Abstract
The extent to which normal (nonmalignant) cells of the body can evolve through mutation and selection during the lifetime of the organism has been a major unresolved issue in evolutionary and developmental studies. On the one hand, stable multicellular individuality seems to depend on genetic homogeneity and suppression of evolutionary conflicts at the cellular level. On the other hand, the example of clonal selection of lymphocytes indicates that certain forms of somatic mutation and selection are concordant with the organism-level fitness. Recent DNA sequencing and tissue physiology studies suggest that in addition to adaptive immune cells also neurons, epithelial cells, epidermal cells, hematopoietic stem cells and functional cells in solid bodily organs are subject to evolutionary forces during the lifetime of an organism. Here we refer to these recent studies and suggest that the expanding list of somatically evolving cells modifies idealized views of biological individuals as radically different from collectives.
Keywords: Cancer, Cell competition, Driver mutation, Mosaicism, Multicellularity, Somatic evolution
Introduction
To enable the emergence of multicellular individuality hundreds of millions of years ago, synergism must have had been reached between cellular and organism levels of selection [1]. This interlevel harmonization was assumed to result from suppression of the autonomous behaviors of individual cells and their subordination to somatic functions [2–4]. To account for this attunement of cellular activities to the greater needs of the organism, specialized “policing” mechanisms have been postulated and suggested to act as conflict modifiers and intercellular cooperation enforcers [5]. In addition, diminished variation between cells and their reliance on extrinsic molecular features were considered to contribute to the stable existence of higher-level units [6]. In short, some form of somatic “de-Darwinization” or suppression of intra-organism evolution has become recognized as a prerequisite for multicellular individuality [5–9].
Despite its appeal, the idea of somatic de-Darwinization has been compromised by the observation that certain forms of intra-organismal variation and somatic competition are actually favored in nature. This includes evolution of somatic cells in plants and clonal selection of lymphocytes in higher vertebrates [10, 11]. Nevertheless, questions have been raised if these instances should be considered as manifestations of widespread selective processes in metazoans or whether they are only rare exceptions of Darwinian processes in selected organisms [6, 10]. Here we refer to recent somatic DNA sequencing and tissue physiology studies to investigate this problem. Pointing at an extraordinary scale of clonal selective processes in multicellular organisms these pioneering studies challenge idealized views of bodily cells as evolutionarily idle and, if affirmed, current understanding of the differentiation of individuals and collectives must be re-considered.
Evolution within the body
Initially considered as mere sequencing errors, observed differences in DNA code between bodily cells appear to be a natural byproduct of accumulating mutations during the lifetime of the organism [12]. Indeed, despite relying on DNA repair and other damage response systems, somatic cells diverge genetically during the lifetime of the organism, contributing to the substantial polyclonality and mosaicism of animal tissues [13, 14]. Passed on from cell generation to cell generation, this post-zygotic variation is most apparent in an embryo whose unstable chromosomes and frequent structural DNA changes leave permanent marks on its genetic makeup [15]. Unequal in their reproductive capacitates, diversified somatic cells are subject to natural selection, which promotes survival of the fitter and extinction of less adapted clones [16, 17]. Not always harmful or pro-malignant, the selective processes influence the dynamics of clonal processes, often contributing to crucial bodily functions [18, 19].
Among the best characterized selective processes in the organism are those involving lymphocytes (B cells and T cells) that, following diversification, can clonally expand or contract depending on the antigen-binding fitness of their receptors [20–22]. To diversify antigen receptor genes, vertebrates, from sharks to humans, rely on the process of V(D)J gene rearrangement, which introduces variation in the initial (naïve) immune repertoire [23]. Varying in their receptor specificities, individual lymphocytes become activated by their antigenic targets to proliferate and increase representation of their progeny in the immune cell pool [24]. Competing for antigen, activated B cells undergo additional cycles of mutation and selection in specialized structures known as germinal centers [25]. As a result of these diversification and selection processes, the repertoire of lymphocytes undergoes adaptive transformations acquiring the potential to handle recurrent pathogenic insults and by constructing its antigenic niche by means of specialized immunosurveillance and microbiota-shaping functions [26–28]. Computational studies help to elucidate this dynamic, shedding light on the diversity and populational structure of the repertoire [29, 30]. Due to their reliance on previous immune encounters and the potential to acquire shared immune receptor signatures, immune repertoires manifest features of contingent as well as convergent evolution [31]. Acting on distinct parts of the B cell receptor molecule, positive, and purifying negative selection are both at play in the repertoire [32]. These findings support the view that immune cells operate as complex systems that evolve, adapt and transform—an observation that validates application of population-based approaches to the lymphocyte repertoire.
Apart from lymphocytes, other cell types undergo selection and adaptation. For example, epithelial cells, to survive in their microenvironment, can differentially proliferate without causing cancer or tissue disfunction. This is evident in mouse esophageal epithelial cells, which by accumulating mutations in NOTCH1, P53 and other so-called “cancer-associated genes,” clonally expand and remodel the tissue [33, 34]. Natural selection also drives changes in the layer of urothelial cells, whose chromatin remodeling genes like KMT2D and KDM6A, confer advantage on selected clones allowing them to proliferate and colonize the tissue [35, 36]. The widespread character of somatic evolution of epithelial cells is further supported by studies of endometrial glands that tend to become dominated by one or a few mutant clones in post-menopausal women without obvious signs of a pathology [37, 38]. Finally, genome sequencing studies of bronchial epithelium in smoking subjects reveal that mutations in NOTCH1, TP53 and ARID2 drive clonal expansion of these cells [39]. A rapid increase in a fraction of less mutated healthy cells following smoking cessation attests to adaptive clonal changes in altered lung environment. Hence, epithelial surfaces of the lungs, uterus, esophagus, and urinary tract are all polyclonal patchworks of evolving cells that, despite accumulating driver mutations in cancer genes, only rarely cause neoplastic changes.
Still another class of cells known to undergo clonal transformations during the lifetime of the organism are hematopoietic stem and progenitor cells, whose pattern of descent, like that of other evolving populations, can be represented as a phylogenetic tree [40]. Based on mutational profiles of these cells, Lee-Six et al. [40] were able to trace their origin back to the most recent common cell ancestor in the same human subject. As this cell ancestor shared characteristic mutations with buccal epithelial cells, it was likely present already in a pre-gastrulation embryo before the separation of germ layers. An aberrant form of hematopoietic stem cell evolution is called clonal hematopoiesis of indeterminate potential (CHIP) and is known to increase hematologic cancer and cardiovascular disease risks in humans [41]. During this process, natural selection rather than neutral genetic drift promotes expansion of one or a few selected clones at the expense of other clones [42]. While pronounced forms of CHIP are pathological, minor forms of clonal hematopoiesis (below 0.02 variant allele fraction) driven by mutations in leukemia-associated genes (DNMT3A and TET2) are widespread, affecting as many as 95% of healthy human subjects aged between 50 and 70 [43]. This ubiquitous, low-grade clonal expansion does not pose a health risk to the affected person and its detection has no prognostic clinical significance. Thus, in addition to adaptive immune cells and epithelial cells, hematopoietic cells also exhibit somatic evolution during the lifetime of the organism.
The expanding list of somatic cells subject to selection also includes post-mitotic neurons that, despite not being able to proliferate, undergo the process of negative selection to eliminate less adapted clones [44]. This process represents differential survival that appears as a Darwinian struggle for neurotrophins that act as survival factors for these cells (indirect competition) and on direct recognition of specialized “fitness markers,” like Flower, which, depending on their expressed isoform, allow fitter cells to induce apoptosis of the suboptimal neurons (direct competition) [45, 46]. Key for eliminating surplus suboptimal cells, the interneural competition not only occurs early in development but also later in life as part of an ongoing process of neurogenesis in the hippocampus [44]. The outcome of this competition depends, among other factors, on genetic characteristics of the involved neurons whose accumulating mutations may gain fitness advantage or disadvantage over other cells [47]. One mechanism for gene diversification in the brain includes an amyloid precursor protein (APP)-based recombination (reminiscent of V(D)J rearrangement in lymphocytes) that generates genetic variation and sets the framework for cell selection in this organ [48, 49]. In sum, an emerging picture shows how joint action of mutation, selection and competition guide development of the nervous system that enables the organism to attain its unique neural circuitry.
In addition to the immune, hematopoietic, neural, and epithelial cells, multiple other types of cells are also subject to somatic evolutionary processes. This includes normal human eyelid epidermis, which, despite the significant burden of mutations bearing characteristic UV exposure signatures in NOTCH1, P53 and other loci, remains histologically normal [50]. Single cell resolution studies of normal melanocytes revealed that these cells have an extremely high mutation burden (especially in sun-exposed areas) and that 20% of these cells, despite being noncancerous, bear alterations in BRAF, NRAS, and other melanoma-associated genes [51]. Beyond the eyelids, cancer-associated genes can promote expansion of epidermal cells in the head, legs, forearms, trunk, and abdomen, shaping clonal skin architecture in these body parts [52]. As revealed by whole genome and targeted DNA sequencing, strong environmental pressures act on the skin, fostering survival of some and extinction of other mutant clones [52]. In addition to environmental factors, such as UV light, mutant selection in the epidermis depends on competitive exchanges between heterogenous clones, which growing laterally, fight for limited space and resources. Hence, clonal dynamic of normal human skin is driven by mutation, selection, and struggle: Darwinian forces influencing changes also in other body parts.
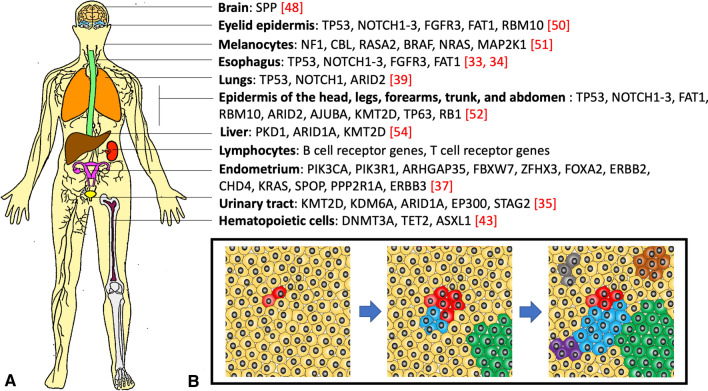
Functional cells of solid organs also undergo somatic evolutionary changes as illustrated by karyotypic adaptive changes of liver cells, which, due to chromosome variation, undergo selection for injury-resistant clones during stress [53]. Indeed, as shown by a model of hereditary tyrosinemia, maladapted euploid hepatocytes can be replaced by clonally expanding fitter aneuploid ones to reconstitute and protect the organ. Further highlighting importance of somatic evolution is the role of genomic diversity of hepatocytes in cirrhotic liver that by hosting a heterogenous set of cells allows the adapted ones to gain proliferative advantage and regenerate the organ [54]. In particular, loss-of-function mutations in PKD1, ARID1A, and KMT2D enhance fitness of hepatocytes to expand and promote liver protection. Complementing the above studies of clonal selection in various body parts is an RNA sequence analysis of 29 tissue types, all of which hosting numerous large clonal cell populations sometimes reaching macroscopic dimensions [55]. Thus, immense in its scale and intensity, clonal evolution appears to be normal and inescapable part of somatic cell dynamics [56] (Fig. 1).
Fig. 1.
Diverse genes modulate fitness of somatic clones in healthy subjects. Characteristic sets of genes in different body parts can confer proliferative advantage on constituent cells in the tissues (A). Somatic evolutionary changes in the organism involve accumulation of mutations and clonal expansion of selected clones, which despite positively selected may cause no pathology (B)
Individuals as collectives
While certain forms of somatic evolution have been recognized, doubts have been raised if bodily cells could be considered as genuine Darwinian population considering that genetic variation and selection are constrained in the tissues [5, 6]. In fact, specialized mechanisms exist to modulate evolution of somatic cells and to prevent uncontrolled expansion of the fittest clones [57]. These mechanisms include targeted elimination of mutated cells by pro-apoptotic and immune effectors as well as maintenance of a stable tissue landscape to prevent rapid alterations of selective pressures in the microenvironment. The importance of these controls is highlighted by the fact that their deterioration later in life greatly increases cancer risk as aging-associated changes in tissue environment can alter selective pressures to favor expansion of malignant clones [58]. Hence, mechanisms are in place to inhibit clonal selective processes in an organism, seemingly supporting the view that these cells have been completely deprived of the capacity to evolve.
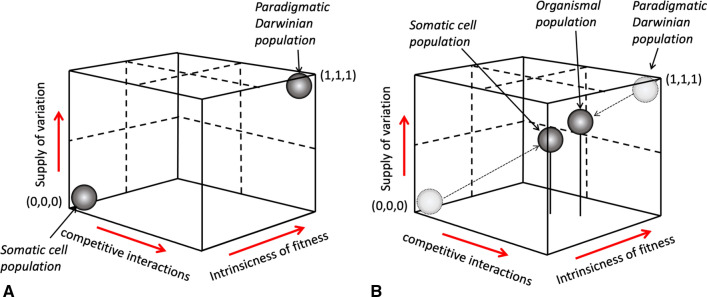
However, as we have seen, clonal selective processes are extremely common in normal healthy tissues. Indeed, instead of blocking clonal evolution, the specialized regulatory mechanisms help to ensure its safe operation reconciling the adaptive interests of the organism and its parts [59]. Minimizing cancer risk, these mechanisms allow cells to operate as semi-autonomous living entities, which can act on their unique features and fitness characteristic much like animals and plants in their natural environment [60]. Indeed, if somatic variation and selection are as ubiquitous as the above recent studies indicate, then the gulf between paradigm Darwinian populations and somatic cell populations is not as wide as often assumed [61] (Fig. 2).
Fig. 2.
Based on criteria such as supply of variation, competitive interactions and dependence on intrinsic characters, somatic cell populations, and organismal populations can no longer be considered as radically opposed. In an idealized model, somatic cell populations were considered to occupy completely different areas in the “individuality space” (A). More realistic model modifies this view, suggesting that somatic cell populations and organismal populations approximate each other with respect to the above three criteria (B). (Adapted with modifications from Godfrey-Smith, 2009)
Studies of lymphocytes help to understand how autonomous cell mechanics and randomness rather than rigorous external controls determine the fate of somatic cells in a population [62]. Suggesting that no two cells are identical in the organism, biological studies blur the divide between individuals and collectives suggesting that organisms operate as “weak individuals”, organized cell populations reminiscent of loosely organized forms of biological coexistence [61]. Compromising the idea of somatic cell de-Darwinization, these recent studies support a view in which bodily cells operate as complex ecosystems rather than as deterministically regulated and integrated wholes [63, 64].
Recognition of the relative autonomy and evolvability of somatic cells opens a framework in which the concepts of habitat, diversity, niche, and population structure are key aspects of normal physiology [63, 65]. So far considered mostly in the context of cancer [66–68], this eco-evolutionary perspective can be expanded also to capture the activity of cells in a healthy organism. Reconceptualizing certain forms of collective cell behavior as equivalent to that of a flock of birds or school of fish such a perspective could help to explain cell coordination in populational terms rather than in top-down regulation categories [64]. This perspective also allows framing interactions between somatic cells and tissue cells in terms of co-evolving predator–prey dynamics, in which selective pressures exerted by self-reactive immune cells promote development of adapted tissue cells to evade immune-related damage [69]. This ecological outlook also allows reframing certain aspects of organogenesis to account for their reliance on tissue-environment interactions rather than on specialized molecular mechanisms [1]. Finally, the environmental vision could advance our efforts to capture the relationship between somatic and microbial cells in the gut in terms of niche construction and ecological equilibrium states [70, 71]. Indeed, when considered in this framework, the internal and external ecologies of the organism cannot be decoupled, meeting at a fluid ecotone at which they interact [72, 73]. All in all, the realization that somatic cells, like other biological populations, mutate, evolve, and adapt demands recognition of their ecological behaviors that encompass a wide spectrum of interactions from cooperation to communication to conflict [74].
Despite varying degrees of autonomy, somatic cells are not unconstrained in their capacity to evolve and engage into ecological relationships in the organism. (Note the gap between somatic cell populations and paradigmatic Darwinian populations in Fig. 2B). Limits imposed on evolving somatic cell populations include regulatory mechanisms that help to target somatic mutagenesis to defined gene regions [75], specialized systems modulating the balance between cell proliferation and cell death [76], and spatial constraints like physical barriers separating liver lobules to prevent uncontrolled expansion of the fittest clones in a cirrhotic liver [54]. The existence of such limitations on somatic evolution does not challenge the ecological and evolutionary perspective on somatic processes in so far as populations of free-living organisms also manifest a variety of “multicellular traits” [77]. Indeed, division of labor, policing controls, regulation of cell proliferation and differentiation can be found not only in somatic cells but also in evolving unicellular populations [78–80]. Thus, following this dynamically informed understanding of the multicellular individual, our understanding of a free-living cell population also changes inasmuch as the latter manifests many features so far attributed mostly to the former and that, accordingly, our idea of paradigmatic Darwinian populations is an idealization that may have no direct counterpart in reality (Fig. 2B).
Implications of the evolutionary framework
While the full implications of these recent findings on understanding somatic evolution and inner ecology of the organism still await to be determined, an outline of the challenges confronting basic assumptions about processes such as cancerogenesis and major evolutionary transitions have emerged.
One of the central tenets of evolutionary studies of cancer is that oncogenic transformation results from an acquired capacity of cells to evolve in the tissues due to somatic mutations in cancer-associated genes [81, 82]. From this point of view, cancerogenesis was often assumed to represent a reversal of the evolved multicellular state and a return to the atavistic state in which cells subjected to selection pursue their own replicative success (see Ref. [78] for the review). In this respect, cancer was considered as an instance of “re-Darwinization” or acquisition of the potential to evolve in the tissues [83]. Studies of somatic evolution in healthy tissues modify this view, highlighting that evolutionary process in the organism are ubiquitous and not exclusive to cancer.
-
The realization that oncogenic transformation does not result simply from an acquisition of evolvability potential by a cell draws attention to contextual factors in the process of oncological transformation [84–86]. The importance of such factors is highlighted, among other things, by the fact that while cells bearing mutations in cancer-associated genes are extremely widespread, their potential to progress into malignancy is exceedingly small due to a network of ecological interactions they make with other cells [87]. For example, despite often affected by oncogenic P53 mutations, epidermal progenitor cells rarely become malignant because of the competitive equilibrium they reach with other mutated cell variants in the basal skin layer [88]. The importance of such Darwinian controls is further supported by studies of esophageal epithelium, where genetically heterogenous, but equally fit clones “collide” to restrain each other’s clonal behavior and to establish a stable state in which no single clone dominates the tissue [89].
In addition to intercellular competition, the organization of tissue landscape helps to prevent mutant clones from progressing into cancer [59]. Indeed, stable microenvironmental niche promotes development of stem cell clones with optimal fitness while favoring elimination of pre-neoplastic mutants [90]. A decline of tissue maintenance systems in the elderly leads to alterations in the tissue environment inducing changes in selective pressures to direct evolution of mutant clones towards malignant phenotypes [57, 91]. Illustrating the impact of environmental pressures on the course of evolutionary changes within an organism is the observation that hematopoietic stem cell mutants that fail to gain proliferative advantage in a young individual, may promote malignancy later in life due to aging-associated alterations in the cytokine milieu [92]. Thus, environmental factors such as the ecology of cell exchanges and organization of a tissue landscape contribute to cancer-protective functions, preventing cells from unlimited growth and invasive spread [93, 94]. It is only when the ecological balances are upset (due to tumor promoting inflammation, age-associated degradation of tissue architecture or noxious substances) that mutated cells can gain competitive superiority over normal cells, a process that mimics a hostile takeover by invasive native species following major environmental alteration like forest fire [95] (Fig. 3).
In addition to elucidating the importance of unperturbed somatic evolutionary and ecological processes in tissue sustainability, studies of intra-organismal variation and selection help to explain the role of stress-induced mutagenic systems in metazoan cells [96]. While existence of such systems in unicellular organisms has obvious benefits, enhancing emergence of fit variants appearing during altered environmental conditions, their persistence in mammalian tissue cells seems to make no adaptive sense beyond their involvement in generating immune and germline diversity [97]. The above-mentioned studies of somatic evolution help to support a hypothesis that stress-induced mutagenic systems persisted in mammals to ensure somatic heterogeneity necessary for tissue resilience, development and normal tissue function [98, 99]. This is confirmed among other things by the fact that the characteristic signatures of APOBEC DNA editing enzymes (a class of deaminases present in somatic cells) are not limited to cancer but also operate in normal colon and bladder linings to increase variation of these cells [35, 100].
Fig. 3.
Positive and negative outcomes of genetic diversity of somatic cells. Genetically homogenous or near-homogenous cells in an embryo (A) acquire mutations during the lifetime of the organism (B) allowing fittest clones not only to expand but also to promote adaptation of the tissue (C). In rare cases, additional mutations and/or ecological perturbations may lead to emergence of malignant cells, which like invasive native pests in a perturbed habitat, monopolize resources and dominate the environment (D)
In conclusion, in contrast to Germain, who argued that natural selection cannot explain cancer progression [101], we maintain that adaptive explanations are applicable to both cancerous as well as noncancerous somatic cells, elucidating their clonal dynamic and development. Indeed, with the progress in our understanding of tissue mosaicism and somatic selection, a broader evolutionary and environmental vision of physiological and pathological processes is emerging.
Finally, an improved understanding of somatic evolution also helps to modify our view of major evolutionary transitions, including those from molecules to cells, cells to multicellular organisms and organisms to societies [102]. Involving integration of lower-level parts into higher levels of organization, and relying on strong cooperation between their parts, these transitions have been considered as well-defined and complete. Instead, the above-mentioned studies suggest that these shifts must have been rather fluid and fragmentary, allowing the lower-level parts to preserve much of their pre-transitional lifestyle and independence. The importance of this realization is underscored by its implications for recent attempts to depict human societies as de-Darwinized populations [6, 9, 103]. While, indeed, human societies departed from paradigm Darwinian populations in their reliance on language, division of labor and cooperation, the involvement of adaptive factors cannot be quite ignored in these communities [104]. A more balanced perspective could help to avoid traps of social Darwinism and its converse, i.e., social collectivism: While the former could lead to eugenics, the latter could embolden implementation of cooperation-enforcing mechanisms in a society.
Conclusions: a return to the concept of inner struggle
Forgotten for over a century now, the idea of inner struggle and adaptation is gaining a new impetus in the light of the above studies [105]. Introduced in 1881 by Wilhelm Roux, this doctrine presupposed that natural selection does not only act on the level of the individual but also on its parts [106]. Indeed, Roux assumed that developmental processes in the organism are guided by competition between cells and other bodily parts, which analogous to autonomous living beings, struggle for resources in their habitats. Finding applications in biology and medicine at the end of the nineteenth century [107], Darwinian ideas infiltrated also bacteriological research helping to lay foundations for adaptive explanations of acquired immunity [108, 109]. Adopted by Elie Metchnikoff to account for immunity, pathology, senescence, and development, the evolutionary approach helped to provide an overarching vision of an organism as internally conflicted and changing [110, 111]. Thus, already considered by developmental biologists, zoologists, bacteriologists, and philosophers, the idea of somatic evolution was considered long before the advancement of the sequencing techniques.
While the Neo-Darwinian synthesis and the immunochemical program led to an abandonment of the somatic evolutionary framework, some of its basic tenets are being revivified in the context of cutting-edge physiological studies. These new advances herald a transition from a framework in which somatic mutations and differential cell expansion are fundamentally pathological, towards a view in which mosaicism and clonal selection are normal parts of organismal physiology. Departing from a view of the adaptive immune system as an encapsulated island of somatically evolving cells in a genetically stable organism, they suggest that the emergence of adaptive immunity in vertebrates was smoother that assumed [112, 113]. While many instances of this ubiquitous somatic variation and selection are detrimental, increasing cancer risk, others are functionally neutral and even beneficial, helping to contribute to developmental, immune, regenerative, and cognitive functions. Challenging the notion that somatic cells had been “de-Darwinized”, these novel studies call for a more nuanced understanding of events that led to evolutionary transitions and open a perspective in which individuals are considered as ecologically balanced collectives [63, 114].
Acknowledgements
I wish to express my most sincere gratitude to Irun R. Cohen and Alfred I. Tauber for their valuable comments on earlier versions of this article. All errors that remain are mine.
Author contributions
Bartlomiej Swiatczak wrote and edited the manuscript.
Funding
This research received no external funding.
Availability of data and material (data transparency)
Not applicable.
Code availability (software application or custom code)
Not applicable.
Declarations
Conflict of interest
The author has no conflicts of interest to declare.
Ethics approval
Not applicable (The study did not involve human or animal participants).
Consent for publication
Not applicable (The study did not involve human or animal participants).
Consent to participate
Not applicable (The study did not involve human or animal participants).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buss LW. The evolution of individuality. Princeton: Princeton University Press; 1987. [Google Scholar]
- 2.Bonner JT. First signals. The evolution of multicellular development. Princeton: Princeton University Press; 2000. [Google Scholar]
- 3.Queller DC, Strassmann JE. Beyond society: the evolution of organismality. Philos Trans R Soc Lond B Biol Sci. 2009;364:3143–3155. doi: 10.1098/rstb.2009.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank SA. Perspective: repression of competition and the evolution of cooperation. Evolution. 2003;57:693–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 5.Michod RE. Darwinian dynamics: evolutionary transitions in fitness and individuality. Princeton: Princeton University Press; 1999. [Google Scholar]
- 6.Godfrey-Smith P. Darwinian populations and natural selection. Oxford: Oxford University Press; 2009. [Google Scholar]
- 7.Godfrey-Smith P. Darwinian individuals. In: Bouchard F, Huneman P, editors. From groups to individuals: evolution and emerging individuality. Cambridge: MIT Press; 2013. pp. 17–36. [Google Scholar]
- 8.Okasha S. Evolution and the levels of selection. Oxford: Oxford University Press; 2006. [Google Scholar]
- 9.Szathmáry E. Toward major evolutionary transitions theory 2.0. Proc Natl Acad Sci USA. 2015;112:10104–10111. doi: 10.1073/pnas.1421398112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke E. Plant individuality and multilevel selection theory. In: Calcott B, Sterelny K, editors. The major transitions in evolution revisited. Cambridge: MIT Press; 2011. pp. 227–250. [Google Scholar]
- 11.Müller V, de Boer RJ, Bonhoeffer S, Szathmáry E. An evolutionary perspective on the systems of adaptive immunity. Biol Rev Camb Philos Soc. 2018;93:505–528. doi: 10.1111/brv.12355. [DOI] [PubMed] [Google Scholar]
- 12.Thorpe J, Osei-Owusu IA, Avigdor BE, Tupler R, Pevsner J. Mosaicism in human health and disease. Annu Rev Genet. 2020;54:487–510. doi: 10.1146/annurev-genet-041720-093403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Vijg J. Somatic mutagenesis in mammals and its implications for human disease and aging. Annu Rev Genet. 2018;52:397–419. doi: 10.1146/annurev-genet-120417-031501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiao YH, Anderson LM. Structural genomic changes during mammalian ontogeny: a new dimension. Epigenomics. 2012;4:1–4. doi: 10.2217/epi.11.100. [DOI] [PubMed] [Google Scholar]
- 15.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, Fryns JP, Verbeke G, D'Hooghe T, Moreau Y, Vermeesch JR. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 16.Bowling S, Lawlor K, Rodríguez TA. Cell competition: the winners and losers of fitness selection. Development. 2019;146:dev167486. doi: 10.1242/dev.167486. [DOI] [PubMed] [Google Scholar]
- 17.Martincorena I. Somatic mutation and clonal expansions in human tissues. Genome Med. 2019;11:35. doi: 10.1186/s13073-019-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease—clones picking up speed. Nat Rev Genet. 2017;18:128–142. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- 19.De S. Somatic mosaicism in healthy human tissues. Trends Genet. 2011;27:217–223. doi: 10.1016/j.tig.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Burnet M. The clonal selection theory of acquired immunity. Cambridge: Cambridge University Press; 1959. [Google Scholar]
- 21.Hodgkin PD, Heath WR, Baxter AG. The clonal selection theory: 50 years since the revolution. Nat Immunol. 2007;8:1019–1026. doi: 10.1038/ni1007-1019. [DOI] [PubMed] [Google Scholar]
- 22.Tauber AI, Podolsky SH. The generation of diversity: clonal selection theory and the rise of molecular immunology. Cambridge: Harvard University Press; 1997. [Google Scholar]
- 23.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershberg U, Luning Prak ET. The analysis of clonal expansions in normal and autoimmune B cell repertoires. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140239. doi: 10.1098/rstb.2014.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Victora GD, Wilson PC. Germinal center selection and the antibody response to influenza. Cell. 2015;163:545–548. doi: 10.1016/j.cell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen IR. Tending Adam's garden: evolving the cognitive immune self. London: Academic Press; 2000. [Google Scholar]
- 27.Cohen IR. Activation of benign autoimmunity as both tumor and autoimmune disease immunotherapy: a comprehensive review. J Autoimmun. 2014;54:112–117. doi: 10.1016/j.jaut.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Huus KE, Petersen C, Finlay BB. Diversity and dynamism of IgA-microbiota interactions. Nat Rev Immunol. 2021 doi: 10.1038/s41577-021-00506-1. [DOI] [PubMed] [Google Scholar]
- 29.Greiff V, Bhat P, Cook SC, Menzel U, Kang W, Reddy ST. A bioinformatic framework for immune repertoire diversity profiling enables detection of immunological status. Genome Med. 2015;7:49. doi: 10.1186/s13073-015-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miho E, Yermanos A, Weber CR, Berger CT, Reddy ST, Greiff V. Computational strategies for dissecting the high-dimensional complexity of adaptive immune repertoires. Front Immunol. 2018;9:224. doi: 10.3389/fimmu.2018.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cobey S, Wilson P, Matsen FA., 4th The evolution within us. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140235. doi: 10.1098/rstb.2014.0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaari G, Benichou JI, Vander Heiden JA, Kleinstein SH, Louzoun Y. The mutation patterns in B-cell immunoglobulin receptors reflect the influence of selection acting at multiple time-scales. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140242. doi: 10.1098/rstb.2014.0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martincorena I, Fowler JC, Wabik A, Lawson ARJ, Abascal F, Hall MWJ, Cagan A, Murai K, Mahbubani K, Stratton MR, Fitzgerald RC, Handford PA, Campbell PJ, Saeb-Parsy K, Jones PH. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama A, Kakiuchi N, Yoshizato T, Nannya Y, Suzuki H, Takeuchi Y, Shiozawa Y, Sato Y, Aoki K, Kim SK, Fujii Y, Yoshida K, Kataoka K, Nakagawa MM, Inoue Y, Hirano T, Shiraishi Y, Chiba K, Tanaka H, Sanada M, Nishikawa Y, Amanuma Y, Ohashi S, Aoyama I, Horimatsu T, Miyamoto S, Tsunoda S, Sakai Y, Narahara M, Brown JB, Sato Y, Sawada G, Mimori K, Minamiguchi S, Haga H, Seno H, Miyano S, Makishima H, Muto M, Ogawa S. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 35.Lawson ARJ, Abascal F, Coorens THH, Hooks Y, O'Neill L, Latimer C, Raine K, Sanders MA, Warren AY, Mahbubani KTA, Bareham B, Butler TM, Harvey LMR, Cagan A, Menzies A, Moore L, Colquhoun AJ, Turner W, Thomas B, Gnanapragasam V, Williams N, Rassl DM, Vöhringer H, Zumalave S, Nangalia J, Tubío JMC, Gerstung M, Saeb-Parsy K, Stratton MR, Campbell PJ, Mitchell TJ, Martincorena I. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science. 2020;370:75–82. doi: 10.1126/science.aba8347. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Du Y, Chen Z, Xu D, Lin T, Jin S, Wang G, Liu Z, Lu M, Chen X, Xu T, Bai F. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science. 2020;370:82–89. doi: 10.1126/science.aba7300. [DOI] [PubMed] [Google Scholar]
- 37.Moore L, Leongamornlert D, Coorens THH, Sanders MA, Ellis P, Dentro SC, Dawson KJ, Butler T, Rahbari R, Mitchell TJ, Maura F, Nangalia J, Tarpey PS, Brunner SF, Lee-Six H, Hooks Y, Moody S, Mahbubani KT, Jimenez-Linan M, Brosens JJ, Iacobuzio-Donahue CA, Martincorena I, Saeb-Parsy K, Campbell PJ, Stratton MR. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580:640–646. doi: 10.1038/s41586-020-2214-z. [DOI] [PubMed] [Google Scholar]
- 38.Lac V, Nazeran TM, Tessier-Cloutier B, Aguirre-Hernandez R, Albert A, Lum A, Khattra J, Praetorius T, Mason M, Chiu D, Köbel M, Yong PJ, Gilks BC, Anglesio MS, Huntsman DG. Oncogenic mutations in histologically normal endometrium: the new normal? J Pathol. 2019;249:173–181. doi: 10.1002/path.5314. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida K, Gowers KHC, Lee-Six H, Chandrasekharan DP, Coorens T, Maughan EF, Beal K, Menzies A, Millar FR, Anderson E, Clarke SE, Pennycuick A, Thakrar RM, Butler CR, Kakiuchi N, Hirano T, Hynds RE, Stratton MR, Martincorena I, Janes SM, Campbell PJ. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature. 2020;578:266–272. doi: 10.1038/s41586-020-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee-Six H, Øbro NF, Shepherd MS, Grossmann S, Dawson K, Belmonte M, Osborne RJ, Huntly BJP, Martincorena I, Anderson E, O'Neill L, Stratton MR, Laurenti E, Green AR, Kent DG, Campbell PJ. Population dynamics of normal human blood inferred from somatic mutations. Nature. 2018;561:473–478. doi: 10.1038/s41586-018-0497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson CJ, Papula AL, Poon GYP, Wong WH, Young AL, Druley TE, Fisher DS, Blundell JR. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–1454. doi: 10.1126/science.aay9333. [DOI] [PubMed] [Google Scholar]
- 43.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergami M, Berninger B. A fight for survival: the challenges faced by a newborn neuron integrating in the adult hippocampus. Dev Neurobiol. 2012;72:1016–1031. doi: 10.1002/dneu.22025. [DOI] [PubMed] [Google Scholar]
- 45.Merino MM, Rhiner C, Portela M, Moreno E. "Fitness fingerprints" mediate physiological culling of unwanted neurons in Drosophila. Curr Biol. 2013;23:1300–1309. doi: 10.1016/j.cub.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 46.Moreno E, Rhiner C. Darwin's multicellularity: from neurotrophic theories and cell competition to fitness fingerprints. Curr Opin Cell Biol. 2014;31:16–22. doi: 10.1016/j.ceb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coelho DS, Moreno E. Emerging links between cell competition and Alzheimer's disease. J Cell Sci. 2019;132:jcs231258. doi: 10.1242/jcs.231258. [DOI] [PubMed] [Google Scholar]
- 48.Lee MH, Siddoway B, Kaeser GE, Segota I, Rivera R, Romanow WJ, Liu CS, Park C, Kennedy G, Long T, Chun J. Somatic APP gene recombination in Alzheimer's disease and normal neurons. Nature. 2018;563:639–645. doi: 10.1038/s41586-018-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaeser G, Chun J. Brain cell somatic gene recombination and its phylogenetic foundations. J Biol Chem. 2020;295:12786–12795. doi: 10.1074/jbc.REV120.009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, Stebbings L, Menzies A, Widaa S, Stratton MR, Jones PH, Campbell PJ. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–996. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang J, Fewings E, Chang D, Zeng H, Liu S, Jorapur A, Belote RL, McNeal AS, Tan TM, Yeh I, Arron ST, Judson-Torres RL, Bastian BC, Shain AH. The genomic landscapes of individual melanocytes from human skin. Nature. 2020;586:600–605. doi: 10.1038/s41586-020-2785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fowler JC, King C, Bryant C, Hall MWJ, Sood R, Ong SH, Earp E, Fernandez-Antoran D, Koeppel J, Dentro SC, Shorthouse D, Durrani A, Fife K, Rytina E, Milne D, Roshan A, Mahububani K, Saeb-Parsy K, Hall BA, Gerstung M, Jones PH. Selection of oncogenic mutant clones in normal human skin varies with body site. Cancer Discov. 2021;11:340–361. doi: 10.1158/2159-8290.CD-20-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu M, Lu T, Jia Y, Luo X, Gopal P, Li L, Odewole M, Renteria V, Singal AG, Jang Y, Ge K, Wang SC, Sorouri M, Parekh JR, MacConmara MP, Yopp AC, Wang T, Zhu H. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177:608–621. doi: 10.1016/j.cell.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yizhak K, Aguet F, Kim J, Hess JM, Kübler K, Grimsby J, Frazer R, Zhang H, Haradhvala NJ, Rosebrock D, Livitz D, Li X, Arich-Landkof E, Shoresh N, Stewart C, Segrè AV, Branton PA, Polak P, Ardlie KG, Getz G. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science. 2019;364:eaaw0726. doi: 10.1126/science.aaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kakiuchi N, Ogawa S. Clonal expansion in non-cancer tissues. Nat Rev Cancer. 2021;21:239–256. doi: 10.1038/s41568-021-00335-3. [DOI] [PubMed] [Google Scholar]
- 57.Laconi E, Marongiu F, DeGregori J. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br J Cancer. 2020;122:943–952. doi: 10.1038/s41416-019-0721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeGregori J. Connecting cancer to its causes requires incorporation of effects on tissue microenvironments. Cancer Res. 2017;77:6065–6068. doi: 10.1158/0008-5472.CAN-17-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeGregori J. Evolved tumor suppression: why are we so good at not getting cancer? Cancer Res. 2011;71:3739–3744. doi: 10.1158/0008-5472.CAN-11-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner RP. Anecdotal, historical and critical commentaries on genetics. Rudolph Virchow and the genetic basis of somatic ecology. Genetics. 1999;151:917–920. doi: 10.1093/genetics/151.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huneman P. Biological individuals as “weak individuals” and their identity. Exploring a radical hypothesis in the metaphysics of science. In: Meincke AS, Dupré J, editors. Biological identity: perspectives from metaphysics and the philosophy of biology. London and New York: Routledge; 2020. pp. 40–62. [Google Scholar]
- 62.Hodgkin PD. Modifying clonal selection theory with a probabilistic cell. Immunol Rev. 2018;285:249–262. doi: 10.1111/imr.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tauber AI. Immunity: the evolution of an idea. New York: Oxford University Press; 2017. [Google Scholar]
- 64.Cohen IR, Efroni S. The immune system computes the state of the body: crowd wisdom, machine learning, and immune cell reference repertoires help manage inflammation. Front Immunol. 2019;10:10. doi: 10.3389/fimmu.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Baalen M, Huneman P. Organisms as ecosystems/ecosystems as organisms. Biol Theory. 2014;9:357–360. doi: 10.1007/s13752-014-0194-7. [DOI] [Google Scholar]
- 66.Thomas F, Roche B, Giraudeau M, Hamede R, Ujvari B. The interface between ecology, evolution, and cancer: more than ever a relevant research direction for both oncologists and ecologists. Evol Appl. 2020;13:1545–1549. doi: 10.1111/eva.13031. [DOI] [Google Scholar]
- 67.Dujon AM, Aktipis A, Alix-Panabières C, Amend SR, Boddy AM, Brown JS, Capp JP, DeGregori J, Ewald P, Gatenby R, Gerlinger M, Giraudeau M, Hamede RK, Hansen E, Kareva I, Maley CC, Marusyk A, McGranahan N, Metzger MJ, Nedelcu AM, Noble R, Nunney L, Pienta KJ, Polyak K, Pujol P, Read AF, Roche B, Sebens S, Solary E, Staňková K, Swain Ewald H, Thomas F, Ujvari B. Identifying key questions in the ecology and evolution of cancer. Evol Appl. 2021;14:877–892. doi: 10.1111/eva.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ujvari B, Roche B, Thomas F. Ecology and evolution of cancer. London: Academic Press; 2017. [Google Scholar]
- 69.Nevo U, Hauben E. Ecoimmunity: immune tolerance by symmetric co-evolution. Evol Dev. 2007;9:632–642. doi: 10.1111/j.1525-142X.2007.00201.x. [DOI] [PubMed] [Google Scholar]
- 70.Chiu L, Gilbert SF. The birth of the holobiont: multi-species birthing through mutual scaffolding and niche construction. Biosemiotics. 2015;8:191–210. doi: 10.1007/s12304-015-9232-5. [DOI] [Google Scholar]
- 71.Swiatczak B. Immune balance: the development of the idea and its applications. J Hist Biol. 2014;47:411–442. doi: 10.1007/s10739-013-9370-z. [DOI] [PubMed] [Google Scholar]
- 72.Gilbert SF, Sapp J, Tauber AI. A symbiotic view of life: we have never been individuals. Q Rev Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 73.Swiatczak B, Tauber AI (2020) Philosophy of immunology. In: Zalta EN (ed) The Stanford encyclopedia of philosophy, Stanford
- 74.Cohen IR. Updating Darwin: information and entropy drive the evolution of life. F1000Research. 2016;5:2808. doi: 10.12688/f1000research.10289.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chandra V, Bortnick A, Murre C. AID targeting: old mysteries and new challenges. Trends Immunol. 2015;36:527–535. doi: 10.1016/j.it.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rué P, Martinez Arias A. Cell dynamics and gene expression control in tissue homeostasis and development. Mol Syst Biol. 2015;11:792. doi: 10.15252/msb.20145549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 78.Nedelcu AM. The evolution of multicellularity and cancer: views and paradigms. Biochem Soc Trans. 2020;48:1505–1518. doi: 10.1042/BST20190992. [DOI] [PubMed] [Google Scholar]
- 79.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci USA. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.West SA, Cooper GA. Division of labour in microorganisms: an evolutionary perspective. Nat Rev Microbiol. 2016;14:716–723. doi: 10.1038/nrmicro.2016.111. [DOI] [PubMed] [Google Scholar]
- 81.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 82.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 83.Lean C, Plutynski A. The evolution of failure: explaining cancer as an evolutionary process. Biol Philos. 2016;31:39–57. doi: 10.1007/s10539-015-9511-1. [DOI] [Google Scholar]
- 84.Soto AM, Sonnenschein C. The tissue organization field theory of cancer: a testable replacement for the somatic mutation theory. BioEssays. 2011;33:332–340. doi: 10.1002/bies.201100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bizzarri M, Cucina A. Tumor and the microenvironment: a chance to reframe the paradigm of carcinogenesis? Biomed Res Int. 2014;2014:934038. doi: 10.1155/2014/934038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laconi E. The evolving concept of tumor microenvironments. BioEssays. 2007;29:738–744. doi: 10.1002/bies.20606. [DOI] [PubMed] [Google Scholar]
- 87.Brown S, Pineda CM, Xin T, Boucher J, Suozzi KC, Park S, Matte-Martone C, Gonzalez DG, Rytlewski J, Beronja S, Greco V. Correction of aberrant growth preserves tissue homeostasis. Nature. 2017;548:334–337. doi: 10.1038/nature23304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Murai K, Skrupskelyte G, Piedrafita G, Hall M, Kostiou V, Ong SH, Nagy T, Cagan A, Goulding D, Klein AM, Hall BA, Jones PH. Epidermal tissue adapts to restrain progenitors carrying clonal p53 mutations. Cell Stem Cell. 2018;23:687–699. doi: 10.1016/j.stem.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colom B, Alcolea MP, Piedrafita G, Hall MWJ, Wabik A, Dentro SC, Fowler JC, Herms A, King C, Ong SH, Sood RK, Gerstung M, Martincorena I, Hall BA, Jones PH. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat Genet. 2020;52:604–614. doi: 10.1038/s41588-020-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rozhok AI, DeGregori J. Toward an evolutionary model of cancer: considering the mechanisms that govern the fate of somatic mutations. Proc Natl Acad Sci USA. 2015;112:8914–8921. doi: 10.1073/pnas.1501713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marongiu F, Serra MP, Doratiotto S, Sini M, Fanti M, Cadoni E, Serra M, Laconi E. Aging promotes neoplastic disease through effects on the tissue microenvironment. Aging (Albany NY) 2016;8:3390–3399. doi: 10.18632/aging.101128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Henry CJ, Casás-Selves M, Kim J, Zaberezhnyy V, Aghili L, Daniel AE, Jimenez L, Azam T, McNamee EN, Clambey ET, Klawitter J, Serkova NJ, Tan AC, Dinarello CA, DeGregori J. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest. 2015;125:4666–4680. doi: 10.1172/JCI83024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tanimura N, Fujita Y. Epithelial defense against cancer (EDAC) Semin Cancer Biol. 2020;63:44–48. doi: 10.1016/j.semcancer.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 94.Baker NE. Emerging mechanisms of cell competition. Nat Rev Genet. 2020;21:683–697. doi: 10.1038/s41576-020-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nackley LL, West AG, Skowno AL, Bond WJ. The nebulous ecology of native invasions. Trends Ecol Evol. 2017;32:814–824. doi: 10.1016/j.tree.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 96.Fitzgerald DM, Hastings PJ, Rosenberg SM. Stress-induced mutagenesis: Implications in cancer and drug resistance. Annu Rev Cancer Biol. 2017;1:119–140. doi: 10.1146/annurev-cancerbio-050216-121919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cisneros L, Bussey KJ, Orr AJ, Miočević M, Lineweaver CH, Davies P. Ancient genes establish stress-induced mutation as a hallmark of cancer. PLoS ONE. 2017;12:e0176258. doi: 10.1371/journal.pone.0176258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.König SG, Nedelcu AM. The genetic basis for the evolution of soma: Mechanistic evidence for the co-option of a stress-induced gene into a developmental master regulator. Proc R Soc Lond B Biol Sci. 2020;287:20201414. doi: 10.1098/rspb.2020.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swiatczak B. Genomic stress responses drive lymphocyte evolvability: an ancient and ubiquitous mechanism. BioEssays. 2020;42:e2000032. doi: 10.1002/bies.202000032. [DOI] [PubMed] [Google Scholar]
- 100.Lee-Six H, Olafsson S, Ellis P, Osborne RJ, Sanders MA, Moore L, Georgakopoulos N, Torrente F, Noorani A, Goddard M, Robinson P, Coorens THH, O'Neill L, Alder C, Wang J, Fitzgerald RC, Zilbauer M, Coleman N, Saeb-Parsy K, Martincorena I, Campbell PJ, Stratton MR. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 101.Germain P-L. Cancer cells and adaptive explanations. Biol Philos. 2012;27:785–810. doi: 10.1007/s10539-012-9334-2. [DOI] [Google Scholar]
- 102.Maynard Smith J, Szathmáry E. The major transitions in evolution. Oxford: W.H. Freeman/Spektrum; 1995. [Google Scholar]
- 103.Dennett DC. From bacteria to Bach and back: the evolution of minds. New York: W.W. Norton; 2017. [Google Scholar]
- 104.Claidière N, Scott-Phillips TC, Sperber D. How Darwinian is cultural evolution? Philos Trans R Soc Lond B Biol Sci. 2014;369:20130368. doi: 10.1098/rstb.2013.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heams T. Selection within organisms in the nineteenth century: Wilhelm Roux's complex legacy. Prog Biophys Mol Biol. 2012;110:24–33. doi: 10.1016/j.pbiomolbio.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 106.Roux W. Der kampf der theile im organismus: Ein beitrag zur vervollständigung der mechanischen zweckmässigkeitslehre. Leipzig: W. Engelmann; 1881. [Google Scholar]
- 107.Méthot P-O, et al. Darwin, evolution, and medicine: historical and contemporary perspectives. In: Heams T, et al., editors. Handbook of evolutionary thinking in the sciences. Bâle: Springer; 2015. pp. 587–617. [Google Scholar]
- 108.Grawitz P. Die theorie der schutzimpfung. Virchow’s Archiv f Patholog Anatomie und Physiologie u f Klin Medicin. 1881;84:87–110. doi: 10.1007/BF01935470. [DOI] [Google Scholar]
- 109.Welch WH. Adaptation in pathological processes. Science. 1897;5:813–832. doi: 10.1126/science.5.126.813. [DOI] [PubMed] [Google Scholar]
- 110.Tauber AI, Chernyak L. Metchnikoff and the origins of immunology. Oxford: Oxford University Press; 1991. [Google Scholar]
- 111.Gourko H, Williamson DI, Tauber AI. The evolutionary biology papers of Elie Metchnikoff. Dordrecht: Kluwer; 2000. [Google Scholar]
- 112.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 113.Flajnik MF. Re-evaluation of the immunological Big Bang. Curr Biol. 2014;24:R1060–R1065. doi: 10.1016/j.cub.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Swiatczak B, Cohen IR. Gut feelings of safety: tolerance to the microbiota mediated by innate immune receptors. Microbiol Immunol. 2015;59:573–585. doi: 10.1111/1348-0421.12318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.