Abstract
Microtubule dynamics ensure multiple cellular events during oocyte meiosis, which is critical for the fertilization and early embryo development. KIF15 (also termed Hklp2) is a member of kinesin-12 family motor proteins, which participates in Eg5-related bipolar spindle formation in mitosis. In present study, we explored the roles of KIF15 in mouse oocyte meiosis. KIF15 expressed during oocyte maturation and localized with microtubules. Depletion or inhibition of KIF15 disturbed meiotic cell cycle progression, and the oocytes which extruded the first polar body showed a high aneuploidy rate. Further analysis showed that disruption of KIF15 did not affect spindle morphology but resulted in chromosome misalignment. This might be due to the reduced stability of the K-fibers, which further induced the loss of kinetochore-microtubule attachment and activated spindle assembly checkpoint, showing with the failed release of Bub3 and BubR1. Based on mass spectroscopy analysis and coimmunoprecipitation data we showed that KIF15 was responsible for recruiting HDAC6, NAT10 and SIRT2 to maintain the acetylated tubulin level, which further affected tubulin acetylation for microtubule stability. Taken together, these results suggested that KIF15 was essential for the microtubule acetylation and cell cycle control during mouse oocyte meiosis.
Keywords: Oocyte, Meiosis, Cell cycle, Microtubule, Aneuploidy
Introduction
Oocyte maturation quality is important for successful fertilization and embryonic development in mammals. Oocyte meiotic maturation is comprised of nuclear maturation and cytoplasmic maturation. Microtubules are involved in multiple cellular processes during nuclear maturation, including assembly of the meiotic spindle and alignment/separation of chromosomes [1, 2]. In metaphase I (MI), homologous chromosomes are correctly connected to the microtubules from the opposite poles of the spindle to ensure alignment of the meiotic spindle at the metaphase plate before the onset of anaphase. Interactions between the microtubules and chromosomes mainly occur at the kinetochores [3, 4]. An unstable attachment between the kinetochores and spindle microtubules causes arrest at MI and activates the spindle assembly checkpoint (SAC). The SAC is active in prometaphase during the microtubule-kinetochore attachment process [5]. Once the correct connection is established between the kinetochores and microtubules, the SAC proteins release from the kinetochores, which triggers chromosome segregation [6]. Defects in kinetochore-microtubule attachment lead to unequal homologous chromosome segregation, which ultimately causes aneuploidy. Several molecules are involved in this process, such as the Mad and Bub families. Bub3 is an essential component of the SAC, which is required for monitoring the cell cycle in mammalian oocytes and ensuring the fidelity of chromosome segregation [7]. BubR1 is also required for almost all surveillance processes to preserve the genomic stability of first meiosis during mammalian oocyte meiotic maturation [8].
Microtubules are cylindrical cytoskeletal polymers composed of α/β-tubulin dimers, which are involved in cell polarity, morphogenesis, and vesicle transport [9]. Post-translational modifications of α/β-tubulin are important for regulating the microtubules [10]. Tubulin acetylation is a post-translational modification that regulates microtubule architecture and maintains microtubule integrity. Changes in the tubulin acetylation level affect stability of the microtubules [11]. N-acetyltransferase 10 (NAT10) is an acetylator that regulates midbody function to mediate cytokinesis by affecting the level of acetylated α-tubulin and maintaining the stability of α-tubulin in Saos-2 cells [12]. NAD-independent histone deacetylase 6 (HDAC6) is an α-tubulin deacetylase co-localized with the microtubule network [13]. HDAC6 possesses intrinsic deacetylase activity towards acetylated α-tubulin, and inhibiting HDAC6 disrupts the progression of mouse oocyte maturation [14]. The NAD-dependent deacetylase Sirtuin 2 (SIRT2) exhibits a substantial degree of functional overlap with HDAC6, which deacetylates the α-tubulin lysine 40 residue to make microtubules dynamic and regulates spindle organization and chromosome alignment during mouse oocyte meiosis [15, 16].
The kinesin superfamily is a group of ATP-driven motor proteins comprised of 14 large families, which play vital roles in intracellular material transport, spindle formation, and chromosome activities [17, 18]. KIF15 (also called Hklp2) is a member of the kinesin superfamily that belongs to the kinesin-12 family of plus-end-directed microtubule-based motor proteins [19, 20]. KIF15 is composed of N-terminal motor domains and a coiled-coil stalk region. Each KIF15 polypeptide chain has two MT-binding domain sites, such as the head and stalk region [21]. KIF15 moves along with the microtubules and binds to the chromosome after walking to the plus end of the microtubule. KIF15 preferentially binds to kinetochore microtubules during mitosis and crosslinks two microtubules through the Coil-1 and motor domains. This ability enables KIF15 to slide microtubules apart in vitro [22]. KIF15 is indispensable for bipolar spindle formation during normal cell division under perturbation by Eg5, which plays a major role on spindle bipolarity [23]. KIF15-inhibited HeLa and RPE1 cells have shorter spindles, suggesting that KIF15 prevents the kinetochore-microtubule from shortening, which is essential to form a bipolar spindle [24]. Moreover, KIF15 is required for assembly of the kinetochore fibers (K-fibers) and aligns the chromosomes to the spindle midzone in HEK293T and HeLa cells [25].
In the present study, we depleted KIF15 expression and inhibited KIF15 activity to explore the potential mechanism for the roles of KIF15 during oocyte maturation. Our results showed that KIF15 recruited deacetylases to stabilize the tubulin acetylation-related microtubules, which further affected microtubule-kinetochore attachment for SAC activity during oocyte meiosis. Our data indicated that KIF15 played distinctive roles during mammalian female meiosis compared with mitosis.
Materials and methods
Collection and culture of mouse oocytes in vitro
ICR mice (4–6-week-old) were used to collect the mouse oocytes. All of our experimental procedures were performed following the guidelines issued by the Animal Research Committee of Nanjing Agriculture University. The mice were fed well, housed at an appropriate temperature, and fed a regular diet. In order to collect the oocytes, the mice were sacrificed by cervical dislocation method, and ovaries were collected and transferred to the pre-warmed M2 medium (Sigma). The germinal vesicle (GV) oocytes were collected under a stereo microscope and cultured in M16 medium under mineral oil at 37 °C in a 5% CO2 atmosphere for in vitro maturation. Generally, mouse oocytes in GV stage were cultured for 8 h to the MI stage, and 10 h of culture for anaphase/telophase I (ATI) stage, and 12 h for metaphase II (MII) stage.
Antibodies and chemicals
The KIF15 inhibitor KIF15-IN-1 (HY-15948) was purchased from MCE (MedChemExpress, Shanghai). Rabbit polyclonal anti-KIF15 antibody [SAB2104083, for immunofluorescence (IF) 1:100, for western blot (WB) 1:1,000], mouse monoclonal anti-acetylated tubulin antibody (T7451, for IF 1:100, for WB 1:1,000), and mouse monoclonal anti-α-tubulin-FITC antibody (F2168, for IF 1:200) were from Sigma (St. Louis, MO, USA). Anti-GAPDH (2118, for WB 1:2000) was purchased from Cell Signaling Technology (Danvers, MA, USA). The rabbit polyclonal anti-SIRT2 antibody (19655-1-AP, for WB 1:500), anti-NAT10 antibody (13365-a-AP, for WB 1:1,000), anti-HDAC6 antibody (12834-1-AP, for WB 1:1,000), and rabbit monoclonal anti-Bub3 antibody (ET7108-82, for IF 1:100) were purchased from Proteintech (Rocky Hill, NJ, USA). Human anti-centromere CREST antibody (15-234, for IF 1:100) was purchased from Antibodies Incorporated (Davis, CA, USA). Hoechst33342, the goat monoclonal anti-BubR1 (SAB1400030, for IF 1:50) antibody, and the mouse monoclonal anti-β-actin antibody (A5441, for WB 1:5000) were purchased from Sigma. TRITC-conjugated rabbit anti-goat IgG antibody (ZF-0317, for IF 1:200), the AlexaFluor 488 goat anti-mouse antibody (ZF-0512, for IF 1:100), and the AlexaFluor 594 goat anti-rabbit antibody (ZF-0516, for IF 1:100) were purchased from Zhongshan Golden Bridge Biotechnology (Beijing, China). Dynabeads Protein G (1003D) was purchased from Thermo Fisher (Waltham, MA, USA). NuPAGE LDS Sample Buffer (NP007) was purchased from Invitrogen (Carlsbad, CA, USA). SDS loading buffer (P2179M) was purchased from Beyotime (Shanghai, China).
KIF15-IN-1 treatment
5 mg of KIF15-IN-1 was dissolved in 232.3 μl DMSO to prepare a 50 mM concentrate for stock. This stock solution was diluted in M16 medium at a ratio of 1:250, and the final concentration of treatment group was 200 μM. The oocytes were cultured for 12 h to determine their maturation status at the MII stage or cultured for 8 h to examine spindle and chromosome morphology at MI stage under 37 °C in an atmosphere of 5% CO2.
KIF15 morpholino injection
KIF15 morpholino 5′-GTC CGA GAT TCG CTT ACG CCA GGAC-3′ (Gene Tools, Philomath, OR, USA) was 300 nmol, diluted with 300 μL water to produce a 1 nmol/μl working concentration, and 5–10 pl of KIF15 morpholino solution were injected into the cytoplasm of fully grown GV oocytes using a Narishige microinjector (Tokyo, Japan) to knockdown KIF15 in mouse oocytes. Correspondingly, 5–10 pl of the non-targeting control were microinjected. After the injections, the oocytes were arrested at the GV stage for 22 h in M16 medium containing 2.5 μM milrinone (475840) from Sigma (St. Louis, MO, USA). The oocytes were completely washed six times with fresh M16 medium for 2 min each. The oocytes were transferred to fresh M16 medium and cultured for experiments at 37 °C in a 5% CO2 atmosphere.
Nocodazole and taxol treatments
Nocodazole (M1404, 10 mg/ml) from Sigma (St. Louis, MO, USA) was added to M16 medium at a ratio of 1:500 and diluted to a final concentration of 20 μg/μl. MI stage oocytes were incubated in M16 medium supplemented with nocodazole at 37 °C for 10 min. Taxol (p875571, 5 mM) from Macklin (Shanghai, China) was added to M16 medium at a ratio of 1:500 and diluted to a final concentration of 10 μM. MI stage oocytes were incubated in M16 medium containing 10 μM taxol for 50 min.
Cold treatment
MI stage oocytes were briefly chilled at 4 °C for 10 min, which depolymerized the unstable microtubules, and then the oocytes were stained with CREST to detect the kinetochores and tubulin antibodies for microtubules. Cold treatment could clearly observe the stable microtubules and kinetochore-microtubule attachment.
Chromosome spread
Oocytes were placed in Tyrode of Sigma (St. Louis, MO, USA), for 10 s to remove the zona pellucida. After the zona pellucida was removed, oocytes were transferred to glass slides and fixed in distilled water solution containing 1% paraformaldehyde, 0.15% Triton X-100 and 3 mM dithiothreitol. The glass slides were dried slowly at room temperature for hours and then the samples were blocked with 1% bovine serum albumin (BSA) prepared in PBS for 1 h at room temperature. The oocytes were incubated with the CREST antibody overnight at 4 °C. After brief washes in PBS, the slides were incubated with secondary antibodies for 1 h at room temperature. The chromosomes on the slides were stained with Hoechst33342 for 15 min, and the samples were examined under a laser scanning confocal microscope. We performed karyotype analysis of MII oocytes by chromosome spreading and counted the number of chromosomes in a single oocyte.
Mass spectroscopy analysis and immunoprecipitation
Ovarian lysates incubated with the KIF15 antibody and the bead complexes were sent to the Beijing Allwegene Technologies Co. (Beijing, China) for mass spectrometry (MS) analysis. Ten ovaries were harvested in 1 ml of lysis buffer containing 20 mM Tris, 150 mM NaCl, 1% Triton X-100, sodium pyrophosphate, β-glycerophosphate, EDTA and leupeptin for co-immunoprecipitation. The rabbit polyclonal anti-KIF15 antibody and rabbit IgG antibody were incubated overnight with the cell lysate and Dynabeads Protein G for 5 h at 4 °C. The tubes were exposed to a magnet. The immune complexes were washed three times and released from the beads by mixing in SDS loading buffer for 20 min at 30 °C. Then, the samples were supplemented with NuPAGE LDS Sample Buffer and heated at 100 °C for 10 min and stored at − 20 °C. These samples were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and blocked in TBST containing 5% non-fat milk for 1 h at room temperature. The membranes were incubated at 4 °C overnight with primary antibodies against KIF15, SIRT2, HDAC6, NAT10. Then, they were washed three times in TBST for 10 min, and the membranes were incubated at 37 °C for 1 h with the appropriate secondary antibody. Finally, the membranes were washed three times with TBST, and specific proteins were visualized using a chemiluminescence reagent (Millipore, Billerica, MA, USA).
Immunofluorescence staining
The oocytes were fixed in 4% paraformaldehyde for 30 min at room temperature and then transferred to permeabilize in PBS supplemented with 0.5% Triton X-100 to for 20 min. Then oocytes were blocked in PBS containing 1% BSA at room temperature for 1 h. The oocytes were incubated with primary anti-KIF15 antibody, anti-acetylated-tubulin antibody, anti-BubR1 antibody, and anti-α-tubulin antibody at room temperature for 6 h or overnight at 4 °C. The oocytes were stained with a human anti-centromere antibody CREST for 48 h at 4 °C. Additionally, PBS containing 0.1% Tween 20 and 0.01% Triton X-100 was used to wash the oocytes three times. The oocytes were incubated in the appropriate secondary antibody at room temperature for 1 h. The samples were mounted on glass slides and detected by confocal laser-scanning microscopy (Zeiss LSM 900 META, Zena, Germany). The fluorescence intensity of tubulin was measured by Image J software. We set up a region of interest (ROI), and the mean fluorescence intensity per unit area of the target region ROI was calculated. For α-tubulin intensity analysis, we calculated the typical certain spindle area; while for ac-tubulin intensity analysis, we measured the whole spindle area. For the chromosome misalignment definition, we also used Image J software to measure the distance between spindle poles and the metaphase plate width. The metaphase plate width exceeding 1/3 of the distance between spindle poles was classified as chromosome misalignment.
Western blot analysis
Approximately 200 live mouse oocytes were lysed in LDS sample buffer at 100 °C for 10 min and stored at − 20 °C. These samples were subjected to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and blocked in TBST containing 5% non-fat milk for 1 h at room temperature. The membranes were incubated at 4 °C overnight with primary antibodies against KIF15, acetylated-tubulin, SIRT2, HDAC6, NAT10, β-actin, and GAPDH. Then, they were washed three times in TBST for 10 min, and the membranes were incubated at 37 °C for 1 h with the appropriate secondary antibody. Finally, the membranes were washed three times with TBST, and specific proteins were visualized using a chemiluminescence reagent and quantified as relative signal intensity using Image J software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Each analysis contained at least three biological replicates. Moreover, each replication was conducted in an independent experiment at different times. The experimental results are presented as mean ± standard error. Using Graph Pad Prism 5 statistical software, the control group and the treatment group were statistically analyzed by t-test method (GraphPad Software Inc., La Jolla, CA, USA). A P value < 0.05 was considered significant.
Results
KIF15 associates with the microtubules during mouse oocyte meiosis
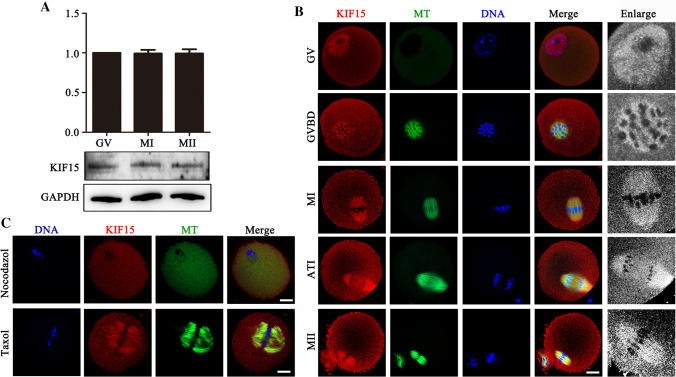
We first examined KIF15 protein expression during mouse oocyte meiosis by western blot analysis. As shown in Fig. 1A, the KIF15 protein was all expressed at the GV, MI and MII stages during oocyte maturation. We next investigated the subcellular localization of KIF15 in oocytes. KIF15 was co-localized with the microtubules at different stages of meiotic maturation after germinal vesicle breakdown (GVBD) (Fig. 1B). To further confirm this localization pattern, we used nocodazole and taxol to detect the change of KIF15 accumulation. The microtubules disassembled and KIF15 was scattered in the cytoplasm after the nocodazole treatment (Fig. 1C). In contrast, the taxol treatment promoted microtubule polymerization, and KIF15 still accumulated at the spindle and asters (Fig. 1C). This distribution pattern indicated that KIF15 had a potential role in microtubule-related cellular processes during oocyte maturation.
Fig. 1.
Expression and localization of KIF15 in mouse oocytes. A Western blot results showed that mouse oocytes expressed KIF15 at different maturation stages (GV, MI and MII). Samples were immunoblotted with anti-KIF15 and anti-GAPDH antibodies. B Oocytes of different stages were stained with anti-KIF15 (red), anti-α-tubulin (green), and chromosomes (blue). KIF15 protein co-localized with the microtubules after GVBD during oocyte meiotic maturation. Scale bars, 20 μm. C Localization of KIF15 after the nocodazole and taxol treatments during mouse oocyte meiosis. Green, α-tubulin; red, KIF15; blue, chormosomes. Scale bars, 20 μm
KIF15 is essential for meiotic progression in mouse oocytes
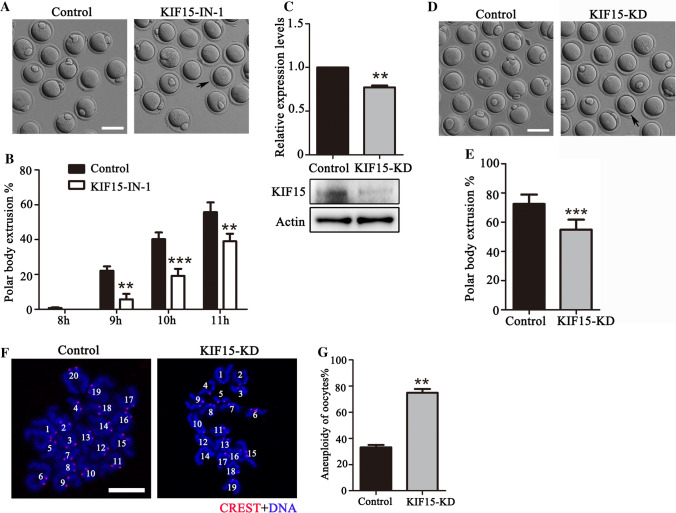
Next, we used the KIF15-specific inhibitor and morpholino to explore the functions of KIF15 in mouse oocytes. We cultured mouse oocytes with 200 μM KIF15-IN-1 for 11 h. As shown in Fig. 2A, inhibiting KIF15 activity led to the decrease of first polar body extrusion compared to the control group. Inhibiting KIF15 delayed the progression of cell cycle beginning at 9 h of culture compared to the control group (8 h: 0.68 ± 0.46%, n = 248 vs. 0 ± 0%, n = 253, P > 0.05; 9 h: 22.16 ± 2.47%, n = 248 vs. 5.68 ± 3.18%, n = 253, P < 0.01; 10 h: 40.20 ± 3.91%, n = 248 vs. 19.18 ± 4.02%, n = 253, P < 0.001; 11 h: 55.80 ± 5.57%, n = 248 vs. 39.00 ± 4.37%, n = 253, P < 0.01) (Fig. 2B). Microinjection of KIF15 morpholino significantly decreased KIF15 expression in oocytes (1 vs. 0.77 ± 0.02, n = 3, P < 0.01) (Fig. 2C), and we observed abnormal polar body extrusion in KIF15-depleted mouse oocytes (Fig. 2D), which was confirmed by the statistical data for the percentage of polar body extrusion (72.43 ± 6.42%, n = 111 vs. 54.87 ± 6.80%, n = 116, P < 0.001) (Fig. 2E). We also performed karyotype analysis of oocytes which reached MII stages by chromosome spread. Depletion of KIF15-induced aneuploid oocytes, which showed incorrect chromosome number in the oocytes (Fig. 2F). The incidence of aneuploidy was significantly higher in the KIF15 knockdown group than in the control group (control: 33.20 ± 1.80%, n = 42 vs. KIF15-KD: 74.93 ± 2.87%, n = 48, P < 0.01) (Fig. 2G). These results showed that inhibiting or depleting KIF15 disturbed cell cycle and induced aneuploidy during mouse oocyte meiotic progression.
Fig. 2.
KIF15 is essential for meiotic progression in mouse oocytes. A Digital image correlation (DIC) images of control oocytes and KIF15-IN-1 treated oocytes after 11 h of culture. The black arrows indicate that the oocyte failed to extrude the polar body after inhibiting KIF15 in mouse oocytes. Scale bars, 100 μm. B The rate of polar body extrusion after 8–11 h of culture in the control group and the KIF15-IN-1 treated group. **, significant difference (P < 0.01); ***, significant difference (P < 0.001). C Western blot analysis of the knockdown efficiency of KIF15 protein expression. Relative intensity of KIF15 and actin were assessed by densitometry. **, significant difference (P < 0.01). D DIC images of control oocytes and KIF15-KD oocytes after 12 h of culture. The black arrows indicate the failed extrusion of the polar body in mouse oocytes after KIF15 depletion. Scale bars, 100 μm. E The rate of polar body extrusion after 12 h of culture in the control group and the KIF15-KD group. ***, significant difference (P < 0.001). F Representative images of chromosome spreads in the control and KIF15-depleted MII oocytes. CREST, red; DNA, blue. Scale bars, 10 μm. G The rate of aneuploidy in the control and KIF15-depleted oocytes. **, significant difference (P < 0.01)
KIF15 prevents chromosome misalignment in mouse oocytes
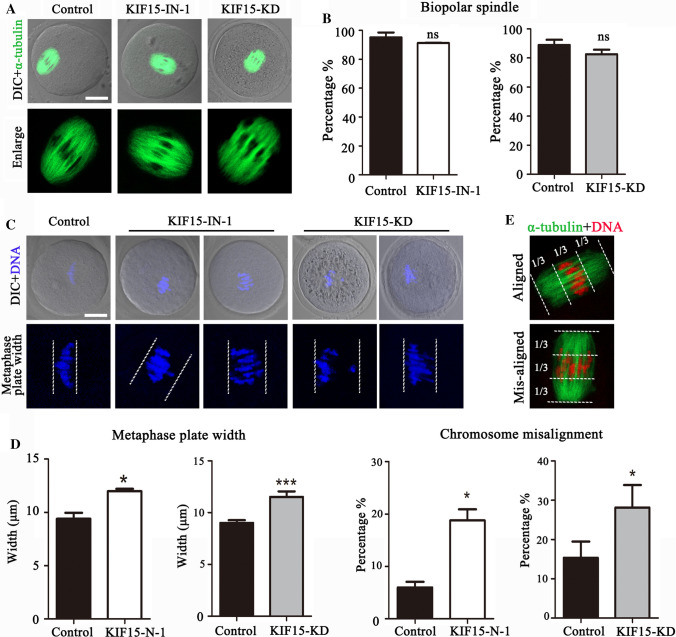
Based on the KIF15 localization pattern, we hypothesized that KIF15 might regulate spindle-related cellular processes. However, most of the oocytes displayed normal spindle morphology after inhibiting or depleting KIF15 compared to the control oocytes (Fig. 3A). The percentage of oocytes with normal spindles was similar between the KIF15 treatment groups and the control group (control: 95.18 ± 3.37%, n = 46 vs. KIF15-IN-1: 91.28 ± 0.28%, n = 48, P > 0.05; control: 88.93 ± 3.59%, n = 52 vs. KIF15-KD: 82.63 ± 3.05%, n = 58, P > 0.05) (Fig. 3B). In contrast to its effects on spindle organization, the chromosomes of most treated oocytes were severely misaligned (we defined that all chromosomes located beyond the central third of the spindle as misalignment, which was based on the general length ratio for chromosomes and spindle in mouse oocytes) (Fig. 3C). We quantified the degree of chromosome misalignment by measuring metaphase plate width, and the KIF15 treatment groups were significantly wider than the control group (control: 9.39 ± 0.55 μm, n = 64 vs. KIF15-IN-1: 11.98 ± 0.21 μm, n = 69, P < 0.05; control: 9.01 ± 0.26 μm, n = 51 vs. KIF15-KD: 11.52 ± 0.52 μm, n = 53, P < 0.001, Fig. 3D). The percentage of misaligned chromosomes in the KIF15 treatment groups were also significantly higher than that in the control group (control: 5.98 ± 1.07%, n = 64 vs. KIF15-IN-1: 18.78 ± 2.14%, n = 69, P < 0.05; control: 15.35 ± 4.13%, n = 51 vs. KIF15-KD: 28.15 ± 5.73%, n = 53, P < 0.05) (Fig. 3E). These results indicated that suppressing KIF15 led to the defects of chromosome alignment in mouse oocytes.
Fig. 3.
KIF15 prevents chromosome misalignment in mouse oocytes. A Representative images of spindle morphology in the control and KIF15-treated oocytes. α-tubulin was shown in green. Scale bars, 20 μm. B The proportion of normal spindles was recorded in the control group and the KIF15 treatment groups. No significant difference was observed between these groups (P > 0.05). C The oocyte metaphase plate width indicated chromosome misalignment in the treated groups. Scale bars, 20 μm. Blue, chromosomes. D The width of the oocyte metaphase plate increased after the KIF15-IN-1 treatment or the morpholino injection. *, significant difference (P < 0.05); ***, significant difference (P < 0.001). E Chromosome misalignment was defined as all chromosomes located beyond the central third of the spindle. The rate of chromosome misalignment in the control and treatment groups. *, significant difference (P < 0.05)
KIF15 maintains K-fiber stability and SAC activity in mouse oocytes
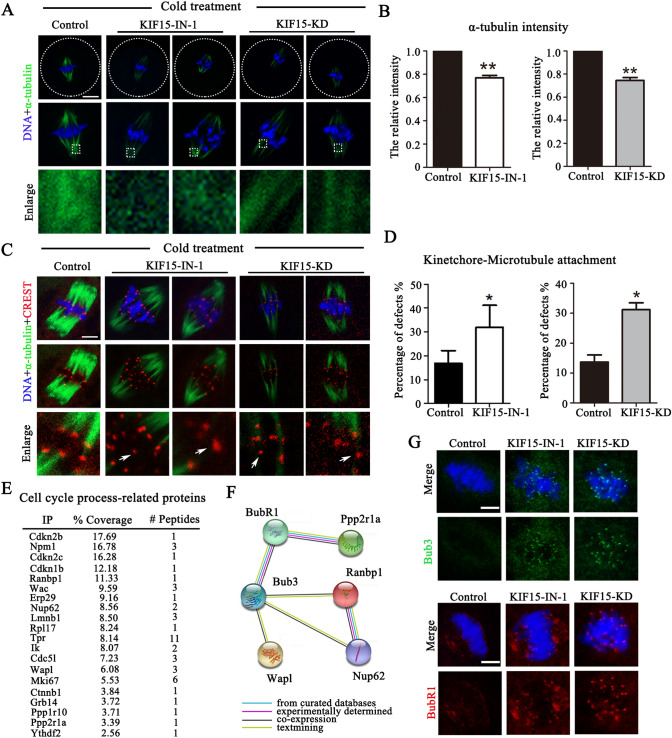
To investigate the causes for the misalignment of chromosomes, we used a cold treatment to depolymerize the unstable microtubules that did not attach to the kinetochores to check K-fiber assembly, since after cold treatment the existed microtubules could stand for the stable K-fiber. The results showed that the fluorescence intensity of tubulin decreased in the KIF15 inhibited or depleted oocytes compared to the control group after cold treatment, which could be observed in the boxed area (Fig. 4A). Fluorescence intensity data also confirmed these results: (control: 1, n = 52 vs. KIF15-IN-1: 0.77 ± 0.02, n = 65, P < 0.01; control: 1, n = 49 vs. KIF15-KD: 0.74 ± 0.03, n = 54, P < 0.01) (Fig. 4B). We next examined kinetochore-microtubule attachment. We defined kinetochore-microtubule de-attachment if the kinetochore did not have microtubules connected, and the results indicated that several kinetochores could not be matched to the corresponding microtubules in KIF15-suppressed oocytes, indicating defects in the kinetochore-microtubule attachment (Fig. 4C). Statistical analysis data showed that the incidence of kinetochore-microtubule attachment deficiency in the KIF15 treatment groups increased significantly compared to the control group (control: 16.82 ± 5.33%, n = 52 vs. KIF15-IN-1: 31.90 ± 9.25%, n = 65, P < 0.05; control: 13.64 ± 2.40%, n = 49 vs. KIF15-KD: 31.19 ± 2.30%, n = 54, P < 0.05) (Fig. 4D). Moreover, we performed MS analysis and found that several cell cycle process-related proteins were associated with KIF15 (Fig. 4E), including the SAC-related protein Wapl, Ppp2r1a, Ranbp1 and Nup62. STRING analysis indicated that these SAC-related proteins were related to BubR1 and Bub3, the essential components of the SAC (Fig. 4F). However, negative Co-IP results were observed for KIF15 with BubR1, Bub3 and Wapl (data not shown), indicating that KIF15 might not directly associate with these proteins but still involves in SAC activity. Indeed, we found that Bub3 and BubR1 still accumulated at the kinetochores in KIF15-treated MI stage oocytes, indicating activation of the SAC (Fig. 4G). These data suggested that KIF15 was critical for the stability of the K-fibers and kinetochore-microtubule attachment, which further regulated the activation of SAC in mouse oocyte meiosis.
Fig. 4.
KIF15 maintains K-fiber stability and SAC activity in mouse oocytes. A K-fiber in the control and KIF15-treated oocytes after the cold treatment. The K-fiber signals decreased after depleting KIF15. DNA and α-tubulin are shown in blue and green, respectively. The boxed area presented the details of α-tubulin signals after cold treatment. Scale bars, 20 μm. B The normalized fluorescence intensity of the K-fibers in control and KIF15 treatment oocytes. **, significant difference (P < 0.01). C The kinetochore-microtubule attachments in the control and KIF15-treated oocytes after the cold treatment. DNA, CREST, and α-tubulin are shown in blue, red and green, respectively. Scale bars, 5 μm. Arrows indicated mis-attachment of microtubules and kinetochores. D The rate of kinetochore-microtubule attachment defects in the control and KIF15 treatment oocytes. *, significant difference (P < 0.05). E Cell cycle process-related proteins which were correlated with KIF15 by mass spectrometry analysis. F Diagram of STRING analysis showed that Bub3 and BubR1 interacted with the molecules listed from mass spectrometry analysis. G. Localization of Bub3 and BubR1 at the MI stage in the control and KIF15-treated oocytes. Scale bar, 5 μm. Green, Bub3; red, BubR1; blue, DNA
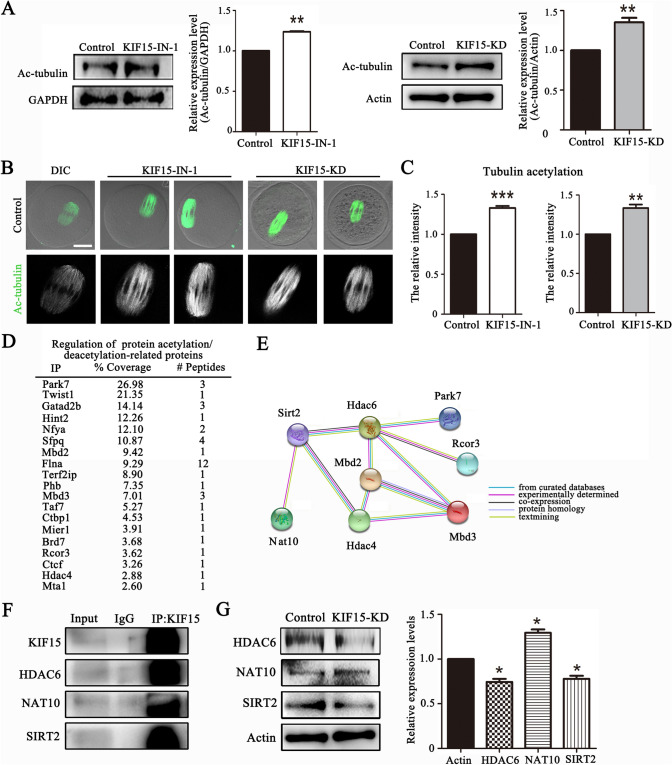
KIF15 regulates tubulin acetylation in mouse oocytes
We next explored the mechanism for the involvement of KIF15 in microtubule stability. The expression level of acetylated tubulin which is crucial for the stability of the microtubules increased in KIF15 inhibited or depleted oocytes according to western blot analysis, which was also confirmed by the band intensity analysis (control: 1 vs. KIF15-IN-1: 1.24 ± 0.01, n = 3, P < 0.01; control: 1 vs. KIF15-KD: 1.35 ± 0.05, n = 4, P < 0.01) (Fig. 5A). Similarly, the acetylated tubulin fluorescence signals at the spindle were also significantly higher in the KIF15 treatment groups than in the control group (Fig. 5B), which was confirmed by the fluorescence intensity data (control: 1, n = 57 vs. KIF15-IN-1: 1.33 ± 0.02, n = 53, P < 0.001; control: 1, n = 43 vs. KIF15-KD: 1.33 ± 0.04, n = 44, P < 0.01) (Fig. 5C). The potential mechanism of the roles of KIF15 in microtubule stability was explored by MS analysis, and the data showed that several acetylation/deacetylation-related proteins were associated with KIF15 (Fig. 5D). STRING analysis data showed that among these molecules, Park7, Rcor3, Hdac4, Mbd3 and Mbd2 were related to HDAC6, SIRT2 and NAT10 (Fig. 5E). Meanwhile, the immunoprecipitation results showed that KIF15 was correlated with the acetylated tubulin-related proteins HDAC6, SIRT2, and NAT10 (Fig. 5F). Western blot densitometry analysis revealed that the expression of HDAC6, a microtubule deacetylase, decreased after the KIF15 depletion (1 vs. 0.74 ± 0.03, n = 3, P < 0.05); while SIRT2 expression decreased after the KIF15 depletion (1 vs. 0.78 ± 0.04, n = 3, P < 0.05); and NAT10 expression, a microtubule acetyltransferase increased after the KIF15 depletion (1 vs. 1.29 ± 0.04, n = 3, P < 0.05) (Fig. 5G). These data indicated that KIF15 recruited acetylase/deacetylase for tubulin acetylation during mouse oocyte meiosis.
Fig. 5.
KIF15 regulates tubulin acetylation in mouse oocytes. A Western blot analysis of Ac-tubulin expression in KIF15-treated and control oocytes at the MI stage, **, significant difference (P < 0.01). B Ac-tubulin fluorescence staining in control and treated MI stage oocytes. Scale bars, 20 μm. Ac-tubulin is shown in green. C The relative fluorescence intensity of Ac-tubulin in KIF15 treatment groups compared with control group, ***, significant difference (P < 0.001); **, significant difference (P < 0.01). D List of acetylation/deacetylation-related proteins which were correlated with KIF15 by mass spectrometry analysis. E Diagram of STRING analysis showed that Nat10, Sirt2 and Hdac6 interacted with the molecules listed from mass spectrometry analysis. F Co-IP results showed that KIF15 was correlated with the acetylase NAT10 and the deacetylases HDAC6/SIRT2. G Quantitative analysis of the relative intensity of NAT10, SIRT2 and HDAC6 in oocytes by western blot. The results indicated that SIRT2/HDAC6 expression decreased and NAT10 expression increased in KIF15-depleted oocytes. *, significant (P < 0.05)
Discussion
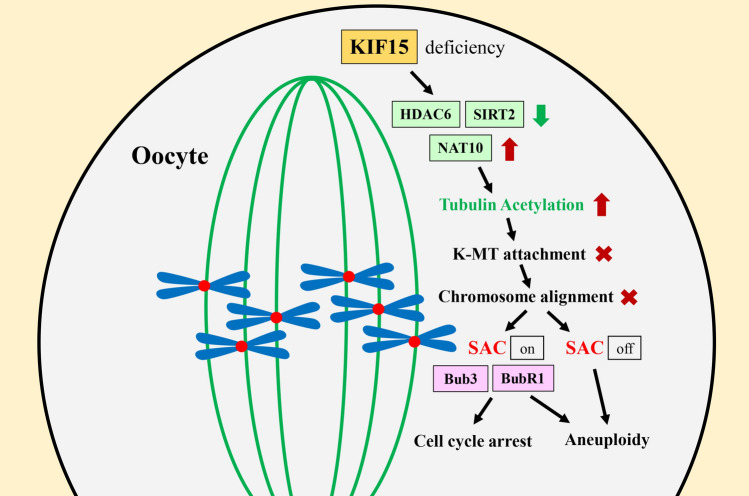
In present study, we tried to determine the roles of KIF15 during mouse oocyte maturation. We perturbed KIF15 expression using a KIF15-IN-1 inhibitor and KIF15 morpholino, and found that the KIF15 was essential for K-fibers stability, the kinetochore-microtubule attachment, chromosomes alignment, and SAC activity during cell cycle progression in oocytes (Fig. 6), which revealed the critical roles and potential regulatory mechanisms of KIF15 during female meiosis.
Fig. 6.
Diagram of the roles of KIF15 in mouse oocyte meiosis. KIF15 regulated tubulin acetylation to stabilize the microtubules, and affected kinetochore-microtubules attachment and SAC activation for chromosome alignment/segregation during mouse oocyte meiosis
KIF15 co-localizes with microtubules after GVBD in mouse oocyte meiosis. This localization pattern is similar to that of somatic cells during mitosis. For example, endogenous KIF15 localizes at the microtubules throughout the spindle and chromosomal area in Hela cells [24, 25]. Functional analysis indicates that KIF15 is essential for meiotic cell cycle progression in mouse oocytes. Previous studies also report the roles of KIF15 on cell cycle regulation. Silencing KIF15 in pancreatic cancer cells results in an inhibited G1/S transition, and suppresses pancreatic cancer cell proliferation [17]. Knockdown of KIF15 in Burkitt lymphoma inhibits proliferation and migration, which promotes apoptosis and cell cycle arrest [26]. Moreover, silencing KIF15 suppresses cell viability, migration, invasion and cell cycle progression in breast cancer [27]. These evidences set up a connection between KIF15 and cell cycle regulation in both mitosis and meiosis.
KIF15 localization pattern prompts us to examine its effects on meiotic spindle organization in mouse oocytes. However, inhibiting and depleting KIF15 all do not affect spindle morphology. A previous study shows that KIF15 cooperates with Eg5 to promote bipolar spindle formation [28], but KIF15 is not essential for spindle bipolarity in U2OS and HeLa cells with full Eg5 activity [29]. This suggests that KIF15 is not essential for spindle bipolarity assembly in both mitosis and meiosis. However, absence of KIF15 induces homologous chromosome misalignment in oocytes. This finding is also supported by several studies showing that KIF15 plays a mechanical role in supporting accurate chromosome alignment and segregation in Ptk2 cells [30]. In addition, KIF15 is involved in efficient chromosome alignment, and KIF15-silenced HEK293T cells specifically delay chromosome alignment during prometaphase [25].
Chromosome misalignment could be due to a failure to generate stable K-fibers, which are the microtubule bundles that attach the spindle poles to the kinetochores [31]. KIF15 is reported to be associated with K-fibers in HeLa cells [32, 33], and KIF15 fortifies and repairs K-fibers, which supports robust K-fiber function and prevents a chromosome segregation error [30]. Furthermore, K-fibers decrease in HeLa cells in the absence of KIF15, indicating that KIF15 is important for K-fiber assembly and stability during mitosis [25]. K-fibers are essential for kinetochore-microtubule attachment, which further guarantee chromosome alignment. The absence of stable kinetochore-microtubule attachment may activate the SAC, and the SAC produces an inhibitory signal that prevents the onset of anaphase I and delays meiosis until all the chromosomes are well aligned [34]. Our results show that KIF15 is essential for K-fibers stability and kinetochore-microtubule attachment in oocytes, which further confirms the role of KIF15 on microtubule stability. Based on MS and STRING analysis, we show that KIF15 is associated with several SAC-related proteins, and depletion of KIF15 causes the accumulation of Bub3 and BubR1 on kinetochores, indicating that the roles of KIF15 on the SAC activation in meiosis. The kinesin-5 motor protein Cut7 (Eg5 homologue) also has been reported to directly interact with SAC protein Mad1, a core SAC component to promote chromosome congression in fission yeast [35]. One of our interesting findings is that besides that part of oocytes show chromosome misalignment, which causes the MI stage arrest by SAC activation, we also observe aneuploidy in the oocytes which develop to the MII stage. There are several reports showing that different from mitosis of somatic cells, oocyte SAC can be inactivated, and meiosis can usually proceed in the presence of a few misaligned chromosomes. Depletion of CENP-E results in polar chromosomal displacement and destabilization of kinetochore-microtubule interactions in oocytes, but SAC is still inactive showing with missing Mad2 at the kinetochores [36]. It is also shown that anaphase onset does not require metaphase alignment of all chromosomes, which means that SAC activity can be silenced in presence of chromosome misalignment, indicating that aneuploidy is inevitable in mouse oocytes [37]. Even further, meiotic maturation can be completed in the presence of SAC activity, producing aneuploid eggs. NEDD1-depleted oocytes disrupt meiotic spindle stabilization and induce chromosome segregation errors even in the presence of SAC [38]. WDR62 involves into oocyte meiotic spindle assembly and K-fiber attachments, and its depletion causes SAC continuous activation but there is still aneuploid generation [39]. Chromosome misalignment causes aneuploidy, and aneuploidy is the main cause of miscarriage, mental retardation, and congenital defects in human oocytes during aging [40]. Whether KIF15 is related aging-related aneuploidy of oocytes needs further study.
Acetylated tubulin level is critical for microtubule stability, and our data show that the KIF15 regulates tubulin acetylation levels in mouse oocytes. Previous studies indicate that decreased tubulin acetylation affects microtubule stability, however, increased tubulin acetylation does not stabilize the microtubules [41]. Instead, recent studies show that increased tubulin acetylation by several kinesins such as KIF17 [42] and KIFC1[43] also affect microtubule stability in oocytes, indicating the general roles of kinesins in microtubule stability during oocyte meiosis. It is possible that moderate increase of tubulin acetylation may protect or does not affect microtubule stability; however, similar with acetylation deficiency, continuous excess of tubulin acetylation level during meiosis may also disturb the microtubule dynamics, which affects K-fiber assembly. Next, we try to explore the mechanism of KIF15 on tubulin acetylation. Several proteins which are related with acetylation or deacetylation are found to be associated with KIF15 according to MS analysis. And based on STRING prediction, these proteins are able to interact with tubulin acetylation-related proteins HDAC6, NAT10 and SIRT2. Acetylation of tubulin is mainly catalyzed by the acetyltransferase NAT10 [44], and deacetylation is mainly performed by the deacetylases HDAC6 and SIRT2 [16, 41]. Our results show that KIF15 is associated with HDAC6, SIRT2 and NAT10 and is essential for the level of tubulin acetylation, which provides a potential mechanism for KIF15 on microtubule stability. One possibility is that since KIF15 is a kinesin which controls intracellular cargo transport, it may transport HDAC6, NAT10 and SIRT2 to the specific destinations to regulate microtubule acetylation in oocytes. While microtubule acetylation is maintained at certain level in dynamics by both acetyl transferase and deacetylase. When KIF15 is deficient, the efficiency of cargo transport for HDAC6 and SIRT2 is reduced, while NAT10 acetylation is depended on SIRT1/SIRT2 [45, 46], which contributes to the increase of NAT10. Then deceased HDAC6/SIRT2 and increased SIRT2 both contribute to the tubulin acetylation elevation. Another possibility is that co-IP results could confirm that KIF15 associates with HDAC6, NAT10 and SIRT2, but it may be not a directly association. There may be some regulators for HDAC6, NAT10 and SIRT2, which directly associate with KIF15, while these regulators may have different roles for the expression of HDAC6, NAT10 and SIRT2. How KIF15 affects both acetyl transferase and deacetylase for tubulin acetylation still needs more studies to clarify.
In summary, our results indicate that KIF15 regulates tubulin acetylation to stabilize the microtubules and affects kinetochore-microtubules attachment and SAC activation for chromosome alignment/segregation during mouse oocyte meiosis.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFC2700100) and the National Natural Science Foundation of China (32170857).
Abbreviations
- GV
Germinal vesicle
- GVBD
Germinal vesicle breakdown
- MI
Metaphase I
- ATI
Anaphase-telophase I
- MII
Metaphase II
- KD
Knock down
- K-fibers
Kinetochore fibers
- SAC
Spindle-assembly checkpoint
- SIRT2
Sirtuin 2
- NAT10
N-acetyltransferase 10
- MAD2
Mitotic arrest deficient 2
- HDAC6
Histone deacetylase 6
- APC
Anaphase promoting complex
Author contribution
YJZ conducted experiments, analyzed the data and wrote the manuscript. MMS, XW conducted experiments. JCL, KHZ, JQJ, CHX contributed to materials and agents. SCS conceived the study, analyzed the data and edited the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2700100) and the National Natural Science Foundation of China (32170857).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declaration
Conflict of interest
There are no competing interests to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coticchio G, Dal Canto M, Mignini Renzini M, Guglielmo MC, Brambillasca F, Turchi D, Novara PV, Fadini R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update. 2015;21:427–454. doi: 10.1093/humupd/dmv011. [DOI] [PubMed] [Google Scholar]
- 2.Severson AF, von Dassow G, Bowerman B. Oocyte meiotic spindle assembly and function. Curr Top Dev Biol. 2016;116:65–98. doi: 10.1016/bs.ctdb.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penttinen R, Kinnula H, Lipponen A, Bamford JK, Sundberg LR. High nutrient concentration can induce virulence factor expression and cause higher virulence in an environmentally transmitted pathogen. Microb Ecol. 2016;72:955–964. doi: 10.1007/s00248-016-0781-1. [DOI] [PubMed] [Google Scholar]
- 4.Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 6.Xie D, Zhang J, Ding J, Yang J, Zhang Y. OLA1 is responsible for normal spindle assembly and SAC activation in mouse oocytes. PeerJ. 2020;8:e8180. doi: 10.7717/peerj.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Li S, Yuan J, Wang ZB, Sun SC, Schatten H, Sun QY. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS ONE. 2009;4:e7701. doi: 10.1371/journal.pone.0007701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homer H, Gui L, Carroll J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science. 2009;326:991–994. doi: 10.1126/science.1175326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 10.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 11.Palazzo A, Ackerman B, Gundersen GG. Cell biology: tubulin acetylation and cell motility. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- 12.Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, Gong Y, Hou L, Zhang B. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Yang XJ. Tubulin acetylation: responsible enzymes, biological functions and human diseases. Cell Mol Life Sci. 2015;72:4237–4255. doi: 10.1007/s00018-015-2000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling L, Hu F, Ying X, Ge J, Wang Q. HDAC6 inhibition disrupts maturational progression and meiotic apparatus assembly in mouse oocytes. Cell Cycle. 2018;17:550–556. doi: 10.1080/15384101.2017.1329067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/S1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Hou X, Ma R, Moley K, Schedl T, Wang Q. Sirt2 functions in spindle organization and chromosome alignment in mouse oocyte meiosis. FASEB J. 2014;28:1435–1445. doi: 10.1096/fj.13-244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Guo X, Xie C, Jiang J. KIF15 promotes pancreatic cancer proliferation via the MEK-ERK signalling pathway. Br J Cancer. 2017;117:245–255. doi: 10.1038/bjc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camlin NJ, McLaughlin EA, Holt JE. Motoring through: the role of kinesin superfamily proteins in female meiosis. Hum Reprod Update. 2017;23:409–420. doi: 10.1093/humupd/dmx010. [DOI] [PubMed] [Google Scholar]
- 19.McHugh T, Drechsler H, McAinsh AD, Carter NJ, Cross RA. Kif15 functions as an active mechanical ratchet. Mol Biol Cell. 2018;29:1743–1752. doi: 10.1091/mbc.E18-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskova A, Knapp B, Matelska D, Reusing S, Arjonen A, Lisauskas T, Pepperkok R, Russell R, Eils R, Ivaska J, Kaderali L, Erfle H, et al. An RNAi screen identifies KIF15 as a novel regulator of the endocytic trafficking of integrin. J Cell Sci. 2014;127:2433–2447. doi: 10.1242/jcs.137281. [DOI] [PubMed] [Google Scholar]
- 21.Reinemann DN, Sturgill EG, Das DK, Degen MS, Voros Z, Hwang W, Ohi R, Lang MJ. Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 kinesin motors. Curr Biol. 2017;27(2810–2820):e2816. doi: 10.1016/j.cub.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle. 2011;10:3533–3544. doi: 10.4161/cc.10.20.17817. [DOI] [PubMed] [Google Scholar]
- 23.Milic B, Chakraborty A, Han K, Bassik MC, Block SM. KIF15 nanomechanics and kinesin inhibitors, with implications for cancer chemotherapeutics. Proc Natl Acad Sci USA. 2018;115:E4613–E4622. doi: 10.1073/pnas.1801242115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malaby HLH, Dumas ME, Ohi R, Stumpff J. Kinesin-binding protein ensures accurate chromosome segregation by buffering KIF18A and KIF15. J Cell Biol. 2019;218:1218–1234. doi: 10.1083/jcb.201806195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouwers N, Mallol Martinez N, Vernos I. Role of Kif15 and its novel mitotic partner KBP in K-fiber dynamics and chromosome alignment. PLoS ONE. 2017;12:e0174819. doi: 10.1371/journal.pone.0174819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Chen M, Fang X, Hong H, Yao Y, Huang H. KIF15 is involved in development and progression of Burkitt lymphoma. Cancer Cell Int. 2021;21:261. doi: 10.1186/s12935-021-01967-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng H, Li T, Zhai D, Bi J, Kuang X, Lu S, Shan Z, Lin Y. ZNF367-induced transcriptional activation of KIF15 accelerates the progression of breast cancer. Int J Biol Sci. 2020;16:2084–2093. doi: 10.7150/ijbs.44204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturgill EG, Norris SR, Guo Y, Ohi R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol. 2016;213:213–227. doi: 10.1083/jcb.201507036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Begley MA, Solon AL, Davis EM, Sherrill MG, Ohi R, Elting MW. K-fiber bundles in the mitotic spindle are mechanically reinforced by Kif15. Mol Biol Cell. 2021;32:br11. doi: 10.1091/mbc.E20-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Regan L, Sampson J, Richards MW, Knebel A, Roth D, Hood FE, Straube A, Royle SJ, Bayliss R, Fry AM. Hsp72 is targeted to the mitotic spindle by Nek6 to promote K-fiber assembly and mitotic progression. J Cell Biol. 2015;209:349–358. doi: 10.1083/jcb.201409151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R. Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol. 2014;24:2307–2313. doi: 10.1016/j.cub.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC, Kim NH. Spindle assembly checkpoint and its regulators in meiosis. Hum Reprod Update. 2012;18:60–72. doi: 10.1093/humupd/dmr044. [DOI] [PubMed] [Google Scholar]
- 35.Akera T, Goto Y, Sato M, Yamamoto M, Watanabe Y. Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat Cell Biol. 2015;17:1124–1133. doi: 10.1038/ncb3219. [DOI] [PubMed] [Google Scholar]
- 36.Gui L, Homer H. Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development. 2012;139:1941–1946. doi: 10.1242/dev.078352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol. 2011;21:651–657. doi: 10.1016/j.cub.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma W, Baumann C, Viveiros MM. NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev Biol. 2010;339:439–450. doi: 10.1016/j.ydbio.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Wang YS, Chen C, Ahmad MJ, Chen F, Ding ZM, Yang SJ, Chen YW, Duan ZQ, Liu M, Liang AX, He CJ, Hua GH, et al. WDR62 regulates mouse oocyte meiotic maturation related to p-JNK and H3K9 trimethylation. Int J Biochem Cell Biol. 2022;144:106169. doi: 10.1016/j.biocel.2022.106169. [DOI] [PubMed] [Google Scholar]
- 40.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 41.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 42.Wang HH, Zhang Y, Tang F, Pan MH, Wan X, Li XH, Sun SC. Rab23/Kif17 regulate meiotic progression in oocytes by modulating tubulin acetylation and actin dynamics. Development. 2019;2019:146. doi: 10.1242/dev.171280. [DOI] [PubMed] [Google Scholar]
- 43.Shan MM, Zou YJ, Pan ZN, Zhang HL, Xu Y, Ju JQ, Sun SC. Kinesin motor KIFC1 is required for tubulin acetylation and actin-dependent spindle migration in mouse oocyte meiosis. Development. 2022;2022:149. doi: 10.1242/dev.200231. [DOI] [PubMed] [Google Scholar]
- 44.Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Cai S, Zhang C, Liu Z, Luo J, Xing B, Du X. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46:9601–9616. doi: 10.1093/nar/gky777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu HY, Liu YY, Yang F, Zhang L, Zhang FL, Hu X, Shao ZM, Li DQ. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.