Abstract
Parthanatos is a form of regulated cell death involved in the pathogenesis of many diseases, particularly neurodegenerative disorders, such as Parkinson's disease, Alzheimer's disease, Huntington’s disease, and amyotrophic lateral sclerosis. Parthanatos is a multistep cell death pathway cascade that involves poly (ADP-ribose) polymerase 1 (PARP-1) overactivation, PAR accumulation, PAR binding to apoptosis-inducing factor (AIF), AIF release from the mitochondria, nuclear translocation of the AIF/macrophage migration inhibitory factor (MIF) complex, and MIF-mediated large-scale DNA fragmentation. All the key players in the parthanatos pathway are pleiotropic proteins with diverse functions. An in-depth understanding of the structure-based activity of the key factors, and the biochemical mechanisms of parthanatos, is crucial for the development of drugs and therapeutic strategies. In this review, we delve into the key players of the parthanatos pathway and reveal the multiple levels of therapeutic opportunities for treating parthanatos-based pathogenesis.
Keywords: Cell death, PARP1, AIF, MIF, Neurodegenerative diseases, Therapeutic target
Introduction
Parthanatos is a form of regulated cell death (RCD) that is initiated by poly (ADP-ribose) polymerase 1 (PARP-1) overactivation, is mediated by apoptosis-inducing factor (AIF), and involves macrophage migration inhibitory factor (MIF)-induced DNA degradation [1]. It was discovered two decades ago [2–4] and has been officially recognized by the Nomenclature Committee on Cell Death (NCCD) [5, 6]. Parthanatos is believed to be involved in the pathogenesis of many disorders, including ischemia reperfusion injury following stroke and myocardial infarction, reactive oxygen species (ROS) -induced injury, glutamate excitotoxicity, and neurodegenerative diseases, such as Parkinson's disease (PD) and Alzheimer's disease (AD) [7–10]. In this review, we elaborate on the pathway of parthanatos by focusing on the key players in the cascade and discuss potential therapeutic targets for parthanatos-related diseases.
Overview of parthanatos
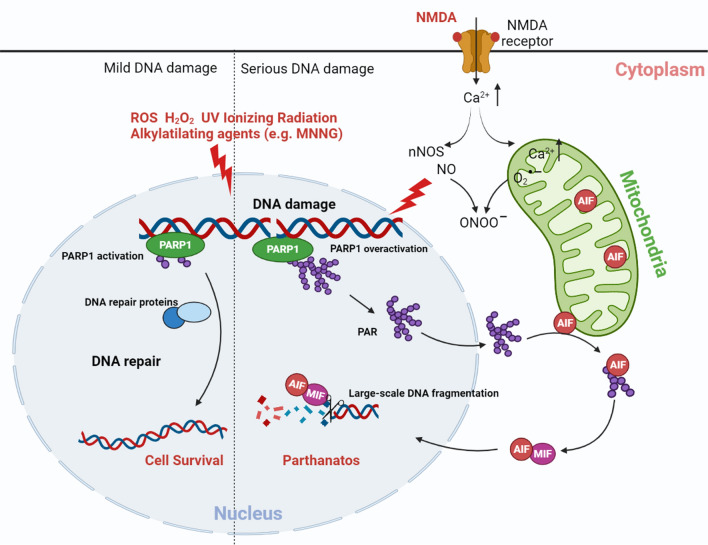
The term “parthanatos” is derived from “PAR” and the Greek word “Thanatos” [11], where “PAR” is “poly (ADP-ribose) (PAR) polymer” and “Thanatos” is the personification of death in Greek mythology. Parthanatos is cell death resulting from the accumulation of PAR polymers [12] and is characterized by a unique pathway, distinct from apoptosis, necroptosis, and any other types of cell death [13]. Oxidative stress, ionizing radiation, or the DNA alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) can cause extensive DNA damage [2–4, 14, 15]. The excitatory activation of N-methyl-D-aspartate (NMDA) receptors can induce calcium influx, neuronal nitric oxide synthases (nNOS) activation, nitric oxide (NO) production, and reactive-oxygen species production. The reaction between NO and superoxide anion (O2•−) produces the potent oxidant peroxynitrite (ONOO−), which leads to extensive DNA damage [2, 14]. This extensive DNA damage induces PARP-1 overactivation and leads to the synthesis of long-chained and branched PAR polymers [2–4]. PAR is subsequently released from the nucleus into the cytosol and mitochondria, where it binds to AIF, resulting in AIF translocation from the mitochondria to cytosol [3, 4]. In the cytosol, AIF in the cytosol binds to and recruits MIF to the nucleus [10]. MIF is a DNA nuclease that can cleave DNA and cause large-scale DNA fragmentation and chromatin condensation [10] (Fig. 1). By eliminating PAR polymers, PAR glycohydrolase (PARG) or ADP-ribosyl-acceptor hydrolase 3 (ARH3) prevents AIF translocation from the mitochondria into the nucleus and, in turn, inhibits cell death [4, 16]. Unlike in apoptosis, parthanatos does not cause the formation of apoptotic bodies, but the results in a loss of membrane integrity that is distinct from cell swelling observed during cell necroptosis [6, 7, 17]. In addition, parthanatos does not depend on the caspase cascade and, thus, pan-caspase inhibitors do not inhibit this cell death process [2, 15]. Briefly, parthanatos is a PARP-1-dependent, caspase-independent, AIF- and MIF-mediated cell death cascade.
Fig. 1.
Schematic model of parthanatos. DNA damage caused by ROS, hydrogen peroxide, UV, ionizing radiation, MNNG, and NMDA. Excitatory activation of NMDA receptors can induce calcium influx, nNOS activation, NO production, and ROS production. The reaction between NO and superoxide anion (O2•−) produces the potent oxidant preoxynitrite (ONOO−), which leads to DNA damage. Other agents such as ROS, hydrogen peroxide, ionizing radiation, and MNNG cause DNA damage, which activates PARP-1. When DNA damage is mild, PARP-1 activates and recruits DNA damage repair proteins to repair the damaged DNA. Widespread DNA damage results in PARP-1 overactivation, which catalyzes PAR polymer formation. PAR polymers bind to AIF and mediate AIF release from the mitochondria. AIF interacts with MIF in the cytoplasm to form the AIF/MIF complex, and nuclear translocation of the AIF/MIF complex. MIF cleaves genomic DNA into large-scale fragments via its nuclease activity, leading to parthanatos. ROS reactive oxygen species, MNNG N-methyl-N'-nitro-N-nitrosoguanidine, NMDA N-methyl-D-aspartate, nNOS neuronal nitric oxide synthases,NO nitric oxide, PARP-1 poly(ADP-ribose) polymerase 1, PAR poly(ADP-ribose), AIF apoptosis-inducing factor, MIF migration inhibitory factor
Although the role of the core pathway involving the PARP1–AIF–MIF axis has been convincingly demonstrated, an AIF/MIF-independent death modality that also requires PARP1 overactivation has drawn attention [18–21]. For instance, AIF proved dispensable for PARP1-dependent programmed necrotic cell death in monocyte-to-macrophage differentiation [19]. The cell death of retinal pigment epithelium exhibits necrotic features, involving PARP1 activation, but lacking a role for AIF [18]. Thus, how cells die in a PARP1-dependent manner if AIF/MIF is not involved has arisen as an intriguing question. The necrotic nature (for instance, increased membrane permeabilization and lack of caspase activity) of PARP1-mediated cell death was recognized in relatively early studies of the historical development of parthanatos [22–24]. In these studies, reductions in ATP and bioenergetic collapse were observed. The nicotinamide adenine dinucleotide (NAD +) content is replenished by a nicotinic acid mononucleotide adenylyl transferase-1 enzyme that synthesizes NAD + from nicotinamide mononucleotide (NMN) and ATP [25, 26]. Thus, it is thought that the consumption of NAD + exhausts ATP, resulting in the loss of ATP; however, studies demonstrated that bioenergetic collapse is not dependent on NAD + depletion and, instead, PARP1 activation initiates a glycolytic defect due to the inhibition of hexokinase activity by PAR polymer binding [27, 28]. It seems that cellular energetic collapse plays a pivotal role in AIF-independent parthanatos, even if it does not seem to be crucial in the classical parthanatos model, which is supported by an earlier observation that pyruvate and α-ketoglutarate supplementation rescues cells from PARP1-mediated cell death [20].
Whether this bioenergetic collapse-driven and AIF/MIF-independent cell death is a new modality of cell death rather than a submodality of parthanatos is still unconfirmed. The controversy suggests the more attention is required in elucidating the molecular mechanisms of parthanatos. It even raises the possibility that energetic collapse is the core driving force for cells undergoing PARP1 activation-mediated cell death because, per the magnitude of PARP1 activation, AIF/MIF nuclear translocation may be a consequence of the extreme activation of PARP1.
Structure and activation of PARP-1
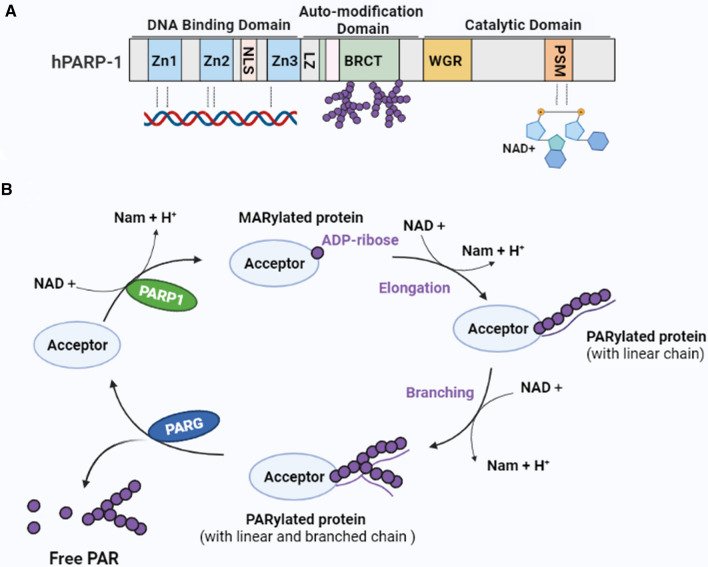
PARP-1 is the most abundant and extensively studied member of the PARP superfamily, comprising 17 related mammalian proteins [29]. PARP-1 functions as a DNA damage sensor to regulate cellular homeostasis and genomic stability. Mammalian PARP-1 is a 116 kDa protein comprising three functional domains [29] (Fig. 2A). The N-terminal DNA-binding domain contains three zinc finger motifs (Zn1, Zn2, and Zn3) and a nuclear localization sequence (NLS). This domain is responsible for identifying and binding damaged DNA. The middle segment is a central, auto-modification domain that contains a leucine zipper (LZ) motif and a BRCA1 C-terminal (BRCT) phosphopeptide-binding motif for protein–protein interaction [29]. In addition, this region contains several glutamate, aspartate, and lysine residues as putative acceptors for self-ADP-ribosylation [29]. The C-terminal catalytic domain contains a tryptophan-, glycine-, and arginine-rich (WGR) domain, and a “PARP signature” sequence [29], which catalyzes PAR synthesis using NAD + as a donor of ADP ribose [30, 31].
Fig. 2.
Structure of PARP-1 and the production and degradation of PAR. A Structure of PARP-1. PARP-1 includes three structural domains: (1) An N-terminal DNA-binding domain, which contains three zinc-binding domains (Zn1, Zn2, and Zn3) and an NLS. The zinc-binding domains are responsible for PARP-1 detection of damaged DNA. (2) A central automodification domain, which contains a leucine zipper (LZ) motif and a BRCA1 C-terminal (BRCT) phosphopeptide-binding motif for protein–protein interaction. (3) A C-terminal catalytic domain that contains a WGR domain and the “PARP signature” sequence (PSM), which catalyzes PAR synthesis using nicotinamide adenine dinucleotide (NAD +) as a donor of ADP-ribose. B Production and degradation of PAR. PARP-1 uses NAD + as a substrate to generate mono(ADP-ribosyl) and nicotinamide (Nam). Mono(ADP-nucleotide) is covalently attached to the target protein, resulting in MARylated protein. Further elongation and branching of the polymer forms PARylated protein. PARG catalyzes the hydrolysis of PAR to free poly(ADP-ribose) or ADP-ribose units through its endoglycosidic or exoglycosidic activity. PARP-1 poly(ADP-ribose) polymerase 1, PAR poly(ADP-ribose), PARG PAR glycohydrolase
In the presence of mild DNA damage, PARP-1 uses its N-terminal DNA-binding domain to identify and bind to the damaged DNA sites, leading to the activation of its catalytic activity. Negatively charged PAR polymers are generated, which covalently attach to PARP-1 itself as well as other nuclear proteins such as histones, transcription factors, DNA helicases, and many other proteins [32]. PARP-1, through the BRCT motif, can recruit DNA repair proteins to DNA damage sites to facilitate DNA repair (Fig. 1) [33]. When DNA damage is severe, PARP-1 overactivation leads to the excessive accumulation of PAR polymers. PAR subsequently translocates from the nucleus to the cytosol and mitochondria, where it stimulates AIF translocation from the mitochondria to the nucleus, leading to cell death [3, 4] (Fig. 1). The inhibition of PARP-1 or the deletion of parp-1 has a significant protective effect in models of many cell injury paradigms, including stroke, diabetes, ischemia–reperfusion injury, and neurodegenerative disease, indicating that PARP-1 overactivation is the first step in the parthanatic cascade [8, 34]. In conclusion, as a cell fate determinant, PARP-1 plays an important role in maintaining genomic stability and protects the host by promoting DNA repair or triggering the cell death pathway [29, 35].
In addition, alternative mechanisms for PARP1 activation in the absence of DNA damage have been identified in various cell types and mouse models [36–41]. They include PARP1 activation by various signal transduction mechanisms, inducing intracellular Ca2+ release, the activation of phosphorylation cascades, and direct protein–protein association [36, 37, 40]. For example, the accumulation of the Parkin substrate aminoacyl-tRNA synthetase complex-interacting multifunctional protein-2 (AIMP2) in vitro and in vivo directly leads to the excessive activation of PARP1 and toxicity in dopaminergic cells [36]. DNA damage-independent PAR formation may fit into the context of chronic diseases, where it takes a long time for the cells to die, and the dysfunction of energy metabolism regulation may be the cue for mediating PARP1-triggered signaling.
Synthesis and hydrolysis of PAR Polymers
Poly(ADP-ribosyl)-ation (PARylation) is a reversible post-translocation modification [42, 43]. PARP-1 catalyzes the transfer of the ADP-ribose (ADPr) unit from NAD + onto the target protein to form a long and branched chain of negatively charged PAR polymers (Fig. 2B) [43]. The first binding of an ADPr moiety to the protein occurs covalently to an aspartate (D), glutamate (E), lysine (K), arginine (R), and serine (S) via an ester bond [42, 44] and leads to the formation of mono-ADP-ribosylated (MARylated) proteins. The next unit of ADPr binds to the previous unit via 2′,1′′-O-glycosidic bonds to form a long PAR chain, forming a branched structure via 2′′,1′′′-O-glycosidic bonds [44], which results in the PARylated protein. Proteins can also noncovalently bind PAR through distinct PAR-binding motifs that include the so-called PAR-binding motifs (PBMs), PAR-binding zinc finger (PBZF) domains, and WWE domains [45]. The negatively charged PAR polymer influences the biochemical properties of the modified or interacting proteins, modulating their structure, function, and localization [29]. Eventually, PARylation affects a number of the cellular biological events involved in chromatin dynamics, genome stability maintenance, transcription, cell metabolism, development, and cell death [29, 42, 44, 46]. Under normal conditions, the level of intracellular PARylation is low, but when PARP-1 is activated, it can increase PARylation levels ranging from 10- to 500-fold [29]. The increase in PARylation depends mainly on the type and degree of DNA damage [47], and is closely linked to inflammation, DNA damage repair, cell senescence, and cell death [29, 42]. Therefore, PARP-1 acts as a “genome guardian” and participates in different cellular biological events by regulating intracellular PAR levels. When cells are exposed to inflammatory stimuli, e.g., TNFα, LPS, etc., they produce low levels of PARylation in response [46, 48, 49]. When DNA damage is mild, PARP-1 activation produces moderate PARylation to participate in the DNA damage repair process [50]. By contrast, severe DNA damage invokes high levels of PARylation to promote cell death [13].
PAR polymers, products of PARP-1 overactivation, are an upstream signal of the parthanatic cascade and directly toxic to neurons [4]. Accompanying the formation of PAR is the reduction in cellular NAD + , and the restoration of NAD + levels requires four adenosine triphosphate (ATP) molecules for every NAD + molecule [27]. Early studies indicated that cell death was caused by the excessive activation of PARP-1, which induced energy collapse via a decrement in NAD + and ATP [24]. However, many studies have since shown that parthanatos is not associated with a loss of cellular energy storage [51, 52] but, rather, depends on PAR signaling [3, 4, 34]. Yu et al. found that either the purified PAR polymer or the PARP-1-activated nuclear supernatant induced AIF release from isolated mitochondria and caused the translocation of AIF from the mitochondria to the nucleus in intact cells [3]. PAR polymers produced primarily in the nucleus can be translocated to the cytoplasm and mitochondria, where they may play a role in mitochondrial AIF release [3, 53]. In addition, the PAR polymer induces cell death in a concentration-dependent way, suggesting AIF release may be a consequence of high levels of PAR accumulation in the cytoplasm. Neutralizing antibodies to PAR abolish its toxicity in either the NMDA excitotoxicity of cortical neurons or MNNG cell toxicity [3]. Therefore, the PAR polymer is aptly described as the “death signal” [4].
The rapid hydrolysis of PAR polymers is, therefore, a similarly essential event in parthanatos, and this hydrolysis is catalyzed by PAR-degrading enzymes, namely, PARG [54, 55]. PARG catalyzes the hydrolysis of PAR to free poly(ADP-ribose) or ADP-ribose units through its endoglycosidic or exoglycosidic activity (Fig. 2B). Thus, PARG inhibits parthanatos through PAR polymer degradation. Previous studies have found that overexpressing PARG significantly protects against focal ischemia, excitotoxicity, and stroke, while reduced levels of PARG result in significantly increased infarct volumes following focal ischemia, and increased sensitivity to NMDA excitotoxicity [4, 56]. In addition, the complete absence of PARG after UV irradiation leads to the activation of AIF-mediated cell death [51]. ARH3, which exhibits PARG activity, can also cleave the ribose–ribose bonds within PAR [57], which protects the cell against hydrogen peroxide (H2O2) exposure by lowering cytosolic and nuclear PAR levels and preventing AIF nuclear translocation [16]. Patients with arh3 mutations that are predicted to produce a truncated, inactive ARH3 protein exhibit a progressive neurodegenerative phenotype [58]. In addition, the fibroblasts of such patients were more sensitive to H2O2 stress-induced PAR accumulation and cell death. Similarly, ARH3-deficient mice demonstrated increased sensitivity to cerebral ischemia/reperfusion-induced PAR accumulation and cell death [58]. In conclusion, parthanatos can effectively be inhibited through the degradation of PAR by PARG or ARH3.
Structure and function of AIF
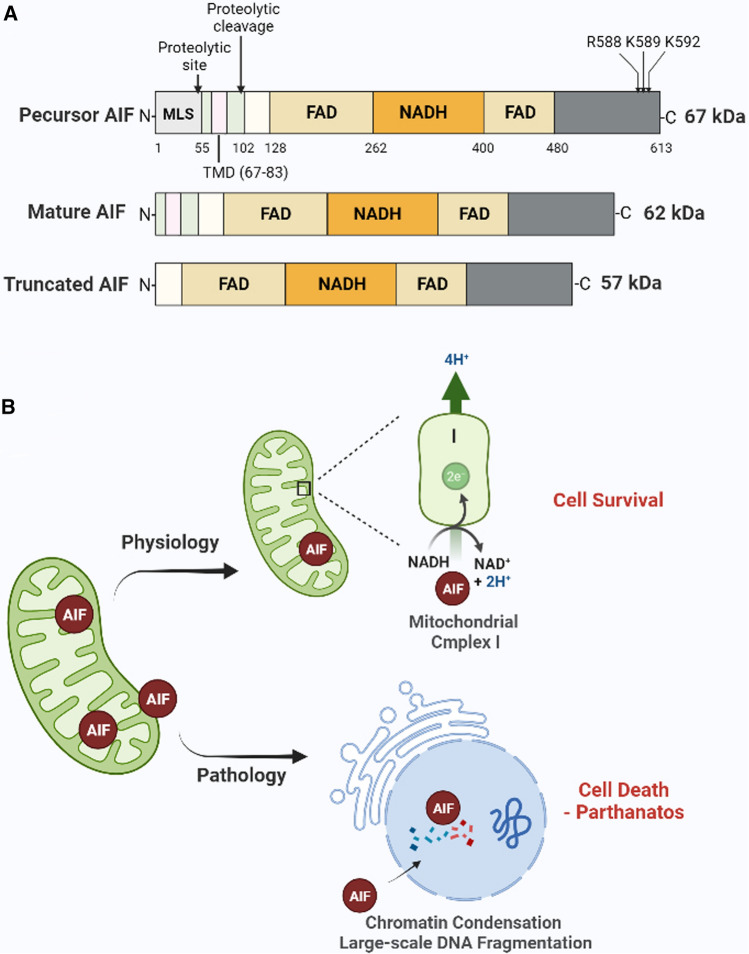
AIF is a mitochondrial flavoprotein that was identified in the late 1990s [59]. AIF is encoded by a single gene on the X chromosome and is transcribed and translated into a 67 kDa precursor protein (Fig. 3A). It is imported into the mitochondria via the mitochondrial localization signal (MLS) located within its N-terminus. Upon import into the mitochondria, the precursor is processed into a mature 62 kDa form via proteolytic cleavage (Fig. 3A). This mature form of AIF is inserted into the inter mitochondrial membrane (IMM) through its N-terminal transmembrane domain (TMD), and the C-terminus is exposed to the mitochondrial intermembrane space (IMS) [60]. In response to a death signal, AIF is further truncated to 57 kDa by calpains and/or cathepsins and released into the cytoplasm (Fig. 3A) [60, 61]. In addition, a small pool of AIF protein is loosely associated with the outer mitochondrial membrane (MOM) on the cytosolic side and can be rapidly released from the mitochondria during parthanatos [62]. The mature form of AIF comprises three structural domains (Fig. 3A); (1) an N-terminal FAD-binding domain, (2) a central reduced NAD(NADH)-binding domain, and (3) a C-terminal domain [63]. As with other flavin proteins, the oxidoreductase component of AIF (FAD-binding domain and NAD-binding domain) confers electron transfer activity to the protein [63–65]. The C-terminal domain is mainly responsible for mediating parthanatos [3, 34]. Therefore, AIF is a bifunctional protein contributing to both cell survival and death.
Fig. 3.
Structure and function of AIF. A The structure of the precursor AIF, mature AIF, and truncated AIF. AIF contains three domains: (1) an N-terminal FAD-binding domain (AA: 128–162 and 400–480), (2) a central reduced NAD(NADH)-binding domain, and (3) a C-terminal domain. The precursor AIF is cleaved at the proteolytic site to form mature AIF. In response to the death signal, the mature AIF is further cleaved at the proteolytic cleavage site to form soluble truncated AIF. R588, K589, and K592 are the binding sites of AIF with PAR. B The function of AIF. Under physiological conditions, AIF spans the intermitochondrial membrane and plays a role in stabilizing mitochondrial complex I and maintaining mitochondrial structure. However, in response to death signals (pathology), AIF translocates from the mitochondria to the nucleus causing chromatin condensation and degradation of large DNA fragments, which leads to parthanatos. AIF apoptosis-inducing factor, MIF migration inhibitory factor, PAR poly(ADP-ribose), MLS mitochondrial localization signal, TMD terminal transmembrane domain
AIF plays an important role in cell survival. Mice with a specific AIF deletion exhibit impaired activity and decreased protein expression of respiratory chain complex I in heart and skeletal muscle. Similarly, mutant animals develop severe dilated cardiomyopathy, heart failure, and skeletal muscle atrophy [66]. During cortical development, AIF is required for neuronal cell survival [67]. Cortical development was impaired and neuronal survival reduced in forebrain-specific AIF ineffective mice due to defects in mitochondrial respiration, which in turn caused death by E17 [67]. AIF plays a role in regulating the mitochondrial structure, namely in determining which AIF-deficient neuronal mitochondria are fragmented with aberrant cristae [67]. Harlequin (Hq) mice, which express only 20% of the regular levels of AIF, exhibit cerebellar degeneration and increased sensitivity to oxidative stress [68], indicating that AIF acts as an oxidative radical scavenger in mitochondria [67, 69]. All these data suggest that AIF promotes cell survival via stabilizing mitochondrial complex I or the maintenance of mitochondrial structure (Fig. 3B).
On the other hand, AIF is also an essential participant in the parthanatos pathway (Fig. 3B). PARP-1 activity leads to calpain activation, which in turn regulates Bax activation. Active bax is required for mitochondrial outer membrane permeabilization (MOMP) and AIF release [70, 71]. The release of AIF from the mitochondria and its translocation into the nucleus result in large-scale DNA fragmentation and cell death [2, 11]. AIF contains a PBM at amino acids 567–592 in the C-terminal domain. In particular, the basic amino acids, arginine 588, lysine 589, and lysine 592, are required for its release from the mitochondria and its ability to induce cell death during parthanatos (Fig. 3A) [3, 34]. Furthermore, the mutation of the PAR-binding site in AIF leads to the failure of its release and translocation, which mediates cell death following PARP-1 activation, whereas the same mutation does not affect its NADH oxidase activity nor ability to bind FAD or DNA [3, 34], providing a further indication that the cell protection and cell death functions of AIF are independent of each other. Either AIF knockdown or its neutralization by anti-AIF antibodies abolish AIF translocation to the nucleus and inhibit MMNG, NMDA, and oxidative stress-triggered cell death [2, 15, 72, 73]. AIF translocation from the mitochondria to the nucleus cannot be inhibited by pan-caspase inhibitors in many cell types. Although AIF transfers from mitochondria, similarly to cytochrome C during caspase-dependent apoptosis [53, 74], it may only represent a by-product of caspase activation and mitochondrial membrane depolarization. Therefore, AIF is a key cell death effector of parthanatos, whose role is independent of caspase activation.
Though the important role of PAR polymers in AIF release from mitochondria and redistribution into nucleus has been validated, it remains to be determined in what form— “naked” polymer or protein-bound—the PAR polymer is released from the nucleus and leads to AIF release from the mitochondria. After DNA damage, 90% of PAR polymers are synthesized by PARP1, with most of the PAR polymers attached to PARP1 itself [75, 76]. Since PARG is involved in the translocation of PAR from the nucleus to the cytoplasm [16, 77], the PAR polymer may work in a soluble form and, thus, diffusion of this “messenger” appears to be an addition layer through which PAR behavior and function is controlled. However, a recent report proposed another possibility—that PAR may be delivered to the cytoplasm and act on AIF in a protein-bound state [76]. PARP1 is cleaved into 89 and 24 kDa fragments upon exposure to staurosporine and actinomycin D. The PAR polymer, attached to the 89 kDa PARP1 fragment, is translocated from nucleus to cytoplasm, where it interacts with AIF, resulting in AIF release and translocation to the nucleus [76]. Although caspase activation-induced PARP1 cleavage sounds controversial for PARP1-dependent cell death, this study evokes a sense of the protein-carried PAR polymer in parthanatos induction.
Structure and function of MIF
Given that AIF itself has no obvious endonuclease activity, AIF-mediated DNA fragmentation likely requires the recruitment of downstream nucleases. A study on Caenorhabditis elegans demonstrated that AIF and endonuclease G (EndoG) together promote DNA degradation [78], yet mammalian EndoG has been shown not to play an essential role in PARP-1-dependent cell death [10, 79]. Kroemer’s group revealed an interaction between AIF and cyclophilin A (CypA) using mass spectrometric methods, protein immunoprecipitation and GST-pulldown assays, demonstrating that CypA is able to degrade plasmid DNA and nuclear DNA extract in vitro [80]. However, the follow-up study found that formation of a complex with CypA is necessary for AIF nuclear translocation in neurons after cerebral hypoxia–ischemia [81]. In 2010, Susin’s group demonstrated that AIF interaction with H2AX [82, 83]. This is an important finding because H2AX (especially phosphorylated H2AX, γH2AX) is closely related to DNA double strand break formation. H2AX genetic ablation, CypA downregulation, or the inhibition of the AIF/CypA complex confers resistance to parthanatos [81–85]. The nuclease that couples with AIF and mediates DNA degradation was discovered by Dawson’s lab. Through a protein chip assay that contains > 16,000 human recombinant proteins, macrophage migration inhibitory factor (MIF) was identified as a PARP-1-dependent AIF-associated nuclease (PAAN) [10].
MIF was identified in 1966 by Bloom and Bennett as a T cell-derived cytokine that inhibits macrophage migration [86, 87]. It is a highly conserved protein, and human MIF consists of 114 amino acids (MW ~ 12.5 kDa) [87, 88]. MIF is a pleiotropic protein, expressed in a number of different cell types such as monocytes, monocytes, fibroblasts, pituitary cells, endothelial cells, neurons, and non-neuronal cells [89]. The most widely studied MIF function is its cytokine activity. In cytokine cascades, MIF is an upstream player that can trigger and amplify cytokine production by stimulating the production of pro-inflammatory mediators, such as tumor necrosis factor α (TNF-α), interleukin-1β (IL- 1β), interferon-γ (IFNγ), and other effector cytokines [90–92]. This function is dependent on its binding with chemokine receptors CXCR2, CXCR4, and CXCR7 [93, 94] and the cell surface receptor CD74, which leads to intramembranous cleavage and signaling or the co-activation of CD44 [95–97]. Following receptor activation, MIF facilitates cell proliferation and inhibition of apoptosis via initiation of the ERK1/2 MAP kinase pathway [96]. Mediated by the non-cognate receptors CXCR2, CXCR4, and CXCR7, MIF inhibits migration and enhances the adhesiveness of immune cells [93, 94]. In addition, MIF is expressed in the central nervous system, where it appears to have multiple beneficial and detrimental effects as a neuroimmunomodulator in neurological disorders [98]. The MIF protein forms a homotrimer and, unlike other cytokine-like molecules, has diverse enzymatic activities including tautomerase, and thiol-protein oxidoreductase (TPOR) activity, which impart MIF with various biological functions [87]. The TPOR activity of MIF catalyzes reductions in insulin and 2-hydroxyethyldisulfide (HED), which are involved in cellular redox protection. This activity is dependent on the cysteines located at positions 57 and 60 in the CXXC motif [99, 100], while the tautomerase activity relies on an N-terminal proline [87]. It is not clear, however, whether the tautomerase enzyme activity of MIF plays a physiological role in mammals, nor is there any known endogenous substrate for this enzyme activity in mammals, or other eukaryotes [89].
In 2016, Dawson’s lab discovered that MIF possesses nuclease activity, which functions in its participation during parthanatos as a downstream effector [10, 101]. The MIF trimer contains a motif characteristic of members of the PD-D/E(X)K superfamily that is also found in many nucleases and highly conserved across mammalian species [10, 102–104]. The glutamic acid residue E22 in the first α helix of MIF is essential for its nuclease activity, and its presence is also consistent with the previously reported active sites of the exonuclease–endonuclease–phosphatase (EEP) domain superfamily nucleases [10, 105, 106]. The nuclease activity of MIF is independent of its oxidoreductase and tautomerase activity. MIF has both 3′ exonuclease and endonuclease activities, preferentially binding to 5′ unpaired bases of ssDNA with a stem–loop structure and cleaving its 3′ unpaired bases [10]. MIF is predominantly localized in the cytosol [10], but in response to various stimuli (NMDA and MNNG), interacts with AIF and is recruited MIF into the nucleus, where it binds to and cleaves genomic DNA into large fragments [10]. Depleting MIF, blocking the AIF/MIF interaction, or mutating the MIF nuclease active site E22 all block MIF nuclease activity and inhibit the chromatinolysis and cell death induced by glutamate excitotoxicity, MNNG, and focal stroke [10]. In conclusion, MIF is an essential nuclease in the parthanatos signaling pathway [101].
Therapeutic targets for parthanatos-based pathogenesis
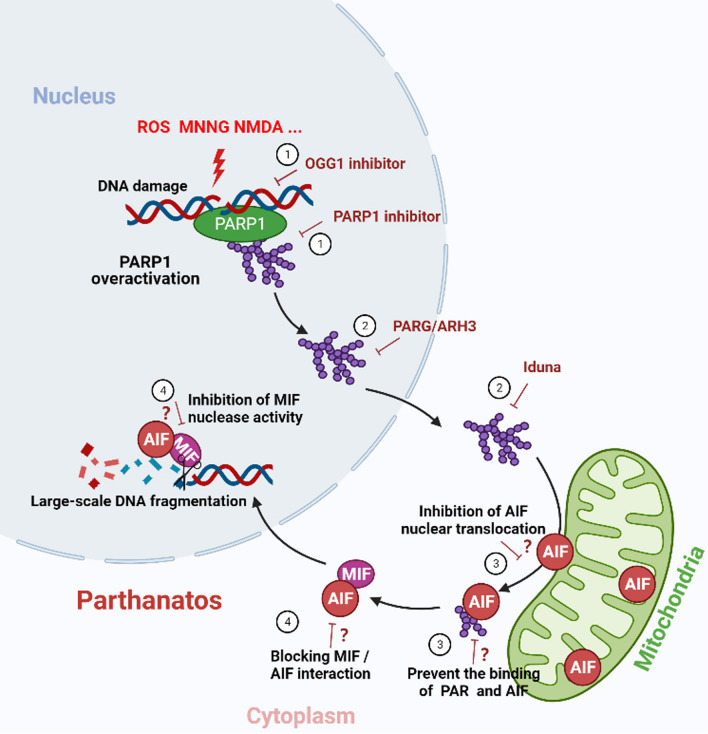
As described above, parthanatos involves a cascade of signaling responses that cause cell death. It is caspase independent, induced by PARP-1 overactivation, and involves AIF and MIF. Hence, targeting any steps of the cascade could serve as a promising therapeutic strategy for parthanatos-based pathologies, such as neurodegenerative diseases, including PD, AD, Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS), and other disorders such as stroke, diabetes, and cerebrovascular diseases [1, 7, 9, 107, 108] (Fig. 4).
Fig. 4.
Potential therapeutic targets for parthanatos-related diseases. ① Inhibition of PARP-1 hyperactivation. Both OGG1 and PARP-1 inhibitors inhibit PARP-1 overactivation and thus inhibit the process of parthanatos. ② Targeted regulation of PARG or ARH3 levels. Promoting PAR degradation, activating Iduna, or mimicking the effects of Iduna binding with PAR to prevent PAR polymer induced parthanatos. ③ Inhibition of AIF nuclear translocation. Research and development agents that inhibit nuclear translocation of AIF, or specifically block the binding of PAR to AIF. ④ Inhibition of the nuclease activity of MIF. On the one hand, the interaction between AIF and MIF can be destroyed to inhibit the transfer of MIF into the nucleus, and on the other hand, targeted inhibition of the nuclease activity of MIF to prevent its cleavage of genomic DNA. “?” refers to a hypothetical therapeutic target to be addressed by joint efforts. PARP-1 poly (ADP-ribose) polymerase 1, OGG1 8-oxo-7,8-dihydroguanine glycosylase1, PARG PAR glycohydrolase, PAR poly(ADP-ribose), ARH3 ADP-ribosyl-acceptor hydrolase 3, AIF apoptosis-inducing factor, MIF migration inhibitory factor
Inhibition of PARP-1 overactivation
As the initial signal of parthanatos, PARP-1 overactivation contributes to the pathological processes of nervous system diseases. Therefore, the effective inhibition of PARP-1 activity is an important target for the treatment of parthanatos-based diseases. Recently, PARP-1 inhibitors have been primarily developed to treat cancer by inhibiting DNA damage repair and promoting cell death [46, 109]. Four PARP inhibitors (olaparib, rucaparib, niraparib, and talazoparib) are currently used in clinical cancer treatment [109–113], and other inhibitors are being tested and used to treat many solid tumors, such as pancreatic and gastric cancer [109, 114]. From a different perspective, these PARP-1 inhibitors could potentially be used to prevent cell death and treat nervous system diseases. PD is an age-related neurodegenerative disease resulting from a pathophysiologic loss or degeneration of dopaminergic neurons in the substantia nigra pars compacta, and the development of neuronal Lewy bodies in intracellular inclusions [115]. Pathological α-synuclein (α-syn) is the main component of Lewy bodies and neurites and contributes to the pathogenesis of PD via various pathways [9]. Dawson’s team found that pathological α-syn activates PARP-1, and PAR generation accelerates the formation of pathological α-syn, resulting in cell death via parthanatos [9]. PARP inhibitors (veliparib, rucaparib, and talazoparib) were shown to prevent the α-syn PFF-mediated PARP activation and cell death in cell and mice models [9]. ALS is a fatal neurodegenerative disease characterized by the accumulation of phosphorylated TAR DNA-binding protein 43 (TDP-43) aggregates in the cytoplasm of affected neurons and glia [108]. Researchers have also reported elevated levels of PAR and the nuclear translocation of AIF in the ALS model [108, 116, 117]. The PARP inhibitor veliparib can inhibit TDP-43-associated neuronal death [108], and olaparib demonstrates remarkable suppression of TDP-43 overexpression-mediated cytotoxicity [118]. AD is the most common type of dementia and is mainly characterized by the aggregation of extracellular amyloid beta peptides (Aβ) and intracellular neurofibrillary tangles [119]. The aggregation of Aβ causes calcium influx, NADPH oxidase activation, and oxidative stress in neuronal cells, which leads to PARP-1 hyperactivation, PAR accumulation, and cell death [120]. The PARP-1 inhibitor PJ34 inhibits Aβ-induced PAR accumulation and thus neuronal death [120–122]. HD is an age-dependent neurodegenerative disease caused by a CAG trinucleotide repeat expansion in exon 1 of the Huntington gene, which leads to the accumulation of mutant huntingtin proteins in the brain [123]. Mutant huntingtin proteins cause the excessive activation of NMDA receptors interacting with postsynaptic density 95 (PSD-95), leading to excitotoxicity [124]. The PARP-1 inhibitor INO-1001 is demonstrated neuroprotection in an HD R6/2 mutant mouse model [125]. Taken together, these data suggest that using PARP-1 inhibitors may represent a promising strategy to treat neurodegenerative diseases in the future.
Recently, Ba’s lab proposed an epistatic determinant of PARP-1 overactivation. After oxidative insult, the 8-oxo-7,8-dihydroguanine glycosylase1 (OGG1)-initiated base excision repair (BER) pathway (OGG1-BER) leads to the generation of excessive BER intermediates, DNA strand breaks, and apurinic/apyrimidinic (AP) sites, which in turn cause PARP-1 overactivation and parthanatos [15]. The lack of OGG1 or repair-deficient OGG1 showed lower levels of DNA strand damage, PARP-1 activation, and AIF nuclear translocation, leading to increased resistance to ROS-induced parthanatos [15]. These results suggest that inhibiting OGG1 activity with specific inhibitors, e.g., TH5487 [126] or O8 [127], may also present a strategy for the treatment of neurodegenerative diseases. However, the inhibition of either PARP-1 or OGG1 activity can also affect DNA damage repair pathways, and these side effects should be considered when developing targeted therapeutics for neurodegenerative diseases.
Control of PARylation
PAR acts as a messenger of death to initiate the parthanatic signaling cascade. Therefore, controlling the level of intracellular PARylation can effectively inhibit this pathway of cell death. Due to the prominent role of PARP-1 activation in DNA repair, interference with PAR signaling may provide a unique opportunity for preventing cell death following PARP-1 activation. As mentioned above, both PARG and ARH3 can regulate PARylation levels through their glycosidic activity [51, 58]. In addition, Dawson’s team found the protein Iduna, a PAR-dependent E3 ligase and PAR-binding protein, to be neuroprotective [128]. Iduna’s protective effects are independent of and do not affect PARP-1 activity[129]. Iduna is protective against NMDA receptor-mediated glutamate excitotoxicity and stroke through interfering with PAR polymer-induced cell death [129]. The PAR polymer binding activity of Iduna is essential for its neuroprotective function. On the one hand, Iduna promotes cell survival by targeting PARylated and PAR binding proteins for ubiquitin proteasomal degradation through PAR binding and ubiquitin E3 ligase activity, and on the other hand, Iduna promotes DNA repair by reducing AP sites [128]. Therefore, the targeted regulation of PARG or ARH3 levels, activating Iduna, or mimicking the effects of Iduna, could represent potential therapeutic strategies to prevent PAR polymer-induced parthanatos.
Inhibition of AIF nuclear translocation
The translocation of AIF from the mitochondria to the nucleus is essential for parthanatos. Therefore, blocking the nuclear transfer of AIF is a further opportunity to prevent cell death following the activation of PARP-1. AIF is a high-affinity PAR-binding protein [34]. The binding of AIF with PAR, particularly through three specific basic amino acids (arginine 588, lysine 589, and lysine 592) found on AIF, is critical for parthanatos [34]. Therefore, developing small-molecule drugs that can inhibit PAR interaction with AIF may be a potential means to protect against parthanatos [34]. Heat shock protein 70 (HSP70) [130] and thioredoxin 1 (Trx1) [131] interact with AIF and inhibit AIF-induced parthanatos in cultured cells. In addition, specifically blocking the interaction between AIF and CypA can prevent AIF nuclear translocation and inhibit cell death. AIF-blocking peptides have been designed to target human CypA, which provides significant neuroprotection as ligand of CypA by blocking AIF binding [132, 133]. Although further validations in animal models is required, these data suggest that agents blocking AIF nuclear translocation may serve as potential drugs to treat neurodegenerative diseases.
In another scenario, the release of AIF from the mitochondria with the concomitant exacerbation of parthanatos may be a further avenue to pursue. For example, atiprimod, a cationic amphiphilic compound, was tested to treat metastatic carcinoid tumors and refractory multiple myeloma. In this study, atiprimod was shown to promote the release of AIF from the mitochondria and subsequent cell death without eliciting PARP-1 activation [134]. Thus, this study highlights that exacerbation of parthanatos, which can be achieved at multiple levels of the cell death pathway, may be an innovative target for cancer therapy.
Inhibition of MIF nuclease activity
MIF is a PARP-1-dependent AIF-associated nuclease that causes large-scale DNA fragmentation during parthanatos [10]. This represents the final step of parthanatos, which ultimately leads to massive DNA damage and irreversible cell death. Various cell death pathways have a “point of no return” [135], and in parthanatos, this is when the nuclease activity of MIF is activated to give rise to large amounts of damaged DNA. The inhibition of MIF’s nuclease activity is thus a potential therapeutic target for neurodegenerative diseases caused by excessive PARP-1 activation [10]. On the one hand, effective inhibition of the nuclear translocation of MIF may insulate its binding to DNA and reduce DNA damage. The transfer of MIF into the nucleus requires interaction with AIF, and effective disruption of the AIF-MIF interaction will inhibit MIF entry into the nucleus. Dawson’s team has resolved the interaction site between AIF and MIF, providing an important reference for the further development of agents as AIF/MIF blockers [10]. MIF’s nuclease activity is independent of its oxidoreductase and tautomerase activity and dependent on the glutamic acid residue E22 in the first α helix of MIF [10]. Thus, it should be possible to develop agents that only inhibit the nuclease activity of MIF without affecting the activity of oxidoreductase and tautomerase [13]. The inhibition of MIF nuclease activity could bypass the need for long-term PARP inhibition, which may impair the detection and repair of DNA damage, and thus offers an opportunity for treating neurodegenerative disease [101].
Conclusion and future prospects
Parthanatos specifically relies on PARP-1 overactivation and is a caspase-independent cell death pathway. It predominantly involves PARP-1 overactivation, PAR accumulation, PAR binding to AIF, AIF release from the mitochondria, nuclear translocation of the AIF/MIF complex, and MIF-mediated large-scale DNA fragmentation. There are still many unsolved questions regarding parthanatos. For example, what is the regulation mechanism of nuclear PAR release, how does AIF release from mitochondria in response to PAR binding, and what chromatin landscape context does MIF recognize? What are the possible molecular determinants of PAR that trigger AIF release in some cell models but not in others? How do cells die in a PARP1-dependent manner if AIF/MIF are not involved? A deeper understanding of the basic biochemical mechanism of parthanatos is a prerequisite for further research on parthanatos-related therapeutics. Parthanatic cell death plays an important role in neurodegenerative disorders such as AD, PD, HD, and ALS, and it may be possible to suppress the development of these neurodegenerative diseases by interfering with key players in the parthanatic cascade, such as PARP-1, the level of PAR signaling, AIF, and MIF. Moreover, inducing parthanatos is emerging as a new strategy to kill tumor cells in cancer therapy, in which PAR/AIF can substitute as a primary mediator of breast cancer cell death after chemotherapy [109, 136]. It is crucial, therefore, that further investigations into the molecular mechanisms of parthanatos are conducted. Strategies to block or exacerbate parthanatos, present promising therapeutic approaches in the treatment of parthanatos-related diseases.
Author contributions
RW and XB had the idea for the article; RW, LL and JL performed the literature search; RW, XB, LL, JL, JG, YK and XZ drafted and critically revised work; RW designed the figures. LL and RW produced the figures.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant number: 31900557 to R.W., 31970686 to X.B. and 31801182 to Y.K.), China Postdoctoral Science Foundation Grant (grant number: 2019M662431 to R.W.).
Availability of data and material
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors consent the publication of the manuscript in CMLS.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Libo Liu and Jiaxiang Li have contributed equally to this work.
Contributor Information
Xueqing Ba, Email: baxq755@nenu.edu.cn.
Ruoxi Wang, Email: 618163@sdnu.edu.cn.
References
- 1.Wang X, Ge P. Parthanatos in the pathogenesis of nervous system diseases. Neuroscience. 2020;449:241–250. doi: 10.1016/j.neuroscience.2020.09.049. [DOI] [PubMed] [Google Scholar]
- 2.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 3.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103(48):18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103(48):18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the nomenclature committee on cell death 2012. Cell Death Differ. 2012;19(1):107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fatokun AA, Dawson VL, Dawson TM. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol. 2014;171(8):2000–2016. doi: 10.1111/bph.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173(1):2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, et al. Poly(ADP-ribose) drives pathologic alpha-synuclein neurodegeneration in Parkinson's disease. Science. 2018 doi: 10.1126/science.aat8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, An R, Umanah GK, Park H, Nambiar K, Eacker SM, Kim B, Bao L, Harraz MM, Chang C, et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016 doi: 10.1126/science.aad6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci (Landmark Ed) 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N Y Acad Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H, Kam TI, Dawson TM, Dawson VL. Poly (ADP-ribose) (PAR)-dependent cell death in neurodegenerative diseases. Int Rev Cell Mol Biol. 2020;353:1–29. doi: 10.1016/bs.ircmb.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24(48):10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Li C, Qiao P, Xue Y, Zheng X, Chen H, Zeng X, Liu W, Boldogh I, Ba X. OGG1-initiated base excision repair exacerbates oxidative stress-induced parthanatos. Cell Death Dis. 2018;9(6):628. doi: 10.1038/s41419-018-0680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashimo M, Kato J, Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc Natl Acad Sci U S A. 2013;110(47):18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan J, Dawson TM, Dawson VL. Cell death mechanisms of neurodegeneration. Adv Neurobiol. 2017;15:403–425. doi: 10.1007/978-3-319-57193-5_16. [DOI] [PubMed] [Google Scholar]
- 18.Jang KH, Do YJ, Son D, Son E, Choi JS, Kim E. AIF-independent parthanatos in the pathogenesis of dry age-related macular degeneration. Cell Death Dis. 2017;8(1):e2526. doi: 10.1038/cddis.2016.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regdon Z, Robaszkiewicz A, Kovacs K, Rygielska Z, Hegedus C, Bodoor K, Szabo E, Virag L. LPS protects macrophages from AIF-independent parthanatos by downregulation of PARP1 expression, induction of SOD2 expression, and a metabolic shift to aerobic glycolysis. Free Radic Biol Med. 2019;131:184–196. doi: 10.1016/j.freeradbiomed.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 20.Erdelyi K, Bai P, Kovacs I, Szabo E, Mocsar G, Kakuk A, Szabo C, Gergely P, Virag L. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 2009;23(10):3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robaszkiewicz A, Erdelyi K, Kovacs K, Kovacs I, Bai P, Rajnavolgyi E, Virag L. Hydrogen peroxide-induced poly(ADP-ribosyl)ation regulates osteogenic differentiation-associated cell death. Free Radic Biol Med. 2012;53(8):1552–1564. doi: 10.1016/j.freeradbiomed.2012.08.567. [DOI] [PubMed] [Google Scholar]
- 22.Virag L, Salzman AL, Szabo C. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J Immunol. 1998;161(7):3753–3759. [PubMed] [Google Scholar]
- 23.Virag L, Scott GS, Cuzzocrea S, Marmer D, Salzman AL, Szabo C. Peroxynitrite-induced thymocyte apoptosis: the role of caspases and poly (ADP-ribose) synthetase (PARS) activation. Immunology. 1998;94(3):345–355. doi: 10.1046/j.1365-2567.1998.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96(24):13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ba X, Garg NJ. Signaling mechanism of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammatory diseases. Am J Pathol. 2011;178(3):946–955. doi: 10.1016/j.ajpath.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32(1):12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Andrabi SA, Umanah GK, Chang C, Stevens DA, Karuppagounder SS, Gagne JP, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc Natl Acad Sci U S A. 2014;111(28):10209–10214. doi: 10.1073/pnas.1405158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, Wheeler D, Redpath P, Li J, Romero G, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8(6):1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26(5):417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. doi: 10.1042/bj3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson BA, Kraus WL. Identification of protein substrates of specific PARP enzymes using analog-sensitive PARP mutants and a "clickable" NAD(+) analog. Methods Mol Biol. 2017;1608:111–135. doi: 10.1007/978-1-4939-6993-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27(16):1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Luo W, Wang Y. PARP-1 and its associated nucleases in DNA damage response. DNA Repair (Amst) 2019;81:102651. doi: 10.1016/j.dnarep.2019.102651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16(10):1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visochek L, Grigoryan G, Kalal A, Milshtein-Parush H, Gazit N, Slutsky I, Yeheskel A, Shainberg A, Castiel A, Seger R, et al. A PARP1-ERK2 synergism is required for the induction of LTP. Sci Rep. 2016;6:24950. doi: 10.1038/srep24950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7):411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 39.Homburg S, Visochek L, Moran N, Dantzer F, Priel E, Asculai E, Schwartz D, Rotter V, Dekel N, Cohen-Armon M. A fast signal-induced activation of Poly(ADP-ribose) polymerase: a novel downstream target of phospholipase c. J Cell Biol. 2000;150(2):293–307. doi: 10.1083/jcb.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju BG, Solum D, Song EJ, Lee KJ, Rose DW, Glass CK, Rosenfeld MG. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119(6):815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, Bendetz-Nezer S, Yao Z, Seger R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell. 2007;25(2):297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Teloni F, Altmeyer M. Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res. 2016;44(3):993–1006. doi: 10.1093/nar/gkv1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen MS, Chang P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol. 2018;14(3):236–243. doi: 10.1038/nchembio.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamaletdinova T, Fanaei-Kahrani Z, Wang ZQ. The enigmatic function of PARP1: from PARylation activity to PAR readers. Cells. 2019 doi: 10.3390/cells8121625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung AK. Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol. 2014;205(5):613–619. doi: 10.1083/jcb.201402114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunze FA, Hottiger MO. Regulating immunity via ADP-ribosylation: therapeutic implications and beyond. Trends Immunol. 2019;40(2):159–173. doi: 10.1016/j.it.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 47.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70(3):789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Ke Y, Jiang X, He F, Pan L, Xu L, Zeng X, Ba X. Lipopolysaccharide activates ERK-PARP-1-RelA pathway and promotes nuclear factor-kappaB transcription in murine macrophages. Hum Immunol. 2012;73(5):439–447. doi: 10.1016/j.humimm.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, Tian X, Ba X, Boldogh I, Zeng X. PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nat Commun. 2017;8:14632. doi: 10.1038/ncomms14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y, Feng X, Koh DW. Activation of cell death mediated by apoptosis-inducing factor due to the absence of poly(ADP-ribose) glycohydrolase. Biochemistry. 2011;50(14):2850–2859. doi: 10.1021/bi101829r. [DOI] [PubMed] [Google Scholar]
- 52.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101(51):17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218(2):193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrision D, Gravells P, Thompson R, Bryant HE. Poly(ADP-ribose) glycohydrolase (PARG) vs. poly(ADP-ribose) polymerase (PARP) - function in genome maintenance and relevance of inhibitors for anti-cancer therapy. Front Mol Biosci. 2020;7:191. doi: 10.3389/fmolb.2020.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Min W, Wang ZQ. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front Biosci (Landmark Ed) 2009;14:1619–1626. doi: 10.2741/3329. [DOI] [PubMed] [Google Scholar]
- 56.Cozzi A, Cipriani G, Fossati S, Faraco G, Formentini L, Min W, Cortes U, Wang ZQ, Moroni F, Chiarugi A. Poly(ADP-ribose) accumulation and enhancement of postischemic brain damage in 110-kDa poly(ADP-ribose) glycohydrolase null mice. J Cereb Blood Flow Metab. 2006;26(5):684–695. doi: 10.1038/sj.jcbfm.9600222. [DOI] [PubMed] [Google Scholar]
- 57.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281(2):705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 58.Mashimo M, Bu X, Aoyama K, Kato J, Ishiwata-Endo H, Stevens LA, Kasamatsu A, Wolfe LA, Toro C, Adams D, et al. PARP1 inhibition alleviates injury in ARH3-deficient mice and human cells. JCI Insight. 2019 doi: 10.1172/jci.insight.124519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Susin SA, Zamzami N, Castedo M, Hirsch T, Marchetti P, Macho A, Daugas E, Geuskens M, Kroemer G. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J Exp Med. 1996;184(4):1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24(7):1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem. 2005;280(8):6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- 62.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009 doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mate MJ, Ortiz-Lombardia M, Boitel B, Haouz A, Tello D, Susin SA, Penninger J, Kroemer G, Alzari PM. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol. 2002;9(6):442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 64.Churbanova IY, Sevrioukova IF. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J Biol Chem. 2008;283(9):5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 65.Sevrioukova IF. Redox-linked conformational dynamics in apoptosis-inducing factor. J Mol Biol. 2009;390(5):924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joza N, Oudit GY, Brown D, Benit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25(23):10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, MacLaurin JG, Rippstein P, Park DS, Shore GC, et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25(17):4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419(6905):367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 69.Lipton SA, Bossy-Wetzel E. Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell. 2002;111(2):147–150. doi: 10.1016/s0092-8674(02)01046-2. [DOI] [PubMed] [Google Scholar]
- 70.Moubarak RS, Yuste VJ, Artus C, Bouharrour A, Greer PA, Menissier-de Murcia J, Susin SA. Sequential activation of poly(ADP-ribose) polymerase 1, calpains, and Bax is essential in apoptosis-inducing factor-mediated programmed necrosis. Mol Cell Biol. 2007;27(13):4844–4862. doi: 10.1128/MCB.02141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cabon L, Galan-Malo P, Bouharrour A, Delavallee L, Brunelle-Navas MN, Lorenzo HK, Gross A, Susin SA. BID regulates AIF-mediated caspase-independent necroptosis by promoting BAX activation. Cell Death Differ. 2012;19(2):245–256. doi: 10.1038/cdd.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Culmsee C, Zhu C, Landshamer S, Becattini B, Wagner E, Pellecchia M, Blomgren K, Plesnila N. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25(44):10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheung EC, Melanson-Drapeau L, Cregan SP, Vanderluit JL, Ferguson KL, McIntosh WC, Park DS, Bennett SA, Slack RS. Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. J Neurosci. 2005;25(6):1324–1334. doi: 10.1523/JNEUROSCI.4261-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost MC, Leber B, Andrews D, Penninger J, et al. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000;14(5):729–739. doi: 10.1096/fasebj.14.5.729. [DOI] [PubMed] [Google Scholar]
- 75.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays. 2004;26(8):882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 76.Mashimo M, Onishi M, Uno A, Tanimichi A, Nobeyama A, Mori M, Yamada S, Negi S, Bu X, Kato J, et al. The 89-kDa PARP1 cleavage fragment serves as a cytoplasmic PAR carrier to induce AIF-mediated apoptosis. J Biol Chem. 2021;296:100046. doi: 10.1074/jbc.RA120.014479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mashimo M, Moss J. Functional role of ADP-ribosyl-acceptor hydrolase 3 in poly(ADP-ribose) polymerase-1 response to oxidative stress. Curr Protein Pept Sci. 2016;17(7):633–640. doi: 10.2174/1389203717666160419144603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298(5598):1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 79.Xu Z, Zhang J, David KK, Yang ZJ, Li X, Dawson TM, Dawson VL, Koehler RC. Endonuclease G does not play an obligatory role in poly(ADP-ribose) polymerase-dependent cell death after transient focal cerebral ischemia. Am J Physiol Regul Integr Comp Physiol. 2010;299(1):R215–221. doi: 10.1152/ajpregu.00747.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cande C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, et al. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23(8):1514–1521. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- 81.Zhu C, Wang X, Deinum J, Huang Z, Gao J, Modjtahedi N, Neagu MR, Nilsson M, Eriksson PS, Hagberg H, et al. Cyclophilin A participates in the nuclear translocation of apoptosis-inducing factor in neurons after cerebral hypoxia-ischemia. J Exp Med. 2007;204(8):1741–1748. doi: 10.1084/jem.20070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Artus C, Boujrad H, Bouharrour A, Brunelle MN, Hoos S, Yuste VJ, Lenormand P, Rousselle JC, Namane A, England P, et al. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J. 2010;29(9):1585–1599. doi: 10.1038/emboj.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baritaud M, Boujrad H, Lorenzo HK, Krantic S, Susin SA. Histone H2AX: the missing link in AIF-mediated caspase-independent programmed necrosis. Cell Cycle. 2010;9(16):3166–3173. doi: 10.4161/cc.9.16.12887. [DOI] [PubMed] [Google Scholar]
- 84.Doti N, Reuther C, Scognamiglio PL, Dolga AM, Plesnila N, Ruvo M, Culmsee C. Inhibition of the AIF/CypA complex protects against intrinsic death pathways induced by oxidative stress. Cell Death Dis. 2014;5:e993. doi: 10.1038/cddis.2013.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez J, Xie C, Li T, Sun Y, Wang Y, Xu Y, Li K, Zhang S, Zhou K, Wang Y, et al. Inhibiting the interaction between apoptosis-inducing factor and cyclophilin A prevents brain injury in neonatal mice after hypoxia-ischemia. Neuropharmacology. 2020;171:108088. doi: 10.1016/j.neuropharm.2020.108088. [DOI] [PubMed] [Google Scholar]
- 86.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153(3731):80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 87.Bloom J, Sun S, Al-Abed Y. MIF, a controversial cytokine: a review of structural features, challenges, and opportunities for drug development. Expert Opin Ther Targets. 2016;20(12):1463–1475. doi: 10.1080/14728222.2016.1251582. [DOI] [PubMed] [Google Scholar]
- 88.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33(47):14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 89.Florez-Sampedro L, Soto-Gamez A, Poelarends GJ, Melgert BN. The role of MIF in chronic lung diseases: looking beyond inflammation. Am J Physiol Lung Cell Mol Physiol. 2020;318(6):L1183–L1197. doi: 10.1152/ajplung.00521.2019. [DOI] [PubMed] [Google Scholar]
- 90.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3(10):791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Onodera S, Nishihira J, Koyama Y, Majima T, Aoki Y, Ichiyama H, Ishibashi T, Minami A. Macrophage migration inhibitory factor up-regulates the expression of interleukin-8 messenger RNA in synovial fibroblasts of rheumatoid arthritis patients: common transcriptional regulatory mechanism between interleukin-8 and interleukin-1beta. Arthritis Rheum. 2004;50(5):1437–1447. doi: 10.1002/art.20190. [DOI] [PubMed] [Google Scholar]
- 92.Schindler L, Dickerhof N, Hampton MB, Bernhagen J. Post-translational regulation of macrophage migration inhibitory factor: basis for functional fine-tuning. Redox Biol. 2018;15:135–142. doi: 10.1016/j.redox.2017.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tarnowski M, Grymula K, Liu R, Tarnowska J, Drukala J, Ratajczak J, Mitchell RA, Ratajczak MZ, Kucia M. Macrophage migration inhibitory factor is secreted by rhabdomyosarcoma cells, modulates tumor metastasis by binding to CXCR4 and CXCR7 receptors and inhibits recruitment of cancer-associated fibroblasts. Mol Cancer Res. 2010;8(10):1328–1343. doi: 10.1158/1541-7786.MCR-10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 95.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25(4):595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, Leng L, Goldenberg DM, Shvidel L, Berrebi A, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci U S A. 2007;104(33):13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leyton-Jaimes MF, Kahn J, Israelson A. Macrophage migration inhibitory factor: a multifaceted cytokine implicated in multiple neurological diseases. Exp Neurol. 2018;301(Pt B):83–91. doi: 10.1016/j.expneurol.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 99.Kleemann R, Kapurniotu A, Frank RW, Gessner A, Mischke R, Flieger O, Juttner S, Brunner H, Bernhagen J. Disulfide analysis reveals a role for macrophage migration inhibitory factor (MIF) as thiol-protein oxidoreductase. J Mol Biol. 1998;280(1):85–102. doi: 10.1006/jmbi.1998.1864. [DOI] [PubMed] [Google Scholar]
- 100.Thiele M, Bernhagen J. Link between macrophage migration inhibitory factor and cellular redox regulation. Antioxid Redox Signal. 2005;7(9–10):1234–1248. doi: 10.1089/ars.2005.7.1234. [DOI] [PubMed] [Google Scholar]
- 101.Jonas E. The MIFstep in parthanatos. Science. 2016;354(6308):36–37. doi: 10.1126/science.aai8756. [DOI] [PubMed] [Google Scholar]
- 102.Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142(1):65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142(1):77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 105.Laganeckas M, Margelevicius M, Venclovas C. Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile-profile alignments. Nucleic Acids Res. 2011;39(4):1187–1196. doi: 10.1093/nar/gkq958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40(15):7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dawson TM, Dawson VL. Mitochondrial mechanisms of neuronal cell death: potential therapeutics. Annu Rev Pharmacol Toxicol. 2017;57:437–454. doi: 10.1146/annurev-pharmtox-010716-105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McGurk L, Mojsilovic-Petrovic J, Van Deerlin VM, Shorter J, Kalb RG, Lee VM, Trojanowski JQ, Lee EB, Bonini NM. Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol Commun. 2018;6(1):84. doi: 10.1186/s40478-018-0586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou Y, Liu L, Tao S, Yao Y, Wang Y, Wei Q, Shao A, Deng Y. Parthanatos and its associated components: promising therapeutic targets for cancer. Pharmacol Res. 2021;163:105299. doi: 10.1016/j.phrs.2020.105299. [DOI] [PubMed] [Google Scholar]
- 110.Matulonis UA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, et al. Olaparib maintenance therapy in patients with platinum-sensitive, relapsed serous ovarian cancer and a BRCA mutation: overall survival adjusted for postprogression poly(adenosine diphosphate ribose) polymerase inhibitor therapy. Cancer. 2016;122(12):1844–1852. doi: 10.1002/cncr.29995. [DOI] [PubMed] [Google Scholar]
- 111.Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 112.Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, Konecny GE, Coleman RL, Tinker AV, O'Malley DM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18(1):75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 113.Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, Fehrenbacher L, Yerushalmi R, Mina LA, Martin M, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh N, Pay SL, Bhandare SB, Arimpur U, Motea EA. Therapeutic strategies and biomarkers to modulate PARP activity for targeted cancer therapy. Cancers (Basel) 2020 doi: 10.3390/cancers12040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beitz JM. Parkinson's disease: a review. Front Biosci (Schol Ed) 2014;6:65–74. doi: 10.2741/s415. [DOI] [PubMed] [Google Scholar]
- 116.Oh YK, Shin KS, Kang SJ. AIF translocates to the nucleus in the spinal motor neurons in a mouse model of ALS. Neurosci Lett. 2006;406(3):205–210. doi: 10.1016/j.neulet.2006.07.044. [DOI] [PubMed] [Google Scholar]
- 117.Shibata N, Kakita A, Takahashi H, Ihara Y, Nobukuni K, Fujimura H, Sakoda S, Sasaki S, Yamamoto T, Kobayashi M. Persistent cleavage and nuclear translocation of apoptosis-inducing factor in motor neurons in the spinal cord of sporadic amyotrophic lateral sclerosis patients. Acta Neuropathol. 2009;118(6):755–762. doi: 10.1007/s00401-009-0580-6. [DOI] [PubMed] [Google Scholar]
- 118.Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019;29(3):233–247. doi: 10.1038/s41422-019-0141-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soria Lopez JA, Gonzalez HM, Leger GC. Alzheimer's disease. Handb Clin Neurol. 2019;167:231–255. doi: 10.1016/B978-0-12-804766-8.00013-3. [DOI] [PubMed] [Google Scholar]
- 120.Angelova PR, Abramov AY. Interaction of neurons and astrocytes underlies the mechanism of Abeta-induced neurotoxicity. Biochem Soc Trans. 2014;42(5):1286–1290. doi: 10.1042/BST20140153. [DOI] [PubMed] [Google Scholar]
- 121.Abeti R, Abramov AY, Duchen MR. Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain. 2011;134(Pt 6):1658–1672. doi: 10.1093/brain/awr104. [DOI] [PubMed] [Google Scholar]
- 122.Kauppinen TM, Suh SW, Higashi Y, Berman AE, Escartin C, Won SJ, Wang C, Cho SH, Gan L, Swanson RA. Poly(ADP-ribose)polymerase-1 modulates microglial responses to amyloid beta. J Neuroinflammation. 2011;8:152. doi: 10.1186/1742-2094-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Labbadia J, Morimoto RI. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci. 2013;38(8):378–385. doi: 10.1016/j.tibs.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sun Y, Savanenin A, Reddy PH, Liu YF. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J Biol Chem. 2001;276(27):24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- 125.Cardinale A, Paldino E, Giampa C, Bernardi G, Fusco FR. PARP-1 inhibition is neuroprotective in the R6/2 mouse model of Huntington's disease. PLoS ONE. 2015;10(8):e0134482. doi: 10.1371/journal.pone.0134482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Visnes T, Cazares-Korner A, Hao W, Wallner O, Masuyer G, Loseva O, Mortusewicz O, Wiita E, Sarno A, Manoilov A, et al. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science. 2018;362(6416):834–839. doi: 10.1126/science.aar8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Donley N, Jaruga P, Coskun E, Dizdaroglu M, McCullough AK, Lloyd RS. Small molecule inhibitors of 8-oxoguanine DNA glycosylase-1 (OGG1) ACS Chem Biol. 2015;10(10):2334–2343. doi: 10.1021/acschembio.5b00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kang HC, Lee YI, Shin JH, Andrabi SA, Chi Z, Gagne JP, Lee Y, Ko HS, Lee BD, Poirier GG, et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc Natl Acad Sci U S A. 2011;108(34):14103–14108. doi: 10.1073/pnas.1108799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andrabi SA, Kang HC, Haince JF, Lee YI, Zhang J, Chi Z, West AB, Koehler RC, Poirier GG, Dawson TM, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17(6):692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Park YH, Seo JH, Park JH, Lee HS, Kim KW. Hsp70 acetylation prevents caspase-dependent/independent apoptosis and autophagic cell death in cancer cells. Int J Oncol. 2017;51(2):573–578. doi: 10.3892/ijo.2017.4039. [DOI] [PubMed] [Google Scholar]
- 131.Shelar SB, Kaminska KK, Reddy SA, Kumar D, Tan CT, Yu VC, Lu J, Holmgren A, Hagen T, Chew EH. Thioredoxin-dependent regulation of AIF-mediated DNA damage. Free Radic Biol Med. 2015;87:125–136. doi: 10.1016/j.freeradbiomed.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 132.Russo L, Mascanzoni F, Farina B, Dolga AM, Monti A, Caporale A, Culmsee C, Fattorusso R, Ruvo M, Doti N. Design, optimization, and structural characterization of an apoptosis-inducing factor peptide targeting human cyclophilin a to inhibit apoptosis inducing factor-mediated cell death. J Med Chem. 2021;64(15):11445–11459. doi: 10.1021/acs.jmedchem.1c00777. [DOI] [PubMed] [Google Scholar]
- 133.Monti A, Sturlese M, Caporale A, Roger JA, Mascanzoni F, Ruvo M, Doti N. Design, synthesis, structural analysis and biochemical studies of stapled AIF(370–394) analogues as ligand of CypA. Biochim Biophys Acta Gen Subj. 1864;12:129717. doi: 10.1016/j.bbagen.2020.129717. [DOI] [PubMed] [Google Scholar]
- 134.Wang M, Zhang L, Han X, Yang J, Qian J, Hong S, Samaniego F, Romaguera J, Yi Q. Atiprimod inhibits the growth of mantle cell lymphoma in vitro and in vivo and induces apoptosis via activating the mitochondrial pathways. Blood. 2007;109(12):5455–5462. doi: 10.1182/blood-2006-12-063958. [DOI] [PubMed] [Google Scholar]
- 135.Kroemer G, El-Deiry WS, Golstein P, Peter ME, Vaux D, Vandenabeele P, Zhivotovsky B, Blagosklonny MV, Malorni W, Knight RA, et al. Classification of cell death: recommendations of the nomenclature committee on cell death. Cell Death Differ. 2005;12(Suppl 2):1463–1467. doi: 10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 136.Feng X, Zhou Y, Proctor AM, Hopkins MM, Liu M, Koh DW. Silencing of apoptosis-inducing factor and poly(ADP-ribose) glycohydrolase reveals novel roles in breast cancer cell death after chemotherapy. Mol Cancer. 2012;11:48. doi: 10.1186/1476-4598-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.