Abstract
Cholesterol biosynthesis plays a critical role in rapidly proliferating tumor cells. X-box binding protein 1 (XBP1), which was first characterized as a basic leucine zipper-type transcription factor, exists in an unspliced (XBP1-u) and spliced (XBP1-s) form. Recent studies showed that unspliced XBP1 (XBP1-u) has unique biological functions independent from XBP1-s and could promote tumorigenesis; however, whether it is involved in tumor metabolic reprogramming remains unknown. Herein, we found that XBP1-u promotes tumor growth by enhancing cholesterol biosynthesis in hepatocellular carcinoma (HCC) cells. Specifically, XBP1-u colocalizes with sterol regulatory element-binding protein 2 (SREBP2) and inhibits its ubiquitination/proteasomal degradation. The ensuing stabilization of SREBP2 activates the transcription of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), a rate-limiting enzyme in cholesterol biosynthesis. We subsequently show that the XBP1-u/SREBP2/HMGCR axis is crucial for enhancing cholesterol biosynthesis and lipid accumulation as well as tumorigenesis in HCC cells. Taken together, these findings reveal a novel function of XBP1-u in promoting tumorigenesis through increased cholesterol biosynthesis in hepatocarcinoma cells. Hence, XBP1-u might be a potential target for anti-tumor therapeutic strategies that focus on cholesterol metabolism in HCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04504-x.
Keywords: XBP1, Unspliced XBP1 (XBP1-u), Cholesterol biosynthesis, Tumorigenesis, SREBP2
Introduction
Metabolic reprogramming is recognized as a key event during tumor initiation and progression [1–3]. Recent studies have revealed that alterations in lipid metabolisms, including cholesterol metabolism, are important hallmarks of tumor metabolic reprogramming [4]. Cholesterol is a 27-carbon tetracyclic lipid molecule and a key constituent of mammalian cell membranes. Its physiological functions range from the maintenance of membrane structure to mediating signal transduction as well as serving as a precursor of steroid hormones, bile acids, vitamin D, and lipoproteins [5–7]. Cholesterol homeostasis is essential for maintaining cellular and body activities [8], and any abnormality is closely related to tumor progression and tumor incidence [9]. To meet the demand for rapid proliferation, tumor cells alter their cholesterol metabolism. This is achieved by enhancing the rate of both de novo cholesterol biosynthesis and cholesterol uptake as well as by deregulating the removal of excess cholesterol and promoting intracellular cholesterol storage [10–12]. Aberrant cholesterol homeostasis increases the proliferation, migration, and invasion potential of various tumors [13–15]; whereas cholesterol-lowering drugs, such as statins, exhibit beneficial effects by reducing the risk and mortality of cancer [16]. Despite the important role of cholesterol metabolism in tumorigenesis, the underlying regulatory mechanism remains largely unknown.
X-box binding protein 1 (XBP1), a member of the basic-region leucine zipper family, was first characterized as a transcription factor that bound to MHC class II promoters [17]. XBP1 is transcribed first in an unspliced form (XBP1-u), which upon exposure to endoplasmic reticulum (ER) stress, is processed to a spliced form (XBP1-s) by activated inositol-requiring enzyme 1 [18, 19]. This splicing causes a codon shift, which excludes 26 bp from + 541 to + 566 of XBP1-u mRNA, leading to completely different amino acid sequences at the C-termini of XBP1-u and XBP1-s proteins. XBP1-s enters the nucleus and regulates the transcription of genes related to the unfolded protein response (UPR), thereby activating the UPR cascade [20]. Instead, XBP1-u, which localizes to the cytoplasm and, thus, does not possess transcriptional activity, has been assumed to be merely a precursor of XBP1-s [21]. However, recent evidence has shown that XBP1-u is the dominant form of XBP1 under non-ER stress conditions [22]. While the function of XBP1-u is still rarely known, previous reports have revealed that unlike XBP1-s who regulates target genes through its activity as a transcriptional factor, XBP1-u regulates its target genes post-translationally. Furthermore, its biological and physiological functions are independent of the UPR. XBP1-u protects endothelial cells from oxidative stress by promoting AKT serine/threonine kinase 1 (AKT) phosphorylation [23], and XBP1-u contributes to tumor cell autophagy by recruiting FoxO1 to the 20S proteasome, and inducing its degradation [24]. Meanwhile, our previous study revealed that XBP1-u, but not XBP1-s, favored tumorigenesis by enhancing the ubiquitination/proteasomal degradation of the tumor suppressor p53 thereby enhancing tumor cells proliferation [22]. Moreover, recent studies showed that XBP1-u could maintain vascular homeostasis by binding with FoxO4 and prevent its nuclearization; and could suppress vascular calcification by inducing β-catenin ubiquitination/proteasomal degradation [25, 26]. However, the specific functions of XBP1-u are still poorly characterized, and it remains to be determined whether XBP1-u could regulate other tumor hallmarks.
In this study, we investigated the correlation between the unspliced form XBP1-u, cholesterol metabolic reprogramming in HCC cells, and tumorigenesis using a xenograft model. We report the post-translational stabilization of sterol regulatory element-binding protein 2 (SREBP2) by XBP1-u, and the consequent effect on cholesterol biosynthesis through the XBP1-u/SREBP2/3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) pathway. This is the first study to link the unspliced form XBP1-u with tumor cell metabolic reprogramming, especially with de novo cholesterol biosynthesis, and to provide novel insights on the regulation of cholesterol metabolism in HCC.
Materials and methods
Cell lines and cell culture
HCC-LM3, MHCC-97H, and HepG2 cells were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), and cultured in Dulbecco's modified Eagle's medium (Gibco, Life Technologies, Grand Island, NY) with 10% FBS (Biological Industries, Beith Haemek, Israel) and 1% penicillin–streptomycin. Cell lines were verified using short-tandem repeat profiling method, and were tested periodically for mycoplasma contamination by using Mycoplasma Detection Kit-QuickTest (Biotool, Houston, TX). For gene-silencing experiments, cells were seeded in 6-well plates, and transfected with 2 µg of indicated shRNA expression vectors. Twenty-four hours after transfection, puromycin selection was performed to eliminate untransfected cells. For overexpression experiments, cells were seeded in 6-well plates and transfected with 2 µg of indicated overexpression vectors. For rescue experiments, cells were transfected with 1 µg of indicated shRNA expression vector and 1 µg of overexpression vector using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA) and subjected to puromycin selection (final concentration: 1.2 μg/ml) for 36 h to eliminate untransfected cells. For establishing XBP1-silenced (HCC-LM3/shXBP1) and XBP1-silenced, SREBP2-overexpressed (HCC-LM3/shXBP1/SREBP2) HCC-LM3 stable cell lines, cells were seeded in 10 cm well-plates, transfected with 12 μg each of shCon or shXBP1 and 6 μg of pcEF9-Puro or pcEF9-Puro-SREBP2 vectors, and subjected to puromycin selection. Stable cell line established from HCC-LM3 cells transfected with shCon and pcEF9-Puro (HCC-LM3/shCon) was used as control.
Vectors and constructs
shRNA expression vectors were constructed as described previously [27]. Target sequences were predicted using algorithm as described previously [28], and the specific target sequences were: 5ʹ-GTA AGA AAT ATT ACT ATA A-3ʹ (shXBP1-1); 5ʹ-AGTAAGAAATATTACTATA-3ʹ (shXBP1-2); 5ʹ-GTT CCA GAA TTT ACG TCA A-3ʹ (shHMGCR-1); 5ʹ-AGG TCA ACA TTA ACA AGA A-3ʹ (shHMGCR-2); 5ʹ-GCC CCA GCC TCA ACC TCA A-3ʹ (shSREBP2-1); and 5ʹ-GGA AGA GCC TTG TCT TCT T-3ʹ (shSREBP2-2). XBP1-u (NM_005080.3), XBP1-s (NM_001079539.1), and ubiquitin overexpression vectors (pcXBP1-u, pcXBP1-s, and pcUbi, respectively) were constructed as described previously [22]. For full-length SREBP2 (NM_004599.4) and HMGCR (NM_000859.3) overexpression vectors (pcSREBP2 and pcHMGCR, respectively), human complementary DNA (cDNA) obtained by reverse-transcribing the total RNA extracted from HCC-LM3 cells using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Dalian, China) was used as the template for amplifying the corresponding regions using the Takara PrimeSTAR Max DNA Polymerase (Takara Bio). The amplicons were then cloned into the BamHI and EcoRI (for pcHMGCR) or into NheI and XhoI (for pcSREBP2) sites of pcEF9-Puro vector [29].
Animal experiment
For the in vivo tumor study, BALB/c-nu/nu mice (male, body weight, 18–22 g; 6 weeks old) were purchase from the Third Military Medical University (Chongqing, China). Animal studies were conducted in the Chongqing University Cancer Hospital, and approved by the Laboratory Animal Welfare and Ethics Committee of Chongqing University Cancer Hospital. All animal experiments conformed to the approved guidelines of the Animal Care and Use Committee of the Chongqing University Cancer Hospital. All efforts to minimize suffering were made. To generate an experimental subcutaneous tumor model, BALB/c-nu/nu mice were randomly divided into three groups (n = 6), and each group was injected subcutaneously with 5 × 106 of indicated cells. Tumor size (V) was evaluated by vernier caliper every 2 days with reference to the following equation: V = a × b2/2, where a and b are the major and minor axes of the tumor, respectively [30]. The investigator was blinded to the group allocation and during the assessment.
Western blotting and quantitative reverse-transcribed PCR (qRT-PCR) analysis
Detailed methods for performing western blotting and qRT-PCR analysis are described in the Supplementary Materials and Methods. The sequences of the primers and the antibodies used are listed in the Tables S1 and S2, respectively.
Statistical analysis
All values of the experimental results were presented as mean ± SD (n = 3; unless further indicated). Statistical analysis was performed using two-tailed unpaired Student’s t-test conducted using SPSS Statistics v. 17.0. For clinical samples and xenograft experiments, one-way ANOVA was performed. A value of P < 0.05 was considered statistically significant.
Results
XBP1-u promotes HCC cholesterol biosynthesis
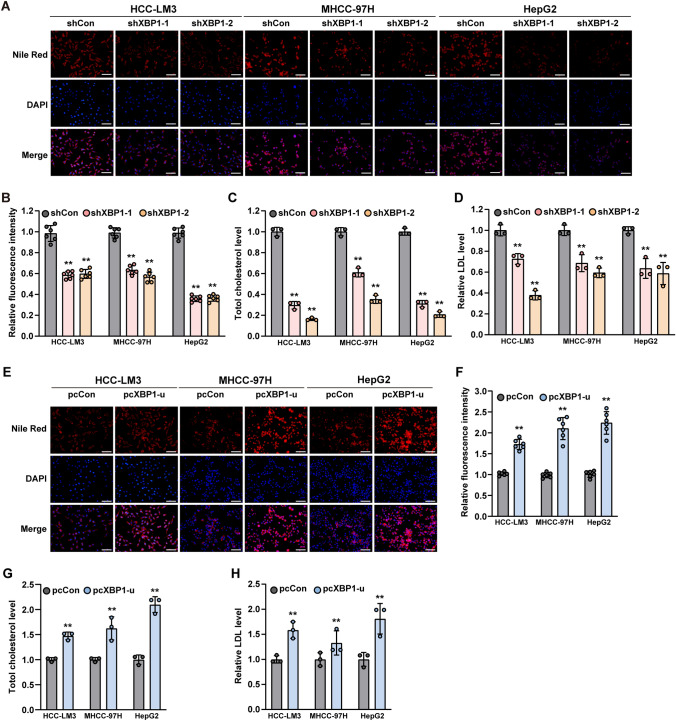
To explore whether XBP1 was involved in lipid metabolism of tumor cells, we first silenced XBP1 in HCC-LM3, MHCC-97H, and HepG2 hepatocarcinoma cells using two shRNA expression vectors targeting XBP1 (Supplementary Fig. S1A) and examined its effect on lipid accumulation. XBP1 silencing significantly decreased the accumulation of lipid droplets in these cells, as indicated by Nile Red-positive cells (Fig. 1A, B). Furthermore, XBP1 silencing also significantly suppressed total cholesterol and low-density lipoprotein (LDL) levels in HCC cells (Fig. 1C, D).
Fig. 1.
XBP1-u regulates HCC cell cholesterol level. A, B Accumulation of lipid droplets in XBP1-silenced HCC cells, as determined using Nile Red staining. Representative images (A; scale bars: 200 μm) and quantification results (B; n = 6) are shown. C Total cholesterol level in XBP1-silenced HCC-cells. D LDL level in XBP1-silenced HCC-cells. E–F Accumulation of lipid droplets in XBP1-u-overexpressed HCC-cells, as determined using Nile Red staining. Representative images (E; scale bars: 200 μm) and quantification results (F; n = 6) are shown. G. Total cholesterol level in XBP1-u-overexpressed HCC-cells. H LDL level in XBP1-u-overexpressed HCC-cells. Cells transfected with shCon or pcCon were used as controls. Total protein was used for normalization of total cholesterol and LDL levels. Quantification data are shown as mean ± SD (n = 3; unless further indicated). pcCon pcDNA3.1(+); **P < 0.01
Similar as reported by previous studies, XBP1-s overexpression significantly increased lipid droplets accumulation as well as the levels of total cholesterol and LDL in HCC cell (Supplementary Fig. S1B–E) [31, 32]. However, as we reported previously, XBP1-u is the major fraction of XBP1 in colorectal cancer cells under non-ER stress condition [22]. The same phenomenon was confirmed in HCC cells (Supplementary Fig. S2A, B). Furthermore, our results showed that under non-ER stress condition, the half-life time of XBP1-u is longer than XBP1-s in these cells (Supplementary Fig. S2C, D), and the shXBP1 vectors silenced mainly XBP1-u (Supplementary Fig. S2E, F). Hence, we next examined the effect of XBP1-u on lipid accumulation and cholesterol level under non-ER stress condition. To this end, first we confirmed the specificity of XBP1-u overexpression vector. Overexpression of XBP1-u did not change the level of XBP1-s (Supplementary Fig. S3A) as well as the expression levels of UPR-related genes ATF4, BIP and CHOP (Supplementary Fig. S3B-D) in HCC-LM3, MHCC-97H, and HepG2 cell lines, indicating that XBP1-u overexpression did not affect XBP1-s level and did not activate the UPR pathway. Surprisingly, overexpressing XBP1-u robustly increased lipid accumulation in HCC-LM3, MHCC-97H, and HepG2 cell lines (Fig. 1E, F). XBP1-u overexpression also conspicuously promoted the levels of total cholesterol and LDL in HCC cells (Fig. 1G, H), suggesting that it is involved in the regulation of cholesterol metabolism. Interestingly, changes in XBP1-u expression did not alter the level of high-density lipoprotein (Supplementary Fig. S3E, F).
To further confirm that XBP1-u could regulate cholesterol metabolism in an XBP1-s-independent manner, we treated HCC-LM3, MHCC-97H, and HepG2 cells overexpressing XBP1-u with 4μ8C, an inhibitor of inositol-requiring enzyme-1α (IRE-1α), the core enzyme that mediates XBP1-u splicing, and confirmed that treatment with 4μ8C further suppressed the amount of XBP1-s in HCC cells under non-stress condition (Fig. S4A–C). However, compared to controls, XBP1-u overexpression still clearly induced the level of total cholesterol even in 4μ8C-treated HCC cells (Fig. S4D–F). In line with this, robust increase of LDL level could also been observed in XBP1-u-overexpressed HCC cells treated with 4μ8C (Fig. S4G–I).
Together, these results show that XBP1-u is involved in lipid metabolism in HCC cells, particularly in the regulation of LDL cholesterol levels, in an XBP1-s- and UPR stress-independent manners.
Cholesterol is critical for XBP1-u-induced HCC cell proliferation and colony formation
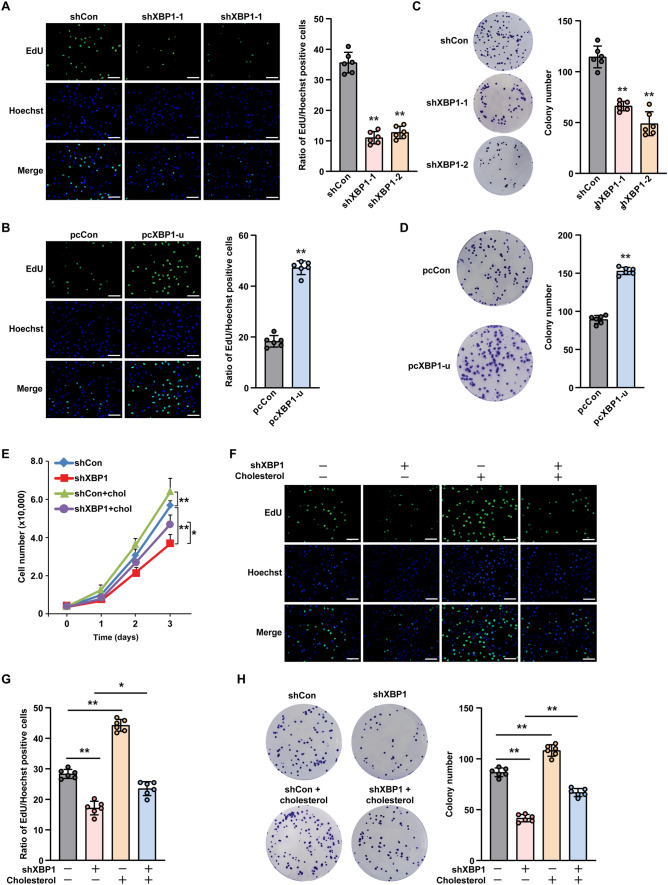
Given that cholesterol is crucial for tumorigenesis, we next examined whether XBP1-u-induced cholesterol biosynthesis could mediate HCC tumorigenesis. To this end, we first assessed the effect of XBP1-u expression on the proliferation and colony formation potential of HCC cells. XBP1 silencing clearly suppressed the viability of HCC-LM3 cells (Supplementary Fig. S5A), as well as the number of 5-ethynyl-2'-deoxyuridine (EdU)-positive cells (Fig. 2A), whereas XBP1-u overexpression significantly increased both parameters (Fig. 2B and Supplementary Fig. S5B). Furthermore, the number of colonies formed by HCC-LM3 cells decreased significantly upon XBP1 silencing (Fig. 2C), but increased significantly following XBP1-u overexpression (Fig. 2D). These results indicate that XBP1-u is crucial for HCC cell proliferation and colony formation.
Fig. 2.
Cholesterol is crucial for XBP1-u regulation on HCC cell tumorigenic potential. A, B Proliferation potential of XBP1-silenced (A) and XBP1-u-overexpressed (B) HCC-LM3 cells, as determined using EdU-incorporation assay. Representative images (left; scale bars: 200 μm) and ratio of proliferative cells (right; n = 6) are shown. C, D Colony formation potential of XBP1-silenced (C) and XBP1-u-overexpressed (D) HCC-LM3 cells. Representative images (left) and the number of the colonies formed (right; n = 6) are shown. E Viability of XBP1-silenced HCC-LM3 cells treated with cholesterol (final concentration: 10 μg/mL) at indicated time points (n = 3). F, G Proliferation potential of XBP1-silenced HCC-LM3 cells treated with cholesterol (final concentration: 10 μg/mL), as determined using EdU-incorporation assay. Representative images (F; scale bars: 200 μm) and ratio of proliferative cells (G; n = 6) are shown. H Colony formation potential of XBP1-silenced HCC-LM3 cells treated with cholesterol (final concentration: 10 μg/mL). Representative images (left) and the number of the colonies formed (right; n = 6) are shown. Cells transfected with shCon or pcCon were used as controls. Quantification data are shown as mean ± SD. pcCon, pcDNA3.1(+); *P < 0.05; **P < 0.01
Next, to understand the role of cholesterol in XBP1-u-induced HCC cell proliferation and colony formation, we added cholesterol to the culture medium of XBP1-silenced HCC-LM3 cells. The addition restored cell viability, the ratio of EdU-positive cells, and colony formation potential (Fig. 2E–H).
To further confirm the role of cholesterol in XBP1-u mediated HCC cells tumorigenic potentials, we treated HCC-LM3 cells overexpressing XBP1-u with statin, a cholesterol lowering compound. Treatment with statin abrogated the increase in lipid droplet accumulation, total cholesterol, and LDL levels in HCC-LM3 cells induced by XBP1-u overexpression (Fig. 3A–C). Furthermore, it cancelled the increase in cell viability, the ratio of EdU-positive cells, and colony formation induced by XBP1-u overexpression (Fig. 3D–G). These results indicate that XBP1-u-regulated cholesterol biosynthesis is crucial for the tumorigenic potential of HCC.
Fig. 3.
Inhibition of cholesterol biosynthesis suppresses XBP1-u-mediated HCC cell tumorigenic potential. A Accumulation of lipid droplets in XBP1-u-overexpressed HCC-LM3 cells treated with statin (final concentration: 15 μM), as analyzed using Nile Red staining. Representative images (left; scale bars: 200 μm) and quantification results (right; n = 6) are shown. B, C Total cholesterol (B) and LDL (C) levels in XBP1-u-overexpressed HCC-LM3 cells treated with statin (final concentration: 15 μM). D Viability of XBP1-u-overexpressed HCC-LM3 cells treated with statin at indicated time points (final concentration: 15 μM). E, F Proliferation potential of XBP1-u-overexpressed HCC-LM3 cells treated with statin (final concentration: 15 μM), as determined using EdU-incorporation assay. Representative images (E; scale bars: 200 μm) and ratio of proliferative cells (F; n = 6) are shown. G Colony formation potential of XBP1-u-overexpressed HCC-LM3 cells treated with statin (final concentration: 15 μM). Representative images (left) and the number of the colonies formed (right; n = 6) are shown. Cells transfected with pcDNA3.1(+) and treated with DMSO were used as controls. Quantification data are shown as mean ± SD (n = 3, unless further indicated). *P < 0.05; **P < 0.01
XBP1-u regulates cholesterol metabolism by modulating HMGCR transcription
It is well known that statin binds and inhibits the activity of HMGCR, the rate-limiting enzyme of the mevalonate pathway in cholesterol biosynthesis. Hence, to elucidate the molecular mechanism of XBP1-u regulation on cholesterol biosynthesis in HCC cells, we first examined the effect of XBP1-u on the expression of HMGCR. XBP1 silencing suppressed HMGCR mRNA expression (Fig. 4A), whereas XBP1-u overexpression significantly enhanced it (Fig. 4B). The same pattern was observed with respect to HMGCR protein expression (Fig. 4C, D). These results indicate that XBP1-u regulates HMGCR at the transcriptional level.
Fig. 4.
XBP1-u regulates HMGCR expression in HCC cells at its transcriptional level. A, B HMGCR mRNA expression level in XBP1-silenced (A) and XBP1-u-overexpressed (B) HCC-LM3, MHCC-97H, and HepG2 cells, as analyzed using qRT-PCR. C, D HMGCR protein expression level in XBP1-silenced (C) and XBP1-u-overexpressed (D) HCC-LM3, MHCC-97H, and HepG2 cells, as analyzed using western blotting. Cells transfected with shCon or pcCon were used as controls. β-actin was used for qRT-PCR normalization and as western blotting loading control. Quantification data are shown as mean ± SD (n = 3). pcCon, pcDNA3.1(+); **P < 0.01
To investigate whether XBP1-u affected cholesterol metabolism in HCC cells and, consequently, proliferation and colony formation, through its regulation of HMGCR, we overexpressed HMGCR in XBP1-silenced HCC-LM3, MHCC-97H, and HepG2 cells (Fig. 5A and Supplementary Fig. S6A, B). Overexpression of HMGCR restored lipid droplet accumulation, total cholesterol, and LDL in HCC-LM3 cells (Fig. 5B–E) as well as in MHCC-97H and HepG2 cells (Supplementary Fig. S6C, D and Supplementary Fig. S7A–D). Meanwhile, silencing HMGCR using shRNA expression vectors targeting HMGCR cancelled the effect of XBP1-u overexpression on promoting the level of HMGCR expression (Supplementary Fig. S7E–G). Accordingly, HMGCR silencing cancelled the positive regulatory effect of XBP1-u overexpression on total cholesterol and LDL levels (Supplementary Fig. S7H, I). HMGCR overexpression abrogated also the suppressive effect of XBP1 silencing on HCC cell viability (Fig. 5F and Supplementary Fig. S8A, B), the number of EdU-positive cells (Fig. 5G, H and Supplementary Fig. S8C, D), and colonies formed (Fig. 5I and Supplementary Fig. S9A, B). Together with the results showing that HMGCR inhibitor, statin, abrogated cholesterol biosynthesis and HCC cell proliferation induced by XBP1-u overexpression, these results strongly indicate that XBP1-u regulation of HMGCR transcription is critical for its tumor cell proliferation-promoting function.
Fig. 5.
HMGCR is crucial for XBP1-u regulation on cholesterol biosynthesis and tumorigenic potential of HCC cells. A Protein expression level of HMGCR in XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells, as analyzed using western blotting. B, C Accumulation of lipid droplets in XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells, as analyzed using Nile Red staining. Representative images (B; scale bars: 200 μm) and quantification results (C; n = 6) are shown. D, E Total cholesterol and LDL levels in XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells. F Viability of XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells at indicated time points. G, H Proliferation potential of XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells, as determined using EdU-incorporation assay. Representative images (G; scale bars: 200 μm) and ratio of proliferative cells (H; n = 6) are shown. I Colony formation potential of XBP1-silenced, HMGCR-overexpressed HCC-LM3 cells. Representative images (left) and the number of the colonies formed (right; n = 6) are shown. Cells transfected with shCon and pcEF9-Puro were used as controls. β-actin was used as western blotting loading control. Total protein was used for normalization of total cholesterol and LDL levels. Quantification data are shown as mean ± SD (n = 3, unless further indicated). *P < 0.05; **P < 0.01
XBP1-u regulates HMGCR expression through SREBP2
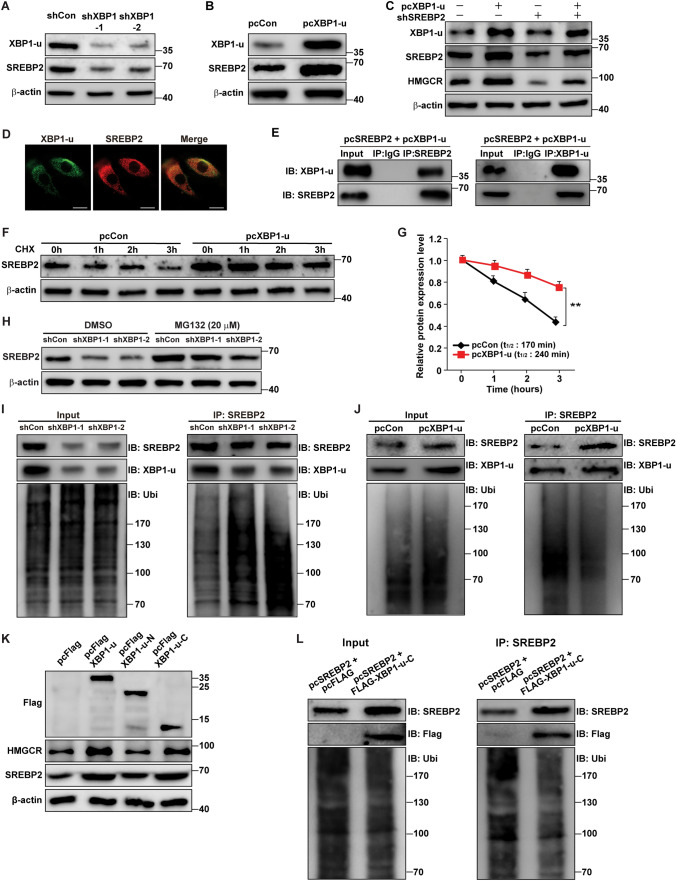
Previous results have shown that unlike XBP1-s, which localizes to the nucleus, XBP1-u possesses a strong nuclear exclusion signal at its C-terminus, localizes to the cytoplasm, and regulates its target genes post-translationally through phosphorylation and ubiquitination [21, 22, 26]. Hence, XBP1-u regulation of HMGCR transcription must occur in an indirect manner, possibly through post-translational protein stabilization. We investigated the effect of XBP1-u on SREBP2 and c-Myc, two transcriptional activators of HMGCR. whose stabilities are controlled post-translationally by ubiquitination [33, 34]. XBP1 silencing significantly suppressed SREBP2 protein levels in HCC-LM3 cells (Fig. 6A); whereas its effect on c-Myc was much lower (Supplementary Fig. S10A). Similarly, while XBP1-u overexpression robustly increased SREBP2 protein levels in HCC-LM3 cells (Fig. 6B), it only slightly affected those of c-Myc (Supplementary Fig. S10B). XBP1-u regulation of SREBP2 levels was confirmed also in MHCC-97H and HepG2 cells (Supplementary Fig. S10C–F). To determine whether XBP1-u regulation of HMGCR occurred through SREBP2, we constructed two shRNA expression vectors and confirmed their silencing effect against SREBP2 and, indirectly, HMGCR (Supplementary Fig. S10G, H). Next, we observed that knockdown of SREBP2 in HCC cells overexpressing XBP1-u abrogated the XBP1-u-mediated increase in HMGCR expression (Fig. 6C). Together, these results indicate that SREBP2 plays a crucial role in XBP1-u regulation of HMGCR expression.
Fig. 6.
XBP1-u stabilizes SREBP2 protein by inhibiting its ubiquitin-proteasomal degradation. A, B SREBP2 protein expression level in XBP1-silenced (A) and XBP1-u-overexpressed (B) HCC-LM3 cells, as analyzed using western blotting. C HMGCR and SREBP2 protein expression levels in SREBP2-silenced, XBP1-u-overexpressed HCC-LM3 cells, as analyzed using western blotting. D Colocalization of endogenous SREBP2 and XBP1-u, as determined by immunofluorescence staining using anti-SREBP2 and anti-XBP1-u antibodies (scale bars: 20 μm). E Physical interaction between XBP1-u and SREBP2, as determined using anti-XBP1-u immunoblotting of cell lysate immunoprecipitated with anti-SREBP2 antibody and vice versa. F, G Degradation rates of SREBP2 protein in XBP1-u-overexpressed HCC-LM3 cells, as analyzed using western blotting at indicated time points after the addition of cycloheximide (final concentration: 200 μg/ml). Western blotting result (F) and the half-life of SREBP2 protein (G) are shown. H SREBP2 protein expression level in XBP1-silenced HCC-LM3 cells treated with MG132 (final concentration: 20 μM) for 8 h, as analyzed using western blotting. I, J SREBP2 ubiquitination levels in HCC-LM3 cells transfected with pcSREBP2, pcUbi, and shXBP1 (I) or pcXBP1-u (J), as analyzed using anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-SREBP2 antibody. K SREBP2 and HMGCR protein expression levels in HCC-LM3 cells transfected with FLAG-XBP1-u, FLAG-XBP1-u-N and FLAG-XBP1-u-C, as determined using western blotting. L SREBP2 ubiquitination level in HCC-LM3 cells transfected with pcFLAG-XBP1-u-C, pcSREBP2, and pcUbi, as analyzed using anti-ubiquitin immunoblotting of cell lysates immunoprecipitated with anti-SREBP2 antibody. Cells transfected with shCon and/or pcCon, or pcFlag, were used as controls. β-actin was used as western blotting loading control. Quantification data are shown as mean ± SD (n = 3). pcCon, pcDNA3.1(+); pcSREBP2: full-length SREBP2 overexpression vector; IB immunoblotting; IP immunoprecipitation; Ubi ubiquitination; **P < 0.01
As XBP1-u did not affect SREBP2 mRNA expression in HCC-LM3, MHCC-97H, and HepG2 cells (Supplementary Fig. S11A), we postulated that XBP1-u might regulate SREBP2 at the protein level. To evaluate this possibility, we first confirmed the subcellular localization of XBP1-u and SREBP2. Immunofluorescent staining using anti-XBP1-u and anti-SREBP2 antibodies showed that endogenous XBP1-u colocalized with SREBP2 in the cytoplasm (Fig. 6D). Upon activation, SREBP2 protein enters the nucleus where it regulates the transcription of its target genes, and where it is ubiquitinated [8]. Indeed, overexpression of SREBP2 and XBP1-u caused their localization to become nuclear, thus coinciding with the site of SREBP2 ubiquitination (Supplementary Fig. S11B). In contrast, SREBP2 knockdown prevented the nuclear colocalization of XBP1-u (Supplementary Fig. S11C). Furthermore, immunoprecipitation assays showed the presence of XBP1-u in the protein fractions immunoprecipitated using anti-SREBP2 antibody and vice versa, revealing a physical interaction between XBP1-u and SREBP2 (Fig. 6E).
Next, we investigated whether XBP1-u was involved in SREBP2 protein stability. We inhibited de novo protein synthesis using cycloheximide and observed that XBP1-u overexpression conspicuously suppressed SREPB2 degradation rate in HCC cells (Fig. 6F, G), whereas XBP1-silencing clearly enhanced it (Supplementary Fig. S11D, E). This finding indicates that XBP1-u is important for maintaining SREBP2 protein stability.
Ubiquitination/proteasomal degradation is essential in SREBP2 post-translational regulation. Treatment with MG132, a proteasomal inhibitor, prevented the drop in SREBP2 level in XBP1-silenced HCC-LM3 cells (Fig. 6H), indicating that XBP1 silencing blocked SREBP2 proteasomal degradation and promoted instead its stability. Moreover, as shown in Fig. 6I, XBP1 silencing robustly enhanced SREBP2 protein ubiquitination, whereas XBP1-u overexpression suppressed it (Fig. 6J). Together, these results clearly show that XBP1-u enhances SREBP2 protein stability by suppressing its ubiquitination/proteasomal degradation.
To determine which region in XBP1-u was crucial for its regulation of SREBP2, we overexpressed the N and C termini of XBP1-u conjugated with the FLAG tag (FLAG-XBP1-u-N and FLAG-XBP1-u-C, respectively; Supplementary Fig. S11F). Only overexpression of the C terminus, but not the N terminus, induced SREBP2 accumulation and increased HMGCR levels in HCC cells (Fig. 6K). Conform with this, C-terminus of XBP1-u, which is different from that of XBP1-s, suppressed SREBP2 ubiquitination (Fig. 6L). Meanwhile, while XBP1-s overexpression promoted the mRNA and protein expression levels of SREBP2 and its downstream target gene HMGCR (Supplementary Fig. S12A–C), it failed to slow down the degradation rate of SREBP2 protein (Supplementary Fig. S12D–E). These results suggest that different from XBP1-s which regulates SREBP2/HMGCR pathway through transcriptional regulation of SREBP2, XBP1-u is required for stabilizing SREBP2 protein. These distinct mechanisms of XBP1 isoforms regulation on SREBP2 are most plausibly due to the difference in their C-termini as the result of the splicing-induced codon shift.
XBP1-u regulates cholesterol metabolism through the SREBP2/HMGCR axis
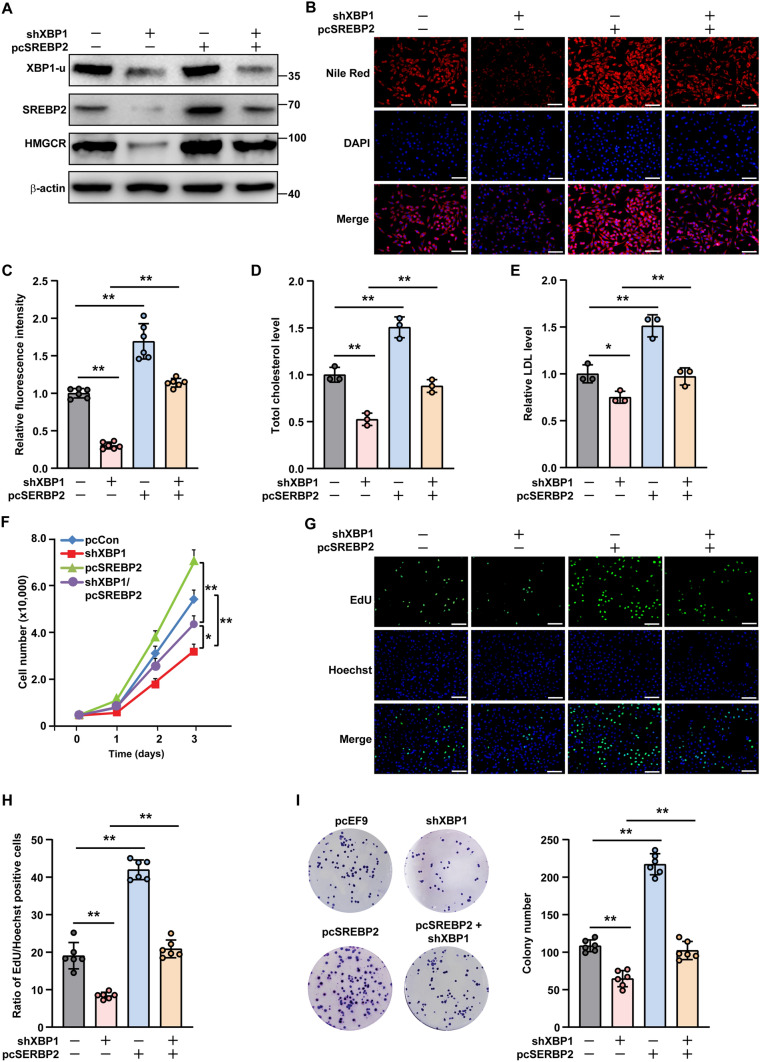
To investigate whether the SREBP2/HMGCR pathway was essential for XBP1-u regulation of cholesterol metabolism in HCC cells, we overexpressed SREBP2 in XBP1-silenced HCC-LM3 cells (Fig. 7A). SREBP2 overexpression restored lipid droplet accumulation previously suppressed by XBP1 silencing (Fig. 7B, C), as well as abrogated the decrease in total cholesterol and LDL levels (Fig. 7D, E). In contrast, silencing of SREBP2 prevented lipid droplet accumulation and canceled the effect of XBP1-u overexpression on total cholesterol and LDL levels (Supplementary Fig. S13A–C). These results suggest that the SREBP2/HMGCR pathway is important for XBP1-u regulation of cholesterol metabolism in HCC cells.
Fig. 7.
SREBP2 is critical for XBP1-u-induced cholesterol biosynthesis and tumorigenic potential of HCC cells. A Protein expression levels of SREBP2 and HMGCR in XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells, as analyzed using western blotting. B, C Accumulation of lipid droplets in XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells, as analyzed using Nile Red staining. Representative images (B; scale bars: 200 μm) and quantification results (C; n = 6) are shown. D, E Total cholesterol (D) and LDL (E) levels in XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells overexpressing SREBP2. F Viability of XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells at indicated time points. G, H Proliferation potential of XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells, as determined using EdU-incorporation assay. Representative images (G; scale bars: 200 μm) and ratio of proliferative cells (H; n = 6) are shown. I Colony formation potential of XBP1-silenced, SREBP2-overexpressed HCC-LM3 cells. Representative images (left) and numbers of the colonies formed (right; n = 6) are shown. Cells transfected with shCon and pcEF9-Puro were used as controls. β-actin was used as western blotting loading control. Total protein was used for normalization of total cholesterol and LDL levels. Quantification data are shown as mean ± SD (n = 3, unless further indicated). pcSREBP2 full-length SREBP2 overexpression vector; *P < 0.05; **P < 0.01
Given that cholesterol is critical for tumorigenesis, we next examined whether the XBP1-u/SREBP2 pathway was involved in HCC cell proliferation and colony formation. As shown in Fig. 7F, SREBP2 overexpression abolished the effect of XBP1 silencing on HCC-LM3 cell viability, while also restoring the number of EdU-positive cells and colonies (Fig. 7G–I). As expected, SREBP2 silencing had the opposite effect and reduced the viability of HCC-LM3 cells, EdU-positive cells, and colonies in XBP1-u-overexpressing cells (Supplementary Fig. S14A–D).
We next investigated whether XBP1-u could act antagonistically to SREBP2 and HMGCR inhibitors. To this end, we evaluated the effect of XBP1-u overexpression on the viability of HCC cells treated with betulin or statin, two cholesterol-lowering compounds that inhibit SREBP2 and HMGCR, respectively. While HCC cell viability decreased in a dose-dependent manner upon betulin addition, XBP1-u overexpression promoted viability at every dose (Supplementary Fig. S15A), resulting in a nearly four-fold increase in the half-maximal inhibitory concentration (IC50) for betulin (Supplementary Fig. S15B). Similarly, XBP1-u overexpression promoted HCC cell viability upon statin treatment, leading to a more than five-fold increase in the IC50 for statin (Supplementary Fig. S15C, D). Together, these results reveal that XBP1-u works antagonistically to the SREBP2/HMGCR pathway, leading to enhanced cholesterol metabolism and, consequently, greater proliferation and colony formation by HCC cells.
XBP1-u mediates the hepatocarcinogenesis potential by positively regulating SREBP2
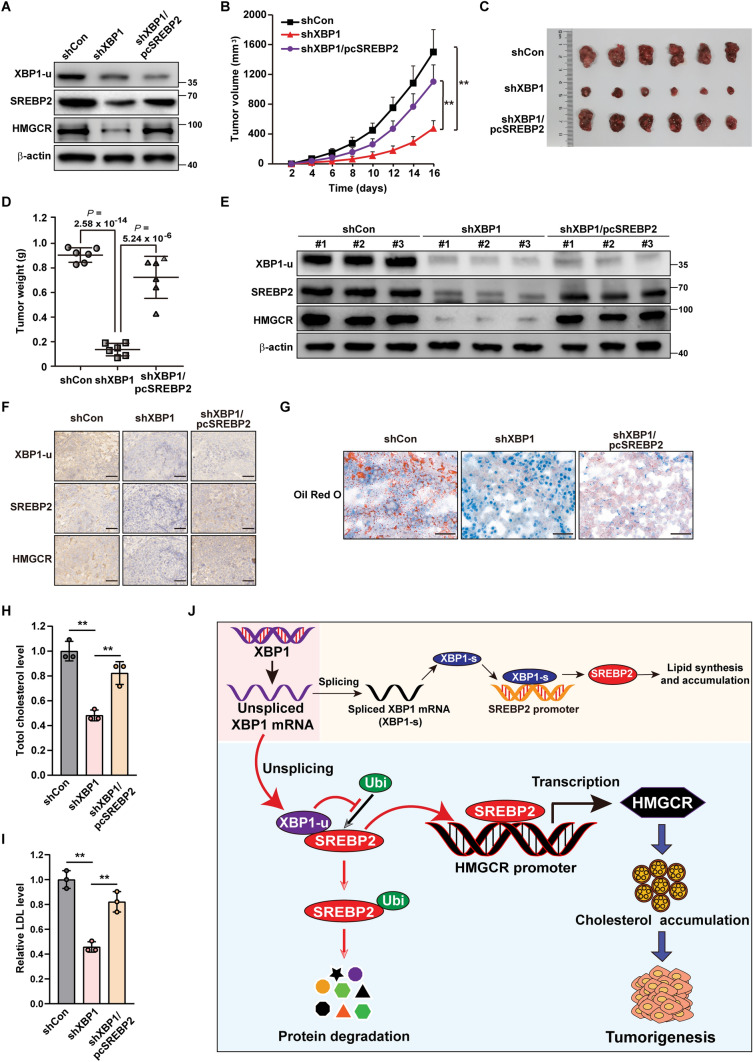
To elucidate the pathological function of the XBP1-u/SREBP2 pathway in vivo, especially during tumorigenesis, we constructed XBP1-silenced (HCC-LM3/shXBP1) and XBP1-silenced, SREBP2-overexpressed (HCC-LM3/shXBP1/pcSREBP2) HCC-LM3 stable cell lines (Fig. 8A). Together with control cells (HCC-LM3/shCon), these test cells were transplanted subcutaneously into BALB/c-nu/nu mice. As shown by the volume and morphological appearance of the tumors (Fig. 8B, C), knockdown of XBP1 significantly suppressed tumor growth, whereas SREBP2 overexpression restored it. These results were further confirmed by the changes in tumor weight (Fig. 8D). As indicated by western blotting (Fig. 8E) and immunohistochemical staining (Fig. 8F), SREBP2 and HMGCR were downregulated in the tumor lesions formed by XBP1-silenced HCC-LM3 cells and restored in those formed by HCC-LM3/shXBP1/pcSREBP2 cells. Hence, SREBP2 is essential for XBP1-induced hepatocarcinogenesis in vivo.
Fig. 8.
XBP1-u mediates hepatocarcinogenesis potential by positively regulates SREBP2. A Protein expression levels of SREBP2 and HMGCR in HCC-LM3/shCon (shCon), HCC-LM3/shXBP1 (shXBP1) and HCC/shXBP1/pcSREBP2 (shXBP1/pcSREBP2) cells, as analyzed using western blotting. B–D Tumorigenesis potentials of HCC-LM3/shCon, HCC-LM3/shXBP1, and HCC-LM3/shXBP1/pcSREBP2 cells, as determined in vivo by subcutaneous injection of these cells into Balb/c-nu/nu mice (n = 6). Tumor volumes at the indicated time points (B), morphological images (C) and tumor weight (D) at day 16 after transplantation are shown. E XBP1-u, SREBP2, and HMGCR protein expression levels in the xenografted tumors formed by the indicated cells, as determined using western blotting. F Immunohistochemistry staining using anti-SREBP2, anti-XBP1-u, and anti-HMGCR antibodies showing the expression levels of SREBP2, XBP1-u, and HMGCR in tissue sections of xenografted tumors in BALB/c-nu/nu mice injected with the indicated cell lines (scale bars: 50 μm). G Accumulation of lipid droplets in the tissue section of xenografted tumors in BALB/c-nu/nu mice injected with the indicated cell lines (scale bars: 50 μm). H, I Total cholesterol (H) and LDL (I) levels in the xenografted tumors in BALB/c-nu/nu mice injected with the indicated cell lines (n = 3). J Schematic diagram showing the mechanism of XBP1-u regulation on the SREBP2/HMGCR axis. β-actin was used as western blotting loading control. Quantification data are shown as mean ± SD. **P < 0.01
Furthermore, XBP1 silencing clearly suppressed lipid accumulation in tumors formed by HCC cells (Fig. 8G), whereas SREBP2 overexpression restored it. Similar results were obtained for total cholesterol and LDL levels (Fig. 8H, I). These findings indicate that XBP1-u/SREBP2 induces hepatocarcinogenesis in vivo by stimulating cholesterol metabolism.
Together, our results reveal a critical role of XBP1-u, the unspliced form of XBP1, in regulating hepatocarcinogenesis via the SREBP2/HMGCR axis. XBP1-u stabilizes SREBP2 by preventing its ubiquitination, thereby activating the SREBP2/HMGCR axis and enhancing cholesterol biosynthesis. Ultimately, together with XBP1-s regulation on SREBP2 transcription, this stimulates the tumorigenic potential of HCC cells (Fig. 8J).
Discussion
Recent studies have revealed that aberrant cholesterol homeostasis is closely related to tumor cell proliferation and tumorigenesis [35]. Excess cholesterol, which is found in HCC, breast cancer, prostate cancer, and colorectal carcinoma, benefits tumor cell proliferation [36–38]. Besides providing tumor cells with energy and major components of the cell membrane, cholesterol directly activates oncogenic cascades through the Hedgehog and Wnt signaling pathways [13, 36, 39, 40]. Cholesterol also contributes to increased tumor incidence, as it downregulates the expression of genes involved in T-cell differentiation, causing a reduction in cancer immunosurveillance [41–43]. This is achieved chiefly through increased cholesterol uptake and endogenous de novo biosynthesis by tumor cells. Upregulation of factors involved in cholesterol uptake, such as LDL receptor as well as those participating in cholesterol biosynthesis, such as HMGCR, oxidative squalene cyclase, and squalene epoxidase, is frequently observed in tumor patients [44–48]. Despite its importance, the detailed molecular mechanism responsible for an altered cholesterol metabolism in tumor cells has not been fully elucidated. In this study, we found that XBP1-u is crucial for de novo cholesterol biosynthesis. XBP1-u, which is upregulated in tumor cells, promotes ubiquitination and destabilization of SREBP2. This, in turn, activates the SREBP2/HMGCR axis, leading to enhanced cholesterol accumulation and tumorigenic potential of HCC cells. Hence, these results reveal a novel regulatory mechanism of cholesterol metabolic reprogramming in HCC cells.
XBP1-u is the unspliced isoform of XBP1 [18, 49]. Under ER stress, XBP1 is spliced into XBP1-u and XBP1-s, whose different C-termini affect the isoform subcellular localization and function [18, 19, 21]. Our results show that endogenous XBP1-u colocalizes with SREBP2 in the cytoplasm and cannot enter the nucleus by itself. Following activation, SREBP2 enters the nucleus, where it transactivates its target genes, and where its ubiquitination takes place. Because XBP1-u can translocate into the nucleus upon overexpression of SREBP2, it likely enters the nucleus together with SREBP2.
XBP1-s possesses a transactivation domain in its C-terminus, enabling it to function as a transcriptional factor [21]; whereas XBP1-u does not possess such a domain and appears to regulate its targets through post-translational modification. Our previous study revealed that XBP1-u, but not XBP1-s, could promote cell cycle progression and proliferation in tumor cells by enhancing the degradation of the tumor suppressor p53 through stabilization of its negative regulator MDM2 [22]. Furthermore, XBP1-u could modulate the autophagic process of tumor cells by recruiting FoxO1 to the 20S proteasome and enhancing FoxO1 degradation [24], and protect endothelial cells from oxidative stress by promoting Akt1 phosphorylation [23]. XBP1-u is also crucial for maintain vascular homeostasis, as it directly associated with the N-terminus of forkhead box protein O4 (FoxO4) in the cytoplasm, thereby preventing FoxO4 nuclear translocation and promoting vascular smooth muscle cells differentiation. Moreover, a very recent study showed that XBP1-u is crucial for suppressing vascular calcification [25]. Under normal physiological conditions, XBP1-u blocks β-catenin nuclear translocation by promoting its ubiquitination/proteasomal degradation, leading to the transcriptional suppression of β-catenin/T cell factor (TCF)-mediated osteogenic markers runt-related transcription factor 2 (Runx2) and msh homeobox2 (Msx2) [26]. Our present results show that XBP1-u enhances de novo cholesterol biosynthesis and, consequently, HCC cell proliferation and tumorigenic potential by inhibiting SREBP2 protein ubiquitination. This evidence indicates for the first time, how alterations in cholesterol metabolism underlie the XBP1-u-mediated tumorigenic potential, linking XBP1-u with tumor cell metabolic reprogramming. Furthermore, they strongly support a specific biological and pathological function for XBP1-u. It is noteworthy that recent study also reported that the stability of the XBP1-u protein was reduced and its proteasomal degradation was accelerated in high phosphate (Pi) condition [26]. Thus, although further investigation is needed, the half-life and major form of XBP1 isoforms might depend on the cell types and other factors such as environment. Nevertheless, together with the fact that XBP1-s could also regulate SREBP2 at transcriptional level, our findings indicate that both XBP1 isoforms could regulate cholesterol metabolism in HCC cells through transcriptional and post-translational regulations.
HMGCR is the first enzyme of the mevalonate pathway, the rate-limiting step in de novo cholesterol biosynthesis [50]. Both HMGCR and its transcription activator SREBP2 correlate positively with cholesterol accumulation in various tumor cells including hepatic, prostate, colorectal, and breast cancers, and in turn promotes their progression [51–54]. Thus, targeting the mevalonate pathway to suppress cholesterol biosynthesis has attracted attention as a potential anti-tumor therapeutic strategy. Recent studies have shown that statins, cholesterol-lowering drugs that inhibit the mevalonate pathway by binding to the active site of HMGCR, and betulin, an inhibitor of SREBP2 activation, could suppress tumor cell growth and promote apoptosis [55, 56]. Our results indicate that XBP1-u acts as an antagonist of these cholesterol-lowering compounds and significantly decreases the sensitivity of HCC cells towards them. Accordingly, elevated XBP1-u levels might block anti-tumor therapeutic strategies targeting the mevalonate pathway. Hence, while further detailed pre-clinical and clinical investigations are needed, our findings show the possibility of employing XBP1-u as a marker to determine the suitability of patients for therapeutic strategies targeting mevalonate pathway, as well as the possibility of potentiating the effect of anti-tumor drugs targeting the mevalonate pathway by suppressing XBP1-u expression.
In summary, we demonstrate a novel regulatory mechanism of XBP1-u on the SREBP2/HMGCR axis in HCC cells, linking the tumorigenic potential of XBP1-u with tumor cell metabolic reprogramming. These findings not only provide new insights on the molecular mechanism of cholesterol metabolic reprogramming in HCC, but highlight also the specific biological and pathological functions of XBP1-u. Furthermore, our findings suggest the possibility of treating tumors by targeting XBP1-u or by combining XBP1-u suppression with drugs targeting the mevalonate pathway in HCC. Finally, as HMGCR is also critical for cholesterol accumulation in non-tumor cells, XBP1-u might be a potential target for other hypercholesterolemia-related diseases.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Professor Xia Zhang (Institute of Pathology, Southwest Hospital, Third Military Medical University) for his helpful comments in preparing this manuscript.
Author contributions
VK and SW conceived the project, designed the experiments, analyzed and interpreted the experimental results, and wrote the manuscript; MW, UN, AH performed most of the experiments and analyzed the experimental data; CH and YL carried out part of the experiments, constructed the vectors and interpreted the data; MM designed the shRNA target sequences and provided part of experimental materials for vector constructions.
Funding
This work was supported by grants from the National Natural Science Foundation of China No. 31871367 (V. Kasim), No. 32070715 (V. Kasim), No. 81872273 (S. Wu), and No. 82173029 (S. Wu).
Availability of data and materials
All data generated or analyzed duringthis study are included in this published article and its supplementary information files.
Declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval and consent to participate
Our studies did not include human participants, human data or human tissue. For the animal studies, a protocol detailing experimental procedures following the Animal Care and Use Committee of the Chongqing University Cancer Hospital guidelines was submitted to—and approved by—the Laboratory Animal Welfare and Ethics Committee of Chongqing University Cancer Hospital.
Consent for publication
We have obtained consent to publish this paper from all the participants of this research.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mankun Wei, Uli Nurjanah and Arin Herkilini contributed equally to this work.
Contributor Information
Shourong Wu, Email: shourongwu@cqu.edu.cn.
Vivi Kasim, Email: vivikasim@cqu.edu.cn.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Kasim V, Yan X, Li L, Meliala ITS, Huang C, Li Z, Lei K, Song G, Zheng XJT. Yin Yang 1 facilitates hepatocellular carcinoma cell lipid metabolism and tumor progression by inhibiting PGC-1β-induced fatty acid oxidation. Theranostics. 2019;9(25):7599. doi: 10.7150/thno.34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat metab. 2020;2(2):132–141. doi: 10.1038/s42255-020-0174-0. [DOI] [PubMed] [Google Scholar]
- 5.Russell DW. Nuclear orphan receptors control cholesterol catabolism. Cell. 1999;97(5):539–542. doi: 10.1016/S0092-8674(00)80763-1. [DOI] [PubMed] [Google Scholar]
- 6.Ikonen E. Mechanisms for cellular cholesterol transport: defects and human disease. Physiol Rev. 2006;86(4):1237–1261. doi: 10.1152/physrev.00022.2005. [DOI] [PubMed] [Google Scholar]
- 7.Baenke F, Peck B, Miess H, Schulze A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis Model Mech. 2013;6(6):1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21(4):225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 9.Silvente-Poirot S, Poirot M. Cholesterol metabolism and cancer: the good, the bad and the ugly. Curr Opin Pharmacol. 2012;12(6):673–676. doi: 10.1016/j.coph.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, Martirosyan A, Hakem A, Hakem R, Jurisica I, Penn LZ. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA. 2010;107(34):15051–15056. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurnher M, Nussbaumer O, Gruenbacher G. Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clin Cancer Res. 2012;18(13):3524–3531. doi: 10.1158/1078-0432.CCR-12-0489. [DOI] [PubMed] [Google Scholar]
- 12.Bathaie SZ, Ashrafi M, Azizian M, Tamanoi F. Mevalonate pathway and human cancers. Curr Mol Pharmacol. 2017;10(2):77–85. doi: 10.2174/1874467209666160112123205. [DOI] [PubMed] [Google Scholar]
- 13.Ding X, Zhang W, Li S, Yang H. The role of cholesterol metabolism in cancer. Am J Cancer Res. 2019;9(2):219–227. [PMC free article] [PubMed] [Google Scholar]
- 14.Chimento A, Casaburi I, Avena P, Trotta F, De Luca A, Rago V, Pezzi V, Sirianni R. Cholesterol and its metabolites in tumor growth: therapeutic potential of statins in cancer treatment. Front Endocrinol. 2018;9:1–14. doi: 10.3389/fendo.2018.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenlee JD, Subramanian T, Liu K, King MR. Rafting down the metastatic cascade: the role of lipid rafts in cancer metastasis, cell death, and clinical outcomes. Cancer Res. 2021;81(1):5–17. doi: 10.1158/0008-5472.CAN-20-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGregor GH, Campbell AD, Fey SK, Tumanov S, Sumpton D, Blanco GR, Mackay G, Nixon C, Vazquez A, Sansom OJ, Kamphorst JJ. Targeting the metabolic response to statin-mediated oxidative stress produces a synergistic antitumor response. Cancer Res. 2020;80(2):175–188. doi: 10.1158/0008-5472.CAN-19-0644. [DOI] [PubMed] [Google Scholar]
- 17.Liou HC, Boothby MR, Finn PW, Davidon R, Nabavi N, Zeleznik-Le NJ, Ting JP, Glimcher LH. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science. 1990;247(4950):1581–1584. doi: 10.1126/science.2321018. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 19.Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331(6017):586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, Yoshida H, Kokame K, Kaufman RJ, Mori K. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem. 2004;136(3):343–350. doi: 10.1093/jb/mvh122. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172(4):565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, Wu S, Ji H, Yan X, Xie Y, Murai S, Zhao H, Miyagishi M, Kasim V. Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci Adv. 2017;3(10):e1701383. doi: 10.1126/sciadv.1701383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin D, Li Y, Yang J, Wang G, Margariti A, Jiang Z, Yu H, Zampetaki A, Hu Y, Xu Q, Zeng L. Unspliced X-box-binding protein 1 (XBP1) protects endothelial cells from oxidative stress through interaction with histone deacetylase 3. J Biol Chem. 2014;289(44):30625–30634. doi: 10.1074/jbc.M114.571984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Li X, Cai MY, Ma K, Yang J, Zhou J, Fu W, Wei FZ, Wang L, Xie D, Zhu WG. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23(4):491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao G, Fu Y, Cai Z, Yu F, Gong Z, Dai R, Hu Y, Zeng L, Xu Q, Kong WJC. Unspliced XBP1 confers VSMC homeostasis and prevents aortic aneurysm formation via FoxO4 interaction. Circ Res. 2017;121(12):1331–1345. doi: 10.1161/CIRCRESAHA.117.311450. [DOI] [PubMed] [Google Scholar]
- 26.Yang L, Dai R, Wu H, Cai Z, Xie N, Zhang X, Shen Y, Gong Z, Jia Y, Yu FJC. Unspliced XBP1 counteracts β-catenin to inhibit vascular calcification. Circ Res. 2022;130(2):213–229. doi: 10.1161/CIRCRESAHA.121.319745. [DOI] [PubMed] [Google Scholar]
- 27.Kasim V, Wu S, Taira K, Miyagishi M. Determination of the role of DDX3 a factor involved in mammalian RNAi pathway using an shRNA-expression library. PLoS ONE. 2013;8(3):e59445. doi: 10.1371/journal.pone.0059445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyagishi M, Taira K. Strategies for generation of an siRNA expression library directed against the human genome. Oligonucleotides. 2003;13(5):325–333. doi: 10.1089/154545703322617005. [DOI] [PubMed] [Google Scholar]
- 29.Wu S, Wang H, Li Y, Xie Y, Huang C, Zhao H, Miyagishi M, Kasim V. Transcription factor YY1 promotes cell proliferation by directly activating the pentose phosphate pathway. Cancer Res. 2018;78(16):4549–4562. doi: 10.1158/0008-5472.CAN-17-4047. [DOI] [PubMed] [Google Scholar]
- 30.Wu S, Kasim V, Kano MR, Tanaka S, Ohba S, Miura Y, Miyata K, Liu X, Matsuhashi A, Chung UI, Yang L, Kataoka K, Nishiyama N, Miyagishi M. Transcription factor YY1 contributes to tumor growth by stabilizing hypoxia factor HIF-1α in a p53-independent manner. Cancer Res. 2013;73(6):1787–1799. doi: 10.1158/0008-5472.CAN-12-0366. [DOI] [PubMed] [Google Scholar]
- 31.So JS, Hur KY, Tarrio M, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, Lichtman AH, Iwawaki T, Glimcher LH, Lee AH. Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16(4):487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casali C, Malvicini R, Erjavec L, Parra L, Artuch A, Fernandez Tomez MC. X-box binding protein 1 (XBP1): a key protein for renal osmotic adaptation. Its role in lipogenic program regulation. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865(4):1–30. doi: 10.1016/j.bbalip.2020.158616. [DOI] [PubMed] [Google Scholar]
- 33.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1(6):379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. Embo J. 1999;18(3):717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickels JT., Jr New links between lipid accumulation and cancer progression. J Biol Chem. 2018;293(17):6635–6636. doi: 10.1074/jbc.H118.002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzu OF, Noory MA, Robertson GP. The role of cholesterol in cancer. Cancer Res. 2016;76(8):2063–2070. doi: 10.1158/0008-5472.CAN-15-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang S, Wang X, Song D, Liu X, Gu Y, Xu Z, Wang X, Zhang X, Ye Q, Tong Z, Yan B, Yu J, Chen Y, Sun M, Wang Y, Gao S. Cholesterol induces epithelial-to-mesenchymal transition of prostate cancer cells by suppressing degradation of EGFR through APMAP. Cancer Res. 2019;79(12):3063–3075. doi: 10.1158/0008-5472.CAN-18-3295. [DOI] [PubMed] [Google Scholar]
- 38.McDonnell DP, Park S, Goulet MT, Jasper J, Wardell SE, Chang C-y, Norris JD, Guyton JR, Nelson ER. Obesity, cholesterol metabolism, and breast cancer pathogenesis. Cancer Res. 2014;74(18):4976–4982. doi: 10.1158/0008-5472.CAN-14-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Xia H, Zhou S, Tang Q, Bi F. Cholesterol activates the Wnt/PCP-YAP signaling in SOAT1-targeted treatment of colon cancer. Cell Death Discov. 2021;7(1):1–13. doi: 10.1038/s41420-021-00421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14(11):683–696. doi: 10.1038/nrurol.2017.144. [DOI] [PubMed] [Google Scholar]
- 41.Shafique K, McLoone P, Qureshi K, Leung H, Hart C, Morrison DS. Cholesterol and the risk of grade-specific prostate cancer incidence: evidence from two large prospective cohort studies with up to 37 years' follow up. BMC Cancer. 2012;12:1–8. doi: 10.1186/1471-2407-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelton K, Freeman MR, Solomon KR. Cholesterol and prostate cancer. Opin Pharmacol. 2012;12(6):751–759. doi: 10.1016/j.coph.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tie G, Yan J, Khair L, Messina JA, Deng A, Kang J, Fazzio T, Messina LM. Hypercholesterolemia Increases colorectal cancer incidence by reducing production of NKT and γδ T cells from hematopoietic stem cells. Cancer Res. 2017;77(9):2351–2362. doi: 10.1158/0008-5472.CAN-16-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villa GR, Hulce JJ, Zanca C, Bi J, Ikegami S, Cahill GL, Gu Y, Lum KM, Masui K, Yang H, Rong X, Hong C, Turner KM, Liu F, Hon GC, Jenkins D, Martini M, Armando AM, Quehenberger O, Cloughesy TF, Furnari FB, Cavenee WK, Tontonoz P, Gahman TC, Shiau AK, Cravatt BF, Mischel PS. An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell. 2016;30(5):683–693. doi: 10.1016/j.ccell.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stopsack KH, Gerke TA, Andrén O, Andersson S-O, Giovannucci EL, Mucci LA, Rider JR. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis. 2017;38(8):806–811. doi: 10.1093/carcin/bgx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu X, Ratliff TL, Cheng JX. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19(3):393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13(3):260–273. doi: 10.1016/j.cmet.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Maione F, Oliaro-Bosso S, Meda C, Di Nicolantonio F, Bussolino F, Balliano G, Viola F, Giraudo E. The cholesterol biosynthesis enzyme oxidosqualene cyclase is a new target to impair tumour angiogenesis and metastasis dissemination. Sci Rep. 2015;5:1–12. doi: 10.1038/srep09054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 50.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9(2):125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 51.Mostaghel EA, Solomon KR, Pelton K, Freeman MR, Montgomery RB. Impact of circulating cholesterol levels on growth and intratumoral androgen concentration of prostate tumors. PLoS ONE. 2012;7(1):e30062. doi: 10.1371/journal.pone.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobs RJ, Voorneveld PW, Kodach LL, Hardwick JC. Cholesterol metabolism and colorectal cancers. Curr Opin Pharmacol. 2012;12(6):690–695. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 53.Ginestier C, Monville F, Wicinski J, Cabaud O, Cervera N, Josselin E, Finetti P, Guille A, Larderet G, Viens P, Sebti S, Bertucci F, Birnbaum D, Charafe-Jauffret E. Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells. 2012;30(7):1327–1337. doi: 10.1002/stem.1122. [DOI] [PubMed] [Google Scholar]
- 54.Xue L, Qi H, Zhang H, Ding L, Huang Q, Zhao D, Wu BJ, Li X. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy. Front Oncol. 2020;10:1–20. doi: 10.3389/fonc.2020.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsson O. HMG-CoA reductase inhibitors: role in normal and malignant cells. Crit Rev Oncol Hematol. 1996;22(3):197–212. doi: 10.1016/1040-8428(96)00193-X. [DOI] [PubMed] [Google Scholar]
- 56.Li N, Zhou ZS, Shen Y, Xu J, Miao HH, Xiong Y, Xu F, Li BL, Luo J, Song BL. Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology. 2017;65(6):1936–1947. doi: 10.1002/hep.29018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed duringthis study are included in this published article and its supplementary information files.