Abstract
In addition to genomic alterations, aberrant changes in post-transcriptional regulation can modify gene function and drive cancer development. RNA-binding proteins (RBPs) are a large class of post-transcriptional regulators that have been increasingly implicated in carcinogenesis. By integrating multi-omics data, we identify LARP1 as one of the most upregulated RBPs in colorectal cancer (CRC) and demonstrate its oncogenic properties. We perform LARP1:RNA interactome profiling and unveil a previously unexplored role for LARP1 in targeting the 3′UTR of oncogenes in CRC. Notably, we identify the proto-oncogenic transcription factor MYC as a key LARP1-regulated target. Our data show that LARP1 positively modulates MYC expression by associating with its 3′UTR. In addition, antisense oligonucleotide-mediated blocking of the interaction between LARP1 and the MYC 3′UTR reduces MYC expression and in vitro CRC growth. Furthermore, a systematic analysis of LARP1:protein interactions reveals IGF2BP3 and YBX1 as LARP1-interacting proteins that also regulate MYC expression and CRC development. Finally, we demonstrate that MYC reciprocally modulates LARP1 expression by targeting its enhancer. In summary, our data reveal a critical, previously uncharacterized role of LARP1 in promoting CRC tumorigenesis, validate its direct regulation of the proto-oncogene MYC and delineate a model of the positive feedback loop between MYC and LARP1 that promotes CRC growth and development.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04093-1.
Keywords: Cancer, Post-transcriptional regulation, RNA-binding protein, LARP1, MYC
Introduction
RNA-binding proteins (RBPs) are a class of key post-transcriptional regulators that have been implicated in multiple steps of the transcript life cycle including RNA maturation, localization, turnover, and translation [1]. Alterations in RBP expression levels and/or binding affinities may lead to the global dysregulation of target gene expression and subsequently contribute to cancer development. With the growing interest in the discovery and characterization of RBPs, 1914 RBPs have been identified, out of which 860 proteins have been validated as mRNA-binding proteins (mRBPs) [2]. However, only a handful of them, such as HuR and eIF4E, have been comprehensively characterized, suggesting that there are still many RBPs which could potentially be involved in diverse physiological and pathophysiological conditions such as cancer [1, 3]. RBPs frequently exhibit tissue- and context-specific expression patterns, necessitating in-depth tissue-specific analysis to accurately characterize their functions.
In this study, we focus on colorectal cancer (CRC), one of the most prevalent cancers globally in terms of both diagnosis and mortality [4]. Despite extensive research efforts pursuing novel treatment options, long-term survival remains limited [5]. This could in part be due to our incomplete understanding of the regulatory networks that are derailed in CRC. We hypothesized that RBPs are key regulators of colorectal cancer (CRC) tumorigenesis and investigated this using a systematic, multi-pronged approach. We anticipate that an unbiased, systematic interrogation of RBP expression in CRC would lead to breakthroughs in our understanding of CRC tumorigenesis, would accelerate the discovery of novel oncogenes and/or tumor suppressors and would provide potential targets for future clinical applications.
Recently, the RBP La-related Protein 1 (LARP1) has attracted attention due to its elevated expression in cancer [6]. LARP1 is a member of the La-related protein (LARP) superfamily of RBPs which are highly conserved throughout eukaryotic evolution [7]. Subsequent studies demonstrated the oncogenic capacity of LARP1 through its regulation of the stability and expression of transcripts such as BCL2 and BIK in ovarian cancer, as well as mTOR in non-small cell lung cancer (NSCLC) and cervical cancer [8, 9]. In addition, LARP1 possesses prognostic potential in several cancer types including CRC, whereby its high expression in patients is associated with poorer prognosis and overall survival [6, 8–10]. Furthermore, LARP1 has been shown to associate with the mTOR complex 1 (mTORC1) and Poly(A)-Binding Protein (PABP) in controlling the stability and translation of mRNAs containing the 5′ terminal oligopyrimidine (TOP) motif, which encode for ribosomal proteins and translational factors [11, 12]. However, a transcriptome-wide mapping of LARP1 target genes in CRC has not been performed to date and the mechanisms driving its oncogenic function remain largely unexplored. Deciphering the detailed function of LARP1 and its targets in CRC could provide important insights into the molecular regulation governing CRC progression and contribute to the development of efficient therapeutic strategies for CRC.
In this study, we performed an integrative, multi-omics analyses of RBPs in CRC, and identified LARP1 as a potential oncogenic mRNA-binding protein (mRBP). Enhanced UV crosslinking and immunoprecipitation (eCLIP)-sequencing analysis confirmed previous reports describing LARP1 regulation of ribosomal proteins and also revealed a previously unexplored association between LARP1 and the 3′UTRs of genes involved in the oncogenic pathways of colon adenocarcinoma (COAD). Among them, we identified and validated the master transcription factor MYC as a target that is positively regulated by LARP1 in CRC. Additionally, we performed quantitative mass spectrometry analysis of LARP1:protein interactions, which revealed IGF2BP3 and YBX1 as RNA-dependent LARP1-interacting proteins that also regulate MYC expression and CRC development. Finally, we demonstrated that MYC directly upregulates LARP1 transcription to form a positive feedback loop that is critical in promoting CRC growth and development.
Materials and methods
Reagents
Reagents are as follows: various antibodies (Table S1); phosphorothioate modified antisense oligonucleotides (ASO) (Integrated DNA Technologies) (Table S2); DharmaFECT 1 (Dharmacon), TRIZOL®, Lipofectamine 3000, Dulbecco’s Modified Eagle Medium (DMEM), Roswell Park Memorial Institute (RPMI) 1640 medium, Eagle Minimum Essential Medium (EMEM), Opti-MEM™ reduced serum media, fetal bovine serum (FBS), Dynabeads® MyOne™ Streptavidin C1 (ThermoFisher Scientific), Dynabeads® Protein G (ThermoFisher Scientific), psiCHECK-2, pGL4.20 and pRL-null (Promega), pcDNA4-Puro (Addgene). Tissue total RNA (5 µg) of ten matched-pairs of human CRC samples were purchased from OriGene.
Clinical samples
Five pairs of matched adjacent normal and CRC tissues were obtained from the National University Hospital (NUH), Singapore. Approximately 10–20 mg of fresh tissue was homogenized using a plastic pestle homogenizer and passed through a 21-gauge needle fitted to an RNase-free syringe. DNA, RNA, and protein were extracted using QIAGEN AllPrep DNA/RNA/Protein Mini Kit according to the manufacturer’s protocol.
Plasmids
The primers used for the amplification of various genes are listed in Table S3. LARP1 was cloned by polymerase chain reaction (PCR) from pCMV6-AC-GFP + LARP1 (OriGene), followed by restriction digest of the PCR products using NotI and XbaI, and ligation into the digested pcDNA4-Puro vector. Primers for the MYC binding sites (Table S3) were annealed by mixing 100 µM of each primer in 5X sequencing and annealing buffer (1 M Tris HCl pH 7.5, 5 M NaCl, 1 M MgCl2). The mixture was heated at 95 °C for 5 min and cooled to room temperature and ligated into the digested vector pGL4.20.
Cell culture and transfection
HCT116 and DLD-1 cells were maintained in DMEM and RPMI, respectively, while CCD-18Co and CCD-841CoN were maintained in EMEM, supplemented with 10% FBS, penicillin/ streptomycin, and L-glutamine. All cell lines were grown at 37 °C in a humidified atmosphere with 5% CO2. Cells were transfected with 50 nM of siRNA using Dharmafect 1 as per the manufacturer’s protocol at a seeding density of 150,000 cells per well in 12-well plates. For plasmid transfection, cells were seeded in 12-well plates at a density of 100,000 cells per well 24 h prior to transfection. Lipofectamine 3000 was used to transfect 500 ng (for HCT116) and 100 ng (for DLD-1) of plasmids and 100 nM of ASOs per well following the manufacturer’s instructions.
CellTiter-Glo® assay
Cells were seeded and transfected at a density of 1000 cells per well in 96-well plates. 48 h post-transfection, 50 µl of CellTiter-Glo® assay reagents were added to each well and incubated for 5 min at room temperature prior to luminescence measurements. For plasmid transfection, cells were seeded at a density of 5000 cells per well in serum-free medium in 12-well plates. 24 h after plasmid transfection, cells were split into 96-well plates at a density of 1000 cells per well and luminescence was measured after 24 h.
Soft agar assay
A 0.6% agarose base was prepared in 6-well plates. 24 h after transfection, cells were trypsinized, resuspended, and diluted to 15,000 cells per well in growth medium. The cells were mixed with agarose to obtain a 0.3% top layer agar which was added above the 0.6% base agar and left to solidify. The cells were maintained at 37 °C and the growth medium was changed twice a week. Images were taken at 4X magnification once every 5 days up to 14 days and quantified using ImageJ.
Xenograft mouse model
Cells were transfected in 12-well plates. 24 h post transfection, cells were trypsinized and collected in the appropriate growth medium. For each injection, 1 million cells were mixed with Matrigel matrix (Corning) in a 1:1 ratio. The cell suspension was injected into the lower flank of immune-deficient nude mice (n = 5 per condition). Xenograft tumor growth was measured every 2 days. After 10–20 days, the mice were killed, and the tumors were extracted and weighed.
Caspase-Glo® 3/7 assay
Cells were transfected in 96-well plates at a density of 5000 cells per well. After 48 h of transfection, 100 μl of the Caspase-Glo® 3/7 assay reagents (Promega) were added to each well and incubated for 1 h at room temperature.
Protein extraction and western blot analysis
Cells were trypsinized and harvested in the appropriate growth medium. Cells were lysed on ice for 10 min in protein lysis buffer (PLB) (10 mM HEPES pH 7.0, 0.1 M KCl, 5 mM MgCl2, 25 mM EDTA pH 8.0, 0.5% (v/v) NP-40, 20 µM DTT, Proteinase Inhibitor). The lysates were centrifuged at 16,000×g for 15 min. The supernatants containing proteins were collected and the concentrations were measured using Bradford Protein Assay (Bio-Rad). 10–12 µg of protein lysates was separated on 4–12% Bis–Tris NuPAGE® Precast gels (ThermoFisher Scientific) and transferred to PVDF membranes using the Mini Trans-Blot® Electrophoretic Transfer Cell (BioRad) in transfer buffer (25 mM Tris HCl, 192 mM glycine and 20% (v/v) methanol). The membranes were probed with specific primary antibodies followed by the corresponding secondary antibodies.
RNA extraction, real-time quantitative PCR (RT-qPCR)
Total RNA was isolated using the TRIZOL® reagent and the PureLink® RNA Mini Kit (Ambion) as per the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). qPCR was performed using the PowerUp™ SYBR® Green Master Mix (Applied Biosystems) on the QuantStudio 5 Real-Time PCR System. The primers used for qPCR are listed in Table S4.
3′ rapid amplification of cDNA ends (3′ RACE)
The 3′ RACE was performed using the RLM-RACE Kit (FirstChoice®) following the manufacturer’s protocol. 1 μg of total RNA from HCT116 and DLD-1 cells was used for the experiment. Primers used for the 3′ RACE are listed in Table S5. The amplified complementary DNAs (cDNAs) were cloned into pcDNA4-TO-Puromycin-mVenus-MAP. Ten clones from each sample were sequenced using Sanger sequencing. The clones were analyzed for the length of the MYC 3′UTR based on a few criteria; the presence of (i) both the forward and reverse inner PCR primers, (ii) both the HindIII and BamHI restriction sites, and (iii) the MYC CDS and stop codon preceding the 3′UTR. The poly(A) at the end of the 3′UTR was included in the calculation of the 3′UTR length.
Luciferase reporter assays
For dual-luciferase reporter assays, cells were seeded at a density of 50,000 cells per well in 24-well plates a day prior to transfection. 5 ng of psiCHECK-2 plasmids was co-transfected with the appropriate transfectants (siRNAs or expression plasmids). At 48 h post-transfection, cells were washed in phosphate-buffered saline (PBS), lysed, and luminescence was measured following the manufacturer’s instructions (Promega). For the promoter luciferase reporter assay, cells were seeded at a density of 40,000 cells per well in 24-well plates one day prior to transfection. 10 ng of each pGL4.20 and pRL-null plasmids were transfected into the cells with the appropriate transfectants. Luminescence of the luciferase reporter was measured after 24 h as described above.
In vitro transcription and pulldown (IVT-PD)
A transcription template for the gene of interest was amplified by PCR using forward primers carrying a T7-tag and untagged reverse primers. For the antisense control, the locations of the tag was reversed. The primers used for IVT are listed in Table S6. 1 µg of PCR product was incubated with the transcription mix (10X transcription buffer, 400 mM NTP mix, 200 U T7 RNA polymerase) at 37 °C for 4 h. The transcription products were precipitated with 100% ethanol at − 20 °C followed by centrifugation at 16,000×g for 1 h at 4 °C. The RNA was purified using Microspin G-50 (GE Healthcare) following the manufacturer’s protocol. For pulldown, 25 µg of the IVT-synthesized RNA was incubated with Dynabeads® MyOne™ Streptavidin C1 (ThermoFisher Scientific) in binding buffer (100 mM NaCl, 10 mM MgCl2, 50 mM HEPES pH 7.4, 0.5% Igepal CA-630) for 30 min at 4 °C. The beads were washed twice in washing buffer (250 mM NaCl, 10 mM MgCl2, 50 mM HEPES pH 7.4, 0.5% Igepal CA-630), followed by incubation with 1 mg of HCT116 whole cell lysate for 1 h at 4 °C. The protein-RNA complexes were dissociated by adding 4X loading buffer and heated at 95 °C for 5 min. The mixtures were centrifuged, and the supernatants were subjected to SDS-PAGE. This protocol was adapted from Butter et al. [13].
Mass spectrometry analysis
The SDS-PAGE gels were fixed in fixing solution (50% methanol, 12% acetic acid, and 0.05% formalin) for 2 h, followed by three 20 min washes in 35% ethanol. Subsequently, the gels were sensitized with 0.02% sodium thiosulphate for 2 min with shaking at 80 rpm, followed by three 5 min washes in water. The gels were stained with the silver stain solution (0.2% silver nitrate and 0.076% formalin). The stain was developed using 6% sodium carbonate, 0.0004% sodium thiosulphate, and 0.05% formalin until the desired intensity was obtained and stopped with 1.46% sodium EDTA. Bands excised from the gels were digested with 0.2 µg of trypsin at 4 °C for 30 min. The excess trypsin was removed and 25 mM ammonium bicarbonate was added to each sample for in-gel tryptic digestion at 37 °C for 16 h. The digested samples were desalted using the Sep-Pak C18 cartridge according to the manufacturer instructions. The eluent which contained the desalted peptides was collected in 3 ml of 80% ACN and dried with Speed Vac. The dried samples were reconstituted in 15 μl of 2% ACN + 0.1% formic acid (FA). Mass spectrometry analysis was performed using a TripleTOF 5600 system (AB SCIEX) at the Protein and Proteomics Centre at the Department of Biological Sciences (DBS), NUS. Peptides were identified using ProteinPilot4.5 software Revision 1656 (AB SCIEX).
RNA immunoprecipitation (RIP)
Protein A-Sepharose beads (Sigma-Aldrich) were coated with 3 µg of LARP1 (Abcam) or Rabbit IgG antibody (Jackson ImmunoResearch Laboratories), followed by incubation with 2 mg of HCT116 or DLD-1 total cell lysates overnight. The RNA-protein-bead complexes were washed once with NT2-crowders (25 mg Ficoll PM400 (GE Healthcare), 75 mg Ficoll PM70 (GE Healthcare), 2.5 mg Dextran Sulphate (Fluka) in 10 ml of NT2 buffer), and five times with NT2 buffer (0.05 M Tris HCl pH 7.0, 0.15 M NaCl, 0.001 M MgCl2, 0.0005% (v/v) NP-40 (Roche) in ultra-pure water). Protein–RNA complexes were collected in 100 μl of NET2 buffer (0.00118 M DTT (Sigma-Aldrich), 0.0176 M EDTA, 200 U RNaseOUT (ThermoFisher Scientific) and 100 U SUPERase• In™ (Ambion) in 1X NT2 crowder), supplemented with 100 μl of 2X SDS-TE (0.04 M Tris HCl pH 7.5, 0.004 M EDTA pH 8.0, 10% SDS). RNA was isolated using the Trizol® reagent and subsequently purified with Phenol:Chloroform:Isoamyl alcohol (25:24:1) (Sigma-Aldrich) and Chloroform:Isoamyl alcohol (24:1) (Sigma-Aldrich). Protocol was adapted from the RIP-ChIP protocol [14].
eCLIP library preparation
DLD-1 cells were cultured to confluency in 15 cm plates. The cells were UV crosslinked (400 mJ/cm2 constant energy) to induce covalent bonds between proteins and nucleic acids in close proximity to detect direct interactions [15]. The cells were immediately pelleted and frozen on dry ice. eCLIP procedure was performed as described [16]. Briefly, cell pellets were lysed in eCLIP lysis buffer and sonicated (BioRuptor). Lysates were treated with RNase I (Ambion) to fragment the RNA and incubated with antibodies against LARP1 (Abcam) to immunoprecipitate LARP1-RNA complexes. 2% of the lysate:antibody mixture was saved as input sample. The remaining immunoprecipitated (IP) samples were subjected to a series of stringent washes, followed by dephosphorylation of RNA by FastAP (ThermoFisher Scientific) and T4 Polynucleotide Kinase (NEB), and ligation of a 3′ RNA adapter with T4 RNA Ligase (NEB). Immunoprecipitates and input samples were separated by SDS-PAGE, transferred to nitrocellulose membrane, and then the region of membrane corresponding to 130–250 kDa was excised for each immunoprecipitate and input sample (Supplementary Fig. S2A). Input samples were referred to as size-matched input (SMInput) controls. RNA was isolated from the membrane, reverse transcribed with AffinityScript (Agilent), free primers were removed (ExoSap-IT, Affymetrix), and a DNA adapter was ligated onto the cDNA product with T4 RNA ligase (NEB). Libraries were then amplified with Q5 PCR mix (NEB), size selected using AMPure XP beads (Beckman Coulter, Inc.), and on a 3% agarose gel, and then quantified with a Bioanalyzer instrument (Agilent). eCLIP-seq for LARP1 was performed in biological duplicates including SMInput duplicates. Paired-end (50 base pair) sequencing of the eCLIP libraries was performed on Illumina HiSeq 3500.
eCLIP data processing and analysis
Processing and analysis of eCLIP sequence reads were performed as previously described [16]. For each cell line, a total of six eCLIP (IP and SMInput) libraries were sequenced to ~ 12–15 million reads for IP libraries and ~ 8–9 million reads for SMInput. Reads were mapped to the human genome (hg19) and only uniquely mapped reads that did not map to repetitive elements were retained for downstream analysis. PCR duplicates were removed with use of a randomer (N10) sequence positioned one of the adapter oligos to obtain ‘usable reads’. For both eCLIP (LARP1 immunoprecipitation) and SMInput samples, usable reads were counted across different genomic regions (i.e. 5′UTR, 3′UTR, coding exons (CDS), and intronic regions) for all coding genes annotated in UCSC Known Gene table (hg19). To identify the reads enrichment above SMInput in certain genomic regions of certain genes, fold-enrichment was calculated as the ratio of read counts within this region in eCLIP versus SMInput. Only regions with at least 10 reads in both of eCLIP and SMInput samples were considered. Sequencing read peaks in both eCLIP and SMInput libraries were identified using CLIPper algorithm [17]. CLIPper-defined peaks were then normalized to SMInput by comparing the read density in LARP1 immunoprecipitation (IP) and SMInput samples. Peaks were considered significant if the number of reads in the IP sample were four-fold greater than the number of reads in the SMInput sample with a P < 0.001. P value was calculated by Yates’ Chi-Square test (Fisher Exact Test when the observed or expected read number was below five).
Peak enrichment analysis
Significantly enriched eCLIP peaks (described above) were assigned to different transcriptome regions (i.e. 5′UTR, 3′UTR, coding exons (CDS), and intronic regions) using BEDTools [18] with genomic annotations from GENCODE. For peaks assigned to multiple regions, peaks were assigned to a single region in the following priority: CDS, 5′UTR, 3′UTR, then proximal or distal introns (as defined as 500 bp or greater from an exon–intron boundary). The fraction of peaks in a given region (FCLIP) was calculated by dividing the number of peaks in a region by the total number of peaks in all regions (5′UTR, 3′UTR, CDS, proximal intron, and distal intron). Fold-enrichment was calculated as , where Fregion is the fractional region size derived by dividing the total number of base pairs in that region relative to the total number of base pairs in all regions. If a base pair associated with multiple regions, this base pair was assigned to regions in the following priority: CDS, 5′UTR, 3′UTR, then proximal or distal introns.
Irreproducibility discovery rate (IDR) analysis
IDR analysis [19] was performed on the SMInput-normalized eCLIP peaks by rank entropy, which is calculated on eCLIP and SMInput read probabilities within each peak for each replicate. The ENCODE IDR Pipeline 2.0.2 was used as documented at https://www.encodeproject.org/software/idr/.
KEGG enrichment analysis
KEGG pathway enrichment analysis was performed using an online toolkit: WebGestalt [20]. The significantly enriched pathways were determined using the false discovery rate (FDR) < 0.05 and affinity propagation was applied for redundancy reduction.
Biotin labelling and pulldown
RNA obtained from IVT was labelled with biotin using the Pierce™ RNA 3′End Desthiobiotinylation Kit (ThermoFisher Scientific). The biotin labelling efficiency was determined using the Chemiluminescent Nucleic Acid Detection Module Kit (ThermoFisher Scientific). The amount of RNA used for pulldown was determined by the efficiency of the biotin labelling. The Pierce™ Magnetic RNA–Protein Pulldown Kit (ThermoFisher Scientific) was used for the pulldown experiments and the eluted protein samples were used for subsequent western blot analysis. All the experiments above were performed according to the corresponding manufacturer’s protocol.
Immunoprecipitation
Cells were lysed with NP-40 lysis buffer (1% (v/v) NP-40, 50 mM Tris HCl pH 7.5, 150 mM NaCl, 50 mM MgCl2, 20% (v/v) glycerol, 1X proteinase inhibitor, 0.5 mM DTT). 1 mg of cell lysate was incubated with 2 µg of antibody overnight with rotation. Protein A Sepharose beads (Abcam) were washed PBS thrice and blocked with 1% w/v BSA for 1 h, followed by twice rinsing in NP-40 lysis buffer before use. The protein–antibody complexes were incubated with 100 µl of beads slurry for 2 h with rotation. The proteins bound to antibody-beads complexes were eluted in 4X sample buffer and heated at 95 °C for 5 min. The eluted samples were used for subsequent immunoblotting or mass spectrometry analysis.
SILAC labeling
For SILAC labelling, cells were incubated in DMEM (-Arg, -Lys) medium containing 10% dialyzed fetal bovine serum (ThermoFisher Scientific) supplemented with 42 mg/l 13C615N4 L-arginine and 73 mg/l 13C615N2 L-lysine (Cambridge Isotope) or the corresponding non-labeled amino acids, respectively.
Mass spectrometry analysis for SILAC labelled cells immunoprecipitation
Samples were boiled at 95 °C prior to separation on a 12% NuPAGE Bis–Tris precast gel (ThermoFisher Scientific) for 10 min at 170 V in 1 × MOPS buffer, followed by gel fixation using the Colloidal Blue Staining Kit (ThermoFisher Scientific). For in-gel digestion, samples were destained in destaining buffer (25 mM ammonium bicarbonate; 50% ethanol), reduced in 10 mM DTT for 1 h at 56 °C followed by alkylation with 55 mM iodoacetamide (Sigma) for 45 min in the dark. Tryptic digest was performed in 50 mM ammonium bicarbonate buffer with 2 μg trypsin (Promega) at 37 °C overnight. Peptides were desalted on StageTips and analyzed by nanoflow liquid chromatography on an EASY-nLC 1200 system coupled to a Q Exactive HF mass spectrometer (ThermoFisher Scientific). Peptides were separated on a C18-reversed phase column (25 cm long, 75 μm inner diameter) packed in-house with ReproSil-Pur C18-AQ 1.9 μm resin (Dr Maisch). The column was mounted on an Easy Flex Nano Source and temperature controlled by a column oven (Sonation) at 40 °C. A 105-min gradient from 2 to 40% acetonitrile in 0.5% formic acid at a flow of 225 nl/min was used. Spray voltage was set to 2.4 kV. The Q Exactive HF was operated with a TOP20 MS/MS spectra acquisition method per MS full scan. MS scans were conducted with 60,000 at a maximum injection time of 20 ms and MS/MS scans with 15,000 resolutions at a maximum injection time of 50 ms. The raw files were processed with MaxQuant [21] version 1.5.2.8 with preset standard settings for SILAC labeled samples and the re-quantify option was activated. Carbamidomethylation was set as fixed modification while methionine oxidation and protein N-acetylation were considered as variable modifications. Search results were filtered with a false discovery rate of 0.01. Known contaminants, proteins groups only identified by site, and reverse hits of the MaxQuant results were removed and only proteins were kept that were quantified by SILAC ratios in both ‘forward’ and ‘reverse’ samples.
Chromatin Immunoprecipitation (ChIP)
Dynabeads® Protein G beads (ThermoFisher Scientific) were blocked using 0.5% BSA in PBS. 5 million cells were crosslinking using 1% formaldehyde and quenched with glycine. Cells were lysed sequentially using lysis buffer 1 (50 mM Hepes–KOH, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1 × protease inhibitor), lysis buffer 2 (10 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1 × protease inhibitor), and lysis buffer 3 (10 mM Tris–HCl, 1 mM EDTA, 0.1% SDS, 1 × protease inhibitor) with rocking at 4 °C. Lysates were sonicated in lysis buffer 3 for 30 × cycles (30 s on, 60 s off) using the Bioruptor Pico sonicator (Diagenode). The lysate was added to blocked beads and incubated overnight with rotation at 4 °C. Beads were washed using wash buffer (50 mM Hepes–KOH, 140 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), high salt wash buffer (50 mM Hepes–KOH, 500 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.2% sodium deoxycholate, 0.1% SDS), LiCl wash buffer (20 mM Tris–HCl, 1 mM EDTA, 250 mM LiCI, 0.5% NP-40, 0.5% sodium deoxycholate), and 10/0.1 TE buffer with 0.1% Triton X-100. Elution buffer (50 mM Tris–HCl, 10 mM EDTA, 1% SDS) was used to elute the DNA at 65 °C followed by reverse crosslinking at 65 °C overnight. Reverse crosslinked DNA was treated with RNase A and protease K, then purified using the Qiaquick™ PCR Purification Kit (Qiagen). This protocol was adapted from Lee et al. [22].
Statistical analysis
All in vitro data were derived from at least three independent experiments. Values calculated from multiple independent experiments were presented as mean ± SEM, while data shown from a representative experiment were presented as mean ± SD. Student’s t-test was used to analyze the statistical significance whereby P values smaller than 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Integrative multi-omics data analysis identifies LARP1 as a potential oncogenic mRNA-Binding Protein (mRBP) in CRC
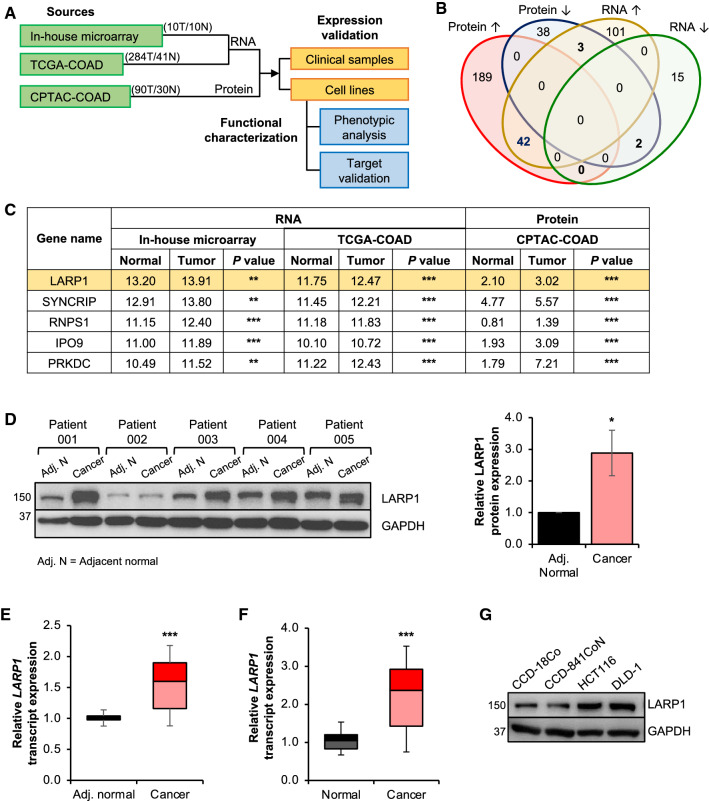
To identify dysregulated RBPs in CRC, we integrated mRNA expression data of ten matched pairs of CRC and normal samples (OriGene) with the COAD dataset (284 COAD versus 41 normal) from The Cancer Genome Atlas (TCGA-COAD), as well as protein expression of 90 COAD clinical samples (versus 30 normal) from the Clinical Proteomic Tumor Analysis Consortium (CPTAC-COAD) (Fig. 1A). By comparing the tumors to adjacent normal tissues, we found that there were more significantly upregulated RBPs (n = 42, adjusted P < 0.05 and fold change > 1.5 in each dataset) than downregulated (n = 2) ones (Fig. 1B), which is consistent with previous findings [23]. Among them, 16 were annotated as mRNA-binding proteins (mRBPs), including LARP1 which ranked as the most highly expressed and dysregulated mRBP (Fig. 1C; Table S7).
Fig. 1.
Integrative multi-omics data analysis identifies LARP1 as a potential oncogenic mRNA binding protein (mRBP) in CRC. A Flowchart outlining the identification and functional characterization of RBPs in colorectal cancer (CRC). Analyses were performed using datasets from an in-house microarray (10 CRC versus 10 normal) (OriGene), The Cancer Genome Atlas colon adenocarcinoma (TCGA-COAD) (284 CRC versus 41 normal) and Clinical Proteomic Tumor Analysis Consortium colon adenocarcinoma (CPTAC-COAD) (90 CRC versus 30 normal). T denotes tumor; N denotes normal. B Venn diagram illustrating the number of overlapping RBPs which have significantly dysregulated expression at the transcript and protein levels. C Table of the top five mRBPs showing consistent upregulation in CRC compared to the adjacent normal tissues at both the transcript and protein levels. **P < 0.01; ***P < 0.001. D and E LARP1 protein expression (left panel) and western blot densitometry quantification (right panel) (D), and LARP1 transcript expression (E) in five pairs matched adjacent normal (Adj. N) and CRC clinical samples. Mean ± STE. *P < 0.05; ***P < 0.001. F LARP1 transcript expression in ten matched pairs of normal and CRC samples (OriGene). Mean ± STE. ***P < 0.001. G Endogenous LARP1 protein expression in immortalized normal colon (CCD-18Co and CCD-841CoN) and CRC (HCT116 and DLD-1) cell lines
We next examined the expression profile of LARP1 in both CRC patient samples and cell lines (Fig. 1D–G). We observed consistent elevated expression of the LARP1 protein and transcript levels in the five matched pairs of CRC patient samples from the National University Hospital (NUH), Singapore (Fig. 1D, E) and tumor samples of the ten matched pairs of CRC patient samples (OriGene) (Fig. 1F). Similarly, LARP1 protein levels were upregulated in CRC cell lines, HCT116, and DLD-1, which were used for subsequent in vitro studies (Fig. 1G). Based on this expression profile, we hypothesized that LARP1 may play an oncogenic role in CRC.
LARP1 possesses oncogenic properties in CRC
Although the tumor-promoting role of LARP1 has been shown in a few cancers, little is known about its role in CRC [6, 8–10]. To date, there is only one study by Ye et al. that demonstrated a correlation between high LARP1 expression and poorer prognosis of CRC patients [10]. To further examine the potential oncogenic properties of LARP1 in CRC, we assessed the effects of altering LARP1 expression in the CRC cell lines.
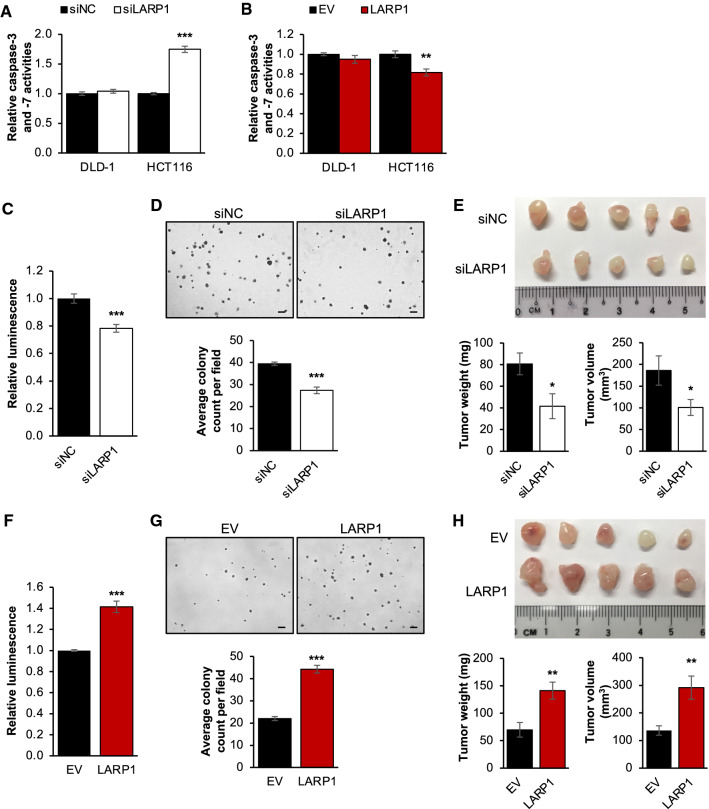
The growth-promoting effect of LARP1 could be in part due to its ability to suppress apoptosis, which has been shown in ovarian cancer [8]. Consistent with the finding, LARP1 knockdown significantly increased, while its overexpression reduced the relative caspase-3 and -7 activities in HCT116 (Fig. 2A, B). However, these effects were not observed in DLD-1. Furthermore, the level of cleaved PARP was significantly upregulated in HCT116, but not in DLD-1, thus demonstrating that the anti-apoptotic effect of LARP1 could be cell line-dependent (Supplementary Fig. S1A).
Fig. 2.
LARP1 possesses oncogenic properties in CRC. A and B Effect of LARP1 knockdown (A) and overexpression (B) on relative caspase-3 and -7 activities. siNC denotes siRNA non-targeting control; EV denotes empty vector. Mean ± STE. **P < 0.01; ***P < 0.001. C – E Effect of LARP1 knockdown on cell viability (C), anchorage-independent growth (D, scale bar: 200 μm) and xenograft tumor formation (E) of DLD-1. Quantification of colony count, tumor weight and volume are shown at the bottom panels. siNC denotes siRNA non-targeting control. Mean ± STE. (E) n = 5 for each group. *P < 0.05; ***P < 0.001. F – H Effect of LARP1 overexpression on cell viability (F), anchorage-independent growth (G, scale bar: 200 μm) and xenograft tumor formation (H) of DLD-1. Quantification of colony count, tumor weight and volume are shown at the bottom panels. EV denotes empty vector. Mean ± STE. (H) n = 5 for each group. **P < 0.01; ***P < 0.001
Nonetheless, we found that the downregulation of LARP1 significantly reduced cell viability (Fig. 2C, Supplementary Fig. S1B) and anchorage-independent growth (Fig. 2D, Supplementary Fig. S1C) in vitro, as well as xenograft tumor growth (Fig. 2E, Supplementary Fig. S1D) of both DLD-1 and HCT116 cells. Consistently, LARP1 overexpression resulted in the reciprocal effect on growth (Fig. 2F–H, Supplementary Fig. S1E–G).
Mapping of transcriptome-wide LARP1-RNA interactions in CRC
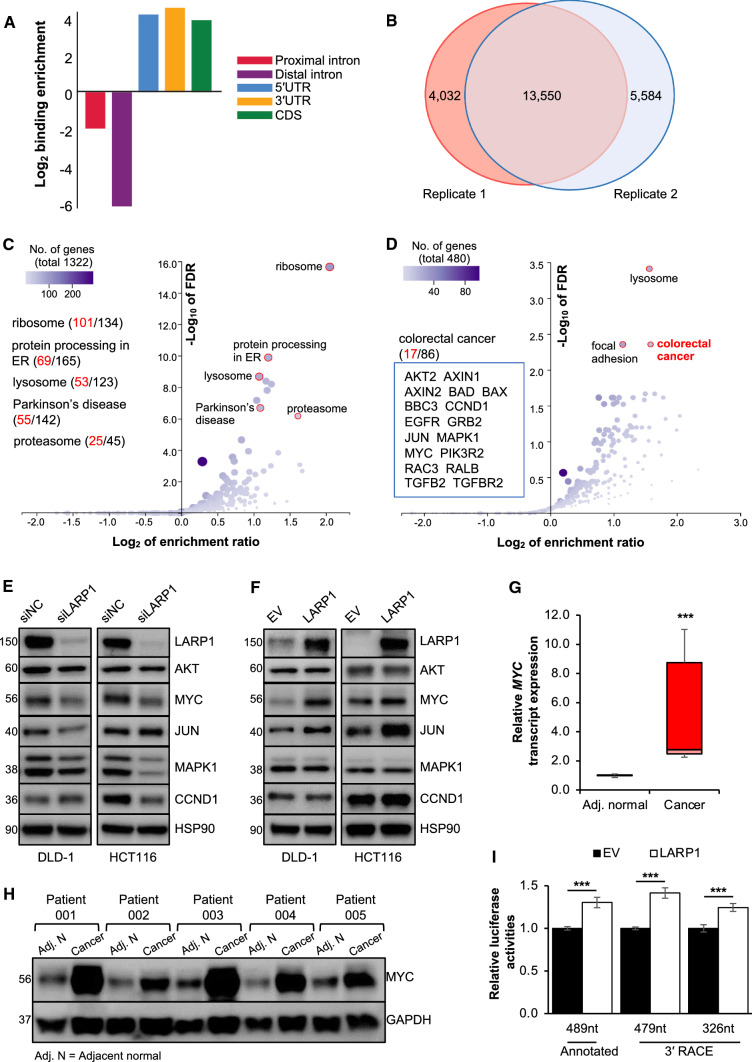
Collectively, our findings suggest that LARP1 possesses some oncogenic properties that are independent of its anti-apoptotic role in CRC. This also highlights the importance of discovering other LARP1 targets which may contribute to CRC progression. Next, we investigated LARP1 binding across the transcriptome by performing enhanced crosslinking and immunoprecipitation (eCLIP) followed by deep sequencing of LARP1-protected RNA fragments in DLD-1 cells (Supplementary Fig. S2A) [16]. LARP1 binding sites were significantly enriched in coding exons (CDS), 5′UTRs and 3′UTRs but not in distal introns (defined as intronic region 500 bp or greater away from an exon–intron boundary) (Fig. 3A). An Irreproducibility Discovery Rate (IDR) analysis revealed highly reproducible LARP1 peaks in replicate datasets (Supplementary Fig. S2B). 17,582 and 19,134 peaks were identified in replicate samples, and they were significantly overlapped (P < 0.001). In total, 13,550 overlapped peaks were used as the putative LARP1 binding sites (Fig. 3B).
Fig. 3.
Mapping of transcriptome-wide LARP1-RNA interactions in CRC. A Binding enrichment of LARP1 normalized to the average length of proximal introns (red), distal introns (purple), 5′UTR (blue), 3′UTR (yellow), coding sequence (CDS, green) in enhanced UV crosslinking and immunoprecipitation (eCLIP) of DLD-1 cells. B Overlap of peaks between the two eCLIP replicates. C Scatter plot illustrating the enriched KEGG pathways for transcripts with LARP1 binding sites at the 5′UTR, CDS and 3′UTR. FDR denotes false discovery rate. D Scatter plot illustrating the enriched KEGG pathways for transcripts with LARP1 binding sites only on their 3′UTRs. E and F Protein expression of candidate LARP1 eCLIP targets upon the knockdown (E) and overexpression (F) of LARP1 in DLD-1 and HCT116 cells. siNC denotes siRNA non-targeting control; EV denotes empty vector. G and H MYC transcript (G) and protein (H) expression in five pairs of matched adjacent normal (Adj. N) and CRC clinical samples. Mean ± STE. ***P < 0.001. I Effect of LARP1 overexpression on the relative luciferase activities of the MYC 3′UTR variants in DLD-1 cells. EV denotes empty vector. Mean ± STE. ***P < 0.001
A few studies have validated the role of LARP1 in regulating protein translation, particularly the translation of 5′ TOP mRNAs which encode for ribosomal proteins [11, 12, 24, 25]. We detected the ribosomal protein synthesis pathway (false discovery rate (FDR) = 0, enrichment ratio = 4.15) as the most significantly enriched KEGG pathway in agreement with previous reports (Fig. 3C). In total, 101 out of 134 transcripts encoding ribosomal proteins contain LARP1 binding sites. Additionally, a LARP1 binding site at the 5′UTR of the RPL32 transcript was observed in our eCLIP data (Supplementary Fig. S2C), consistent with a previous report [11].
Interestingly, we observed more than 400 transcripts that contain LARP1 binding sites only at their 3′UTRs. These transcripts were not enriched for the ribosomal protein synthesis pathway, but were significantly enriched (FDR < 0.004, enrichment ratio = 2.99) for the KEGG colorectal cancer (CRC) pathway (Fig. 3D). Among the 17 genes in the CRC pathway that contain LARP1 binding sites at their 3′UTR regions, many are well-known oncogenes. These results suggested that LARP1 plays a pivotal role in CRC by directly binding to the 3′UTRs of key regulators of tumorigenesis and provide support for an additional mechanism of LARP1 function beyond binding to 5′TOP mRNAs.
MYC is a LARP1 target in CRC
A few candidates from the KEGG CRC pathway (AKT2, MYC, JUN, MAPK1, and CCND1) were selected for further experimental validation. The upregulation of AKT2 [26], MYC [27, 28], JUN [29], and CCND1 [30] has been independently verified in CRC patients, whilst the activation of MAPK signalling is implicated in the induction of CRC growth [31]. To validate the LARP1-mediated regulation of these targets, we performed LARP1 knockdown and overexpression in both DLD-1 and HCT116 cells. There were no uniform trends between treatments and cell lines for most of the targets apart from MYC, whose expression was consistently and significantly decreased upon LARP1 knockdown and increased upon LARP1 overexpression in both cell lines (Fig. 3E, F, Supplementary Fig. S2D, S2E). We thus focused on MYC, a key proto-oncogenic transcription factor which has been extensively studied over the past 40 years.
Numerous reports have highlighted the importance of MYC in promoting growth of multiple cancers, including CRC. Remarkably, a study found that almost 100% of CRC cells of different mutational status and anatomical origin have deregulated expression of MYC target genes, including CCND2, CCNA, CDK4, p21Cip1, and p27Kip1, implicating it in CRC development [32, 33]. Although the MYC transcript is overexpressed by 5 to 400-fold in 70% of primary CRC, this increase is not due to genetic rearrangement, which only accounts for 10% of primary CRC [27, 28, 34]. These findings point towards the possibility that MYC dysregulation in primary CRC may occur at the post-transcriptional level, which is a critical step in the complex process of gene regulation. Indeed, we observed the upregulation of the MYC transcript (Fig. 3G) as well as protein expression (Fig. 3H, Supplementary Fig. S2F) in CRC tumors compared to the adjacent normal colon tissues. The positive trend between MYC and LARP1 expression (Fig. 1D–G) suggests that LARP1 could be a positive regulator of MYC in CRC.
The importance of 3′-untranslated regions (3′UTR) in translational regulation is well-documented, including that of MYC. Recent studies demonstrated frequent 3′UTR shortening of oncogene transcripts, which facilitates their escape from post-transcriptional regulation mediated by regulatory molecules such as RBPs and microRNAs. These events resulted in the upregulation of oncogene expression and subsequent cancer progression [35, 36]. To examine the possibility of MYC 3′UTR shortening eliminating LARP1 binding and regulation, we performed 3′ rapid amplification of cDNA end (3′ RACE) of the MYC transcript in DLD-1 and HCT116. We found two additional MYC 3′UTR variants which correspond to known polyadenylation or poly(A) signals and contribute to the heterogenous population of MYC transcripts (Supplementary Fig. S2G). Subsequently, we cloned the annotated MYC 3′UTR (489nt) and both MYC 3′UTR variants (479 nucleotides (nt) and 326nt) into a luciferase reporter vector. The overexpression of LARP1 significantly and consistently increased the relative luciferase activities of all three constructs (Fig. 3I, Supplementary Fig. S2H), suggesting the shortening of MYC 3′UTR does not affect LARP1-mediated regulation of MYC.
LARP1 associates with the MYC transcript to regulate its expression
To verify the interaction between LARP1 and the annotated MYC 3′UTR, we performed in vitro transcription and pulldown (IVT-PD) using HCT116 cells, followed by mass spectrometry analysis. Candidates were shortlisted based on three criteria: (i) the proteins matched at least two unique peptides with > 95% confidence interval (C.I.), (ii) the unused score (total score of distinct peptides detected for a protein) was > 1.5, and (iii) the proteins were not detected in the antisense control pulldown (Supplementary Fig. S3A). IGF2BP2, an RBP known to bind to the MYC 3′UTR [37], was enriched in our MYC 3'UTR pulldown (Supplementary Fig. S3A). This suggests that its interaction with MYC is conserved in different cell types and demonstrates the robustness of our pulldown experiment.
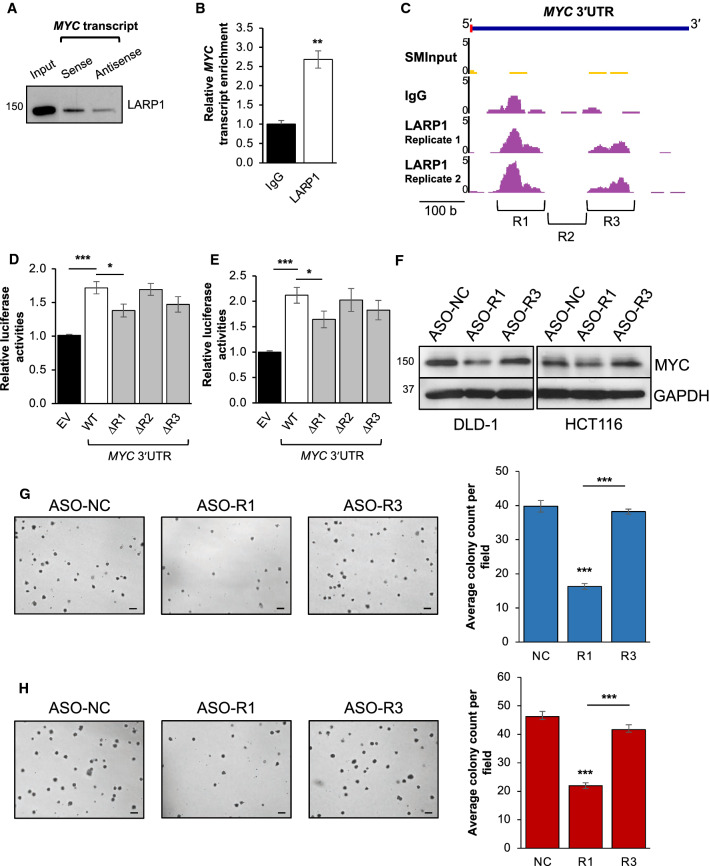
Most importantly, we were able to detect and validate the enrichment of LARP1 in the MYC pulldown (Fig. 4A, Supplementary Fig. S3A-E). Two paralogues exist for human LARP1: LARP1, and LARP2 (also known as LARP1a and LARP1b). LARP1 and LARP2 are located on chromosomes 5q34 and 4q28, respectively, and they encode two proteins with 60% sequence homology but of different sizes [38]. Despite their similarity, mass spectrometry of the MYC 3′UTR pulldown detected peptides which were distinctly LARP1, indicating that the MYC 3′UTR specifically interacts with LARP1, but not LARP2 (Supplementary Fig. S3B, C). The LARP1:MYC interaction was also validated by Western blot analysis that showed LARP1 protein enrichment in the MYC pulldown (Fig. 4A, Supplementary Fig. S3D–E). To further confirm this interaction, we performed a complementary experiment in the form of an RNA immunoprecipitation (RIP) using the LARP1 protein as bait, which significantly enriched for the MYC transcript (Fig. 4B, Supplementary Fig. S3F). The corroborative data from these bidirectional studies reaffirm the interaction between the LARP1 protein and the MYC transcript.
Fig. 4.
LARP1 associates with the MYC transcript to regulate its expression. A LARP1 protein enrichment upon MYC transcript pulldown in DLD-1 cells. The antisense pulldown was used as a negative control. B MYC transcript enrichment upon LARP1 RNA immunoprecipitation in DLD-1. Mean ± STD. **P < 0.01. C Genome browser track of LARP1 peaks indicating the binding of LARP1 to the MYC 3′UTR and size-matched input (SMInput) from the LARP1 enhanced UV crosslinking and immunoprecipitation (eCLIP) of DLD-1 cells. Inverted brackets indicate regions of MYC 3′UTR that were deleted to create the MYC 3′UTR ΔR1, ΔR2, ΔR3 mutant luciferase reporters. D, E Effect of LARP1 overexpression on the relative luciferase activities of the wild-type (WT) and mutant (ΔR1, ΔR2, ΔR3) MYC 3′UTR-reporters in DLD-1 (D) and HCT116 (E). EV denotes empty vector. Mean ± STE. *P < 0.05; ***P < 0.001. F–H Effect of the antisense oligonucleotide (ASO) targeting the MYC 3′UTR R1 region (ASO-R1) and MYC 3′’UTR R3 region (ASO-R3) compared to ASO non-targeting control (ASO-NC) on MYC protein expression (F left: DLD-1, F right: HCT116) and anchorage-independent growth (G: DLD-1, H: HCT116, scale bar: 200 μm). Quantification of anchorage-independent growth is shown at the right panels. Mean ± STE. ***P < 0.001
Our LARP1 eCLIP of DLD-1 cells identified two distinct LARP1 binding sites (referred to as R1 [chr8:128,753,264–128,753,359] and R3 [chr8:128,753,457–128,753,557]) separated by a stretch of sequence termed R2 [chr8:128,753,360–128,753,456] on the MYC 3′UTR (Fig. 4C). To validate these putative LARP1 binding sites, we deleted regions R1, R2 or R3 from the MYC 3′UTR-luciferase reporter construct (∆R1, ∆R2, ∆R3). LARP1 overexpression significantly upregulated the luciferase activity of the WT MYC 3′UTR reporter, while R1 deletion resulted in a marked decrease in the MYC 3′UTR luciferase signal compared to WT (Fig. 4D, E). On the other hand, deletion of the R2 and R3 regions did not have a significant effect. We further validated the importance of the R1 region in regulation of MYC using a steric blocking antisense oligonucleotide (ASO) targeting this region (ASO-R1). ASO-R1 significantly decreased MYC protein expression (Fig. 4F) and reduced the anchorage-independent growth of CRC cells (Fig. 4G, H) compared to a non-targeting control (ASO-NC) and the negative ASO-R3 region (ASO-R3). However, ASO-R1 treatment appears to increase the MYC mRNA levels, suggesting a potential translation inhibition mechanism that results in accumulation of MYC mRNA (Supplementary Fig. S3G, H). Taken together, these results suggest that the LARP1 potentially regulates MYC through the R1 region. Moreover, the R1 region is present in all the MYC 3′UTR isoforms, hence suggesting its suitability for ASO-mediated targeting to control MYC expression in cancer.
Quantitative mass spectrometry analysis of LARP1:protein interactions reveals IGF2BP3 and YBX1 as LARP1-interacting proteins
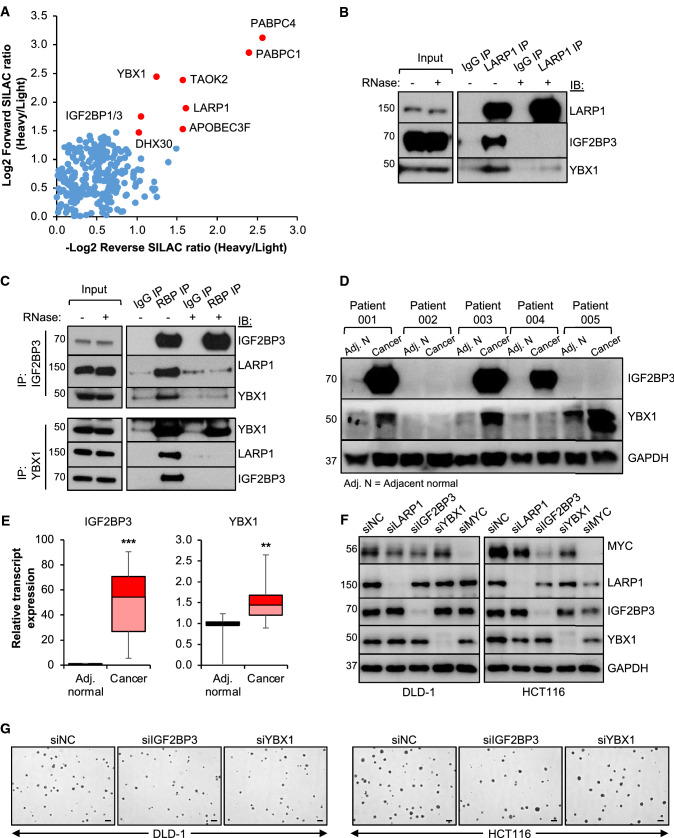
To better understand the mechanism underlying LARP1 action in CRC, we performed a systematic analysis of LARP1-interacting proteins by performing LARP1 immunoprecipitation (IP) using SILAC labelled cells, followed by quantitative mass spectrometry analysis (Data File S1). These were shortlisted based on two criteria; (i) forward SILAC ratio > 2.5 and (ii) reverse SILAC ratio < 0.5 (Fig. 5A). Among the shortlisted candidates, IGF2BP3 and YBX1 were validated as LARP1-interacting partners (Fig. 5B, Supplementary Fig. S4A). The association among these three RBPs was further verified by independent IGF2BP3 and YBX1 IPs (Fig. 5C, Supplementary Fig. S4B). Interestingly, the association between LARP1, IGF2BP3, and YBX1 appeared to be RNA-dependent as it was lost upon RNase treatment (Fig. 5B, C, Supplementary Fig. S4A, S4B).
Fig. 5.
Quantitative mass spectrometry analysis of LARP1:protein interactions reveals IGF2BP3 and YBX1 as LARP1-interacting proteins. A Quantitative mass spectrometry detection of proteins enriched upon LARP1 immunoprecipitation (IP) in HCT116 cells grown in stable isotope labelling by amino acids in cell culture (SILAC) media. B Western blot validating the enrichment of LARP1 and its interacting proteins upon LARP1 IP compared to immunoglobulin (IgG) IP in DLD-1 cells. C Western blot validating the enrichment of LARP1 and YBX1 in IGF2BP3 IP (top panel) and the enrichment of LARP1 and IGF2BP3 in YBX1 IP (bottom panel) compared IgG IP in DLD-1 cells. D and E IGF2BP3 and YBX1 protein (D) and transcript (E) expression in five pairs of matched adjacent normal (Adj. N) and CRC patient samples. Mean ± STE. **P < 0.01; ***P < 0.001. F and G Effect of IGF2BP3 and YBX1 knockdown on MYC protein expression (F) and anchorage-independent growth (G, scale bar: 200 μm) of DLD-1 and HCT116. siNC denotes siRNA non-targeting control
Both YBX1 and IGF2BP3 have been shown to possess oncogenic properties in CRC [39, 40]. Our analysis also identified IGF2BP3 as one of the 16 annotated mRBPs which are significantly upregulated in CRC (Table S7), supporting its importance in CRC. In addition, these RBPs have been shown to regulate MYC expression in other cancers: YBX1 was found to regulate MYC translation in multiple myeloma [41], while IGF2BP3 increased MYC expression in mixed lineage leukemia–rearranged (MLL-rearranged) B-acute lymphoblastic leukemia (B-ALL) [42]. However, their potential regulation of MYC in CRC remains unexplored to date. We observed upregulation of IGF2BP3 and YBX1 protein and transcript expression in CRC samples relative to adjacent normal tissue (Fig. 5D, E, Supplementary Fig. S4C). Moreover, we found that knockdown of IGF2BP3 and YBX1 significantly decreased MYC protein expression (Fig. 5F, Supplementary Fig. S4D) and anchorage-independent growth of CRC cells (Fig. 5G, Supplementary Fig. S4E). Collectively, these data validate the oncogenic properties of IGF2BP3 and YBX1 in CRC, which is at least in part through their regulation of MYC. We also postulate that LARP1, IGF2BP3, and YBX1 may work in tandem with each other to regulate MYC expression and CRC development.
MYC positively regulates LARP1 transcription
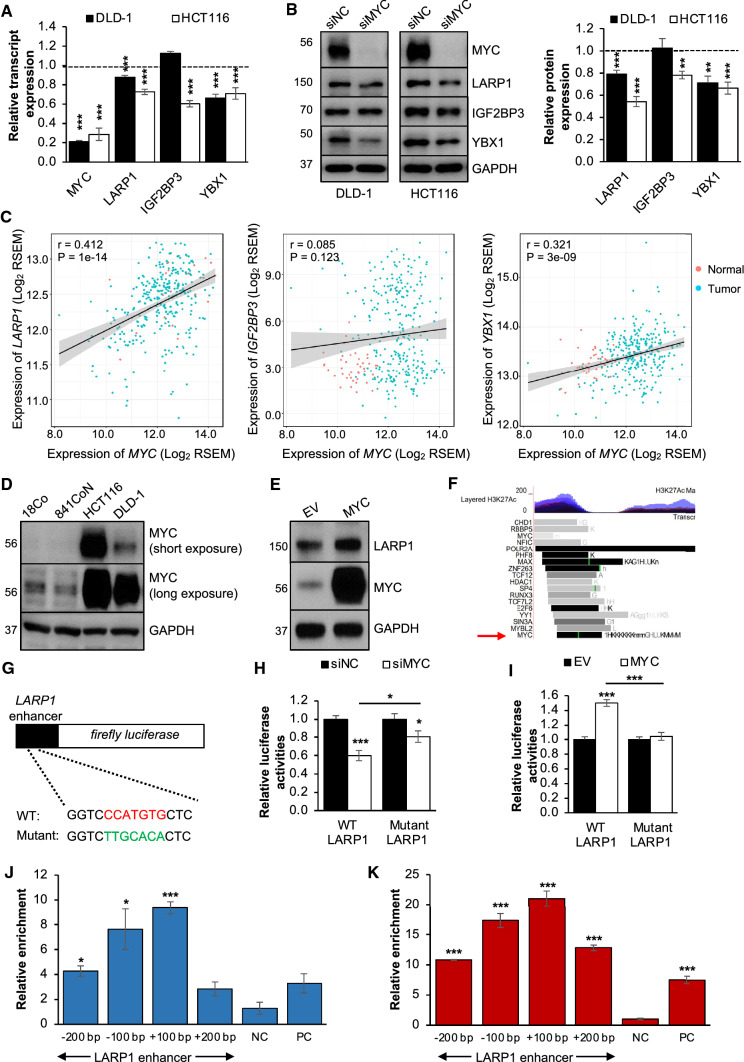
Given that MYC is a well-established transcription factor that governs the expression of numerous genes [43], we explored the hypothesis that MYC could regulate the expression of its RBPs in a positive feedback loop. In line with this hypothesis, MYC knockdown resulted in a significant reduction in the transcript and protein expression of LARP1 and YBX1 in both cell lines (Fig. 6A, B). However, the effects of MYC knockdown on the IGF2BP3 transcript and protein expression were only observed in HCT116, suggesting a cell line-specific regulation of IGF2BP3 expression. Next, we observed that MYC expression was positively correlated with the expression of LARP1 and YBX1, but not IGF2BP3, across 325 TCGA-COAD samples (Fig. 6C). This was consistent with our in vitro findings and supported the potential reciprocal relationship between MYC and its RBPs, LARP1, and YBX1, in maintaining their high levels in CRC.
Fig. 6.
MYC positively regulates LARP1 transcription. A and B Effect of MYC knockdown on the transcript (A) and protein (B) expression of LARP1, IGF2BP3 and YBX1 in DLD-1 and HCT116. Western blot densitometry quantification of RBP expression upon MYC knockdown is shown at the right panel. siNC denotes siRNA non-targeting control. Mean ± STE. **P < 0.01; ***P < 0.001. C Scatter plots demonstrating the correlation of transcript expression between MYC and LARP1 (left panel), IGF2BP3 (middle panel) and YBX1 (right panel) in the TCGA-COAD samples (284 CRC versus 41 normal). RSEM denotes RNA-Seq by Expectation Maximization. D Endogenous MYC protein expression in immortalized normal colon (CCD-18Co and CCD-841CoN) and CRC (HCT116 and DLD-1) cell lines. E Effect of MYC overexpression on LARP1 protein expression in DLD-1. EV denotes empty vector. F Schematic representation of the LARP1 enhancer cloned into the pGL4.20 promoterless vector. G Schematic representation of the LARP1 enhancer cloned into the pGL4.20 promoterless vector. H and I Relative luciferase activities of the WT and mutant LARP1 enhancer-reporters upon MYC knockdown (H) and overexpression (I) in DLD-1. siNC denotes siRNA non-targeting control; EV denotes empty vector. Mean ± STE. *P < 0.05; ***P < 0.001. J and K ChIP-qPCR showing the relative enrichment of the LARP1 enhancer region in DLD-1 (J) and HCT116 (K). A chromosome region upstream of the LARP1 promoter was used as a negative control. The MYU promoter was used as a positive control. NC denotes negative control; PC denotes positive control. Mean ± STE. *P < 0.05; ***P < 0.001
MYC has been shown to regulate YBX1 at the transcriptional level in multiple myeloma [41]; however, its transcriptional regulation of LARP1 is unknown. To validate the regulatory effect of MYC on LARP1, we performed a reciprocal MYC overexpression experiment in DLD-1 cells, which have lower levels of endogenous MYC compared to HCT116 (Fig. 6D) and observed a significant increase in LARP1 transcript and protein levels (Supplementary Fig. S5A, S5B; Fig. 6E). Furthermore, we searched for potential MYC binding sites on LARP1 using the UCSC genome browser. We observed high levels of the H3K27 acetylation mark at the enhancer region of LARP1 (Supplementary Fig. S5C), signifying increased transcriptional activity in this region. Upon further analysis, we found a MYC binding site at position chr5:154,134,296–154,134,812 of the LARP1 enhancer region, which is present in multiple cell lines (Fig. 6F), suggesting that MYC could be modulating LARP1 transcription through its enhancer region. To evaluate this, we generated a luciferase reporter construct containing the LARP1 enhancer sequence in a promoterless plasmid. We also designed a mutant enhancer construct by altering the identified MYC-binding sequence based on purine-to-purine and pyrimidine-to-pyrimidine substitutions to ensure that the DNA structure was minimally disrupted compared to the wild-type (WT) (Fig. 6G). Furthermore, the WT and mutant sequences were cloned in the reverse orientation as well.
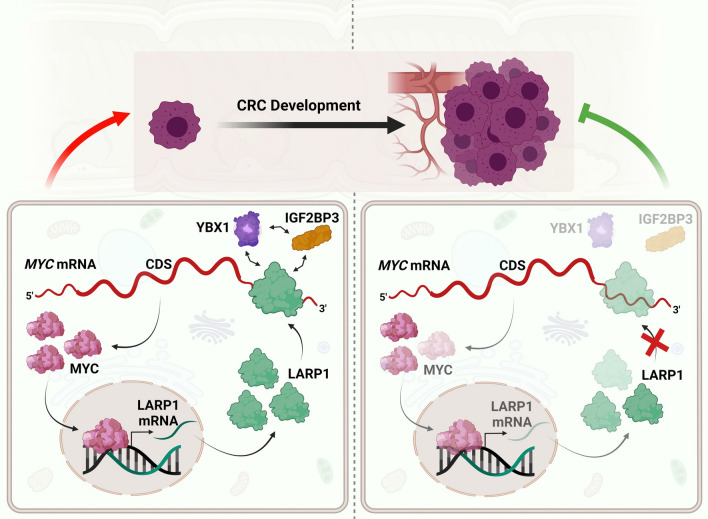
Relative to the promoterless vector, both the forward and reverse LARP1 enhancer reporters showed significantly increased luciferase activity, suggesting this region is a functional enhancer (Supplemental Fig. S5D, E). Following MYC knockdown, we detected a significant decrease in the relative luciferase activity of the WT LARP1 enhancer-reporter, with little to no effect on the mutant enhancer-reporter (Fig. 6H, Supplemental Fig. S5F). Consistently, MYC overexpression significantly increased the luciferase activity of the WT reporter with no significant change in the mutant variant (Fig. 6I, Supplementary Fig. S5G). MYC ChIP-qPCR detected significant enrichment of the LARP1 enhancer region in both CRC cell lines, suggesting that MYC binds to the enhancer sequence in its native chromatin context (Fig. 6J, K). Together, our findings establish a novel positive feedback loop between RBPs and MYC that could promote tumorigenesis in CRC (Fig. 7).
Fig. 7.
Model of the feedforward loop between LARP1 and MYC in CRC. In the steady state of CRC cells (left panel), the association of LARP1 with the MYC 3′UTR increases MYC protein expression. In turn, MYC acts as a transcriptional activator of LARP1 to induce LARP1 transcription and subsequently increase LARP1 transcript and protein levels. Cumulatively, this positive feedback loop promotes CRC progression. In addition, LARP1 interacts with other RBPs, IGF2BP3 and YBX1 which also regulate MYC expression, in an RNA-dependent manner. Disruption of LARP1 binding to the MYC 3′UTR reduces MYC protein expression and inhibits CRC progression (right panel)
Discussion
CRC is one of the most prevalent cancers in terms of diagnosis and mortality; hence, there is an urgency for deeper insights into the molecular basis underlying CRC development [4]. It has been suggested that a deeper understanding of the global transcriptomic landscape may provide a link between molecular underpinnings and cellular phenotypes. The global transcriptomic balance is frequently disrupted in cancers, and this could be in part due to the dysregulation of RNA-binding proteins (RBPs) which modulate a broad range of target transcripts.
In this study, we identify LARP1 as an RBP which is upregulated in CRC. In addition to being a robust prognostic marker in several cancer types, [6, 8–10] our data suggest that LARP1 possesses strong oncogenic properties in CRC. We demonstrate that LARP1 associates with the 3′UTRs of key oncogenes in CRC. Specifically, we show that LARP1 binds to and positively regulates MYC expression and that disruption of the association between LARP1 and the MYC 3′UTR significantly decreases MYC expression. Additionally, we uncover a positive feedback loop whereby MYC regulates LARP1 at the transcriptional level.
Our data suggest that the oncogenic role of LARP1 could be partly attributed to its positive regulation of MYC expression. However, the existing literature postulates that LARP1 can both promote and repress growth. Both phenomena can be attributed to LARP1 binding to the cap structure of 5′ TOR mRNAs, which encode for ribosomal proteins and translational factors required for growth. Binding to mRNA cap can result in competition with either de-capping factors or cap-binding translation factors, stabilizing the mRNA or inhibiting translation, respectively, in turn promoting or suppressing growth. Furthermore, other functions of LARP1, such as regulating mRNAs through their 3′ UTRs, are comparatively unexplored. While we, and others, have demonstrated LARP1’s oncogenic role in various cancers, a coherent function of LARP1 in cancer has yet to be established [11, 12, 24, 25, 44]. Indeed, the function of LARP1 could be cancer- and tissue-specific, leading to different functions depending on cell type or cell state [44].
We also identify and validate LARP1-interacting RBPs, IGF2BP3 and YBX1, which associate with LARP1 and each other in an RNA-dependent manner (Fig. 7). The interactions between these RBPs have been studied independently, however, their association and effect on a common target transcript have not been explored until now. The interaction between YBX1 and LARP1 has been shown to prevent Hepatitis C virus (HCV) particle production in the HCV-infected human hepatoma cells [45]. On the other hand, the association between YBX1 and IGF2BP3 has been shown in the renal cell carcinoma [46]. We postulate that a comprehensive study of the interaction of RBPs on common target transcripts will lead to deeper insights into their roles in post-transcriptional regulation and cancer progression.
MYC is overexpressed in more than half of human cancers, and its key regulatory functions in CRC as well as other cancers is well established [43]. Our study uncovers the reciprocal relationship between MYC and LARP1 which may in part explain the maintenance of their elevated expression and oncogenic capacity in CRC (Fig. 7). While the oncogenic effects of MYC protein are well-studied, the MYC mRNA itself is important to cellular processes, such as regulation of general transcription based on glutamine levels [47]. This highlights the importance of considering the effect of post-transcriptional regulators on both the MYC mRNA and protein levels when investigating such complex, interconnected regulatory networks.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all past and present YT lab members for their constructive feedback on this project. We also thank Teck Kwang Lim from Protein and Proteomics Centre at the Department of Biological Sciences (DBS), NUS, and Charlene Chan from Cancer Science Institute of Singapore, for their assistance in performing the mass spectrometry analyses. In addition, we thank Avencia Sanchez-Mejias and Xiao Hong Chew for their technical assistance. Furthermore, we thank Shi Hao Tan from Cancer Science Institute of Singapore for his assistance in the design of the ChIP-qPCR experiments.
Author contributions
YT, ND, and JJC conceptualized and designed the project. ND and QYT designed and performed experiments, analyzed and interpreted data and generated figures. VT, JJC, BZ, HT, ZHK, and CYL designed and performed experiments and analyzed data. HQT, KK, WJ, and GWY performed, analyzed and interpreted the eCLIP experiments. BZ and HY performed bioinformatics analysis and interpretation. CYL, SW, PCLL, BES, KCL, CSC, and KKT obtained and processed the patient samples. HTT, MCMC and DK performed mass spectrometry experiments and analysis. ND, QYT, JJC, YT, KK, BZ, HTT, and DK wrote and edited the manuscript.
Funding
Y.T. is funded by a Singapore National Research Foundation Fellowship, a National Medical Research Council Open Fund—Individual Research Grant and a National University of Singapore President’s Assistant Professorship. Y.T., K.K. and D.K. are supported by the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s AcRF Tier 3 Grant (MOE2014-T3-1-006). G.W.Y. is funded by the National University of Singapore and National Research Foundation of Singapore. N.D. is supported by the NUS Research Scholarship. QYT is supported by a Cancer Science Institute of Singapore Research Scholarship. This research is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative, as well as the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s AcRF Tier 3 Grants, Grant number MOE2014-T3-1-006.
Availability of data and material
The authors confirmed that the data supporting the findings of this study are available within the article and its supplementary material.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ng Desi and Qing Yun Tong contributed equally to this work.
Velda Teh, Jia Jia Chan and Bin Zhang contributed equally to this work.
References
- 1.Wurth L. Versatility of RNA-binding proteins in cancer. Comp Funct Genomics. 2012;2012:178525. doi: 10.1155/2012/178525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze MW, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 3.Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol Cell. 2004;96(7):479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Arnold M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 5.Kuipers EJ, et al. Colorectal cancer. Nat Rev Dis Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie C, et al. LARP1 predict the prognosis for early-stage and AFP-normal hepatocellular carcinoma. J Transl Med. 2013;11:272. doi: 10.1186/1479-5876-11-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousquet-Antonelli C, Deragon JM. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15(5):750–764. doi: 10.1261/rna.1478709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins TG, et al. The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res. 2016;44(3):1227–1246. doi: 10.1093/nar/gkv1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mura M, et al. LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene. 2015;34(39):5025–5036. doi: 10.1038/onc.2014.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye L, et al. Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumour Biol. 2016;37(11):14585–14594. doi: 10.1007/s13277-016-5332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca BD, et al. La-related Protein 1 (LARP1) represses terminal Oligopyrimidine (TOP) mRNA translation downstream of mTOR Complex 1 (mTORC1) J Biol Chem. 2015;290(26):15996–16020. doi: 10.1074/jbc.M114.621730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tcherkezian J, et al. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev. 2014;28(4):357–371. doi: 10.1101/gad.231407.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butter F, et al. Unbiased RNA–protein interaction screen by quantitative proteomics. Proc Natl Acad Sci USA. 2009;106(26):10626–10631. doi: 10.1073/pnas.0812099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon JH, Srikantan S, Gorospe M. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012;58(2):81–87. doi: 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meisenheimer KM, Koch TH. Photocross-linking of nucleic acids to associated proteins. Crit Rev Biochem Mol Biol. 1997;32(2):101–140. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- 16.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20(12):1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high throughput experiments. Ann Appl Stat. 2011;5:1752–1779. [Google Scholar]
- 20.Liao Y, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 22.Lee TI, et al. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protocol. 2006;1(2):729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, et al. A comprehensive expression landscape of RNA-binding proteins (RBPs) across 16 human cancer types. RNA Biol. 2020;17(2):211–226. doi: 10.1080/15476286.2019.1673657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahr RM, et al. The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence. Nucleic Acids Res. 2015;43(16):8077–8088. doi: 10.1093/nar/gkv748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lahr RM, et al. La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife. 2017 doi: 10.7554/eLife.24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy HK, et al. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23(1):201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- 27.Rochlitz CF, Herrmann R, de Kant E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology. 1996;53(6):448–454. doi: 10.1159/000227619. [DOI] [PubMed] [Google Scholar]
- 28.Erisman MD, et al. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol Cell Biol. 1985;5(8):1969–1976. doi: 10.1128/mcb.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Birkenbach M, Hart J. Expression of Jun family members in human colorectal adenocarcinoma. Carcinogenesis. 2000;21(7):1313–1317. [PubMed] [Google Scholar]
- 30.Arber N, et al. Increased expression of cyclin D1 is an early event in multistage colorectal carcinogenesis. Gastroenterology. 1996;110(3):669–674. doi: 10.1053/gast.1996.v110.pm8608874. [DOI] [PubMed] [Google Scholar]
- 31.Sebolt-Leopold JS, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat Med. 1999;5(7):810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coller HA, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97(7):3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DR, Myint T, Goh HS. Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br J Cancer. 1993;68(2):407–413. doi: 10.1038/bjc.1993.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andres SF, et al. IMP1 3' UTR shortening enhances metastatic burden in colorectal cancer. Carcinogenesis. 2019;40(4):569–579. doi: 10.1093/carcin/bgy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, et al. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23(6):1176–1188. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stavraka C, Blagden S. The la-related proteins, a family with connections to cancer. Biomolecules. 2015;5(4):2701–2722. doi: 10.3390/biom5042701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim A, et al. Inhibition of Y box binding protein 1 suppresses cell growth and motility in colorectal cancer. Mol Cancer Ther. 2020;19(2):479–489. doi: 10.1158/1535-7163.MCT-19-0265. [DOI] [PubMed] [Google Scholar]
- 40.Xu W, et al. Increased IGF2BP3 expression promotes the aggressive phenotypes of colorectal cancer cells in vitro and vivo. J Cell Physiol. 2019;234(10):18466–18479. doi: 10.1002/jcp.28483. [DOI] [PubMed] [Google Scholar]
- 41.Bommert KS, et al. The feed-forward loop between YB-1 and MYC is essential for multiple myeloma cell survival. Leukemia. 2013;27(2):441–450. doi: 10.1038/leu.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palanichamy JK, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest. 2016;126(4):1495–1511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berman AJ, et al. Controversies around the function of LARP1. RNA Biol. 2021;18(2):207–217. doi: 10.1080/15476286.2020.1733787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatel-Chaix L, et al. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J Virol. 2013;87(21):11704–11720. doi: 10.1128/JVI.01474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. C1QBP negatively regulates the activation of oncoprotein YBX1 in the renal cell carcinoma as revealed by interactomics analysis. J Proteome Res. 2015;14(2):804–813. doi: 10.1021/pr500847p. [DOI] [PubMed] [Google Scholar]
- 47.Dejure FR, et al. The MYC mRNA 3'-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 2017;36(13):1854–1868. doi: 10.15252/embj.201796662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirmed that the data supporting the findings of this study are available within the article and its supplementary material.