Abstract
Chromatin remodeler complexes regulate gene transcription, DNA replication and DNA repair by changing both nucleosome position and post-translational modifications. The chromatin remodeler complexes are categorized into four families: the SWI/SNF, INO80/SWR1, ISWI and CHD family. In this review, we describe the subunits of these chromatin remodeler complexes, in particular, the recently identified members of the ISWI family and novelties of the CHD family. Long non-coding (lnc) RNAs regulate gene expression through different epigenetic mechanisms, including interaction with chromatin remodelers. For example, interaction of lncBRM with BRM inhibits the SWI/SNF complex associated with a differentiated phenotype and favors assembly of a stem cell-related SWI/SNF complex. Today, over 50 lncRNAs have been shown to affect chromatin remodeler complexes and we here discuss the mechanisms involved.
Keywords: ATP-dependent helicase, Nucleosome, Histone and epigenetic regulation
Introduction
Long non-coding RNAs (lncRNAs) are a heterogeneous class of long RNAs without a large open reading frame encoding proteins. To better understand the role of lncRNAs in the formation and recruitment of chromatin remodeler complexes, we will briefly discuss the organization of DNA into chromatin, and then review the chromatin remodeler subfamilies with the involved subunit proteins.
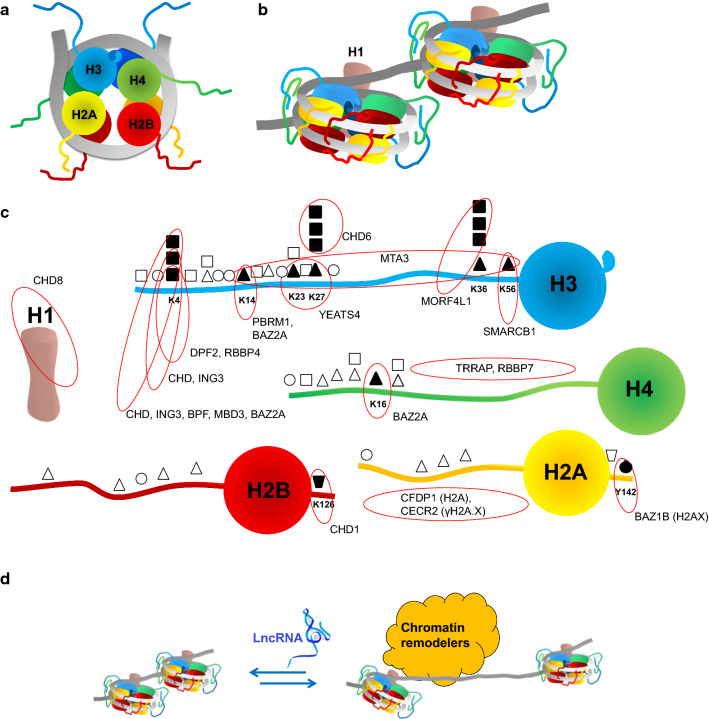
Chromatin is the state of organized DNA condensation in the nucleus. DNA is wrapped into nucleosome structures that are further packed into chromatin fibers and condensed into either “euchromatin” or highly condensed “heterochromatin”. Chromatin condensation is a dynamic process resulting in different nuclear sub‐compartments, including topologically lamina-associating domains, nucleoli, Cajal bodies, nuclear stress bodies, paraspeckles and non-chromatin bodies like nuclear speckles and PML bodies (reviewed by [1]). In the fundamental nucleosome unit, ~ 146 base-pairs (bp) of the negatively charged DNA helix are folded ~ 1.7 times around an octamer of positively charged histone proteins. A tetramer consisting of two H3 and two H4 histones binds to the DNA, where after two H2A-H2B dimers join the complex. The first ~ 20 bp of DNA that sort the fold are hold together by the histone H1 protein and a “beads on a string” structure is formed with strings of ~ 15–70 bp free DNA (Fig. 1a, b, [2]).
Fig. 1.
Organization of chromatin. Depicted is a DNA helix (in grey) with histone proteins H2A, H2B, H3, H4 and H1 (different colors). a Schematized top view of a nucleosome with tetramer (H3-H4)2 and two dimers (H2A-H2B). b Nucleosomes side view with histone H1 stabilizing the DNA linker. Methylated histone tails have stronger DNA interactions and then acetylated tails. c Presented are common histones modifications, such as methylation (squares), acetylation (triangles), phosphorylation (circles) and ubiquitination (trapeziums). Histones modifications are frequently recognized by chromatin readers. Modifications that are implicated in chromatin remodeling complexes are depicted by solid symbols and their corresponding readers are annotated (red circles). d LncRNAs (in blue) can either inhibit or stimulate chromatin remodeling via interference with remodeler complexes
Nucleosome positioning is part of epigenetic regulation since the condensed DNA is inaccessible for transcription, replication, and repair [3]. Positioning of nucleosomes is not arbitrary; firstly, it depends on the thermodynamic bending properties of different DNA sequences. In particular, the properties of poly(deoxyAdenylic(dA): deoxyThymidylic(dT)) DNA stretches disfavoring nucleosome formation. DNA sequence motifs can also be recognized by sequence-specific factors that may lead to nucleosome remodeling or depletion. Secondly, DNA modifications, such as 5-methylation and 5-hydroxymethylation of cytosines, affect DNA flexibility and nucleosome stability. Thirdly, the DNA-interactions with histone tails may vary, as these are subjected to several posttranslational modifications, which may change electric charges and/or evoke steric hindrance (Fig. 1c). In addition, acetylation of lysine residue 16 of histone 4 (H4K16) weakens its protein–protein interaction with residues from the H2A-H2B dimer of the adjacent nucleosome and significantly reduces nucleosome stacking [4]. Moreover, modifications like methylation and acetylation of lysine residues are important for the recognition by chromatin reader proteins [5]. These chromatin readers are known to recruit complexes with writers and erasers of chromatin modifications, as well as chromatin remodelers (Fig. 1c).
Rather than modifying DNA or histones to modulate chromatin (like writers/erasers), the remodeler complexes change nucleosome positioning (Fig. 1d). They affect spacing of nucleosomes, regulate nucleosomes transfer and evoke histone variant switching. The kinetic energy needed to modulate the stable nucleosome organization comes from ATP hydrolysis. All chromatin remodeling complexes contain a catalytic unit from the superfamily 2 helicases, which categorizes them into four types of chromatin remodeling complexes: (1) the SWI/SNF, (2) the INO80/SWR1, (3) the ISWI and (4) the CHD family. The complexes of each chromatin remodeler family have distinct subunits and different functionalities that are listed in Table 1. Today, 34 lncRNAs have been described to interact with diverse subunits. Another 18 lncRNAs were reported to act as competing endogenous (ce) RNA with miRNA targets. We will summarize the current knowledge of the effect that lncRNAs have on the function of each chromatin remodeler complex family.
Table 1.
Proteins of human chromatin remodeling complexes
| Hugo name | Alternative names | Complex family | Bound complexes | Function | References | Implicated lncRNAs |
|---|---|---|---|---|---|---|
| SWI/SNF subunits (Histone dimer/nucleosome removal, nucleosome sliding) | ||||||
| 1. Catalytic subunit (SMARC subfamily A) | ||||||
| SMARCA4 | BRG1 | SWI/SNF | BAF, PBAF | ATP-dependent helicase | [6] |
Evf2, UCA1, TUG1, Myheart, CPS1‐IT1, NEAT1, MALAT1, LincRNA Cox2 IL-7–AS, LncFZD6, lncTCF7 |
| SMARCA2 | BRM | SWI/SNF | BAF | NEAT1, lncBRM | ||
| 2. SMARC subfamily B subunit | ||||||
| SMARCB1 | SNF5, BAF47 | SWI/SNF | BAF, PBAF | Constitutive Core Subunit, chromatin reader binding to H3K56Ac, Enhances DNA integration | [5] | SWINGN, HOTAIR |
| 3–4. SMARC subfamily C dimer (BAF155-155 or 155-170) | ||||||
| SMARCC1 | BAF155 | SWI/SNF | BAF, PBAF | Constitutive Core Subunit | [6] | |
| SMARCC2 | BAF170 | SWI/SNF | BAF | Core Subunit; mono-ADP-ribosylated by SIRT6 | [113] | |
| 5. AT-rich interaction domain subunit | ||||||
| ARID2 | BAF200 | SWI/SNF | PBAF | DNA and transcription factor binding | [6] | |
| ARID1A | BAF250A | SWI/SNF | BAF | HOTAIR, MVIH, DGCR5, LINC00163, CASC15 | ||
| ARID1B | BAF250B | SWI/SNF | BAF | |||
| 6. SMARC subfamily E subunit | ||||||
| SMARCE1 | BAF57 | SWI/SNF | cBAF, PBAF | Binding to '4-way' branched DNA (Holliday junction) | [6] | |
| 7–8. Actin-related Subunits | ||||||
| ACTL6A | BAF53A, Arp4, INO80K | General | BAF,PBAF, INO80, p400/TRRAP | SMARCA/TRRAP binding | [6] | uc.291 |
| ACTL6B | BAF53B | SWI/SNF | ncBAF | |||
| ACTB | (β-Actin) | General | BAF,PBAF, p400/TRRAP | |||
| 9. Poly-bromo subunit | ||||||
| PBRM1 | BAF180 | SWI/SNF | PBAF | chromatin reader binding to H3K14ac, phosphorylated by ATM | [11, 114] | |
| 10. B-cell lymphoma 11 subunit | ||||||
| BCL11A | General | BAF/PBAF, NuRD, SIN3A, PRC2 [115] | Enhances DNA integration | [6] | CDKN2B-AS1, uc.57 | |
| BCL11B | SWI/SNF | BAF/PBAF | ||||
| 11. Bromodomain subunit | ||||||
| BRD7 | SWI/SNF | PBAF | Bromo-domain interaction with acetylated lysine. | [116] | ||
| BRD9 | LAVS3040 | SWI/SNF | ncBAF | [117] | ||
| 12. B-cell lymphoma 7 subunit | ||||||
| BCL7A | SWI/SNF | BAF | (B-cell CLL/Lymphoma 7) | [6] | ||
| BCL7B | SWI/SNF | npBAF | ||||
| BCL7C | SWI/SNF | npBAF | ||||
| 13. SMARC subfamily D subunit | ||||||
| SMARCD1 | BAF60A | SWI/SNF | BAF/PBAF | Transcription factor binding [118] | DLEU1 | |
| SMARCD2 | BAF60B | SWI/SNF | PBAF | [118] | ||
| SMARCD3 | BAF60C | General | BAF, MyoD | [119] | ||
| 14. PHD-zinc finger subunit | ||||||
| PHF10 | BAF45A | SWI/SNF | PBAF | Phosphorylated protein, complex stability | [6] | |
| DPF1 | BAF45B, NEUD4 | SWI/SNF | BAF | (Double Plant homeodomain Finger) | [6] | |
| DPF2 | BAF45C | SWI/SNF | BAF | Chromatin reader binding to H3K4me1 | [14] | |
| DPF3 | BAF45D | SWI/SNF | BAF | [5] | ||
| 15. BRD-interacting subunit | ||||||
| BICRA | GLTSCR1 | SWI/SNF | ncBAF | (Glioma tumor suppressor candidate region 1), BRD interacting protein | [6] | |
| BICRAL | GLTSCR1L | SWI/SNF | ncBAF | [6] | ||
| 16. Synovial Sarcoma translocation protein Chr18 subunit | ||||||
| SS18 | SSXT/SYT | SWI/SNF | ncBAF | Interaction with BAF47 | [120] | |
| SS18L1 | CREST | SWI/SNF | ncBAF | Calcium-responsive transactivator | [120] | |
| INO80/SWR1 subunits (nucleosome replacement) | ||||||
| 1. Catalytic subunit (ATPase) | ||||||
| INO80 | INO80A, INOC1, | INO80/SWR1 | INO80 | ATPase | [121] | LCTS5, HAND2‐AS1 |
| SRCAP | EAF1, FLHS, SWR1, DOMO1 | INO80/SWR1 | SRCAP | [121] | LncKdm2b | |
| EP400 | p400 | INO80/SWR1 | p400/TRRAP | [121] | ||
| 2–3. Catalytic subunit dimer (helicase) | ||||||
| RUVBL1 | INO80H, RVB1, TIP49 | INO80/SWR1 | INO80, SRCAP, p400/TRRAP | ATP-dependent helicase | [121] | |
| RUVBL2 | INO80J, RVB2, TIP48 | INO80/SWR1 | INO80, SRCAP, p400/TRRAP | |||
| 4–6. Effector subunit | ||||||
| YY1 | INO80S | General | INO80 , p400/TRRAP, PRC2 | Transcription factor (GLI-Krüppel zinc finger), Polycomb proteins recruitment | [122] | ANRIL, Linc-YY1, TUG1, SPAG5-AS1, HOTAIR, LINC00899, LINC00668, Linc01134, |
| KAT5 | TIP60 | INO80/SWR1 | p400/TRRAP | Lysine (K) acetyltransferase 5 | [121] | |
| TRRAP | Tra1 | INO80/SWR1 | p400/TRRAP | Histone acyltransferase (H4) | [121] | |
| 7–9. Actin-related subunits | ||||||
| ACTR5 | ARP5, INO80M | INO80/SWR1 | INO80 | INO80 binding | [121] | |
| ACTR8 | ARP8, INO80N | INO80/SWR1 | INO80 | |||
| ACTL6A | Arp4, INO80K, BAF53A | General | INO80, SRCAP, p400/TRRAP, BAF, PBAF, | TRRAP/SMARCA binding | ||
| ACTB | (β-Actin) | General | p400/TRRAP, BAF, PBAF | |||
| ACTR6 | ARP6, CDA12; hARPX, HSPC281, MSTP136 | INO80/SWR1 | SRCAP | SRCAP binding | ||
| 10–17. INO80 subunits | ||||||
| INO80B | IES2, PAPA-1 | INO80/SWR1 | INO80 | INO80 binding | [121] | |
| INO80C | IES6 | INO80/SWR1 | INO80 | |||
| TFPT | INO80F, FB1, amida | INO80/SWR1 | INO80 | Modulated by SUMOylation (increases interaction with INO80E), transcription factor? | [123] | |
| INO80E | CCDC95 | INO80/SWR1 | INO80 | Binding to TFPT | [121] | |
| INO80D | INO80/SWR1 | INO80 | Has two putative DNA-binding domains | CR933609 | ||
| MCRS1 | INO80Q, MSP58, P78 | INO80/SWR1 | INO80 | Has a nuclear and nucleolar localization signal | ||
| NFRKB | INO80G | INO80/SWR1 | INO80 | NF (nuclear factor) binding to kappa B regulatory elements/winged-helix domains involved in protein-protein interactions, recruitment of EXO1 enhances resection, stimulating homologous recombinase DNA repair (non-essential for the in vitro nucleosome sliding) | [30] | |
| UCHL5 | UCH37 | INO80/SWR1 | INO80, RPN13 | Deubiquitinating enzyme, but inhibited by NFRKB, recruitment of EXO1 (non-essential for the in vitro nucleosome sliding) | [124], | DRAIC |
| 9–12. SRCAP/TRAPP subunits | ||||||
| VPS72 | YL1; CFL1; Swc2; YL-1; TCFL1 | INO80/SWR1 | SRCAP, p400/TRRAP | Histone chaperone | [125] | |
| DMAP1 | SWC4, MEAF2, EAF2, DNMTAP1 | General | SRCAP, p400/TRRAP, DNMT1/HDAC complexes | Regulation of transcription by binding to DNMT1, HDAC2 and HAT complexes | [121] | |
| BRD8 | p120 | INO80/SWR1 | SRCAP, p400/TRRAP | Bromodomain interaction with acetylated lysine (histone H4) and nuclear receptors (TR, RXR) | [121] | RNCR3 |
| YEATS4 | GAS41; NUBI-1 | INO80/SWR1 | SRCAP, p400/TRRAP | Chromatin reader, binding to H3K23acK27ac binding, H3K122suc, homology to transcription factors MLLT 1, 3 | [126, 127] | lncAKHE |
| 13–14. SRCAP specific subunits | ||||||
| CFDP1 | Swc5, p97, BCNT | INO80/SWR1 | SRCAP | Interacting with H2A | [121] | |
| ZNHIT1 | ZNFN4A1 | INO80/SWR1 | SRCAP | Interacting with H2AZ | [128] | |
| 13–16. P400/TRAPP specific subunits | ||||||
| MORF4L1 | MRG15 | General | p400/TRRAP (MRG15-MRGBP), HDAC Rpd3S/ Sin3S (MRG15-Pf1) | Chromatin reader that binds to H3K36me3, H3K4me1 and H3K4me3 | [121] | |
| MRGBP | Eaf7, MRG15BP, URCC4 | INO80/SWR1 | p400/TRRAP | Interaction with BDR8, MORF4L1 | [121] | |
| ING3 | ING2, EAF4 | General | p400/TRRAP, | Chromatin reader, binding to H3K4me3, interaction with TP53 | [121] | CASC7 |
| MEAF6 | EAF6 | General | p400/TRRAP, HAT complexes (eg., MOZ/MORF, NuA3) | Interaction with EAF4 | [121] | |
| ISWI (nucleosome spacing, DNA repair) | ||||||
| 1. Catalytic subunit (ATP-dependent helicase) | ||||||
| SMARCA5 | SNF2H | ISWI | WICH, NoRC, RSF, ACF/CHRAC | ATP-dependent helicase | [46] | |
| SMARCA1 | SNF2L | ISWI | NURF, CERF | [46] | DLEU1 | |
| SMARCAD1 | ADERM, BASNS, ETL1, HEL1, HRZ | ? | “CUE” |
ATP-dependent helicase, binds to H2A-ubiquitin, CUE-domain binds KAP1; maintaining H3K9me3 marks. |
[129] | |
| HELLS | SMARCA6 | ? | CHIRRC | ATP-dependent helicase, binds to CDCA7 | [48] | BlackMamba |
| 2. Signature subunit | ||||||
| BAZ1B | WSTF | ISWI | WICH | Histone phosphorylation (H2A.X-pY142), interacts with RNA polymerase II, stimulates transcription-coupled homologous recombination | [52] | |
| BAZ2A | Tip5, NoRC | ISWI | NoRC | Chromatin reader that binds to H3K4, H4K16ac, H3K14ac. Binds also to TCF7L2 and ncRNAs. Responsible for heterochromatin formation at major clusters of repetitive elements | [130, 131] | Lnc pRNA |
| RSF | HBXAP | ISWI | RSF | Remodeling and spacing factor 1; histone chaperone, involved in homologous recombination DNA repair | [132] | |
| BAZ1A | ACF1 | ISWI | ACF/CHRAC | Dimer ACF complex, RNF20-mediated chromatin relaxation, and KU70 interaction, mediating DNA repair | [55] | NEXN-AS1 |
| BPTF | NURF301 | ISWI | NURF | Chromatin reader binding to H3K4me3 and H4K16ac, interacts with AT rich DNA sequences | [57] | NMR (LINC01672) |
| CECR2 | CERF | ISWI | CERF | Chromatin reader binding to acetyl-lysine residues of histone H2A and H3, stimulating γH2A.X formation | [53] | |
| TRIM28 | KAP1 | ? | “CUE” | Maintaining H3K9me3 marks. | [129] | Paupar |
| CDCA7 | ICF3, JPO1 | ? | CHIRRC | Nucleosome sliding | [48] | FGD5-AS1 |
| 3–4 CHRAC subunits | ||||||
| CHRAC1 | YCL1, CHRAC15 | ISWI | ACF/CHRAC | Chromatin accessibility complex subunit 1; histone‐fold protein that form a stable complex with POLE3 and binds naked DNA | [76] | |
| POLE3 | CHRAC17, p17, YBL1 | General | ACF/CHRAC, DNA POL ϵ complex, HAT GCN5/PCAF complex | DNA polymerase epsilon 3; accessory subunit; histone H3, H4 chaperone | [133] | |
| 3–4 RB binding subunits | ||||||
| RBBP4 | NURF55, RBAP48 | General | NURF, NuRD , PRC2 complex | RB binding protein 4; histone tetramer deposition; histone H3 and BCL11A binding | [76] | |
| RBBP7 | RbAp46 | General | NURF, NuRD, PRC2 complex | Binding to histone H4, histone acetyltransferase HAT1 | [76] | |
| CHD (chromodomain helicase DNA-binding, DNA repair; NuRD, histone deacetylation/demethylase complex) | ||||||
| 1. Catalytic subunit (Chromodomain Helicase DNA-binding) | ||||||
| CHD1 | PILBOS | CHD1/ PAF1, CHD1/ SSRP1, CHD1/ NCoR CHD1/HAT complexes ( SAGA/SLIK/GCN5) | Chromatin reader that binds to H3K4me2/3, stabilization of H2AX at DSBs, stabilization of H2B(tail)K123ub | [67–70, 134] | MATN1-AS1 | |
| CHD2 | EEOC | CHD2/PARP-1 | Deposition of histone variant H3.3 in NHEJ DNA-repair | [72] | CHASERR | |
| CHD6 | CHD5, RIGB | CHD6/Polycomb units | Chromatin reader that binds to H3K27me3 | [135] | ||
| CHD7 | CRG, KAL5 | CHD7/SOX2, CHD7/SOX9, CHD7/8, PBAF | SOX2, SOX9, SMARCC2 binding | [79] | LINC01410 | |
| CHD8 | AUTS18 | CHD8/β-catenin, CHD8/CHD7, CHD8/MT complexes (BRD4-NSD3, BACH1-MAFG-DNMT3, KMT2) | Binds to histone H1, H3K36 methyltransferase (MT) complex | [80, 81] | ||
| CHD9 | KISH2 | CHD2/PPARα, CHD2/PPARγ, BAF |
Chromatin reader that binds to H3 (K9me2/3,K27me3), also interaction with PPARα, γ, SMARCA2. Ribosomal gene regulation |
[76] | ||
| 1. Catalytic subunit NuRD | ||||||
| CHD3 | Mi-2a | NuRD | NuRD, PRC2 complex | Generation of compacted chromatin after DNA replication; isoforms CHD3.1 and CHD3.3: SUMOylated KAP-1 binding; transcription factor interaction | [55] | |
| CHD4 | Mi-2b | NuRD | NuRD, PRC2 complex | Transcription factor interaction, interaction with RNF8 | [76, 136] | |
| CHD5 | NuRD | NuRD, PRC2 complex | H3K27me3 binding | [135] | ||
| 2. Interacting subunit | ||||||
| FAM124B | CHD7/8 | [86] | ||||
| 3. GATA-type Zn-finger subunit | ||||||
| GATAD2A | p66a | NuRD | NuRD/CHD5 | CHD and MBD2 binding | [75] | |
| GATAD2B | p68 | NuRD | NuRD/CHD5 | Protein-protein interaction, deacetylating, methylated nucleosomes | ||
| 4–5 Histone deacetylase Subunit | ||||||
| HDAC1 | general | NuRD, PRC2 complex | Histone deacetylase (Kac) | [76] | ANRIL | |
| HDAC2 | general | NuRD, PRC2 complex | SNHG15, ARAP1-AS1 | |||
| 6–7 RB binding subunits | ||||||
| RBBP4 | NURF55, RBAP48 | general | NURF, NuRD , PRC2 complex | RB binding protein 4; histone tetramer deposition, histone H3 and BCL11A binding | [76] | |
| RBBP7 | RbAp46 | general | NURF, NuRD, PRC2 complex | Binding to histone H4 and histone acetyltransferase HAT1 | ||
| 2. Interacting subunit | ||||||
| CTBP2 | NuRD | NuRD, PRC2 complex | C-terminal binding protein 2; NADH-dependent interaction with p300-Runx2 complex, decoy inhibiting transcriptional activation of this complex | [137] | ||
| 3. Methyl-CpG binding subunit | ||||||
| MBD3 | NuRD | NuRD, PRC2 complex | methyl-CpG binding domain protein 3; no binding to methylated DNA, mediates the association of MTA2 with HDAC, H3K4me3 binding | [136] | ||
| MBD2 | DMTase | NuRD | NuRD, PRC2 complex | methyl-CpG binding domain protein 2; reposition nucleosomes away from unmethylated CpG-rich regions of the genome | [136] | |
| 4. Metastasis-associated subunit | ||||||
| MTA1 | NuRD | NuRD, PRC2 complex | Metastasis-associated protein 1 | [76] | ||
| MTA2 | NuRD | NuRD, PRC2 complex | Metastasis-associated protein 2 | [76] | HOTAIR | |
| MTA3 | NuRD | NuRD, DMT complex LSD1/SIX3, PRC2 complex | Binds to pan-acetylated H3, histone demethylase, opposing action to MTA1, binding to BCL6 | [76] | ||
| 5. Associated subunit | ||||||
| CDK2AP1 | DOC1 | NuRD | NuRD/CHD3/4 | NuRD recruitment to displace SWI/SNF complex | [75] | |
ARID AT-rich interactive domain, ARP actin-related protein, BAF BRG1-associated factor, BAZ bromodomain adjacent to zinc finger domain, BRD bromodomain containing, BPTF bromodomain PHD finger transcription factor, CHD Chromodomain Helicase DNA-binding, CHIRRC CDCA7–HELLS ICF-related nucleosome remodeling complex, EAF Esa1- associated factor, HAT histone acetyltransferase, HDAC histone deacetylase, ISWI Imitation SWI, Kac acetyl-lysine, Ksuc succinyl-lysine, Kub ubiquitin-lysine, NURF nucleosome remodeling factor, NuRD Nucleosome remodeling and histone deacetylation, SMARC SWI/SNF related, matrix associated, actin-dependent regulator of chromatin, SNF Sucrose Non-Fermentable, SWI SWItch [128]
Chromatin remodeling complexes of the SWI/SNF family
Subunits of the SWI/SNF complexes
The SWI/SNF proteins were discovered in yeast as important factors regulating mating-type switching (SWI = SWItch) and regulating the use of different energy sources (SNF = Sucrose Non-Fermentable) [6]. The catalytic subunit in these complexes is either the ATP-dependent helicase Brahma (SMARCA2, BRM) or Brahma-related gene-1 (SMARCA4, BRG1). The core subunits are SMARCB1 (SNF5), SMARCC1 (BAF155) and SMARCC2 (BAF170) (Table 1). SWI/SNF remodeler complexes anchors to histone proteins and translocate 1–2 bp of DNA along the surface of the nucleosome, depending on the ATPase activity, this results in nucleosome sliding, or in destabilizing and removal of H2A–H2B dimers or entire histone cores [7], thus creating open chromatin structures.
Depending on subunit composition the SWI/SNF complexes are classified as PBAF (PolyBromo-Associated Factors) and BAF (BRG1- or BRM-Associated Factors). Exclusive subunits of the PBAF complex are ARID2, PBRM1 (BAF180), PHF10, and BRD7, in combination with the helicase SMARCA4. In the embryonic stem cell, the PBAF complex is important for maintaining the stem cell transcriptome [8]. Indeed, SMARCA4 plays an important role in the NANOG signaling pathway [9]. The PBAF complex inhibits growth through down-regulation of cell cycle genes CDK2, CDK4 and CCND1 and interference with the phosphorylated RB-pathway [10]. In addition, the PBAF complex is implicated in transcriptional repression at DNA double-strand breaks [11].
The BAF complex harbors the exclusive subunits ARID1A or ARID1B, BRD9 and SS18. They contain either the helicase SMARCA4 or SMARCA2. The difference between the PBAF and BAF complexes also results in different functionality [10, 12]. The SMARCA4- and SMARCA2-containing complexes have been assigned to ‘actively marked’ and ‘repressively marked’ chromatin binding complexes, respectively [13]. In line with this, both complexes have differential affinity for histone modifications (PBAF complexes binds to H3K4-monomethylated, BAF binds to H3K4-trimethylated; [14]).
Whereas the PBAF complex is associated with an embryonic stem cell phenotype, BAF complexes seem to be tissue- and cell-type specific [15]. In particular, SMARCD3 subunits defines cardiac progenitors (cBAF complex) and SMARCD1 neuronal progenitors (npBAF complex), and the association with SS18L1 is associated with post-mitotic neurons (nBAF complex) [15].
Long non-coding RNAs and the SWI/SNF family
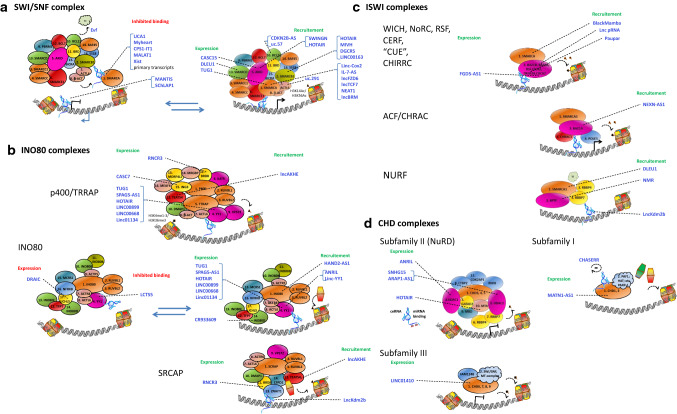
Amongst the chromatin remodelers, the SWI/SNF complex has frequently been shown to interact with lncRNAs (Table 2, Fig. 2a). In particular, SMARCA4 physically associates with primary transcribed RNA [16], suggesting a cis-acting function as shown for the lncRNAs lncFZD6 and lncTCF7. Some lncRNAs (e.g., MANTIS, SChLAP1) do not bind the SWI/SNF complex, but compete for DNA-binding at a particular chromosomal promoter gene region (e.g., MANTIS at the ICAM-1 gene). SMARCA4-lncRNA interaction may also interfere with recruitment of SWI/SNF complexes at multiple gene regions (trans-acting) (e.g. LincRNA Cox2 and IL-7–AS). Recently, also SMARCB1 (SNF5) has been shown to interact with a pool of lncRNAs [17], including SWINGN and HOTAIR. Interestingly, lncBRM seem to inhibit SWI/SNF-SMARCA2 BAF complexes by its association with SMARCA2 (BRM), which in turn favors the assembly of SWI/SNF-SMARCA4 BAF complexes in liver cancer stem cells [18]. The BAF-specific subunit ARID1A has also been shown to bind lncRNAs (HOTAIR, MVIH, DGCR5 and LINC00163) thereby interfering with the transcription of several genes (Table 2 and Fig. 2a). Binding of lncRNA uc.291 with ACTL6A competes with BAF-ACTL6A binding and affects the expression of several differentiation genes in skin keratinocytes [19]. In addition, the MAPK pathway is affected by the interaction of SWI/SNF subunit BCL11A with CDKN2B-AS1 in lymphocytes and lncRNA uc.57 in breast cancer cells [20, 21]. Lots of lncRNAs have been shown to regulate gene expression by competing for miRNA binding [22], and for the SWI/SNF family lncRNA CASC15 and DLEU1 were reported to act as ceRNA for ARID1A and SMARCD1 transcripts, respectively [23, 24]. In addition, lncRNA DSCAM-AS1 may upregulate BCL11A expression [25].
Table 2.
LncRNAs known to interact with human chromatin remodeling complexes
| lncRNA (name UCSC) | Binding to | Type of lncRNA | Action | Refs. |
|---|---|---|---|---|
| SWI/SNF subunits (Histone dimer/nucleosome removal, nucleosome sliding) | ||||
| Evf2 (DLX6-AS1) | SMARCA4 | Antisense of DLX6 | Competing with transcription factor DLX1 for binding to BRG1 | [138] |
| UCA1 | SMARCA4 | Intergenic | Prevents binding to p21 promoter UCA1 expression regulated by ARID1A | [139, 140]$ |
| TUG1 | SMARCA4 | Intergenic, sharing promoter with MORC2 | Inhibiting BRG1 degradation (MORC2 has PARP1-mediated ATPase and chromatin remodeling activities important in the DNA-damage response [141]). | [142] |
| Myheart (Mhrt) | SMARCA4 | Antisense of MYH7 | Binding to BRG1 prevents DNA binding of SWI/SNF complex | [143] |
| CPS1‐IT1 | SMARCA4 | Intronic of CPS1 gene | Prevents binding to Cyr61 promoter region | [144]$ |
| NEAT1 | SMARCA2, SMARCA4 | Intergenic | Facilitates the organization of paraspeckle proteins (independent of SMARCA catalytic activity) | [145] |
| MALAT1 | SMARCA4 | intergenic | Complex with HDAC9 that represses vascular smooth muscle cell genes | [146] |
| LincRNA Cox2 | SMARCA4 | Antisense | Recruiting the SWI/SNF complex to inflammatory-response genes, e.g., Saa3 and Ccl5 | [147] |
| IL-7–AS | p300/SMARCA4 | Antisense | Recruiting HAT p300 followed by recruitment of SWI/SNF complex to activate inflammatory genes, (e.g., CCL2, CCL5, and IL-6) | [148] |
| LncFZD6 (BAALC-AS1) | SMARCA4 | Antisense, sharing promoter with FZD6 | Recruits SWI/SNF complex to FZD6 promoter, | [149]$ |
| lncTCF7 | SMARCA4 | antisense | Recruiting the SWI/SNF complex to the TCF7 promoter | [143] |
| lncBRM | SMARCA2 | Antisense | Associates with BRM and favors assembly of BRG1- BAF complexes in liver cancer stem cells | [18]$ |
| SWINGN (LINC00565/LINC00452) | SMARCB1 | Intergenic, sharing promoter with GAS6 | Transcriptional activation of several genes (e.g., GAS6, PDGFRB and COL1A1). | [17]$ |
| HOTAIR | SMARCB1, ARID1A | Intergenic, sharing promoter of HOXC11 | Affecting expression of transcriptional repressor gene SNAIL | [150]$ |
| MVIH (AK094613) | ARID1A | Intragenic, overlapping RPS24 exons | Affects CDKN1A transcription | [151]$ |
| DGCR5 | ARID1A | Intergenic | Promotes p21 expression | [152]$ |
| LINC00163 | ARID1A | Intergenic | Stimulation of TCF21 expression | [153]$ |
| uc.291* | ACTL6A | Intragenic at LRMDA gene | Prevents inhibition by ACTL6A on epidermal differentiation genes | [19] |
| CDKN2B-AS1 | BCL11A | Antisense | Inhibition of MAP4K1 | [20] |
| uc.57* | BCL11A | Intergenic upstream of BCL11A | Inhibition of PI3K/AKT and MAPK signaling pathways | [21]$ |
| Xist | SWI/SNF | Intergenic | Binding to SWI/SNF complex inhibits binding to Xi-genes regions | [154] |
| MANTIS | DNA | Intragenic, ANXA4 gene | Preventing SWI/SNF complex binding at ICAM-1 promoter region | [155] |
| SChLAP1 | DNA | intergenic | SMARCA4 associates frequently with primary transcripts including SChLAP1, preventing SWI/SNF complex binding | [16, 102, 156]$ |
| CASC15 | miR-221 | Intergenic | ceRNA ARID1A | [23]$ |
| DLEU1 | miR-490-3p | Intergenic sharing promoter of DLEU2 | ceRNA CDK1, CCND1 and SMARCD1 | [24]$ |
| INO80/SWR1 subunits (nucleosome replacement) | ||||
| LCTS5 (AC008610.1) | INO80 | Intergenic | Binding to INO80 inhibits binding to enhancer regions near lung cancer associated genes. | [31] |
| HAND2‐AS1 (lncHand2) | INO80 | Antisense, sharing promoter with SCRG1 | Recruiting the INO80 complex to e.g. the BMP-R1A and Nkx1-2 promoter | [32, 33]$ |
| ANRIL | YY1 | Antisense in INK4 Locus | Recruitment of YY1 to promoter loci of IL6 and IL8 | [34] |
| Linc-YY1 | YY1 | Intragenic of YY | evict YY1/Polycomb repressive complex (PRC2) from target promoters | [35] |
| lncAKHE (AK056594) | YEATS4 | Intragenic of TRIM55 | activation of NOTCH2 signaling in hepatocellular carcinoma | [36]$ |
| DRAIC | UCHL5 | Intergenic | Promotes NFRKB (INO80G) ubiquitination-mediated degradation | [37]$ |
| CASC7 | miR-21 | Intergenic (in between CHRAC1-Ago2) | ceRNA ING3 | [41]$ |
| RNCR3 (LINC00599) | miR-185-5p | Intergenic | ceRNA BRD8 | [40]$ |
| TUG1 | miR-145 and YY1 | Intergenic, sharing promoter with MORC2 | Interference PRC2 complex | [157]$ |
| CR933609 | miRNA-5096 | Overlapping INO80D 3’UTR | ceRNA INO80D | [39]$ |
| SPAG5-AS1 | miR-769-5p | Antisense | ceRNA YY1 | [42] |
| HOTAIR | miR-1, miR-206 | Intergenic, sharing promoter of HOXC11 | ceRNA YY1 | [43]$ |
| LINC00899 | miR-744-3p | Intergenic | ceRNA YY1 | [44]$ |
| LINC00668 | miR-532-5p | Intergenic | ceRNA YY1 | [45]$ |
| Linc01134 | miR-324-5p | Intergenic | ceRNA IGF2BP1, increased stability YY1 expression | [158]$ |
| LncKdm2b (KDM2B-DT) | SRCAP | Antisense to Kdm2b | SRCAP complex recruitment to Zbtb3 gene, activating expression of this transcription factor | [59] |
| ISWI (nucleosome spacing, DNA repair) | ||||
| LncKdm2b (KDM2B-DT) | SATB1-ISWI | Antisense to Kdm2b | NURF complex directed to Zfp292 promoter via recruitment of chromatin organizer SATB1 (also capable of recruiting SWI/SNF complexes) | [58] |
| NMR (LINC01672) | BPTF | Intergenic | NURF complex recruitment promoting ERK1/2 signaling pathway | [60]$ |
| DLEU1 | SMARCA1 | Intergenic sharing promoter of DLEU2 | NURF complex recruitment activating of KPNA3 | [61]$ |
| NEXN-AS1 | BAZ1A | Antisense | Upregulation of NEXN expression, which is associated with atherosclerosis-related diseases | [62] |
| Lnc pRNA | BAZ2A | At intergenic spacer (IGS) sequences separating rDNA regions | Establishment of heterochromatin at ribosomal RNA genes (repression) e.g. RNA45SN2 (45S pre-rRNA) | [63, 159] |
| Paupar | TRIM28 | Intragenic of PAX6-AS1 | promotes KAP1 (H3K9me3 deposition) recruitment at a subset of distal targets, through formation Paupar-KAP1-PAX6 complex | [64] |
| BlackMamba (–) | HELLS | Intergenic upstream of KCNMA1 | Gene expression (e.g., RGS1, CCL17, CCL22, PAK2, and KCNMA1) in anaplastic large cell lymphoma | [65]$ |
| FGD5-AS1 | miR-302e | Antisense | ceRNA CDCA7 | [66]$ |
| CHD (chromodomain helicase DNA-binding, DNA repair; NuRD, histone deacetylation/demethylase complex) | ||||
| CHASERR (LINC01578) | CHD2 | Intergenic, upstream of CHD2 | Transcriptional interference between Chaserr and CHD2 results in negative feedback loop | [87] |
| MATN1-AS1 | miR-200b/c/429 | Antisense | ceRNA CHD1 | [91]$ |
| LINC01410 | miR-23c | Intergenic | ceRNA CHD7 | [92]$ |
| ANRIL | miR-34a | Antisense in INK4 Locus | ceRNA HDAC1 | [93]$ |
| SNHG15 | miR-490-3p | miRNA host gene | ceRNA HDAC2 | [94]$ |
| ARAP1-AS1 | miR-2110 | antisense | ceRNA HDAC2 | [95]$ |
| HOTAIR | miR-326 | Intergenic, sharing promoter of HOXC11 | ceRNA MTA2 | [96]$ |
*UltraConserved elements gene location at https://genome-test.gi.ucsc.edu/~hiram/hubs/GillBejerano/hg19/hg19.ultraConserved.bb. $Studies showing the role of lncRNA chromatin remodeling in cancer
Fig. 2.
Effects of long non-coding RNAs on chromatin remodeling complexes. LncRNAs (in blue) affect chromatin remodeling by changing subunit expression (transcription and/or competing for miRNA binding (ceRNAs)), by inhibiting complex binding to or by stimulating the recruitment to a genomic region. a Interaction of SWI/SNF complexes with lncRNA results in nucleosome sliding either further apart or closer together. b Histone replacement (H2A.Z) and modifications in nucleosomes are affected by lncRNAs that regulate the INO80 complexes. c LncRNAs known to regulate the different ISWI complexes are mainly recruiting these complexes. d Generally, lncRNAs that function as ceRNAs are known for the CHD family
Chromatin remodeling complexes of the INO80/SWR1 family
Subunits of the INO80/SWR1 complexes
The INO80/SWR1 factors were first identified in budding yeast in a screen for regulators of phospholipid biosynthesis (INOsitol requiring) and for switching from glycolytic to oxidative metabolism (SWR1) [26]. This family includes three major complexes, each having a unique ATPase unit (INO80, SRCAP or p400) in combination with the two ATP-driven helicases RUVBL1 and RUVBL2. The “effector” sub-units may recruit other enzymes, such as transcription factors and histone acetyltransferases. In particular, the p400-TRRAP-TIP60 complex mediates acetylation of histone H4 and H2A-tails. This acetylation stimulates the INO80 and SRCAP complexes to replace histone H2A with the 60% homologous histone H2A.Z. The H2A.Z diverges with H2A at two loops interacting with the nucleosomal DNA and at the tail section interacting with histone H3, which may result in a reduced stability or in nucleosome sliding [27]. Nucleosomes with histone H2A.Z are enriched at flanking regions of transcription start sites. In addition, H2A.Z can also be modified by acetylation, methylation, phosphorylation, SUMOylation and ubiquitination [28]. Removal of H2A.Z from DNA by the histone chaperone ANP32E and INO80 is a primary step in DNA repair [29]. The INO80 complex may recruit additional subunits, e.g., NFKRP, implicated in double strand break DNA repair [30].
Long non-coding RNAs and the INO80/SWR1 family
Both the lncRNA LCTS5 and HAND2‐AS1 interact with the INO80 subunit, however, they have opposite effects (Fig. 2b). LCTS5 inhibits and HAND2‐AS1 stimulates INO80 complex recruitment [31–33]. Similarly, ANRIL and Linc-YY1 both interact with transcription factor YY1 resulting, respectively, in recruitment to promoter loci of IL6/IL8 and eviction of polycomb repressive complex (PRC2) regulated promoters [34, 35]. Interaction of lncAKHE with YEATS4 from the histone acetylation p400-TRRAP complex results in activation of NOTCH2 signaling in hepatocellular carcinomas [36].
Several lncRNAs regulate the expression of INO80 subunits. UCHL5 mediates de-ubiquitination of NFRKB (INO80G), which is prevented by its interaction with lncRNA DRAIC [37]. The subsequent NFRKB ubiquitination-mediated degradation affects resection of DNA in double strand breaks DNA-repair [30]. In addition, lncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3 [38]. The lncRNAs CR933609, RNCR3, and CASC7 were reported to act as ceRNAs targeting INO80D, BRD8 and ING3, respectively [39–41]. SPAG5-AS1, HOTAIR, LINC00899 and LINC00668 were reported to act as ceRNAs targeting YY1 [42–45].
Chromatin remodeling complexes of the ISWI family
Subunits of the ISWI complexes
Initially discovered in a search for SWI/SNF genes in Drosophila [46], ISWI complexes regulate nucleosome spacing and are implicated in DNA repair. These chromatin remodelers are rather small complexes with 2–4 sub-units. The best-known ATP-helicases in these complexes are SMARCA5 and SMARCA1. Both SMARCAD1 and SMARCA6 (HELLS) are SWI/SNF genes that can also be classified in this family. SMARCAD1-TRIM28 binds to H2A-ubiquitin and stimulates acetylation [47]. HELLS-CDC7A re-modulates the nucleosome to facilitate access to DNA for DNMT3B [48, 49]. SMARCA3 and SMARCAL1 are also ATP-driven DNA-binding helicases, but they are implicated, respectively, in DNA unwinding, and in the stabilization of the replication fork and they do not interact with nucleosomes [50, 51].
The WICH (SMARCA5-BAZ1B) and CERF (SMARCA1-CECR2) complexes affect the phosphorylation of histone H2A.X (γH2A.X) [52, 53]. This histone variant is highly homologous to H2A, but has a 13 amino acid extended C-tail that includes three phosphorylation sites (T136, S139 and Y142) [54]. Phosphorylation and ubiquitination of H2A.X are key events in the detection and response to double strand breaks of DNA damage.
The ACF (SMARCA5-BAZ1A) complex can either function as a heterodimer complex playing a role in double strand break repair [55], or as the CHRAC complex (ACF interacting with CHRAC1 and POLE3). Both complexes have very similar nucleosome sliding activity [56]. The NURF (SMARCA1-BPTF-RBBP4-RBBP7) complex also mediates nucleosome sliding and enhances recruitment of transcription factors and chromatin insulator protein elements [57].
Long non-coding RNAs and the ISWI family
The LncKdm2b (KDM2B-DT) seems to have a general function in recruiting chromatin remodeler complexes, as it was reported to direct both SRCAP (INO80 family) and NURF (ISWI family) complexes to two different zinc finger protein genes (Zbtb3 and Zfp292) [58, 59] (Fig. 2b, c). The NURF complex is also recruited through binding of lncRNA NMR to BPTF [60] and of DLEU1 to SMARCA1 [61]. Interestingly, DLEU1 has a double function as it seems to inhibit other complexes via miRNA-mediated decay of SMARCD1 (subunit of SWI/SNF complex). The NEXN-AS1-mediated recruitment of BAZ1A (ACF complex) to NEXN was shown to upregulate its expression [62], which may suggest a cis-acting function similar to the recruitment of the SWI/SNF complex to primary transcribed RNA. The “lnc pRNA” has a specific action on ribosomal RNA genes as its interaction with PARP1 and BAZ2A (NoRC complex) represses these genes in particular [63].
Recently, it was shown that Pauper interacts with KAP1 to promote H3K9me3 deposition at a subset of distal targets, through formation of a Paupar-KAP1-PAX6 complex [64]. If the ATP-dependent helicase SMARCAD1, also interacting with KAP1, is part of this complex was not assessed in this study. The CHIRRC complex was reported to be affected by lncRNA BlackMamba via interaction with HELLS and by FGD5-AS1 as ceRNA inhibiting CDCA7 degradation [65, 66].
Chromatin remodeling complexes of the CHD family
Subunits of the CHD complexes
The chromodomain helicase DNA-binding (CHD) proteins that were initially identified as mammalian DNA-binding factors with a SWI-like helicase domain [67], are implicated in DNA repair and in recruitment of histone deacetylase/demethylase enzymes. CHD1 and CHD2 belong to the same subclass I. CHD1 has been shown to promote stabilization of H2A.X and the efficient repair of double strand breaks through homologous recombination [68]. CHD1 has no specific subunit, but has been described to interact with several factors, such as SSRP1 and the NCoR complex [69, 70]. CHD1 is a critical regulator of transcription initiation and elongation stimulating androgen receptor (AR)-mediated regulation and pluripotency gene expression (Oct 4 and Nanog) [67, 71].
The CHD2 protein has been shown to interact with PARP1 complex stimulating histone variant H3.3 deposition in non-homologous end joining (NHEJ) DNA-repair regions [72]. The histone H3.3 has only four amino acid differences compared to H3, introducing a posttranslational phosphorylation site (S31). Phosphorylated H3.3S31p stimulates p300-mediated histone H3K27 acetylation of neighboring nucleosomes [73]. In addition, the combination of histone H3.3 with H2A.Z in nucleosomes enriched near promoters and enhancers also evokes a loose nucleosomal packaging [74].
In the NuRD complex, one of the helicase from sub-family II, CHD3, CHD4 or CHD5 is present where they interact with GATAD2A/B. Interaction of the GATAD2 proteins with MDB2/3 results in recruitment of the NuRD histone deacetylase sub-complex (HDAC1/2, RBBP4/7, CTBP2, MTA1/2/3) [75]. The NuRD complex may also interact with several additional proteins, such as CDK2AP1, SALL1/4 and ZMYND8 that may direct the NuRD complex to specific regions [75]. CHD4 is also binding to RNF8, which is implicated in histone H1-ubiquitinylation and loading of the BRCA1 complex [76].
Subfamily III of CHD remodelers includes CHD6–CHD9. They interact with diverse transcription factors and posttranslationally modified histone H3. For CHD6, CHD7 and CHD8 it has been shown that they bind to the DNA strings in between nucleosomes. CHD6 and CHD7 both bind to short linker DNA, whereas CHD8 requires longer DNA sequences for binding and thus slides nucleosomes further apart [77]. CHD6 rather disrupts nucleosomes and was recently shown to relocalize to sites of DNA damage, suggesting a role in DNA repair [77, 78]. CHD7 and CHD9 have been shown to interact with chromatin remodelers from the SWI/SNF family (PBAF/BAF) [79]. CHD8 can directly bind β-catenin mediating interaction with histone H1 or other factors like the Gfi1b-complex regulating Wnt/β-catenin-dependent gene expression [80, 81]. CHD8 was further shown to interact with diverse methyltransferase complexes, such as BRD4-NSD3 [82], BACH1-MAFG-DNMT3 [83], SET1 (KMT2) [84] and WDR5-ASH2L-RbBP5 [85]. In addition, CHD7 was shown to interact with CHD8/FAM124B [86]. Both CHD7 and CHD8 are implicated in neuronal and developmental disorders (e.g., CHARGE, autism [77]).
Long non-coding RNAs and the CHD family
So far, only a few lncRNAs have been described in the context of CHD complexes (Fig. 2d). Recently, a negative regulation loop beween lncRNA CHASERR and CHD2 has been described. The CHASERR gene is located on the same strand (1.7 kb apart) as the CHD2 gene at chr15q26. The CHASERR transcript can be bound by CHD2 protein and regulate transcription of the CHD2 gene [87]. Some lncRNAs have been shown to affect transcriptional regulation of CHD subunits. The lncRNA MTA2TR transcriptionally upregulates MTA2 expression [88]. LUCAT1 upregulates MTA1 in cervical cancer and HDAC1 in papillary thyroid cancer [89, 90]. Six lncRNAs have been described as ceRNAs for CHD subunits (MATN1-AS1 ceRNA CHD1 [91], LINC01410 ceRNA CHD7 [92], ANRIL ceRNA HDAC1 [93], SNHG15 and ARAP1-AS1 ceRNA HDAC2 [94, 95] and HOTAIR ceRNA MTA2 [96]).
Concluding remarks
The lncRNAs described in this review often recruit chromatin remodeler complexes at promoter and enhancer regions by binding a particular subunit. Some other lncRNAs, not discussed herein, are related to the chromatin remodeler complex through their position, but they seem to have unrelated functions (lnc-Arid2-IR, lnc-SMARCC2, lnc-MDB2, lnc-Dpf3) [97–100].
A recurrent chromatin regulatory mechanism is recruitment of remodeler complexes by interaction with primary transcribed RNAs, either by antisense lncRNA encoded at genomic proximity, or by trans-acting lncRNAs (e.g., LncKdm2b-mediated recruitment of SRCAP and NURF complexes [58, 59]). The interaction of chromatin remodeler subunits with some lncRNAs results in a distinct cellular function. For example, interaction of lncBRM with BRM inhibits the SWI/SNF complex associated with a differentiated phenotype and favors assembly of a stem cell-related SWI/SNF complex [18]. Also lnc-pRNA regulates primarily ribosomal RNA gene expression via the NoRC complex [63]. The interaction between chromatin remodeler subunits, in particular, the flexible and/or successive nature of complex formation in the different processes like NHEJ or HR-mediated DNA repair will be an important asset to further define the role of lncRNAs in these processes. The role of the chromatin remodelers in the different stages of DNA repair is closely related to cancer (e.g., [67, 101–103]). Indeed, from the lncRNAs that affect the chromatin remodeling complexes (Table 2) at least 35 are associated with cancer. Cellular capture-based approaches can be employed to further analyze the interactions of lncRNAs with the chromatin remodeler complexes, using either RNA-specific, DNA-locus specific or genome wide approaches (reviewed by [104]), although non-specific background interaction signals and sequential complex formation remains a major issue. The CRISPR-Cas9 engineering revolution now enables in situ capture of RNA-chromatin interactions by biotinylated dCas9 [104], which may also reveal implication of lncRNAs in the cancer-related nucleosome remodeling evidenced by Druliner et al. [105]. In addition, analyzing CRISPR/Cas9-based gene perturbations (e.g., lncRNAs invalidation/activation) combined with single cell sequence analysis (recently reported by [106–108]), may unravel the specific actions of lncRNAs, that are measured as low expressed in bulk-analyzing experiments but can be abundantly expressed in individual, and phenotype-specific cells (e.g., shown in neuronal cells and cardiomyocytes [109, 110]). Although today still presenting a huge technical challenge, these analyses could be combined with studies of chromatin organization and protein interactions in single cells [111]. Identifying the lncRNAs that contribute to epigenetic regulation by controlling the specific chromatin modifications associated with disease, may result in interesting novel targets for e.g. oligonucleotide-based therapy [112]. In particular, DNA-Gapmer antisense oligonucleotides, of which several have received therapeutic FDA-approval, are suitable for targeting lncRNAs as they can evoke RNASEH1-mediated transcript degradation both within the nucleus and cytosol.
In conclusion, the description of lncRNAs interacting with chromatin remodeler complexes in this review, intends to highlight the importance of lncRNA-mediated chromatin remodeling via remodeler complexes in physiological and carcinogenic conditions.
Acknowledgements
This work was supported by grants from “Institut National de la Santé et de la Recherche Médicale” (Inserm), “Centre National de la Recherche Scientifique” (CNRS) and the “Cancéropôle Nord-Ouest” (Bourse Emergence 2019) and “Ligue Nationale contre le Cancer” (Comité Départemental CD59, CD80). The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish this review.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Audrey Vincent and Isabelle Van Seuningen are last co-auteurs.
References
- 1.Shah FR, Bhat YA, Wani AH. Subnuclear distribution of proteins: links with genome architecture. Nucleus. 2018;9(1):42–55. doi: 10.1080/19491034.2017.1361578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annunziato AT (2008) DNA packaging: nucleosomes and chromatin. Nat Educ 1(26):310. https://www.nature.com/scitable/topicpage/dna-packaging-nucleosomes-and-chromatin-310
- 3.Chereji RV, Clark DJ. Major determinants of nucleosome positioning. Biophys J. 2018;114(10):2279–2289. doi: 10.1016/j.bpj.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang R, Erler J, Langowski J. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys J. 2017;112(3):450–459. doi: 10.1016/j.bpj.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfert A, Moreno N, Kerl K. The BAF complex in development and disease. Epigenetics Chromatin. 2019;12(1):19. doi: 10.1186/s13072-019-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18(7):407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, Zhu J, Fan G, Archer TK, Wang Y. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3(3):460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27(12):2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez-Vilendrer SB, Rai SK, Gramling SJ, Lu L, Reisman DN. BRG1 and BRM loss selectively impacts RB and P53, respectively: BRG1 and BRM have differential functions in vivo. Oncoscience. 2016;3(11–12):337–350. doi: 10.18632/oncoscience.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakarougkas A, Ismail A, Chambers AL, Riballo E, Herbert AD, Kunzel J, Lobrich M, Jeggo PA, Downs JA. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol Cell. 2014;55(5):723–732. doi: 10.1016/j.molcel.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Zhang J, Chen X. The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem. 2007;282(52):37429–37435. doi: 10.1074/jbc.M706039200. [DOI] [PubMed] [Google Scholar]
- 13.Giles KA, Gould CM, Du Q, Skvortsova K, Song JZ, Maddugoda MP, Achinger-Kawecka J, Stirzaker C, Clark SJ, Taberlay PC. Integrated epigenomic analysis stratifies chromatin remodellers into distinct functional groups. Epigenetics Chromatin. 2019;12(1):12. doi: 10.1186/s13072-019-0258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Local A, Huang H, Albuquerque CP, Singh N, Lee AY, Wang W, Wang C, Hsia JE, Shiau AK, Ge K, Corbett KD, Wang D, Zhou H, Ren B. Identification of H3K4me1-associated proteins at mammalian enhancers. Nat Genet. 2018;50(1):73–82. doi: 10.1038/s41588-017-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raab JR, Smith KN, Spear CC, Manner CJ, Calabrese JM, Magnuson T. SWI/SNF remains localized to chromatin in the presence of SCHLAP1. Nat Genet. 2019;51(1):26–29. doi: 10.1038/s41588-018-0272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grossi E, Raimondi I, Goni E, Gonzalez J, Marchese FP, Chapaprieta V, Martin-Subero JI, Guo S, Huarte M. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat Commun. 2020;11(1):936. doi: 10.1038/s41467-020-14623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, Du Y, Gao G, Tian Y, He L, Fan Z. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panatta E, Lena AM, Mancini M, Smirnov A, Marini A, Delli Ponti R, Botta-Orfila T, Tartaglia GG, Mauriello A, Zhang X, Calin GA, Melino G, Candi E. Long non-coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex. EMBO Rep. 2020;21(3):e46734. doi: 10.15252/embr.201846734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei JJ, Li HQ, Mo ZH, Liu KJ, Zhu LJ, Li CY, Chen WL, Zhang L. Long noncoding RNA CDKN2B-AS1 interacts with transcription factor BCL11A to regulate progression of cerebral infarction through mediating MAP4K1 transcription. FASEB J. 2019;33(6):7037–7048. doi: 10.1096/fj.201802252R. [DOI] [PubMed] [Google Scholar]
- 21.Zhang CH, Wang J, Zhang LX, Lu YH, Ji TH, Xu L, Ling LJ. Shikonin reduces tamoxifen resistance through long non-coding RNA uc.57. Oncotarget. 2017;8(51):88658–88669. doi: 10.18632/oncotarget.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Gao S, Zheng Y, Yao M, Ruan F. LncRNA CASC15 functions as an unfavorable predictor of ovarian cancer prognosis and inhibits tumor progression through regulation of miR-221/ARID1A axis. Onco Targets Ther. 2019;12:8725–8736. doi: 10.2147/ott.S219900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LL, Sun KX, Wu DD, Xiu YL, Chen X, Chen S, Zong ZH, Sang XB, Liu Y, Zhao Y. DLEU1 contributes to ovarian carcinoma tumourigenesis and development by interacting with miR-490-3p and altering CDK1 expression. J Cell Mol Med. 2017;21(11):3055–3065. doi: 10.1111/jcmm.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao J, Xie N. Long noncoding RNA DSCAM-AS1 functions as an oncogene in non-small cell lung cancer by targeting BCL11A. Eur Rev Med Pharmacol Sci. 2019;23(3):1087–1092. doi: 10.26355/eurrev_201902_16998. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Gao S, Peng X, Wu K, Yang S. Roles of the INO80 and SWR1 Chromatin Remodeling Complexes in Plants. Int J Mol Sci. 2019 doi: 10.3390/ijms20184591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16(2):166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Giaimo BD, Ferrante F, Herchenrother A, Hake SB, Borggrefe T. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin. 2019;12(1):37. doi: 10.1186/s13072-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alatwi HE, Downs JA. Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 2015;16(8):986–994. doi: 10.15252/embr.201540330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, Clague MJ, Urbe S, Jackson SP. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16(10):1016–1026. doi: 10.1038/ncb3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Wang Y, Ma D, Wang L, Yang M. Long noncoding RNA LCTS5 inhibits non-small cell lung cancer by interacting with INO80. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117680. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W, Du Y, Ye B, Wang D, He L, Ren W, Wang J, Sun X, Chen R, Tian Y, Fan Z. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019;38(17):e101110. doi: 10.15252/embj.2018101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu P, Wang J, Zhu X, Luo J, Meng S, Wu J, Ye B, He L, Du Y, He L, Chen R, Tian Y, Fan Z. Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J Hepatol. 2018;69(4):861–872. doi: 10.1016/j.jhep.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X, Gan LM, Cao H, Liang Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016;13(1):98–108. doi: 10.1080/15476286.2015.1122164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L, Sun K, Zhao Y, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, Zhu X, Wang L, Tang LY, Esteban MA, Wang CC, Jauch R, Sun H, Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 36.Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi B, Luo Y, Jian Z, Zhou X. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis. 2018;9(5):487. doi: 10.1038/s41419-018-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Hu X, Kuang J, Liao J, Yuan Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett. 2020;25:29. doi: 10.1186/s11658-020-00221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang XM, Liu Y, Fan YX, Liu Z, Yuan QL, Jia M, Geng ZS, Gu L, Lu XB. LncRNA PTCSC3 affects drug resistance of anaplastic thyroid cancer through STAT3/INO80 pathway. Cancer Biol Ther. 2018;19(7):590–597. doi: 10.1080/15384047.2018.1449610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CC, Liu TY, Lee YT, Chen YC, Yeh KT, Lee CC, Chen YL, Lin PC, Chang YS, Chan WL, Liu TC, Chang JG. Genome-wide analysis of lncRNAs in 3'-untranslated regions: CR933609 acts as a decoy to protect the INO80D gene. Int J Oncol. 2018;53(1):417–433. doi: 10.3892/ijo.2018.4398. [DOI] [PubMed] [Google Scholar]
- 40.Tian C, Deng Y, Jin Y, Shi S, Bi H. Long non-coding RNA RNCR3 promotes prostate cancer progression through targeting miR-185-5p. Am J Transl Res. 2018;10(5):1562–1570. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Fu C, Xu Q, Wei X. Long non-coding RNA CASC7 inhibits the proliferation and migration of colon cancer cells via inhibiting microRNA-21. Biomed Pharmacother. 2017;95:1644–1653. doi: 10.1016/j.biopha.2017.09.052. [DOI] [PubMed] [Google Scholar]
- 42.Xu J, Deng Y, Wang Y, Sun X, Chen S, Fu G. SPAG5-AS1 inhibited autophagy and aggravated apoptosis of podocytes via SPAG5/AKT/mTOR pathway. Cell Prolif. 2020;53(2):e12738. doi: 10.1111/cpr.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Li N, Fu J, Zhou W. Long noncoding RNA HOTAIR promotes medulloblastoma growth, migration and invasion by sponging miR-1/miR-206 and targeting YY1. Biomed Pharmacother. 2020;124:109887. doi: 10.1016/j.biopha.2020.109887. [DOI] [PubMed] [Google Scholar]
- 44.Dong X, Xu X, Guan Y. LncRNA LINC00899 promotes progression of acute myeloid leukaemia by modulating miR-744-3p/YY1 signalling. Cell Biochem Funct. 2020 doi: 10.1002/cbf.3521. [DOI] [PubMed] [Google Scholar]
- 45.You G, Zhou C, Xuan W. LncRNA LINC00668 promotes cell proliferation, migration, invasion ability and EMT process in hepatocellular carcinoma by targeting miR-532-5p/YY1 axis. Biosci Rep. 2020 doi: 10.1042/bsr20192697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aydin OZ, Vermeulen W, Lans H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle. 2014;13(19):3016–3025. doi: 10.4161/15384101.2014.956551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kazakevych J, Denizot J, Liebert A, Portovedo M, Mosavie M, Jain P, Stellato C, Fraser C, Correa RO, Celestine M, Mattiuz R, Okkenhaug H, Miller JR, Vinolo MAR, Veldhoen M, Varga-Weisz P. Smarcad1 mediates microbiota-induced inflammation in mouse and coordinates gene expression in the intestinal epithelium. Genome Biol. 2020;21(1):64. doi: 10.1186/s13059-020-01976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kollarovic G, Topping CE, Shaw EP, Chambers AL. The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin. Nucleic Acids Res. 2020;48(4):1872–1885. doi: 10.1093/nar/gkz1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenness C, Giunta S, Muller MM, Kimura H, Muir TW, Funabiki H. HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex defective in ICF syndrome. Proc Natl Acad Sci USA. 2018;115(5):E876–E885. doi: 10.1073/pnas.1717509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng E, Batenburg NL, Walker JR, Ho A, Mitchell TRH, Qin J, Zhu XD. CSB cooperates with SMARCAL1 to maintain telomere stability in ALT cells. J Cell Sci. 2020 doi: 10.1242/jcs.234914. [DOI] [PubMed] [Google Scholar]
- 51.Oh YS, Gao P, Lee KW, Ceglia I, Seo JS, Zhang X, Ahn JH, Chait BT, Patel DJ, Kim Y, Greengard P. SMARCA3, a chromatin-remodeling factor, is required for p11-dependent antidepressant action. Cell. 2013;152(4):831–843. doi: 10.1016/j.cell.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ji JH, Min S, Chae S, Ha GH, Kim Y, Park YJ, Lee CW, Cho H. De novo phosphorylation of H2AX by WSTF regulates transcription-coupled homologous recombination repair. Nucleic Acids Res. 2019;47(12):6299–6314. doi: 10.1093/nar/gkz309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SK, Park EJ, Lee HS, Lee YS, Kwon J. Genome-wide screen of human bromodomain-containing proteins identifies Cecr2 as a novel DNA damage response protein. Mol Cells. 2012;34(1):85–91. doi: 10.1007/s10059-012-0112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corujo D, Buschbeck M. Post-translational modifications of H2A histone variants and their role in cancer. Cancers (Basel) 2018 doi: 10.3390/cancers10030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klement K, Luijsterburg MS, Pinder JB, Cena CS, Del Nero V, Wintersinger CM, Dellaire G, van Attikum H, Goodarzi AA. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol. 2014;207(6):717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scacchetti A, Brueckner L, Jain D, Schauer T, Zhang X, Schnorrer F, van Steensel B, Straub T, Becker PB. CHRAC/ACF contribute to the repressive ground state of chromatin. Life Sci Alliance. 2018;1(1):e201800024. doi: 10.26508/lsa.201800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alkhatib SG, Landry JW. The nucleosome remodeling factor. FEBS Lett. 2011;585(20):3197–3207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, Du Y, Wu J, Qin X, Chen R, Tian Y, Fan Z. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol. 2017;18(5):499–508. doi: 10.1038/ni.3712. [DOI] [PubMed] [Google Scholar]
- 59.Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W, Zhu P, Wang Y, Wang S, Xia P, Du Y, Meng S, Huang G, Wu J, Chen R, Tian Y, Fan Z. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018 doi: 10.15252/embj.201797174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Li J, Luo M, Zhou C, Shi X, Yang W, Lu Z, Chen Z, Sun N, He J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57–66. doi: 10.1016/j.canlet.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 61.Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17(1):118. doi: 10.1186/s12943-018-0873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu YW, Guo FX, Xu YJ, Li P, Lu ZF, McVey DG, Zheng L, Wang Q, Ye JH, Kang CM, Wu SG, Zhao JJ, Ma X, Yang Z, Fang FC, Qiu YR, Xu BM, Xiao L, Wu Q, Wu LM, Ding L, Webb TR, Samani NJ, Ye S. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J Clin Invest. 2019;129(3):1115–1128. doi: 10.1172/jci98230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell. 2012;45(6):790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 64.Pavlaki I, Alammari F, Sun B, Clark N, Sirey T, Lee S, Woodcock DJ, Ponting CP, Szele FG, Vance KW. The long non-coding RNA Paupar promotes KAP1-dependent chromatin changes and regulates olfactory bulb neurogenesis. EMBO J. 2018 doi: 10.15252/embj.201798219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fragliasso V, Verma A, Manzotti G, Tameni A, Bareja R, Heavican TB, Iqbal J, Wang R, Fiore D, Mularoni V, Chan WC, Lhoumaud P, Skok J, Zanetti E, Merli F, Ciarrocchi A, Elemento O, Inghirami G. The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK(-) anaplastic large cell lymphoma by regulating the DNA helicase HELLS. Leukemia. 2020 doi: 10.1038/s41375-020-0754-8. [DOI] [PubMed] [Google Scholar]
- 66.Li D, Jiang X, Zhang X, Cao G, Wang D, Chen Z. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. Vitro Cell Dev Biol Anim. 2019;55(8):577–585. doi: 10.1007/s11626-019-00376-x. [DOI] [PubMed] [Google Scholar]
- 67.Mills AA. The chromodomain helicase DNA-binding chromatin remodelers: family traits that protect from and promote cancer. Cold Spring Harb Perspect Med. 2017 doi: 10.1101/cshperspect.a026450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Li J, Serafim RB, Ketchum S, Ferreira CG, Liu JC, Coe KA, Price BD, Yusufzai T. Human CHD1 is required for early DNA-damage signaling and is uniquely regulated by its N terminus. Nucleic Acids Res. 2018;46(8):3891–3905. doi: 10.1093/nar/gky128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelley DE, Stokes DG, Perry RP. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999;108(1):10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- 70.Tai HH, Geisterfer M, Bell JC, Moniwa M, Davie JR, Boucher L, McBurney MW. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun. 2003;308(1):170–176. doi: 10.1016/s0006-291x(03)01354-8. [DOI] [PubMed] [Google Scholar]
- 71.Skene PJ, Hernandez AE, Groudine M, Henikoff S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, van Attikum H. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61(4):547–562. doi: 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, Tastemel M, Banaszynski LA. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet. 2019;51(6):941–946. doi: 10.1038/s41588-019-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21(12):1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leighton G, Williams DC., Jr The methyl-CpG-binding domain 2 and 3 proteins and formation of the nucleosome remodeling and deacetylase complex. J Mol Biol. 2019 doi: 10.1016/j.jmb.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rother MB, van Attikum H. DNA repair goes hip-hop: SMARCA and CHD chromatin remodellers join the break dance. Philos Trans R Soc Lond B Biol Sci. 2017 doi: 10.1098/rstb.2016.0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manning BJ, Yusufzai T. The ATP-dependent chromatin remodeling enzymes CHD6, CHD7, and CHD8 exhibit distinct nucleosome binding and remodeling activities. J Biol Chem. 2017;292(28):11927–11936. doi: 10.1074/jbc.M117.779470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moore S, Berger ND, Luijsterburg MS, Piett CG, Stanley FKT, Schrader CU, Fang S, Chan JA, Schriemer DC, Nagel ZD, van Attikum H, Goodarzi AA. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat Commun. 2019;10(1):241. doi: 10.1038/s41467-018-08111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463(7283):958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishiyama M, Skoultchi AI, Nakayama KI. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol. 2012;32(2):501–512. doi: 10.1128/MCB.06409-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shooshtarizadeh P, Helness A, Vadnais C, Brouwer N, Beauchemin H, Chen R, Bagci H, Staal FJT, Cote JF, Moroy T. Gfi1b regulates the level of Wnt/β-catenin signaling in hematopoietic stem cells and megakaryocytes. Nat Commun. 2019;10(1):1270. doi: 10.1038/s41467-019-09273-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60(6):847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol Cell. 2014;55(6):904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao C, Dong C, Frah M, Deng Y, Marie C, Zhang F, Xu L, Ma Z, Dong X, Lin Y, Koenig S, Nait-Oumesmar B, Martin DM, Wu LN, Xin M, Zhou W, Parras C, Lu QR. Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev Cell. 2018;45(6):753–768. doi: 10.1016/j.devcel.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yates JA, Menon T, Thompson BA, Bochar DA. Regulation of HOXA2 gene expression by the ATP-dependent chromatin remodeling enzyme CHD8. FEBS Lett. 2010;584(4):689–693. doi: 10.1016/j.febslet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 86.Batsukh T, Schulz Y, Wolf S, Rabe TI, Oellerich T, Urlaub H, Schaefer IM, Pauli S. Identification and characterization of FAM124B as a novel component of a CHD7 and CHD8 containing complex. PLoS ONE. 2012;7(12):e52640. doi: 10.1371/journal.pone.0052640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rom A, Melamed L, Gil N, Goldrich MJ, Kadir R, Golan M, Biton I, Perry RB, Ulitsky I. Regulation of CHD2 expression by the Chaserr long noncoding RNA gene is essential for viability. Nat Commun. 2019;10(1):5092. doi: 10.1038/s41467-019-13075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, Zhong JX, Deng SJ, Liu ML, Huang K, Li Q, Li W, Hu YH, Wang F, Wang CY, Zhao G. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics. 2019;9(18):5298–5314. doi: 10.7150/thno.34559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang AH, Zhao JM, Du J, Pang QX, Wang MQ. Long noncoding RNA LUCAT1 promotes cervical cancer cell proliferation and invasion by upregulating MTA1. Eur Rev Med Pharmacol Sci. 2019;23(16):6824–6829. doi: 10.26355/eurrev_201908_18721. [DOI] [PubMed] [Google Scholar]
- 90.Luzón-Toro B, Fernández RM, Martos-Martínez JM, Rubio-Manzanares-Dorado M, Antiñolo G, Borrego S. LncRNA LUCAT1 as a novel prognostic biomarker for patients with papillary thyroid cancer. Sci Rep. 2019;9(1):14374. doi: 10.1038/s41598-019-50913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu J, Gu W, Yu C. MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif. 2020;53(1):e12700. doi: 10.1111/cpr.12700. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Lu M, Ding N, Zhuang S, Li Y. LINC01410/miR-23c/CHD7 functions as a ceRNA network to affect the prognosis of patients with endometrial cancer and strengthen the malignant properties of endometrial cancer cells. Mol Cell Biochem. 2020 doi: 10.1007/s11010-020-03723-9. [DOI] [PubMed] [Google Scholar]
- 93.Wang CH, Li QY, Nie L, Ma J, Yao CJ, Chen FP. LncRNA ANRIL promotes cell proliferation, migration and invasion during acute myeloid leukemia pathogenesis via negatively regulating miR-34a. Int J Biochem Cell Biol. 2020;119:105666. doi: 10.1016/j.biocel.2019.105666. [DOI] [PubMed] [Google Scholar]
- 94.Dai W, Dai JL, Tang MH, Ye MS, Fang S. lncRNA-SNHG15 accelerates the development of hepatocellular carcinoma by targeting miR-490-3p/histone deacetylase 2 axis. World J Gastroenterol. 2019;25(38):5789–5799. doi: 10.3748/wjg.v25.i38.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu C, Wang X, Zhao X, Xin Y, Liu C. Long non-coding RNA ARAP1-AS1 accelerates cell proliferation and migration in breast cancer through miR-2110/HDAC2/PLIN1 axis. Biosci Rep. 2020 doi: 10.1042/bsr20191764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tao D, Zhang Z, Liu X, Zhang Z, Fu Y, Zhang P, Yuan H, Liu L, Cheng J, Jiang H. LncRNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through metastasis-associated gene 2. Mol Carcinog. 2020;59(4):353–364. doi: 10.1002/mc.23159. [DOI] [PubMed] [Google Scholar]
- 97.Zhou Q, Huang XR, Yu J, Yu X, Lan HY. Long noncoding RNA Arid2-IR is a novel therapeutic target for renal inflammation. Mol Ther. 2015;23(6):1034–1043. doi: 10.1038/mt.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji R, Guo Q, Li Q, Ye Y, Wu J, Zhou Y, Qiao L. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett. 2018;418:84–96. doi: 10.1016/j.canlet.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 99.Ge Y, Zhang R, Feng Y, Li H. Mbd2 mediates retinal cell apoptosis by targeting the lncRNA Mbd2-AL1/miR-188-3p/Traf3 axis in ischemia/reperfusion injury. Mol Ther Nucleic Acids. 2020;19:1250–1265. doi: 10.1016/j.omtn.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, Chen Y, Zhu H, Li Z, Cao X. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50(3):600–615.e615. doi: 10.1016/j.immuni.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 101.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21(3):396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee RS, Roberts CW. Linking the SWI/SNF complex to prostate cancer. Nat Genet. 2013;45(11):1268–1269. doi: 10.1038/ng.2805. [DOI] [PubMed] [Google Scholar]
- 103.Hasan N, Ahuja N. The emerging roles of ATP-dependent chromatin remodeling complexes in pancreatic cancer. Cancers (Basel) 2019 doi: 10.3390/cancers11121859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet. 2019;20(9):503–519. doi: 10.1038/s41576-019-0135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Druliner BR, Vera D, Johnson R, Ruan X, Apone LM, Dimalanta ET, Stewart FJ, Boardman L, Dennis JH. Comprehensive nucleosome mapping of the human genome in cancer progression. Oncotarget. 2016;7(12):13429–13445. doi: 10.18632/oncotarget.6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell. 2016;167(7):1853–1866.e1817. doi: 10.1016/j.cell.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell. 2016;167(7):1883–1896.e1815. doi: 10.1016/j.cell.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 108.Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14(3):297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA, Lim DA. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016 doi: 10.1186/s13059-13016-10932-13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.See K, Tan WLW, Lim EH, Tiang Z, Lee LT, Li PYQ, Luu TDA, Ackers-Johnson M, Foo RS. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8(1):225. doi: 10.1038/s41467-017-00319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ortiz V, Yu M. Analyzing circulating tumor cells one at a time. Trends Cell Biol. 2018;28(10):764–775. doi: 10.1016/j.tcb.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rezazadeh S, Yang D, Tombline G, Simon M, Regan SP, Seluanov A, Gorbunova V. SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res. 2019;47(15):7914–7928. doi: 10.1093/nar/gkz528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Porter EG, Dykhuizen EC. Individual bromodomains of polybromo-1 contribute to chromatin association and tumor suppression in clear cell renal carcinoma. J Biol Chem. 2017;292(7):2601–2610. doi: 10.1074/jbc.M116.746875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moody RR, Lo MC, Meagher JL, Lin CC, Stevers NO, Tinsley SL, Jung I, Matvekas A, Stuckey JA, Sun D. Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes. J Biol Chem. 2018;293(6):2125–2136. doi: 10.1074/jbc.M117.811463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci USA. 2010;107(32):14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gatchalian J, Malik S, Ho J, Lee DS, Kelso TWR, Shokhirev MN, Dixon JR, Hargreaves DC. A non-canonical BRD9-containing BAF chromatin remodeling complex regulates naive pluripotency in mouse embryonic stem cells. Nat Commun. 2018;9(1):5139. doi: 10.1038/s41467-018-07528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang RR, Pan R, Zhang W, Fu J, Lin JD, Meng ZX. The SWI/SNF chromatin-remodeling factors BAF60a, b, and c in nutrient signaling and metabolic control. Protein Cell. 2018;9(2):207–215. doi: 10.1007/s13238-017-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harada A, Ohkawa Y, Imbalzano AN. Temporal regulation of chromatin during myoblast differentiation. Semin Cell Dev Biol. 2017;72:77–86. doi: 10.1016/j.semcdb.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McBride MJ, Pulice JL, Beird HC, Ingram DR, D'Avino AR, Shern JF, Charville GW, Hornick JL, Nakayama RT, Garcia-Rivera EM, Araujo DM, Wang WL, Tsai JW, Yeagley M, Wagner AJ, Futreal PA, Khan J, Lazar AJ, Kadoch C. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell. 2018;33(6):1128–1141. doi: 10.1016/j.ccell.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Willhoft O, Wigley DB. INO80 and SWR1 complexes: the non-identical twins of chromatin remodelling. Curr Opin Struct Biol. 2019;61:50–58. doi: 10.1016/j.sbi.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40(8):3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cox E, Hwang W, Uzoma I, Hu J, Guzzo CM, Jeong J, Matunis MJ, Qian J, Zhu H, Blackshaw S. Global analysis of SUMO-binding proteins identifies SUMOylation as a key regulator of the INO80 chromatin remodeling complex. Mol Cell Proteomics. 2017;16(5):812–823. doi: 10.1074/mcp.M116.063719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell. 2015;57(5):887–900. doi: 10.1016/j.molcel.2014.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liang X, Shan S, Pan L, Zhao J, Ranjan A, Wang F, Zhang Z, Huang Y, Feng H, Wei D, Huang L, Liu X, Zhong Q, Lou J, Li G, Wu C, Zhou Z. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat Struct Mol Biol. 2016;23(4):317–323. doi: 10.1038/nsmb.3190. [DOI] [PubMed] [Google Scholar]
- 126.Cho HJ, Li H, Linhares BM, Kim E, Ndoj J, Miao H, Grembecka J, Cierpicki T. GAS41 recognizes diacetylated Histone H3 through a bivalent binding mode. ACS Chem Biol. 2018;13(9):2739–2746. doi: 10.1021/acschembio.8b00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Y, Jin J, Chung MWH, Feng L, Sun H, Hao Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc Natl Acad Sci USA. 2018;115(10):2365–2370. doi: 10.1073/pnas.1717664115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hota SK, Bruneau BG. ATP-dependent chromatin remodeling during mammalian development. Development. 2016;143(16):2882–2897. doi: 10.1242/dev.128892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ding D, Bergmaier P, Sachs P, Klangwart M, Ruckert T, Bartels N, Demmers J, Dekker M, Poot RA, Mermoud JE. The CUE1 domain of the SNF2-like chromatin remodeler SMARCAD1 mediates its association with KRAB-associated protein 1 (KAP1) and KAP1 target genes. J Biol Chem. 2018;293(8):2711–2724. doi: 10.1074/jbc.RA117.000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Anosova I, Melnik S, Tripsianes K, Kateb F, Grummt I, Sattler M. A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes. Nucleic Acids Res. 2015;43(10):5208–5220. doi: 10.1093/nar/gkv365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tallant C, Valentini E, Fedorov O, Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S, Ciulli A. Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC. Structure. 2015;23(1):80–92. doi: 10.1016/j.str.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle. 2013;12(18):3070–3082. doi: 10.4161/cc.26033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bellelli R, Belan O, Pye VE, Clement C, Maslen SL, Skehel JM, Cherepanov P, Almouzni G, Boulton SJ. POLE3-POLE4 is a Histone H3–H4 chaperone that maintains chromatin integrity during DNA replication. Mol Cell. 2018;72(1):112–126. doi: 10.1016/j.molcel.2018.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee JS, Garrett AS, Yen K, Takahashi YH, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26(9):914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Egan Chris M, Nyman U, Skotte J, Streubel G, Turner S, O’Connell David J, Rraklli V, Dolan Michael J, Chadderton N, Hansen K, Farrar Gwyneth J, Helin K, Holmberg J, Bracken Adrian P. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell. 2013;26(3):223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 136.Torchy MP, Hamiche A, Klaholz BP. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci. 2015;72(13):2491–2507. doi: 10.1007/s00018-015-1880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang W, Duan N, Zhang Q, Song T, Li Z, Chen X, Wang K. The intracellular NADH level regulates atrophic nonunion pathogenesis through the CtBP2-p300-Runx2 transcriptional complex. Int J Biol Sci. 2018;14(14):2023–2036. doi: 10.7150/ijbs.28302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cajigas I, Leib DE, Cochrane J, Luo H, Swyter KR, Chen S, Clark BS, Thompson J, Yates JR, 3rd, Kingston RE, Kohtz JD. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142(15):2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang X, Gong Y, Jin BO, Wu C, Yang J, Wang LE, Zhang Z, Mao Z. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol Rep. 2014;32(3):1281–1290. doi: 10.3892/or.2014.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guo X, Zhang Y, Mayakonda A, Madan V, Ding LW, Lin LH, Zia S, Gery S, Tyner JW, Zhou W, Yin D, Lin DC, Koeffler HP. ARID1A and CEBPalpha cooperatively inhibit UCA1 transcription in breast cancer. Oncogene. 2018 doi: 10.1038/s41388-018-0371-4. [DOI] [PubMed] [Google Scholar]
- 141.Zhang L, Li DQ. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019;47(16):8502–8520. doi: 10.1093/nar/gkz545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ming N, Na HST, He JL, Meng QT, Xia ZY. Propofol alleviates oxidative stress via upregulating lncRNA-TUG1/Brg1 pathway in hypoxia/reoxygenation hepatic cells. J Biochem. 2019;166(5):415–421. doi: 10.1093/jb/mvz054. [DOI] [PubMed] [Google Scholar]
- 143.Chang CP, Han P. Epigenetic and lncRNA regulation of cardiac pathophysiology. Biochim Biophys Acta. 2016;1863(7 Pt B):1767–1771. doi: 10.1016/j.bbamcr.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou X, Rao Y, Sun Q, Liu Y, Chen J, Bu W. Long noncoding RNA CPS1-IT1 suppresses melanoma cell metastasis through inhibiting Cyr61 via competitively binding to BRG1. J Cell Physiol. 2019;234(12):22017–22027. doi: 10.1002/jcp.28764. [DOI] [PubMed] [Google Scholar]
- 145.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci USA. 2015;112(14):4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lino Cardenas CL, Kessinger CW, Cheng Y, MacDonald C, MacGillivray T, Ghoshhajra B, Huleihel L, Nuri S, Yeri AS, Jaffer FA, Kaminski N, Ellinor P, Weintraub NL, Malhotra R, Isselbacher EM, Lindsay ME. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat Commun. 2018;9(1):1009. doi: 10.1038/s41467-018-03394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, Su CJ, Shibata A, Strauss-Soukup JK, Drescher KM, Chen XM. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J Immunol. 2016;196(6):2799–2808. doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu X, Lu Y, Zhu J, Liu M, Xie M, Ye M, Li M, Wang S, Ming Z, Tong Q, Liu F, Zhou R. A long noncoding RNA, antisense IL-7, promotes inflammatory gene transcription through facilitating histone acetylation and switch/sucrose nonfermentable chromatin remodeling. J Immunol. 2019;203(6):1548–1559. doi: 10.4049/jimmunol.1900256. [DOI] [PubMed] [Google Scholar]
- 149.Chen Z, Gao Y, Yao L, Liu Y, Huang L, Yan Z, Zhao W, Zhu P, Weng H. LncFZD6 initiates Wnt/beta-catenin and liver TIC self-renewal through BRG1-mediated FZD6 transcriptional activation. Oncogene. 2018;37(23):3098–3112. doi: 10.1038/s41388-018-0203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Imai-Sumida M, Dasgupta P, Kulkarni P, Shiina M, Hashimoto Y, Shahryari V, Majid S, Tanaka Y, Dahiya R, Yamamura S. Genistein represses HOTAIR/chromatin remodeling pathways to suppress kidney cancer. Cell Physiol Biochem. 2020;54(1):53–70. doi: 10.33594/000000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cheng S, Wang L, Deng CH, Du SC, Han ZG. ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem Biophys Res Commun. 2017;491(1):178–182. doi: 10.1016/j.bbrc.2017.07.072. [DOI] [PubMed] [Google Scholar]
- 152.Fang C, He W, Xu T, Dai J, Xu L, Sun F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J Cell Physiol. 2019;234(5):6254–6262. doi: 10.1002/jcp.27356. [DOI] [PubMed] [Google Scholar]
- 153.Guo X, Wei Y, Wang Z, Liu W, Yang Y, Yu X, He J. LncRNA LINC00163 upregulation suppresses lung cancer development though transcriptionally increasing TCF21 expression. Am J Cancer Res. 2018;8(12):2494–2506. [PMC free article] [PubMed] [Google Scholar]
- 154.Jegu T, Blum R, Cochrane JC, Yang L, Wang CY, Gilles ME, Colognori D, Szanto A, Marr SK, Kingston RE, Lee JT. Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat Struct Mol Biol. 2019;26(2):96–109. doi: 10.1038/s41594-018-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Leisegang MS, Bibli SI, Gunther S, Pfluger-Muller B, Oo JA, Hoper C, Seredinski S, Yekelchyk M, Schmitz-Rixen T, Schurmann C, Hu J, Looso M, Sigala F, Boon RA, Fleming I, Brandes RP. Pleiotropic effects of laminar flow and statins depend on the Kruppel-like factor-induced lncRNA MANTIS. Eur Heart J. 2019;40(30):2523–2533. doi: 10.1093/eurheartj/ehz393. [DOI] [PubMed] [Google Scholar]
- 156.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Katsushima K, Natsume A, Ohka F, Shinjo K, Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, Shibata T, Naito M, Kim HJ, Miyata K, Kataoka K, Kondo Y. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rong Z, Wang Z, Wang X, Qin C, Geng W. Molecular interplay between linc01134 and YY1 dictates hepatocellular carcinoma progression. J Exp Clin Cancer Res. 2020;39(1):61. doi: 10.1186/s13046-020-01551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, Cinelli P, Santoro R. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15(6):720–734. doi: 10.1016/j.stem.2014.10.00. [DOI] [PubMed] [Google Scholar]