Abstract
Glutamine is essential for maintaining the TCA cycle in cancer cells yet they undergo glutamine starvation in the core of tumors. Cancer stem cells (CSCs), responsible for tumor recurrence are often found in the nutrient limiting cores. Our study uncovers the molecular basis and cellular links between glutamine deprivation and stemness in the cancer cells. We showed that glutamine is dispensable for the survival of ovarian and colon cancer cells while it is required for their proliferation. Glutamine starvation leads to the metabolic reprogramming in tumor cells with enhanced glycolysis and unaltered oxidative phosphorylation. Production of reactive oxygen species (ROS) in glutamine limiting condition induces MAPK-ERK1/2 signaling pathway to phosphorylate dynamin-related protein-1(DRP1) at Ser616. Moreover, p-DRP1 promotes mitochondrial fragmentation and enhances numbers of CD44 and CD117/CD45 positive CSCs. Besides the established features of cancer stem cells, glutamine deprivation induces perinuclear localization of fragmented mitochondria and reduction in proliferation rate which are usually observed in CSCs. Treatment with glutaminase inhibitor (L-DON) mimics the effects of glutamine starvation without altering cell survival in in vitro as well as in in vivo model. Interestingly, the combinatorial treatment of L-DON with DRP1 inhibitor (MDiVi-1) reduces the stem cell population in tumor tissue in mouse model. Collectively our data suggest that glutamine deficiency in the core of tumors can increase the cancer stem cell population and the combination therapy with MDiVi-1 and L-DON is a useful approach to reduce CSCs population in tumor.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03818-6.
Keywords: Glutaminase, Glutamine metabolism, ROS, Tumor growth, Mitochondrial fission
Introduction
The most prominent feature of cancer is the rapid proliferation of abnormal cells that grow beyond their usual boundaries and metastasize to other parts of the body. In the current scenario, cancer metastasis and its recurrence are the main clinical problems for the scientific community. The recurrence occurs after a short break of treatment due to the chemoresistance property acquired in the tumor cells. A tiny population of CSCs within the tumor is responsible for this chemoresistance [1].
One of the emerging hallmarks of cancer is metabolic reprogramming or plasticity, which is generated in the cancer cells to acclimatize in the nutrient-deprived and hypoxic environment. For their survival and proliferation, cancer cells depend upon two important nutrients glucose and glutamine. In tumor cells, glucose can supply rapid energy and produce lactate from pyruvate under the aerobic condition, known as the “Warburg effect” [2]. This pyruvate cannot enter into the TCA cycle as pyruvate dehydrogenase (PDH) remains inactive due to its phosphorylation by pyruvate dehydrogenase kinase (PDK). In such cells, glutamine plays a crucial role to fuel the TCA cycle and act as an anaplerotics agent to synthesize lipids, nucleotides, and hexosamines [3]. The normal physiological concentration of glutamine in blood is 0.6–0.9 mM and the level may reach up to 20 mM in some tissues [4]; however, in tumor tissue the glutamine level is reduced drastically. This is due to the fact that the tumor cells are highly glutamine-dependent and they utilize the glutamine rigorously leading to its reduction in the tumor microenvironment (TME). To meet the high demand for glutamine in tumor cells, its synthesis increases in cancer-associated fibroblasts (CAF) in TME [5]. In mammalian cell, de novo glutamine synthesis occurs through the glutamine synthetase (GS). However, GS does not express significantly in all tumor cells and hence they mostly depend on the extracellular sources [6]. In addition, glutamine transporters, such as ASCT2 (SLC1A5), LAT1 (SLC7A5), ATB0,+ (SLC6A14), are overexpressed in tumor cells which help in glutamine uptake [7].
Glutaminase (GLS) is involved in the conversion of glutamine to glutamate and ammonium [8] and the glutamate is converted into α-ketoglutarate by glutamate dehydrogenase (GDH), which acts as a substrate for TCA cycle. Glutamine is also a constituent of the tripeptide glutathione (GSH), an antioxidant and there by maintains oxidative stress within the cell. Glutamine derived glutamate is exported out of the cell in exchange for cysteine through xCT(SLC7A11), where cysteine is the rate-limiting amino acid for GSH [3].
Mitochondria are the prime site for TCA cycle and glutamine metabolism. ROS also has been reported to be beneficial for cancer progression [9]. Moreover, mitochondrial dynamics and their specific localization inside the cell play an important role in oncogenesis. Recent reports suggest that mitochondrial fragmentation is increased in cancer cells to support actin polymerization and cell migration [10]. In some cells, after fragmentation, perinuclear localization of mitochondria increases regional ROS level in the vicinity of the nucleus and induces stem-like properties [11]. The cellular requirement of glutamine varies extensively among different cancer types as some are glutamine-dependent, whereas some cells can survive and proliferate in its absence [12]. Glutamine itself acts as a signaling molecule to activate STAT3 and mTORC1 for cancer proliferation, which is independent of glutamine metabolism. Some reports suggest that glutamine dependency increases with their aggressiveness [12–14]. Hence, the modulation of glutamine metabolism is associated with tumor progression, and regulation of this might be important from the therapeutic point of view. Various small molecules such as BPTES, compound 968, CB-839 that target GLS are presently in clinical trial. Though BPTES specifically targets GLS, it happens to be a poor drug candidate owing to its solubility and bioavailability. EGCG and R162, inhibitors of GDH have been shown to attenuate tumor growth in preclinical trial [15]. It is evident that the tumor core contains a population of CSCs and has limited nutrient supply. This regional glutamine deprivation leads to the de-differentiation of the cancer cells followed by chemoresistance [16, 31]. These stem-like cells can be identified by the expression of various cell surface antigens (CDs) and pluripotent stemness markers like Sox2, Oct4, Nanog, and high aldehyde dehydrogenase (ALDH) activity [1].
Considering this background information, we aimed to study the mechanism of survival of cancer cells in glutamine-deprived environment. We have mainly used epithelial ovarian cancer cell line, PA1 and OAW42 and the colorectal cancer cell line HCT116. Our study indicated that glutamine deprivation promotes mitochondrial fragmentation, which in turn enhances stem-like characteristics and chemoresistance. We also found that GLS1 inhibition enhances the expression of the stemness markers. Therefore, inhibitors of both GLS1 and DRP1 might be effective as promising therapeutic agent to treat the tumor growth and disease recurrence.
Materials and methods
Cell culture and glutamine deprivation
Human ovarian cancer cell lines PA1 (ATCC;Manassas, Virginia, United State; Cat# CRL-1572, RRID:CVCL_0479) and OAW42 (SIGMA-ALDRICH;St. Louis, Missouri, United States; Cat# 85073102, RRID:CVCL_1615) were grown in Minimal essential media alpha (MEM α, Gibco) and Dulbecco’s modified Eagle’s medium (DMEM, Gibco), respectively. Another ovarian carcinoma cell line OVCAR3 (RRID:CVCL_0465, a gift from Dr. Asima Mukhopadhyay, CNCI, Kolkata, India) was grown in RPMI-1640 media (Gibco). Human colorectal carcinoma cell line HCT116 (ATCC Cat# CCL-247, RRID:CVCL_0291), ID8 (MERCK Cat# SCC145,RRID:CVCL_IU14) and human non-small cell lung carcinoma H1299 (RRID: CVCL_0060, a gift from Prof. Samit Chattopadhyay, CSIR-IICB, Kolkata, India) were cultured in DMEM. Human cervical carcinoma cell lines, HeLa (RRID:CVCL_0058) and SiHa (RRID:CVCL_0032) (were gifts from Dr. Chinmay Kumar Panda, CNCI, Kolkata, India) were maintained in DMEM and MEM α respectively. All the media were supplemented with 10% FBS (Cat #16000–044, Gibco) and the cells were grown at 37 °C in a humified 5% CO2 incubator. Glutamine deprivation studies were performed in glutamine-free medium (MEM α, Cat#AL080, DMEM, Cat#AL066, RPMI-1640, Cat#AL060, HIMEDIA) for respective cell lines with 10% FBS reconstitution. The cells were starved for at least 6 h prior to the glutamine deprivation.
Treatments of cells and siRNA transfection
The cells were starved for at least 6 h and the respective treatments were given. As mentioned, the cells were treated with 14 µM of MDiVi-1(Cat#M0199, Sigma-Aldrich), 2 mM of glutathione reduced ethyl ester (GSH, Cat#G1404, Sigma-Aldrich), 0.5 mM, 1 mM, 3 mM and 5 mM of N-acetyl cysteine (NAC, Cat#A7250, Sigma-Aldrich), 10 µM of ERK inhibitor (Cat#328006, Calbiochem), 10 µM of U0126 (Cat#662005, Calbiochem), 30 µM of H2O2 (Cat#107209, Merck), 20 µM, 40 µM, 100 µM of L-DON (Cat#D2141, 6-Diazo-5-oxo-L-norleucine, Sigma-Aldrich), 250 µM of SSZ (Sulphasalazine, Cat#S0883, Sigma-Aldrich). The siRNA against DRP1 (Cat#sc-43732), xCT (Cat#sc-76933) and PDHE1α (Cat#sc-91064) (Santa Cruz Biotechnology; Dallas, Texas,United State) were used at 20 nM concentration along with lipofectamine 2000 (Cat#11668019, Invitrogen) in the cell lines. Scrambled (SCR) siRNA (Cat#sc-37007, Santa Cruz Biotechnology) was used as control for knockdown studies. The transfection was done at 50–60% confluency and the transfection reagents were added in Opti-MEM (Cat#31985062, Gibco) medium and after 4 h of transfection the media was changed to respective media of treatment (either glutamine containing or glutamine free medium).
Quantitative real-time PCR
Total RNA was isolated from the cells using RNAiso Plus reagent (Cat#9109, Takarabio; Kyoto, Japan) following the standard protocol. iScript cDNA synthesis kit (Cat#1708891, Bio-Rad; Hercules, California, United States) was used for synthesizing cDNA. Quantitative real time-PCR (Q-PCR) was performed with iTaq universal SYBR green supermix (Cat#1725120, Bio-Rad) on the ABI 7500 Fast Real-Time PCR system (Applied Biosystems; Waltham, Massachusetts, USA; 7500 Real-Time PCR Software, RRID:SCR_014596). Fold change in gene expression was calculated against 60S ribosomal protein L19 mRNA expression (considered as endogenous control). The primers were designed using Primer Express software (Primer Express Software, RRID:SCR_017376) and verified using Primer-Blast (Primer-BLAST, RRID:SCR_003095). Custom oligos were obtained from IDT (Integrated Technology Enterprise Inc, RRID:SCR_012186) and the sequences are given in Table 1.
Table 1.
Primer details used for checking the respective mRNA expression
| Gene name | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon size |
|---|---|---|---|
| OCT4 | GCAGCTTAGCTTCAAGAACATGTG | TCAGCTTCCTCCACCCACTT | 66 |
| SOX2 | TGCGAGCGCTGCACAT | GCAGCGTGTACTTATCCTTCTTCA | 93 |
| NANOG | GCATCCGACTGTAAAGAATCTTCA | CATCTCAGCAGAAGACATTTGCA | 90 |
| ABCG2 | GATGTCTAAGCAGGGACGAACAA | GGTGAGGCTATCAAACAACTTGAA | 82 |
| CD44s | TCCAACACCTCCCAGTATGACA | GGCAGGTCTGTGACTGATGTACA | 83 |
| CD44 epithelial | GAAAGGAGCAGCACTTCAGG | GAGGTCCTGTCCTGTCCAAA | 202 |
| L19 | GCGGATTCTCATGGAACACA | GGTCAGCCAGGAGCTTCTTG | 68 |
Western blot analysis
The cells were lysed using radioimmunoprecipitation assay (RIPA) buffer and the protein was extracted. All the proteins were then subjected to western blot analysis using the standard protocol [17]. Each blot was stripped with stripping buffer (Cat#T7135A, TAKARA) and re-probed for checking corresponding α-tubulin as endogenous control (Cell signaling Technology (CST); Danvers, Massachusetts; Cat# 2125, RRID:AB_2619646). The primary antibodies such as p-DRP1 (Ser616) (CST Cat# 3455, RRID:AB_2085352), DRP1 (CST Cat# 8570, RRID:AB_10950498), HK2 (CST Cat# 2867, RRID:AB_2232946), SDHA (CST Cat# 5839, RRID:AB_10707493), VDAC (CST Cat# 4661, RRID:AB_10557420), MFN1 (CST Cat# 14739BC), MFN2 (CST Cat# 9482), p-AKT (CST Cat# 4060, RRID:AB_2315049), AKT (CST Cat# 4691, RRID:AB_915783), p38MAPK (CST Cat# 9218, RRID:AB_10694846), p-p38MAPK (T180/Y182) (CST Cat# 4631, RRID:AB_331765), p-p44/42MAPK (CST Cat# 9101, RRID:AB_331646), OCT4 (CST Cat# 2750, RRID:AB_823583), SOX2 (CST Cat# 3579, RRID:AB_2195767), xCT/SLC7A11 (CST Cat# 12691, RRID:AB_2687474), α-tubulin (CST Cat# 2125) were purchased from CST and used at 1:2000 dilution. Antibodies of PDHE1α (Santa Cruz Biotechnology Cat# sc-377092, RRID:AB_2716767), ABCG2 (Santa Cruz Biotechnology Cat# sc-58222, RRID:AB_630828) were obtained from Santa Cruz Technology and used at 1:1000 dilution. Antibodies against p-PDHE1α (Ser2193) (Millipore; Burlington, Massachusetts, United States; Cat# ABS204, RRID:AB_11213668) and total OXPHOS (Abcam; Cambridge, United Kingdom; Cat# ab110411, RRID:AB_2756818) complex were procured from Merck (1:10,000 dilution) and Abcam (1:3000 dilution), respectively. The chemiluminescent bands were detected in the blot using ECL detection reagent (Cat# 1705062, Bio-Rad).
Extracellular flux analysis
Extracellular acidification rate (ECAR) was measured using Agilent seahorse extracellular flux analyzer (XFe24) as described earlier [17]. Briefly, PA1 and HCT116 cells were seeded at 3000 cells/well and 10,000 cells/well were plated for OAW42. Each treatment was given in triplicates. Primarily basal oxygen consumption rate (OCR) and ECAR were measured under glucose-free condition. After that, ECAR was measured using Glycostress assay kit with three consecutive injections of glucose, oligomycin and 2-deoxyglucose (2-DG) (Agilent; Santa Clara, CA; Cat#103344-100). Mitostress assay kit (Agilent, Cat# 103015-100) was used for analyzing mitochondrial bioenergetics using three consecutive injections of oligomycin, FCCP (Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone) and antimycin/rotenone. The ECAR and OCR values were normalized by estimating protein concentration and cell concentration in each well. Analysis was done by WAVE (Seahorse Wave, RRID:SCR_014526) software.
Lactate assay
Amount of lactate was measured in both the condition media of glutamine-deprived condition and control media using lactate assay kit (Sigma-Aldrich, Cat# MAK064) following manufacturer’s protocol. Lactate production was normalized using cell numbers and represented as 'fold change' with respect to control.
Analysis of metabolites using NMR
After completion of the treatment the media was aspirated, centrifuged in 12000 g for 5 min at 4 °C for the isolation of cell debris. From here, 1 ml media was withdrawn and mixed thoroughly with 300 μl of methanol-chloroform mixture (2:1) and centrifuged at 12000g for 10 min at 4 °C. Then the upper aqueous layer was extracted, mixed with five times of HPLC grade water and freezed at − 80 °C. Then it was lyophilized and re-suspended in 600 μl of D2O (Sigma-Aldrich, Cat#151882) mixture (10% D2O, 90% H2O). TSP (Cat #269913, Sigma-Aldrich) was used as a control for reference peak. The samples were analyzed in 600 MHz proton NMR instrument (Brucker; Billerica, Massachusetts, US). The.fid file was uploaded in MetaboAnalyst 3 (MetaboAnalyst, RRID:SCR_015539) web application and output data was exported in.xlsx format. ClustVis (ClustVis, RRID:SCR_017133) was used to create a heatmap of the acquired metabolites after PCA analysis.
Confocal microscopy and live-cell video microscopy
To visualize the mitochondrial morphology and the fission, MitoTracker Red CMXRos (50 nM, Invitrogen; Waltham, Massachusetts, USA; Cat#M7512) was used for 20 min at 37 °C. Immunofluorescence microscopy was performed with DRP1 Antibody (CST Cat# 8570, RRID:AB_10950498), followed by Alexa flour 488 conjugated secondary antibody (Thermo Fisher Scientific; Waltham, Massachusetts, US; Cat# A32731, RRID:AB_2633280) as described previously [18]. The nuclei were stained with DAPI (0.25 µg/ml) for 5 min. The images were visualized and acquired by Leica confocal microscope SP8 (Leica Microsystems; Wetzlar, Germany; RRID:SCR_008960) with 63X oil immersion objective lens, Zeiss LSM 800 microscope (Zeiss; acquisition software: LAS X; Oberkochen, Germany) with 63X oil immersion objective lens (NA 1.4) and Olympus BX51 microscope (Olympus; acquisition software: cellSens; Shinjuku, Tokyo, Japan; RRID:SCR_017564) with 40X (NA 0.75) objective lens. 3D reconstruction of the images was done by LAS X software (LAS X, RRID:SCR_013673). Images were taken in room temperature.
For time-lapse video microscopy, the cells were grown in 35 mm cover glass-bottom dish (SPL Lifesciences; Korea), and the treatment was done for glutamine-deprived conditions. After completion of the treatment, the cells were incubated with MitoTracker Red (50 nM, Invitrogen, Cat#M7512) and Hoechst stain (5 µg/ml, Sigma-Aldrich, cat#14533) for 20 min at 37 °C in 5% CO2 incubator. Then the images were taken in every 13.75 s for 7 min 6 s in case of control cells. The images were taken in every 5.061 s for 3 min 7.25 s for the cells grown in glutamine-deprived condition with 63X objective lens (NA 1.4). The video reconstruction was done by LAS X software (LAS X, RRID:SCR_013673).
Cellular ROS localization study using DCFDA and Mitotracker Red
The cells were grown in 35 mm cover glass-bottom dish and treated with respective glutamine-containing media and glutamine-deprived media. At 24 h the cells were stained with 2 μM DCFDA (CM-H2DCFDA, Invitrogen, Cat# C6827) and MitoTracker Red CMXRos (50 nM, Invitrogen, Cat#M7512) in the respective culture medium for 30 min in humified 37 °C incubator (5% CO2). After that, the cells were washed with PBS and observed under Leica SP8 microscope (Leica Microsystems, RRID:SCR_008960) under 63X oil immersion objective [11].
Intracellular and extracellular antigen staining and measurement by flow cytometry
For intracellular antigen staining, the cells were grown in 60 mm dish. After the completion of the treatment, the media was discarded and cells were scraped with phosphate buffered saline (PBS). Then the cells were fixed with 4% formaldehyde, which was added drop-wise with vigorous mixing in vortex and kept at 37 °C for 10 min followed by a brief incubation in ice for 1 min. Then the cells were centrifuged at 3000g for 5 min and the supernatant was discarded. The cells were then resuspended in PBS and permeabilized by adding drop-wise 90% chilled methanol under stirring condition and incubated in ice and then kept it overnight at − 20 °C. Next day, the cells were washed with PBS and incubated with anti–GSH antibody (Cloud clone Corp. Cat# PAA294Ge01) at 37 °C for 2 h followed by washing with PBS and then incubated with FITC-tagged secondary antibody (Thermo Fisher Scientific Cat# A32731, RRID:AB_2633280) for 1 h. Flow cytometric analysis was performed using LSR Fortessa cell analyzer (BD Biosciences, RRID:SCR_013311).
Extracellular antigen staining was done in live cells. Cells from 60 mm dish were scrapped with PBS and incubated with Fluorophore tagged primary antibody CD44-PE (BD Biosciences Cat# 555479, RRID:AB_395871), CD117-APC (BD Biosciences; Franklin Lakes, New Jersey, U.S.; Cat# 550412, RRID:AB_398461), CD31-FITC (BD Biosciences Cat# 555445, RRID:AB_395838) and CD45-PE-Cy7 (BD Biosciences Cat# 557748, RRID:AB_396854) (used in 1:400 dilution) at 37 °C for 30 min followed by washing and analysis by flowcytometry [1]. Unstained control cells were used as experimental control.
Aldehyde dehydrogenase (ALDH) activity assay
Aldehyde dehydrogenase (ALDH) assay was done using the ALDEFLUOR kit (Stem Cell Technologies; Vancouver, British Columbia, Canada; Cat# 01700) following the manufacturer's protocol. Fluorescence intensity was measured by flow cytometer using FACSDiva (BD FACSDiva Software, RRID:SCR_001456). In the dot plot, gating was done on stained ‘CONTROL + DEAB’ tube keeping 1% cells in P3 gate.
Spheroid formation
Initially cells were culture in 35 mm dish and treated as mentioned. From these dishes, cells were trypsinized and 5×103 cells/well were seeded in low attachment 6-well plates (Cat#CLS3471, Corning, New York, NY). They were allowed to grow in serum-free media (SFM) supplemented with B27 supplement (Cat#17504044, Invitrogen), rhEGF (Cat# F0291, Sigma-Aldrich) and insulin (Cat#I0320000, Sigma-Aldrich) up to 4 days following the previous standard protocol [1]. The spheroids were visualized, counted, and their images were acquired using a phase-contrast microscope with 20X objective (EVOS, Invitrogen). The ImageJ (ImageJ, RRID:SCR_003070) software was used for measuring the size of the spheroids. The scale length of acquired images was fed to the software and after thresholding the image, the volume of every spheroid was acquired.
Rhodamine efflux assay
Cells were grown in 6-well plate. After treatment is over, Rh123 (200 ng/ml, Cat# R302, Invitrogen), a substrate for efflux transporter, was given in the medium and incubated in 37 °C CO2 incubator for 1 h [1]. Then the cells were washed and again replenished with respective media under the optimum condition for 24 h. The cells that refluxed or retained drug, were measured through FACS analysis on the basis of unstained control cells.
Cell cycle analysis
The cells were grown in 6-well plate and after completion of the treatment, cells were trypsinized and washed with PBS. Fixation was done with 70% ethanol with gentle vortexing and were kept overnight at − 20 °C. Next day, after centrifugation, the cells were washed again with PBS and re-suspended in PBS containing Propidium Iodide (PI) (1.5 mM, Cat# P1304MP, Invitrogen), RNaseA (1 mg/ml, Cat# EN0531, Fermentas; Waltham, Massachusetts, USA). They were incubated for 15 min at room temperature and analyzed by flow cytometer with BD FACSDiva Software, (RRID:SCR_001456) (493/636 nm). Gating of flow cytometric data were done using unstained control cells. Histogram overlay were prepared using FlowJo software (FlowJo, RRID:SCR_008520).
Cell proliferation study
3000 cells were seeded per well in a 12-well plate. The cell count was taken at 24 h, 48 h, and 72 h of glutamine deprivation. The media was discarded and then the cells were trypsinized with 100 μl of trypsin-ETDA and mixed with 100 μl of complete medium. From this cell solution, 10 μl was added to the TC-10 cell counting slide and cell concentration was measured using TC-10 automated cell counter (Bio-Rad, USA).
Annexin-V-FITC/PI apoptosis assay
Annexin-V-FITC/PI assay was done to check the apoptosis in glutamine-deprived condition by flow cytometric analysis following the manufacturer’s protocol (Invitrogen). The results have been represented as dot plot using BD FACSDiva Software ( RRID: SCR_001456) [18]. Using unstained control cells, gating was done in dot plot data of flow cytometry.
Estimation of mitochondrial ROS generation
1X106 cells were scraped and washed with PBS. To determine the mitochondrial ROS generation, cells were resuspended with 100 μl of PBS containing MitosoxTMred (5 µM) mitochondrial superoxide indicator (Invitrogen, Cat#M36008) and incubated for 20 min at 37 °C. Then the cells were washed with PBS and the amount of ROS generation was determined using flow cytometer (510/580 nm) (BD FACSDiva Software, RRID:SCR_001456) [17].
Estimation of mitochondrial membrane potential
Approximately, 1×106 cells were scraped and washed with PBS. They were resuspended in PBS containing JC-1 (5 μg/ml, Invitrogen) followed by incubation for 15 min at 37 °C. The ratio of cells showing red/green (485/535 nm)/(514/529 nm) fluorescence was calculated from the dot plot of the flow cytometric data using BD FACSDiva Software (RRID:SCR_001456) [18].
Glucose oxidation
The cells were kept in serum-starved condition for 2 h prior to the addition of 0.5 µCi of U-C14-glucose (BRIT) to the media and incubated at 37 °C for 2 h. During incubation, Whatman chromatography paper soaked with 3 M NaOH was placed on the lid of the plate, such that each well is covered. After incubation, 500 µL of perchloric acid was added to release the dissolved CO2, which was then trapped in Whatman paper for 1 h. The filter paper was dried overnight and then put in the cocktail-T solution. The levels of C14 were measured using the scintillation counter (Tri-Carb 2810TR, Perkin Elmer) and the CPM value was normalized with the cell numbers [19].
Glucose uptake assay
0.5 µCi of 2-DG (C14, BRIT; Navi Mumbai, India) was added to each well of a 6-well plate following 30 min of incubation in 37 °C. Then the media was discarded and the cells were trypsinized with 200 µl of trypsin–EDTA solution (0.25%). 10 µl cell solution was used to count the cells and the rest was pelleted down. After discarding the supernatant, the pellet was re-suspended and incubated for 20 min at room temperature in 10% NP40 for cell lysis. This lysed cell solution was then mixed with 5 ml of cocktail-T solution and the scintillation count was taken in Tri-Carb 2810TR counter (Perkin Elmer; Waltham, Massachusetts, US).
Determination of mitochondrial load using MitoTracker green and nonyle acridine orange
MitoTracker green (100 nM, Invitrogen, Cat# M7514) was added in the cell suspension in 1X PBS and incubated at 37 °C for 10–15 min. The green fluorescence (490⁄516 nm) intensity of the cells was determined using flow cytometer using BD FACSDiva Software (RRID:SCR_001456). Similarly 1 μM NOA (Nonyle acridine orange, Invitrogen, Cat# A1372) staining was performed where the incubation period is 30 min [20].
Analysis of mitochondrial morphology using Atomic force microscope (AFM)
Mitochondria were isolated from the treated cells using mitochondrial isolation kit (QIAGEN; Venlo, Netherlands; Cat#37612) following the manufacturer’s protocol. The mitochondrial pellet was washed with PBS for two times and the suspension in PBS was fixed with 4% paraformaldehyde with gentle shaking. The fixed cells were washed twice with 1×PBS and then with deionized water and the pellet was resuspended in deionized water. 5 μl of the mitochondrial sample was deposited onto Ruby mica sheet (ASTM V1 Grade Ruby Mica from MICAFAB; Chennai, India) or coverslips for 5–10 min. Then the sample was gently washed with 0.5 ml Milli-Q water to remove the loosely attached molecule.
Contact mode AFM was performed using a Pico plus 5500 AFM (Agilent Technologies USA) with a piezo scanner having a maximum range of 100 μm. Image processing has been done through Pico Image Advanced version software (Agilent Technologies, USA).
Pyruvate dehydrogenase (PDH) activity assay
PDH activity was measured using the PDH assay kit (Sigma-Aldrich, Cat#MAK183) as instructed in the manufacturer’s protocol. Briefly, mitochondria were isolated from both control and glutamine-deprived cells using mitochondria isolation kit (QIAGEN, Cat#37612) and the protein concentration was estimated in each sample. Using the NADH standard, the PDH activity of the unknown sample was measured.
Immunohistochemistry
Primary ovarian tumor tissue samples were collected from the Saroj Gupta Cancer Centre & Research Institute, Kolkata after appropriate approval from the Institutional Ethics Committee (approval number-IEC SGCCRI REF NO.-16/2/2018/Non-Reg/SSR/3) and informed written consent from the patients in accordance with the 1964 Helsinki declaration. The nature of the tissue and stage of the disease was validated by the concerned clinician and respective histology experts of the source institute. Tissue sections were processed and fluorescently stained against CD44 (PE tagged) (BD Biosciences Cat# 555479, RRID:AB_395871) and p-DRP1 (S616) (CST Cat# 3455,RRID:AB_2085352) (stained with Alexa-fluor 488 secondary antibody, Thermo Fisher Scientific Cat# A32731, RRID:AB_2633280). Mouse tumor tissue sections of 5 μm thickness were stained with Cleaved Caspase 3 (Cell Signaling Technology Cat# 9661, RRID:AB_2341188), Ki67 (Abcam Cat# ab15580, RRID:AB_443209), ABCG2 antibody, p-DRP1 (Ser616), OCT4 (CST Cat# 2750, RRID:AB_823583), SOX2 (CST Cat# 3579, RRID:AB_2195767), and PE tagged CD44 antibody. Anti-mouse AF555 tagged antibody (Thermo Fisher Scientific Cat# A28180, RRID:AB_2536164) was used against ABCG2 antibody and AF488 tagged anti rabbit antibody was used to counterstain other primary antibodies. All the tissue sections were counter-stained with DAPI (0.25 µg/ml) for staining the nucleus. The images were acquired with Leica confocal microscope SP8 (Leica Microsystems, RRID:SCR_008960) with 20× objective lens (NA 0.4) in room temperature.
Co-immunoprecipitation assay
Proteins were extracted from whole cell lysate using immunoprecipitation lysis buffer and the protein concentration was estimated. Then 200 μg of protein was incubated with CD44 (BD Biosciences Cat# 555479, RRID:AB_395871) primary antibody for the formation of protein-antibody conjugation complex, which was then pulled down using ProteinA magnetic beads (Bio-Rad, Cat# 1614013). This protein-antibody conjugation complex was released from the beads by boiling in the SDS-electrophoresis sample buffer (1X), which was then subjected to SDS-PAGE followed by western blot using xCT antibody (CST Cat# 12691, RRID:AB_2687474) [17].
Image analysis
Image analysis was done with ImageJ software (ImageJ, RRID:SCR_003070). For measuring the mitochondrial distance from the nucleus, the red channel (MitoTracker red) images were first converted into 8-bit images, and then a straight line is drawn from the center of the nucleus up to the region of mitochondrial fluorescence within the cell boundary. The fluorescent intensity throughout the line was extracted.
For 'line scan' analysis, first, a freehand line was drawn over the mitochondrial length in the red channel image after converting them into 8-bit image and the intensity profile was extracted with the “Plot profile” option. Then the ROI was copied from that image of MitoTracker channel to green channel (8-bit image of the Drp-1) of the same image and the Drp-1 intensity throughout the ROI was plotted with “Plot Profile” option.
The relative mean fluorescence intensity was measured with ImageJ software after selecting the region of interest.
The mitochondrial length analysis and co-localization analysis were done with LAS X software (LAS X, RRID:SCR_013673) as done previously [21] and the co-localization was analyzed with ImageJ (ImageJ, RRID:SCR_003070) software.
TUNEL assay
The tissues were deparaffinized using xylene and then hydrated using serial ethanol gradation. After hydrating, the tissue sections were washed with 0.9% NaCl and digested with Proteinase-K. For positive control, the tissue was incubated with DNase1 (Invitrogen, Cat#AM2224) and TUNEL assay was performed according to the manufacturer’s protocol (Invitrogen, Cat#A35125). DAPI was used for counter-staining the nucleus. The images were acquired using Leica SP8 confocal microscope.
Animal model
All animal procedures were approved by institutional animal ethics committee following the guidelines of CPCSEA, Govt. of India. The ID8 cells were cultured in DMEM supplemented with 10% FBS. About 3×107 cells/ml in PBS was mixed with Matrigel (19 mg/ml) (BD, Cat no. 356234) solution in 1:1 ratio prior to injection C57BL/6 female mice (3–4 weeks old) were injected subcutaneously with 300 μl cell-matrigel suspension at the lower right quadrant using a 1 ml insulin syringe (30G 5/16 in). After 4 days of injection, the animals which have detectable tumors were included in the study. When the tumors reached about 100 mm3 size, they were randomly grouped in control and treatment set. Control mice were injected with 125 μl PBS and treated mice were injected with 125 μl L-DON solution (0.6 mg/kg body weight), 125 μl MDiVi-1 (5 mg/kg body weight) and 125 μl of MDiVi-1 + L-DON (5 mg/kg and 0.6 mg/kg of body weight), respectively, through tail vein for three consecutive days. Each day body weight and the tumor size were measured and after completion of the treatment the tumors were isolated and weighed. The tumors were then fixed with 10% formalin and paraffin blocks were prepared for histological analysis. Bright field images of Hematoxylin and Eosin stained histological sections were acquired under EVOS microscope (Applied Biosystems) with 20× objective lens (NA 0.45).
Two-tailed paired t test was performed when the number of groups was two and the one-way ANOVA (followed by Bonferroni post-test analysis) was done, where the number of groups was more than two. P values < 0.05 were considered to be statistically significant.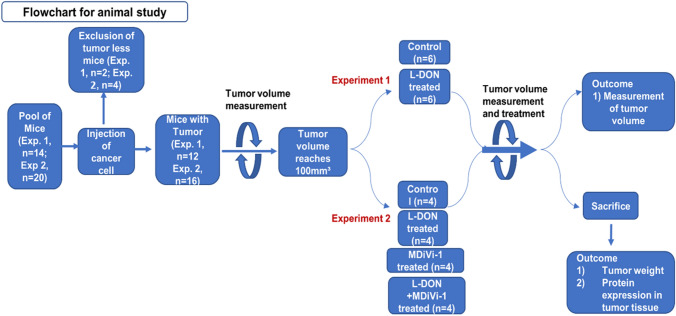
Statistical analysis
All statistical analyses were performed using GraphPad Prism-5 (GraphPad Prism, RRID:SCR_002798) software. The data were represented as ± SEM. Two-tailed paired t test was performed when the number of groups was two and the one-way ANOVA (followed by Bonferroni post-test analysis) was done, where the number of groups was > 2. P values < 0.05 were considered to be statistically significant. All the experiments were performed thrice unless otherwise stated. Graphs were made with excel software (Microsoft Excel, RRID:SCR_016137).
Results
Tumor cells can survive in glutamine deficient condition
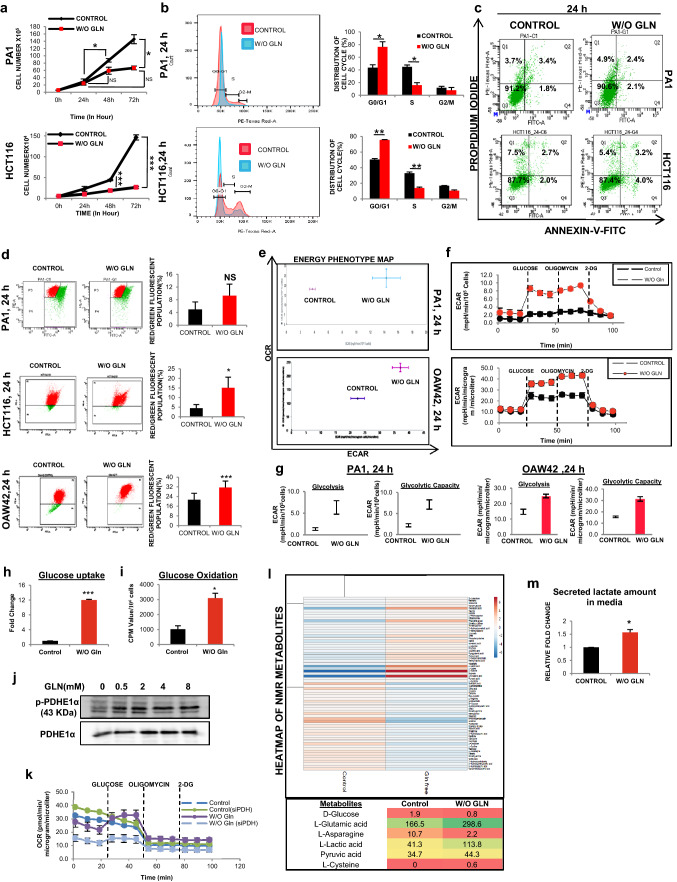
To understand whether glutamine harmonizes cell growth and proliferation in cancer, we first examined cellular proliferation of PA1 and HCT116 cells cultured with or without glutamine. Although cell number did not change significantly with time when grew in glutamine-starved condition (Fig. 1a), but cell population in S-phase was significantly reduced with concomitant enhancement of control as observed in the cell cycle (Fig. 1b). To test whether the reduced cell count was due to cell death upon glutamine deprivation, we performed annexin-V/PI apoptosis assay and observed that there was no significant change in the live cell population (Fig. 1c, Supplementary Fig. S1A). Mitochondrial membrane potential known to be decreased in cells undergoing apoptosis [18], was also found to be increased whenever glutamine is depleted from the culture medium (Fig. 1d). Collectively, these data suggest that PA1 and HCT116 cells can sustain in glutamine-deprived condition, however, their proliferation is delayed.
Fig. 1.
Cancer cells can survive and rearrange their metabolic phenotype in absence of glutamine. a Cell proliferation assay was done by counting cell number in 3 consecutive time point (24 h, 48 h, and 72 h) in glutamine-deprived condition (indicated as W/O GLN) in PA1 (n = 3) and HCT116 (n = 2) cells. b Histogram of cell cycle analysis and corresponding bar diagram of the histogram depicts that the cells were arrested in G0 phase upon glutamine deprivation in both PA1 and HCT116 (n = 3). c Apoptosis assay conducted in glutamine-deprived condition (24 h) and did not find any change in the live-cell population of PA1, and HCT116 (n = 3). d Red/green fluorescent population (%) was quantified for mitochondrial membrane potential using JC-1 dye in PA1, HCT116, and OAW42 upon 24 h of glutamine limiting condition, and presented in bar diagram (n = 3). e, f Extracellular flux analysis dictates that the cells became more glycolytic in nature while OXPHOS remained unchanged at 24 h time point in PA1 and OAW42 cells. g At 24 h of glutamine deprivation, glycolysis rate and glycolytic capacity increased in both PA1 and OAW42 cells. h Glucose uptake was increased in 24 h of glutamine deprivation in PA1 (n = 3). i Glucose oxidation through TCA cycle was increased in absence of glutamine at 24 h in PA1 (n = 3). j PDH phosphorylation increased in absence of glutamine at 24 h shown by Western blot in PA1 cells. k Glycostress assay kit was used to check the change in OCR after glucose injection in glutamine-deprived condition and silencing the PDHE1α in PA1 cells. l Heatmap of metabolites measured through NMR spectroscopy at 24 h of glutamine-starved condition in PA1. m Lactate level was increased in the medium when the cells are cultured in 24 h of glutamine limiting condition in PA1 (n = 3). Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001). NS non-significant
Glutamine starvation leads to altered cellular bioenergetics
As cancer cells endure glutamine-restricted condition, we focused on bioenergetics profile to understand their survival strategy. Although the in vitro cellular oxygen consumption rate (OCR) was changed insignificantly at the basal condition at 24 h (Supplementary Fig. S1B), the extracellular acidification rate (ECAR) started increasing at 18 h and was increased at 24 h and 48 h in PA1 and OAW42 cells (Fig. 1e, f, Supplementary Fig. S1C-F). Similar result was obtained for HCT116 (Supplementary Fig. S5L). The glycolytic reserve, which is the level of full potential of the cell to perform glycolysis was also observed to be increased at 48 h, but not at 24 h of glutamine starvation (Supplementary Fig. S1F-G). Apart from the unaltered basal OCR, the coupling efficiency, which is the percentage of basal OCR used for the ATP synthesis remained unchanged and the glutamine-starved cells produced similar amounts of ATP compared to control cells (Supplementary Fig. S1B). Further, the enhancement of glycolysis and glycolytic capacity in glutamine starvation (Fig. 1g) correlated with the enhanced glucose uptake and glucose oxidation (Fig. 1h, i). In contrast, the protein level of Hexokinase 2 (HK2), a rate-limiting enzyme of glycolysis, remained unchanged (Supplementary Fig. S1H). Glucose is known to be converted into lactate in aerobic condition (Warburg effect) and glutamine can fuel TCA cycle in cancer cells [3]. To rescue the oxidative phosphorylation in glutamine-restricted condition, glucose can provide pyruvate to enter into the TCA cycle to maintain OCR. Therefore, we checked the PDH activity, which was modestly increased (Supplementary Fig. S1I) with a concomitant reduction of its phosphorylation in glutamine-starved condition (Fig. 1j). In addition, we observed that when glucose was injected during extracellular flux analysis, OCR was increased in absence of glutamine, but increment of OCR was reduced when PDHE1α (a subunit of PDH) was silenced (Fig. 1k). These data clearly indicated that in absence of glutamine, glucose takes part in TCA cycle and increases the OCR in PDH dependent manner. As shown by NMR spectroscopy data, glucose produces a higher amount of lactate apart from being the fuel of TCA cycle to maintain OCR (Fig. 1l). Similar result was obtained when extracellular lactate from the medium was quantified in glutamine-restricted condition (Fig. 1m, Supplementary Fig. S1J). Altogether, these first sets of data indicate that in glutamine-restricted condition, cancer cells maintain their cellular bioenergetics by utilizing glucose as the major fueling source for TCA cycle as well as for glycolysis.
Glutamine deprivation promotes mitochondrial fragmentation
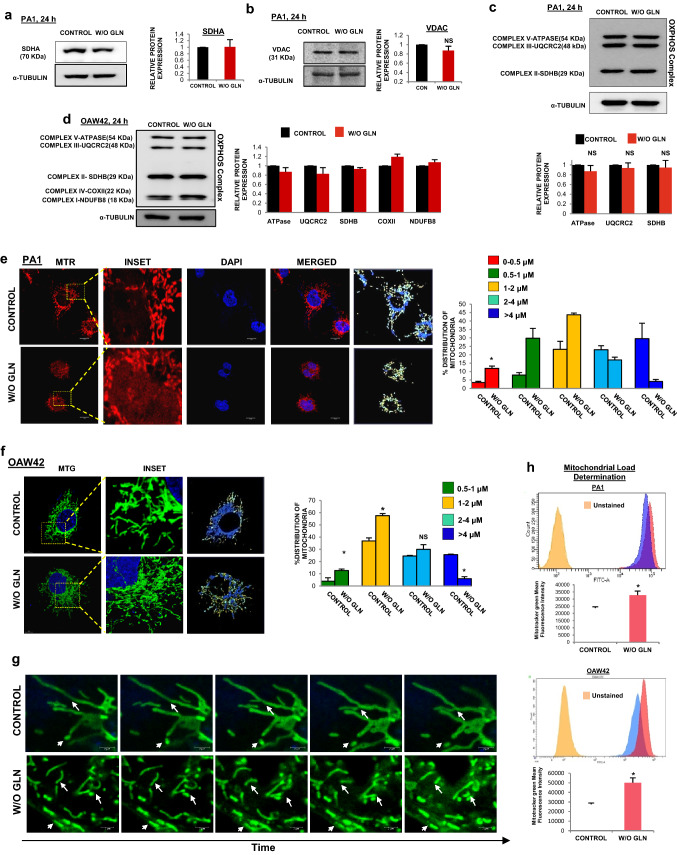
Mitochondria are the major site for glutamine metabolism and OXPHOS. In glutamine-restricted condition the tumor cells adapt themselves to maintain an uninterrupted OXPHOS. Hence, we wanted to connect the mitochondrial dynamics and modulation of cellular bioenergetics in glutamine-deprived tumor cells. We did not find any significant change in the expression of mitochondrial proteins such as voltage-dependent anion channel (VDAC), succinate dehydrogenase A (SDHA), and total OXPHOS complex (complex I-V), which indicate unaltered mitochondrial mass (Fig. 2a-d). Surprisingly, we observed that mitochondria are fragmented in the glutamine-starved state. In PA1 and OAW42 cells, the relative number of short mitochondria (≤ 2 µm) was increased with a concomitant depletion of long mitochondria (> 4 µm) (Fig. 2e, f, Supplementary Vid. S1-4). Similar results have been obtained for HCT116 cell line (Supplementary Fig. S2A-B). In live-cell videography of PA1 under glutamine limiting condition, shorter mitochondria were observed due to the rapid occurrence of ‘kiss-n-run’ phenomenon. In contrast, elongated mitochondria were present in the untreated cells with ubiquitous fission and fusion phenomenon (Fig. 2g, Supplementary Vid. S5-6). Interestingly, mitochondria fragmentation was found to be time-dependent, which started appearing at 18 h post-starvation and increased at 24 h (Supplementary Fig. S2C-D). Fragmentation was also increased in non-small cell lung cancer cell H1299 and in another epithelial ovarian cancer cell line OVCAR3, at 24 h of glutamine starvation (Supplementary Fig. S2E-F). However, we did not find any difference in mitochondrial fragmentation in other cervical cancer cell line HeLa and SiHa (Supplementary Fig. S2G-H). Mitochondrial fragmentation has been reported to occur during apoptosis and mitophagy [18, 22]. Since the cells did not undergo apoptosis in glutamine limiting condition (Fig. 1c), we focused on the status of mitophagy using mitotracker green by flow cytometry. Mitochondrial amount is known to decrease due to mitophagy as well as mitochondrial degradation [23]. We found enhanced mitotracker green intensity, suggesting increased mitochondrial load in glutamine starved condition both in PA1 and OAW42 cells (Fig. 2h). As mitotracker green intensity depends upon mitochondrial membrane potential and ROS, we further analyzed the mitochondrial amount by flow cytometry, using nonyl-acridine orange (NAO) which binds to cardiolipin present in inner mitochondrial membrane irrespective of mitochondrial metabolism through flow cytometric analysis [20]. No significant change was found in mitochondrial amount (Supplementary Fig. S2I) and hence we ruled out the possibility of mitophagy. Atomic force microscopic images also depicted that mitochondria were fragmented and their surface roughness remained unchanged in glutamine limiting condition (Supplementary Fig. S2J). Convincingly all these envisioned that glutamine restriction could promote mitochondrial fission in cancer cells.
Fig. 2.
Glutamine starvation leads to the fragmentation of mitochondrion. a–d Western blot analysis of SDHA (n = 2), VDAC, OXPHOS complex proteins (PA1 (n = 3) and OAW42 (n = 2) revealed that there was no change in the functional protein amount upon 24 h of glutamine starvation. The quantitative fold change of each protein is represented in the bar diagram along with the respective blot. Microscopic observation with Mitotracker red (MTR) and Mitotracker green (MTG) showing increased fragmentation of mitochondria upon 24 h of glutamine starvation in PA1 (e) and OAW42 (f) respectively (Being a 3D image, this image does not contain scale bar). Different sized mitochondria are indicated with different colour and their percentage was quantified and expressed in bar diagram for respective cell line g Sequential images of live (time lapse) videography of mitochondria undergoing fission and fusion at 24 h of glutamine limiting condition in PA1. The time interval between two consecutive frames for control was 13.75 s and for glutamine-starved cells was 7.25 s. h Mitochondrial load determination by Mitotracker Green (MTG) showed a significant increase in the mitochondrial amount upon 24 h of without glutamine condition in PA1 and OAW42 which represented in both histogram and bar diagram (n = 3). Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t-test (*p < 0.05). NS non-significant. Scale bars 10 μm (e), 2 μm (g)
Stemness and chemoresistance properties also promoted upon glutamine limitation in cancer cells
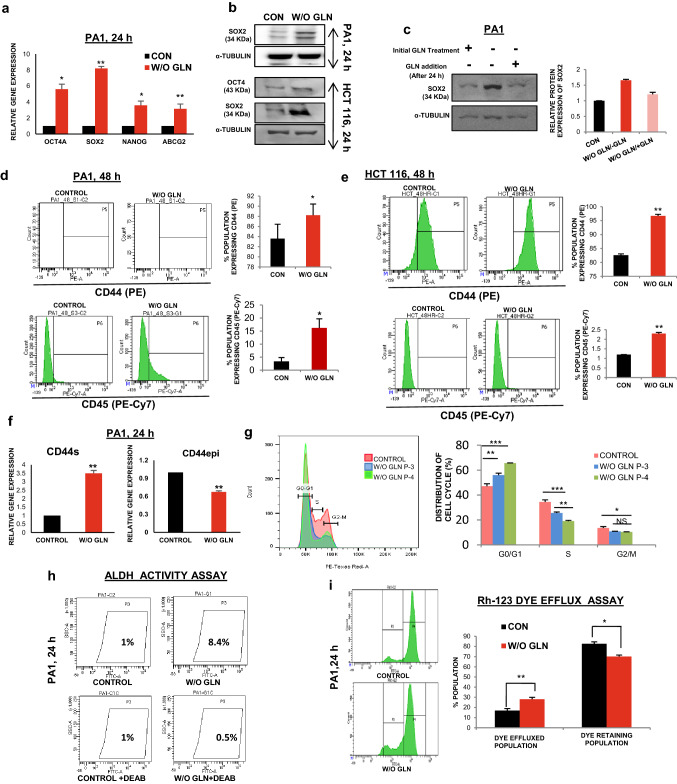
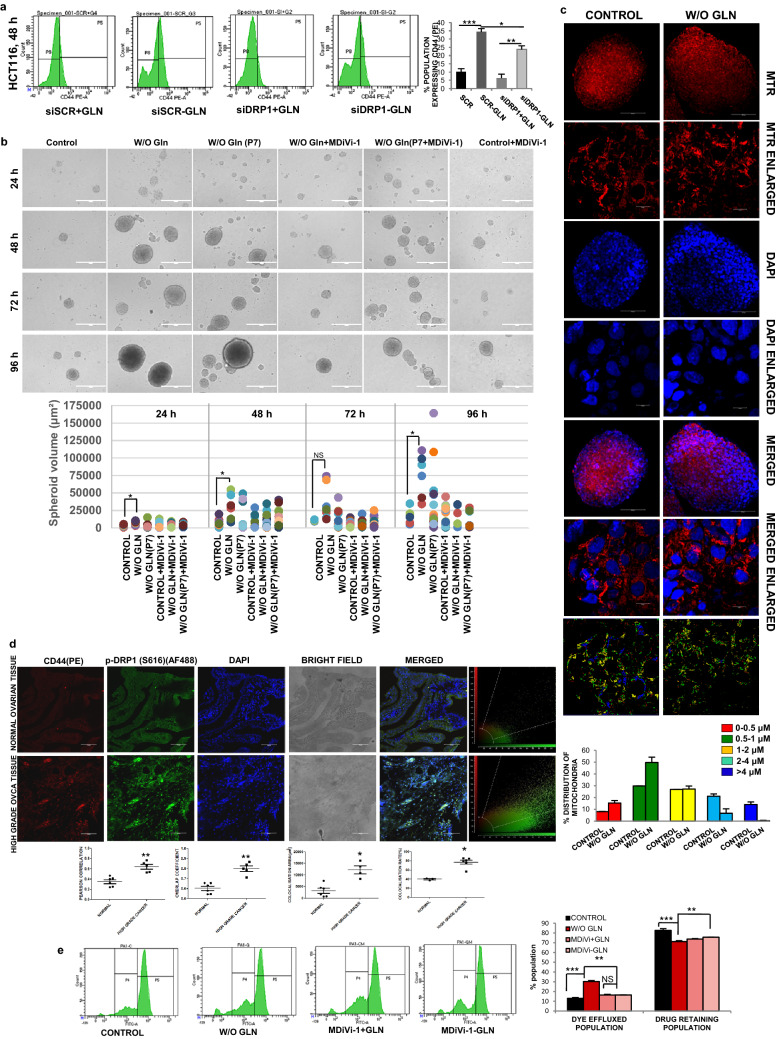
CSCs are a small population of cells within tumors with self-renewal capacity. They are quiescent in nature having reduced cell proliferation and prevalence of perinuclear localization of fragmented mitochondria [24]. We observed the perinuclear localization of fragmented mitochondria in glutamine limiting state (Supplementary Fig. S3A). The pluripotent stem cell markers like SOX2, OCT4, NANOG are highly expressed in CSCs along with the cell surface antigens, like CD44, CD117, CD45, CD31 [25, 26]. As expected, the expression of pluripotent cell markers was found to be upregulated both at the mRNA (Fig. 3a) and protein level (Fig. 3b, Supplementary Fig. S3B) in glutamine-deprived condition. Moreover, when glutamine was added to the medium at 24 h of glutamine starvation, the expression of SOX2 (as a representative marker for stemness) was reduced to the normal level after 24 h of glutamine supplementation (Fig. 3c). These results suggest that glutamine deprivation is responsible for promoting stem-like traits in cancer cells. When the status of different cell surface markers was observed at different hours of glutamine starvation, CD44 and CD117 were increased at 24 h in PA1 (Supplementary Fig. S3C), whereas the expression of CD44 and CD45 was elevated in PA1 and HCT116 at 48 h of glutamine deprivation (Fig. 3d-e). We did not find any significant change in the level of CD31 up to 48 h and similarly, CD45 remains unaltered at 24 h in PA1 cells (Supplementary Fig. S3C-D). However, CD117 was enhanced at 48 h in PA1 (Supplementary Fig. S3D). It was previously reported that deregulated alternative splicing of CD44 enhances CSC traits in tumor cells [27]. We observed increased gene expression of CD44 standard (CD44s) isoform, whereas its epithelial variant (CD44e) was downregulated due to glutamine starvation in PA1 cells (Fig. 3f). To test the ultimate effect of glutamine deprivation, we made continuous culture of cells in glutamine-depleted media up to four passages. The cells were found to be arrested at the G0/G1 phase of the cell cycle irrespective of their passage number as evident from the flow cytometric analysis (Fig. 3g). Interestingly, the enhanced ALDH activity, a signature of CSCs, is also found in glutamine-starved PA1 cells (Fig. 3h).
Fig. 3.
Glutamine deprivation promotes stemness. a The markers of stemness like OCT4, SOX2, NANOG, and ABCG2 mRNA expression was increased as shown by quantitative RT-PCR at 24 h glutamine-starved condition in PA1 (n = 3). (CON indicates the control cell). b Protein expression of different stem markers (SOX2 and OCT4) was increased upon 24 h of glutamine deprivation in both PA1 and HCT116 cells. c Re-supplementation of glutamine after 24 h, in the glutamine-starved cell reduced the SOX2 protein level in PA1 and fold change is depicted in the bar diagram. Flow cytometry data reveals that expression of CD44 and CD45 increased in absence of glutamine at 48 h (n = 3) in PA1 (d) and HCT116 (e) and % population of expressing cells represented in bar diagram. f Gene expression of CD44 variants (CD44epi) was decreased, whereas, CD44s expression was enhanced upon 24 h of glutamine deprivation in PA1 cells (n = 3). g Continuous culture of cells without glutamine at passage 3 (P-3) and passage 4 (P-4) showed an increased population in G1 stage with a reduced population in S phase in PA1 (n = 3) as shown by flowcytometry. These data are represented in both the histogram and bar diagram. h ALDH activity was increased in PA1 cells grew in glutamine-free medium for 24 h. i Drug efflux capacity was increased in glutamine-starved PA1 cells at 24 h. Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post-hoc analysis) (*p < 0.05, **p < 0.01, ***p < 0.001). NS non-significant
Chemoresistance in CSCs is marked by the upregulation of efflux transporters of the ABC family, like ABCG2,which export drugs out of the cells [1]. Gene and protein expression analysis revealed that ABCG2 was elevated after glutamine deficiency (Fig. 3a, Supplementary Fig. S3B). Rhodamine-123 (Rh123) efflux assay was performed to study the efflux property of glutamine-deprived cells, which export out Rh123 more efficiently compared to the cells grown in glutamine-supplemented condition. There was also a significant reduction of cells that retain Rh123 in glutamine limiting condition as shown by flow cytometry (Fig. 3i, Supplementary Fig. S3E). Therefore, these results indicate that glutamine deprivation might promote stemness and chemoresistance in cancer cells.
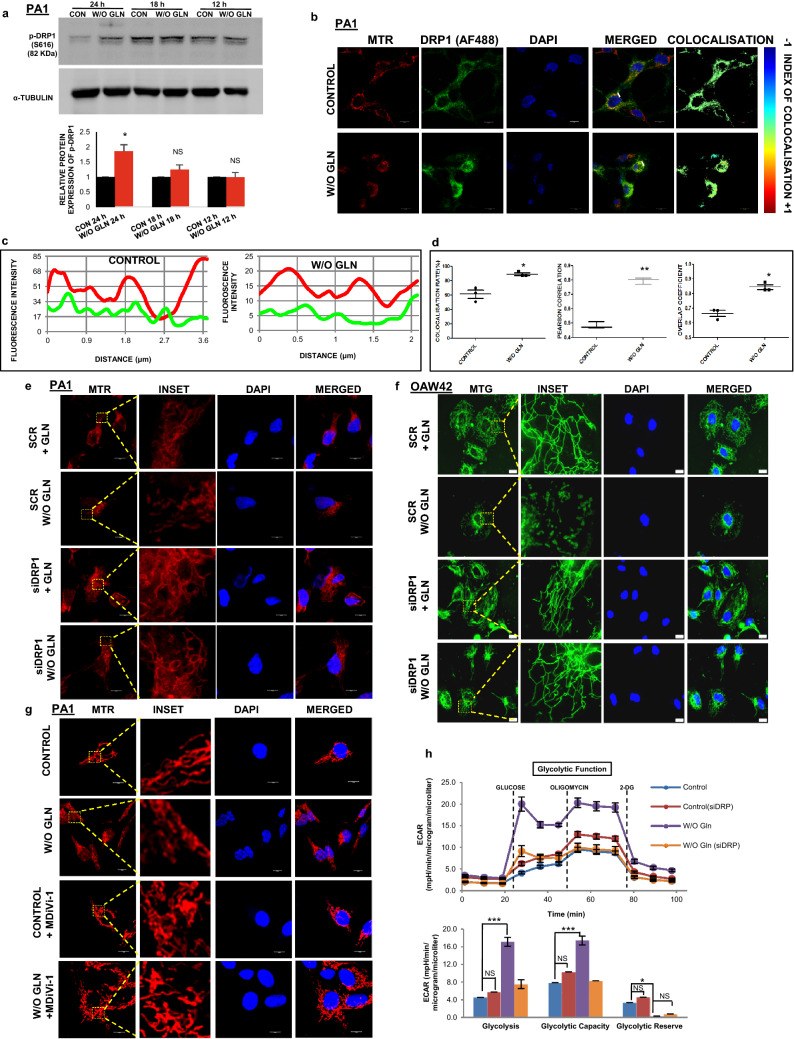
Mitochondrial fragmentation is induced through enhanced DRP1 phosphorylation in glutamine-restricted condition
DRP1 is a cytosolic protein that shuttles between cytosol and mitochondrial outer membrane to promote mitochondrial fission. After phosphorylation of DRP1 at S616, it can bind to the mitochondrial outer membrane and constricts the membrane using its GTPase activity [28]. We observed that S616 phosphorylation was elevated upon glutamine deprivation for 24 h in PA1 and HCT116, although there was no significant change of p-DRP1 till 18 h of starvation (Fig. 4a, Supplementary Fig. S4A). We did not observe any change of expression in the mitochondrial fusion protein such as Mitofusin-1 (MFN1) and Mitofusin-2 (MFN2) (Supplementary Fig. S4B). Glutamine deprivation showed mitochondria-specific localization of DRP1 in PA1 cells (Fig. 4b). The colocalization was established qualitatively using line-scan analysis and intensity correlation analysis (ICA). The line scan graph analysis suggested that the mitochondrial localization of DRP1 was very specific in glutamine-restricted condition (Fig. 4c). The ICA statistical data showed that the colocalization rate, Pearson's correlation, and overlap coefficient were enhanced in glutamine deficiency (Fig. 4d). Further, the silencing of DRP1 using siRNA inhibited mitochondrial fission in glutamine depleted PA1 and OAW42 cells (Fig. 4e, f). Similar results were observed when DRP1 inhibitor MDiVi-1 was used (Fig. 4g). In contrast, extracellular flux analysis indicates that ECAR, total glycolysis, glycolytic capacity as well as the glycolytic reserve were decreased after treating the cells with MDiVi-1 and siDRP1 individually in glutamine-deprived condition (Fig. 4h, Supplementary Fig. S4C-E). These results thus point DRP1 as the potential component for mitochondrial fragmentation in glutamine-restricted condition.
Fig. 4.
DRP1 phosphorylation and localization promote mitochondrial fragmentation. a The cells were grown for 12 h, 18 h, and 24 h in glutamine-free medium and cellular protein was isolated followed by SDS-PAGE and western blotting. The phosphorylation of DRP1 (S616) was increased at 24 h (n = 3), as shown by bar diagram. b Microscopy-based colocalization analysis in PA1 revealed the significant enhancement of association between DRP1 and mitochondria in absence of glutamine at 24 h (n = 3). c Line scan analysis of colocalization microscopy depicted significant overlapping of mitochondrial (RED) and DRP1 (GREEN) increased in absence of glutamine. d Intensity correlation analysis of colocalization through fluorescence microscopy of mitochondria and DRP1 revealed that colocalization rate, Pearson's correlation, and overlap coefficient are enhanced upon glutamine deprivation. Knockdown of DRP1 protected the mitochondria from fragmentation in PA1 (e) and OAW42 (f) as depicted in confocal microscopy. g Similarly, inhibition of DRP1 with MDiVi-1 significantly inhibits mitochondrial fragmentation upon 24 h of glutamine starvation in PA1 as observed by confocal microscopy. h Extracellular Flux analysis revealed that DRP1 knockdown was capable of restoring the glycolysis, glycolytic capacity similar to the control cells of PA1 upon 24 h of glutamine starvation (n = 3). Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post hoc analysis) (*p < 0.05, **p < 0.01, ***p < 0.001). NS non-significant. Scale bars 10 μm (b, e, g), 20 μm (f)
DRP1 has a role in stimulating stem-like characteristics and chemoresistance
To test the impact of inhibition of mitochondrial fission, we used MDiVi-1 as an inhibitor of fission in glutamine-deprived condition. We observed a significant reduction of CD44 (a stem cell marker) after silencing and inhibiting DRP1 in HCT116 and PA1, respectively, in glutamine limiting condition (Fig. 5a, Supplementary Fig. S4F-G). Another remarkable trait of CSCs is to form spheroid in non-adherent condition. Glutamine-starved cells gave rise to larger spheroids when cultured in non-adherent condition for 4 days and its volume became the same as that of untreated cells, when treated with MDiVi-1 in absence of glutamine (Fig. 5b). We have also performed this assay with the cells continuously cultured for seven passages in absence of glutamine. They formed larger spheroids and could able to generate a greater number of spheroids than the other sets. It also gave rise to new smaller spheroids even after 4 days of the assay, which suggests that continuous culture in absence of glutamine could significantly increase the stemness potential. To check if the cells maintain the fragmented mitochondrial phenotype in spheroids, we have used mitotracker red (MTR) for confocal microscopy and observed the presence of fragmented mitochondria in the spheroid (Fig. 5c). In human high-grade ovarian cancer tissue sample, colocalization of CD44 and p-DRP1 (S616) was increased as compared to the normal ovarian tissue section (Fig. 5d). Taken together, we can conclude that glutamine deprivation can enhance stem-like characteristics through DRP1 signaling in cancer.
Fig. 5.
DRP1 inhibition reduced stem-like features. a Flow cytometric data showed that the reduced expression of CD44 in HCT116 cells upon DRP1 silencing when grown in glutamine-starved condition for 48 h. The % population of CD44 expressing cells are quantified and represented in the bar diagram (n = 2). b MDiVi-1 treatment reduced spheroid size in absence of glutamine in PA1 as shown in bright field microscopy suggesting DRP1 dependency in spheroid formation. The number and volume of spheroid at different time points are shown in the bar diagram (n = 3). c In the spheroid cultures of PA1, the cells retained their fragmented morphology of mitochondria as shown in confocal microscopy and quantified the percentage of different mitochondrial length (n = 2). d CD44 expression and colocalization with p-DRP1 was high in high-grade ovarian cancer compared to normal ovarian tissue in IHC. Intensity correlation analysis of this colocalization was shown in the bar diagram in the form of co-localization rate, colocalization area, Pearson's correlation co-efficient, overlap co-efficient (e) MDiVi-1 treatment also reduced drug efflux capacity of cells at 24 h of glutamine starvation in PA1 (n = 2). Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post hoc analysis) (*p < 0.05, **p < 0.01, ***p < 0.001). NS non-significant. Scale bars 400 μm (b), 100 μm (c, d), 10 μm (Enlarged images of c)
We then wanted to determine whether DRP1 mediated stem-like characteristics are associated with chemoresistance. Both MDiVi-1 and siDRP1 independently can markedly enhanced the Rh123 dye retaining population, which was decreased in glutamine-starved condition, suggesting a role of DRP1 in chemoresistance (Fig. 5e, Supplementary Fig. S4H-I). These results suggest that DRP1 promotes stem-like characteristics coupled with chemoresistance in cancer cells in glutamine limiting condition.
Glutamine-restricted cells augment ERK1/2 induced DRP-1 phosphorylation through ROS
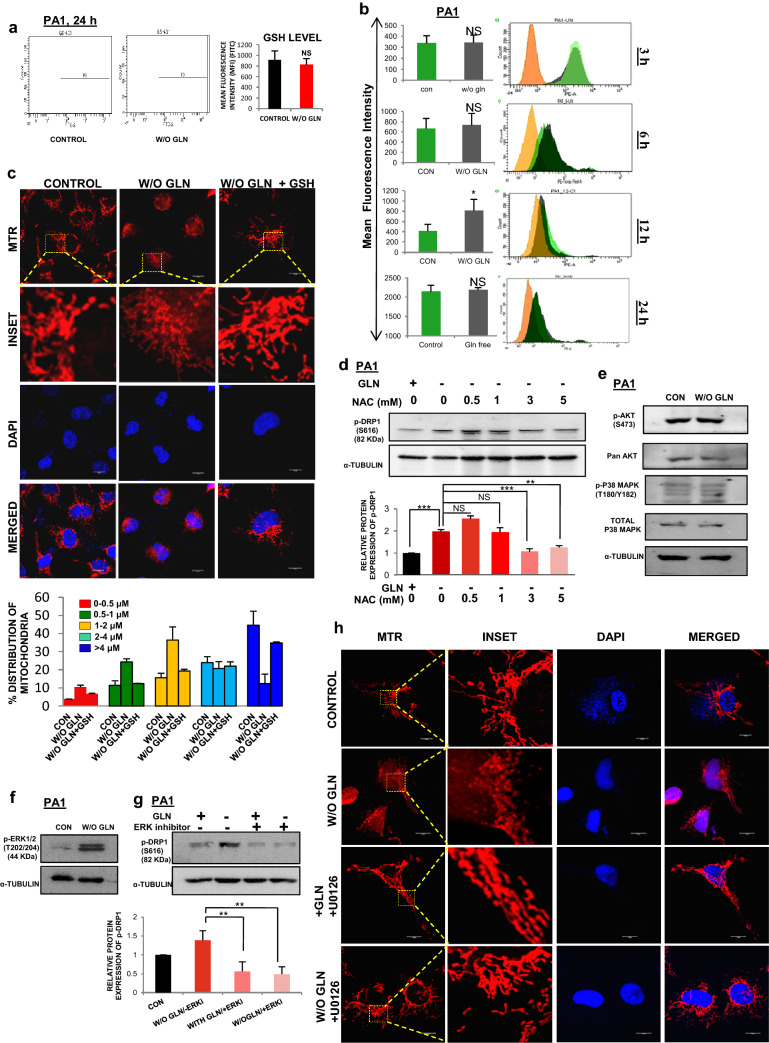
Glutamine also plays a role in synthesizing GSH via glutamate. GSH is a major endogenous antioxidant and linked with the ROS level [29]. As glutamine restriction was associated with reduced GSH synthesis, we focused on the status of mitochondrial ROS and tried to find a causal link between the ROS and DRP1 phosphorylation at S616. To validate this, we found that there was no significant difference in cellular GSH at 24 h of glutamine starvation (Fig. 6a). Next, we measured mitochondrial ROS generation at 3, 6, 12 and 24 h of glutamine deprivation (Fig. 6b) and found its significant elevation only at 12 h. By H2O2 treatment, we have shown that ROS independently induces mitochondrial fragmentation which is reduced by NAC treatment, an exogenous ROS scavenger (Supplementary Fig. S5A-B). In glutamine-deprived condition, another exogenous ROS scavenger, cell-permeable glutathione (GSH) reduces ROS-induced mitochondrial fragmentation (Fig. 6c). Similarly, NAC also reduces the phosphorylation of DRP1 at S616 (Fig. 6d) and reduced mitochondrial fragmentation (Supplementary Fig. S5C) in glutamine-depleted situation. Thus, glutamine depletion generates ROS, which might be responsible for the phosphorylation of DRP1 followed by mitochondrial fragmentation.
Fig. 6.
DRP1 phosphorylation occurs through ROS driven ERK1/2 signaling pathway (a) Total internal GSH was measured through flow cytometry at 24 h of glutamine starvation in PA1 and MFI was quantified and represented in the bar diagram (n = 3). b Mitochondrial ROS was measured upon glutamine deprivation at different time points (3 h,6 h,12 h, and 24 h) by flow cytometry in PA1. At 12 h only, there was a significant increment in the ROS level which decreased eventually (n = 3). The results were represented as histograms along with the bar diagram. c GSH treatment in absence of glutamine at 24 h reduced mitochondrial fragmentation in PA1 as depicted by confocal microscopy. d NAC treatment for 24 h in glutamine depleted media significantly reduced the DRP1 phosphorylation in PA1, as observed in western blot analysis (n = 3). Fold change was measured by densitometric analysis using ImageJ software and represented as bar diagram. e As shown by western blot analysis, there was no change in phosphorylation of AKT and p38 MAP kinase upon 24 h of glutamine starvation in PA1 cell. f There was a significant change in phosphorylated ERK1/2 at 24 h of glutamine starvation in PA1. g ERK inhibitor is sufficient to inhibit DRP1 phosphorylation at glutamine starvation in PA1 at 24 h. Densitometric analysis of this blot quantified the fold change of protein expression and represented in bar diagram. h Treatment of U0126 also inhibited mitochondrial fragmentation at 24 h of glutamine-starved condition in PA1 cell as shown in confocal microscopy. Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post hoc analysis) (*p < 0.05, **p < 0.01, ***p < 0.001). NS is for non-significant. Scale bars 10 μm (c, h)
To explore the pathways associated with mitochondrial fragmentation and DRP1 phosphorylation, we further checked the phosphorylation level of AKT (S473) and p38MAPK (T180/Y182) through Western blot analysis and found no alteration of their phospho- and total forms at 24 h of glutamine deprivation (Fig. 6e). The ERK1/2 phosphorylation on T202/204 was increased at 24 h of glutamine depletion (Fig. 6f). Moreover, DRP1 phosphorylation (S616) was found to be dependent on the ERK1/2 signaling pathway as ERK inhibitor prevented this phosphorylation (Fig. 6g). The number of longer mitochondria was significantly increased when U0126 (inhibitor of MEK, the upstream kinase of ERK) was added in the glutamine-depleted media (Fig. 6h). The above results convincingly show that DRP1 was phosphorylated through ROS-dependent activation of the ERK1/2 signaling pathway and promote mitochondrial fragmentation in tumor cells.
It is well known that xCT (antiporter of glutamate and cysteine) colocalizes with CD44 protein for maintaining the redox balance in the cell. Its expression also increased along with its enhanced colocalization with CD44 (Supplementary Fig. S5D-G). Since the ROS induced phosphorylation of DRP1, we checked the role of xCT in maintaining the ROS level in glutamine-deprived condition; however, no significant increase in the total cellular and mitochondrial ROS was observed upon inhibition of xCT by siRNA and its inhibitor sulfasalazine (Supplementary Fig. S5H-J). It suggested that xCT is not involved in the maintenance of ROS in glutamine-restricted condition. Perinuclear localization of mitochondria is known to increase nuclear ROS accumulation and activate various transcription factors for their downstream functioning. Therefore, we performed microscopy with DCFDA and MTR-stained cells and found that the perinuclear localization of mitochondria led to the accumulation of ROS in the nucleus in glutamine limiting condition (Supplementary Fig. S5K). Considering these results, we speculated that ROS accumulation within the nucleus is responsible for promoting stemness in glutamine-deprived condition.
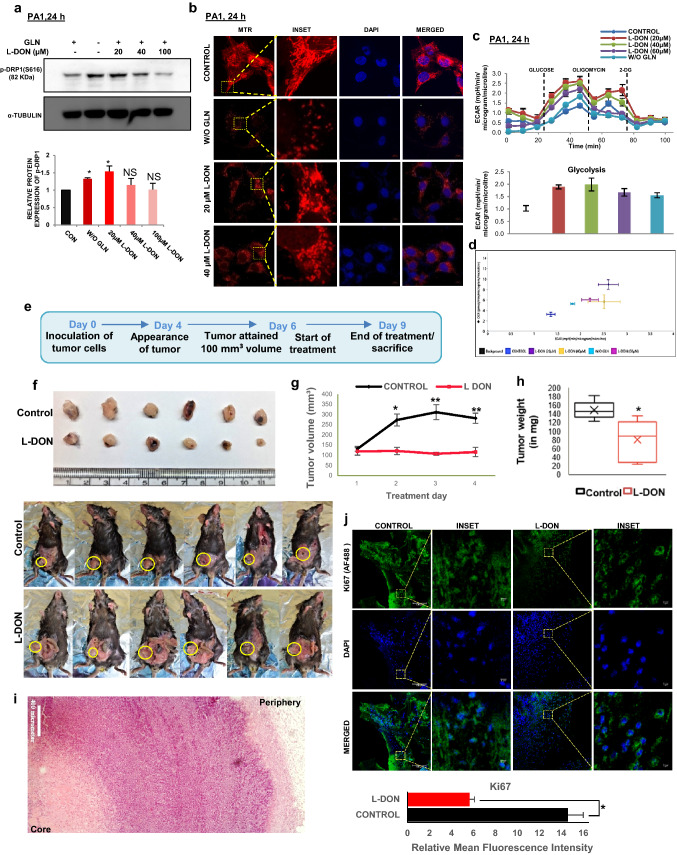
Combinatorial treatment of glutaminase inhibitor (L-DON) and MDiVi-1inhibit tumor growth and reduce CSCs population
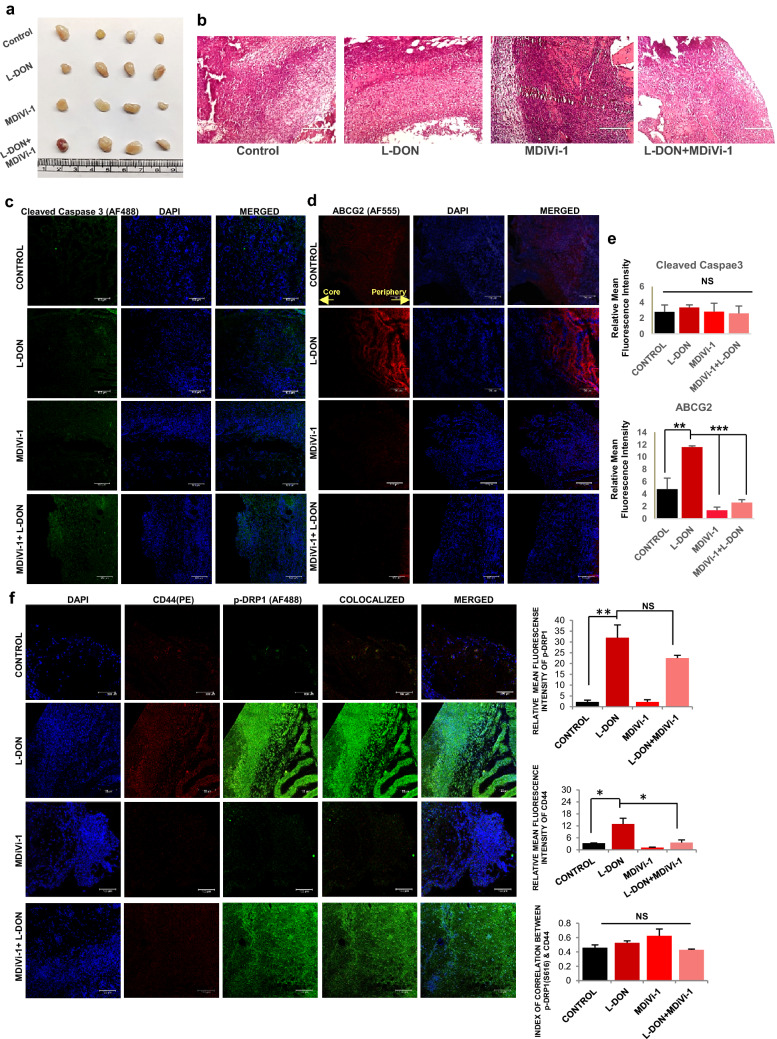
Glutamine metabolism is therapeutically targeted for killing aggressive cancer cells. GLS1 inhibition by L-DON can stop the usage of glutamine by the cancer cells and hence, it can mimic a situation of glutamine deprivation. We observed that L-DON treatment can significantly increase the level of p-DRP1 at S616 (Fig. 7a) and consequently, the number of fragmented mitochondria (Fig. 7b). Like glutamine-starved condition, L-DON treatment also increases ECAR (Fig. 7c, d, Supplementary Fig. S5L) and protein expression of SOX2 (Supplementary Fig. S5M). Further, to establish the results in in-vivo, we studied the parameters in syngeneic mouse model (C57BL/6) using ID8 (mouse ovarian epithelial cancer cell) cells. After the appearance of 100 mm3 tumor volume, mice were injected intravenously with L-DON consecutively for 3 days, whereas the animals of the control set were injected with equal volume of PBS (Fig. 7e). We found that L-DON treatment significantly inhibited tumor growth and weight gain as compared to the control set (Fig. 7f–h). Histological analysis with hematoxylin and eosin staining demarcated the tumor core area with lower number of cells, implying that glutamine starvation in the tumor core could be responsible for lowered cell number (Fig. 7i). To check if there is any necrotic cell population in the core of those tumors, we performed TUNEL assay and found no significantly increased TUNEL positive cells (Supplementary Fig. S6A). There was reduced Ki67 protein in the nucleus of L-DON treated cells, depicting that L-DON treatment ceased cell proliferation (Fig. 7j). To further validate the effect of MDiVi-1 in glutamine-starved condition, we injected it with or without L-DON in tumor-bearing mice (Fig. 8a, b). We found no significant increase in cell death as shown by Cleaved caspase-3 staining (Fig. 8c, e). L-DON treatment even in presence of MDiVi-1 significantly inhibited tumor growth (Supplementary Fig. S6B). There was increased expression of ABCG2, SOX2 and OCT4 after L-DON treatment that was significantly reduced by MDiVi-1 in in vivo model (Fig. 8d, e, Supplementary Fig. S6C-D). L-DON-treated tumors showed increased DRP1 phosphorylation (S616) and CD44 expression along with increased co-localization of these proteins, while dual treatment of L-DON and MDiVi-1 significantly reduced the CD44 expression (Fig. 8f). These results point towards a possibility that simultaneous pharmacological inhibition of GLS1 and mitochondrial fission might inhibit tumor growth with reduction in CSC population.
Fig. 7.
L-DON mimics glutamine starvation in tumor cells. a L-DON treatment increased DRP1 phosphorylation at the concentration of 20 µM in PA1 cell. Densitometric analysis was done using ImageJ software and the fold change is presented as bar diagram (n = 3). b L-DON treatment increased mitochondrial fragmentation as observed in confocal microscopy. c L-DON treatment for 24 h also increased the ECAR value along with glycolysis like glutamine starvation in PA1. d The energy phenotype graph also showed increased ECAR after L-DON treatment in PA1. e Timeline of development of tumor in mouse model and L-DON treatment. f Representative images of tumors formed in C57BL/6 injected with PBS or L-DON. g Tumor volume measured during 3 days of L-DON treatment course and represented graphically (n = 6) and h tumor weight was measured at the end of treatment course (Day-4) and represented in bar diagram. i Histological section stained with Hematoxylin and Eosin showed meagre amount of cell in the core of the tumor. j Fluorescence-immunohistochemistry depicted that amount of Ki67 in tumors was decreased in L-DON treated set (n = 5). In the control tissue, Ki67 localized in the nucleus whereas extranuclear localization of Ki67 was observed in L-DON treated set. Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post hoc analysis) (*p < 0.05, **p < 0.01). NS non-significant. Scale bars 10 μm (b), 400 μm (i), 100 μm (j) and 5 μm (INSET of j)
Fig. 8.
Pharmacological inhibitor of glutaminase (L-DON) and DRP1 (MDiVi-1) can together reduce CSCs population in in-vivo mouse model. a Representative images of tumors formed in C57BL/6 injected with PBS, L-DON, MDiVi-1 and MDiVi-1 + L-DON. b Hematoxylin and eosin stained histology sections of the tumor. c There is minimal expression of cleaved caspase 3 showing no change in control and L-DON treated tumors whereas (d) ABCG2 expression was increased in tumors treated with L-DON. as depicted in immunofluorescence microscopy images. MDiVi-1 treatment significantly reduced the ABCG2 expression. e Relative fluorescence intensity of cleaved caspase 3 (n = 3), Ki67 (n = 3) and ABCG2 (n = 5) were presented in bar diagram. f Both the expression and colocalization of CD44 with phosphorylated DRP1 (Ser616) was increased. Data are expressed in ± SEM, and statistical significance was calculated using two-tailed Student’s t test and one-way ANOVA (followed by Bonferroni post hoc analysis) (*p < 0.05, **p < 0.01, ***p < 0.001). NS is for non-significant. Scale bars 400 μm (b), 100 μm (c, d, f)
Discussion
Growing tumors require and consume plenty of glutamine for their survival and this addiction for glutamine is used to diagnose the growing tumors through 18F-FGln based PET scanning systems [30]. It is well established that the core of a solid tumor is glutamine-deprived and cancer cells survive in this nutrient limiting condition [16]. Further, this regional glutamine deficiency is supposed to be a major contributor of chemoresistance property that tumor cells acquire subsequently [31]. In this report, we showed that glutamine deficiency generates stem-like characteristics to induce chemoresistance property in the tumor cells by inducing mitochondrial fragmentation.
Glutamine dependency is acquired by tumor cells and is a crucial phenotype for aggressive cancer cell. Glutamine is required for their survival, proliferation, and metastasis [32]. In contrast, our present findings suggest that glutamine is crucial for the proliferation of cancer cells, but is not essential for their survival (Fig. 1a–c, Supplementary Fig. S1A). However, in absence of glutamine, the cell division potency of cancer cells did not cease, but significantly reduced as they still continued to proliferate even after several passages of culture in glutamine-free medium (Fig. 1b, 3g). This addiction towards glutamine is varied in different cancer cell types and their metabolic phenotype changes with their glutamine dependency level [32, 33]. In some cervical and breast cancer cells, basal oxygen consumption rate and glycolytic efficiency decreased in the absence of glutamine independently of their metabolic phenotype [12]. However, we identified that upon glutamine starvation, glucose uptake and glucose oxidation is enhanced to maintain both glycolysis and TCA cycle (Fig. 1e–m, Supplementary Fig. S1B-G, I-J). In tumor core, there is a possibility of both glucose and glutamine scarcity. In that condition, it is interesting to understand how glutamine-starved cells maintain high glycolysis rate. The possible explanation could be addressed by two different mechanisms. Firstly, in the periphery of the tumor, the glucose consumption is less than the core, rendering high availability and consumption of glucose by the glutamine-starved cells in the core [34]. Second, when the cells in the core perform higher glycolysis, they produce extracellular lactate, which can relay signal to the peripheral cells to reduce their glucose uptake [35]. Thus, it ensures that the core cells get adequate glucose in absence of glutamine. To further confirm the effect of lactate, we have treated the PA1 cells with it in presence of glutamine and observed reduced glucose uptake and glycolysis. Further, by blocking the lactate receptor with 3-OBA (3-hydroxy-butyrate) and using its transporter (MCT1) blocker the lactate signaling was found inhibited in glutamine supplemented cells (data not shown).
Mitochondrial fragmentation is an essential phenotype for various types of tumors and in melanoma cells, it is found to increase glycolytic rate [36]. We convincingly showed that glutamine deficiency facilitates mitochondrial fragmentation through phosphorylation of DRP1 at S616 (Fig. 4). DRP1 level is directly linked to the mitochondrial network (length) as silencing or inhibiting DRP1 reduces the mitochondrial fragmentation in absence of glutamine (Fig. 4e-g) and this DRP1 phosphorylation also linked with cell metabolism as glycolytic function remain same as control when DRP1 was silenced or inhibited (Fig. 4h, Supplementary Fig. S4C–E). We did not find any difference in mitochondrial protein level (Fig. 2a–d) and the level of mitochondrial OXPHOS (Supplementary Fig. S1B) and hence, we have not linked mitochondrial function with DRP1 level upon glutamine starvation. Treatment with MDiVi-1 also inhibited the retrograde transport of the mitochondria, i.e., the perinuclear localization of the mitochondria, indicating that the retrograde movement is fission dependent as reported previously [37].
Mitochondrial ROS act as an important second messenger to drive intracellular signaling pathways [38] and it is associated with mitochondrial fragmentation in malignant cells through ROS-RAS-RAF-ERK1/2-DRP1 signaling axis [39, 40]. It is reported that high glucose treatment in primary liver cells increased ROS production with concomitant enhancement in the mitochondrial fragmentation followed by perinuclear localization [41]. According to this report, ROS was increased at 15 min of incubation and decreased after 60 min, but the mitochondrial fragmentation persisted. Similarly, in the present study, we have also found a very dynamic and variable pattern of ROS level, which increases at 12 h of glutamine deprivation and surprisingly reduced again at 24 h, but the mitochondria remain fragmented (Fig. 6b, 2e). Treatment with cell-permeable anti-oxidant GSH and NAC in glutamine-deprived conditions can rescue the mitochondria from fragmentation (Fig. 6c, Supplementary Fig. S5C). Therefore, the rise in the mitochondrial ROS may be responsible for the acute activation of ERK1/2 and DRP1. Then perinuclear accumulation of fragmented mitochondria increases the local ROS accumulation in the nucleus (Supplementary Fig. S3A, S5K). This local ROS accumulation in the nucleus has been reported as a regulatory factor of gene expression through guanidine oxidation [11]. Moreover, we also found nuclear accumulation of NRF2, which is a well-established transcriptional regulator of ABCG2 in glutamine limiting condition (data not given). We suggested that DRP1-driven mitochondrial fragmentation and its perinuclear localization may promote stemness via nuclear accumulation of NRF2 in glutamine-deprived condition. Therefore, it is an interesting area to follow-up in detail and future studies are needed to determine the actual cause of ROS generation that leads to mitochondrial fragmentation in glutamine-starved cells.
In gynecological cancer, DRP1 and mitochondrial dynamics play a vital role in promoting chemoresistance [42, 43]. According to recent evidence, mitochondrial fragmentation and clustering around the nucleus is an important feature associated with stem cells [11, 28, 44]. Some reports have demonstrated that this "mitochondrial flirtation" with the nucleus induces the ROS level in the nucleus and this micro milieu leads to the activation of various transcription factors that can regulate gene associated with stemness [11]. ROS also can regulate stemness by suppressing Wnt/β-catenin signaling pathway in glutamine-dependent pancreatic ductal carcinoma, hepatocellular carcinoma, and non-small cell lung carcinoma [45, 46]. In contrast, glutamine deprivation inhibits Wnt signaling pathway and induces stemness in colorectal cancer. This effect can be reversed by the addition of α-ketoglutarate which may take over the function of glutamine in the downstream signaling [47]. In this report, we show that glutamine starvation enhances perinuclear localization of fragmented mitochondria, which leads to the promotion of stem-like features and chemoresistance in cancer cells (Fig. 3, Supplementary Fig. S3). The CD44-xCT colocalization has been reported to maintain the redox balance in cancer cells. xCT is an antiporter for glutamate and cysteine and is involved in the synthesis of glutathione and thereby preserves the redox homeostasis within the cell. Therefore, the interaction between CD44 and xCT is important for maintaining the ROS balance in CSCs [48]. Glutamine starvation also leads to the enhanced expression as well as colocalization of xCT with CD44 (Supplementary Fig. S5D-G). However, we did not find any significant increase in the cellular or mitochondrial ROS level after inhibiting xCT, which indicates that it is not involved in reducing the mitochondrial ROS after 12 h of glutamine starvation (Supplementary Fig. S5H-J). In high-grade ovarian cancer patient samples, we found enhanced colocalization of p-DRP1 (S616) and CD44, which again suggests that mitochondrial fragmentation is associated with stem-like feature in cancer (Fig. 5d).
Targeting glutaminolysis has become one of the most emerging therapeutic strategies to reduce tumor growth [49]. In contrast, a recent report suggested that blocking of glutamine metabolism affects the glycolysis and OXPHOS of tumor cells, but enhances the oxidative metabolism of effector T cells, which helps the tumor cells to overcome immune supervision [50]. Our observation suggests that glutaminase inhibitor L-DON is capable of mimicking glutamine-deprived conditions in the tumor as it enhances glycolysis and mitochondrial fragmentation (Fig. 7a–d, Supplementary Fig. S5L). L-DON treatment in mouse tumor model attenuated tumor growth without hampering the cell survival (Fig. 7f–h, 8c). It is interesting that, there was increased expression of ABCG2 in the core region than the periphery of the same tissue. This result indicated that glutamine starvation in the tumor core region may lead to increased expression of ABCG2. The MDiVi-1 treatment successfully reduced CD44 expression without affecting the p-DRP1 level. However, the co-treatment with L-DON and MDIVi-1 significantly reduced the expression of the stemness markers, like OCT4, SOX2, CD44 and ABCG2 (Supplementary Fig. S6C–D, Fig. 8d–f). Considering the importance of glutamine metabolism in cancer, it has been suggested that combinatorial therapy of glutaminase inhibitor along with the blocking of specific signaling pathway might have high anti-proliferative activity [51]. In addition, MDiVi-1-mediated blocking of mitochondrial fragmentation has also been used as therapeutics to combat cancer stem cells [52]. Altogether, this study shows a promising approach to inhibit tumor cell-specific growth and reduce CSCs population using a dual treatment of L-DON and MDiVi-1. Future studies including clinical investigation will strengthen the possibilities for the usage of a combination of such metabolic inhibitors as therapeutic agent.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thankfully acknowledge Mr. Sounak Bhattacharya (confocal microscopy), Mr. Tanmoy Dalui, & Mrs. Debalina Chakraborty (flow cytometry), Dr. E. Padmanaban (NMR spectroscopy), and Mr. T. Muruganandan (AFM) of Central Instrumentation Facility of IICB. Prof. Pijush K. Das (CSIR-IICB) and Dr. Partha Chakrabarti (CSIR-IICB) are gratefully acknowledged for their valuable suggestions in preparing the manuscript. We are also thankful to Prof. Susanta Roychoudhury and Dr. Damayanti Das Ghosh (both from Saroj Gupta Cancer Centre & Research Institute, Kolkata, India) for providing human patient tissue samples and also for their valuable suggestions. Technical assistance of Mr. Prabir Kumar Dey is acknowledged. Other members of SSR laboratory are acknowledged for their co-operation.
Author contributions
PP, SG and SSR conceptualized the study; PP and SG investigated, performed analysis and validation of the data and wrote the original draft. SSR provided the financial support, supervised the work, reviewed, and approved the final manuscript.
Funding
This work was supported by grants from Science and Engineering Research Board (SERB) project GAP-360 (EMR/2016/002578) and the Council of Scientific and Industrial Research (CSIR) in house projects.
Availability of data
Data are available on request from the corresponding author.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval for animal studies
All animal experiments were approved by the institutional animal ethics committee (IAEC), CSIR-Indian Institute of Chemical Biology, India, (Registration no. 147/GO/ReBi/S/99/CPSCEA) following the guidelines of Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Govt. of India.
Ethical approval for human studies
The human patients' tissue samples were collected from Saroj Gupta Cancer Centre and Research Institute (SGCCRI), Kolkata, India with proper human ethics clearance from the Institutional Ethics Committee (approval number -IEC SGCCRI REF NO.- 16/2/2018/Non-Reg/SSR/3).
Consent to participate
Informed written consent was taken from the patients in accordance with the 1964 Helsinki declaration.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Parash Prasad and Sampurna Ghosh contributed equally to this work.
References
- 1.Mitra T, Prasad P, Mukherjee P, et al. Stemness and chemoresistance are imparted to the OC cells through TGFβ1 driven EMT. J Cell Biochem. 2018;119:5775–5787. doi: 10.1002/jcb.26753. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Pavlova NN, Thompson CB. The Emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Souba WW. Glutamine and cancer. Ann Surg. 1993;218:715–728. doi: 10.1097/00000658-199312000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Achreja A, Yeung TL, et al. Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metab. 2016;24:685–700. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. Glutamine metabolism in cancer: understanding the heterogeneity. Trends in Cancer. 2017;3:169–180. doi: 10.1016/j.trecan.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scalise M, Pochini L, Galluccio M, et al. Glutamine transport and mitochondrial metabolism in cancer cell growth. Front Oncol. 2017;7:1–9. doi: 10.3389/fonc.2017.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Choi W, Chen Y, et al. A proposed role for glutamine in cancer cell growth through acid resistance. Cell Res. 2013;23:724–727. doi: 10.1038/cr.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Zhang J, Yu M, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Mehdi AB, Pastukh VM, Swiger BM, et al. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:1–10. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cacace A, Sboarina M, Vazeille T, Sonveaux P. Glutamine activates STAT3 to control cancer cell proliferation independently of glutamine metabolism. Oncogene. 2017;36:2074–2084. doi: 10.1038/onc.2016.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Moss T, Mangala LS, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:1–23. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan L, Sheng X, Willson AK, et al. Glutamine promotes ovarian cancer cell proliferation through the mTOR/S6 pathway. Endocr Relat Cancer. 2015;22:577–591. doi: 10.1530/ERC-15-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y, Park K. Targeting glutamine metabolism for cancer treatment. Biomol Ther. 2018;26:19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 17.Das N, Mandala A, Naaz S, et al. Melatonin protects against lipid-induced mitochondrial dysfunction in hepatocytes and inhibits stellate cell activation during hepatic fibrosis in mice. J Pineal Res. 2017;62:1–21. doi: 10.1111/jpi.12404. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury SR, Ray U, Chatterjee BP, Roy SS. Targeted apoptosis in ovarian cancer cells through mitochondrial dysfunction in response to Sambucus nigra agglutinin. Cell Death Dis. 2017;8:1–12. doi: 10.1038/cddis.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan M, Biswas D, Ghosh M, et al. mTORC2 controls cancer cell survival by modulating gluconeogenesis. Cell Death Discov. 2015;1:1–12. doi: 10.1038/cddiscovery.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maftah A, Petit JM, Ratinaud MH, Julien R. 10-N nonyl-acridine orange: a fluorescent probe which stains mitochondria independently of their energetic state. Biochem Biophys Res Commun. 1989;164:185–190. doi: 10.1016/0006-291X(89)91700-2. [DOI] [PubMed] [Google Scholar]
- 21.De R, Sarkar S, Mazumder S, et al. Macrophage migration inhibitory factor regulates mitochondrial dynamics and cell growth of human cancer cell lines through CD74-NF-κB signaling. J Biol Chem. 2018;293:19740–19760. doi: 10.1074/jbc.RA118.003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordi M, Nazio F, Campello S. The close interconnection between mitochondrial dynamics and mitophagy in cancer. Front Oncol. 2017;7:1–9. doi: 10.3389/fonc.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao B, Deng X, Zhou W, Tan EK. Flow cytometry-based assessment of mitophagy using mitotracker. Front Cell Neurosci. 2016;10:1–4. doi: 10.3389/fncel.2016.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song IS. Mitochondria as therapeutic targets for cancer stem cells. World J Stem Cells. 2015;7:418. doi: 10.4252/wjsc.v7.i2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parte SC, Batra SK, Kakar SS. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J Ovarian Res. 2018;11:1–16. doi: 10.1186/s13048-018-0439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akhter Z, Sharawat SK, Kumar V, et al. Aggressive serous epithelial ovarian cancer is potentially propagated by EpCAM + CD45 + phenotype. Oncogene. 2018 doi: 10.1038/s41388-017-0106-y. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya R, Mitra T, Ray Chaudhuri S, Roy SS. Mesenchymal splice isoform of CD44 (CD44s) promotes EMT/invasion and imparts stem-like properties to ovarian cancer cells. J Cell Biochem. 2018;119:3373–3383. doi: 10.1002/jcb.26504. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Chan DC. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metab. 2017;26:39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cetinbas NM, Sudderth J, Harris RC, et al. Glucose-dependent anaplerosis in cancer cells is required for cellular redox balance in the absence of glutamine. Sci Rep. 2016;6:1–12. doi: 10.1038/srep32606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venneti S, Dunphy MP, Zhang H, et al. Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci Transl Med. 2015;7:1–10. doi: 10.1126/scitranslmed.aaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan M, Reid MA, Lowman XH, et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol. 2016;18:1090–1101. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zacharias NM, McCullough C, Shanmugavelandy S, et al. Metabolic differences in glutamine utilization lead to metabolic vulnerabilities in prostate cancer. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-16327-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu L, Ploessl K, Zhou R, et al. Metabolic imaging of glutamine in cancer. J Nucl Med. 2017;58:533–537. doi: 10.2967/jnumed.116.182345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonveaux P, Végran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trotta AP, Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci. 2017;74:1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campello S, Scorrano L. Mitochondrial shape changes: Orchestrating cell pathophysiology. EMBO Rep. 2010;11:678–684. doi: 10.1038/embor.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao RZ, Jiang S, Zhang L, Bin YuZ. Mitochondrial electron transport chain, ROS generation and uncoupling (review) Int J Mol Med. 2019;44:3–15. doi: 10.3892/ijmm.2019.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashatus JA, Nascimento A, Myers LJ, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afanas’ev I Reactive oxygen species signaling in cancer: comparison with aging. Aging Dis. 2011;2:219–230. [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong B, Tsuyoshi H, Orisaka M, et al. Mitochondrial dynamics regulating chemoresistance in gynecological cancers. Ann N Y Acad Sci. 2015;1350:1–16. doi: 10.1111/nyas.12883. [DOI] [PubMed] [Google Scholar]
- 43.Kingnate C, Charoenkwan K, Kumfu S, et al. Possible roles of mitochondrial dynamics and the effects of pharmacological interventions in chemoresistant ovarian cancer. EBioMedicine. 2018;34:256–266. doi: 10.1016/j.ebiom.2018.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao J, Liu PP, Hou G, et al. Regulation of stem-like cancer cells by glutamine through β-catenin pathway mediated by redox signaling. Mol Cancer. 2017;16:1–13. doi: 10.1186/s12943-017-0623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Cao Y, Meng G, et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine. 2019;39:239–254. doi: 10.1016/j.ebiom.2018.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tran TQ, Hanse EA, Habowski AN, et al. α-Ketoglutarate attenuates Wnt signaling and drives differentiation in colorectal cancer. Nat Cancer. 2020;1:345–358. doi: 10.1038/s43018-020-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ju HQ, Lu YX, Chen DL, et al. Redox regulation of stem-like cells though the CD44v-xCT axis in colorectal cancer: mechanisms and therapeutic implications. Theranostics. 2016;6:1160–1175. doi: 10.7150/thno.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin L, Alesi GN, Kang S. Glutaminolysis as a target for cancer therapy. Oncogene. 2016;35:3619–3625. doi: 10.1038/onc.2015.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leone RD, Zhao L, Englert JM, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matés JM, Di Paola FJ, Campos-Sandoval JA, et al. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin Cell Dev Biol. 2020;98:34–43. doi: 10.1016/j.semcdb.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 52.Peiris-Pagès M, Bonuccelli G, Sotgia F, Lisanti MP. Mitochondrial fission as a driver of stemness in tumor cells: mDIVI1 inhibits mitochondrial function, cell migration and cancer stem cell (CSC) signalling. Oncotarget. 2018;9(17):13254–13275. doi: 10.18632/oncotarget.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request from the corresponding author.