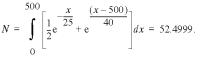Abstract
The vast body of Expressed Sequence Tag (EST) data in the public databases provide an important resource for comparative and functional genomics studies and an invaluable tool for the annotation of genomic sequences. We have developed a rigorous protocol for reconstructing the sequences of transcribed genes from EST and gene sequence fragments. A key element in developing this protocol has been the evaluation of a number of sequence assembly programs to determine which most faithfully reproduce transcript sequences from EST data. The TIGR Gene Indices constructed using this protocol for human, mouse, rat and a variety of other plant and animal models have demonstrated their utility in a variety of applications and are freely available to the scientific research community.
INTRODUCTION
Our efforts to catalog the collection of human genes are progressing rapidly. Although both public and private efforts have greatly accelerated the pace of human genome sequencing, annotation of the genome, including identification of the gene sequences, remains a significant challenge. Expressed Sequence Tag (EST) sequences represent the most extensive available survey of the transcribed portion of the genome. ESTs are single pass, partial sequencing reads generated from either the 5′- or the 3′-end of a cDNA clone (1). There are >4 000 000 ESTs in GenBank, nearly two-thirds of which are human (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html ). ESTs have proven to be an indispensable tool for the identification of expressed genes (2) and for genomic mapping (3,4).
There have been a number of attempts to identify unique genes represented by EST data (5). UniGene (6) uses pairwise sequence comparisons at various levels of stringency to group related sequences, placing closely related and alternatively spliced transcripts into clusters. The TIGR Gene Indices described here use assembly algorithms, rather than clustering, to produce tentative consensus (TC) sequences that represent the underlying mRNA transcripts (7). This has several advantages: it separates closely related genes into distinct consensus sequences; it separates splice variants; it produces longer representations of the underlying gene sequences. The resulting TCs can be used for eukaryotic genome sequence annotation (8,9), integration of complex mapping data and identification of orthologous genes (I.Holt, F.Liang, G.Pertea, S.Karamycheva and J.Quackenbush, submitted for publication). However, the quality and utility of the assembled sequences relies on the ability of sequence assembly programs to effectively generate high fidelity consensus sequences from the available EST data.
Among the assembly programs that have been developed for genomic sequencing projects, the most extensively used are Phrap (http://www.phrap.org/phrap.docs/phrap.html ), CAP3 (10) and TIGR Assembler (11). The version of TIGR Assembler now in use for genomic sequence assembly has been modified significantly from the original, which was optimized for assembly of EST sequences. In this manuscript we refer to the original version as TA-EST and the modified version, optimized for genomic assembly, as TIGR Assembler. While all of these have proven their utility in assembling genomic shotgun sequencing data, EST sequences present a number of distinct computational problems for an assembler. In genomic shotgun sequencing, which typically uses a single clone for the source DNA, sequences sharing <98% identity can be assumed to come from different copies of a repetitive sequence element. In contrast, EST data are derived from a wide variety of sources representing the spectrum of polymorphisms in the original samples. This is compounded by sequencing errors inherent in single pass sequencing, including a relatively high rate of insertions and deletions, contamination by vector and linker sequences and the non-random distribution of sequence start sites in oligo(dT)-primed libraries. Therefore, the degree of identity in overlapping sequences from the same gene will often be lower than in genomic projects. In addition, the patterns of overlapping sequences caused by alternative transcripts are different from that observed in a genomic shotgun project. Finally, gene sequences in GenBank and the ESTs in dbEST lack the base call quality values that most assembly programs now use as part of the assembly process. Sequence chromatograms can be obtained for approximately half of the nearly 2 000 000 human ESTs from the Washington University ftp site and quality values can be derived for these sequences. This information is not available for the remaining ESTs and for all of the gene sequences. Of the four programs we evaluated, only TIGR Assembler is capable of assembling a mix of sequences with and without quality values.
Using the >118 000 rat ESTs in dbEST as a model, we evaluated Phrap, CAP3, TA-EST and TIGR Assembler to determine which program most faithfully assembles ESTs to produce TC sequences, to compare the number of TCs and singletons produced and to evaluate the relative performance of the algorithms. In this comparison we have focused on a number of known genes. As there are a number of potential difficulties in working with available EST data, including the presence of undetected gene families and variable error rates, we augmented our studies using simulated sequences designed to model known sequencing errors (12) or ESTs transcribed from closely related genes. Finally, we validated our simulation results by assembling ESTs derived from 73 known, annotated genes in GenBank. In our analysis we have found that CAP3 consistently out-performs the other programs, producing the fewest high quality assemblies from single genes while being tolerant to random errors yet maintaining the ability to discriminate between related genes; we have adopted CAP3 as the assembler for the TIGR Gene Indices.
We used CAP3 to construct the most recent release of the Human Gene Index (HGI) (9), which is based on 1 610 947 human ESTs, 47 283 human sequences derived from CDS features in GenBank (we refer to these as NP, for NucProt, sequences) and 7223 curated expressed transcript (ET) sequences from the TIGR EGAD database (http://www.tigr.org/tdb/egad/egad.html ). Using the 52 825 ESTs that have been mapped by the International Radiation Hybrid Mapping consortium (13), we were able to assign map locations to >40% of the tentative human consensus (THC) sequences. While adding significant value to the HGI, this mapping information also serves to validate the assemblies, as THCs containing multiple, independently mapped markers almost invariably map to consistent locations within the genome.
MATERIALS AND METHODS
Rat Gene Index assembly
Rat EST sequences were downloaded from dbEST. These were trimmed to remove vector sequences, poly(A/T) tails, adaptor sequences and contaminating bacterial sequences. The cleaned ESTs were clustered by comparing all pairs using WU-BLAST (http://blast.wustl.edu ) (14) and collecting those with ≥95% identity over regions at least 40 bp in length with unmatched overhangs <20 bp. The sequences comprising each cluster were assembled using Phrap (v.990315), CAP3, TIGR Assembler and TA-EST and the results from the independent assemblies were compared.
Modeling error rates for EST sequence assembly
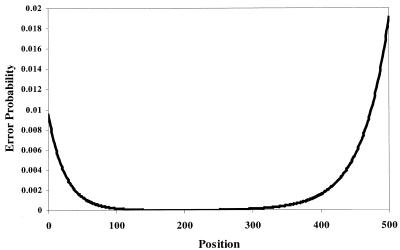
Errors produced during automated DNA sequencing are non-uniformly distributed and tend to be concentrated at the beginning and end of the sequence read (12). To assess the effects of sequencing errors we used a 600 base segment of a reference sequence (ECA1, GenBank accession no. U96455) to model the distribution of sequence start positions and errors in EST data. From the reference sequence a set of fragment sequences ranging from 450 to 550 bases in length was generated. Sequencing errors with a pattern similar to that previously reported (12) were introduced as substitutions, insertions or deletions with a 3:1:1 ratio at positions selected using the normalized probability density model of the form (see Fig. 1):
Figure 1.
DNA sequencing base call error probability. Error probability distribution adapted from Ewing and Green (12) used to simulate systematic base call errors.
where x is the position along the length of the sequence read and N is a normalization constant equal to:
Total sequencing error rates ranging from 1 to 8% (5–40 errors/sequence) and sequence coverages ranging from 5- to 50-fold were simulated. Model sequences were generated and each assembly program was used to independently assemble the sequences. The numbers of contiguous assemblies and singletons were recorded. The best consensus sequence produced by each program was compared with the original sequence and its fidelity was assessed using an assembly score (A-score) defined by
A-score = (2 × sequence length) – (15 × no. of insertions) – (15 × no. of deletions) – (5 × no. of substitutions),
where a perfect assembly would have an A-score of 1200 for our 600 base test sequence.
Assembly of gene families
Gene families were modeled by taking an 1800 bp segment of the ECA1 gene and introducing substitutions at random positions, generating eight sequences that were 99, 98, 97, 96, 95, 94 and 90% identical to the original. For each of the eight family members, the gene sequence was artificially shotgunned, creating 5-fold coverage of EST fragments 450–550 bp in size. Two pools of EST sequences were created, one with all eight sequences and a six sequence family containing only those ≥95% identical to the reference sequence. Each family was assembled using the four assembly programs; the consensus assemblies were evaluated by comparing them with each member of the gene family. Six independent simulations were conducted.
Assembly and evaluation of representative human genes
A set of 73 representative human genes was selected based on their EST content. EST sequences representing each gene were assembled using Phrap, CAP3, TA-EST and TIGR Assembler; the parent gene sequences were not included in the assembly process. For each gene the longest consensus sequence produced by each assembler was compared to the original gene sequence in order to assess consensus quality; the errors in the consensus sequences were tabulated and classified and a normalized A-score was calculated for each program by summing the A-scores for each gene and dividing by the total sequence length. For a perfect sequence reconstruction the normalized A-score would be 2.0.
HGI assembly and analysis
Human EST and coding gene sequences were downloaded from dbEST and GenBank records and cleaned using the same filters as were used for rat. Cleaning eliminated 82 228 (5.1%) of the original 1 610 947 ESTs and trimmed an additional 8 350 769 bases of contaminating sequence. A total of 54 506 human gene sequences were included: 47 283 human transcripts (NP sequences) parsed through Entrez from the CDS and CDS-join features in GenBank records and 7223 curated ET sequences from the TIGR EGAD database (http://www.tigr.org/tdb/egad/egad.html ). Sequences were compared using FLAST, a rapid sequence comparison program based on DDS (15) in which query sequences are first concatenated and then searched against a nucleotide database. Sequences were placed in clusters using criteria identical to those used for rat EST clustering. Sequences in each cluster were assembled using CAP3. A THC sequence containing a known gene was assigned the function of that gene; THCs without assigned functions were searched using DPS (15) against a non-redundant protein database; high scoring hits were assigned a putative function. The THC sequences were assigned map locations using the most recent data from the International Radiation Hybrid Mapping Consortium (13). EST mapping information was downloaded via ftp from the NCBI (ftp://ftp.ncbi.nlm.nih.gov/repository/genemap ) and map locations assigned using Greg Schuler’s e-PCR program (16).
RESULTS
Incorporation of EST sequences into TC assemblies
Construction of the TIGR Gene Indices relies on faithfully clustering and assembling sequences, so ESTs from the same transcript are properly assembled while ESTs from distinct but closely related transcripts are appropriately placed into separate assemblies. Sequences that do not fit into any assemblies are called singletons. The number of singletons provides an estimate of the number of rare transcripts represented in the data; to avoid overestimates, assemblers must be fairly tolerant of sequencing errors so as to not produce an excessive number of singletons.
Following sequence cleaning, pairwise comparisons placed 118 473 Rat ESTs in 16 183 clusters. The sequences comprising each cluster were assembled using Phrap, CAP3, TA-EST and TIGR Assembler, respectively, using each program’s default parameters. Following assembly, the number of consensus sequences and singletons was tabulated, as shown in Table 1. While each program produces approximately the same number of assemblies, TA-EST gives nearly 20 times the number of singletons produced by CAP3 or Phrap, suggesting that it is much less tolerant of sequence discrepancies. This observation is further supported by the slightly larger number of TCs generated from high coverage clusters, suggesting that it is also more likely to split sequence contigs when sequencing errors occur.
Table 1. Summary of TC and singletons of rat EST clusters using Phrap, CAP3 and TA-EST.
| Phrap | CAP3 | TA-EST | TIGR Assembler | |
|---|---|---|---|---|
| TCs | 16 635 | 16 647 | 16 977 | 17 653 |
| Singletons | 121 | 138 | 2751 | 7540 |
| Total | 16 756 | 16 785 | 19 728 | 25 193 |
Much of the difference between Phrap and CAP3 can be attributed to large clusters containing tens or hundreds of sequences. These present a unique challenge to the assembly programs because they contain many closely related sequences that must be correctly assembled, despite sequencing errors and polymorphisms inherent in the data. The results for four representative clusters are presented in Table 2. Again, we can see clear differences between the programs, indicating that TA-EST and TIGR Assembler are far less error tolerant than Phrap and CAP3. This analysis at first glance suggests that Phrap may be better at assembling ESTs containing errors. However, as described below, Phrap tends to misassemble sequences and to produce low fidelity consensuses containing many insertions and miscalled bases.
Table 2. Contigs and singletons produced by Phrap, CAP3, TA-EST and TIGR Assembler for four representative ‘large’ clusters of rat sequences.
| Cluster (no. of ESTs) | Contigs/singletons produced | |||
|---|---|---|---|---|
| Phrap | CAP3 | TA-EST | TIGR Assembler | |
| 1. (135) | 11/0 | 17/0 | 21/43 | 25/41 |
| 2. (270) | 25/1 | 35/1 | 37/125 | 42/126 |
| 3. (540) | 1/0 | 1/0 | 2/2 | 3/12 |
| 4. (1791) | 15/0 | 18/7 | 28/62 | 71/229 |
| Total | 52/1 | 61/8 | 88/232 | 141/408 |
Consensus assessment
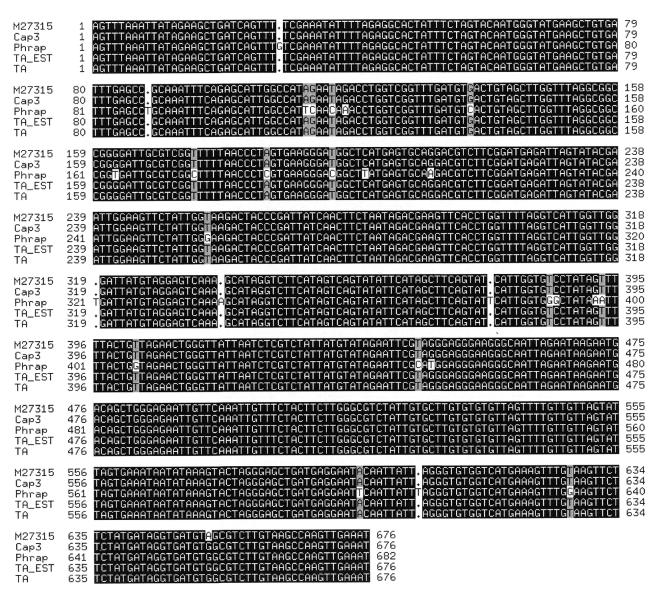
While clustering alone can provide an estimate of the number of genes represented in an EST database, the construction of TCs has a number of advantages. For example, each TC sequence tends to be longer than its component ESTs, facilitating functional assignment, transcript mapping and genomic sequence annotation. The utility of TC sequences depends critically on the fidelity of the consensus produced. To evaluate the quality of this consensus for each assembly program, we used ESTs from known, annotated genes and compared the consensus sequences produced by each program to the reference sequence. An example, representing the single copy cytochrome c oxidase subunit II gene of the rat mitochondrial genome (GenBank accession no. M27315), is shown in Figure 2 (this corresponds to Cluster 3 in Table 2). Analysis of the alignment in Figure 2 shows that CAP3, TA-EST and TIGR Assembler were all able to accurately reproduce the reference sequence (modulo one consistent difference that suggests an error or polymorphism in the GenBank sequence). However, while CAP3 was able to use all the sequence data to produce a single consensus, TA-EST and TIGR Assembler used some of the lower quality sequences to produce a second consensus that spans only part of the reference sequence and that differs from it by 9.3% (data not shown). Phrap assembled all of the ESTs into a single consensus, but the resulting sequence contains a large number of insertions and other errors, representing a 5% error rate. While Phrap has been shown to produce accurate consensus sequences for genomic sequencing projects, the lack of quality values for EST sequences appears to have a significant adverse effect on its output. Further analysis, described below, suggests that Phrap also over-assembles sequences, combining ESTs from distinct transcribed genes into single consensus sequences. However, in genomic sequence assembly using quality values CAP3 has been demonstrated to produce fewer errors than Phrap (10).
Figure 2.
CLUSTAL W (17) alignment of consensus sequence assemblies for the rat cytochrome c oxidase gene produced by Phrap, CAP3, TA-EST and TIGR Assembler.
Effects of sequencing errors on EST assembly
The errors generated in automated DNA sequencing are known to be concentrated at the start and end of the sequence read (12). In genomic sequence assembly this is mitigated by the random distribution of sequence start points. The situation is quite different for ESTs. cDNA clones are constructed from polyadenylated mRNA using oligo(dT) to prime reverse transcription first-strand DNA synthesis. Consequently, clone ends and 3′-EST sequences start from the same position (or nearly so) and errors, while independent, are positionally clustered. The existence of these correlated errors can have a significant impact on EST assembly; assembly programs must effectively handle this in order to generate high fidelity contigs and an accurate estimate of the number of transcripts represented within the data.
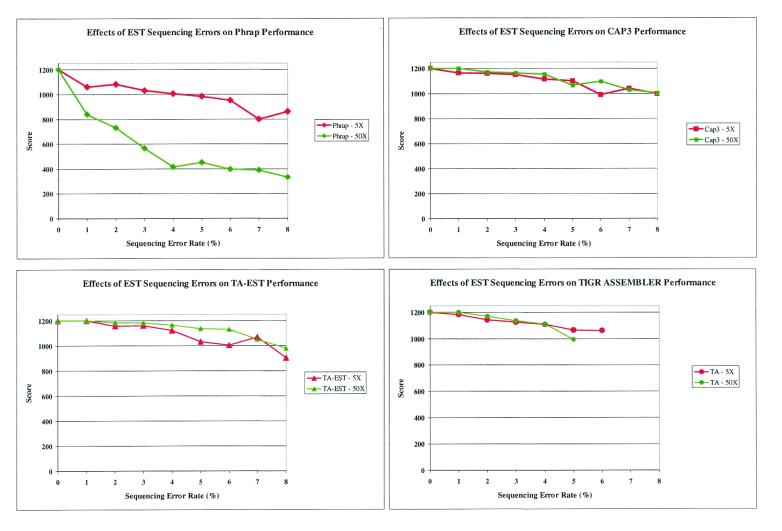
To systematically assess the relative performance of the various assembly programs we generated model EST data with lengths of 450–550 bp, error rates ranging from 1 to 8% and various levels of coverage spanning a 600 base segment of the ECA1 gene (GenBank accession no. U96455). These were assembled using each of the programs: both the number of contigs and singletons and the quality of the consensus sequences were compared (Figs 3 and 4). In each instance, Phrap and CAP3 produced a single consensus sequence. In contrast, TA-EST and TIGR Assembler split sequences into singletons or separate contigs as the error rate increased (data not shown). We also assessed the quality of the consensus sequences. For each program we calculated an A-score (see Materials and Methods) for the consensus sequences produced by each program at 5- and 50-fold EST coverage. Figure 3 shows the A-score for the best consensus sequence produced by each program as a function of EST error rates. Although both CAP3 and Phrap produced a single consensus in all of the situations analyzed, the fidelity of the Phrap consensus sequence was consistently worse than that generated by CAP3. The best consensus assemblies produced by TA-EST and TIGR Assembler score similarly to CAP3, although this must be considered in the light of the fact that the former programs tend to generate additional consensus sequences and singletons and fail to produce a consensus if the error rate is sufficiently high.
Figure 3.
Consensus sequence errors. Plot of A-scores for the best consensus assemblies produced by Phrap, CAP3, TA-EST and TIGR Assembler (TA) using simulated data for various error rates at 5× and 50× sequence coverage.
Figure 4.
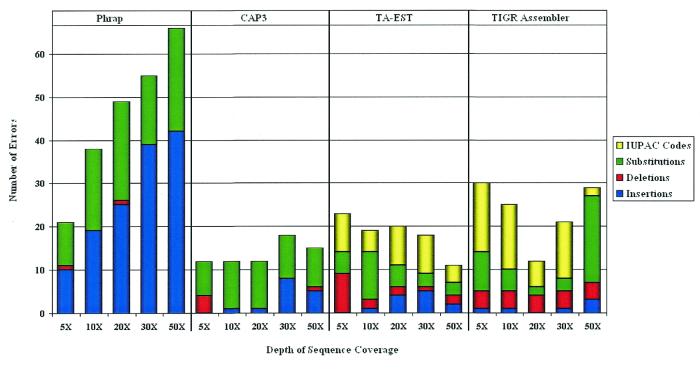
Error source distribution and normalized A-score for assemblies of 73 known genes. Consensus sequence error classification for Phrap, CAP3, TA-EST and TIGR Assembler using EST sequences containing 5% errors at various depths of coverage.
The consensus sequence discrepancies generated by each program were classified as insertions, deletions, substitutions or IUPAC codes (an IUPAC code represents an ambiguous nucleotide, e.g. Y represents one of C or T) and tabulated. Figure 4 shows the distribution of consensus errors for each program at various depths of coverage. While the total error rates for CAP3, TA-EST and TIGR Assembler are relatively constant and independent of depth of coverage, the errors produced by Phrap, particularly the insertions and substitutions, increase as the depth of coverage increases. This is consistent with the data in Figure 3, where the A-score for Phrap at 50-fold coverage is lower than that for 5-fold coverage. Phrap has a tendency to retain the errors in the EST sequences, introducing them as insertions in the consensus.
Assembly of ESTs from a family of genes
While software used for EST assembly should be relatively tolerant of random errors, it should be capable of separating ESTs from distinct but closely related transcripts. To assess the performance of the assembly programs to handle data from gene families we generated model ESTs from sequences sharing 90% or greater identity (see Materials and Methods) and measured the number of contigs generated by each of the programs. TA-EST was generally unable to separate the gene family members, always grouping the six member family into a single consensus and the eight member family into an average of 1.38. Phrap did only slightly better, generating an average of 2 and 4.33 consensus sequences from the six and eight member families, respectively. CAP3 did a good job of discriminating between closely related but distinct transcripts, however, it too failed with sequences that share >96% identity, producing an average of 4.5 and 6.67 for the two families. TIGR Assembler provided the greatest discrimination, generating an average of 5.83 and 8.5 consensus sequences, respectively, for the two families.
Evaluation of EST assembly algorithms using highly represented human genes
To further validate the results from our simulation studies, we examined 73 human genes with EST sequences spanning their lengths. The length of these genes was 1881 ± 1037 bp and the sequences had an average coverage of 203 ± 183 ESTs. Ideally, the ESTs from each gene should assemble to a single contig without singletons. However, without the gene sequence to serve as a reference, regions of low coverage and errors in the ESTs may cause multiple contigs and singletons to be formed. We examined the performance of each assembler, tallying the number of contigs and singletons produced for each gene. As summarized in Table 3, CAP3 was able to produce an average of 1.26 ± 0.58 contigs with a single contig in 59 of 73 cases (81%). The performance of Phrap was nearly as good, with 46 (63%) of the genes producing a single consensus and an average of 1.56 ± 0.88 assemblies. Neither CAP3 nor Phrap generated a significant number of singletons. Based on these measures, both programs performed significantly better that either TA-EST or TIGR Assembler.
Table 3. Performance of the four assemblers under evaluation for ESTs representing 73 known genes with an average coverage of 196 ± 180 sequences.
| Phrap | CAP3 | TA-EST | TIGR Assembler | |
|---|---|---|---|---|
| (A) | ||||
| No. of single assemblies | 46 | 59 | 15 | 2 |
| Mean no. of assemblies | 1.56 | 1.26 | 2.85 | 17.26 |
| Standard deviation | 0.88 | 0.58 | 1.79 | 17.20 |
| (B) | ||||
| Mean no. of singletons | 0.07 | 0.10 | 8.05 | 38.55 |
| Standard deviation | 0.35 | 0.45 | 10.09 | 47.14 |
For each of these genes, one would expect the assembler to produce a single consensus without singletons. (A) The number of single contigs produced by each assembler and the mean and standard deviation of the number of assemblies. (B) The mean and standard deviation of the number of singletons produced by each of the assemblers.
For each gene we assessed the fidelity of the longest consensus sequence produced by each of the four programs. As for our simulation studies, the best assemblies produced by CAP3, TIGR Assembler and TA-EST were all significantly better than those produced by Phrap. Figure 5 shows the number of errors, classified by type, generated by each program. Phrap produced considerably more insertions, deletions and substitutions than did the other assemblers. As a measure of the fidelity of the best assembly produced by each gene we normalized the total A-score for all assemblies by the total length of the assembled sequence; in this case, perfect assemblies would produce a value of 2. The normalized A-score for CAP3 was 1.59, while those for TA-EST, TIGR Assembler and Phrap were 1.49, 1.25 and 0.55, respectively.
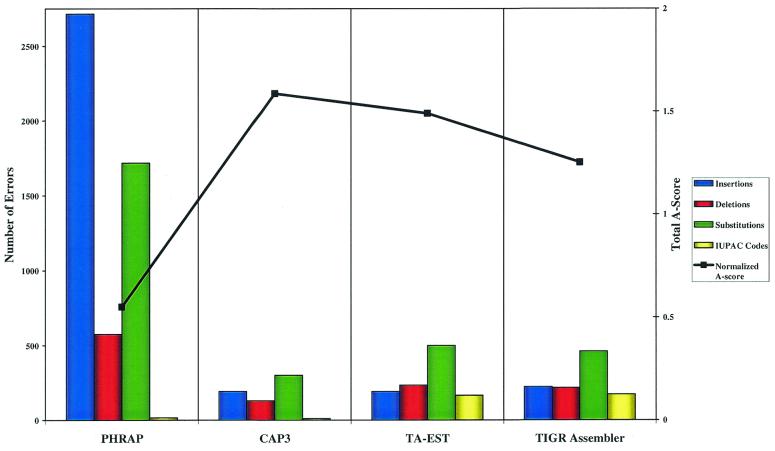
Figure 5.
DNA sequencing base call error probability. The total number of errors, classified by type, in the best assembly produced by the four assemblers and the normalized A-score for 73 known genes.
As expected, based on our previous results, CAP3 generated the highest quality assemblies of the corresponding gene sequences. Further, CAP3 exceeded our expectations, generating fewer consensus sequences than even Phrap, which had produced the fewest assemblies in both our simulations and our analysis of the Rat Gene Index. While these results may have been different if sequence quality values had been used in the assemblies, the gene sequence and EST data in GenBank do not include these data. For the available data our results clearly indicate that CAP3 has the best balance of error tolerance and error resolution.
Assembling the HGI and assessing THC fidelity by e-PCR
The HGI was assembled using CAP3 from 1 524 335 ESTs, 47 283 NPs and 7223 ET sequences, producing 75 424 THCs and 338 999 singletons. Of the 52 825 EST-based markers placed on radiation hybrid maps, we were able to assign 28 577 markers to one or more THCs. In all, 32 404 map assignments were made, suggesting a redundancy in the THC data set of 1.13-fold (32 404/28 577). Of 20 731 THCs assigned map locations, 7328 contained two or more independently mapped markers; of these, 7104 THCs (97%) contained multiple markers that mapped to nearby chromosomal locations, suggesting that the assemblies properly reconstructed the gene sequence.
DISCUSSION
EST data have proven to be an important resource for gene discovery and mapping and promise to be invaluable for the annotation of the eukaryotic genomes soon to be completed. However, the large number of EST sequences have made working with this data a challenge. The TIGR Gene Indices are an attempt to reduce those data to a manageable, well-defined collection of high fidelity consensus sequences. Central to this process is the use of a sequence assembly program that provides an accurate representation of the gene sequences from which the ESTs were derived. We have conducted an extensive analysis of the performance of four of the most widely known DNA sequence assembly programs—Phrap (http://www.phrap.org/phrap.docs/phrap.html ), CAP3 (10) and two versions of TIGR Assembler (11)—and used a variety of measures to assess the fidelity of the consensus sequences produced by this process.
Evaluation of sequence assembly programs
While none of the assembly programs performed perfectly, CAP3 consistently provided the highest fidelity assemblies, accurately assembling ‘dirty’ EST data without introducing an inordinate number of errors into the consensus or generating unnecessary singletons. TIGR Assembler and TA-EST proved slightly more sensitive to subtle yet consistent differences in sequence, such as those present in closely related members of a gene family. However, this sensitivity, combined with the naturally occurring errors inherent in ESTs, causes both to split transcripts, generating an over-representation of some genes. In contrast, Phrap is insufficiently sensitive to sequence differences, causing it to over-assemble ESTs and sacrifice the fidelity of the consensus sequences it produces by generating a significantly higher number of insertions and incorrect base assignments. CAP3 incorporates the best features of these other programs, producing high fidelity consensus sequences and maintaining a high level of sensitivity to gene family members while effectively handling sequencing errors. Based on our analysis we have selected CAP3 for assembly of the TIGR Gene Indices (http://www.tigr.org/tdb/tgi.org ).
Factors that influence consensus sequence fidelity
Most EST sequences in the public repositories do not have quality values assigned to each base. Quality values indicate how accurate the base call is; values >20 (99% confidence) represent high confidence calls (12). Without these, Phrap assigns a default quality of 15 to each base. The construction by Phrap of a consensus relies heavily on the quality value; when several input sequences disagree, it often resolves the problem by inserting two different bases in the final consensus, producing an insertion error. In contrast, CAP3, TA-EST and TIGR Assembler use a ‘majority rule’ scheme that tends to resolve disagreements correctly. [This result is consistent with that reported by Miller and Powell (18), although that study considered an earlier generation of assembly algorithms.] CAP3 uses only the majority base for its consensus; TA-EST and TIGR Assembler use an IUPAC code to represent possible ambiguities.
Although each of the assembly programs uses internal checks to discriminate between sequences, all include a user-definable parameter that specifies how similar two sequences must be to initially be considered identical. The default values of this parameter for the four assemblers are 95, 65, 94.5 and 97.5% for Phrap, CAP3, TA-EST and TIGR Assembler, respectively. This explains in part why TA-EST and Phrap, and to a lesser extent CAP3, could not separate ESTs from genes sharing >95% DNA sequence identity. One could increase the discrimination of these programs by selecting a higher stringency, but this has other unwanted effects, including increasing the number of consensus sequences and singletons.
Human sequence mapping and validation
The most extensive collection of EST and genomic mapping and sequence data is available for humans. This provides a unique opportunity to assess the fidelity of the consensus sequences contained within the TIGR Gene Indices. Radiation hybrid mapping does not provide precise map locations, but rather bins markers into approximate chromosomal locations. The likelihood that two markers from independently mapped ESTs fall into the same or adjacent bins is extremely small, unless the ESTs were derived from the same gene. The 97% (7104/7328) concordance between map locations for the THCs containing multiple, independent radiation hybrid markers suggests that the consensus sequences faithfully reconstruct the genes from which the ESTs were derived. In many cases the mapped markers fall into distinct, non-overlapping regions of the THCs. If there were a large number of chimeric or misassembled sequences in the THCs one would expect a discordance rate significantly higher than the 3% observed; this rate is not significantly different than that expected due to mapping errors at the various radiation hybrid laboratories (13). The fact that these discrete markers map to consistent locations within the genome provides an independent, experimental validation for the clustering and assembly process used to create the TIGR Gene Indices. Representative data for 14 of the 7328 THCS containing multiple mapped ESTs can be found in Table 4; radiation hybrid map locations for the HGI are available at http://www.tigr.org/tdb/hgi/searching/rh_map.html
Table 4. Radiation hybrid mapping data for a representative sample of THCs from HGI5.0 that contain multiple, independently mapped markers.
| THC ID | Marker position | RHDB ID | Chromosome | Location | Panel | Score |
|---|---|---|---|---|---|---|
| THC403868 | 766–890 | RH53683 | 14 | 281.46 | GB4 | P = 1.70 |
| THC403868 |
766–890 |
RH14883 |
14 |
4460 |
G3 |
F |
| THC403877 | 71–335 | RH26593 | 6 | 105.5 | GB4 | P > 3.00 |
| THC403877 | 74–212 | RH46761 | 6 | 105.5 | GB4 | P > 3.00 |
| THC403877 | 1224–1344 | RH46705 | 6 | 105.5 | GB4 | P > 3.00 |
| THC403877 |
1414–1661 |
RH26034 |
6 |
105.5 |
GB4 |
P > 3.00 |
| THC403892 | 552–748 | RH24987 | 9 | 404 | GB4 | P > 3.00 |
| THC403892 | 594–798 | RH11721 | 9 | 403.9 | GB4 | P > 3.00 |
| THC403892 |
660–798 |
RH13861 |
9 |
4806 |
G3 |
F |
| THC403910 | 830–989 | RH44275 | 10 | 547.83 | GB4 | P = 2.44 |
| THC403910 |
916–1036 |
RH16755 |
10 |
548.62 |
GB4 |
P = 1.28 |
| THC403911 | 50–149 | RH51822 | 11 | 247.67 | GB4 | P = 2.48 |
| THC403911 | 86–185 | RH27455 | 11 | 247.67 | GB4 | P = 2.48 |
| THC403911 |
86–185 |
RH10295 |
11 |
242.54 |
GB4 |
P = 0.00 |
| THC403912 | 2–101 | RH51822 | 11 | 247.67 | GB4 | P = 2.48 |
| THC403912 | 38–137 | RH27455 | 11 | 247.67 | GB4 | P = 2.48 |
| THC403912 |
38–137 |
RH10295 |
11 |
242.54 |
GB4 |
P = 0.00 |
| THC403923 | 597–759 | RH15921 | 19 | 216.04 | GB4 | P = 0.01 |
| THC403923 |
1179–1280 |
RH16797 |
19 |
214.04 |
GB4 |
P = 1.26 |
| THC403929 | 1637–1761 | RH12769 | 2 | 574.71 | GB4 | P > 3.00 |
| THC403929 | 1773–1903 | RH56254 | 2 | 573.36 | GB4 | P = 1.03 |
| THC403929 | 1908–2241 | RH14021 | 2 | 8064 | G3 | F |
| THC403929 | 1918–2049 | RH70825 | 2 | 572.14 | GB4 | P = 0.84 |
| THC403929 |
1929–2217 |
RH56929 |
2 |
557.16 |
GB4 |
P = 0.96 |
| THC403934 | 755–883 | RH39372 | 17 | 295.52 | GB4 | P = 0.76 |
| THC403934 |
793–998 |
RH76470 |
17 |
293.11 |
GB4 |
P = 0.02 |
| THC403947 | 22–122 | RH49734 | 1 | 145.4 | GB4 | P = 1.33 |
| THC403947 |
1431–1555 |
RH50152 |
1 |
145.91 |
GB4 |
P = 1.12 |
| THC403950 | 1724–1872 | RH78931 | 11 | 18.46 | GB4 | P = 0.36 |
| THC403950 | 1745–1920 | RH32214 | 11 | 17 | G3 | P = 1.62 |
| THC403950 |
1766–1877 |
RH27310 |
11 |
4.63 |
GB4 |
P = 2.39 |
| THC403956 | 2300–2430 | RH91675 | 16 | 194.86 | GB4 | F |
| THC403956 |
2300–2430 |
RH79175 |
16 |
193.96 |
GB4 |
P = 0.33 |
| THC403987 | 25–174 | RH55229 | 17 | 329.2 | GB4 | P = 1.52 |
| THC403987 | 25–174 | RH14431 | 17 | 2298 | G3 | P = 0.47 |
| THC403987 |
353–473 |
RH70694 |
17 |
319.86 |
GB4 |
P = 2.12 |
| THC404053 | 354–470 | RH44831 | 12 | 401.81 | GB4 | P > 3.00 |
| THC404053 | 557–686 | RH52825 | 12 | 400.21 | GB4 | P > 3.00 |
| THC404053 | 1146–1390 | RH75162 | 14 | 140.79 | GB4 | P = 0.17 |
The first column is the THC ID, the second column represents the position of the marker within the THC sequence, the third contains the RHDB ID (http://corba.ebi.ac.uk/RHdb ) for the marker, the fourth contains the chromosome associated with the marker, the fifth is the location of the marker on the chromosome expressed in CentiRays (CR), the sixth is the radiation hybrid panel on which the marker was mapped and the final column is the score associated with the marker position (F for G3 means that this is a ‘framework’ marker). It should be noted that the G3 and GB4 panels were constructed using different radiation dosages. Consequently, the ‘size’ of the chromosome in CR is different. Single lines separate distinct THCs; of the THCs shown, only the last, THC404053, has a discrepancy in its map location.
Conclusions
We have conducted a careful analysis of sequence assembly programs in order to determine their performance in assembling EST sequences and developed a refined process of EST sequence cleaning, clustering, assembly and annotation that provides a faithful representation of the gene sequences from which the ESTs were derived. With the imminent completion of the sequence of the human and other genomes, our challenge will be to use all our available resources to accurately catalog and characterize the encoded genes. Finding genes in a genomic sequence is a significant challenge; the vast body of EST data represents a tremendous resource that can be applied to this problem. The TIGR Gene Indices provide a reliable reduction of the EST data and can simplify annotation by providing fewer, accurate sequences that can be searched against genomic sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge Phil Green for providing Phrap and Xiaoqui Huang for CAP3. This work could not have been accomplished without the remaining members of the TIGR Gene Index Team, Thomas Hansen and Jonathan Upton. The authors are indebted to Anna Glodek for her database development efforts. The authors also wish to thank Michael Heaney and Susan Lo for database support, Vadim Sapiro, Billy Lee, Sonja Gregory, Rajeev Karamchedu, Corey Irwin, Lily Fu and Eddy Arnold for computer system support and Cathy Ronning, Robin Buell, Joseph White and Claire M. Fraser for thoughtful comments and suggestions. This work was supported by a grant from the US Department of Energy. S.L.S. was supported in part by NIH Grant R01 LM06845-01 and NSF Grant IIS-9902923. S.L.S., J.Q. and S.K. were supported in part by NSF Grant KDI-9980088. F.L., I.E.H., G.P. and J.Q. were supported in part by grant DE-FG02-99ER62852 from the US Department of Energy
REFERENCES
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Science, 252, 1651–1661. [DOI] [PubMed] [Google Scholar]
- 2.Adams M.D., Kerlavage,A.R., Fleischmann,R.D., Fuldner,R.A., Bult,C.J., Lee,N.H., Kirkness,E.F., Weinstock,K.G., Gocayne,J.D., White,O. et al. (1995) Nature, 377, 3–174. [PubMed] [Google Scholar]
- 3.Hudson T.J., Stein,L.D., Gerety,S.S., Ma,J., Castle,A.B., Silva,J., Slonim,D.K., Baptista,R., Kruglyak,L., Xu,S.H. et al. (1995) Science, 270, 1945–1954. [DOI] [PubMed] [Google Scholar]
- 4.Schuler G.D., Boguski,M.S., Stewart,E.A., Stein,L.D., Gyapay,G., Rice,K., White,R.E., Rodriguez-Tome,P., Aggarwal,A., Bajorek,E. et al. (1996) Science, 274, 540–546. [PubMed] [Google Scholar]
- 5.Bouck J., Yu,W., Gibbs,R. and Worley,K. (1999) Trends Genet., 15, 159–162. [DOI] [PubMed] [Google Scholar]
- 6.Boguski M.S. and Schuler,G.D. (1995) Nature Genet., 10, 369–371. [DOI] [PubMed] [Google Scholar]
- 7.Quackenbush J. Liang,F., Holt,I., Pertea,G. and Upton,J. (2000) Nucleic Acids Res., 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin X., Kaul,S., Rounsley,S., Shea,T.P., Benito,M.I., Town,C.D., Fujii,C.Y., Mason,T., Bowman,C.L., Barnstead,M. et al. (1999) Nature, 402, 761–768. [DOI] [PubMed] [Google Scholar]
- 9.Liang F., Holt,I., Pertea,G., Karamycheva,S., Salzberg,S.L. and Quackenbush,J. (2000) Nature Genet., 25, 239–240. [DOI] [PubMed] [Google Scholar]
- 10.Huang X. and Madan,A. (1999) Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton G., White,O., Adams,M.D. and Kerlavage,A.R. (1995) Genome Sci. Technol., 1, 9–18. [Google Scholar]
- 12.Ewing B. and Green,P. (1998) Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 13.Deloukas P., Schuler,G.D., Gyapay,G., Beasley,E.M., Soderlund,C., Rodriguez-Tome,P., Hui,L., Matise,T.C, McKusick,K.B., Beckmann,J.S. et al. (1998) Science, 282, 744–746. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Huang X., Adams,M.D., Zhou,H. and Kerlavage,A.R. (1997) Genomics, 46, 37–45. [DOI] [PubMed] [Google Scholar]
- 16.Schuler G.D. (1997) Genome Res., 7, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller M.J. and Powell J.I. (1994) J. Comp. Biol., 1, 257–269. [DOI] [PubMed] [Google Scholar]