Abstract
Pancreatic ductal adenocarcinoma (PDAC) is correlated with poor outcomes because of limited therapeutic options. Laminin-5 gamma-2 (LAMC2) plays a critical role in key biological processes. However, the detailed molecular mechanism and potential roles of LAMC2 in PDAC stay unexplored. The present study examines the essential role and molecular mechanisms of LAMC2 in the tumorigenesis of PDAC. Here, we identified that LAMC2 is significantly upregulated in microarray cohorts and TCGA RNA sequencing data of PDAC patients compared to non-cancerous/normal tissues. Patients with higher transcript levels of LAMC2 were correlated with clinical stages; dismal overall, as well as, disease-free survival. Additionally, we confirmed significant upregulation of LAMC2 in a panel of PDAC cell lines and PDAC tumor specimens in contrast to normal pancreatic tissues and cells. Inhibition of LAMC2 significantly decreased cell growth, clonogenic ability, migration and invasion of PDAC cells, and tumor growth in the PDAC xenograft model. Mechanistically, silencing of LAMC2 suppressed expression of ZEB1, SNAIL, N-cadherin (CDH2), vimentin (VIM), and induced E-cadherin (CDH1) expression leading to a reversal of mesenchymal to an epithelial phenotype. Interestingly, co-immunoprecipitation experiments demonstrated LAMC2 interaction with epidermal growth factor receptor (EGFR). Further, stable knockdown of LAMC2 inhibited phosphorylation of EGFR, ERK1/2, AKT, mTOR, and P70S6 kinase signaling cascade in PDAC cells. Altogether, our findings suggest that silencing of LAMC2 inhibited PDAC tumorigenesis and metastasis through repression of epithelial–mesenchymal transition and modulation of EGFR/ERK1/2/AKT/mTOR axis and could be a potential diagnostic, prognostic, and therapeutic target for PDAC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04392-1.
Keywords: Pancreatic carcinoma, Epithelial-mesenchymal transition, LAMC2, EGFR, AKT, mTOR, Xenograft model, Migration
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a fatal malignancy with an average 5-year survival rate of ≤ 8%, and median survival of 3–6 months [1–4]. PDAC is the fourth and seventh common cause of cancer-related mortalities in North America and worldwide, respectively [5]. Surgical resection is the most effective treatment, especially in patients with early stages of the disease. However, only approximately 20% of patients can qualify for this treatment, while the rest of the patients are often diagnosed with advanced stages, making treatment very difficult [6, 7]. However, even with the advancement in technologies in the field of medicine, PDAC is still resistant to conventional chemotherapies and other therapies [4, 8–10]. Therefore, an urgent need exists for the identification of new therapeutic molecular targets to foster clinical care of pancreatic cancer patients.
Laminin proteins are a group of evolutionarily conserved proteins [11]. Laminin-5 belongs to the family of basement membrane proteins and is crucial for several biological processes including tissue development, differentiation, cell adhesion, wound healing, proliferation, migration, and metastasis [12–15]. Laminin-5 is a heterotrimeric glycoprotein and is composed of three non-identical chains of laminin [alpha (α3), beta (β3), and gamma (ϒ2)] [12, 15]. Several studies demonstrated robust expression of laminin ϒ2 subunit (known as LAMC2) in a variety of human malignancies including carcinomas of the pancreas, colorectal, cholangiocarcinoma, gastric, tongue, lung, cervix, esophagus (squamous), and thyroid [10, 15–23]. Growing evidence suggests that the highly invasive potential of the tumor cells and distant metastasis depends on the strong cytoplasmic expression of the LAMC2 and is mostly correlated with poor outcome, recurrence, and metastasis [17, 21, 24]. Xue and colleagues have used the LASSO regression method on the TCGA and GEO datasets and discovered a network of six genes including LAMC2 to predict the metastatic status of patients with PDAC [25]. Our previous study revealed that elevated expression of LAMC2 was associated with aberrant proliferation, migration, invasion, and tumor growth in anaplastic thyroid carcinoma (ATC) [26]. Moreover, LAMC2 interacts with epidermal growth factor receptor (EGFR) and activates downstream signaling in ATC [26]. Interestingly, another study has shown the importance of LAMC2 as a promising and reliable serum biomarker for early detection of pancreatic carcinoma along with CA19.9 using proteomic analysis on serum samples obtained from patients with pancreatic adenocarcinoma [27]. In the present investigation, we evaluated expression levels of LAMC2 in a panel of PDAC cell lines and tumor samples compared to normal pancreatic tissue and cell line (control). Moreover, we explored the potential role of LAMC2 in proliferation, migration, invasion, epithelial–mesenchymal transition (EMT), tumor growth, and metastasis using the in-vitro and in-vivo models of PDAC. At molecular levels, we identified the role of LAMC2 in the EGFR/ERK1/2/AKT/mTOR signaling cascade in PDAC.
Materials and methods
Cell culture and antibodies
Pancreatic cancer cell lines (HPAC, AsPC-1, BxPC-3, Capan-2, Panc0203, Panc0327, Panc0403, Panc0504, Panc0813, PL45, SW1990, CFPAC-1, Panc10.05 and hTERT-HPNE, human pancreatic normal epithelial cells were obtained from American Type Culture Collection, USA. HPAC, AsPC-1, BxPC-3, Capan-2, Panc0203, Panc0327, Panc0403, Panc0504, Panc0813, PL45, Panc10.05, SW1990, CFPAC-1 were cultured and maintained in RPMI-1640 (Gibco), Leibovitz's L-15 (Gibco), McCoy's 5A (Gibco), DMEM/F12 medium with 10% FBS and 1% penicillin–streptomycin (Invitrogen, Carlsbad, CA) at 37 °C in a humidified atmosphere with 5% CO2. hTERT-HPNE cells were grown in 75% high glucose DMEM (Sigma-Aldrich, Merck), 25% M3 medium (INCELL), 10 ng per ml EGF (Sigma-Aldrich, Merck), and puromycin (Merck).
Antibodies against LAMC2 and GAPDH were purchased from Santa Cruz Biotechnologies (Dallas, TX, USA). Antibodies against ZEB1, SNAIL, Vimentin, N-cadherin, E-cadherin, EGFR, p-EGFR, ERK1/2, p-ERK1/2, AKT, p-AKT, mTOR, p-mTOR, P70S6K, p-P70S6K, Ki-67, and PECAM1 were procured from Cell Signaling Technology (Danvers, MA, USA). Details of the antibodies are provided in Supplementary Table 1. Total RNA from normal human pancreas tissues was purchased from Invitrogen (Cat. # QS0621; Thermo-Fisher Scientific).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was isolated from HPAC, AsPC-1, BxPC-3, Capan-2, Panc0203, Panc0327, Panc0403, Panc0504, Panc0813, PL45, SW1990, CFPAC-1, Panc10.05 and hTERT-HPNE cells using NucleoSpin RNA kit (MACHEREY-NAGEL, Duren, Germany). Two micrograms of total RNA were reverse transcribed to complementary DNA using RevertAid First Strand cDNA Synthesis Kit (Thermo-Fisher Scientific, USA). qRT-PCR was performed using PowerUp SYBR Green PCR Master Mix kit (Thermo-Fisher Scientific, USA) using the ABI StepOnePlus real-time PCR System (Thermo-Fisher Scientific, USA). GAPDH mRNA levels were used to normalize the levels of target genes. The primers used for qRT-PCR are provided in Supplementary Table 2.
Immunoblot analysis
Whole-cell lysates were prepared from HPAC, AsPC-1, BxPC-3, Capan-2, Panc0203, Panc0327, Panc0403, Panc0504, Panc0813, PL45, SW1990, CFPAC-1, Panc10.05, and hTERT-HPNE cells using M-PER (Mammalian protein extraction reagent; Thermo-Fisher Scientific) with 1X protease inhibitor cocktail (Roche Molecular Biochemicals, Pleasanton, CA). Briefly, the proteins were transferred to a polyvinylidene fluoride membrane (Merck-Millipore), blocked with 3% milk, and incubated overnight with respective primary antibodies. Blots were washed and incubated with HRP-conjugated secondary antibodies for 1 h. Either chemiluminescent substrate West Dura or SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Biotechnology, Rockford, IL) was used for protein detection.
Immunofluorescence assay
HPAC, BxPC-3, and Panc0504 cells were grown in chamber slides (SPL Life Science, Korea) for 24 h. Cells were fixed with ice-cold methanol and incubated with murine monoclonal LAMC2 antibody (1:250 dilution) for 2 h. Cells were washed and incubated with a secondary antibody conjugated with Alexa Fluor 488 (Thermo-Fisher Scientific, USA). 4´,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO) was used for nuclear staining. The slides were mounted with an Antifade gold medium (Life Technologies, USA) and imaged on a light microscope (Nikon, Japan).
Immunohistochemistry (IHC) assay
All the patient tissues and data on the PDAC TMA were obtained and evaluated in accordance with prior institutional review board approval as previously described in Manuyakorn A et al., in 2010 [28]. For LAMC2 IHC, the University of California Los Angeles (UCLA), USA tissue microarray (TMA) for pancreatic adenocarcinoma was used as described earlier [28] and is comprised of 145 treatment-naïve AJCC stage I and II PDA tumors resected at UCLA from 1991–2005. Paraffin-embedded sections were deparaffinized with xylene and rehydrated through graded ethanol. Antigen retrieval was performed with citrate buffer (pH 6.0) in a vegetable steamer for 20 min. After antigen retrieval and peroxidase blocking step, slides were incubated with anti-LAMC2 antibody diluted 1:200 in DAKO protein blocking solution overnight at 4 °C. For Ki-67 and PECAM1 staining, sodium citrate buffer (pH 6.0) was used for 5 min at 95–100 °C [29, 30]. The signals were detected using the Dakocytomation Envision ⊕ System Labelled Polymer HRP anti-murine antibody and visualized by diaminobenzidine reaction as per manufacturer’s instructions. Counterstaining was done using hematoxylin, and slides were mounted with DPX (Sigma-Aldrich, St. Louis, MO). Three representative cores per tumor on the TMA were evaluated by a practicing gastrointestinal pathologist (DWD) using a histoscore (range 0–200). The histoscore is the product of the staining intensity (0-absent, 1-weak, 2-strong) multiplied by percent tumor cell staining (0–100). The average of the histoscores for the three cores of each tumor was used for statistical analysis. Tumors were dichotomized into two groups with either low (histoscore ≤ 100, N = 58) or moderate/strong (histoscore > 100, N 62) staining. 11 patients had either no or insufficient tumor for histologic evaluation.
Small interfering RNA
To generate stable knockdown of LAMC2 in PDAC cell lines for functional studies. LAMC2 specific shRNA or scramble shRNA were transfected in 293 T cells with jetPrime transfection reagent (Polyplus, France) to generate lentiviral particles. PDAC cells were then transduced with either LAMC2 shRNA or scramble shRNA lentiviral particles as described earlier [26, 31]. Stable cells were selected using puromycin for 2–3 weeks. LAMC2 knockdown was confirmed with qRT-PCR and western blotting in PDAC cells. Each experiment was performed at least three times in three biological replicates.
Cell proliferation assay
PDAC cells (104 cells per well) were plated in 96-well plates, and MTT (3-(4, 5-dimethylthiazol-2-yl)-2,5diphenyl tetrazolium bromide) assays were performed at 24, 48, 72, and 96 h. Cells were incubated with MTT (0.5 mg/ml) for 2 h at 37 °C in a CO2 incubator. Formazan crystals were dissolved in 100 ul of stop solution (SDS-HCl). Absorbance was measured at 570 nm using a Multiskan GO Microplate Spectrophotometer (Thermo Scientific, USA). Experiments were carried out in triplicate and repeated at least three times.
Cell migration and invasion assays
Cell migration assays were performed using transwell inserts with 8 µm pores (Corning Life Sciences, NY, USA) as per manufacturer protocol. Briefly, cells were placed in the top chamber and 500 µl medium with 10% FBS was added to the lower chamber for chemotaxis. After 48 h, cells migrated to the lower chamber were fixed and stained with crystal violet. For invasion assay, transwell inserts were coated with Matrigel (Corning Life Sciences, NY, USA), the rest of the protocol was similar to the migration assays. All assays were performed in triplicates and repeated at least in three biological replicates.
Tumor growth in PDAC murine xenograft
Specific Pathogen Free male Ncr Nude (NCRNU) mice (4–6 weeks age) were procured from Vivo Bio-Tech Ltd., Hyderabad, India, an authorized Breeder & Distributor of Taconic Biosciences, Inc. These mice were maintained in the individually ventilated caging system and housed in cleanroom facility (Class 10,000). Mice were fed with an irradiated Altromin 1324P (Phytoestrogen poor) diet and maintained in autoclaved corn cob bedding. Animal experiments were carried out with prior approval from the Institutional Animal Ethics Committee of Cancer Institute (W.I.A), Adyar, Chennai, India (project approval number # CIWIA1016LAMC2). Animal housing conditions were maintained as reported [32]. Four to six weeks old NCRNU mice were used for developing PDAC xenografts. HPAC and BxPC-3 (2 × 106 cells/mice) having either stable LAMC2 knockdown or scramble shRNA were subcutaneously injected in the right and left flanks of the nude mice, respectively. Tumor diameters were assessed with a caliper and calculated by V = π/6 × Dl × Ds2 (Dl, largest diameter; Ds, smallest diameter) [33]. Once the tumor volume reached > 1000 mm3, mice were sacrificed, and tumor tissues were dissected and weighed. Half portion of each tumor was fixed in 10% formaldehyde for IHC, and the remaining tumor tissue was preserved at − 80 °C for RT-PCR and western blotting.
Immunoprecipitation assay
PDAC cells were harvested and washed with 1X cold Phosphate Buffered Saline (PBS), and lysates were prepared using M-PER solution (Pierce, Thermo-Fisher Scientific) containing 1X protease inhibitor (Roche, Germany). PDAC cell lysates were incubated with LAMC2 mouse mAb (Santa Cruz Biotechnology) overnight at 4 °C. The next day, A/G Agarose beads (Santa Cruz Biotechnology) were washed and added to the lysate and allowed to form immune complexes at rocker in the cold room for 1 h. The immune complexes were washed with PBS, followed by centrifugation, and eluted by heating the beads with SDS-sample buffer at 95 °C for 5 min. Western blotting was carried out in 10% SDS-PAGE to separate the proteins. Anti-EGFR monoclonal antibody (1:1000) was used as the primary antibody followed by incubation with secondary HRP conjugated anti-rabbit IgG (1:10,000) for immuno-detection.
Statistical analysis
Statistical significance was determined by either a two-tailed unequal variance or unpaired Student t-test; ANOVA using statistical software package SPSS (version 16.0; SPSS Inc., Chicago, IL) and GraphPad Prism (San Diego, CA). Kaplan–Meier plot analyses were used for survival and the Log-rank test was used for statistical analysis. Data were represented as mean ± standard deviation. All experiments were repeated thrice and performed in biological triplicates.
Results
LAMC2 is highly upregulated and associated with worse survival in patients with PDAC
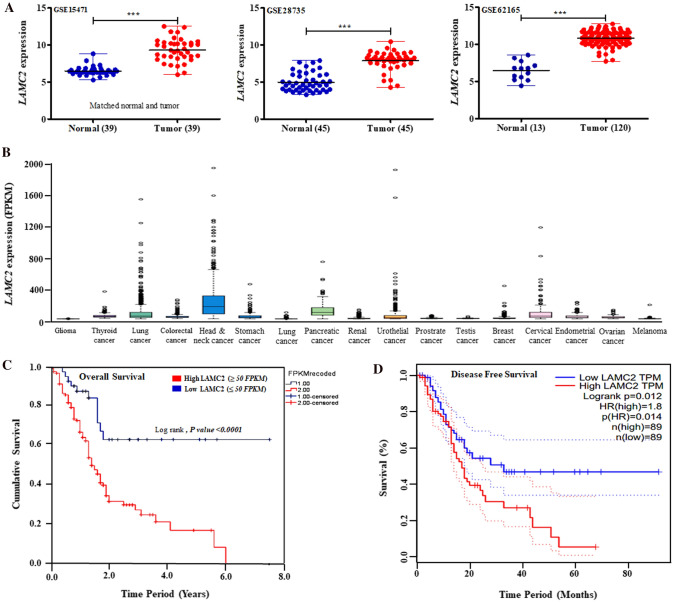
Initially, we analyzed the expression of LAMC2 transcript in normal and tumor pancreatic tissue samples using different microarray datasets (GSE15471, GSE28735, GSE62165, GSE60980, GSE62452, and GSE16515) available at NCBI-Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). Pancreatic tumor specimens displayed significantly (P-value < 0.0001) higher expression of LAMC2 transcript compared to normal pancreatic specimens in GSE15471, GSE28735, GSE62165, GSE60980, GSE62452, and GSE16515 (Fig. 1A & Supplementary Fig. 1A). Further, TCGA RNA sequencing data available at The Human Protein Atlas showed significant and much higher LAMC2 expression in pancreatic and head and neck cancers than other cancer subtypes (Fig. 1B). Interestingly, we performed a survival analysis of the RNA sequencing data which displayed that higher LAMC2 mRNA expression was correlated with poor overall and disease-free survival in patients with pancreatic carcinoma (Fig. 1C, D). A comparison of 179 PDAC and 171 normal/non-cancerous pancreatic samples in the GTEX and GEPIA databases [https://www.gtexportal.org, http://gepia.cancer-pku.cn/] also displayed significant upregulation of LAMC2 mRNA in PDAC samples (Supplementary Fig. 1B). Furthermore, increased expression of LAMC2 was positively correlated with pathological stages in PDAC samples (Supplementary Fig. 1C).
Fig. 1.
LAMC2 is highly upregulated and associated with worse survival in patients with PDAC. A LAMC2 transcript is overexpressed in PDAC tumor specimens compared to normal or adjacent non-cancerous pancreatic samples in the microarray data (GSE15471, GSE28735, and GSE62125). Statistical significance was calculated using GraphPad Prism (***P-value < 0.0001; two-tailed t-test). B RNA sequencing data analysis displayed higher LAMC2 transcript in PDAC and other human malignancies in TCGA Pan-Cancer datasets. C, D Kaplan–Meier plots analysis displayed elevated levels of LAMC2 transcripts were linked with overall (P-value < 0.0001) and disease-free survival (P-value < 0.012) in the patients with PDAC (TCGA data). Statistical significance was calculated using the Log-rank test
Overexpression of LAMC2 transcript and protein in PDAC cell lines and clinical samples
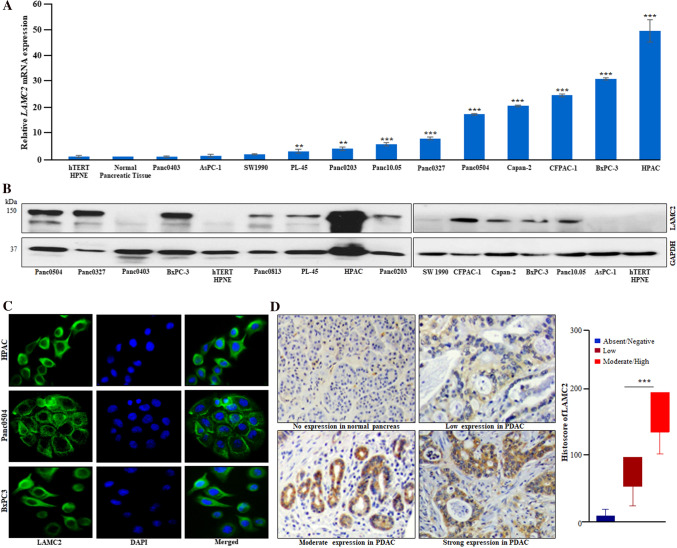
LAMC2 mRNA expression was demonstrated in a panel of 12 PDAC cell lines by qRT-PCR. Our data revealed that LAMC2 was significantly upregulated in most of the PDAC cell lines (except Panc0403, and AsPC-1) as compared to normal human pancreatic tissue and hTERT-HPNE (human pancreatic normal epithelial) cells (Fig. 2A). Concurrently, elevated levels of LAMC2 protein were observed in most of the PDAC cell lines in comparison to hTERT-HPNE. Further, our densitometric analysis of the western blot data demonstrated higher endogenous expression of LAMC2 protein in HPAC, BxPC-3, CFPAC-1, Panc0327, and Panc0504; whereas lower expression occurred in Panc0203, Panc0813, PL45, Capan-2, Panc10.05 and either undetectable or low expression in AsPC-1, Panc0403, SW1990 and hTERT-HPNE cells (Fig. 2B & Supplementary Fig. 2A). Additionally, the results of the immunofluorescence staining displayed cytoplasmic localization of LAMC2 protein in HPAC, Panc0504, and BxPC-3 (Fig. 2C). However, no staining was observed in Panc0403, AsPC-1, and SW1990 (data not shown). Importantly, immunohistochemistry of the pancreatic cancer tissue microarrays (TMAs) confirmed cytoplasmic staining of LAMC2 protein in PDAC tissue samples; either weak or negative staining was found in normal or adjacent non-cancerous pancreatic tissue (Fig. 2D & Supplementary Fig. 2B). Also, we observed that PDAC TMA sections displayed varying levels of immunostaining for LAMC2 protein. Moreover, our histoscore analysis revealed that 43% (58/134) of the PDAC samples demonstrated low LAMC2 staining with a histoscore ≤ 100, and 46% (62/134) of PDAC samples showed moderate/strong LAMC2 staining with a histoscore > 100. Interestingly, a significant difference between the patients with lower LAMC2 staining and the moderate/high staining group was noticed (Fig. 2D).
Fig. 2.
Overexpression of LAMC2 transcript and protein in PDAC cell lines and clinical samples. A Quantitative real-time PCR (qPCR) displayed upregulation of LAMC2 transcript in PDAC cells compared to normal human pancreatic tissue and cell line (hTERT-HPNE). Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value < 0.0001; two-tailed t-test. B Western blot analysis displayed higher LAMC2 protein expression in PDAC cell lines compared to hTERT-HPNE (normal human pancreatic epithelial) cells. C Immunofluorescence experiments demonstrated LAMC2 protein expression in the cytoplasm of PDAC cells. DAPI (4′,6′-diamino-2-phenylindole) was used for nuclear staining. D IHC (immunohistochemistry) data showed strong, moderate, and low staining for LAMC2 in the representative PDAC clinical tissue sections. No/negative staining was noticed in normal pancreatic tissue sections (original magnification, X200). Histoscore analysis displayed significant difference in the staining intensity among low and moderate/high groups. Results represent mean ± SD. ***P-value < 0.0001
Inhibition of LAMC2 significantly decreased the growth of PDAC cells in vitro
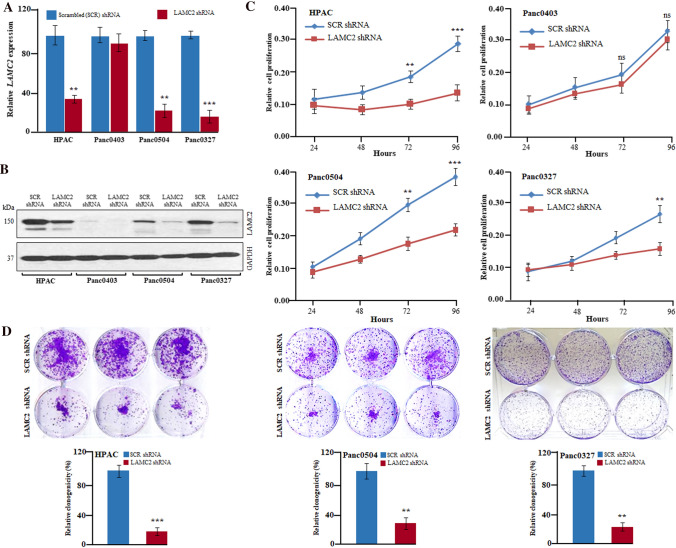
To study the potential role of LAMC2 in pancreatic carcinoma, stable clones were generated either with LAMC2 shRNA or scramble shRNA in PDAC cell lines (HPAC, Panc0403, Panc0504, Panc0327, Panc0203, and BxPC-3). LAMC2 shRNA caused approximately 80–90% silencing of LAMC2 both at mRNA and protein levels in HPAC, Panc0504, Panc0327, Panc0203, and BxPC-3 cells (Figs. 3A, B; Supplementary Fig. 3A). The silencing of LAMC2 led to a significant decrease in cellular growth of HPAC, Panc0504, Panc0327, Panc0203, and BxPC-3 cells compared to scramble shRNA. No significant effect by LAMC2 shRNA on the growth of Panc0403 cells (no LAMC2 expression) was observed, suggesting that LAMC2 shRNA did not produce an off-target effect (Fig. 3C & Supplementary Fig. 3B). Moreover, the knockdown of LAMC2 also significantly reduced the clonogenic growth of PDAC cells (HPAC, Panc0504, Panc0327, Panc0203, and BxPC-3) (Fig. 3D, Supplementary Fig. 3C).
Fig. 3.
Inhibition of LAMC2 significantly decreased the growth of PDAC cells in-vitro. A, B. LAMC2 shRNA stable clones of HPAC, Panc0403, Panc0504, and Panc0327 displayed depletion of LAMC2 both at the transcript and protein levels than scramble (SCR) shRNA. GAPDH primers and antibodies were used for normalization and loading controls in qRT-PCR and western blotting experiments, respectively. Experiments were performed thrice in biological triplicates and the results represent mean ± SD. **P-value < 0.001; ***P-value < 0.0001; two-tailed t-test. C. LAMC2 shRNA displayed reduced growth of HPAC, Panc0504, and Panc0327 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value < 0.0001; two-tailed t-test. D LAMC2 depletion showed decreased clonogenic growth of HPAC, Panc0504, and Panc0327 cells. Data represent mean ± SD; n = 3. ***P-value < 0.0001;**P-value < 0.001 two-tailed t-test
Inhibition of LAMC2 suppressed migration and invasion of PDAC cells through Epithelial-Mesenchymal Transition (EMT)
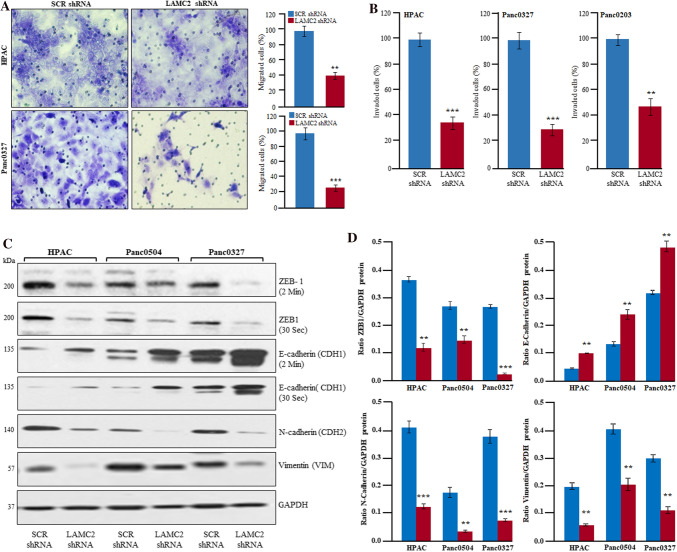
Cell migration and invasion of cancer cells through the extracellular matrix is an essential feature in the process of metastasis. In this regard, EMT has been shown to play a crucial role during tumor progression by promoting metastasis and invasiveness of cancer cells [34]. Therefore, the effect of LAMC2 knockdown was evaluated on the migration and invasion of PDAC cells. Silencing of LAMC2 displayed a significantly decreased motility of HPAC, Panc0327, Panc0203, and BxPC-3 cells through 8 µm inserts compared to scramble shRNA cells (Fig. 4A & Supplementary Fig. 4A). Silencing of LAMC2 showed a significant decrease in the invasiveness of HPAC, Panc0327, and Panc0203 cells through transwell insert (8-µm) coated with Matrigel in comparison to scrambled shRNA (Fig. 4B). Moreover, we evaluated the expression of several crucial EMT markers. Our data displayed that suppression of LAMC2 expression resulted in the inhibition of ZEB1 and SNAIL expression, the master regulators of EMT in HPAC, Panc0504, Panc0327, and Panc0203 cells. Further, inhibition of LAMC2 showed significantly decreased expression of N-cadherin (CDH2), and Vimentin (VIM) (mesenchymal markers) along with an increased expression of E-cadherin (CDH1) (epithelial marker) (Fig. 4C, D & Supplementary Fig. 4C). Additionally, we have observed increased expression of E-cadherin (CDH1) on the surface of LAMC2 knockdown PDAC cells with a marked decrease in the cytoplasmic expression of Vimentin (VIM) (Fig. 5A). Our qRT-PCR data also confirms the above findings at transcriptional levels, indicating the reversal of mesenchymal to epithelial phenotype in PDAC cells (Supplementary Fig. 4D).
Fig. 4.
Inhibition of LAMC2 suppressed migration and invasion of PDAC cells through Epithelial-Mesenchymal Transition (EMT). A, B LAMC2 knockdown inhibited migration and invasion of HPAC, Panc0327, and Panc0203 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value < 0.0001; two-tailed t-test. C, D Western blotting and densitometry data displayed significantly decreased protein expression of ZEB1, N-cadherin, vimentin, and increased expression of E-cadherin in LAMC2 depleted PDAC cells
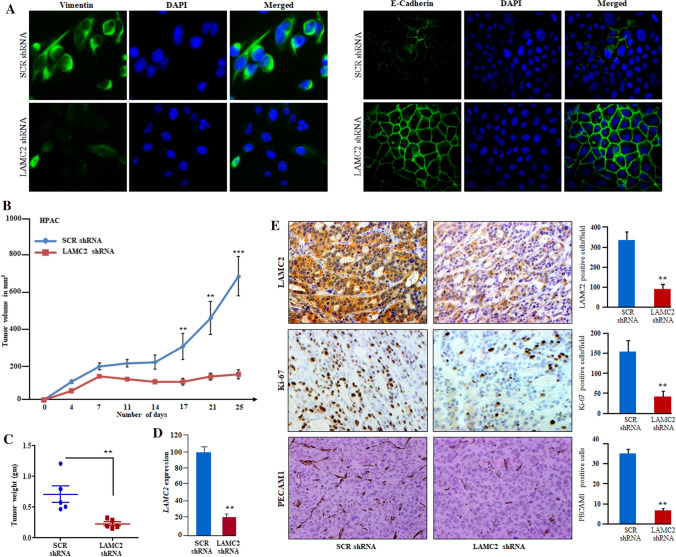
Fig. 5.
Inhibition of LAMC2 suppressed the tumor growth in PDAC xenografts. A Immunofluorescence staining of LAMC2 silenced cells displayed expression of E-cadherin on the cell surface and suppressed the expression of vimentin in PDAC cells. DAPI was used for nuclear staining. B Graph displayed decreased tumor volumes and growth in the LAMC2 silenced group compared to the scramble shRNA group (n = 6). C The weight of the tumors in the LAMC2 silenced group was significantly lower (**P-value < 0.001; t-test) than the scramble shRNA group. D qPCR confirmed LAMC2 knockdown in tumor samples. E IHC on tumor section displayed decreased reactivity for LAMC2, Ki-67, and PECAM1 in LAMC2 silenced tumors sections compared to scramble shRNA Tumor sections. Data represent mean ± SD; n = 3. **P-value < 0.001; two-tailed t-test. Original magnification, X200
Inhibition of LAMC2 suppressed the tumor growth in PDAC xenografts
As our results displayed that knockdown of LAMC2 suppresses the growth of pancreatic cancer cells in liquid culture. Next, we studied the ability of stably LAMC2 silenced HPAC and BxPC-3 cells on tumor growth in the xenograft murine model. Silencing of LAMC2 resulted in tumor growth inhibition leading to smaller tumors, whereas scramble shRNA flanks were noticed with bigger tumors in both HPAC and BxPC-3 xenograft models. Also, we observed a significant decrease in the mean tumor volumes and weights in LAMC2 silenced tumors compared to the scramble knockdown tumors (Fig. 5B, C; Supplementary Fig. 5A, B). However, no significant change in the bodyweight of the xenografts was observed among scrambled and LAMC2 knockdown groups. As expected, we observed reduced expression of LAMC2 both at mRNA and protein levels in the LAMC2 shRNA xenograft tumors section. Furthermore, immunohistochemistry analysis confirmed the stable knockdown of LAMC2 in PDAC xenografts tumors sections along with decreased expression of Ki-67 and, PECAM1 (CD31) (blood vessels marker) in the LAMC2 shRNA tumors than scramble shRNA tumors (Fig. 5D, E). Interestingly, the qRT-PCR data demonstrated a significant decrease in the expression of ZEB1, SNAIL, CDH2 (N-Cadherin), VIM (Vimentin) transcripts along with an increased expression of CDH1 (E-Cadherin) in the LAMC2 knockdown tumor tissues compared to scrambled tumor tissues (Supplementary Fig. 5C).
Inhibition of LAMC2 suppressed EGFR/ERK1/2/AKT/mTOR/P70S6K cascade in PDAC
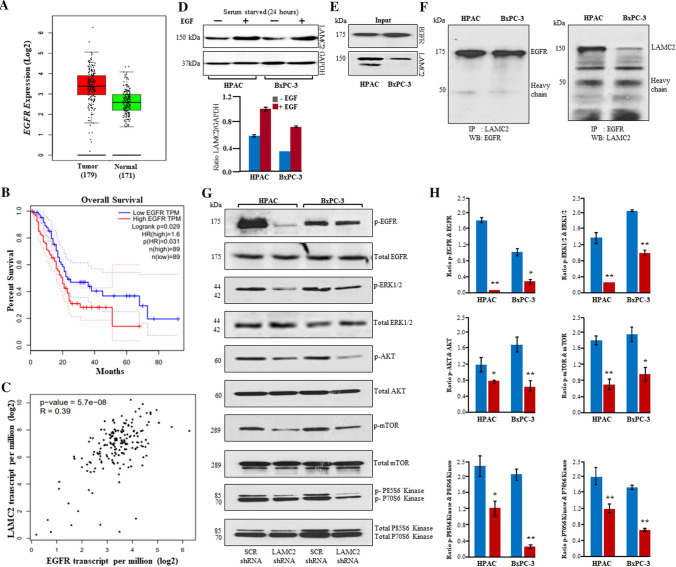
LAMC2 has been shown to have EGF-like motif and is displayed to modulate EGFR signaling cascade in human malignancies [15, 35]. To understand whether depletion of LAMC2 can inhibit the downstream targets of the EGFR cascade to suppress the progression of PDAC, we analyzed EGFR expression in TCGA RNA sequencing data for PDAC patients samples. The EGFR expression was significantly higher in patients with PDAC and correlated with worse overall and disease-free survival. We were more interested to know the co-relation between LAMC2 and EGFR. Therefore, the Pearson correlation analysis was performed and the results displayed a significant and positive correlation between LAMC2 and EGFR expression in PDAC (Fig. 6A–C). Additionally, we have evaluated the effect of EGF ligand on the expression of LAMC2 in HPAC and BxPC-3 cell lines. HPAC and BxPC-3 cells were starved for 24 h followed by EGF treatment (100 ng/mL) for 24 h. Interestingly, EGF treatment showed increased expression of LAMC2 protein in HPAC and BxPC-3 cells as compared to unstimulated cells. This suggested that EGF can induce the expression of LAMC2 in PDAC (Fig. 6D). Further, we observed an interaction between LAMC2 and EGFR proteins in our coimmunoprecipitation experiments in HPAC and BxPC-3 cells (Fig. 6E, F). Moreover, the inhibition of LAMC2 significantly inhibited the phosphorylation of EGFR, ERK1/2, AKT, mTOR, P70S6K, and P85S6K upon starvation and EGF stimulation as compared to scrambled shRNA cells (Fig. 6G, H). These data suggested that LAMC2 can modulate the EGFR/ERK1/2, AKT, mTOR, P70S6K, and P85S6K cascade in PDAC cells (Fig. 7).
Fig. 6.
Inhibition of LAMC2 suppressed EGFR/ERK1/2/AKT/mTOR/P70S6K cascade in PDAC. A Upregulation of EGFR transcripts in 179 PDAC samples compared to 171 non-cancerous pancreatic samples in TCGA data. B Higher expression of EGFR displayed inferior overall survival. C Pearson analysis revealed a positive correlation between LAMC2 and EFGR expression. D Epidermal growth factor (100 ng/mL) treatment induced the expression of LAMC2 in HPAC and BxPC-3 cells. E Western blotting experiments displayed robust expression of LAMC2 and EGFR in protein lysates (input) of HPAC and BxPC-3. F LAMC2 antibodies were used for immunoprecipitation in the cell lysates of HPAC and BxPC-3 and immunoprecipitated complexes were immunoblotted with EGFR antibodies. EGFR antibodies were used for immunoprecipitation and immunoprecipitated complexes were subjected to western blotting with LAMC2 antibodies. G HPAC and BxPC-3 cells (either with LAMC2 shRNA or scramble shRNA) were starved for at least 24 h followed by stimulation with EGF (100 ng/mL) for 20 min. Phosphorylation of EGFR, ERK1/2, AKT, mTOR, P70S6K, and P85S6K was demonstrated using western blotting experiments in HPAC and BxPC-3 cells. H Densitometric analysis revealed a significant decrease in phosphorylation of EGFR, ERK1/2, AKT, mTOR, P70S6K, and P85S6K
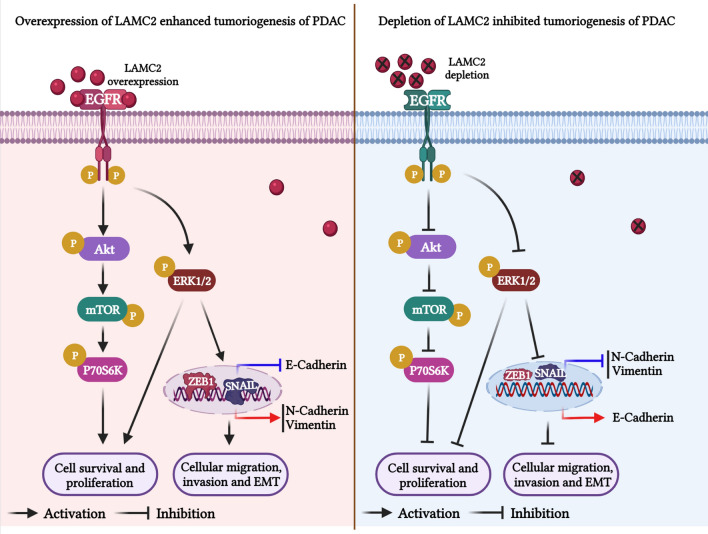
Fig. 7.
Graphical representation of the molecular mechanism/mode of action of LAMC2 in the tumorigenesis of PDAC. Overexpression of LAMC2 activates EGFR signaling pathway and its downstream signaling by phosphorylating and activating ERK1/2, and AKT/mTOR/P70S6K cascade to enhance the proliferation, survival, migration, invasion, and EMT of PDAC cells. On the other hand, depletion of LAMC2 expression suppressed the phosphorylation of ERK1/2 and AKT/mTOR/P70S6K proteins in HPAC and BxPC-3 cells and reduces the tumorigenesis of PDAC
Discussion
Pancreatic carcinoma has a grave prognosis compared to all other human malignancies [3]. PDAC will become the second leading cause of cancer-related deaths because of the sharp increase in PDAC cases by the year 2030 [36]. Hence, dissecting the key molecular mechanisms related to the tumorigenesis of pancreatic cancer is crucial for the treatment of this devastating disease.
LAMC2 is involved in various key physiological functions like tissue development, differentiation, cell adhesion, wound healing, proliferation, and migration in normal cells [15, 24, 37]. In the past decade, emerging pieces of evidence, including our previous study on thyroid cancer, suggest that aberrant expression of LAMC2 plays an essential role in the progression, metastasis, drug resistance, and survival of human malignancies [15, 24]. In this current study, we demonstrated robust expression of LAMC2 transcripts in microarray data containing either matched normal/tumor or unmatched normal/tumor samples. Further, significantly higher transcript and protein expression of LAMC2 occurred in a panel of pancreatic cancer cell lines and PDAC patient samples compared to normal pancreatic tissue, and cell line (hTERT-HPNE). Our histoscore analysis on PDAC TMAs verified the LAMC2 protein expression in ~ 90% of the PDAC patients. Noticeably, histoscore results displayed a significant difference in the low and moderate/high LAMC2 expressing patients. Interestingly, overexpression of LAMC2 transcripts was positively linked with pathological stages, worse overall, and disease-free survival in the PDAC. Taken together, these data indicated that elevated expression of LAMC2 is crucial to progression, worse prognosis, and survival corresponding to elevated LAMC2 levels in PDAC patients in discovery and validation cohorts. Next, we established stable knockdown in several PDAC cell lines either with high, low, or no LAMC2 expression to understand the biological/functional relevance of high endogenous LAMC2 expression in PDAC cells through several functional experiments. Stable silencing of LAMC2 demonstrated decreased proliferation in liquid culture, tumor growth, and tumor volumes in a murine PDAC xenograft model in the cells that robustly expressed LAMC2. Additionally, the tumor sections displayed lower proliferation and lesser blood vessels (Ki-67 and PECAM1) in LAMC2-depleted tumors. To the best of our knowledge, these data first time convincingly displayed the role of LAMC2 in cellular proliferation and tumor progression in the murine model.
Moreover, the suppression of LAMC2 expression caused a significant reduction in the migration and invasion of PDAC cells in Boyden Chamber assays. These results are consistent with our previous findings in thyroid carcinoma and other reports on several other tumor types [23, 26]. These findings might be associated with the epithelial–mesenchymal transition (EMT) in PDAC cells. EMT is essential for maintaining aggressive, invasive, and metastatic phenotypes while changing the expression of various key proteins involved in this process [38]. As anticipated, our data demonstrated that depletion of LAMC2 markedly suppressed EMT of PDAC cells by decreasing their levels of master transcription factors related to EMT including ZEB1, Snail, as well as mesenchymal markers (for example, Vimentin, and N-cadherin). Simultaneously, an increase occurred in the levels of E-cadherin at molecular levels. Additionally, our immunostaining data confirmed the re-expression of the E-cadherin on the surface of PDAC cells and a decrease in the cytoplasmic localization of vimentin. These data suggest that LAMC2 expression is associated with EMT in PDAC. Kosanam H et al., demonstrated the higher expression of LAMC2 in the serum samples obtained from patients with PDAC from different countries compared to normal serum samples through proteomic analysis [27]. Their study concluded that LAMC2 might be used as a diagnostic biomarker along with CA19.9. Interestingly, we also confirmed the secretion of LAMC2 protein in the conditioned medium of PDAC cells and found that secreted LAMC2 induced the EMT phenotype in AsPC-1. AsPC-1 cells had no endogenous LAMC2 expression both at transcript and protein levels. These data reveal the functional relevance of LAMC2 in PDAC for acquiring aggressive, migratory, and invasive phenotypes. Taken together, our data strongly suggest that LAMC2 has an essential role in the progression and metastasis of PDAC.
Furthermore, we were interested to understand the molecular mechanism underlying the mode of action of LAMC2. Mechanistically, we have observed a significant correlation between LAMC2 and EGFR. Also, EGFR expression is one of the prognostic factors in PDAC. We observed the interaction between EGFR and LAMC2 proteins. The first evidence was provided by Schenk S and colleagues that Domain III of LAMC2 is composed of EGF-like repeats, and binding of a recombinant DIII fragment to EGFR can stimulate downstream MAPK signaling, resulting in cell migration in breast carcinoma [35]. Recently, Daisuke H et al. have used the purified LAMC2 protein containing EGF-like motifs and showed the activation of the EGFR signaling cascade and its downstream molecules in SKOV3 cells [39] and was comparable EGF stimulation which was used as a positive control. Other studies in cholangiocarcinoma and laryngeal carcinoma including our previous study in thyroid cancers have displayed the correlation between EGFR and LAMC2 and cetuximab markedly suppressed the expression of LAMC2 [22, 26, 40]. Interestingly, our data showed that EGF stimulation can activate the expression of LAMC2 in PDAC cells. Also, coimmunoprecipitation experiments confirm the interaction between LAMC2 and EGFR. Altogether these data suggested that the EGF-like motifs in LAMC2 can activate the EGFR signaling cascade [35, 39]. Furthermore, the present study demonstrated that depletion of LAMC2 markedly suppressed both EGFR-ERK1/2 and AKT/mTOR/P70S6K/P85S6K kinase pathway to decrease the tumorigenesis, invasiveness, and migration ability in PDAC. Based on our findings, LAMC2 could be used as a diagnostic, prognostic, and potential target for designing therapeutic strategies for PDAC. Our studies provide the foundation for further investigation of LAMC2 as a promising target for the treatment of PDAC.
Conclusions
Altogether, our findings highlight that LAMC2 is highly expressed in PDAC and associated with worse overall and disease-free survival in patients with PDAC. Silencing of LAMC2 significantly inhibited PDAC growth, invasive and metastatic potential in vitro, and murine PDAC xenografts. Mechanistically, LAMC2 inhibition caused the suppression of epithelial–mesenchymal transition by modulating the expression of ZEB1, SNAIL, E-cadherin, Vimentin, N-cadherin in both in vitro and xenograft models of PDAC. Moreover, the silencing of LAMC2 showed inhibition of key oncogenic signaling cascade including EGFR/ERK1/2/AKT/mTOR, a new perspective in PDAC (Fig. 7). Taken together, LAMC2 can be utilized as a potential biomarker for diagnostic and prognostic purposes as well as a target for designing therapeutic strategies for PDAC.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure-1: LAMC2 is significantly overexpressed and positively linked with pathological stages in PDAC. A. LAMC2 expression in GSE60980, GSE62452, and GSE16515. B. Elevated expression of LAMC2 in PDAC (n=179) versus normal pancreatic specimens (n=171) in the GTEX and GEPIA databases. C. LAMC2 transcripts were positively associated with pathological stages in PDAC samples. HNSC, head and neck squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; LUSC, lung squamous cell carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; CESC, cervical squamous cell carcinoma; BLCA, bladder urothelial carcinoma; KIRP, kidney renal papillary cell carcinoma; CHOL, cholangiocarcinoma
Supplementary Figure-2: LAMC2 protein expression in PDAC cell lines and tissue samples. A. Densitometric analysis showed LAMC2 expression in a panel of PDAC cell lines. B. Strong, moderate, and weak LAMC2 protein reactivity was detected in PDAC tissues
Supplementary Figure-3: Knockdown of LAMC2 inhibited proliferation and clonogenic growth in PDAC. A. Panc0203, BxPC-3 cells showed LAMC2 knockdown both at the transcript and protein levels in LAMC2 shRNA stable clones than scramble shRNA. GAPDH was used for normalization and internal control. All the experiments were performed thrice in biological triplicates and the results represent mean ± SD. ***P-value <0.0001; two-tailed t-test. B. LAMC2 shRNA suppressed the growth of Panc0203, BxPC-3 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test. C. LAMC2 knockdown inhibited the clonogenic ability of Panc0203, BxPC-3 cells. Data represents mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Supplementary Figure-4: knockdown of LAMC2 inhibited migration, invasion of Epithelial-Mesenchymal (EMT) in PDAC. A and B. LAMC2 knockdown inhibited migration of BxPC-3, and Panc0203 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; two-tailed t-test. C. LAMC2 knockdown decreased protein expression of LAMC2, Snail, ZEB1, N-cadherin, vimentin, and increased E-cadherin protein expression in Panc0203. D. qPCR results displayed inhibition of LAMC2, Snail, ZEB1, N-cadherin, vimentin, and induction of E-cadherin in BxPC-3 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Supplementary Figure-5: Knockdown of LAMC2 suppressed the tumor growth in PDAC xenografts. A. Photographs of dissected tumors displayed decreased tumor size and growth in the LAMC2 silenced group compared to scramble shRNA group. B. Tumor weights were lowered in LAMC2 silenced tumors than scramble shRNA tumors. ***P-value <0.0001; C. qPCR displayed a marked decrease in transcripts of mesenchymal markers and increased transcripts of the epithelial marker. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Acknowledgements
Not applicable
Abbreviations
- ATC
Anaplastic thyroid carcinoma
- AKT
Ak strain transforming
- ANOVA
Analysis of variance
- CD31
Cluster of differentiation 31
- DAPI
4´,6-Diamidino-2-phenylindole
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial–mesenchymal transition
- ERK1/2
Extracellular signal-regulated kinase ½
- FBS
Fetal bovine serum
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- GEO
Gene expression omnibus
- HRP
Horseradish peroxide
- LAMC2
Laminin-5 gamma-2
- M-PER
Mammalian protein extraction reagent
- m-TOR
Mammalian target of rapamycin
- PDAC
Pancreatic ductal adenocarcinoma
- PIER
Proteolytic induced epitope retrieval
- P70S6K
Ribosomal protein S6 kinase
- qRT-PCR
Quantitative real-time polymerase chain reaction
- shRNA
Short hairpin ribonucleic acid
- SCR
Scramble
- TCGA
The Cancer Genome Atlas
- TMA
Tissue microarray
- ZEB1
Zinc finger E-box-binding homeobox-1
Author contributions
MG conceived the idea and designed the study. AK, AKP, BR, DWD, MG, performed the experiments, acquired and analyzed the data. AK, DPM, GS, TSG helped and performed in silico data analysis. AK, MG prepared the final figures and wrote the manuscript. HPK, MG assisted with the final preparation of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Science and Engineering Research Board, Department of Science and Technology, Government of India under its ECRA scheme (SERB File No. ECR/2016/001519) awarded to Dr. Manoj Garg. We acknowledge the Department of Biotechnology (DBT), Government of India for the Ramalingaswami Fellowship (BT/RLF/Re-entry/24/2014). We acknowledge the Indian Council of Medical Research (ICMR), Government of India for providing Senior Research Fellowship (No.3/2/5/59/2020-NCD-III) to Anuradha Kirtonia under Dr. Manoj Garg.
Availability of data and materials
Microarray data on the PDAC and normal/non-cancerous pancreatic tissue samples are available at NCBI Gene Expression Omnibus (GSE15471, GSE28735, GSE62165, GSE60980, GSE62452, and GSE1651).
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
This study was approved by the Ethics Committee of Amity University, Uttar Pradesh. Animal experiments were carried out with prior approval from the Institutional Animal Ethics Committee of Cancer Institute (W.I.A), Adyar, Chennai, India (project approval number # CIWIA1016LAMC2).
Consent for publication
The authors have agreed to publish this manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tempero MA, Malafa MP, Behrman SW, et al. Pancreatic adenocarcinoma, version 2.2014: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2014;12:1083–1093. doi: 10.6004/jnccn.2014.0106. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 3.Pandya G, Kirtonia A, Singh A, et al. A comprehensive review of the multifaceted role of the microbiota in human pancreatic carcinoma. Semin Cancer Biol. 2021;26:S1044-579X(21)00157-7. doi: 10.1016/j.semcancer.2021.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Pandya G, Kirtonia A, Sethi G, Pandey AK, Garg M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim Biophys Acta Rev Cancer. 2020;1874:188423. doi: 10.1016/j.bbcan.2020.188423. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 7.Chien W, Sudo M, Ding LW, et al. Functional genome-wide screening identifies targets and pathways sensitizing pancreatic cancer cells to dasatinib. J Cancer. 2018;9:4762–4773. doi: 10.7150/jca.25138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao H, Le D, Yang LX. Current status in chemotherapy for advanced pancreatic adenocarcinoma. Anticancer Res. 2013;33:1785–1791. [PubMed] [Google Scholar]
- 9.Tiriac H, Belleau P, Engle DD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada Y, Nishiwada S, Yamamura K, et al. Identification of laminin gamma2 as a prognostic and predictive biomarker for determining response to gemcitabine-based therapy in pancreatic ductal adenocarcinoma. Eur J Cancer. 2021;146:125–134. doi: 10.1016/j.ejca.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sztal T, Berger S, Currie PD, Hall TE. Characterization of the laminin gene family and evolution in zebrafish. Dev Dyn. 2011;240:422–431. doi: 10.1002/dvdy.22537. [DOI] [PubMed] [Google Scholar]
- 12.Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell. 1991;65:599–610. doi: 10.1016/0092-8674(91)90092-D. [DOI] [PubMed] [Google Scholar]
- 13.Rousselle P, Lunstrum GP, Keene DR, Burgeson RE. Kalinin: an epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J Cell Biol. 1991;114:567–576. doi: 10.1083/jcb.114.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan MC, Christiano AM, Engvall E, et al. The functions of laminins: lessons from in vivo studies. Matrix Biol. 1996;15:369–381. doi: 10.1016/S0945-053X(96)90157-2. [DOI] [PubMed] [Google Scholar]
- 15.Garg M, Braunstein G, Koeffler HP. LAMC2 as a therapeutic target for cancers. Expert Opin Ther Targets. 2014;18:979–982. doi: 10.1517/14728222.2014.934814. [DOI] [PubMed] [Google Scholar]
- 16.Sordat I, Bosman FT, Dorta G, et al. Differential expression of laminin-5 subunits and integrin receptors in human colorectal neoplasia. J Pathol. 1998;185:44–52. doi: 10.1002/(SICI)1096-9896(199805)185:1<44::AID-PATH46>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 17.Hlubek F, Jung A, Kotzor N, Kirchner T, Brabletz T. Expression of the invasion factor laminin gamma2 in colorectal carcinomas is regulated by beta-catenin. Cancer Res. 2001;61:8089–8093. [PubMed] [Google Scholar]
- 18.Koshikawa N, Moriyama K, Takamura H, et al. Overexpression of laminin gamma2 chain monomer in invading gastric carcinoma cells. Cancer Res. 1999;59:5596–5601. [PubMed] [Google Scholar]
- 19.Ono Y, Nakanishi Y, Ino Y, et al. Clinocopathologic significance of laminin-5 gamma2 chain expression in squamous cell carcinoma of the tongue: immunohistochemical analysis of 67 lesions. Cancer. 1999;85:2315–2321. doi: 10.1002/(SICI)1097-0142(19990601)85:11<2315::AID-CNCR3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 20.Skyldberg B, Salo S, Eriksson E, et al. Laminin-5 as a marker of invasiveness in cervical lesions. J Natl Cancer Inst. 1999;91:1882–1887. doi: 10.1093/jnci/91.21.1882. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto H, Itoh F, Iku S, Hosokawa M, Imai K. Expression of the gamma(2) chain of laminin-5 at the invasive front is associated with recurrence and poor prognosis in human esophageal squamous cell carcinoma. Clin Cancer Res. 2001;7:896–900. [PubMed] [Google Scholar]
- 22.Pei YF, Liu J, Cheng J, Wu WD, Liu XQ. Silencing of LAMC2 reverses epithelial-mesenchymal transition and inhibits angiogenesis in cholangiocarcinoma via inactivation of the epidermal growth factor receptor signaling pathway. Am J Pathol. 2019;189:1637–1653. doi: 10.1016/j.ajpath.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Takahashi N, Takayama T, Goel A. LAMC2 promotes cancer progression and gemcitabine resistance through modulation of EMT and ATP-binding cassette transporters in pancreatic ductal adenocarcinoma. Carcinogenesis. 2021;42:546–556. doi: 10.1093/carcin/bgab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousselle P, Scoazec JY. Laminin 332 in cancer: When the extracellular matrix turns signals from cell anchorage to cell movement. Semin Cancer Biol. 2020;62:149–165. doi: 10.1016/j.semcancer.2019.09.026. [DOI] [PubMed] [Google Scholar]
- 25.Xue K, Zheng H, Qian X, et al. Identification of key mRNAs as prediction models for early metastasis of pancreatic cancer based on LASSO. Front Bioeng Biotechnol. 2021;9:701039. doi: 10.3389/fbioe.2021.701039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garg M, Kanojia D, Okamoto R, et al. Laminin-5gamma-2 (LAMC2) is highly expressed in anaplastic thyroid carcinoma and is associated with tumor progression, migration, and invasion by modulating signaling of EGFR. J Clin Endocrinol Metab. 2014;99:E62–72. doi: 10.1210/jc.2013-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosanam H, Prassas I, Chrystoja CC, et al. Laminin, gamma 2 (LAMC2): a promising new putative pancreatic cancer biomarker identified by proteomic analysis of pancreatic adenocarcinoma tissues. Mol Cell Proteomics. 2013;12:2820–2832. doi: 10.1074/mcp.M112.023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manuyakorn A, Paulus R, Farrell J, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J Clin Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garg M, Kanojia D, Mayakonda A, et al. Molecular mechanism and therapeutic implications of selinexor (KPT-330) in liposarcoma. Oncotarget. 2017;8:7521–7532. doi: 10.18632/oncotarget.13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanojia D, Garg M, Saini S, Agarwal S, Kumar R, Suri A. Sperm associated antigen 9 expression and humoral response in chronic myeloid leukemia. Leuk Res. 2010;34:858–863. doi: 10.1016/j.leukres.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Garg M, Kanojia D, Mayakonda A, et al. Selinexor (KPT-330) has antitumor activity against anaplastic thyroid carcinoma in vitro and in vivo and enhances sensitivity to doxorubicin. Sci Rep. 2017;7:9749. doi: 10.1038/s41598-017-10325-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Ramachandran B, Jayavelu S, Murhekar K, Rajkumar T. Repeated dose studies with pure Epigallocatechin-3-gallate demonstrated dose and route dependant hepatotoxicity with associated dyslipidemia. Toxicol Rep. 2016;3:336–345. doi: 10.1016/j.toxrep.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg M, Kanojia D, Suri S, Suri A. Small interfering RNA-mediated down-regulation of SPAG9 inhibits cervical tumor growth. Cancer. 2009;115:5688–5699. doi: 10.1002/cncr.24658. [DOI] [PubMed] [Google Scholar]
- 34.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Schenk S, Hintermann E, Bilban M, et al. Binding to EGF receptor of a laminin-5 EGF-like fragment liberated during MMP-dependent mammary gland involution. J Cell Biol. 2003;161:197–209. doi: 10.1083/jcb.200208145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 37.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7:370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 38.Thiery JP. EMT: an update. Methods Mol Biol. 2021;2179:35–39. doi: 10.1007/978-1-0716-0779-4_6. [DOI] [PubMed] [Google Scholar]
- 39.Daisuke H, Kato H, Fukumura K, et al. Novel LAMC2 fusion protein has tumor-promoting properties in ovarian carcinoma. Cancer Sci. 2021;112:4957–4967. doi: 10.1111/cas.15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang QY, Liu YC, Zhou SH, Chen HH. LAMC2 acts as a novel therapeutic target of cetuximab in laryngeal cancer. Neoplasma. 2021;68:1257–1264. doi: 10.4149/neo_2021_210421N549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure-1: LAMC2 is significantly overexpressed and positively linked with pathological stages in PDAC. A. LAMC2 expression in GSE60980, GSE62452, and GSE16515. B. Elevated expression of LAMC2 in PDAC (n=179) versus normal pancreatic specimens (n=171) in the GTEX and GEPIA databases. C. LAMC2 transcripts were positively associated with pathological stages in PDAC samples. HNSC, head and neck squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma; LUSC, lung squamous cell carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; CESC, cervical squamous cell carcinoma; BLCA, bladder urothelial carcinoma; KIRP, kidney renal papillary cell carcinoma; CHOL, cholangiocarcinoma
Supplementary Figure-2: LAMC2 protein expression in PDAC cell lines and tissue samples. A. Densitometric analysis showed LAMC2 expression in a panel of PDAC cell lines. B. Strong, moderate, and weak LAMC2 protein reactivity was detected in PDAC tissues
Supplementary Figure-3: Knockdown of LAMC2 inhibited proliferation and clonogenic growth in PDAC. A. Panc0203, BxPC-3 cells showed LAMC2 knockdown both at the transcript and protein levels in LAMC2 shRNA stable clones than scramble shRNA. GAPDH was used for normalization and internal control. All the experiments were performed thrice in biological triplicates and the results represent mean ± SD. ***P-value <0.0001; two-tailed t-test. B. LAMC2 shRNA suppressed the growth of Panc0203, BxPC-3 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test. C. LAMC2 knockdown inhibited the clonogenic ability of Panc0203, BxPC-3 cells. Data represents mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Supplementary Figure-4: knockdown of LAMC2 inhibited migration, invasion of Epithelial-Mesenchymal (EMT) in PDAC. A and B. LAMC2 knockdown inhibited migration of BxPC-3, and Panc0203 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; two-tailed t-test. C. LAMC2 knockdown decreased protein expression of LAMC2, Snail, ZEB1, N-cadherin, vimentin, and increased E-cadherin protein expression in Panc0203. D. qPCR results displayed inhibition of LAMC2, Snail, ZEB1, N-cadherin, vimentin, and induction of E-cadherin in BxPC-3 cells. Results represent mean ± SD; n = 3. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Supplementary Figure-5: Knockdown of LAMC2 suppressed the tumor growth in PDAC xenografts. A. Photographs of dissected tumors displayed decreased tumor size and growth in the LAMC2 silenced group compared to scramble shRNA group. B. Tumor weights were lowered in LAMC2 silenced tumors than scramble shRNA tumors. ***P-value <0.0001; C. qPCR displayed a marked decrease in transcripts of mesenchymal markers and increased transcripts of the epithelial marker. **P-value < 0.001; ***P-value <0.0001; two-tailed t-test
Data Availability Statement
Microarray data on the PDAC and normal/non-cancerous pancreatic tissue samples are available at NCBI Gene Expression Omnibus (GSE15471, GSE28735, GSE62165, GSE60980, GSE62452, and GSE1651).