Abstract
Oxaliplatin is the first-line regime for advanced gastric cancer treatment, while its resistance is a major problem that leads to the failure of clinical treatments. Tumor cell heterogeneity has been considered as one of the main causes for drug resistance in cancer. In this study, the mechanism of oxaliplatin resistance was investigated through in vitro human gastric cancer organoids and gastric cancer oxaliplatin-resistant cell lines and in vivo subcutaneous tumorigenicity experiments. The in vitro and in vivo results indicated that CD133+ stem cell-like cells are the main subpopulation and PARP1 is the central gene mediating oxaliplatin resistance in gastric cancer. It was found that PARP1 can effectively repair DNA damage caused by oxaliplatin by means of mediating the opening of base excision repair pathway, leading to the occurrence of drug resistance. The CD133+ stem cells also exhibited upregulated expression of N6-methyladenosine (m6A) mRNA and its writer METTL3 as showed by immunoprecipitation followed by sequencing and transcriptome analysis. METTTL3 enhances the stability of PARP1 by recruiting YTHDF1 to target the 3′-untranslated Region (3′-UTR) of PARP1 mRNA. The CD133+ tumor stem cells can regulate the stability and expression of m6A to PARP1 through METTL3, and thus exerting the PARP1-mediated DNA damage repair ability. Therefore, our study demonstrated that m6A Methyltransferase METTL3 facilitates oxaliplatin resistance in CD133+ gastric cancer stem cells by Promoting PARP1 mRNA stability which increases base excision repair pathway activity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04129-0.
Keywords: Digestive system tumors, Chemotherapy resistance, Epigenetic modulation, DNA repair
Introduction
Gastric cancer (GC) still remains among the most common malignant tumors being the third leading cause of cancer-related deaths worldwide [1]. Moreover, GC is primarily found in Asian populations, and China has the largest population of gastric cancer patients around the world [2]. Chemotherapies such as the ones based on oxaliplatin are commonly used postoperatively in the clinics for treatment of GC patients [3]. However, it is still important to study the mechanism of congenital and acquired oxaliplatin resistance in GC treatment [4]. Different tumorigenic cell subpopulations are thought to be the sources of cancer heterogeneity [5]. Cancer initiating cells (CICs) are known to present enhanced chemoresistance, metastatic potential, and ability to develop cell heterogeneity [6]. However, the role and function of these subpopulations in drug resistance and metastasis are still poorly characterized [6]. Currently, there are only very few studies exploring in depth the role of CSCs in GC [7]. Previous studies demonstrated that CD44+ and CD133+ cells show the characteristics of cancer stem cells and drug resistance properties [7–9]. The acquisition of chemotherapeutic resistance can be mediated by tumor cell dormancy, increased DNA damage repair and drug efflux capacity, reduced cell apoptosis, and the interaction between CSC and its supporting microenvironment (CSC niche) [10]. At present, CSC is closely related to chemotherapy resistance of a variety of tumors, including breast cancer, pancreatic cancer, liver cancer, and colorectal cancer [11–14]. However, there are very few relevant studies on platinum resistance of CSC in gastric cancer, and our understanding is still at the early stage, which requires further study [15].
The powerful DNA damage repair ability of chemotherapy-resistant cancer stem cells has been described in many studies [16, 17]. However, the question of how cancer stem cells exert their strong DNA damage repair ability has puzzled us for many years. The N6-methyladenosine (m6A) RNA modification is a common internal mRNA modification that modulates the outcome of gene expression by regulating RNA processing, localization, translation and final attenuation, all of which can be modulated by the “writer”, “eraser” and “reader” of this epigenetic marker [18]. How tumor stem cells exert the function of m6A has been studied in glioma, colorectal cancer, liver cancer and leukemia, but not yet in gastric cancer [19–23]. In fact, regulation of m6A mRNA modification is an important strategy to overcome drug-induced cell death for cancer cells [24–26]. However, the mechanism on how m6A modification affects the drug resistance of tumor cells is still not fully understood and needs to be explore in depth. In particular, there is currently no relevant research on whether tumor stem cells can overcome drug resistance by exerting the effect of m6A in gastric cancer.
The purpose of this study is to find out which CSC subpopulation is the key player for the acquired oxaliplatin resistance in GC, and explore the possible modulation mechanism of m6A on the oxaliplatin resistance of this subpopulation, to facilitate our clinical efforts to overcome oxaliplatin resistance. The mechanism of oxaliplatin resistance was investigated by means of using in vitro human gastric cancer organoid models and gastric cancer oxaliplatin-resistant cell lines, as well as in vivo subcutaneous tumorigenicity studies.
Methods
Cell culture
GC cell lines used in this study, including the MKN74, HEK293T and AGS, were provided by The Francis Crick Institute Cell Services. SNU719 cell line was obtained from Keygen Biotechnology Co., Ltd. A fluorescence-based mycoplasma detection assay was performed, and subsequently confirmed with agar culture. The cells were cultured until passage 20. The AGS, MKN45 and SNU719 cells were cultured in RPMI medium containing 10% FBS. The HEK293T cells were grown in DMEM medium containing 10% FBS.
A stable oxaliplatin-resistant GC cell lines was maintained in culture for long term (No. S1224, Selleckchem), AGS, SNU719 and MKN74 cells were grown in RPMI with the initial oxaliplatin concentration of 1 μmol/L and 10% FCS. When the density of surviving cells reached 80%, the cells were passed twice within 9 days to ensure optimal growth. The procedure was repeated until the cells were tolerant to oxaliplatin at a concentration of 200 μmol/L. Finally, IC50 and colony formation tests were used to verify their resistance (Figure S1A-E).
Human tissue and organoids
GC Tissue used for organoid culture was derived from GC patients after surgery in the First Affiliated Hospital of Sun Yat-sen University (FAHSYSU), with signed consent form from the patients. The study was approved and supervised by the Ethics Committee of Clinical Research and Animal Experiments of the FAHSYSU (No. 2017-208). This research conforms to the ethics of all animal and human tissue research. Specimens that met the standards were then screened through the Scientific Research Center of the Seventh Affiliated Hospital of Sun Yat-sen University, and finally organoid specimens of four patients were finally selected in this study.
After gastric cancer surgery, the tissues were obtained by dissection, and the tumor samples were placed in a 50 U/mL penicillin–streptomycin (Thermo Fisher Scientific, Waltham, MA, USA) freezing solution. Then, the GC tissue samples were cut into small pieces within a sterile cell culture hood and then the minced tissue was digested in a 1 mg/mL collagenase V DMEM solution (Sigma-Aldrich, USA) at 37 °C for 1 h. To stop the digestion, large amount of ice-cold PBS was added to the mixture which was subsequently centrifuged at 4 °C (300 G, 5 min). The GC tissue samples were further digested using TrypLE (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C (5 min), and then stopped with plenty of cold PBS. The suspension was filtered by means of using 40 nylon meshes. Then, the cells were cultured in the culture medium and passaged every 2 weeks. The medium used for establishment and cultivation of human GC organoids was as described elsewhere [27].
Lentivirus production and infection of organoids
The control plasmid and the PLKO plasmid expressing shRNA_PARP1, the PLKO vector expressing shRNA_METTL3, and the PLKO vector expressing shRNA_YTHDF1 were purchased from Sigma-Aldrich (St. Louis, MS, USA). To generate the lentivirus, METTL3 and PARP1 overexpression vectors were designed by Shanghai Genechem Co., Ltd. (Shanghai, China). The HEK293T cells were used to produce the lentiviral particles following standard procedure. For transfection, the organoids were firstly extracted and then re-suspended and mixed with the virus solution for 6 h within an incubator. The cells were re-seeded into Matrigel and divided when the antibiotic selection initiated after 3 up to 7 days.
Quantitative real-time PCR
For RNA extraction, a MagMAX-96 Kit (Ambion) was used according to the standard procedures. The cDNA synthesis was performed using the iScript cDNA synthesis kit (BioRad) and following the manufacturer's instructions. The cDNA was diluted fivefold with distilled water, diluted to 2 µM/L to use each RT-qPCR reaction, and measured on the expression of SYBR GreenER (Thermo Fisher Scientific, Waltham, MA, USA) by ABI7500 (Applied Biosystems). The Universal Probabilistic Analysis and Design Center (Roche) were employed for the design of primers and to make sure the exon-exon junction was covered. The transcription level of the family gene (actin) is used for standardization. The sequenced of the RT-qPCR primers can be found in Table S1.
m6A quantitative measurement
Quantitative detection of m6A was described elsewhere [28]. In brief, the total RNA of GC cells was isolated by means of using Trizol according to manufacturer's instructions. Following, the level of m6A was quantified using a m6A RNA Methylation Kit (Abcam).
Methylated RNA immune-precipitation qPCR (MeRIP-qPCR)
The MeRIP-qPCR assay was carried out as described elsewhere [28]. Total RNA obtained from the GC cells binds to protein A/G magnetic beads in an IP buffer containing anti-m6A antibody (ABCAM) and anti-IgG. RNase inhibitors and protease inhibitors were added to the mixture and stored overnight at 4 °C to precipitate the m6A-modified PARP1 grade. The relative enrichment was quantified using 2−ΔΔCt and compared with the input samples. Primers to m6A negative region of EEF1A as the negative control and primers to m6A positive region of EEF1A as the positive control.
Dual luciferase plasmid system
A fragment containing the predicted m6A modification site in human PARP1 3'UTR was amplified by PCR and cloned into the psiCheck2 plasmid (Promega) to form a double luciferase reporter vector. The vector construction was carried out by Genechem. Luciferase activity was measured using a dual luciferase reporter gene assay kit (Promega) and an Infinite F Plex microreader (Tecan). The results were calculated by normalizing the luciferase activity of fireflies.
RNA stability
The RNA of GC cells was extracted using Trizol and subsequently treated with actinomycin D (1 μg/mL). The level of PARP1 mRNA at a specific time point was detected by qRT-PCR.
Western blot (WB)
The extraction of total cell protein was performed by means of using our group's standard cocktail method [5]. The NE-PER™ Extraction Reagent (Thermo Fisher Scientific, MA, USA) was used to extract the nuclear proteins. The pre-determined BCA (Pierce, Rockford, IL, USA) and WB procedures were used as described above [29]. Antibodies used included Anti-GAPDH antibody (1:10,000, Proteintech, 60,004–1-Ig), Anti-gamma H2A.X (phospho S139) antibody (1:1000, Abcam, ab2893), PARP-1 antibody (F-2) (1:500, Santa Cruz, sc-8007), METTL3 Polyclonal Antibody (Catalog number: 15073-1-AP), YTHDF1 Polyclonal Antibody (Catalog number: 17479-1-AP) and XRCC1 (1:1000, Abcam, ab44830).
Flow cytometry and FACS
The FACS method used was the same as the standard method we published [5]. Cellular apoptosis was tested by means of using the Annexin V-FITC Apoptosis Detection Kit (Sigma-Aldrich, St. Louis, MS, USA), and following the protocol provided by the manufacturer. The obtained data was evaluated using the FlowJo 10 software.
The stained CD133+ and CD133− cells were sorted by flow cytometry. GC Oxaliplatin resistance cell lines, PT1 and PT2 organoids were analyzed through a flow analyzer and the sorted cells were used for subsequent experiments.
Colony formation assay and cell viability
For the colony formation assay, 500 cells were seeded into each well of the 6-well plate. Control (DMSO) and oxaliplatin (10 µM/mL) were added into the cell culture medium. Two weeks later, obvious colonies or giant spheres were formed. The cell colonies were fixed, stained by crystal violet, and counted. This procedure was repeated three times.
For cell viability test, a total of 3,000 cells were seeded in each well of a 96-well transparent bottom blackboard (organoids were embedded within the Matrigel). To each well, it was added the oxaliplatin according to a tenfold concentration gradient. After 48 h, the level of ATP was measured using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA).
Spheroid colony formation assay
The specific method of the spherical colony formation test was as described previously [8]. Human GC cells were sown in wells (1000 cells/or indicated) in ultra-low-attachment 6-well plates (Corning Life Sciences, lot: 08,817,006) supplemented with 2 mL DMEM/F12 medium (Glico) and 10 mM HEPES, recombinant human epidermal growth factor (EGF) (Invitrogen) at a concentration of 20 ng/mL, and human recombinant basic fibroblast growth factor (Invitrogen) at a concentration of 10 ng/mL. After 3 to 4 weeks, each well was inspected with a light microscope, and the spherical colonies were counted in 5 random fields of view.
Immunofluorescent staining
Immunofluorescence staining of organoids, tumor tissues and cell lines was performed using method we previously published [5]. Fluorescence staining was imaged on a Zeiss LSM780 confocal microscope. Antibodies used included Anti-CD133 (Prominin-1) Monoclonal Antibody (1:200, Invitrogen, 17-1331-81), PARP-1 antibody (F-2) (1: 500, Santa Cruz, sc-8007), METTL3 Polyclonal Antibody (1:1000, Anti-gamma H2A.X (phospho S139) antibody, Abcam, ab2893).
The patient-derived organoid xenograft (PDOX) mouse model
In vivo experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) regulations and approved by the clinical research and animal experiment ethics committee of the FAHSYSU. To study the tumorigenic ability and drug resistance of CD133+ cell subset (previously screened by flow cytometry), we inoculated 100,000 CD133+ and CD133− cells embedded in Matrigel (BD, 354,230) into BALB/C naked mice body. After 25 days, 6 organoid transplanted tumor (CD133+ and CD133−) mice received oxaliplatin (Selleckchem, s1224) treatment at a dose of 5 mg/Kg twice a week for 4 weeks. The remaining 6 mice were injected intraperitoneally with PBS. After 4 weeks, the tumor-bearing BALB/C nude mice were sacrificed, and the tumors were harvested for measuring the size and weight.
To study the influence of PARP1 and METTL3 expressions on drug resistance, 100,000 pLKO and PARP1-sh1 (PT1 and PT2) cells were mixed with matrix gel and inoculate into BALB/C nude mice, respectively. After 25 days, 6 organoid transplanted tumor mice were treated with oxaliplatin (Sellekchem, s1224) twice a week for 4 weeks at a dose of 5 mg/kg. After 4 weeks, the tumor-bearing BALB/C nude mice were sacrificed, and the tumors were taken out for measuring the size and weight.
To study the influence of PARP1 and METTL3 expression on drug resistance, we used Matrigel (BD, 354,230) to inoculate 200,000 pLKO and PARP1-sh1 (PT3 and PT4) cells into BALB/C nude mice. After 25 days of implantation, 6 organ transplanted tumor mice received oxaliplatin (Selleckchem, s1224) at a dose of 5 mg/kg, twice a week for 4 weeks. After 4 weeks, the tumor-bearing BALB/C nude mice were sacrificed, and the tumors were excised for measuring the size and weight.
The size and weight of the tumors were measured every three days. One month later, the mice were sacrificed and the tumor tissues were processed for histological examination. Tumor volume (mm3) = 0.5 × Width2 × Length. All animal experiments were carried out in accordance with the health guidelines, and the protocol was formulated by the Animal Protection and Use Committee of Sun Yat-sen University. When the mice reached the endpoint, the tumors were photographed and weighed.
RNA isolation, microarray and Illumina sequencing methods
Total RNA was extracted from tissue samples, and the concentration and purity of RNA were detected with Nanodrop2000. Agarose gel electrophoresis was used to detect RNA integrity, and Agilent 2100 was used to determine the RIN value. The construction of a single library requires that the total amount of RNA is not less than 5 μg, the concentration is 0.5 μg ≥ 200 ng/μL, and the OD260/280 is between 1.8 and 2.2. The mRNA capture and library preparation were completed using the KAPA-mRNA-HyperPrep kit (Roche) and the advanced sequencing equipment of Shanghai Origingene Biomedical Technology Co., LTD. The biological triple library was sequenced on the facility’s Illumina Truseq TM RNA sample preparation kit platform, and each sample produced an average of 25 million single-ended reads of 75 bp. Align the high-quality sequence after quality control with the designated reference genome. First compare the PDOX sample with the mouse reference genome. After removing the mouse-related data, it is compared with the human reference genome. The human reference genome comes from the Ensembl database, the genome version is GRCh38, and the gene annotation information is Ensemble92. Before comparison, Cutadapt (version 1.9.1) was used to perform quality control and adaptor trimming of the original readings. Use annotation version 86, use RSEM 1.3.0 and STAR 2.5.2 to sort the read sequence of human genome GRCh38, and calculate the subsequent gene level. In version 3.6.1 of the R package, the DESeq2 package (version 1.24.0) is used for the standardization of raw count data and differential expression analysis. Perform regularized logarithmic transformation on the rlog function.
Patient information in public databases
Transcriptome data of patients with gastric adenocarcinoma confirmed by pathology can be downloaded from the TCGA website (https://portal.gdc.cancer.gov/) in June 2020, including data on 416 patients with gastric adenocarcinoma and general information on the corresponding cases. Excluding data that did not list survival time, 416 cases of gastric cancer and 33 cases of adjacent tissues. Inclusion criteria: (a) Diagnosis age ≥ 8 years (b) Tumor location: stomach (3) Clear pathological cases. The exclusion criteria are as follows: (a) Multiple tumors (b) Carcinoma in situ (c) Incomplete follow-up data (d) Death within 30 days. The stem cell index is an index that describes the similarity between tumor cells and stem cells. mRNAsi is an index calculated based on expression data. Through machine learning, stem cell epigenetic regulation of the expression of related genes is trained to obtain the EREG-mRNAsi index. The index is between 0 and 1. The closer to 1, the lower the degree of cell differentiation and the stronger the characteristics of stem cells. Malta et al. used a computer to calculate the TCGA gastric cancer stem cell index [30].
Bioinformatics
GSEA is performed by the software (GSEA V4.0.3). We mainly compared the enrichment analysis of CD133 positive and negative cells from TCGA gastric cancer dataset and our data.
Screening of differently expressed genes (DEGs), are normalized by quantiles and are normally distributed [31]. In this study, the R software package limma program v3.28.14 was used to analyze the differential genes in the gene expression data. The mRNA satisfies P < 0.01, false discovery rate (FDR) < 0.01, |log2 fold change (FC)|> 1.5, where P < 0.05 indicates that the hypothesis test is statistically significant. Use R language to construct heat maps and volcano maps of differential genes for visual comparison. PPI network construction of key module gene, use the string data set is an online biological resource that can decode the interaction between protein and protein to obtain the actual precise function of the protein [32]. P < 0.05 was considered as statistically significant. For the scRNA-seq data analysis, we use single-cell sequencing data of early gastric cancer tissue which was obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134520). The scRNA-seq data analysis performed in R version 4.0.3 was as follows: (1) Seurat R package was used to convert scRNA-seq data into Seurat objects [33]. (2) After data quality control, the “FindVariableFeatures” function was used to find the first 1,500 highly variable genes; (3) Based on these 1500 genes, principal component analysis (PCA) and Uniform Manifolds Approximation and Projection (UMAP) analysis was performed to find the top 10 scRNA seq data; (4) Identification of cell type specific activation pathways (CTSAPs). A single cell based on the PAS matrix, multiple non-parametric statistical methods and fold-change analysis can be used to identify CTSAPs. These distinguishable cell populations are the cell type of interest and other cell types composed of the case control group, and CTSAPs with statistically significant activation in different cell types were identified [34]. Cells that met one of the following conditions were excluded: (1) the number of expressed genes was less than 101 or more than 6000; (2) 10% or more of its unique molecular identifiers (UMIs) were mapped to mitochondria. The m6A public data from RMVar M6a methylation database can be found in this website: https://rmvar.renlab.org/result_level_1.html?input_type=Gene&input_text=parp1.
Statistical analysis
The images and graphs shown represent several experiments repeated on different individuals at different times. Each experiment is repeated independently. All statistical data are carried out using SPSS and R software. The Student’s T test was used for the comparison between two groups and one-way analysis of variance (ANOVA) was employed for the comparison of multiple groups. The results were presented as Mean ± SD, and P < 0.05 was considered as statistically significant.
Results
Oxaliplatin-resistant primary human gastric cancer and patient-derived organoids (PDOs) enriched for CD133+ cells that show highly stem-like properties
To find the cause of chemotherapy resistance, four PDOs (PT1, PT2, PT3, PT4) were used. PT1 and PT2 were obtained from patients whose GC recurred after postoperative chemotherapy. By its turn, PT3 and PT4 were from patients without recurrence after postoperative chemotherapy (PDOs picture see Fig. S1A–E). In a viability assay, PT1 (IC50 = 32.06 µmol/L) and PT2 (IC50 = 61.66 µmol/L) were found to be more resistant to oxaliplatin as compared to PT3 (IC50 = 3.26 µmol/L) and PT4 (IC50 = 1.55 µmol/L) (Fig. 1A). The four PDOs were then sequenced. From the result, it was found that CD133 expression was higher in the oxaliplatin resistant PT1 and PT2 than the non-resistant PT3 and PT4 (Fig. 1B). However, there were no significant differences in the expressions of CD44, LGR5, ALDH1, EPCAM, and SOX2 among the four groups. Next, flow analysis allowed investigation of ratio of CD133+ cells in the organoids. The results showed that the ratios of CD133+ cell subpopulation in organoids PT1 (14.5%) and PT2 (28.0%) were higher compared to that for PT3 (3.89%) and PT4 (0.31%) (Fig. 1C). Oxaliplatin-resistant cell lines showed stronger ability in sphere formation as compared to non-resistant cell lines (Fig. S2A, B). Not only that, the expression of CD133 of the oxaliplatin resistant cells lines were higher than that of the wild-type cell line (Fig. S2C). The expression of LGR5 in the AGS and MKN74 oxaliplatin resistant strains was higher than that of the wild-type cell line, while there were no significant differences in CD44 expression (Fig. S2D,E). The CD133+ cell subpopulation of the resistant strain was significantly higher than the wild-type cell line (Fig. S2F), suggesting the importance of CD133+ subpopulation in the formation of oxaliplatin resistance. To verify the drug resistance of CD133+ cells and their CSC potential, CD133+ and CD133− cells from the organoids of PT1 and PT2 patients were sorted by flow analysis then cultured, and then implanted under the skin of BALB/C NUDE mice. After studying the sorted organoid spheroidization and in vivo tumorigenesis situations, it was found that CD133+ cells have a strong spheroidization ability in in vitro organoid formation and in vivo tumorigenesis (Fig. 1D–I). The tolerance of CD133+ cells to oxaliplatin was also significantly higher than that of CD133- cells (Fig. 1D–I). Flow cytometry experiments on the spheroidization of CD133+ and CD133− oxaliplatin resistant cell lines showed that the ability of CD133+ cells to spheroidize was significantly higher than that observed for CD133- cells (Fig. S3A–D). However, the expression of LGR5 and CD44 of CD133+ cells was not significantly different from that of negative cells (Fig. S2C, D). Through in vitro experiments of organoids and oxaliplatin resistant cell lines, and in vivo tumorigenesis experiments, CD133+ cells subset was proven to have obvious CSC characteristics and oxaliplatin tolerance. We used a machine-learning methodology to verify oncogenic dedifferentiation-related stemness characteristics against stemness indices calculated from TCGA patients to analyze gastric CSC markers from Fig. S4A, it can be observed that the expression of CD133 in patients with high stemness index was higher than that in patients with low stemness index. The enrichment analysis also showed that patients with high stemness index were enriched in the JAK STAT signaling pathway, MAPK signaling pathway, TGF-β signaling pathway, and WNT signaling pathway (Fig. S4 B–E). This indicates that the stemness index is a reliable indicator, and also confirmed that CD133 indeed can be used as an important marker for gastric CSCs.
Fig. 1.
CD133+ cells are main cells responsible for oxaliplatin-resistance in human primary gastric cancer-patient derived organoids. A Compared with PT3 and PT4, PT1 and PT2 have obvious tolerance to oxaliplatin. The abscissa represents the concentration of oxaliplatin, and the ordinate represents cell viability. B The statistical significance of the fold change in gene expression (x-axis, log2 transformation) between oxaliplatin-resistant (PT1 and PT2) and sensitive (PT3 and PT4) populations (n = 3) determined by RNA sequencing (y-axis, log2 transformation) volcano map. FC, fold change; padj, adjust the p value for false discovery rate. Red dots indicate differentially expressed genes with padj < 0.05 of PT1 and PT2. Green dots indicate differentially expressed genes with padj < 0.05 of PT3 and PT4. C Flow cytometry analysis and comparison of CD133+ cells in PT1, PT2, PT3, and PT4 organoids. Isotype is no antibody control. The abscissa represents CD133, and the ordinate represents cells. D Representative images of organoid culture based on cells sorted by flow cytometry. The scale represents 200 μm. E Number of organoids in (D) after organoid formation. F Size of organoids in (D) after organoid formation. G A representative image of size change of tumor post BALB/C NUDE mice implantation with a dose of Oxaliplatin with its vehicle. The scale represents 1 cm. H Statistics of tumor volume after tumorigenesis in (G). I Statistics of tumor quality after tumorigenesis in (G). PT1 gastric cancer (GC) patient 1. PT2 GC patient 2. PT3 GC patient 3. PT4 GC patient 4. OXA Oxaliplatin. CON Solvent group. *p < 0.05, **p < 0.01, ***p < 0.001
The CD133 expression in individual groups with the GSE134520 single cell sequencing data set was analyzed. From Figure S5A and B, it can be observed that the GC tissue was mainly composed of 7 groups of cells (C1–7). It was also found that C1 and C4 mainly participated in the metabolism, C2 was mainly related to oxidative stress, C3 was primarily related with ribosome, C5 was mainly related with cell cycle and DNA damage repair ability, C6 was associated to the cellular proliferation, and C7 was associated with immunology (Fig. S5C). In terms of CD133 expression of each group, C5 and C6 showed the highest CD133+ ratios (Fig. S5D) suggesting that CD133+ cells not only have strong proliferation and self-renewal ability (TGF beta signaling pathway), but also have significant correlation with DNA repair (Fig. S5E).
PARP1 is the central gene of CD133+ cells responsible for oxaliplatin resistance in gastric cancer
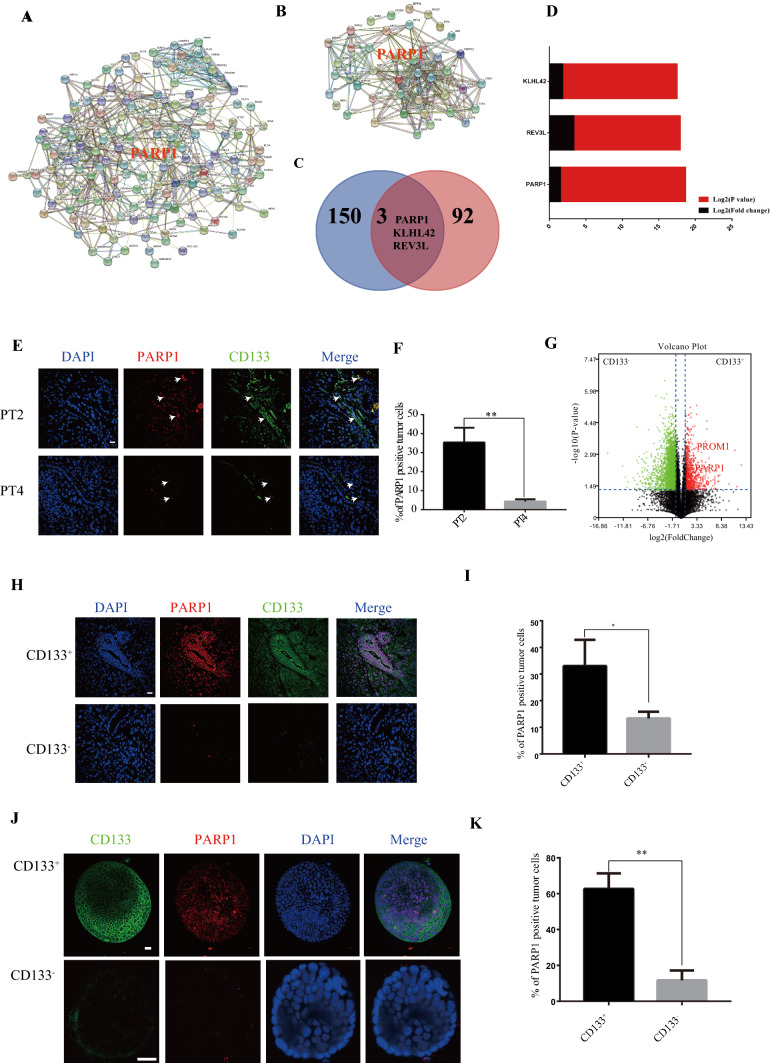
Through the above experiments, it was determined that CD133+ gastric cancer cells had the characteristics of CSCs and could mediate oxaliplatin resistance. To find out the mechanism of the drug resistance in CD133+ CSCs, we first compared the differential genes expressions between clinical tissues from oxaliplatin resistance patients (PT1 and PT2) and chemotherapy sensitive patients (PT3 and PT4), then compared the differential genes expression between CD133+ and CD133− cells after in vivo tumorigenesis in mice. After that, the protein interaction network was used to further screen the core functional genes of tissue from oxaliplatin resistance patients (PT1 and PT2) and chemotherapy sensitive patients (PT3 and PT4) (Fig. 2A), and the core functional genes of CD133+ and CD133− cells after in vivo tumorigenesis in mice (Fig. 2B). There were three common core functional genes, namely PARP1, KLHL42 and REV3L (Fig. 2C). Finally, PARP1 was identify to be the gene with the most significant difference amongst the common core genes by comparison (Fig. 2D).
Fig. 2.
PARP1 is the central gene of CD133+ cells responsible for oxaliplatin resistance in gastric cancer. A Compared with PT3 and PT4 tumors, the protein interaction network diagram of STING database of mRNA differential expression in PT1 and PT2 tumors. The edges represent protein–protein associations. Cambridge Blue: from the curatorial database. Violet: measured experimentally. Green: The Jean community. Red: gene fusion. Blue: Co-occurrence of genes. Reseda: text mining. Black: Common expression. Clove: protein homology. B STING database CD133+ and CD133− tumor mRNA differentially expressed protein interaction network diagram. The edges represent protein–protein associations. Cambridge Blue: from the curatorial database. Violet: measured experimentally. Green: The Jean community. Red: gene fusion. Blue: co-occurrence of genes. Reseda: text mining. Black: common expression. Clove: protein homology. C There are 3 common core genes in (A) and (B), including PARP1, KLHL42 and REV3L. D Comparison of LOG2 (P value) and LOG2 (Fold change) of two gene sets in core genes of (C). E CD133 and PARP1 immunofluorescence staining of PT2 and PT4 tumor. Scale bar: 20 μm. F Proportion of PARP1+ cells in tumor in (E). G RNA-seq (n = 3) measures the fold change (y-axis, log2 transformation) and statistical significance (y-axis, log2 transformation) of gene expression (x-axis, log2 transformation) between CD133+ and CD133− cells populations. FC: fold change. Padj: adjust the p value for false discovery rate. The red dots represent the significantly differentially expressed genes of CD133− with padj < 0.05. Green dots indicate differentially expressed genes with CD133+ padj < 0.05. PROM1 (log2FC = 5.40, P = 0.0047), PARP1 (log2FC = 1.10, P = 0.000589). H Representative images of PARP1 and CD133 levels stained by immunofluorescence in CD133+ and CD133− tumors. The scale represents 20 μm. I Proportion of PARP1+ cells in tumor in (H). J Representative images of PARP1 and CD133 levels stained by immunofluorescence CD133+ and CD133− organoids. The scale represents 20 μm. K Proportion of PARP1 + cells in organoids in (J). *p < 0.05, **p < 0.01, ***p < 0.001
The number of CD133+ and PARP1 positive cells in PT2 tumor tissue (oxaliplatin resistant) was higher than that of PT4 tumor tissue (oxaliplatin sensitive) through immunofluorescence staining (Fig. 2E, Fig. S6A). Moreover, from their localization, CD133+ cells significantly overexpressed PARP1 (Fig. 2E, F). The sequencing data of CD133+ and CD133− cells after tumorigenesis in organoids, showed that the expression of PARP1 in CD133+ cells significantly higher than that in CD133− cells (Fig. 2G). Additionally, it was also confirmed that the expression of PARP1 in CD133+ cells was significantly higher than the CD133− cells in PDOX tumor immunofluorescence (Fig. 2H, I, Fig. S6B) and organoid immunofluorescence after tumorigenesis in BALB/C NUDE mice (Fig. 2J, K, Fig. S6C). The expression of PARP1 in CD133+ cells in oxaliplatin resistance GC strains was also significantly higher than that in CD133- cells, and the expression of PARP1 in oxaliplatin resistance GC strains was also significantly higher than that of wild-type cell lines (Fig. S7 A–C). Finally, through the analysis of the TCGA database, PARP1 was found to be highly expressed in patients with high stemness index (Fig. S4A).
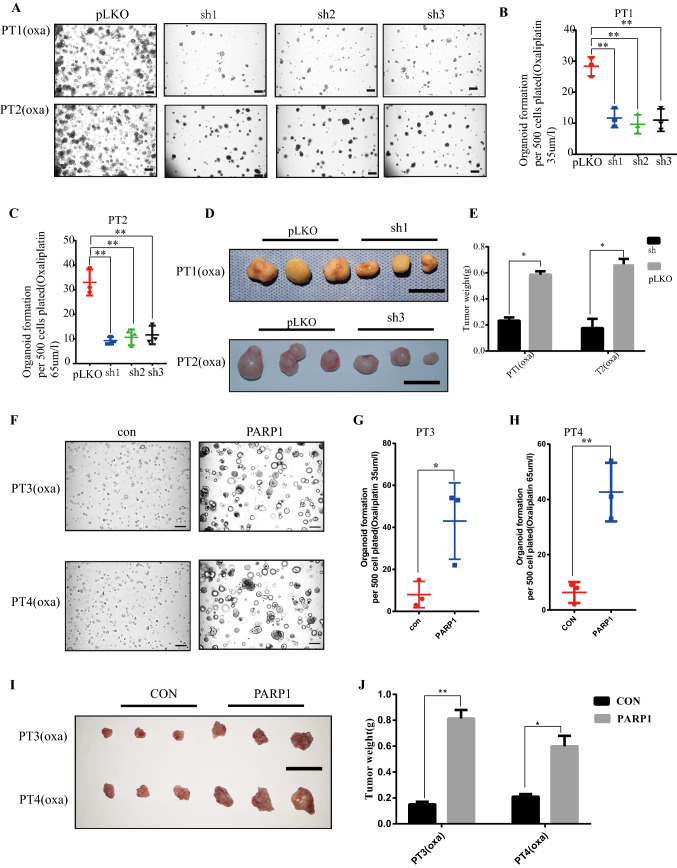
PARP1 is required for oxaliplatin resistance development
To verify the role of PARP1 in oxaliplatin resistance, PARP1 was knocked down in resistant organoids PT1 and PT2. Through in vitro and in vivo experiments, we found that the tolerance to oxaliplatin decreased significantly after PARP1 was knocked down (Fig. 3A–E, Fig. S8A–B). PARP1 was overexpressed in oxaliplatin sensitive organoids PT3 and PT4. It was found that the tolerance to oxaliplatin increased significantly after PARP1 was overexpressed (Fig. 3F–J, Fig. S8C). Furthermore, based on our analysis of GSE134520 single cell sequencing data set, it was found that CD133+ tumor stem cells were enriched in the Base Excision Repair (BER) pathway (Fig. S5E). To study whether PARP1 can mediate BER to repair DNA damage and thereby induce the occurrence of oxaliplatin resistance, the following analysis was conducted.
Fig. 3.
PARP1 is required for Oxaliplatin resistance development. (A) Organoid formation assay (organoid number) of PT1 and PT2 organoids treat with oxaliplatin following PARP1 knockdown using shRNAs 1–3 The scale represents 200 μm. (B, C) Number of organoids in A after organoid formation. PT1 organoids treat with oxaliplatin 35 μm/L. PT2 organoids treat with oxaliplatin 65 μm/L. (D) Representative image of the tumors. Scale bar, 1 cm. (E) The weight of the subcutaneous grafts at the end point after injection of 10,000 cells (n = 3 biologically independent animals). (F) Organoid formation assay (organoid number) of PT3 and PT4 organoids treat with oxaliplatin following PARP1 overexpression. The scale represents 200 μm. (G, H) Number of organoids in F after organoid formation. PT3 organoids treat with oxaliplatin 2 μm/L. PT4 organoids treat with oxaliplatin 4 μm/L. (I) Representative image of the tumors. Scale bar, 1 cm. (J) The weight of the subcutaneous grafts at the end point after injection of 10,000 cells (n = 3 biologically independent animals). * p < 0.05, ** p < 0.01, *** p < 0.001
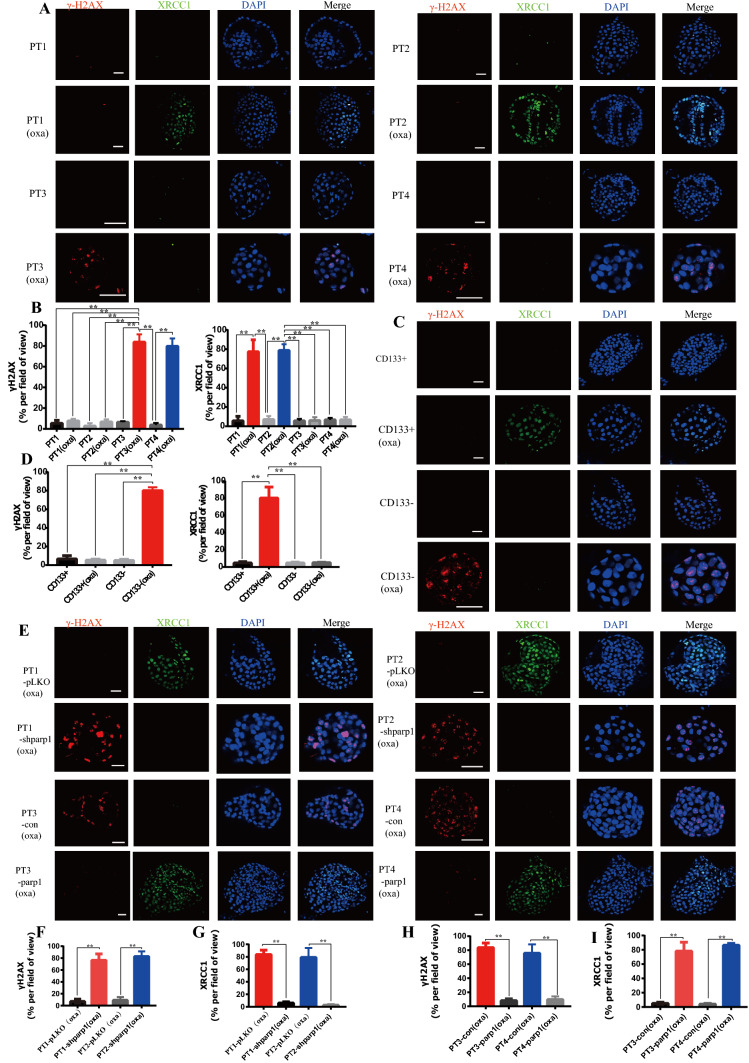
PARP1 can mediate oxaliplatin resistance through Base Excision Repair (BER)
Since BER is an important signaling pathway leading to oxaliplatin resistance, we studied the DNA damage caused by oxaliplatin and the expression changes in BER pathway marker XRCC1 in both oxaliplatin resistant and sensitive patients. DNA damage marker γH2AX was not significantly changed in oxaliplatin resistant PT1 and PT2 organoids with or without oxaliplatin treatment, but the level of γH2AX in oxaliplatin sensitive organoid PT3 and PT4 was significantly increased. This indicates that oxaliplatin resistant organoids have stronger ability to repair DNA damage compared to oxaliplatin sensitive ones (Fig. 4A, B). When oxaliplatin treatment was applied to oxaliplatin resistant organoid, the BER pathway marker XRCC1 was significantly increased compared to sensitive organoid. However, there was no significant change without oxaliplatin treatment (Fig. 4A, B). This indicates that BER plays a role in the repair of DNA damage during DNA damage process caused by oxaliplatin, leading to the survival of cells. Due to the weak BER pathway, sensitive organoids cannot effectively repair DNA damage, resulting in DNA damage and thereby leading to cell apoptosis. At the same time, it was also found that the CD133+ cells underwent DNA damage repair through BER pathway, thus leading to cell survival (Fig. 4C, D). These results indicated that CD133+ tumor stem cells mainly acted through BER pathway to acquired oxaliplatin resistance.
Fig. 4.
PARP1 can mediate oxaliplatin resistance through base excision repair. A Representative images of γ-H2AX and XRCC1 levels stained by immunofluorescence of PT1, PT1 OXA, PT2, PT2 OXA, PT3, PT3 OXA, PT4, PT4 OXA organoids. The scale represents 20 μm. B Proportion of γ-H2AX + and XRCC1 + cells in organoids in (A). C Representative images of γ-H2AX and XRCC1 levels stained by immunofluorescence CD133 ± and CD133 ± OXA organoids. The scale represents 20 μm. D Proportion of γ-H2AX + and XRCC1 + cells in organoids in (C). E Representative images of γ-H2AX and XRCC1 levels stained by immunofluorescence PT1 pLKO, PT1 sh-parp1, PT2 pLKO, PT2 sh-parp1, PT3 con, PT3 parp1, PT4 con, PT4 parp1 organoids. The scale represents 20 μm. F, G Proportion of γ-H2AX + cells and XRCC1 + in organoids in (E). * p < 0.05, ** p < 0.01, *** p < 0.001. H, I Proportion of γ-H2AX+ cells and XRCC1+ in PDX in (E)
Since PARP1 is an important gene affecting BER pathway, we knocked down PARP1 to see its influence on DNA damage repair and BER pathway. It was found that oxaliplatin resistant organoids were significantly less able to repair DNA damage after PARP1 knockdown. Additionally, PARP1 functioned by influencing BER pathway (Fig. 4E–G). However, after PARP1 was overexpressed in sensitive organoids, DNA damage repair can be activated by powerful BER effect (Fig. 4E, H, I). These results suggested that PARP1 could exert DNA damage repair by mediation of BER, leading to oxaliplatin resistance in GC organoids.
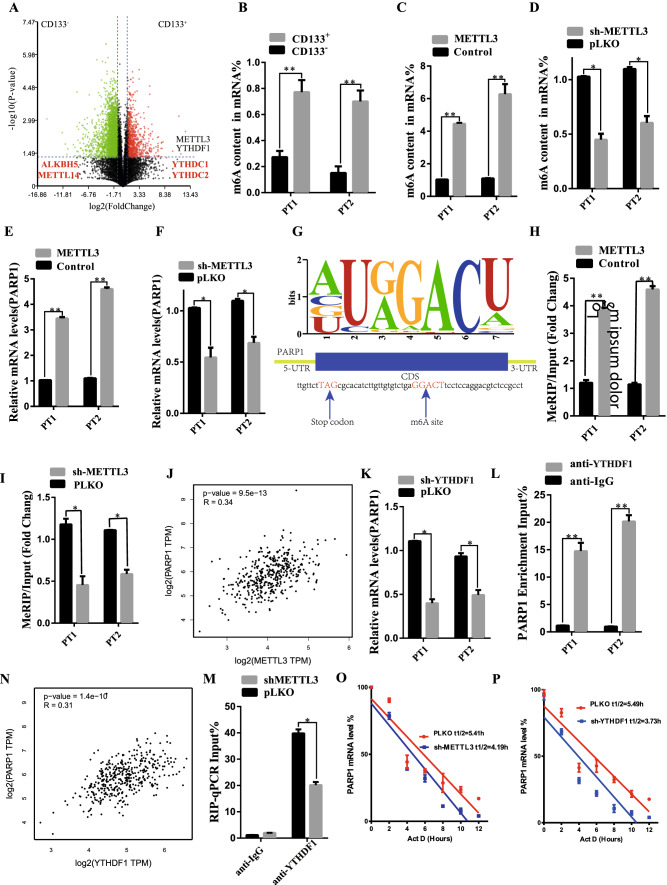
N6-methyladenosine METTL3 maintains PARP1 mRNA stability in CD133+ GC stem cells
PARP1 is an important factor in the function maintenance of CD133+ tumor stem cells, but by which mechanism CD133+ cells modulate PARP1 is still unknown. By studying the sequencing results of CD133+ and CD133− cells for enrichment analysis, it was found that CD133+ CSCs subpopulation was not only enriched in the powerful proliferative pathways (including the Wnt signaling pathway, BMP signaling pathway, Transforming growth factor beta-activated receptor activity and DNA damage repair pathway—base-excision repair). In addition to cellular response to DNA damage stimulus, CD133+ CSCs were also associated with RNA modification (mRNA binding, translation regulator activity, N6-methyladenosine-containing RNA binding) (Fig. S9A, B). Moreover, through differential gene analysis, it was found that the expression levels of METTL3 and YTHDF1 in CD133+ cells were significantly higher than those in CD133− cells (Fig. 5A). Moreover, the CD133+ cells showed stronger m6A modification function compared to the CD133- cells, via quantitative analysis of m6A content (Fig. 5B). We further studied whether CD133+ cells regulate m6A level through METTL. The results showed that METTL3 knockdown inhibited m6A content (Fig. 5C, Figure S9 C), while METTL3 overexpression increased m6A content (Fig. 5D, Figure S9 C). RT-PCR results showed that METTL3 knockdown decreased PARP1 mRNA expression (Fig. 5E), while METTL3 overexpression promoted PARP1 mRNA expression (Fig. 5F). By performing m6A sequencing, we identified the m6A motif of METTL3 and the m6A site of PARP1 mRNA in coding sequence (CDS) (Fig. 5G). MeRIP-qPCR results showed that METTL3 knockdown decreased PARP1 mRNA level precipitated by m6A antibody (Fig. 5H, Fig. S10A), while METTL3 overexpression increased PARP1 mRNA level (Fig. 5I, Fig. S10B). Spearman rank correlation coefficient (GEPIA, http://gepia.cancer-pku.cn/) was used for correlation analysis, indicating that METTL3 was positively correlated with PARP1 expression in GC tissue samples (Fig. 5J). Through RMVar methylation database, we also found that PARP1 and we had related M6A methylation modification sites in HEK293T cells (see supplement Table 2). In conclusion, these findings support that METTL3 can act on PARP1 through m6A modification to enable the stable expression and function of PARP1.
Fig. 5.
N6-Methyladenosine METTL3 maintains expression of PARP1 in CD133+ gastric cancer stem cells. A RNA-seq measures the statistical significance (y-axis, log2 transformation) volcano plot of the fold change of gene expression (x-axis, log2 transformation) between CD133+ and CD133− cells populations. FC, fold change; padj, adjust the p value for false discovery rate. Red dots indicate significantly differentially expressed genes of CD133+ with padj < 0.05. The green dots indicate a significant difference between padj < 0.05 and CD133- gene expression. The m6A genes of METTL3 and YTDHF1 are highly expressed in CD133+ . B m6A quantitative analysis showed the percentage of m6A content in CD133+ and CD133−. C m6A quantitative analysis showed the percentage of m6A content in PT1 and PT2 organoid transfected with METTL3 overexpression. D m6A quantitative analysis showed the percentage of m6A content in PT1 and PT2 organoid transfected with METTL3 knockdown. E, F RT-PCR indicated the PARP1 mRNA in the transfection of METTL3 knockdown or METTL3 Overexpression. G Schematic diagram demonstrated the m6A motif of METTL3 and the m6A site in the 3’-UTR of PARP1 mRNA (near stop codon). H, I MeRIP-qPCR indicated the PARP1 mRNA enrichment precipitated by m6A antibody. J Correlation analysis by Spearman’s rank correlation coefficient (GEPIA, http://gepia.cancer-pku.cn/) showed the correlation within METTL3 and PARP1 in the GC tissue specimens. K RT-PCR demonstrated the PARP1 mRNA expression in PT1 and PT2 organoid transfected with YTHDF1 Knock down. L RNA immunoprecipitation (RIP)-PCR indicated the direct binding within YTHDF1 and PARP1 mRNA. M Correlation analysis by Spearman’s rank correlation coefficient showed the positive correlation within PARP1 expression and YTHDF1 in the GC tissue specimens. N RIP-qPCR indicated the enrichment of HK2 mRNA in PT1 and PT2 organoid, using anti-YTHDF1 antibody, with METTL3 knockdown. O, P RNA decay rate followed by RT-PCR assay demonstrated the PARP1 mRNA half-lives upon the METTL3 knockdown and YTHDF1 knockdown. Data were detected at indicated timepoint with actinomycin D (Act D, 5 μg/mL) treatment. *p < 0.05, **p < 0.01, ***p < 0.001
The results of existing studies indicated that PARP1 is the target protein of METTL3. However, the underlying mechanism by which METTL3 promotes PARP1 expression remains unclear. The RT-PCR analysis showed that YTHDF1 knockdown reduced PARP1 mRNA expression, suggesting that YTHDF1 may be involved in the epigenetic regulation of PARP1 (Fig. 5K, Fig. S9D). By its turn, the RNA immunoprecipitation (RIP)-PCR studies showed that YTHDF1 directly bound to PARP1 mRNA (Fig. 5L, Fig. S10C). Spearman's rank Correlation coefficient (GEPIA, http://gepia.cancer-pku.cn/) showed a positive Correlation between the expression of YTHDF1 and PARP1 in GC tissue samples (Fig. 5M). In addition, RNA immunoprecipitation and qPCR showed that METTL3 knockdown reduced the direct interaction between YTHDF1 and PARP1 mRNA (Fig. 5N). RNA decay rate analysis showed that the PARP1 mRNA half-life was significantly shortened after METTL3 knockdown and YTHDF1 silencing (Fig. 5O, P and Fig. S10D, E, F). Taken together, these findings confirm that methylated PARP1 mRNA is recognized by YTHDF1 and that METTL3/YTHDF1 may enhance its mRNA stability.
N6-Methyladenosine METTL3 maintains the function of PARP1 in oxaliplatin resistance
Above results showed that CD133 tumor stem cells can regulate the expression of PARP1 through RNA N 6-methyladenosine methyltransferase METTL3. However, the effect of METTL3 on oxaliplatin resistance is not clear. To this end, we will observe the effect of oxaliplatin on oxaliplatin-resistant organoids by knockdown of METTL3. Herein, we found that knockdown of METTL3 can significantly reduce the resistance ability of oxaliplatin-resistant organoids to oxaliplatin (Fig. 6A–E). Additionally, through the immunofluorescence staining results, it was found that METTL3 can affect the expressions of PARP1 and DNA repair marker γ-H2AX, indicating that METTL3 can affect the ability of oxaliplatin-resistant organoids to repair damaged DNA (Fig. 6F–I and Figure S6 D). Through the verification of CD133+ and CD133− GC cell lines cells, as well as oxaliplatin-resistant and sensitive strains, it was found that METTL3 was significantly increased in CD133+ tumor stem cells and oxaliplatin-resistant strains (Fig. S7C–F).
Fig. 6.
METTL3 knockdown significantly reduced oxaliplatin tolerance in drug-resistant GC. A Organoid formation assay (organoid number) of PT1 and PT2 organoids following METTL3 knockdown using shRNAs 1–3 The scale represents 200 μm. B, C Number of organoids in (A) after organoid formation. PT1 organoids treat with oxaliplatin 35 μm/L. PT2 organoids treat with oxaliplatin 65 μm/L. D Representative image of the tumors. Scale bar, 1 cm. E The weight of the subcutaneous grafts at the end point after injection of 10,000 cells (n = 3 biologically independent animals). F Representative images of γ-H2AX, PARP1, METTL3 levels stained by immunofluorescence METTL3 knockdown organoids. The scale represents 20 μm. G Proportion of γ-H2AX, PARP1, METTL3 + cells in organoids in (F). H Representative images of γ-H2AX, PARP1, METTL3 levels stained by immunofluorescence METTL3 knockdown tumor. The scale represents 200 μm. I Proportion of γ-H2AX, PARP1, METTL3 + cells in tumor in (H). *p < 0.05, **p < 0.01, ***p < 0.001
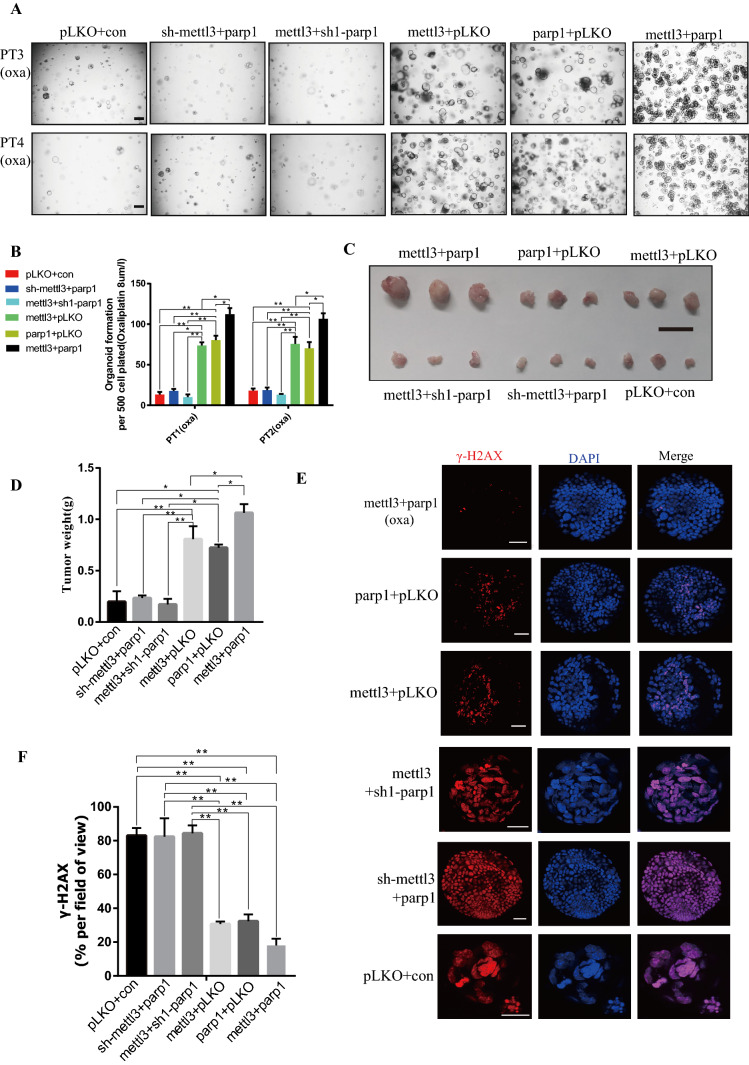
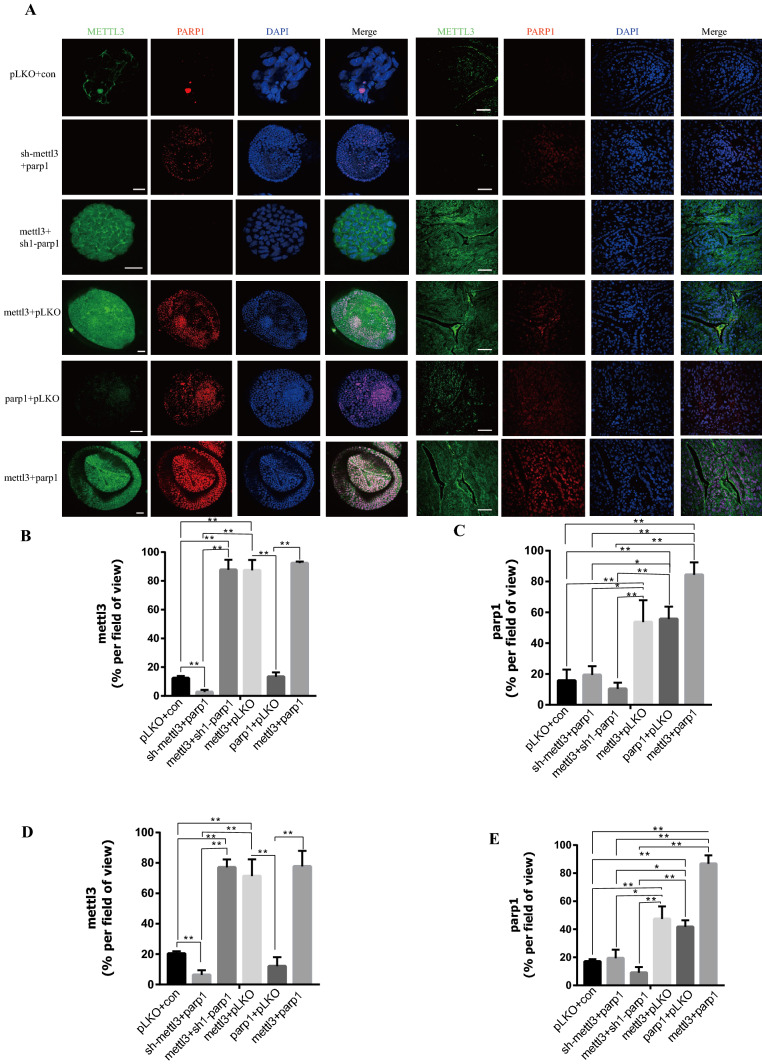
To study whether PARP1 plays a role through the regulation of METTL3, METTL3 knockdown combined with PARP1 overexpression and PARP1 knockdown combined with METTL3 overexpression in oxaliplatin-sensitive organoids (PT3 and PT4) were performed. In in vitro and in vivo experiments, it was found that the effects of oxaliplatin on them were not significantly different from those in the control group (Fig. 7A–E). Moreover, there was no significant change in their effect on DNA damage (Fig. 7E–F), suggesting that they work through interdependence. According to immunofluorescence and WB, there was no significant difference in the expression of PARP1 among the three groups. Although PARP1 was overexpressed, the expression of PARP1 was affected due to the loss of METTL3 function (Fig. 8A–E, Fig. S9E). This indicates that PARP1 can be stabilized and expressed only under the condition of normal function of METTL3. Next, overexpressions of METTL3 and PARP1 in oxaliplatin-sensitive organoids were conducted (PT3 and PT4), respectively. Results showed that the tolerance of sensitive organoids to oxaliplatin was significantly improved after overexpressions of PARP1 of METTL3 (Fig. 7A–E). Oxaliplatin sensitive organoids after PARP1 and METTL3 overexpression showed significantly higher DNA repair ability than that of the control group (Fig. 7E–G). Moreover, through immunofluorescence and WB, it was observed that the overexpression of METTL3 can significantly affect the expression of PARP1 (Fig. 8A–E, Fig. S9F). This result suggests that METTL3 can play a role by stabilizing and promoting the expression of PARP1. Finally, through the simultaneous overexpression of METTL3 and PARP1, we found that their tolerance to oxaliplatin was significantly improved (Fig. 7A–E). According to the above research results, we can know that METTL3 plays an important role in maintaining and stabilizing the expression of PARP1 (see Fig. 9).
Fig. 7.
N6-Methyladenosine METTL3 maintains the function of PARP1 in oxaliplatin resistance. A Organoid formation assay (organoid number) of PT3 and PT4 organoids following METTL3 knockdown and PARP1 overexpression, PARP1 knockdown and METTL3 overexpression, METTL3 overexpression, PARP1 overexpression, METTL3 overexpression and PARP1 overexpression. The scale represents 200 μm. B Number of organoids in (A) after organoid formation. PT3 organoids treat with oxaliplatin 8 μmol/L. PT2 organoids treat with oxaliplatin 8 μmol/L. C Representative image of the tumors. Scale bar, 1 cm. D The weight of the subcutaneous grafts at the end point after injection of 20,000 cells (n = 3 biologically independent animals). E Representative images of γ-H2AX levels stained by immunofluorescence of organoids of METTL3 and PARP1 overexpressions, PARP1 overexpression, METTL3 overexpression, METTL3 overexpression and PARP1 knockdown, METTL3 knockdown and PARP1 overexpression. The scale represents 20 μm. F Proportion of γ-H2AX + cells in organoids in (E). * p < 0.05, ** p < 0.01, *** p < 0.001
Fig. 8.
N6-Methyladenosine METTL3 maintains the expression of PARP1. A Representative images of PARP1 and METTL3 levels stained by immunofluorescence of organoids of METTL3 overexpression, PARP1 overexpression, METTL3 overexpression and PARP1 overexpression in organoid and tumor. The scale represents 20 μm of organoid. The scale represents 200 μm of tumor. B Proportion of METTL3 + cells in organoids in (A). C Proportion of PARP1 + cells in organoids in (A). D Proportion of METTL3 + cells in tumor in (A). E Proportion of PARP1 + cells in tumor in (D). * p < 0.05, ** p < 0.01, *** p < 0.001
Fig. 9.
The schematic model of METTL3 in regulating PARP1 mediated oxaliplatin resistance through DNA damage repair
Discussion
Platinum chemotherapy drugs play an important role in the treatment of advanced gastric cancer [35]. But unfortunately, drug resistance of platinum chemotherapeutic drugs is still a challenging problem [36]. It is now found that tumors are composed of different subsets of cells, and CSCs are important cells for drug resistance, proliferation and differentiation of tumors [37]. Through our study, we found that CD133+ CSCs are an important subset in oxaliplatin resistance of GC. However, how CD133+ CSCs mediate oxaliplatin resistance has not been studied. Through sequencing, we found that CD133+ CSCs have a strong ability of DNA damage repair and RNA modification. Oxaliplatin can kill tumor cells mainly by mediating DNA damage of tumor cells [38]. Many studies have found that CSCs have a strong ability to repair DNA damage. Park et al. studied the effect of CSCs on radiotherapy tolerance and found that CSCs can play a powerful role in DNA damage repair by mediating the JAK2/STAT3 pathway, leading to the tolerance to radiotherapy [39]. In our study, it was found that CD133+ CSCs can mediate oxaliplatin resistance by expressing PARP1. The PARP protein family mediates the PARylation of the translated substrate proteins involved in transcription and DNA damage repair, and PARP1 is a particularly important protein for detecting single-stranded DNA breaks (SSBs) [40]. A recent study showed that PARP1 can condenses DNA via loop stabilization [41]. The increased expression of PARP1 is also an important factor that mediates the drug resistance of tumor chemotherapy, especially the drug resistance related to DNA loss [42]. Hu et al. studied the mechanism of chemotherapeutic drug resistance and found that PARP1 induced the degradation of BRD7, leading to the resistance of cancer cells to DNA damage agents [43]. Avitabile et al. found that PARP1 was one of the main DNA damage sensors involved in DNA repair system and genomic stability, and PARP1 expression is closely related to chemotherapy resistance of neuroblastoma [44]. BER is a very important pathway leading to oxaliplatin resistance [45]. The above-mentioned studies showed that PARP1 was the main target gene that mediates BER pathway. Ronson et al. found that PARP1 repaired the alkylation DNA base damage through BER redundancy [46]. Reynolds et al. confirmed the repair of SSBS and purinyl damage was through BER pathway and requires the participation of XRCC1 and PARP1 [47]. Our study revealed that CD133+ GC CSCs could affect the BER pathway by expressing PARP1, thus leading to the occurrence of DNA damage repair and subsequent oxaliplatin resistance.
In this study, we found that CD133+ CSCs can mediate a strong DNA damage repair ability through the action of PARP1. However, in addition to the ability to repair DNA damage, our sequencing data also showed that CD133+ CSCs were able to modify RNA through the action of m6A. m6A modification is the methylation of the sixth nitrogen site of mRNA adenosine. The modification of m6A on mRNA is dynamically reversible and is operated by methyltransferase or methyltransferase [48]. Mettl3-, Mettl14- and WTAP mediated mRNA m6A modification can affect mRNA stability and expression [48]. Our study found that CD133+ CSCs can upregulate METTL3 to play a role in regulating m6A. Zhang et al. found that tumor stem cells can regulate and stabilize the expression of miR-25-3p by upregulating METTL3 to mediate m6A methylation, thereby causing the progression of pancreatic cancer [49]. Zhang et al. studied the effect of CSCs on tumor progression and found that THDF2 regulates OCT4 expression through m6A RNA methylation and promotes HCC stem cell phenotype and tumor metastasis [22]. Hao et al. found that m6A-YTHDF1-mediated upregulation of TRIM29 promoted the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells [50]. The correlation between M6A methylation and drug resistance has also been reported in relevant literature, which further indicates that M6A plays an important role in regulating stem cells and drug resistance [51, 52]. Our study also found that CD133+ CSCs had stronger m6A methylation function, and mediated the drug resistance of tumor cells to oxaliplatin through the expression stability and transcription of PARP1 by METTL3.
In conclusion, using GC patient derived oxaliplatin resistant and sensitive organoid models, we found that CD133+ CSCs acquired oxaliplatin resistance by regulated PARP1 at the RNA level via m6A modification through METTL3. This METTL3 mediated m6A modification led to the stabilization of PARP1 expression and subsequent strong DNA damage repair which finally mediated the occurrence of oxaliplatin resistance. These findings significantly contribute to our understanding of the mechanism of oxaliplatin-resistance in GC and other cancers.
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1. Verifying the drug resistance level in GC resistant strains. (A) Representative images of colony formation of SNU719, MKN74 and AGS resistant strains and their corresponding wild-type cell lines in the same dose of oxaliplatin. (B) Comparison of the number of colonies in (A). (C-E) Indicate the comparison of cell viability between AGS, SNU719 and MNK74 Oxaliplatin resistance strains and wild-type cell strains under the effects of oxaliplatin respectively. OXA: Oxaliplatin. OLP: Olaparib. CON: Control group. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001 (PDF 1686 KB)
Figure S2. Oxaliplatin-resistant human gastric cancer cell lines enrich for CD133+ cells. (A) Representative image of the comparison of the spheroidizing ability of SNU719, MKN74, AGS-resistant strains and their wild-type cell lines. (B) Proportion of spheroids in (A). (C-E) RT-qPCR was used to compare the mRNA expression levels of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines LGR5, CD133, and CD44 respectively. (F) Flow cytometric analysis and comparison of CD133+ cells in SNU719, AGS and MKN74 resistant strains and their corresponding wild-type cell lines. Isotype is a control without antibody. The abscissa represents CD133, and the ordinate represents cells. * p < 0.05, ** p < 0.01, *** p < 0.001. (PDF 1870 KB)
Figure S3. CD133+ cell show highly stem-like properties. (A) A representative image of the spheroidization experiment based on the cells processed in flow cytometry. (B) The proportion of spheroids in (A). (C, D) RT-qPCR was performed on the cells sorted in H and their mRNA differences were analyzed. OXA: Oxaliplatin. OLP: Olaparib. CON: Control group. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001 (PDF 4920 KB)
Figure S4. CD133 and PARP1 are elevated in CSCs. (A) Volcano plot for the statistical significance (y-axis, log2 transformation) versus the fold change of gene expression (x-axis, log2 transformation) between populations with high and low stemness indices determined by RNA-seq (n = 3). FC, fold change; padj, false-discovery-rate adjusted p value. The red dots denote significantly differentially expressed genes with padj < 0.05 of low stemness. The green dots denote significantly differentially expressed genes with padj < 0.05 of high stemness. (B-E) Stemness index is highly enriched in JAK STAT signaling pathway, MAPK Signaling pathway, TGF BETA signaling pathway, WNT Signaling pathway (PDF 3013 KB)
Figure S5. CD133+ cells are a subgroup of cells capable of self-renewal and DNA repair. (A, B) Nonlinear dimension reduction analysis of pathway activity score using t-SNE and UMAP method. (C) The heatmap of cell-type-specific activation pathways. (D) The ratio of other groups of CD133+ cells. (E) The first eight sites of C6 and C5 enrichment pathways (PDF 3788 KB)
Figure S6. The expression of PARP1 and CD133 in PT2, PT4, CD133+, CD133-, pLKO and SH-mettl3 were verified by WB. (A) comparison of PARP1 expression between PT2 and PT4. (B) comparison of expression of CD133+ and CD133-organoidPARP1. (C) CD133+ and CD133- PARP1 expression in subcutaneous tumors. (D) comparison of PARP1 expression in pLKO and SH-MettL3 organs and subcutaneous tumors (PDF 973 KB)
Figure S7. PARP1 and METTL3 were elevated in CSCs and oxaliplatin resistance cells. (A) RT-qPCR was used to compare the expression of PARP1 mRNA in SNU719, MKN74 and AGS-resistant CD133+ and CD133- cells. (B) RT-qPCR was used to compare PARP1 mRNA expression in SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (C) RT-qPCR was used to compare the expression of METTL3 mRNA in SNU719, MKN74 and AGS-resistant CD133+ and CD133- cells. (D) RT-qPCR was used to compare METTL3 mRNA expression in SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (E) By comparing the expression levels of PARP1 of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (F) By comparing the expression levels of METTL3 of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001. (PDF 1137 KB)
Figure S8. PARP1 knockdown and overexpression were verified by WB. (A) Comparing the expression levels of PARP1 of PT1 pLKO, PT1 sh1, PT1 sh2, PT1 sh3. (B) Comparing the expression levels of PARP1 of PT2 pLKO, PT2 sh1, PT2 sh2, PT2 sh3. (C) Comparing the expression levels of PARP1 of PT3 CON, PT3 PARP1, PT4 CON, PT4 PARP1 (PDF 1305 KB)
Figure S9. CD133+ cells were enriched in tumor stem cells and DNA damage repair related pathways and RNA modification. (A, B) CD133+ CSCs were enriched in tumor stem cells and DNA damage repair related pathways and RNA modification. (C, D) WB analysis of METTL3 and YTHDF1 in successfully transfected stable strains. (E, F) WB analysis of PT3 and PT4 organoids following METTL3 knockdown and PARP1 overexpression, PARP1 knockdown and METTL3 overexpression, METTL3 overexpression, PARP1 overexpression, METTL3 overexpression and PARP1 overexpression (PDF 631 KB)
Figure S10. m6A positive and negative controls and Dual Luciferase plasmid system validation. (A, B, C) MerIP-QPCR positive and negative controls indicated that the experimental results were reliable. (D) RNA decay rate positive and negative controls indicated that the experimental results were reliable. (E, F) RNA decay rate followed by Dual luciferase plasmid system assay demonstrated the PARP1 mRNA half-lives upon the METTL3 knockdown and YTHDF1 knockdown. Data were detected at indicated timepoint with actinomycin D (Act D, 5 μg/mL) treatment (PDF 488 KB)
Acknowledgements
The authors would like to thank Professor Antonio Tedeschi (Adult Stem Cell Laboratory, The Francis Crick Institute) for valuable comments on this article.
Author contributions
All authors participated in the writing of the manuscript. AB, YH, CZ, HL and RLR designed the study. HL, CW and LL worked on the methodologies. HL, CW, LL, IE, WL, QS and ZZ performed data collection (providing animals, collection of patients samples, providing facilities, etc.). HL, CW, LL, ER conducted data analysis and interpretation (for example, statistical analysis, biostatistics, computational analysis). HL, CW, WC, ZH, LY were responsible for cultivation and development of organoids. LY, JMO and RLR supervised the study and revised the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant No. 82073148), the Sanming Project of Medicine in Shenzhen (SZSM201911010), the Shenzhen Key Medical Discipline Construction Fund (SZXK016), the Shenzhen Sustainable Project (KCXFZ202002011010593), Shenzhen-Hong Kong-Macau Technology Research Programme (Type C) (Grant No. SGDX2020110309260100) and the Guangdong Provincial Key Laboratory of Digestive Cancer Research (No. 2021B1212040006).
Availability of data and materials
The tumor data of BALB/C NUDE mice have been deposited in the NCBI BioProject database (www.ncbi.nlm.nih.gov/bioproject) under BioProject accession no. PRJNA669425, PRJNA669419. The RNA-Seq data in gastric cancer in the TCGA database was downloaded from https://cptac-data-portal.georgetown.edu/study-summary/S025. Obtain single-cell sequencing data of early gastric cancer tissue from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134520). All relevant data could be obtained from the corresponding author.
Declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (Ethical Review [2018] No. 087). And the clinical research and animal experiment ethics committee of the First Affiliated Hospital of Sun Yat-sen University (Ethical Review [2017] No. 208).
Consent for publication
We understand that journals may be available in both print and on the internet, and will be available to a broader audience through marketing channels and other third parties. I understand that readers may include not only medical professionals and scholarly researchers but also journalists and general members of the public.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Huafu Li, Chunming Wang, Linxiang Lan and Leping Yan are joint first authors.
Contributor Information
Axel Behrens, Email: Axel.Behrens@crick.ac.uk.
Rui L. Reis, Email: rgreis@i3bs.uminho.pt
Changhua Zhang, Email: zhchangh@mail.sysu.edu.cn.
References
- 1.Arnold M, Ferlay J, van Berge Henegouwen MI, Soerjomataram I. Global burden of oesophageal and gastric cancer by histology and subsite in 2018. Gut. 2020;69(9):1564–1571. doi: 10.1136/gutjnl-2020-321600. [DOI] [PubMed] [Google Scholar]
- 2.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet (London, England) 2016;388(10060):2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 3.Boku N, Ryu MH, Kato K, Chung HC, Minashi K, Lee KW, et al. Safety and efficacy of nivolumab in combination with S-1/capecitabine plus oxaliplatin in patients with previously untreated, unresectable, advanced, or recurrent gastric/gastroesophageal junction cancer: interim results of a randomized, phase II trial (ATTRACTION-4) Ann Oncol Off J Eur Soc Med Oncol. 2019;30(2):250–258. doi: 10.1093/annonc/mdy540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harada K, Sakamoto N, Ukai S, Yamamoto Y, Pham QT, Taniyama D, et al. Establishment of oxaliplatin-resistant gastric cancer organoids: importance of myoferlin in the acquisition of oxaliplatin resistance. Gastric Cancer 2021. [DOI] [PubMed]
- 5.Wang VM, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, et al. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol. 2019;21(11):1425–1435. doi: 10.1038/s41556-019-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16(3):225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu M, Gong A, Yang H, George SK, Jiao Z, Huang H, et al. Sonic hedgehog-glioma associated oncogene homolog 1 signaling enhances drug resistance in CD44(+)/Musashi-1(+) gastric cancer stem cells. Cancer Lett. 2015;369(1):124–133. doi: 10.1016/j.canlet.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells (Dayton, Ohio) 2009;27(5):1006–1020. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yano S, Tazawa H, Hashimoto Y, Shirakawa Y, Kuroda S, Nishizaki M, et al. A genetically engineered oncolytic adenovirus decoys and lethally traps quiescent cancer stem-like cells in S/G2/M phases. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19(23):6495–6505. doi: 10.1158/1078-0432.CCR-13-0742. [DOI] [PubMed] [Google Scholar]
- 10.Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. 2018;53:156–167. doi: 10.1016/j.semcancer.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Bai X, Ni J, Beretov J, Graham P, Li Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat Rev. 2018;69:152–163. doi: 10.1016/j.ctrv.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu Y, et al. The epithelial to mesenchymal transition (EMT) and cancer stem cells: implication for treatment resistance in pancreatic cancer. Mol Cancer. 2017;16(1):52. doi: 10.1186/s12943-017-0624-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L, Cai J, Guo H, Gu J, Tong Y, Qiu B, et al. ID3 promotes stem cell features and predicts chemotherapeutic response of intrahepatic cholangiocarcinoma. Hepatol (Baltimore, MD) 2019;69(5):1995–2012. doi: 10.1002/hep.30404. [DOI] [PubMed] [Google Scholar]
- 14.Lucena-Cacace A, Otero-Albiol D, Jiménez-García MP, Muñoz-Galvan S, Carnero A. NAMPT is a potent oncogene in colon cancer progression that modulates cancer stem cell properties and resistance to therapy through Sirt1 and PARP. Clin Cancer Res Off J Am Assoc Cancer Res. 2018;24(5):1202–1215. doi: 10.1158/1078-0432.CCR-17-2575. [DOI] [PubMed] [Google Scholar]
- 15.Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018;11(1):17. doi: 10.1186/s13045-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 17.Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell. 2017;66(3):306–319. doi: 10.1016/j.molcel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, et al. m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature. 2021;591(7849):312–316. doi: 10.1038/s41586-020-03135-1. [DOI] [PubMed] [Google Scholar]
- 19.Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37(4):522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 20.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205.e199. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, et al. YTHDF2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating OCT4 expression via m6A RNA methylation. Oncogene. 2020;39(23):4507–4518. doi: 10.1038/s41388-020-1303-7. [DOI] [PubMed] [Google Scholar]
- 23.Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A Reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11(2):480–499. doi: 10.1158/2159-8290.CD-20-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39(12):e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Gonzalez G, Dai X, Miao W, Yuan J, Huang M, et al. Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m6A-based epitranscriptomic mechanism. Mol Ther. 2020;28(12):2593–2604. doi: 10.1016/j.ymthe.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seidlitz T, Merker SR, Rothe A, Zakrzewski F, von Neubeck C, Grützmann K, et al. Human gastric cancer modelling using organoids. Gut. 2019;68(2):207–217. doi: 10.1136/gutjnl-2017-314549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz EJ, Diefenbacher ME, Nelson JK, Sancho R, Pucci F, Chakraborty A, et al. LUBAC determines chemotherapy resistance in squamous cell lung cancer. J Exp Med. 2019;216(2):450–465. doi: 10.1084/jem.20180742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338–354.e315. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang Y, Hu J, Zhang J, Guo F, Zhou M, et al. scTPA: a web tool for single-cell transcriptome analysis of pathway activation signatures. Bioinf (Oxf, Engl) 2020;36(14):4217–4219. doi: 10.1093/bioinformatics/btaa532. [DOI] [PubMed] [Google Scholar]
- 35.Al-Batran SE, Homann N, Pauligk C, Goetze TO, Meiler J, Kasper S, et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet (Lond, Engl) 2019;393(10184):1948–1957. doi: 10.1016/S0140-6736(18)32557-1. [DOI] [PubMed] [Google Scholar]
- 36.Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Balibrea E, Martínez-Cardús A, Ginés A, Ruizorras V, Moutinho C, Layos L, et al. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol Cancer Ther. 2015;14(8):1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- 39.Park SY, Lee CJ, Choi JH, Kim JH, Kim JW, Kim JY, et al. The JAK2/STAT3/CCND2 axis promotes colorectal cancer stem cell persistence and radioresistance. J Exp Clin Cancer Res CR. 2019;38(1):399. doi: 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noordermeer SM, van Attikum H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol. 2019;29(10):820–834. doi: 10.1016/j.tcb.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Bell NAW, Haynes PJ, Brunner K, de Oliveira TM, Flocco MM, Hoogenboom BW, et al. Single-molecule measurements reveal that PARP1 condenses DNA by loop stabilization. Sci Adv. 2021;7(33):3641. doi: 10.1126/sciadv.abf3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh MP, Cho HJ, Kim JT, Baek KE, Lee HG, Kang SC (2019) Morin hydrate reverses cisplatin resistance by impairing PARP1/HMGB1-dependent autophagy in hepatocellular carcinoma. Cancers 11(7) [DOI] [PMC free article] [PubMed]
- 43.Hu K, Wu W, Li Y, Lin L, Chen D, Yan H et al (2019) Poly(ADP-ribosyl)ation of BRD7 by PARP1 confers resistance to DNA-damaging chemotherapeutic agents. EMBO Rep 20(5) [DOI] [PMC free article] [PubMed]
- 44.Avitabile M, Lasorsa VA, Cantalupo S, Cardinale A, Cimmino F, Montella A, et al. Association of PARP1 polymorphisms with response to chemotherapy in patients with high-risk neuroblastoma. J Cell Mol Med. 2020;24(7):4072–4081. doi: 10.1111/jcmm.15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slyskova J, Sabatella M, Ribeiro-Silva C, Stok C, Theil AF, Vermeulen W, et al. Base and nucleotide excision repair facilitate resolution of platinum drugs-induced transcription blockage. Nucleic Acids Res. 2018;46(18):9537–9549. doi: 10.1093/nar/gky764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronson GE, Piberger AL, Higgs MR, Olsen AL, Stewart GS, McHugh PJ, et al. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat Commun. 2018;9(1):746. doi: 10.1038/s41467-018-03159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reynolds P, Cooper S, Lomax M, O'Neill P. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 2015;43(8):4028–4038. doi: 10.1093/nar/gkv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Can Res. 2019;79(7):1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Et Biophys Acta Mol Cell Res. 2021;1868(1):118878. doi: 10.1016/j.bbamcr.2020.118878. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Meng Q, Ma B. Characterization of the prognostic m6A-Related lncRNA signature in gastric cancer. Front Oncol. 2021;11:630260. doi: 10.3389/fonc.2021.630260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garbo S, Zwergel C, Battistelli C. m6A RNA methylation and beyond—the epigenetic machinery and potential treatment options. Drug Discov Today. 2021;26(11):2559–2574. doi: 10.1016/j.drudis.2021.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Verifying the drug resistance level in GC resistant strains. (A) Representative images of colony formation of SNU719, MKN74 and AGS resistant strains and their corresponding wild-type cell lines in the same dose of oxaliplatin. (B) Comparison of the number of colonies in (A). (C-E) Indicate the comparison of cell viability between AGS, SNU719 and MNK74 Oxaliplatin resistance strains and wild-type cell strains under the effects of oxaliplatin respectively. OXA: Oxaliplatin. OLP: Olaparib. CON: Control group. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001 (PDF 1686 KB)
Figure S2. Oxaliplatin-resistant human gastric cancer cell lines enrich for CD133+ cells. (A) Representative image of the comparison of the spheroidizing ability of SNU719, MKN74, AGS-resistant strains and their wild-type cell lines. (B) Proportion of spheroids in (A). (C-E) RT-qPCR was used to compare the mRNA expression levels of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines LGR5, CD133, and CD44 respectively. (F) Flow cytometric analysis and comparison of CD133+ cells in SNU719, AGS and MKN74 resistant strains and their corresponding wild-type cell lines. Isotype is a control without antibody. The abscissa represents CD133, and the ordinate represents cells. * p < 0.05, ** p < 0.01, *** p < 0.001. (PDF 1870 KB)
Figure S3. CD133+ cell show highly stem-like properties. (A) A representative image of the spheroidization experiment based on the cells processed in flow cytometry. (B) The proportion of spheroids in (A). (C, D) RT-qPCR was performed on the cells sorted in H and their mRNA differences were analyzed. OXA: Oxaliplatin. OLP: Olaparib. CON: Control group. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001 (PDF 4920 KB)
Figure S4. CD133 and PARP1 are elevated in CSCs. (A) Volcano plot for the statistical significance (y-axis, log2 transformation) versus the fold change of gene expression (x-axis, log2 transformation) between populations with high and low stemness indices determined by RNA-seq (n = 3). FC, fold change; padj, false-discovery-rate adjusted p value. The red dots denote significantly differentially expressed genes with padj < 0.05 of low stemness. The green dots denote significantly differentially expressed genes with padj < 0.05 of high stemness. (B-E) Stemness index is highly enriched in JAK STAT signaling pathway, MAPK Signaling pathway, TGF BETA signaling pathway, WNT Signaling pathway (PDF 3013 KB)
Figure S5. CD133+ cells are a subgroup of cells capable of self-renewal and DNA repair. (A, B) Nonlinear dimension reduction analysis of pathway activity score using t-SNE and UMAP method. (C) The heatmap of cell-type-specific activation pathways. (D) The ratio of other groups of CD133+ cells. (E) The first eight sites of C6 and C5 enrichment pathways (PDF 3788 KB)
Figure S6. The expression of PARP1 and CD133 in PT2, PT4, CD133+, CD133-, pLKO and SH-mettl3 were verified by WB. (A) comparison of PARP1 expression between PT2 and PT4. (B) comparison of expression of CD133+ and CD133-organoidPARP1. (C) CD133+ and CD133- PARP1 expression in subcutaneous tumors. (D) comparison of PARP1 expression in pLKO and SH-MettL3 organs and subcutaneous tumors (PDF 973 KB)
Figure S7. PARP1 and METTL3 were elevated in CSCs and oxaliplatin resistance cells. (A) RT-qPCR was used to compare the expression of PARP1 mRNA in SNU719, MKN74 and AGS-resistant CD133+ and CD133- cells. (B) RT-qPCR was used to compare PARP1 mRNA expression in SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (C) RT-qPCR was used to compare the expression of METTL3 mRNA in SNU719, MKN74 and AGS-resistant CD133+ and CD133- cells. (D) RT-qPCR was used to compare METTL3 mRNA expression in SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (E) By comparing the expression levels of PARP1 of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. (F) By comparing the expression levels of METTL3 of SNU719, MKN74 and AGS resistant strains and their wild-type cell lines. AGSRE: AGS oxaliplatin resistance. SNU719RE: SNU719 oxaliplatin resistance. MKN74RE: MKN74 oxaliplatin resistance. * p < 0.05, ** p < 0.01, *** p < 0.001. (PDF 1137 KB)
Figure S8. PARP1 knockdown and overexpression were verified by WB. (A) Comparing the expression levels of PARP1 of PT1 pLKO, PT1 sh1, PT1 sh2, PT1 sh3. (B) Comparing the expression levels of PARP1 of PT2 pLKO, PT2 sh1, PT2 sh2, PT2 sh3. (C) Comparing the expression levels of PARP1 of PT3 CON, PT3 PARP1, PT4 CON, PT4 PARP1 (PDF 1305 KB)
Figure S9. CD133+ cells were enriched in tumor stem cells and DNA damage repair related pathways and RNA modification. (A, B) CD133+ CSCs were enriched in tumor stem cells and DNA damage repair related pathways and RNA modification. (C, D) WB analysis of METTL3 and YTHDF1 in successfully transfected stable strains. (E, F) WB analysis of PT3 and PT4 organoids following METTL3 knockdown and PARP1 overexpression, PARP1 knockdown and METTL3 overexpression, METTL3 overexpression, PARP1 overexpression, METTL3 overexpression and PARP1 overexpression (PDF 631 KB)
Figure S10. m6A positive and negative controls and Dual Luciferase plasmid system validation. (A, B, C) MerIP-QPCR positive and negative controls indicated that the experimental results were reliable. (D) RNA decay rate positive and negative controls indicated that the experimental results were reliable. (E, F) RNA decay rate followed by Dual luciferase plasmid system assay demonstrated the PARP1 mRNA half-lives upon the METTL3 knockdown and YTHDF1 knockdown. Data were detected at indicated timepoint with actinomycin D (Act D, 5 μg/mL) treatment (PDF 488 KB)
Data Availability Statement
The tumor data of BALB/C NUDE mice have been deposited in the NCBI BioProject database (www.ncbi.nlm.nih.gov/bioproject) under BioProject accession no. PRJNA669425, PRJNA669419. The RNA-Seq data in gastric cancer in the TCGA database was downloaded from https://cptac-data-portal.georgetown.edu/study-summary/S025. Obtain single-cell sequencing data of early gastric cancer tissue from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134520). All relevant data could be obtained from the corresponding author.