Abstract
Glioblastoma (GBM) is the most aggressive cancer of central nervous system with worst patient outcome. Telomere maintenance is a crucial mechanism governing GBM initiation and progression making it an attractive target. microRNAs (miRNAs) have shown therapeutic potential in GBM. Earlier, we showed miR-490 is downregulated in GBM patients and plays a tumor suppressive role. Here, we show that miR-490 regulates telomere maintenance program in GBM by directly targeting Telomeric Repeat-binding Factor 2 (TERF2) of the shelterin complex, Tankyrase 2 (TNKS2) and Serine/Threonine-protein kinase, SMG1. Overexpression of miR-490 resulted in effects characteristic to hampered telomere maintenance via TERF2 inhibition. These include induction of telomere dysfunction-induced foci and global DNA damage (53BP1 foci), along with an increase in p-γH2AX levels. Further, it led to inhibition of telomere maintenance hallmarks via reduced stemness (SOX2 and SOX4 downregulation) and induction of senescence (H3K9me3 marks gain and SIRT1 downregulation). It also initiated downstream DNA damage response (DDR) leading to p53 pathway activation. Moreover, microarray data analysis highlighted an overlap between miR-490 expression and REST-inhibition responses in GBM. Thus, miR-490-mediated targeting of telomere maintenance could be therapeutically important in GBM.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03644-2) contains supplementary material, which is available to authorized users.
Keywords: miR-490, Glioblastoma, Telomeres, TERF2, REST, DNA damage
Introduction
Glioblastoma remains the most aggressive tumour of the central nervous system. Despite the advances in conventional therapeutic strategies, the prognostic outcome of GBM patients has improved marginally [1]. However, the evaluation of the molecular aberrations involved in gliomagenesis and progression has advanced our understanding about gliomas while simultaneously enabling the identification of promising druggable targets. One of the key factors regulating crucial processes like aging as well as carcinogenesis and cancer progression is the regulation of telomere maintenance mechanisms (TMMs) that include activity of telomerase, shelterin complex and other factors [2]. Telomeres are the ends of linear eukaryotic chromosomes composed of tandem 5’-TTAGGG-3’ repeats. Although telomere length has been directly correlated to organismal aging, recent evidence suggests that the rate of telomere shortening in an organism is a better indicator of its biological age [3]. Replication of linear chromosomes is eventually met with a hindrance called the end replication problem where the DNA replication machinery is unable to replicate the linear overhangs which causes recession of these ends. If unchecked, this leads to loss of vital genetic information. Cells solve this issue by initial activation of telomerase to elongate the telomeres via addition of the repeats, which are then replicated by DNA replication machinery. However, telomeres get progressively shortened with each round of replication, which eventually leads to cellular senescence or apoptosis. Telomere maintenance is, thus, especially important to malignant transformation for evading apoptosis and senescence and maintaining proliferative potential [4]. Cancer cells, unlike normal cells, frequently harbor TERT promoter mutations (TPMs) to reactivate the telomerase and exhibit enhanced genomic instability, both of which collectively contribute to tumorigenesis [5]. Telomere maintenance has been implicated in increased cancer stemness [6, 7], proliferation [8] and resistance to radio/chemotherapy [9–11], and hence tumour recurrence in glioma. However, evaluating the therapeutic potential of targeting telomere maintenance in GBM remains poorly explored.
MicroRNAs (miRNAs) are the non-coding RNAs that mediate post-transcriptional gene regulation. miRNAs bind to discrete binding sites on the 3’UTR of cognate target mRNA transcripts. This leads to the formation of RNA-induced silencing complex (RISC) which eventually leads to the degradation of the transcript or repression of its translation [12]. Interestingly, the expression of miRNAs themselves is deregulated in various malignancies including GBM [13, 14]. Generally, the downregulated miRNAs are known to play a tumour-suppressive function and upregulated miRNAs are oncogenic in nature (oncomiRs) [15]. Thus, miRNAs have emerged as novel regulators of gene expression and crucial therapeutic targets/agents.
Earlier, we showed that miR-490 is an epigenetically suppressed miRNA in GBM and that it functions as a tumour-suppressor miRNA by inhibiting the TGF-β pathway-mediated epithelial-to-mesenchymal transition (EMT) [16]. The hallmarks suppressed by miR-490 and recent evidence about the role of TGF-β pathway in telomere dynamics prompted us to investigate its participation in regulation of telomere maintenance [17]. In that context, we report here that miR-490 regulates telomere maintenance in GBM cells. miR-490 was also shown to target various genes (TERF2 of the shelterin complex, TNKS2 and SMG1) that are known to regulate TMM in GBM. Overexpression of miR-490 leads to telomere dysfunction-induced foci formation and DNA damage in GBM cells. miR-490 induces DNA-damage response and the senescence program in GBM cells as well as inhibits stemness. It also leads to the activation of p53 signaling and induction of REST-inhibition response. Thus, overexpression of miR-490 inhibits key hallmarks of telomere maintenance in GBM characteristic of TERF2 inhibition leading to impaired telomere maintenance and activation of corresponding tumour-suppressive events in GBM cells. Thus, targeting TMMs in GBM cells to inhibit its progression via miR-490-mediated TERF2 inhibition could be a viable therapeutic strategy.
Materials and methods
Cell culture
Since the dysregulation of telomere maintenance culminates in apoptosis or senescence mostly regulated by the p53 pathway, GBM cell lines U87MG (wild-type p53) and T98G (p53 mutant c.711G > T) both differing in their p53 status were chosen for analysis. U87MG and T98G cell lines were maintained in DMEM medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin and incubated at 37 ºC and 5% CO2 in a humidified atmosphere in an incubator. The U87MG cell line was procured from the cell line repository at the National centre for cell science (NCCS, Pune, India). The T98G cell line was a kind gift from Dr. Ellora Sen (National brain research centre, Manesar, India).
Transient transfections
Cells were seeded such that they were ~ 80% confluent 24 h post seeding. Cells were transfected with miR-490 overexpressing plasmid PC-490 or its control PCDNA (1.5 µg or 3 µg for 12-well and 6-well plate format, respectively), using Lipofectamine 2000 transfection reagent (Invitrogen, USA) as per manufacturer’s instructions. The cells were processed after 48 h for the quantification of transcript levels using qPCR or performing the cellular assays.
miRNA target prediction and TCGA data analysis
Potential targets of miR-490-3p were predicted using the miRWalk 2.0 database that uses 12 different miRNA target prediction programs [18]. Genes predicted by at least 5 programs were considered as potential targets for further analysis (Supplementary Table 1). Further, shortlisting of the targets was done on the basis of their novelty and involvement in different modes of telomere maintenance. Project Betastasis (https://www.betastasis.com) was used for gene set correlation analysis from the cancer genome atlas (TCGA) GBM patient cohort.
Luciferase assay
The binding sites of miR-490 in the 3’UTR of the target transcripts were obtained using targetscan [19]. The primers flanking the miR-490 binding sites were designed using the gene runner program. The region of 3’UTRs of the selected target genes containing miR-490 binding sites was amplified by PCR. It was then cloned into the luciferase reporter vector (pMIR-REPORT) in the sites for SacI and SpeI downstream of a firefly luciferase gene. pMIR-REPORT vector was a kind gift from Dr. Dipanjan Chowdhury, Dana Farber Cancer Institute, USA. To perform PCR-based site-directed mutagenesis, primers incorporating the indicated mutations in the miR-490-3p-binding site on TERF2 mRNA were designed. The primer sequences are available in Supplementary Table 2. PCR was then performed using these primers and the luciferase construct harboring wild-type binding site as a template. This was followed by DpnI digestion and purification of PCR products to eventually yield a luciferase construct with the mutant binding site. Cells (3 × 104 cells/well of a 24-well plate) were co-transfected with 3’UTR luciferase constructs and miR-490 overexpression vector or its control using Lipofectamine 2000 (Invitrogen). Luciferase activity was quantified with the Dual Luciferase reporter assay (Promega) 48 h post transfection as per the manufacturer’s instructions using FB 12 Luminometer (Berthold).
Total RNA isolation
Total RNA isolation from cell lines either from transfected groups or non-transfected cells was performed 48 h post transfection using an RNA extraction kit (Qiagen) according to the manufacturer’s instructions. The concentration and purity of RNA samples was determined using a NanoDrop spectrophotometer (Thermo fisher scientific).
Reverse transcription and quantitative polymerase chain reaction (qPCR)
Stem-loop Reverse Transcription-PCR was performed using miRNA-specific stem-loop primers to reverse-transcribe miR-490-3p and RNU6-6P (used for normalization). qPCR for miRNAs was performed using miRNA-specific forward primer and a universal reverse primer (Supplementary Table 2). Reverse transcription of mRNAs was performed using Oligo dT primer from iScript cDNA synthesis kit (BioRad). The cDNA prepared as above was further quantified using respective primers in CFX96 real-time system (Biorad) using SsoFast Evagreen Supermix (Biorad). Beta-Actin and RNU6-6P were used as controls for the normalization of mRNAs and miRNAs, respectively. Data were plotted using the 2-ΔΔCt method.
Protein isolation and western blotting
The cells were collected and lysed in non-denaturing lysis buffer at the end of the treatments 48 h post transfection. Protein estimation was done by Bradford assay. For western blot analysis, 30 µg protein was denatured in Laemmli sample buffer and subjected to SDS-PAGE on 8 or 10% Tris–glycine gel. The resolved proteins were transferred on a nitrocellulose membrane and blocked for 2 h at room temperature in 5% skim milk. The membranes were probed with the primary antibodies for TERF2 (Cat. No. 2645, cell signaling technology; 1:1000), phospho-γH2AX (Cat. No. 05–636, Upstate; 1:1000), TP53 (Cat no. 2524S, Cell Signaling Technology; 1:1000), p21 (Cat. No. 2947 T, cell signaling technology; 1:1000), H3k9me3 (Cat. No. 07–442, Upstate; 1:500), SIRT1 (Cat. No. AB12193, Abcam; 1:2000), SOX2 (Cat. No. AB97959, Abcam; 1:1000), SOX4 (Cat. No. AB10537, Abcam; 1:500), SMC1 (Cat. No. A300-055A, Bethyl Laboratories; 1:4000) and Beta-actin (Cat. No. A5316; Sigma-Aldrich; 1:4000) overnight at 4 °C followed by incubation in respective HRP-conjugated secondary antibodies at room temperature for an hour. The ECL (Millipore) detection system was employed for visualization. Beta-actin was used as a loading control.
Preparation of metaphase spreads
For metaphase preparation, cells were treated for 4 h with 0.1 μg/ml Colcemid after respective treatment 48 h post transfection. Cells were then trypsinized and re-suspended slowly in pre-warmed (37 °C) hypotonic solution (0.03 M sodium citrate) for 25 min. Then, cells were fixed in a low volume of fixative solution (3:1 methanol/acetic acid), centrifuged, collected and supernatant was aspirated. Fixative solution was again added while gently vortexing. This was repeated 3 times. Finally, the cells were re-suspended in a small volume of fixative. Glass slides were wet with 45% acetic acid and drained. Cell suspension was added onto glass slides from a maximum distance possible. The slides were dried overnight.
Quantitative fluorescence in situ hybridization (Q-FISH) on metaphase spreads
Slides were fixed with 4% formaldehyde solution in PBS and treated with Pepsin/HCl solution. After washing with PBS, slides were dehydrated with increasing ethanol concentrations (70%, 90%, and 100%) and were dried overnight. Slides were then incubated with Tel-Cy3 PNA telomeric probe mix for 3.5 min at 85 °C followed by 2 h RT incubation in a wet chamber. After the incubation, the slides were washed with wash solution (70% formamide, 10% BSA, 1% 1 M Tris pH 7.2 in water) and then in 0.08% TBS‐Tween. The slides were then incubated in a DAPI bath for 5 min at room temperature followed by Vectashield application. A coverslip was added to each slide and the slides were sealed with nail polish. Slides were stored at 4 °C in the dark before confocal microscope imaging. Six images of the metaphase spreads (random) per slide were obtained using a confocal microscope (Leica TCSSP5) followed by quantification of the telomere probe fluorescent signal.
Preparation of slides for immunofluorescence
Cells were trypsinized 48 h post transfection and the pellet was re-suspended in 10% formaldehyde solution in PBS (pH 7.0) for 20 min at room temperature. Porcine type A gelatin solution (10% gelatin in PBS) was made and added to an equal volume of cell suspension. The solution was gently poured into an Eppendorf tube and allowed to solidify for 5 min at 4 °C. 1 mL of formaldehyde solution was added on top and tubes were stored at 4 °C overnight. The gelatin block was extruded into histopathology cassettes the following day and processed for paraffinization and sectioning.
Immunofluorescence staining
After deparaffinization and citrate antigen retrieval, the slides with gelatin sections were washed in PBS and permeabilized with 0.5% Triton in PBS for 3 h. Followed by washing in PBS again, slides were blocked with 100% FBS for 1 h at room temperature. Respective antibodies were diluted using antibody diluent with background reducing agents and applied onto the slides followed by incubation overnight at 4 °C in a wet chamber. The slides were washed with 0.1% Tween 20 in PBS followed by 1 h incubation with appropriate secondary antibodies. After washing slides again with PBS, the slides were incubated in DAPI bath for 5 min followed by washing with PBS. A coverslip was added after the Vectashield was applied and slides were sealed with nail polish and stored at 4 °C in the dark before imaging.
Telomere dysfunction-induced foci (TIFs) and DNA damage foci analysis
To identify the incidence of DNA damage at telomeres, immunofluorescence staining was performed in cells transfected with either PCDNA or PC-490 48 h post transfection. The slides with gelatin sections were processed for immunofluorescence as described in the immunofluorescence staining section. Antibodies against TERF1 (Cat. No. AB10579, Abcam; 1:100) and 53BP1 (Cat. No. NB100-304, Novus Biologicals; 1:1000) were used as telomere and DNA-damage markers, respectively. Images were obtained using a confocal microscope (Leica TCSSP5). To quantify TIFs, images were analyzed using ImageJ. The fluorescent spots positive for DAPI, TERF1, and 53BP1 were considered as TIFs. The same slides were used to quantify 53BP1 foci. Cells with at least 3 distinct 53BP1 foci against a DAPI background were considered for analysis.
Microarray data acquisition
Two separate datasets based on gene expression by microarray with Accession IDs GSE4290 and GSE118983 were obtained from NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). Data for 77 GBM patients were obtained from GSE4290. Data for differentially expressed genes (DEGs) in response to miR-490 overexpression in GBM cells were obtained from GSE118983.
Downstream analysis of DEGs
GBM patients from GSE4290 were stratified into two groups REST-High and REST-Low based on the expression profile of REST. Gene expression data were downloaded using GEO2R tool and were used to identify significantly altered DEGs among REST-Low vs. REST-High (GSE4290) and miR-490 overexpression vs. control (GSE118983) groups based on p value (p < 0.05). Further, the Panther pathways tool [20] was used to perform the pathway enrichment analysis in the respective DEGs. The top 5 pathways based on number of representative genes were analyzed by intersecting the DEG profiles obtained from these groups. AmiGO gene ontology [21] analysis tool was used to obtain gene entries associated with GO terms ‘senescence’, ‘apoptosis’ and ‘stem cell population maintenance’.
Statistical analysis
All in vitro experiments were performed in three biological replicates with a minimum of two technical replicates. Two-tailed Student’s t test was applied for the calculation of p values using Microsoft excel. Data with p value < 0.05 were considered significant.
Results
miR-490 targets multiple genes involved in telomere maintenance in GBM
Previously, we showed that miR-490 acts as a tumour-suppressor miRNA in GBM by inhibiting GBM proliferation and inducing apoptosis among other hallmarks. Interestingly, these features are characteristic to telomere maintenance as well. First, to investigate the mode of action of miR-490 in the context of telomere maintenance, we analyzed miR-490 targets predicted by at least 5 target prediction algorithms using miRWalk [18] (Supplementary Table 1). We intersected this list with the genes with GO term ‘telomere maintenance’. Out of the 16 genes identified (Supplementary Table 1), we decided to further focus on four genes TERF2, SMG1, TNKS2 and PML (Fig. 1a). These were shortlisted with an attempt to gauge the effect of miR-490 on different modes of telomere maintenance, for instance, direct maintenance (via TERF2 of the shelterin complex) or indirect maintenance (via TNKS2 and SMG1) and based on their novelty. To study the effect of miR-490 on the expression of these genes, we transfected GBM cells with either control PCDNA (mammalian expression vector pcDNA 3.1( +) (without insert)) or PC-490 (pre-miR-490 cloned in pcDNA 3.1( +) for overexpression of miR-490). A qPCR analysis upon miR-490 overexpression revealed significant downregulation of TERF2, TNKS2, and SMG1 indicating that these genes could be direct targets of miR-490 (Fig. 1b). Using Targetscan, we predicted the binding sites of miR-490 in their 3’UTR (Fig. 1c). To investigate direct binding of miR-490 to these predicted sites, we cloned these sites in pMIR-REPORT vector downstream to the firefly luciferase gene. A significant inhibition of luciferase activity was observed upon miR-490 overexpression when TNKS2 and SMG1 WT 3’UTR luciferase constructs were used indicating that TNKS2 and SMG1 could be direct targets of miR-490 (Fig. 1d). Owing to the direct involvement of TERF2 at the telomeres, we further focused on TERF2. To that end, we also performed PCR-based site-directed mutagenesis on miR-490-binding site in TERF2 3’UTR to yield TERF2 Mut UTR luciferase construct. Subsequent luciferase assay showed that miR-490 overexpression significantly downregulated the luciferase activity in the case of TERF2 UTR luciferase construct which was restored significantly when TERF2 mut UTR luciferase construct was used, suggesting that miR-490 brings about downregulation of TERF2 levels through direct binding on its binding site in its 3’UTR (Fig. 1e). We further observed downregulation of TERF2 expression at the protein level after overexpressing miR-490 by western blotting, confirming that miR-490 inhibited the expression of TERF2 in GBM (Fig. 1f). Thus, miR-490 targeted the telomere maintenance program in GBM cells by affecting the levels of TERF2, TNKS2, and SMG1 transcripts. Given the positive correlation in the expression of TERF2, TNKS2, and SMG1 in GBM (Supplementary Fig. 1), miR-490 appears to regulate the telomere maintenance program by multifaceted targeting.
Fig. 1 .
miR-490 targets multiple genes involved in telomere maintenance in GBM. a Pipeline to identify miR-490-3p targets involved in telomere maintenance. b qPCR analysis of target genes TERF2, TNKS2 and SMG1 upon miR-490 overexpression in U87MG and T98G cell lines. c Prediction of binding sites of miR-490 in 3’UTR of candidate genes using Targetscan. d 3’UTR Luciferase dual-reporter assay for constructs harboring 3’UTR of TNKS2 and SMG1 with wild-type miR-490-3p-binding sites. e 3’UTR Luciferase dual-reporter assay for construct harboring 3’UTR of TERF2 with wild-type or mutated miR-490-3p-binding site. f Western blotting for TERF2 upon overexpression of miR-490 in U87MG and T98G cells. Images are representative of western blotting performed as three replicates. For quantitative PCR analysis, actin was used as a control for normalization of mRNA data. Data was plotted using 2-ΔΔCt method. Beta-actin was used as a loading control for western blotting. The graphs represent mean ± SD of at least three independent experiments (*p < 0.05, **p < 0.01)
miR-490 induces telomere fragility in GBM cells
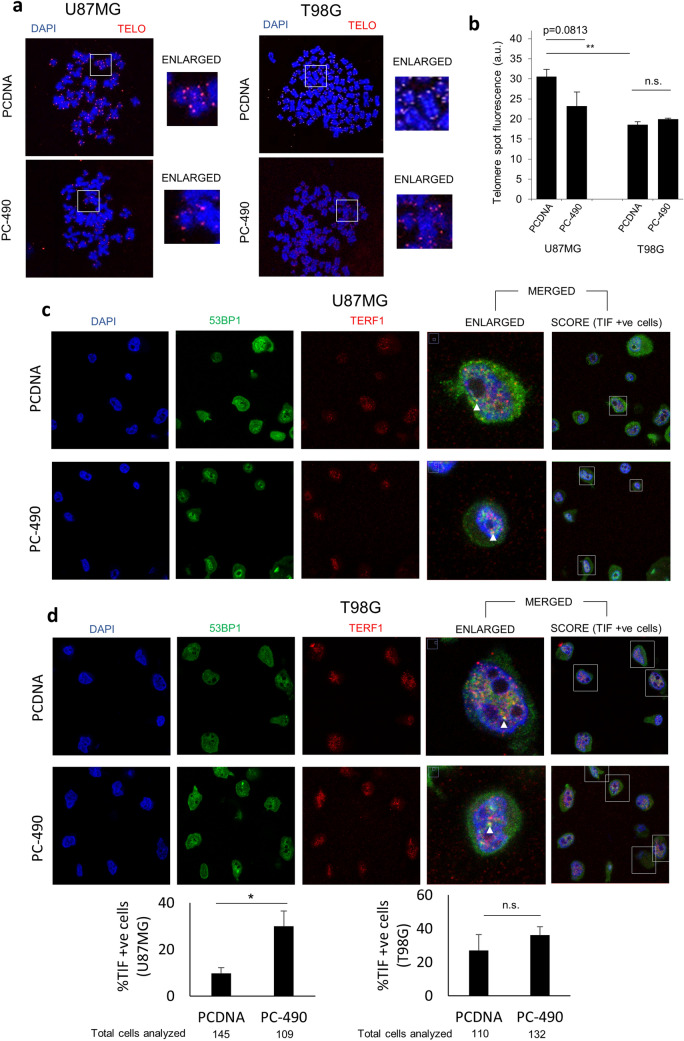
One of the consequences of hampered telomere maintenance is the introduction of telomere fragility. To investigate the effect of miR-490 overexpression on telomere maintenance, we performed quantitative fluorescence in situ hybridization (Q-FISH) on metaphase preparations to measure the relative telomere length in GBM cells transfected with either PCDNA or PC-490 in T98G and U87MG cell lines. A decrease in relative mean fluorescent signal intensity (corresponding to decrease in average telomere length) was observed in U87MG cells overexpressing miR-490. Although the data were not significant (p = 0.08 for U87MG), the trend seems to indicate progressive shortening of telomere length in the U87MG GBM cell line (Fig. 2a). The quantification of the intensities is plotted in Fig. 2b. However, no decrease in relative mean fluorescent signal intensity was observed in the T98G cell line. A comparison between control groups in each cell line indicates shorter telomere length in T98G as compared to U87MG (Fig. 2b). Next, to gauge the effect of miR-490 on telomere DNA damage, we performed telomere DNA dysfunction-induced foci (TIFs) analysis using immunofluorescence staining on interphase samples. We used antibodies against TERF1 (telomere probe) and 53BP1 (pan-nuclear DNA-damage marker) and checked for co-localization of their respective fluorescent signals against a DAPI (nuclear stain) background. It was observed that a higher proportion of cells overexpressing miR-490 exhibited an incidence of TIFs (percentage of cells with TIFs) with statistically significant differences in U87MG cell line but not in T98G cell line (Fig. 2c, d). This could be so owing to the higher incidence of TIFs in the cells with mutated p53 like T98G. Thus, miR-490 overexpression could result in telomere fragility as seen from the incidence of TIF formation and telomere length shortening in GBM cells with wild-type p53 (U87MG cells).
Fig. 2 .
miR-490 induces telomere fragility in GBM cells. a Representative images of quantitative fluorescent in situ hybridization (Q-FISH) using metaphase spreads of U87MG and T98G cells transfected with either PCDNA or PC-490. DAPI and cy3-labeled telomere PNA probe were used as nucleus and telomere stains, respectively. The region highlighted in insets is shown separately as an enlarged image. b Quantification of Q-FISH data represented as average telomere spot fluorescence. Number of spots analyzed for each sample is as follows: U87MG PCDNA (4975 spots), U87MG PC-490 (8257 spots), T98G PCDNA (19,247 spots) and T98G PC-490 (14,812 spots). Representative images for the incidence of telomere DNA damage as telomere dysfunction-induced foci (TIFs) in (c) U87MG and (d) T98G cells upon miR-490 overexpression by immunofluorescence microscopy, with the quantification for both cell lines shown in (d). DAPI was used as a nucleus stain, 53BP1 was used as a pan-nuclear DNA damage marker, and TERF1 was used as a telomere probe. Cells with colocalized fluorescent spots for all three markers were considered as positive for TIFs (highlighted in the insets). Sample TIFs analyzed are marked. Data was represented as the percentage of cells with at least one TIF in each sample group. The graphs represent mean ± SD of at least three independent experiments (*p < 0.05, **p < 0.01)
miR-490 induces DNA damage in GBM
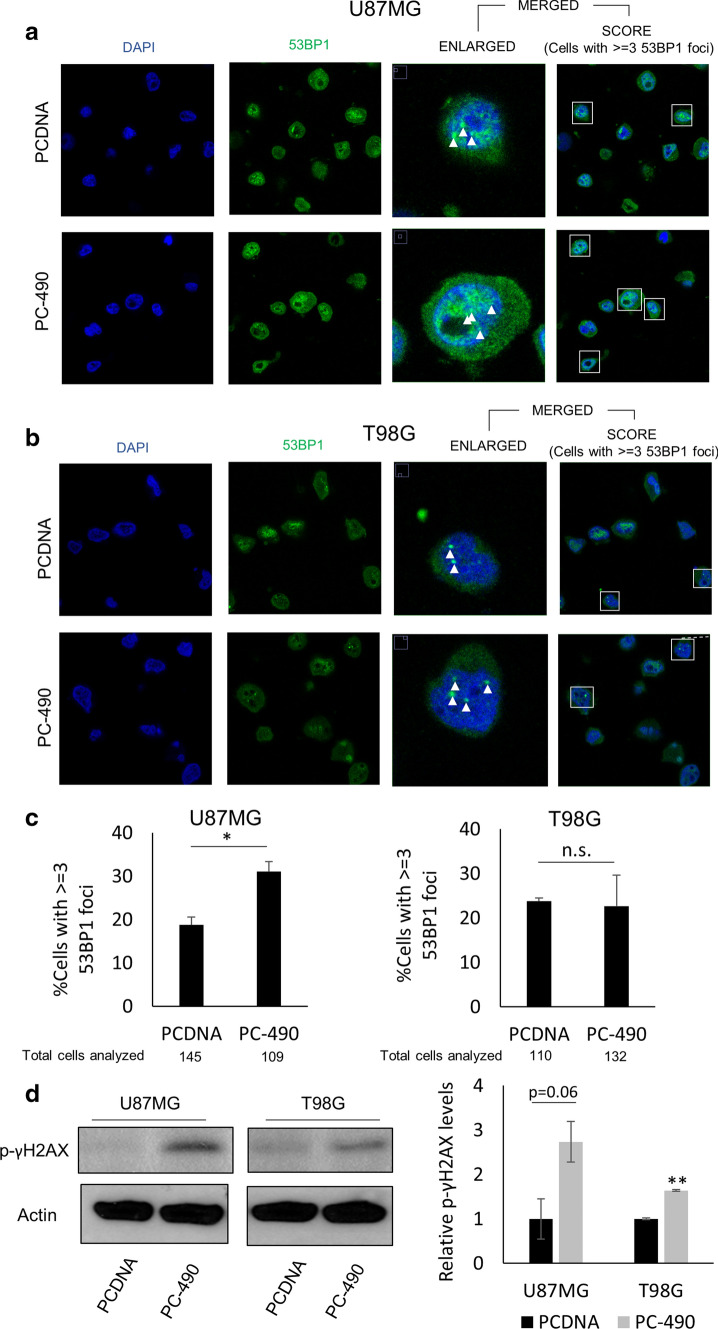
One of the prime functions of shelterin complex proteins apart from maintaining telomere integrity is to prevent the activation of DNA-damage response (DDR) pathways. For instance, it has been shown that upon depletion of the shelterin component TERF2, ATM kinase-mediated DDR signaling is activated, culminating in apoptosis, cell cycle arrest, or senescence via activation of p53 signaling [22]. We checked the onset of DDR by counting DNA damage foci by immunofluorescence staining using 53BP1 as a pan-nuclear DNA-damage marker post transfection with either PCDNA or PC-490. We observed that miR-490 overexpression led to an increase in the proportion of U87MG cells having more 53BP1 foci as compared to the cells transfected with PCDNA (Fig. 3a). Interestingly, T98G cells did not exhibit such response (Fig. 3b). The quantification of the percentage of cells with 3 or more 53BP1 foci for both the U87MG and T98G cell line is shown in Fig. 3c. Further, to validate the significant induction of global DNA damage as seen from 53BP1 staining of single cells, we performed western blotting using bulk cells for another pan-nuclear DNA-damage marker p-γH2AX. Interestingly, it was observed that miR-490 overexpressing cells showed an accumulation of p-γH2AX in both the cell lines (Fig. 3d). Overexpression of miR-490, thus, induced DNA damage in these cells.
Fig. 3 .
miR-490 induces DNA Damage in GBM. Representative images of incidence of global DNA damage in a U87MG and b T98G cells upon miR-490 overexpression using immunofluorescence. DAPI and 53BP1 were used as nucleus stain and pan-nuclear DNA damage marker respectively. Cells with at least three distinct colocalized fluorescent spots for both markers were considered as positive for DNA damage (highlighted in the insets). Sample foci analyzed are marked with a white arrow. c Quantification of DNA damage foci was represented as percentage of cells with at least three 53BP1 foci in each sample group. d Western blotting for pan-nuclear DNA damage marker p-γH2AX upon miR-490 overexpression in U87MG and T98G cell lines. Beta-actin was used as a loading control for western blotting. The graphs represent mean ± SD of at least three independent experiments (*p < 0.05, **p < 0.01)
miR-490 inhibits hallmarks of telomere maintenance
Cellular senescence is the prolonged arrest of cell cycle progression that arises due to various cellular insults including telomere damage. Tri-methylation of Lys9 in Histone H3 (H3K9me3) is considered as an epigenetic marker of senescence in GBM [23, 24]. Also, a member of the sirtuin family of proteins, namely SIRT1, has been shown to suppress senescence in GBM via suppression of CHK2 activation in the ATM kinase-dependent DDR pathway [25]. To investigate the onset of senescence, we thus performed western blotting for H3K9me3 and SIRT1in GBM cells and qPCR analysis for SIRT1 and P53β (P53BETA) after transfection with either PCDNA or PC-490. We observed that upon miR-490 overexpression, a senescence program was activated in the cells as seen by increased H3K9me3 marks and downregulation of SIRT1 (Fig. 4a). qPCR analysis showed downregulation of SIRT1 in T98G cells with no change in P53β transcript levels (Fig. 4b). This indicated that miR-490 overexpression could induce senescence in GBM cells. Interestingly, in support of our observation, the expression of TERF2 is negatively correlated with senescence-promoting genes-D-amino acid oxidase (DAO), Epsin-3 (EPN3) and positively correlated with senescence-inhibiting SIRT1 [26] which prompts towards the regulation of senescence program by miR-490 via TERF2 inhibition (Supplementary Fig. 2).
Fig. 4 .
miR-490 inhibits hallmarks of telomere maintenance. a Western blotting for senescence markers H3K9me3 and SIRT1 upon miR-490 overexpression in U87MG and T98G cell lines. b qPCR analysis of SIRT1 and P53BETA upon miR-490 overexpression in U87MG and T98G cell lines. c Western blotting for stemness markers SOX2 and SOX4 upon miR-490 overexpression in U87MG and T98G cell lines. d qPCR analysis of SOX2, SOX4 and CD133 upon miR-490 overexpression in U87MG and T98G cell lines. For quantitative PCR analysis, actin was used as a control for normalization of mRNA data. Data was plotted using 2-ΔΔCt method. Beta actin was used as a loading control for western blotting. The graphs represent mean ± SD of at least three independent experiments (*p < 0.05, **p < 0.01)
Telomere maintenance in GBM has been well associated with enhanced stemness and targeting shelterin proteins has shown promise in its inhibition [8]. Recent research in the context of stemness in GBM has identified the crucial factors that regulate cell stemness, which is implied in enhanced chemoresistance and tumour recurrence. For instance, SOX2 and SOX4 were identified to promote glioma self-renewal and are widely implicated in stemness [27]. We performed western blotting for stemness genes SOX2 and SOX4 and qPCR analysis for SOX2, SOX4, and CD133 after transfection with either PCDNA or PC-490 to evaluate the link between miR-490-mediated telomere damage and stemness. Interestingly, we observed that miR-490 overexpression brought about downregulation of SOX2 in T98G and SOX4 levels in both T98G and U87MG cells at the protein levels with minimal effects at transcript levels indicating that miR-490 affected the translation of these proteins or their stability (Fig. 4c). qPCR analysis showed inhibition of SOX2 and CD133 in U87MG and T98G cell lines, respectively (Fig. 4d). The more pronounced effect of miR-490 overexpression on stemness genes was observed at the protein level and not at transcript levels, possibly indicating that miR-490 affected the translation or stability of these proteins via other effectors rather than by post-transcriptional regulation. We also compared the genes associated with GO terms ‘senescence’, ‘apoptosis’ and ‘stem cell population maintenance’ with significantly altered DEGs representing miR-490 overexpression obtained from GSE118983 and found many overlapping entries (Supplementary Fig. 3). miR-490 could, thus, be crucial in regulation of these hallmarks of telomere maintenance in GBM.
miR-490 activates downstream p53 signaling pathway
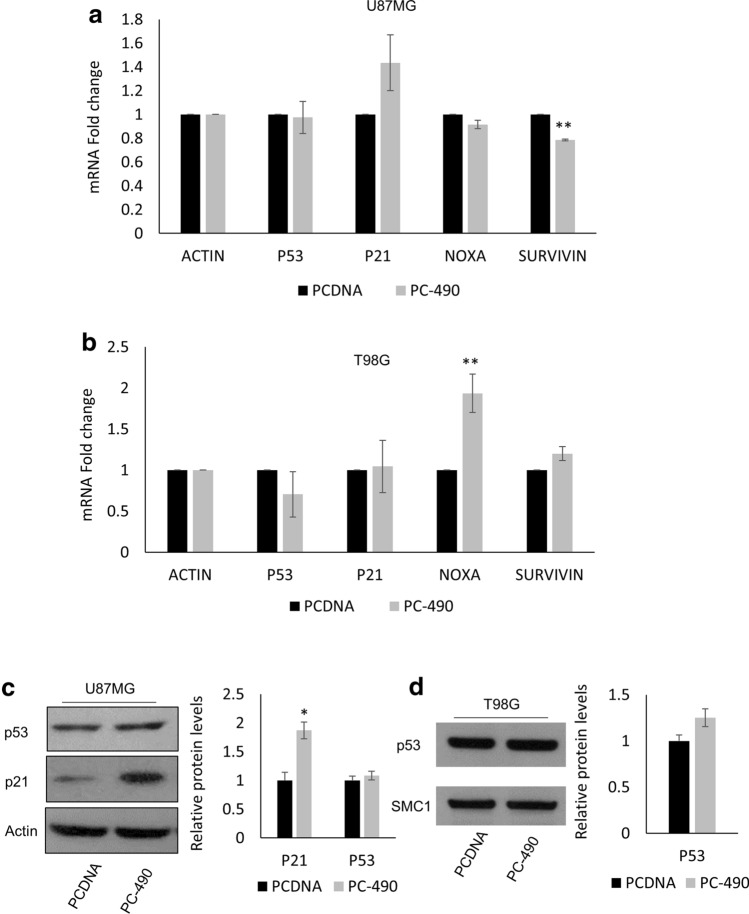
To investigate the pathway activated upon TERF2 depletion and telomere DNA damage instigated by miR-490 overexpression, we hypothesized that miR-490 would activate the p53 signaling pathway since p53 is a well-known master regulator of DNA-Damage Response. To that end, we performed qRT-PCR analysis post transfection with either PCDNA or PC-490. We studied the expression of genes in the p53 pathway including TP53, p21, Survivin and NOXA. Upon miR-490 overexpression in U87MG, we observed a moderate induction in p21 levels (p = 0.1) and a downregulation in Survivin indicating that downstream p53 signaling was activated (Fig. 5a). On the other hand, upon miR-490 overexpression in T98G cells with defective p53 signaling, we did not observe induction in p53 pathway genes with the exception of NOXA (Fig. 5b). Next, we performed western blotting for p53 and p21 after transfection with either PCDNA or PC-490 in the U87MG cell line. Although we did not observe an induction in total p53 protein levels, a significant upregulation was seen in p21 protein levels after miR-490 overexpression in line with the qPCR data (Fig. 5c). Also, in accordance with qRT-PCR data, we did not observe an induction in p53 levels in the T98G cell line (Fig. 5d). Thus, miR-490 overexpression-induced DNA damage and DDR signaling in GBM cells, culminating in the activation of the p53 pathway in cells with wild-type p53.
Fig. 5 .
miR-490 activates downstream p53 signaling. a, b qPCR analysis of p53 pathway genes P53, P21, NOXA, SURVIVIN, upon miR-490 overexpression in U87MG and T98G cell lines, respectively. c Western blotting for p53 and p21 in U87MG cell line upon miR-490 overexpression. d Western blotting for p53 in T98G cell line upon miR-490 overexpression. For quantitative PCR analysis, actin was used as a control for normalization of mRNA data. Data was plotted using 2-ΔΔCt method. Beta-actin or SMC1 was used as a loading control for western blotting. The graphs represent mean ± SD of at least three independent experiments (*p < 0.05, **p < 0.01)
miR-490 mediates a cellular response similar to REST downregulation in GBM
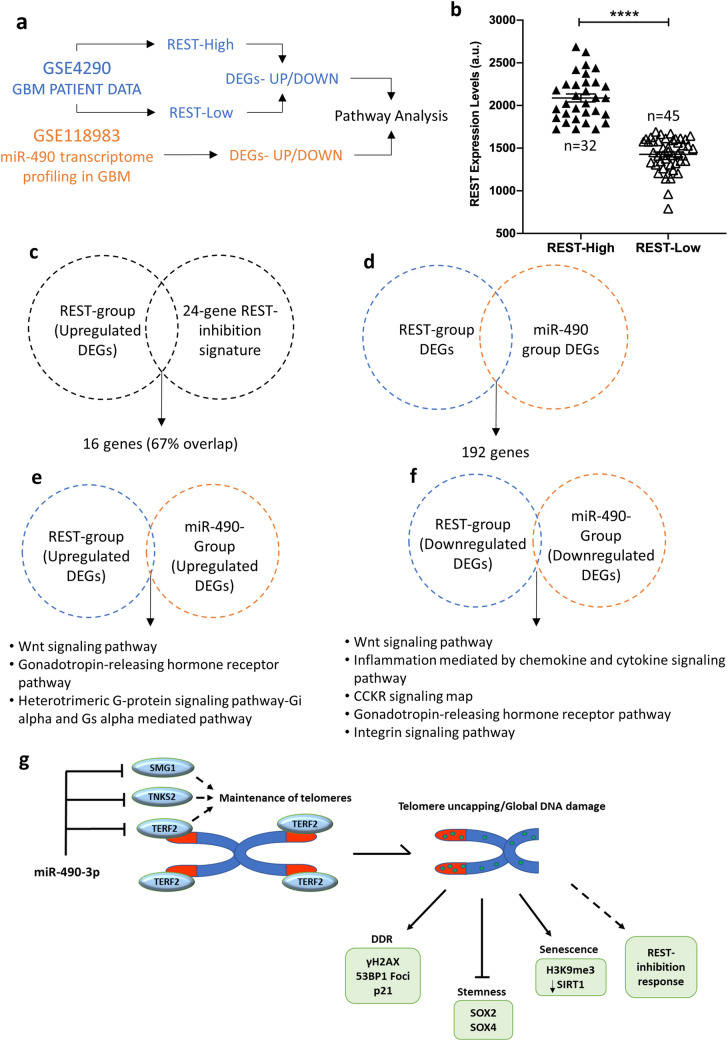
RE1 Silencing Transcription Factor (REST) is a known oncogene that promotes glioma aggressiveness and is associated with a poor prognostic outcome [28]. The association of TERF2 with REST has been shown to promote REST stability and repression of its target genes [29]. We, thus, hypothesized that miR-490, via TERF2 inhibition, led to the activation of transcriptional response similar to REST downregulation. We performed gene expression data analysis using publicly available microarray datasets. A schematic representation of the approach followed is shown in Fig. 6a. First, we downloaded the gene expression microarray data for REST in 77 GBM samples from GEO dataset GSE4290 from NCBI Gene Expression Omnibus database using GEO2R tool. To study differentially expressed genes (DEGs) pertaining to REST, we divided the GBM samples into two groups, namely REST-High (n = 32) and REST-Low (n = 45) on the basis of mean expression of REST. We then compared the expression of REST in these groups (REST-Low vs. REST-High) and found significant difference in REST expression, which validated the stratification approach (Fig. 6b). As a further step of validation, we compared the 24-gene REST signature [30] (genes upregulated upon REST inhibition) with the upregulated DEGs (representative of REST downregulation) from our analysis. Interestingly, we observed a significant overlap (~ 67%; 16 out of 24 genes) indicating the activation of transcriptional response for REST-inhibition (Fig. 6c). Significantly altered DEGs representing REST downregulation (referred to as REST-group; p < 0.05) were, thus, obtained from comparison of REST-Low vs. REST-High groups using GEO2R (Supplementary Table 3). Similarly, we obtained significantly altered DEGs representing miR-490 upregulation (referred to as miR-490 group; p < 0.05) in response to miR-490 overexpression in GBM cells from our GEO dataset GSE118983 (Supplementary Table 3). Importantly, we found 192 common DEGs among REST and miR-490 groups which underlined the regulation of the REST pathway by miR-490 (Fig. 6d). Based on our hypothesis that miR-490 led to REST inhibition via targeting TERF2, we then performed pathway enrichment analysis for these groups of DEGs using the Panther pathways analysis tool and compared the enriched pathways (Supplementary Table 4). The pathways were ranked based on the number of genes represented. Interestingly, we observed that the DEGs represented by miR-490 overexpression were enriched in pathways overlapping with those represented by REST downregulation in GBM (Fig. 6e, f).
Fig. 6 .
miR-490 mediates a cellular response similar to REST downregulation in GBM. a Schematic representation of the approach used for microarray data analysis. b Validation of group stratification (REST-High n = 32 and REST-Low n = 45 groups) based on REST expression analysis in GBM patient data from GSE4290. c Validation of upregulated DEGs against an established 24-gene REST-inhibition signature. d Intersection of significantly altered DEGs among REST and miR-490 groups. e, f Panther Pathway enrichment analysis for upregulated and downregulated DEGs, respectively, in the indicated groups. g Cartoon summarizing the role of miR-490 in regulation of telomere maintenance in GBM by direct targeting of TERF2, TNKS2, and SMG1. miR-490 overexpression lead to telomere and global DNA damage and suppression of telomere maintenance associated hallmarks like senescence induction and inhibition of stemness including activation of p53 signaling and REST-inhibition response
This indicated that miR-490 overexpression in GBM cells activated a cellular response profile that significantly matches that of REST downregulation which could be a viable approach in GBM therapy. A diagram of the proposed model for how miR-490 plays a role in telomere maintenance and senescence is shown in Fig. 6g.
Discussion
Telomere maintenance has been widely implicated in cancer progression. Targeting telomere maintenance has, thus, been suggested as an alternative therapeutic option for GBM management. For instance, targeting shelterin protein TERF1 was shown to inhibit tumour progression in mouse models and patient-derived xenografts [8] whereas targeting adrenocortical dysplasia protein homolog (ACD) enhanced radiosensitivity of GBM cells [9]. Also, downregulation of stemness in glioblastoma stem cells was achieved via inhibition of TERF2 [6]. Likewise, inhibition of hTERT by the means of drugs or RNA-interference has also shown promise by downregulating various hallmarks of glioblastoma [31, 32]. However, the role of miRNAs in the regulation of telomere maintenance is beginning to be explored with promising reports indicating that miRNAs could have therapeutic potential [33].
Several miRNAs have been shown to regulate telomere maintenance via various mechanisms. For instance, miR-155 and miR-23a were shown to induce telomere DNA damage by targeting shelterin components TERF1 and TERF2, respectively [34, 35]. Likewise, miR-21-mediated regulation of hTERT was identified as a mechanism promoting GBM cell growth [36]. Here, we show that miR-490 targeted the telomere maintenance program and its associated hallmarks in GBM via regulating TERF2, TNKS2, and SMG1. Forced expression of miR-490 in vitro in GBM cells induced telomere uncapping and damage along with global DNA damage and downstream p53-related DDR signaling. The miR-490, thus, challenged the telomere maintenance program in GBM cells via downregulation of effectors participating directly (TERF2) or indirectly (TNKS2 and SMG1) in the maintenance of telomeres. However, the role of miR-490 in the regulation of telomere maintenance mechanisms, namely alternative lengthening of telomeres (ALT) and telomerase activation (TA), needs further evaluation [37]. Since this study focuses on the role of miR-490 in vitro, further studies concerning the role of miR-490 in the regulation of telomere maintenance in pre-clinical in vivo mouse models need to be performed.
Recent reports have highlighted the link between the mechanisms of genomic instability/telomere maintenance and EMT in cancer progression [17, 38]. Interestingly, along with other oncogenic effects of REST in glioblastoma [39], it has also been shown to promote EMT [40]. Thus, extra-telomeric binding of TERF2 to stabilize REST [29] could be instrumental in the maintenance of telomeres and EMT. Also, our microarray data analysis highlighted the significant overlap in the transcriptional profiles for miR-490 overexpression and REST-inhibition strategies which further unravels the mode of action of miR-490 through regulation of REST signaling in GBM. However, these findings would need further experimental validation. Apparently, the suppression of miR-490 in GBM could be of prime importance leading to enhanced GBM aggressiveness via the miR-490|TERF2|REST axis. Our earlier observations regarding miR-490-mediated regulation of EMT [16] together with the present study regarding miR-490-mediated telomere maintenance make miR-490 a central miRNA exerting a multi-pronged effect in inhibiting GBM progression. From a therapeutic intervention point of view, activation of miR-490 could be achieved either by targeted delivery of miR-490 mimics or overexpression vector or using a targeted demethylation approach employing CRISPR-dCas9-mediated recruitment of TET1 (ten–eleven translocation methylcytosine dioxygenase), a DNA demethylase. A similar approach has been shown to be effective in the activation of target genes suppressed via DNA methylation [41].
Our observations indicated that miR-490 overexpression leads to the downregulation of TERF2 levels and an increased incidence of telomere uncapping and dysfunction (TIFs) and global DNA damage (53BP1 foci) in U87MG cells. This is consistent with the findings reported earlier regarding the incidence of TIFs and 53BP1 foci upon RNAi-mediated inhibition of TERF2 [42]. The similarity in the percentage of cell populations exhibiting TIFs and 53BP1 foci incidence further highlights the efficacy of miR-490 in initiating telomere specific DNA damage and concurrent formation of 53BP1 recruitment to DNA damage lesions (global DNA damage) in GBM cells. Furthermore, we did not observe significant induction of TIFs or 53BP1 foci in the T98G cell line. One of the factors responsible for these observations could be the p53 mutation in T98G cells, as such cells are known to exhibit an inherently higher incidence of TIFs [43] and an improper recruitment of 53BP1 to DNA damage sites [44, 45]. However, 53BP1 recruitment in the p53 mutant background of T98G cells should be further explored. The resilience of T98G cells to miR-490 overexpression-mediated telomere length shortening could also be in part attributed to shorter telomere lengths. Nevertheless, the induction of the DNA-damage response was observed in these cells after miR-490 overexpression as seen by an accumulation of p-γH2AX. This observation reiterates the shift in the choice of DNA damage pathway even in the presence of the p53 defect in such cells [45].
In glioma, the inhibition of TERF2 has been shown to suppress tumorigenicity, chemoresistance, and proliferative ability of glioblastoma stem cells (GSCs) [6]. Also, treatment of GSCs with the G-quadruplex (G4s)-stabilizing agent telomestatin leads to the dissociation of TERF2 from telomeres, the induction of TIFs, and the consequent DNA damage response and growth inhibition of GSCs [46]. miR-490 replacement could, thus, be of vital importance in the GBM stem cell niche owing to the importance of TERF2 in GBM stem cells. Other than shelterin proteins, SMG1 and TNKS2 also regulate telomere maintenance. SMG1 has been shown to participate in the maintenance of telomeres and regulate the recruitment of TElomeric Repeat-containing RNA (TERRA) [47, 48]. Interestingly, TERRA levels show grade-dependent expression in glioma with the highest expression in GBM, highlighting the relevance of SMG1 in telomere maintenance in GBM [49]. Depletion of SMG1 promotes spontaneous DNA damage and activation of the ATM pathway [50], and induction of senescence [51]. TNKS2, on the other hand, is involved in the maintenance of telomere length [52]. Also, TNKS2, along with TNKS1, has been shown to mediate the DNA damage response (DDR), and depletion of TNKS2 resulted in hampered recruitment of BRCA1 to DNA damage lesions and faulty repair [53]. Thus, in line with these observations, the tumour-suppressive effects of miR-490 in the context of telomere maintenance could be in part via direct targeting of TERF2, SMG1, and TNKS2.
Overall, we show that telomere fragility and the consequent DNA damage response inflicted in GBM cells upon miR-490 overexpression result in the inhibition of aggressive GBM traits. The miR-490 highlights the sensitivity and resistance of GBM cells harboring wild-type and mutant p53, respectively, for the induction of DNA damage. Epigenetic suppression of miR-490 in early grades of glioma could be coinciding with activation of telomere maintenance during gliomagenesis and progression. miR-490, thus, appears to regulate the telomere maintenance program in GBM owing to its potential to target multiple genes involved. This feature could be harnessed to develop a novel therapeutic approach for GBM by targeting telomere maintenance via forced expression of miR-490. Overall, miR-490 overexpression may serve as a therapeutic strategy for GBM by not only preventing cell proliferation and migration as previously reported [16], but also by promoting DNA damage and limiting telomere maintenance, leading to increased apoptosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
RK is thankful to the IIT Delhi internal grant for funding this study. OSV thanks the Ministry of Human Resource and Development (MHRD), Govt. of India for Senior Research Fellowship and European Molecular Biology Organization (EMBO) for a Short-Term Fellowship. The authors also thank Miguel Jiménez for help with cell culture and the CNIO histopathology and confocal microscopy cores.
Author contributions
RK and MAB conceived the project. RK supervised the whole study. MAB hosted OSV for an EMBO fellowship and supervised the study pertaining to telomere research at CNIO, Spain. OSV and RK designed the experiments. OSV performed all the experiments. DK assisted in preparation of luciferase assay constructs. KW mentored OSV in performing experiments pertaining to telomere research at CNIO, Spain. RK and OSV wrote the manuscript. KW and MAB reviewed the manuscript.
Funding
IIT Delhi Internal Research Grant, Senior Research Fellowship, EMBO Short term fellowship.
Data availability
Upon reasonable request.
Compliance with ethical stanadrds
Conflicts of interest
The authors declare no conflict of interest).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 2.Gaspar TB, Sá A, Lopes JM, et al. Telomere maintenance mechanisms in cancer. Genes (Basel) 2018;9(5):241. doi: 10.3390/genes9050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whittemore K, Vera E, Martínez-Nevado E, et al. Telomere shortening rate predicts species life span. Proc Natl Acad Sci USA. 2019;116:15122–15127. doi: 10.1073/pnas.1902452116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Lange T. Activation of telomerase in a human tumor. Proc Natl Acad Sci USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba K, Lorbeer FK, Shain AH, et al. Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science. 2017;80(357):1416–1420. doi: 10.1126/science.aao0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai Y, Lathia JD, Zhang P, et al. Molecular targeting of TRF2 suppresses the growth and tumorigenesis of glioblastoma stem cells. Glia. 2014;62:1687–1698. doi: 10.1002/glia.22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki T, Pan Y, Joshi K, et al. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin Cancer Res. 2012;18:1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bejarano L, Schuhmacher AJ, Méndez M, et al. Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell. 2017;32:590–607.e4. doi: 10.1016/j.ccell.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Polito F, Cucinotta M, Abbritti RV, et al. Silencing of telomere-binding protein adrenocortical dysplasia (ACD) homolog enhances radiosensitivity in glioblastoma cells. Transl Res. 2018;202:99–108. doi: 10.1016/j.trsl.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Gao K, Li G, Qu Y, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7(8):8712–8725. doi: 10.18632/oncotarget.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo Y, Kondo S, Tanaka Y, et al. Inhibition of telomerase increases the susceptibility of human malignant glioblastoma cells to cisplatin-induced apoptosis. Oncogene. 1998;16:2243–2248. doi: 10.1038/sj.onc.1201754. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.ZHANG Y, CRUICKSHANKS N, PAHUSKI M, et al (2017) Noncoding RNAs in Glioblastoma. In: Glioblastoma. Codon Publications, pp 95–130 [PubMed]
- 14.Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem Biophys Res Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs–MicroRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Vinchure OS, Sharma V, Tabasum S, et al. Polycomb complex mediated epigenetic reprogramming alters TGF-β signaling via a novel EZH2/miR-490/TGIF2 axis thereby inducing migration and EMT potential in glioblastomas. Int J Cancer. 2019;145(5):1254–1269. doi: 10.1002/ijc.32360. [DOI] [PubMed] [Google Scholar]
- 17.Mazzolini R, Gonzàlez N, Garcia-Garijo A, et al. Snail1 transcription factor controls telomere transcription and integrity. Nucleic Acids Res. 2018;46:146–158. doi: 10.1093/nar/gkx958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dweep H, Gretz N. MiRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carbon S, Ireland A, Mungall CJ, Shu ShengQiang, Marshall B, Lewis S, the AmiGO Hub and the WPWG, AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlseder J, Broccoli D, Yumin D, et al. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;80(283):1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 23.Sidler C, Kovalchuk O, Kovalchuk I. Epigenetic regulation of cellular senescence and aging. Genet: Front; 2017. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeon HY, Kim JK, Ham SW, et al. Irradiation induces glioblastoma cell senescence and senescence-associated secretory phenotype. Tumor Biol. 2016;37:5857–5867. doi: 10.1007/s13277-015-4439-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Feng Y, Guo Q, et al. SIRT1 modulates cell cycle progression by regulating CHK2 acetylation−phosphorylation. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmeer K, Wengerodt, , et al. Dissecting aging and senescence—current concepts and open lessons. Cells. 2019;8:1446. doi: 10.3390/cells8111446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikushima H, Todo T, Ino Y, et al. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Wang Z, Tang X, et al. Molecular mechanisms and potential prognostic effects of REST and REST4 in glioma (Review) Mol Med Rep. 2017;16:3707–3712. doi: 10.3892/mmr.2017.7071. [DOI] [PubMed] [Google Scholar]
- 29.Zhang P, Pazin MJ, Schwartz CM, et al. Nontelomeric TRF2-REST interaction modulates neuronal gene silencing and fate of tumor and stem cells. Curr Biol. 2008;18:1489–1494. doi: 10.1016/j.cub.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagoner MP, Gunsalus KTW, Schoenike B, et al. The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 2010;6:1–12. doi: 10.1371/journal.pgen.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallini R, Sorrentino A, Pierconti F, et al. Telomerase inhibition by stable RNA interference impairs tumor growth and angiogenesis in glioblastoma xenografts. Int J cancer. 2006;118:2158–2167. doi: 10.1002/ijc.21613. [DOI] [PubMed] [Google Scholar]
- 32.Lavanya C, Venkataswamy MM, Sibin MK, et al. Down regulation of human telomerase reverse transcriptase (hTERT) expression by BIBR1532 in human glioblastoma LN18 cells. Cytotechnology. 2018;70:1143–1154. doi: 10.1007/s10616-018-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santambrogio F, Gandellini P, Cimino-Reale G, et al. MicroRNA-dependent regulation of telomere maintenance mechanisms: a field as much unexplored as potentially promising. Curr Pharm Des. 2014;20:6404–6421. doi: 10.2174/1381612820666140630095918. [DOI] [PubMed] [Google Scholar]
- 34.Dinami R, Ercolani C, Petti E, et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014;74:4145–4156. doi: 10.1158/0008-5472.CAN-13-2038. [DOI] [PubMed] [Google Scholar]
- 35.Luo Z, Feng X, Wang H, et al. Mir-23a induces telomere dysfunction and cellular senescence by inhibiting TRF2 expression. Aging Cell. 2015;14:391–399. doi: 10.1111/acel.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang YY, Sun G, Luo H, et al. Mir-21 modulates htert through a stat3-dependent manner on glioblastoma cell growth. CNS Neurosci Ther. 2012;18:722–728. doi: 10.1111/j.1755-5949.2012.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naderlinger E, Holzmann K. Epigenetic regulation of telomere maintenance for therapeutic interventions in gliomas. Genes (Basel) 2017;8(5):145. doi: 10.3390/genes8050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Badawy A, Ghoneim NI, Nasr MA, et al. Telomerase reverse transcriptase coordinates with the epithelial-to-mesenchymal transition through a feedback loop to define properties of breast cancer stem cells. Biol Open. 2018;7(7):bio034181. doi: 10.1242/bio.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamal MM, Sathyan P, Singh SK, et al. REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells. 2012;30:405–414. doi: 10.1002/stem.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang J, Meng Q, Zhao W, et al. An expression based REST signature predicts patient survival and therapeutic response for glioblastoma multiforme. Sci Rep. 2016;6:34556. doi: 10.1038/srep34556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Tao Y, Gao X, et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016;2:16009. doi: 10.1038/celldisc.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takai H, Smogorzewska A, De Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/S0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 43.Tutton S, Lieberman PM. A role for p53 in telomere protection. Oncol: Mol Cell; 2017. p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suchánková J, Legartová S, Ručková E, et al. Mutations in the TP53 gene affected recruitment of 53BP1 protein to DNA lesions, but level of 53BP1 was stable after γ-irradiation that depleted MDC1 protein in specific TP53 mutants. Histochem Cell Biol. 2017;148:239–255. doi: 10.1007/s00418-017-1567-3. [DOI] [PubMed] [Google Scholar]
- 45.Moureau S, Luessing J, Harte EC, et al. A role for the p53 tumour suppressor in regulating the balance between homologous recombination and non-homologous end joining. Open Biol. 2016;6(9):160225. doi: 10.1098/rsob.160225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa D, Okabe S, Okamoto K, et al. G-quadruplex ligand-induced DNA damage response coupled with telomere dysfunction and replication stress in glioma stem cells. Biochem Biophys Res Commun. 2016;471:75–81. doi: 10.1016/j.bbrc.2016.01.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azzalin CM, Reichenbach P, Khoriauli L, et al. Telomeric repeat-containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;80(318):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 48.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2009;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 49.Sampl S, Pramhas S, Stern C, et al. Expression of telomeres in astrocytoma WHO grade 2 to 4: TERRA level correlates with telomere length, telomerase activity, and advanced clinical grade. Transl Oncol. 2012;5:56–65. doi: 10.1593/tlo.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brumbaugh KM, Otterness DM, Geisen C, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Crutchley J, Zhang D, et al. Identification of a DNA damage–induced alternative splicing pathway that regulates p53 and cellular senescence markers. Cancer Discov. 2017;7:766–781. doi: 10.1158/2159-8290.CD-16-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhardwaj A, Yang Y, Ueberheide B, Smith S. Whole proteome analysis of human tankyrase knockout cells reveals targets of tankyrase-mediated degradation. Nat Commun. 2017;8(1):2214. doi: 10.1038/s41467-017-02363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy Z, Kalousi A, Furst A, et al. Tankyrases promote homologous recombination and check point activation in response to DSBs. PLoS Genet. 2016;12(2):e1005791. doi: 10.1371/journal.pgen.1005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Upon reasonable request.