Abstract
Since the emergence of the first case of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2), the viral genome has constantly undergone rapid mutations for better adaptation in the host system. These newer mutations have given rise to several lineages/ variants of the virus that have resulted in high transmission and virulence rates compared to the previously circulating variants. Owing to this, the overall caseload and related mortality have tremendously increased globally to > 233 million infections and > 4.7 million deaths as of Sept. 28th, 2021. SARS-CoV-2, Spike (S) protein binds to host cells by recognizing human angiotensin-converting enzyme 2 (hACE2) receptor. The viral S protein contains S1 and S2 domains that constitute the binding and fusion machinery, respectively. Structural analysis of viral S protein reveals that the virus undergoes conformational flexibility and dynamicity to interact with the hACE2 receptor. The SARS-CoV-2 variants and mutations might be associated with affecting the conformational plasticity of S protein, potentially linked to its altered affinity, infectivity, and immunogenicity. This review focuses on the current circulating variants of SARS-CoV-2 and the structure–function analysis of key S protein mutations linked with increased affinity, higher infectivity, enhanced transmission rates, and immune escape against this infection.
Keywords: Lineages, S protein, SARS-CoV-2, Structure-functional analysis, Variant of concern (VOC)
Introduction
Novel coronavirus, also termed Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of Corona virus disease (COVID-19), continuously imposes serious challenges to both global health and economic conditions [1]. The virus was first reported from Wuhan city, China among patients suffereing from pneumonia with unknown etiology. Subsequently, it spread and became a serious health problem worldwide. World Health Organization (WHO) declared SARS-CoV-2 infection as a pandemic in March 2020 [2]. As of September 2021, more than 233,158,434 infections and 4,771,151 deaths have been reported globally [3].
To date, more than 1.2 million SARS-CoV-2 genome sequences are available at Global Initiative on Sharing Avian Influenza Data (GISAID), permitting real-time surveillance of the variants identified globally [4]. The deposited data enable comprehension of epidemiology related to evolving SARS-CoV-2 variants and related mutations associated with modified viral properties. The emergence of multiple variants of SARS-CoV-2, represents its consistent evolutionary fitness in terms of virus adaptability to evade host immune response and confer enhanced infectivity [5]. Various nomenclature systems have been adopted for determining the emerging variants of SARS-CoV-2. The recently approved WHO classification system distinguishes new emerging variants into the variant of concern (VOC) and the variant of interest (VOI) niche [6]. SARS-CoV-2 is known to recognize the human angiotensin-converting enzyme 2 (hACE2) receptor through its Spike (S) protein. Mutations in the S protein of SARS-CoV-2 have been linked with high transmissibility, increased virulence, enhanced infectivity, poor diagnostic sensitivity, and viral immune escape. Therefore, S protein has been considered as an attractive immunogenic target for vaccine design [7]. Several vaccines are in phase III clinical trials, wherein some of them have been approved globally for emergency use. Due to the continuously evolving nature of SARS-CoV-2, the efficacy of these vaccines against the emerging variants is still largely unknown [8]. As the virus is rapidly mutating its genome, the design of a specific drug or vaccine against a particular variant of SARS-CoV-2 is difficult. This review summarizes the structural, functional, and antigenic aspects of the interactions involving S protein from SARS-CoV-2 with its host hACE2 receptor and the current emerging variants associated with the rapid resurgence of SARS-CoV-2 infections. The emphasis is on the structural and functional aspects of the key S protein mutations, associated with WHO-defined variants linked with increased affinity, transmission, and immune escape. This review will help to develop newer strategies for the design of drugs and immunotherapies to combat viral infection.
Genomic organization of SARS-CoV-2
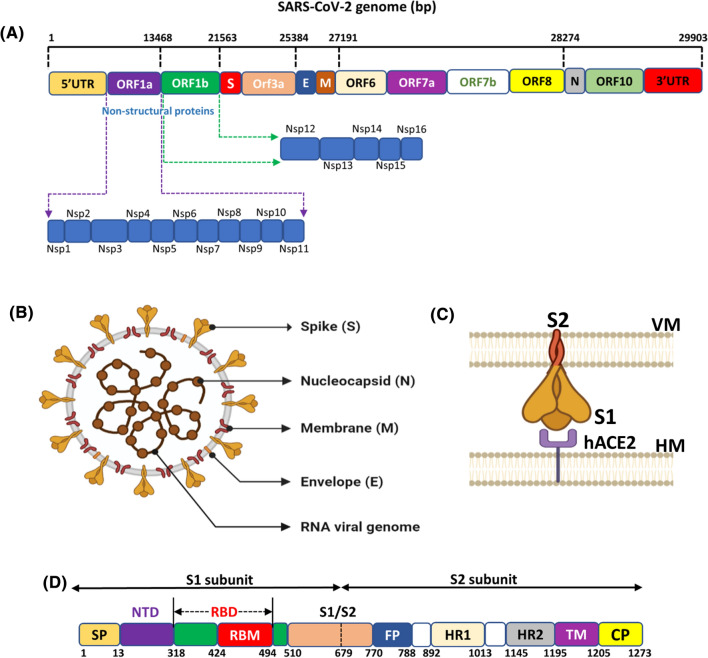
The SARS-CoV-2 virus belongs to the beta coronavirus family that contains single positive-strand RNA as a genetic material [9]. Its viral genome contains approximately 29,881 nucleotides (Gene ID—MN908947) which encode 9860 amino acids [10]. The viral genome of SARS-CoV-2 bound to nucleocapsid consists of a 5′- cap and a 3′- poly-acetylated tail (A) bordered by 5′ and 3′ untranslated region (UTRs) [11]. The 5′UTR and 3′UTR comprise 265 and 229 nucleotides, respectively. These UTRs play an essential role in viral gene transcription and regulation. The viral genome includes several open reading frames (ORFs) that translate into multiple proteins [12]. The 5’-end consists of the ORF1a and ORF1b genes that harbor most of the genome (~ 21 kb). The tail portion towards the 3’-end contains the gene for four structural proteins (envelope (E), matrix (M), nucleocapsid (N), S protein and nine accessory proteins [13]. The functions of the accessory genes ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c and ORF10 [14] are not clear yet (Fig. 1A). ORF1a and ORF1b genes encode for 2 polypeptides, pp1a and pp1ab, which are then chopped into 16 non-structural proteins (nsps) by viral chymotrypsin-like protease (3CLpro) or main protease (Mpro) and papain-like proteases (PLpro) [15]. PLpro cleaves the site between nsp1/2, nsp2/3, nsp3/4 proteins while the 3CLpro cleaves at the Q/SAG site to produce the non-structural proteins, nsp4 to nsp16. Nsp12 (known as RNA-dependent RNA polymerase) along with nsp7, nsp8, nsp13 (helicase), and nsp14 (exonuclease) are the key enzymes that mediate the replication and transcription machinery of SARS-CoV-2 in the host cell. These nsps also react to regulate the host immune system and act as a cofactor for other nsps to activate them and amplify their functions [16]. Both nsp3 and nsp16 hamper the natural immune responses of the host cells to promote viral proliferation [17].
Fig. 1.
The structural organization of SARS-CoV-2 genome. (A) Schematic representation of SARS-CoV-2 genome organization where, bp: base pair; ORF: Open reading frame; S: Spike protein; E: Envelope protein; M: Membrane protein; N: Nucleocapsid protein. (B) The structural features of SARS-CoV-2 (C) S1 and S2 subunits of SARS-CoV-2 S protein. (C) Block representation of SARS-CoV-2 S protein with its different domains namely, NTD, N-terminal domain; RBD, receptor-binding domain; FP, fusion peptide; HR1, heptad repeat 1, HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic domain
Structural proteins of SARS-CoV-2
Structurally, the virus has a double-layered lipid envelope that contains four structural proteins namely, S, M, E, and N (Fig. 1B) [18]. The S protein has been recognized to contribute significantly to both cell recognition as well as the fusion to the cell membrane. The S protein comprises two monomeric subunits S1 and S2, which assemble into trimers on the surface of the virion. The S1 monomeric subunit forms the head region while the S2 subunit encompasses the stalk region of S protein that together impart the visual effect of the crown shape virus [14] (Fig. 1C). S protein incorporates the receptor-binding domain (RBD) that participates in binding viral particles to the host cell receptors. Therefore, this domain is widely used as a target for neutralizing antibodies generated after infection, vaccination and monoclonal antibodies and as a drug target [19].
The M protein spans the viral membrane bilayer and is responsible for maintaining the shape of the virion. The highly conserved E protein is a hydropathic transmembrane protein, rich in valine and leucine residues that are essential for viral pathogenesis [20]. It plays a vital role in the budding process of viral particles from infected macrophages. The E and M proteins collectively facilitate the viral entry into the host cell, replication, and assembly of viruses within human host cells. Together with the viral RNA, the N protein constitutes the nucleocapsid and functions to maintain the genome structure and facilitate viral assembly, replication, and interaction with host cells [21].
Structural organization of S protein
The S protein contains 1273 amino acid residues, distributed into S1 and S2 subunits, spanning (14–685) and (686–1273) residues respectively, preceeded by a 1–13 amino acid long signal polypeptide chain towards the N-terminal end. The S1 subunit represents the variable part of the protein, while the S2 subunit is much more conserved. The first 13 amino acid residues constituting the signal peptide (SP) of S1 are required to guide its transportation to its membrane destination [22]. The S1 subunit contains the N-terminal domain (NTD) (14–305) and RBD spanning 319–541 residues. The S2 subunit incorporates the fusion peptide (FP) (788–806), heptapeptide repeat sequence 1 (HR1) (912–984), heptapeptide repeat sequence 2 (HR2) (1163–1213), transmembrane domain (TM) (1213–1237), and the cytoplasm domain (CP) (1237–1273) [20] (Fig. 1D)
NTD is responsible for binding to carbohydrates. RBD of S protein permits the virus entry into the host cell receptor. RBD has extensively been studied as an anti-SARS-CoV-2 drug target and vaccine candidate [23]. The FP, located in the S2 subdomain, comprising 15–20 residues, plays a vital role in mediating membrane fusion with the hACE2. HR1 and HR2 contain a heptapeptide repeat sequence, HPPHCPC (where H stands for hydrophobic residue, P for polar and C for charged), and interact with each other through hydrophobic (non-polar) interactions to form a six helical bundled (6-HB) fusion core structure [24]. This coiled-coil core conformation is mainly responsible for bringing viral and human cells in close vicinity for fusion, thus permitting the viral attachment to the host cell. The TM domain of the S2 subunit is highly conserved in SARS-CoV-2 and, along with the CP domain, contributes to the adherence of S protein to the host cells.
SARS-CoV-2 entry into host cells
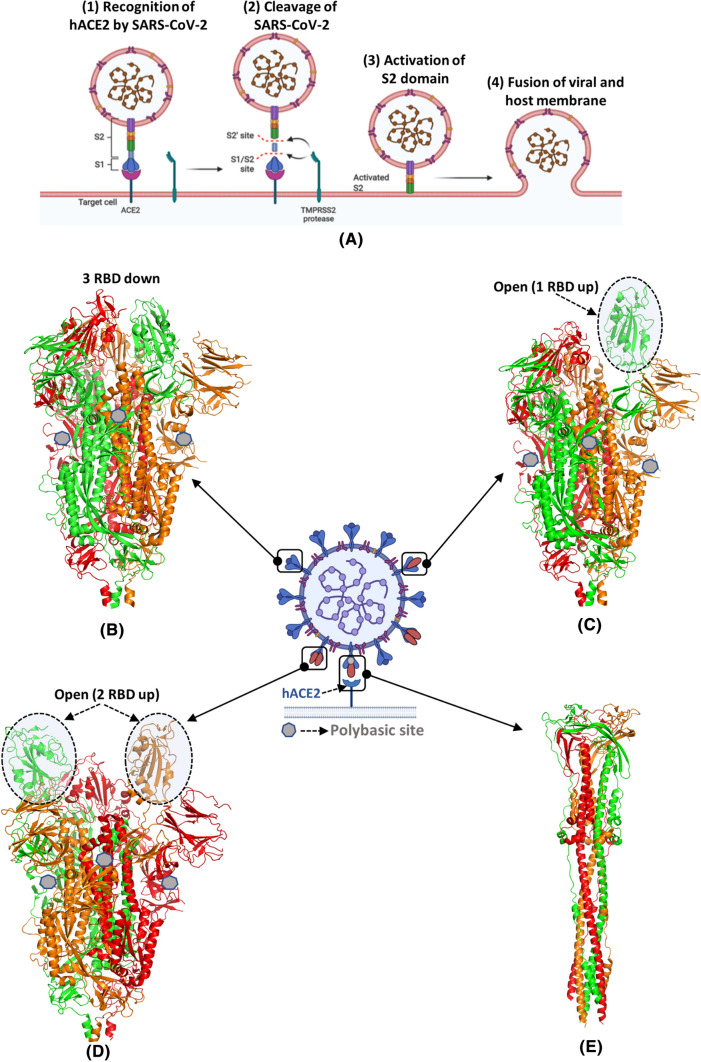
SARS-CoV-2 invades the host cell by establishing direct contact with the host cell receptor. Within the S1 subunit, RBD specifically recognizes the integral membrane protein, hACE2, as its receptor [25] (Fig. 2A: Step (1)). The conformation of S protein alters after its association with the hACE2 receptor and facilitates the availability of the S1 and S2 cleavage sites to the transmembrane protease serine kinase 2 (TRMPSS2) (Fig. 2A: Step(2)) [26]. The cleavage of S1 and S2 enables the fusion of S protein with the host by facilitating the insertion of the FP domain of the S2 subunit into the host cell membrane (Fig. 2A: Step (3)). This induces another conformational change in S2 subunit, which facilitates the viral and host membranes to attain sufficient proximity. Consequently, this triggers membrane fusion resulting in viral-host cellular fusion and viral entrance into host cells (Fig. 2A: Step (4)).
Fig. 2.
Interactions of SARS-CoV-2 with hACE2 receptor. (A) Diagrammatic representation of the entry of SARS-CoV-2 in the human cell through angiotensin-converting enzyme type 2 receptor (hACE2). (B-E) Cryo-EM structures of trimeric S protein representing structural plasticity in its prefusion and postfusion states (B) three RBD down closed prefusion state (PDB id: 6VXX) (C) one RBD up prefusion states (PDB id: 6VYB) (D) two RBD up (PDB id: 6X2B) prefusion state (E) postfusion state (PDB id: 6M3W) of the S protein. Three protomers of S protein are colored by chain using red, green, and orange colors. The polybasic cleavage site is represented in the centre in grey heptagon
Cryo-EM structures of SARS-CoV-2 S protein
The cryo-electron microscopy structure reveals that the SARS-CoV-2 S protein adopts two isoforms, the prefusion and postfusion [27]. The prefusion isoform is further classified into three conformations, closed (PDB id: 6VXX), open RBD up (PDB id: 6VYB), and two RBD up (PDB id: 6X2B) (Fig. 2 BCD) [28]. In the closed conformation of the prefusion isoform, all the three RBDs are covered by the NTD of S protein. While, in the open conformation, the RBD is dissociated from its apical central axis of the S2 subdomain. The two up conformations of RBD (both one RBD and two RBD up) have the ability to recognize the hACE2 receptor. S protein undergoes conformational plasticity during its transition from prefusion to the postfusion state upon binding to the hACE2 receptor. The postfusion state is characterized by the formation of a needle-like structure in which both FP and TM associate together (PDB id: 6M3W) (Fig. 2E). The postfusion trimer state is possibly linked with immune escape mechanism of SARS-CoV-2 as this state masks the prefusion trimers with neutralizing antibodies and thus cannot be used as a vaccine target [19, 27, 29].
Three-dimensional structure of RBD-hACE2 complex
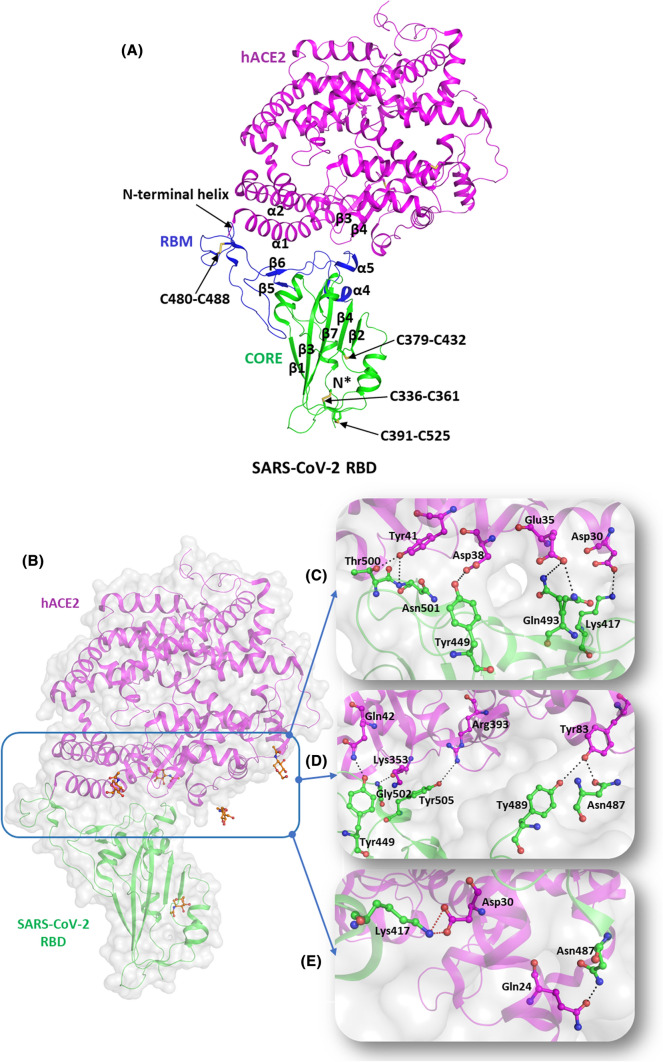
The hACE2 is a membrane protein, which primarily functions as a hormone. It is distributed in the alveolar epithelial type II cells of the lungs, kidney, heart, and intestine [30]. hACE2 is the main receptor responsible for the binding of SARS-CoV-2 to host cells [31]. The N terminal region of hACE2 receptor contains the N-terminal protease domain and a collectrin-like C-terminal domain. The RBD is housed on the N-terminal peptidase domain of the hACE2 receptor. At the same time, the contact regions are located in the two helices (α1 and α2) as well as the loop region between two antiparallel strands (β3 and β4) (Fig. 3A).
Fig. 3.
Molecular mechanisms of interaction of SARS-CoV-2 S protein with hACE2 receptors. (A) Structural representation of SARS-CoV-2 RBD (green, cartoon) with RBM (blue, cartoon) bound to hACE2 (magenta, cartoon) (PBD id: 6M0J). The disulfide bridges are highlighted in yellow (stick representation). (B) The interaction interface of hACE2 (magenta, cartoon) and SARS-CoV-2 (green, cartoon) with NAG (orange, ball and stick). Several hydrogen bonds interactions and salt bridges stabilize the RBD-hACE2 complex. (C) Hydrogen-bonded interactions involving residues Tyr449, Gln493, Thr500, and Asn501 of RBD (igreen, ball and stick) with Asp38, Glu35, and Tyr41 of hACE2 receptor, respectively. (D) Hydrogen-bonded interactions between the residues Tyr487 and Asn489 of RBD (green, ball and stick) and Tyr83 of hACE2 (magenta, ball and stick). It also depicts the involvement of Tyr449, Gly502, and Tyr505 residues of RBD (green, ball and stick) with Gln42, Lys353, and Arg393 of hACE2 (magenta, ball and stick) through H bonds. (E) Hydrogen-bonded interactions of Asp30 (RBD, green ball and stick) with Gln24 (hACE2, magenta, ball and stick) and salt bridge interactions between Lys 417 (RBD, green ball and stick) and Asp30 (hACE2, magenta, ball and stick). Hydrogen bonds are marked as blacked dotted lines and salt bridges as red dotted lines
RBD is one of the antigenic determining components of the viral S protein and encompasses most of the recognition sites to facilitate SARS-CoV-2 interaction with the hACE2 receptor. The three-dimensional structural analysis of the RBD-hACE2 complex revealed the presence of five β strands (β1, β2, β3, β4, and β7) arranged in antiparallel mode with short connecting loops and helices to constitute the RBD core (PDB id: 6M0J) [32] (Fig. 3A). Between the β4 and β7 strands of the core protein, an extended insertion referred to as the receptor-binding motif (RBM) is present, which includes the short β5 and β6 sheets, α4 and α5 helices and loop regions. Most of the contacting residues of S protein lie in this region. Three pairs of disulfide linkages present in the core RBD (Cys336-Cys361, Cys379-Cys432, and Cys391-Cys525) stabilize the β sheet region of the core S protein while one pair (Cys480- Cys488) connects the loop regions present at the end of RBM [33].
The RBD of S protein interacts with N-terminal helix of hACE2 receptor through hydrophilic interactions (Fig. 3B). RBD contains the RBM (residues 424–494) that directly interact with the N terminal region of the hACE2 receptor. The residues Tyr449, Gln493, Thr500, and Asn501 of RBM make hydrogen-bonded interactions with Asp38, Glu35, and Tyr41 of hACE2 receptor, respectively (Fig. 3C). However, Lys417, present outside the RBM, is uniquely situated to form both hydrogen-bonded and salt bridge interaction with Asp30 residue of RBD (Fig. 3C). The polar residues, Asn487 and Tyr489, of RBM make hydrogen-bonded interactions with Tyr83 of hACE2 receptor while Gly502 and Tyr505 of RBM form hydrogen bonds with Lys353 and Arg393 of hACE2 receptor (Fig. 3D). Further, the RBD-hACE2 interaction is stabilized by hydrogen-bonded interactions mediated by Asn487 with Gln24 of hACE2 receptor (Fig. 3E).
Emerging SARS-CoV-2 variants
Throughout the COVID-19 pandemic, large number of sequences of the SARS-CoV-2 viral genomes have been deposited to public databases [6]. There are presently more than 38,63,286 publicly available complete or nearly complete SARS-CoV-2 genome sequences (28th Sept. 2021), and the number continues to grow [6]. These new SARS-CoV-2 variants harbour mutations at nsps as well as the structural proteins of SARS-Cov-2 that might impact viral fitness and transmissibility [34, 35].
There are various terms like variant, strain, and lineage currently being used to describe different forms of SARS-CoV-2. A new variant is said to be formed when specific mutations get selected through several cycles of viral replication. If these sequence variations yield a virus that noticeably has specific phenotypical characteristics, then it is designated as a new strain [36]. The variants can be grouped into larger groups like lineages and clades [37]. A new lineage is formed when the novel variant can be distinguished as a separate branch on a phylogenetic tree [36].
There are three main nomenclature systems for the classification of SARS-CoV-2 at the species level, as proposed by GISAID [6], Nextstrain [37], and Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) [38].
GISAID was established as a global science initiative and primary source in 2008 and provides an open-access platform to upload and retrieve information related to influenza viruses and the coronavirus [39]. The GISAID nomenclature system is based on the large clades formed by the variants [40]. The variants are segregated into eight different clades termed S, O, L, V, G, GH, GR, and GV based on global data available on GISAID [41]. Nextstrain is an open-source pipeline for phylogenetic and phylodynamic analysis to track the pathogen evolution [37]. The SARS-CoV-2 lineages are referred to as the mutation accumulated with the year of detection followed by a successive alphabetic letter. Nextstrain classified the SARS-CoV-2 genome in 13 clades, including 19A to 19B, 20A to 20 J, and 21A [42]. PANGOLIN has proposed a dynamic nomenclature for the appellation of SARS-CoV-2 lineages [38]. In this classification scheme, the viral genome is divided into lineages like A, B, and C. These lineages are further subdivided into sub-lineages by adding the numeric numbers up to four different levels arranged hierarchically, each level is considered as a descendant from a former level [38].
Public health England (PHE) has categorized viral lineages into three classes: VOC, VOI, and a variant of high consequence [43]. Any SARS-CoV-2 strain is assigned as a VOI if, it harbors new mutations with recognized or supposed phenotypic consequences compared to the reference strain. Simultaneously, it should be responsible for transmission in a specific area, causing multiple COVID-19 cases/clusters, or have been reported in multiple countries. VOC is a SARS-CoV-2 variant that meets the definition of a VOI and is associated with at least one change with a public health implication like upsurge in transmission or specific epidemiological changes, enhanced virulence or modification in its exhibition of clinical illness; declined efficacy of preventive or therapeutic measures. Each variant is named by year, month and one number in the format [YY]-[MMM]-[NN]. A suffix like 'VOI' and 'VOC' is included to specify a variant under investigation and a variant of concern, respectively [13].
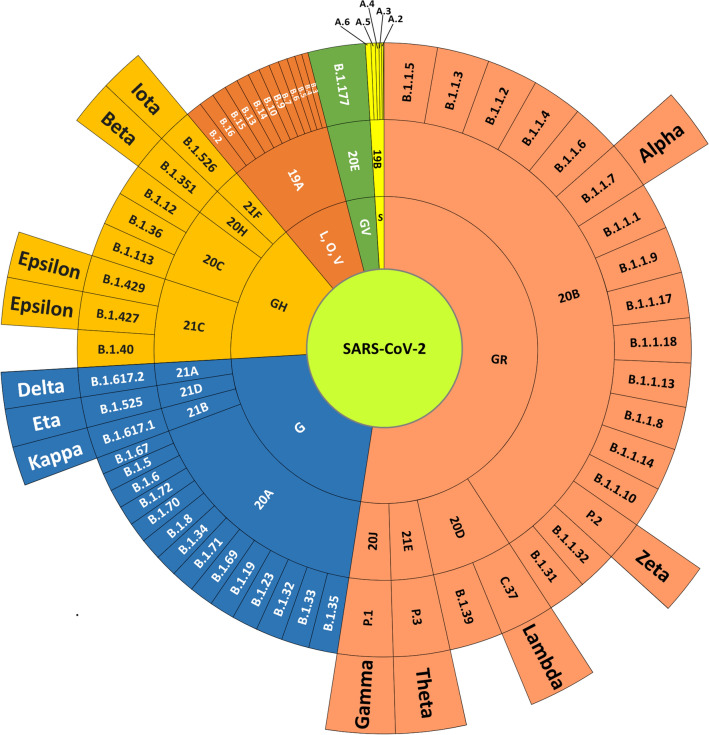
WHO has recently proposed the nomenclature for VOIs and VOCs using the Greek alphabets (Table 1 and Fig. 4) [44]. This nomenclature is not intended to replace the phylogenetic labels but provides consistent names to identify VOCs and VOIs present across the globe [45]. Phylogenetic classification schemes for SARS-CoV-2 nomenclature are given in Table 1.
Table 1.
Phylogenetic classification schemes for SARS-CoV-2 nomenclature
| WHO (Label) | Pangolin (Lineage) | GISAID (Clade/Lineage) | Nextstrain (Clade) | Mutations | First Isolated (Location and Date) | |
|---|---|---|---|---|---|---|
| VOC | Alpha (α) |
B.1.1.7 |
GRY (formerly GR/501Y.V1) | 20I/501Y.V1 | H69-70, Y144-145, N501Y, P681H, L452R, E484Q [48–50] |
United Kingdom, September, 2020 |
| Beta (β) |
B.1.351 [51] |
GH/501Y.V2 [51] |
20H/501Y.V2 |
L18F, D80A D215G, (Del 241–243), K417N, E484K, N501Y, D614G, A701V [52, 53] |
South Africa, May,2020 |
|
| Gamma (γ) | P.1 | GR/501Y.V3 | 20 J/501Y.V3 | L18F, T20N, P26S, D138Y, R190S, K417N/T, E484K, N501Y, D614G, H655Y, T1027I, V1176F [54, 55] |
Brazil, November, 2020 |
|
| Delta (δ) | B.1.617.2 | G/478 K.V1 | 21A/S:478 K |
T19R, G142D, 156del, 157del, R158G, L452R, T478K, D614G, P681R, D950N |
India, October, 2020 |
|
| VOI | Epsilon (ε) | B.1.427/B.1.429 | GH/452R.V1 | 21C |
S13I, W152C, L452R, D614G |
United States of America, March, 2020 |
| Zeta (ζ) | P.2 | GR/484 K.V2 | 20B/S.484 K | E484K, D614G, T859I, V1176F [64] | Brazil, April, 2020 | |
| Eta (η) | B.1.525 | G/484 K.V3 | 21D | Q52R, E484K, D614G, Q677H, F888L [65] |
Multiple countries, December, 2020 |
|
| Theta (θ) | P.3 | GR/1092 K.V1 | 21E | E484K, N501Y, D614G, P681H, E1092K, H1101Y, V1176F [66] |
Philippines, January, 2021 |
|
| Iota (ι) | B.1.526 | GH/253G.V1 | 21F | L5F, T95I, D253G, S477N, E484K, D614G, A701V [67] |
United States of America, November, 2020 |
|
| Kappa (κ) | B.1.617.1 | G/452R.V3 | 21B | L452R, E484Q, D614G, P681R [58] | India, October, 2020 | |
| Lambda (λ) | C.37 | GR/452Q.V1 | 20D | G75V, T76I, DEL247/253, L452Q, F490S, D614G, T859N [68] |
Peru, August, 2020 |
*References are provided in square brackets
Fig. 4.
Nomenclature systems of SARS-CoV-2. The schematic representation of the phylogenetic association between different nomenclature systems: The sunburst diagram represents the annotations from GISAID, Nextstrain, PANGOLIN, and WHO from inner to outer circles
Alpha (α)
The alpha variant was first detected in September 2020 from the United Kingdom. It belongs to lineage B.1.1.7 and is notated as a VOC-202012/01 and 20I/501Y.V1 (Next strain) [46, 47]. As of May 2021, this lineage has been reported in 120 countries. Genome analysis revealed 23 mutations, out of which 17 are non-synonymous. The most common mutations in the S protein, E484K, N501Y, A570D, D614G, T716I, S982A, D1118H, and P681H [48] and deletions (ΔH69, ΔV70, ΔY144), are associated with the evading ability of corona virus against host immune responses [49, 50].
Beta (β)
The B.1.351 lineage in PANGOLIN system is designated as beta variant and referred to as H/501Y.V2 and VOC-20DEC-02 by GISAID and PHE, respectively [51]. The first sequence of this variant was reported in South Africa in May 2020 [52]. The most frequent mutations present in beta variant include L18F, D80A, D215G, Δ242, Δ243, Δ244, R246I, K417N, E484K, N501Y, D614G, and A701V in the S protein region, which is responsible for increased binding affinity for human cells and ineffectiveness against host immune system [53].
Gamma (γ)
This variant first reported from Brazil belongs to P.1 lineage and is termed as a variant of concern (January, 2021) by PHE and 20 J/501Y.V3 by Nextstrain [6, 37]. As per PANGOLIN nomenclature, P.1 lineage is an alias form of the lineage B.1.1.28.1. The Gamma variant possesses 17 unique amino acid substitutions, out of which 10 are harboured in the S region [54]. The most common mutations observed are L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, and T1027I [55].
Delta (δ)
The new coronavirus variant belonging to lineage B1.617.2 is labeled as the Delta variant by WHO. This variant is also termed as G/478 K.V1 by GISAID and 21A/S: 478 K by Nextstrain nomenclature system. This variant was first reported in October 2020 in India and has also been annotated as the double mutant variant by INSACOG (Indian SARS-CoV-2 Consortium on Genomics) [56]. The Delta variant harbors a double mutation in S protein, that yields increased infectivity and immune response evasion. The most prevalent substitutions present in the Delta variant are T19R, Δ157-158, L452R, T478K/T478R, D614G, P681R, and D950N [57]. A sub-variant of Delta housing the substitution at K417N (also present in Beta variant) was nicknamed "Delta plus K417N" (AY.1) on 14 June 2021 [135]. The emergence of this variant has raised concern about the likelihood of immunity evasion and reinfection [23].
Kappa (κ)
The Kappa variant of SARS-CoV-2, also recognized as lineage B.1.617.1, is one of the three sub-lineages of Pango lineage B.1.617 [45]. It was firstly reported from India in December, 2020 and accounted for more than half of the variants identified in India. Public Health England designated it as a VOI (VUI-21APR-01) on April 1, 2021. This variant contains 13 S protein mutations, the majority of which include the amino acid substitutions L452R, E484Q, D614G, P681R, and Q1071H [58]. This variant is also reported to be resistant against neutralizing X593 and P2B-2F6 monoclonal antibodies [59, 60].
Epsilon (ε)
The WHO has assigned the lineage B.1.429 and B.1.427 as the Epsilon variants. This lineage first reported in California is also known as GH/452R.V1 and 21C as per Nextstrain and GISAID nomenclature, respectively [45]. The lineage B.1.427 of Epsilon variant harbors the mutants, D614G and L452R, which have reduced affinities with monoclonal antibodies and polyclonal sera. The lineage B.1.429 contains five distinct mutations, including S13I, W152C, L452R in the S region and I4205V and D1183Y in the region, ORF1ab [35]. The signature substitution, L452R, present in other unrelated lineages might be responsible for high transmissibility [61, 62].
Zeta (ζ)
The P.2 variant, a derivative of B.1.1.28, like P.1, is categorized as a variant of concern, Zeta, by WHO [45]. It was first reported in the state of Rio de Janeiro, Brazil [63]. Other mutations included in the zeta lineage are D614G, V1176F, P314L, L3468V, L3930F, A119S, R203K, G204R, and M234I [64].
Eta (η)
The Eta variant belongs to the PANGOLIN lineage B.1.525, and is also recognized as VUI-21FEB-03 and 21D. It was first reported in December 2020 in the United Kingdom and Nigeria simultaneously [65]. This variant harbors the E484K substitution detected in Gamma, Zeta, and Beta variants, and ΔH69/ΔV70 as present in the Alpha variant. It also has been reported to contain an additional mutation, F888L [35].
Theta (θ)
The PANGOLIN lineage P.3 is designated as Theta by WHO [45]. This lineage belongs to GR/1092 K.V1 and 21E clade of GISAID and Nextstrain classification systems, respectively [6, 37]. The Theta variant was first reported in the Philippines on 18 February 2021. This variant is derived from the lineage B.1.1.28, and like the Gamma variant, it comprises two amino acid substitutions, E484K and N501Y [66].
Iota (ι)
The lineage B.1.526 is assigned to the VOI category by WHO and called the Iota variant [45]. This lineage belongs to GH/253G.V1 clade of GISAID and 21F clade of Nextstrain [6, 37]. This variant was first reported in November 2020 in the USA and since spread to 18 countries by April 11, 2021 [67].
Lambda (λ)
The Lambda variant is designated as C.37, GR/452Q.V1, and 20D in PANGOLIN, GISAID, and Nextstrain nomenclatures, respectively [6, 37, 38]. The first case was reported in Peru in August 2020 and, by April 2021, reached a frequency of over 80% of the total cases reported. WHO called it a VOC on Jun 24, 2021 [68].
Mutation of the S protein
Several structural, biophysical and bioinformatics studies have identified S protein as a critical target for protective antibodies, wherein NTD and RBD (S1subunit) have been identified as the major immunogenic regions of S protein. However, limited data are available on the antigenicity of the S2 subunit. The interaction of RBD with the host cell has been extensively studied as an attractive immunogenic target for vaccine design. The structure–function relationship of the key frequently occurring mutations has been discussed here and is provided in Table 2.
Table 2.
Key S protein mutations with their location and significance
| Serial Number |
Mutation | Spike protein region | Significance |
|---|---|---|---|
| 1 | L18F | S1-NTD | Compromises the binding of neutralizing antibody [128]* |
| 2 | Δ69 /70 | S1 -NTD | Role in immune escape and diagnostic failure [159] |
| 3 | L54F | S1-NTD | Increases infectivity and transmissibility [160] |
| 4 | Δ144 | S1-NTD | Alters immunogenicity [161] |
| 5 | Δ156 | S1-NTD | Alters immunogenicity [34] |
| 6 | Δ157 | S1-NTD | Alters immunogenicity [34] |
| 7 | D215G | S1-NTD | Alters immunogenicity [34] |
| 8 | A222V | S1-NTD | Alters immunogenicity [34] |
| 9 | K417N/K417T | S1-NTD-RBD | Helps the virion to bind with hACE2 in a more efficient way [84, 131] |
| 10 | L452R | S1-RBD (RBM) | Increases S protein affinity towards hACE2 and linked with reduce neutralization [90, 102] |
| 11 | N439K |
S1-RBD (RBM) |
Increases binding affinity towards hACE2 receptor and linked with immune evasion towards neutralization activity [88] |
| 12 | N440K | S1-RBD (RBM) | High infectivity, transmission, and immune escape [107, 108] |
| 13 | Y453F | S1-RBD (RBM) | Enhanced binding affinity to hACE2 receptor and can likely evade neutralizing antibodies [114, 117] |
| 14 | S477G/N | S1-RBD (RBM) | Increases binding affinity towards hACE2 receptor [89] |
| 15 | T478K | S1-RBD (RBM) | Increases infectivity and reduce neutralizing activity [158] |
| 16 | E484K/Q | S1-RBD (RBM) | Reduces neutralization activity [104] |
| 17 | N501Y | S1-RBD (RBM) | Increase binding affinity to hACE2 receptor and increased transmission and infectivity [75] |
| 18 | A570D | S1-spike | Alters the balance of closed to open conformation [157] |
| 19 | D614G | S1-spike | Predominant mutation linked with increase affinity towards hACE2 receptor and increased transmission [70–73] |
| 20 | H655Y | S1-spike part of S1/S2 junction | Furin cleavage [162] |
| 21 | P681H | S1- spike part of S1/S2 junction | Proximal to furin cleavage site [162] |
| 22 | H1101D/Y | S2- spike | Increases the stability of spike protein by surface alteration [163] |
| 33 | G1124V | S2-spike | Imparts structural rigidity to S2 subunit [164] |
*References are provided in square brackets
Predominant mutations of S protein
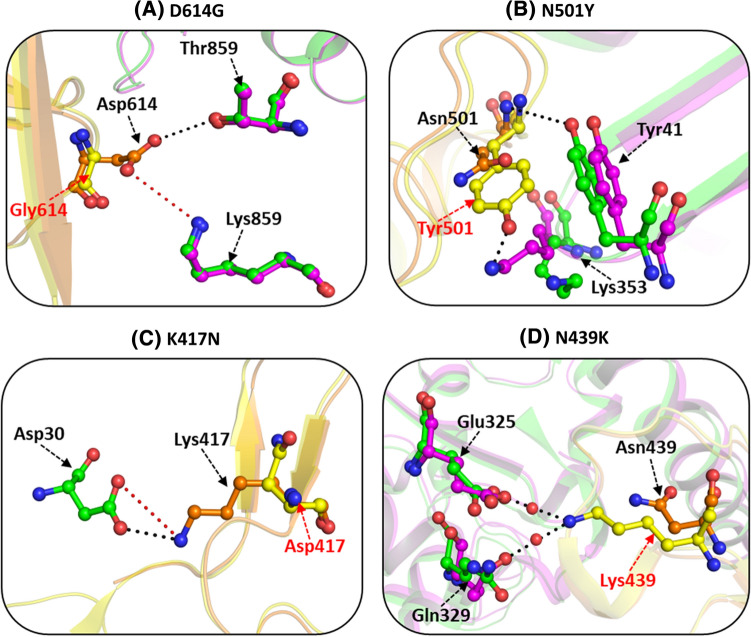
D614G
The D614G mutation is one of the predominant mutations across the globe that presents fitness survival to the virus in terms of its higher infectivity, transmissibility, and stability. This mutation was initially recognized as the Wuhan reference strain in early March 2020. Later on, this mutation was identified in almost all B.1 lineages of SARS-CoV-2 (B.1.17, B.1.351, P1, P2, B.1.425, and B.1.617.2, etc.). In the wild-type trimeric structure of S protein, the Asp614 residue (chain A) forms a hydrogen-bonded interaction with Thr859 (chain B) and a salt bridge interaction with Lys854 residue of adjacent subunit (chain B) [69] (Fig. 5A). The cryo-EM structure of D614G mutant (PDB id: 6XS6) revealed that these interactions were lost in the mutant and the lack of acetyl group in the D614G mutant is primarily responsible for the loss of these polar interactions. Experimental studies conducted in retroviruses pseudotyped with D614G mutation-infected hACE2-expressing cells revealed that the D614G mutation enhances the viral infectivity by diminishing the S1 shedding of viral S protein [70–73]. It has also been associated with reduced neutralization against monoclonal antibodies and increased transmissibility.
Fig. 5.
Structural effect of the mutations of SARS-CoV-2 S protein on interactions with hACE2. Intra- and inter- molecular non-covalent interactions of wild and mutant spike SARS-CoV-2 RBD with hACE2 receptor. in (A) D614G [PDB id: 7DWZ (W), 7DX1 (M)] (B) N501Y: [PDB id: 6MOJ (W), 7MJM (M)] (C) K417N [PDB id: 6MOJ (W), 7NX7 (M] (D) N439K [PDB id: 6MOJ (W), 7LON (M)].Wild type (W) SARS-CoV-2 residues are shown in orange and mutated residues (M) are in yellow. Interacting hACE2 with wild type RBD is shown in green and the correspnding residues on interaction with the mutant are in magenta. Hydrogen bonds are marked as black dashed lines and salt bridges as red dashed lines. Mutated residues are marked in red font
Mutation at RBD of S protein
N501Y
This mutation has been majorly reported in A.27, A.28, B.1.1.7, B.1.351, and B.1.621 lineages. The crystal structure of the native RBD-hACE2 complex (PDB id: 6MOJ) reveals that the hydrophilic residue, Asn501, is situated at the RBD-hACE2 interface and makes critical contacts with several hACE2 residues [32]. In the wild-type S protein, Asn 501 forms a hydrogen bond with Tyr41 of the hACE2 receptor and weak hydrophobic contacts with Tyr41 and Lys353 alkane side chain of hACE2 to further stabilize the complex (Fig. 5B). The cryo-EM structure of N501Y mutant ((PDB id: 7MJM) revealed that this mutation strengthens the intermolecular hydrogen-bonding between the RBD-hACE2 complex and thus results in higher infectivity and affinity [74]. Replacement of Asn501 residue with the aromatic Tyr residue in the S protein results in stronger π-π stacking interaction of the two aromatic benzene rings, Tyr501 in the mutant and Tyr 41 of hACE2 protein [75]. Additionally, a hydrogen bond is formed between Tyr501 and Lys353 residues of RBD and hACE2, respectively. This substitution increases the binding propensity of S protein for the hACE2 protein and is functionally also associated with immune escape against antibodies. N501Y mutation causes a 9 fold increase in binding affinity to the hACE2 receptor as compared to the wild type protein [53, 66, 76–79].
K417N
The Lys to Asn mutation at position 417 has been observed most frequently in the B.1.351/beta and B.1.617.2/delta variants. K417N mutation is positioned at one of the key determining sites of RBD that is associated with reduced binding with hACE2 receptor. The residue Lys417 forms hydrogen-bonded and salt bridge interactions with Gln24 and Asp30 residues, respectively, of hACE2 receptor [80] (Fig. 5C). The K417N mutation is responsible for destabilizing the salt bridge interaction as the shorter asparagine side chain (compared to the longer side chain of lysine) cannot interact with Asp30. K417N moderately decreases the binding affinity of hACE2 receptor and has been linked with reduced neutralizing activity against monoclonal antibodies. K417N mutation exists mainly in combination with N501Y mutation and thus has a more pronounced effect in terms of infectivity and neutralization reaction [53, 81–84].
N439K
This substitution was reported in B.1.1.7, B.1.351, B.1.258, and B.1.617 variants. N439K is one of the most common RBD mutations occurring independently worldwide. However, its prevalence in India is lower in frequency as compared globally. This mutation has been linked with an increased affinity towards hACE2 receptor, viral fitness, and immune escape. X-ray crystal structure of N439K (PDB id: 7L0N) reveals that this mutation leads to a higher affinity of SARS-CoV-2 towards the hACE2 receptor than the wild type [85]. The presence of the longer and more positively charged Lys residue (in place of shorter Asn) in mutant structure enhances binding affinity via the formation of additional water-mediated hydrogen bonds with residues Gln325 and Glu329 of hACE2 receptor (Fig. 5D). In contrast, the shorter Asn439 residue in the wild-type protein is positioned distantly and cannot form hydrogen-bonded interactions. N439K mutation has also been associated with resistance towards REGN10987 antibodies [82, 85–88].
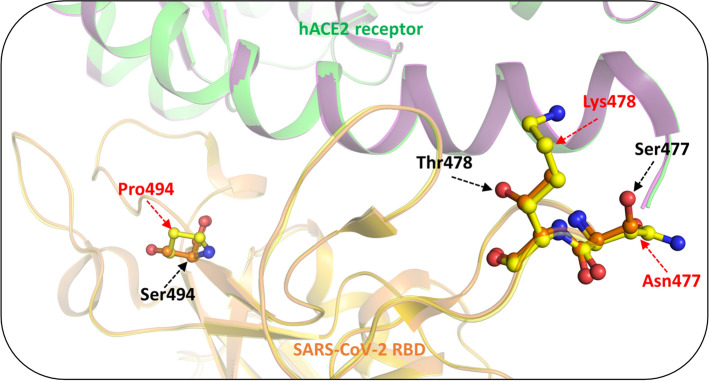
S477N
The S477N mutation has been reported in B.1.620. This frequently observed mutation occurs in the flexible loop region (473–490) of S protein that is responsible for the stronger binding of the mutant with hACE2 [89]. This mutation involves the replacement of the small charged Ser residue with the longer branched Asn residue (Fig. 6). This substitution mediates local conformational changes in the protein loop region housing this mutation that enhances its binding ability with the hACE2 receptor [89]. Co-occurrence of S477N with N439K and N501Y has been linked with an increased viral affinity towards the hACE2 receptor and immune escape. [82, 89, 90]. BioNTech has defined S477N mutation as a relevant mutation for future designing of vaccines against COVID-19 [91]. The co-occurrence of another mutation, D614G, though structurally situated far away from the S477N mutation, in most frequent cases, is linked with high viral load [87, 89, 92].
Fig. 6.
Conformational changes in SARS-CoV-2 S protein mutants and their interactions with hACE2. Representation of conformational changes in the tertiary structure of S protein mediated by S477N, T478K and S494P mutation. PDB id: 6LZG was used for wild-type protein while mutant types were prepared by PyMOL [156] and energy minimized for further studies. Wild type SARS-CoV-2 residues are indicated in orange and mutated residues are depicted in yellow. Mutated residues are marked in red font. hACE2 is represented in green and magenta
T478K
T478K mutation has been frequently observed in B.1.617.2 and B.1.1.519 lineages. This mutation resides at the RBD—hACE2 interface. The substitution of the shorter and neutral Thr by the longer basic Lys residue introduces a high charge potential which disrupts the S protein–hACE2 interaction (Fig. 6). T478K mutation occurring along with D614G, P681H, and T732A mutations are associated with increased resistance to neutralizing antibodies and increased infectivity [90, 93–95].
S494P
Ser494 residue sits at the β6 strand of RBD, an interface where the protein interacts with the hACE2 receptor. S494P mutation reported in B.1.1.7 contributes to the increased binding affinity of S protein with the hACE2 receptor through the strong interfacial complementarity between the S protein and hACE2. In this mutation, the charged Ser amino acid is replaced by an imino Pro. (Fig. 6) [90, 96–98]. However, this mutation is a milder substitution that causes a three to fivefold reduction in neutralization activity [99].
Mutations Alterating Immunogenicity of S protein
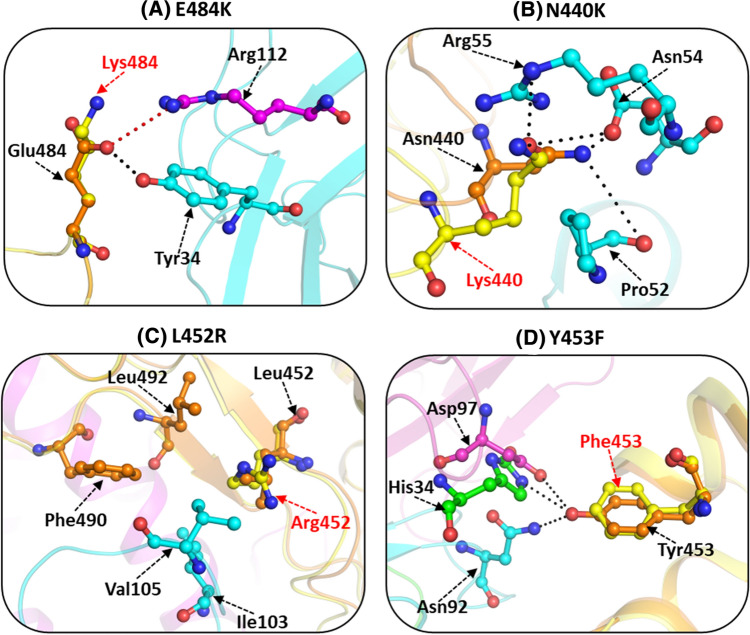
E484K/Q
This mutation was reported in B.1.351 (South Africa), P.1 (Brazil), and B.1.617 variants. E484K mutation is responsible for the fast transmission and higher infectivity of this SARS-CoV-2 variant [53, 66, 82]. The Glu residue present in the RBM of S protein has not been observed to be involved in any direct interaction with the hACE2 receptor. However, it disrupts the hydrogen-bonded and salt bridge interaction associated with binding to the neutralizing antibody. This mutant serves as an escape mutant against SARS-CoV-2 infection as it interferes with the ability of the antibodies to bind with the RBD region [53, 82, 90, 100, 101]. Glu484 residue makes a hydrogen bond with Tyr34 residue of the light chain and salt bridge interaction with the Arg112 residue of the heavy chain of monoclonal antibody P2B-2F6 (Fig. 7A). E484K mutation produces a local conformational perturbation wherein the resultant substituted positively charged residue, Lys484, is pushed away from another positively charged, Arg112 residue, located in the heavy chain. Consequently, this leads to the disruption of the intermolecular hydrogen-bonded interaction as well as salt bridge interaction formed in the wild-type protein. In the E484K mutant, the Lys residue forms an intra-molecular H-bond with the carbonyl oxygen of Gly485 residue, thus further ascertaining the inability of the mutant to interact with the hACE2 receptor.
Fig.7.
Interactions of SARS-CoV-2 S protein mutants with neutralizing antibodies. Intermolecular non-covalent interactions of wild-type and mutant S protein with neutralizing antibodies, (A) N440K with antibody C135 (residues in cyan). S protein complex with Fab portion of C135 neutralizing antibody (PDB id: 7K8Z) was used as wild type while for mutant type PDB id: 6VXX was taken (B) L452R with REGN10933. RBD complex with REGN10933 neutralizing antibody (PDB id: 6XDG) was used as wild type while for mutant PDB id 6LZG was used (C) Y453F with CC12.1 (light chain and heavy chain residues are shown in cyan and magenta, respectively). RBD complex with CC12.1 neutralizing antibody (PDB id: 6XC2) was used as wild type while for creating mutant type PDB id: 6LZG was employed. (D) E484K with P2B-2F6 (light chain and heavy chain residues shown in cyan and magenta, respectively). RBD complex with P2B-2F6 neutralizing antibody (PDB id: 7BWJ) was utilized as wild type while for mutant (PDB id: 6LZG) was considered. Mutant structures were generated with PyMOL and were energy minimized. Wild-type S protein residues are shown in orange and mutated residues in yellow. Hydrogen bonds are marked as black dotted lines and salt bridges as red dotteded lines. Mutated residues are marked by red font
E484Q mutant, like E484K, is found in several lineages, including B.1.351, B.1.17, B.1.617 as well as P1 and P2 variants [58, 97, 102]. E484Q has been seen very frequently in the Indian subcontinent along with L452R and P681R mutations [58, 90, 103]. This mutation enhances the viral S protein affinity towards the hACE2 receptor and improves viral ability to evade immune escape [104]. Similar to E484K, the E484Q mutations disrupt the interaction between the RBD and the neutralizing antibody. Residue Glu484 interacts with two residues of P2B-2F6 monoclonal antibody (Asn33 and Tyr34) to form hydrogen-bonded interaction with the light chain of the antibody. It is also associated with salt bridge formation with the side chain of Arg112 residue located on the light chain of P2B-2F6. E484Q mutation disrupts the hydrogen-bonded interaction formed by Arg112 residue.
N440K
N440K mutation found in B.1.36, was first perceived in Visakhapatnam and other parts of South India [105, 106]. This substitution has been linked with higher transmission and infectivity as well as immune escape across the south-eastern part of India [107, 108]. Asn440 residue in the wild-type protein resides in the proximity of the antiparallel β-sheet core of RBD, which lies adjacent to the hACE2 receptor (Fig. 7B). In the N440K mutant, the negatively charged Asn is replaced by the positively charged Lys. Hence, this substitution causes a charge reversal that contributes to distinct conformational perturbations in the RBM loop region facing the hACE2 receptor. Cell-based infectivity assay also validates the immune escape from monoclonal antibodies C135 anti-SARS-CoV-2 antibody [108]. This mutation generally co-occurs with the D614G mutation [109]. Structural–functional analysis of RBD with human monoclonal antibody C135 anti-SARS-CoV-2 antibody can explain the immune evasion effect of N440K mutation. Asn440 in the native protein forms a strong hydrogen-bonded interaction with Asp54 residue and weaker hydrogen bond with Pro52 and Arg55 residues of C135 antibody, while the mutant is capable of only weak hydrogen-bonded interactions with Asp54 residue.
L452R
The L452R mutation frequently observed in B.1.427/429, B.1.1.7, B.1.526, and B.1.617 lineages favors the fitness adaptability of SARS-CoV-2. This mutation has been linked with higher affinity, immune evasion, higher infectivity, and transmission [57, 61, 62, 90, 102]. The higher affinity can be attributed to the replacement of the smaller hydrophobic Leu residue with the hydrophilic basic Arg residue. In the wild-type protein, Leu452 makes a hydrophobic contact with Leu492 residue of the RBD, which further forms hydrophobic contact with the Phe490 residue (Fig. 7C). These interactions are responsible for its binding with monoclonal antibodies (REGN10933). Replacement of Leu residue with Arg residue results in the loss of this sequential chain of hydrophobic interactions in the mutant.
This mutation has also been associated with disruption of interfacial interaction with monoclonal antibodies (REGN10933) [58, 82, 110]. Leu452 residue interacts with the heavy chain of REGN10933 through hydrophobic contacts with the residues Ile103 and Val105 [111]. The presence of a longer charged residue Arg in place of the smaller hydrophobic Leu in the L452R mutant leads to disruption of the hydrophobic interactions mediated through Ile103 and Val105 residues. Co-occurrence of E484Q and L452R mutations in the B.1.617 variant has been linked with a more pronounced effect with enhanced infectivity and immune escape as compared to wild-type SARS-CoV-2 [112].
Y453F
This mutation was reported in B.1.1.298 lineage and has been related to an increased affinity towards hACE2 receptor in both humans and mink [113, 114]. Structural studies illustrate that in the wild-type, Tyr453 makes hydrogen-bonded interaction with His34 of hACE2. This mutation is responsible for the reduced neutralization with monoclonal antibodies. Tyr453 residue interacts with monoclonal antibody CC12.1 and forms a hydrogen-bonded interaction with Asn92 of the light chain, and Asp97 of the heavy chain of monoclonal antibody CC12.1, respectively [115] (Fig. 7D). In the mutant, the polar Tyr is substituted with the hydrophobic non-polar Phe residue. Consequently, the mutant can no longer participate in hydrogen bond formation leading to weak interaction with the hACE2 receptor. Moreover, the mutant is unable to interact with Asn92 and Asp97 residues of monoclonal antibody CC12.1 resulting in partial escape from monoclonal antibodies [82, 110, 116, 117].
Apart from RBD mutations, two key mutations H655Y and P681H/R are situated near the poly-cleavage site.
Mutation at the poly cleavage site of S protein
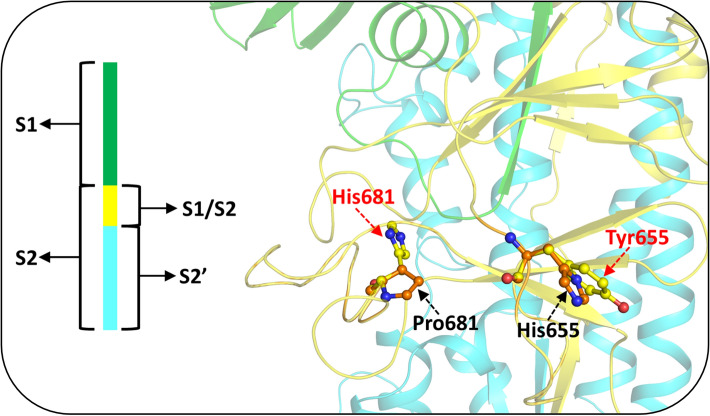
H655Y
This mutation was reported in P.2 lineage and has been associated with conferring resistance to monoclonal antibodies. The residue His655 is located close to the polybasic cleavage site, between RBD and the FP of S protein (Fig. 8). The substitution of the five-member imidazole ring of His residue with the benzene-containing aromatic Tyr residue that includes a polar group modifies the existing hydrophobic local environment to a comparatively more charged environment in this region. Therefore, this mutant has been hypothesized to play a role in regulating the efficiency of S protein fusion and host cell entry in mammals [118–120].
Fig. 8.
Mutations affecting the cleavage site of SARS-CoV-2 S protein. Ball and stick representation of the wild-type (PDB id: 6VYB) and mutant S protein residues at S1-S2 furin cleavage site affecting the fusion, facilitation, and efficient entry in mammal host cells. Wild-type SARS-CoV-2 residues are represented in orange and mutated residues in yellow. Mutated residues are marked in red font. Mutations were generated in the same model and structure was stabilized through energy minimization
P681H/R
The P681H variant is the most common mutation in B.1.1.7, B.1.620, and B.1.621 lineages [121–123], while the P681R mutation is present in B.1.617.1, B.1.617.2, and B.1.617.3 variants [93, 102, 124]. It occurs at the S1-S2 furin cleavage site. The replacement of the imino Pro residue by the much larger and charged basic amino acid residue, Arg, or the substitution with imidazole ring containing a bigger Trp residue, introduces a larger side chain in the region of mutation, resulting in a comparatively more hydrophilic environment (Fig. 8). This mutation enhances the basicity of this polybasic stretch and impairs the functioning or processing of proteases that facilitate the efficient spread and infection of SARS-CoV-2 [125–127]. The P681R mutation along with L452R and E484Q leads to convergent evolution success of SARS-CoV-2 in better adaption to its human hosts [58, 84, 103].
Mutation at N terminal domain of S protein
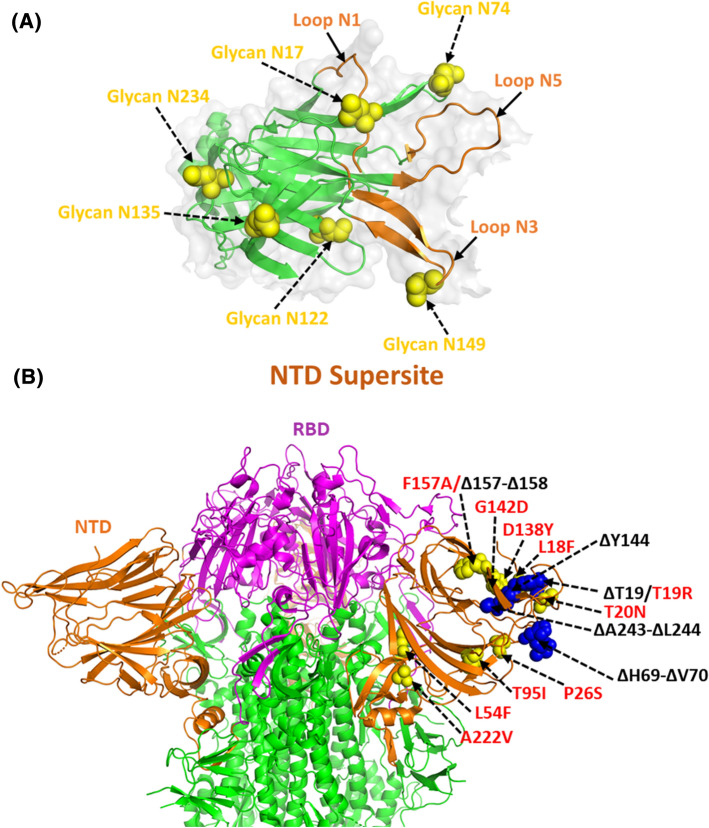
Several epitope mapping, antibody footprinting, and structural studies also include the NTD as the antigenic domain of the S1 subunit of S protein. Both the cryo-EM (PDB Ids: 7C2L, 7JJI, 6M17, 7LXY, 7LY0, 7LY2, 7LXX) and crystal structures (PDB id: 7LY3) of the potent NTD neutralizing antibodies reveal that all antibodies target the common region lined by N17, N74, N122 and N149 glycans (Fig. 9A). Epitope binding studies of 41 NTD-specific monoclonal antibodies define the existence of 6 common antigenic binding sites known as “NTD supersites” on S1 subunit encompassing residues 14–20, 140–158, and 245–264 (Fig. 9A). These NTD supersites possess strong positive electrostatic potential, while the recognizing antibodies have an electronegative potential [128–130].
Fig.9.
NTD of SARS-CoV-2 S protein as a supersite for interaction. (A) Structural representation of antigenic supersite of NTD (PBD id: 7L2C) surrounded by loop regions where the glycans on NTD are highlighted as yellow spheres. (B) Cartoon representation of SARS-CoV-2 trimeric S protein (PBD id: 6ZGE) rendering the position of key substitutions (red font) and deletion mutations (black font) in yellow and blue spheres, respectively
Cryo-EM structures of NTD-complex with neutralizing antibodies (PDB ids: 7L2C, 7L2D, 7L2E, 7L2F, 7LQV, and 7LQW and EMDB Ids: EMD-23150 and EMD-23151) reveal that the antibodies target the overlapping surface enclosed by several glycan moieties (at positions N17, N74, N122, and N149) known as NTD antigenic supersite [130]. This supersite is composed of N1 (14–26), N3 β-hairpin (141–156), and N5 loop (246–260) regions that impart functional plasticity to it. Some of the deletion mutations detected in the N-terminal region which alter the immunogenicity of S protein are ΔT19, ΔH69 -70, ΔY144 (B.1.617, B.1.1.7, B.1.1.318, B.1.525 lineages), Δ157-158, and Δ243–244 [65, 87, 131–133]. These deletion mutations may disrupt the binding sites of neutralizing anti-NTD antibodies and decrease neutralization reactions mediated by the anticipated antibody. Some of the most common substitution mutations seen in NTD are L18F (B.1.351, B.1.1.7), T19R (B.1.617), T20N (P.1), P26S (P1), L54F (B.1.617), T95I (B.1.617, B.1.526, B.1.1.318), D138Y (P1), G142D (B.1.617), F157A (A.23.1) and A222V (B.1.17) [87, 98, 119, 132–138] (Fig. 9B). However, several less common prevalent substitutions involve the mutants L5F, R78M, V483A, Q675H, and S939F [72, 98, 120, 139–142]. Substitution mutations like L18F, T20N, and P26S situated in the NTD supersite have been linked with reduced binding with monoclonal sera. Leu is a non-polar amino acid. Its replacement in the L54F mutant with the aromatic benzene side chain containing Phe can stiffen the overall structure by mediating stacking interactions or aromatic-aromatic interaction. The T19R mutation involves a change from a smaller polar, non-charged Thr residue to a much longer polar positively charged Arg, which increases the interaction capability between the S and the hACE2 proteins. The replacement of Thr by the similar but comparatively longer Asn at position 20 (T20N mutant) does not alter the environment of the binding region but influences the overall binding ability. The P26S mutation involves the replacement of the imino Pro with the polar, non-charged Ser residue, whereas in the L54F mutant a non-polar, aliphatic residue is replaced by an aromatic residue. In the T95I mutation, the polar, non-charged Thr residue is substituted by the non-polar, aliphatic Ile residue. These mutations thus alter the overall milieu of the protein in the region of replacement. A significant change in the local environment occurs in the D138Y mutant where a shorter negatively charged, hydrophilic residue is substituted by a bulkier aromatic Tyr residue. The replacement of Gly residue which lacks a side chain by the polar acidic Asp residue in the G142D mutant increases its hydrogen bond forming capability. In the F157A mutant, the small non-polar Ala is present in place of the bulky aromatic ring containing Phe and consequently results in loss of interactions. The A222V mutant comprises the similar nature of the substituted residue where both Ala and Val are non-polar and aliphatic wherein the side chain in the wild type is slightly shorter than that in the mutant. All these NTD mutations along with RBD have been linked with a decrease in neutralizing activity of antibodies [61, 87, 128, 144, 145].
Mutation at S2 domain
The S2 subunit forms a six-helical bundle (6-HB) structure via two HR repeat sequences that contribute to the merging of the viral membrane with the hACE2 receptor. The antigenicity of the S2 domain is found to be comparatively lower than the S1 domain. It has been hypothesized that this may be due to the extensive glycan shielding of this domain. Based on the protein’s conformational epitope, the key antigenic site where the antibodies interact with the S2 domain have been hypothesized in several loop regions of the S2 domain (793–794, 808–812, 850–854, 978–984, 1,099–1,100, and 1,139–1,146) [34]. However, the immunogenic significance of these regions is not well defined. Several mutations of the S2 domain include D769H, D950N (B.1.617) [146], D982A (B.1.17), T1027I (P1), Q1071H ((B.1.617.1), D1118H (B.1.1.7), H1101D (B.1.617), and V1176F (P.1) [66, 94, 103, 123, 136, 143, 147–151] (Fig. 10). In the D769H mutation, the acidic Asp residue is changed to the longer polar His. In D950N mutant, the acidic Asp is replaced by a polar, non-charged Asn, whereas the S982A mutation mediates the change of a polar residue by a shorter non-polar Val. The D1118H mutation involves the replacement of the negatively charged polar Asp with the positively charged imidazole-ring containing His residue, and the reverse happens in the case of the H1101D mutant. The T1027I mutation involves the substitution of the polar, non-charged Thr residue to the non-polar, aliphatic Ile residue. The polar, non-charged Gln in the Q1071H mutant is replaced by the imidazole ring containing His residue. The V1176F mutant involves the substitution of the non-polar aliphatic Val to the aromatic ring containing Phe residue. All these substitutions possibly perturb the local environment of the region of replacement as well as influence the interaction propensities due to a change in the nature and size of the substituted amino acid residue. However, the exact role of these mutations has not been defined yet.
Fig.10.
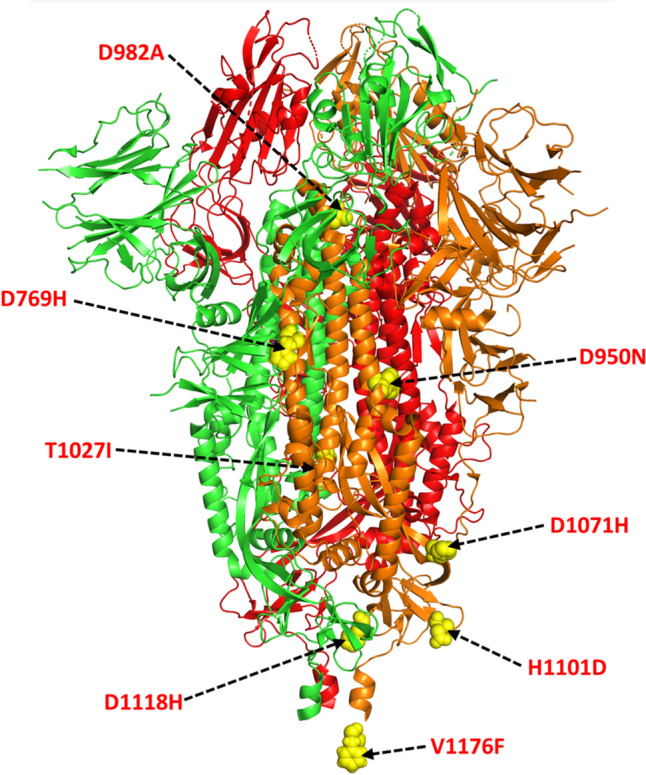
Mutations hotspots in SARS-CoV-2 S protein in S2 region Cartoon representation of trimeric S protein (PDB id: 6VXX) depicting single amino acid substitution mutation sites of S2 subdomain in yellow spheres with red font. Each monomers is coloured differently
Conclusion and prospectives
SARS-CoV-2 is continuously evolving in the form of new variants by accumulating the mutations to escape the host immunity [34]. The present review discusses the novel mutations in SARS-CoV-2, S protein, as well as their influence on transmissibility and vaccine efficacy along with their frequency mutations in VOC and VOI. Additionally, these mutations are contextualized with the conformational changes in protein structure.
It is worth mentioning that S protein is solely responsible for viral entry into the hACE2 receptor and has also been referred to as a key antigenic target against SARS-CoV-2 infection. Several structural studies have elucidated the atomic-level view of the molecular pathway through which viral S protein undergoes different topological conformations during its cellular entry [27]. Data analysis of genomic data obtained since last year revealed that the virus is rapidly evolving its genome through positive selection for better adaptability in changing environmental conditions [152]. Thus genome sequencing at a higher rate is required to track the emergence of new variants that will assist the execution of targeted control measures as well as the development of new vaccination measures, diagnostic techniques, and therapeutic agents [153, 154]. However, the mutations in the immunogenic region of S protein can impair vaccine effectiveness and lead to developing an unadorned illness [155]. Due to the ever changing nature of the virus to constantly adapt itself, the development of vaccine candidates is required to be accordingly modified regularly. This review has discussed the role of S protein mutations in modulating changes in protein structure that can enable more efficient and prolonged transmission in the human host. It, thus provides an overview that may assist in the development of new therapeutics against emerging variants of SARS-CoV-2.
Acknowledgements
DG thanks DHR/HRD for providing grants and fellowship under the young scientist scheme. Figures and illustrations were created using the collective resources of Biorender.com, Microsoft Office Powerpoint, and PyMOL.
Abbreviations
- Covid-19
Coronavirus disease 2019
- CP
Cytoplasm domain
- E
Envelope
- FP
Fusion peptide
- GISAID
Global Initiative on Sharing All influenza data
- HB
Helical bundle
- HR1
Heptapeptide repeat sequence 1
- HR2
Heptapeptide repeat sequence 2
- hACE2
Human angiotensin-converting enzyme 2
- M
Matrix
- Nsps
Non-structural proteins
- NTD
N-terminal domain
- ORF
Open reading frame
- PANGOLIN
Phylogenetic Assignment of Named Global Outbreak Lineages
- RBD
Receptor-binding domain
- RBM
Receptor-binding motif
- S
Spike protein
- SARS-CoV-2
Severe acute respiratory syndrome Corona Virus 2
- SP
Signal peptide
- TM
Transmembrane domain
- VOC
Variant of concern
- VOI
Variant of interest
- WHO
World Health Organization
Author contributions
PK and ASE conceptualized the article. DG designed the workflow, wrote and edited the major portions of the manuscript. PS wrote and edited the SARS-CoV-2 variants section. MS discussed the S protein mutations and drew the figures and illustrations. MK edited and revised the manuscript. DG, MS, PS, MK,ASE, and PK co-revised the manuscript.
Funding
Not applicable.
Availability of data and material
Not applicable.
Declarations
Conflict of interest
The authors report no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The manuscript was reviewed and approved by all authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Zheng J. SARS-coV-2: An emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cucinotta D, Vanelli M (2020) WHO declares COVID-19 a pandemic. Acta Biomed 91:157–160. 10.23750/abm.v91i1.9397 [DOI] [PMC free article] [PubMed]
- 3.COVID Live Update: 233,158,434 Cases and 4,771,151 Deaths from the Coronavirus - Worldometer. https://www.worldometers.info/coronavirus/. Accessed 28 Sep 2021.
- 4.Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 27 Sept 2021.
- 5.Lo Presti A, Rezza G, Stefanelli P. Selective pressure on SARS-CoV-2 protein coding genes and glycosylation site prediction. Heliyon. 2020;6:e05001. doi: 10.1016/j.heliyon.2020.e05001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GISAID - Initiative. https://www.gisaid.org/. Accessed 27 Sept 2021
- 7.Salvatori G, Luberto L, Maffei M, et al. SARS-CoV-2 spike protein: An optimal immunological target for vaccines. J Transl Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Zhou L, Ma Z, et al. Promising Immunotherapies against COVID-19. Adv Ther. 2021 doi: 10.1002/adtp.202100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai C, Zhong Q, Gao GF. Overview of SARS-CoV-2 genome-encoded proteins. Sci China Life Sci. 2021 doi: 10.1007/S11427-021-1964-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman R, Patel KJ, Ranjan K. COVID-19: unmasking emerging SARS-CoV-2 variants vaccines and therapeutic strategies. Biomolecules. 2021;11:993. doi: 10.3390/biom11070993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastogi M, Pandey N, Shukla A, Singh SK. SARS coronavirus 2: from genome to infectome. Respir Res. 2020;21:318. doi: 10.1186/s12931-020-01581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Pandey R, Sharma P, et al (2020) Integrated genomic view of SARS-CoV-2 in India. Wellcome open research. 5:184. 10.12688/wellcomeopenres.16119.1 [DOI] [PMC free article] [PubMed]
- 13.Yadav R, Chaudhary JK, Jain N, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells. 2021;10:821. doi: 10.3390/cells10040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemm T, Ebert G, Calleja DJ, et al (2020) Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 39(18): e106275. 10.15252/embj.2020106275 [DOI] [PMC free article] [PubMed]
- 16.Jauregui AR, Savalia D, Lowry VK, et al. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS ONE. 2013;8:e62416. doi: 10.1371/journal.pone.0062416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson BS, Kolte V, Khan A, Sharma G. Dynamic tracking of variant frequencies depicts the evolution of mutation sites amongst SARS-CoV-2 genomes from India. J Med Virol. 2020;93:2534–2537. doi: 10.1002/jmv.26756. [DOI] [PubMed] [Google Scholar]
- 18.Boyton RJ, Altmann DM. Risk of SARS-CoV-2 reinfection after natural infection. Lancet. 2021;397:1161–1163. doi: 10.1016/S0140-6736(21)00662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi MK, Singh P, Sharma S, et al. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J Biomol Struct Dyn. 2020;39:1–14. doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu G, Zhu C, Zhu Y, Sun F. Minireview of progress in the structural study of SARS-CoV-2 proteins. Curr Res Microb Sci. 2020;1:53–61. doi: 10.1016/j.crmicr.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tripathi MK, Nath A, Singh TP, et al. Evolving scenario of big data and Artificial Intelligence (AI) in drug discovery. Mol Divers. 2021;25:1439–1460. doi: 10.1007/s11030-021-10256-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shrestha UK. SARS-CoV-2 vaccines and their challenges against the variants. J Adv Intern Med. 2021;10:1–3. doi: 10.3126/jaim.v10i1.37080. [DOI] [Google Scholar]
- 24.Xu Y, Lou Z, Liu Y, et al. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ismail AM, Elfiky AA. SARS-CoV-2 spike behavior in situ: a Cryo-EM images for a better understanding of the COVID-19 pandemic. Signal Transduct Target Ther. 2020;5:252. doi: 10.1038/s41392-020-00365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, Zhang J, Xiao T, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W (2020) Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pacific J Allergy Immunol. 38:10–18. 10.12932/AP-200220-0773 [DOI] [PubMed]
- 30.Perlot T, Penninger JM. ACE2 - From the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013;15:866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 33.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janik E, Niemcewicz M, Podogrocki M, et al. The emerging concern and interest sars-cov-2 variants. Pathogens. 2021;10:633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikhra V (2021) Evolving Patterns in COVID-19: The Virus, its Variants and Infectivity-cum-Virulence. Biomed J Sci Tech Res 33:25712–25722: 10.26717/bjstr.2021.33.005382
- 37.Hadfield J, Megill C, Bell SM, et al. NextStrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu Y, McCauley J (2017) GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 22:30494. 10. 2807/1560–7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed]
- 40.Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamed SM, Elkhatib WF, Khairalla AS, Noreddin AM. Global dynamics of SARS-CoV-2 clades and their relation to COVID-19 epidemiology. Sci Rep. 2021;11:8435. doi: 10.1038/s41598-021-87713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodcroft, E. CoVariants (2021): SARS-CoV-2 mutations and variants of interest (NextStrain). CoVariants https://covariants.org/2021.
- 43.Public Health England (2020), Investigation of novel SARS-COV-2 variant: Variant of Concern 2020/12/01. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201.
- 44.Embrace the WHO’s new naming system for coronavirus variants (2021). Nature 594:149. 10.1038/d41586-021-01508-8 [DOI] [PubMed]
- 45.Konings F, Perkins MD, Kuhn JH, et al. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6:821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 46.González-Candelas F, Shaw MA, Phan T, et al. One year into the pandemic: Short-term evolution of SARS-CoV-2 and emergence of new lineages. J Infect Genet Evol. 2021;92:104869. doi: 10.1016/j.meegid.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Public Health England (2020) Investigation of novel SARS-COV-2 variant: Variant of Concern 2020/12/01 Technical Briefing 3. GovUk 1–11; https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/950823/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3_-_England.pdf.
- 48.B.1.1.207 Overview, Nomenclature, Criteria for notability, Notable variants, The Free Encyclopedia (2021). https://www.mobilewiki.org/en/B.1.1.207-6666680484. Accessed 11 May 2021.
- 49.Galloway SE, Paul P, MacCannell DR, et al (2021) Emergence of SARS-CoV-2 B.1.1.7 Lineage. Mmwr 70:95–99. 10.15585/mmwr.mm7003e2 [DOI] [PMC free article] [PubMed]
- 50.Frampton D, Rampling T, Cross A, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis. 2021;21:1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Toole Á, Kraemer MUG, Hill V, et al (2021) Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res 6:121. 10.12688/wellcomeopenres.16661.1 [DOI] [PMC free article] [PubMed]
- 52.Feder KA, Pearlowitz M, Goode A, et al (2021) Linked Clusters of SARS-CoV-2 Variant B.1.351 - Maryland, January–February 2021. MMWR Morb Mortal Wkly Rep 70:627–631. 10.15585/mmwr.mm7017a5 [DOI] [PMC free article] [PubMed]
- 53.Kim YJ, Jang US, Soh SM, et al (2021) The impact on infectivity and neutralization efficiency of sars-cov-2 lineage b.1.351 pseudovirus. Viruses 13:633. 10.3390/v13040633 [DOI] [PMC free article] [PubMed]
- 54.Oliveira MDL, Oliveira KMT, Silva JN, et al (2021) Theoretical causes of the Brazilian P.1 and P.2 lineages of the SARS-CoV-2 virus through molecular dynamics. bioRxiv 2021.04.09.439181. doi: 10.1101/2021.04.09.439181
- 55.Hirotsu Y, Omata M. Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu. Japan J Infect. 2021;82:276–316. doi: 10.1016/j.jinf.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava S, Banu S, Singh P, et al. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J Biosci. 2021;46:22. doi: 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar V, Singh J, Hasnain SE, Sundar D. Possible link between higher transmissibility of B.1.617 and B.1.1.7 variants of SARS-CoV-2 and increased structural stability of its spike protein and hACE2 affinity. Int J Mol Sc. 2021;22:9131. doi: 10.3390/ijms22179131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cherian S, Potdar V, Jadhav S, et al (2021) Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021.04.22.440932. 10.1101/2021.04.22.440932 [DOI] [PMC free article] [PubMed]
- 59.Singh J, Rahman SA, Ehtesham NZ, et al. SARS-CoV-2 variants of concern are emerging in India. Nat Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 60.Yadav PD, Sapkal GN, Abraham P, et al (2021) Neutralization potential of Covishield vaccinated individuals sera against B.1.617.1. Clin Infect Di, ciab483. 10.1093/cid/ciab483 [DOI] [PubMed]
- 61.McCallum M, Bassi J, Marco A De, et al (2021) SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv 2021.03.31.437925. 10.1101/2021.03.31.437925
- 62.Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184:3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voloch CM, da Silva FR, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol. 2020;95:e00119–e121. doi: 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goes LR, Siqueira JD, Garrido MM, et al. New infections by SARS-CoV-2 variants of concern after natural infections and post-vaccination in Rio de Janeiro. Brazil Infect Genet Evol. 2021;94:104998. doi: 10.1016/j.meegid.2021.104998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCormick KD, Jacobs JL, Mellors JW. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/science.abg4493. [DOI] [PubMed] [Google Scholar]
- 66.Andrew N, Bascos D, Mirano-Bascos D, Saloma CP (2021) Structural Analysis of Spike Protein Mutations in the SARS-CoV-2 P.3 Variant. bioRxiv 2021.03.06.434059. 10.1101/2021.03.06.434059
- 67.Annavajhala MK, Mohri H, Zucker JE, et al (2021) A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv 2021.02.23.21252259v4. 10.1101/2021.02.23.21252259
- 68.Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Accessed 2 Jun 2021
- 69.Isabel S, Graña-Miraglia L, Gutierrez JM, et al. Evolutionary and structural analyses of SARS-CoV-2 D614G spike protein mutation now documented worldwide. Sci Rep. 2020;10:14031. doi: 10.1038/s41598-020-70827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Jackson CB, Mou H, et al (2020) The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020.06.12.148726. 10.1101/2020.06.12.148726
- 71.Raghav S, Ghosh A, Turuk J, et al. Analysis of Indian SARS-CoV-2 Genomes Reveals Prevalence of D614G Mutation in Spike Protein Predicting an Increase in Interaction With TMPRSS2 and Virus Infectivity. Front Microbiol. 2020;11:594928. doi: 10.3389/fmicb.2020.594928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu J, He CL, Gao QZ, et al (2020) D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. bioRxiv 2020.06.20.161323. 10.1101/2020.06.20.161323
- 73.Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu X, Mannar D, Srivastava SS, et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19:e3001237. doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veeramachaneni GK, Thunuguntla VBSC, Bobbillapati J, Bondili JS. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J Biomol Struct Dyn. 2020;39:4015–4025. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ali F, Kasry A, Amin M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng L, Song S, Zhou B, et al. Impact of the N501Y substitution of SARS-CoV-2 Spike on neutralizing monoclonal antibodies targeting diverse epitopes. Virol J. 2021;18:87. doi: 10.1186/s12985-021-01554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luan B, Wang H, Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Santos JC, Passos GA (2021) The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv 2020.12.29.424708.10.1101/2020.12.29.424708
- 80.Koirala RP, Thapa B, Khanal SP, et al. Binding of sars-cov-2/sars-cov spike protein with human ace2 receptor. J Phys Commun. 2021;5:035010. doi: 10.1088/2399-6528/abea27. [DOI] [Google Scholar]
- 81.Villoutreix BO, Calvez V, Marcelin AG, Khatib AM. In silico investigation of the new UK (B.1.1.7) and South African (501y.v2) SARS-CoV-2 variants with a focus at the ace2-spike rbd interface. Int J Mol Sci. 2021;22:1–13. doi: 10.3390/ijms22041695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Focosi D, Maggi F. Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines. Rev Med Virol. 2021 doi: 10.1002/rmv.2231.10.1002/rmv.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 84.Khan A, Zia T, Suleman M, et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J Cell Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu J, Zhang L, Zhang Y, et al. The antigenicity of epidemic SARS-CoV-2 variants in the United Kingdom. Front Immunol. 2021;12:687869. doi: 10.3389/fimmu.2021.687869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou W, Xu C, Wang P, et al. N439K variant in spike protein may alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. Front cell dev bio. 2020;9:697035. doi: 10.3389/fcell.2021.697035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh A, Steinkellner G, Köchl K, et al. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci Rep. 2021;11:4320. doi: 10.1038/s41598-021-83761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang R, Chen J, Gao K, Wei GW. Vaccine-escape and fast-growing mutations in the United Kingdom, the United States, Singapore, Spain, India, and other COVID-19-devastated countries. Genomics. 2021;113:2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meo SA, Bukhari IA, Akram J, et al (2021) COVID-19 vaccines: Comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna vaccines. Eur Rev Med Pharmacol Sci 25:1663–1679. 10.26355/eurrev_202102_24877 [DOI] [PubMed]
- 92.Mejdani M, Haddadi K, Pham C, Mahadevan R. SARS-CoV-2 receptor binding mutations and antibody contact sites. Antib Ther. 2021;4:149–158. doi: 10.1093/abt/tbab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shrestha UK. B.1.617.2 variant and increasing surge of COVID-19 in Nepal. Nepal Mediciti Med J. 2021;2:1–4. doi: 10.3126/nmmj.v2i1.37212. [DOI] [Google Scholar]
- 94.Barona-Gomez F, Delaye L, Diaz-Valenzuela E, et al (2021) Phylogenomics and population genomics of SARS-CoV-2 in Mexico reveals variants of interest (VOI) and a mutation in the Nucleocapsid protein associated with symptomatic versus asymptomatic carriers. medRxiv 2021.05.18.21256128v2. 10.1101/2021.05.18.21256128
- 95.Di Giacomo S, Mercatelli D, Rakhimov A, Giorgi FM. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J Med Virol. 2021;93:5638–5643. doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem Biophys Res Commun. 2021;534:374–380. doi: 10.1016/j.bbrc.2020.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen J, Gao K, Wang R, Wei G-W (2021) Revealing the threat of emerging SARS-CoV-2 mutations to antibody therapies. bioRxiv 2021.04.12.439473. 10.1101/2021.04.12.439473 [DOI] [PMC free article] [PubMed]
- 98.Grabowski F, Preibisch G, Giziński S, et al. Sars-cov-2 variant of concern 202012/01 has about twofold replicative advantage and acquires concerning mutations. Viruses. 2021;13:392. doi: 10.3390/v13030392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greaney AJ, Loes AN, Crawford KHD, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laffeber C, de Koning K, Kanaar R, Lebbink JHG. Experimental evidence for enhanced receptor binding by rapidly spreading SARS-CoV-2 variants. J Mol Biol. 2021;433:167058. doi: 10.1016/j.jmb.2021.167058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu Z, VanBlargan LA, Bloyet LM, et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou H, Dcosta BM, Samanovic MI, et al (2021) The Spike Proteins of SARS-CoV-2 B.1.617 and B.1.618 Variants Identified in India 1 Provide Partial Resistance to Vaccine-elicited and Therapeutic Monoclonal 2 Antibodies. bioRxiv 2021.05.14.444076. 10.1101/2021.05.14.444076
- 103.Cherian S, Potdar V, et al (2021) Convergent evolution of SARS-CoV2 spike mutations, L452R, E484Q and P681R, in the second wave of Covid-19 in Maharashtra, India. bioRxiv 2021.04.22.440932. 10.1101/2021.04.22.440932 [DOI] [PMC free article] [PubMed]
- 104.Devendran R, Kumar M, Chakraborty S. Genome analysis of SARS-CoV-2 isolates occurring in India: Present scenario. Indian J public health. 2020;64:S147–S155. doi: 10.4103/ijph.IJPH_506_20. [DOI] [PubMed] [Google Scholar]
- 105.Pattabiraman C, Prasad P, George AK, et al (2021) Importation, circulation, and emergence of variants of SARS-CoV-2 in the South Indian state of Karnataka. Wellcome Open Res 6:110. 10.12688/wellcomeopenres.16768.1 [DOI] [PMC free article] [PubMed]
- 106.Yadav PD, Nyayanit DA, Majumdar T, et al. An epidemiological analysis of sars-cov-2 genomic sequences from different regions of India. Viruses. 2021;13:925. doi: 10.3390/v13050925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rani PR, Imran M, Lakshmi JV, et al. Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape. J Med Virol. 2021;93:4163–4165. doi: 10.1002/jmv.26997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tandel D, Gupta D, Sah V, Harinivas Harshan K (2021) N440K variant of SARS-CoV-2 has Higher Infectious Fitness. bioRxiv. 2021.04.30.441434. 10.1101/2021.04.30.441434
- 109.Singh J, Samal J, Kumar V, et al (2021) Structure-function analyses of new sars-cov-2 variants b.1.1.7, b.1.351 and b.1.1.28.1: Clinical, diagnostic, therapeutic and public health implications. Viruses 13:439. 10.3390/v13030439 [DOI] [PMC free article] [PubMed]
- 110.Tada T, Dcosta BM, Zhou H, et al (2021) Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv 2021.02.18.431897. 10.1101/2021.02.18.431897
- 111.Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Verghese M, Jiang B, Iwai N, et al. A SARS-CoV-2 Variant with L452R and E484Q Neutralization Resistance Mutations. J Clin Microbiol. 2021;59:e0074121. doi: 10.1128/jcm.00741-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ortuso F, Mercatelli D, Guzzi PH, Giorgi FM. Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1886175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bayarri-Olmos R, Rosbjerg A, Johnsen LB, et al. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J Biol Chem. 2021;296:100536. doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuan M, Liu H, Wu NC, et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Upadhyay V, Lucas A, Panja S, Mallela K (2021) Interplay between protein stability, binding to ACE2 and escape from neutralizing antibodies determines the natural selection of SARS-CoV-2 receptor binding domain variants. bioRxiv 2021.05.23.445348. 10.1101/2021.05.23.445348
- 117.Takuma H, Yaegashi. N, Ikuo K (2021) Effect of RBD (Y453F) mutation in spike glycoprotein of SARS-CoV-2 on neutralizing IgG affinity. medRxiv 2021.01.28.21250577v2. 10.1101/2021.01.28.21250577
- 118.Colson P, Levasseur A, Delerce J, et al. Spreading of a new SARS-CoV-2 N501Y spike variant in a new lineage. Clin Microbiol Infect. 2021;27:1352.e1–1352.e5. doi: 10.1016/j.cmi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mallm J-P, Bundschuh C, Kim H, et al (2021) Local emergence and decline of a SARS-CoV-2 variant with mutations L452R and N501Y in the spike protein. medRxiv 2021.04.27.21254849v1. 10.1101/2021.04.27.21254849
- 120.Manickavasagam P (2020) Spike protein of SARS-CoV-2: Impact of single amino acid mutation and effect of drug binding to the variant-in silico analysis. Preprints.10.20944/preprints202008.0447.v1
- 121.Zuckerman NS, Fleishon S, Bucris E, et al (2021) A unique SARS-CoV-2 spike protein P681H strain detected in Israel Israel National Consortium for SARS-CoV-2 sequencing. medRxiv 2021.03.25.21253908v1. 10.1101/2021.03.25.21253908
- 122.Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, et al (2021) Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect Genet Evol 95:105038. 10.1016/j.meegid.2021.10503 [DOI] [PMC free article] [PubMed]
- 123.Meng B, Kemp SA, Papa G, et al. (2021) Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep 35:109292. 10.1016/j.celrep.2021.109292 [DOI] [PMC free article] [PubMed]
- 124.Peacock TP, Sheppard CM, Brown JC, et al (2021) The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin. bioRxiv 2021.05.28.446163. 10.1101/2021.05.28.446163
- 125.Yadav PD, Mohandas S, Shete AM, et al (2021) SARS CoV-2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. bioRxiv 2021.05.05.442760. 10.1101/2021.05.05.442760
- 126.Shirakawa K, Sadamasu K, Kimura I, et al (2021) SARS-CoV-2 spike P681R mutation enhances and accelerates viral fusion. bioRxiv 2021.06.17.448820. 10.1101/2021.06.17.448820
- 127.Ferreira I, Datir R, Papa G, et al (2021) SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv 2021.05.08.443253. 10.1101/2021.05.08.443253.
- 128.McCallum M, De Marco A, Lempp FA, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lok SM. An NTD supersite of attack. Cell Host Microbe. 2021;29:744–746. doi: 10.1016/j.chom.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cerutti G, Guo Y, Zhou T, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vega-Magaña N, Sánchez-Sánchez R, Hernández-Bello J, et al. RT-qPCR Assays for Rapid Detection of the N501Y, 69–70del, K417N, and E484K SARS-CoV-2 Mutations: A Screening Strategy to Identify Variants With Clinical Impact. Front Cell Infect Microbiol. 2021;11:672562. doi: 10.3389/fcimb.2021.672562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hacisuleyman E, Hale C, Saito Y, et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N Engl J Med. 2021;384:2212–2218. doi: 10.1056/nejmoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dhar MS, Marwal R, Ponnusamy K, et al (2021) Genomic characterization and Epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. medRxiv 2021.06.02.21258076. 10.1101/2021.06.02.21258076 [DOI] [PMC free article] [PubMed]
- 134.Laha S, Chakraborty J, Das S, et al. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Osina NA, Krasnov YM, Guseva NP, et al (2021) Molecular-genetic monitoring of sARs-CoV-2 genovariants in the territory of the volga federal district of the Russian Federation. Communication. Probl Osobo Opasnykh Infektsii 122–127. 10.21055/0370-1069-2021-1-122-127
- 136.Toovey OTR, Harvey KN, Bird PW, Tang JWTWT. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J Infect. 2021;82:e23–e24. doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cai HY, Cai A. SARS-CoV2 spike protein gene variants with N501T and G142D mutation–dominated infections in mink in the United States. J Vet Diagnostic Investig. 2021;33(5):939–942. doi: 10.1177/10406387211023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bartolini B, Rueca M, Gruber CEM, et al (2020) The newly introduced SARS-CoV-2 variant A222V is rapidly spreading in Lazio region, Italy. medRxiv 2020.11.28.20237016v1. 10.1101/2020.11.28.20237016
- 139.Gunadi WH, Hakim MS, et al. Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters. BMC Med Genomics. 2021;14:144. doi: 10.1186/s12920-021-00990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ashwaq O, Manickavasagam P, Haque SM. V483A: an emerging mutation hotspot of SARS-CoV-2. Future Virol. 2021;16:419–429. doi: 10.2217/fvl-2020-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li Q, Wu J, Nie J, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;82:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ahamad S, Hema K, Gupta D. Structural stability predictions and molecular dynamics simulations of RBD and HR1 mutations associated with SARS-CoV-2 spike glycoprotein. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1889671. [DOI] [PubMed] [Google Scholar]
- 143.Aleem A, Akbar Samad AB, Slenker AK. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19) In StatPearls: Treasure Island (FL) StatPearls Publishing; 2021. [PubMed] [Google Scholar]
- 144.Cristina Resende P, Naveca FG, Lins RD, et al (2021) The ongoing evolution of variants of concern and interest of SARS-CoV-2 in Brazil revealed by convergent indels in the amino (N)-terminal domain of the Spike protein. Virus evolution 7:veab069. 10.1093/ve/veab069 [DOI] [PMC free article] [PubMed]
- 145.Winger A, Caspari T. The spike of concern-the novel variants of Sars-cov-2. Viruses. 2021;13:1002. doi: 10.3390/v13061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kemp SA, Collier DA, Datir R, et al. Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation. Preprint medRxiv. 2020 doi: 10.1101/2020.12.05.20241927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Franceschi VB, Caldana GD, de Menezes MA, et al. Genomic epidemiology of SARS-CoV-2 in Esteio, Rio Grande do Sul. Brazil BMC Genomics. 2021;22:371. doi: 10.1186/s12864-021-07708-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ramírez-Soto MC, Arroyo-Hernández H, Ortega-Cáceres G. Sex differences in the incidence, mortality, and fatality of COVID-19 in Peru. PLoS ONE. 2021;16:e0253193. doi: 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jungreis I, Sealfon R, Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat Commun. 2021;2:2642. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hirotsu Y, Omata M (2021) Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLoS Pathog. 17:e1009619. 10.1371/journal.ppat.1009619 [DOI] [PMC free article] [PubMed]
- 151.Chrysostomou AC, Vrancken B, Koumbaris G, et al. A Comprehensive Molecular Epidemiological Analysis of SARS-CoV-2 Infection in Cyprus from April 2020 to January 2021: Evidence of a Highly Polyphyletic and Evolving Epidemic. Viruses. 2021;13:1098. doi: 10.3390/v13061098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rochman, Nash D et al. (2020). Ongoing Global and Regional Adaptive Evolution of SARS-CoV-2. bioRxiv 2020.10.12.336644. 10.1101/2020.10.12.336644
- 153.V’kovski P, Kratzel A, Steiner S,, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guo S, Liu K, Zheng J. The genetic variant of sars-cov-2: Would it matter for controlling the devastating pandemic? Int J Biol Sci. 2021;17:1476–1485. doi: 10.7150/ijbs.59137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Om BB, N W, DF N,, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med. 2021;27:1518–1524. doi: 10.1038/S41591-021-01472-W. [DOI] [PubMed] [Google Scholar]
- 156.PyMOL | pymol.org. https://pymol.org/2/.
- 157.Warwicker J. A model for pH coupling of the SARS-CoV-2 spike protein open/closed equilibrium. Brief Bioinform. 2021;22:1499–1507. doi: 10.1093/bib/bbab056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Starr TN, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kemp SA, Collier DA, Datir RP, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jakhmola S, Indari O, Kashyap D, et al. Mutational analysis of structural proteins of SARS-CoV-2. Heliyon. 2021;7:e06572. doi: 10.1016/j.heliyon.2021.e06572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang P, Casner RG, Nair MS, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Laha S, et al. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. J Infect Genet Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Andrew, N., Bascos, D., Mirano-Bascos, D., & Saloma, C. P. (2021) Structural Analysis of Spike Protein Mutations in an Emergent SARS-CoV-2 Variant from the Philippines. bioRxiv 2021.03.06.434059. 10.1101/2021.03.06.434059
- 164.Maitra A, et al. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: Possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci. 2020;45:76. doi: 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.