Abstract
Strains of Saccharomyces cerevisiae were constructed in which chromosomal rDNA repeats are completely deleted and their growth is supported by a plasmid carrying a single rDNA repeat, either a plasmid carrying the 35S rRNA gene transcribed from the native promoter by RNA polymerase I or a plasmid carrying the 35S rRNA gene fused to the GAL7 promoter for transcription by RNA polymerase II. This system has made it possible to assess the expression of rDNA by measuring the ability of synthesized rRNA to support cell growth as well as by measuring the actual rRNA synthesized rather than by the use of reporter mini-rDNA genes. Using this system, deletion analysis of the rDNA promoter confirmed the presence of two elements, the upstream element and the core promoter, and showed that basal transcription from the core promoter, if it takes place in vivo as was observed in vitro, is not sufficient to allow cell growth. We have also succeeded in integration of a rDNA repeat and its copy number expansion at the original chromosomal locus, which will allow future mutational analysis not only of rRNA but also other DNA elements involved in rRNA transcription, rDNA replication and recombination within a repeated rDNA structure.
INTRODUCTION
In most eukaryotic organisms the DNA encoding the large precursor rRNA (rDNA) is tandemly repeated at one or a few chromosomal loci and is present in many copies. Structures of repeating rDNA units have been studied in several organisms, ranging from the yeast Saccharomyces cerevisiae to human, with respect to DNA elements implicated in rRNA transcription, DNA replication and recombination. In addition to the promoter (gene promoter) for the large precursor rRNA, there are several DNA elements within the intergenic spacer region that are postulated to stimulate or regulate rRNA transcription, such as spacer promoters and enhancers (for reviews see 1,2). In S.cerevisiae (called yeast in this article), which carries ∼150 copies of tandemly repeated rDNA on chromosome XII, DNA elements in the intergenic spacer region are somewhat different from those of higher eukaryotes (Fig. 1). First, there is the gene for 5S RNA, which is transcribed by RNA polymerase III. Second, there are no spacer promoter or repeating enhancer elements resembling those found in Xenopus or mouse. However, there is a DNA element which is located just distal to the transcription termination site or 2.1 kb upstream of the initiation site (indicated as E in Fig. 1) which was reported to show a large stimulation of transcription in experiments using reporter mini-gene systems (3,4). This element is called the enhancer of yeast rDNA. In addition, some other DNA elements are universally present from yeast to human reflecting common features of repeating rDNA structures. For example, at the 3′-end of each rRNA gene there is a replication fork barrier (RFB) in all eukaryotes studied which allows progression of the DNA replication fork in the direction of 35S rRNA transcription, but not in the opposite direction (for yeast see 5–7; for other eukaryotes see 8 and papers cited therein). RFB has been shown to stimulate recombination within rDNA repeats, allowing expansion and contraction of rDNA repeats and contributing to sequence homogenization within rDNA repeats (9,10).
Figure 1.
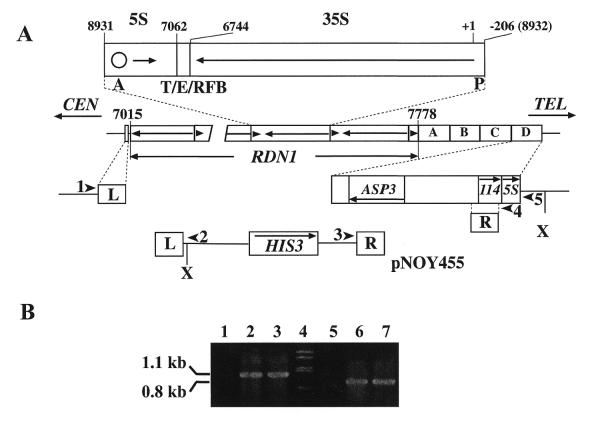
Complete deletion of chromosomal rDNA repeats. (A) The structure of the RDN1 locus on chromosome XII is shown in the middle. The RDN1 locus contains ∼150 tandem repeats of rRNA genes. A single repeat unit is expanded above RDN1, showing the 35S rRNA coding region (long arrow) and the 5S RNA coding region (short arrow, not to scale) as well as the locations of the 35S rRNA promoter (P), ARS (A, shown as a circle), the terminator of 35S rRNA transcription (T), the enhancer (E) and replication fork blocking element (RFB). One repeat unit of rDNA is 9137 bp and is numbered with respect to the Pol I transcription start site (+1). By convention, one repeat unit is shown as the fragment obtained after digestion of rDNA with SmaI. T, E and RFB overlap and are present within the fragment between the EcoRI (6744) and HpaI (7062) sites. The left and right end repeats of the RDN1 locus are not complete and the rDNA base pairs adjacent to the non-rDNA are indicated by their position numbers (7015 and 7778, respectively). The positions of flanking L and R sequences used for complete deletion of rDNA repeats are indicated. R is a 556 bp DNA sequence that is present in each of the four non-rDNA repeats, A–D, at the telomere-proximal end of the RDN1 locus. Each of these repeats is 3652 bp in length and contains gene ASP3, a 114 amino acid ORF (114) and the 5S RNA gene. L is a 565 bp DNA sequence located 297 bp from the centromere-proximal end of the RDN1 locus. Complete deletion of rDNA repeats was by standard gene replacement using an integration plasmid, pNOY455, linearized by digestion with BamHI as indicated. The linearized form of pNOY455 is shown. It should be noted that recombination using the R sequence homology could take place at any one of the four chromosomal R sequences. Strain NOY890 obtained was found to have used the R at repeat D as shown. Southern blot analysis of XhoI (indicated as X) digested DNA from NOY890 using the R sequence as probe yielded a signal with a size of ∼5.3 kb (data not shown). If recombination using the R sequence homology took place at A, B, C or D, one would expect to observe a 16.4, 12.7, 9.0 or 5.3 kb signal. Thus, the results support recombination at repeat D. Locations of PCR primers used to confirm the gene replacement (1–5) are indicated by arrowheads. (B) PCR analysis of DNA isolated from rdnΔΔ deletion strains NOY890 (lanes 2, 6) and NOY891 (lanes 3, 7) as well as control strain NOY505 (lanes 1 and 5). Products of PCR using primers 1 and 2 (lanes 1–3) and those using primers 3 and 5 (lanes 5–7) were analyzed together with molecular weight standards (lane 4) by agarose gel electrophoresis.
Despite extensive studies of DNA elements in rDNA repeats, our understanding of their function is limited. For example, gene promoters in all eukaryotes studied consist of two cis elements, the upstream element and the core. The former is required for high level transcription, but is dispensable, whereas the latter is essential for accurate transcription initiation. This conclusion was made using in vitro rRNA transcription systems and various in vivo reporter systems, but has never been confirmed by demonstrating their actual function in synthesizing rRNA in vivo. Much less is known about the mechanism(s) by which enhancer elements (including the yeast enhancer) stimulate rRNA synthesis in vivo. As in the case of mutational analysis of rRNA in ribosome function in vivo, the presence of many copies of rRNA genes in the chromosomal rDNA repeats has been an obvious obstacle to mutational analysis of the physiological importance of these DNA elements in rRNA synthesis as well as the mechanisms involved in their function. To overcome this obstacle we have constructed yeast strains in which the chromosomal rDNA repeats were completely deleted and growth was supported by the presence of a single kind of rDNA repeat on a plasmid. Using plasmid shuffling, this system can be used to examine the effects of any mutational changes of rDNA on growth of yeast cells, on rRNA transcription by RNA polymerase I (Pol I) and on rRNA structures involved in ribosome assembly and/or function. Using this system, we were able to confirm the importance of the promoter elements defined in previous studies and the results will be described in this paper. We have also succeeded in integrating a new rDNA unit at the original chromosomal locus, increasing the numbers of the rDNA copy by repeat expansion and thus constructing a new rDNA repeat structure, which will allow future mutational studies of DNA elements within a repeated rDNA structure.
MATERIALS AND METHODS
Media, strains, plasmids and genetic methods
YEPD medium contains 1% yeast extract, 2% BactoPeptone (Difco Laboratories, Detroit, MI) and 2% glucose. YEPGal medium is the same as YEPD except that 2% galactose is substituted for glucose. Synthetic glucose (SD) medium (2% glucose, 0.67% yeast nitrogen base) (Difco Laboratories) was supplemented with tryptophan and required bases as described by Sherman et al. (11). Synthetic galactose (SGal) is the same as SD but 2% galactose is substituted for glucose. To make solid media, 2% agar was added. Cells were grown at 30°C unless otherwise stated.
The yeast strains and plasmids used in this study are described in Table 1 and Figures 1 and 2. All genetic and cloning techniques were standard procedures (11,12). NOY505 was the parent strain to all the rdnΔΔ strains constructed. Complete deletion of rDNA repeats was done in two steps. The first step was construction of strain NOY758 by deleting most of the rDNA repeats according to the method described by Chernoff et al. (13) and was described previously (14). NOY505 was first transformed with pRDN-hyg1 using URA3 for selection. The transformants were directly plated on YEPD medium containing 300 µg/ml hygromycin (Calbiochem-Novabiochem, La Jolla, CA). NOY758 was isolated as one of the ‘stably’ hygromycin-resistant mutants with most (but not all) of the rDNA repeats deleted. The second step was to remove the remaining rDNA repeats in NOY758 by a standard gene replacement technique as described in Figure 1. For this purpose we constructed plasmid pNOY455, a derivative of pRS303 (15) containing the L and R sequences flanking the rDNA repeats (see Fig. 1). The 567 bp L sequence was isolated from clone λ5611 (ATCC no. 70595) by HpaI and EcoRI digestion and cloned into pRS304 (15) through SmaI and EcoRI sites to make pHW51. The 554 bp R sequence was isolated from clone λ2786 (ATCC no. 70081) by SpeI and PmlI digestion and cloned into pRS304 through SpeI and SmaI sites to make pHW52. The SacI (blunt-ended)–SalI fragment containing the L sequence from pHW51 was cloned between the EcoRI (blunt-ended) and SalI sites of pHW52 to make pHW53. The 1.5 kb PvuII fragment from pHW53 was cloned into the PvuII sites of pRS303 to make pNOY455. pNOY455 was linearized with BamHI (shown in Fig. 1A) and NOY758 was transformed with this linear fragment using HIS3 for selection, yielding the rdnΔΔ strain NOY890. NOY890 grows by transcription of the rRNA gene on plasmid pRDN-hyg1 by Pol I and we call this plasmid, as well as similar plasmids carrying the native 35S rRNA gene, the Pol I plasmid. NOY891 was constructed from NOY890 by replacing plasmid pRDN-hyg1 with pNOY353 (Fig. 2; 14). NOY891 synthesizes rRNA for growth by transcription of the GAL7–35S rDNA hybrid gene on the plasmid by RNA polymerase II (Pol II). We call plasmid pNOY353 and similar plasmids carrying the GAL7–35S rDNA hybrid gene the Pol II plasmid. [In our previous work on nucleolar structures of rdnΔΔ strains (14) we used a rdnΔΔ strain, NOY770, carrying a Pol I plasmid, which was obtained by the same method as described here, but details of the method were not described. After publication of the paper (14) we noted that strain NOY770 and other strains derived from NOY770 (NOY773 and NOY780) were unable to utilize mannose or glycerol, a phenotype which was different from the parent, NOY758. Since this phenotype was unexpected, we repeated the second step, transformation of NOY758 with the linearized pNOY455 DNA, and isolated NOY890. This strain, NOY890, grew normally on mannose and glycerol. Therefore, it is likely that the observed defect of the previous strains, NOY770 and strains derived from it, in utilizing mannose and glycerol was caused by a mutation which took place fortuitously during strain construction. Nucleolar structures of the new rdnΔΔ strains constructed from NOY890, NOY908 (which carries Pol I plasmid pNOY373) and NOY891 (which carries Pol II plasmid pNOY353) were very similar to those published previously using NOY770 (which carries a Pol I plasmid) and NOY773 (which was derived from NOY770 and carries a Pol II plasmid), respectively.]
Table 1. Yeast strains and plasmids used.
| Designation (strain/plasmid) | Description |
|---|---|
| Strain | |
| NOY505 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 |
| NOY556 | MATα ade2-1 ura3-1 trp1-1 leu2-3, 112 his3-11 can1-100 carrying pNOY103 |
| NOY758 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔ carrying pRDN-hyg1 |
| NOY890 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pRDN-hyg1 |
| NOY891 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY353 |
| NOY892 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 |
| NOY903 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and pNOY373 |
| NOY904 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and pNOY444 |
| NOY905 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and pNOY445 |
| NOY907 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY130 and YEp351 |
| NOY908 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY373 |
| NOY909 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY444 |
| NOY910 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::HIS3 carrying pNOY373 and pNOY353 |
| NOY989 | MATa ade2-1 ura3-1 trp1-1 leu2-3,112 his3-11 can1-100 rdnΔΔ::URA3 carrying pNOY353 |
| Plasmid | |
| YEp351 | High copy number plasmid carrying LEU2, 2µ, amp (32) |
| pNOY103 | High copy number plasmid carrying GAL7–35S rDNA, ADE3, URA3, 2µ, amp (20) |
| pNOY130 | High copy number plasmid carrying GAL7–35S rDNA, 5S rDNA, URA3, 2µ, amp |
| pRDN-hyg1 (pNOY290) | High copy number plasmid carrying hygromycin-resistant rDNA with promoter starting from –206, leu2-d, URA3, 2µ, amp (13; Fig. 2) |
| pNOY353 | A derivative of high copy number plasmid pTV3 (16) carrying GAL7–35S rDNA, 5S rDNA, TRP1, 2µ, amp (14; Fig. 2) |
| pNOY373 | A derivative of high copy number plasmid YEp351carrying rDNA with promoter starting from –206 with XhoI–NotI flanked enhancer, LEU2, 2µ, amp (Fig. 2) |
| pNOY444 | A derivative of high copy number plasmid YEp351carrying rDNA with promoter starting from –100 with XhoI–NotI flanked enhancer, LEU2, 2µ, amp |
| pNOY445 | A derivative of high copy number plasmid YEp351 carrying rDNA with promoter starting from +9 with XhoI–NotI flanked enhancer, LEU2, 2µ, amp |
| pNOY455 | A derivative of pRS303 carrying the R segment inserted between SpeI and EcoRV and the L segment inserted between SmaI and EcoRI (see Fig. 1A for the structure after linearization with BamHI) |
| pNOY500 | A derivative of pRS314 (15) carrying the L and R segments in addition to a single rDNA repeat carrying hyg1 (see Fig. 6 for the structure of the insert; see text for details of construction) |
Figure 2.
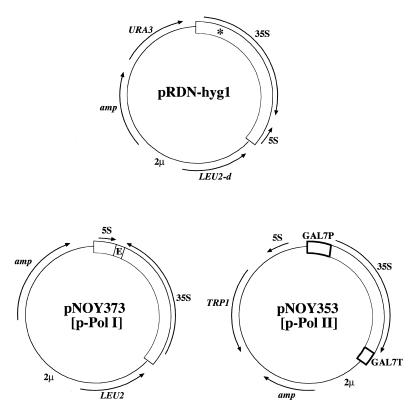
Plasmids carrying rDNA which were used to support growth of rdnΔΔ strains. Structures of plasmids pRDN-hyg1, pNOY373 (p-Pol I) and pNOY353 (p-Pol II) are shown. Plasmid pRDN-hyg1 carries a T→C mutation at the site (indicated by an asterisk) encoding position 1756 of 18S rRNA (13). E in the rDNA carried by pNOY373 is the segment containing T/E/RFB (see Fig. 1) and two flanking lines indicate the two restriction sites created by mutagenesis.
Plasmid pNOY373 (Fig. 2), used as a Pol I plasmid, was constructed in the following way. First, a single 9.1 kb rDNA unit, which corresponds to that carried by pRDN-hyg1 but without the hyg1 mutation, was obtained by cutting pRDN-wt (13) at the flanking BamHI sites and then cloned into the BamHI site of pTV3, a TRP1, ARS, 2µ plasmid vector (16). Because the rDNA derived from pRDN-wt contained a XhoI site in its 5′-proximal region, the 1.5 kb SmaI–SacII segment containing this site was then replaced by the corresponding segment without the XhoI site derived from a different rDNA clone, prib105 (17), leading to plasmid pNOY372. In order to make a deletion of the terminator/enhancer/RFB region (the region from the EcoRI site at 6743 to the HpaI site at 7060; see Fig. 1) for some other studies not described in this paper, the 2.6 kb PflMI–BamHI fragment containing this region was replaced by the corresponding DNA fragment derived from pHW73, a plasmid with rDNA which contains mutations changing TA at 6740 and 6741 to CG, thus creating a XhoI site CTCGAG (6738–6743), and mutations changing GACAT at 7068–7072 to CGGCC, thus creating a NotI site CGGCCG (7068–7073). The resulting plasmid, pNOY374, thus contains a XhoI–NotI flanked terminator/enhancer/RFB region. The creation of these XhoI and NotI sites did not affect the expression of rDNA. Oligonucleotide-directed mutagenesis was used to make these mutations (18). Finally, the 9.1 kb PstI–BamHI rDNA repeat from pNOY374 was inserted into the PstI and BamHI sites of YEp351, leading to pNOY373.
Plasmids carrying deletions of the rDNA promoter from upstream regions were derivatives of pNOY373 and constructed by replacing the 2.15 kb PstI–SacI fragment of pNOY373 by the corresponding PstI–SacI fragments derived from deletion derivatives of plasmid pSIRTΔ described previously (19); these derivatives were called 5′Δ–208/–155, 5′Δ–208/–101, 5′Δ–208/–76, 5′Δ–208/–52, 5′Δ–208/–38 and 5′Δ–208/+8, respectively (19).
Integration of a new rDNA repeat and its expansion were carried out using a DNA fragment obtained from plasmid pNOY500. This fragment consists of a single rDNA unit carrying the hyg1 mutation with the L sequence added at the left end and the L plus R sequences at the right end (see Fig. 5). Plasmid pNOY500 is a derivative of pRS314 (15) and was constructed as follows. The first step was to amplify the L and R sequences by PCR from yeast genomic DNA. The L sequence was amplified in two different ways using two sets of primer, 101/102 and 103/104. Primer 101 (CCCCCTGCAGTAATTTTCAGTTAATGGATGGTATCAGACG) hybridized to the 5′-end sequence of L with the additional PstI restriction sequence (underlined) added just 5′ to L. Primer 102 (CCCCGAATTCTTTTTACTATAAATCCAATTACTTATATGT) hybridized to the 3′-end sequence of L with the additional EcoRI restriction sequence (underlined) added just 3′ to L. Primer 103 (CCCCACTAGTATTTCTGTTTGTCAGTAATTTTCAGTTAATGG) hybridized to the 5′-end sequence of L with the additional SpeI restriction sequence (underlined) added just 5′ to L. Primer 104 (CCCCTCTAGATATGTGTATTACGTGATATACAGTGACAGCC) hybridized to the 3′-end sequence of L with the additional XbaI restriction sequence (underlined) added just 3′ to L. The 101/102 primer set specifically amplified the PstI–L–EcoRI 0.54 kb DNA fragment. The 103/104 primer set specifically amplified the SpeI–L–XbaI 0.53 kb DNA fragment. The R sequence was amplified by the 105/106 primer set. Primer 105 (CCCCATCGATGGTCGCATTCATTGCTACTCATCTGCGAGTG) hybridized to the 5′-end sequence of R with the additional ClaI restriction sequence (underlined) added just 5′ to R. Primer 106 (CCCCGTCGACGCAAAGTGCAGGAGCITTTGGAAAGAGCGTGC) hybridized to the 3′-end sequence of R with the additional SalI restriction sequence (underlined) added to the 3′-end of R. PCR using the 105/106 primer set specifically amplified the ClaI–R–SalI 0.56 kb DNA fragment. Then, the SpeI–L–XbaI DNA fragment was cloned into the SpeI site on pRS314 with orientation of the XbaI end adjacent to the BamHI site on the plasmid, followed by insertion of the PstI–L–EcoRI and ClaI–R–SalI fragments at their respective restriction sites on the same plasmid. Finally, the rDNA-hyg1 fragment (obtained as a PstI–BamHI fragment from a hyg1 derivative of pNOY373) was inserted between the two L sequences utilizing the PstI and BamHI sites, yielding pNOY500. The complete ∼10.7 kb reintegration fragment was isolated after digestion of pNOY500 with SalI and SpeI.
Figure 5.
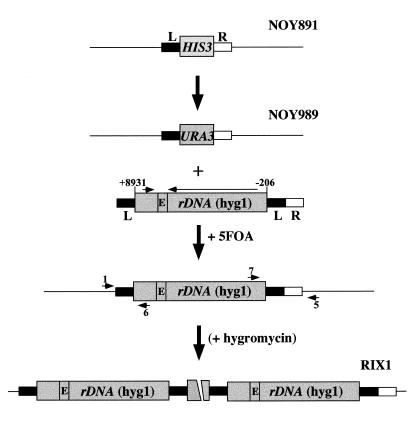
Integration and expansion of a new rDNA repeat. The L (filled box) and R (open box) sequences used for integration of rDNA are those described in Figure 1. Locations of PCR primers used for confirmation of integration (1 and 6; 7 and 5) are indicated by arrowheads. E indicates the location of E/T/RFB shown in Figure 1. As mentioned in the text, hygromycin selection was used in construction of the RIX1-1 and RIX1-2 strains analyzed in Figures 6 and 7, but repeat expansion takes place without hygromycin selection and, hence, this selection step is parenthesized.
PCR primers used to confirm complete deletion and integration of new rDNA repeats
Complete deletion of rDNA repeats was confirmed by PCR using primers 1–5 as indicated in Figure 1. Integration of new rDNA repeats was confirmed by PCR using primers 1 and 6 and 7 and 5, as indicated in Figure 5. The sequences of these primers were: primer 1, 5′-GCTGAGACCTTTCCATTGGG-3′; primer 2, 5′-GGAAACAGCTATGACCATG-3′; primer 3, 5′-GTAATACGACTCACTATAGGGC-3′; primer 4, 5′-GAAGCCGGGCGATGGCTCCGCC-3′; primer 5, 5′-GACGGGGTCCGGGTAAC-3′; primer 6, 5′-TCCGTATTTTCCGCTTCCGC-3′; primer 7, 5′-CTAAACCCCCCCTCCC-3′.
Other methods
Analysis of RNA synthesis by [3H]uridine incorporation was carried out as described previously (20) and the amounts of radioactive RNA were determined by scintillation counting after cutting RNA bands from the gel. Analysis of chromosome XII by contour-clamped homogeneous electric field (CHEF) electrophoresis was done as described previously (21). Isolation of DNA and Southern hybridization were carried out as described by Maniatis et al. (22).
RESULTS
Construction of rdnΔΔ strains with complete deletions of chromosomal rDNA
The structure of yeast rDNA repeats on chromosome XII is shown in Figure 1A. We used two steps to delete chromosomal rDNA repeats completely. The first step was to follow the method originally devised by E. Morgan and described by Chernoff et al. (13). Strain NOY505 was transformed with pRDN-hyg1 (Fig. 2) using URA3 for selection. Plasmid pRDN-hyg1 carries, in addition to URA3, a single copy of rDNA repeats with a hygromycin-resistant mutation. As in the case of many other aminoglycoside antibiotics acting on ribosomes, the hygromycin-resistant mutation is recessive to the wild-type allele. NOY758 was isolated as a stable hygromycin-resistant mutant and its rDNA repeats were mostly but not completely deleted (the mutation called rdnΔ; see 14). The second step was a standard gene replacement method using HIS3 for selection to remove the remaining rDNA repeats completely (the deletion called rdnΔΔ), as schematically shown in Figure 1A. Knowing the DNA sequences flanking the rDNA repeats, we selected a centromere-proximal 565 bp DNA segment, called L, and a telomere-proximal 556 bp DNA segment, called R, for homologous recombination and constructed plasmid pNOY455 for this purpose. Complete deletion of the chromosomal rDNA repeats was achieved. One of the resulting rdnΔΔ strains, NOY890, as well as a derivative, NOY891 (see below), were used to confirm complete deletion of the rDNA repeats. DNA from these strains was examined by PCR. PCR using primers 1 and 2 (shown in Fig. 1A) gave the expected 1.1 kb product (Fig. 1B, lanes 2 and 3), confirming recombination at the L segment, which must have led to deletion of 297 bp of non-rDNA on the centromere-proximal side of the rDNA repeats. On the telomere-proximal side of the rDNA repeats there are four 3652 bp repeats, each of which contains ASP3, an uncharacterized open reading frame encoding 114 amino acids and a single copy of the 5S RNA gene (see Fig. 1A; the four repeats are indicated as A–D). The rdnΔΔ strains NOY890 and NOY891 were analyzed with respect to the site of homologous recombination within these 3.7 kb repeats. First, PCR using primers 3 and 4 (shown in Fig. 1A) gave the expected 0.7 kb product, demonstrating that recombination took place using the R sequence of the plasmid DNA and the R sequence on one of the 3.7 kb repeats (data not shown). Second, the results of Southern analysis of XhoI digested DNA from strain NOY890 were consistent with those expected from recombination at the R sequence in the last copy (copy D in Fig. 1A) (data not shown; for further explanation see the legend to Fig. 1). Finally, primer 5, which hybridizes to a sequence outside the 3.7 kb repeats, was used in combination with primer 3 for PCR and a 0.8 kb reaction product was obtained for both NOY890 and NOY891 (Fig. 1B, lanes 6 and 7), confirming recombination at the last copy of the 3.7 kb repeats. As expected from complete deletion of chromosomal rDNA repeats, Southern hybridization designed to detect chromosomal rDNA repeats showed no signal above background for genomic DNA obtained from the rdnΔΔ strain (NOY890), but detected a small number of residual rDNA repeats (corresponding to a few percent of control strain NOY505) for genomic DNA from the rdnΔ strain (NOY758) (data not shown). Analysis of chromosome XII of the rdnΔΔ strain NOY891 by CHEF electrophoresis showed a chromosome band with a homogeneous size, which migrated to a position close to that corresponding to the size (1.1 Mb) calculated for complete deletion of rDNA repeats (23) and was in clear contrast to the size of ∼2.6 Mb with heterogeneous size distribution of chromosome XII from the control strain (NOY505) (see below and Fig. 6). All these results demonstrate that the chromosomal rDNA repeats, the RDN1 locus, have been completely deleted in NOY890 (and other rdnΔΔ strains derived from it) and growth of this strain is dependent on a single rDNA repeat carried on plasmid pRDN-hyg1.
Figure 6.
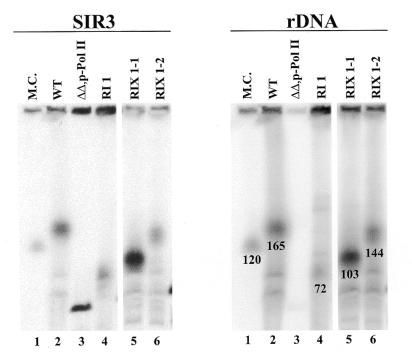
Analysis of chromosome XII by CHEF electrophoresis. Chromosomal DNA was isolated from the following strains and analyzed: NOY505 (WT), lane 2; NOY891 (ΔΔ,p-Pol II), lane 3; a clone with new integrated rDNA repeats obtained without the hygromycin selection step (RI1), lane 4; two clones with new integrated rDNA repeats obtained after the hygromycin selection step (RIX1-1 and RIX1-2), lanes 5 and 6; marker yeast chromosomes (M.C.) (obtained from Bio-Rad, Richmond, CA) were also run, lane 1. The left and right panels show autoradiograms obtained after hybridization with a SIR3 probe and a rDNA probe, respectively. (In this experiment hybridization with the SIR3 probe was done first, followed by hybridization with the rDNA probe. The faint radioactive band seen for the ΔΔ,p-Pol II strain in the right panel corresponds to chromosome XII and the radioactivity detected on this chromosome appears to be due to incomplete stripping of the SIR3 probe or background. In other experiments no hybridization with the rDNA probe was detected for chromosome XII in this strain.) The size of chromosome XII was determined from a plot of the relative migration of other chromosomes versus chromosome size (for each sample) and comparing the plot with that obtained for marker chromosomes. The size of XII in the marker chromosomes is 2.2 Mb according to the supplier and that carried by NOY891 (lane 3) should be 1.1 Mb because of complete deletion of the rDNA repeats. (The estimated values for the latter ranged from 1.1 to 1.3 Mb.) The sizes of chromosome XII in lanes 2, 4, 5 and 6 were estimated to be 2.6, 1.8, 2.1 and 2.5 Mb, respectively, and they correspond to rDNA repeat numbers of 165, 72, 103 and 144, respectively. The number of rDNA repeats was calculated by subtracting 1.1 Mb for non-rDNA from the values for the size of chromosome XII and then dividing by the repeat length of 9.1 kb for the native rDNA or 9.7 kb for the reintegrated and expanded rDNA. The values thus obtained are indicated under the relevant chromosome XII bands in the right panel. In a separate experiment the size of the rDNA repeats was estimated after digestion with a restriction enzyme (FspI) that cuts outside rDNA repeats. Similar values were obtained.
Either Pol I-driven or Pol II-driven 35S rRNA genes can support growth of rdnΔΔ strains
We have previously demonstrated that the GAL7 promoter-driven 35S rRNA gene (GAL7–35S rDNA) on a multi-copy plasmid (such as pNOY103) allows growth of mutants defective in Pol I or Pol I-specific transcription factors on galactose (20,24). We constructed plasmid pNOY353, which carries the 5S RNA gene in addition to GAL7–35S rDNA (14; Fig. 2). Through plasmid shuffling on galactose medium we constructed strain NOY891, which is identical to NOY890 but carries plasmid pNOY353 (called the helper Pol II plasmid or p-Pol II) instead of pRDN-hyg1. As expected, this strain was galactose dependent and did not grow on glucose. The doubling time of this strain in YEPGal medium at 30°C was ∼6.5 h. We also constructed a strain which is identical to NOY890 but carries a single rDNA repeat without the hyg1 mutation on plasmid pNOY373 (Fig. 2; called the helper Pol I plasmid or p-Pol I). This strain, NOY908, grew on both galactose and glucose and had a doubling time of ∼3 h in YEPGal and ∼2 h in YEPD at 30°C. Thus, growth defects observed in the rdnΔΔ strain carrying a Pol I plasmid are rather small compared to control strains with the intact rDNA repeats (e.g. NOY505 showing doubling times of 2.2 and 1.7 h in YEPGal and YEPD, respectively.).
It was previously shown that in the presence of certain mutations yeast variants are produced which are able to transcribe chromosomal rDNA repeats from a cryptic promoter using Pol II (21,25,26). Transcription of rDNA from Pol I plasmids in rdnΔΔ strains is carried out by Pol I and not by Pol II. This was demonstrated in the following way. We first constructed a rdnΔΔ strain (NOY911) carrying both a Pol II plasmid (pNOY353) and a Pol I plasmid (pNOY373). This strain grew on both glucose and galactose. The RPA135 gene, which encodes the second largest subunit of Pol I, was then disrupted by the LEU2 gene as was done previously (24). The resultant strain became galactose dependent in its growth, demonstrating that the 35S rRNA gene on the Pol I plasmid is in fact transcribed by Pol I in these rdnΔΔ strains.
Deletion analysis of the Pol I promoter
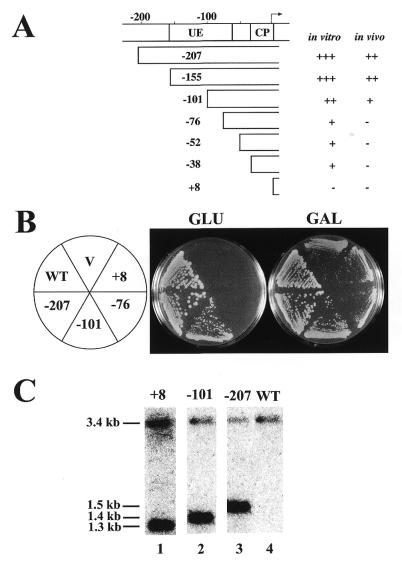
A series of 5′ promoter deletions was constructed (Fig. 3A) and the effects of the deletions on Pol I transcription of rDNA were examined by introducing Pol I plasmids with these deletions into rdnΔΔ strain NOY892, which carries a helper Pol II plasmid (pNOY130). Plasmid pNOY130 is similar to pNOY353 in allowing rdnΔΔ strains to grow on galactose but not on glucose. When control Pol I plasmid pNOY373 with the intact promoter was introduced into this strain it allowed the strain to grow on glucose (NOY903; Fig. 3B, –207), whereas no growth on glucose was observed when the vector, YEp351, was introduced (NOY907; Fig. 3B, V). A plasmid with a deletion to –155 allowed growth on glucose like the control Pol I plasmid, but a plasmid with a deletion to –101 allowed only weak growth on glucose relative to the control strain NOY903 (NOY904; Fig. 3B, –101). Plasmid constructs with further deletions to –76, –52, –38 or +8, allowed no growth on glucose (Fig. 3A and B and data not shown). The partial reduction in growth on glucose caused by deletion to –101 relative to the control strain (NOY903) was also confirmed by measuring doubling times in liquid cultures. The doubling times of the control and –101 deletion strains in YEPD were 2.0 and 3.4 h, respectively, at 30°C (see doubling time, DT, in Table 2).
Figure 3.
Deletion analysis of the 35S rDNA promoter using rdnΔΔ strains. (A) The promoter region and various deletion constructs. The names of deletion constructs indicate the last upstream base pair deleted. Numbering is with respect to the transcription start site (+1). Locations of the upstream element (UE) and the core promoter (CP) within the promoter are indicated. A summary of transcription activities of the various promoters in vitro (19) and in vivo [as judged by the extent of rRNA synthesis to allow cell growth (B in this figure and other data not shown) and by rRNA accumulation shown in Fig. 4] is also given. (B) Growth of rdnΔΔ strains on YEPD (Glu) and YEPGal (Gal) plates incubated at 30°C for 5 days. All of these strains carry a helper Pol II plasmid (pNOY353) and deletion derivatives (–101, –76 and +8 as indicated) of a Pol I plasmid (pNOY373, indicated as –207) or a control vector (V). NOY505 (WT) was also included. (C) Southern blot analysis of plasmid copy numbers. DNA was prepared from NOY505 (lane 4, WT), NOY905 (lane 1, +8), NOY909 (lane 2, –101) and NOY908 (lane 3, –207). The DNA preparations were digested with BamHI and EcoRI, subjected to agarose gel electrophoresis and analyzed using a radioactive probe containing LEU2. A picture obtained with a PhosphorImager is shown. The fragment containing the chromosomal LEU2 gene has a calculated size of ∼3.4 kb and that from pNOY373 (in NOY908) has a calculated size of ∼1.5 kb. The sizes of those from the other two plasmids are shorter due to the deletions. Quantification of plasmid copy numbers relative to chromosomal LEU2 gave the following values: pNOY445 (in NOY905), 12; pNOY444 (in NOY909) and pNOY373 (in NOY908), 70–100.
Table 2. Relative accumulation of 18S and 25S rRNA in rdnΔΔ strains carrying various Pol I plasmids.
| Strain | Glucose | Galactose | ||||||
|---|---|---|---|---|---|---|---|---|
| RDN1 | p-Pol IIa | p-Pol I | rRNA | rRNA – bg | DT (h) | rRNA | DT (h) | |
| NOY556 | WT | + | 1.14 | 1.12 | 1.7 | 1.07 | 2.2 | |
| NOY903 | ΔΔ | + | –207 | 0.48 | 0.46 | 2.0 | 0.50 | 2.7 |
| NOY904 | ΔΔ | + | –101 | 0.15 | 0.13 | 3.4 | 0.31 | 3.9 |
| NOY905 | ΔΔ | + | +8 | 0.015 | (0) | ND | 0.31 | 6.5 |
| NOY907 | ΔΔ | + | (vector) | 0.020 | ND | 0.39 | 6.5 |
In experiments similar to those shown in Figure 4, [25S + 18S]/[5S + tRNA] values were calculated and averages obtained from four experiments are given under rRNA. For the values obtained for glucose medium, corrected values obtained after subtraction of the background value (0.020) are given under rRNA – bg. No such corrections are shown for galactose medium because of the substantial rRNA synthesis from the GAL7–35S rDNA fusion gene (see text). Growth rates of these strains in galactose and glucose media (YEPGal and YEPD, respectively) are also shown as doubling time (DT) in hours.
aNOY556 carries pNOY103 and all other strains carry pNOY130 as the Pol II plasmid, thus making these strains Ura+.
ND, not determined. NOY904 and NOY907 do not grow on glucose plates and hence no measurements were performed.
The abilities of various Pol I deletion plasmids to allow growth on glucose are also reflected in the growth rates of these rdnΔΔ strains on galactose. The rdnΔΔ strains without Pol I plasmids (NOY892 and NOY907) had doubling times of ∼6.5 h on YEPGal. The three strains with extra Pol I plasmids, the control plasmid (–207), the partially active plasmid (–101) or an inactive plasmid (+8), showed doubling times of 2.7, 3.9 and 6.5 h, respectively (see DT in Table 2). The differences in growth rates on galactose among these three strains were also evident by comparing colony sizes of these strains on galactose plates (Fig. 3B).
The copy number of the two plasmids with –101 and +8 promoter deletions and the control plasmid (–207) was determined by Southern blot analysis using DNA from cells grown in SGal (leucine drop-out) medium. DNA digested with BamHI and EcoRI was analyzed using a LEU2 DNA probe and the radioactive signals produced by the fragments derived from the plasmids (1.5 kb or smaller due to deletions) were quantified by comparing those produced by a single copy chromosomal 3.4 kb LEU2 fragment. The results showed that both the –207 and –101 plasmids were present in approximately the same number (70–100 copies; Fig. 3C, lanes 2 and 3). In contrast, the +8 deletion plasmid showed only ∼12 copies (Fig. 3C, lane 1), which was about the same as the copy number of the YEp351 vector plasmid (data not shown). The elevated level of the –207 and –101 plasmids is probably due to selection for faster growth while the +8 plasmid did not show such an increase because it did not have any growth promoting activity. It is interesting to note, however, that the –101 plasmid did not show a higher copy number than that of the control –207 plasmid, even though the growth rate of the rdnΔΔ strain carrying the former was definitely slower than that carrying the latter. Perhaps, the copy numbers (70–100) achieved by both strains represents the upper limit attainable by selection under the growth conditions used in these experiments, i.e. the amounts of Pol I and/or transcription factors may become limiting and further increases in copy numbers would not increase rRNA synthesis rates.
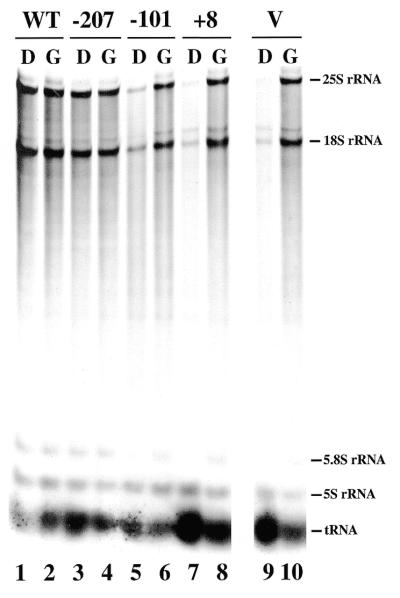
Effects of partial and complete deletions of the promoter sequence were then studied by measuring the rate of rRNA accumulation in the rdnΔΔ strains carrying the –207, –101, +8 or YEp351 plasmids. These yeast strains were grown in SGal (uracil drop-out) medium to a cell density of A600 = 0.2–0.3. After harvesting and washing of cells, each of the cultures was divided into two parts, one incubated in the same SGal (uracil drop-out) medium and the other in SD (uracil drop-out) medium for 1 h. 3H-labeled uridine was then added to each culture and incubation was continued for another hour to label RNA. RNA samples prepared from these cultures were then analyzed by electrophoresis on polyacrylamide–agarose composite gels followed by autoradiography (Fig. 4). The amounts of 3H-labeled RNA in 25S rRNA, 18S rRNA, 5S RNA and tRNA were determined and the sum of the values for 25S rRNA and 18S rRNA (Pol I transcripts) was normalized to the sum of the values for 5S RNA and tRNA (Pol III transcripts). The results obtained in this way are shown in Table 2.
Figure 4.
Polyacrylamide–agarose gel electrophoresis of RNA synthesized in rdnΔΔ strains carrying a Pol II helper plasmid and a Pol I helper plasmid with various deletions. Strains NOY556 (WT), NOY903 (–207), NOY904 (–101), NOY905 (+8) and NOY907 (V) were grown in SGal medium. At a cell density (A600) of 0.2 the cultures were divided: one part was shifted to SD medium (D) and the other was kept in the same medium (G). One hour after the shift, cells were labeled with [3H]uridine for 1 h. RNA was isolated from each culture and samples containing approximately equal radioactivity were subjected to electrophoresis. An autoradiogram of the gel is shown.
Transcription from the GAL7–35S rDNA fusion gene by Pol II is repressed by glucose. Only a very low level of rRNA accumulation (0.020) was observed in glucose medium for the strain (NOY907) not carrying a Pol I plasmid (Table 2). The residual incorporation into rRNA may be the result of read-through transcription from an unknown promoter located upstream of the GAL7 promoter on pNOY130. As expected from the absence of growth on glucose, the strain carrying the +8 deletion plasmid (NOY905) showed rRNA accumulation similar to the background seen for the above strain without a Pol I plasmid (NOY907). Thus, as was found in in vitro experiments, deletion to +8 completely abolished Pol I transcription in vivo. The strain carrying the –207 Pol I plasmid (NOY903) showed a value (0.46) which was 41% of the control strain (NOY556) with intact chromosomal rDNA repeats. (Doubling times of NOY903 and NOY556 in YEPD were 2.0 and 1.7 h, respectively. Thus, the lower rRNA accumulation in NOY903 may reflect the slower growth rate relative to NOY556.) The strain carrying the –101 Pol I plasmid (NOY904) showed a value for rRNA accumulation (0.13) which was 28% of the control strain (NOY903) with the intact Pol I plasmid. Thus, in agreement with the previous in vitro results (19,27; see Discussion), deletion from upstream to –155 did not cause any decrease in transcription, but further deletion to –101 caused a partial decrease (3.6-fold) in rDNA transcription. This partial decrease in the efficiency of Pol I transcription caused by the –101 deletion is consistent with the slower growth of strain NOY904 carrying the Pol I plasmid with this deletion. We conclude that the upstream element defined in vitro as the region which is required for stable binding of UAF and is stimulatory for rRNA transcription (28) plays an important role in rRNA transcription and cell growth in vivo.
Table 2 also shows rRNA accumulation data in galactose medium. Since the rRNA accumulation in galactose is the sum of that synthesized by Pol II using the Pol II plasmid (pNOY130) and that synthesized by Pol I using the Pol I plasmid (pNOY373) or its deletion derivatives, accurate quantification of rRNA accumulation due to Pol I transcription may be difficult because of a high degree of rRNA production from the GAL7 promoter. Although we detected stimulation by the intact (Pol I) plasmid, we failed to detect the expected (small) stimulation by the –101 deletion plasmid compared to the control, the inactive +8 Pol I plasmid or the vector plasmid.
Plasmids with deletion beyond –101, i.e. deletions –76, –52 and –38, did not support growth of rdnΔΔ strains on glucose (Fig. 3A and B and data not shown). Although we did not measure rDNA transcription rates in the strains carrying these plasmids, we conclude that basal transcription from the core promoter, if it takes place in vivo as was observed in vitro, is not sufficient to allow cell growth.
Integration and copy number expansion of a new rDNA repeat
We initially used the method outlined in Figure 5 in order to integrate a new rDNA repeat into the original locus of rdnΔΔ strains. For this purpose we used a DNA fragment obtained from a plasmid (pNOY500) which carries a single rDNA repeat with the hyg1 mutation and the L region and the L plus R regions flanking the rDNA unit as shown in Figure 5. The presence of the L element on the right side of the rDNA was designed to initiate expansion by an unequal crossing-over or a DNA breakage–repair process. The HIS3 gene that replaced RDN1 in the rdnΔΔ strain NOY891 was first replaced with URA3 and the resulting strain (NOY989) was transformed with the DNA fragment from pNOY500, selecting transformants by the loss of URA3. Colonies formed on plates containing 5-fluoroorotic acid (5-FOA) were screened for integration of the rDNA fragment by PCR using primers 1 and 6 for the left junction and primers 7 and 5 for the right junction, as indicated in Figure 5. Approximately 40% of colonies formed on 5-FOA plates showed the expected integration. Several independent colonies were then grown on YEPGal containing hygromycin. The original rdnΔΔ strain carried a helper Pol II plasmid and could grow only on galactose-containing media. In the initial experiments the concentration of hygromycin was gradually increased on the assumption that hygromycin treatment would facilitate expansion of the integrated rDNA carrying the recessive hyg1 mutation, since the original cells were growing using multiple copies of the helper Pol II plasmid carrying the wild-type rDNA. Independent clones grown on YEPGal containing hygromycin were then tested for their ability to grow on glucose. All of them were found to be able to grow on glucose and had lost the helper Pol II plasmid.
Integration of the rDNA unit and its expansion within chromosome XII were confirmed by CHEF electrophoresis followed by hybridization to detect chromosome XII using a reference probe for SIR3 (which is on chromosome XII) and a rDNA probe (Fig. 6). Chromosome preparations from the original rdnΔΔ strain with a helper Pol II plasmid (NOY891) showed a sharp band on chromosome XII with the SIR3 probe, but not with the rDNA probe. Its size was close to 1.1 Mb, which is the size calculated for complete deletion of the rDNA repeats (Fig 6, lane 3). Two independent clones, RIX1-1 and RIX1-2 (RIX for rDNA integration and expansion), showed broad chromosome XII bands detected with both the SIR3 and rDNA probes with sizes of ∼2.1 and 2.5 Mb, respectively. These sizes correspond to estimated rDNA repeat numbers of ∼103 and 144, respectively (Fig. 6, lanes 5 and 6). We also analyzed a clone (RI1, for rDNA integration) obtained after 5-FOA selection which was not subjected to hygromycin selection. Somewhat unexpectedly, we found that expansion of the rDNA copy had taken place without hygromycin selection (Fig. 6, lane 4). The size of the chromosome was estimated to be ∼1.8 Mb, which corresponds to an rDNA repeat number of ∼72. [In a subsequent experiment this particular clone was further analyzed after additional growth on YEPGal plates without hygromycin (∼40 more generations). The size of chromosome XII estimated by CHEF electrophoresis was larger, indicating a further expansion of rDNA to a repeat number of ∼103 (data not shown).] It appears that expansion of the native rDNA copy (which is transcribed by Pol I) on chromosome XII may give a growth advantage relative to the original strain whose growth is dependent on transcription of GAL7–35S rDNA on the helper plasmid by Pol II, leading to repeat expansion without hygromycin selection.
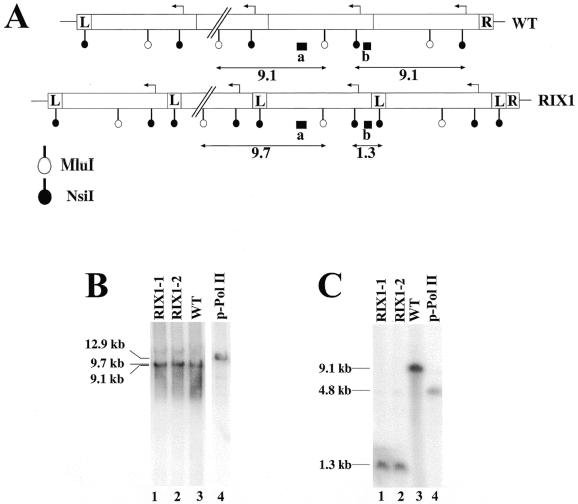
The expanded rDNA repeats on chromosome XII in the RIX1 strains are expected to have the structure shown in Figure 7A, namely each repeating unit has an extra sequence derived from the 565 bp L segment in addition to the native 9.1 kb rDNA repeat. Southern blot analysis using MluI confirmed the repeating structure (Fig. 7B, lanes 1 and 2) and that using NsiI confirmed the presence of the expected L element adjacent to the native 9.1 kb rDNA unit, the feature which clearly distinguishes it from the native rDNA repeats (Fig. 7C, compare lanes 1 and 2 with lane 3).
Figure 7.
Comparison of the structures of integrated new rDNA repeats and that of native rDNA repeats. (A) The structure of integrated new rDNA repeats (RIX1) and that of native rDNA repeats (WT) are schematically shown. The sizes of the L and R segments are not to scale. Expected sizes of fragments detected after digestion of genomic DNA with MluI or NsiI and hybridized with radioactive probe a or b (indicated as a filled box) are indicated (in kb). (B and C) Southern analysis of genomic DNA using MluI and probe a (B) and that using NsiI and probe b (C). DNA was isolated and analyzed from the following strains: two independent clones (RIX1-1 and RIX1-2) with integrated new rDNA repeats obtained after hygromycin selection, lanes 1 and 2; NOY505 (WT), lane 3; plasmid pNOY353 (p-Pol II), lane 4. The sizes of bands estimated from size markers run in parallel are indicated.
The presence of the extra 565 bp L sequence in rDNA repeats does not appear to be significantly harmful to cell growth. Growth rates of five independent RIX1 clones in YEPD were measured and all of them showed doubling times of ∼2.5 h. Although this doubling time is significantly longer than the 1.7 h observed for wild-type strain NOY505 (i.e. the growth rate of RIX1 was 68% of the wild-type), it is known that the hygromycin resistance mutation causes slower growth. We observed that rdnΔΔ strain NOY758 carrying pRDN-hyg1 grows with a doubling time of 3.3 h in YEPD, whereas the doubling time of rdnΔΔ strain NOY903 carrying a helper Pol I plasmid with the wild-type rDNA (pNOY373) is ∼2.0 h in the same medium (i.e. the growth rate achieved with a Pol I plasmid with the hyg1 mutation was 61% of that achieved with a similar Pol I plasmid without the mutation).
DISCUSSION
We have constructed yeast strains in which chromosomal rDNA repeats are completely deleted and rRNA is synthesized from a DNA template on a plasmid, either rRNA genes transcribed by Pol I or the GAL7–35S rDNA fusion gene transcribed by Pol II. The growth rate of rdnΔΔ strains with a standard wild-type Pol I plasmid (e.g. NOY908) was only ∼20% slower than the strain without such a chromosomal rDNA deletion (e.g. NOY505) in YEPD. We have not determined the reason for this small decrease in growth rate, but speculate that it is caused by disruption of the intact nucleolar structure. We have previously reported nucleolar structures of rdnΔΔ strains carrying a helper Pol I plasmid (NOY770) or helper Pol II plasmid (NOY773) and found alterations of nucleolar structures in these rdnΔΔ strains (14). The rdnΔΔ strains used in the current work showed the same alterations, namely the crescent-shaped organization of the nucleolus as seen for the wild-type (RDN1) yeast strain was absent and the rdnΔΔ strains carrying a helper Pol I plasmid showed fragmented mini-nucleoli localized primarily at the nuclear periphery, whereas the rdnΔΔ strains carrying a helper Pol II plasmid contained a round nucleolus that often lacked extensive contact with the nuclear periphery (M.Oakes and M.Nomura, unpublished results; see 14). Altered nucleolar structures might have caused the inefficiency in rDNA transcription which we observed (Fig. 4 and Table 2). However, we cannot conclude that the reduction in rRNA synthesis is a direct consequence of the disrupted nucleolar structure rather than an indirect consequence of some other defect, e.g. inefficient rRNA processing/ribosome assembly or possible defects in regulation of the cell cycle (for the involvement of nucleolar structures in cell cycle regulation see for example 29). If disrupted nucleolar structures in rdnΔΔ strains are responsible for the small reduction in growth rate one might expect that strains with a new integrated rDNA in an expanded form may have a growth rate similar to the wild-type, since these strains contain a crescent-shaped nucleolar structure similar to that in wild-type strains as judged by immunofluorescence microscopy (M.Oakes and M.Nomura, unpublished results). However, as mentioned in Results, we constructed these strains with a new integrated rDNA using an rDNA fragment carrying the hyg1 mutation, which is somewhat harmful to normal growth. Repeating rDNA repeat integration experiments using the wild-type rDNA without the hygromycin selection step is necessary to confirm the expected wild-type growth rate of strains with integrated new rDNA repeats.
Mutational analysis of rRNA in relation to ribosome function was previously carried out (13) using a system corresponding to the rdnΔ system described in this and a previous paper (14). In rdnΔ strains there are residual copies of rDNA repeats and these strains are not completely stable, although deletion of the FOB1 gene may help to prevent an increase in the residual rDNA repeats (see 10). The rdnΔΔ system described in this paper, which utilizes a helper Pol II plasmid in addition to a Pol I plasmid with a single native rDNA repeat, may represent an improved system to carry out mutational analysis of the structure–function relationship of rRNAs. Similarly, the present system could also be used for mutational analysis of processing of the 35S precursor rRNA to mature rRNA products. Although we have not done such experiments, we have used the system to study nucleolar structures (14; unpublished results) and to analyze promoter elements as described in this paper. We also used this system to study the significance of the enhancer element in rDNA transcription, the results of which will be reported elsewhere.
Two cis elements of the yeast Pol I promoter were previously defined by in vivo experiments using a reporter gene on a plasmid (30) and by in vitro experiments using crude or partially fractionated extracts (19,27,28) or using a system consisting of purified Pol I and transcription factors (31). In the present mutational analysis of the promoter we measured the synthesis of actual rRNAs as well as the growth-promoting activity of these rRNA products in rdnΔΔ strains which have the chromosomal rDNA repeats completely deleted. The results generally support the previous conclusions on the presence of two elements, the upstream element and core promoter, for the Pol I promoter. The upstream element was previously demonstrated to be the region required for stable binding of transcription factor UAF, as judged by template competition experiments. Deletion analysis showed that its upstream end is between –155 and –119 and its downstream end is between –70 and –41 (28). As observed in previous in vitro transcription experiments using crude extracts (19,27), partial deletion from upstream to –101 partially decreased transcription in the present in vivo experiments. However, transcription experiments using all purified components gave a different result (31). Deletion to –101 still allowed a high level of transcription, almost comparable with the intact (–207) promoter. It appears that there are some components present in vivo or in crude extracts, but not in the purified transcription system, which may interact, specifically or non-specifically, with the upstream element and that such interactions might be responsible for the apparent discrepancy in the effects of partial deletion of the upstream element between the present in vivo analysis (or in vitro experiments using crude extracts) and the in vitro analysis using purified components.
Deletion of the promoter from upstream to –76 and farther led to inviability of rdnΔΔ strains, indicating a further reduction in rRNA transcription and suggesting that the basal transcription observed in vitro, if it does operate in vivo in the rdnΔΔ strains, is insufficient to sustain growth. We have previously observed that in mutants defective for a Pol I-specific subunit of UAF there is residual transcription of rRNA genes (a few percent of the control), but it is not transcription by Pol I, rather transcription by Pol II using several different upstream start sites, and that in a majority of cells this weak transcription is insufficient to sustain growth to form colonies (21). Because of the upstream deletions we do not expect any transcription by Pol II in deletions (such as –56) which remove the upstream element completely but leave the core promoter intact. We have not studied the question of whether weak basal transcription by Pol I takes place with such promoters in the rdnΔΔ strains as in the case of in vitro transcription.
Integration of a new rDNA unit at the original locus in rdnΔΔ strains and its expansion were achieved using a rDNA unit with the hyg1 mutation and the attached L sequence. As mentioned above, hygromycin selection was not necessary for rDNA repeat expansion. We should also be able to devise a scheme to eliminate the use of attached sequences, such as the L sequence, for repeat expansion, so that the reconstituted rDNA repeats would be identical to the original one. Regardless of this question, mutational analyses now appear to be applicable to studies of rDNA elements involved not only in rDNA transcription but also in rDNA replication, recombination and nucleolar organization in the context of rDNA repeat structures. In addition, integration of rDNA repeats in other chromosomal loci should also be possible. This would then answer the question of the significance of the chromosomal location of rDNA repeats in relation to the structural organization and functions of the nucleolus.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr S. Liebman for providing plasmids pRDN-hyg1 and pRDN-wt. We thank Dr S. M. Arfin for reading the manuscript and C. Carmen for help in preparation of the manuscript. This work was supported by Public Health Service grant GM 35949.
REFERENCES
- 1.Paule M.R. (1998) In Paule,M.R. (ed.), Transcription of Ribsomal RNA Genes by Eukaryotic RNA Polymerase I. Springer-Verlag, Germany and R.G. Landes Co., Georgetown, TX, pp. 1–8.
- 2.Reeder R.H. (1999) Prog. Nucleic Acid Res. Mol. Biol., 62, 293–327. [DOI] [PubMed] [Google Scholar]
- 3.Elion E.A. and Warner,J.R. (1984) Cell, 39, 663–673. [DOI] [PubMed] [Google Scholar]
- 4.Elion E.A. and Warner,J.R. (1986) Mol. Cell. Biol., 6, 2089–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer B.J. and Fangman,W.L. (1988) Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- 6.Linskens M.H.K. and Huberman,J.A. (1988) Mol. Cell. Biol., 8, 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi T., Hidaka,M., Nishizawa,M. and Horiuchi,T. (1992) Mol. Gen. Genet., 233, 355–362. [DOI] [PubMed] [Google Scholar]
- 8.Gerber J.-K., Gogel,E., Berger,C., Wallisch,M., Muller,F., Grummt,I. and Grummt,F. (1997) Cell, 90, 559–567. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi T. and Horiuchi,T. (1996) Genes Cells, 1, 465–474. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi R., Heck,D.J., Nomura,M. and Horiuchi,T. (1998) Genes Dev., 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Guthrie C. and Fink,G.R. (1991) Methods Enzymol., 194, 1–183. [DOI] [PubMed] [Google Scholar]
- 13.Chernoff Y.O., Vincent,A. and Liebman,S.W. (1994) EMBO J., 13, 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakes M., Aris,J.P., Brockenbrough,J.S., Wai,H., Vu,L. and Nomura,M. (1998) J. Cell Biol., 143, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sikorski R.S. and Hieter,P. (1989) Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose M.D. and Broach,J.R. (1991) Methods Enzymol., 194, 195–230. [DOI] [PubMed] [Google Scholar]
- 17.Swanson M.E., Yip,M. and Holland,M.J. (1985) J. Biol. Chem., 260, 9905–9915. [PubMed] [Google Scholar]
- 18.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 19.Kulkens T., Riggs,D.L., Heck,J.D., Planta,R.J. and Nomura,M. (1991) Nucleic Acids Res., 19, 5363–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogi Y., Vu,L. and Nomura,M. (1991) Proc. Natl Acad. Sci. USA, 88, 7026–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oakes M.L., Siddiqi,I., Vu,L., Aris,J. and Nomura,M. (1999) Mol. Cell. Biol., 19, 8559–8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual, 1st Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Goffeau A., Barrell,B.G., Bussey,H., Davis,R.W., Dujon,B., Feldmann,H., Gailbert,F., Hoheisel,J.D., Jacq,C., Johnston,M., Louis,E.J., Mewes,H.W., Murakami,Y., Philippsen,P., Tettelin,H. and Oliver,S.G. (1996) Science, 274, 563–567. [DOI] [PubMed] [Google Scholar]
- 24.Nogi Y., Yano,R. and Nomura,M. (1991) Proc. Natl Acad. Sci. USA, 88, 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vu L., Siddiqi,I., Lee,B.-S., Josaitis,C.A. and Nomura,M. (1999) Proc. Natl Acad. Sci. USA, 96, 4390–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad-Webb H. and Butow,R.A. (1995) Mol. Cell. Biol., 15, 2420–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choe S.Y., Schultz,M.C. and Reeder,R.H. (1992) Nucleic Acids Res., 20, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keys D.A., Lee,B.-S., Dodd,J.A., Nguyen,T.T., Vu,L., Fantino,E., Burson,L.M., Nogi,Y. and Nomura,M. (1996) Genes Dev., 10, 887–903. [DOI] [PubMed] [Google Scholar]
- 29.Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W.S., Jang,J., Shevchenko,A., Charbonneau,H. and Deshaies,R.J. (1999) Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- 30.Musters W., Knol,J., Maas,P., Dekker,A.F., van Heerikhuizen,H. and Planta,R.J. (1989) Nucleic Acids Res., 17, 9661–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keener J., Josaitis,C.A., Dodd,J.A. and Nomura,M. (1998) J. Biol. Chem., 273, 33795–33802. [DOI] [PubMed] [Google Scholar]
- 32.Hill J.E., Myers,A.M., Koener,T.J. and Tzagoloff,A. (1986) Yeast, 2, 163–167. [DOI] [PubMed] [Google Scholar]