Abstract
Both adipose tissue and skeletal muscle are highly dynamic tissues and interact at the metabolic and hormonal levels in response to internal and external stress, and they coordinate in maintaining whole-body metabolic homeostasis. In our previous study, we revealed that adipocyte-specific Rnf20 knockout mice (ASKO mice) exhibited lower fat mass but higher lean mass, providing a good model for investigating the adipose-muscle crosstalk and exploring the effect of the adipocyte Rnf20 gene on the physiology and metabolism of skeletal muscle. Here, we confirmed that ASKO mice exhibited the significantly increased body weight and gastrocnemius muscle weight. Fiber-type switching in the soleus muscle of ASKO mice was observed, as evidenced by the increased number of fast-twitch fibers and decreased number of slow-twitch fibers. Serum metabolites with significant alteration in abundance were identified by metabolomic analysis and the elevated lysophosphatidylcholine 16:0 [LysoPC (16:0)] was observed in ASKO mice. In addition, lipidome analysis of gonadal white adipose tissue revealed a significant increase in LysoPCs and LysoPC (16:0) in ASKO mice. Furthermore, knockdown of Rnf20 gene in 3T3-L1 cells significantly increased the secretion of LysoPC, suggesting that LysoPC might be a critical metabolite in the adipose-muscle crosstalk of ASKO mice. Furthermore, in vitro study demonstrated that LysoPC (16:0) could induce the expression of fast-twitch muscle fibers related genes in differentiated C2C12 cells, indicating its potential role in adipose-muscle crosstalk. Taken together, these findings not only expand our understanding of the biological functions of Rnf20 gene in systemic lipid metabolism, but also provide insight into adipose tissue dysfunction-induced physiological alterations in skeletal muscle.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-04896-4.
Keywords: RNF20, Adipose-muscle crosstalk, Fiber-type switching, LysoPC (16:0)
Introduction
Skeletal muscle is the most abundant tissue in mammalian body, and is essential for various physiological activities, including movement, thermogenesis and metabolic homeostasis etc. [1]. It is a heterogeneous tissue in nature comprised of muscle fibers, which can be classified as slow-twitch (type I) and fast-twitch (type IIa, IIx and IIb) fibers according to the characteristics of contraction. Based on their metabolic properties, these four fiber types can be divided into oxidative and glycolytic myofibers [2, 3]. Numerous studies have indicated that under the influence of internal and external factors, such as heredity, nutritional factors, exercise, and environmental factors, myofibers can be transformed from slow-twitch to fast-twitch or vice versa [2, 4, 5].
Numerous studies revealed that skeletal muscle exert their biological functions by communicating with other organs, including adipose tissue, liver, pancreas, bone and brain [6]. As an important endocrine organ, adipose tissue can express and subsequently release a wide range proteins and other molecules into the circulation, thereby communicating with skeletal muscle [7]. Adipokines, including adiponectin and resistin, have been reported to inhibit glucose uptake in rat skeletal muscle by decreasing GLUT4 translocation and regulating insulin receptor substrate-1 (IRS-1) function [8]. Leptin or adiponectin administration resulted in increased fatty acid oxidation and decreased TAG content in mice skeletal muscle [9, 10]. Tumor necrosis factor α, which is released from adipose tissue, has been reported to induce the atrophy of C2C12 cells by increasing myofibrillar protein degradation [11]. In addition to adipokines, other factors released by adipose tissue, such as lipids, metabolites, noncoding RNAs, and exosomes, have also been shown to participate in adipose-muscle conversation [12]. Free fatty acids (FFAs), both palmitate and oleate, released from fat tissue inhibit protein degradation by suppressing IRS-1/AKT signaling in C2C12 skeletal muscle cells [13]. Mice with lower serum lysophosphatidic acids (LPAs) exhibited the enhanced glucose utilization, glucose tolerance and insulin sensitivity in skeletal muscle [14]. Sphingosine-1-phosphate, a lipid mediator formed by the phosphorylation of sphingosine, triggers skeletal muscle cells to increase glucose uptake [15]. Moreover, the clinical data demonstrated that lipodystrophic patients are normally associated with muscular hypertrophy [16]. In Lipin1-deficient and Bscl2/Seipin-deficient lipodystrophic mice, the phenotypes of skeletal muscle fibers were also observed to be altered [17, 18]. Nonetheless, the effect of dysfunction in adipose tissue on cellular and molecular alterations of skeletal muscle is still largely incomplete and remain to be elucidated.
RING finger 20 (RNF20) is an E3 ligase and crucial for multiple biological processes, such as cancer development and progression, spermatogenesis, brain development and so on [19]. Accumulating evidence demonstrated that RNF20 also plays a critical role in fat metabolism. Ren et al. has been demonstrated that RNF20 could increase C/EBPα expression through stimulating the polyubiquitination and proteasome-dependent of AP-2α [20]. It has been reported that RNF20 acts as a negative regulatory of hepatic fatty acid metabolism through degradation sterol regulatory element binding protein 1c (SREBP1c), a key transcription factor for de novo lipogenesis [21]. Studies with RNF20+/− mice and 3T3-L1 cells revealed that RNF20 stimulates the transcriptional activity of PPARγ via promoting the proteasomal degradation of nuclear receptor corepressor 1 (NCoR1) [22]. In addition, it has been reported that knockdown the expression of RNF20 impaired the porcine adipogenesis via the mitotic clonal expansion, indicating its essential role in adipocyte differentiation [23]. Our previous study found that mice with adipocyte-specific deletion of Rnf20 (ASKO mice) exhibited significantly decreased fat mass but markedly increased body weight and lean mass [24], suggesting the potential crosstalk between fat and skeletal muscle. It is of interest to explore the mechanisms by which adipocyte Rnf20 gene affects the growth and metabolism of skeletal muscle.
In the present study, we analyzed the mass and fiber composition of skeletal muscles (soleus and gastrocnemius) from 6-month-old WT and ASKO mice. Untargeted serum metabolomic analysis and gonadal white adipose tissue (gWAT) lipidomic analysis were performed with two groups of mice, and the potential candidate crosstalk molecules were screened. Furthermore, in vitro experiments were used to validate the effect of the candidate metabolite on the fiber type transition.
Materials and methods
Animals
The Rnf20flox/flox adiponectin-Cre+ mice (referred to as ASKO mice) and Rnf20flox/flox adiponectin-Cre− mice (referred to as WT mice) were obtained as previously described [24]. All male mice (C57BL/6 background) were housed on a 12 h (h) light/dark cycle with free access to water and food throughout the experiment. All procedure involving animals were in compliance with the Animal Research Panel of the Committee on Research Practice of the University of Chinese Academy of Sciences (IOZ20190077).
Immunofluorescence staining and imaging
For staining of muscle sections, we collected the soleus and gastrocnemius muscle samples frozen by liquid nitrogen-cooled isopentane in Tissue-Tek OCT (SAKURA, Japan) and then sliced muscles into 5-μm sections by a cryostat (CM1850, Leica, Germany). Sections were dried at room temperature for 5 min, fixed with cold acetone for 10 min at 4 °C, washed with PBS and blocked with 10% goat serum (Beyotime Biotechnology, China) in PBST at room temperature for 1 hour (h). After blocking, sections were incubated at 4 °C overnight in primary antibodies, including anti-slow skeletal myosin heavy chain (MYH7, 1:500, Abcam Cat#ab11083, RRID:AB_297734), anti-fast skeletal myosin heavy chain (MYH4, 1:500, Abcam Cat#ab91506, RRID:AB_10714690) and laminin (1:500, Abcam Cat#ab11575, RRID:AB_298179). The next day, the sections were washed three times with PBST and incubated with goat anti-mouse IgG H&L Alexa Fluor 555 (1:200, Abcam Cat#ab150114, RRID:AB_2687594) or goat anti-rabbit IgG H&L Alexa Fluor 488 (1:200, Abcam Cat#ab150113, RRID:AB_2576208) antibodies at room temperature for 1 h. Finally, the sections were mounted with aqueous mounting medium (with DAPI) (Thermo Fisher Scientific, USA) and photographed under a fluorescence microscope (Nikon, Japan). The number of total muscle fibers, MYH7-positive fibers (slow-twitch fibers), and MYH4-positive fibers (fast-twitch fibers) was quantified using Image J (National Institute of Health, RRID:SCR_003070). The slow-twitch percentage was calculated as follows: slow-twitch fiber (%) = MYH7-positive fibers/total muscle fibers × 100%. The fast-twitch percentage was expressed as follows: fast-twitch fiber (%) = MYH4-positive fibers/total muscle fibers × 100%.
For immunocytochemistry staining, the differentiated C2C12 myotubes were washed three times with DPBS (HyClone, USA) and fixed with 4% paraformaldehyde (PFA, Solarbio, China) for 20 min at room temperature. Cells were permeabilized with PBST (0.1% Triton-X 100) for 15 min and then blocked with 5% bovine serum albumin (BSA, Solarbio, China) for 30 min with slow shaking at room temperature. Then, the cells were incubated with anti-MyHC (1:200, Santa Cruz Biotechnology Cat#sc-376157, RRID: AB_10989398) antibody at 4 °C overnight, and the corresponding secondary antibodies were supplied for use. Cell images were observed and captured under a fluorescence microscope (Nikon, Japan). At least five randomly selected areas in three independent cultures were examined.
Quantification of muscle cross-sectional area
In cryosectioned muscle preparations, we used immunofluorescence for laminin staining. To determine the relative size of muscle fibers, we measured muscle fiber cross-sectional area (CSA) in the soleus and gastrocnemius muscles of 6-month-old WT and ASKO mice. In addition, the sizes of MYH4 + and MYH7 + fibers were measured respectively. Three independent samples per group were analyzed. Each group included at least one hundred fifty myofibers. Twenty randomly selected nonoverlapping images were acquired for each cross-section, and the CSAs of almost all muscle fibers in each image were measured by Image J software (National Institute of Health, RRID:SCR_003070), except for the fibers with blurred outlines that could not be recognized by the software.
Serum sample collection and untargeted metabolomic analysis
Serum obtained by centrifugation of clotted blood, snap-frozen in liquid nitrogen and stored at − 80 °C for untargeted metabolomics. LC‒MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher Scientific, USA) coupled with an Orbitrap Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific, USA) in both positive and negative modes. Quality control (QC) samples were the mixture prepared from an equal amount in each sample supernatant and analyzed with the same procedure as that for the experimental samples. Untargeted metabolomics profiling was performed on Shanghai Applied Protein Technology Co. Ltd (China). A complete list of all metabolites is provided in Table S1.
Principal component analysis (PCA) was calculated from relative abundances and performed with R package ‘gmodels’. Univariate analysis included the two-tailed student’s t test (P value) and fold change (FC) analysis. Metabolomic data were examined at an annotation cutoff of 0.5 and analyzed following log2 transformation with P value of the metabolite ratios in positive and negative ion mode. The different metabolites with FC larger than 1.5 and P value less than 0.05 were considered as significant. Functional enrichment analyses of differential metabolites were analyzed by MetaboAnalyst 5.0 online software (https://www.metaboanalyst.ca/faces/home.xhtml). Kyoto Encyclopedia of Genes and Genomes (KEGG) with P < 0.05 were considered as significantly altered.
Lipid sample preparation and lipidomic analysis
For lipidomic analysis, gWAT was collected from 6-month-old WT and ASKO male mice (6 samples per group). Lipids were extracted from approximately 50 mg gWAT as previously described [25]. Mass spectrometry-based lipid detection and data analysis were performed by Lipidall Technologies Co., Ltd (China). Molecular lipids were analyzed in both positive and negative ion modes using multiple reaction monitoring (MRM)-based methods. The molecular lipid species were identified and quantified in absolute (TAG, DAG, CE, PC, PE, PS, LysoPC, LysoPE, LysoPS and SM) amounts and normalized to their respective internal standard and the sample amount. Statistical significantly changed lipids were screening using of the criteria P < 0.05 and FC > 2.0. Lipids were presented as μmol/g wet tissues, full list of the lipidomics data is provided in Table S2.
RNA extraction, reverse transcription, and qPCR
Quantitative reverse-transcription PCR (qRT-PCR) was performed as described previously [23]. Total RNA was extracted from tissues or cells using RNAiso reagent (Takara, Japan) according to the manufacturer’s instructions. Then, 1 μg of total RNA was reverse transcribed with the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Japan) using random hexamer primers according to the manufacturer’s instructions. Real-time PCR was performed with the Applied Biosystems Quant Studio 3 Real-time PCR System (Thermo Fisher Scientific, USA). Gene-specific primers were designed using the online website Primer 3 (v. 0. 4. 0) (http://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by Shanghai Generay Biotech Co., Ltd. The primer sequences are listed in Table S3. Relative RNA expression was calculated using the 2−ΔΔCt method. Gapdh or 18s was used as a candidate housekeeping gene in the skeletal muscle or adipose tissue, respectively.
Protein extraction and Western blotting analysis
Total proteins were extracted from frozen tissues or cell samples by using T-PER™ Tissue Protein Extraction Reagent (Thermo Fisher Scientific, USA) supplemented with protease and phosphatase inhibitor mini tablets (Roche, Switzerland) per 10 mL and then centrifuged for 20 min at 12,000 rpm at 4 °C to remove cell debris. Total protein concentrations were determined using a BCA protein assay reagent (Beyotime Biotechnology, China). Western blotting was performed as previously described [23]. The following primary antibodies were used: GAPDH (1:5000, Cell Signaling Technology Cat#2118, RRID: AB_561053), RNF20 (1:2000, Proteintech Cat#21625-1-AP, RRID:AB_10734436), anti-MYH7 (1:2000, Abcam Cat#ab11083, RRID: AB_297734), anti-MYH4 (1:2000, Abcam Cat#ab91506, RRID:AB_10714690), anti-CTα (1:2000, Abcam Cat#ab109263, RRID:AB_10859965), MyoD1 (1:2000, Cell Signaling Technology Cat#13812, RRID: AB_2798320), Myogenin (1:1000, Santa Cruz Biotechnology Cat#sc-12732, RRID: AB_627980), MyHC (1:1000, Santa Cruz Biotechnology Cat#sc-376157, RRID:AB_10989398) and β-Tubulin (1:2000, Cell Signaling Technology Cat#15115, RRID:AB_2798712). Horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2000, Cell Signaling Technology Cat#7074, RRID: AB_2099233) or goat anti-mouse IgG (1:5000, Cell Signaling Technology Cat#91196, RRID: AB_321928) was used as a secondary antibody, and blots were detected with Tanon™ High-sig ECL Western Blotting Substrate (Tanon Science & Technology Co., Ltd., China). The densitometric quantification of the blots was performed using Image J software (National Institute of Health, RRID:SCR_003070), and the relative expression of protein was normalized to the housekeeping protein (GAPDH or β -Tubulin).
Cell transfection and differentiation
3T3-L1 cells (RRID:CVCL_0123) were seeded in 6-well plate with culture media DMEM (Gibco, USA) containing fetal bovine serum (FBS, PAN, Germany) and 1% penicillin streptomycin (PS, Gibco, USA). To explore whether Rnf20 knockdown affect LysoPC secretion, 3T3-L1 cells were transfected with siRNF20 (siRNA specific for mouse Rnf20 gene) or siNC (siRNA for negative control) and then differentiated to mature adipocytes as described previously [24]. Cells were then treated with serum-free DMEM for 24 h. Two groups of cells were harvested for knockdown efficiency detection, and the culture media was collected for LysoPC measurement.
LysoPC measurement
Serum from 6-month-old WT and ASKO mice and the culture media that we described above were collected for LysoPC measurement by enzyme-linked immunosorbent assays (ELSA) kit (CEK621Ge; Cloud-Clone Corp., China). Briefly, 50 μL samples and 50 μL detection reagent were mixed and incubated for 1 h at 37 °C. After washing three times and adding 100 μL detection reagent B to each well, the mixture was then incubated for 30 min at 37 °C. The samples were aspirated five times, 90 μL 3,3′, 5,5′-tetramethylbenzidine (TMB) substrate solution was added, and the mixture was incubated for 15 min at 37 °C. Last, 50 μL stop solution was added to each well. Absorbance was detected immediately on SpectraMax M5 (Molecular Devices, San Jose, USA) at 450 nm. The exact LysoPC concentration of the sample was determined by a standard curve created by measuring the absorbance of a dilution series of LysoPC standards.
C2C12 cell culture and treatment
Mouse C2C12 myoblasts (RRID: CVCL_0188) were cultured in high-glucose DMEM (HyClone, USA) supplemented with 10% FBS (FBS, PAN, Germany) and 1% PS (Gibco, USA) under a 5% CO2 atmosphere at 37 °C. Upon reaching nearly confluence (80–90%), the cells were switched to differentiation medium [DMEM containing 2% horse serum (HS, Gibco, USA)] for 8 days to form mature myotubes, and fresh differentiation medium was provided every 2 days.
1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine (LysoPC (16:0), 855675P, ≥ 99%) and 1-stearoyl-2-hydroxy-sn-glycero-3-phosphocholine (LysoPC (18:0), 855775P, ≥ 99%) were purchased from Avanti Polar Lipids (Alabama, USA) and dissolved in methanol as a 50 mM stock. Lipid stocks were stored at −20 °C as single-use aliquots and used within 6 months. LysoPC (16:0) or LysoPC (18:0) was conjugated to 0.5% BSA (fatty acid-free grade, Applygen, China) dissolved in serum-free DMEM. To illuminate the mechanism of LysoPC (16:0) on muscle fiber-type transition, differentiated myotubes were cultured in different concentrations (0, 1, 5, 10, 20 and 50 μM) of the LysoPC (16:0)-0.5% BSA complex for 24 h, and vehicle-0.5% BSA was used as a control.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 software (RRID:SCR_002798, USA). Significant differences were evaluated through an unpaired two-tailed Student’s t test (P value). All data are expressed as the mean ± standard error mean (SEM). In all cases, the statistical significance of differences between groups was denoted as. P < 0.05 was considered significant. *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Increased fiber size in skeletal muscle of ASKO mice
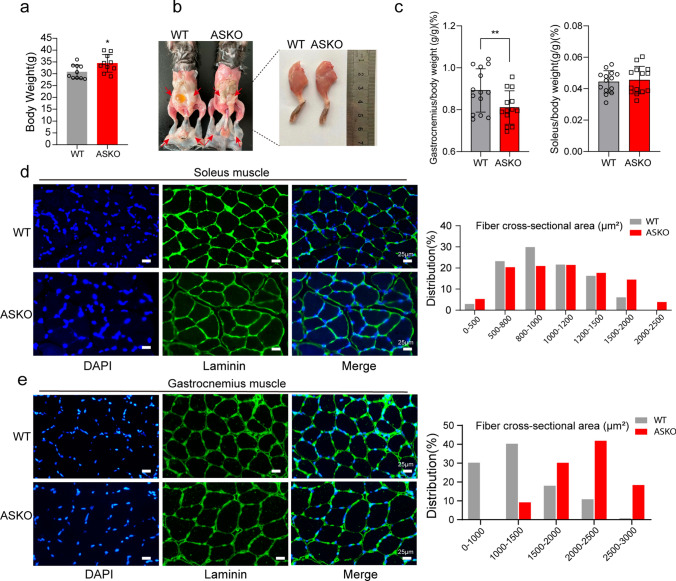
In our previous study, we found that the adipocyte-specific deletion of Rnf20 gene in mice dramatically decreased fat mass but significantly increased both body weight and lean mass [24]. It is of interest to investigate how Rnf20 ablation in adipose tissue affects the physiology and metabolism of skeletal muscle. First, we checked the expression levels of Rnf20 in the soleus and gastrocnemius muscles from both WT and ASKO mice, and the unchanged expression of Rnf20 at both the RNA and protein levels excluded the RNF20 leakage effect on muscle (Fig. S1). Certainly, as we observed before, the body weight of ASKO mice was significantly larger than that of littermate controls (Fig. 1a, + 11.62%, P < 0.05). Figure 1b shows a representative photo of WT and ASKO mice. Notably, the leg was larger in the ASKO mice. In addition, we found that the relative weight of the gastrocnemius muscle to body weight in the 6-month-old ASKO mice was significantly higher than that in control mice (Fig. 1c, left panel, + 9.53%, P < 0.01), while no significant differences were observed in the soleus muscle (Fig. 1c, right panel). Next, we investigated whether the muscle fiber size was changed in ASKO mice by laminin immunofluorescence staining. Our data revealed enlarged muscle fibers in both the soleus (Fig. 1d, left panel) and gastrocnemius muscles (Fig. 1e, left panel) from ASKO mice. The distribution of muscle fiber sizes of 6-month-old WT and ASKO mice was further quantitatively calculated by at least 1000 fibers, and the proportion of larger muscle fibers (> 1500 μm2) was increased in both the soleus and gastrocnemius muscles of ASKO mice (Fig. 1d, right panel; Fig. 1e, right panel). In particular, the changes in fiber size were mild for the soleus muscle but stronger in the gastrocnemius muscle from ASKO mice. Together, these results suggest that the ablation of Rnf20 in adipose tissue altered skeletal muscle fiber size and thereby potentially affected metabolic characteristics.
Fig. 1.
The muscle mass and muscle fiber size were significantly increased in ASKO mice. a The body weight of ASKO mice was significantly heavier than that of control littermates (n = 10 mice/group). b Representative photographs of the body (left panel) and whole left leg muscles (right panel). c Relative skeletal muscle weights of 6-month-old WT and ASKO mice (n = 14 mice/group). All data were normalized to the body weight (g/g) × 100%. d Representative images (left panel) and quantification (right panel) of laminin immunochemistry staining showing the fiber size in the soleus muscle from 6-month-old WT and ASKO mice (n = 5 mice/group). e Representative images (left panel) and quantification (right panel) of laminin immunochemistry staining showing the size of fibers in the gastrocnemius muscle from 6-month-old WT and ASKO mice (n = 5 mice/group). The cross-sectional areas of 1200 muscle fibers were measured by Image J software. The results are presented as the mean ± SEM. Scale bar = 25 μM. *P < 0.05 and **P < 0.01
Slow-twitch to fast-twitch switching of skeletal muscle fibers in ASKO mice
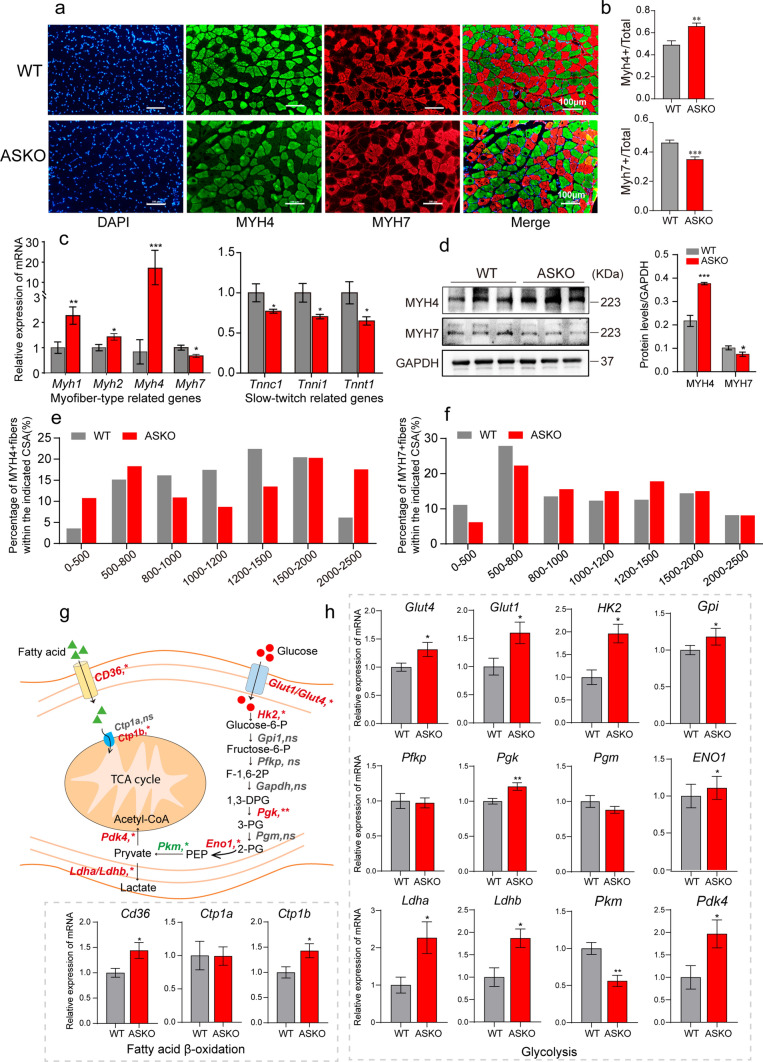
Skeletal muscle fibers can be classified as slow-twitch and fast-twitch types based on the twitch speed and primary ATP production pathway [3]. Slow-twitch fibers are oxidative and express myosin heavy chain 7 (MYH7), while fast-twitch fibers are glycolytic and express myosin heavy chain 4 (MYH4). To test whether the skeletal muscle fiber types were changed in ASKO mice, we stained the myofiber-specific markers (MYH4 and MYH7) in skeletal muscle. In soleus muscle, the immunofluorescence data demonstrated increased MYH4 staining (+ 34.69% for MYH4 + fibers, P < 0.01) and decreased MYH7 staining (−24.61% for MYH7 + fibers, P < 0.001) in ASKO mice (Fig. 2a). Quantitative data of the images from biological replicates confirmed this finding (Fig. 2b). This observation was further confirmed by the significantly decreased expression level of Myh7 and the significantly increased expression level of Myh4 at both the RNA (Fig. 2c, left panel) and protein levels (Fig. 2d). The RNA levels of the other two fast-twitch fiber-related genes, Myh1 and Myh2, were also observed to be significantly increased in soleus of ASKO mice (Fig. 2c, left panel). The expression levels of the slow-twitch troponin genes (Tnnc1, Tnni1 and Tnnt1) were also observed to be significantly decreased in soleus muscle of ASKO mice (Fig. 2c, right panel). The proportion of larger MYH4 + fibers (> 2500 μM2) was increased in soleus muscle of ASKO mice (Fig. 2e), while the size of MYH7 + fibers were not changed significantly in soleus muscle of both mice (Fig. 2f). Our data indicated the increased in both CSA and the number of fast-twitch muscle fibers.
Fig. 2.
The fiber types of skeletal muscles are transformed in ASKO mice. a, b Representative images (a) and quantification (b) of MYH4 and MYH7 staining in the soleus muscle (n = 5 mice/group). Scale bar = 100 μm. Blue: DAPI; Green: MYH4; Red: MYH7. c The expression levels of fiber type-related genes (Myh1, Myh2, Myh4 and Myh7) and slow-twitch troponin related genes (Tnnc1, Tnni1 and Tnnt1) were detected by qPCR in the soleus muscle from 6-month-old mice (n = 6 mice/group). d Immunoblots (left panel) and quantification (right panel) of MYH4 and MYH7 in the soleus show the increased MYH4 but decreased MYH7. GAPDH was used as a loading control. e, f Comparison of the percentage of MYH4 + (e) and MYH7 + (f) fibers in the indicated cross-sectional area in soleus muscle of WT and ASKO mice. g Schematic diagram of the processes of fatty acid oxidation and glycolysis in the skeletal muscle. h The mRNA expression levels of genes in (g) were detected by qPCR in the soleus from WT and ASKO mice (n = 6 mice/group). Values are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001
Skeletal muscle fiber-type switching is closely related to lipid and glucose metabolism. In slow-twitch fibers, energy is provided mainly by mitochondrial oxidative metabolism, while in fast-twitch fibers there is a greater contribution of glycolysis to ATP formation [26]. We further examined the expression levels of genes involved in glycolysis and fatty acid oxidation metabolism. We found that the genes involved in glycolysis (Glut1, Glut4, Hk2, Eno1 and Pgk) were significantly increased in the soleus muscle from ASKO mice (Fig. 2g; h), suggesting the enhanced glycolysis metabolic processes. In addition, parallel experiments were performed in the gastrocnemius muscle of the two groups of mice. We found that the level of MYH7 was also decreased in the mixed gastrocnemius muscle of ASKO mice (Fig. S2a; S2b). The proportion of larger MYH4 + and MYH7 + fibers (> 1500 μM2) was increased in gastrocnemius muscle of ASKO mice (Fig. S2c, S2d). However, the expression levels of MyHC-specific genes and glycolytic genes were not significantly changed in the gastrocnemius muscle of the two groups (Fig. S2e; S2f). Overall, these results indicated that ablation of the Rnf20 gene in adipose tissue leads to increased fast-twitch muscle fibers but decreased slow-twitch fibers in skeletal muscle.
LysoPCs were significantly increased in serum of ASKO mice
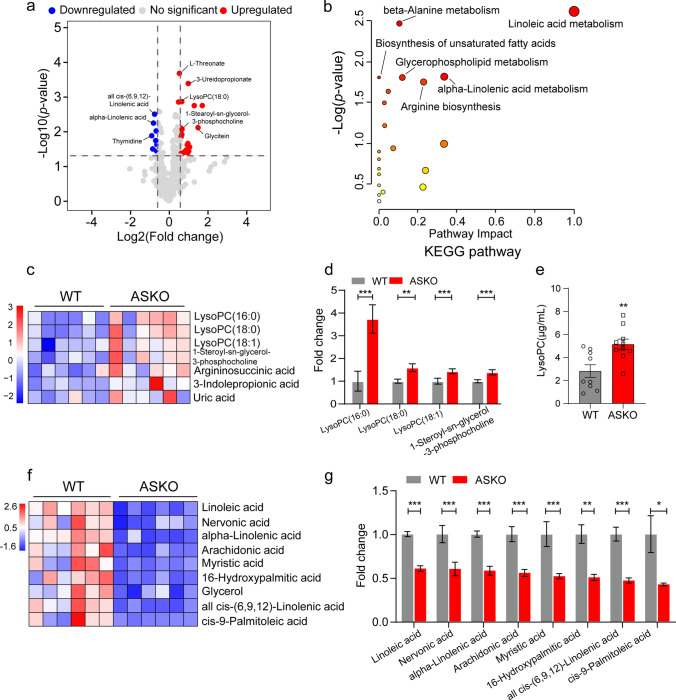
We speculate that the crosstalk between adipose tissue and skeletal muscle might be mediated by a molecule produced by fat tissues and secreted into the bloodstream. Thus, untargeted metabolomic analysis of the serum samples from 6-month-old WT and ASKO mice was performed, and a total of 484 metabolites were identified in distinct metabolic profiles (Table S1). PCA clearly distinguished two clusters of serum from both groups (Fig. S3). To obtain a broad overview of changes in metabolite profiles, volcano plots were built based on the P value and FC of all identified metabolites. With the criteria P < 0.05 and FC > 1.5, 40 differential metabolites were identified, of which 21 were significantly increased and 19 were markedly decreased in the circulation of ASKO mice (Fig. 3a). Pathway enrichment analysis of differential metabolites highlighted several metabolic pathways, in particular, linoleic acid metabolism, glycerophospholipid metabolism, alpha-linolenic acid metabolism and biosynthesis of unsaturated fatty acids (Fig. 3b), which are critical pathways for lipid metabolism. Notably, the significantly increased levels of LysoPC (16:0) (+ 273.7%, P < 0.001), LysoPC (18:0) (+ 59.8%, P < 0.01), LysoPC (18:1) (+ 43.8%, P < 0.001), and 1-stearoyl-sn-glycerol 3-phosphocholine (+ 39.3%, P < 0.001) were found in the serum from ASKO mice compared with those from WT mice (Fig. 3c, d). The significant increased serum LysoPC level in ASKO mice was further confirmed by ELISA analysis (Fig. 3e). In addition, heatmap analysis also showed noticeably decreased levels of polyunsaturated fatty acids (PUFAs), including linoleic acid (−39.5%, P < 0.001), nervonic acid (−40.0%, P < 0.001), alpha-linolenic acid (−42.0%, P < 0.001), arachidonic acid (−44.5%, P < 0.001), myristic acid (−48.3%, P < 0.001), 16-hydroxypalmitic acid (−49.7%, P < 0.01), all cis-(6,9,12)-linolenic acid (−53.3%, P < 0.001) and cis-9-palmitoleic acid (−57.6%, P < 0.05), in the serum of ASKO mice (Fig. 3f; 3g). Mechanistically, this decrease might be the result of an almost 50% reduction in the expression of stearoyl-CoA desaturase-1 (SCD1) in gWAT of ASKO mice (Fig. S4), which is the main enzyme responsible for desaturation of palmitic and stearic acid. Taken together, our data showed that loss of Rnf20 in adipose tissue altered the serum metabolite profiles in mice.
Fig. 3.
A significantly increased LysoPCs were found in serum of ASKO mice. a Volcano plots were built based on the P value and FC for overall metabolites, which were measured by metabolomics in serum of 6-month-old WT and ASKO mice (n = 6 mice/group). With the criteria of P < 0.05 and |FC|> 1.5, 21 metabolites (red dots) were significantly increased, and 19 metabolites (blue dots) were markedly decreased. Gray dots represent the no significant differences in metabolites. b KEGG pathway analysis of the differential metabolites in serum of the two groups of mice. c Heatmap indicating several upregulated LysoPCs in serum of ASKO mice. d The intensity of the upregulated LysoPCs in serum of WT and ASKO. e The concentration of serum LysoPC in two groups was detected by ELISA (n = 10 mice/group). f Heatmap indicating several downregulated PUFAs in serum of ASKO mice. g The intensity of downregulated PUFAs. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001
PCs and LysoPCs were highly induced in gWAT of ASKO mice
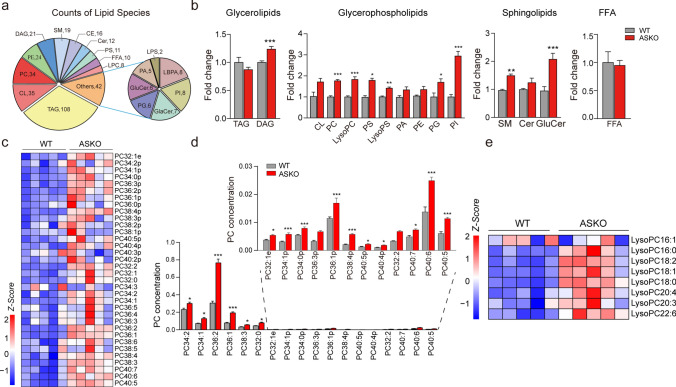
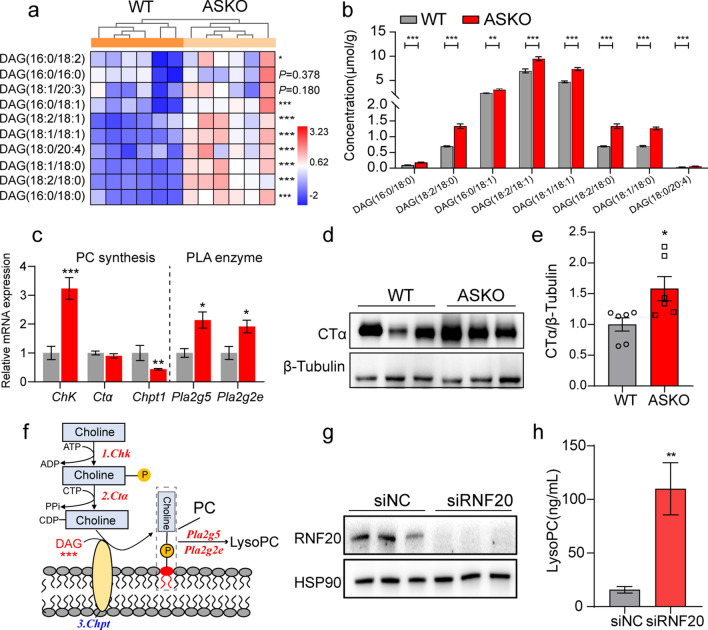
We speculate that the increased serum LysoPCs in ASKO mice might be derived from fat tissue. To elucidate the effects of Rnf20 gene ablation on the overall lipid composition in adipose tissue, gWAT from WT and ASKO mice were collected and subjected to mass spectrometry-based lipidomic analysis (Table S2). The detected lipid classes and their abbreviations are shown in Table S4. We examined over 361 different lipid species in the gWAT, consisting of 108 TAGs, 21 DAGs, 34 PCs, 24 PEs, 35 CLs and other lipid classes (Fig. 4a). PCA showed distinct clusters of gWAT lipidomic data from WT and ASKO mice (Fig. S5). Using a P value of 0.05 as a cutoff, a total of 109 species were identified as significantly altered in the gWAT from ASKO mice compared with control mice. Relative quantification results showed that adipocyte-specific Rnf20 disruption in mice significantly increased the contents of diglyceride (DAG) (+ 23.7% for total DAG content, P < 0.001) in glycerolipids; phosphatidylcholine (PC) (+ 76.7%, P < 0.001), lysophosphatidylcholine (LysoPC) (+ 83.1%, P < 0.001), phosphatidylserine (PS) (+ 81.4%, P < 0.05), lysophosphatidylserine (LysoPS) (+ 41.1%, P < 0.01), phosphatidylinositol (PI) (+ 194.6%, P < 0.001), and phosphatidylglycerol (PG) (+ 69.8%, P < 0.05) in glycerophospholipids; and sphingomyelin (SM) (+ 54.4%, P < 0.01), ceramides (Cer) (+ 23.9%, P > 0.05) and glucosylceramide (GluCer) (+ 100.0%, P < 0.001) in sphingolipids (Fig. 4b). These findings suggest that the depletion of Rnf20 induces considerable alterations in the content of lipid species in fat tissue.
Fig. 4.
PCs and LysoPCs were significantly increased in the gWAT of ASKO mice. a Distribution of lipid classes considered for subsequent analysis in all samples detected by lipidomic. b The fold change of glycerolipids, glycerophospholipids, sphingolipids and free fatty acids in gWAT (n = 8 mice/group). c Heatmap was built with the upregulated PCs (long-chain fatty acids C32-C40). d The concentrations of PCs in gWAT of 6-month-old WT and ASKO mice. e Heatmap was constructed from the increased LysoPCs associated with different acyl chains. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001
As increased LysoPCs were observed in the serum of ASKO mice, considering the close anatomical and functional relationship between blood vessels and fat tissue, we profiled the PCs and LysoPCs in the gWAT of two groups of mice. Eighteen PC species with long-chain fatty acids (C32–C40) in adipose tissue clearly exhibited increased levels in the gWAT of ASKO mice (Fig. 4c; d). In addition, most LysoPCs with different chains were found to be significantly induced in the gWAT of ASKO mice, except for LysoPC (16:1) and LysoPC (20:3) (Fig. 4e). In summary, the Rnf20 gene influences the PC and LysoPC profiles of white adipose tissue.
PC and LysoPC biosynthesis-related genes were upregulated in gWAT of ASKO mice
The increased concentrations of PCs and LysoPCs in Rnf20−/− fat tissue suggest that the PC and LysoPC biosynthesis pathways may be enhanced in ASKO mice. It has been reported that de novo PC biosynthesis relies on two substrates, DAG and cytidine diphosphate-choline [27]. As we described above, the total level of DAG was significantly elevated in gWAT of ASKO mice (Fig. 4b, left panel). Thus, we profiled the significantly elevated DAG species based on lipidomic data of gWAT from both mice, including DAG (16:0/18:0), DAG (18:2/18:0), DAG (16:0/18:1), DAG (18:2/18:1), DAG (18:1/18:1), DAG (18:1/18:0) and DAG (18:0/20:4) (Fig. 5a, b). In addition, we also measured the expression levels of choline kinase (Chk), phosphocholine cytidylyltransferase α (Ctα), choline phosphotransferase (Chpt) and phospholipase A2 (Pla2g2e and Pla2g5) in gWAT from both mice. Our data revealed the significantly elevated expression of Chk, Pla2g2e, and Pla2g5 in gWAT of ASKO mice (Fig. 5c). Although Ctα was not altered at the RNA level (Fig. 5c), its protein level was increased in gWAT of ASKO mice (Fig. 5d, e). These results indicate that the increased PC and LysoPC in gWAT of Rnf20−/− mice appears to be due to the accumulated DAG content and the activated PC and LysoPC biosynthesis-related genes. The diagram figure presented the PC and LysoPC biosynthesis pathway for ASKO mice (Fig. 5f).
Fig. 5.
Significantly increased DAG content and PC synthesis-related genes were observed in gWAT of ASKO mice. a Heatmap of the extensively changed DAGs in gWAT from WT and ASKO mice (n = 6 mice/group). b The concentration of the increased DAGs is shown. c The mRNA expression levels of PC synthesis-related genes and phospholipase A2 genes in gWAT of two groups of mice (n = 6 mice/group). 18s was used as an internal control. d Western blotting analysis of CTα protein level in gWAT from 6-month-old WT and ASKO mice (n = 3 mice/group). e The relative protein level of CTα was normalized to β-Tubulin. f Schematic representation of the process of PC and LysoPC synthesis. The red words display the upregulated genes, and the blue words display the downregulated genes in gWAT of ASKO mice. g Western blotting analysis of RNF20 protein level in siNC- and siRNF20-transfected mature adipocytes. h The level of LysoPC in culture media derived from siNC and siRNF20 cells was detected by ELISA (n = 6 samples/group). Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001
To prove that the increase in serum LysoPC in ASKO mice is due to adipose tissue-derived LysoPC, the in vitro experiment was performed. In detail, 3T3-L1 cells were transfected with siRNF20 or siNC and were differentiated into mature adipocytes. Both cells were then treated with serum-free media for 24 h. Cells were collected for knockdown efficiency detection and the culture media was collected for LysoPC measurement. Our data revealed the decreased protein level of RNF20 (Fig. 5g) and the significant increased LysoPC in siRNF20 group (Fig. 5h). This data indicated that adipose tissue-derived LysoPC may contribute to the elevation of the serum LysoPC in ASKO mice.
LysoPC (16:0) increased the expression of fast fiber-related genes in C2C12 myotubes
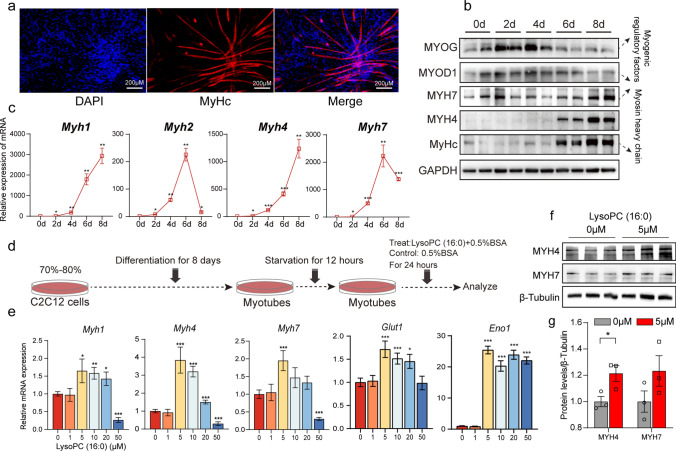
Extensively increased LysoPC (16:0) and LysoPC (18:0) were found in both circulating and fat tissue of ASKO mice, and we speculated that LysoPC (16:0), LysoPC (18:0), or both might participate in the connection of adipose tissue and skeletal muscle. To test whether fiber switching was regulated in a LysoPC (16:0)/(18:0)-dependent manner, C2C12 cells were differentiated in vitro and then treated with LysoPC (16:0) or LysoPC (18:0). The successful myogenic differentiation of C2C12 myoblasts was proven by MyHC immunofluorescence staining (Fig. 6a) and the significantly induced myogenesis markers MYOG and MYOD1 at the protein level (Fig. 6b). The expression levels of myosin heavy chain genes were also robustly elevated during differentiation at both the mRNA and protein levels (Fig. 6c; b). Next, the differentiated cells were starved for 12 h and incubated with different concentrations of LysoPC (16:0) for 24 h (Fig. 6d). We found that LysoPC (16:0) treatment significantly increased the expression levels of fast-specific MyHC (Myh1 and Myh4) and glycolysis genes (Glut1 and Eno1), and 5 μM LysoPC (16:0)-0.5% BSA treatment achieved the best effect (Fig. 6e). However, only 5 μM LysoPC (16:0) showed a slight increase in Myh7 gene expression (Fig. 6e). The protein level of MYH4 but not MYH7 was significantly increased in cells treated with 5 μM LysoPC (16:0) (Fig. 6f, g), but not with LysoPC (18:0) (Fig. S6). Taken together, these results demonstrated that LysoPC (16:0) might be the mediator that induces the expression of fast fiber-type genes and leads to the fiber-type switching in the skeletal muscle of ASKO mice.
Fig. 6.
LysoPC (16:0) promotes the expression of fast-twitch muscle fiber-related genes in C2C12 myotubes. a MyHC staining of C2C12 nucleic myotubes at day 8 after differentiation. Fused myotubes were positive for MyHC (red), and cell nuclei were positive for DAPI (blue). b Western blotting analysis of myogenic regulatory factors (MYOG and MYOD1) and myosin heavy chain (MYH7, MYH4 and MyHC) during C2C12 myoblast differentiation (0, 2, 4, 6, and 8 days after the induction of differentiation). GAPDH was used as a protein loading control. c The expression levels of myosin heavy chain-related genes (Myh1, Myh2, Myh4 and Myh7) during C2C12 myoblast differentiation. d Schematic diagram of the effect of LysoPC (16:0) on skeletal muscle fiber remodeling. Myotubes were incubated with various concentrations of LysoPC (16:0)-0.5% BSA (treat groups) and vehicle-0.5% BSA (control group) for 24 h. e qPCR analyses of myosin heavy chain-related genes (Myh1, Myh4 and Myh7) and glycolysis-related genes (Glut1 and Eno1) (n = 6). f Western blotting of MYH4 and MYH7 in the treat and control groups. g The relative protein levels were normalized to β-Tubulin. Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001
Discussion
This study demonstrates the effects of adipocyte Rnf20 on the alteration of skeletal muscle growth and metabolism. Previously, we found the significantly increased lean mass in adipocyte-specific Rnf20 knockout mice [24]. Here, we showed that the heavier muscle mass in ASKO mice is partially due to the larger muscle fiber size (Fig. 1). Therefore, we examined several critical genes which have been shown to stimulate skeletal muscle growth, including Myod, Myog, Mstn, Igf2, Bmp4, Murf1, Igf2r and Zbed6. Our data showed that only Myog was significantly induced in the skeletal muscle of ASKO mice (Fig. S7), suggesting its potential role in skeletal muscle hypertrophy of ASKO mice. It has been reported that the AKT signaling pathway plays a critical role in the regulation of skeletal muscle growth. For example, overexpression of constitutively active forms of protein AKT1 induced muscle hypertrophy in mice [28]. Enhanced phosphorylation of AKT (p-AKT) could promote protein synthesis and inhibit protein degradation in skeletal muscle [28, 29]. Thus, we detected p-AKT in skeletal muscle from both mice and found that the level of p-AKT was significantly inhibited in the skeletal muscle of ASKO mice (Fig. S8), excluding the alteration of muscle hypertrophy via the AKT signaling pathway. In addition, p-AKT was also an indicator signaling of insulin response, thus the decreased p-AKT in skeletal muscle might explain the slight decreased insulin sensitivity phenotype that we reported previously [24].
It has been reported that altered fiber size potentially affects metabolic characteristics and that fast-twitch fibers have a larger fiber size than slow-twitch fibers [30]. Our results showed that the ASKO mice exhibited an increased fast-twitch fiber percentage and a decreased slow-twitch fiber percentage (Fig. 2), implying that the increased muscle fiber size might be caused by the remodeling of muscle fibers. Activation of ERK1/2 signaling induced upregulation of the fast-twitch fiber program in soleus muscle [31]; in contrast, the activation of AMPK promoted the formation of slow-twitch fibers [32]. Here, we also tried to explore whether these pathways are involved in fiber type switching in ASKO mice. Our data demonstrated that the levels of ERK1/2 and p-ERK1/2 were not significantly changed in the soleus muscle of ASKO mice, but the protein AMPK was significantly decreased (Fig. S8). These data indicated that the decreased slow-twitch fibers in ASKO mice might be caused by the inactivation of AMPK. Together, the detailed mechanisms underlying the regulation of muscle fiber size and type by adipocyte Rnf20 ablation need to be further investigated.
In general, adipose tissue has been recognized as a major endocrine organ that produces, releases and conveys biological signals (especially adipokines) to regulate whole-body metabolic homeostasis via communication with other metabolically active organs [33]. The adipokines, such as adiponectin and leptin, were observed to be decreased in serum of ASKO mice [24], and consistently, their decreased mRNA expression levels were also observed in skeletal muscle (Fig. S9), suggesting the small amount of organ signaling by adipokines. Indeed, lipids released by adipocytes can act as messenger molecules to mediate organ crosstalk. For example, LPA, which is secreted from adipocytes, has been shown to antagonize insulin signaling and inhibit mitochondrial respiration in muscle tissues and C2C12 cells [34]. The monounsaturated fatty acid palmitoleate is also an adipose-secreted lipid metabolite that improves systemic insulin sensitivity by acting on both liver and muscle [35]. The lipid 12,13-dihydroxy-9Z-octadecenoic acid, which is secreted by brown fat tissues, exhibits metabolically beneficial bioactivates by promoting fatty acid uptake in skeletal muscle [36]. N-acyl-amino acids exhibited autocrine/paracrine and endocrine activities as direct stimulators of cellular respiration in non-adipocytes such as C2C12 muscle cells [37].
Here, combination data of serum metabolomic analysis and gWAT lipidomic analysis revealed that adipocyte Rnf20 depletion significantly increased the level of LysoPC (16:0) in serum and gWAT, indicating its potential role in mediating the conversion of both organs. Consistent with our observation, significantly lower serum PC and LysoPC levels were found in obese mice than in lean mice [38]. Indeed, LysoPCs, which are released by adipose tissue, have been reported to act as messenger molecules that reduce skeletal muscle contractile force-generating capacity [39]. It also revealed a robust positive correlation between serum LysoPC-acyl C16:0 levels and brown adipose tissue (BAT) activity [40]. Our data revealed that LysoPC (16:0) treatment increased the expression of fast-specific MyHC genes (Myh1 and Myh4) in C2C12 myotubes, implying that LysoPC (16:0) promoted the formation of fast-twitch fibers in vitro.
Our previous study showed that BAT of ASKO mice displayed a significantly decreased total content of PCs and LysoPCs [24], together with increased PCs and LysoPCs in gWAT, indicating the distinct roles of Rnf20 in different fat depots. Adipose tissue and liver are the major organs for PC and LysoPC biogenesis [41, 42] and the unchanged LysoPC level in liver of ASKO mice (Fig. S10) suggested that the increased serum LysoPC level was probably affected by adipose tissue. Our in vitro data revealed that RNF20 knockdown in 3T3 cells could induce the secretion of LysoPC, suggesting that adipose tissue-derived LysoPC may contribute to the elevation of the serum LysoPC in ASKO mice.
In addition, we investigated the molecular mechanisms of increased DAG levels in gWAT of ASKO mice. Surprisingly, all DAG biogenesis related genes that we examined, including Mgat1, Dgat1, Mgll, Atgl, Pnpla3 and Hsl, were dramatically suppressed in gWAT of ASKO mice (Fig. S11), suggesting the impaired DAG metabolism was occurred in ASKO mice. It has been reported that tissue levels of DAG were not increased in Dgat1−/− mice, although DGAT1 converts DAG to TAG [43]. However, another in vitro study showed that Dgat1−/− cells exhibited the elevated DAG and phospholipid metabolites [44], suggesting that the decreased Dgat1 may contribute to increased DAG in our study. For Mgat1, it has been reported that Mgat1 inhibition also increased both membrane and cytosolic compartment DAG levels [45] and our data is consistent with this trend. In addition, stable overexpression of Atgl resulted in DAG accumulation in myotubes [46]. Taken together, we speculated that the decrease of Mgat1, Dgat1 and Hsl might partially explain the increase of DAG level in the gWAT of ASKO mice. However, the detailed molecular mechanisms by which knockout of Rnf20 decreases the expression of these genes remain unclear and need to be further explored.
In summary, our data reveal a previously unappreciated role of adipocyte Rnf20 expression in the regulation of the fiber type switching of skeletal muscle via LysoPC (16:0)-mediated adipose-muscle dialog (Fig. 7). This study not only identifies a new messenger molecule for regulating the metabolism of skeletal muscle, but also provides novel evidence for a better understanding of adipose tissue dysfunction-induced physiological alterations in skeletal muscle.
Fig. 7.
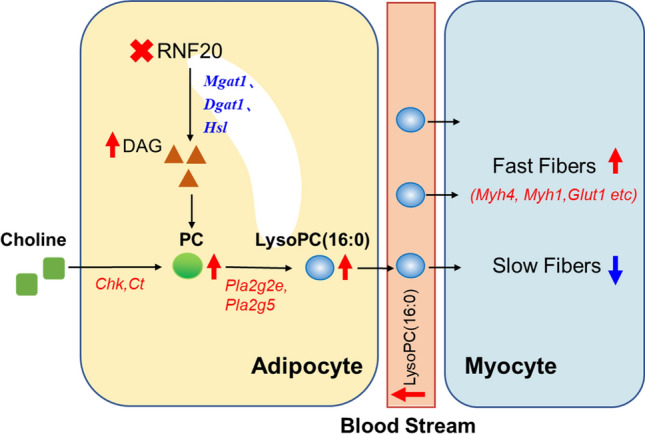
LysoPC (16:0) mediated the crosstalk between adipose tissue and skeletal muscle in the mice with the adipocyte-specific deletion of Rnf20 gene.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank our lab members for critical reading of the manuscript and helpful discussions.
Abbreviations
- RNF20
Ring finger 20
- WT mice
Wild-type mice
- ASKO mice
Adipocyte-specific Rnf20 knockout mice
- ERK1/2
Extracellular regulated protein kinase 1/2
- GLUT4
Glucose transporter type 4
- IRS-1
Insulin receptor substrate-1
- SREBP1c
Sterol regulatory element binding protein 1c
- NCoR1
Nuclear receptor corepressor 1
- FFA
Free fatty acid
- LPA
Lysophosphatidic acid
- gWAT
Gonadal white adipose tissue
- PBST
Phosphate buffered solution
- BSA
Bovine serum albumin
- CSA
Cross-sectional area
- TAG
Triacylglycerol
- DAG
Diacylglycerol
- CE
Cholesteryl esters
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PS
Phosphatidylserine
- LysoPC
Lysophosphatidylcholine
- LysoPE
Lysophosphatidylethanolamine
- LysoPS
Lysophosphatidylserine
- SM
Sphingomyelin
- qRT-PCR
Quantitative reverse transcription PCR
- FBS
Fetal bovine serum
- PCA
Principal component analysis
- FC
Fold change
- PUFA
Polyunsaturated fatty acids
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- SCD-1
Stearoyl-CoA desaturase-1
- PLA2
Phospholipase A2
- Chk
Choline kinase
- Ctα
Phosphocholine cytidylyltransferase α
- Chpt
Choline phosphotransferase
Author contributions
All authors contributed to the study conception and design. The project was designed by YW, JZ and NY. Material preparation, data collection and analysis were performed by YZ, CC, JP, SY, TW and CT. Lipid measurement were performed by GS and SML. The first draft of the manuscript was written by YZ and YW, and all authors commented on previous versions of manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by Lingnan Modern Agriculture Project (NT2021005), the National Key R & D Program of China (2020YFA0509500 and 2021YFA0805903), National Natural Science Fundation for Distinguished Young Scholars (32025034 and 31925036) and the Agricultural Science and Technology Innovation Program (ASTIP).
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was performed in line with the principles of Animal Research Panel of the Committee on Research Practice. Approval was granted by Animal Ethics Committee of the Institute of Animal Science (No. IOZ20190077).
Consent for publication
Human experiments are not involved in this article.
Consent to participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianguo Zhao, Email: zhaojg@ioz.ac.cn.
Yanfang Wang, Email: wangyanfang@caas.cn.
References
- 1.Relaix F, Bencze M, Borok MJ, Der Vartanian A, Gattazzo F, Mademtzoglou D, Perez-Diaz S, Prola A, Reyes-Fernandez PC, Rotini A, Taglietti T. Perspectives on skeletal muscle stem cells. Nat Commun. 2021;121:692. doi: 10.1038/s41467-020-20760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaauw B, Schiaffino S, Reggiani C. Mechanisms modulating skeletal muscle phenotype. Compr Physiol. 2013;34:1645–1687. doi: 10.1002/cphy.c130009. [DOI] [PubMed] [Google Scholar]
- 3.Zierath JR, Hawley JA. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2004;210:e348. doi: 10.1371/journal.pbio.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;914:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Xu YQ, Yuan YX, Xu PW, Zhang C, Li F, Wang LN, Yin C, Zhang L, Cai XC, Zhu CJ, Xu JR, Liang BQ, Schaul S, Xie PP, Yue D, Liao ZR, Yu LL, Luo L, Zhou G, Yang JP, He ZH, Du M, Zhou YP, Deng BC, Wang SB, Gao P, Zhu XT, Xi QY, Zhang YL, Shu G, Jiang QY. Succinate induces skeletal muscle fiber remodeling via Suncr1 signaling. EMBO Rep. 2019;209:e47892. doi: 10.15252/embr.201947892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol. 2012;88:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 7.Ukropec J, Ukropcova B, Kurdiova T, Gasperikova D, Klimes I. Adipose tissue and skeletal muscle plasticity modulates metabolic health. Arch Physiol Biochem. 2008;1145:357–368. doi: 10.1080/13813450802535812. [DOI] [PubMed] [Google Scholar]
- 8.Fan HQ, Gu N, Liu F, Fei L, Pan XQ, Guo M, Chen RH, Guo XR. Prolonged exposure to resistin inhibits glucose uptake in rat skeletal muscles. Acta Pharmacol Sin. 2007;283:410–416. doi: 10.1111/j.1745-7254.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- 9.Muoio DM, Dohm GL, Fiedorek FT, Jr, Tapscott EB, Coleman RA. Leptin directly alters lipid partitioning in skeletal muscle. Diabetes. 1997;468:1360–1363. doi: 10.2337/diab.46.8.1360. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, Eto K, Akanuma Y, Froguel P, Foufelle F, Ferre P, Carling D, Kimura S, Nagai R, Kahn BB, Kadowaki T. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating amp-activated protein kinase. Nat Med. 2002;811:1288–1295. doi: 10.1038/nm788. [DOI] [PubMed] [Google Scholar]
- 11.Fong Y, Moldawer LL, Marano M, Wei H, Barber A, Manogue K, Tracey KJ, Kuo G, Fischman DA, Cerami A, et al. Cachectin/Tnf or Il-1 alpha induces cachexia with redistribution of body proteins. Am J Physiol. 1989;2563(Pt 2):R659–R665. doi: 10.1152/ajpregu.1989.256.3.R659. [DOI] [PubMed] [Google Scholar]
- 12.Funcke JB, Scherer PE. Beyond adiponectin and leptin: adipose tissue-derived mediators of inter-organ communication. J Lipid Res. 2019;6010:1648–1684. doi: 10.1194/jlr.R094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, Du J, Hu Z, Walsh K, Wang XH. Evidence for adipose-muscle cross talk: opposing regulation of muscle proteolysis by adiponectin and fatty acids. Endocrinology. 2007;14812:5696–5705. doi: 10.1210/en.2007-0183. [DOI] [PubMed] [Google Scholar]
- 14.Rancoule C, Attane C, Gres S, Fournel A, Dusaulcy R, Bertrand C, Vinel C, Treguer K, Prentki M, Valet P, Saulnier-Blache JS. Lysophosphatidic acid impairs glucose homeostasis and inhibits insulin secretion in high-fat diet obese mice. Diabetologia. 2013;566:1394–1402. doi: 10.1007/s00125-013-2891-3. [DOI] [PubMed] [Google Scholar]
- 15.Samad F, Hester KD, Yang G, Hannun YA, Bielawski J. Altered adipose and plasma sphingolipid metabolism in obesity: a potential mechanism for cardiovascular and metabolic risk. Diabetes. 2006;559:2579–2587. doi: 10.2337/db06-0330. [DOI] [PubMed] [Google Scholar]
- 16.Vantyghem MC, Pigny P, Maurage CA, Rouaix-Emery N, Stojkovic T, Cuisset JM, Millaire A, Lascols O, Vermersch P, Wemeau JL, Capeau J, Vigouroux C. Patients with familial partial lipodystrophy of the dunnigan type due to a Lmna R482w mutation show muscular and cardiac abnormalities. J Clin Endocrinol Metab. 2004;8911:5337–5346. doi: 10.1210/jc.2003-031658. [DOI] [PubMed] [Google Scholar]
- 17.Sellers RS, Mahmood SR, Perumal GS, Macaluso FP, Kurland IJ. Phenotypic modulation of skeletal muscle fibers in Lpin1-deficient lipodystrophic ( Fld) mice. Vet Pathol. 2019;562:322–331. doi: 10.1177/0300985818809126. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Zhou H, Xuan H, Saha P, Wang G, Chen W. Novel metabolic disorders in skeletal muscle of lipodystrophic Bscl2/seipin deficient mice. Mol Cell Endocrinol. 2019;482:1–10. doi: 10.1016/j.mce.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs G, Oren M. Writing and reading H2b monoubiquitylation. Biochim Biophys Acta. 2014;18398:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Ren P, Sheng Z, Wang Y, Yi X, Zhou Q, Zhou J, Xiang S, Hu X, Zhang J. Rnf20 promotes the polyubiquitination and proteasome-dependent degradation of Ap-2alpha protein. Acta Biochim Biophys Sin (Shanghai) 2014;462:136–140. doi: 10.1093/abbs/gmt136. [DOI] [PubMed] [Google Scholar]
- 21.Lee JH, Jeon YG, Lee KH, Lee HW, Park J, Jang H, Kang M, Lee HS, Cho HJ, Nam DH, Kwak C, Kim JB. Rnf20 suppresses tumorigenesis by inhibiting the Srebp1c-Pttg1 axis in kidney cancer. Mol Cell Biol. 2017 doi: 10.1128/MCB.00265-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon YG, Lee JH, Ji Y, Sohn JH, Lee D, Kim DW, Yoon SG, Shin KC, Park J, Seong JK, Cho JY, Choe SS, Kim JB. Rnf20 functions as a transcriptional coactivator for ppargamma by promoting Ncor1 degradation in adipocytes. Diabetes. 2020;691:20–34. doi: 10.2337/db19-0508. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Pan J, Cao C, Liang X, Yang S, Liu L, Tao C, Zhao J, Wang Y. Rnf20 affects porcine adipocyte differentiation via regulation of mitotic clonal expansion. Cell Prolif. 2021;5412:e13131. doi: 10.1111/cpr.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang X, Tao C, Pan J, Zhang L, Liu L, Zhao Y, Fan Y, Cao C, Liu J, Zhang J, Lam SM, Shui G, Jin W, Li W, Zhao J, Li K, Wang Y. Rnf20 deficiency in adipocyte impairs adipose tissue development and thermogenesis. Protein Cell. 2021;126:475–492. doi: 10.1007/s13238-020-00770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan J, Tao C, Cao C, Zheng Q, Lam SM, Shui G, Liu X, Li K, Zhao J, Wang Y. Adipose lipidomics and Rna-Seq analysis revealed the enhanced mitochondrial function in Ucp1 knock-in pigs. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;186410:1375–1383. doi: 10.1016/j.bbalip.2019.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Pereyra AS, Lin CT, Sanchez DM, Laskin J, Spangenburg EE, Neufer PD, Fisher-Wellman K, Ellis JM. Skeletal muscle undergoes fiber type metabolic switch without myosin heavy chain switch in response to defective fatty acid oxidation. Mol Metab. 2022;59:101456. doi: 10.1016/j.molmet.2022.101456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Zhou M. Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis. Nat Commun. 2023;141:2753. doi: 10.1038/s41467-023-38003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/Mtor pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;311:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 29.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of Igf-1-induced skeletal myotube hypertrophy by Pi(3)K/Akt/Mtor and Pi(3)K/Akt/Gsk3 pathways. Nat Cell Biol. 2001;311:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 30.Ryu YC, Choi YM, Lee SH, Shin HG, Choe JH, Kim JM, Hong KC, Kim BC. Comparing the histochemical characteristics and meat quality traits of different pig breeds. Meat Sci. 2008;802:363–369. doi: 10.1016/j.meatsci.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 31.Shi H, Scheffler JM, Pleitner JM, Zeng C, Park S, Hannon KM, Grant AL, Gerrard DE. Modulation of skeletal muscle fiber type by mitogen-activated protein kinase signaling. FASEB J. 2008;228:2990–3000. doi: 10.1096/fj.07-097600. [DOI] [PubMed] [Google Scholar]
- 32.Koh HJ, Brandauer J, Goodyear LJ. Lkb1 and Ampk and the regulation of skeletal muscle metabolism. Curr Opin Clin Nutr Metab Care. 2008;113:227–232. doi: 10.1097/MCO.0b013e3282fb7b76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frayn KN. Adipose tissue as a buffer for daily lipid flux. Diabetologia. 2002;459:1201–1210. doi: 10.1007/s00125-002-0873-y. [DOI] [PubMed] [Google Scholar]
- 34.D'Souza K, Nzirorera C, Cowie AM, Varghese GP, Trivedi P, Eichmann TO, Biswas D, Touaibia M, Morris AJ, Aidinis V, Kane DA, Pulinilkunnil T, Kienesberger PC. Autotaxin-Lpa signaling contributes to obesity-induced insulin resistance in muscle and impairs mitochondrial metabolism. J Lipid Res. 2018;5910:1805–1817. doi: 10.1194/jlr.M082008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;1346:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, Takahashi H, Hirshman MF, Schlein C, Lee A, Baer LA, May FJ, Gao F, Narain NR, Chen EY, Kiebish MA, Cypess AM, Bluher M, Goodyear LJ, Hotamisligil GS, Stanford KI, Tseng YH. The cold-induced lipokine 12,13-dihome promotes fatty acid transport into brown adipose tissue. Nat Med. 2017;235:631–637. doi: 10.1038/nm.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H, Long JZ, Roche AM, Svensson KJ, Dou FY, Chang MR, Strutzenberg T, Ruiz C, Cameron MD, Novick SJ, Berdan CA, Louie SM, Nomura DK, Spiegelman BM, Griffin PR, Kamenecka TM. Discovery of hydrolysis-resistant isoindoline N-acyl amino acid analogues that stimulate mitochondrial respiration. J Med Chem. 2018;617:3224–3230. doi: 10.1021/acs.jmedchem.8b00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer N, Yu Z, Wagener A, Millrose MK, Reissmann M, Bortfeldt R, Dieterich C, Adamski J, Wang-Sattler R, Illig T, Brockmann GA. Changes in metabolite profiles caused by genetically determined obesity in mice. Metabolomics. 2014;103:461–472. doi: 10.1007/s11306-013-0590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrara PJ, Verkerke ARP, Maschek JA, Shahtout JL, Siripoksup P, Eshima H, Johnson JM, Petrocelli JJ, Mahmassani ZS, Green TD, McClung JM, Cox JE, Drummond MJ, Funai K. Low lysophosphatidylcholine induces skeletal muscle myopathy that is aggravated by high-fat diet feeding. FASEB J. 2021;3510:e21867. doi: 10.1096/fj.202101104R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon MR, Bakker LEH, Prehn C, Adamski J, Vosselman MJ, Jazet IM, Arias-Bouda LMP, van Lichtenbelt WDM, van Dijk KW, Rensen PCN, Mook-Kanamori DO. Lysopc-Acyl C16:0 is associated with brown adipose tissue activity in men. Metabolomics. 2017;135:48. doi: 10.1007/s11306-017-1185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekas G, Patton GM, Lincoln EC, Robins SJ. Origin of plasma lysophosphatidylcholine: evidence for direct hepatic secretion in the rat. J Lab Clin Med. 1985;1052:190–194. [PubMed] [Google Scholar]
- 42.Mehedint MG, Zeisel SH. Choline’s role in maintaining liver function: new evidence for epigenetic mechanisms. Curr Opin Clin Nutr Metab Care. 2013;163:339–345. doi: 10.1097/MCO.0b013e3283600d46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L, Yu S, Khan RS, Ables GP, Bharadwaj KG, Hu Y, Huggins LA, Eriksson JW, Buckett LK, Turnbull AV, Ginsberg HN, Blaner WS, Huang LS, Goldberg IJ. Dgat1 deficiency decreases Ppar expression and does not lead to lipotoxicity in cardiac and skeletal muscle. J Lipid Res. 2011;524:732–744. doi: 10.1194/jlr.M011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlegel C, Lapierre LA, Weis VG, Williams JA, Kaji I, Pinzon-Guzman C, Prasad N, Boone B, Jones A, Correa H, Levy SE, Han X, Wang M, Thomsen K, Acra S, Goldenring JR. Reversible deficits in apical transporter trafficking associated with deficiency in diacylglycerol acyltransferase. Traffic. 2018;1911:879–892. doi: 10.1111/tra.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall AM, Soufi N, Chambers KT, Chen Z, Schweitzer GG, McCommis KS, Erion DM, Graham MJ, Su X, Finck BN. Abrogating monoacylglycerol acyltransferase activity in liver improves glucose tolerance and hepatic insulin signaling in obese mice. Diabetes. 2014;637:2284–2296. doi: 10.2337/db13-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watt MJ. Storing up trouble: does accumulation of intramyocellular triglyceride protect skeletal muscle from insulin resistance? Clin Exp Pharmacol Physiol. 2009;361:5–11. doi: 10.1111/j.1440-1681.2008.05075.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.