Abstract
Class I PI3K are heterodimers composed of a p85 regulatory subunit and a p110 catalytic subunit involved in multiple cellular functions. Recently, the catalytic subunit p110β has emerged as a class I PI3K isoform playing a major role in tumorigenesis. Understanding its regulation is crucial for the control of the PI3K pathway in p110β-driven cancers. Here we sought to evaluate the putative regulation of p110β by SUMO. Our data show that p110β can be modified by SUMO1 and SUMO2 in vitro, in transfected cells and under completely endogenous conditions, supporting the physiological relevance of p110β SUMOylation. We identify lysine residue 952, located at the activation loop of p110β, as essential for SUMOylation. SUMOylation of p110β stabilizes the protein increasing its activation of AKT which promotes cell growth and oncogenic transformation. Finally, we show that the regulatory subunit p85β counteracts the conjugation of SUMO to p110β. In summary, our data reveal that SUMO is a novel p110β interacting partner with a positive effect on the activation of the PI3K pathway.
Keywords: p110β, p85β, Stability, SUMO, Transformation
Introduction
Class IA phosphatidylinositol-4,5,bisphosphate 3-kinases (PI3Ks) are heterodimeric proteins composed of one p110 catalytic subunit (p110α, β, or δ) and one p85 regulatory subunit (p85α, p55α, p50α, p85β, and p55γ) [1]. Mutational activation or altered expression of class IA PI3K results in enhanced PI3K signaling, which is associated with oncogenic cellular transformation and cancer [2–6]. The p85 subunit inhibits the p110 basal activity, allows its activation downstream RTKs, and is essential for its thermal stability [7–9]. Therefore, optimal signaling through the PI3K pathway depends on a critical molecular balance between the regulatory and the catalytic subunits [10]. The stabilization of p110 by p85 is not totally understood. Interaction between the iSH2 domain of p85 and the N-terminus of p110 stabilizes p110 in mammalian cells [11–14]. However, stabilization of p110 can be also achieved by protein synthesis at low temperature or the linkage of epitope tags to the N-terminus of p110 [8].
The regulatory activity of p85 on p110 is modulated by different p85 post-translational modifications such as the phosphorylation at tyrosine residues or its conjugation to the small ubiquitin-related modifier (SUMO) [15–17]. Conjugation of SUMO to the iSH2 domain on p85 reduces its tyrosine phosphorylation and reinforces its inhibitory effect on p110 [17]. Covalent interaction with SUMO is also a critical event in the regulation of other key PI3K signaling components such as AKT or PTEN [18–22]. So far, no direct post-translational modification of p110 by SUMO has been reported.
Here, we evaluated the potential regulation of the PI3K catalytic subunit p110β by SUMO. In vitro and in vivo SUMOylation assays show that p110β is an authentic SUMO substrate, that lysine residue K952, located at the activation loop in p110β, is essential for SUMO conjugation, and that SUMOylation increases the stability of p110β. In addition, functional studies reveal that mutation of the SUMOylation site in p110β downmodulated the p110β-mediated PI3K activation downstream pathway, its cell proliferation-promoting activity, and the oncogenic capability. Finally, our data show that the regulatory subunit p85β negatively modulates the interaction of p110β with SUMO. These results indicate that SUMO conjugation to p110β has a positive impact on the activation of the PI3K pathway, and its modulation could represent a novel strategy for PI3K regulation.
Materials and methods
Cell culture, plasmids and transfections
HEK-293, PC3, Rat-2 and NIH-3T3 cells were purchased from ATCC. Primary chicken embryo fibroblasts (CEF) were obtained from specific-pathogen-free (SPF) 10-day-old eggs (Intervet, Salamanca, Spain) following the protocol previously described [23]. HEK-293, PC3, and CEF cells were cultured in complete medium (DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin). Rat-2 and NIH-3T3 were cultured in DMEM supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin. The cells were transiently transfected using PEI (Polysciences, Inc), according to the manufacturer’s instructions. pcDNA-His6-SUMO1, pcDNA-His6-SUMO2, and pcDNA-SV5-Ubc9 plasmids were previously described [24, 25]. The vectors encoding the HA-tagged form of p85β (HA-p85β) and p85β SUMOylation mutant (HA-p85β K535RK592R) were previously described [17]. The myc-tagged p110β expression plasmid [26] was kindly provided by Jonathan M Backer. The HA-tagged p110α expression plasmid (Addgene #1397) was kindly provided by Lewis Cantley. Myc-p110β construct was mutated using Quickchange site directed mutagenesis kit (Stratagene) and the oligonucleotides p110β -K647R-F-5′-GTGCAAGTGTTAAGATATGAGCCTTTTC-3′, p110β-K647R-R-5′-GAAAAGGCTCATATCTTAACACTTGCAC-3′, p110β-K952R-F-5′-CTAAGTTTGGCATTAGAAGGGAGCGAGTG-3′, and p110β-K952R-R-5′-CACTCGCTCCCTTCTAATGCCAAACTTAG-3′. All mutations were confirmed by sequencing. For generation of stably expressing cell lines, NIH-3T3 or Rat-2 cells were co-transfected with pcDNA, myc-p110β WT or myc-p110β K952R and a vector with a puromycin resistance gene (ratio 5:1) and selected by incubating in 2.5 µg/ml puromycin.
Focus formation assay
Stably transfected NIH-3T3 cells expressing p110β or p110β K952R were plated in six-well plates. Cells were grown for 20 days with changes of medium every 3 days. The cells were stained with crystal violet and the number of transformed foci per well was counted. In addition, NIH-3T3 cells stably transfected with empty vector or p110β K952R were transduced with a lentiviral vector encoding an empty vector or p110β WT, seeded to six-well plates, and after 20-day incubation, focus formation was observed.
Antibodies
Anti-HA antibody (901513) was from Biolegend. Anti-SUMO2 (4971S), anti-phospho-Ser-473 of AKT (9271S), anti-phospho-FoxO1 (Thr24)/FoxO3 (Thr32) (9464S), anti-FoxO3a (2497S), anti-lamin A/C (4C11), and anti-AKT (9272S) were from Cell Signaling. Anti-B23 antibody (B0556) was from Sigma. Anti-myc antibodies were from Cell Signaling (9B11) and Santa Cruz Biotechnology (sc-40). Anti-GAPDH antibody (sc-32233) was from Santa Cruz Biotechnology. Antibodies against p110β were purchased from Invitrogen (PA5-22152) and Cell Signaling (3011S). Anti-Histidine antibody (MA1-21315) was from Invitrogen.
Nuclear-cytoplasm fractionation
The nuclear and cytoplasmic fractions were isolated using a nuclear-cytosol fractionation kit (BioVision), following manufacturer´s instructions.
In vitro SUMOylation assays
In vitro SUMO conjugation assays were performed on [35S]methionine-labeled in vitro-transcribed/translated proteins as described previously [27], using recombinant E1 (Biomol), Ubc9, and SUMO1 or SUMO2. The in vitro transcription/translation of proteins was performed using 1 µg of plasmid DNA and a rabbit reticulocyte-coupled transcription/translation system according to the instructions provided by the manufacturer (Promega). Briefly, [35S]methionine-labeled in vitro-transcribed/translated proteins were incubated with E1 in a 10µl reaction including an ATP regenerating system (50 mM Tris (pH7.5), 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate, 3.5 U of creatine kinase/ml, and 0.6 U of inorganic pyrophosphatase/ml), 600 ng of Ubc9 and 10 µg of SUMO1 or SUMO2, at 37 °C for 90 min. Reactions were stopped by adding SDS-PAGE loading buffer, boiled for 5 min at 100 °C, analyzed by SDS-PAGE and detected by autoradiography.
In vitro deSUMOylation assay
In vitro deSUMOylation assay with recombinant GST-SENP1 (Biomol) was performed on p110β-SUMO1 or p110β-SUMO2 as described previously [27]. p110β-SUMO1 or p110β-SUMO2 proteins obtained after in vitro SUMOylation assay were incubated with GST-SENP1. Reactions were incubated at 37 °C for 1 h, terminated by adding SDS-PAGE loading buffer, boiled for 5 min at 100 °C, analyzed by SDS-PAGE and detected by autoradiography.
Immunofluorescence staining
Immunofluorescence staining and confocal analysis were performed as described previously [27]. Briefly, cells cultured on coverslips were fixed with 2% paraformaldehyde, permeabilized with 0.25% Triton X-100 in PBS and blocked in 2% bovine serum albumin (BSA). Preparations were incubated with primary antibodies overnight in a moist chamber at 4 °C. Coverslips were washed with PBS and incubated with Alexa conjugated secondary antibody for 1 h at room temperature. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole), and coverslips were mounted with ProLong. Mouse anti-myc antibody was used at a dilution of 1:200. Secondary Alexa 488-conjugated antibody was obtained from Invitrogen. Analysis of the samples was carried out on a Leica TCS SP5 confocal laser microscope. Images were exported using Adobe Photoshop.
Purification of His-tagged conjugates
Cells were lysed in G-buffer (6 M Guanidine hydrocloride, 100 mM sodium phosphate pH 8.0, 50 mM Tris–HCl pH8.0) and incubated with pre-equilibrated Ni2+-NTA-agarose beads for 2 h at room temperature. Beads were washed four times with U-buffer (8 M urea, 100 mM sodium phosphate, 10 mM Tris–HCl pH8.0), resuspended in SDS-PAGE loading buffer and analyzed by SDS-PAGE followed by Western-blot.
Immunoprecipitation assay
Immunoprecipitation of endogenous SUMO1 or SUMO2 modified proteins was performed using denaturing cell lysis as described previously [28]. Briefly, cells were lysed with lysis buffer (PBS, 1% (wt/vol) SDS, 5 mM EDTA, 5 mM EGTA, 10 mM NEM) containing a protease inhibitor cocktail. Cell lysates were sonicated to reduce viscosity. DTT (50 mM final concentration) was added and cell lysates were boiled at 97 °C for 10 min, diluted (1:10) in cold RIPA buffer and supplemented with NEM (10 mM final concentration). Lysates were then cleared by centrifugation at 16,000 g for 15 min, and the supernatant was incubated overnight at 4 °C with SUMO1 or SUMO2 antibody-coupled beads. Control lysate was mixed with normal mouse IgG and beads. The next day, the beads were then washed three times with RIPA buffer, immunoprecipitated proteins were resuspended in SDS-PAGE loading buffer, boiled for 5 min at 100 °C and subjected to immunoblot analysis with the indicated antibodies.
Coimmunoprecipitation of p110β with p85β subunit of PI3K was evaluated in cells lysed in BC-100 buffer (20 mM Tris, pH 8, 0.1 M KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 0.1% Nonidet P40) containing 10 mM NEM and a protease inhibitor cocktail. Cell lysates were cleared by centrifugation at 20,000 g for 10 min, and the supernatant was incubated overnight at 4 °C with the primary antibody. The next day, lysates were incubated with protein A-sepharose beads for 2 h at 4 °C. Control lysate was mixed with normal mouse IgG and beads. The beads were then washed four times with BC-100 buffer, resuspended in SDS-PAGE loading buffer, boiled for 5 min at 100 °C and subjected to immunoblot analysis with the indicated antibodies.
Stability assay
HEK-293 cells were transfected with the indicated plasmids and 24 h after transfection cells were treated with cycloheximide (100 µg/ml). At different times after treatment, cells were analyzed by Western-blot with the indicated antibodies. Bands intensity was measured using ImageJ software. p110β bands intensity was normalized to a housekeeping protein.
Statistical analysis
Statistical analysis was performed using Student’s t test for comparison between two groups. We used the one-way Analysis of Variance (ANOVA) followed by Dunnett’s test using the GraphPad Prism 8.0.2 software for comparisons of more than two groups. All data are presented as mean ± standard deviation (SD). The significance level chosen for the statistical analysis was p < 0.05.
Results
Modification of p110β by SUMO
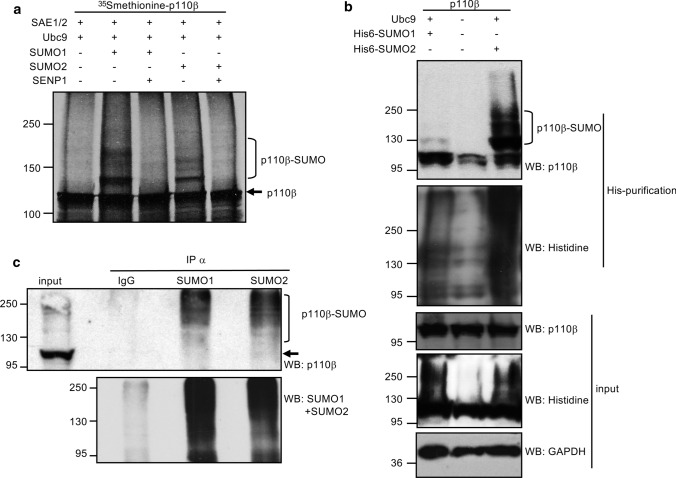
We carried out an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2 using in vitro translated [35S] methionine-labeled p110β protein as a substrate. The p110β protein was detected as a single band of the expected 110 kDa molecular weight. When the reaction was incubated with SUMO1 or SUMO2 we observed a strong additional band of around 130 kDa and fainter higher bands of around 150 and 170 kDa, which disappeared after incubating the reaction with the recombinant SUMO specific protease 1 (SENP1) (Fig. 1a). These results demonstrate that p110β is SUMOylated by SUMO1 and SUMO2 in vitro. In addition, the presence of several bands corresponding to p110β-SUMO1 in the in vitro assay indicates that SUMO1 can conjugate to more than one lysine residue, although the much stronger intensity of the 130 kDa band in comparison with the higher ones also suggests that the predominant form of SUMOylated p110β has only one SUMO1 molecule conjugated at a time. Then, to determine whether p110β also conjugates to SUMO within the cell, HEK-293 cells were co-transfected with myc-tagged p110β together with pcDNA or Ubc9 and His6-tagged SUMO1 or SUMO2 plasmids. At 48 h after transfection, His6-tagged proteins were purified in denaturing conditions using nickel beads. Western-blot analysis of the purified extracts with anti-p110β antibody revealed bands of the expected size corresponding to p110β-SUMO1 and p110β-SUMO2 only in those cells co-transfected with His6-SUMO1 and His6-SUMO2, respectively, indicating that p110β is modified by SUMO1 and SUMO2 in transfected cells (Fig. 1b). Finally, we decided to evaluate the SUMOylation of endogenous p110β protein using a method that allows high enrichment of SUMO targets under physiological conditions [28]. Protein extracts from PC3 cells were lysed under denaturing conditions, diluted in non-denaturing buffer and subjected to immunoprecipitation with validated anti-SUMO1 or anti-SUMO2 antibodies (SUMO1 21C7 or SUMO2 8A2 Hybridoma Bank). Finally, the immunoprecipitated extracts were evaluated by Western-blot analysis with anti-p110β antibody. As shown in Fig. 1c, we detected SUMOylated p110β bands in the SUMO-immunoprecipitated extracts but not in the anti-IgG immunoprecipitated extracts, indicating that endogenous p110β is SUMOylated in the cell without overexpression of the SUMO machinery. Altogether these results demonstrate that p110β protein is modified by SUMO.
Fig. 1.
p110β protein is modified by SUMO1 and SUMO2 in vivo and in vitro. a Modification of in vitro translated p110β by SUMO1 or SUMO2. [35S]methionine labeled in vitro translated p110β was subjected to an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2, as indicated. SUMOylated p110β protein was then subjected to a deSUMOylation assay in the presence of SENP1. b SUMOylation of p110β in transfected cells. HEK-293 cells were co-transfected with myc-p110β together with pcDNA or Ubc9 and His6-SUMO1 or His6-SUMO2, and 48 h after transfection whole protein extracts (input) and histidine-tagged purified proteins were analyzed by Western-blot using the indicated antibodies. c SUMOylation of endogenous p110β under denaturing conditions. PC3 cells were lysed under denaturing conditions, immunoprecipitated using anti-SUMO1 or anti-SUMO2 antibodies and analyzed by Western-blot with anti-p110β antibody
Lysine residue K952 in p110β is a SUMO acceptor site
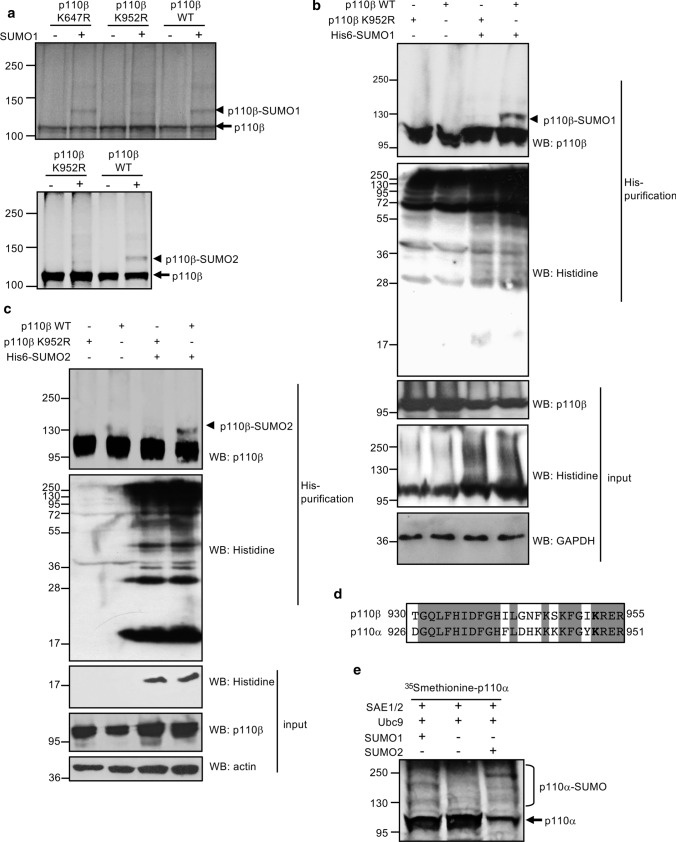
To map the SUMO acceptor lysine of p110β, we constructed single mutants in two candidate lysines (K647 and K952) identified by the GPS-SUMO software as present within a type I SUMO consensus motif, and analyzed their capability to conjugate to SUMO in vitro. We did not observe differences in the SUMO1 conjugation between the WT and the K647R mutant p110β proteins (Fig. 2a, upper panel). However, SUMO1 modification was reduced after mutation of lysine K952 to arginine (Fig. 2a, upper panel), thus identifying this residue, located at the activation loop of p110β, as the main SUMO acceptor site. Similarly, a reduction in the in vitro SUMO2 modification of the K952R mutant relative to the WT p110β protein was observed (Fig. 2a, lower panel). To verify the involvement of this residue in p110β SUMOylation, we evaluated the SUMOylation of WT and K952R p110β proteins in cells. We did not observe modification of p110β K952R with SUMO1 (Fig. 2b) or SUMO2 (Fig. 2c), confirming that lysine residue K952 in p110β is a SUMO acceptor site.
Fig. 2.
Lysine residue K952 in p110β is the main SUMO conjugation site. a Modification of in vitro translated p110β WT, p110β K647R or p110β K952R by SUMO1 or SUMO2 in vitro. 35S-methionine-labeled in vitro translated WT or mutant p110β proteins were subjected to an in vitro SUMOylation assay in the presence of SUMO1 (upper panel) or SUMO2 (lower panel). b SUMO1 modification of p110β WT or p110β K952R in transfected cells. HEK-293 cells were co-transfected with myc-p110β or myc-p110β K952R together with pcDNA or Ubc9 and His6-SUMO1, and 48 h after transfection whole protein extracts and histidine-tagged purified proteins were analyzed by Western-blot with the indicated antibodies. c SUMO2 modification of p110β WT or p110β K952R in transfected cells. HEK-293 cells were co-transfected with myc-p110β or myc-p110β K952R together with pcDNA or Ubc9 and His6-SUMO2, and 48 h after transfection whole protein extracts and histidine-tagged purified proteins were analyzed by Western-blot with the indicated antibodies. d Amino acid sequence homology between the activation loop of p110α and p110β. Lysine residue involved in SUMO conjugation to p110β and the corresponding lysine residue in p110α are labeled in bold. e Modification of in vitro translated p110α by SUMO1 or SUMO2. [35S]methionine labeled translated p110α was subjected to an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2, as indicated
The lysine residue that corresponds to K952 of p110β is conserved in the activation loop of p110α (Fig. 2d). In addition, residues 941 to 950 in p110α are disordered [29], a feature that has been reported to facilitate SUMO conjugation [30]. Therefore, we hypothesized that p110α could be also SUMOylated. To test this hypothesis, we carried out an in vitro SUMOylation assay in the presence of SUMO1 or SUMO2 using in vitro translated [35S] methionine-labeled p110α protein as a substrate. We observed that p110α is SUMOylated by SUMO1 and SUMO2 in vitro (Fig. 2e), suggesting that p110β is not the only PI3K catalytic subunit modulated by SUMO.
SUMO modulates the stability of p110β
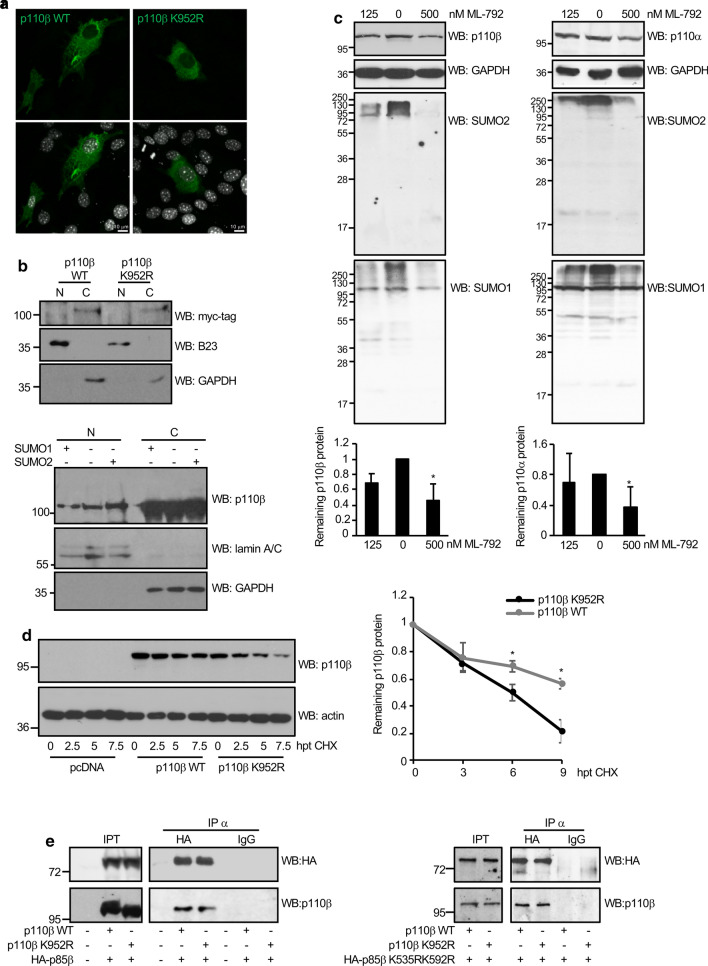
SUMOylation can have varied effects on the target protein including the regulation of its subcellular localization, stability, transcriptional activity and interaction with other proteins [31]. We first studied whether mutation of the SUMOylation site in p110β altered its subcellular localization. Confocal analysis of NIH-3T3 cells transfected with myc-tagged p110β WT or K952R mutant plasmids revealed that both proteins, the WT and the SUMOylation mutant, were detected mainly at the cell cytoplasm (Fig. 3a). Similar results were observed after Western-blot analysis of nuclear and cytoplasm fractions obtained from cells transfected with myc-tagged p110β WT or K952R mutant using anti-myc antibody (Fig. 3b, upper panel). In addition, we did not observe a change in the subcellular distribution of endogenous p110β in HEK-293 cells, detected using a cell fractionation assay, after transfection of SUMO1 or SUMO2 (Fig. 3b, lower panel), suggesting that SUMO does not affect the subcellular localization of p110β.
Fig. 3.
SUMOylation of p110β enhances its protein stability. a Subcellular localization of p110β WT or p110β K952R in NIH-3T3 cells. NIH-3T3 cells were transfected with p110β WT or p110β K952R. At 36 h after transfection, cells were stained with mouse anti-myc-tag antibody and Alexa-488 conjugated donkey anti-mouse secondary antibodies. Nuclei were visualized by DAPI staining (white). b Subcellular localization of p110β WT or p110β K952R in HEK-293 cells (upper panel). HEK-293 cells were transfected with p110β WT or p110β K952R. At 48 h cells were subjected to subcellular fractionation and then analyzed by Western-blot with the indicated antibodies. Subcellular localization of endogenous p110β protein in HEK-293 cells transfected with SUMO1 or SUMO2 (lower panel). Transfected cells were subjected to subcellular fractionation and then analyzed by Western-blot with the indicated antibodies. c PC3 cells were treated with the indicated concentrations of the SUMOylation inhibitor ML-792 and 12 h after treatment cells were harvested for Western-blot analysis with the indicated antibodies (upper panel). Quantification of endogenous p110β or p110α protein levels relative to GAPDH expression from at least three independent experiments is shown in the lower panel. Data are mean ± SD of three experiments. Statistical analysis was assessed by one-way ANOVA followed by Dunnett’s test. *p < 0.05. d HEK-293 cells transfected with p110β WT or p110β K952R were incubated in the presence of cycloheximide (CHX). At the indicated times after CHX treatment protein extracts were analyzed by Western-blot analysis with the indicated antibodies (left panel). The stability of each protein (mean ± SD) from three independent experiments is shown (right panel). *p < 0.05, Student’s paired t test. e Interaction between p110β and p85β is not altered by p110β SUMOylation. HEK-293 cells were co-transfected with myc-p110β WT or myc-p110β K952R together with HA-p85β WT or HA-p85β K535RK592R, as indicated. At 48 h after transfection, cells were lysed, immunoprecipitated using anti-HA antibodies and analyzed by Western-blot with anti-p110β antibody
To determine whether SUMO had an impact on p110β stability, we first treated cells with two different concentrations of the SUMOylation inhibitor ML-792 and evaluated by Western-blot the levels of the endogenous p110β protein. We observed a significant decrease in p110β levels after treatment with 500 nM of the inhibitor (Fig. 3c, left panel). Similar trends were observed on p110α, with a statistically significant reduction in protein levels at the highest dose of ML-792 (Fig. 3c, right panel). Therefore, we speculated that SUMO may modulate the stability of the protein. Cycloheximide chase experiments were then carried out to determine the stability of p110β WT and K952R mutant. p110β K952R exhibited a shorter half-life than the WT protein (Fig. 3d). These results suggest that SUMO conjugation stabilizes p110β protein.
The p85 regulatory subunit interacts with and stabilizes p110 [8]. Therefore, we decided to evaluate whether this interaction is modulated by SUMO conjugation. HEK-293 cells were co-transfected with p110β WT or K952R together with HA-p85β WT or the p85β SUMOylation mutant, and 48 h after transfection p110β-p85β interaction was analyzed by co-immunoprecipitation. We did not observe differences in the interaction between p110β WT or p110β K952R, and p85β WT or the p85β SUMOylation mutant (Fig. 3e), which suggests that an alteration in the p110β-p85β interaction is not responsible for the stabilization of p110β by SUMO.
Mutation of the SUMOylation site in p110β has a negative impact on the activation of the PI3K pathway mediated by the protein
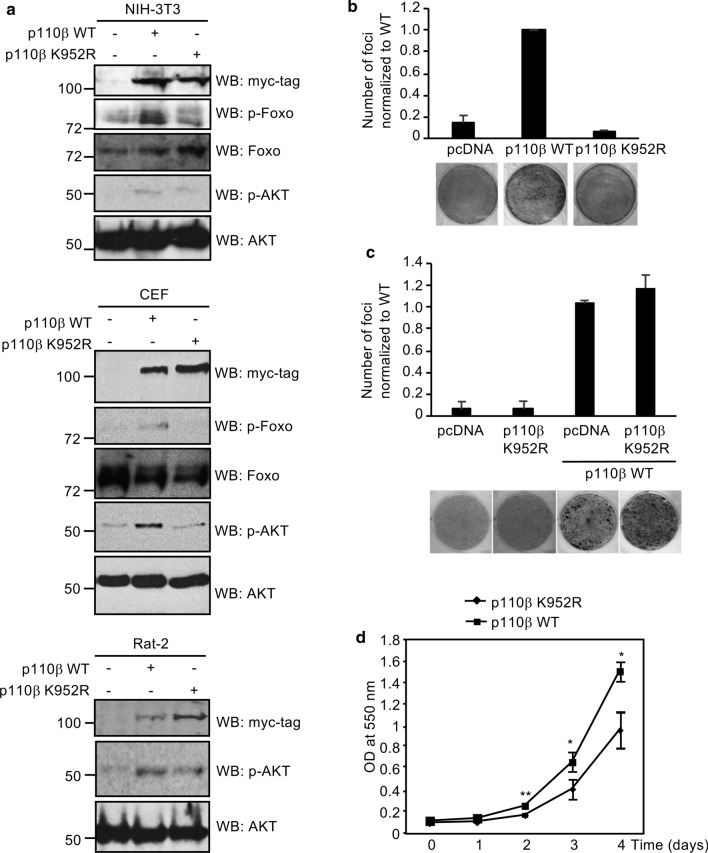
To determine the consequences of p110β SUMOylation on signaling, NIH-3T3 cells transfected with pcDNA, p110β WT or K952R were serum deprived for 16 h, and activation of the AKT pathway was analyzed by Western-blot with a specific anti-phospho Ser473-AKT antibody or with anti-phospho-FoxO-specific antibody. Overexpression of p110β led to phosphorylation of FoxO and to AKT activation, as previously reported [3, 32]. The levels of phosphorylated AKT or FoxO proteins in cells expressing p110β K952R protein were higher than in pcDNA transfected control cells but lower than those detected in p110β WT transfected cells (Fig. 4a, upper panel). Similar results were observed after analysis of phosphorylated AKT or FoxO proteins in both CEF and Rat-2 cells transfected with p110β WT or the SUMOylation mutant (Fig. 4a, middle and lower panel, respectively), indicating that mutation of the SUMOylation site in p110β has a negative impact on the activation of the PI3K pathway mediated by the protein.
Fig. 4.
Lysine residue K952 in p110β is important for PI3K pathway activation and p110β-induced transformation and proliferation. a Analysis of AKT or FoxO phosphorylation in NIH-3T3, CEF or Rat-2 cells expressing the indicated constructs and incubated for 16 h in serum-free media. b Focus formation assay. NIH-3T3 cells stably expressing p110β WT or p110β K952R protein were plated and left to grow to confluence for 20 days when foci were stained and counted (upper panel). Data are mean ± SD of triplicate samples for one experiment. Experiments were repeated twice. A representative image is shown (lower panel). c NIH-3T3 cells stably transfected with empty vector or p110β K952R were transduced with lentivirus expressing empty vector or p110β WT, plated and left to grow to confluence for 20 days when foci were stained and counted (upper panel). Data are mean ± SD of triplicate samples for one experiment. A representative image is shown (lower panel). d Rat-2 cells stably expressing p110β WT or p110β K952R protein were plated in 96-well plates, incubated for the indicated times and assayed using the MTT assay. Data are mean ± SD of triplicate samples. *p < 0.05, **p < 0.005, Student’s paired t test
p110β induces oncogenic transformation in cell culture [3]. Therefore, we decided to analyze the transforming activity of the p110β SUMOylation mutant. We generated NIH-3T3 cells that stably overexpress WT or the K952R mutant p110β protein and tested their ability to form foci of transformed cells. As shown in Fig. 4b, we detected focus formation in cells transfected with p110β WT, as previously reported [3, 32]. However, p110β K952R was not able to induce foci (Fig. 4b). In addition, we evaluated the formation of transformed foci of NIH-3T3 cells stably transfected with an empty vector or with p110β K952R as the result of transduction with a lentivirus expressing p110β WT. The number of transformed foci was similar in both conditions (Fig. 4c). These results suggest that SUMOylation of p110β is required for cellular transformation. Finally, we also generated Rat-2 cells that stably overexpress p110β WT or p110β K952R protein and proliferation of the two cell lines was compared by the MTT assay. We observed that the cells expressing p110β WT grew at a higher rate than the K952R mutant cells (Fig. 4d). Altogether, these results suggest that SUMO conjugation to p110β positively modulates its oncogenic capability, its cell proliferation-promoting activity, and the PI3K downstream pathway activation.
The PI3K regulatory subunit p85β negatively modulates the SUMOylation of p110β
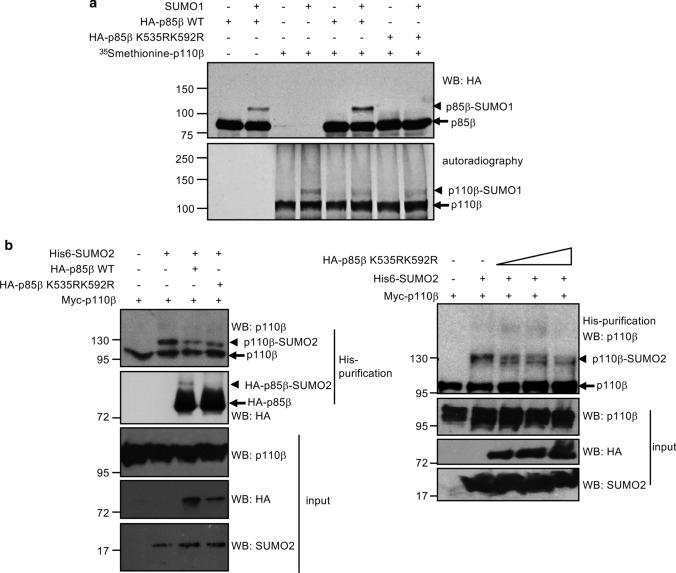
Our data indicate that SUMO conjugates to a lysine residue located within the second basic box of the activation loop of p110β. Interaction of p85 nSH2 domain with p110 has been proposed to confine the activation loop into a collapsed conformation [33, 34]. Therefore, we speculated that p85 may modulate the accessibility of SUMO to conjugate to p110β. To evaluate this possibility, we first carried out an in vitro SUMOylation assay with SUMO1 using in vitro translated [35S] methionine-labeled p110β protein as a substrate, in the presence or absence of in vitro translated p85β protein. In addition, since p85β protein can also be SUMOylated [17], and to discard that any alteration in the SUMOylation of p110β is due to a competition for limiting amounts of SUMO in the reaction, we also carried out the experiment in the presence of a p85β SUMOylation mutant. We observed a down-modulation of p110β SUMOylation by p85β and this effect was independent of the p85β SUMOylation (Fig. 5a). To confirm these results, we studied the SUMOylation of p110β in cells co-transfected with pcDNA, p85β WT or the p85β SUMOylation mutant. Co-expression of p85β WT or p85β SUMOylation mutant led to a reduction in p110β SUMOylation (Fig. 5b, left panel). In addition, we also observed a reduction in the SUMOylation of p110β after transfection of increasing amounts of the p85β SUMOylation mutant (Fig. 5b, right panel). These results suggest that p85β modulates the covalent interaction of SUMO with p110β.
Fig. 5.
Regulation of p110β SUMOylation by p85β. a p85β negatively modulates the SUMOylation of p110β in vitro. [35S]methionine labeled in vitro translated p110β protein was subjected to an in vitro SUMOylation assay with SUMO, in presence or absence of in vitro translated HA-tagged p85β WT or p85β SUMOylation mutant (p85β K535RK592R) proteins. b HEK-293 cells were co-transfected with myc-p110β and pcDNA or myc-p110β, Ubc9, His6-SUMO2, and pcDNA, HA-p85β WT or a HA-tagged p85β SUMOylation mutant (HA-p85β K535RK592R) (left panel). 48 h after transfection whole protein extracts and histidine-tagged purified proteins were analyzed using the indicated antibodies. HEK-293 cells were co-transfected with myc-p110β together with Ubc9, His6-SUMO2, and increasing amounts of the HA-tagged p85β SUMOylation mutant (HA-p85β K535RK592R) plasmid (right panel). 48 h after transfection whole protein extracts and histidine-tagged purified proteins were analyzed using the indicated antibodies
Discussion
Here we demonstrate that the PI3K catalytic subunit p110β can be modified by SUMO in vitro, in transfected cells and under completely endogenous conditions, indicating that SUMOylation of p110β is physiologically relevant. We identified lysine residue K952, located at the activation loop in p110β, as the SUMO-acceptor site. The lysine residue that corresponds to K952 of p110β is conserved in a disordered region of the activation loop of p110α [29], suggesting that p110α may be also SUMOylated. This hypothesis is supported by the detection of p110α protein conjugated to SUMO by in vitro SUMOylation analysis.
SUMOylation can regulate the stability of the target proteins [31]. Here we show that the endogenous p110β and p110α protein levels decrease after SUMOylation inhibition and that mutation of the SUMOylation site in p110β reduces the stability of the transfected protein but did not affect its interaction with p85β. These results are in agreement with the observed reduction in the levels of PI3K in response to knockdown of SUMO in microglia [35] and led us to propose that SUMO increases the stability of p110β. The PI3K catalytic subunit p110 is thermally unstable and can be conformational stabilized by different mechanisms including the interaction with the regulatory subunit p85, the synthesis at low temperature or the linkage of an epitope tag to the N-terminus of the protein [8]. Based on these data, we propose that SUMO conjugation may stabilize the overall conformation of the catalytic subunit. Interestingly, SUMOylation has been recently suggested as a mechanism that might promote protein thermal stability during mitosis [36]. In agreement with the role of SUMO in the stability of p110β protein, mutation of the SUMOylation site in p110β reduced its ability to activate the AKT signaling pathway, promote cell proliferation and induce transformation of NIH-3T3 cells, suggesting that SUMO positively modulates the activity of the p110β protein and that the full oncogenic activity of p110β may require SUMO conjugation. Supporting this hypothesis, it has been previously reported that the proliferation of the prostate cancer cell line PC3 depends on both, the SUMO conjugating enzyme Ubc9 [19] and p110β [37].
Interaction of p85 with p110 inhibits its activity [8]. Here we show that p85 also inhibits the SUMOylation of p110β. Recently, Zhang et al. proposed that the interaction of the nSH2 motif of p85 with the second basic box (KRER) of the activation domain, confines the activation loop into a collapsed conformation, inhibiting p110 [33]. The restriction in the flexibility of the activation loop exerted by p85 may then explain the inhibition of the SUMOylation of p110β mediated by p85β.
In summary, we have identified SUMOylation as a new mechanism of regulation of p110β. Our data indicate that SUMO conjugation to p110β has a positive impact on the activation of the PI3K pathway likely through increasing its stability. These results together with previous data demonstrating that SUMOylation of p85β negatively regulates PI3K [17], indicate that SUMO can positively or negatively modulate the signaling pathway, depending on the targeted protein, and suggest that the interaction of SUMO with each PI3K component must be tightly regulated. Moreover, the results shown here suggest that p85 may be one of these regulators. This information could be useful for designing PI3K inhibitors.
Acknowledgements
We thank Dr, Jonathan M Backer for kindly providing the myc-p110β plasmid and Dr. Lewis Cantley for the HA-tagged p110α expression plasmid. Funding at the laboratory of CR is provided by Ministry of Science, Innovation and Universities and FEDER (BFU-2017–88,880-P) and Xunta de Galicia (ED431G 2019/02). SV and RS are predoctoral fellows funded by Xunta de Galicia-Consellería de Cultura, Educación y Ordenación Universitaria (ED481A-2018/110 and ED481A-2020/160, respectively).
Abbreviations
- CHX
Cycloheximide
- PI3Ks
Phosphatidylinositol-4,5,bisphosphate 3-kinases
- SUMO
Small ubiquitin-related modifier
- SENP1
SUMO specific protease 1
Authors contributions
AEM, CFC-H, SV, RS, YHB, and MB-M conducted the experiments; ME and MSR generated reagents; AV, MSR, ME, MC, EL, and CR analyzed the results; CR, designed the experiments and wrote the paper.
Funding
Funding at the laboratory of CR is provided by Ministry of Science, Innovation and Universities and FEDER (BFU-2017-88880-P) and Xunta de Galicia (ED431G 2019/02). The laboratory of MC is funded by grant RTI2018-095818-B-100 (MCIU/AEI/ FEDER, UE). SV and RS are predoctoral fellows funded by Xunta de Galicia-Consellería de Cultura, Educación y Ordenación Universitaria (ED481A-2018/110 and ED481A-2020/160, respectively).
Availability of data and materials
All data generated or analysed during this study are included in this published article. The materials used in this study are available from the corresponding authors, CR, upon reasonable request.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interest to disclosure.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethics approval
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ahmed El Motiam and Carlos F de la Cruz-Herrera, equal contribution.
References
- 1.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85–p110 heterodimers. Proc Natl Acad Sci USA. 2007;104:7809–7814. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Oncogenic transformation induced by the p110beta, -gamma, and -delta isoforms of class I phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3'-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3'-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/MCB.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-E. [DOI] [PubMed] [Google Scholar]
- 10.Ueki K, Fruman DA, Brachmann SM, Tseng YH, Cantley LC, Kahn CR. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol Cell Biol. 2002;22:965–977. doi: 10.1128/MCB.22.3.965-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt KH, Olson L, Moye-Rowley WS, Pessin JE. Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol Cell Biol. 1994;14:42–49. doi: 10.1128/MCB.14.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu P, Schlessinger J. Direct association of p110 beta phosphatidylinositol 3-kinase with p85 is mediated by an N-terminal fragment of p110 beta. Mol Cell Biol. 1994;14:2577–2583. doi: 10.1128/MCB.14.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu P, Mondino A, Skolnik EY, Schlessinger J. Cloning of a novel, ubiquitously expressed human phosphatidylinositol 3-kinase and identification of its binding site on p85. Mol Cell Biol. 1993;13:7677–7688. doi: 10.1128/MCB.13.12.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klippel A, Escobedo JA, Hu Q, Williams LT. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol Cell Biol. 1993;13:5560–5566. doi: 10.1128/MCB.13.9.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuevas BD, Lu Y, Mao M, Zhang J, LaPushin R, Siminovitch K, Mills GB. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J Biol Chem. 2001;276:27455–27461. doi: 10.1074/jbc.M100556200. [DOI] [PubMed] [Google Scholar]
- 16.Mellor P, Furber LA, Nyarko JN, Anderson DH. Multiple roles for the p85α isoform in the regulation and function of PI3K signalling and receptor trafficking. Biochem J. 2012;441:23–37. doi: 10.1042/BJ20111164. [DOI] [PubMed] [Google Scholar]
- 17.de la Cruz-Herrera CF, Baz-Martínez M, Lang V, El Motiam A, Barbazán J, Couceiro R, Abal M, Vidal A, Esteban M, Muñoz-Fontela C, Nieto A, Rodríguez MS, Collado M, Rivas C. Conjugation of SUMO to p85 leads to a novel mechanism of PI3K regulation. Oncogene. 2016;35:2873–2880. doi: 10.1038/onc.2015.356. [DOI] [PubMed] [Google Scholar]
- 18.González-Santamaría J, Campagna M, Ortega-Molina A, Marcos-Villar L, de la Cruz-Herrera CF, González D, Gallego P, Lopitz-Otsoa F, Esteban M, Rodríguez MS, Serrano M, Rivas C. Regulation of the tumor suppressor PTEN by SUMO. Cell Death Dis. 2012;3:e393. doi: 10.1038/cddis.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz-Herrera CF, Campagna M, Lang V, del Carmen G-S, Marcos-Villar L, Rodriguez MS, Vidal A, Collado M, Rivas C. SUMOylation regulates AKT1 activity. Oncogene. 2015;34:1442–1450. doi: 10.1038/onc.2014.48. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, Zhu C, Chen C, Liu X, Cheng J, Mustelin T, Feng GS, Chen G, Yu J. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73:5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- 22.Risso G, Pelisch F, Pozzi B, Mammi P, Blaustein M, Colman-Lerner A, Srebrow A. Modification of Akt by SUMO conjugation regulates alternative splicing and cell cycle. Cell Cycle. 2013;12:3165–3174. doi: 10.4161/cc.26183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotter CA, Earl PL, Wyatt LS, Moss B. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Protein Sci. 2017;89:5.12.1–5.12.18. doi: 10.1002/cpps.34. [DOI] [PubMed] [Google Scholar]
- 24.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/S1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 25.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Dbouk HA, Backer JM. Novel approaches to inhibitor design for the p110β phosphoinositide 3-kinase. Trends Pharmacol Sci. 2013;34:149–153. doi: 10.1016/j.tips.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campagna M, Marcos-Villar L, Arnoldi F, de la Cruz-Herrera CF, Gallego P, Gonzalez-Santamaria J, Gonzalez D, Lopitz-Otsoa F, Rodríguez MS, Burrone OR, Rivas C. Rotavirus viroplasm proteins interact with the cellular SUMOylation system: implications for viroplasm-like structure formation. J Virol. 2012;87:807–817. doi: 10.1128/JVI.01578-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barysch SV, Dittner C, Flotho A, Becker J, Melchior F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 2014;9:896–909. doi: 10.1038/nprot.2014.053. [DOI] [PubMed] [Google Scholar]
- 29.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 30.Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA, Jensen LJ, Nielsen ML. Site-specific characterization of endogenous SUMOylation across species and organs. Nat Commun. 2018;9:2456. doi: 10.1038/s41467-018-04957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 32.Pazarentzos E, Giannikopoulos P, Hrustanovic G, St John J, Olivas VR, Gubens MA, Balassanian R, Weissman J, Polkinghorn W, Bivona TG. Oncogenic activation of the PI3-kinase p110β isoform via the tumor-derived PIK3Cβ(D1067V) kinase domain mutation. Oncogene. 2016;35:1198–1205. doi: 10.1038/onc.2015.173. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Jang H, Nussinov R. Structural features that distinguish inactive and active PI3K lipid kinases. J Mol Biol. 2020 doi: 10.1016/j.jmb.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, Stephens LR, Williams RL. Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol Cell. 2011;41:567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saw G, Krishna K, Gupta N, Soong TW, Mallilankaraman K, Sajikumar S, Dheen ST. Epigenetic regulation of microglial phosphatidylinositol 3-kinase pathway involved in long-term potentiation and synaptic plasticity in rats. Glia. 2020;68:656–569. doi: 10.1002/glia.23748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becher I, Andrés-Pons A, Romanov N, Stein F, Schramm M, Baudin F, Helm D, Kurzawa N, Mateus A, Mackmull MT, Typas A, Müller CW, Bork P, Beck M, Savitski M. pervasive protein thermal stability variation during the cell cycle. Cell. 2018;173:1495–1507. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci USA. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article. The materials used in this study are available from the corresponding authors, CR, upon reasonable request.
Not applicable.