Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system provides a groundbreaking genetic technology that allows scientists to modify genes by targeting specific genomic sites. Due to the relative simplicity and versatility of the CRISPR/Cas system, it has been extensively applied in human genetic research as well as in agricultural applications, such as improving crops. Since the gene editing activity of the CRISPR/Cas system largely depends on the efficiency of introducing the system into cells or tissues, an efficient and specific delivery system is critical for applying CRISPR/Cas technology. However, there are still some hurdles remaining for the translatability of CRISPR/Cas system. In this review, we summarized the approaches used for the delivery of the CRISPR/Cas system in mammals, plants, and aquacultures. We further discussed the aspects of delivery that can be improved to elevate the potential for CRISPR/Cas translatability
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-020-03725-2.
Keywords: CRISPR/Cas, Gene editing, Gene delivery, Gene therapy
Introduction
The innovation of gene editing has enabled the precise modification of specific genomic regions in a wide variety of organisms. Gene editing is mainly accomplished using programmable nucleases that are highly specific. These nucleases create double-strand breaks (DSBs) in regions of interest of the genome. These DSBs are then repaired by nonhomologous end-joining (NHEJ), which is error-prone, or homology-directed repair (HDR), which is error-free; specific changes, such as insertions or deletions (indels), are thus introduced into desired regions of the genome [1–3]. By introducing HDR repair template, the defects in genes may be corrected, thus providing hope for correcting inherent errors in DNA.
A recent new programmable nuclease technology; clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)-type II RNA-guided nucleases system [4], has revolutionized the scientific field of gene editing. Although the versatility and ease of construction and target design make CRISPR/Cas extremely attractive for breakthrough gene therapy achievements and crop improvement, there are still important limitations to consider [5, 6]. One of the obstacles is the immune response in animal systems; since the components of the CRISPR/Cas system are bacterially derived, this system is expected to trigger host immune responses. Another obstacle is the size of the components in the system, which are all macromolecules; thus, they are unable to spontaneously enter the cytosol and then the nucleus [6], which are essential for successful gene modification [7]. In addition, the large size of the CRISPR/Cas system may also make it difficult to package into delivery vehicles such as viral vectors. Another aspect of the difficulty of the CRISPR/Cas system is its stability. The CRISPR/Cas system needs to be highly stable and functional; otherwise, it will be degraded or eliminated during circulation in the targeted organs or tissues. Efficient delivery is one of the last major hurdles to overcome in CRISPR/Cas-mediated gene editing. As such, developing stable and effective delivery approaches is critical for its application. In this review, we summarized the approaches used for the delivery of the CRISPR/Cas system in different biological systems, including mammals, aquacultures, and plants (Fig. 1). We also discussed the aspects of delivery that can be improved to elevate the potential translatability the CRISPR/Cas system.
Fig. 1.
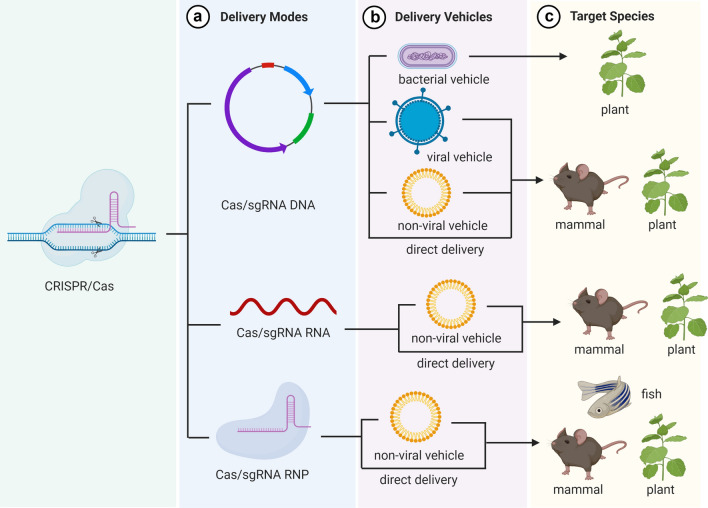
Schematic diagrams of in vivo CRISPR/Cas delivery modes and vehicles in different biological systems. Systems used for delivery of CRISPR/Cas components (a) can be separated into two major categories, CRISPR/Cas delivery mode (b) and delivery vehicle (c). Three CRISPR/Cas delivery models including DNA (plasmid encoding both the Cas protein and the gRNA), RNA (mRNA for Cas protein translation and a separate gRNA) and protein (Cas protein with gRNA as a ribonucleoprotein complex, RNP) can be delivered in to mammalians, aquacultures or plants via bacterial or viral vectors, non-viral carriers and physically direct delivery (d). The figure was created with BioRender.com
CRISPR/Cas gene-editing system
The CRISPR/Cas system was first identified as a prokaryotic adaptive immune system. It was the first programmable nuclease system that was found to function as ribonucleoprotein particles that utilized base pairing to recognize its targets [8]. This system for gene editing has been widely adopted since it is relatively easy to redesign to produce target specificity. Scientists have engineered and modified this system to allow CRISPR/Cas to act as a successful gene-editing tool [4].
There are three key components in the CRISPR/Cas9 system: the tracrRNA, Cas9 protein, and pre-crRNA. The tracrRNA forms a complex with pre-crRNA after transcription. The Cas9 protein stabilizes the complex, and the pre-crRNA is then processed by RNase III to generate crRNA [9]. The Cas9/gRNA (made up of crRNA and tracrRNA) complex recognizes the protospacer adjacent motif (PAM), which is a short motif that is located adjacent to the target DNA sequence [10]. Then, the complex unwinds the target DNA beginning at the seed region (10–12 nucleotides) [11]. When the DNA sequence corresponds to the gRNA, two nuclease domains of Cas9 cleave the target strands [12–14]. The Cas9/gRNA complex can tolerate single or sometimes multiple mismatches, with mismatches downstream of the seed region typically being more frequently tolerated [13, 15].
There are six types of CRISPR/Cas systems (type I–VI) that are further classified into two classes: the class 1 CRISPR/Cas system and the class 2 CRISPR/Cas system [16, 17]. The main feature of the class 1 CRISPR/Cas system, which is subclassified into types I, III, and IV, is that they have multisubunits of effector nuclease complexes. The class 2 CRISPR/Cas system differs from class 1 because it requires only a single effector nuclease; class 2 is subclassified into types II, V, and VI. Their programmable single effector nucleases enable nucleic acid detection and genome engineering [3, 18–21]. Types II, V, and VI are based on Cas9, Cas12, and Cas13 effectors [22–24]. Among them, the CRISPR/Cas9 system is the most commonly used system to date.
Application of the CRISPR/Cas system for gene editing
Strategies based on Cas nuclease activity
Based on the nuclease activity of CRISPR/Cas, there are various gene editing strategies that have been developed for DNA (gene disruption, precise repair, targeted insertion, large-scale DNA editing) and RNA modification (Supplementary Table S1; Fig. 2a, b).
Fig. 2.
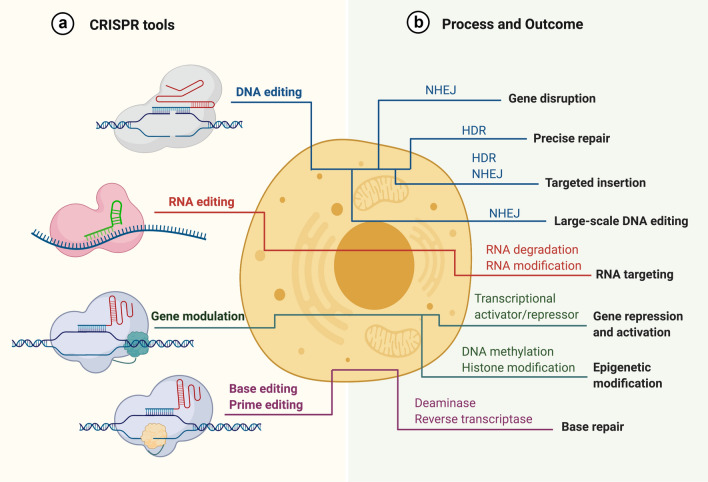
CRISPR/Cas-mediated gene-editing strategies. The versatile CRISPR/Cas system is a powerful tool for DNA, RNA editing, gene modulation and base, prime editing by leveraging different approaches (a) to achieve numerous gene-editing outcomes (b). The figure was created with BioRender.com
DNA editing
For gene disruption, NHEJ ligates DSBs introduced by Cas endonuclease, and this break and repair pattern takes place repeatedly until the target sequence is altered and an indel occurs [25]. An indel can cause frameshifting or exon skipping and subsequent gene disruption. Gene disruption can also silence dominant negative mutations by disrupting the mutant allele while preserving the normal allele. In addition, HDR inserts a donor template, which has homology arms to match the target locus to the genome and cause a deletion [26].
For precise repair, HDR uses a donor template that has the desired insertion or modification. This donor template has homology arms that enable it to match the target locus and insert the desired genetic material or modify the genome with high precision [25].
For targeted insertion, HDR allows precise insertion of exogenous DNA sequences into the genomes of dividing cells, while homology-independent targeted integration (HITI) allows insertion of exogenous DNA sequences into the genomes of nondividing cells using an NHEJ-based homology-independent strategy [27].
For large-scale DNA editing (editing a size of up to several megabase pairs [Mbp]), DNA fragments can be deleted by introducing the CRISPR/Cas system with two guide RNAs that target different sites. In addition, allelic exchange can correct recessive compound heterozygous mutations. This is achieved by generating homologous DNA breaks in both chromosomes, and the allelic exchange between mutated alleles can rescue the disease phenotype [28].
RNA editing
The CRISPR/Cas system acts not only on DNA but also RNA. Previous studies have identified an RNA-targeting CRISPR/Cas effector complex, termed the psiRNA-Cmr protein complex, which comprise prokaryotic silencing (psi)RNAs and Cmr Cas proteins. This complex cleaves target RNAs at a predetermined site, indicating that prokaryotes have their own unique RNA silencing system [29]. Cas endonuclease has also been shown to bind to and cleave ssRNA targets [30]. Strutt et al. showed that type II-A and II-C Cas9 endonucleases are capable of recognizing and cleaving ssRNA without a PAM. [31]. Recently, scientists have discovered the RNA-editing Cas13 family. The Cas13 family has been shown to be a programmable RNA-editing CRISPR/Cas system. Compared to other RNA targeting approaches, this system is more specific and efficient [32–34]. Recently, Konermann et al. discovered a Cas13d in Ruminococcus flavefaciens XPD3002 (CasRx), and it possesses high activity in human cells. CasRx is small, consisting of 930 amino acids, and it can be flexibly packaged into an adeno-associated virus (AAV), making it suitable for delivery by AAV vectors. In addition to the knockdown activity, catalytically inactivated CasRx can be utilized to regulate pre-mRNA splicing by acting as a splice effector [35].
Strategies based on Cas-effector fusion protein activity
Since CRISPR/Cas possesses DNA-binding properties, it may play a crucial role in important applications other than site-specific gene editing. A catalytically dead Cas9 enzyme (dCas9) has been developed to control gene expression [36]. dCas9, like Cas9, is capable of recognizing and binding to a target DNA sequence. However, instead of cleaving the target DNA sequence, dCas9 has been used for transcriptional repression, transcriptional activation, introducing epigenetic modifications, and base editing. These functions are achieved by fusing dCas9 to gene activators, repressors, acetyltransferases or adenosine deaminases. Since dCas9 is in a catalytically inactive form, it is used here for precise targeting instead of its catalytic activity (Supplementary Table S2; Fig. 2c, d).
Transcriptional regulation
dCas9 can be fused with transcriptional repressors or activators to target the promoter region of the gene of interest and result in transcriptional repression (CRISPR interference), or activation (CRISPR activation) without changing the genome; this activity has been demonstrated in Escherichia coli as well as in plant and mammalian cells [36–38]. In addition, studies have shown that a modifying sgRNA can also enhance the specificity of transcriptional regulation. For example, using the Cas9-VP64 transcriptional activator together with an sgRNA that has two MS2 RNA aptamer hairpin sequences added to it can successfully induce sequence-specific transcriptional activation [39].
The CRISPR/Cas system can also be utilized for epigenetic modification. Hilton et al. fused dCas9 with acetyltransferase that catalyze the acetylation of histone H3 at lysine 27. This modulation has been shown to strongly activate specific gene expression. Not only acetylation but also methylation may be accomplished using this approach [40].
Base and prime editing
CRISPR/dCas has been utilized for precise DNA base editing. The CRISPR/nickase Cas9 (nCas9)-based base editor was first developed by Komor et al. and was used to convert a targeted C–G base pair to T–A by a DNA cytosine deaminase [41]. Gaudelli et al. subsequently developed a transfer RNA adenosine deaminase that, when fused to nCas9, can convert A–T base pairs to G–C base pairs [42]. This kind of CRISPR/Cas-mediated editing is powerful since single point mutations account for a large category of genetic diseases.
Recently, a more powerful and versatile gene-editing method, prime editing, was discovered as a way to introduce indels and enable base conversions in both transitions and transversions [43]. The editor used in prime editing is termed prime editor. The prime editor is composed of nCas9 fused with reverse transcriptase. The prime editor is guided by a prime editing gRNA (pegRNA). After nCas9 nicks the target site, the pegRNA binds to a single strand DNA (ssDNA) and initiates reverse transcription. The reverse transcribed pegRNA is then incorporated into the target site.
Current approaches for delivering the CRISPR/Cas system in mammals
CRISPR/Cas can be delivered using different modalities, including DNA, RNA and protein. When it is delivered in a DNA mode, Cas and gRNA are delivered as a single plasmid. For the RNA mode, Cas mRNA is delivered with a separate gRNA. For the protein mode, Cas protein is delivered with gRNA as a ribonucleoprotein complex (RNP). Each mode exhibits overall effectiveness but also includes some limitations. Packaging Cas9 and gRNA in the same plasmid makes the delivered cargo more stable than that of other methods; however, the large size of the plasmid increases the difficulty of delivery, and the integration of plasmids into the host genome and prolonged expression are potential limitations of this delivery method. Delivery of Cas mRNA enables faster gene editing; however, RNA is fragile, and the degradation of gRNA may initiate before Cas9 mRNA is successfully translated. The RNP is the most direct and fastest mode for gene editing. However, compared to plasmids or mRNAs, it is much more challenging to obtain a pure protein. In addition, the sudden introduction of bacterial proteins may induce an immune response in the host.
The delivery vehicles can be separated into two groups: viral and nonviral vectors. For in vivo delivery of CRISPR/Cas, viral vectors are the preferred vehicle. To date, nonviral vector delivery has not been as commonly used as viral-based delivery. However, nonviral vectors are comparable to viral vectors and are a topic of intense research. The delivery vehicles for the in vivo CRISPR/Cas system discussed below are summarized in Fig. 3 and Table 1.
Fig. 3.
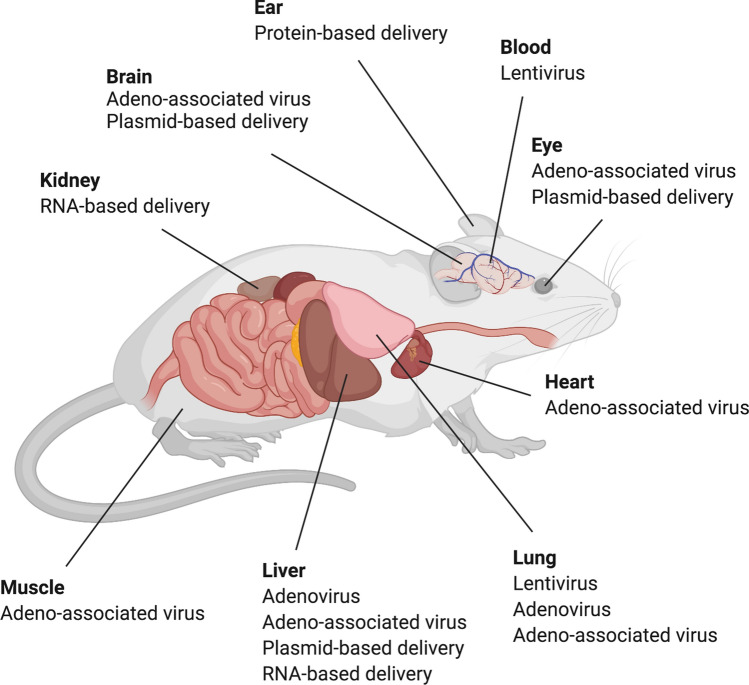
Representation of different delivery methods of the CRISPR/Cas system to target organs in the rodent. Delivery methods including virus-based (lentivirus, adenovirus and adeno-associated virus) and non-virus-based (plasmid-, RNA- or protein-based) delivery have been used to deliver CRISPR/Cas system to different organs in the rodent. The figure was created with BioRender.com
Table 1.
Delivery methods for CRISPR/Cas system in mammals
| Model-target tissue (disease) | Delivery methods | Editing gene | References | |
|---|---|---|---|---|
| Viral delivery system | No viral delivery system | |||
| Mouse blood (myeloid malignancy) | Lentivirus | Tet2, Runx1, Dnmt3a, Ezh2, Nf1, Smc3, p53 and Asxl1 | [46] | |
| Mouse lung (lung cancer) | Lentivirus | Pten and Nkx2-1 | [47] | |
| Mouse liver (NASH) | Adenovirus | Pten | [56] | |
| Mouse liver (cardiovascular disease) | Adenovirus | Pcsk9 | [57] | |
| Mouse lung (lung cancer) | Adenovirus | Eml4 and Alk | [58] | |
| Mouse brain | AAV | Mecp2, Dnmt1, Dnmt3a and Dnmt3b | [62] | |
| Mouse eye (LCA) | AAV | Yfp | [63] | |
| Mouse muscle (DMD) | AAV | Dmd | [68, 69] | |
| Mouse lung (lung cancer) | AAV | Kras, p53 and Lkb1 | [74] | |
| Mouse liver (OTC deficiency) | AAV | Otc | [72] | |
| Mouse liver (cardiovascular disease) | AAV | Pcsk9 | [78] | |
| Mouse brain (Huntington disease) | AAV | Htt | [192] | |
| Mouse brain (GBM) | AAV | Trp53, Nf1 and Rb1 | [76] | |
| Mouse brain | AAV | Camk2a, Erk2 and Actb | [193] | |
| Mouse muscle (DMD) | AAV | Dmd | [67] | |
| Mouse muscle (MDC1A) | AAV | Lama2 | [194] | |
| Mouse eye (retinal degeneration) | AAV | Nrl | [64] | |
| Mouse spleen, lungs, heart, colon, and brain (HIV/AIDS) | AAV | HIV-1 provial DNA | [195] | |
| Mouse liver (Hemophilia B) | AAV | F9 | [196] | |
| Mouse liver (cancer) | AAV | Tsgs | [77] | |
| Mouse liver | AAV | HBV cccDNA | [197] | |
| Mouse eye (X-linked retinitis pigmentosa) | AAV | Rpgr | [65] | |
| Mouse liver (tyrosinemia) | AAV | Fah | [198] | |
| Mouse liver (OTC deficiency) | AAV | Otc | [71] | |
| Mouse muscle (DMD) | AAV | Dmd | [70] | |
| Mouse brain (schizophrenia) | AAV | Mir137 | [75] | |
| Mouse muscle (ALS) | AAV | Igf1 | [66] | |
| Mouse liver (tyrosinemia) | Plasmid | Fah | [84] | |
| Mouse liver | Plasmid | HBsAg | [85] | |
| Rat eye (retinitis pigmentosa) | Plasmid | (Rho(S334)) | [86] | |
| Mouse eye (retinitis pigmentosa) | Plasmid | Rho | [87] | |
| Mouse brain (MB and GBM) | Plasmid | Trp53, Pten and Nf1 | [199] | |
| Mouse brain | Plasmid | Satbs | [89] | |
| Mouse eye (IRDs) | Plasmid | Rho-P23H | [88] | |
| Mouse liver (tyrosinemia) | mRNA | Fah | [90] | |
| Mouse liver (cardiovascular disease) | mRNA | Pcsk9 | [91, 93] | |
| Mouse liver (tyrosinemia) | mRNA | Ttr | [92] | |
| Mouse liver, kidney, and lung | mRNA | floxed tdTomato | [200] | |
| Mouse ear | Protein | Egfp | [94] | |
| Mouse muscle (DMD) | Protein | Dmd | [100] | |
| Mouse brain (FXS) | Protein | Grm5 | [101] | |
| Mouse brain (Alzheimer’s disease) | Protein | Th, Bace1 | [96] | |
| Mouse liver and spleen | Protein | Pten | [99] | |
| Mouse liver and spleen (HT1) | Protein | Hpd | [95] | |
AAV adeno-associated virus, ALS amyotrophic lateral sclerosis, DMD Duchenne muscular dystrophy, FXS fragile X syndrome, GBM glioblastoma, HIV/AIDS human immunodeficiency virus/acquired immunodeficiency syndrome, HT1 hereditary tyrosinemia type I, IRDs inherited retinal degenerations, LCA Leber congenital amaurosis, MB medulloblastoma, MDC1A congenital muscular dystrophy type 1A, NASH non-alcoholic steatohepatitis, OTC ornithine transcarbamylase
Viral-based CRISPR/Cas gene editing and delivery
Viral vectors are commonly used vehicles for introducing gene-editing materials such as DNA. Lentivirus, adenovirus, and AAV are three major types of viral vectors widely used for the gene delivery of CRISPR/Cas system. Though viral delivery has high efficiency in vivo, there are some disadvantages, including safety issues. These viruses work by releasing the viral genome into host cells after infection. This means that the interactions between the virus and host cells must be strong; thus, viral delivery methods are more complicated than most of the nonviral methods under in vivo conditions.
Lentivirus
Lentiviruses are RNA viruses with the capability to integrate into dividing and nondividing cells. Lentiviruses are an excellent delivery vehicle for cells that are hard to transfect by chemical methods. Furthermore, it has a large packaging capacity of ~ 10.7 kb [44]. This property allows it to carry multiple sgRNA sequences that can induce multiple gene edit at once [45]. Due to these advantages, lentiviruses have been used in many initial gene-editing studies. Mouse models of myeloid malignancy [46] and lung cancer [47] have been generated using lentivirus delivery. However, there are some disadvantages of using lentivirus, including the integration of the viral genome, which may be carcinogenic [48].
To overcome these issues, lentiviral vectors have been further developed into integration-deficient lentiviral vectors (IDLVs) to reduce the undesired integration of the viral genome into the host cell genome [49, 50]. IDLVs retain the property of being able to edit genes in hard-to-transfect cells [51, 52]. Although IDLVs have been found to cause unwanted gene modifications, the study also showed that IDLVs have effective site-specific gene repair activity due to their active recruitment of host HDR proteins [53]. Therefore, pairing IDLVs with safer endonucleases such as SpCas9-HF or eSpCas9 may improve its application [54, 55].
Adenovirus
Adenoviruses are double-stranded DNA (dsDNA) viruses. Similar to a lentivirus, an adenovirus can infect both dividing and nondividing cells. However, since they do not generally induce genome integration in the host DNA, adenoviruses do not cause potential off-target effects the way a lentivirus does. It has been shown that adenovirus-based delivery of the CRISPR/Cas system can result in the efficient editing of the Pcsk9 (proprotein convertase subtilisin/kexin type 9) and Pten (phosphatase and tensin homolog) genes in adult mouse liver [56, 57]. Moreover, adenovirus-based delivery also has been successfully used to induce specific chromosomal rearrangements to generate echinoderm microtubule-associated protein like 4-anaplastic lymphoma kinase (EML4-ALK)-driven lung cancer in vivo [58]. However, adenoviruses can elicit a significant immune response. Adenoviruses are also costly and difficult to produce in high volumes. These shortcomings set a limit for the applications of adenovirus-mediated delivery in clinical gene therapy [56].
Adeno-associated virus
AAVs are small ssDNA viruses. Compared to lentivirus- and adenovirus-based delivery, AAV-based delivery is safe and efficient since it results in only minor cytotoxicity and immune responses [59, 60]. AAVs have a wide range of serotypes, which helps to achieve a broad range of tissue tropisms and are used for efficient gene editing [61]. For example, Swiech et al. reported a first successful AAV-based CRISPR/Cas9 gene editing in the mouse brain [62]. A similar approach was used by Hung et al. for retinal gene editing and achieved high editing effects in the adult mouse retina [63]. Studies have also demonstrated successful AAV-based CRISPR/Cas9 gene editing in the retina of retinal degeneration mouse models [64, 65]. AAV-based delivery of CRISPR/Cas components have also been used to knockdown IGF in the central nervous system [66]. In addition, studies have also demonstrated that muscle tissue-specific delivery of CRISPR/Cas components using AAV vectors can correct the mutated dystrophin gene in Duchenne muscular dystrophy (DMD), and functional recovery was observed in vivo [61, 67–69]. Zhang et al. recently demonstrated improved CRISPR-Cas9-mediated gene-editing efficiency in DMD mouse model using self-complementary AAV (scAAV) system [70]. AAV-based delivery of CRISPR/Cas9 has also been used to achieve effective gene correction in metabolic liver disease in newborn mice [71, 72]. Moreover, delivery of sgRNAs using AAVs in a tissue-specific SpCas9 transgenic mouse can be employed to generate the disease animal model such as cardiomyopathy [73] and lung adenocarcinoma [74]. Also, Murlidharan et al. used chimeric AAV (AAV2g9) to deliver gRNAs targeting the schizophrenia risk gene MIR137 into the brain of a CRISPR/Cas9 knock-in mouse model, to achieve brain-specific gene deletion of the gene [75]. Furthermore, delivery of sgRNAs using AAVs into CRISPR/Cas9 knock-in mice can be used to perform high-throughput mutagenesis to generate autochthonous mouse models of cancer [76, 77]. Despite progress in using AAVs for CRISPR/Cas-based gene editing, the small cargo capacity (< 4.7 kb) of AAVs can limit its application. Thus, when combining conventional SpCas9, which has a size of 4.2 kb, with the addition of sgRNA, another vector system is usually required. Later on, several smaller Cas9 orthologs (such as Staphylococcus aureus (SaCas9) [78], Campylobacter jejuni (CjCas9) [79], Streptococcus thermophilus (StCas9) [80] and Neisseria meningitidis (NmCas9) [79]) were developed by scientists to enable the in vivo gene editing by a single AAV vector.
Nonviral-based CRISPR/Cas gene editing and delivery
DNA-based delivery
DNA-based delivery is commonly used for introducing the CRISPR/Cas system into cells because it is more stable than RNA. CRISPR/Cas-encoding DNA facilitates greater gene-editing efficiency than other methods [81–83]. For example, the CRISPR/Cas9 components were delivered in the form of DNA by tail-vein hydrodynamic injection to a mouse model of tyrosinemia and achieved > 6% gene correction in the liver cells after a single application [84]. Furthermore, Zhen et al. also reported that hydrodynamic injection of CRISPR/Cas9-encoding DNA can effectively disable the hepatitis B virus replication by creating mutations in virus DNA [85]. Apart from systemic administration, subretinal injection of CRISPR/Cas components in a plasmid form in combination with electroporation has also been reported to enable an allele-specific gene editing in the retina of a rat model of retinitis pigmentosa [86]. A similar effect also found by Latella et al. in a mouse model of retinitis pigmentosa, which significantly reduced mutated protein levels and prevented major visual dysfunction [87]. In addition, Li et al. demonstrated an allele-specific gene editing in the retinas of Rho-P23H knock-in mice which selectively targeting the P23H allele that has a single-nucleotide mutation [88]. Moreover, Shinmyo et al. introduced a plasmid containing CRISPR/Cas components into the mouse brain using in utero electroporation for effective brain-specific gene editing in vivo [89]. These works demonstrated the applicability of DNA-based delivery of CRISPR/Cas9 in vivo.
RNA-based delivery
RNA-based delivery methods largely decrease the risk of host genome integration. However, the effective time of RNA-based delivery methods is relatively fast, and there are some additional shortcomings of such delivery methods. For example, the stability of RNA, and the need to deliver the components (Cas mRNA and sgRNA) separately are the two main concerns of this method. Yin et al. demonstrated a delivery method that utilized different vehicles for introduction of the CRISPR/Cas9 components, lipid nanoparticles delivered the Cas9 mRNA, and an AAV delivered the sgRNA/HDR template. By utilizing this strategy, they showed an efficient correction of Fah (fumarylacetoacetate hydrolase) gene in a mouse model of hereditary tyrosinemia [90]. However, it is important to note that this combination approach still requires viral codelivery to achieve certain efficacy, and compared to DNA and protein, RNA is unstable. Moreover, the degradation of sgRNA may significantly affect editing efficiency. Future research into increasing sgRNA stability is required to improve the efficiency of these methods. Studies have showed that modifying sgRNA has beneficial effects on the stability of sgRNA. Yin et al. modified sgRNA by switching the 2′OH group of RNA to 2′OMe and 2′F and added phosphorothioate bonds [91]. This study reported that a single injection induced more than 80% efficiency in editing Pcsk9 in the livers of mice, demonstrating a potential modified method for improving the stability of RNA to overcome the obstacles of RNA-based delivery. In addition, other researchers reported a similar study in which modified sgRNA and Cas9-encoding mRNA were packaged into a lipid nanoparticle vehicle. With a single administration, a more than 97% reduction in the mouse Ttr (transthyretin) gene was shown in the serum protein levels of the liver. This study demonstrated efficient gene editing that could persist for at least 1 year [92]. Another study has also demonstrated a high editing efficacy (~ 80%) by unitizing a lipid nanoparticle with disulfide bonds (BAMEA-O16B) to deliver Cas9 mRNA and sgRNA in vivo [93].
Protein-based delivery
Delivering Cas protein with gRNA as a Cas9 RNP is the fastest and most direct pathway for gene editing, and it is suitable for in vivo therapeutic applications. To facilitate the delivery of Cas9 RNPs into target cells, a fusion protein of Cas9 and negatively supercharged proteins was created to enable the delivery by cationic lipid formulated transfection reagents such as RNAiMAX [94]. Delivery of the Cas9 RNP/RNAiMAX complex via injection into the cochlea of transgenic Atoh1 (atonal bHLH transcription factor 1)-GFP mice caused a 13% reduction in GFP in the ears of the transgenic mice. Mangeot et al. designed a vector based on murine leukemia virus (MLV), termed nanoblades, to deliver Cas9 RNPs for in vivo gene editing [95]. Moreover, an amphiphilic nanocomplex has also been developed to deliver Cas9 RNPs in vivo and showed effective gene editing in the brain of mouse model of Alzheimer’s disease [96]. Furthermore, to enhance endosomal escape, PEI polymers or combined PEI polymers with liposomes were used for Cas9 RNP delivery in vivo. Sun et al. coated a DNA nanoclew with PEI polymers to deliver Cas9 RNPs into the nuclei of human cells. Using this vehicle, target gene disruption can be achieved with negatively impacting cell viability [97]. The study also noted that the modification of DNA nanoclew to partially complementary with the sgRNA can further enhance the editing efficacy. In addition, the modification of Cas9 protein can also improve the efficacy of direct cytoplasmic/nuclear delivery of Cas9 RNP. Mout et al. developed the Cas9En protein, in which the N-terminus of Cas9 protein has an attached oligo glutamic acid tag that is negatively charged [98]. Cas9En RNPs were delivered using arginine-functionalized gold nanoparticles (Arg-AuNPs), which are positively charged. With the NLS attached, Cas9 RNPs were delivered directly to the cytosol, accumulated in the nucleus, and provided ~ 30% editing efficiency. Recently, this nano-assembled platform has been used for Cas9 RNP delivery in vivo and achieved > 8% gene editing efficiency [99].
AuNPs have also been used to deliver Cas9 RNPs in vivo for gene editing and correction in the disease models. AuNPs can be conjugated with donor DNA, Cas9 RNPs and the endosomal disruptive polymer poly[N-[N-(2-aminoethyl)-2-aminoethyl]aspartamide] (PAsp(DET) to form a vehicle termed CRISPR-Gold. Lee et al. reported that CRISPR-Gold-based Cas9 RNPs delivery can achieve 5.4% correction of the dystrophin gene in the muscle tissue of DMD mice [100]. Another study also showed that intracranial injection of CRISPR-Gold in the brain rescued mice from abnormal behaviors caused by fragile X syndrome [101]. CRISPR-Gold may offer the opportunity in the development of therapeutic approaches targeting the muscle and brain diseases, while effective endosomal escape is still required for higher delivery efficiency.
Overall, protein-based delivery offers reduced off-target effects and a low immune response compared to DNA and RNA-based delivery [102]. Cas9 RNPs increase efficacy by avoiding the degradation of sgRNA. However, transport of Cas9 RNPs into the cytosol or the nucleus is critical for therapeutic effects. Thus, endosomal entrapment is still a crucial obstacle to overcome [103].
Current approach of delivering the CRISPR/Cas system in aquaculture
Genomes of several aquaculture species, including zebrafish, Atlantic salmon, Nile tilapia, sea bream, catfish, carp, rainbow trout, Northern Chinese lamprey and Pacific oyster, have been successfully modified with the CRISPR/Cas system (Table 2). CRISPR/Cas protocols developed in model species such as zebrafish have been followed for gene editing in aquaculture species [104]. The standard gene transfer method used in aquaculture species is microinjection. Microinjection is performed using special equipment to inject the CRISPR/Cas complex into newly fertilized eggs; this method has high gene-editing efficiency [105]. In most cases, NHEJ was used to induce mutations, while HDR has been successfully used in rohu carp [106]. However, if gene editing continues at different stages of embryonic development, mosaicism could occur. These concerns are the focus of current research, which aim to enable more widespread adoption of CRISPR/Cas techniques in aquaculture. CRISPR/Cas techniques have been used to address characteristics such as sterility, growth, and disease resistance of aquaculture species. The reason for inducing sterility in fish is to preserve the domesticated strains by preventing gene flow. For example, CRISPR/Cas techniques have been used to induce sterility in Atlantic salmon [107]. Several papers have demonstrated gene editing of the myostatin gene using the CRISPR/Cas approach to enhance the growth of fish, including channel catfish and common carp [108, 109]. The CRISPR/Cas approach has also been used to investigate immunity and disease resistance in channel catfish, rohu carp, and grass carp [106, 110, 111]. Disruption of the tlr22 gene in rohu carp resulted in a model for studying immunology, demonstrating the capability of CRISPR/Cas to aid in the development of effective treatments for aquaculture. By understanding the underlying pathways of transcription and translation through CRISPR/Cas-based mechanisms, it is possible to strengthen disease resistance, decrease disease incidence, and improve species resilience in aquaculture. Aquaculture is highly suited for the application of CRISPR/Cas gene editing for numerous reasons. Sample sizes can be large without generating cumbersome costs; thousands of externally fertilized embryos enable microinjection by hand. The large sample size is impartial and useful for comparisons of successfully edited samples with controls and for the assessment of pathogen resistance. Furthermore, a large sample size enables the development of well-developed disease challenge models since extensive phenotypes are practical. With the technology becoming mature in aquaculture species, it is becoming easier to study gene function, improve disease resistance, and generate new strains with selected characteristics that can improve economic value.
Table 2.
Delivery methods for CRISPR/Cas system in aquacultures
| Species | Applications | References |
|---|---|---|
| Zebrafish (Danio rerio) | Gene editing of multiple genes | [104] |
| Rohu carp (Labeo rohita) | Gene editing of tlr22 gene | [106] |
| Atlantic salmon (Salmo salar L.) | Gene editing of dnd gene | [107] |
| Gene editing of tyr and slc45a2 genes | [201] | |
| Gene editing of elov12 gene | [202] | |
| Channel catfish (Ictalurus punctatus) | Gene editing of mstn gene | [108] |
| Gene editing of ticam and rb1 gene | [110] | |
| Common carp (Cyprinus carpio) | Gene editing of sp7 and mstn genes | [109] |
| Grass carp (Ctenopharyngodon idella) | Gene editing of gcjam-a gene | [111] |
| Nile tilapia (Oreochromis niloticus) | Gene editing of nanos2, nanos3, dmrt1 and fox12 genes | [203] |
| Gene editing of gsdf gene | [204] | |
| Gene editing of aldh1a2 and cyp26a1 genes | [205] | |
| Gene editing of sf-1 gene | [206] | |
| Gene editing of dmrt6 gene | [207] | |
| Gene editing of amhy gene | [208] | |
| Gene editing of wt1a and wt1bgenes | [209] | |
| Southern catfish (Silurus meridionalis) | Gene editing of aldh1a gene | [210] |
| Sea bream (Pagrus major) | Gene editing of mstn gene | [211] |
| Rainbow trout (Oncorhynchus mykiss) | Gene editing of igfbp2b1 and igfbp2b2 genes | [212] |
| Pacific oyster (Crassostrea gigas) | Gene editing of mstn and twist genes | [213] |
| Northern Chinese lamprey (Lethenteron morii) | Gene editing of multiple gene | [214] |
Current approach of delivering the CRISPR/Cas system in plants
As shown above, the CRISPR/Cas system is highly adept at modifying animal genomes. Studies have also demonstrated its ability to modify plant genomes. Conventionally, a mixed dual promoter system is used to express CRISPR/Cas system in plants. In mixed dual promoter systems, RNA polymerase II promoters are used to express Cas protein and RNA polymerase III promoters specifically expressed in plants, such as AtU6 for Arabidopsis or tomato, TaU6 for wheat, and OsU6 or OsU3 for rice, are used to express gRNA [112–115]. However, to utilize CRISPR/Cas9 technology in creating new traits in plants, efficient delivery of the CRISPR/Cas system into cells is essential. The two delivery methods utilized in plants are indirect and direct methods. Indirect methods (such as agroinfiltration, agroinfection, and viral infection/agroinfection) use plant bacteria or viruses to mediate the introduction of DNA constructs into target plant cells. By contrast, no biological organisms are used as mediators for direct delivery. Protoplast transfection and biolistic particle delivery are the most commonly used direct methods. Agroinfiltration is usually used as a transient assay and has been widely used for its versatility and simplicity [116–120]. Agroinfection, biolistic particle delivery, and viral infection are usually used for stable editing. Protoplast transfection can be used for both transient and stable editing. The delivery methods used in plant gene editing (Fig. 4 and Table 3) will be summarized in the following sections.
Fig. 4.
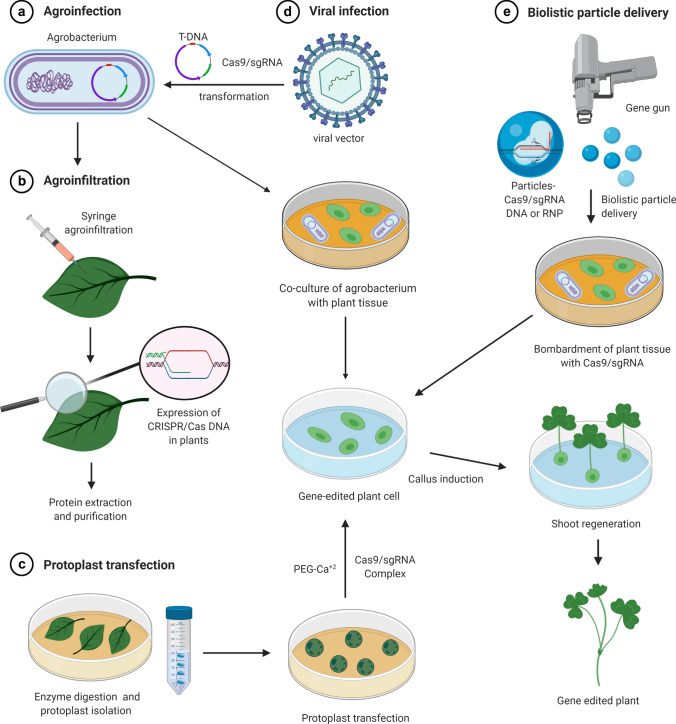
Schematic representation of main methods used to modify plant genome by CRISPR/Cas system. A schematic diagram showing major steps involved in the generation of gene-edited plants using direct and indirect methods including agroinfiltration (a), protoplast transfection (b), agroinfection (c), and virus infection (d) and biolistic particle delivery (e)
Table 3.
Delivery methods for CRISPR/Cas system in plants
| Species | Delivery methods | Edited gene | References | |
|---|---|---|---|---|
| Stable | Transient | |||
| Arabidopsis thaliana | Protoplast, Agroinfiltration | PDS3, FLS2, RACK1b and RACK1c | [122] | |
| Arabidopsis thaliana | Agroinfiltration | GFP | [215] | |
| Arabidopsis thaliana | Agrobacterium | GFP | [216] | |
| Arabidopsis thaliana | Agrobacterium | Protoplast | BRI1, JAZ1, GAI, and YFP | [122] |
| Arabidopsis thaliana | Agrobacterium | BRI1, JAZ1, GAI, CHLI1/2, AP1, TT4 and GUUS | [217] | |
| Arabidopsis thaliana | Agrobacterium | Protoplast | CHLI1, CHLI2 and TT4 | [218] |
| Arabidopsis thaliana | Agrobacterium | ADH1 | [219, 220] | |
| Arabidopsis thaliana | Agrobacterium | TRY, CPC and ETC2 | [221] | |
| Arabidopsis thaliana | Agrobacterium | At5g55580 | [145] | |
| Arabidopsis thaliana | Agrobacterium | ADH1 and TT4 | [222] | |
| Arabidopsis thaliana | Agrobacterium | BRI1 | [224] | |
| Arabidopsis thaliana | Agrobacterium | ALS | [225] | |
| Arabidopsis thaliana | Agrobacterium | ETC2, TRY, CPC and CHLI1/2 | [226] | |
| Arabidopsis thaliana | Agrobacterium | FT and SPL4 | [227] | |
| Arabidopsis thaliana | Agrobacterium | AP1, TT4 and GL2 | [228] | |
| Arabidopsis thaliana | Agrobacterium | PDS3, AG, DUO1 and ADH1 | [229] | |
| Arabidopsis thaliana | Agrobacterium | At3g04220 | [230] | |
| Nicotiana benthamiana | Agrobacterium | Agroinfiltration | PDS | [121] |
| Nicotiana benthamiana | Agroinfiltration | PDS | [123, 262, 231] | |
| Nicotiana benthamiana | Virus | PCNA and PDS | [136] | |
| Nicotiana benthamiana | Agroinfiltration | GFP | [215] | |
| Nicotiana benthamiana | Virus | PDS, IspH and fsGUS | [140] | |
| Nicotiana tabacum | Protoplast | PDS and PDR6 | [232] | |
| Nicotiana tabacum | Virus | SurA and SurB | [138] | |
| Nicotiana tabacum | Protoplast | PDS | [132] | |
| Nicotiana tabacum | Agrobacterium | PDS and STF1 | [233] | |
| Nicotiana tabacum | Protoplast | AOC | [234] | |
| Populus trichocarpa | Agrobacterium | PDS | [235] | |
| Oryza sativa | Agrobacterium | ROC5, SPP and YSA | [122] | |
| Oryza sativa | Protoplast | SWEET11 and SWEET14 | [215] | |
| Oryza sativa | Agroinfiltration | PDS | [121] | |
| Oryza sativa | Biolistic | Protoplast | PDS-SP1, BADH2, Os02g23823 and MPK2 | [144] |
| Oryza sativa | Agrobacterium | Protoplast | MYB1 | [218] |
| Oryza sativa | Protoplast | MPK5 | [236] | |
| Oryza sativa | Agrobacterium | CAO1 and LAZY1 | [237] | |
| Oryza sativa | Agrobacterium | PTG1, PTG2, PTG3, PTG4, PTG5, PTG6, PTG7, PTG8 and PTG9 | [236] | |
| Oryza sativa | Agrobacterium | OsBEL | [238] | |
| Oryza sativa | Agrobacterium | FTL, GSTU, MRP15 and Waxy | [145] | |
| Oryza sativa | Agrobacterium | Protoplast | SWEET1a, SWEET1b, SWEET11, SWEET13 and CYP76 | [239] |
| Oryza sativa | Agrobacterium | PDS, PMS3, EPSPS, DERF1, MSH1, MYB5, MYB1, ROC5, SPP and YSA | [240] | |
| Oryza sativa | Agrobacterium | DMC1A | [241] | |
| Oryza sativa | Protoplast | PDS, DEP1, ROC5 and miR159b | [242] | |
| Oryza sativa | Agrobacterium | DL and ALS | [233] | |
| Oryza sativa | Protoplast | PDS | [132] | |
| Triticum aestivum | Protoplast | Mlo | [144] | |
| Triticum aestivum | Biolistic | Protoplast | Gw2 | [147] |
| Triticum aestivum | Agroinfiltration | Inox and Pds | [231] | |
| Zea mays | Protoplast | zm ipk | [128] | |
| Zea mays | Agrobacterium | Protoplast | zm hkt1 | [221] |
| Zea mays | Biolistic | lig1, ms26, ms45, als1 and als2 | [146] | |
| Zea mays | Biolistic | lig, ms26, ms45 and als2 | [148] | |
| Zea mays | Biolistic | argos8 | [243] | |
| Glycine max | Agrobacterium | GFP, Glyma07g14530, 01gDDM1, 11gDDM1, Met1-04g, Met1-06g, miR1514 and miR1509 | [244] | |
| Glycine max | Biolistic | DD20, DD43 and ALS1 | [261] | |
| Glycine max | Agrobacterium | Glyma06g14180, Glyma08g02290 and Glyma12g37050 | [246] | |
| Glycine max | Agrobacterium | bar, FEI1, FEI2 and SHR | [245] | |
| Glycine max | Agrobacterium | PDS11 and PDS18 | [247] | |
| Glycine max var | Protoplast | FAD2-1A and FAD2-1B | [234] | |
| Solanum tuberosum | Agrobacterium | StIAA2 | [223] | |
| Solanum tuberosum | Agrobacterium | StALS1 | [248] | |
| Solanum tuberosum | Protoplast | GBSS | [142] | |
| Hordeum vulgare | Agrobacterium | HvPM19 | [249] | |
| Marchantia polymorpha | Agrobacterium | ARF1 | [250] | |
| Solanum lycopersicum | Agrobacterium | SlAGO7 | [251] | |
| Solanum lycopersicum | Virus | ANT1 | [127] | |
| Solanum lycopersicum | Agrobacterium | RIN | [252] | |
| Brassica oleracea | Agrobacterium | BolC.GA4.a | [249] | |
| Papaver somniferum | Agroinfiltration | 4′OMT2 | [253] | |
| Papaver somniferum | Agrobacterium | eIF4E | [254] | |
| Xanthomonas citri subsp. citri | Agrobacterium | Agroinfiltration | PDS | [255] |
| Xanthomonas citri subsp. citri | Agrobacterium | Agroinfiltration | LOB1 | [256] |
| Vitis vinifera | Agrobacterium | IdnDH | [257] | |
| Chardonnay | Protoplast | MLO-7 | [258] | |
| Golden delicious | Protoplast | DIPM-1, DIPM-2 and DIPM-4 | [258] | |
Transient events
Indirect method
Agroinfiltration. Agrobacterium spp. are plant pathogens. When infecting plants, Agrobacterium tumefaciens causes tumor-like growth on aerial parts of the plant (crown gall), while Agrobacterium rhizogenes induces root tumors. Agrobacteria contain a large plasmid (exceeding 200 kb), which is named Ti in the case of A. tumefaciens or Ri in the case of A. rhizogenes, and it can transfer a specific DNA segment (transfer DNA or T-DNA) into the infected plant cells, enabling the T-DNA to integrate into the host genome. These two strains of agrobacterium have been modified to contain a disarmed Ti/Ri plasmid where tumor-inducing genes have been deleted. The essential parts of the T-DNA, border repeats (25 bp), are needed for plant transformation and are used to generate transgenic plants. Agroinfiltration is a transient assay in which an A. tumefaciens culture containing modified T-DNA is directly injected into plant leaves (Fig. 4a) [121–123]. For root hair transformation, A. rhizogenes is specifically used to evaluate editing efficiency in plant root hairs, and this method has mainly been used in legume species such as Medicago and soybean [124–126].
Direct method
Protoplast transfection. A method for transfection and transient assays is protoplast transfection. This method enzymatically digests the cell walls of plant tissues and uses polyethyleneglycol (PEG) for transfection or electroporation for delivery (Fig. 4b). The same protoplasts can deliver several DNA constructs. Protoplast transfection has been proven to successfully deliver the CRISPR/Cas system and result in gene editing in Arabidopsis thaliana, Nicotiana benthamiana, rice, wheat, and maize, among others [113, 127–132].
Stable events
Indirect method
Agroinfection. Agrobacterium-mediated DNA delivery is the most commonly used method for almost all model plant species, main crop species, vegetable and fruit crops and forest crops. Similar to agroinfiltration, Agrobacterium can also create transgenic plants by genome integration in the plant nuclear DNA [133] (Fig. 4c).
Viral infection. The first viral vector used in plants was tobacco mosaic virus (TMV). Researchers used TMV to silence a gene in N. benthamiana [134]. The majority of plant viruses are RNA viruses whose genomes are ssRNAs, as such they can be synthesized in vitro and used to inoculate plants, or they can be synthesized in vivo as DNA viruses from a plasmid introduced directly to plants by mechanical means for gene delivery [135]. To accelerate the delivery process, the viral genome can be inserted as a cDNA fragment into a binary vector and then can be used for agroinfection-mediated delivery into a plant cell (Fig. 4d).
Tobacco rattle virus (TRV) is an ssRNA virus that has two genome components, TRV1 (or RNA1) and TRV2 (or RNA2). Both genome components are required for inoculation. Plants edited using RNA viruses do not exhibit germline transmission of edits. For instance, Ali et al. used agroinfection to deliver the RNA1 genomic component of TRV and a vector derived from TRV RNA2 containing targeting gRNA into the leaves of N. benthamiana overexpressing Cas9 for gene editing in plant cells [136].
Geminiviruses, unlike TRV, do not require in vitro transcription prior to inoculation. Geminiviruses have a circular ssDNA genome [137]. Geminiviruses do not have a gene encoding DNA polymerase; therefore, their ssDNA genomes are converted into dsDNA genomes by host DNA polymerases in the nucleus. The dsDNA genome is then used as a template for virus transcription and rolling circle replication. Replication initiator protein (Rep) is essential for the initiation of rolling-circle replication. Rolling circle replication can either convert ssDNA genomes into dsDNA genomes or package ssDNA genomes into virions. Plant plasmodesmata pathways facilitate the transport of virions to adjacent cells [138, 139]. Bean yellow dwarf virus (BeYDV), which is a geminivirus, has been used to deliver the CRISPR/Cas system [138]. Studies have demonstrated gene editing using BeYDV in tomato (anthocyanin mutant 1 gene, ANT1), and a modified cabbage leaf curl virus (CaLCuV) has been used in tobacco [127, 140]. Such approaches have also been applied in wheat, and researchers have enhanced the efficiency of this method by developing an optimized wheat dwarf virus (WDV) system [141].
Direct method
Protoplast transfection. Unlike the transient method of protoplast transfection, the stable transformation method generated targeted genome modifications in whole plants that were regenerated from gene-edited protoplasts [130, 131]. Two advantages of protoplast transfection are the ability to deliver multiple components and to do so at a high quantity. This method is highly suitable for gene editing using donor template repair. A high quantity of transfected cells can promote the recovery of gene editing via donor template repair. However, a disadvantage of protoplast transfection is the rate of plant regeneration in monocot plants. Protoplast transfection has been used for gene editing in potato [142], tobacco, and lettuce [131].
Biolistic particle delivery. Biolistic particle delivery is accomplished by transfecting cells via bombardment. Gene guns can penetrate the cell wall of plant cells with physical force to deliver DNA (Fig. 4e). This method is common in transforming plants due to its efficiency and its ability to deliver multiple DNA constructs simultaneously [143]. Most importantly, there is no plant species restriction to biolistic particle-based delivery. The main disadvantage of this method is that by introducing multiple copies of the DNA in the target plants, undesired effects such as gene suppression might occur in the recovered transgenic plants. Biolistic particle delivery has been used for gene editing in rice and wheat, soybean and maize using the CRISPR/Cas system [144–146]. In addition, this method is also used to deliver CRISPR/Cas9 RNPs for gene editing in crops, such as hexaploid wheat and maize [147, 148].
Future prospects in CRISPR/Cas delivery
The CRISPR/Cas system is simple but versatile. The CRISPR/Cas system has great potential for gene editing, but the delivery of CRISPR/Cas into cells dramatically impacts editing efficiency. There are still some aspects of delivery that can be improved to elevate the potential for translatability.
Immunity to the CRISPR/Cas system and its delivery vehicle
It is known that the Cas gene must be delivered into cells to express the Cas protein, and the long-term and robust expression of bacterially derived protein is expected to activate the host immune system. One solution to this problem is to use a protein-based delivery of the CRISPR/Cas system, which may have less immunogenicity, as the Cas protein would only be present in the target cell for a short period of time [98]. When combined with immunogenic effects caused by certain delivery vehicles, the level of immunogenicity might make negligible the efficiency of the CRISPR/Cas system. It has been reported that exogenous RNA delivered by lipid nanoparticles might activate Toll-like receptors and subsequent immune responses [149]. Therefore, the type of delivery vector should be carefully chosen. Moreover, it is especially important to consider the side effects of viral vectors. When compared to lentiviruses, AAVs and adenoviruses can avoid the risk of undesired DNA integration into the host genome. Producing viral DNA or protein within the cells of host can generate a risk for clinical applications [150, 151].
Engineered biomaterials in improving the delivery efficiency
Among the delivery vectors, the most suitable vectors for in vivo delivery may be nonviral vectors rather than viral vectors. Nonviral delivery, compared with viral delivery, exhibits potential advantages. It reduces the risk of off-target effects by decreasing the expression period of nuclease and enables better control of dosing duration [90]. The emergence and development of nanotechnology and material sciences have produced versatile applications in gene editing. It has been shown that gold-based nanoparticles enable effective delivery of RNP both in vitro and in vivo [100]. In addition, polymeric-based and lipid-based nanoparticles exhibit low immunogenicity, especially in their ability to encapsulate large cargos [152]. Additionally, it has been demonstrated recently that PEI-magnetic nanoparticles can improve the delivery of CRISPR/Cas9 constructs in vitro with low cell toxicity and have been shown to be a promising delivery system that can improve the safety and utility of gene editing [153, 154]. Moreover, researchers have demonstrated the delivery of the Cas9 RNP complex directly into cells using the nanoneedle array system and showed approximately 32% and 16% gene disruption efficiencies in HeLa cells and mouse breast cancer cells, respectively. Although the efficiency needs to be improved, researchers were able to successfully demonstrate gene editing by the direct delivery of Cas9/sgRNA using a nanoneedle array, and this method of delivery may be applied to gene knock-in via HDR [155]. Recently, Chen et al. demonstrated a platform comprised of vertically aligned silicon nanotube (VA-SiNT) arrays for gene editing. They successfully delivered Cas9 RNP to the target gene and demonstrated more than 80% efficiency of SiNT-facilitated biocargo internalization. This indicated that the nanotube-facilitated molecular delivery platform has great potential to propel gene-editing technologies [156]. However, nanoparticle-mediated protein delivery still has challenges, including the difficult process of packaging into designed materials and the prevention of RNP degradation before it enters the nucleus. Therefore, biocompatible, well-tolerated, high capability, and nonimmunogenic delivery vehicles are required to deliver cargos to the nucleus for effective gene editing, and these characteristics are essential when designing any nonviral delivery material.
Spatial and temporal regulation of Cas9 activity
As previously discussed, the unintended off-target effect of the CRISPR/Cas system is a major concern. Regulating delivery of the components of the CRISPR/Cas system to specific target sites before Cas9 is turned on and delivery of certain factors that switch on this machinery at a specific time point is critical. A number of teams have identified Cas9 endonuclease inhibitors. These anti-CRISPR (Acr) proteins, such as AcrIIA4, can shut off Cas9 activity [157, 158]. Moreover, anti-CRISPRs could be used to limit editing activity to particular cells and tissues in the body. Researchers designed miRNA-responsive Acr switches, and delivery of this machinery with Cas9 or dCas9 enabled tissue-specific editing [159]. In a recent study, researchers generated Cas9 variants called ProCas9s that enabled the CRISPR/Cas9 system to be turned on only in target cells [160]. ProCas9 senses the type of cell it is in based on proteases. This machinery enables the safer translational application of CRISPR/Cas9 gene editing, and this technology could be used to help plants defend against viral pathogens.
Several strategies to control the activity or expression of Cas9 have also been demonstrated (Table 4). It has been reported that Cas9 can be expressed in a split [161–164] or inactive form [165, 166]. In addition, an inducible system enabled Cas9 to be activated only when stimulated by a chemical inducer [167–171] or by exposure to certain types of light [164]. Studies have engineered a split-Cas9 system in which the activity of Cas9 is induced only when the two domains, recognition domain and nuclease domain, are assembled [172]. This split-Cas9 system is also utilized for gene editing using inteins. Inteins are protein introns that excise themselves out of host polypeptides to generate a functional protein [173]. The intein-based split-Cas9 system is composed of the split Cas9 domains, each of which is fused to intein sequences. Upon dimerization, these intein sequences will be spliced out, and fully active Cas9 can be generated [161]. Truong et al. demonstrated that Cas9 domains can be delivered by AAV vectors separately and retain comparable editing efficiencies as full-length Cas9 [161]. Cas9 can also be chemically inducible by exposure to rapamycin, which induces FK506-binding protein (FKBP)-FKBP rapamycin binding (FRB) dimerization [174]. Rapamycin-inducible split-Cas9 is composed of split Cas9 fragments each fused with FRB and FKBP fragments. In the presence of rapamycin, a fully active Cas9 is formed. Researchers have also demonstrated a photoactivatable Cas9 (paCas9) system that utilized photoinducible dimerizing protein domains termed Magnets [164]. This optically controlled split-Cas9 system was generated by fusing each Cas9 fragment with magnet fragments (pMagnet and nMagnet) and triggering magnet dimerization upon blue light treatment [175]. Several other optically controlled systems have also been reported to enable CRISPR/Cas-based transcriptional activation and gene editing [175–178]. Nihongaki et al. developed a light-inducible system. They fused integrin binding protein 1 (CIB1) with dCas9 and fused cryptochrome 2 (CRY2) with a transcriptional activator domain, and then they used blue light to trigger dimerization of CIB1 and CRY2, resulting in subsequent expression of downstream targets [175]. Shao et al. developed a optogenetic far-red light (FRL)-activated CRISPR/dCas9 effector (FACE) system based on dCas9 [179–181] and the bacterial phytochrome BphS [182] that induced transcription of target genes in the presence of FRL [178].
Table 4.
Summary of regulatory CRISPR/Cas systems
| Type of system | Split-Cas9 | Light-inducible | Destabilizing domain | NS3 domain | ||
|---|---|---|---|---|---|---|
| Intein-inducible | Rapamycin-inducible | Photoactivatable | ||||
| In vivo studies | [259] | n/a | n/a | [178] | [260] | n/a |
| Delivery vehicle | Viral-based delivery: AAV | n/a | n/a | DNA-based delivery: electroporation | DNA-based delivery: tail vein hydrodynamic injection | n/a |
Other strategies can also enable tunable regulation of CRISPR/Cas9 systems. Wandless and colleagues used small cell-permeable molecules to regulate protein stability. This chemical-genetic approach allowed rapid and tunable expression of a specific protein by fusing the molecules to a destabilizing domain [183]. The destabilizing domain acts as a degron that directs the fusion protein to proteasome-dependent degradation without the presence of a small molecule ligand, which allows tunable control of protein function. Ligand binding to the destabilizing domain protects the fusion protein from degradation and allows the protein of interest to function normally. Thus far, several ligand-destabilizing domain pairs have been discovered, including Shield-1 with mutant K506-binding protein (FKBP) 12 destabilized domain (FKBP[DD]), trimethoprim with mutant dihydrofolate reductase (DHFR) destabilized domain (DHFR[DD]), and CMP8 with the 4-OHT-estrogen receptor destabilized domain (ER50[DD]) [183–185]. This concept can be utilized for switchable gene editing and activation [186–188]. FKBP[DD], DHFR[DD], and ER50[DD] were fused to Cas9 for drug inducible gene editing [187, 188]. DHFR[DD] or ER50[DD] were fused to PP7-activation domain [179], and DHFR[DD] can be fused directly to dCas9 activator [186] for drug inducible gene activation. Multidimensional control can be achieved by pairing different ligand-destabilizing domain pairs with different aptamers [187]. Another platform utilizes the hepatitis C virus (HCV) nonstructural protein 3 (NS3) protease domain and its various inhibitors and has also been used to regulate CRISPR/Cas activity [189]. Tague et al. integrated the NS3 protease domain into dCas9–VPR to form a ligand-inducible CRISPR activation platform [189, 190]. The NS3 protease domain was inserted between the DNA binding scaffold and the C-terminal region, which is where NLS and VPR are located, to form a dCas9–NS3–NLS–VPR complex. NS3 protease can separate VPR from dCas9 and subsequently inhibit transcriptional activation, while in the presence of protease inhibitor, transcriptional activation is achieved. Recently, Cas9 has been fused with small molecule-assisted shut-off tag (SMASh), which consists of the HCV NS3 and nonstructural protein 4a (NS4A, acting as a degron). Cas9 stability can be controlled by SMASh via asunaprevir, an HCV protease inhibitor. Cas9 protein is degraded when NS3–NS4A is inhibited in the presence of asunaprevir, while in the absence of asunaprevir, the gene editing activity of Cas9 was restored [191].
Unfortunately, there are still some obstacles to progressing with the application of the regulatory approach to the CRISPR/Cas system. Chemical inducers may elicit cytotoxicity, which would make application of this approach in vivo more difficult. Additionally, light-induced systems may be limited to in vitro studies since activating such a system with light in vivo would be invasive, and penetration of light into tissue may cause other problems. Further investigation, optimization, and development are needed to overcome these challenges to advance the clinical translation of the CRISPR/Cas system.
Conclusion
The discovery and application of the CRISPR/Cas system offers great hope for the human disease treatment as well as revolutionize plant breeding. Although research on the CRISPR/Cas system in the life sciences community is well underway, there are still substantial barriers to efficient delivery that need to be overcome to achieve effective gene editing. Factors related to specificity, efficacy and regulatable expression are important to consider when selecting an approach. The development of new delivery methods has overcome many disadvantages that severely impede the translatability of the CRISPR/Cas system. With the rapid development of delivery methods, the successful translation of CRISPR/Cas technology into medical and agricultural applications is imperative and major improvements can be anticipated.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Health and Medical Research Council of Australia under Grant 1185600 and 1123329; the Ophthalmic Research Institute of Australia; and the Shenzhen Key Laboratory of Biomimetic Materials and Cellular Immunomodulation under Grant ZDSYS20190902093409851.
Author contribution
Conceptualization: Y.F.C. and G.S.L. Writing (original draft): Y.F.C. and G.S.L. Writing (review and editing): A.J.P., F.L.L., V.H., A.W.H., and P.Y.W. Visualization: Y.F.C., A.J.P., and G.S.L. Funding acquisition: P.Y.W. and G.S.L.
Funding
This work was supported by the National Health and Medical Research Council of Australia under Grant 1185600 and 1123329; the Ophthalmic Research Institute of Australia; and the Shenzhen Key Laboratory of Biomimetic Materials and Cellular Immunomodulation under Grant ZDSYS20190902093409851.
Availability of data and material
All data relevant to the study are available upon reasonable request from the corresponding author.
Code availability
Not applicable.
Compliance with ethical standards
Conflict of interest
The authors report no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peng-Yuan Wang, Email: py.wang@siat.ac.cn.
Guei-Sheung Liu, Email: rickliu0817@gmail.com.
References
- 1.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387. doi: 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belting M, Sandgren S, Wittrup A. Nuclear delivery of macromolecules: barriers and carriers. Adv Drug Deliv Rev. 2005;57(4):505–527. doi: 10.1016/j.addr.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, Liu Z, Doudna JA, Tjian R. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350(6262):823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- 8.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders AP, Dickman MJ, Doudna JA, Boekema EJ, Heck AJ, van der Oost J, Brouns SJ. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 9.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10(5):726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111(27):9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimasu H, Cong L, Yan WX, Ran FA, Zetsche B, Li Y, Kurabayashi A, Ishitani R, Zhang F, Nureki O. Crystal Structure of Staphylococcus aureus Cas9. Cell. 2015;162(5):1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164(1–2):29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 18.Barrangou R, Horvath P. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017;2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 19.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 20.Klompe SE, Sternberg SH. Harnessing “A Billion Years of Experimentation”: the ongoing exploration and exploitation of CRISPR-Cas immune systems. CRISPR J. 2018;1(2):141–158. doi: 10.1089/crispr.2018.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 26.Norris AD, Kim HM, Colaiacovo MP, Calarco JA. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics. 2015;201(2):449–458. doi: 10.1534/genetics.115.180679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nunez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JC. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540(7631):144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Li J, Song CQ, Tran K, Mou H, Wu PH, Tai PWL, Mendonca CA, Ren L, Wang BY, Su Q, Gessler DJ, Zamore PD, Xue W, Gao G. Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat Biotechnol. 2018;36(9):839–842. doi: 10.1038/nbt.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strutt SC, Torrez RM, Kaya E, Negrete OA, Doudna JA. RNA-dependent RNA targeting by CRISPR-Cas9. Elife. 2018 doi: 10.7554/eLife.32724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, Wang Y. The Molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170(4):714–726.e710. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 35.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676.e614. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowder LG, Paul JW, 3rd, Qi Y. Multiplexed transcriptional activation or repression in plants using CRISPR-dCas9-based systems. Methods Mol Biol. 2017;1629:167–184. doi: 10.1007/978-1-4939-7125-1_12. [DOI] [PubMed] [Google Scholar]
- 38.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33(5):510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rittiner JE, Moncalvo M, Chiba-Falek O, Kantor B. Gene-editing technologies paired with viral vectors for translational research into neurodegenerative diseases. Front Mol Neurosci. 2020;13:148. doi: 10.3389/fnmol.2020.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42(19):e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32(9):941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, Jacks T. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou S, Fatima S, Ma Z, Wang YD, Lu T, Janke LJ, Du Y, Sorrentino BP. Evaluating the safety of retroviral vectors based on insertional oncogene activation and blocked differentiation in cultured thymocytes. Mol Ther. 2016;24(6):1090–1099. doi: 10.1038/mt.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joglekar AV, Hollis RP, Kuftinec G, Senadheera S, Chan R, Kohn DB. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol Ther. 2013;21(9):1705–1717. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wanisch K, Yanez-Munoz RJ. Integration-deficient lentiviral vectors: a slow coming of age. Mol Ther. 2009;17(8):1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 52.Sessa M, Lorioli L, Fumagalli F, Acquati S, Redaelli D, Baldoli C, Canale S, Lopez ID, Morena F, Calabria A, Fiori R, Silvani P, Rancoita PM, Gabaldo M, Benedicenti F, Antonioli G, Assanelli A, Cicalese MP, Del Carro U, Sora MG, Martino S, Quattrini A, Montini E, Di Serio C, Ciceri F, Roncarolo MG, Aiuti A, Naldini L, Biffi A. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: an ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet. 2016;388(10043):476–487. doi: 10.1016/S0140-6736(16)30374-9. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Wang Y, Chang T, Huang H, Yee JK. Integration-defective lentiviral vector mediates efficient gene editing through homology-directed repair in human embryonic stem cells. Nucleic Acids Res. 2017;45(5):e29. doi: 10.1093/nar/gkw1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Mou H, Li S, Li Y, Hough S, Tran K, Li J, Yin H, Anderson DG, Sontheimer EJ, Weng Z, Gao G, Xue W. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther. 2015;26(7):432–442. doi: 10.1089/hum.2015.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, Lowe SW, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15(7):445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, Della Peruta M, Lheriteau E, Patel N, Raj D, Riddell A, Pie J, Rangarajan S, Bevan D, Recht M, Shen YM, Halka KG, Basner-Tschakarjan E, Mingozzi F, High KA, Allay J, Kay MA, Ng CY, Zhou J, Cancio M, Morton CL, Gray JT, Srivastava D, Nienhuis AW, Davidoff AM. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371(21):1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson CE, Gersbach CA. Engineering delivery vehicles for genome editing. Annu Rev Chem Biomol Eng. 2016;7:637–662. doi: 10.1146/annurev-chembioeng-080615-034711. [DOI] [PubMed] [Google Scholar]
- 62.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hung SS, Chrysostomou V, Li F, Lim JK, Wang JH, Powell JE, Tu L, Daniszewski M, Lo C, Wong RC, Crowston JG, Pebay A, King AE, Bui BV, Liu GS, Hewitt AW. AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest Ophthalmol Vis Sci. 2016;57(7):3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 64.Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, Ataeijannati Y, Sun X, Dong L, Li T, Swaroop A, Wu Z. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu S, Du J, Chen N, Jia R, Zhang J, Liu X, Yang L. In vivo CRISPR/Cas9-mediated genome editing mitigates photoreceptor degeneration in a mouse model of X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2020;61(4):31. doi: 10.1167/iovs.61.4.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin H, Hu H, Duan W, Liu Y, Tan G, Li Z, Liu Y, Deng B, Song X, Wang W, Wen D, Wang Y, Li C. Intramuscular delivery of scAAV9-hIGF1 prolongs survival in the hSOD1(G93A) ALS mouse model via upregulation of D-amino acid oxidase. Mol Neurobiol. 2018;55(1):682–695. doi: 10.1007/s12035-016-0335-z. [DOI] [PubMed] [Google Scholar]
- 67.Bengtsson NE, Hall JK, Odom GL, Phelps MP, Andrus CR, Hawkins RD, Hauschka SD, Chamberlain JR, Chamberlain JS. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, Cong L, Zhang F, Vandenberghe LH, Church GM, Wagers AJ. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351(6271):407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Li H, Min YL, Sanchez-Ortiz E, Huang J, Mireault AA, Shelton JM, Kim J, Mammen PPA, Bassel-Duby R, Olson EN. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv. 2020;6(8):eaay6812. doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Yang Y, Breton C, Bell P, Li M, Zhang J, Che Y, Saveliev A, He Z, White J, Latshaw C, Xu C, McMenamin D, Yu H, Morizono H, Batshaw ML, Wilson JM. A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci Adv. 2020;6(7):eaax5701. doi: 10.1126/sciadv.aax5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, Batshaw ML, Wilson JM. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34(3):334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carroll KJ, Makarewich CA, McAnally J, Anderson DM, Zentilin L, Liu N, Giacca M, Bassel-Duby R, Olson EN. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–343. doi: 10.1073/pnas.1523918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murlidharan G, Sakamoto K, Rao L, Corriher T, Wang D, Gao G, Sullivan P, Asokan A. CNS-restricted transduction and CRISPR/Cas9-mediated gene deletion with an engineered AAV vector. Mol Ther Nucleic Acids. 2016;5(7):e338. doi: 10.1038/mtna.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, Renauer P, Bilguvar K, Gunel M, Sharp PA, Zhang F, Platt RJ, Chen S. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20(10):1329–1341. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang G, Chow RD, Ye L, Guzman CD, Dai X, Dong MB, Zhang F, Sharp PA, Platt RJ, Chen S. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR-mediated direct in vivo screening. Sci Adv. 2018;4(2):eaao5508. doi: 10.1126/sciadv.aao5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther. 2016;24(3):636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patsali P, Kleanthous M, Lederer CW. Disruptive technology: CRISPR/Cas-based tools and approaches. Mol Diagn Ther. 2019;23(2):187–200. doi: 10.1007/s40291-019-00391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, Jin S. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci (Weinh) 2020;7(6):1902312. doi: 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22(5):404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 86.Bakondi B, Lv W, Lu B, Jones MK, Tsai Y, Kim KJ, Levy R, Akhtar AA, Breunig JJ, Svendsen CN, Wang S. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24(3):556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Latella MC, Di Salvo MT, Cocchiarella F, Benati D, Grisendi G, Comitato A, Marigo V, Recchia A. In vivo editing of the human mutant rhodopsin gene by electroporation of plasmid-based CRISPR/Cas9 in the mouse retina. Mol Ther Nucleic Acids. 2016;5(11):e389. doi: 10.1038/mtna.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li P, Kleinstiver BP, Leon MY, Prew MS, Navarro-Gomez D, Greenwald SH, Pierce EA, Joung JK, Liu Q. Allele-specific CRISPR-Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 2018;1:55–64. doi: 10.1089/crispr.2017.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shinmyo Y, Tanaka S, Tsunoda S, Hosomichi K, Tajima A, Kawasaki H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci Rep. 2016;6:20611. doi: 10.1038/srep20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, Gupta A, Bolukbasi MF, Walsh S, Bogorad RL, Gao G, Weng Z, Dong Y, Koteliansky V, Wolfe SA, Langer R, Xue W, Anderson DG. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, Mou H, Oberholzer A, Ding J, Kwan SY, Bogorad RL, Zatsepin T, Koteliansky V, Wolfe SA, Xue W, Langer R, Anderson DG. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol. 2017;35(12):1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finn JD, Smith AR, Patel MC, Shaw L, Youniss MR, van Heteren J, Dirstine T, Ciullo C, Lescarbeau R, Seitzer J, Shah RR, Shah A, Ling D, Growe J, Pink M, Rohde E, Wood KM, Salomon WE, Harrington WF, Dombrowski C, Strapps WR, Chang Y, Morrissey DV. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22(9):2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 93.Liu J, Chang J, Jiang Y, Meng X, Sun T, Mao L, Xu Q, Wang M. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mater. 2019;31(33):e1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33(1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mangeot PE, Risson V, Fusil F, Marnef A, Laurent E, Blin J, Mournetas V, Massourides E, Sohier TJM, Corbin A, Aube F, Teixeira M, Pinset C, Schaeffer L, Legube G, Cosset FL, Verhoeyen E, Ohlmann T, Ricci EP. Genome editing in primary cells and in vivo using viral-derived nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat Commun. 2019;10(1):45. doi: 10.1038/s41467-018-07845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Park H, Oh J, Shim G, Cho B, Chang Y, Kim S, Baek S, Kim H, Shin J, Choi H, Yoo J, Kim J, Jun W, Lee M, Lengner CJ, Oh YK, Kim J. In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat Neurosci. 2019;22(4):524–528. doi: 10.1038/s41593-019-0352-0. [DOI] [PubMed] [Google Scholar]
- 97.Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mout R, Ray M, Yesilbag Tonga G, Lee YW, Tay T, Sasaki K, Rotello VM. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11(3):2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee YW, Mout R, Luther DC, Liu YC, Castellanos-García L, Burnside AS, Ray M, Tonga GY, Hardie J, Nagaraj H, Das R, Phillips EL, Tay T, Vachet RW, Rotello VM. In vivo editing of macrophages through systemic delivery of CRISPR-Cas9-ribonucleoprotein-nanoparticle nanoassemblies. Adv Ther. 2019;2(10):7. doi: 10.1002/adtp.201900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee K, Conboy M, Park HM, Jiang F, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, Park HM, Brenner R, Murthy N, Lee HY. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2(7):497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: progress and challenges. Bioconjug Chem. 2017;28(4):880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev. 2007;59(8):748–758. doi: 10.1016/j.addr.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110(34):13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goto R, Saito T, Matsubara T, Yamaha E. Microinjection of marine fish eggs. Methods Mol Biol. 2019;1874:475–487. doi: 10.1007/978-1-4939-8831-0_27. [DOI] [PubMed] [Google Scholar]
- 106.Chakrapani V, Patra SK, Panda RP, Rasal KD, Jayasankar P, Barman HK. Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. Dev Comp Immunol. 2016;61:242–247. doi: 10.1016/j.dci.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 107.Wargelius A, Leininger S, Skaftnesmo KO, Kleppe L, Andersson E, Taranger GL, Schulz RW, Edvardsen RB. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci Rep. 2016;6:21284. doi: 10.1038/srep21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khalil K, Elayat M, Khalifa E, Daghash S, Elaswad A, Miller M, Abdelrahman H, Ye Z, Odin R, Drescher D, Vo K, Gosh K, Bugg W, Robinson D, Dunham R. Generation of myostatin gene-edited channel catfish (Ictalurus punctatus) via zygote injection of CRISPR/Cas9 system. Sci Rep. 2017;7(1):7301. doi: 10.1038/s41598-017-07223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhong Z, Niu P, Wang M, Huang G, Xu S, Sun Y, Xu X, Hou Y, Sun X, Yan Y, Wang H. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci Rep. 2016;6:22953. doi: 10.1038/srep22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Elaswad A, Khalil K, Ye Z, Liu Z, Liu S, Peatman E, Odin R, Vo K, Drescher D, Gosh K, Qin G, Bugg W, Backenstose N, Dunham R. Effects of CRISPR/Cas9 dosage on TICAM1 and RBL gene mutation rate, embryonic development, hatchability and fry survival in channel catfish. Sci Rep. 2018;8(1):16499. doi: 10.1038/s41598-018-34738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ma J, Fan Y, Zhou Y, Liu W, Jiang N, Zhang J, Zeng L. Efficient resistance to grass carp reovirus infection in JAM-A knockout cells using CRISPR/Cas9. Fish Shellfish Immunol. 2018;76:206–215. doi: 10.1016/j.fsi.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 112.Kim JS. Precision genome engineering through adenine and cytosine base editing. Nat Plants. 2018;4(3):148–151. doi: 10.1038/s41477-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 113.Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9(10):2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- 114.Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T, Kondo A. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol. 2017;35(5):441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 115.Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6(6):1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 116.Bhaskar PB, Venkateshwaran M, Wu L, Ane JM, Jiang J. Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS ONE. 2009;4(6):e5812. doi: 10.1371/journal.pone.0005812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Circelli P, Donini M, Villani ME, Benvenuto E, Marusic C. Efficient Agrobacterium-based transient expression system for the production of biopharmaceuticals in plants. Bioeng Bugs. 2010;1(3):221–224. doi: 10.4161/bbug.1.3.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Figueiredo JF, Romer P, Lahaye T, Graham JH, White FF, Jones JB. Agrobacterium-mediated transient expression in citrus leaves: a rapid tool for gene expression and functional gene assay. Plant Cell Rep. 2011;30(7):1339–1345. doi: 10.1007/s00299-011-1045-7. [DOI] [PubMed] [Google Scholar]
- 119.Kim MJ, Baek K, Park CM. Optimization of conditions for transient Agrobacterium-mediated gene expression assays in Arabidopsis. Plant Cell Rep. 2009;28(8):1159–1167. doi: 10.1007/s00299-009-0717-z. [DOI] [PubMed] [Google Scholar]
- 120.Zheng L, Liu G, Meng X, Li Y, Wang Y. A versatile Agrobacterium-mediated transient gene expression system for herbaceous plants and trees. Biochem Genet. 2012;50(9–10):761–769. doi: 10.1007/s10528-012-9518-0. [DOI] [PubMed] [Google Scholar]
- 121.Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9(1):39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13(4):578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 124.Zhang H, Cao Y, Zhang H, Xu Y, Zhou C, Liu W, Zhu R, Shang C, Li J, Shen Z, Guo S, Hu Z, Fu C, Sun D. Efficient generation of CRISPR/Cas9-mediated homozygous/biallelic Medicago truncatula mutants using a hairy root system. Front Plant Sci. 2020;11:294. doi: 10.3389/fpls.2020.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM. CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food. 2015;6(4):243–252. doi: 10.1080/21645698.2015.1106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang S, Kondorosi E, Kereszt A. An anthocyanin marker for direct visualization of plant transformation and its use to study nitrogen-fixing nodule development. J Plant Res. 2019;132(5):695–703. doi: 10.1007/s10265-019-01126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41(2):63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 129.Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao C. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38(5):582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- 130.Park J, Choe S. DNA-free genome editing with preassembled CRISPR/Cas9 ribonucleoproteins in plants. Transgenic Res. 2019;28(Suppl 2):61–64. doi: 10.1007/s11248-019-00136-3. [DOI] [PubMed] [Google Scholar]
- 131.Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 132.Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HC, Zhang Y, Zhang R, Chang WJ, Yu CT, Wang W, Liao LJ, Gelvin SB, Shih MC. Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J. 2018;16(7):1295–1310. doi: 10.1111/pbi.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hwang HH, Yu M, Lai EM. Agrobacterium-mediated plant transformation: biology and applications. Arabidopsis Book. 2017;15:e0186. doi: 10.1199/tab.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kumagai MH, Donson J, Della-Cioppa G, Harvey D, Hanley K, Grill LK. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci U S A. 1995;92(5):1679–1683. doi: 10.1073/pnas.92.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nagyova A, Subr Z. Infectious full-length clones of plant viruses and their use for construction of viral vectors. Acta Virol. 2007;51(4):223–237. [PubMed] [Google Scholar]
- 136.Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, Dinesh-Kumar S, Mahfouz MM. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8(8):1288–1291. doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 137.Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Biochem Mol Biol. 2000;35(2):105–140. [PubMed] [Google Scholar]
- 138.Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26(1):151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wawrzyniak P, Plucienniczak G, Bartosik D. The different faces of rolling-circle replication and its multifunctional initiator proteins. Front Microbiol. 2017;8:2353. doi: 10.3389/fmicb.2017.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep. 2015;5:14926. doi: 10.1038/srep14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sanchez-Leon S, Baltes NJ, Starker C, Barro F, Gao C, Voytas DF. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89(6):1251–1262. doi: 10.1111/tpj.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Andersson M, Turesson H, Nicolia A, Falt AS, Samuelsson M, Hofvander P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017;36(1):117–128. doi: 10.1007/s00299-016-2062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kikkert JR, Vidal JR, Reisch BI. Stable transformation of plant cells by particle bombardment/biolistics. Methods Mol Biol. 2005;286:61–78. doi: 10.1385/1-59259-827-7:061. [DOI] [PubMed] [Google Scholar]
- 144.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 145.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 146.Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, Gao C. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;8:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31(26):6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 150.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17(3):295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shirley JL, de Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28(3):709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 153.Hryhorowicz M, Grzeskowiak B, Mazurkiewicz N, Sledzinski P, Lipinski D, Slomski R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol Biotechnol. 2019;61(3):173–180. doi: 10.1007/s12033-018-0145-9. [DOI] [PubMed] [Google Scholar]
- 154.Rohiwal SS, Dvorakova N, Klima J, Vaskovicova M, Senigl F, Slouf M, Pavlova E, Stepanek P, Babuka D, Benes H, Ellederova Z, Stieger K. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci Rep. 2020;10(1):4619. doi: 10.1038/s41598-020-61465-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yamagishi A, Matsumoto D, Kato Y, Honda Y, Morikawa M, Iwata F, Kobayashi T, Nakamura C. Direct delivery of Cas9-sgRNA ribonucleoproteins into cells using a nanoneedle array. Appl Sci. 2019;9:965. doi: 10.3390/app9050965. [DOI] [Google Scholar]
- 156.Chen Y, Aslanoglou S, Murayama T, Gervinskas G, Fitzgerald LI, Sriram S, Tian J, Johnston APR, Morikawa Y, Suu K, Elnathan R, Voelcker NH. Silicon-nanotube-mediated intracellular delivery enables Ex vivo gene editing. Adv Mater. 2020 doi: 10.1002/adma.202000036. [DOI] [PubMed] [Google Scholar]
- 157.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell. 2017;168(1–2):150–158.e110. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dong GM, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z. Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature. 2017;546(7658):436–439. doi: 10.1038/nature22377. [DOI] [PubMed] [Google Scholar]
- 159.Hoffmann MD, Aschenbrenner S, Grosse S, Rapti K, Domenger C, Fakhiri J, Mastel M, Borner K, Eils R, Grimm D, Niopek D. Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 2019;47(13):e75. doi: 10.1093/nar/gkz271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oakes BL, Fellmann C, Rishi H, Taylor KL, Ren SM, Nadler DC, Yokoo R, Arkin AP, Doudna JA, Savage DF. CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell. 2019;176(1–2):254–267.e216. doi: 10.1016/j.cell.2018.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Truong DJ, Kuhner K, Kuhn R, Werfel S, Engelhardt S, Wurst W, Ortiz O. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 2015;43(13):6450–6458. doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol. 2015;11(5):316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33(7):755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 165.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, Gale M, Xu Q, Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44(19):e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dow LE, Fisher J, O'Rourke KP, Muley A, Kastenhuber ER, Livshits G, Tschaharganeh DF, Socci ND, Lowe SW. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33(4):390–394. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zetsche B, Volz SE, Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol. 2015;33(2):139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu KI, Ramli MN, Woo CW, Wang Y, Zhao T, Zhang X, Yim GR, Chong BY, Gowher A, Chua MZ, Jung J, Lee JH, Tan MH. A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat Chem Biol. 2016;12(11):980–987. doi: 10.1038/nchembio.2179. [DOI] [PubMed] [Google Scholar]
- 171.Lu J, Zhao C, Zhao Y, Zhang J, Zhang Y, Chen L, Han Q, Ying Y, Peng S, Ai R, Wang Y. Multimode drug inducible CRISPR/Cas9 devices for transcriptional activation and genome editing. Nucleic Acids Res. 2018;46(5):e25. doi: 10.1093/nar/gkx1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci U S A. 2015;112(10):2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shah NH, Muir TW. Inteins: nature’s gift to protein chemists. Chem Sci. 2014;5(1):446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 175.Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol. 2015;22(2):169–174. doi: 10.1016/j.chembiol.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 176.Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical control of CRISPR/Cas9 gene editing. J Am Chem Soc. 2015;137(17):5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol. 2015;11(3):198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, Ye H. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc Natl Acad Sci U S A. 2018;115(29):E6722–E6730. doi: 10.1073/pnas.1802448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1–2):339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Du D, Roguev A, Gordon DE, Chen M, Chen SH, Shales M, Shen JP, Ideker T, Mali P, Qi LS, Krogan NJ. Genetic interaction mapping in mammalian cells using CRISPR interference. Nat Methods. 2017;14(6):577–580. doi: 10.1038/nmeth.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gao Y, Xiong X, Wong S, Charles EJ, Lim WA, Qi LS. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods. 2016;13(12):1043–1049. doi: 10.1038/nmeth.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ryu MH, Gomelsky M. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol. 2014;3(11):802–810. doi: 10.1021/sb400182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17(9):981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Miyazaki Y, Imoto H, Chen LC, Wandless TJ. Destabilizing domains derived from the human estrogen receptor. J Am Chem Soc. 2012;134(9):3942–3945. doi: 10.1021/ja209933r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Rep. 2015;5(3):448–459. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Maji B, Moore CL, Zetsche B, Volz SE, Zhang F, Shoulders MD, Choudhary A. Multidimensional chemical control of CRISPR-Cas9. Nat Chem Biol. 2017;13(1):9–11. doi: 10.1038/nchembio.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Senturk S, Shirole NH, Nowak DG, Corbo V, Pal D, Vaughan A, Tuveson DA, Trotman LC, Kinney JB, Sordella R. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat Commun. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tague EP, Dotson HL, Tunney SN, Sloas DC, Ngo JT. Chemogenetic control of gene expression and cell signaling with antiviral drugs. Nat Methods. 2018;15(7):519–522. doi: 10.1038/s41592-018-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, Ter-Ovanesyan D, Braff JL, Davidsohn N, Housden BE, Perrimon N, Weiss R, Aach J, Collins JJ, Church GM. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wu Y, Yang L, Chang T, Kandeel F, Yee JK. A small molecule-controlled Cas9 repressible system. Mol Ther Nucleic Acids. 2020;19:922–932. doi: 10.1016/j.omtn.2019.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther. 2017;25(1):12–23. doi: 10.1016/j.ymthe.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nishiyama J, Mikuni T, Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96(4):755–768.e755. doi: 10.1016/j.neuron.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kemaladewi DU, Maino E, Hyatt E, Hou H, Ding M, Place KM, Zhu X, Bassi P, Baghestani Z, Deshwar AG, Merico D, Xiong HY, Frey BJ, Wilson MD, Ivakine EA, Cohn RD. Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat Med. 2017;23(8):984–989. doi: 10.1038/nm.4367. [DOI] [PubMed] [Google Scholar]
- 195.Yin C, Zhang T, Qu X, Zhang Y, Putatunda R, Xiao X, Li F, Xiao W, Zhao H, Dai S, Qin X, Mo X, Young WB, Khalili K, Hu W. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol Ther. 2017;25(5):1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Singh K, Evens H, Nair N, Rincon MY, Sarcar S, Samara-Kuko E, Chuah MK, VandenDriessche T. Efficient in vivo liver-directed gene editing using CRISPR/Cas9. Mol Ther. 2018;26(5):1241–1254. doi: 10.1016/j.ymthe.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kayesh MEH, Amako Y, Hashem MA, Murakami S, Ogawa S, Yamamoto N, Hifumi T, Miyoshi N, Sugiyama M, Tanaka Y, Mizokami M, Kohara M, Tsukiyama-Kohara K. Development of an in vivo delivery system for CRISPR/Cas9-mediated targeting of hepatitis B virus cccDNA. Virus Res. 2020;290:198191. doi: 10.1016/j.virusres.2020.198191. [DOI] [PubMed] [Google Scholar]
- 198.Krooss SA, Dai Z, Schmidt F, Rovai A, Fakhiri J, Dhingra A, Yuan Q, Yang T, Balakrishnan A, Steinbruck L, Srivaratharajan S, Manns MP, Schambach A, Grimm D, Bohne J, Sharma AD, Buning H, Ott M. Ex vivo/in vivo gene editing in hepatocytes Using “All-in-One” CRISPR-adeno-associated virus vectors with a self-linearizing repair template. Science. 2020;23(1):100764. doi: 10.1016/j.isci.2019.100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, Largaespada D, Roussel MF, Korshunov A, Reifenberger G, Pfister SM, Lichter P, Kawauchi D, Gronych J. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 2017;56(4):1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS ONE. 2014;9(9):e108622. doi: 10.1371/journal.pone.0108622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Datsomor AK, Zic N, Li K, Olsen RE, Jin Y, Vik JO, Edvardsen RB, Grammes F, Wargelius A, Winge P. CRISPR/Cas9-mediated ablation of elovl2 in Atlantic salmon (Salmo salar L.) inhibits elongation of polyunsaturated fatty acids and induces Srebp-1 and target genes. Sci Rep. 2019;9(1):7533. doi: 10.1038/s41598-019-43862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Li M, Yang H, Zhao J, Fang L, Shi H, Li M, Sun Y, Zhang X, Jiang D, Zhou L, Wang D. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics. 2014;197(2):591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jiang DN, Yang HH, Li MH, Shi HJ, Zhang XB, Wang DS. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol Reprod Dev. 2016;83(6):497–508. doi: 10.1002/mrd.22642. [DOI] [PubMed] [Google Scholar]
- 205.Feng R, Fang L, Cheng Y, He X, Jiang W, Dong R, Shi H, Jiang D, Sun L, Wang D. Retinoic acid homeostasis through aldh1a2 and cyp26a1 mediates meiotic entry in Nile tilapia (Oreochromis niloticus) Sci Rep. 2015;5:10131. doi: 10.1038/srep10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xie QP, He X, Sui YN, Chen LL, Sun LN, Wang DS. Haploinsufficiency of SF-1 causes female to male sex reversal in nile tilapia. Oreochromis niloticus Endocrinology. 2016;157(6):2500–2514. doi: 10.1210/en.2015-2049. [DOI] [PubMed] [Google Scholar]
- 207.Zhang X, Wang H, Li M, Cheng Y, Jiang D, Sun L, Tao W, Zhou L, Wang Z, Wang D. Isolation of doublesex- and mab-3-related transcription factor 6 and its involvement in spermatogenesis in tilapia. Biol Reprod. 2014;91(6):136. doi: 10.1095/biolreprod.114.121418. [DOI] [PubMed] [Google Scholar]
- 208.Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K, Jiang D, Zhou L, Sun L, Tao W, Nagahama Y, Kocher TD, Wang D. A tandem duplicate of anti-Mullerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015;11(11):e1005678. doi: 10.1371/journal.pgen.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jiang D, Chen J, Fan Z, Tan D, Zhao J, Shi H, Liu Z, Tao W, Li M, Wang D. CRISPR/Cas9-induced disruption of wt1a and wt1b reveals their different roles in kidney and gonad development in Nile tilapia. Dev Biol. 2017;428(1):63–73. doi: 10.1016/j.ydbio.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 210.Li M, Feng R, Ma H, Dong R, Liu Z, Jiang W, Tao W, Wang D. Retinoic acid triggers meiosis initiation via stra8-dependent pathway in Southern catfish, Silurus meridionalis. Gen Comp Endocrinol. 2016;232:191–198. doi: 10.1016/j.ygcen.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 211.Kishimoto KWY, Yoshiura Y, Toyoda A, Ueno T, Fukuyama H, Kato K, Kinoshita M. Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture. 2018;495:415–427. doi: 10.1016/j.aquaculture.2018.05.055. [DOI] [Google Scholar]
- 212.Cleveland BM, Yamaguchi G, Radler LM, Shimizu M. Editing the duplicated insulin-like growth factor binding protein-2b gene in rainbow trout (Oncorhynchus mykiss) Sci Rep. 2018;8(1):16054. doi: 10.1038/s41598-018-34326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yu H, Li H, Li Q, Xu R, Yue C, Du S. Targeted gene disruption in Pacific oyster based on CRISPR/Cas9 ribonucleoprotein complexes. Mar Biotechnol (NY) 2019;21(3):301–309. doi: 10.1007/s10126-019-09885-y. [DOI] [PubMed] [Google Scholar]
- 214.Zu Y, Zhang X, Ren J, Dong X, Zhu Z, Jia L, Zhang Q, Li W. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system. Sci Rep. 2016;6:23496. doi: 10.1038/srep23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jiang W, Yang B, Weeks DP. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE. 2014;9(6):e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111(12):4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6(6):2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schiml S, Fauser F, Puchta H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014;80(6):1139–1150. doi: 10.1111/tpj.12704. [DOI] [PubMed] [Google Scholar]
- 220.Steinert J, Schiml S, Fauser F, Puchta H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 2015;84(6):1295–1305. doi: 10.1111/tpj.13078. [DOI] [PubMed] [Google Scholar]
- 221.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79(2):348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 223.Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015;34(9):1473–1476. doi: 10.1007/s00299-015-1816-7. [DOI] [PubMed] [Google Scholar]
- 224.Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant. 2015;8(12):1820–1823. doi: 10.1016/j.molp.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 225.Wolter F, Klemm J, Puchta H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018;94(4):735–746. doi: 10.1111/tpj.13893. [DOI] [PubMed] [Google Scholar]
- 226.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hyun Y, Kim J, Cho SW, Choi Y, Kim JS, Coupland G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta. 2015;241(1):271–284. doi: 10.1007/s00425-014-2180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu JK. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2016;14(2):519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tsutsui H, Higashiyama T. pKAMA-itachi vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 2017;58(1):46–56. doi: 10.1093/pcp/pcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wu R, Lucke M, Jang YT, Zhu W, Symeonidi E, Wang C, Fitz J, Xi W, Schwab R, Weigel D. An efficient CRISPR vector toolbox for engineering large deletions in Arabidopsis thaliana. Plant Methods. 2018;14:65. doi: 10.1186/s13007-018-0330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3(12):2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol. 2015;87(1–2):99–110. doi: 10.1007/s11103-014-0263-0. [DOI] [PubMed] [Google Scholar]
- 233.Endo A, Masafumi M, Kaya H, Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep. 2016;6:38169. doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci Rep. 2015;5:12217. doi: 10.1038/srep12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112(11):3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J. Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N Y) 2014;7(1):5. doi: 10.1186/s12284-014-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 241.Mikami M, Toki S, Endo M. Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 2016;57(5):1058–1068. doi: 10.1093/pcp/pcw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants. 2017;3:17103. doi: 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- 243.Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15(2):207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15:16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS ONE. 2015;10(8):e0136064. doi: 10.1371/journal.pone.0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2016;217:90–97. doi: 10.1016/j.jbiotec.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 248.Butler NM, Atkins PA, Voytas DF, Douches DS. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS ONE. 2015;10(12):e0144591. doi: 10.1371/journal.pone.0144591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015;16:258. doi: 10.1186/s13059-015-0826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55(3):475–481. doi: 10.1093/pcp/pcu014. [DOI] [PubMed] [Google Scholar]
- 251.Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166(3):1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467(1):76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- 253.Alagoz Y, Gurkok T, Zhang B, Unver T. Manipulating the biosynthesis of bioactive compound alkaloids for next-generation metabolic engineering in opium poppy using CRISPR-Cas 9 genome editing technology. Sci Rep. 2016;6:30910. doi: 10.1038/srep30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17(7):1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Jia H, Wang N. Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014;33(12):1993–2001. doi: 10.1007/s00299-014-1673-9. [DOI] [PubMed] [Google Scholar]
- 256.Jia H, Orbovic V, Jones JB, Wang N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccDeltapthA4:dCsLOB1.3 infection. Plant Biotechnol J. 2016;14(5):1291–1301. doi: 10.1111/pbi.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ren C, Liu X, Zhang Z, Wang Y, Duan W, Li S, Liang Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.) Sci Rep. 2016;6:32289. doi: 10.1038/srep32289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Malnoy M, Viola R, Jung MH, Koo OJ, Kim S, Kim JS, Velasco R, Nagamangala Kanchiswamy C. DNA-Free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng. 2020;4(1):97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Bao J, Liu W, Xie J, Xu L, Guan M, Lei F, Zhao Y, Huang Y, Xia J, Li H. Nix Co3-x O4 nanoneedle arrays grown on Ni Foam as an efficient bifunctional electrocatalyst for full water splitting. Chem Asian J. 2019;14(3):480–485. doi: 10.1002/asia.201801710. [DOI] [PubMed] [Google Scholar]
- 261.Li Z, Liu Z-B, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM. Cas9-Guide RNA directed genome editing in soybean. Plant Physiol. 2015;169:960–970. doi: 10.1104/pp.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Komatsu H, Abdellatif IMY, Yuan S, Ono M, Nonaka S, Ezura H, Ariizumi T, Miura K. Genome editing in PDS genes of tomatoes by non-selection method and of Nicotiana benthamiana by one single guide RNA to edit two orthologs. Plant Biotechnol. 2020;37(2):213–221. doi: 10.5511/plantbiotechnology.20.0527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study are available upon reasonable request from the corresponding author.
Not applicable.