Abstract
TNF-α-induced NF-κB pathway is an essential component of innate and adaptive immune pathway, and it is tightly regulated by various post-translational modifications including ubiquitination. Oscillations in NF-κB activation and temporal gene expression are emerging as critical determinants of inflammatory response, however, the regulators of unique outcomes in different patho-physiological conditions are not well understood. Tripartite Motif-containing proteins (TRIMs) are RING domain-containing E3 ligases involved in the regulation of cellular homeostasis, metabolism, cell death, inflammation, and host defence. Emerging reports suggest that TRIMs are recruited at different steps of TNF-α-induced NF-κB pathway and modulate via their E3 ligase activity. TRIMs show synergy and antagonism in the regulation of the NF-κB pathway and also regulate it in a feedback manner. TRIMs also regulate pattern recognition receptors (PRRs) mediated inflammatory pathways and may have evolved to directly regulate a specific arm of immune signalling. The review emphasizes TRIM-mediated ubiquitination and modulation of TNF-α-regulated temporal and NF-κB signaling and its possible impact on unique transcriptional and functional outcomes.
Keywords: TNF-α, TRIMs, Oscillatory NF-κB, Feedback regulation, Inflammation
TNF-α: a pleiotropic cytokine and essential regulator of pathophysiology
TNF-α is a pleiotropic cytokine physiologically important for normal response against bacterial, viral, parasitic infections and inherent cellular stress. It has both pro-inflammatory and immunomodulatory functions. Primarily macrophages and monocytes, as well as other cell types in stress conditions, expresses TNF-α as a 26 kDa type II transmembrane protein. It is further processed as a 17-kDa soluble form upon the proteolytic cleavage by TNF-α converting enzyme (TACE) [1]. Soluble TNF-α is released as 51 kDa homotrimer and exerts its biological function upon binding to tumor necrosis factor receptor superfamily (TNFSF) receptors: TNFR1 and TNFR2.
Beyond its immune and inflammatory functions, TNF-α also plays an essential role in several homeostatic functions. It is required for establishing lymphoid-organ architecture, formation of germinal-center formation, and maturation of humoral immune response [2–4]. Interestingly, TNF-α knockout mice show delayed response to heat-killed Corynebacterium parvum and aberrant inflammatory response leading to death [4], suggesting that TNF-α-induced signalling may not only activate inflammatory response, but also help in the coordination of its regulation. Furthermore, the role of TNF-α in tissue regeneration and wound healing [5–7], neuronal remyelination [8], and cardiac remodelling [9] are emerging. Physiologically, TNF-α regulates lipid metabolism in adipose tissue and protein catabolism in muscle [10, 11]. During normal physiology, TNF-α protects against the expansion of the adipose tissue depots, increases lipolysis, and free fatty acid oxidation [12, 13]. Additionally, TNFSF also activates immune cell, produce tissue inflammatory proteins and molecules that induce immune suppression or immune cell death [14, 15]. Given the implication of TNF-α in many pathophysiological conditions, TNF-α-regulated pathway is one of the most explored biological mechanisms, however, the mechanism of unique outcome in a given patho-physiological condition is still not well understood.
TNF-α: an activator of pro-inflammatory transcription factors
TNF-α is a prototypic activator of the NF-κB transcription factors; essential mediators of pro-inflammatory response and cell survival [16–18]. The binding of TNF-α to its receptors: TNFR1 or TNFR2 receptor subtype, leads to all its different cellular and pathological effects. TNF receptors are single transmembrane glycoproteins and show 28% homology mostly in their extracellular domain and have distinct four tandemly repeated cysteine-rich motifs. Interestingly, intracellular domains of either receptor show almost no homology. Both membrane-bound and soluble TNF-α activates signalling pathways and its binding with TNFR1 and R2 receptors have different implications. TNFR1 is constitutively expressed in most tissues and acts as the key mediator of TNF-α-mediated signalling [14, 19–21]. TNF-α–TNFR1-mediated signalling promotes the production of stress and inflammatory response factors by activation of two major transcription factors NF-κB and activator protein 1 (AP-1) [14, 19].
On the other hand, TNFR2 expression is highly regulated and specifically expressed on immune cells and preferentially promotes cell survival and proliferation [22]. The binding of TNF-α to TNFR2 induces assembly of the TNFR2–Etk–VEGFR2 (vascular endothelial growth factor receptor 2) complex which promotes cell adhesion, migration, survival, and proliferation of human umbilical vein endothelial cells (HUVEC) and bovine aorta endothelial cells (BAEC) [23]. TNF-α–TNFR2 can also activate both STAT5 and NF-κB signalling pathways and play a major role in the differentiation of Th cells [24, 25].
In this review, we majorly focus on TNF-α binding to TNFR1 which activates the NF-κB pathway by dynamic assembly of several distinct protein complexes and had been implicated in many acute and chronic patho-physiological conditions. Many of these complexes require post-translational modification (PTMs) for activation or optimal function. Hence, PTM’s role in activation and temporal regulation of TNF-α-induced NF-κB pathway has been discussed further.
Regulation of TNF-α-induced NF-κB pathway: multifaceted roles of ubiquitination
PTM of a substrate via ubiquitin show various outcomes including its stabilization or degradation. The seven lysine residues of the ubiquitin (K6, K11, K27, K29, K33, K48, and K63) had been now shown to be utilized for the formation of polyubiquitin chain of different topologies, which is now being explored for their different role in cellular functions. Moreover, an eighth chain type classified as Met1-linked or 'linear' chains, is generated when ubiquitin is attached to the N-terminus of a second ubiquitin. Mono or polyubiquitination of proteins increases their structural diversity and determines their fate. Modification of substrates by specific chain linkages can determine their fate and functional outcomes. K48-linked chains are well known for their role in substrate degradation, whereas, K63-linked chains result in the assembly of complexes and non-proteolytic functions [26]. Among the atypical ubiquitin chains, K11-linked chains regulate different functions in cell cycle regulation [27] and ERAD pathway [28]. A recent review by Tracz and Bialek discusses the atypical ubiquitin chains and their functional outcomes [29]. Mechanistically, the addition of specific chain linkages depends on the combinations of ubiquitin-conjugating E2 enzymes (UBE2s) and E3 ligases, and also on the side chain of the acceptor lysine residue [30, 31]. TNF-α binding to TNFR1 leads to sequential assembly of three major dynamic complexes: membrane-bound TRADD–TRAF complex, cytosolic TAB–TAK, and IKK kinase complex and nuclear translocation and regulation of NF-κB heterodimers. Ubiquitination of critical proteins and regulation at these distinct steps may not only affect NF-κB activation but also determine the functional outcome. In the following sections, we discuss various aspects of ubiquitin signalling in the regulation of the TNF-α induced NF-κB pathway at discreet levels.
Regulation of NF-κB pathway at membrane-bound receptor complex: ubiquitin chains as assembly platforms
The binding of TNF-α to its receptor TNFR1 initiates assembly of adapter proteins TRADD, TRAF2, or TRAF5, receptor-interacting protein 1(RIP1), and E3 ubiquitin ligases cIAP1 and cIAP2 at the cytosolic face of the receptor. Initially, TRAF2 is recruited to the N-terminal TRAF-binding domain of the TRADD through its C-terminal TRAF domain [17, 32]. TNF-α-induced Protein Kinase C (PKC) phosphorylates TRAF2 at Thr117 leading to its K63 linked ubiquitination at Lysine (K) 31 which serves as a site for recruitment of other E3 ligases and kinases essential for activation of NF-κB pathway [33]. Further, TRAF2 recruits RIP1 to the TRADD complex and promotes K63 linked ubiquitination of RIP1 [34]. TRAF2 also recruits E3 ligases cIAP1 and 2 to the membrane-bound TNFR1 complex, which further contributes to RIP1 ubiquitination and stabilization of the membrane-bound TRADD–TRAF complex.
The recruitment of cIAPs catalyse K11-, K48- and K63-linked ubiquitin chains to different components of the TNFR1 complex including K11-linked ubiquitination of RIP1 [35]. The E3 ligase activity of cIAPs and not TRAF2 is essential for HOIL-1 [Linear ubiquitin chain assembly complex (LUBAC) component] recruitment to the TNFR1 complex [36]. Consequently, LUBAC (Composed of SHARPIN, HOIL-1L, and HOIP) increases the stability of membrane-bound complex by promoting linear ubiquitination of TRADD and RIPK1. Additionally, it promotes linear ubiquitination of NEMO, a regulatory subunit of cytosolic IKK kinase complex which enhances its recruitment to TNFR1 [36, 37].
Regulation of NF-κB activating kinase complexes in the cytosol: ubiquitin as a switch for activation
Ubiquitination of RIP1 on Lys377 residue is critical for the recruitment of both IKK and TAB–TAK complex at TNFR1 [18, 35, 38]. TAB–TAK complex is composed of TAB2, TAB3, and TGFβ (transforming growth factor β)-activated kinase-1 (TAK1) and binds to K63-linked RIP1 ubiquitin chains through their ubiquitin-binding domains (UBDs) [18, 38]. Importantly, the kinase activity of RIP1 is not required for activation of TNF-α induced NF-κB or p38 MAP kinase pathway, suggesting it primarily acts as an assembly factor. Later TRAF2 ubiquitinates TAK1 at lysine 158 which induces a conformational change and promotes its auto-phosphorylation at Thr178, Thr184, Thr187, and Ser192 positions [18, 39]. The other downstream kinase complex IKK, composed of IKKα, IKKβ, and regulatory IKKγ/NEMO, is also recruited to the K63 ubiquitinated RIP1 in a TRAF2 dependent manner. NEMO binds to K63 linked Ub chains and is essential for stabilization of RIP1 at the TRADD–TRAF complex in TNF-α stimulated cells [40]. The presence of both TRAF2 and RIP1 is indispensable for the kinase activity of the IKK complex [41]. NEMO, the regulatory subunit of IKK complex had been shown to be ubiquitinated by different topologies in different conditions and activates downstream NF-κB pathway. cIAP1 mediated NEMO ubiquitination is indispensable for TNF-α-induced NF-κB activation [42], and its ubiquitination at Lys277 and Lys309 is important for genotoxic stress-induced NF-κB activation [43]. Likewise, polymethylmethacrylate (PMMA) particles released from joint implants induce NF-κB pathway leading to inflammatory osteoclastogenesis and osteolysis [44] via K63-linked ubiquitination of NEMO at Lys392. In TNF-α stimulated cells, LUBAC adds linear polyubiquitin chains on NEMO at Lys285 and Lys309 residue [45] and promotes downstream pathway for NF-κB activation [36, 37].
It is important to note that other than NEMO, K63 linked ubiquitination of IKKβ at Lys147 play role in IL-6 mediated STAT3 activation. Lys147 of IKKβ is a highly conserved site in many ortholog and paralogs of IKKβ. Intriguingly, Lys171 mutation of IKKβ enhances K63 linked ubiquitination of Lys147 and increased activation of STAT3 by both TNF-α and IL6 which is associated with multiple myeloma (MM), spleen marginal zone lymphoma (SMZL), and mantle cell lymphoma (MCL) [46]. In addition to this physiologically relevant ubiquitination site, several ubiquitination sites in IKKα and IKKβ have been predicted/identified in high throughput proteomics studies [47, 48]. However, none of these sites had been validated hence requires further study for determining their role in physiological outcomes and pathological conditions.
The IκBα bound NF-κB heterodimer remains in the cytosol, and stimuli induced IκBα degradation unmasks the nuclear localization signal of NF-κB heterodimer; promoting its nuclear translocation [49]. The SCFβ−TrCP (containing substrate recognizing F-box protein) E3 ubiquitin ligase complex is recruited at the IκBα bound NF-κB heterodimers. SCFβ−TrCP recognizes IκBα phosphorylation at Ser32 and Ser36 by IKK complex, promotes its K48 linked ubiquitination and degradation by the ubiquitin–proteasome system (UPS) [17, 32]. The NF-κB heterodimers bind to the target DNA sequence and activate transcription of target genes which may play a critical role in inflammatory response, cell survival, and proliferation, depending on the patho-physiological cues. Recently, it had been shown that higher levels of SCF component β-TrCP correlate with constitutive expression of NF-κB signalling in colorectal cancer [50], skin tumors [51], and lymphocytic leukemia [52]. F-box and WD repeat domain-containing 7 (FBW7); a member of the F-box protein family, is one of the subunits of the Skp1, Cul1, and F-box protein (SCF) ubiquitin ligase complex. High levels of FBW7 lead to NF-κB mediated intestinal inflammation in Crohn's Disease (CD) and Ulcerative Colitis (UC) [53]. These evidence suggests that the regulation of IκBα degradation by SCF complex may affect chronic NF-κB activation and associated pathological conditions.
Regulation of NF-κB heterodimers in the nucleus: ubiquitination in control of transcriptional regulation
p65-p50 heterodimer is the most abundant Rel dimers found in almost all cell types. Besides, other Rel dimers like p65/p65, p65/c-Rel, p65/p52, c-Rel/c-Rel, p52/c-Rel, p50/c-Rel, p50/p50, RelB/p50, and RelB/p52 have been identified in limited subsets of cells. Moreover, Rel dimers lacking TAD domains (p50 or p52) also bind to DNA and inhibit transcription of target genes unless bound to secondary proteins (Fig. 1).
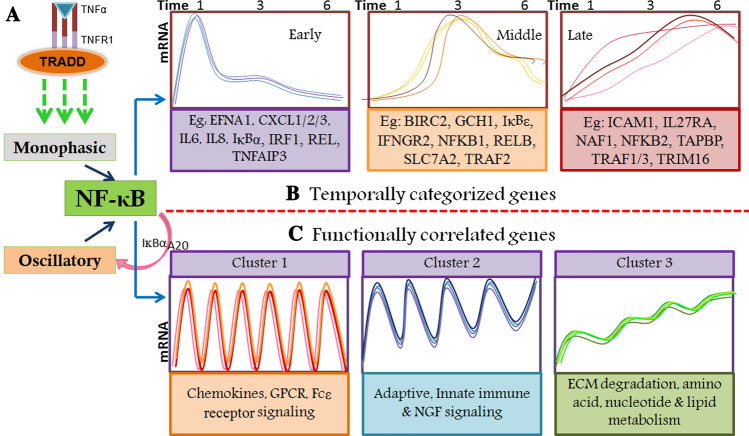
Fig. 1.
TNF-α-induced temporal and functionally correlated genes. A Activation of monophasic or oscillatory NF-κB pathway. Under distinct physiological conditions, TNF-α-induces NF-κB pathway in a monophasic manner which ends with resynthesis of early response feedback inhibitory genes like IκBα and TNFAIP3/A20. The long-term presence of TNF-α leads to translocation cycles of NF-κB heterodimers to nucleus and IκBα/A20-resynthesis dependant nuclear exit leading to oscillatory signalling. B TNF-α-induced NF-κB-dependant gene expression had been categorized temporally. Based on their peak expression these genes show a unique pattern. The figure is
adapted from Tian et al. [64]. C TNF-α-induced NF-κB pathway activates the expression of functionally correlated genes. Cluster 1 which also corroborates with early response genes shows a highly oscillatory pattern and includes genes related to proinflammatory. Cluster 2 shows expression patterns like middle response genes and are functionally correlated to innate and adaptive immune response. Cluster 3 shows temporally increasing expression and includes genes related to metabolism and ECM modification. The figure is adapted from Zambrano et al. and modified [65]
Once in the nucleus, the NF-κB heterodimers bind to the consensus sequences termed as κB DNA elements within the enhancer or promoter region of target genes. NF-κB subunits are regulated by several post-translational modifications including phosphorylation, acetylation, and methylation. These PTMs not only affect the DNA binding ability of NF-κB but also modulate its interactions with transcriptional co-activators, hence affect discreet transcriptional outcomes. For instance, Ser276 phosphorylation of p65 induces structural changes to promote its interaction with the cofactors p300/CBP and lead to enhanced p65 transcriptional activity [54, 55]. Furthermore, nuclear cofactors like p53 and RPS3 have been shown to enhance (> 50 fold) the DNA binding ability of p65 [56]. The effect of other PTMs like acetylation and methylation has been thoroughly reviewed earlier [57].
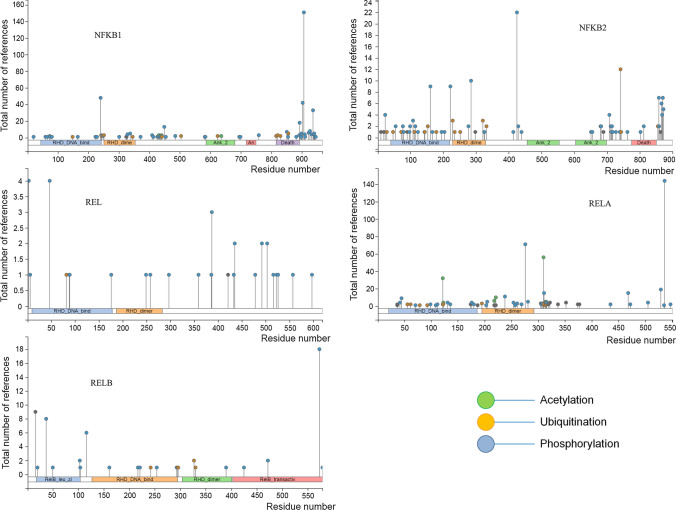
Additionally, ubiquitination of different NF-κB subunits is also emerging as nuclear regulation of the TNF-α-induced NF-κB pathway. EC2S (Elongin BC-CUL2-SOCS-box protein) ubiquitin ligase complex component Suppressor of cytokine signalling (SOCS-1) promotes proteasomal degradation of p65 however did not affect protein levels of p50 [58]. Other E3 ligases like EC5S, PDLIM2, PPAR-gamma, and ING4 also promote UPS-mediated degradation of p65 [59]. Recently, it has been shown that PDLIM7 heterodimerizes with PDLIM2 and promotes K63 linked ubiquitination of PDLIM2. Further, p62/Sqstm1 binds to polyubiquitinated PDLIM2 for shuttling of PDLIM2–p65 complex to nuclear proteasome; leading suppression of NF-κB-mediated inflammatory responses [60]. Furthermore, a phosphorylation-dependant ubiquitination switch has been identified to regulate the selective elimination of p65 on promoter of target genes. TNF-α induced p65 phosphorylation at Ser468 by IKKε promotes its binding with COMMD1 and CUL2, promoting UPS-mediated degradation of p65 [61]. Interestingly, UPS-mediated degradation of nuclear p65/RelA is observed in both IκBα + / + and IκBα −/− cells, suggesting independent control over nuclear NF-κB response through its proteasomal degradation. Additionally, p65 mutant that fails to bind to the κB DNA elements showed reduced polyubiquitination, suggesting UPS-mediated degradation of nuclear p65/RelA requires its DNA binding ability and ubiquitinates promoter bound NF-κB. Furthermore, the report also shows that both IκBα resynthesis and nuclear NF-κB degradation cooperates in termination of the NF-κB response [62]. In addition to p65 ubiquitination, other NF-κB subunits also possess lysine residues that can be ubiquitinated (Fig. 2), and many of these possible ubiquitination sites are conserved in humans, mice, and rats. Identification of ubiquitination-mediated regulation of other NF-κB subunits by novel E3 ligases may provide cues about transcription of specific sets of target genes in a unique physiological response.
Fig. 2.
PTM sites in NF-κB subunits. The NF-κB transcription factors are
modified by different PTMs. PTM of NF-κB subunits provide an additional layer of regulation and bring stringency in their transcriptional outcomes. NF-κB transcription factors possess multiple acetylation, phosphorylation, and ubiquitination sites across various domains. Reversible phosphorylation and acetylation of RELA have been extensively studied but PTMs of other NF-κB subunits are less known. NFKB1, NFKB2, and RELA possess several lysine residues that can be ubiquitinated, whereas REL and RELB have fewer ubiquitination sites. Recruitment of signal specific ubiquitin ligases at NF-κB heterodimers and their ubiquitination may affect stability and structural changes to modulate transcriptional outcomes. In addition to a different combination of NF-κB dimers, combinatorial PTMs of these subunits may provide stringent control and discrete transcriptional outcomes in response to various pathophysiological cues. The figure was generated using PhosphoSitePlus® (https://www.phosphosite.org/homeAction)
TNF-α-induced oscillatory NF-κB activation and temporal gene expression
TNF-α-induced NF-κB can be transient as nuclear translocation and transcriptional response of NF-κB leads to re-synthesis of IĸBα protein, resulting in feedback inhibition of the pathway. Conversely, the constitutive presence of TNF-α leads to NF-κB oscillations due to prolonged IKK complex activation resulting in continued proteolytic degradation of IĸBα and several rounds of NF-κB heterodimer nucleo-cytoplasmic translocations [63, 64]. The existence of transient and oscillatory phases suggests biphasic activation of NF-κB.
Regulation of oscillatory NF-κB activation
The resolution of the transient phase and activation of NF-κB oscillations depend on the IκBα transcription which is NF-κB target gene [63, 64]. The other NF-κB target gene TNFAIP3 (A20) also regulate NF-κB oscillations [65, 66]. More importantly, another NF-κB activator, IL-1, promotes the expression of A20 and limits inflammation but surprisingly does not activate a biphasic response [67, 68]. LPS can activate a biphasic response in vivo by inducing platelet-activating factor (PAF) mediated induction of TNF-α, which in turn activates the second phase of NF-κB activation [69].
Further, the functional outcome of biphasic response depends on the number, period, and amplitude of oscillations [63] and the feedback regulators IĸBα and A20 [66]. It had been observed that these oscillations translate into functionally related proteins suggesting NF-κB oscillations may have a critical role in the execution of TNF-α-induced temporal gene expression and effector response [65].
TNF-α-induced temporal gene expression
Recorded temporal expression of TNF-α-induced genes in response to biphasic/oscillatory NF-κB activation had been classified into three distinct classes of genes. These genes show expression peaks at 1, 3, and 6 h, termed as ‘Early’, ‘Mid’, and ‘Late’ response genes. Further, the expressions of ‘Late’ response genes are temporarily dependent on oscillatory NF-κB activation [64]. The ‘Early’ response genes show functions related to cytokine, chemokines signalling; 'Mid' response genes show sugar and protein binding, whereas the ‘Late’ response genes show peptide transporter activity and protein binding functions [64]. Both of the biphasic response regulatory proteins, IκBα and A20, are ‘Early’ response genes, and very less is known about the possible role of ‘Mid’ and ‘Late’ response genes in feedback regulation of TNF-α-induced NF-κB pathway and biphasic response.
Genome-wide expression of TNF-α upregulated genes have been grouped in 3 clusters. Interestingly, cluster 1 (broadly related to chemokines and chemokine receptors; early genes response gene) contained genes related to GPCR ligand binding and downstream signalling, cytokine signalling in the immune system, Fc epsilon receptor signalling and RIG-I mediated IFNα/β pathways.
Cluster 2 (broadly related to the innate and adaptive immune system; mid/intermediate response genes) includes genes related to the adaptive immune system, TLR signalling, collagen formation, and activation of matrix metalloproteinases. Further, cluster 3 (late response genes) comprised genes related to the metabolism of amino acids and nucleotides, degradation of the extracellular matrix, and S-phase proteins showed distinct dynamics that resemble the pattern of slowly increasing dynamics of Ccl5 [65].
Notably, a comparison of transcript dynamics of cluster 1 genes showed highly similar dynamics compared to the dynamics of early response gene IκBα, whereas, cluster 3 genes showed transcript dynamics comparable to slowly increasing dynamics of a late response gene Ccl5. Moreover, genes categorized in these 3 clusters were also enriched with known NF-κB target genes [65]. TNF-α-induced temporally and functionally correlated genes are shown as a schematic in Fig. 1. These observations connect the temporal and oscillatory response with functionally related transcriptional outcomes, however, their functional relevance in pathophysiological conditions remains elusive. Hence, investigation of novel regulators involved in NF-κB oscillations may answer major questions including how a unique transcriptional response is achieved by NF-κB transcription factors activated under different stimuli.
TNF-α-mediated secondary inflammation
Here we discuss the unique regulatory modules in some pathophysiological conditions regulated by TNF-α-induced NF-κB gene regulation. TNF-α enhances the expression of antiviral cytokines in response to virus infection and activates lower but sustained type I IFNs in macrophages. Furthermore, increased hepatitis C virus (HCV) virus replication, reactivation of hepatitis B virus infections, and increased frequency of viral infection is observed in many cases of patients receiving TNF-α blockade therapy [70, 71]. TNF-α has been shown to promote expression of the IFN response gene by stimulating IFNR1 expression for controlling viral infection [72, 73]. Interferon-stimulated genes (ISGs) are the primary effectors of antiviral signalling activated by IFNs. Interestingly, TNF-α-mediated IFN-stimulated response element (ISRE) activation requires TNFR1-induced NF-κB activation, and it cooperates with IFN-α to activate antiviral response against HCV and hepatitis E virus (HEV) [74]. The usage of anti-TNF-α antibodies reduced the levels of pro-inflammatory cytokines in the blood and the brains of the murine CMV-infected mice model [75]. These reports indicate the role of TNF-α in augmenting physiologically essential innate immune response.
Contrary to the above-mentioned reports, few clinical studies suggest that TNF-α blockade dysregulates type I IFN response leading to paradoxical psoriasis [76, 77]. In support of these anti-inflammatory roles, it has been observed that type I IFNs are essential mediators of TNF-α-induced lethal inflammatory shock. The study also pointed out that type I IFN deficiency reduced the expression of TNF-α-dependant early genes as well as genes that are direct NF-κB targets, further strengthening the cooperation between TNF-α and IFNs [78]. Additionally, it was observed that TNF-α induces the expression of TNFR2. This can further augment the inflammatory response through TNFR2 mediated NF-κB activation and also results in apolipoprotein-A4 expression mediated kidney injury [79]. In a different physiological context, an elevated level of salivary TNF-α was observed in cases of pregnancy-induced periodontal inflammation [80].
Interestingly, in monocytes, TNF-α induces selective hypo-responsiveness and reduced cytokine production on secondary TLR challenge, representing a state of tolerance. It is achieved by GSK3-mediated suppression of chromatin accessibility and termination of NF-κB signalling by inducing negative feedback regulatory proteins A20 and IκBα [15]. TNF-α optimizes inflammatory response to prevent toxicity by tolerizing inflammatory genes through selective chromatin remodelling in macrophages [81]. Another interesting study exploring temporal gene expression in macrophages and fibroblast-like synoviocytes (FLS) showed that TNF-α-induced inflammatory genes show different kinetics in these cell types. More importantly, several TNF-α-induced inflammatory genes that are transiently expressed in macrophages (with peak expression at 1–6 h, ‘tolerized’ genes) show sustained expression in FLS (up to at least 24–72 h). The presence of TNF-α shows increased chromatin accessibility in regulatory elements of NF-κB, interferon-regulatory factors (IRFs), and activating protein-1 (AP-1) [82]. More recent reports have confirmed the role of TNF-α-mediated activation of NF-κB pathway through p-38 and JNK pathway contributing to the expression of inflammatory factors and apoptosis-related proteins ultimately leading to endothelial cell injury and atherosclerosis [83].
Collectively, these reports suggest that TNF-α activates the “feed-forward” loop for sustaining secondary inflammation, and play a pivotal role in disease progression and organismal survival. PTMs specifically ubiquitination may play important role in the regulation of temporal gene expression leading to either subverted or persistent inflammation. The forthcoming sections discuss the role of TRIM E3 ligases-mediated regulation of TNF-α-activated NF-κB signaling.
TRIMs: an unique class of RING domain-containing E3 ligases
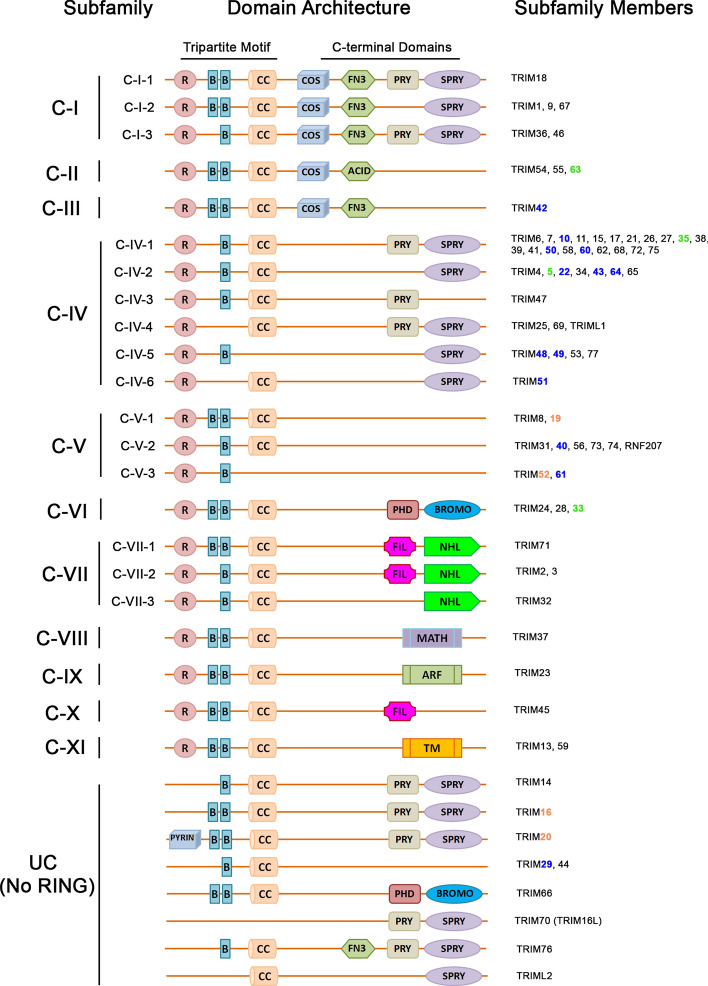
Tripartite motif-containing proteins (TRIMs) are a member of the RING family of ubiquitin E3 ligases [84, 85]. TRIMs possess a distinctive motif composed of N-terminal RING, one or two B-Box, and coiled-coil domain [84–86]. TRIMs also possess additional C-terminal domains of various lengths and compositions, hence further sub-classified based on their c-terminal domain composition (Fig. 3). The majority of TRIMs possess PRY or/and SPRY domain in their C-terminal region. C‐terminal subgroup One Signature (COS), fibronectin type 3 (FN3), plant homeodomain (PHD), bromodomain (BR), bromodomain, filamin type Ig (FIL), NCL-1, HT2A and LIN-41 (NHL) repeats, meprin and TRAF homology (MATH), ADP ribosylation factor-like (ARF) and transmembrane domains (TMs) are among the additional C-terminal domain possessed by members of TRIM proteins [86–88].
Fig. 3.
Classification of TRIMs. Based on their variable C-terminal domains TRIMs are classified into 11 sub-classes. Additionally, 9 RING-less TRIMs had been identified which are grouped as unclassified (UC). TRIMs highlighted in green are validated NF-κB target genes; highlighted in blue areTRIMs induced by TNF-α in THP-1 cells, and orange color TRIMs show TNF-α-induced expression in other cell types. The figure is adapted from Tomar and Singh [87] and had been modified
These proteins are modular in nature and specific functions can be ascribed to each domain. E3 ligase activity is attributed to the RING and B-box domain of TRIMs, whereas the coiled-coil (CC) domain is required for homo and heteromeric interactions leading to oligomers and complexes of higher-order structures [85, 89]. The B30.2 domain (also known as RFP-like or PRY/SPRY) is defined by the presence of highly conserved sequence motifs (LDP, WEVE, and LDYE). Proteins containing PRY/SPRY domain have been shown to have functions related to innate immune response [90].
Interestingly, TRIMs are known to interact with a variety of E2 enzymes (UBE2) with a preference for the D and E classes of UBE2 enzymes. Some of these TRIMs show unique, whereas some members show redundant interaction with different E2 enzymes [91, 92]. Therefore, TRIMs can play an important role in the identification of substrate and directing the assembly of specific ubiquitin chains to their substrates and regulate functional outcomes [91, 93]. TRIM21-mediated Ube2N (a UBE2 enzyme) interaction results in catalysis and association with K63-linked chains essential for its own function [91]. Similarly, other TRIMs also mediate catalysis of different ubiquitin chains on diverse proteins in NF-κB and other inflammatory pathways. For example, TRIM6-mediated unanchored K48-linked ubiquitin chains promote IKKε activation and antiviral response, whereas unanchored K63 linked chains promoted by TRIM5 induces TAK1 activation and HIV restriction [94, 95]. TRIM41 promotes K48-linked ubiquitination of Influenza A virus (IAV)’s Nucleoprotein (NP) and inhibits IAV infection. Similarly, TRIM32 inhibits IAV infection by promoting the degradation of PB1 protein [96]. TRIM14, 22, and 52 also inhibit viral infections by ubiquitin proteasome-dependent target substrate degradation [97–100].
Recent evidence also suggests that TRIMs assemble novel signalosomes in different pathophysiological conditions, hence modulate different cellular pathways [101–106]. Beyond its role in the regulation of antiviral and innate immune response, TRIMs can also regulate autophagy, cell death, and cellular homeostasis by ubiquitin-dependent or independent mechanisms [107–109].
TRIMs modulate TNF-α-induced NF-κB pathway: stimuli-specific expression, recruitment, and E3 ligase activity
Several TRIMs show tissue-specific or stimulus-specific expression and turnover [110–114]. Isolated reports have confirmed the role of TRIMs in the regulation of the TNF-α-induced NF-κB pathway. Reports from our lab and others suggest that individual TRIMs regulate the TNF-α-induced NF-κB pathway at distinct steps by ubiquitin-dependent and independent signalling.
TNF-α modulates the expression of TRIMs
Several TRIMs—TRIM5, TRIM33, TRIM35, TRIM63—have been postulated as NF-κB target genes [115]. Previously, TRIM16 had been shown as TNF-α-induced ‘late’ response gene [64]. The expression of promyelocytic leukemia protein (PML/TRIM19) is induced by TNF-α in an internal ribosome entry site (IRES) dependant manner in HUVEC cells [116]. Expression of another nuclear TRIM protein TRIM52 is also induced by TNF-α in 293 T cells [117]. Gene responsible for familial Mediterranean fever ‘MEFV’also known as TRIM20/PYRIN is increased in peripheral blood leukocytes treated with TNF-α [118] as well as in HeLa and HEK293 cells [119]. Apart from these isolated reports, TNF-α-induced temporally expressed TRIM genes had not been investigated systematically. Since the feedback regulator of the TNF-α-induced NF-κB pathway can affect NF-κB oscillations and unique transcriptional outcomes, more focused investigations are required to identify and validate TRIMs in the regulation of the NF-κB pathway and inflammation.
TRIMs modulate TNF-α-induced NF-κB pathway: interplay between site-specific recruitment and ubiquitin chain topology
In recent years TRIMs have emerged as crucial regulators of innate and inflammatory response. Emerging evidence suggest that TRIMs are selectively recruited at discreet steps of TNF-α-induced NF-κB pathway, which we further discuss in the following sections.
TRIMs: at membrane-bound TRADD–TRAF complex
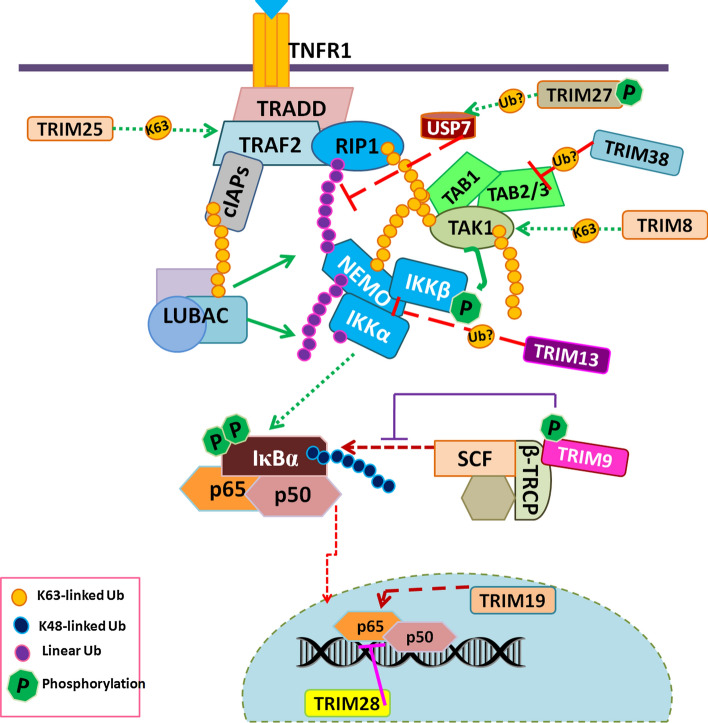
As discussed earlier, the membrane-bound TRADD–TRAF complex requires the recruitment of several E3 ligases to stabilize the complex and promote the assembly of sequential activation nodes. A recent report had identified TRIM25 as an activator of the TNF-α-induced NF-κB pathway in various cell types. TRIM25 promotes K63-linked ubiquitination of TRAF2 and enhances the interaction between TRAF2 and TAK1 or IKKβ (Fig. 4). TRIM25 also promotes NF-κB activation induced by IL-1β and LPS [120].
Fig. 4.
Recruitment of TRIMs at distinct steps of the pathway. Several TRIMs are recruited at different steps of the TNF-α-induced NF-κB pathway. TRIMs either share the same substrate or distinct target and show functional synergy and antagonism in regulation of the TNF-α-induced NF-κB pathway. TRIM38 is induced by TNF-α, hence act as a feedback negative regulator of the TNF-α-induced NF-κB pathway. PTM of TRIM9 and 27 by phosphorylation provides an additional layer of control over the TRIM-mediated regulation TNF-α-induced NF-κB pathway
Ubiquitination of RIP1 at the membrane-bound TRADD–TRAF complex is a critical switch between TNF-α-induced NF-κB and cell death activation [121].TRIM27 also known as RFP inhibits NF-κB and type 1 IFN response activated by TNF-α, IL-1, poly I:C (TLR3 ligand), and viral infections [122]. Interestingly, TRIM27 along with its substrate; USP7 promotes deubiquitylation of RIP1 [123, 124], consequently, TNF-α-induced apoptosis. This strongly suggests that TRIM27 may act as an important node regulating RIP1-dependant cell survival or death switch.
TRIMs regulate the activity of cytosolic kinase complexes
TRIM8 and TRIM38 are positive and negative regulators of the TNF-α-induced NF-κB pathway, respectively. Interestingly, both are recruited at the cytosolic kinase complex and specifically at the TAB–TAK complex (Fig. 4) [114, 125, 126]. TRIM8; predominantly a nuclear protein, is translocated to cytosol in presence of TNF-α where it promotes K63 linked TAK1 ubiquitination and NF-κB activation [114, 125]. Conversely, TRIM38 destabilizes the TAB2/3 complex and promotes lysosome-dependent degradation of TAB2 hence inhibition of NF-κB activity [126]. These evidence suggest, that TAB–TAK complex is an important hub for TRIM-mediated regulation of the TNF-α-induced NF-κB pathway.
TRIM13 is one of the two transmembrane domain-containing TRIMs, anchored to ER membrane; involved in the regulation of ER stress-induced autophagy [112]. TRIM13 is also a negative regulator of the TNF-α-induced NF-κB pathway and acts at the IKK kinase complex (Fig. 4). TRIM13 promotes ubiquitination-mediated degradation of NEMO to inhibit TNF-α-induced NF-κB activation [127]. In contrast, another group shows decreased TRIM13 expression results in inhibition NF-κB pathway specifically in Multiple myeloma (MM) [128].
Besides its role in modulation of RIP1 ubiquitination, TRIM27 interacts with canonical (IKKβ and IKKα) as well as noncanonical (IKKε and TBK1) kinases and potently inhibits signalling activated by these kinases [122]. Notably, TRIM27 is phosphorylated by these aforementioned kinases [122], but its implication in functional outcomes of TNF-α-induced NF-κB is not known.
TRIMs: emerging regulators of nuclear NF-κB activity
TRIM9 is a brain-specific TRIM protein identified as a negative regulator of NF-κB which blocks both canonical and non-canonical NF-κB activation. TRIM9 inhibits TNF-α-induced NF-κB activation and pro-inflammatory IL-6 cytokine production. Mechanistically, phosphorylation-dependant interaction of TRIM9 with SCF-cullin F-box E3 ligase complex component, β-TrCP, blocks IκBα degradation, and inflammatory response (Fig. 4). Physiologically TRIM9 is expressed in the peri-infarct region of mice brain after ischemic stroke to protect the ischemic region from consequences of neuroinflammation [129, 130]. In consonance, molluscan TRIM9 had been shown to inhibit NF-κB activation [131]. On the contrary, a recent report demonstrated that TRIM9 promotes the formation and development of uterine leiomyoma (UM) by regulating NF-κB signalling [132].
Several nuclear-localized TRIMs had been shown to modulate TNF-α-induced NF-κB activation. One of the most studied nuclear TRIM proteins, PML/TRIM19 forms subnuclear structures known as PML nuclear bodies (NBs) and regulates inflammatory response and tumorigenesis. It has been shown that PML promotes TNF-α-induced expression of several NF-κB target genes. Furthermore, the report shows decreased NF-κB p65 phosphorylation and defective NF-κB transcriptional activity in PML −/− cells [133]. Nuclear localized TRIM28/KAP1 is a transcriptional co-repressor and a well-known oncogene. It inhibits TNF-α-induced NF-κB activation and IL-6 production (Fig. 4) [134–136]. Importantly TRIM28 directly interacts with NF-κB heterodimers and p300, resulting in decreased NF-κB acetylation and nuclear retention in TNF-α treated cells [137]. Contrary to its role in the inhibition of inflammatory response, evidence suggests that TRIM28-dependant expression of TNFR1 and R2 receptors in endothelial cells (ECs) contributes to enhanced inflammation in ECs [138]. TRIM39 is also localized in nucleus and negatively regulates the TNF-α-induced NF-κB activation. It has been shown that TRIM39 promotes the stabilization of TNF-α-indued gene Cactin to inhibit NF-κB activation [139].
Post-translational regulation of TRIMs is also emerging as an additional layer of control over cellular pathways. For example, phosphorylation of TRIM9 and TRIM27 has a functional impact on NF-κB activation, whereas N-terminal cleavage of TRIM20/MEFV leads to its interaction with IκBα and potentiates its degradation thus leading to activation of NF-κB and inflammatory response [140]. Together this evidence suggests that individual TRIM proteins are recruited at different steps of the TNF-α-induced NF-κB pathway and regulate the NF-κB pathway. However, their temporal recruitment and role in modulation of inflammatory outcomes remain elusive.
TRIMs: vanguards of PRR induced inflammation
Other than cytokines, PRRs can induce pro-inflammatory response. Persistent infection or stress can result in chronic inflammation and contribute to pathological conditions [141]. Toll-like receptors (TLRs) are a major class of PRRs and contribute to initiating inflammatory response by sensing various pathological macromolecules [142]. TRIMs are also emerging as modulators of PRR-induced NF-κB and inflammatory pathways.
PRR-induced TRIMs
TRIMs have been identified as crucial regulators of innate immune pathways, inflammatory response, and antiviral response [111, 143]. During stress conditions and infection, TRIMs regulate PRR-induced signalling and majorly act as innate immune modulators [111]. We have catalogued the PRR induced expression of TRIMs in Table 1.
Table 1.
PRR-induced TRIMs
| PRR | TRIMs | Cell type |
|---|---|---|
| TLR1/2 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [144] |
| TRIM13 | BMDM, RAW264.7 [154] | |
| TRIM38 | RAW264.7 [150] | |
| TLR3 | TRIM5, 6, 10, 14, 15, 19, 20, 21, 22, 25, 25, 29, 31, 34, 38, 40, 42, 43, 48, 49, 50, 51, 56, 60, 61, 64, 69, 77 | THP-1-derived macrophages [144] |
| TRIM13 | RAW264.7 [154] | |
| TRIM30α | BMDC, J744 macrophages[145] | |
| TRIM38 | RAW264.7 [150] | |
| TRIM25 | PBMCs[174] | |
| 7TLR4 | TRIM5, 6, 10, 14, 15, 19, 20, 21, 22, 25, 25, 29, 31, 34, 38, 40, 42, 43, 48, 49, 50, 51, 56, 60, 61, 64, 69, 77 | THP-1-derived macrophages [144] |
| TRIM13 | RAW264.7 [154] | |
| TRIM20/MEFV | Peripheral blood leukocytes [118] | |
| TRIM30α | BMDC, J744 macrophages[145] | |
| TRIM38 | RAW264.7 [150] | |
| TLR5 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1 derived macrophages [144] |
| TRIM13 | RAW264.7 [154] | |
| TLR6/2 | TRIM13 | RAW264.7 [154] |
| TLR7 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [144] |
| TRIM35 | RAW264.7 [175] | |
| TRIM38 | RAW264.7 [150] | |
| TRIM25 | PBMCs[174] | |
| TRIM32 | Mouse skin[176] | |
| TLR8 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [144] |
| TRIM13 | RAW264.7 [154] | |
| TLR9 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [144] |
| TRIM13 | RAW264.7 [154] | |
| TRIM30α | BMDC, J744 macrophages [145] | |
| TRIM35 | RAW264.7 [175] | |
| TRIM25 | PBMCs [174] | |
| TLR1/2 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [138] |
| TRIM13 | BMDM, RAW264.7 [139] | |
| TRIM38 | RAW264.7 [140] | |
| TLR3 | TRIM5, 6, 10, 14, 15, 19, 20, 21, 22, 25, 25, 29, 31, 34, 38, 40, 42, 43, 48, 49, 50, 51, 56, 60, 61, 64, 69, 77 | THP-1-derived macrophages [138] |
| TRIM13 | RAW264.7 [139] | |
| TRIM30α | BMDC, J744 macrophages [141] | |
| TRIM38 | RAW264.7 [140] | |
| TRIM25 | PBMCs [142] | |
| TLR4 | TRIM5, 6, 10, 14, 15, 19, 20, 21, 22, 25, 25, 29, 31, 34, 38, 40, 42, 43, 48, 49, 50, 51, 56, 60, 61, 64, 69, 77 | THP-1-derived macrophages [138] |
| TRIM13 | RAW264.7 [139] | |
| TRIM20/MEFV | Peripheral blood leukocytes [114] | |
| TRIM30α | BMDC, J744 macrophages [141] | |
| TRIM38 | RAW264.7 [140] | |
| TLR5 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [138] |
| TRIM13 | RAW264.7 [139] | |
| TLR6/2 | TRIM13 | RAW264.7 [139] |
| TLR7 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [138] |
| TRIM35 | RAW264.7 [143] | |
| TRIM38 | RAW264.7 [140] | |
| TRIM25 | PBMCs [142] | |
| TRIM32 | Mouse skin[144] | |
| TLR8 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [138] |
| TRIM13 | RAW264.7 [139] | |
| TLR9 | TRIM10, 15, 29, 31, 40, 42, 43, 48, 49, 50, 51, 60, 61, 64, 77 | THP-1-derived macrophages [138] |
| TRIM13 | RAW264.7 [139] | |
| TRIM30α | BMDC, J744 macrophages [141] | |
| TRIM35 | RAW264.7 [143] | |
| TRIM25 | PBMCs [142] |
Interestingly, expression of TRIM14 and 34 increases in THP-1 cells in a dose- and time-dependent manner [144] in the presence of TLR3 and TLR4 ligands. TRL3, 4, and 9 induced the expression of TRIM30α in BMDCs, whereas, TLR2, 3, 4, and 7 induced expression of TRIM38 in RAW264.7. Interestingly most of the TLR-induced expression of TRIMs is NF-κB dependant [145]. The majority of TLR-induced TRIMs expression data come from innate immune cells and limited data from cells of different tissues and organs is available. Therefore, TRIMs expression in response to ligands-specific PRRs may vary from tissue to tissue which may have an important role in the regulation of associated cellular functions.
TRIMs: emerging regulators of TLR-induced signalling
Several of the aforementioned TLR-induced TRIMs regulate NF-κB-dependant inflammatory pathways. A series of investigations by Hong-Bing Shu group identified that TRIM8, TRIM32, and TRIM38 act as feedback negative regulators of TLR3/4 induced signalling [146, 147]. TRIM8 and TRIM38 modify TIR-domain-containing adapter-inducing interferon-β (TRIF) by K6/K33 and K48-linked ubiquitin chains, respectively, to abolish TLR-induced signalling [146, 147], whereas TRIM32 promotes autophagy-mediated TRIF degradation without affecting its ubiquitination [148]. TRIM8 inhibits TLR3/4-induced signalling by disrupting TRIF–TBK1 interaction. TRIM38 inhibits the TLR3/4 induced pathway by promoting UPS-mediated degradation of NAP1 [149], TNF-α receptor-associated factor 6 (TRAF6) [150], and TRIF [147, 151]. Both TRIM8 and TRIM38 knockout mice are more susceptible to poly(I:C), LPS, and S. Typhimurium-induced death. TRIM72/MG53 shows a protective effect in LPS-induced neurotoxicity and neuroinflammation by inhibiting the TLR4-induced NF-κB pathway both in vitro and in vivo [152]. In a different patho-physiological condition, TRIM72 leads to idiopathic inflammatory myopathies (IIM) [153] suggesting TRIM72 broadly has an anti-inflammatory function. Conversely, TRIM13 acts as a feedback positive regulator of TLR2-induced signalling by promoting K29-linked ubiquitination of TRAF6 [154] to potentiate inflammatory response.
TRIM-mediated ubiquitination regulates MAVS/STING-mediated inflammation
Intracellular pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DMAPs) can be recognized by intracellular receptors like RIG-I and MDA5/AIMs which through recruitment of specific adaptor proteins can activate downstream NF-κB and IRF3 mediated inflammatory pathways [155, 156]. These adaptor proteins: mitochondrial antiviral signaling protein (MAVS) and Stimulator of interferon genes (STING) are localized on mitochondria or ER and mitochondria contact site, respectively. Interestingly, intracellular dsRNA either from viral or bacterial origin or released from mitochondria can be recognized by RIG-I which recruits MAVS and activate downstream NF-κB and IFN pathways. Interestingly, pathogen-derived dsDNA or intracellular DNA released from nucleus or mitochondria in cytosol can be recognized by cyclic GMP–AMP synthase (cGAS) which can recruit STING to activate downstream pathway [155, 156]. Recent evidence suggests that TRIMs are important regulators of STING-mediated inflammatory signaling. TRIM56 expression is induced by dsDNA and poly (I:C) stimulation and it ubiquitinates STING through K63-linked poly-ubiquitin chains at Lys150. The ubiquitination of STING is required for its dimerization, recruitment of TBK1, and downstream IFN response [157]. Both TRIM29 and TRIM30α are expressed during DNA virus infection and negatively regulate the STING-mediated innate immune signaling. TRIM30α interacts with STING in mice dendritic cells, adds K48-linked polyubiquitin chains to Lys275 residue, and enhances its turnover [158]. Similarly, TRIM29 also promotes STING turnover by promoting its K48 linked ubiquitination at Lys370 [159].Moreover, TRIM56 monoubiquitinates cGAS at Lys335 position and regulates HSV-1 infection [160]. Interestingly, TRIM41 also monoubiquitinates cGAS to promote innate antiviral response [161].
Similarly, RIG-I-MAVS-mediated signaling is also modulated by TRIMs. TRIM31 is recruited to mitochondria after viral infection and interacts with MAVS. Further, it catalyzes K63-linked polyubiquitination of MAVS at Lys10, Lys311, and Lys461. The specific ubiquitination pattern on MAVS promotes the formation of prion-like aggregates of MAVS after viral infection which modulates IFN response [162]. Specific TRIMs may have acquired ubiquitin chain editing either through deubiquitinase (DUB) like activity or recruitment of some novel DUBs. TRIM44 does not have the typical RING finger domain, instead, the N-terminal region of TRIM44 contains a ZF UBP domain, which inhibits MAVS ubiquitination and turnover hence positively regulate NF-κB and IFN activity [163]. Interestingly, TRIM38 acts as E3 SUMO1 ligase for MDA5, RIG-I [164], cGAS, and STING [165]. This antagonizes their K48-linked polyubiquitination and degradation, hence positively regulates the cGAS- and RLR-mediated innate immune signalling [164]. Human Cytomegalovirus (HMCV) uses a unique molecular mimicry-based mechanism to evade immune surveillance [166]. HMCV encoded UL144 protein recruits host cellular protein TRIM23 to activate NF-κB pathway [167], however, TRIM23 does not affect NF-κB activation by dsRNA or TNF-α (168). Contrarily, another report shows that TRIM23 mediated atypical K27-linked ubiquitination of NEMO is essential for TLR3- and RIG-I/MDA5-mediated antiviral innate and inflammatory response [168]. It is important to note that these TRIMs may act synergistically or antagonistically to regulate the STING/MAVS-dependent innate and inflammatory signaling. Systemic screening of TRIMs and other ubiquitin ligases in specific patho-physiological conditions is required for better understanding the role of these critical adaptors and inflammatory singnaling.
Conclusions and perspectives
The literature reviewed here till date suggests that different TRIMs are recruited at distinct steps for regulation of TNF-α-induced NF-κB pathway to regulate it, however functional, and substrate redundancy is observed in the case of many TRIMs. It will be interesting to explore if the temporal recruitment of TRIMs at distinct steps of the NF-κB pathway can alter the transcriptional outcomes and overall inflammatory response. Moreover, TRIMs homo/heterodimerize and their interaction may dictate the functional outcomes. Additionally, their stimuli-specific expression may have important regulatory roles in regulation of various cellular pathways including inflammation. Furthermore, TRIMs via their E3 ligase activity-dependent UPS-mediated degradation of target substrates but promote lysosome dependant degradation.
TNF-α-induced TRIM38 and TRIM13 can regulate NF-κB oscillations. The role of these TRIMs requires in-depth studies to decipher the largely unknown aspects of temporally unique transcriptional outcomes achieved by the NF-κB pathway under different stimuli. The selectivity and coordination between TRIMs for the regulation of the TNF-α-induced NF-κB pathway remain elusive and further investigations may identify their role in optimization of inflammatory response. Functional redundancy and antagonism have been observed for TRIM family proteins in regulation of innate immune pathways, however, the impact of this aspect on regulation of proinflammatory NF-κB pathway needs further investigation.
In addition to their dynamic localization, their translocation to specific cellular compartments also enhances the landscape of their functional outcomes. Mitochondria are emerging as an important signalling hub that controls inflammation [169–171]. Dynamic interaction of TRIMs (TRIM4 and TRIM32) and mitochondria has been reported [172, 173], however, whether these TRIMs can modulate inflammatory pathways associated with mitochondria remains elusive.
In conclusion, further studies in this direction will help decipher the mystery of higher eukaryotes having several layers of ubiquitin-mediated regulation of NF-κB pathway which may be a critical determinant of the activation or resolution of inflammation indifferent patho-physiological conditions.
Acknowledgements
The work in the area of TRIMs supported by the grant (BT/PR20692/BRB/10/1538/2016) from the Department of Biotechnology (DBT), Govt. of India, and grant (CRG/2019/000316) from DST-SERB, Govt. of India to Prof. Rajesh Singh. This work constitutes a part of the Ph.D. thesis of Milton Roy.
Author contributions
Authors MR and RS conceptualized the review. MR complied, wrote the first draft, and edited the review. RS acquired the funding, edited and supervised the writing of the review.
Funding
This work is supported by the grant (BT/PR20692/BRB/10/1538/2016) from the Department of Biotechnology (DBT), Govt. of India to Prof. Rajesh Singh and grant (CRG/2019/000316) from DST-SERB, Govt. of India to Department of Biochemistry, The M.S. University of Baroda.
Declarations
Conflict of interest
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moss ML, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385(6618):733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 2.Furtado GC, et al. TNFalpha-dependent development of lymphoid tissue in the absence of RORgammat(+) lymphoid tissue inducer cells. Mucosal Immunol. 2014;7(3):602–614. doi: 10.1038/mi.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasparakis M, et al. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J Exp Med. 1996;184(4):1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marino MW, et al. Characterization of tumor necrosis factor-deficient mice. Proc Natl Acad Sci USA. 1997;94(15):8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Macrophages induce AKT/beta-catenin-dependent Lgr5(+) stem cell activation and hair follicle regeneration through TNF. Nat Commun. 2017;8:14091. doi: 10.1038/ncomms14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heo SC, et al. Tumor necrosis factor-alpha-activated human adipose tissue-derived mesenchymal stem cells accelerate cutaneous wound healing through paracrine mechanisms. J Invest Dermatol. 2011;131(7):1559–1567. doi: 10.1038/jid.2011.64. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen-Chi M, et al. TNF signaling and macrophages govern fin regeneration in zebrafish larvae. Cell Death Dis. 2017;8(8):e2979. doi: 10.1038/cddis.2017.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brambilla R, et al. Astrocytes play a key role in EAE pathophysiology by orchestrating in the CNS the inflammatory response of resident and peripheral immune cells and by suppressing remyelination. Glia. 2014;62(3):452–467. doi: 10.1002/glia.22616. [DOI] [PubMed] [Google Scholar]
- 9.Sun M, et al. Tumor necrosis factor-alpha mediates cardiac remodeling and ventricular dysfunction after pressure overload state. Circulation. 2007;115(11):1398–1407. doi: 10.1161/CIRCULATIONAHA.106.643585. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Martinez C, Lopez-Soriano FJ, Argiles JM. Acute treatment with tumour necrosis factor-alpha induces changes in protein metabolism in rat skeletal muscle. Mol Cell Biochem. 1993;125(1):11–18. doi: 10.1007/BF00926829. [DOI] [PubMed] [Google Scholar]
- 11.Ryden M, et al. Targets for TNF-alpha-induced lipolysis in human adipocytes. Biochem Biophys Res Commun. 2004;318(1):168–175. doi: 10.1016/j.bbrc.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Laurencikiene J, et al. NF-kappaB is important for TNF-alpha-induced lipolysis in human adipocytes. J Lipid Res. 2007;48(5):1069–1077. doi: 10.1194/jlr.M600471-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Porter MH, et al. Effects of TNF-alpha on glucose metabolism and lipolysis in adipose tissue and isolated fat-cell preparations. J Lab Clin Med. 2002;139(3):140–146. doi: 10.1067/mlc.2002.121552. [DOI] [PubMed] [Google Scholar]
- 14.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12(1):49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park SH, et al. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat Immunol. 2011;12(7):607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi K, Karin M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 17.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Wajant H, Scheurich P. TNFR1-induced activation of the classical NF-kappaB pathway. FEBS J. 2011;278(6):862–876. doi: 10.1111/j.1742-4658.2011.08015.x. [DOI] [PubMed] [Google Scholar]
- 19.Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10(1):45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 20.Croft M, Siegel RM. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat Rev Rheumatol. 2017;13(4):217–233. doi: 10.1038/nrrheum.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holbrook J, et al. Tumour necrosis factor signalling in health and disease. F100Res. 2019;8:F1000. doi: 10.12688/f1000research.17023.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye LL, et al. The significance of tumor necrosis factor receptor type II in CD8(+) regulatory T cells and CD8(+) effector T cells. Front Immunol. 2018;9:583. doi: 10.3389/fimmu.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R, et al. Etk/Bmx transactivates vascular endothelial growth factor 2 and recruits phosphatidylinositol 3-kinase to mediate the tumor necrosis factor-induced angiogenic pathway. J Biol Chem. 2003;278(51):51267–51276. doi: 10.1074/jbc.M310678200. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, et al. TNF-alpha enhances Th9 cell differentiation and antitumor immunity via TNFR2-dependent pathways. J Immunother Cancer. 2019;7(1):28. doi: 10.1186/s40425-018-0494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller PG, Bonn MB, McKarns SC. Transmembrane TNF-TNFR2 impairs Th17 differentiation by promoting Il2 expression. J Immunol. 2015;195(6):2633–2647. doi: 10.4049/jimmunol.1500286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579–586. doi: 10.1038/ncb3358. [DOI] [PubMed] [Google Scholar]
- 27.Jin L, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133(4):653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tracz M, Bialek W. Beyond K48 and K63: non-canonical protein ubiquitination. Cell Mol Biol Lett. 2021;26(1):1. doi: 10.1186/s11658-020-00245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liwocha J, et al. Linkage-specific ubiquitin chain formation depends on a lysine hydrocarbon ruler. Nat Chem Biol. 2021;17(3):272–279. doi: 10.1038/s41589-020-00696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickliffe KE, et al. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144(5):769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2(3):a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Wang L, Dorf ME. PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell. 2009;33(1):30–42. doi: 10.1016/j.molcel.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez SE, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465(7301):1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertrand MJ, et al. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1–4) PLoS ONE. 2011;6(9):e22356. doi: 10.1371/journal.pone.0022356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36(5):831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Spit M, Rieser E, Walczak H. Linear ubiquitination at a glance. J Cell Sci. 2019;132(2):jcs208512. doi: 10.1242/jcs.208512. [DOI] [PubMed] [Google Scholar]
- 38.Ea CK, et al. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22(2):245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Fan Y, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J Biol Chem. 2010;285(8):5347–5360. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu CJ, et al. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8(4):398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 41.Devin A, et al. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12(4):419–429. doi: 10.1016/S1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 42.Tang ED, et al. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J Biol Chem. 2003;278(39):37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 43.Jin HS, et al. cIAP1, cIAP2, and XIAP act cooperatively via nonredundant pathways to regulate genotoxic stress-induced nuclear factor-kappaB activation. Cancer Res. 2009;69(5):1782–1791. doi: 10.1158/0008-5472.CAN-08-2256. [DOI] [PubMed] [Google Scholar]
- 44.Alhawagri M, et al. Lysine392, a K63-linked ubiquitination site in NEMO, mediates inflammatory osteoclastogenesis and osteolysis. J Orthop Res. 2012;30(4):554–560. doi: 10.1002/jor.21555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11(2):123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 46.Gallo LH, et al. Novel Lys63-linked ubiquitination of IKKbeta induces STAT3 signaling. Cell Cycle. 2014;13(24):3964–3976. doi: 10.4161/15384101.2014.988026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akimov V, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 2018;25(7):631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- 48.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fagerlund R, et al. NF-kappaB is transported into the nucleus by importin α-3 and importin α-4. J Biol Chem. 2005;280(16):15942–15951. doi: 10.1074/jbc.M500814200. [DOI] [PubMed] [Google Scholar]
- 50.Ougolkov A, et al. Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. J Natl Cancer Inst. 2004;96(15):1161–1170. doi: 10.1093/jnci/djh219. [DOI] [PubMed] [Google Scholar]
- 51.Bhatia N, et al. Mouse homologue of HOS (mHOS) is overexpressed in skin tumors and implicated in constitutive activation of NF-kappaB. Oncogene. 2002;21(10):1501–1509. doi: 10.1038/sj.onc.1205311. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, et al. Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of NF-kappaB and beta-catenin/TCF signaling pathways. Cell Death Dis. 2018;9(4):427. doi: 10.1038/s41419-018-0440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng Q, et al. miRNA-129/FBW7/NF-kappaB, a novel regulatory pathway in inflammatory bowel disease. Mol Ther Nucleic Acids. 2020;19:731–740. doi: 10.1016/j.omtn.2019.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Naumann M, Scheidereit C. Activation of NF-kappa B in vivo is regulated by multiple phosphorylations. EMBO J. 1994;13(19):4597–4607. doi: 10.1002/j.1460-2075.1994.tb06781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann M, et al. RelA/p65 is a molecular target for the immunosuppressive action of protein kinase A. EMBO J. 1995;14(9):1991–2004. doi: 10.1002/j.1460-2075.1995.tb07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulero MC, et al. Protein cofactors are essential for high-affinity DNA binding by the nuclear factor kappaB RelA Subunit. Biochemistry. 2018;57(20):2943–2957. doi: 10.1021/acs.biochem.8b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giridharan S, Srinivasan M. Mechanisms of NF-kappaB p65 and strategies for therapeutic manipulation. J Inflamm Res. 2018;11:407–419. doi: 10.2147/JIR.S140188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryo A, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12(6):1413–1426. doi: 10.1016/S1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 59.Xu H, et al. Ubiquitin-mediated NFkappaB degradation pathway. Cell Mol Immunol. 2015;12(6):653–655. doi: 10.1038/cmi.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jodo A, et al. PDLIM7 synergizes with PDLIM2 and p62/Sqstm1 to inhibit inflammatory signaling by promoting degradation of the p65 subunit of NF-kappaB. Front Immunol. 2020;11:1559. doi: 10.3389/fimmu.2020.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geng H, et al. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009;10(4):381–386. doi: 10.1038/embor.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saccani S, et al. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J Exp Med. 2004;200(1):107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 64.Tian B, Nowak DE, Brasier AR. A TNF-induced gene expression program under oscillatory NF-kappaB control. BMC Genomics. 2005;6:137. doi: 10.1186/1471-2164-6-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zambrano S, et al. NF-kappaB oscillations translate into functionally related patterns of gene expression. Elife. 2016;5:e09100. doi: 10.7554/eLife.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werner SL, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22(15):2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladner KJ, Caligiuri MA, Guttridge DC. Tumor necrosis factor-regulated biphasic activation of NF-kappa B is required for cytokine-induced loss of skeletal muscle gene products. J Biol Chem. 2003;278(4):2294–2303. doi: 10.1074/jbc.M207129200. [DOI] [PubMed] [Google Scholar]
- 68.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145(7):1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han SJ, et al. Molecular mechanisms for lipopolysaccharide-induced biphasic activation of nuclear factor-kappa B (NF-kappa B) J Biol Chem. 2002;277(47):44715–44721. doi: 10.1074/jbc.M202524200. [DOI] [PubMed] [Google Scholar]
- 70.Pompili M, et al. Tumor necrosis factor-alpha inhibitors and chronic hepatitis C: a comprehensive literature review. World J Gastroenterol. 2013;19(44):7867–7873. doi: 10.3748/wjg.v19.i44.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murdaca G, et al. Infection risk associated with anti-TNF-alpha agents: a review. Expert Opin Drug Saf. 2015;14(4):571–582. doi: 10.1517/14740338.2015.1009036. [DOI] [PubMed] [Google Scholar]
- 72.Yarilina A, et al. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9(4):378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 73.Matikainen S, et al. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J Virol. 2006;80(7):3515–3522. doi: 10.1128/JVI.80.7.3515-3522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang W, et al. Convergent transcription of interferon-stimulated genes by TNF-alpha and IFN-alpha augments antiviral activity against HCV and HEV. Sci Rep. 2016;6:25482. doi: 10.1038/srep25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seleme MC, et al. Tumor necrosis factor alpha-induced recruitment of inflammatory mononuclear cells leads to inflammation and altered brain development in murine cytomegalovirus-infected newborn mice. J Virol. 2017 doi: 10.1128/JVI.01983-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conrad C, et al. TNF blockade induces a dysregulated type I interferon response without autoimmunity in paradoxical psoriasis. Nat Commun. 2018;9(1):25. doi: 10.1038/s41467-017-02466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vasconcellos JB, et al. Paradoxical psoriasis after the use of anti-TNF in a patient with rheumatoid arthritis. An Bras Dermatol. 2016;91(5 suppl 1):137–139. doi: 10.1590/abd1806-4841.20164456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huys L, et al. Type I interferon drives tumor necrosis factor-induced lethal shock. J Exp Med. 2009;206(9):1873–1882. doi: 10.1084/jem.20090213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee HH, et al. TNF-alpha-induced inflammation stimulates apolipoprotein-A4 via activation of TNFR2 and NF-kappaB signaling in kidney tubular cells. Sci Rep. 2017;7(1):8856. doi: 10.1038/s41598-017-08785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lasisi TJ, Abdus-Salam RA. Pregnancy-induced periodontal inflammation: Influence of salivary cytokines and antimicrobial proteins. Saudi Dent J. 2018;30(4):306–311. doi: 10.1016/j.sdentj.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park SH, et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol. 2017;18(10):1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loh C, et al. TNF-induced inflammatory genes escape repression in fibroblast-like synoviocytes: transcriptomic and epigenomic analysis. Ann Rheum Dis. 2019;78(9):1205–1214. doi: 10.1136/annrheumdis-2018-214783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou P, et al. Attenuation of TNF-alpha-induced inflammatory injury in endothelial cells by ginsenoside Rb1 via inhibiting NF-kappaB, JNK and p38 signaling pathways. Front Pharmacol. 2017;8:464. doi: 10.3389/fphar.2017.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of single protein RING finger E3 ubiquitin ligases. BioEssays. 2005;27(11):1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 85.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sardiello M, et al. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomar D, Singh R. TRIM family proteins: emerging class of RING E3 ligases as regulator of NF-kappaB pathway. Biol Cell. 2015;107(1):22–40. doi: 10.1111/boc.201400046. [DOI] [PubMed] [Google Scholar]
- 88.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Li Y, et al. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24(6):762–765. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Cruz AA, et al. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 2013;22(1):1–10. doi: 10.1002/pro.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kiss L, et al. A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases. Nat Commun. 2019;10(1):4502. doi: 10.1038/s41467-019-12388-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Napolitano LM, et al. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J. 2011;434(2):309–319. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- 93.Koliopoulos MG, et al. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35(11):1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Campbell EM, et al. TRIM5alpha-mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J Virol. 2016;90(4):1849–1857. doi: 10.1128/JVI.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu B, et al. TRIM32 senses and restricts influenza a virus by ubiquitination of PB1 Polymerase. PLoS Pathog. 2015;11(6):e1004960. doi: 10.1371/journal.ppat.1004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S, et al. TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci Rep. 2016;6:32336. doi: 10.1038/srep32336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fan W, et al. TRIM52 inhibits Japanese encephalitis virus replication by degrading the viral NS2A. Sci Rep. 2016;6:33698. doi: 10.1038/srep33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang C, et al. Interferon alpha (IFNalpha)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell Mol Immunol. 2016;13(1):94–102. doi: 10.1038/cmi.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eldin P, et al. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J Gen Virol. 2009;90(Pt 3):536–545. doi: 10.1099/vir.0.006288-0. [DOI] [PubMed] [Google Scholar]
- 101.Rajsbaum R, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014;40(6):880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kumar S, et al. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy. 2017;13(6):1086–1087. doi: 10.1080/15548627.2017.1307487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kimura T, Mandell M, Deretic V. Precision autophagy directed by receptor regulators—emerging examples within the TRIM family. J Cell Sci. 2016;129(5):881–891. doi: 10.1242/jcs.163758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kimura T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210(6):973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mandell MA, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30(4):394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tan P, et al. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes RIG-I-mediated antiviral signaling. Mol Cell. 2017;68(2):293–307. doi: 10.1016/j.molcel.2017.09.035. [DOI] [PubMed] [Google Scholar]
- 107.Di Rienzo M, et al. TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020;27(3):887–902. doi: 10.1038/s41418-020-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mandell MA, Saha B, Thompson TA. The tripartite nexus: autophagy, cancer, and tripartite motif-containing protein family members. Front Pharmacol. 2020;11:308. doi: 10.3389/fphar.2020.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lionnard L, et al. TRIM17 and TRIM28 antagonistically regulate the ubiquitination and anti-apoptotic activity of BCL2A1. Cell Death Differ. 2019;26(5):902–917. doi: 10.1038/s41418-018-0169-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajsbaum R, Stoye JP, O'Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38(3):619–630. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- 111.Versteeg GA, et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38(2):384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tomar D, et al. TRIM13 regulates ER stress induced autophagy and clonogenic ability of the cells. Biochim Biophys Acta. 2012;1823(2):316–326. doi: 10.1016/j.bbamcr.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 113.Roy M, et al. TRIM8 regulated autophagy modulates the level of cleaved Caspase-3 subunit to inhibit genotoxic stress induced cell death. Cell Signal. 2018;48:1–12. doi: 10.1016/j.cellsig.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 114.Tomar D, et al. Nucleo-cytoplasmic trafficking of TRIM8, a novel oncogene, is involved in positive regulation of TNF induced NF-kappaB pathway. PLoS ONE. 2012;7(11):e48662. doi: 10.1371/journal.pone.0048662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang Y, Wu J, Wang J. A database and functional annotation of NF-κB target genes. Int J Clin Exp Med. 2016;9(5):7986. [Google Scholar]
- 116.Hsu KS, et al. Translational control of PML contributes to TNFalpha-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs. Cell Death Differ. 2016;23(3):469–483. doi: 10.1038/cdd.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fan W, et al. TRIM52: A nuclear TRIM protein that positively regulates the nuclear factor-kappa B signaling pathway. Mol Immunol. 2017;82:114–122. doi: 10.1016/j.molimm.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 118.Centola M, et al. The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood. 2000;95(10):3223–3231. doi: 10.1182/blood.V95.10.3223. [DOI] [PubMed] [Google Scholar]
- 119.Papin S, et al. The tumor necrosis factor alpha-dependent activation of the human mediterranean fever (MEFV) promoter is mediated by a synergistic interaction between C/EBP beta and NF kappaB p65. J Biol Chem. 2003;278(49):48839–48847. doi: 10.1074/jbc.M305166200. [DOI] [PubMed] [Google Scholar]
- 120.Liu Y, et al. TRIM25 promotes TNF-alpha-induced NF-kappaB activation through potentiating the K63-linked ubiquitination of TRAF2. J Immunol. 2020;204(6):1499–1507. doi: 10.4049/jimmunol.1900482. [DOI] [PubMed] [Google Scholar]
- 121.Delanghe T, Dondelinger Y, Bertrand MJM. RIPK1 kinase-dependent death: a symphony of phosphorylation events. Trends Cell Biol. 2020;30(3):189–200. doi: 10.1016/j.tcb.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 122.Zha J, et al. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J Immunol. 2006;176(2):1072–1080. doi: 10.4049/jimmunol.176.2.1072. [DOI] [PubMed] [Google Scholar]
- 123.Zaman MM, et al. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2013;33(24):4971–4984. doi: 10.1128/MCB.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cai J, et al. USP7-TRIM27 axis negatively modulates antiviral type I IFN signaling. FASEB J. 2018;32(10):5238–5249. doi: 10.1096/fj.201700473RR. [DOI] [PubMed] [Google Scholar]
- 125.Li Q, et al. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA. 2011;108(48):19341–19346. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hu MM, et al. TRIM38 inhibits TNFalpha- and IL-1beta-triggered NF-kappaB activation by mediating lysosome-dependent degradation of TAB2/3. Proc Natl Acad Sci USA. 2014;111(4):1509–1514. doi: 10.1073/pnas.1318227111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tomar D, Singh R. TRIM13 regulates ubiquitination and turnover of NEMO to suppress TNF induced NF-kappaB activation. Cell Signal. 2014;26(12):2606–2613. doi: 10.1016/j.cellsig.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 128.Gatt ME, et al. TRIM13 (RFP2) downregulation decreases tumour cell growth in multiple myeloma through inhibition of NF Kappa B pathway and proteasome activity. Br J Haematol. 2013;162(2):210–220. doi: 10.1111/bjh.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shi M, et al. Negative regulation of NF-kappaB activity by brain-specific TRIpartite Motif protein 9. Nat Commun. 2014;5:4820. doi: 10.1038/ncomms5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zeng J, et al. TRIM9-mediated resolution of neuroinflammation confers neuroprotection upon ischemic stroke in mice. Cell Rep. 2019;27(2):549–560. doi: 10.1016/j.celrep.2018.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Y, et al. The first molluscan TRIM9 is involved in the negative regulation of NF-kappaB activity in the Hong Kong oyster Crassostrea hongkongensis. Fish Shellfish Immunol. 2016;56:106–110. doi: 10.1016/j.fsi.2016.06.057. [DOI] [PubMed] [Google Scholar]
- 132.Yang F, et al. TRIM9 overexpression promotes uterine leiomyoma cell proliferation and inhibits cell apoptosis via NF-kappaB signaling pathway. Life Sci. 2020;257:118101. doi: 10.1016/j.lfs.2020.118101. [DOI] [PubMed] [Google Scholar]
- 133.Ahmed A, et al. Regulation of NF-kappaB by PML and PML-RARalpha. Sci Rep. 2017;7:44539. doi: 10.1038/srep44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen L, Munoz-Antonia T, Cress WD. Trim28 contributes to EMT via regulation of E-cadherin and N-cadherin in lung cancer cell lines. PLoS ONE. 2014;9(7):e101040. doi: 10.1371/journal.pone.0101040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pineda CT, Potts PR. Oncogenic MAGEA-TRIM28 ubiquitin ligase downregulates autophagy by ubiquitinating and degrading AMPK in cancer. Autophagy. 2015;11(5):844–846. doi: 10.1080/15548627.2015.1034420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li J, et al. TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene. 2017;36(21):2991–3001. doi: 10.1038/onc.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kamitani S, et al. Kruppel-associated box-associated protein 1 negatively regulates TNF-alpha-induced NF-kappaB transcriptional activity by influencing the interactions among STAT3, p300, and NF-kappaB/p65. J Immunol. 2011;187(5):2476–2483. doi: 10.4049/jimmunol.1003243. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y, et al. Tripartite motif-containing 28 bridges endothelial inflammation and angiogenic activity by retaining expression of TNFR-1 and -2 and VEGFR2 in endothelial cells. FASEB J. 2017;31(5):2026–2036. doi: 10.1096/fj.201600988RR. [DOI] [PubMed] [Google Scholar]
- 139.Suzuki M, et al. TRIM39 negatively regulates the NFkappaB-mediated signaling pathway through stabilization of Cactin. Cell Mol Life Sci. 2016;73(5):1085–1101. doi: 10.1007/s00018-015-2040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chae JJ, et al. The familial Mediterranean fever protein, pyrin, is cleaved by caspase-1 and activates NF-kappaB through its N-terminal fragment. Blood. 2008;112(5):1794–1803. doi: 10.1182/blood-2008-01-134932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cassell GH. Infectious causes of chronic inflammatory diseases and cancer. Emerg Infect Dis. 1998;4(3):475–487. doi: 10.3201/eid0403.980339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.El-Zayat SR, Sibaii H, Mannaa FA. Toll-like receptors activation, signaling, and targeting: an overview. Bull Natl Res Centre. 2019;43(1):187. doi: 10.1186/s42269-019-0227-2. [DOI] [Google Scholar]
- 143.Uchil PD, et al. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol. 2013;87(1):257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jiang MX, et al. Expression profiling of TRIM protein family in THP1-derived macrophages following TLR stimulation. Sci Rep. 2017;7:42781. doi: 10.1038/srep42781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shi M, et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9(4):369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 146.Ye W, et al. TRIM8 negatively regulates TLR3/4-mediated innate immune response by blocking TRIF-TBK1 interaction. J Immunol. 2017;199(5):1856–1864. doi: 10.4049/jimmunol.1601647. [DOI] [PubMed] [Google Scholar]
- 147.Hu MM, et al. TRIM38 negatively regulates TLR3/4-mediated innate immune and inflammatory responses by two sequential and distinct mechanisms. J Immunol. 2015;195(9):4415–4425. doi: 10.4049/jimmunol.1500859. [DOI] [PubMed] [Google Scholar]
- 148.Yang Q, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13(9):e1006600. doi: 10.1371/journal.ppat.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao W, et al. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J Immunol. 2012;188(11):5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- 150.Zhao W, et al. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J Immunol. 2012;188(6):2567–2574. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 151.Xue Q, et al. TRIM38 negatively regulates TLR3-mediated IFN-beta signaling by targeting TRIF for degradation. PLoS ONE. 2012;7(10):e46825. doi: 10.1371/journal.pone.0046825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guan F, et al. MG53 attenuates lipopolysaccharide-induced neurotoxicity and neuroinflammation via inhibiting TLR4/NF-kappaB pathway in vitro and in vivo. Prog Neuropsychopharmacol Biol Psychiatry. 2019;95:109684. doi: 10.1016/j.pnpbp.2019.109684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McElhanon KE, et al. Autoantibodies targeting TRIM72 compromise membrane repair and contribute to inflammatory myopathy. J Clin Invest. 2020;130(8):4440–4455. doi: 10.1172/JCI131721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang B, Baek SH. Trim13 potentiates toll-like receptor 2-mediated nuclear factor kappaB activation via K29-linked polyubiquitination of tumor necrosis factor receptor-associated factor 6. Mol Pharmacol. 2017;91(4):307–316. doi: 10.1124/mol.116.106716. [DOI] [PubMed] [Google Scholar]
- 155.Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol. 2020;21(9):501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 156.Decout A, et al. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021 doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tsuchida T, et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33(5):765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 158.Wang Y, et al. TRIM30alpha is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 2015;11(6):e1005012. doi: 10.1371/journal.ppat.1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xing J, et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun. 2017;8(1):945. doi: 10.1038/s41467-017-00101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Seo GJ, et al. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun. 2018;9(1):613. doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu ZS, et al. RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 2018;8:35. doi: 10.1186/s13578-018-0233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu B, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18(2):214–224. doi: 10.1038/ni.3641. [DOI] [PubMed] [Google Scholar]
- 163.Yang B, et al. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190(7):3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 164.Hu MM, et al. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. 2017;214(4):973–989. doi: 10.1084/jem.20161015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hu MM, et al. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45(3):555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 166.Poole E, et al. The UL144 gene product of human cytomegalovirus activates NFkappaB via a TRAF6-dependent mechanism. EMBO J. 2006;25(18):4390–4399. doi: 10.1038/sj.emboj.7601287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Poole E, et al. Identification of TRIM23 as a cofactor involved in the regulation of NF-kappaB by human cytomegalovirus. J Virol. 2009;83(8):3581–3590. doi: 10.1128/JVI.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arimoto K, et al. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci USA. 2010;107(36):15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhong Z, et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature. 2018;560(7717):198–203. doi: 10.1038/s41586-018-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sandhir R, Halder A, Sunkaria A. Mitochondria as a centrally positioned hub in the innate immune response. Biochim Biophys Acta Mol Basis Dis. 2017;1863(5):1090–1097. doi: 10.1016/j.bbadis.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 171.Missiroli S, et al. The role of mitochondria in inflammation: from cancer to neurodegenerative disorders. J Clin Med. 2020;9(3):740. doi: 10.3390/jcm9030740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tomar D, et al. TRIM4; a novel mitochondrial interacting RING E3 ligase, sensitizes the cells to hydrogen peroxide (H2O2) induced cell death. Free Radic Biol Med. 2015;89:1036–1048. doi: 10.1016/j.freeradbiomed.2015.10.425. [DOI] [PubMed] [Google Scholar]
- 173.Prajapati P, et al. TRIM32 regulates mitochondrial mediated ROS levels and sensitizes the oxidative stress induced cell death. Cell Signal. 2020;76:109777. doi: 10.1016/j.cellsig.2020.109777. [DOI] [PubMed] [Google Scholar]
- 174.Wei Y, et al. TRIM25 identification in the Chinese goose: gene structure, tissue expression profiles, and antiviral immune responses in vivo and in vitro. Biomed Res Int. 2016;2016:1403984. doi: 10.1155/2016/1403984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang Y, et al. TRIM35 negatively regulates TLR7- and TLR9-mediated type I interferon production by targeting IRF7. FEBS Lett. 2015;589(12):1322–1330. doi: 10.1016/j.febslet.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 176.Liu Y, et al. Trim32 deficiency enhances Th2 immunity and predisposes to features of atopic dermatitis. J Invest Dermatol. 2017;137(2):359–366. doi: 10.1016/j.jid.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]