10 in the intracellular trafficking of neosynthesised SHH, we went back to our SHH-RUSH system and measured the amount of SHH at the cell surface by flow cytometry as in Fig. 2E. We observed a significant decrease in SHH surface levels in siTMED10-treated cells compared to control cells at early timepoints, while the delay was compensated at later timepoints (Fig. 3D). This decrease was even more severe than the one observed for Rab6a, underlying the importance of TMED10 during SHH secretion. As a control, we also showed that TMED10 silencing decreased significantly the secretion of a neosynthesized GPI-anchored protein using the RUSH system (Supplemental Fig. S4), consistent with previous report [40]. Interestingly, in stark contrast to SHH-RUSH and GPI-RUSH, the trafficking of TfR-RUSH to the cell surface was not perturbed in TMED10-depleted cells (Fig. 3E), highlighting that TMED10 is important for the biosynthetic pathway secretion of a specific subset of cargoes.
Next, to better define at which stage TMED10 could intervene in SHH trafficking, we labelled the cis-Golgi (GM130) and the trans-Golgi (TGN46) compartments at different timepoints after biotin addition. Colocalisation analyses revealed that silencing TMED10 led to significant decrease in colocalisation between SHH-RUSH and GM130 at 15 min post biotin addition, and with TGN46 at 45 min post biotin addition compared to control cells (Fig. 3F, G and Supplemental Fig. S3B–E), suggesting that SHH-RUSH trafficking is delayed in reaching and passing through the Golgi stacks.
Due to a lack of reliable antibodies against endogenous TMED10, we decided to co-express GFP-tagged TMED10 [41] alongside SHH-RUSH in SVG-A cells to observe TMED10 dynamics in the context of SHH secretion (Fig. 3E, left panel and Video S5). We observed that a large fraction of TMED10 was enriched in a structure reminiscent of the Golgi complex, with a small subset of TMED10 present in distinct puncta reminiscent of ERES. These puncta were also positive for SHH-RUSH. At 60 s post-biotin addition, we observed double-positive puncta (GFP-TMED10 and SHH-RUSH) trafficking towards the perinuclear region together (Fig. 3H, right panels and Video S6).
Thus, our data indicates that TMED10 is an important regulator of SHH biosynthetic secretion and may play a role of ER-to-Golgi trafficking of SHH.
TMED10 associates with SHH-RUSH at ERES and aids in its exit from ER
TMED10 has been shown to play an important role in loading lipidated cargo proteins (such as GPI-anchored proteins and Wnt) in the ER into COPII vesicles en route to the cis-Golgi [40]. As SHH is also a lipidated protein, and lacks the necessary transmembrane motifs to interact with the COPII machinery, we hypothesised that TMED10 could act as an ER cargo receptor for SHH.
To assess whether there was an association between SHH and TMED10, we setup a proximity ligation assay (PLA). Cells were co-transfected with SHH-RUSH and GFP-TMED10. The number of PLA puncta per cell (indicative of SHH and TMED10 proximity) was quantified through confocal imaging and automated image analysis (Supplemental Fig. S5A). At 0 min post-biotin addition, we saw no/few PLA puncta per cell, suggesting that SHH-RUSH is not in proximity with GFP-TMED10. However, at 5 min and 10 min post-biotin, we observed PLA puncta, suggesting that at these timepoints, TMED10 and SHH were in close proximity with each other, likely in the same subcellular structures. At 15 min post biotin, fewer PLA puncta were observed, while the majority of SHH-RUSH was localised to the Golgi. These results suggest that TMED10 may associate with SHH at pre-Golgi stages.
Our numerous attempts to co-immunoprecipitate TMED10 and SHH were inconclusive and irreproducible, which prevents us to definitely conclude on the potential interactions between the two proteins. However, the p24 complex is composed of four subfamilies, and the stoichiometry of p24 subfamily members in the p24 complex is key for efficient cargo transport. Hence it is possible that other partners interact more directly with SHH. On this line, we found that TMED2, another subfamily member of the p24 complex, is also required for efficient SHH arrival to the plasma membrane (Supplemental Fig. S5B). Moreover, TMED10 knock-down results in the slight upregulation of TMED7 mRNA (Supplemental Fig. S5C). These data suggest that the different p24 members are likely cooperating to allow SHH exit from the ER.
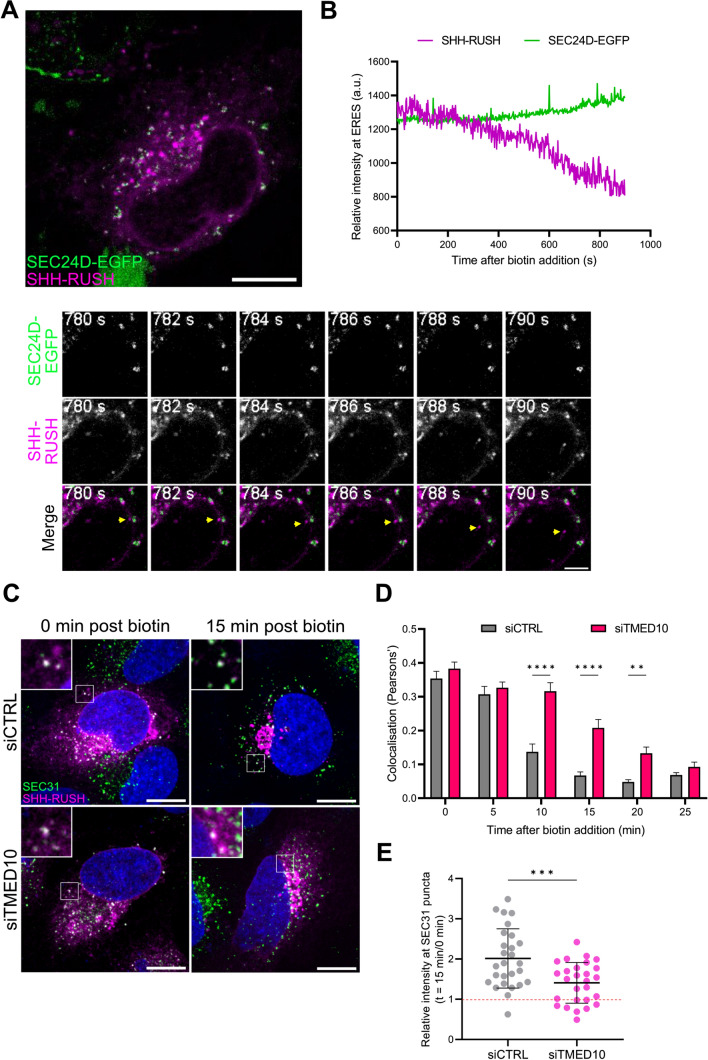
As TMED10 interacts with the inner COPII coat protein SEC24D [42], we next investigated the dynamics of SHH-RUSH at ERES using live confocal time-lapse microscopy. Even before the addition of biotin, a significant subset of SHH-RUSH was localised at SEC24D-puncta (Fig. 4A and associated Video S7). Fluorescent intensity analysis of this structure containing both SHH-RUSH and GFP-SEC24D revealed that shortly after biotin addition, the intensity of SHH-RUSH dropped whereas the intensity of SEC24D remains the same (Fig. 4B), suggesting that SHH exited the ER through an ERES. Of note, given the recent advances on the structure of ERES [43], super-resolution microscopy could help investigating further the sublocation of TMED10 and SHH in ERES.
Fig. 4.
TMED10 associates with SHH-RUSH at ERES and aids in its exit from ER. A Live confocal imaging of ER exit site marker SEC24D (green) and SHH-RUSH (purple) associated to Video S7. The left panel shows a still image of a cell co-transfected with SEC24D-EGFP and SHH-RUSH at 0 s post biotin addition. Scale bar = 10 µm. The right panel highlights zoomed images of a SHH-RUSH vesicle leaving a SEC24D-positive ERES. Scale bar = 2 µm. B Quantification of SHH-RUSH at SEC24D-positive puncta over time post-biotin addition. Data is plotted as mean fluorescence intensity of SHH-RUSH (magenta) and SEC24D-EGFP (green) and is representative of 5 individual cells. C Representative confocal images from a single panel in a z-stack of cells treated with siCTRL or siTMED10 and transfected with SHH-RUSH, fixed at either 0 min or 15 min post biotin addition, permeabilized and stained with antibodies against SEC31. Scale bar = 10 µm. D Quantification of SHH-RUSH colocalisation with SEC31 over time in siCTRL and siTMED10 treated cells. Data is plotted as mean ± SD (n = 30 cells). Unpaired t-test p value < 0.01 (**) or < 0.0001 (****). E Quantification of the ratio of SHH-RUSH intensity at SEC31-positive puncta at 15 min versus 0 min post biotin addition in siCTRL or siTMED10 treated cells. Dotted line (red) represents relative SHH-RUSH intensity at 0 min post biotin addition. Data is plotted as mean ± SD (n = 30 cells). Unpaired t-test p value < 0.001 (***).
To investigate the role of TMED10 in the ER exit of SHH-RUSH, SHH-RUSH expressing cells were treated with siCTRL or siTMED10 and immunolabelled with antibodies against SEC31, an outer COPII coat protein known to localise to ERES [18]. Before the addition of biotin, SHH-RUSH was distributed to the ER in both siCTRL and siTMED10 treated cells. Colocalisation analysis demonstrated that SHH-RUSH colocalised with SEC31 for a longer period of time in siTMED10 cells compared to siCTRL (Fig. 4C, D), suggesting that TMED10 depletion delayed the exit of SHH-RUSH from ER exit sites. To assess whether TMED10 could help SHH to reach ERES, we measured the fluorescence intensity of SHH-RUSH at SEC31-positive puncta over time in siCTRL and siTMED10 cells and found that indeed, SHH-RUSH intensity at SEC31 puncta was significantly reduced in TMED10-depleted cells compared to control cells (Fig. 4E).
Taken together, these results suggest that TMED10 promotes the recruitment of SHH to ERES, thereby favouring its trafficking towards the Golgi and subsequent secretory pathways.
Loss of baiser function affects Hh secretion and signalling in vivo
Given that TMED10 depletion leads to a delay in neosynthesised SHH trafficking from the ER to the cell surface and reduced SHH secretion from producing cells, we investigated whether similar phenotypes could be observed in an established in vivo model such as Drosophila.
Hedgehog (Hh) is known to play a vital role in axial patterning of the epithelial wing imaginal disc cells (WIDs) in Drosophila larvae [3]. The WID is mainly composed of a monolayer of two populations of epithelial cells (columnar cells), posterior cells expressing Hh and anterior cells responding to Hh signalling. Hh induces differential responses in the first proximal 6–8 anterior cells located at the antero-posterior (A/P) border [44, 45]. This readout, and also the failure to respond, can be directly analysed by measuring the expression level of Hh target genes which give specific response to different levels of Hh concentration: short-range target genes, such as the transcription factor Engrailed (En) and the Hh receptor Patched (Ptc), and long-range target genes, such as Decapentaplegic (dpp), the Drosophila homologue of TGF-β [46, 47].
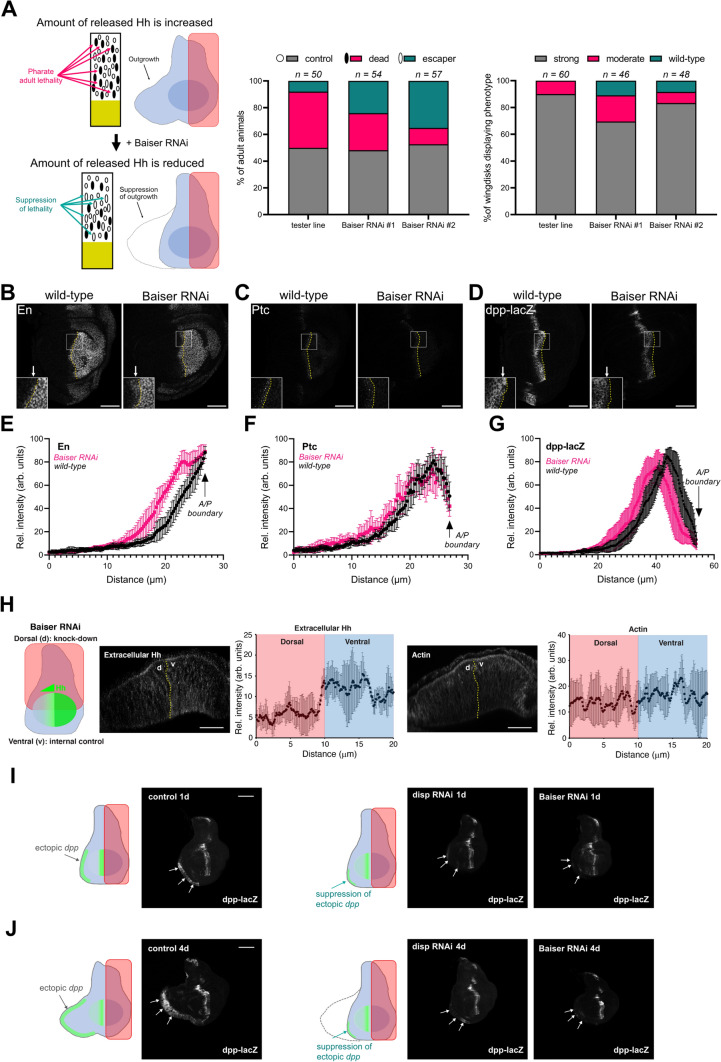
First, we used a ‘tester line’ in which Hh is conditionally overexpressed in the posterior compartment of the WID, leading to distal ectopic expression of the long-range Hh target gene dpp, resulting in an outgrowth of the anterior compartment at the opposite site of the source of Hh [48] and post-larvae lethality during pupal stage (Fig. 5A, I–J). The expression of two different RNAi targeting baiser (bai), the Drosophila homologue of TMED10, in the posterior cells induced a moderate rescue of Hh-induced pupal lethality, as well as dpp-dependent anterior outgrowth (Fig. 5A). Because the RNAi, which is expressed only in posterior cells, induces a non-cell autonomous effect on Hh-dependent activity in anterior cells, our data suggest that bai may play a role in Hh secretion in vivo.
Fig. 5.
Loss of baiser function affects Hh secretion and signalling in vivo. A Left panel: cartoon illustrating pupal lethality rescue assay and larval wing imaginal disc (WID) outgrowth assay. In the “tester line”, expression of Hh is increased in the WID posterior domain (in pink) leading to distal anterior tissue outgrowth. This can be rescued upon decreasing Hh level or Hh extracellular secretion. Middle panel: lethality rescue assay—percentage of escaper animals in”tester line” control, and overexpressing bai RNAi-s in the posterior domain of the “tester line”. Right panel: WID outgrowth assay—percentage of outgrown, moderately and fully rescued discs with respect to their genotypes. Sample numbers (n) are indicated above the graphs. B–D Hh target gene expression in wild-type and bai RNAi wing discs. B Engrailed, C Patched (Ptc) and D anti-β-gal (dpp-lacZ). The anterior/posterior (A/P) compartment boundaries are marked with yellow dotted lines. Placement of insets are indicated by white rectangles. White arrows point to the areas in the anterior compartment, where proximal Hh target gene expression is different between wild-type and bai RNAi conditions. Scale bar – 50 µm. E–G Quantification of Engrailed (E, n = 8), Patched (F, n = 10) and dpp-lacZ (G, n = 10) in the anterior compartment of wild-type control (black) and bai RNAi (magenta) discs. The A/P boundary is indicated by black arrows. H Left panel: Cartoon illustrating the dorsal depletion of bai using RNAi, with ventral area serving as an internal control. Middle and right panels: Extracellular staining of endogenous Hh (middle) and filamentous Actin (right) in discs with bai RNAi. Dotted yellow line labels boundary between dorsal and ventral compartments. Scale bar – 50 µm. Quantification of extracellular Hh and Actin intensity levels between the dorsal and ventral compartments are shown next to the corresponding immunostainings. Data plotted as mean ± SD, n = 10. I, J Distal ectopic dpp-lacZ gene expression measured in discs overexpressing Hh in producing cells for 1 day (I) or 4 days (J). Cartoons next to the fluorescent panels illustrate dpp-lacZ expression pattern (green). The pink rectangles indicate the domain of expression of UAS-Hh together with GFP RNAi (control), disp RNAi (positive control) and bai RNAi. White arrows point to ectopic dpp-lacZ expression (or in case of disp and bai RNAi the lack of it) in cuboidal cells of the WID. Note, that 1-day induction of Hh overexpression does not lead to changes in WID morphology, and that bai RNAi in Hh producing cells is capable of suppressing ectopic distal dpp-lacZ expression. Additional examples can be found on Supplementary Fig. S6. Scale bar – 100 µm.
To verify the role of bai in the establishment of Hh gradient activity, we examined the expression levels of Hh target genes in bai RNAi treated WIDs expressing a physiological level of Hh (Fig. 5B–D). We observed an increased number of cells expressing the high-level Hh target En in anterior Hh-receiving cells (3–4 rows, instead of 1–2 in the wild-type discs, Fig. 5B, E). We did not detect significant changes in Ptc expression (Fig. 5C, F). In contrast, the range of dpp-lacZ expression covered a smaller territory in posterior bai-depleted discs compared to wild-type (Fig. 5D, G). This change was very apparent in cells close to the A/P border, where, since En is a transcriptional repressor for dpp, the increased ectopic En expression is known to suppressed dpp transcription.
To determine whether bai depletion plays a direct role on Hh secretion, we stained extracellular Hh and actin as a control, and found that within the same animal, the bai RNAi region had decreased extracellular Hh compared to non-RNAi treated areas, while actin staining remained similar regardless of the region observed (Fig. 5H). Together, the data suggests that in vivo depletion of bai decreases Hh secretion, likely differentially affecting Hh targets, and perturbs the physiological Hh gradient involved in WID morphogenesis.
Finally, ectopic distal dpp-lacZ patterns were observed in the WID of the UAS-Hh tester line together with either control GFP RNAi or with RNAi targeting bai or dispatched (disp; a strong regulator of Hh secretion in both flies and mammals [49] (Fig. 5I–J and Supplemental Fig. S6A, B). In control samples, ectopic dpp-lacZ was found at the distal tip of the outgrowing WID at 1- and 4-days post induction. In contrast, we found that bai RNAi results in decreased distal dpp-lacZ expression with a reduced outgrowth area at 4 days post-induction. This phenotype was very reminiscent of the one observed for disp RNAi, used here as a positive control. These data suggest that bai regulates Hh and is particularly important for long-range Hh signalling in flies.
Discussion
The present study aimed to provide a better understanding of the complex mechanisms governing the intracellular transport of SHH by using a novel SHH-RUSH reporter system. Our results demonstrate that the SHH-RUSH system enables efficient transport of SHH through the classical biosynthetic secretory pathway while maintaining its full signalling capability. This system allowed to identify two novel players involved in the trafficking of neosynthesised SHH: TMED10 and Rab6. Moreover, we have uncovered TMED10 as a novel regulator of SHH biosynthetic secretory pathway. We show that TMED10 mediates ER-to-Golgi translocation of SHH by contributing to its loading onto ERES. Furthermore, TMED10 plays a significant role in Hh/SHH secretion and downstream signalling in vitro and in vivo, highlighting its physiological relevance during embryonic development.
Recently, a suite of sophisticated tools has been developed to elucidate the intricate spatial and temporal mechanisms underlying the secretory pathway. Early investigations into protein secretion were conducted using the thermosensitive vesicular stomatitis virus glycoprotein (ts045VSV-G) [50]. However, to retain physiological conditions and monitor the trafficking of cargoes other than VSV-G, the RUSH system was developed [31]. Here, the cells we used do not express endogenous SHH and thus the CRISPR approach could not be employed. The RUSH system may also perturb intracellular trafficking as the RUSH-tagged protein is released as a single wave, potentially causing an overload of the system. Although this is important to take into account, we investigated SHH-RUSH kinetics in various cell lines (Thauvin et al. [32] and this study), testing its signalling activity, and confirmed our main findings with non-RUSH approaches, allowing us to further consolidate our conclusions. Nonetheless, it is imperative to exercise caution when utilizing the RUSH system to study the normal trafficking and function of cargoes, taking into account their size and post-translational modifications [51]. These findings suggest that our approach would be suitable for future investigations into other aspects of Hh/SHH biology.
We identified Rab6-positive carriers as a mean of transport of neosynthesised SHH from the Golgi apparatus to the cell surface in non-polarised mammalian cells. This finding is consistent with previous reports on the importance of Rab6 for the biosynthetic secretory pathway of other proteins [52]. Rab6 has also been implicated in cargo transport to other subcellular compartments, such as the recycling endosome, late endosomes/lysosomes, and Golgi stacks, highlighting its versatility in intracellular transport [53–55]. However, silencing of Rab6a induces only a mild (although significant) decrease of SHH arrival at the PM, which suggest either that other Rab6 isoforms are participating in SHH secretion, or that other parallel pathways are at play. Future studies should be undertaken to explore the regulatory partners involved in post-Golgi transport of SHH. Such investigations would significantly deepen our understanding of the interwoven dynamics of the intracellular trafficking machinery that governs protein secretion.
In our study, the association of TMED10 with SHH at ERES provides insight into the delayed SHH secretion observed upon TMED10 depletion. Of note however, given the recent advances on the structure of ERES [43], super-resolution microscopy could help investigating further the sublocation of TMED10 and SHH in ERES. We found that the depletion of one p24 family member results in abnormal expression of other members and that TMED2 silencing also decreases SHH secretion. These observations argue in favour of the requirement for a mature p24 complex, composed of the four subfamily members. Yet, only TMED10 was found in SHH-containing EVs [56], suggesting that TMED10 could play additional roles that remain to be characterized.
The p24 family proteins have been extensively studied as cargo receptors for proteins such as GPI-anchored and Wnt proteins in mammals, flies and plants [57, 42, 58]. As lipidated cargo proteins have been identified as requiring p24 family proteins for efficient ER-to-Golgi trafficking, we hypothesize that such proteins utilise common mechanisms of ER egress. Recently, SURF4 was identified as another cargo receptor for SHH in the ER [59]. The SURF4-SHH interaction was found in HeLa and 293T cells, while we used an astrocytic cell line to study SHH secretion as well as in vivo models. One can hypothesize that SURF4 and TMED10 are differentially recruited according to the producer cell type investigated, and thus, further work is needed to decipher the interplay of the various SHH partners in the ER. Whether SURF4 and TMED10 are redundant or playing different roles during SHH secretion remains unclear at this stage. SURF4 is a major player in the role of maintaining the architecture of the ERGIC and Golgi apparatus [60], and thus, it may be possible that SURF4 has a broad effect on protein secretion. In contrast, TMED10 silencing had no effect on the TfR neosynthetic pathway in mammalian cells, suggesting that TMED10 is cargo-specific. Yet, the molecular determinants governing specificity remain to be identified. Interestingly, TMED10 was recently shown to be involved in SMO secretion, another component of SHH signalling cascade [61]. It would be interesting to determine whether TMED10 plays a role on other components of the hedgehog pathway, as well as on Desert Hedgehog and Indian Hedgehog secretion. Finally, TMED10 has been involved in various neurodevelopmental defects [62–66] and found associated to SHH-containing extracellular vesicles in ART-EVs, a subtype of vesicles found in the cerebrospinal fluid of a glioblastoma patient [56].
In this study, we highlight that bai/TMED10 participates in the secretion of Hh/SHH in Drosophila and in mammalian cells, and revealed its pivotal role during ER egress. Bai has been shown to be required for the secretion of other morphogens such as Wingless, the Drosophila homologue of Wnt [67, 68], providing further evidence supporting our hypothesis on the role of TMEDs in selective cargo secretion. Hh is secreted in various forms: a form that is involved in short-range signalling (monitored through En staining), and a form more potent for long-range signalling mediated by extracellular vesicles (monitored through Dpp-LacZ expression). We have often observed a tradeoff between the long-range and the short-range Hh pathway targets, in other words that defects in long-range Hh signalling was associated to increased short-range signalling [69, 28, 29]. This is observed specifically when trafficking of Hh in its secreting cells is perturbed, suggesting that apical and basolateral trafficking of Hh in the polarized producing cells are not independent. Thus, the increased En staining observed may solely result from a defective apical secretion induced by bai RNAi. This trade-off is extremely difficult to investigate because little is known about polarized trafficking in wing imaginal discs.
Intriguingly, the low, yet significant and consistent, impact of bai depletion in the WID outgrowth assays closely resembles the effect of TMED10 depletion that we observed on SHH secretion under steady-state conditions. As previously stated, steady-state condition is not an ideal system to decipher the biosynthetic secretory pathway, and thus, to further elucidate the role of bai in Hh secretion and downstream signalling, it will be beneficial to employ the RUSH system, which has recently been implemented in Drosophila [70].
In conclusion, our study lays the groundwork for future research on the intricate workings of SHH biology and showcases the potential of the SHH-RUSH reporter system as a potent instrument for delineating the molecular mechanisms underlying the intracellular trafficking and subsequent secretion of SHH. Notably, our discovery of the pivotal role of TMED10 as an important modulator of SHH secretion and its therapeutic potential as a target against abnormal SHH signalling should be further considered.
Methods
Cell culture and transfections
Human brain astroglial cell line SVG-A [71] was cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM-GlutaMAX, Gibco), supplemented with 10% foetal bovine serum (FBS, Dominique Dutscher), 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco). Transient transfections were performed on 70% confluent cells using JetPRIME (Polyplus) according to the manufacturer’s instructions. Briefly, a known concentration of DNA was diluted in JetPRIME buffer in a microcentrifuge tube and vortexed. A volume of JetPRIME reagent was added to the solution at a 1:2 DNA/JetPRIME ratio. The tube was briefly vortexed, centrifuged and incubated for 10 min at room temperature (RT). The transfection mix was added dropwise to cells in complete growth media and incubated for 24 – 48 h at 37 °C and 5% CO2. For siRNA transfections, single siRNA sequences targeting a gene of interest were purchased from Ambion (ThermoFisher Scientific). 50,000 cells were seeded in 24-well plates, and the next day, 11 pmol of siRNA was transfected using Lipofectamine 3000 (ThermoFisher Scientific) following the manufacturer’s instructions. For RUSH experiments, cells were maintained in avidin-rich media (2 µg/ml, Sigma) to prevent leakage of the reporter.
Plasmids
The SHH-RUSH construct was previously described [32]. The GFP-Rab6A cDNA construct cloned into the pBS vector under control of the weak promoter L30 was generated by the Montpellier Genomic Collection (MGC). The TfR-RUSH construct (pStrepKDEL-TfR-SBP-EGFP) was a kind gift from Dr. Franck Perez. The Nanoluciferase (NLuc) plasmid (pNL2.1) was purchased from Promega. SHH-NLuc was a kind gift from Dr. Adrian Salic (Harvard Medical School) [39]. Hsp70-NLuc was a kind gift from Dr. Gregory Lavieu (Institut Curie) [72]. GFP-TMED10 was a kind gift from Dr. Robert Blum (Uni. Wurzburg) [41].
Antibodies and reagents
For immunofluorescence, primary antibodies used were rabbit anti-Calnexin (1:500; Elabscience), rabbit anti-Rab6 (1:1000; Cell Signalling Technology), rabbit anti-Rab7 (1:250; Cell Signalling Technology), mouse anti-GM130 (1:1000; BD Biosciences), sheep anti-TGN46 (1:1000; Bio-Rad), mouse anti-SEC31 (1:250; BD Biosciences), rabbit anti-SHH (1:250; Abcam), mouse anti-GFP (1:250; Invitrogen), guinea-pig anti-Engrailed (1:100; Pizette et al. [29]), mouse anti-Ptc (1:400,kind gift from P. Ingham), chicken anti-β-gal (1:1000; GeneTex) and rabbit anti-Hh (1:500; Matusek et al. [48]). Secondary antibodies used were Alexa Fluor 488 donkey anti-mouse PLUS (1:5000, ThermoFisher Scientific), Alexa Fluor 488 donkey anti-rabbit PLUS (1:5000; ThermoFisher Scientific), Alexa Fluor 647 donkey anti-rabbit PLUS (1:5000; ThermoFisher Scientific), Alexa Fluor 647 donkey anti-sheep (1:1000; ThermoFisher Scientific), Cy3-conjugated donkey anti- chicken (1:200; Jackson ImmunoResearch), goat anti-mouse Alexa 488 (1:500; Life Technologies), Cy3-conjugated anti-guinea pig (1:200; Jackson ImmunoResearch), goat anti-rabbit Cy5 (1:200; Jackson ImmunoResearch), and Rhodamine phalloidin (1:100; Sigma-Aldrich). Probes used for immunofluorescence were Duolink In Situ PLA Probe donkey anti-rabbit PLUS, donkey anti-mouse MINUS and Detection Reagents Far-Red (Sigma-Aldrich). DAPI (4,6-diamidino-2-phenylindole) (1:1000; Sigma-Aldrich) was used to stain the nucleus. For flow cytometry, rabbit anti-SHH antibody (1:1000; Abcam) and Alexa Fluor 647 donkey anti-rabbit PLUS (1:5000; ThermoFisher Scientific) were used. Antibodies used for immunoblotting were rabbit anti-Rab6 (1:1000; Cell Signalling Technology), rabbit anti-Rab7 (1:1000; GeneTex), rabbit anti-TMED10 (1:1000; Sigma-Aldrich), mouse anti-GAPDH (1:5000, GeneTex), mouse anti-GFP (1:1000, Invitrogen), goat anti-SHH (1:1000, Santa Cruz) and rabbit anti-SHH (1:1000; Abcam).Secondary antibodies used for immunoblotting were goat anti-mouse immunoglobulin G (IgG) horseradish peroxidase (HRP) antibody (1:10,000; Jackson ImmunoResearch), goat anti-rabbit IgG HRP antibody (1:10,000; Jackson ImmunoResearch) and donkey anti-goat IgG HRP antibody (1:10,000; Invitrogen).
Retention using selective hook (RUSH) procedure
Cells were cultured with 2 µg/ml avidin (Sigma-Aldrich) 24 h prior experiments started to quench endogenous biotin from the media and/or produced by the cells. To release RUSH constructs, media was replaced with complete medium containing 40 µM biotin (Sigma-Aldrich) and incubated for the indicated amount of time at 37 °C and 5% CO2. Cells were then placed on ice, and media was replaced with ice-cold phosphate-buffered saline (PBS) and cells were maintained on ice for 10 min.
Cell surface staining and flow cytometry
To measure the amount of protein at the cell surface, cells were incubated for 1 h with a primary antibody, washed three times with ice-cold PBS then incubated for 30 min with a secondary antibody coupled to Alexa Fluor 647 (abbreviated as AF-647 on the corresponding figures). Cells were then washed twice with cold PBS and detached with pre-warmed 0.5 mM EDTA solution. Cells were collected and centrifuged at 500×g for 5 min at 4°C. Cell fixation was performed using 4% PFA for 15 min at RT, and after two washes, they were resuspended in FACS buffer (PBS, 0.5% BSA (w/v), 0.5 mM EDTA). Samples were run on a NovoCyte (ACEA Biosciences) flow cytometer equipped with 561- and 640-nm lasers and four-filter set. Data analysis was performed using FlowJo software (FlowJo LLC, v10.8.1).
Nanoluciferase secretion assay
SVG-A cells were seeded in 6-well plates and transfected with plasmids encoding NLuc-tagged proteins. At 16–24 h post-transfection, cells were washed with PBS and media was replaced with serum-free DMEM. After 24 h, cell culture supernatant was collected and centrifuged at 10,000×g for 5 min. Cells were also trypsinised and pelleted at 500×g for 3 min before lysis using NanoGlo Luciferase Assay Reagent (Promega). NLuc activity was measured on an Infinite200 PRO Plate Reader (TECAN) using the NanoGlo Luciferase Assay System kit (Promega) according to the manufacturer’s instructions.
Immunofluorescence and proximity ligation assay
Cells were seeded onto 12 mm glass coverslips (EMS) in 24-well plates and cultured overnight in complete medium. For RUSH assays, cells were cultured in complete medium supplemented with avidin (2 µg/ml). At 16–24 h post-transfection, cells were incubated at indicated timepoints in complete medium containing 40 µM biotin. Cells were washed twice with 1X PBS and then fixed in 4% PFA for 15 min at RT. Cells were then simultaneously permeabilised and blocked for 15 min at RT with PBS containing 0.1% Triton X-100 (v/v) (Sigma-Aldrich) and 0.5% BSA (w/v) (Euromedex). Cells were subsequently incubated with primary antibodies diluted in perm/block buffer for 2 h at RT in a humidified chamber. For immunofluorescence, cells were incubated for 1 h at RT in a humidified chamber with the corresponding secondary antibodies and DAPI diluted in the same buffer as the primary antibodies. Secondary antibodies were washed three times for 5 min in PBS at RT and coverslips were then mounted onto glass slides using Mowiol 4–88 (Sigma-Aldrich). For PLA, after incubation with primary antibodies, cells were washed twice for 5 min with 1X Wash Buffer A at RT. Cells were then incubated for 1 h at 37 °C in a pre-heated humidity chamber with Sigma PLUS and MINUS PLA probes in the 1X Sigma dilution solution. Sigma probes were washed twice for 5 min with 1X Wash Buffer A. The ligation step was performed at 37 °C for 30 min followed by two washes with 1X Wash Buffer A. Amplification was performed for 100 min at 37°C in dark conditions. Cells were then washed twice at RT for 10 min each with 1X Wash Buffer B and finally for 1 min in 0.01X Wash Buffer B. Coverslips were then mounted onto glass slides using Mowiol 4–88 (Sigma-Aldrich).
Image acquisition
For fixed imaging, confocal microscopy was performed using an Eclipse Ti inverted microscope (Nikon) mounted with a CSU-X1 spinning disk head (Yokogawa), a back-illuminated EMCCD Andor camera (Oxford Instruments), and a 100X (1.45 NA) oil objective lens controlled by the Andor iQ3 software (Oxford Instruments).
For live cell imaging, cells were seeded onto 25 mm glass coverslips (EMS) and transfected using JetPRIME (Polyplus) according to the manufacturer’s instructions. Coverslips with transfected cells were then transferred into an Attofluor Cell Chamber (Invitrogen) and placed onto the microscope stage and maintained in a dark atmosphere-controlled chamber at 37 °C and 5% CO2. Live-cell confocal imaging was performed using either an inverted IX83 microscope (Olympus) coupled to a CSU-W1 spinning disk unit (Yokogawa), a sCMOS Fusion camera (Hamamatsu) and a 60X (1.3 NA) oil objective lens controlled by the CellSens software (Olympus), or an Eclipse Ti2 inverted microscope (Nikon) coupled to the Andor Dragonfly spinning disk system (Oxford Instruments), an EMCCD Andor camera (Oxford Instruments), and a 100X APO (1.45 NA) oil objective lens controlled by Andor Fusion software (Oxford Instruments). For TIRF microscopy, live imaging was performed with an Eclipse Ti inverted microscope (Nikon), a back-illuminated EMCCD camera (Evolve 512, Photometrics) and a 100X APO (1.49 NA) oil objective lens controlled by MetaMorph. The TIRF angle was chosen to obtain a calculated evanescent field depth of < 100 nm. Acquisition was performed from 30 min after biotin addition.
Image analyses
Image processing was performed using either the Fiji upgrade of ImageJ or Imaris software v9.2 (Bitplane, Oxford Instruments). Quantifications for mean fluorescence intensities and colocalisation measurements were performed using Imaris software v9.2 (Bitplane, Oxford Instruments). For colocalisation analysis of the different Rab GTPases, cells were stained with a Golgi marker (Mannosidase-II-EGFP) and the Golgi staining was masked prior colocalization measurements to assess colocalization in non-Golgi area. The fluorescence intensities of SHH and given Rab proteins were then measured within each manually delimited cell.
Drosophila stocks and experimental conditions
Flies were cultured on standard Drosophila medium. In rescue experiments of Hh-dependent wing imaginal disc outgrowth and lethality, “tester line” virgins with the genotype of w1118; tubGal80ts<dpp-LacZ; hhGal4 < UAS-Hh(M4)/TM6b-Tb [48] were crossed to control w1118 and bai RNAi lines (UAS-bai330020SH: Vienna Drosophila Resource Center, VDRC#330,020, UAS-baiser100612KK: Vienna Drosophila Resource Center, VDRC#100,612). Parents were transferred to new vials every 12 h at 25 °C. This allowed sufficient number of eggs to be laid without considerable overcrowding. Early 2nd instar larvae were shifted to 29 °C to inactivate tubGal80ts thus inducing UAS-Hh(M4) expression and the UAS transgenes tested in the system. Following 3 days of incubation at 29 °C, wandering 3rd instar larvae were dissected and their WIDs were scored into categories (with wild-type like, moderately outgrown, fully outgrown morphologies). To assess lethality rescue, in parallel, vials with the same progeny were kept at 25 °C till hatching, and then scored for pharate adult viability (empty pupae). In 50% of the progeny Tb animals (represented by empty small circles on Fig. 5A) do not contain the1 hhGal4 < UASHh (M4) recombinant chromosome, expected to be viable, therefore serve as an internal control. Non-Tb animals carry the hhGal4 < UASHh (M4) recombinant chromosome and either a neutral GFP RNAi (VDRC#60,102) or bai RNAi-s. To overexpress bai RNAi in wild-type background, UAS-bai330020SH UAS-bai100612KK and were crossed to tubGal80ts < dpp-lacZ; hhGal4/TM6b-Tb virgins. As a control we crossed tubGal80ts < dpp-lacZ; hhGal4/TM6b-Tb virgins to w1118 as well. Egg laying was allowed at 25 °C and vials were then shifted up to 29 °C until dissection.
Immunostaining and imaging of WIDs
Conventional and extracellular immunolabeling was performed as previously described [27, 48]. For extracellular Hh labelling without detergent but with initial fixation, discs were dissected in ice-cold PBS, and then fixed in 2% PFA (v/v) in PBS for 15 min at RT. After fixation, discs were rinsed in PBS three times for 5 min at RT, and then incubated with anti-Hh antibody for 1 h in 2% BSA (w/v) in PBS. Discs were then rinsed three times in 0.3% Triton-X100 (v/v) in PBS and blocked again for an hour in 0.1% Triton X-100 (v/v), 1% BSA (w/v) in PBS. Following this step, anti-Ptc was added in the detergent-containing block solution and the samples were incubated at 4 °C overnight. The next day, samples were washed three times for 10 min in 0.3% Triton-X100 (v/v) in PBS before incubation with secondary antibodies and phalloidin. For imaging, wing imaginal discs were mounted in VECTASHIELD mounting medium (Vector Laboratories). All confocal images were recorded with Leica TCS SP5 confocal laser scanning microscope. dpp-lacZ intensity was measured on single slices on images recorded in 1024 × 1024 resolution, with 40 × objective (HCX PLAN APO; NA = 1.3) at 1,3 × zoom level, with 400Hz speed, within a 200 × 100 pixel rectangle. For En and Ptc range measurements, the same imaging conditions, and a 100 × 100 pixel square was used. Optical Z sections within the posterior compartment along the dorsoventral axis were taken at 1µm thickness in 1024 × 1024 resolution, with 40 × objective (HCX PLAN APO; NA = 1.3) at 3.52 × zoom level, with 400Hz speed. Images presented in the manuscript are a stack of 5 single sections. To image whole discs single confocal sections were collected with 20 × objective (HCX PL APO CS 20.0 × 0.70 IMM UV NA = 0.7) without zoom at 1024 × 1024 resolution with 400Hz speed. To analyze ectopic dpp-lacZ patterns, expression of UAS-Hh (tester line) together with either control UAS-GFP RNAi (VDRC #60,102), UAS-disp RNAi (Bloomington Drosophila Stock Center, BDSC#27,247) or with UAS-bai RNAi (VDRC #100,612) was induced for either 1 or 4 days.
Western blotting
Cells were seeded in 6-well plates in complete growth media. After 16–24 h post-transfection, cells were washed twice with ice-cold PBS and lysed with NP40 buffer [150 mM NaCl, 1% NP-40, 50 mM Tris (pH 8.0), and protease inhibitor (Promega)]. Cells were rotated on a carousel for 10 min at 4 °C then centrifuged at 15,000 × g for 20 min at 4 °C. The supernatant was collected and protein concentration was determined using the Pierce BCA Assay kit (ThermoFisher Scientific). A total of 20 µg of protein lysates was run on Bolt 4–12% Bis–Tris Mini Protein gels (Invitrogen) and proteins were transferred onto nitrocellulose membranes using the iBlot2 Transfer system (Invitrogen) according to the manufacturer’s instructions. Membranes were blocked with 5% low-fat milk (w/v) in TBS-T [Tris-buffered saline (TBS) at pH 7.4 and 0.05% Tween20] for 30 min at RT prior to incubation with primary antibodies overnight at 4 °C in TBS-T containing 0.5% milk. Secondary antibodies were incubated for 1 h at RT in TBS-T containing 0.5% milk. After several washes with TBS-T, membranes were revealed by chemiluminescence using Clarity Western ECL Blotting Substrates (Bio-Rad) and images were acquired using the ChemiDoc Touch System (Bio-Rad). Relative intensities of western blot bands were quantified using ImageLab software (Bio-Rad, v6.0.1) using Volume Tools, global background subtraction and linear regression method.
SHH-ELISA
SVG-A cells transfected with SHH-RUSH were incubated in complete growth medium supplemented with 40 µM biotin at indicated timepoints. The media was collected and centrifuged at 10,000 × g for 5 min and used in the SHH-ELISA kit (R&D Systems) according to the manufacturer’s instructions.
SHH signalling and RT-qPCR
NIH 3T3 cells were seeded into 48-well plates and cultured in complete growth media until confluent. Cells were then serum-starved for 24 h then incubated with cell culture supernatants taken from SVG-A cells expressing SHH or SHH-RUSH in absence or presence of biotin. After 48 h, cells were lysed and prepared for RT-qPCR analysis. For all RT-qPCR experiments, cells were lysed and RNA immediately extracted using the Luna Cell Ready Lysis Module (New England Biolabs) according to the manufacturer’s instructions. RT-qPCR on extracted RNA was performed using the Luna One-Step RT-qPCR kit (New England Biolabs) and samples were analysed using a LightCycler 480 instrument (Roche) using the following thermal cycle: reverse transcription step at 55 °C for 15 min, initial denaturation at 95 °C for 2 min; then, 40 cycles were programmed consisting of 15 s of denaturation at 95 °C followed by 30 s of annealing/extension at 60 °C. Fluorescent data collection was performed at the end of each cycle.
| Oligonucleotides | Sequence (5′–3′) |
|---|---|
| Gli1 qPCR F (Ms) a | CCTTTCTTTGAGGTTGGGATGA |
| Gli1 qPCR R (Ms) a | TTGGATTGAACATGGCGTCT |
| Gli1 qPCR F (Ms) b | CTTCAAGGCCCAATACATGC |
| Gli1 qPCR R (Ms) b | GTGAATAGGACTTCCGACAGC |
| Gapdh qPCR F (Ms) | AATGGTGAAGGTCGGTGTG |
| Gapdh qPCR R (Ms) | GTGGAGTCATACTGGAACATGTAG |
| RAB6A qPCR F | CGTTTCCGTAGCCTCATTC |
| RAB6A qPCR R | ACATCACTTCCTCTTTCTGTTC |
| RAB7A qPCR F | CTCACACACACACACACA |
| RAB7A qPCR R | GGGAAAGGGGGGGTAAAAAG |
| TMED10 qPCR F | GAGATGCGTGATACCAACGA |
| TMED10 qPCR R | TTCTTGGCCTTGAAGAAGCG |
| TMED9 qPCR F | GCGCTCTACTTTCACATCGG |
| TMED9 qPCR R | CACCTCCACAAACATGCCAA |
| TMED7 qPCR F | TTGGAGAAGACCCACCTTTG |
| TMED7 qPCR R | GCCCTATGCTAACCACCAGA |
| TMED2 qPCR F | GTAAAGCACGAACAGGAATACATG |
| TMED2 qPCR R | GTCATGGCAACTAGAACAAGAGC |
| GAPDH qPCR F | GAGTCAACGGATTTGGTCGT |
| GAPDH qPCR R | TTGATTTTGGAGGGATCTCG |
Statistical analysis
All statistical analyses were performed using GraphPad Prism (v9.5.1). Student’s t-test was performed for comparisons between two groups of data that seemed to be normally distributed with similar variances. Multiple group comparisons were performed using one-way ANOVA.
Supplementary Information
Below is the link to the electronic supplementary material.
Video S1: Dual-colour live-cell imaging of SHH-RUSH trafficking through the Golgi complex, related to Figure 1. SVG-A cell co-transfected with SHH-RUSH (magenta) and the medial Golgi marker, Mannosidase-II-EGFP (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started 5 mins after the addition of 40 µM biotin. Frames were taken every 5 mins for 90 mins. Scale bar – 10µm
Video S2: Zoomed inset of Video S1, related to Figure 1. SHH-RUSH (magenta) transported into and exits from the Golgi (green). Frames taken every 5 mins for 90 min. Scale bar – 5 µm
Video S3: Two colour TIRF imaging of SHH-RUSH and GFP-Rab6a, related to Figure 2. SVG-A cell co-transfected with SHH-RUSH (magenta) and GFP-Rab6a (green). Acquisition started 30 mins after the addition of 40 µM biotin. Frames were taken every 2 seconds for an additional 30 mins. Scale bar – 10 µM
Video S4: Zoomed inset of Video S3, related to Figure 2. Video corresponds to 1890s to 1920s post biotin addition and demonstrates a double-positive vesicle of SHH-RUSH (magenta) and GFP-Rab6a (green) arriving at the plasma membrane. Scale bar – 2 µm
Video S5: Dual-colour live-cell imaging of SHH-RUSH and GFP-TMED10, related to Figure 3. SVG-A cell co-transfected with SHH-RUSH (magenta) and GFP-TMED10 (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started immediately after the addition of 40 µM biotin. Frames were taken every 500 ms for 10 mins. Scale bar – 10 µM
Video S6: Zoomed inset of Video S5, related to Figure 3. A double-positive vesicle of SHH-RUSH (magenta) and GFP-TMED10 (green) trafficking towards a perinuclear region. Video corresponds to 49s to 99s post biotin addition. Scale bar – 5 µm
Video S7: Dual colour live-cell imaging of SHH-RUSH and SEC24D-EGFP, related to Figure 4. SVG-A cells co-transfected with SHH-RUSH (magenta) and SEC24D-EGFP (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started immediately after the addition of 40 µM biotin. Frames were taken every 2 s for 15 min. Scale bar – 10 µm
Acknowledgements
We acknowledge the MRI imaging facility, member of the national infrastructure France-BioImaging, for advice and training. We thank Drs. Adrian Salic, Franck Perez, Gregory Lavieu and Robert Blum for the gift of constructs used in this study
Author contributions
Conceptualisation: YB, RG. Methodology: YB, TM, SV, PT, MSD, RG. Investigation: YB, TM, PT, RG. Analysis: YB, TM, PT, RG. Supervision: PT, RG. Writing—original draft: YB, RG. Writing—review & editing: YB, TM, SV, PT, MSD, RG.
Funding
This work was supported by the Agence Nationale de la Recherche (ANR-18-CE13-0003–01) to RG and PT. YB obtained a post-doctoral fellowship from Fondation pour la Recherche Médicale (FRM, SPF202110014043).
Data and materials availability
All data are available in the main text, the supplementary materials or will be made available on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wolpert L (2016). Positional information and pattern formation. In: Current topics in developmental biology. Vol. 117. Elsevier Inc. 10.1016/bs.ctdb.2015.11.008. [DOI] [PubMed]
- 2.Álvarez-Buylla A, Ihrie RA. Sonic hedgehog signaling in the postnatal brain. Semin Cell Dev Biol. 2014;33:105–111. doi: 10.1016/j.semcdb.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briscoe J, Thérond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):418–431. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 4.Dahmane N, Ruiz I Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development (Cambridge, England) 1999;126(14):3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 5.Groves I, Placzek M, Fletcher AG. Of mitogens and morphogens: modelling Sonic Hedgehog mechanisms in vertebrate development: modelling Shh mechanisms in development. Philos Trans R Soc B Biol Sci. 2020 doi: 10.1098/rstb.2019.0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palma V, Lim DA, Dahmane N, Sánchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Álvarez-Buylla A, Ruiz I Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132(2):335–344. doi: 10.1242/dev.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straface G, Aprahamian T, Flex A, Gaetani E, Biscetti F, Smith RC, Pecorini G, et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med. 2009;13(8b):2424–2435. doi: 10.1111/j.1582-4934.2008.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrales JMD, Blaess S, Mahoney EM, Joyner AL. The Level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133(9):1811–1821. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- 9.Melamed JR, Morgan JT, Ioele SA, Gleghorn JP, Sims-Mourtada J, Day ES. Investigating the role of hedgehog/GLI1 signaling in glioblastoma cell response to temozolomide. Oncotarget. 2018;9(43):27000–27015. doi: 10.18632/oncotarget.25467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin LL, de Sauvage FJ. Targeting the hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5(12):1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 11.Brady MV, Vaccarino FM. Role of SHH in patterning human pluripotent cells towards ventral forebrain fates. Cells. 2021 doi: 10.3390/cells10040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill SA, Blaeser AS, Coley AA, Xie Y, Shepard KA, Harwell CC, Gao W-J, Denise A, Garcia R. Sonic hedgehog signaling in astrocytes mediates cell type-specific synaptic organization. Elife. 2019;8:1–23. doi: 10.7554/elife.45545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yam PT, Charron F. Signaling mechanisms of non-conventional axon guidance cues: the Shh, BMP and Wnt morphogens. Curr Opinion Neurobiol Dev Neurons Glia. 2013;23(6):965–973. doi: 10.1016/j.conb.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Maity T, Fuse N, Beachy PA. Molecular Mechanisms of sonic hedgehog mutant effects in holoprosencephaly. Proc Natl Acad Sci. 2005;102(47):17026–17031. doi: 10.1073/pnas.0507848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClelland K, Li W, Rosenblum ND. Pallister-Hall syndrome, GLI3, and kidney malformation. Am J Med Genet C Semin Med Genet. 2022;190(3):264–278. doi: 10.1002/ajmg.c.31999. [DOI] [PubMed] [Google Scholar]
- 16.Hutchings J, Zanetti G. Coat flexibility in the secretory pathway: a role in transport of bulky cargoes. Curr Opin Cell Biol. 2019;59(August):104–111. doi: 10.1016/j.ceb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14(1):20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 18.Peotter J, Kasberg W, Pustova I, Audhya A. COPII-mediated trafficking at the ER/ERGIC interface. Traffic. 2019;20(7):491–503. doi: 10.1111/tra.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saraste J, Marie M. Intermediate compartment (IC): from Pre-Golgi vacuoles to a semi-autonomous membrane system. Histochem Cell Biol. 2018;150(5):407–430. doi: 10.1007/s00418-018-1717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borchers A-C, Langemeyer L, Ungermann C. Who’s in control? Principles of Rab GTPase activation in endolysosomal membrane trafficking and beyond. J Cell Biol. 2021;220(9):e202105120. doi: 10.1083/jcb.202105120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deffieu MS, Cesonyte I, Delalande F, Boncompain G, Dorobantu C, Song E, Lucansky V, et al. Rab7-harboring vesicles are carriers of the transferrin receptor through the biosynthetic secretory pathway. Sci Adv. 2021;7(2):eaba7803. doi: 10.1126/sciadv.aba7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goud B, Liu S, Storrie B. Rab proteins as major determinants of the Golgi complex structure. Small GTPases. 2018;9(1–2):66–75. doi: 10.1080/21541248.2017.1384087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-Palmitoylation of Sonic Hedgehog. J Biol Chem. 2008;283(32):22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293(5537):2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 25.Porter JA, Young KE, Beachy PA. Cholesterol modification of hedgehog signaling proteins in animal development. Science. 1996;274(5285):255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 26.Resh MD. Palmitoylation of hedgehog proteins by hedgehog acyltransferase: roles in signalling and disease. Open Biol. 2021;11(3):rsob.200414. doi: 10.1098/rsob.200414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Angelo G, Matusek T, Pizette S, Thérond PP, D’Angelo G, Matusek T, Pizette S, et al. Endocytosis of hedgehog through dispatched regulates long-range signaling. Dev Cell. 2015;32(3):290–303. doi: 10.1016/j.devcel.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Gore T, Matusek T, D’Angelo G, Giordano C, Tognacci T, Lavenant-Staccini L, Rabouille C, Thérond PP. The GTPase Rab8 differentially controls the long- and short-range activity of the hedgehog morphogen gradient by regulating hedgehog apico-basal distribution. Development. 2021;148(5):dev191791. doi: 10.1242/dev.191791. [DOI] [PubMed] [Google Scholar]
- 29.Pizette S, Matusek T, Herpers B, Thérond PP, Rabouille C. Hherisomes, hedgehog specialized recycling endosomes, are required for high level hedgehog signaling and tissue growth. J Cell Sci. 2021 doi: 10.1242/jcs.258603. [DOI] [PubMed] [Google Scholar]
- 30.Sandoval Li, Labarca M, Retamal C, Sánchez P, Larraín J, González A. Sonic hedgehog is basolaterally sorted from the TGN and transcytosed to the apical domain involving dispatched-1 at Rab11-ARE. Fronti Cell Dev Biol. 2022 doi: 10.3389/fcell.2022.833175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9(5):493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 32.Thauvin M, Amblard I, Rampon C, Mourton A, Queguiner I, Li C, Gautier A, Joliot A, Volovitch M, Vriz S. Reciprocal regulation of Shh trafficking and H2O2 levels via a noncanonical BOC-Rac1 pathway. Antioxidants (Basel, Switzerland) 2022;11(4):841–856. doi: 10.3390/antiox11040718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamberlain CE, Jeong J, Guo C, Allen BL, McMahon AP. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135(6):1097–1106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- 34.Aber R, Chan W, Mugisha S, Jerome-Majewska LA. Transmembrane Emp24 domain proteins in development and disease. Genet Res. 2019;101:e14. doi: 10.1017/S0016672319000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strating JRPM, Martens GJM. The P24 family and selective transport processes at the ER-Golgi interface. Biol Cell. 2009;101(9):495–509. doi: 10.1042/bc20080233. [DOI] [PubMed] [Google Scholar]
- 36.D’Arcangelo JG, Crissman J, Pagant S, Čopič A, Latham CF, Snapp EL, Miller EA. Traffic of P24 proteins and COPII coat composition mutually influence membrane scaffolding. Curr Biol. 2015;25(10):1296–1305. doi: 10.1016/j.cub.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuiki I, Volchuk A. Diverse Roles for the P24 family of proteins in eukaryotic cells. Biomol Concepts. 2012;3(6):561–570. doi: 10.1515/bmc-2012-0028. [DOI] [PubMed] [Google Scholar]
- 38.Pastor-Cantizano N, Montesinos JC, Bernat-Silvestre C, Marcote MJ, Aniento F. P24 family proteins: key players in the regulation of trafficking along the secretory pathway. Protoplasma. 2016;253(4):967–985. doi: 10.1007/s00709-015-0858-6. [DOI] [PubMed] [Google Scholar]
- 39.Petrov K, Wierbowski BM, Liu J, Salic A. Distinct cation gradients power cholesterol transport at different key points in the hedgehog signaling pathway. Dev Cell. 2020;55(3):314–327.e7. doi: 10.1016/j.devcel.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tashima Y, Hirata T, Maeda Y, Murakami Y, Kinoshita T. Differential Use of P24 family members as cargo receptors for the transport of glycosylphosphatidylinositol-anchored proteins and Wnt1. J Biochem. 2021;171(1):75–83. doi: 10.1093/jb/mvab108. [DOI] [PubMed] [Google Scholar]
- 41.Blum R, Lepier A. The luminal domain of P23 (Tmp21) plays a critical role in P23 cell surface trafficking. Traffic. 2008;9(9):1530–1550. doi: 10.1111/j.1600-0854.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- 42.Lopez S, Perez-Linero AM, Manzano-Lopez J, Sabido-Bozo S, Cortes-Gomez A, Rodriguez-Gallardo S, Aguilera-Romero A, Goder V, Muñiz M. Dual independent roles of the P24 complex in selectivity of secretory cargo export from the endoplasmic reticulum. Cells. 2020;9(5):1295. doi: 10.3390/cells9051295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weigel AV, Chang C-L, Gleb Shtengel C, Shan Xu, Hoffman DP, Freeman M, Iyer N, et al. ER-to-golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 2021;184(9):2412–2429.e16. doi: 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 44.Hatori R, Kornberg TB. Hedgehog produced by the Drosophila wing imaginal disc induces distinct responses in three target tissues. Development (Cambridge, England) 2020;147(22):dev195974. doi: 10.1242/dev.195974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell. 1994;76(1):89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 46.Tanimoto H, Itoh S, ten Dijke P, Tabata T. Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol Cell. 2000;5(1):59–71. doi: 10.1016/s1097-2765(00)80403-7. [DOI] [PubMed] [Google Scholar]
- 47.Torroja C, Gorfinkiel N, Guerrero I. Mechanisms of Hedgehog Gradient Formation and Interpretation. J Neurobiol. 2005;64(4):334–356. doi: 10.1002/neu.20168. [DOI] [PubMed] [Google Scholar]
- 48.Matusek T, Wendler F, Polès S, Pizette S, D’Angelo G, Fürthauer M, Therond PP. The ESCRT machinery regulates the secretion and long-range activity of hedgehog. Nature. 2014;516(729):99–103. doi: 10.1038/nature13847. [DOI] [PubMed] [Google Scholar]
- 49.Burke R, Nellen D, Bellotto M, Hafen E, Senti K-A, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99(7):803–815. doi: 10.1016/S0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 50.Balch WE, Michael McCaffery J, Plutner H, Farquhar MG. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76(5):841–852. doi: 10.1016/0092-8674(94)90359-X. [DOI] [PubMed] [Google Scholar]
- 51.Barlowe C, Helenius A. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol. 2016;32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- 52.Fourriere L, Kasri A, Gareil N, Bardin S, Bousquet H, Pereira D, Perez F, Goud B, Boncompain G, Miserey-Lenkei S. RAB6 and microtubules restrict protein secretion to focal adhesions. J Cell Biol. 2019;218(7):2215–2231. doi: 10.1083/jcb.201805002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson LJ, Liu S, Storrie B. Rab6 Is required for rapid, cisternal-specific, intra-Golgi cargo transport. Sci Rep. 2020;10(1):16604. doi: 10.1038/s41598-020-73276-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huber ME, Kurapova R, Heisler CM, Karamooz E, Tafesse FG, Harriff MJ. Rab6 regulates recycling and retrograde trafficking of MR1 molecules. Sci Rep. 2020;10(1):20778. doi: 10.1038/s41598-020-77563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patwardhan A, Bardin S, Miserey-Lenkei S, Larue L, Goud B, Raposo G, Delevoye C. Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes. Nat Commun. 2017 doi: 10.1038/ncomms15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coulter ME, Dorobantu CM, Lodewijk GA, Delalande F, Cianferani S, Ganesh VS, Smith RS, et al. The ESCRT-III protein CHMP1A mediates secretion of sonic hedgehog on a distinctive subtype of extracellular vesicles. Cell Rep. 2018;24(4):973–986.e8. doi: 10.1016/j.celrep.2018.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernat-Silvestre C, De Sousa V, Vieira J-S, Pastor-Cantizano N, Hawes C, Marcote MJ, Aniento F. P24 family proteins are involved in transport to the plasma membrane of GPI-anchored proteins in plants. Plant Physiol. 2020;184(3):1333–1347. doi: 10.1104/pp.20.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mañuel M, Howard R. Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J Lipid Res. 2016;57(3):352–360. doi: 10.1194/jlr.R062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang X, Chen R, St Dollente Mesias V, Wang T, Wang Y, Poljak K, Fan X, et al. A SURF4-to-proteoglycan relay mechanism that mediates the sorting and secretion of a tagged variant of Sonic Hedgehog. Proc Natl Acad Sci USA. 2022;119(11):e2113991119. doi: 10.1073/pnas.2113991119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri H-P. ‘The Cargo Receptors Surf4, Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC)-53, and P25 Are Required to Maintain the Architecture of ERGIC and Golgi. Edited by Benjamin Glick. Mol Biol Cell. 2008;19(5):1976–1990. doi: 10.1091/mbc.e07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Minin G, Holzner M, Grison A, Dumeau CE, Chan W, Monfort A, Jerome-Majewska LA, Roelink H, Wutz A. TMED2 binding restricts SMO to the ER and Golgi compartments. PLoS Biol. 2022 doi: 10.1371/journal.pbio.3001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim P, Scott MR, Meador-Woodruff JH. Abnormal ER quality control of neural GPI-anchored proteins via dysfunction in ER export processing in the frontal cortex of elderly subjects with schizophrenia. Transl Psychiatry. 2019;9(1):1–9. doi: 10.1038/s41398-018-0359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiu K, Zhang X, Wang S, Li C, Wang X, Li X, Wu Y. TMP21 in Alzheimer’s disease: molecular mechanisms and a potential target. Front Cell Neurosci. 2019 doi: 10.3389/fncel.2019.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin JH, Park SJ, Jo DS, Park NY, Kim JB, Bae JE, Jo YK, et al. Down-regulated TMED10 in Alzheimer disease induces autophagy via ATG4B activation. Autophagy. 2019;15(9):1495–1505. doi: 10.1080/15548627.2019.1586249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vetrivel KS, Kodam A, Gong P, Chen Y, Parent AT, Kar S, Thinakaran G. Localization and regional distribution of P23/TMP21 in the brain. Neurobiol Dis. 2008;32(1):37–49. doi: 10.1016/j.nbd.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie J, Yang Y, Li J, Hou J, Xia K, Song W, Liu S. Expression of Tmp21 in normal adult human tissues. Int J Clin Exp Med. 2014;7(9):2976–2983. [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Yihui Wu, Shen C, Belenkaya TY, Ray L, Lin X. Drosophila P24 and Sec22 regulate wingless trafficking in the early secretory pathway. Biochem Biophys Res Commun. 2015;463(4):483–489. doi: 10.1016/j.bbrc.2015.04.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Port F, Hausmann G, Basler K. A Genome-wide RNA interference screen uncovers two P24 proteins as regulators of wingless secretion. EMBO Rep. 2011;12(11):1144–1152. doi: 10.1038/embor.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ayers KL, Gallet A, Staccini-Lavenant L, Thérond PP. The Long-Range activity of hedgehog is regulated in the apical extracellular space by the glypican dally and the hydrolase notum. Dev Cell. 2010;18(4):605–620. doi: 10.1016/j.devcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 70.Glashauser J, Camelo C, Hollmann M, Backer W, Jacobs T, Sanchez JI, Schleutker R, et al. Acute manipulation and real-time visualization of membrane trafficking and exocytosis in Drosophila. bioRxiv. 2023 doi: 10.1101/2022.03.25.483021. [DOI] [PubMed] [Google Scholar]
- 71.Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci. 1985;82(4):1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonsergent E, Grisard E, Buchrieser J, Schwartz O, Théry C, Lavieu G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat Commun. 2021;12(1):1864. doi: 10.1038/s41467-021-22126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: Dual-colour live-cell imaging of SHH-RUSH trafficking through the Golgi complex, related to Figure 1. SVG-A cell co-transfected with SHH-RUSH (magenta) and the medial Golgi marker, Mannosidase-II-EGFP (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started 5 mins after the addition of 40 µM biotin. Frames were taken every 5 mins for 90 mins. Scale bar – 10µm
Video S2: Zoomed inset of Video S1, related to Figure 1. SHH-RUSH (magenta) transported into and exits from the Golgi (green). Frames taken every 5 mins for 90 min. Scale bar – 5 µm
Video S3: Two colour TIRF imaging of SHH-RUSH and GFP-Rab6a, related to Figure 2. SVG-A cell co-transfected with SHH-RUSH (magenta) and GFP-Rab6a (green). Acquisition started 30 mins after the addition of 40 µM biotin. Frames were taken every 2 seconds for an additional 30 mins. Scale bar – 10 µM
Video S4: Zoomed inset of Video S3, related to Figure 2. Video corresponds to 1890s to 1920s post biotin addition and demonstrates a double-positive vesicle of SHH-RUSH (magenta) and GFP-Rab6a (green) arriving at the plasma membrane. Scale bar – 2 µm
Video S5: Dual-colour live-cell imaging of SHH-RUSH and GFP-TMED10, related to Figure 3. SVG-A cell co-transfected with SHH-RUSH (magenta) and GFP-TMED10 (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started immediately after the addition of 40 µM biotin. Frames were taken every 500 ms for 10 mins. Scale bar – 10 µM
Video S6: Zoomed inset of Video S5, related to Figure 3. A double-positive vesicle of SHH-RUSH (magenta) and GFP-TMED10 (green) trafficking towards a perinuclear region. Video corresponds to 49s to 99s post biotin addition. Scale bar – 5 µm
Video S7: Dual colour live-cell imaging of SHH-RUSH and SEC24D-EGFP, related to Figure 4. SVG-A cells co-transfected with SHH-RUSH (magenta) and SEC24D-EGFP (green) and imaged using 3D spinning disk confocal microscopy. Acquisition started immediately after the addition of 40 µM biotin. Frames were taken every 2 s for 15 min. Scale bar – 10 µm
Data Availability Statement
All data are available in the main text, the supplementary materials or will be made available on reasonable request.