Abstract
Since the initial report of V (D) J “allelic exclusion/inclusion” (allelic exclusion rearrangement or allelic inclusion rearrangement) and the concept of the “dual B cell receptor (BCR)” in 1961, despite ongoing discoveries, the precise proportion and source mechanism of dual BCR under physiological conditions have been puzzling immuologists. This study takes advantage of the single cell B cell receptor sequencing (scBCR-seq) technology, which can perfectly match the heavy and light chains of BCR at the level of a single B cell, and obtain the full length mRNA sequence of the complementary determining region 3 (CDR3). Through analyzing the pairing of functional IGH (immunoglobulin heavy chain) and IGL (immunoglobulin light chain) in single B cell from both human and mouse bone marrow and peripheral blood, it was observed that dual BCR B cells exhibit stable and high levels of expression. Among them, the human bone marrow and peripheral blood contain about 10% dual (or multiple) BCR B cells, while in mouse peripheral blood and bone marrow memory B cells, this proportion reaches around 20%. At the same time, we innovatively found that in each research sample of humans and mice, there are three (or more) functional rearrangements (mRNA level) of a single chain in a single B cell. By analyzing the position, direction and other compositional characteristics of the V(D)J gene family, we found that at least two (or more) of them are derived from over two (or more) specific allelic inclusion rearrangements of a single chromosome (mRNA molecular level evidence), our findings also highlighted the necessity of classified single cell sequencing data based on single, dual (or multiple) and cannot be assembled into BCR when analyzing the B cell repertoire. The results of this article provides new methods and modeling references for evaluating the proportion and source mechanisms of dual BCR B cells, as well as potential significance of allelic inclusion (exclusion escape) of V(D)J rearrangement.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-023-04973-8.
Keywords: Single cell B cell receptor sequencing, Allelic inclusion (exclusion escape), V(D)J rearrangement, Dual BCR
Introduction
The classical clonal selection theory of “one B cell-one antibody” [1] is the basis of B cell development, tolerance, and response, which relies on the V(D)J allelic exclusion rearrangement, the B cells did not conform to the rearrangement rules would undergo apoptosis, that is, the mature B cells express only one functional Heavy chain and Light chain. Allelic exclusion was proposed in 1957 and has been studied for nearly 60 years, but the detailed mechanism has not been clarified [2–4]. V(D)J allelic inclusion (incomplete allelic exclusion or allelic escape) rearrangement and dual BCR B cells have been widely supported by experiments since it was reported in 1961 [5]. Nonetheless, owing to the limited scope of single B cell studies, the exact proportions, mechanisms, and implications of dual BCR B cells within individuals remain ambiguous. [6–9]. scBCR-seq enables the analysis of a large number of single B cells with one (or more) Heavy chain and light chain mRNA expressing, providing an opportunity for the intensive investigation of dual BCR proportion and mechanism. In 2019, Qiu et al. [10] pioneered the use of scVDJ(BCR)-seq technology to analyze peripheral blood samples from five healthy individuals. They observed the occurrence of two or more V-D-J (heavy chain) and V-J (light chain) recombinations in a certain number of single B cells. By combining scRNA-seq and Sanger sequencing of single B cells using PCR, the authors examined the presence of two or more instances of class switching events in single B cells that expressed dual (or multiple) BCR. Additionally, they also investigated the distribution of V-D-J (heavy chain) and V-J (light chain) in B cells expressing dual (or multiple) BCR within cell clusters classified as naïve B cells, memory B cells, and plasma B cells. This work has established the groundwork for conducting research on the dual receptor expression in B cells using scRNA + BCR-seq analysis. In 2022, Pelanda et al. [11] compared the rearrangement of dual K (Kappa-Light Chain, κ), dual L (Lambda-Light Chain, λ), and K + L in three healthy individuals and three SLE patients. Expanding on the groundbreaking research conducted by Qiu et al. [10] and Pelanda et al. [11] who employed scRNA-seq technology to explore dual BCR B cells, our study provides a comprehensive analysis of the proportion and underlying mechanisms governing the origin of these dual BCR B cells. According to the BCR rearrangement rule, even if both chromosomes are involved in rearrangement, a single B cell can only rearrange two types of Heavy chains, two types of K and two types of L chains [3, 4, 9]. We systematically analyzed human and mouse bone marrow and peripheral blood samples from multiple laboratories using published and shared “B cell single cell sequencing V(D)J repertoire”, it is interesting to note that in the BCR sequence data of single B cells, we found three or more types of complete mRNAs of heavy or light chains in each sample [10–13]. This raises the question of how the V(D)J allelic escape rearrangement occurs in these B cells. Meanwhile, for the first time, we found a high proportion of B cells expressing dual (or multiple) BCR in single cell B cell receptor sequencing of human and mouse central and peripheral B cells (approximately 10% in human bone marrow and peripheral blood, approximately 20% in mouse memory B cells from peripheral blood and bone marrow), the detailed mechanism of formation and the biological significance of these B cells still require further research and exploration. Furthermore, this study revealed a high proportion of B cells theoretically “cannot be assembled into a BCR” based on the complete mRNA (V-D-J-C) data obtained from single cell B cell receptor sequencing. These findings suggest that investigations into BCR repertoire through single cell sequencing should assess the proportion and significance of B cells that can be assembled into “functional BCR” while also analyzing the potential sources and mechanisms of complete IGH and IGL mRNA.
Materials and methods
The detail information of all samples
The scBCR-seq datasets were download from 10 Company and Gene Expression Omnibus (GEO) data repository. Among them, human bone marrow sample (BM01) and peripheral blood sample (HB08), mouse peripheral blood sample (PBMC_BALB/C, PBMC_C57BL/6) were all provided by 10Company. Human peripheral blood samples (HB01, HB02, HB03) were healthy female volunteers aged 41, 36 and 68. (HB04, HB05, HB06) were all healthy volunteers aged 65 years. Mouse bone marrow samples (bm1, bm2): single cell suspensions from bone marrow of 3 × NP-CGG immunized 8–12 weeks female C57BL/6 mice were prepared and CD19+ cells were enriched by magnetic cell sorting using anti-CD19 microbeads (Miltenyi Biotech). Ex vivo IgG1+/IgG2b+CD19+CD38+GL7−CD138−IgM−IgD− memory B cells were isolated by FACS (Influx cell sorter (BD Bioscience)) and applied to the 10 Genomics platform using the Single Cell 5′ Library & Gel Bead Kit (10 Genomics) following the manufacturer’s instructions.
Definition of functional and non-functional B cells
IGH (Functional) and IGL (Functional) sequences refer to in-frame recombination of functional V(D)J genes to form mRNA that can be completely transcribed, and then expressed as polypeptide chain sequences; functional B cells refer to a single B cell contains at least one IGH (Functional) sequence and one IGL (Functional) sequence; non-functional B cells refers to the single B cells(1) no IGH (Functional) and IGL (Functional) sequences; (2) Only IGH (Functional) sequences were detected; (3) Only IGL (Functional) sequences were detected.
Functional H and L chain expression and combination proportion analysis process
One type of chain "cannot be assembled into BCR" B cells: H; K; L.
Two types of chains "can be assembled into a single BCR"B cells: H + K; H + L; Two types of chains "cannot be assembled into BCR" of B cells: H1 + H2; K1 + K2; L1 + L2; K + L.
Three types of chains “can be assembled into dual BCR” B cells: H1 + H2 + K; H1 + H2 + L; H + K1 + K2; H + L1 + L2; H + K + L. Three types of chains “cannot be assembled into BCR” B cells: H1 + H2 + H3; K1 + K2 + K3; L1 + L2 + L3; K1 + K2 + L; K + L1 + L2.
Four (or more) types of chains “can be assembled into three or more BCR”: H1 + H2 + H3 + K; H1 + H2 + H3 + L; H1 + H2 + K1 + K2; H1 + H2 + L1 + L2; H + K1 + K2 + K3; H + L1 + L2 + L3.
It is rare for five or more types of chains to pair in B cells and form multiple BCR. Direct analysis does not show combinations such as H1 + H2 + H3 + H4 + H5, count to others.
Statistical analysis
Data are presented as Mean ± SEM, statistical analysis with SPSS 29.0 (IBM Corp., Armonk, NY, USA, Version 29.0) software was used to evaluate significant differences between the groups, and p < 0.05 was considered statistically significant. GraphPad Prism software and Adobe Illustrator software were used for data visualization.
Results
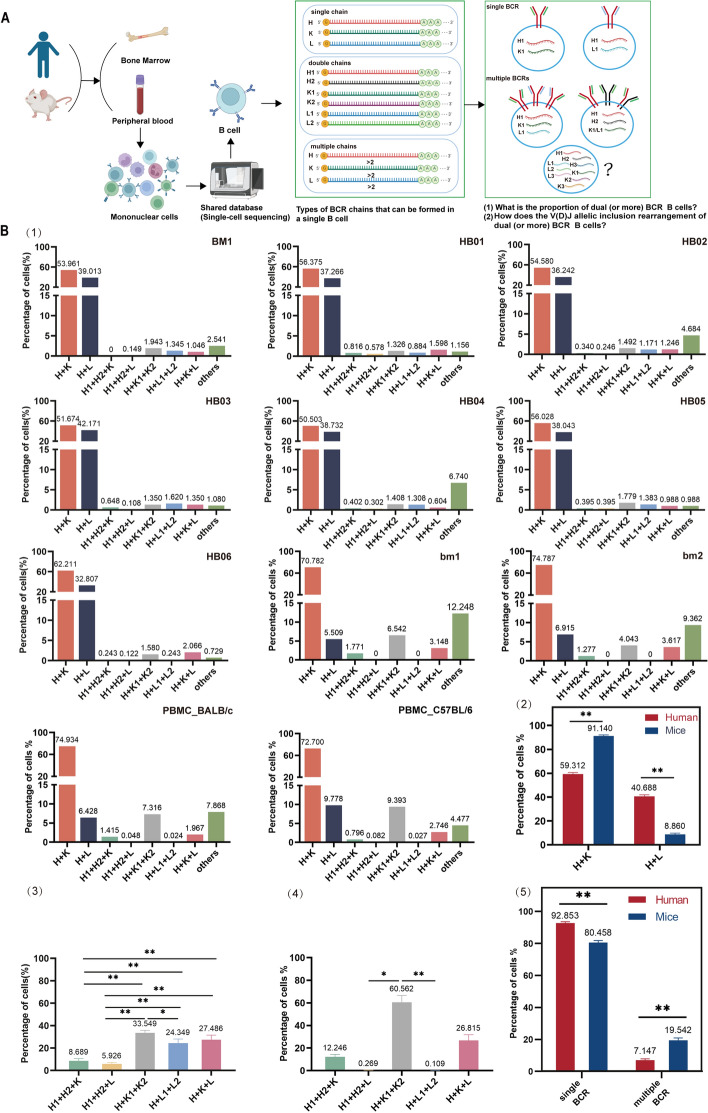
We analyzed the single cell B cell receptor sequencing data of bone marrow and peripheral blood in the physiological condition of human and mice (Fig. 1, Table S1). The main results are:
H + K pairing was significantly higher than H + L in the single BCR B cells, and the proportion of mouse H + L pairing was much lower than that of human, the results consistent with the classical K superiority over Lambda utilization reported (Fig. 1B, Fig S2). In addition, we found the dual (or multiple) BCR was mainly H + K + K, H + K + L, that is, K is also advantage utilization than L in dual BCR. In each sample, there are some H + H + n K/L and H + H + H + n K/L B cells (details examples in Fig. 2). For the first time, a high proportion of dual (or multiple) BCR B cells (about 10% in human bone marrow and peripheral blood, about 20% in mouse peripheral blood and bone marrow memory B cells) were found (Fig. 1B, Fig S2). It were much higher than normal subjects (0.2–2%) [14, 15] and normal mice (0.1–4%) [6, 16] reported by FCM and other technical studies.
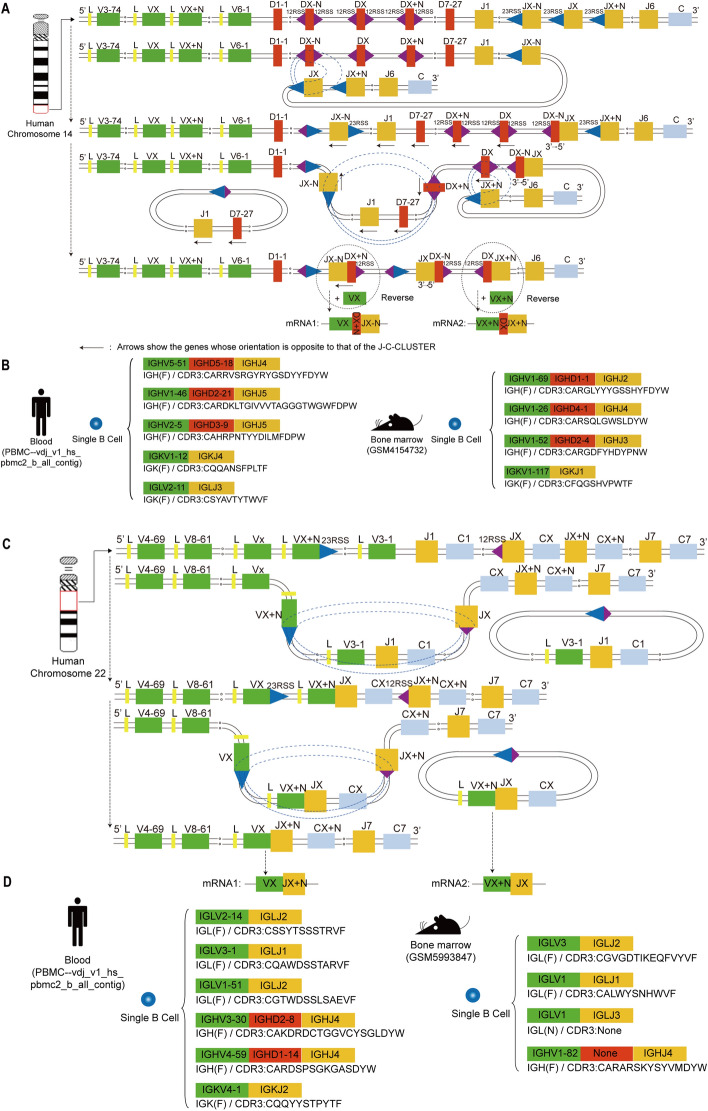
A high proportion of single B cells containing three or more H (or K or L) chain V(D)JC mRNAs were found in both human and mouse samples. In this study, according to the position of a large number of V(D)J pairing families, V(D)J order on human and mouse H locus, direction of RSS, 12/23 deletional (looped out) and inversional rearrangement rules, and H chain D to J recombination preceding V to DJ recombination rules. We found the first time that three (or more) V(D)J mRNAs in a single B cell required a “single chromosome with specific allelic inclusion rearrangement”. BCR H chain required twice D-J inversional recombination on a single chromosome to form two types VDJC mRNAs transcripts (Fig. 3A, B: the rearrangement mechanism and examples). And the L chain V-J required twice deletional rearrangement to form two types VJC mRNAs transcripts on a single chromosome (Fig. S3). If a single B cell can express four (or more) mRNAs only when both paternal and maternal chromosomes undergo twice (or more) rearrangements. The RSS direction of V(D)J in IG gene locus is different from TR, both V and J of IGH can undergo deletional and inversional rearrangement with IGHD (V3′ with 7-23-9 RSS, D5′ and D3′ with 7-12-9 RSS, J5′ with 7-23-9 RSS), which greatly increases the accessibility of secondary or more rearrangements of the H chain in single chromosome. Individuals initiate rearrangements of alleles of the heavy (or light) chain of both paternal and maternal chromosomes, even twice and more rearrangement on one chromosome which has very positive implications for increasing the in-frame rearrangements of effective BCR and the diversity of BCR repertoire. However, the proportion, mechanism and significance of its occurrence remain to be studied in more depth by single cell sequencing, such as the heavy and light chain pairing rule of dual BCR B cells, and functional studies at the protein level.
In the process of analyzing the V(D)J sequences of all single B cell samples, we found that a certain proportion of B cells can only detect IGH (Functional) sequences or only IGL (Functional) sequences, and these cells cannot express a functional BCR (Fig. S1) and should not be included in the analysis of B cell outcomes.
Fig. 1.
The high proportion dual (or multiple) BCR expressing B cells. A Graphical abstract. single cell B cell receptor sequencing samples from human and mouse bone marrow and peripheral blood were used to analyze the proportion and rearrangement mechanism of single BCR and multiple BCR. B (1) The proportion of Heavy and Light chains assembled into single and dual (or multiple) BCR expressing B cells in human and mice bone marrow and peripheral blood. B (2) Statistical comparison of the proportions of H + K (mice > human) and H + L (human > mice) pairing of single B cells in human and mice. B (3) Statistical comparison of the proportions of three chains (H, K, L) assembled into different types of dual BCR in human. The results was H + K1 + K2 > H + K + L > H + L1 + L2 > H + H + K > H + H + L. B(4) Statistical comparison of the proportions of three chains (H, K, L) assembled into different types of dual BCR in mice. The results was H + K1 + K2 > H + K + L > H1 + H2 + K > H1 + H2 + L > H + L1 + L2. B(5) Statistical comparison of the proportions of single BCR and multiple BCR expressing B cells between human and mice. The figure depicts a statistical analysis of single BCR (H + K and H + L) B cells and multiple BCR B cells (H1 + H2 + K, H1 + H2 + L, H + K1 + K2, H + L1 + L2, H + K + L, others) from seven human samples and four mouse samples. The results indicate that the proportion of single BCR B cells in humans is 92.853%, which is higher than the proportion in mice, 80.458%. Meanwhile, the proportion of multiple BCR B cells in mice is 19.542%, significantly higher than that in humans, 7.147%. (1) The proportion of single and dual (or multiple) BCR B cells was based on the number of cells of the clone type in each sample. (2) Data in bar graphs are shown as Mean ± SEM. Statistical analysis was performed with unpaired t test, One-way Anova test, *p < 0.05, **p < 0.01. (3) H = Heavy Chain, K = Kappa-Light Chain, L = Lambda-Light Chain
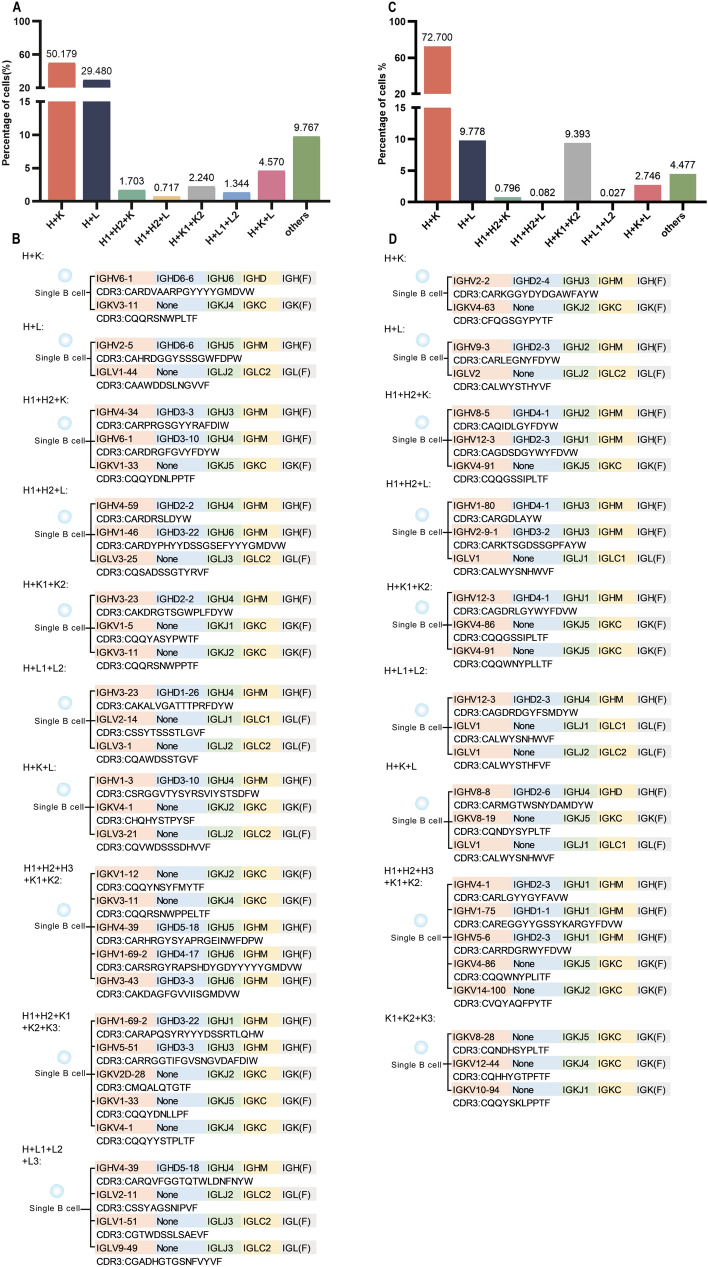
Fig. 2.
Examples of CDR3 sequences in a single B cell that can assemble into single BCR or multiple BCR in both human (HB08) and mouse(PBMC_C57BL/6). A Pairing of heavy and light chains that can assemble into different types of single and dual (or multiple) BCR in human (HB08) sample. B Examples of CDR3 sequences and the V(D)J recombination in a single B cell of human (HB08) sample. C Pairing of Heavy and Light chains that can assemble into different types of single and dual (or multiple) BCR in mouse (PBMC_C57BL/6) sample. D Examples of CDR3 sequences and the V(D)J recombination in a single B cell of mouse (PBMC_C57BL/6) sample. The proportion of H + K pairing was significantly higher than H + L in the single BCR B cells of both human (HB08) and mouse (PBMC_C57BL/6) samples. However, the proportion of H + L pairing was much lower in mouse compared to human, dual or multiple BCR in both species are mainly H + K + K and H + K + L, indicating that Kappa is utilized more frequently than Lambda in single or dual BCR. There are cells expressing three H chains (three K chains or three L chains, etc.) in “others”, examples of the VH(D)JH recombination patterns and CDR3 sequences in single cells expressing different types of single and dual (or multiple) BCR B cells in both human peripheral blood (HB08) and mouse (PBMC_C57BL/6) samples
Fig. 3.
The Special rearrangement mechanisms for three types of IGH and IGL mRNA in a single B cell and an example of a single B cell CDR3 sequence in human and mouse samples. A The H chain on a single human chromosome experiences two (or more) D-J inversional rearrangements. A single IGH chain on human chromosome 14, where DX−N initiates the "first inversion rearrangement" with JX (depicted by dashed thin trajectory). The 5′ end of DX−N with its 12RSS, binds to the 23RSS of JX (indicated by dashed bold trajectory). Subsequently, the 23RSS of JX−N at the 5′ end engages with the 12RSS of DX+N to execute a “deletion (loop out) rearrangement,” resulting in the JX−N-DX+N combination. Following this, the JX−N-DX+N combination undergoes an "inversion rearrangement" with VX to form VX-DX+N-JX−N (the first mRNA transcript). The intermediate genes “J1 and D7-27” loop out from the chromosome. At the 3′ end, Dx participates in a “second inversion rearrangement” with JX+N, resulting in the DX-JX+N combination, which then undergoes an “inversion rearrangement” with VX+N to yield the VX+N-DX-JX+N combination (the second mRNA transcript). B Examples of CDR3 sequences with three H chains from single B cells in both humans and mouse samples. Based on the VDJ rearrangement pattern illustrated in A, provide examples of the composition of single B cell mRNA sequences for both human and mouse samples. C The L chain on a single human chromosome experiences two (or more) V-J deletional rearrangements. A single IGL chain on human chromosome 22, the 23RSS of VX+N binds to the 12RSS of JX, leading to a “deletion (loop-out) rearrangement” that generates the VX+N-JX combination. Subsequently, the 5′ end of Vx and the 3′ end of JX+N engage in another “deletion (loop-out) rearrangement,” resulting in the formation of VX-JX+N(the first mRNA transcript). This process leads to the loop-out of the previously rearranged VX+N-JX (the second mRNA transcript). D Examples of CDR3 sequences with three L chains from single B cells in both human and mouse samples. Based on the VJ rearrangement pattern illustrated in C, provide examples of the composition of single B cell mRNA sequences for both human and mouse samples. The H chain required two (or more) D-J inversional recombination on a single chromosome to form two (or more) VDJC mRNAs transcripts (among the three types of H in single human and mouse B cells as shown in the figure, at least two of them are derived from over two specific allelic inclusion rearrangements of a single chromosome). The L chain required twice deletional rearrangement to form two types VJC mRNAs transcripts on a single chromosome (among the three types of IGL in single human and mouse B cells as shown in the figure, at least two types of IGL are derived from specific allelic inclusion rearrangements in a single chromosome)
Discussion
Extensive experimental evidence has verified the widely accepted concept of the “clonal selection theory”, which proposes that “each lymphocyte expresses only a single type of antigen receptor” [1]. This process is mainly attributed to “allelic exclusion”. However, the exact mechanisms responsible for allelic exclusion have yet to be fully elucidated, despite over 60 years of invesimmune diseases and plasmacytomas [8, 9, 11, 17], since the first report of their existence in 1961. Nevertheless, the proportion and source of these B cells under normal physiological conditions have been unresolved problems. This study takes advantage of the single cell B cell receptor sequencing technology, we systematically analyzed the proportion and potential mechanisms of IGH and IGL allelic inclusion rearrangement in B cells from human and mouse bone marrow and peripheral blood samples across multiple laboratories using published and shared “single B cell sequencing data” [10–12]. Qiu et al. [10] were the first to utilize scRNA-seq to report the presence of B cells expressing dual (or multiple) BCR in peripheral blood samples from healthy individuals, while Pelanda et al. [11] were the first to employ scRNA-seq to report B cells expressing dual (or multiple) BCR in peripheral blood samples from SLE patients. We have reported, for the first time, a high and stable proportion of B cells that express dual (or multiple) BCR in physiological states, with approximately 10% in both human bone marrow, as well as peripheral blood and approximately 20% in memory B cells in both mouse bone marrow and peripheral blood. while both single and dual (multiple) BCR B cells showed the expression of K chain is superior to that of the L chain. Additionally, our findings reveal that in both human and mouse central and peripheral, multiple BCR B cells exhibit expression of five types of chains (μ, γ, α, δ, ε), the evidence suggests that multiple BCR B cells all possess the capability to undergo class switching. These observations align with those made in peripheral blood by Qiu et al. [10]. Furthermore, they conducted further analysis of the co-expression of types such as IGM + IGA; IGA + IGD; IGM + IGA + IGG; IGM + IGD + IGG in single B cells expressing dual (or multiple) BCR. Considering the analysis of a limited set of 13 samples in this study, along with the restricted number of sequenced single B cells per sample, no significant differences were observed in VDJ usage and pairing between single and multiple BCR B cells. This implies that the VDJ rearrangements in both single and multiple BCR B cells are the result of random recombination processes.
Meanwhile, during the analysis process of this study, we innovatively found that in each sample, there are V(D)JC mRNA sequences containing three or more H (or K or L chains) in a single B cell, which are derived from single chromosome with specific allelic inclusion rearrangement. That is, the BCR H chain required twice D-J inversional recombination, K chain V-J required twice deletional or inversional rearrangements, and L chain V-J required twice deletional rearrangement to form two types of V(D)JC mRNA transcripts on a single chromosome (mRNA molecular level evidence). We have identified a novel mechanism of “allelic exclusion escape” involving “dual (or multiple) rearrangements” on a single chromosome, which provides new technical approaches for modeling “allelic exclusion escape” and related studies. However, In physiological conditions, it remains to be further studied why B cells undergo a high proportion of “three (or more) times” functional rearrangement of heavy and light chains. The results of this article provides a more accurate and comprehensive understanding of B cell diversity and function, which may help address potential biases in the application of single cell sequencing in BCR repertoire studies.
Currently, in the analysis of single cell B cell receptor sequencing results, many laboratories primarily conduct comparative statistical analyses of total B cells based on the “barcode” of each cell. In our study, we performed separate statistical analyses, comparing the total number of B cells (Fig. 1) and number of clone type cells (Fig. S2) for each individual BCR sequencing sample. Among these 13 samples, it has been observed that the vast majority of single BCR B cells and dual BCR B cells are monoclonal. However, B cells expressing two distinct types of BCR exhibit instances of clonal expansion with clone counts equal to or greater than two. It is worth noting that under normal physiological conditions, the clonal expansion proportion of dual BCR B cells remains very low. Nevertheless, our study has found a higher proportion of clonal expansion among dual BCR B cells in autoimmune pathologies closely associated with them [11, 17]. By considering both the total number of B cells and number of clone type cells, analyzing the VDJ rearrangement mechanisms and usage biases of dual BCR B cells could unveil novel targets for immunodiagnosis and therapeutic interventions in diseases.
In single cell B cell receptor sequencing results, the presence of single IGH or IGL mRNA chain that “cannot assemble into BCR,” prompted us to conduct a causal analysis. Throughout the procedure of scBCR-seq, it is possible that there was a challenge in effectively tracking the IGH and IGL mRNA within single B cells at an equivalent level, or the V(D)J genes could not be sequenced completely. In bone marrow B cells, there is a subset known as developing B cells, which may have undergone successful rearrangement of only one chain. Moreover, in the repertoire preparation process for single BCR sequencing samples, there is a possibility of the two chains becoming asynchronous, leading to the sequencing of only one of the chains in the end. Based on the current applications of single cell BCR (or TCR) sequencing research, it is commonly observed that a significant proportion of lymphocytes cannot be assembled into BCR or TCR. The theoretical possibility of partial lymphocytes expressing only a single chain cannot be entirely dismissed [18–21]. Therefore, our results suggest that the scBCR-seq should be analyzed separately by single BCR, dual (or multiple) BCR B cells, and cells that cannot be assembled into BCR, it is crucial not to include them in relevant physiological or pathological correlation analyses solely due to their functional IGH (or IGL) mRNA, as this could lead to an amplification and distortion of BCR information.
In both human and mouse samples, the presence of single cells expressing three or more functional IGH or IGL chain mRNA is evident. This suggests a widespread occurrence of V(D)J allelic inclusion rearrangements in BCR, yet the intricate mechanisms behind this phenomenon remain to be further validated. Although this allelic inclusion rearrangement pattern theoretically holds the potential to broaden an individual's BCR repertoire, it also concurrently escalates the risks of individual B cell tumors and autoimmune diseases. Currently, whether this observed rearrangement pattern in humans and mice exists in other mammals remains uncertain. Investigating whether this phenomenon correlates with the distinct immune response capabilities or disease occurrences among different mammalian species represents an entirely novel research avenue worthy of pursuit. However, despite the utilization of extensive sequencing data generated by scBCR-seq technology in studies conducted by Qiu et al. [10], Pelanda et al. [11] and our research, which included preliminary analyses of the characteristics of single B cells expressing dual (or multiple) BCR, further exploration of the functional aspects of these B cells at the protein level, as well as the investigation into the physiological and pathological significance of B cells expressing dual (or multiple) BCR, requires the collaboration and participation of more research laboratories.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to the 10×Company, GEO and IMGT databases for providing the availability of the data. In addition, thanks to Shi, Z, Peterson, J.N, Wang P and Riedel R for sharing the single cell sequencing data.
Author contributions
XY completed the experimental design and paper writing, LZ, QP, and YW completed the data analysis and chart making.
Funding
This study was supported by the National Natural Science Foundation of China (82160279) and the Guizhou Provincial Hundred level Talent Fund [No. (2018)5637].
Data and materials availability
All data are available in the main text or the supplementary materials.
Declarations
Conflict of interest
The authors declare no conflict of interests.
Ethics declarations
This study does not involve ethical requests or approvals.
Consent for publication
All authors have read the manuscript and agreed to give their consent for the publication of information in the Journal of Cellular and Molecular Life Sciences.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lanwei Zhu, Qi Peng, and Yingjie Wu are joint first authors.
References
- 1.Burnet SFM. The clonal selection theory of acquired immunity. Nashville: Vanderbilt University Press; 1959. [Google Scholar]
- 2.Casellas R, Shih TAY, Kleinewietfeld M, Rakonjac J, Nemazee D, Rajewsky K, Nussenzweig MC. Contribution of receptor editing to the antibody repertoire. Science. 2001;291(5508):1541–1544. doi: 10.1126/science.1056600. [DOI] [PubMed] [Google Scholar]
- 3.Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237(1):22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Outters P, Jaeger S, Zaarour N, Ferrier P. Long-range control of V (D) J recombination & allelic exclusion: modeling views. Adv Immunol. 2015;128:363–413. doi: 10.1016/bs.ai.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Mäkelä O, Nossal GJV. Study of antibody-producing capacity of single cells by bacterial adherence and immobilization. J Immunol. 1961;87:457–463. doi: 10.4049/jimmunol.87.4.457. [DOI] [PubMed] [Google Scholar]
- 6.Ten Boekel E, Melchers F, Rolink AG. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity. 1998;8(2):199–207. doi: 10.1016/S1074-7613(00)80472-0. [DOI] [PubMed] [Google Scholar]
- 7.Rezanka LJ, Kenny JJ, Longo DL. 2 BCR or NOT 2 BCR–Receptor dilution: a unique mechanism for preventing the development of holes in the protective B cell repertoire. Immunobiology. 2005;210(10):769–774. doi: 10.1016/j.imbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Xu D. Dual surface immunoglobulin light-chain expression in B-cell lymphoproliferative disorders. Arch Pathol Lab Med. 2006;130(6):853–856. doi: 10.5858/2006-130-853-DSILEI. [DOI] [PubMed] [Google Scholar]
- 9.Pelanda R. Dual immunoglobulin light chain B cells: Trojan horses of autoimmunity? Curr Opin Immunol. 2014;27:53–59. doi: 10.1016/j.coi.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Z, Zhang Q, Yan H, Yang Y, Wang P, Zhang Y, Deng Z, Yu M, Zhou W, Wang Q, Yang X, Mo X, Zhang C, Huang J, Dai H, Sun B, Zhao Y, Zhang L, Yang YG, Qiu X. More than one antibody of individual B cells revealed by single-cell immune profiling. Cell Discov. 2019;5(1):1–13. doi: 10.1038/s41421-019-0137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson JN, Boackle SA, Taitano SH, Sang A, Lang J, Kelly M, Rahkola JT, Miranda AM, Sheridan RM, Thurman JM, Rao VK, Torres RM, Pelanda R. Elevated detection of dual antibody B cells identifies lupus patients with B cell-reactive VH4–34 autoantibodies. Front Immunol. 2022;13:795209. doi: 10.3389/fimmu.2022.795209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Luo M, Zhou W, Jin X, Xu Z, Yan S, Li Y, Xu C, Cheng R, Huang Y, Lin X, Yao L, Nie H, Jiang Q. Global characterization of peripheral B cells in Parkinson's disease by single-cell RNA and BCR sequencing. Front Immunol. 2022;13:814239. doi: 10.3389/fimmu.2022.814239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riedel R, Addo R, Ferreira-Gomes M, Heinz GA, Heinrich F, Kummer J, Greiff V, Schulz D, Klaeden C, Cornelis R, Menzel U, Kröger S, Stervbo U, Köhler R, Haftmann C, Kühnel S, Lehmann K, Maschmeyer P, McGrath M, Naundorf S, Hahne S, Sercan-Alp Ö, Siracusa F, Stefanowski J, Weber M, Westendorf K, Zimmermann J, Hauser AE, Reddy ST, Durek P, Chang HD, Mashreghi MF, Radbruch A. Discrete populations of isotype-switched memory B lymphocytes are maintained in murine spleen and bone marrow. Nat Commun. 2020;11(1):2570. doi: 10.1038/s41467-020-16464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giachino C, Padovan E, Lanzavecchia A. κ+λ+ dual receptor B cells are present in the human peripheral repertoire. J Exp Med. 1995;181(3):1245–1250. doi: 10.1084/jem.181.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser LD, Zhao Y, Lutalo PM, D'Cruz DP, Cason J, Silva JS, Dunn-Walters DK, Nayar S, Cope AP, Spencer J. Immunoglobulin light chain allelic inclusion in systemic lupus erythematosus. Eur J Immunol. 2015;45(8):2409–2419. doi: 10.1002/eji.201545599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rezanka LJ, Kenny JJ, Longo DL. Dual isotype expressing B cells [κ+/λ+ ] arise during the ontogeny of B cells in the bone marrow of normal nontransgenic mice. Cell Immunol. 2005;238(1):38–48. doi: 10.1016/j.cellimm.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Sang A, Danhorn T, Peterson JN, Rankin AL, O’Connor BP, Leach SM, Torres RM, Pelanda R. Innate and adaptive signals enhance differentiation and expansion of dual-antibody autoreactive B cells in lupus. Nat Commun. 2018;9(1):1–14. doi: 10.1038/s41467-018-06293-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JE, Botting RA, Domínguez Conde C, Popescu DM, Lavaert M, Kunz DJ, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367(6480):eaay3224. doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo OJ, Lei W, Zhu G, Ren Z, Xu Y, Xiao C, et al. Multidimensional single-cell analysis of human peripheral blood reveals characteristic features of the immune system landscape in aging and frailty. Nat Aging. 2022;2(4):348–364. doi: 10.1038/s43587-022-00198-9. [DOI] [PubMed] [Google Scholar]
- 20.Stubbington MJT, Lönnberg T, Proserpio V, Clare S, Speak AO, Dougan G, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13(4):329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu L, Peng Q, Li J, Wu Y, Wang J, Zhou D, Ma L, Yao X. scRNA-seq revealed the special TCR β & α V(D)J allelic inclusion rearrangement and the high proportion dual (or multiple) TCR expressing cells. Cell Death Dis. 2023;14(7):487. doi: 10.1038/s41419-023-06004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.