Abstract
Breast cancer is the leading cause of cancer death in female. Until now, advanced breast cancer is still lack effective treatment strategies and reliable prognostic markers. In the present article, we introduced the physiologic and pathologic functions and regulation mechanisms of ZBTB28, a tumor suppressor gene, in breast cancer. ZBTB28 is frequently silenced in breast cancer due to promoter CpG methylation, and its expression is positively correlated with breast cancer patient survival. The antineoplastic effect of ZBTB28 in breast cancer was elucidated through a series of in vitro and in vivo measurements, including cell proliferation, apoptosis, cell cycle, epithelial mesenchymal transition (EMT), and growth of xenografts. Furthermore, ZBTB28 can directly regulate IFNAR to activate interferon-stimulated genes and potentiate macrophage activation. Ectopic ZBTB28 expression in breast cancer cells was sufficient to downregulate CD24 and CD47 to promote phagocytosis of macrophages, demonstrating that ZBTB28 was beneficial for the combination treatment of anti-CD24 and anti-CD47. Collectively, our results reveal a mode of action of ZBTB28 as a tumor suppressor gene and suggest that ZBTB28 is an important regulator of macrophage phagocytosis in breast cancer, holding promise for the development of novel therapy strategies for breast cancer patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04124-x.
Keywords: ZBTB28, Breast cancer, IFNAR, CD24, CD47, Phagocytosis
Introduction
Breast cancer is a public-health issue that affects women, and in particular, triple-negative breast cancer (TNBC) is the most common cause of death [1]. Although there are several methods for detecting this kind of cancer at the earliest stage and effective treatments for patients after diagnosis, the multiple factors that affect complex and heterogeneous of mammary cancer appear to be poorly understood [2, 3]. During the development and progression of breast cancer, a great many of tumor suppressors or oncogenes are regulated by cancer-related transcription factors [4]. The latest study suggested that the inactivation of the tumor suppressor gene is not only involved in tumorigenesis, but also contributed to tumor-immune escape [5].
Transcription factor ZBTB28 (also known as BCL6B, BAZF or ZNF62), a novel suppressor for tumor, is a paralog of BCL6 [6]. We have been identified that ZBTB28 is expressed widely in normal tissues, and CpG methylation of its promoter leads to downregulation of ZBTB28 in various tumors such as colorectal cancer, gastric cancer, hepatocellular carcinoma, as well as cervical cancer [7–10]. While the role and mechanism of ZBTB28 in tumorigenesis for mammary cancer remain to be elucidated.
With the development of molecular biology, pharmacogenetics, and tumor immunology, immunotherapy has become a prospective new field among clinical cancer therapies [11]. As a large class of proteins that regulate immune response, cytokines can directly activate immune effector cells or stimulate tumor stromal cells to enhance tumor cell susceptibility to immune attack, as well as exert different anti-tumor responses according to different tumor microenvironments [12]. Interferons (IFNs) are among the most important cytokines in response to anti-tumor response, facilitating macrophage activation and anti-tumor immunity [13, 14]. About 20 IFNs bind to 3 kinds of cell-surface receptors, IFNAR (the type I IFN receptor), IFNGR (the type II IFN receptor), and IFNLR (the type III IFN receptor), to exert different biological effects [15]. Evidence suggests that damaged IFN receptor on tumor cells are associated with reduction of anti-tumor effect [16]. Furthermore, the latest studies implicated that CD24, a mucin-like cell-surface protein, can interact with the inhibitory receptor sialic-acid-binding Ig-like lectin 10 (Siglec-10), which located on the surface of macrophages, to compose novel immune checkpoint [17, 18]. CD24 also is known as small-cell lung carcinoma cluster 4 antigen or heat-stable antigen, which is overexpressed in nearly 70% of human cancers [19, 20]. The binding of CD24 to Siglec-10 elicits an inhibitory signaling cascade, damages macrophage phagocytosis, evades immune surveillance, and allows tumor growth [18, 21]. Intriguingly, upon dual blockade of CD24 and CD47 with antibodies further augmented phagocytosis ability of macrophage, suggesting the cooperativity of combinatorial blockade of CD24 and CD47 in breast cancer cells [18]. CD47 (integrin-associated protein), another widely expressed transmembrane protein, is correlated with a decreased probability of survival for multiple types of cancer [22]. In solid tumor cell-surface, CD47 interacts with its receptor on macrophages, SIRPα, to transmit a “don’t eat me” signal for phagocytic cells [23, 24]. As a ‘self-labeling’ protein which is overexpressed broadly across tumor types, it is emerging as a novel and effective macrophage immune checkpoint for cancer immunotherapy [25].
Cancer-associated transcription factors are key players in cancer development and tumor-immune processes, orchestrating gene expression networks in cancers [26]. In this study, we identified the inhibitory effect of ZBTB28 on the occurrence and development of breast cancer. In vitro and in vivo experiments proved that ZBTB28 inhibited the growth and movement of breast cancer cell. In addition, we also found that ZBTB28 regulated IFNAR to activate downstream ISGs, as well as to promote macrophages polarization. Mechanistic studies have shown that ZBTB28 favored the phagocytosis of macrophages by simultaneously down-regulating CD24 and CD47, and enhanced the sensitivity of CD24 and CD47 antibody combination effect. These results indicated that ZBTB28 plays an important role in regulating the biological functions of breast cancer and has the potential to act as an effective indicator of breast cancer treatment and prognosis.
Materials and methods
Cell lines and human tumor samples
MCF7, MB231, BT549, MB468, SK-BR3, YCCB1, T47D, ZR75-1, THP-1, HEK-293 T, MCF10A, and 4T1 cells were obtained from the American Type Culture Collection (ATCC) or collaborators. Cells were routinely maintained at 37 ℃ with 5% CO2 in RPMI-1640 (Gibco-BRL, Karlsruhe, Germany) or DMEM (Gibco-BRL, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (Biological Industries). Primary human tumor tissues, which were assessed by pathologists that the percentage of tumor cells was no less than 70%, were obtained from the First Affiliated Hospital of Chongqing Medical University. The clinical and pathological studies were followed by Institutional Ethics Committees of the First Affiliated Hospital of Chongqing Medical University (Approval notice: # 2015-01-24).
Methylation-specific PCR (MSP) and 5-Aza-2’-deoxycytidine (5-Aza) treatment
Bisulfite modification of DNA and how to conduct MSP was described previously, as well as the primers of MSP [27]. MSP products were electrophoresis with 2% agarose gels and were recorded on a gel imaging system (Bio-RAD Gel Doc XR + , CA). Breast cancer cell lines were treated with 10 μmol/L 5-Aza (Sigma-Aldrich, Steinheim, Germany), a demethylation agent, for 4 days. Then, harvested cells for qRT-PCR analysis.
Real-time quantitative PCR and RT-polymerase chain reaction
qRT-PCR was performed using an SYBR-Green ER Kit (Life Technologies, USA) and the HT7500 System (Applied Biosystems). RT-PCR was performed using Go-Taq (Promega, Madison, WI), and the reaction conditions were as previously described [28]. All the indicated primers are listed in supplementary Table 1 and supplementary Table 2. The mRNA expression of ZBTB28 in normal breast cells or breast carcinoma was analyzed from an online cancer database (http://www.Oncomine.org).
CCk8, colony formation assays, wound healing, and Transwell® assays
Cells were cultured in 96-well plates at a density of 3000 cells/well. The absorbance (at 450 nm) in each well was measured with CCK8 kit (C0037, Beyotime) at 0 h, 24 h, 48 h, 72 h. For colony formation assays, cells were plated in 6-well plates (200 cells/well) with/without soft-agar for 14 days [27]. Cell colonies were photographed with a phase contrast microscope (Leica DMI4000B, Milton Keynes, Buckinghamshire, UK); colonies with more than 50 cells were manually counted with Photoshop software. Wound healing and Transwell® assays were performed as previously described [10, 27].
Immunofluorescence staining
Immunofluorescence staining was performed following the protocol that we previously described [27]. In short, cells were incubated with whole goat serum containing primary antibody against vimentin (#2707-1; Epitomics, Cambridge, MA), and E-cadherin (#1702-1; Epitomics, Cambridge, MA) overnight at 4 °C. Then, the cells were incubated with biotin-labeled secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) for an additional 20–30 min. At the same time, nuclei were counterstained with DAPI (Roche, Palo Alto, CA, USA). The staining was observed under fluorescence microscopy.
Western blot assay
Western blotting was performed following the protocol which we previously described [29]. The primary antibodies used included HA-tag (#3724, Cell Signaling Technology), vimentin (sc-6260, Santa Cruz), Occludin (TA306787, OriGene Technologies), Snail (#3897; Cell Signaling Technology), iNOS (ab15323, Abcam), CD206 (ab8918, Abcam), CD24 (sc-19585, Santa Cruz), CD47 (sc-12730, Santa Cruz), IFN-γRβ (sc-377291, Santa Cruz), IFN-γ (sc-8423, Santa Cruz), GAPDH (sc-47724, Santa Cruz), and β-actin (sc-8432, Santa Cruz). The protein bands were clarified with a chemiluminescence kit (Amersham Pharmacia Biotech, Piscataway, NJ).
Immunohistochemistry (IHC)
Samples from mice were studied following a previously published protocol [10]. The sections were incubated in blocking solution (Reagent 2) for 15 min, followed by overnight incubation at 4 °C with PCNA (#13,110, Cell Signaling Technology), Ki-67 (ab16667, Abcam), CD24 (sc-19585, Santa Cruz), and CD47 (sc-12730, Santa Cruz) antibody in immunohistochemical wet box. After the final staining with DAB (ZSGB-BIO, ZLI-9018) and counterstaining with hematoxylin (BL702B, Biosharp, China), the samples were scanned using an IHC scanner (PANNNORAMIC MIDI, Budapest, Hungary).
Dual-Luciferase® assays
Luciferase reporter assay was performed following the protocol which we previously described [10]. The dual-luciferase reporter assay kit and the Luminometer (Infinite M200 PRO, Tecan, Austria) were used to quantify the light emission of luciferase.
Chromatin immunoprecipitation (ChIP) analysis
ChIP-PCR analysis was performed using the SimpleChIP® Enzymatic Chromatin IP Kit (#9003, Cell Signaling Technology) and followed the protocol that we previously described [10]. The antibodies used included HA-tag antibody (#3274, Cell Signaling Technology), histone H3 antibody (#4620, Cell Signaling Technology), and normal rabbit IgG (#2729, Cell Signaling Technology). Primers are listed in supplementary Table 3.
In vivo assay
BALB/c nude mice, 4–6 weeks of age, were injected with 5 × 106 different cells into their lower backs. Tumors’ volumes (mm3) were calculated as follows: volume = length × width2 × 0.52. For the macrophage infiltration assay, BALB/c mice were injected with 2 × 105 4-T1 cells. Tumor tissues were acquired fresh on the day of resection and dissociated. Cells were analyzed by flow cytometry on an LRSFortessa Analyzer (BD Biosciences), and measured the number of CD11b+F4/80+ macrophages were measured and quantified as a percentage of the total cells. The experiments were following by Institutional Ethics Committees of the First Affiliated Hospital of Chongqing Medical University (Approval notice: # 2016–75).
Macrophages co-culture with breast cancer cells
For macrophages testing, we derived human macrophages from monocytic cell THP-1, and seeded 1 × 106 THP-1 cells with RPMI-1640 medium-containing 10 ng/mL 12-myristate 13-acetate (PMA, Sigma) at the bottom chamber of the non-contact co-culture Transwell® system (#3450, Corning, USA). After 24 h, the medium was replaced by RPMI-1640 medium with 10% fetal calf serum. At the same time, the breast tumor cells with or without ZBTB28 overexpression were seeded at the upper chamber and co-cultured with THP-1 macrophages for 5 days. Then, the macrophage cells were harvested for the next experiments.
When detecting cancer cells, 5 × 105 THP-1 cells were seeded in upper chambers then treated with PMA for 3 days. The breast cancer cells were seeded in the bottom chambers with/without IFN alpha-IFNAR-IN-1 hydrochloride (HY-12836A, MCE) or IFN-γRβ (sc-377291, Santa Cruz) for 8 h. The tumor cells were collected for the next experiments after 24 h.
Phagocytosis assay
Monocytic line THP-1 cells were seeded with RPMI-1640 medium-containing 10 ng/mL PMA for 4 days to generate macrophages. Next, the macrophages were harvested via gentle scraping and then staining with Cslcein, AM (Yeasen, China). Next, plated 5 × 104 cells per well in a 6-well culture plate (3471, Corning) with serum-free media for 4 h. Then, 2.5 × 105 target cells with DIL (USEVERBRIGHT® INC) staining were added to the macrophage containing wells and incubated overnight and then imaged by confocal laser scanning microscopy.
When using flow cytometry to observe phagocytosis, cancer cells were blocked using anti-IgG (#2729, Cell Signaling Technology), anti-CD24 (sc-19585, Santa Cruz), or anti-CD47 (sc-12730, Santa Cruz) for 4 h before co-culture. Then, THP-1 macrophages were incubated with breast tumor cells overnight in 24-well culture plate (3473, Corning) with serum-free media. Before testing, cells were washed 3 times with PBS thoroughly. Prepare single-stained THP-1 macrophages and single-stained tumor cells for gating. Phagocytosis was calculated as the percentage of PI+FITC+ cells among FITC+ cells (1 × 104 cells per sample).
Statistical analyses
The NCBI Blast program (http://www.ncbi.nlm.nih.gov/BLAST/) was used for cDNA sequences analyses. SPSS Statistics software (SPSS, Chicago, IL, USA) was used for the statistical analyses. An ANOVA test, two-tailed Fisher’s exact test, and Chi-square test also were applied for the statistical analyses. For all experiments, p values < 0.05 indicated statistically significance.
Results
ZBTB28 expression is repressed by CpG methylation in mammary carcinoma
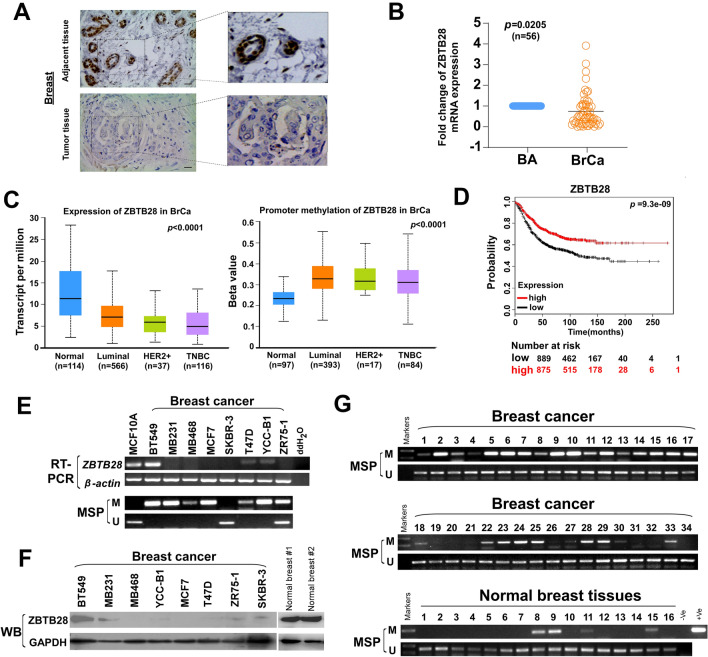
As reported in our previous online bioinformatics analysis, expression of ZBTB28 was decreased in a large number of cancer types [10, 27]. Immunohistochemical staining labeled ZBTB28 in breast cancer tissue and adjacent tissue, and showed low expression levels of ZBTB28 in tumor tissues (Fig. 1A). The reduced ZBTB28 expressions in breast cancer tissues (BrCa) compared to adjacent non-tumor specimens (BA) were detected by qRT-PCR (Fig. 1B). We further analyzed ZBTB28 expression and methylation in breast cancer tissues form online database (http://ualcan.path.uab.edu/index.html). The data illustrated that expression of ZBTB28 was lower and methylation of ZBTB28 was higher in luminal, human epidermal growth factor receptor type-2 (HER2) positive and triple-negative breast cancer (TNBC) subtype of breast cancer tissues, compared to normal mammary tissues (Fig. 1C). Next, we confirmed a strong positive correlation between high ZBTB28 expression and survival advantage for patients with breast cancer (Fig. 1D). Also, the correlation between ZBTB28 methylation levels and survival times of breast cancer patients is shown in supplementary Fig. 1E. We found that ZBTB28 expression was silenced in 7/8 breast cancer cell lines, and further methylation-specific PCR (MSP) analysis has revealed that the methylation of the ZBTB28 promoter was correlated with its downregulation in most breast cancer cell lines (Fig. 1E). Meanwhile, from The Cancer Genome Atlas databases, increased ZBTB28 promoter methylation was accompanied by a decrease in ZBTB28 expression in breast cancer specimens (Supplementary Fig. 1A, B). Then, we detected the ZBTB28 expression with western bolt in breast cancer cell lines and found that ZBTB28 was maintained at a relatively low level in most breast cancer cells (Fig. 1F). Furthermore, ZBTB28 promoter methylation was detected in 73.6% (128/174) of primary breast tumors, but rarely in normal mammary tissues (Fig. 1G). In addition, MethylTarget® detection revealed that there are higher degrees of methylation at almost CpG sites of ZBTB28 promoter CpG island in 3/5 breast cancer samples (Supplementary Fig. 1C). After treatment with a demethylating agent, 5-aza-2’-deoxycytidine (Aza), ZBTB28 expression was restored and its methylation level was reduced (Supplementary Fig. 1D). To further evaluate the clinical significance of ZBTB28 in breast cancer, the clinicopathological features of 174 BrCa patients were analyzed. The methylation of ZBTB28 showed a positive correlation with the patients’ age (Supplementary Table 4). However, no significant association of the methylation status with other clinical variables was observed. All of results suggested that ZBTB28 is frequently down-regulated or silenced in breast cancer through promoter methylation, and that this is related to prognosis.
Fig. 1.
CpG methylation and expression about ZBTB28 in mammary carcinoma. A Immunohistochemical staining labeled ZBTB28 for adjacent and paired breast tumor tissue. B Expression of ZBTB28 in breast cancer tissues (BrCa) and paired adjacent tissues (BA). C Data
available at UALCAN databases showing ZBTB28 expression and methylation in Luminal, HER2 positive and TNBC types of breast cancer tissues compared to normal mammary tissues. D The relationship between ZBTB28 expression and survival of patients with breast cancer were illustrated through Kaplan–Meier plots database. E The expression and CpG methylation level of ZBTB28 in normal breast cell line and several kinds of breast cancer cell lines. β-actin expression as control. F Western blot analysis confirmed the exogenous expression of ZBTB28 in normal breast cells and breast cancer cell lines, with GAPDH as a control. G ZBTB28 methylation in primary breast cancer tissues (n = 34) and normal breast tissues (n = 16) were measured by MSP (M methylated; U unmethylated)
ZBTB28 suppresses tumorigenesis of breast cancer
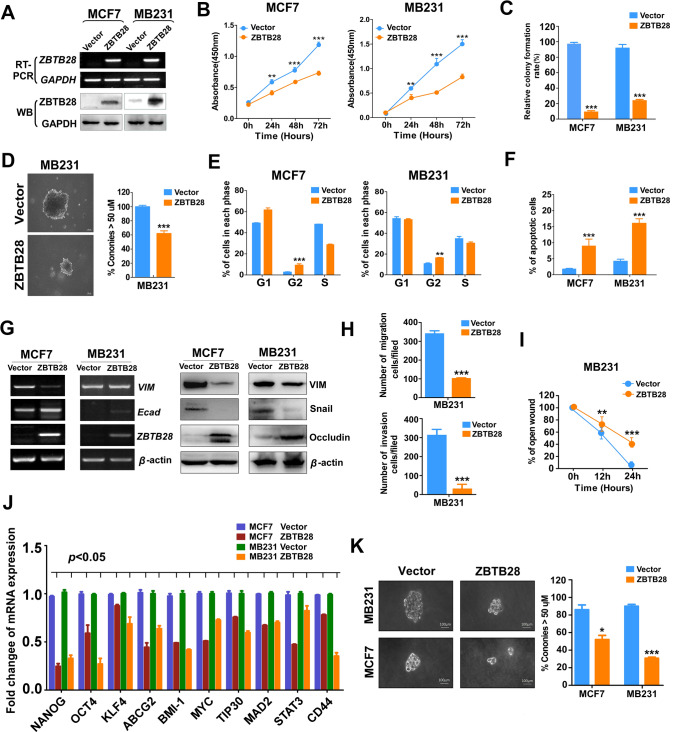
To explore the role of ZBTB28 for breast cancer via gain-of-function cell models: first, RT-PCR and western blotting verified the overexpression levels of ZBTB28 (Fig. 2A). The exogenous expression of ZBTB28 was involved in the prevention of cell viability and proliferation, and both CCK8 and colony formation assays correctly reflected the phenomenon of the inhibition (Fig. 2B, C and Supplementary Fig. 2A). Furthermore, a soft-agar colony formation assay was performed to assess the proliferation capacity of breast cancer cells under three-dimensional culture conditions. The results revealed that ZBTB28 suppressed colony formation of MB231 cells in soft-agar (Fig. 2D). Cell cycle arrest was determined by flow cytometry analysis, and cells with ZBTB28 displayed increased G2/M fractions compared to the control group (Fig. 2E and Supplementary Fig. 2C). AO/EB assay revealed the cellular effects of ZBTB28 on cell apoptosis (Fig. 2F and Supplementary Fig. 2B). Meanwhile, increased epithelial markers (E-cadherin, occludin) and decreased mesenchymal markers (vimentin, snail) were found in stable gain-of-function cell lines (Fig. 2G). With immunofluorescent staining, we confirmed that ZBTB28 up-regulated the expression of the epithelial markers E-cadherin, and down-regulated vimentin (Supplementary Fig. 2D). Moreover, we observed that ectopic ZBTB28 reversed EMT from scattered invasive properties to tightly growth structures (Supplementary Fig. 2E).
Fig. 2.
ZBTB28 acts as a tumor suppressor to inhibit tumorigenesis of breast cancer. A Ectopic ZBTB28 expression at transcription and post-transcription level was detected by RT-PCR and Western blot. GAPDH was used as negative control. B Ectopic ZBTB28 expression impacts breast cancer cell growth was analyzed by the CCK-8 kit. ZBTB28 overexpression can suppress proliferation of breast cancer cells which were assessed through colony formation assay without soft-agar (C) and with soft-agar (D). E Flow cytometry analysis of cell cycle of MCF7 and MB231 which was overexpression pcDNA 3.1 or pcDNA-ZBTB28 by PI staining. F Ectopically expressed ZBTB28-induced apoptosis in breast cancer cells were tested by AO/EB staining assay. G Epithelial markers (E-cadherin, occludin) and mesenchymal markers (N-cadherin, snail) were detected via RT-PCR and Western blot, β-actin expression as control. H, I Transwell assay and wound-healing assay were used to investigate cell motility and invasiveness. Stem cell biomarker expression and growth of floating spheroid colonies were determined by RT-PCR (J) and spheroid formation assay (K) (independent experiment = 3, and a representative data was shown; ‘*’means p < 0.05; ‘**’means p < 0.01; ‘***’means p < 0.001)
As we know, EMT is the key process of metastasis and invasion of tumors [30]. Over-expression of ZBTB28 significantly inhibited the ability of migration and invasion of MB231 cells compared with control cells (Fig. 2H and Supplementary Fig. 2F). Furthermore, ZBTB28-overexpressing breast cancer cells reduced the ability of motility (Fig. 2I and Supplementary Fig. 2G). Because EMT plays pivotal role in self-renewal process of cells, and thus, we measured cancer stem cells biomarkers by RT-PCR analysis. In result, ZBTB28 inhibited NANOG, OCT4, KLF4, ABCG2, BMI-1, MYC, TIP30, MAD2, STAT3, and CD44 at the mRNA level (Fig. 2J). Furthermore, a spheroid formation assay showed that ZBTB28 prevented growth of floating spheroid colonies (Fig. 2K). Next, we chose BT549, a breast cancer cell line which express ZBTB28, to further evaluated the biological effects of ZBTB28. Knock-down of ZBTB28 in BT549 cell line was detected by RT-PCR (Supplementary Fig. 3A). Meanwhile, we found that downregulation of ZBTB28 promoted cell proliferation, migration, and invasion (Supplementary Fig. 3B–D). Altogether, ZBTB28 exerted the capacity to suppress proliferation, induce cell cycle arrest and apoptosis, and restrain EMT and cell stemness which is a general feature of the tumor suppressor gene in breast tumor cells.
ZBTB28 directly regulated IFNAR to stimulate interferon-stimulated genes (ISGs)
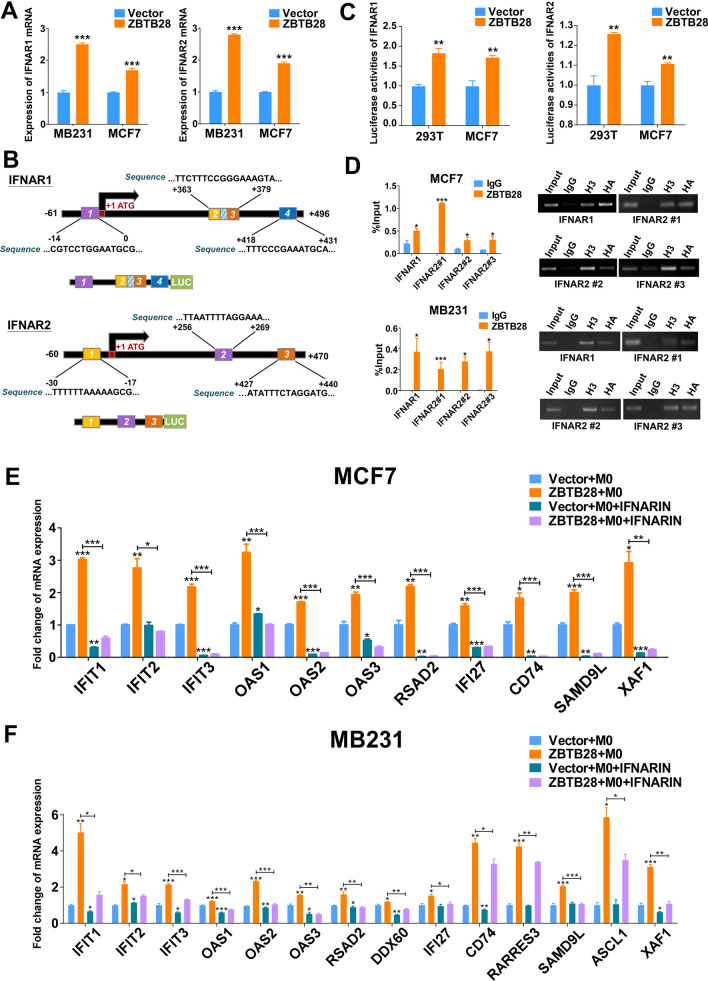
Interestingly, we have previously shown that ZBTB28 could exert its anti-tumor efficacy by activating ISGs through RNA sequencing (RNA-seq) analysis in KYSE150 cells. Activation of IFN receptors will trigger the transcription of hundreds of different ISGs, and this attracted us to determine whether ZBTB28 could regulate IFN receptors to take part in anti-tumor function in breast cancer [15]. We next used global transcriptional profiling of RNA isolated from MCF7. RNA sequencing analysis identified IFNAR1, IFNAR2, IFNGR1, and IFNGR2 as differentially expressed genes in ZBTB28-expressing MCF7 cells. qRT-PCR confirmed that IFNAR1, IFNAR2, IFNGR1, and IFNGR2 were up-regulated by ZBTB28 in both MCF7 and MB231 cells (Fig. 3A and Supplementary Fig. 4C). The online database JASPAR was used to search potential ZBTB28 transcription factor-binding sites (TFBSs) on promoter region of these four genes. According to the prediction results, IFNAR1 has four suitable binding sites at positions -14 to 0, + 363 to + 376, + 366 to + 379, and + 418 to + 431. IFNAR2 has three binding sites at positions − 30 to − 17, + 256 to + 269, and + 427 to + 440 (Fig. 3B). However, no suitable predicted binding sites were found on the IFNGR1 and IFNGR2 promoter regions. Furthermore, a dual-luciferase reporter assay revealed that ZBTB28 up-regulated the promoter activity of IFNAR1 and IFNAR2 in 293 T and MCF7 cell lines (Fig. 3C). Moreover, the direct interaction of the IFNAR1 or IFNAR2 with ZBTB28 promoter was confirmed by ChIP-qPCR analysis in MCF7 and MB231 cells lines, and ZBTB28 was enriched at the predicted regions of the IFNAR1 and IFNAR2 promoter (Fig. 3D). To make sure upregulation of ISGs was caused by ZBTB28, we used an IFNAR inhibitor to block IFNAR1 and IFNAR2, which are the subunits of the type I IFN receptor (IFNAR). The cancer cells were harvested after co-culturing with macrophages to detect ISGs mRNA levels. The results thus far demonstrated that ZBTB28 could upregulate IFNAR as well as downstream signaling pathway activation (Fig. 3E, F). In addition, RT-PCR and qRT-PCR illustrated that interferon-γ (IFN-γ) was up-regulated by ZBTB28; also, the expression of IFNGR2, which encodes the IFN-γ receptor to management IFN-γ, was up-regulated by ZBTB28 in MCF7 and MB231 cell lines (Supplementary Fig. 4A–C). Western blot results indicated that after blocking IFNGR2, the level of IFN-γ induced by ZBTB28 was decreased in the co-culture system (Supplementary Fig. 4D). This suggested that ZBTB28 may enhance IFN-γ by regulating IFNGR2 to strengthen its anti-tumor immunity. These data suggested that ZBTB28 has the potential to stimulate both IFNAR and IFNGR to incite effective anti-tumor immunity to maintain durable tumor control.
Fig. 3.
ZBTB28 regulated IFNAR to activate ISGs. A qRT-PCR results of IFNAR1 and IFNAR2 mRNA expression in stable transfected breast cancer cells. B Structure of wild-type IFNAR1 and IFNAR2 reporter plasmid. The sequence of the predicted site has been displayed. C Promoter luciferase activity of IFNAR1 and IFNAR2 in 293 T and MCF7 cells. D ChIP analysis with HA-tag for ZBTB28 binding to the IFNAR1 or IFNAR2 promoter. Then, products of ChIP-PCR were used for electrophoresis. E, F Cancer cells were pretreatment with/without IFNAR inhibitor (1 μM) for 8 h and then co-cultured with THP-1 macrophages for another 24 h later. Expressions of ISGs in the indicated groups were detected by qRT-PCR
Over-expression of ZBTB28 inhibits tumor growth in vivo and regulates macrophage polarization
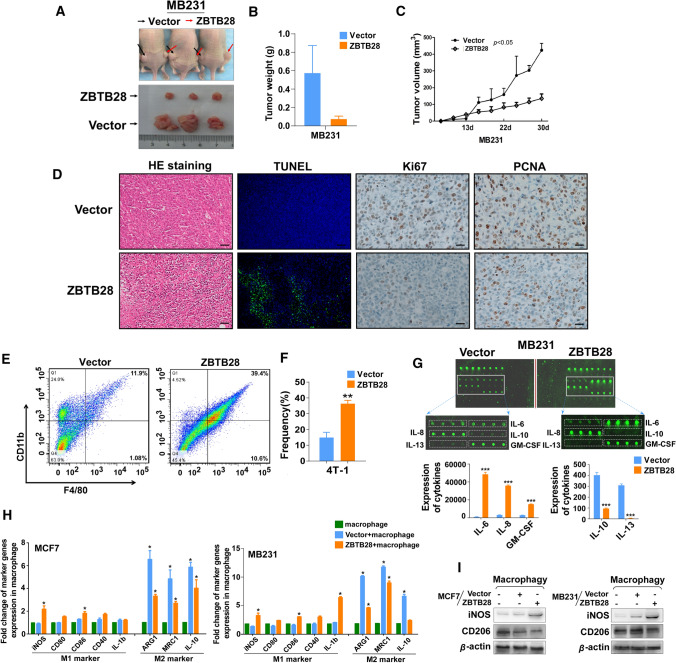
Having observed that ZBTB28 inhibited breast cancer cells in vitro, we hypothesized that overexpression of ZBTB28 might exert suppressive effects on tumor growth in vivo. To test this hypothesis, we used ZBTB28-transfected MB231 cells for in vivo studies. The ZBTB28-transfected MB231 cells grew slower than the control-transfected MB231 cells when they were subcutaneously implanted into nude mice (Fig. 4A–C). And compared with the control group, the xenografts with ZBTB28 overexpression exhibited a significantly reduced weight (Fig. 4B). Then, we performed histological analysis to assess the anti-tumor features of ZBTB28 in vivo. Hematoxylin and eosin (H&E) and TUNEL staining confirmed the effective inhibition of MB231 cell proliferation by ZBTB28 (Fig. 4D). Stronger Ki-67 and PCNA expression was found in tumors derived from the control group than in those from the ZBTB28 group, indicating that the ectopic expression of ZBTB28 slowed the tumor proliferative activity (Fig. 4D). It has been reported that interferon can activate macrophages and induce polarization [15]. Thus, infiltrating macrophages from mice bearing 4-T1 cells stably overexpressing Vector or m-ZBTB28 were stained and assessed via flow cytometry to determine whether ZBTB28 could influence anti-tumor immunity in vivo. The results showed that the levels of macrophages (CD11b+F4/80+) were higher in the ZBTB28 high expression group than in the control group (Fig. 4E, F). These findings suggested that ZBTB28 inhibited breast cancer growth in vivo and was associated with macrophage infiltration.
Fig. 4.
In vivo tumorigenicity was suppressed by ZBTB28. A Representative picture showing xenografts growth 30 days after subcutaneously implanted MB231 cells. B, C The weight of tumors was measured, respectively (n = 3). Tumors’ volume was measured every 3 days and then draw it in a line graph. D Typical Images of HE staining, TUNEL assays, Ki-67, and PCNA expression. E, F Representative FACS dot plots of tumor tissue single-cell suspensions stained with CD11b and F4/80. Bar graph reports the mean ± SEM of CD11b+ F4/80+ subset from combined independent experiments. G An antibody array was used for cytokines detection of medium from MB231 cells. Fluorescence imaging (upper) and analysis of extracted data (lower) were shown. H Expression of markers associated with M1 and M2 polarization in macrophages of the indicated groups. I WB results of iNOS (M1 marker) and CD206 (M2 marker) after co-cultivation
It is known that tumor cells secrete several cytokines (autocrines), thereby inducing the recruitment of immune cells to the tumor [31]. Thus, cancer cell supernatants were collected to test cytokine production. MB231 cells that expressed ZBTB28 exhibited a significant increase in the proinflammatory cytokines, such as IL-6 (markers associated with M1 macrophage polarization), IL-8, and granulocyte–macrophage colony-stimulating factor (GM-CSF) (Fig. 4G) [32]. We also observed a reduction in the expression of Th2 cytokines, for example, IL-10, as well as markers associated with M2 macrophage polarization, for example, IL-13, in ZBTB28 overexpression tumors (Fig. 4G). Macrophages can quickly change their status and function in response to local immune microenvironmental signals which makes the state of macrophages determine their role in tumor biology [33]. Thus, we explored whether there was an association between ZBTB28 expression and polarization of the macrophages. First, we established an in vitro co-culture system that cultured BrCa cells (with/without ZBTB28 expression) with PMA-treated THP-1 cells. qRT-PCR results showed that the enhanced expression of M1 markers, iNOS, CD80, CD86, and IL-1b was accompanied by the decreased expression of M2 markers, ARG1, MRC1, and IL-10 in THP-1 macrophages (Fig. 4H). Western blotting also showed that ZBTB28 increased the expression of iNOS and suppressed CD206, which are associated with M1 and M2 macrophages, respectively (Fig. 4I). This reflected more specifically the highly complex in vivo immune environments in which macrophages could acquire an activation status due to the expression of ZBTB28 in breast cancer cells.
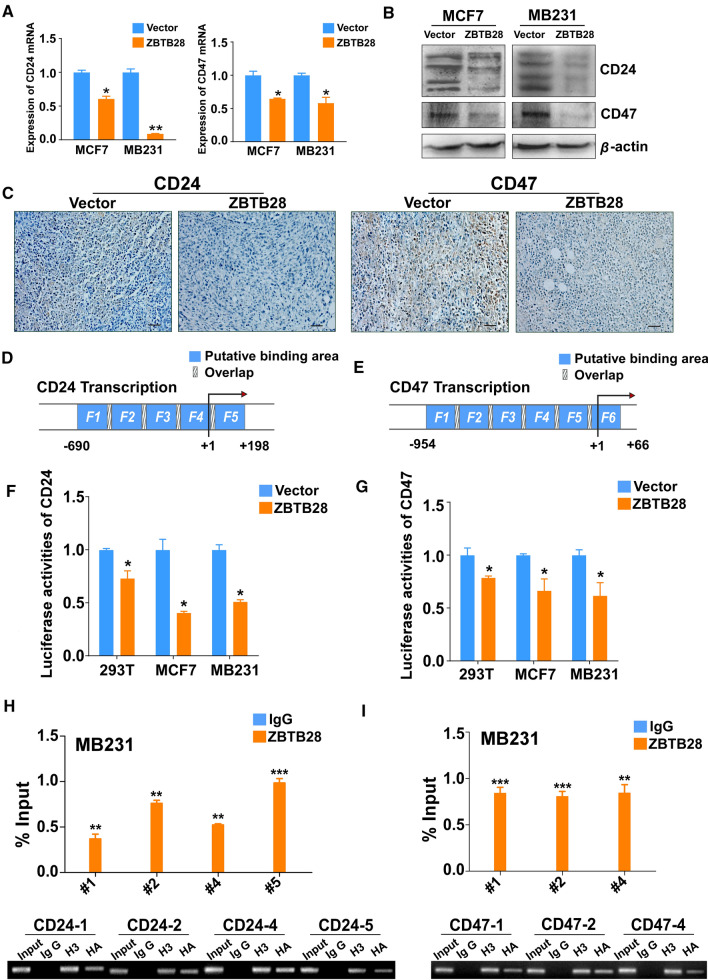
ZBTB28 downregulates the expressions of CD24 and CD47
It is noteworthy that the RNA sequencing results also showed that the expression of CD24 and CD47 were down-regulated by ZBTB28. Next, qRT-PCR was performed to confirm that ZBTB28 down-regulated the expressions of CD24 and CD47 in both MCF7 and MB231 cell lines (Fig. 5A). Western blotting proved that ZBTB28 down-regulated CD24 and CD47 at the protein level (Fig. 5B). IHC results from tumor tissue sections of nude mice also showed that ZBTB28 down-regulated the expressions of CD24 and CD47 in vivo (Fig. 5C). Furthermore, we searched potential ZBTB28 TFBSs on CD24 and CD47 promoter regions through the open-access database JASPAR (Fig. 5D, E). The promoter regions of CD24 and CD47 which contain different predicted TFBSs of ZBTB28 were cloned into the pGL3-Basic reporter vector, respectively. Luciferase reporter assay showed that ZBTB28 suppressed the promoter activities of CD24 and CD47 in 293 T, MCF7 and MB231 cell lines (Fig. 5F, G). Then, we conducted the ChIP assay to confirm the putative binding sites. The results revealed that ZBTB28 was enriched at the 4/5 predicted regions of CD24 promoter and 3/6 predicted regions of CD47 promoter, while a control IgG antibody showed no significant enrichment over the entire surveyed region (Fig. 5H, I). In our previous studies, we found that in lung cancer and esophageal cancer, ZBTB28 and BCL6 inhibited each other’s transcription [27]. Therefore, we tried to explore whether there is any interaction between them in breast cancer. Analysis from the TCGA online database showed that the relationship between the expression of ZBTB28 and BCL6 is negatively correlated in breast cancer (Supplementary Fig. 5A). We examined further whether a ZBTB28 and BCL6 interaction could affect the expressions of CD24 and CD47. Knock-down of BCL6 with siRNA could reduce the expressions of CD24 and CD47 in both MCF7 and MB231 cells. Furthermore, knock-down BCL6 after overexpression of ZBTB28 strengthened the inhibitory effect of ZBTB28 on CD24 and CD47 (Supplementary Fig. 5B, C). In K562 cell line, with no BCL6 expression, exogenous expression of BCL6 up-regulated CD24 and CD47. Meanwhile, ZBTB28 down-regulated CD24 and CD47 expression [34]. Over-expression of BCL6 partly offseted the inhibitory effect of ZBTB28 on CD24 and CD47 (Supplementary Fig. 5D). This suggested that the BCL6 might be involved in the regulation of CD24 and CD47 by ZBTB28. The above results proved that CD24 and CD47 were indeed the target genes of ZBTB28.
Fig. 5.
Expression of CD24 and CD47 was down-regulated by ZBTB28. A, B qRT-PCR and Western blot results of CD24 and CD47 expression in stable transfected breast cancer cells, respectively. C Representative images of CD24 and CD47 immunostaining in MB231 xenografts sections of vector and overexpression of ZBTB28. (D) Locations of ChIP-PCR primers (Fragement1 (− 690 ~ -465), 2 (− 486 ~ -323), 3 (− 342 ~ − 122), 4 (− 143 ~ + 18), and 5 (− 2 ~ + 198)) at the CD24 promoter, transcription start site (TSS) was designated as nucleotide + 1. (E) Locations of ChIP-PCR primers (Fragement1 (− 954 ~ − 783), 2 (− 804 ~ − 620), 3 (− 638 ~ − 489), 4 (− 510 ~ − 334), 5 (− 351 ~ − 95), and 6 (− 112 ~ + 66)) at the CD47 promoter, TSS was designated as nucleotide + 1. F, G The effect of ZBTB28 on CD24 and CD47 promoter activity was detected by dual-luciferase reporter system in 293 T, MCF7 and MB231 cells. H, I %Input of CD24 DNA or CD47 DNA by anti-HA antibody was determined by ChIP-PCR. Then, products of ChIP-PCR were used for electrophoresis
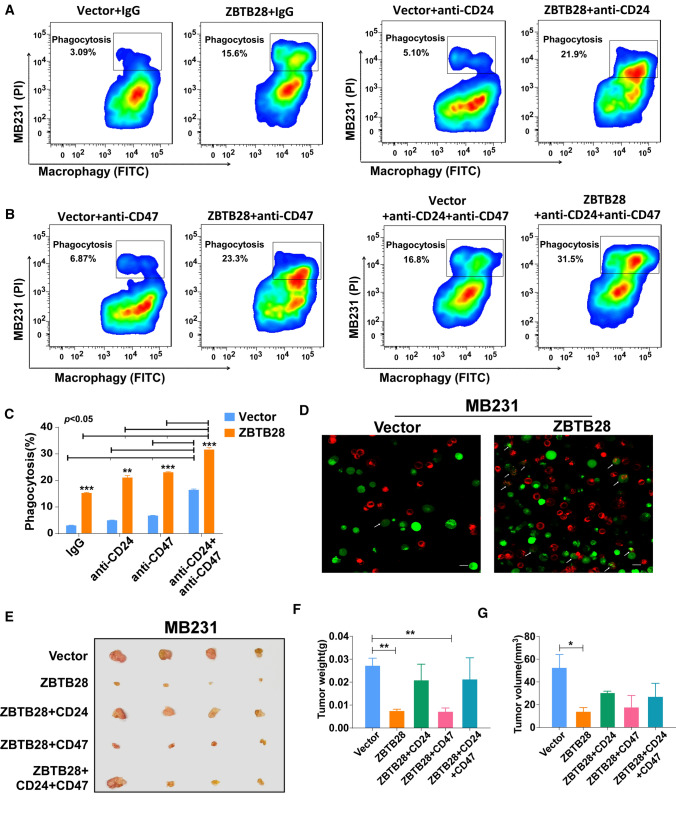
ZBTB28 favors macrophages to increase phagocytosis
To explore the impact of ZBTB28 on the ability of macrophages to engulf cancer cells. THP-1 cells could differentiate from a monocytic phenotype into macrophage-like cells by stimulation with phorbol 12-myristate 13-acetate (PMA). Therefore, this is a widely used model cell line for human monocytes (M0) and macrophage-like cells (MΦ) [35]. We developed an in vitro co-culture system to investigate phagocytosis. A cell tracking assay was established by labeling BrCa cells with the red fluorescent dye DIL and THP-1 macrophages with the green fluorescent dye Calcein-AM. After co-culturing, the flow cytometry assay clearly showed that, compared with that of vector MCF7 and MB231 cells, the phagocytosis of these cancer cells with stable overexpression of ZBTB28 by THP-1 macrophages was increased (Fig. 6A, C and Supplementary Fig. 6A, B). Also, confocal microscopy visually displayed that MCF7 and MB231 cells with red fluorescence were engulfed by THP-1 macrophages with green fluorescence (Fig. 6D and Supplementary Fig. 6C). According to the reports, the presence of CD24-Siglec10 or CD47-SIRPα mediated inhibition could govern the occurrence of phagocytosis [18, 25]. To assess the role of CD24-Siglec10 and CD47-SIRPα signaling in regulating the macrophage-mediated anti-tumor-immune response in BrCa, we first made sure that PMA-treated THP-1 was still expressing Siglec10 and SIRPα (Supplementary Fig. 5E). The blockade of CD24 and CD47 augmented the phagocytosis of BrCa cells by macrophages, which was more obvious in the ZBTB28 overexpression group. Similarly, FACS-based measurements showed a robust increase in ZBTB28 induced phagocytosis upon the addition of anti-CD24 mAb and anti-CD47 mAb as compared to the vector control, which was greater than the effect observed with only CD24 or CD47 blockade (Fig. 6A–C and Supplementary Fig. 6A, B). This implied that ZBTB28 can enhance the cooperativity of combinatorial blockade of CD24 and CD47 in vitro. To confirm that the anti-tumor immunity of ZBTB28 is indeed related to CD24 and CD47, we restored the expression of CD24, CD47 in MB231 cells with stable overexpression of ZBTB28 cells. Then, these cells were subcutaneously implanted into nude mice. We found that the inhibitory effect of ZBTB28 on tumor growth was offset by the restoration of CD24, whereas in these cases, restoration of CD47 had little effect on ZBTB28’s anti-tumor effects. Furthermore, we observed that the simultaneous restoration of CD47 and CD24 was insufficient to completely abrogate the anti-tumor effect of ZBTB28 in vivo (Fig. 6E–G). However, in vitro flow cytometry analysis showed that the recovery of CD24 and CD47 successfully inhibited the phagocytosis induced by ZBTB28, and exhibited a combined effect (Supplementary Fig. 7A–C). Therefore, considering in vitro and in vivo studies, our results revealed that ZBTB28 directly inhibited the expressions of CD24 and CD47 to result in the increased phagocytosis of BrCa cells by macrophages.
Fig. 6.
Over-expression of ZBTB28 promotes phagocytosis of tumor cells by macrophages. A, B Representative flow-cytometry plots portraying the phagocytosis of MB231 cells treated with anti-CD24 mAb, anti-CD47 mAb or dual treatment, compared with the IgG control. C Phagocytosis efficiency was shown as a bar graph. D Representative phagocytosis images of THP-1 macrophages engulfing MB231 cells with vector and ZBTB28 overexpression. THP-1 macrophages were stained green (Calcein-AM); cancer cells were labeling with red (DIL). The white arrows indicate macrophages that engulfed cancer cells. (E) Representative pictures showing tumor growth 14 days after subcutaneously implanted different MB231 cells. (F, G) Tumors’ volume and weight were measured, respectively (n = 4). All statistical data were shown as mean ± SEM.*p < 0.05, **p < 0.01, ***p < 0.001
Discussion
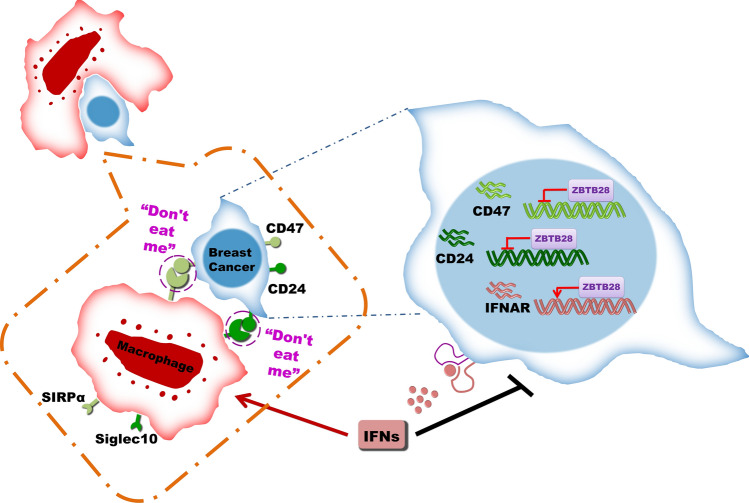
Tumor suppressor genes are well recognized of mainly related to cancer cell proliferation and metastasis. Interestingly, a recent finding has suggested that inactivation of tumor suppressor gene also exerts a major role in tumor-immune escape [5]. In the present study, we investigated the transcription factor, ZBTB28, to determine its functional role in breast cancer and its relationship to anti-tumor immunity and phagocytosis. We found that ZBTB28 inhibited breast cancer cell malignant functions in vivo and in vitro, and demonstrated the involvement of phagocytosis by macrophages and immune checkpoint molecules in the process (Fig. 7).
Fig. 7.
Molecular mechanism of ZBTB28 strengthen anti-tumor immunity to inhibiting breast cancer
In vitro co-culture systems, we detected that ZBTB28 primed the type I interferon system and activated ISGs. IFNs have regulatory functions, which include inhibition of tumors growth and strengthen anti-tumor-immune activity through wake immune cells (such as dendritic cells, macrophages, or NK cells) [36, 37]. Nevertheless, the wide distribution of interferon receptors makes the treatment of type I IFN is often highly toxic [38]. The ectopic expression of ZBTB28 up-regulates IFNAR in breast cancer cells, theoretically guaranteeing the more binding of type I interferon, which optimized the auto-immunity control of tumors, and provided a new strategy for reducing the side effects of high-dose interferon therapy.
A recent study demonstrated a role for tumor-expressed CD24 and CD47 in promoting immune evasion by breast cancer through their respective interactions with Siglec10 and SIRPα, which are expressed by tumor-associated macrophages. It is worth noticing that the expression of CD24 was significantly higher in TNBC or ER + PR + breast cancers than in normal breast cells. More interestingly, “lower PD-L1 expressors” were showed to be associated with higher CD24 expression [18]. Herein, the present investigation showed that the expression of CD24 and CD47, at mRNA and protein levels, was down-regulated by ZBTB28. Mechanism research ascribed this downregulation to ZBTB28 transcription factors directly binding the promoter of CD24 and CD47. Thus, the reduction of CD24 and CD47 on cancer cells endued ZBTB28 the ability to induced macrophage phagocytosis.
Blockade of the CD24-Siglec10 or CD47-SIRPα interaction using CD24 blocking mAb (clone SN3) or CD47 blocking mAb (clone Hu5F9-G4), respectively, augments phagocytosis by macrophages and inhibits tumor growth [18, 39]. In breast cancer, combination inhibited the immune checkpoints CD24-Siglec10 and CD47-SIRPα significantly improves the ability of macrophages to phagocytize cancer cells, indicating a synergistic effect between immune checkpoint molecules CD24 and CD47 [18]. In our study, we determined that ZBTB28 enhanced the phagocytic effect of anti-CD24 or anti-CD47 treatment alone in vitro. Moreover, ZBTB28 made the dual blockade of CD24 and CD47 further augmented the phagocytic ability of macrophages, thereby confirming a synergistically role for CD24 and CD47 in inhibiting phagocytosis, which suggested that ZBTB28 could regulate anti-tumor immunity by targeting CD24 and CD47 in BrCa. This provides a new mechanism by which ZBTB28 can regulate the CD24 and CD47 immune checkpoint, so that targeting of CD24 and CD47 by ZBTB28 may be a potential novel therapy approach to the treatment of BrCa. That may potentially increase the efficacy and decrease toxic side effects compared with the traditional radiotherapy and chemotherapy.
To investigate the impact of ZBTB28 on the tumor-immune microenvironment in vivo, we restored the expressions of CD24 and CD47 in cells overexpressing ZBTB28. The ability of ZBTB28 to inhibit tumor growth is severely diminished due to the restoration of CD24 expression. Restoration of CD47 was insufficient to abrogate the anti-tumor effect of ZBTB28 in vivo; however, in vitro phagocytosis assays showed that CD47 indeed influenced ZBTB28 induced phagocytosis. It is likely that differences in the amino acid sequences of human and mouse SIRPα may bring about inconsistent results for in vivo and in vitro experiments [40].
The tumor suppressor gene ZBTB28 can not only activate the IFNAR to enhance ISGs expression and boost macrophage but also reduce the expression of CD24 and CD47 to allow breast cancer cells phagocytosis by macrophages. Eventually, the exacerbated immune response driven by ZBTB28 makes it an important target for a potential “synergistic” immune therapy against breast cancer.
In conclusion, we demonstrated that ZBTB28 acts as a functional tumor suppressor in breast cancer. The silencing of ZBTB28 due to methylation of its promoter associated with poor survival of patients. By upregulating IFNAR to activate ISGs and macrophages, targeting CD24 and CD47 on breast cancer cells to increased macrophage-mediated phagocytosis, and enhancing the synergistic effect of dual treatment with anti-CD24 and anti-CD47, ZBTB28 exerts its anti-tumor effects. This shows that assessing the methylation and expression status of ZBTB28 in breast cancer might be new prognostic markers for risk stratification and lead to better treatment strategies for breast cancer patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by National Natural Science Foundation of China (#81872380, #81572769, #82003135), Natural Science Foundation of Chongqing (2020ZYO13799), and Hong Kong-RGC (GRF# 14115019). The authors thank Prof. Qian Tao (the Chinese University of Hong Kong, Hong Kong, China) for generously providing primers and plasmids.
Abbreviations
- 5-Aza
5-Aza-2’-deoxycytidine
- BrCa
Breast cancer
- ChIP
Chromatin immunoprecipitation
- EMT
Epithelial–mesenchymal transition
- IFNAR
The type I IFN receptor
- ISGs
Interferon-stimulated genes
- MSP
Methylation-specific PCR
- PMA
12-Myristate 13-acetate
- Siglec-10
Sialic-acid-binding Ig-like lectin 10
- SIRPα
Signal regulatory protein alpha
- TFBSs
Transcription factor-binding sites
- ZBTB28
Zinc-finger and BTB/POZ (Poxvirus and Zinc-finger) domain-containing family protein 28
Author contributions
LL and XT: conception and design. LL, GY, and TJ performed majority of experiments. PW, CZ, YR, DC, and LL performed experiments and analyzed data. XQ, MJ, YC, XJ, and WY collected samples. LL and GY drafted the manuscript. XT, LL, and GY reviewed data and manuscript. XT, LL, GY, and LX reviewed data and finalized the manuscript. All authors reviewed and approved the final version.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Ethical approval and consent to participate
The study was authorized by the Institutional Ethics Committees of The First Affiliated Hospital of Chongqing Medical University and conformed to the principles of the Declaration of Helsinki.
Consent for publication
We have obtained consent to publish from the participant to report individual patient data.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Li, Yijia Gong, and Jun Tang have contributed equally to this work.
References
- 1.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6(12):718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 2.Bilimoria MM, Morrow M. The woman at increased risk for breast cancer: evaluation and management strategies. CA Cancer J Clin. 1995;45(5):263–278. doi: 10.3322/canjclin.45.5.263. [DOI] [PubMed] [Google Scholar]
- 3.Tichy JR, Lim E, Anders CK. Breast cancer in adolescents and young adults: a review with a focus on biology. J Nat Compr Cancer Netw JNCCN. 2013;11(9):1060–1069. doi: 10.6004/jnccn.2013.0128. [DOI] [PubMed] [Google Scholar]
- 4.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12(6):488–496. doi: 10.1245/aso.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Martin TD, Patel RS, Cook DR, Choi MY, Patil A, Liang AC, et al. The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science. 2021;373(6561):1327–1335. doi: 10.1126/science.abg5784. [DOI] [PubMed] [Google Scholar]
- 6.Hartatik T, Okada S, Okabe S, Arima M, Hatano M, Tokuhisa T. Binding of BAZF and Bc16 to STAT6-binding DNA sequences. Biochem Biophys Res Commun. 2001;284(1):26–32. doi: 10.1006/bbrc.2001.4931. [DOI] [PubMed] [Google Scholar]
- 7.Sakashita C, Fukuda T, Okabe S, Kobayashi H, Hirosawa S, Tokuhisa T, Miyasaka N, Miura O, Miki T. Cloning and characterization of the human BAZF gene, a homologue of the BCL6 oncogene. Biochem Biophys Res Commun. 2002;291(3):567–573. doi: 10.1006/bbrc.2002.6481. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Huang P, Panyisha W, Kong R, Jiang X, Zhang L, Yang Q, Xie Q, et al. BCL6B expression in hepatocellular carcinoma and its efficacy in the inhibition of liver damage and fibrogenesis. Oncotarget. 2015;6(24):20252–20265. doi: 10.18632/oncotarget.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu S, Cao B, Zhang M, Linghu E, Zhan Q, et al. Epigenetic silencing BCL6B induced colorectal cancer proliferation and metastasis by inhibiting P53 signaling. Am J Cancer Res. 2015;5(2):651–662. [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Gong Y, Xu K, Chen W, Xia J, Cheng Z, et al. ZBTB28 induces autophagy by regulation of FIP200 and Bcl-XL facilitating cervical cancer cell apoptosis. J Exp Clin Cancer Res. 2021;40(1):150. doi: 10.1186/s13046-021-01948-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu LY, Tang J, Zhang CM, Zeng WJ, Yan H, Li MP, et al. new immunotherapy strategies in breast cancer. Int J Environ Res Public Health. 2017 doi: 10.3390/ijerph14010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger A, Colpitts SJ, Seabrook MSS, Furlonger CL, Bendix MB, Moreau JM, et al. Interleukin-15 in cancer immunotherapy: IL-15 receptor complex versus soluble IL-15 in a cancer cell-delivered murine leukemia model. J Immunother Cancer. 2019;7(1):355. doi: 10.1186/s40425-019-0777-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H, Qiao J, Fu YX. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370(1):85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salvagno C, Ciampricotti M, Tuit S, Hau CS, van Weverwijk A, Coffelt SB, et al. Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response. Nat Cell Biol. 2019;21(4):511–521. doi: 10.1038/s41556-019-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesev EV, LeDesma RA, Ploss A. Decoding type I and III interferon signalling during viral infection. Nat Microbiol. 2019;4(6):914–924. doi: 10.1038/s41564-019-0421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xuan C, Steward KK, Timmerman JM, Morrison SL. Targeted delivery of interferon-alpha via fusion to anti-CD20 results in potent antitumor activity against B-cell lymphoma. Blood. 2010;115(14):2864–2871. doi: 10.1182/blood-2009-10-250555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirruccello SJ, LeBien TW. The human B cell-associated antigen CD24 is a single chain sialoglycoprotein. J Immunol. 1986;136(10):3779–3784. [PubMed] [Google Scholar]
- 18.Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572(7769):392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006;66(4):1917–1922. doi: 10.1158/0008-5472.Can-05-3855. [DOI] [PubMed] [Google Scholar]
- 20.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35(3):255–262. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. doi: 10.1038/ncomms6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11(3):130–135. doi: 10.1016/s0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 23.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci USA. 2012;109(17):6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138(2):271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng R, Zhao H, Xu J, Shen C. CD47: the next checkpoint target for cancer immunotherapy. Crit Rev Oncol Hematol. 2020;152:103014. doi: 10.1016/j.critrevonc.2020.103014. [DOI] [PubMed] [Google Scholar]
- 26.Qu M, Zou X, Fang F, Wang S, Xu L, Zeng Q, et al. Platelet-derived microparticles enhance megakaryocyte differentiation and platelet generation via miR-1915-3p. Nat Commun. 2020;11(1):4964. doi: 10.1038/s41467-020-18802-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang T, Tang J, Li L, Peng W, Du Z, Wang X, et al. Tumor suppressive BTB/POZ zinc-finger protein ZBTB28 inhibits oncogenic BCL6/ZBTB27 signaling to maintain p53 transcription in multiple carcinogenesis. Theranostics. 2019;9(26):8182–8195. doi: 10.7150/thno.34983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Fan J, Fan Y, Li L, He X, Xiang Q, et al. The new 6q27 tumor suppressor DACT2, frequently silenced by CpG methylation, sensitizes nasopharyngeal cancer cells to paclitaxel and 5-FU toxicity via beta-catenin/Cdc25c signaling and G2/M arrest. Clin Epigenetics. 2018;10(1):26. doi: 10.1186/s13148-018-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16(11):2949–2958. doi: 10.1158/1078-0432.ccr-09-3178. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49(3):361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20(5):576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Y, Yang J, Xu D, Gao XM, Zhang Z, Hsu JL, et al. Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade. Gut. 2019;68(9):1653–1666. doi: 10.1136/gutjnl-2019-318419. [DOI] [PubMed] [Google Scholar]
- 33.Casazza A, Mazzone M. Altering the intratumoral localization of macrophages to inhibit cancer progression. Oncoimmunology. 2014;3(1):e27872. doi: 10.4161/onci.27872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, et al. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86(1):45–53. doi: 10.1182/blood.V86.1.45.bloodjournal86145. [DOI] [PubMed] [Google Scholar]
- 35.Herdoiza Padilla E, Crauwels P, Bergner T, Wiederspohn N, Förstner S, Rinas R, et al. mir-124-5p regulates phagocytosis of human macrophages by targeting the actin cytoskeleton via the ARP2/3 complex. Front Immunol. 2019;10:2210. doi: 10.3389/fimmu.2019.02210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borden EC. Interferons α and β in cancer: therapeutic opportunities from new insights. Nat Rev Drug Discov. 2019;18(3):219–234. doi: 10.1038/s41573-018-0011-2. [DOI] [PubMed] [Google Scholar]
- 37.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16(3):131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 38.Aricò E, Castiello L, Capone I, Gabriele L, Belardelli F. Type I interferons and cancer: an evolving story demanding novel clinical applications. Cancers (Basel) 2019 doi: 10.3390/cancers11121943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkarni A, Chandrasekar V, Natarajan SK, Ramesh A, Pandey P, Nirgud J, et al. A designer self-assembled supramolecule amplifies macrophage immune responses against aggressive cancer. Nat Biomed Eng. 2018;2(8):589–599. doi: 10.1038/s41551-018-0254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murata Y, Saito Y, Kotani T, Matozaki T. CD47-signal regulatory protein α signaling system and its application to cancer immunotherapy. Cancer Sci. 2018;109(8):2349–2357. doi: 10.1111/cas.13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.