Abstract
Numerous studies have established the critical roles of microRNAs in regulating post-transcriptional gene expression in diverse biological processes. Here, we report on the role and mechanism of miR-24-3p in skeletal muscle differentiation and regeneration. miR-24-3p promotes myoblast differentiation and skeletal muscle regeneration by directly targeting high mobility group AT-hook 1 (HMGA1) and regulating it and its direct downstream target, the inhibitor of differentiation 3 (ID3). miR-24-3p knockdown in neonatal mice increases PAX7-positive proliferating muscle stem cells (MuSCs) by derepressing Hmga1 and Id3. Similarly, inhibition of miR-24-3p in the tibialis anterior muscle prevents Hmga1 and Id3 downregulation and impairs regeneration. These findings provide evidence that the miR-24-3p/HMGA1/ID3 axis is required for MuSC differentiation and skeletal muscle regeneration in vivo.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04168-7.
Keywords: Skeletal muscle stem cell, Myoblast, Differentiation, Development, Regeneration, microRNA, miR-24-3p, HMGA1, Id3
Introduction
During postnatal skeletal muscle development and regeneration, quiescent MuSCs are activated to re-enter the cell cycle, followed by proliferation to form a pool of myoblasts, which then differentiate and fuse into newly formed or existing myofibers [1]. This extensive process of making new muscle is known as myogenesis and is essential for maintaining normal physiological function. Dysregulation of the critical molecular and cellular events linked to skeletal myogenesis is associated with muscle degenerative diseases. Most of the understanding of the critical myogenic processes, including myoblast differentiation and skeletal muscle regeneration, is based on the regulation of myogenic transcription factors and signaling molecules [1–8]. However, it is not well understood how these factors of the myogenic network are regulated to maintain normal myogenesis and skeletal muscle function. In this context, an emerging research area is exploring microRNAs’ role in post-transcriptional gene regulation during skeletal myogenesis.
Recently we have begun to elucidate the role of microRNAs in skeletal muscle differentiation, development, and muscle degenerative diseases. The microRNAs involved in myogenesis have recently been reviewed [9]. Most studies have demonstrated the roles of microRNAs in muscle differentiation by overexpression and knockdown experiments using the C2C12 myoblast cell line [10–25]. miR-1 and miR-206, which share identical target binding seed sequences, promote myoblast differentiation by regulating several targets, including Hdac4, Pax7, Smarcd2, and Smarcb1 [14, 17, 26]. miR-26a promotes myoblast differentiation by regulating the TGFβ/BMP pathway transcription factors SMAD1 and SMAD4 [27]. miR-181 induces muscle differentiation by repressing HOXA11 [28]. In addition, we have demonstrated that several microRNAs, including miR-26a, miR-206, miR-322/424, miR-378, miR-486, and miR-503, are induced during myoblast differentiation and promote myoblast differentiation by regulating critical myogenic regulatory factors and signaling molecules [14, 21–24, 27, 29]. Intriguingly, we have shown that H19 non-coding RNA generates miR-675-3p and miR-675-5p to promote muscle differentiation [29]. Through ChIP-Seq analyses, we have revealed how crucial myogenic transcription factors, including MYOD and MEF2C, repress their repressors by inducing myogenic microRNAs [14, 24], suggesting that microRNAs tightly regulate many critical factors of the myogenic network and maintain the balance between myoblast proliferation and differentiation. However, only a few studies have revealed microRNA function in skeletal muscle development and regeneration in animals [26, 27, 29–36]. Therefore, it is crucial to uncover microRNA function in myogenesis in vivo to identify new therapeutic targets for muscle degenerative diseases.
Here, we report that miR-24-3p is abundantly expressed in skeletal muscles and upregulated during myoblast differentiation and muscle regeneration. Consistent with our findings, an earlier study showed that miR-24-3p promotes muscle differentiation in vitro [37]. However, it has remained unknown how miR-24-3p promotes myoblast differentiation and whether it plays any role during myogenesis in vivo. We show that miR-24-3p promotes myoblast differentiation by directly targeting Hmga1and regulating it and its direct downstream target, Id3. HMGA1 is a prominent member of the high mobility group A (HMGA) proteins preferentially expressed in the proliferating embryonic tissues but usually absent in differentiated cells [38, 39]. HMGA1 downregulation is crucial for myoblast differentiation [40]. Sustained expression of HMGA1 in myoblast cells represses promyogenic genes, including Myod [40]. HMGA1’s direct target ID3 represses MYOD activity by binding and sequestering E12/47 away from the MYOD binding sites of the promyogenic genes, thereby repressing myogenesis. Our findings suggest that miR-24-3p promotes myogenesis by increasing MYOD activity through the downregulation of HMGA1 and ID3. Furthermore, knockdown of miR-24-3p in a neonatal mouse model increases the PAX7-positive proliferating MuSCs by derepressing Hmga1 and Id3. In addition, we show that miR-24-3p is required for skeletal muscle regeneration after injury. Thus, our study establishes a role for miR-24-3p in skeletal muscle function in vivo and elucidates the molecular mechanism by which this myogenic microRNA affects muscle differentiation and regeneration: inhibiting a well-established repressor of myogenesis.
Materials and methods
Cell culture
Mouse C2C12 myoblast cell line was purchased from the American Type Culture Collection (ATCC) [41]. These cells were cultured subconfluently in a growth medium (GM) consisting of DMEM (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (ATCC) and 1X Antibiotic–Antimycotic (Thermo Scientific). These cells were differentiated into myocytes or myotubes in a differentiation medium (DM) [42]. The DM consisted of DMEM containing 2% heat-inactivated horse serum (Hyclone) and 1X Antibiotic–Antimycotic (Thermo Scientific). Mouse primary myoblast cells were isolated from C57BL/6 J mice following a standard protocol described earlier [43]. Mouse primary myoblast cells were differentiated as described [14]. U2OS cells were cultured in DMEM (ATCC) supplemented with 10% heat-inactivated FBS (ATCC) and 1X Antibiotic–Antimycotic (thermo scientific). Muscle stem cells (MuSCs) were isolated from mouse TA muscle using a satellite cell isolation kit (Miltenyi Biotec) following the manufacturer's instructions.
Isolation of total RNA and performance of qRT–PCR
We isolated total RNAs using Trizol reagent (Invitrogen) or RNeasy mini kit (Qiagen) following the manufacturer’s instructions. We carried out cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad) as instructed. We performed qRT-PCR using the SYBR Green PCR master mix (Bio-Rad) in a Bio-Rad thermal cycler using specific primers for Myogenin (Myog) (Forward: AGCGCAGGCTC AAGAAAGTGAATG; Reverse: CTGTAGGCGCTCAATGTACTGGAT), Myosin Heavy Chain (Mhc) (forward: TCCAAACCGTCTCTGCACTGTT; Reverse: AGCGTACAAAGTGTGGGTGTGT), Hmga1 (Forward: CGACCAAAGGGAAGCAAGAATAA; Reverse: TCCTCTTCC TCCTTCTCCAGTTTC), and Id3 (Forward: CTCTTAGCCTCTTGGACGACATGA; Reverse: TGT AGTCTATGACACGCTGCAGGA). We used Rsp13 (Forward: TGACGACGTGAAGGAACAG ATTT; Reverse: ATTTCCAGTCACAAAACGGACCT) or Gapdh (Forward: ATGACATCAAGAAG GTGGTGAAGC; Reverse: GAAGAGTGGGAGTTGCTGTTGAAG) primer pairs as a housekeeping gene for normalizing the values of Myog, Mhc, Hmga1, and Id3. We carried out qRT-PCR for miR-24-3p using the miRCURY LNA kit as described (Qiagen). The primer sequences used for U6Sn, miR-24-3p, and miR-192 were CTGCGCAAGGATGACACGCA, TGGCTCAGTTCAGCAGGAACAG, and CTGACCTATGAATTGACAGCC, respectively.
Plasmid construction
Mouse Hmga1 3′UTR was amplified by PCR from C2C12 myoblast genomic DNA and cloned into pRL-CMV. Specific point mutations in Hmga1 3′UTR (shown in Fig. 3A) cloned into pRL-CMV vector were generated using site-directed mutagenesis kit (Stratagene). In addition, Hmga1 ORF without or with its 3′UTR was subcloned into pMSCV retroviral expression vector as described [14]. We made retroviruses using these constructs in HEK-293 T cells by cotransfecting helper plasmids using a standard protocol.
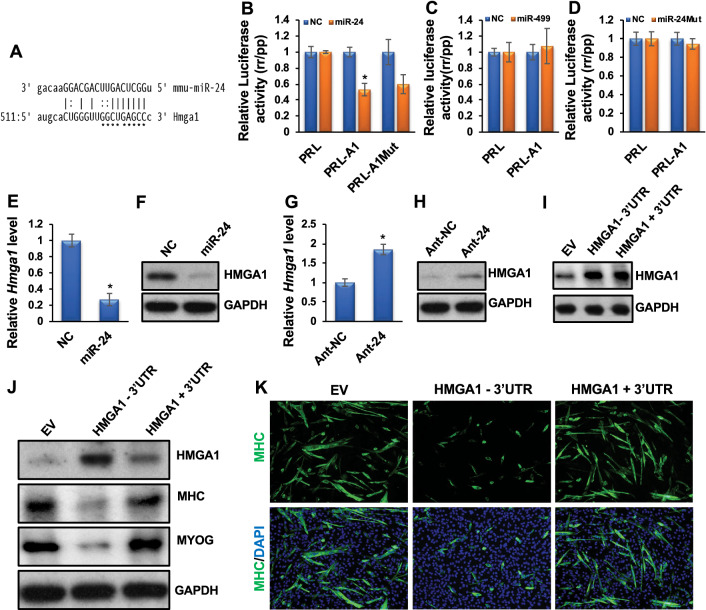
Fig. 3.
miR-24-3p promotes myoblast differentiation by directly targeting and regulating Hmga1. A The microRNA target prediction program miRanda predicts a miR-24-3p binding site on the Hmga1 3′UTR. The stars indicate the bases that were mutated (PRL-A1Mut). B Luciferase assays were performed to measure the effect of miR-24-3p transfection on a Renilla (rr) luciferase reporter fused to the wild-type and mutated Hmga1 3′UTRs (PRL-A1 and PRL-A1-Mut), respectively. A firefly (pp) luciferase plasmid was co-transfected as a transfection control. The rr/pp value was normalized with a control rr luciferase plasmid without an Hmga1 3′UTR (PRL) and is expressed relative to the normalized rr/pp value in GL2-transfected cells (NC). C The luciferase activity was similarly determined when NC or miR-499 was co-transfected with the PRL-A1 construct. D An NC or a mutant form of miR-24-3p (miR-24Mut) was co-transfected with PRL or PRL-A1, and luciferase activity was similarly determined. E, F Exogenous expression of miR-24-3p downregulated Hmga1 mRNA and protein levels. G, H Ant-24 sustained the expression of Hmga1 mRNA and protein levels. I HMGA1 levels in an empty retroviral vector (EV) or the retroviral vectors stably expressing HMGA1 in myoblasts lacking its 3′UTR (HMGA1-3′UTR) or the 3′UTR attached to it (HMGA1 + 3′UTR). J Myoblast cells stably expressing EV and the constructs lacking or containing the Hmga1 3’UTR were differentiated for 3 days. HMGA1, MYOG, MHC, and GAPDH protein levels are shown. GAPDH served as a loading control. (K) MHC immunostaining on these cells. The values are expressed as Mean ± SD of biological triplicates. *P < 0.001
Luciferase assays
We first transfected U2OS cells with miR-24-3p (UGGCUCAGUUCAGCAGG AACAG), miR-24-3p mutant (UCGCACUGUUCUGCAGGAACAG), or GL2 (UCGAAGUAUUCC GCGUACG), and after 24 h, we transfected the same cells with Renilla luciferase (rr) plasmids containing 3′UTR. pGL3 firefly Photinus pyralis (pp) (Promega) was co-transfected as an internal transfection control. Then we harvested the cells after another 48 h and performed luciferase assays with a dual-luciferase reporter assay system using a GloMax luminometer (Promega) following our established protocol [14, 27]. We first normalized rr values to the co-transfected pp luciferase values. Then we normalized each rr/pp value in the miR-24-3p-transfected samples with the rr/pp values obtained from GL2-transfected samples.
Western blotting and antibodies
We harvested the cells, washed them with 1X PBS, lysed them in NP40 lysis buffer (50mMTris-HCl, 150mMNaCl, 0.1% NP-40, 5 mM EDTA, 10% glycerol) with protease inhibitors cocktail (Sigma), and sonicated using a Bioruptor (Diagenode). We separated proteins on SDS-PAGE, transferred, and immunoblotted with various antibodies. The antibodies used were anti-HMGA1 (dilution 1:500; Cell Signaling), anti-MYOG (dilution 1:500; Santa Cruz), anti-MHC (dilution 1:3000; Sigma), and anti-GAPDH (dilution 1:10,000; Sigma).
Intraperitoneal injection of neonatal mice
All animal work was done following our Institutional Animal Care and Use Committee (IACUC) protocol. An antagomir specific to miR-24-3p (Ant-24) and a control GL2 antagomir (Ant-NC) was designed following the procedure, as described previously [44] and were synthesized by Horizon. The sequences for Ant-24 and Ant-NC were mU(*)mG(*)mGmCmUmCmAmGmUmUmCmAmG mCmAmGmGmAmAmCmA(*)mG(*)(3′-Chl) and mU(*)mA(*)mUmCmGmCmGmAmGmUmAmC mGmUmCmGmAmG(*)mG(*)mC(*)mC(*)(3′-Chl), respectively. Here, “m” represents a 2′-O-methyl-modified nucleotide, “*” indicates a phosphorothioate linkage, and “Chl” indicates cholesterol moiety linked to the oligos through a hydroxyprolinol linkage. On postnatal day 3, C57BL/6 J pups were injected intraperitoneally (IP). We followed NIH guidelines of sex as biological variables (SABV) and used both male and female pups for our experiments. Ant-24 and Ant-NC were administered twice at a dose of 100 mg/kg body weight in 10–15 μl per injection volume. BrdU was injected 4 h before harvesting the hind leg skeletal muscles on day 6 for qRT–PCR and fresh-frozen (OCT) samples.
Skeletal muscle regeneration model and TA muscle injection
We developed an injury-mediated mouse model of skeletal muscle regeneration by intramuscular injection of cardiotoxin (CTX) from Naja nigricollis (EMD Millipore) following our established procedure as described earlier [27, 29]. Briefly, about 10-week-old male C57BL/6 J mice were injected into TA muscles with 100ul of 10 μM CTX. This high volume of injection materials, pressure, and post-injection massages spread the injected material throughout the TA muscle compartment. However, we analyzed the middle two-thirds of the TA muscle closest to the injection site. We injected 100 μl (15 μg) of Ant-24 into the TA muscle of one leg and 100 μl (15 μg) of Ant-NC into the contralateral leg 3 days after the CTX injection. Mice were euthanized by CO2 and sacrificed by cervical dislocation before harvesting the muscle samples at different time intervals.
Immunocytochemistry and immunohistochemistry
We carried out immunocytochemistry following our standard protocol described previously [14, 27]. Briefly, we grew the cells on sterile glass coverslips and fixed them with 2% formaldehyde in PBS for 15 min. Next, we permeabilized the cells with 0.2% Triton X-100 and 1% normal goat serum (NGS) in ice-cold PBS for 5 min and blocked them with 1% NGS in PBS twice for 15 min. We incubated the fixed and blocked cells with primary antibodies (MYOG 1:50; Santa Cruz Biotechnology and MHC 1:400; Sigma) in 1% NGS for 1 h. Next, we washed the cells twice with 1X PBS and incubated them with FITC-conjugated anti-mouse IgG (dilution 1:500; Dako Cytomation) for 1 h. Finally, we rewashed the cells twice and counterstained the nuclei with DAPI while mounting the coverslips on a glass slide (H-1200, Vector Laboratories). Images were captured with an Olympus Hi-Mag microscope. The immunostaining using the skeletal muscle tissue sections was carried out with slight modifications of the protocols described by others: anti-BrdU and LAMININ [45], and anti-DESMIN [34]. The primary antibodies, mouse BrdU (1:50; Roche), rat anti-LAMININ (1:100; Millipore), mouse anti-DESMIN (1:200; Dako), rat anti-F4/80 (1:200; Biolegend), rat anti-CD45 (1:200; Invitrogen), and rat anti-CD4 (1:100; Invitrogen) were used. The secondary antibodies, Alexa Fluor 594 goat anti-mouse IgG1 (1:400; Invitrogen), and Alexa Fluor 488 goat anti-rabbit IgG (1:400; Invitrogen), were used. Images were captured using Zeiss LSM-710 confocal microscope. H&E staining was carried out using a standard protocol, and bright-field images were captured using an Olympus microscope.
Statistical analyses
Data are presented as the mean ± standard deviation of three or more biological replicates. A two-tailed Student’s t-test was employed to determine P-values.
Results
miR-24-3p is abundantly expressed in adult skeletal muscle and is regulated during myoblast differentiation and skeletal muscle regeneration
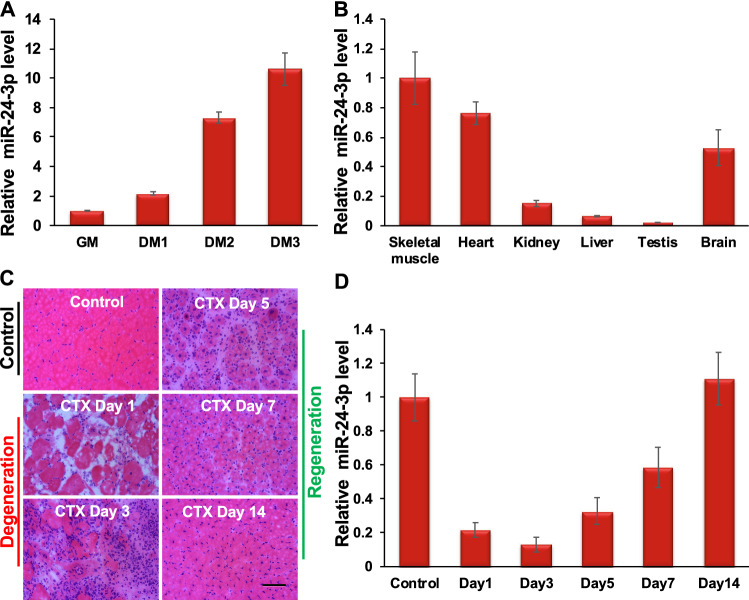
An earlier study showed that miR-24-3p induces myoblast differentiation in vitro [37]. However, the molecular mechanism of miR-24-3p in myoblast differentiation and its function in skeletal myogenesis in vivo remained undetermined. Therefore, we focused on miR-24-3p to reveal its function and molecular mechanism in skeletal myogenesis using primary myoblast cells, neonatal mice, and a mouse model of skeletal muscle regeneration. We first confirmed that miR-24-3p was upregulated during C2C12 myoblast differentiation (Suppl. Figure 1). Next, we determined that miR-24-3p was upregulated during mouse primary myoblast differentiation, a physiologically more relevant system for studying myogenesis (Fig. 1A). We also examined the expression level of miR-24-3p in various mouse tissues and found that it is abundantly expressed in adult skeletal muscle (Fig. 1B). We then examined the expression of miR-24-3p during skeletal muscle degeneration and regeneration. We generated a well-accepted cardiotoxin (CTX)-induced mouse skeletal muscle regeneration model [46] (Fig. 1C). We observed a degeneration phase after the CTX injury when quiescent MuSCs are activated and give rise to myoblast cells (Fig. 1C). This is followed by a regeneration phase when myoblasts differentiate and fuse to the existing myofibers or make new myofibers (Fig. 1C). miR-24-3p decreased rapidly on days 1 through 3 (degeneration phase) and increased on days 5 through 14 (regeneration phase) (Fig. 1D). These findings suggest the role of miR-24-3p in myoblast differentiation and skeletal muscle regeneration.
Fig. 1.
miR-24-3p is abundantly expressed in adult skeletal muscle and upregulated during myoblast differentiation and muscle regeneration. A qRT-PCR analyses show that miR-24-3p is upregulated during mouse primary myoblast differentiation. miR-24-3p values were first normalized to the U6sn values, and the fold-change of miR-24-3p was determined relative to the undifferentiated myoblasts (GM). DM1, DM2, and DM3 indicates the number of days myoblasts cultured in differentiation medium (DM). B miR-24-3p is abundantly expressed in adult skeletal muscle. qRT-PCR was performed using various mouse tissues to measure miR-24-3p. The values were normalized to the respective U6sn values and again to the values of skeletal muscle. C Hematoxylin and eosin (H&E) staining of tibialis anterior (TA) muscle cross-sections from noninjected control and on days 1, 3, 5, 7, and 14 post-injury caused by intramuscular injection of CTX. D miR-24-3p is downregulated on days 1–3 post-injury and upregulated on days 5–14 post-injury. The values are expressed as mean ± SD of three (A–B) and five (D) biological replicates
miR-24-3p promotes myoblast differentiation
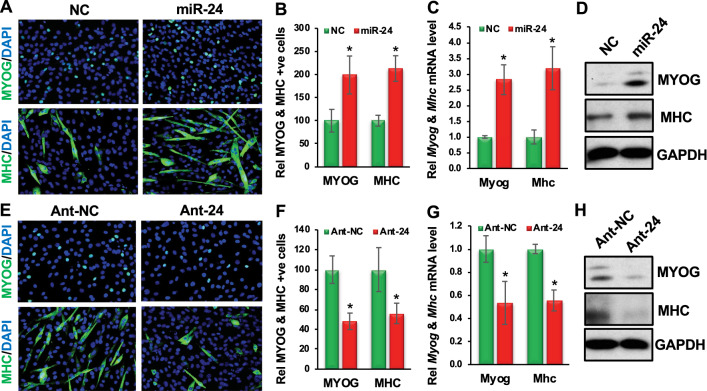
Since myoblast differentiation is a critical step in postnatal muscle development and regeneration, we assessed the function of miR-24-3p in primary myoblast differentiation. We first confirmed an earlier report that miR-24-3p promotes C2C12 myoblast differentiation (Suppl. Figure 2). Next, we examined its function during mouse primary myoblast differentiation. We transfected primary myoblast cells with mature miR-24-3p or GL2, an RNA duplex of the identical length of miR-24-3p from the luciferase gene, as a negative control (NC) in the growth medium (GM). We transfected the cells twice at 24 h intervals in GM and replaced the GM with the differentiation medium (DM). We carried out immunostaining of the cells for an early myogenic marker, myogenin (MYOG), and a late myogenic marker, myosin heavy chain (MHC), 24 and 48 h after adding the DM, respectively. The exogenous miR-24-3p increased differentiation, as seen by the increased number of MYOG- and MHC-positive cells with elongated and multinucleated cellular morphology (Fig. 2A, B; Suppl. Table 1). We also observed increased Myog and Mhc mRNA and protein levels in these cells when maintained throughout in GM (Fig. 2C, D). These results demonstrate that miR-24-3p indeed promotes myoblast differentiation.
Fig. 2.
miR-24-3p promotes and is required for primary myoblast differentiation. A Primary myoblast cells were transfected twice at 24 h intervals with GL2 or miR-24-3p in GM, and then the GM was replaced with DM. Immunostaining of these cells was carried out for MYOG and MHC 24 and 48 h after replacing the GM with DM, respectively. B Fractions of MYOG- and MHC-positive cells are presented relative to the GL2 (NC) as 100%. C Myog and Mhc mRNA levels when the cells were maintained throughout in GM. qRT-PCR was carried out for Myog and Mhc, and the values were normalized to Gapdh, then again to the NC values. D MYOG, MHC, and GAPDH protein levels. GAPDH served as a loading control. E Primary myoblast cells were transfected twice at 24 h intervals with antagomirs specific to GL2 (Ant-NC) or miR-24-3p (Ant-24), then the GM was replaced with DM. Immunostaining of these cells was carried out for MYOG and MHC 24 and 48 h after replacing the GM with DM, respectively. F Fractions of MYOG- and MHC-positive cells are presented relative to the Ant-NC control as 100%. G qRT-PCR was carried out for Myog and Mhc, and the values were normalized as C. H MYOG, MHC, and GAPDH protein levels. GAPDH served as a loading control. The values are expressed as mean ± SD of biological triplicates. *P < 0.001
We also investigated whether miR-24-3p is required for myoblast differentiation. First, we examined the silencing efficiency of antagomirs specific to miR-24-3p (Ant-24). We transfected primary myoblast cells twice with Ant-24 or control antagomir (Ant-NC) at 24 h intervals in GM and harvested the cells after another 48 h. We determined by qRT-PCR analyses that the endogenous miR-24-3p level was downregulated five-fold in the Ant-24-transfected samples (Suppl. Figure 3). Next, we transfected the cells twice with Ant-24 or Ant-NC at 24 h intervals in GM and replaced the GM with the DM. We carried out immunostaining of the cells for MYOG and MHC 24 and 48 h after adding the DM, respectively. Ant-24 decreased the MYOG- and MHC-positive cell numbers and differentiation morphology (Fig. 2E, F; Suppl. Table 1). We also found that both Myog and Mhc mRNA and protein levels were significantly decreased in these samples (Fig. 2G, H). We followed up these cells for five days (Suppl. Figure 4). Whereas Ant-NC-treated cells differentiate normally and form multinucleated myotubes (Suppl. Figure 4A, D), the Ant-24-treated cells don’t differentiate following the normal kinetics (Suppl. Figure 4B, E) and the majority of the cells looks like proliferating myoblasts (Suppl. Figure 4C, F). These findings establish that miR-24-3p is required for myoblast differentiation.
miR-24-3p promotes muscle differentiation by directly targeting and regulating Hmga1
To determine how miR-24-3p promotes myoblast differentiation, we revealed its direct target relevant to myogenesis. The microRNA target prediction algorithm miRanda revealed Hmga1 as a potential target for miR-24-3p (Fig. 3A). HMGA1 is a known myogenic repressor, and its downregulation is required for myoblast differentiation [40]. Therefore, we chose Hmga1 to investigate whether miR-24-3p promotes muscle differentiation by directly targeting and regulating Hmga1. To determine if Hmga1 3′UTR has a valid target site(s) for miR-24-3p, we fused the Hmga1 3′UTR to a luciferase reporter gene driven by the cytomegalovirus (CMV) promoter. Transfection of miR-24-3p with the Hmga1 3′UTR luciferase construct repressed luciferase activity (Fig. 3B). However, mutations in the seed sequence of the predicted target site in the Hmga1 3′UTR did not relieve the repression significantly (Fig. 3B), suggesting a non-canonical (non-seed-match) target site(s) for miR-24-3p in the Hmga1 3′UTR. Non-seed-match target sites have previously been reported by us and others [14, 47]. Consistent with this notion, the RNA hybrid target prediction program revealed several non-canonical targets sites of miR-24-3p in the Hmga1 3′UTR (Suppl. Figure 5).
In addition to the conventional experiments as described above, we carried out additional control experiments where we co-transfected Hmga1 3′UTR with a control miR-499 (similarly upregulated during myoblast differentiation but does not contain Hmga1 target sites) and miR-24Mut (randomly mutated miR-24-3p at the seed nucleotide positions 2, 5, and 7 and a non-seed nucleotide position 12). We changed one base outside the seed sequence as non-seed sequences also influence microRNA binding to the target sites. Co-transfection of the 3′UTR of Hmga1 either with miR-499 or mutated form of miR-24-3p did not suppress the luciferase activity (Fig. 3C, D), further suggesting that Hmga1 is specifically targeted by miR-24-3p.
To demonstrate that Hmga1 is indeed a valid target of miR-24-3p during myogenesis, we transfected primary myoblasts with GL2 or miR-24-3p twice at 24 h intervals in GM. We harvested the cells for qRT-PCR and Western blotting for HMGA1 48 h after the last transfection. miR-24-3p drastically downregulated Hmga1 mRNA and protein levels in these samples (Fig. 3E, F). We also observed downregulation of HMGA1’s direct target Id3 transcripts in these samples (Suppl. Figure 6A). HMGA1 is known to bind to the Id3 promoter and regulate its expression positively [48]. Consistent with this finding, knockdown of Hmga1 in myoblasts decreased the Id3 level (Suppl. Figure 6B, C). In a reciprocal experiment, inhibiting miR-24-3p in DM caused derepression of endogenous Hmga1 mRNA and protein (Fig. 3G, H) and Id3 transcript (Suppl. Figure 6D) levels during primary myoblast differentiation. These findings demonstrate that miR-24-3p is responsible for the repression of Hmga1 mRNA and protein levels during myoblast differentiation.
We further examined if the 3′UTR of Hmga1 containing the miR-24-3p target site was necessary for downregulation of the Hmga1 mRNA and protein levels during myoblast differentiation. We used retroviral vectors stably expressing Hmga1 lacking its 3′UTR (HMGA1-3′UTR) or the 3′UTR attached to it (HMGA1 + 3′UTR) during myoblast differentiation. We also used myoblasts stably expressing empty vectors (EV). We chose stable cells containing either HMGA1-3′UTR or HMGA1 + 3′UTR constructs expressing a similar basal level of HMGA1 for differentiation assays (Fig. 3I). After three days in DM, HMGA1 protein persisted in the cells expressing Hmga1 without its 3′UTR, in contrast to the cells expressing Hmga1 containing the 3′UTR or an EV (Fig. 3J). As expected, the myoblast cells expressing Hmga1 attached to its 3′UTR or an EV differentiate normally, as found by immunoblotting for MYOG and MHC and immunofluorescence for MHC (Fig. 3J, K). However, the cells expressing Hmga1 without its 3′UTR repress differentiation (Fig. 3J, K). Together, these findings confirm that Hmga1 is a bonafide target for miR-24-3p and that miR-24-3p promotes myoblast differentiation by directly targeting and regulating Hmga1.
miR-24-3p regulates proliferation of PAX7-positive MuSCs in vivo
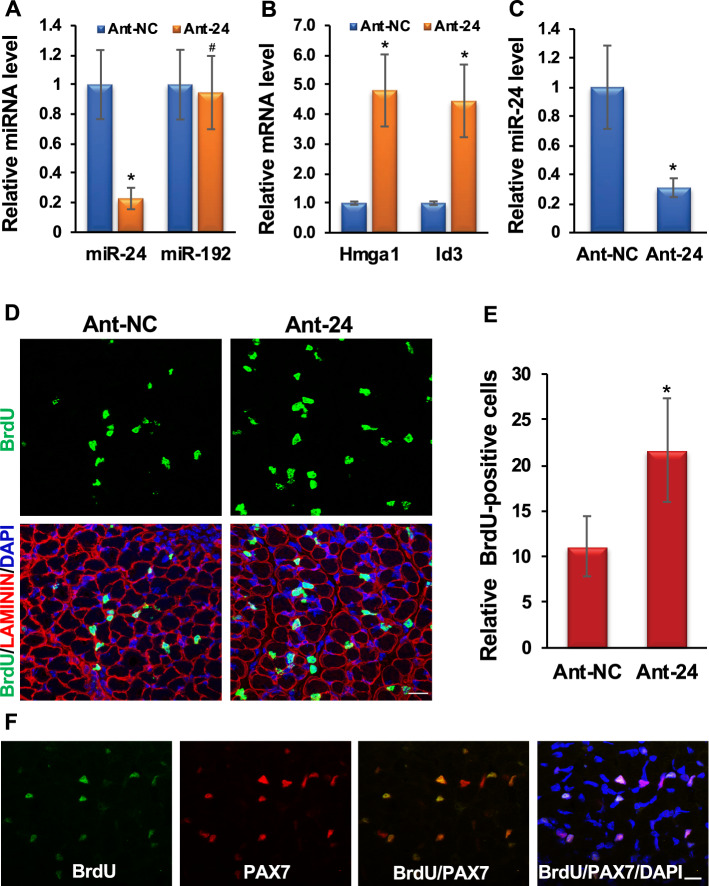
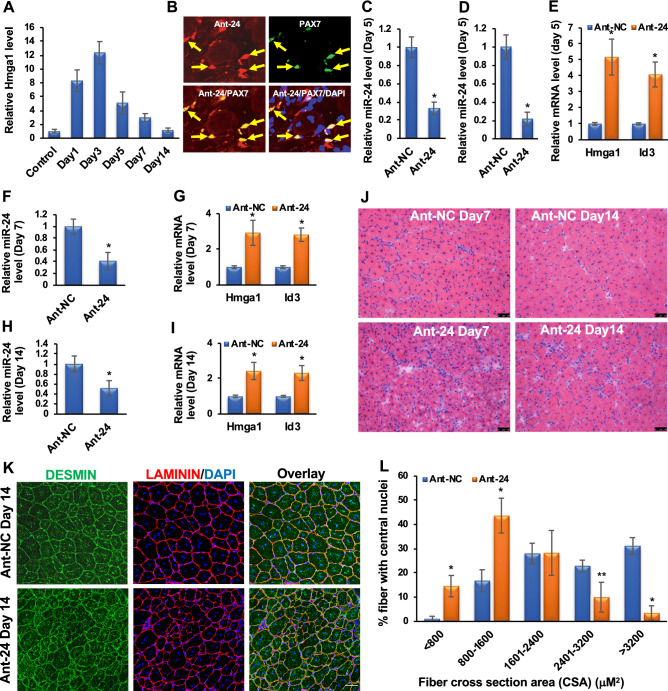
To determine the role of miR-24-3p in vivo, we knocked down endogenous miR-24-3p in the skeletal muscle of neonatal mice using antagomirs [44]. We injected 3-day-old neonatal mice intraperitoneally with antagomirs specific to miR-24-3p (Ant-24) or NC (Ant-NC) twice at 24 h intervals and harvested the muscles on day 6. Ant-24 knocked down miR-24-3p in the hind leg skeletal muscles but had no impact on control miR-192 (Fig. 4A). The Hmga1 mRNA level was derepressed in the miR-24-3p knockdown skeletal muscles (Fig. 4B). HMGA1 activity was upregulated in these muscles, as indicated by an increase in Id3 transcripts (Fig. 4B). Ant-24 decreased the endogenous miR-24-3p level in the MuSCs isolated from these samples (Fig. 4C). Consequently, Myog and Mhc levels were decreased in the undifferentiated and differentiated MuSCs isolated from these samples (Suppl. Figure 7A, B). To examine the effects of miR-24-3p knockdown on MuSC proliferation, we injected BrdU into these mice to label the proliferating cells 4 h before harvesting the muscles. We observed a significant increase in BrdU-positive cells in Ant-24 skeletal muscles (Fig. 4D, E). The BrdU-positive cells were located at the periphery of the muscle fibers, and 90.87% of these cells also expressed the MuSC marker PAX7 in Ant-24 samples (Fig. 4F). Based on these results, we conclude that miR-24-3p restricts the proliferative potential of MuSCs and induces differentiation in vivo. Thus, our findings suggest that the miR-24-3p/HMGA1/ID3 axis plays an essential role during neonatal skeletal muscle development by regulating MuSC proliferation.
Fig. 4.
Knockdown of miR-24-3p increases PAX7-positive proliferating MuSCs in vivo. A Intraperitoneal injection of Ant-24 decreases endogenous miR-24-3p in neonatal skeletal muscle. qRT-PCR values of miR-24-3p normalized to the U6sn values are expressed relative to Ant-NC values. miR-192 was used as an NC. B Ant-24 derepresses Hmga1 and Id3 transcript levels. qRT-PCR of Hmga1 and Id3 on hind leg skeletal muscle normalized to Gapdh, then again to the respective Ant-NC samples. C miR-24-3p level is decreased in the MuSCs isolated from Ant-24-injected neonatal mice. D Confocal images of skeletal muscle harvested 4 h after BrdU labeling from Ant-24- or Ant-NC-injected neonates. Cell proliferation was determined by anti-BrdU antibody (green), the cell surface was marked by LAMININ (red), and nuclei were counterstained with DAPI (blue). E Relative BrdU-positive nuclei per field. Mean ± SD of 10 random fields from five mice. F Confocal microscopy images of Ant-24-injected neonatal skeletal muscles from D immunostained for BrdU (green), PAX7 (red), DAPI (blue), BrdU and PAX7 (orange), and BrdU, Pax7, and DAPI (magenta). Mean ± SD of the samples from five neonatal mice. *P < 0.001. Scale bar: 50 μM
miR-24-3p is essential for adult skeletal muscle regeneration
We used a well-established mouse model of skeletal muscle regeneration by intramuscular injection of CTX into the tibialis anterior (TA) muscle. This model first induces injury (degeneration) and later regenerates spontaneously, allowing us to study muscle regeneration (Fig. 1C). As described earlier, miR-24-3p decreased rapidly on days 1–3 after CTX injury, then gradually increased as new muscles were formed (Fig. 1D). We examined the Hmga1 levels in these samples and found its expression pattern anti-correlated to miR-24-3p, increasing on days 1–3 after injury and decreasing steadily on days 5–14 after injury (Fig. 5A). These findings suggest that miR-24-3p regulates Hmga1 during regeneration.
Fig. 5.
miR-24-3p is essential for skeletal muscle regeneration. A Hmga1 is gradually upregulated in TA muscles on days 1–3 post-injury and downregulated on days 5–14 post-injury. Hmga1 values were normalized to the Rps13 values. B We injected a fluorescence (DyLight594) conjugated Ant-24 into TA muscle of adult mice on day 3 after CTX injury and harvested the samples day 5 post-injury. Ant-24 incorporates into TA muscle, including MuSCs on day 5 post-injury. Arrows indicate a subset of PAX7-positive MuSCs overlapped with Ant-24 fluorescence. C miR-24-3p level was also decreased in the MuSCs isolated from these samples. qRT-PCR was carried out for miR-24-3p, and its values were first normalized to the U6sn values and again relative to NC values. D, E Ant-NC and Ant-24 were injected in TA muscle on day 3 post-injury. The miR-24-3p level was repressed, and Hmga1 and Id3 transcript levels were derepressed in the TA muscle on day 5 post-injury. Hmga1 and Id3 values were normalized to the respective Rps13 values and then again to the Ant-NC values. F–I The miR-24-3p level remained repressed on days 7 and 14 post-injury, and Hmga1 and Id3 transcript levels remained deprepressed in the Ant-24 samples. J Representative images of H&E staining showing that the Ant-24 impairs regeneration on days 7 and 14 post-injury. K, L DESMIN and LAMININ staining and cross-section areas (CSA) of the regenerating fibers on day 14 using the Fiji software. More than 600 fibers were counted from ten random sections of five mice in both groups. Scale bar: 50 μM. Mean ± SD of the samples from five mice. *P < 0.001; **P < 0.01
Next, we confirmed the role of miR-24-3p during skeletal muscle regeneration by knocking it down using Ant-24. We first determined whether Ant-24 incorporated into MuSCs after injury and decreased endogenous miR-24-3p level by injecting a fluorescence conjugated Ant-24 into TA muscle of adult mice on day 3 post-injury. Ant-24 incorporates into TA muscle, including MuSCs on day 5 post-injury (Fig. 5B), and indeed decreased miR-24-3p level in these samples (Fig. 5C, D). Furthermore, decreased miR-24-3p level in TA muscle was associated with derepression of Hmga1 and Id3 transcript levels (Fig. 5E).
Since miR-24-3p was upregulated on days 5–14 after CTX injury, we injected Ant-24 into the TA muscles of one leg and Ant-NC into the TA muscles of the contralateral leg on day 3 post-injury. On day 5 post-injury, the regeneration process was early, when new myofibers were just beginning to form. Therefore, we examined the impact of Ant-24 on days 7 and 14 after injury. The miR-24-3p level remained repressed while the Hmga1 and Id3 transcript levels remained derepressed in the Ant-24 samples (Fig. 5F–I). In Ant-NC mice, muscle regenerated normally, and the morphology was restored on day 14, except for the presence of the central nuclei, a signature of regenerating muscles (Fig. 5J). In contrast, the regeneration process was impaired in the Ant-24 muscles (Fig. 5J). In addition, we observed the presence of numerous nuclei, indicating increased inflammatory cells among other cell types [49–51] (Suppl. Figure 8A–L), and the persistence of smaller myofibers, indicating impaired regeneration (Fig. 5J). We further characterized the regenerating skeletal muscle sections on day 14 after injury by DESMIN and LAMININ immunostaining and measuring myofiber cross-section areas (CSA). DESMIN is an intermediate filament protein abundantly expressed in newly generated muscle fibers, while LAMININ labels the muscle fiber boundaries. DESMIN was expressed in the myofibers of regenerating muscles in both Ant-24 and Ant-NC samples (Fig. 5K; Suppl. Figure 9). However, the muscle fibers were significantly smaller in the Ant-24 muscles than in the Ant-NC muscles (Fig. 5L). In addition, interfiber spaces were larger and filled with nuclei, further supporting abnormal regeneration in the Ant-24 muscles. These findings suggest that the miR-24-3p/HMGA1/ID3 axis plays a critical role in normal skeletal muscle regeneration in vivo.
Discussion
Most of the understanding of skeletal muscle development and regeneration is based on the regulation of myogenic transcription factors and signaling molecules [1–8]. However, it remains elusive how these critical factors of the fundamental myogenic processes are themselves regulated. Furthermore, molecular mechanisms underlying the regulation of gene expression during skeletal muscle development and regeneration are not entirely understood, hindering the development of therapeutic interventions for muscle degenerative diseases. To fill this knowledge gap, we have examined the role of a myogenic microRNA in myoblast differentiation and skeletal muscle regeneration. Thus far, most of the studies have demonstrated the role of microRNAs in C2C12 myoblast differentiation in vitro [10–25]. However, only a limited number of studies, including ours, have revealed the function of microRNAs using animal models of muscle development and regeneration [26, 27, 29, 34–36]. In this study, we demonstrated the role of miR-24-3p in muscle differentiation and regeneration in mice. An earlier study also suggests that this microRNA is upregulated during myoblast differentiation and promotes differentiation in vitro [37]. However, it remains unknown how miR-24-3p promotes myoblast differentiation and whether it has any role in myogenesis in vivo. Here we demonstrated that miR-24-3p promotes myoblast differentiation by directly targeting and regulating HMGA1, a well-known repressor of myogenesis [40]. More importantly, we have shown that miR-24-3p is essential for skeletal muscle regeneration. Our study establishes the role of a myogenic microRNA in skeletal muscle function in vivo. Also, it provides mechanistic insights into how a myogenic microRNA sculpts myogenesis by regulating a well-established repressor of myogenesis.
HMGA1 is an important member of the high mobility group A (HMGA) proteins predominantly expressed in the proliferating embryonic tissues but mostly absent in differentiated cells [38, 39]. HMGA1 downregulation is essential for myogenic differentiation [40]. Sustained expression of HMGA1 in myoblast cells represses promyogenic genes, including Myod [40]. HMGA1’s direct target ID3 represses MYOD activity by binding and sequestering E12/47 away from the MYOD binding sites of the critical promyogenic genes, thereby repressing myogenesis. Therefore, our findings suggest that miR-24-3p promotes myogenesis by increasing MYOD activity through the downregulation of HMGA1 and ID3. Consistent with this idea, HMGA1 is involved in the differentiation of several cell types, including embryonic lymphohematopoietic cells and adipocytes [52, 53]. These findings indicate that the strictly regulated expression of HMGA1 is critical for normal cellular differentiation, including myogenic differentiation. Our study reveals how a myogenic microRNA regulates this crucial factor during myoblast differentiation. Our studies suggest that miR-24-3p/HMGA1/ID3 axis is essential for normal myogenesis. Since the loss of Hmga1 gene function affects cellular differentiation processes [39] and Hmga1 knockout mice develop diseases including type 2 diabetes [54], cardiac hypertrophy [55], and myelolymphoproliferative disorders [55], we envisage that dysregulation of miR-24-3p/HMGA1/ID3 axis may lead to muscle degenerative diseases.
Only a handful of studies have implicated microRNAs in skeletal muscle function in animal development [26, 27, 29–36]. This gap prompted us to study the function of miR-24-3p in skeletal muscle differentiation and regeneration in vivo using mouse models. Though defective skeletal muscle development in the muscle-specific deletion of Dicer suggests a critical role for microRNAs during skeletal muscle development in animals [30], there have been conflicting findings in the literature on whether specific microRNAs are essential for these processes. In one study, the germline deletion of miR-206 did not result in any significant defects in skeletal muscle development and exerted only a mild effect on skeletal muscle innervation following injury [31]. Although miR-1 is expressed in skeletal muscles, miR-1 knockout mice showed defects only in cardiac muscle development; no visible phenotypes were observed in skeletal muscles [56]. These discrepancies might be due to the functional redundancy between miR-1 and miR-206, as they share identical seed sequences. In another study, miR-206 knockout delayed skeletal muscle regeneration following CTX injury [34]. We demonstrated earlier that miR-26a and H19-derived miR-675-3p and miR-675-5p play essential roles in skeletal muscle development and regeneration [27, 29]. Here, we have demonstrated a critical role of miR-24-3p in myogenesis in vivo. It’s important to elucidate the role of the repertoire of microRNAs in myogenesis in vivo because their concerted actions maintain the normal skeletal muscle function. Future studies will explore its role in muscle degenerative diseases. Nonetheless, our findings elucidate an essential function of miR-24-3p in skeletal muscle differentiation and regeneration in vivo.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the inputs of the Dey lab members. They also acknowledge Dr. Melinda Larsen for providing F4/80 and CD45 antibodies.
Author contributions
BKD conceived the project; PD and BKD designed the study; PD, MAS, and BKD performed the experiments; PD and BKD analyzed the data and prepared the manuscript.
Funding
This work was supported by SUNY startup and American Heart Association (AHA 17SDG33670339) grants to BKD.
Availability of data and material
Data generated for this study are included in the manuscript and available from the corresponding author.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The animal work of this manuscript was approved by the Institutional Animal Care and Use Committee (IACUC).
Consent for publication
The authors give consent for publication in CMLS.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4(2):a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asfour HA, Allouh MZ, Said RS. Myogenic regulatory factors: the orchestrators of myogenesis after 30 years of discovery. Exp Biol Med (Maywood) 2018;243(2):118–128. doi: 10.1177/1535370217749494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(4–5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Bi P, Yue F, Sato Y, Wirbisky S, Liu W, Shan T, et al. Stage-specific effects of Notch activation during skeletal myogenesis. Elife. 2016 doi: 10.7554/eLife.17355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrichs M, Wirsdöerfer F, Flohé SB, Schneider S, Wuelling M, Vortkamp A. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 2011;12:26. doi: 10.1186/1471-2121-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paris ND, Soroka A, Klose A, Liu W, Chakkalakal JV. Smad4 restricts differentiation to promote expansion of satellite cell derived progenitors during skeletal muscle regeneration. Elife. 2016 doi: 10.7554/eLife.19484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 8.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132(12):2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Chen M, Lian D, Li Y, Li Y, Wang J, et al. Non-coding RNA regulates the myogenesis of skeletal muscle satellite cells. Injury Repair Dis Cells. 2019 doi: 10.3390/cells8090988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou A, Mastroyiannopoulos NP, Uney JB, Phylactou LA. miR-186 Inhibits muscle cell differentiation through myogenin regulation. J Biol Chem. 2014;289(7):3923–3935. doi: 10.1074/jbc.M113.507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, et al. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS ONE. 2009;4(10):e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng X, Du J, Shen L, Tan Z, Jiang D, Jiang A, et al. MiR-204-5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed Pharmacother. 2018;101:528–535. doi: 10.1016/j.biopha.2018.02.096. [DOI] [PubMed] [Google Scholar]
- 14.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31(1):203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gan M, Du J, Shen L, Yang D, Jiang A, Li Q, et al. miR-152 regulates the proliferation and differentiation of C2C12 myoblasts by targeting E2F3. In Vitro Cell Dev Biol Anim. 2018;54(4):304–310. doi: 10.1007/s11626-017-0219-1. [DOI] [PubMed] [Google Scholar]
- 16.Ge G, Yang D, Tan Y, Chen Y, Jiang D, Jiang A, et al. miR-10b-5p regulates C2C12 myoblasts proliferation and differentiation. Biosci Biotechnol Biochem. 2019;83(2):291–299. doi: 10.1080/09168451.2018.1533805. [DOI] [PubMed] [Google Scholar]
- 17.Goljanek-Whysall K, Pais H, Rathjen T, Sweetman D, Dalmay T, Munsterberg A. Regulation of multiple target genes by miR-1 and miR-206 is pivotal for C2C12 myoblast differentiation. J Cell Sci. 2012;125(Pt 15):3590–3600. doi: 10.1242/jcs.101758. [DOI] [PubMed] [Google Scholar]
- 18.Huang Z, Chen X, Yu B, He J, Chen D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem Biophys Res Commun. 2012;423(2):265–269. doi: 10.1016/j.bbrc.2012.05.106. [DOI] [PubMed] [Google Scholar]
- 19.Katase N, Terada K, Suzuki T, Nishimatsu S-I, Nohno T. miR-487b, miR-3963 and miR-6412 delay myogenic differentiation in mouse myoblast-derived C2C12 cells. BMC Cell Biol. 2015 doi: 10.1186/s12860-015-0061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren R-M, Liu H, Zhao S, Cao J. Targeting of miR-432 to myozenin1 to regulate myoblast proliferation and differentiation. Genet Mol Res GMR. 2016 doi: 10.4238/gmr15049313. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21(13):2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagan J, Dey BK, Dutta A. MicroRNAs regulate and provide robustness to the myogenic transcriptional network. Curr Opin Pharmacol. 2012 doi: 10.1016/j.coph.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286(22):19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 MicroRNAs in differentiating myoblasts. J Biol Chem. 2012;287(48):40360–40370. doi: 10.1074/jbc.M112.378414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191(2):347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J-F, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190(5):867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26(19):2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8(3):278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 29.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28(5):491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, et al. Essential role for dicer during skeletal muscle development. Dev Biol. 2007;311(2):359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mok GF, Lozano-Velasco E, Maniou E, Viaut C, Moxon S, Wheeler G, et al. miR-133-mediated regulation of the Hedgehog pathway orchestrates embryo myogenesis. Development. 2018 doi: 10.1242/dev.159657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vechetti IJ, Jr, Wen Y, Chaillou T, Murach KA, Alimov AP, Figueiredo VC, et al. Life-long reduction in myomiR expression does not adversely affect skeletal muscle morphology. Sci Rep. 2019;9(1):5483. doi: 10.1038/s41598-019-41476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Invest. 2012;122(6):2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galimov A, Merry TL, Luca E, Rushing EJ, Mizbani A, Turcekova K, et al. MicroRNA-29a in adult muscle stem cells controls skeletal muscle regeneration during injury and exercise downstream of fibroblast growth factor-2. Stem Cells (Dayton, Ohio) 2016;34(3):768–780. doi: 10.1002/stem.2281. [DOI] [PubMed] [Google Scholar]
- 36.Wu R, Li H, Zhai L, Zou X, Meng J, Zhong R, et al. MicroRNA-431 accelerates muscle regeneration and ameliorates muscular dystrophy by targeting Pax7 in mice. Nat Commun. 2015;6:7713. doi: 10.1038/ncomms8713. [DOI] [PubMed] [Google Scholar]
- 37.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, et al. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36(8):2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murani E, Muraniova M, Ponsuksili S, Schellander K, Wimmers K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol. 2007;7:109. doi: 10.1186/1471-213X-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17(2):72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocher J, Vogel B, Hock R. HMGA1 down-regulation is crucial for chromatin composition and a gene expression profile permitting myogenic differentiation. BMC Cell Biol. 2010;11(1):64. doi: 10.1186/1471-2121-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270(5639):725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 42.Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132(4):657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125(6):1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138(17):3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, et al. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem. 2003;278(10):8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 47.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez Hoyos J, Fedele M, Battista S, Pentimalli F, Kruhoffer M, Arra C, et al. Identification of the genes up- and down-regulated by the high mobility group A1 (HMGA1) proteins: tissue specificity of the HMGA1-dependent gene regulation. Cancer Res. 2004;64(16):5728–5735. doi: 10.1158/0008-5472.CAN-04-1410. [DOI] [PubMed] [Google Scholar]
- 49.Doyonnas R, LaBarge MA, Sacco A, Charlton C, Blau HM. Hematopoietic contribution to skeletal muscle regeneration by myelomonocytic precursors. Proc Natl Acad Sci USA. 2004;101(37):13507–13512. doi: 10.1073/pnas.0405361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang W, Hu P. Skeletal muscle regeneration is modulated by inflammation. J Orthop Translat. 2018;13:25–32. doi: 10.1016/j.jot.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kharraz Y, Guerra J, Mann CJ, Serrano AL, Munoz-Canoves P. Macrophage plasticity and the role of inflammation in skeletal muscle repair. Mediators Inflamm. 2013;2013:491497. doi: 10.1155/2013/491497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, Croce CM, et al. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. FASEB J. 2003;17(11):1496–1498. doi: 10.1096/fj.02-0977fje. [DOI] [PubMed] [Google Scholar]
- 53.Melillo RM, Pierantoni GM, Scala S, Battista S, Fedele M, Stella A, et al. Critical role of the HMGI(Y) proteins in adipocytic cell growth and differentiation. Mol Cell Biol. 2001;21(7):2485–2495. doi: 10.1128/MCB.21.7.2485-2495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Foti D, Chiefari E, Fedele M, Iuliano R, Brunetti L, Paonessa F, et al. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat Med. 2005;11(7):765–773. doi: 10.1038/nm1254. [DOI] [PubMed] [Google Scholar]
- 55.Fedele M, Fidanza V, Battista S, Pentimalli F, Klein-Szanto AJ, Visone R, et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006;66(5):2536–2543. doi: 10.1158/0008-5472.CAN-05-1889. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated for this study are included in the manuscript and available from the corresponding author.