Abstract
Among all reactive oxygen species (ROS), hydrogen peroxide (H2O2) takes a central role in regulating plant development and responses to the environment. The diverse role of H2O2 is achieved through its compartmentalized synthesis, temporal control exerted by the antioxidant machinery, and ability to oxidize specific residues of target proteins. Here, we examine the role of H2O2 in stress acclimation beyond the well-studied transcriptional reprogramming, modulation of plant hormonal networks and long-distance signalling waves by highlighting its global impact on the transcriptional regulation and translational machinery.
Keywords: Redox signalling, Chromatin remodeling, Oxidative posttranslational modifications, Stress priming
Introduction
Crops grown in the field rarely achieve their genetically encoded yield potential. Fluctuating environmental conditions and severe or mild abiotic and biotic stress episodes are the main drivers of reduced plant productivity [1, 2]. Their amplitude, intervals of occurrences throughout the growing season, and plant developmental stage at which they occur are inextricably intertwined with plant yield. Any changes in growth conditions are immediately perceived and transduced via signalling pathways ultimately remodelling the epigenetic landscape, gene expression, proteome, and metabolome. This ensures plant survival in a changing environment. The dynamics and amplitude of those responses are a function of stress severity and duration. Creating further complexity, plants often experience a combination of stress factors which can occur either simultaneously or separate in time [3–5].
Even when stress conditions subside, many molecular processes are not immediately reset to their prestress levels and create a new base level that underlies a conceptually new response to future environmental fluctuations. Similarly, plants experiencing mild stress will react differently to a subsequent harsher stress than naïve plants. The new response is often faster and stronger and has a lesser negative impact on plant physiology and growth compared to plants that have not been previously exposed to any stress condition. This concept is often referred to as priming, acclimation, or hardening [6–10]. Depending on the nature and duration of the initial stress exposure, those terms can often be used interchangeably but in general, acclimation denotes processes that develop over prolonged mild stress periods [10, 11].
Plants experiencing adverse environmental conditions accumulate reactive oxygen species (ROS) that are partially reduced (superoxide radicals, hydrogen peroxide and hydroxyl radicals) or excited (singlet oxygen) forms of O2 [12, 13]. Historically, excessive ROS levels associated with abiotic stresses has been exclusively seen as a trigger of oxidative stress characterized by indiscriminate oxidative attack on proteins, DNA, and lipids [14]. This paradigm has led to numerous attempts to engineer stress resilience by overexpression of components of the antioxidant machinery [15, 16]. Despite certain successful outcomes, this approach has revealed that boosting the antioxidant capacity can have far more outreaching effects than minimizing oxidative impact. This largely stems from the fact that apart from their damaging nature, ROS also initiate, integrate, and fine-tune numerous signalling cascades involved in growth, development, and defense [17–19].
The highly reactive nature of ROS, their interconversion, and extremely short lifetime have been a serious barrier to understanding the precise roles of individual ROS types [13, 20–22]. Among them, hydrogen peroxide (H2O2) has attracted significant attention due to its relative stability, transmembrane mobility, and direct sensing by receptor proteins [23–25]. Apart from the fact that H2O2 accumulation has been associated with various stress conditions, exogenously applied H2O2 can also prime plants against a range of adverse environmental conditions [11, 26, 27]. Here, we take a critical look at the role of H2O2 in stress acclimation. The origins of H2O2, its signalling role, and crosstalk with various phytohormonal pathways have been extensively reviewed previously [23, 24, 28, 29] and will not be the focus of this review. Instead, we summarize and explore less researched but equally important and intriguing areas related to the impact of H2O2 on the transcriptional regulation and translational machinery that are likely to have a global regulatory effect during stress acclimation. Moreover, we revisit the use of H2O2 as a priming agent and discuss its potential as a solution against adverse environmental stresses.
Effect of H2O2 on plant signaling and metabolism
Priming against adverse environmental conditions upon H2O2 exposure
Plants cannot only be primed against adverse environmental conditions after experiencing a mild stress episode but can also be chemically primed upon exposure to natural and synthetic small molecules [30–32]. The scientific literature abounds with examples of various chemical compounds that improve stress tolerance in model and crop species [26, 27, 30, 33–36]. Although many of these reports are anecdotic and the diverse range of stress conditions, concentrations, application modes, and plant developmental stages makes it difficult to draw outreaching conclusions for the efficacy and applied potential of most compounds, stress protective agrochemicals are an exciting alternative to climate resilient crops. Several small molecules have been shown to protect crops in the field and a substantial effort fueled by the latest developments in chemical biology is underway to discover and commercialize novel agrochemicals with stress protective effects [37, 38].
The priming effect of exogenously applied H2O2 has been documented in various stress scenarios (salt, drought, heat, cold, and heavy metal stress) and model plant and crop species [35, 39–41]. Seed treatment, spraying or addition to the growth medium was used to administer a wide range of H2O2 concentrations. The effect of H2O2 priming was assessed by monitoring plant growth parameters, photosynthetic efficiency, photosynthetic pigments and chloroplast structure, osmolytes, ion leakage, lipid peroxidation, levels of enzymatic and non-enzymatic antioxidants, and endogenous ROS content [26, 33, 42–45]. Not all H2O2 concentrations provoke an equal response. In fact, H2O2 application is often employed as a proxy for oxidative stress with a negative impact on Arabidopsis rosette and root growth and germination rate observed when plants are germinated and grown on 0.5–2.5 mM H2O2 [46]. Tobacco plants primed by spraying with 5 mM H2O2 displayed improved performance under high light and aminotriazole, a catalase inhibitor, whereas concentrations above 50 mM were lethal [47]. The concentrations of H2O2 used to successfully prime plants against subsequent stresses have been reported in the range from 0.05 µM to 200 mM. This vast range likely reflects the modes of application, their duration, and plant-specific morphological and physiological features. For example, a high H2O2 concentration (200 mM) was needed to successfully prime 7-day-old Vigna radiata seedlings by spraying them 12 h before exposure to chilling stress (4 °C for 36 h) [48]. In contrast, pretreatment of tomato seedling roots with only 1 mM H2O2 for an hour enhanced plant tolerance to chilling (3 °C for 16 h) 4 days after the priming [49]. Even lower amounts of H2O2 (10 µM) were sufficient to improve the resistance of hydroponically grown rice plants to salinity and heat when H2O2 was present in the medium for 2 days [50].
H2O2 production and signalling
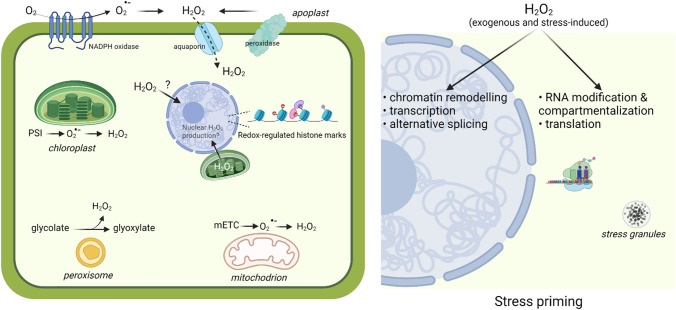
The negative effects observed at high H2O2 concentrations are largely a reflection of a general oxidative stress response that leads to indiscriminate damage of cellular constituents. Separating the damaging and signalling aspects of externally applied H2O2 would prove to be extremely difficult not least because exogenous H2O2 is likely to activate and/or perturb other ROS-producing mechanisms. Multiple sources contribute to H2O2 production which is highly compartmentalized between different cellular compartments (Fig. 1). Their contribution to the overall H2O2 content depends on the particular cell type and physiological state. For example, in illuminated leaves, chloroplasts, together with peroxisomes, are major sources of H2O2 [51, 52]. H2O2 is actively produced in the apoplast by dismutation of NADPH oxidase-derived superoxide and cell wall peroxidases [13, 23, 53]. Plant NADPH oxidases (NOXs) have been implicated in many developmental processes, such as root hair formation and cell expansion, stomatal closure, as well as defense responses [54–56]. Propagation of long-distance ROS signals that alert systemic tissues to mount a coordinated response upon abiotic and biotic stresses are also dependent on NOX activity [57].
Fig. 1.
Sources of H2O2 and its possible role in stress acclimation. H2O2 is produced in different subcellular compartments in response to environmental stimuli and during normal metabolism. Elevated H2O2 levels can impact key cellular processes such as gene expression, chromatin remodeling, alternative splicing, RNA modification and compartmentalization, and translation that can ultimately lead to stress priming. Created with BioRender.com
The exact molecular mechanisms that underlie the improved response to stress after H2O2 treatment remain largely unclear. Increased H2O2 levels and perturbation of cellular redox homeostasis is a common theme among many environmental stresses. In this respect, exogenous H2O2 application is likely to mimic naturally occurring redox processes that are integral to plant stress responses since elevated ROS levels have also been observed upon H2O2-induced priming. Priming of maize with H2O2 increased the endogenous levels of superoxide radicals, but not H2O2, prior to stress exposure and led to attenuated accumulation of ROS during a subsequent salt stress exposure [45]. Similarly, a peak of H2O2 was detected during priming of maize with heat (42 °C for 4 h) which correlated with improved subsequent performance to salinity, chilling, drought, and heat stresses [58]. Due to its relatively long half-life and physicochemical properties that resemble water, H2O2 can migrate significant distances and enter the cell from its extracellular production site via aquaporin membrane proteins [51]. Aquaporins might play a role in internalizing externally applied H2O2. Whether the increase in endogenous ROS content is simply due to uptake of the applied H2O2 and its conversion to other ROS types or exogenous H2O2 triggers in planta ROS production is not immediately clear but most likely, it involves a combination of both. The subcellular production sites that might be involved in exogenous H2O2-triggered ROS production are not yet resolved. Among the direct targets of externally applied H2O2 might be the plasma membrane localized H2O2 sensor HPCA1 that is activated via covalent modification of extracellular cysteine residues [59]. HPCA1 is a leucine-rich-repeat receptor kinase that mediates Ca2+ influx into the cytosol by activating Ca2+ channels. Since calcium spikes are closely interacting with ROS production and amplify each other, apoplastic H2O2 production through calcium-stimulated NOXs activity is likely to contribute to the observed ROS increase upon H2O2 priming [59, 60].
Impact of H2O2 on cellular redox homeostasis
In many reports, the beneficial effect of H2O2 priming has been found to correlate with increased activities of antioxidant enzymes, higher content of small molecule antioxidants, diminished oxidative damage, and lower ROS levels under stress [26, 41, 42]. Whereas some of these parameters are not so technically demanding to assess, the reported content of H2O2 and other ROS types like superoxide radicals is prone to misinterpretation due to the difficulties in measuring specific ROS [21, 61]. The outlines of these studies and their conclusions largely reflect the predominant view that has dominated the redox field in the last decades. Namely that adverse environmental conditions lead to oxidative stress which if counteracted by the antioxidant machinery would result in improved stress tolerance [42, 49, 62]. Thus, the common theme among the vast majority of studies reporting priming with H2O2 is enhanced tolerance to stress correlated with activation of the antioxidant machinery. Overexpression of various key antioxidant enzymes and boosting the levels of ascorbate and glutathione does not necessarily translate into enhanced tolerance to oxidative stress-promoting conditions suggesting that other mechanisms are likely to contribute to the beneficial effects of H2O2 priming. For example, accumulation of osmoprotectants (proline and soluble carbohydrates) correlated with improved tolerance to osmotic stress in H2O2 primed plants [63]. H2O2 pretreatment regulated ion uptake thus enhancing salinity tolerance [27, 64]. Reduced uptake of heavy metals and increased vacuolar sequestration was observed after H2O2 priming as well [65].
H2O2-induced posttranslational modifications
The signalling roles of H2O2 are especially crucial in establishing a primed state due to the profound effect H2O2 has on gene expression, hormonal pathways, developmental and defense responses [23]. H2O2-induced oxidation of Cys residues is recognized as an important molecular switch that can regulate numerous cellular processes and signalling pathways [66–68]. Cysteine residues with low pKa that reside in specialized protein environments and exist as thiolate anions can be selectively oxidized by H2O2 [69]. The first oxidation product of a cysteine thiol is sulfenic acid (-SOH) that is highly unstable and can be further oxidized to sulfinic (-SO2H) and sulfonic acid (-SO3H). Whereas, sulfinic and sulfonic acid are largely seen as irreversible oxidative modifications of damaged proteins, sulfenic acid formation is specific and reversible. Sulfenic acid can react with proximal thiols to form functionally important intramolecular and intermolecular disulfides. Alternatively, sulfenylated cysteine residues can form mixed disulfides with glutathione which is often regarded as a way to protect proteins from further oxidation [68, 70]. Cysteine oxidation can ultimately lead to functionally significant conformational changes that modulate protein function and interaction with protein partners and DNA. H2O2 can potentially oxidize thousands of cellular proteins, many of which have been identified in large-scale proteomics approaches [67, 71–73]. Nevertheless, a biological role has been attributed to only a fraction of these redox posttranslational modifications. It is very likely that not all protein oxidation events have a functional relevance, and the future challenge will lie in systematically analyzing their biological roles.
Impact of H2O2 on post-transcriptional regulation
Elevated H2O2 levels either as a result of exogenous chemical treatment, genetic perturbation, or adverse environmental conditions trigger extensive transcriptional reprogramming [74, 75]. Accumulating evidence supports the notion that oxidative stress can also regulate gene expression at the post-transcriptional level that includes mRNA cleavage, pre-mRNA splicing (alternative splicing; AS) and translation initiation. For instance, the Cu/Zn superoxide dismutase genes CSD1 and CSD2 in Arabidopsis are post-transcriptionally induced under oxidative stress. Under normal growth conditions, miR398 cleaves the CSD1 and CSD2 transcripts or causes their translational repression which is lifted upon oxidative stress-induced suppression of miR398 expression [76, 77]. Oxidative stress caused by methyl viologen treatment in the human neuroblastoma cells induces extensive changes in the AS [78]. Direct evidence on the impact of H2O2 in modifying AS landscape is currently not available in plants, however, oxidative stress-promoting conditions such as salinity stress and temperature variations are known to cause such modifications [79, 80]. Under salt stress, 10% of the total intron-containing genes show significantly differential AS and these genes are related to stress responses and RNA splicing [79]. For example, an E3-ligase encoding gene Salt-Responsive Alternatively Spliced gene 1 (SRAS1) in Arabidopsis has two splicing variants SRAS1.1 and SRAS1.2 with opposing functions. Under salinity, SRAS1.1 is accumulated in higher amount to support growth during salt stress by SRAS1.1-mediated degradation of COP9 signalosome 5A (CSN5A), an important regulator of plant development and growth [81]. Similarly, spliceosomal protein AtU1A controls alternative splicing of ACONITASE 1 (ACO1;[82]) under salt stress. ACO1 was proposed to affect the transcript levels of antioxidant enzyme CSD2 by binding to its 5’UTR [83]. Interestingly, mutant plants lacking AtU1A are sensitive to exogenous H2O2 suggesting that alternative splicing is implicated in maintenance of the cellular redox homeostasis [82]. While heat stress is known to widely repress pre-mRNA splicing across eukaryotes in general, thermopriming leads to derepression of splicing upon subsequent exposure to lethal temperatures [84]. The possibility of existence of such a ‘splicing memory’ in response to H2O2 priming cannot be denied. AS provides another layer of control in response to environmental perturbations, and the functional relevance of these differential AS is to preferentially accumulate the splice variants which participate in the stress-response pathways owing to their different biochemical and cellular properties.
Impact of H2O2 on translation
Gene expression regulation at the translational level allows quick acclimatory response without a need for de novo mRNA synthesis, splicing, and mRNA export from the nucleus. Translation in plants occurs in three distinct compartments i.e., cytoplasm, mitochondria, and chloroplasts. In this review, we focus on regulation of cytoplasmic translation catalyzed by eukaryotic type 80S ribosomes. The impact of oxidative stress on translation is likely to have a global regulatory effect on gene expression and is emerging as an important mechanism to modulate plant responses to adverse environmental conditions [85]. H2O2 can influence translation at multiple levels including the structure and posttranslational modifications of the ribosome, activation of specific signalling pathways, regulation of tRNA levels, and mRNA modification and compartmentalization.
Can ribosome heterogeneity play a role during oxidative stress?
Cytoplasmic ribosomes, apart from rRNAs, contain around 80 nuclear-encoded ribosomal proteins (RPs). The Arabidopsis ribosomal proteins genes have two to seven paralogs each and encode all together more than 230 RPs that could theoretically lead to 1034 different ribosome conformations [86]. The differential paralog usage can regulate ribosome composition. Ribosomes can also display heterogeneity due to posttranslational modifications of RPs, binding to ribosome-associated proteins, and variations in rRNA sequences [87]. Ribosome specialization can provide an additional layer of gene expression control that regulates the translation of specific mRNAs in a spatio-temporal manner [88]. Nevertheless, currently, it is not clear whether ribosome heterogeneity exists and has translational consequences in plant cells. The expression of paralogous ribosomal proteins genes under oxidative stress, however, is regulated at the transcriptional level suggesting that it can influence ribosome composition [86, 89]. Moreover, decrease of ribosome abundances and increase of average ribosome age were observed in Arabidopsis cells exposed to H2O2. [90]. Interestingly, the ribosomal protein RPS14C showed increased turnover rate and degraded more rapidly upon oxidative stress. Since RPS14C is relatively stable under control conditions, this might implicate its involvement in ribosome repair and/or increased susceptibility to H2O2 [90].
Can TOR kinase modulate translation in H2O2-dependent manner?
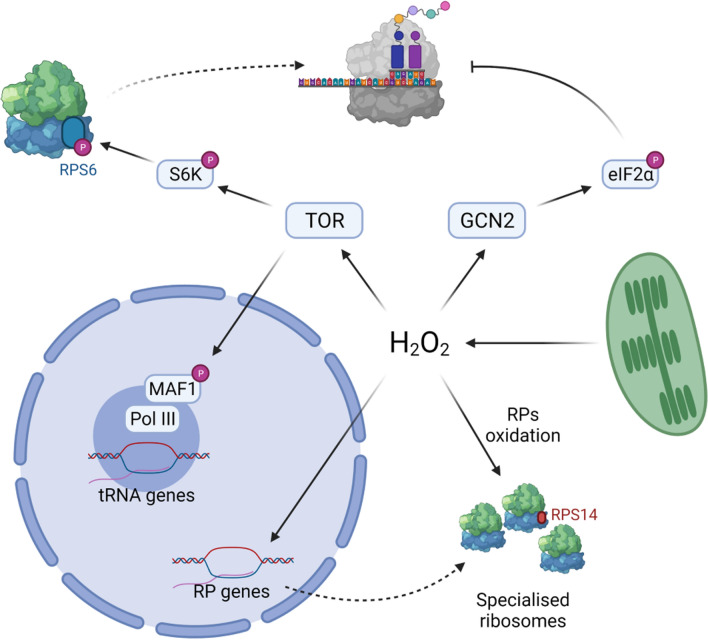
Post-translational modifications of RPs, such as phosphorylation, can introduce an additional source of ribosome heterogeneity. One of the best characterized signaling components affecting translation is target of rapamycin (TOR) kinase which regulates growth and development of eukaryotic organisms. Interestingly, TOR kinase plays a central role in plant responses to stress. TOR signalling regulates translation on multiple levels and promotes protein translation in plants [91]. For example, it activates 40S ribosomal protein kinase which phosphorylates ribosomal S6 protein (RPS6) in so-called TOR-RPS6 pathway, activates translation initiation factor eIF3h, and stimulates transcription of rRNAs and tRNAs through phosphorylation of RNA polymerase III repressor MAF1 (Fig. 2) [92].
Fig. 2.
Possible effects of H2O2 on the translational apparatus. H2O2 can influence gene expression through TOR and GCN2 signalling cascades by promoting or repressing translation and regulating tRNA transcription. H2O2 can also influence ribosome composition and redox posttranslational modifications on ribosomal proteins leading to the generation of a pool of specialized ribosomes. Created with BioRender.com
Holistic description of TOR signalling events in response to H2O2 in plants is still missing but recent works suggest that TOR pathway is activated by H2O2 and might influence translational output. TOR is likely activated by chloroplast-derived H2O2 [93]. Arabidopsis plants treated with methyl viologen, which leads to superoxide-mediated production of H2O2 in chloroplasts, has a higher activity of TOR-RPS6 pathway combined with increased expression of TOR complex genes [93]. In yeast, H2O2 causes widespread translational reprogramming [94] and likely similar mechanism can be observed in plants since excess light which promotes generation of chloroplast H2O2 significantly influences translational output in Arabidopsis [95]. Moreover, the biological significance of TOR–RPS6 pathway in H2O2 signalling was shown in mutant plants lacking components of TOR complex, as such they were more susceptible to oxidative stress and had altered levels of H2O2 [93]. These works suggest that a specific signalling pathway H2O2–TOR–RPS6 might exist in plants.
One of the roles of the TOR pathway is inhibition of MAF1 which acts as RNA polymerase III repressor. As such, TOR activates transcription of diverse tRNAs, 5S rRNA and small RNAs, and likely influences translational output. Inactivation of MAF1, in maf1 mutant, leads to hypersensitivity to H2O2 [96]. Thus, it is proposed that MAF1 regulates tRNA biogenesis in response to environmental cues and influences translation. However, it would be interesting to test whether such regulation is global or rather specific, and leads to the selective mRNA translation. In yeast, tRNAs’ abundance is regulated in response to H2O2 which results in selective translation [97]. It is not clear how expression of tRNAs is regulated but MAF1 can play a key role in this process.
tRNA-derived RNA fragments (tRFs) are products of tRNA cleavage and likely influence translation [98]. They were found in all eukaryotes and in plants, tRFs were identified for the first time in 2008 in response to H2O2 [99]. Notably, tRFs arise not only from cytosolic but also from organellar (i.e. chloroplastic and mitochondrial) tRNAs [100]. Organellar tRFs do not accumulate inside chloroplasts or mitochondria but rather are found outside these organelles suggesting that tRFs can play a regulatory role and/or can be involved in signalling.
Role of GCN2 kinase in translation response during oxidative stress
One of the best characterized pathways involved in translational regulation in all eukaryotes is phosphorylation of the translation initiation factor eIF2α by General Control Nonderepressible 2 (GCN2) kinase. In plants, GCN2 is activated by diverse conditions including abiotic stresses, pathogens, and photosynthetic electron transport-derived H2O2 [101]. Whereas in animals and yeast, activation of GCN2 leads to global repression of translation, it is not clear whether the same regulation exists in plants. In Arabidopsis, activation of the GCN2–eIF2α pathway by bacterial infection, promotes translational derepression of transcription factor TBF1 and expression of ABA-related genes [102]. In other work, activation of GCN2 led to significant inhibition of translation of many mRNAs. It is likely that this pathway specifically regulates translation because a pool of mRNAs (enriched for kinase and E3 ligase encoding genes) were not repressed after GCN2 activation [101]. These results together with the above-mentioned regulation of TOR activity suggest that chloroplast, might exert control over cytosol translation. Additionally, H2O2 can influence translation not only through changes of ribosome composition or posttranslational modifications of RPs but also via tRNA biogenesis and generation of tRFs.
Impact of H2O2 on RNA modification and compartmentalization
mRNA modifications influence translation, and recently their role in response to H2O2 has started to be elucidated. The importance of RNA methylation (i.e. incorporation of 5-methylcytosine, m5C) in response to oxidative stress was shown in Arabidopsis [103]. As such, plants lacking the methyltransferase TRM4B, crucial for m5C catalysis, were sensitive to H2O2. In addition, the trm4b mutant plants had reduced tRNA stability suggesting the role of m5C in translation.
Another example is a direct RNA oxidation by ·OH, being a result of H2O2 decay and leading to the formation of 8-hydroxyguanosine (8-oxo-G) [104]. Since the 8-oxo-G can pair with A and C, its presence in mRNA can cause the formation of aberrant proteins and ribosome stalling leading to the premature translation termination and accumulation of short polypeptides. Oxidation of the mRNA is widespread and it is estimated that on average at least one 8-oxo-G is present in each mRNA [105]. This process seems to be non-random since experiments on sunflower seeds showed that only selected mRNAs are oxidized [106]. Additionally, compartmentalization of oxidized mRNA seems to be crucial for RNA quality control. In unicellular alga, Chlamydomonas reinhardtii, 8-oxo-G was detected in pyrenoid (chloroplast CO2 assimilation micro-compartment found in most algae) where large subunit of RuBisCo was involved in the control of the oxidized mRNA levels [107]. Apart from mRNA, rRNA and tRNA can be also oxidized leading to the decrease of protein production rate and tRNA degradation. In higher plants, chloroplast is a key source of H2O2 during light stress. Thus, it is likely that RNA oxidation may influence RNA metabolism and translation in this organelle; however, such regulation has not been so far described.
Capped and polyadenylated mRNA in cytoplasm is generally ready for translation. However, it can be sequestered in membrane-less condensates such as stress granules (SGs) [108, 109]. SGs transiently form in response to diverse stresses leading to translation inhibition, and disassemble after the stress is over to release translation-competent mRNA. In mammalian cells, the SGs can form in response to H2O2 [110], however, so far in plants, SGs were only shown to accumulate in cytoplasm in response to hypoxia [111] and heat [112]. Although hypoxia and heat shock are related to the H2O2 accumulation and signalling in plants, it is not clear if SGs formation can be induced by H2O2. Intriguingly, SG-like condensates were also observed in Arabidopsis chloroplasts in response to heat shock [113] and in C. reinhardtii exposed to H2O2 [114]. Such granules in C. reinhardtii contained chloroplast encoded mRNAs, SGs marker proteins and large subunit of RuBisCo which might function in mRNA metabolism. These results suggest the existence of an elegant mechanism of translation regulation during oxidative stress.
Hydrogen peroxide in the nucleus
Nuclear sources of H2O2
In contrast to cellular compartments such as chloroplasts, mitochondria, and peroxisomes which are well explored with regards to H2O2 synthesis and signalling, our understanding of the redox homeostasis in the nucleus remains enigmatic. The majority of H2O2 detected in the nucleus likely diffuses from other cellular compartments like cytosol and chloroplasts through nuclear pores [115, 116]. Direct physical association between chloroplasts and nuclei facilitated by the formation of stromules has been suggested to be instrumental for H2O2 accumulation in the nucleus [117]. Intriguingly, exogenously applied H2O2 can stimulate stromule induction [118]. Elicitor-induced H2O2 accumulation visualized with 2′, 7′-dichlorofluorescein diacetate was observed in the nucleolus of tobacco BY-2 cells suggesting even more granular distribution of H2O2 at the subnuclear level [119]. Interestingly, the nucleolus harbours the rRNA biosynthetic machinery that can potentially be subjected to redox regulation. The elevated H2O2 levels in the nucleolus can be further activated to hydroxyl radicals in a reaction with ferrous ions which are abundant in this subnuclear compartment [120].
Active nuclear ROS production might also contribute to the overall ROS content in the nucleus since isolated nuclei can generate H2O2 upon treatment with Ca2+ [119]. Blue light-induced ROS synthesis mediated by cryptochromes represents another exciting mechanism that is involved in H202 production and signalling in the nucleus [121]. Manipulation of cryptochrome responses through blue light illumination has been proposed as a chemical-free approach to prime plants against abiotic stresses.
Different components of the antioxidant machinery have been found in the nucleus supporting the notion that the nuclear H2O2 homeostasis is actively regulated. The two major non-enzymatic antioxidants ascorbate and glutathione have been consistently detected in the nucleus [122, 123]. Moreover, several antioxidant enzymes that use GSH as a reductant, such as glutathione peroxidases (GPXs), glutaredoxins (GRXs), and glutathione S-transferase (GSTs) can also be targeted to the nucleus [124]. For example, GPX8 is partitioned between the cytosol and the nucleus [125]. Glutathione reductase, a component of the Halliwell-Asada-Foyer cycle that detoxifies H2O2, is also observed in the nucleus [126]. Several thioredoxins that are either exclusively localized in the nucleus or shuffle between the nucleus and the cytosol have been also characterized [127, 128]. Nuclear partitioning of antioxidant enzymes is often promoted by oxidative stress conditions further corroborating the idea that the nuclear redox homeostasis is dynamically fine-tuned [129].
Redox signalling to chromatin
Adverse environmental conditions trigger extensive transcriptional reprogramming that is partially regulated at the chromatin level which adds critical context for the activity of transcription factors. Chromatin features such as posttranslational histone modifications (acetylation, methylation, sumoylation, phosphorylation and ubiquitinoylation) and DNA methylation are dynamically altered by histone modifying enzymes and DNA (de)methyltransferases in response to abiotic and biotic stimuli [130]. The persistence of these epigenetic marks can vary widely and some of them are reverted back as soon as the stress episode is over while others can last throughout the whole life of the plant or even in the next generations [131–133]. Epigenetic mechanisms are likely at the core of stress priming and acclimation with specific histone marks such as H3K4 di- and tri-methylation implicated in the process [134]. Interestingly, epigenetic variations are attracting interest in plant breeding and represent a novel source of diversity in crop improvement [135].
Redox regulation of chromatin remodeling is emerging as an important mechanism to systematically control gene expression. Numerous studies have demonstrated that adverse environmental conditions linked to oxidative stress in plants trigger profound epigenetic changes [136]. Similarly, oxidative stress in animal systems associated with pathogenicity, such as cancer and cardiovascular disorders, impacts the epigenetic landscape [59, 137, 138].
Among the four main groups of epigenetic regulators, i.e. (1) histone modifying enzymes (methyltransferase, acetyltransferase, demethylase, deacetylase, kinase), (2) chromatin remodelers (CHD, SWI/SNF, INO80/SWR1 and IMITATION SWITCH (ISWI)), (3) DNA (de)-methylation enzymes (CHG/CG/CHH methyltransferase and demethylase), and (4) ncRNAs (miRNA, small-interfering RNA), histone modifying enzymes and DNA methyltransferases are most likely to be directly redox regulated according to the currently available evidence [139]. Such processes have been predominantly described in animal systems but are likely to be evolutionary conserved and occur in plants as well. Nevertheless, the identification and particularly, the functional characterization of specific oxidation events remain technically challenging due to their transient nature.
H2O2 treatment has been shown to recruit DNA methyltransferase 1 to specific genome regions and promote the formation and re-localization of protein complexes with other epigenetic modifiers [140]. The ROS-sensitive histone methyltransferases of the H3K4-trimethylating protein complex (COMPASS) in Caenorhabditis elegans deplete H3K4me3 marks upon transient ROS increase that ultimately enhances stress resistance and prolongs lifespan [141]. Some nuclear enzymes such as Repressor of Silencing1 and Demeter-like family members involved in removal of methylation marks from the DNA backbone are suspected to be redox sensitive due to the presence of Fe-S clusters [142]. Enzymes that use Fe2+ as a factor can be also potentially susceptible to oxidation. Among them, the activity of the demethylation enzymes from the TET and JmjC families which demethylate DNA and histones, respectively, might reflect changes in the cellular redox status [143].
Histones are highly decorated by PTMs which provides an additional layer of dynamic regulation for the chromatin structure. Histone marks can have a direct effect on the chromatin landscape and activity or influence epigenetic readers that target adaptor proteins and chromatin remodelling complexes to certain genome regions [144]. In cardiac muscle cells exposed to ROS-generating stimuli, the histone deacetylase HDAC4 forms an intramolecular disulfide bridge that promotes its nuclear exit [145]. Interestingly, NOX4-generated H2O2 was also able to oxidize HDAC4 in endothelial cells which resulted in increased HDAC4 phosphorylation and disturbance of the complex between HDAC4 and Mef2A, an important transcription factor involved in the activation of stress-induced genes [146]. The histone deacetylase activity of Arabidopsis HDACs can be inhibited by NO ultimately leading to genome-wide hyperacetylation of stress-induced genes [147]. Among them, HDA6 has been experimentally validated to be directly inhibited by NO [148]. Interestingly, its closest human ortholog, HDAC2, is also modified by NO, which has a direct effect on chromatin remodelling [149, 150]. Genome-wide distribution analysis of the histone acetylation mark H3K9ac which was unchanged in Arabidopsis hda6 mutants upon GSNO treatment, but accumulated in the wild type, further positioned HDA6 as an important player in the deacetylation of growth responsive genes [148].
Interestingly, mammalian histone H3 contains a redox-active conserved Cys residue which can be glutathionylated during cell proliferation and deglutathionylated during aging thus affecting nucleosome stability [151]. The highly reactive lipid oxidation products α, β-unsaturated aldehydes have been shown to commonly target histones in animal systems which could destine them for removal from the chromatin. Even though most α, β-unsaturated aldehydes would react with a wide range of cellular proteins, 4-oxo-2-nonenal (4-ONE) preferentially targets histones [152]. Intriguingly, adduct between 4-ONE and H3K27 was suggested as a redox-mediated histone mark that could stimulate transcription [153]. Because histones are the most abundant chromatin proteins, any changes that impact their structure and distribution are likely to have a global effect on gene expression and genome stability (Fig. 1). Whereas direct evidence for redox PTMs on plant histones is currently lacking, deposition of histone variants that are often decorated with distinct histone marks is crucial for establishment and reprogramming of plant chromatin landscapes [154].
Transcriptional expression of components involved in maintenance of ROS homeostasis might be equally subjected to regulation by dynamic changes in chromatin accessibility. The bread wheat histone acetyltransferase TaHAG1 directly targets three NADPH oxidases and facilitates their expression under salt stress by increased histone H3 acetylation [155]. Plants overexpressing TaHAG1 were more tolerant to salt stress which was modulated by H2O2 production. Salt tolerance was also associated with the ploidy level where hexaploid wheat was more tolerant than its tetraploid wheat progenitor. In animal cells, upregulation of transcription of the Nox4 gene was also associated with enrichment of activating acetylated histone marks, whereas silencing of the HAT that acetylates H4K16 negatively impacted Nox4 expression [156]. The relationship between histone acetylation and transcriptional activation of H2O2 producing enzymes might not be that straight forward since reports where the expression of Nox4 has been shown to be inversely correlated with histone acetylation also exist [157]. For example, treatment of smooth muscle cells with a range of HDAC inhibitors led to decreased levels of Nox4 expression [158]. Regulation of NADPH oxidases by histone methylation marks and DNA methylation have also been shown in animal models [156, 159] but currently, there is no experimental evidence for such regulation in plants.
Redox-regulated transcription factors
Apart from the genome-wide effect on gene expression that H2O2 might exert by modulating certain epigenetic regulators, numerous transcription factors can be governed by redox regulation causing either conformational changes that alter their association with DNA, protein interaction partners, or partitioning to the nucleus [160, 161]. The following examples are not meant to give a comprehensive view on this exciting area of redox signalling which has been excellently summarized previously [161, 162] but to illustrate the potential of H2O2 and redox homeostasis to orchestrate gene expression through its effect on transcription factors. Many TFs reside in the cytosol and are partitioned to the nucleus upon stress. A notable example is NONEXPRESSOR OF PATHOGENESIS-RELATED GENE 1 (NPR1), a master transcriptional regulator of pathogen-induced gene expression, that is retained in the cytosol as inactive oligomers maintained by intermolecular disulfide bonds involving Cys82 and Cys216 [163]. SA-induced perturbation of redox homeostasis triggers the thioredoxin H3/H5-dependent reduction of these disulfide bonds. The resulting monomeric NPR1 migrates to the nucleus and activates gene expression including that of PATHOGENESIS-RELATED (PR) genes [164].
The effect of heat stress is closely intertwined with H2O2 accumulation, and several heat shock transcription factors (HSFs) are activated by oxidation [165, 166]. The Arabidopsis HSFA8 is destined to the nucleus under H2O2 treatment, and its nuclear partitioning is dependent on two redox-sensitive Cys residues [167]. HSFA1A is similarly activated by H2O2 through trimerization which induces its binding to heat shock elements in target promoters [168].
H2O2 has a drastic impact on gene expression and thousands of induced or repressed transcripts have been identified in various model systems [75, 165]. Functioning alongside redox-sensitive TFs, numerous other TFs that recognize specific cis-regulatory DNA sequences have been implicated in the transcriptional response to H2O2 [162, 169]. Nevertheless, a gene regulatory network that provides a complete set of regulatory interactions between TFs and their target genes in the response to H2O2 is still missing. Systematically identifying TF-binding sites through yeast one-hybrid (Y1H) screens, chromatin immunoprecipitation (ChIP) experiments, and information about open chromatin profiling is instrumental in constructing gene regulatory networks, but the challenge is to extract information about functionally significant interactions. To address this, a network-based approach based on supervised learning for large-scale functional data integration was used to capture and validate regulators of ROS transcriptional regulation. This network covering 1,491 TFs and 31,393 target genes (1.7 million interactions) contained 124 ROS-related TFs that target core ROS responsive marker genes [170]. Five of them (WRKY15, WRKY28, ERF6, JAM1, and JUB1) have been previously reported to function in plant responses to H2O2 [171–177]. Moreover, newly predicted ROS regulators were experimentally validated using gain- or loss-of-function lines. Among them, Arabidopsis mutants lacking WRKY45 displayed enhanced sensitivity to 1 µM 3-amino triazole (3-AT), a catalase inhibitor used to increase endogenous H2O2 content, whereas WRKY45 overexpression resulted in improved performance. In contrast, transgenic lines overexpressing ERF115 displayed reduced growth and bleaching in comparison to the wild type when grown at 1.5 µM 3-AT [170]. Such integrative networks combining different experimental types are the next step in studying the systems biology of H2O2-induced stress acclimation.
Conclusions and perspectives
The versatile roles of H2O2 in plant growth, development, and stress responses reflect the enormous importance of redox chemistry in living systems. Myriad of cellular constituents can be oxidized by H2O2 potentially changing their physicochemical properties and functional roles. This seemingly indiscriminate impact of H2O2 and other ROS types on proteins, lipids, DNA, and small molecules, together with their accumulation under adverse environmental conditions have historically attracted a negative connotation and fueled a long line of research aimed to counteract the damaging effects of ROS and/or prevent their buildup. Ample evidence now demonstrates that H2O2 is much more than a harmful molecule and plants have integrated numerous H2O2-induced oxidation events in their signalling networks. H2O2 synthesis and signalling sustain plant growth and development not only under favourable conditions but also ensure mounting of timely and effective responses to abiotic and biotic stresses. Here, we discussed that H2O2 can impact key cellular processes like translation and chromatin remodelling which integrate various intracellular and extracellular cues. Adding these effects to the already wide repertoire of H2O2 target proteins and signalling networks impacted by H2O2 makes the mechanistic understanding of the systems biology of H2O2-induced stress acclimation even more challenging. The largely empirical effect of H2O2 priming is an excellent example that the regulatory roles of H2O2 can be harnessed without a complete knowledge of the underlying molecular mechanisms. Nevertheless, H2O2 priming has not found a place in agricultural practices not least because of the many variables associated with its application and unfavourable physicochemical properties. Chemicals that can produce H2O2 upon contact with plant tissues or trigger H2O2 synthesis in planta are viable alternatives to exploit the acclimation potential of H2O2 priming. Arguably, the most important implication of identifying crucial regulatory components mediating H2O2 signalling will be the engineering of climate resilient crops. Among the approaches that have the potential to dominate the field in the future are genome-editing technologies aimed at fine-tuning the signalling properties of key proteins by informed substitution of redox-sensitive Cys residues and/or modulation of their local environment. Epigenetic engineering of histone marks that are under redox control is another exciting avenue to capitalize on the rapidly growing information about redox signalling to chromatin.
Author contributions
PK and PG conceived and wrote the manuscript. MQ, SM, and SJ took part in writing the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Czech Science Foundation (Grant number 22-17092S to PK), Polish National Science Centre (SONATA12 UMO-2016/23/D/NZ3/02491 given to PG), and Ministry of Education, Youth and Sports of the Czech Republic (European Regional Development Fund-Project “Centre for Experimental Plant Biology” (Grant no. CZ.02.1.01/0.0/0.0/16_019/0000738 to SJ).
Availability of data and materials
Not applicable.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mills G, Sharps K, Simpson D, Pleijel H, Frei M, Burkey K, Emberson L, Uddling J, Broberg M, Feng Z, Kobayashi K, Agrawal M. Closing the global ozone yield gap: Quantification and cobenefits for multistress tolerance. Glob Chang Biol. 2018;24:4869–4893. doi: 10.1111/gcb.14381. [DOI] [PubMed] [Google Scholar]
- 2.Bailey-Serres J, Parker JE, Ainsworth EA, Oldroyd GED, Schroeder JI. Genetic strategies for improving crop yields. Nature. 2019;575:109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Sinha R, Fritschi FB, Zandalinas SI, Mittler R. The impact of stress combination on reproductive processes in crops. Plant Sci. 2021;311:111007. doi: 10.1016/j.plantsci.2021.111007. [DOI] [PubMed] [Google Scholar]
- 5.Rivero RM, Mittler R, Blumwald E, Zandalinas SI. Developing climate-resilient crops: improving plant tolerance to stress combination. Plant J. 2021 doi: 10.1111/tpj.15483. [DOI] [PubMed] [Google Scholar]
- 6.Walter J, Jentsch A, Beierkuhnlein C, Kreyling J. Ecological stress memory and cross stress tolerance in plants in the face of climate extremes. Environ Exp Bot. 2013;94:3–8. doi: 10.1016/J.ENVEXPBOT.2012.02.009. [DOI] [Google Scholar]
- 7.Godwin J, Farrona S. Plant Epigenetic Stress Memory Induced by Drought: A Physiological and Molecular Perspective. Methods Mol Biol. 2020;2093:243–259. doi: 10.1007/978-1-0716-0179-2_17. [DOI] [PubMed] [Google Scholar]
- 8.Serrano N, Ling Y, Bahieldin A, Mahfouz MM (2019) Thermopriming reprograms metabolic homeostasis to confer heat tolerance. Sci Rep 2019 9:1 9:1–14. 10.1038/s41598-018-36484-z [DOI] [PMC free article] [PubMed]
- 9.Crisp PA, Ganguly D, Eichten SR, et al. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv. 2016 doi: 10.1126/SCIADV.1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leuendorf JE, Frank M. Schmülling T (2020) Acclimation, priming and memory in the response of Arabidopsis thaliana seedlings to cold stress. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-019-56797-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiszniewska A, Muszyńska E, Kołton A, Kamińska I, Hanus-Fajerska E. In vitro acclimation to prolonged metallic stress is associated with modulation of antioxidant responses in a woody shrub Daphne jasminea. Plant Cell Tiss Organ Cult. 2019;139:339–357. doi: 10.1007/s11240-019-01688-2. [DOI] [Google Scholar]
- 12.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Mittler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 15.Gómez R, Vicino P, Carrillo N, Lodeyro AF. Manipulation of oxidative stress responses as a strategy to generate stress-tolerant crops. From damage to signaling to tolerance. Crit Rev Biotechnol. 2019;39:693–708. doi: 10.1080/07388551.2019.1597829. [DOI] [PubMed] [Google Scholar]
- 16.Kerchev PI, Van Breusegem F. Improving oxidative stress resilience in plants. Plant J. 2021 doi: 10.1111/tpj.15493. [DOI] [PubMed] [Google Scholar]
- 17.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 19.Mhamdi A, Van Breusegem F. Reactive oxygen species in plant development. Development. 2018;145:164376. doi: 10.1242/dev.164376. [DOI] [PubMed] [Google Scholar]
- 20.Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- 21.Dikalov SI, Harrison DG. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid Redox Signal. 2014;20:372–382. doi: 10.1089/ars.2012.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noctor G, Mhamdi A, Foyer CH. Oxidative stress and antioxidative systems: recipes for successful data collection and interpretation. Plant Cell Environ. 2016;39:1140–1160. doi: 10.1111/pce.12726. [DOI] [PubMed] [Google Scholar]
- 23.Petrov VD, Van Breusegem F. Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants. 2012 doi: 10.1093/aobpla/pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foyer CH. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ Exp Bot. 2018;154:134–142. doi: 10.1016/j.envexpbot.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang J, Li JL, Ma XR. Exogenous hydrogen peroxide enhanced the thermo tolerance of Festuca arundinacea and Lolium perenne by increasing the antioxidative capacity. Acta Physiol Plant. 2014;36:2915–2924. doi: 10.1007/s11738-014-1661-2. [DOI] [Google Scholar]
- 27.Ashfaque F, Iqbal M, Khan R, Khan NA. Exogenously Applied H2O2 promotes proline accumulation, water relations, photosynthetic efficiency and growth of wheat (Triticum aestivum L.) under salt stress. Annu Res Rev Biol. 2014;4:105–120. doi: 10.9734/ARRB/2014/5629. [DOI] [Google Scholar]
- 28.Phua SY, De Smet B, Remacle C, Chan KX, Van Breusegem F. Reactive oxygen species and organellar signaling. J Exp Bot. 2021;72:5807–5824. doi: 10.1093/jxb/erab218. [DOI] [PubMed] [Google Scholar]
- 29.Foyer CH. How plant cells sense the outside world through hydrogen peroxide. Nature. 2020;578:518–519. doi: 10.1038/d41586-020-00403-y. [DOI] [PubMed] [Google Scholar]
- 30.Aranega-Bou P, de la O Leyva M, Finiti I, García-Agustín P, González-Bosch C (2014) Priming of plant resistance by natural compounds Hexanoic acid as a model. Front Plant Sci 5:488. 10.3389/fpls.2014.00488 [DOI] [PMC free article] [PubMed]
- 31.Balmer A, Pastor V, Gamir J, Flors V, Mauch-Mani B. The ‘prime-ome’: Towards a holistic approach to priming. Trends Plant Sci. 2015;20:443–452. doi: 10.1016/j.tplants.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Vincent C, Rowland D, Schaffer B, Bassil E, Racette K, Zurweller B. Primed acclimation: A physiological process offers a strategy for more resilient and irrigation-efficient crop production. Plant Sci. 2020;295:110240. doi: 10.1016/j.plantsci.2019.110240. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Zhou M, Zhou H, et al. Hydrogen sulfide, a signaling molecule in plant stress responses. J Integr Plant Biol. 2021;63:146–160. doi: 10.1111/JIPB.13022. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Ji F, Zhang Y, et al. Interactions between hydrogen sulphide and nitric oxide regulate two soybean citrate transporters during the alleviation of aluminium toxicity. Plant Cell Environ. 2019;42:2340–2356. doi: 10.1111/PCE.13555. [DOI] [PubMed] [Google Scholar]
- 35.Ishibashi Y, Yamaguchi H, Yuasa T, Iwaya-Inoue M, Arima S, Zheng SH. Hydrogen peroxide spraying alleviates drought stress in soyabean plants. J Plant Physiol. 2011;168:1562–1567. doi: 10.1016/j.jplph.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Conrath U, Beckers GJ, Flors V, García-Agustín P, Jakab G, Mauch F, Newman MA, Pieterse CM, Poinssot B, Pozo MJ, Pugin A, Schaffrath U, Ton J, Wendehenne D, Zimmerli L, Mauch-Mani B. Priming: getting ready for battle. Mol Plant Microbe Interact. 2006;19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- 37.Vaidya AS, Peterson FC, Eckhardt J, Xing Z, Park SY, Dejonghe W, Takeuchi J, Pri-Tal O, Faria J, Elzinga D, Volkman BF, Todoroki Y, Mosquna A, Okamoto M, Cutler SR. Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo. Proc Natl Acad Sci USA. 2021;118:e2108281118. doi: 10.1073/pnas.2108281118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerchev P, van der Meer T, Sujeeth N, Verlee A, Stevens CV, Van Breusegem F, Gechev T. Molecular priming as an approach to induce tolerance against abiotic and oxidative stresses in crop plants. Biotechnol Adv. 2020;40:107503. doi: 10.1146/10.1016/j.biotechadv.2019.107503. [DOI] [PubMed] [Google Scholar]
- 39.Gondim FA, Gomes-Filho E, Costa JH, Alencar NLM, Priso JT. CAT plays a key role in salt stress acclimation induced by hydrogen peroxide pretreatment in maize. J Plant Physiol Biochem. 2012;56:62–71. doi: 10.1016/j.plaphy.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 40.Habib N, Ali Q, Ali S, Javed MT, Zulqurnain Haider M, Perveen R, Shahid MR, Rizwan M, Abdel-Daim MM, Elkelish A, Bin-Jumah M. Use of nitric oxide and hydrogen peroxide for better yield of wheat (Triticum aestivum L.) under water deficit conditions: growth, osmoregulation, and antioxidative defense mechanism. Plants. 2020;9:285. doi: 10.3390/plants9020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silva PCC, Azevedo Neto ADD, Gheyi HJ, Ribas RF, Silva CRDR, Cova AMD. Salt tolerance induced by hydrogen peroxide priming on seed is related to improvement of ion homeostasis and antioxidative defense in sunflower plants. J Plant Nutr. 2021;44:1207–2121. doi: 10.1080/01904167.2020.1862202. [DOI] [Google Scholar]
- 42.de Azevedo Neto AD, Prisco JT, Enéas-Filho J, Medeiros JV, Gomes-Filho E. Hydrogen peroxide pre-treatment induces salt-stress acclimation in maize plants. J Plant Physiol. 2005;162:1114–1122. doi: 10.1016/j.jplph.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Wahid A, Perveen M, Gelani S, Basra SM. Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J Plant Physiol. 2007;164:283–294. doi: 10.1016/j.jplph.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Xu FJ, Jin CW, Liu WJ, Zhang YS, Lin XY. Pretreatment with H2O2 alleviates aluminum-induced oxidative stress in wheat seedlings. J Integr Plant Biol. 2010;53:44–53. doi: 10.1111/j.1744-7909.2010.01008.x. [DOI] [PubMed] [Google Scholar]
- 45.Dos Santos AG, de Oliveira P-M, de Paiva Pinheiro SK, de Castro ME, de Sousa LL, Camelo Marques E, de Carvalho HH, Gomes-Filho E. H2O2 priming promotes salt tolerance in maize by protecting chloroplasts ultrastructure and primary metabolites modulation. Plant Sci. 2021;303:110774. doi: 10.1016/j.plantsci.2020.110774. [DOI] [PubMed] [Google Scholar]
- 46.Claeys H, Van Landeghem S, Dubois M, Maleux K, Inzé D. What Is Stress? Dose-Response Effects in Commonly Used in Vitro Stress Assays. Plant Physiol. 2014;165:519–527. doi: 10.1104/pp.113.234641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gechev T, Gadjev I, Van Breusegem F, Inzé D, Dukiandjiev S, Toneva V, Minkov I. Hydrogen peroxide protects tobacco from oxidative stress by inducing a set of antioxidant enzymes. Cell Mol Life Sci. 2002;59:708–714. doi: 10.1007/s00018-002-8459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu CW, Murphy TM, Lin CH. Hydrogen peroxide-induced chilling tolerance in mung beans mediated through ABA-independent glutathione accumulation. Funct Plant Biol. 2003;30:955–963. doi: 10.1071/FP03091. [DOI] [PubMed] [Google Scholar]
- 49.İşeri ÖD, Körpe DA, Sahin FI, Haberal M. Hydrogen peroxide pretreatment of roots enhanced oxidative stress response of tomato under cold stress. Acta Physiol Plant. 2013;35:1905–1913. doi: 10.1007/s11738-013-1228-7. [DOI] [Google Scholar]
- 50.Uchida A, Jagendorf AT, Hibino T, Takabe T, Takabe T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002;163:515–523. doi: 10.1016/S0168-9452(02)00159-0. [DOI] [Google Scholar]
- 51.Mubarakshina Borisova MM, Kozuleva MA, Rudenko NN, Naydov IA, Klenina IB, Ivanov BN. Photosynthetic electron flow to oxygen and diffusion of hydrogen peroxide through the chloroplast envelope via aquaporins. Biochim Biophys Acta. 2012;1817:1314–1321. doi: 10.1016/j.bbabio.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 52.Del Río LA, López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol. 2016;57:1364–1376. doi: 10.1093/pcp/pcw076. [DOI] [PubMed] [Google Scholar]
- 53.Segal AW. NADPH oxidases as electrochemical generators to produce ion fluxes and turgor in fungi, plants and humans. Open Biol. 2016;6:160028. doi: 10.1098/rsob.160028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal. 2009 doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 55.Marino D, Dunand C, Puppo A, Pauly N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012;17:9–15. doi: 10.1016/j.tplants.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, Chen S, Zhou JM. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014;15:329–338. doi: 10.1016/j.chom.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 58.Gong M, Chen B, Li ZG, Guo LH. Heat-shock-induced cross adaptation to heat, chilling, drought and salt stress in maize seedlings and involvement of H2O2. J Plant Physiol. 2001;158:112–1130. doi: 10.1078/0176-1617-00327. [DOI] [Google Scholar]
- 59.Wu F, Chi Y, Jiang Z, et al. Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature. 2020;578:577–581. doi: 10.1038/s41586-020-2032-3. [DOI] [PubMed] [Google Scholar]
- 60.Gilroy S, Białasek M, Suzuki N, Górecka M, Devireddy Amith R, Karpinski S, Mittler R. ROS, calcium and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 2016;171:1606–1615. doi: 10.1104/pp.16.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winterbourn CC. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochem Biophys Acta. 2014;1840:730–738. doi: 10.1016/j.bbagen.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Li J, Wang J, Li Z. Exogenous H2O2 improves the chilling tolerance of manilagrass and mascarenegrass by activating the antioxidative system. Plant Growth Regul. 2010;61:195–204. doi: 10.1007/s10725-010-9470-0. [DOI] [Google Scholar]
- 63.Marthandan V, Geetha R, Kumutha K, Renganathan VG, Karthikeyan A, Ramalingam J. Seed Priming: A feasible strategy to enhance drought tolerance in crop plants. Int J Mol Sci. 2020;21:8258. doi: 10.3390/ijms21218258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu T, Chen K, Hu L, Amombo E, Fu J. H2O2 and Ca2+-based signaling and associated ion accumulation, antioxidant systems and secondary metabolism orchestrate the response to NaCl stress in perennial ryegrass. Sci Rep. 2016;6:36396. doi: 10.1038/srep36396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y, Ge Y, Zhang C, Ju T, Cheng W. Cadmium toxicity and translocation in rice seedlings are reduced by hydrogen peroxide pretreatment. Plant Growth Regul. 2009;59:51–61. doi: 10.1007/s10725-009-9387-7. [DOI] [Google Scholar]
- 66.Couturier J, Chibani K, Jacquot JP, Rouhier N. Cysteine-based redox regulation and signaling in plants. Front Plant Sci. 2013;4:105. doi: 10.3389/fpls.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacques S, Ghesquière B, De Bock PJ, Demol H, Wahni K, Willems P, Messens J, Van Breusegem F, Gevaert K. Protein methionine sulfoxide dynamics in Arabidopsis thaliana under oxidative stress. Mol Cell Proteomics. 2015;14:1217–1229. doi: 10.1074/mcp.M114.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang J, Willems P, Van Breusegem F, Messens J. Pathways crossing mammalian and plant sulfenomic landscapes. Free Radic Biol Med. 2018;122:193–201. doi: 10.1016/j.freeradbiomed.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Roos G, Foloppe N, Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 70.Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 71.Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Van Molle I, De Smet B, Vertommen D, Gevaert K, De Jaeger G, Van Montagu M, Messens J, Van Breusegem F. Sulfenome mining in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2014;111:11545–11550. doi: 10.1073/pnas.1411607111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, Sun R, Liu K, Yang J, Carroll KS. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat Chem Biol. 2018;14:995–1004. doi: 10.1038/s41589-018-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Smet B, Willems P, Fernandez-Fernandez AD, Alseekh S, Fernie AR, Messens J, Van Breusegem F. In vivo detection of protein cysteine sulfenylation in plastids. Plant J. 2019;97:765–778. doi: 10.1111/tpj.14146. [DOI] [PubMed] [Google Scholar]
- 74.Gadjev I, Vanderauwera S, Gechev TS, et al. Transcriptomic Footprints Disclose Specificity of Reactive Oxygen Species Signaling in Arabidopsis. Plant Physiol. 2006;141:436–445. doi: 10.1104/PP.106.078717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willems P, Mhamdi A, Stael S, Storme V, Kerchev P, Noctor G, Gevaert K, Van Breusegem F. The ROS wheel: Refining ROS transcriptional footprints. Plant Physiol. 2016;171:1720–1733. doi: 10.1104/pp.16.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beauclair L, Yu A, Bouché N. microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 2010;62:454–462. doi: 10.1111/J.1365-313X.2010.04162.X. [DOI] [PubMed] [Google Scholar]
- 77.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional Induction of Two Cu/Zn Superoxide Dismutase Genes in Arabidopsis Is Mediated by Downregulation of miR398 and Important for Oxidative Stress Tolerance. Plant Cell. 2006;18:2051. doi: 10.1105/TPC.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vivarelli S, Lenzken SC, Ruepp MD, et al. Paraquat Modulates Alternative Pre-mRNA Splicing by Modifying the Intracellular Distribution of SRPK2. PLoS ONE. 2013;8:e61980. doi: 10.1371/JOURNAL.PONE.0061980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding F, Cui P, Wang Z, et al. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014;15:1–14. doi: 10.1186/1471-2164-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John S, Olas JJ, Mueller-Roeber B. Regulation of alternative splicing in response to temperature variation in plants. J Exp Bot. 2021;72:6150–6163. doi: 10.1093/JXB/ERAB232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Y, Li XH, Guo QH, et al. Salt responsive alternative splicing of a RING finger E3 ligase modulates the salt stress tolerance by fine-tuning the balance of COP9 signalosome subunit 5A. PLoS Genet. 2021 doi: 10.1371/JOURNAL.PGEN.1009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu J, Xia Z, Luo Y, et al. Spliceosomal protein U1A is involved in alternative splicing and salt stress tolerance in Arabidopsis thaliana. Nucleic Acids Res. 2018;46:1777. doi: 10.1093/NAR/GKX1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moeder W, del Pozo O, Navarre DA, et al. Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana. Plant Mol Biol. 2007;63:273–287. doi: 10.1007/S11103-006-9087-X. [DOI] [PubMed] [Google Scholar]
- 84.Ling Y, Serrano N, Gao G, et al. Thermopriming triggers splicing memory in Arabidopsis. J Exp Bot. 2018;69:2659–2675. doi: 10.1093/JXB/ERY062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Urquidi Camacho RA, Lokdarshi A, von Arnim AG. Translational gene regulation in plants: A green new deal. Wiley Interdiscip Rev RNA. 2020 doi: 10.1002/WRNA.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martinez-Seidel F, Beine-Golovchuk O, Hsieh YC, Kopka J. Systematic review of plant ribosome heterogeneity and specialization. Front Plant Sci. 2020;11:948. doi: 10.3389/fpls.2020.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Genuth NR, Barna M. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat Rev Genet. 2018;19:431–452. doi: 10.1038/s41576-018-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Lan P, Gao H, Zheng L, Li W, Schmidt W. Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics. 2013;14:783. doi: 10.1186/1471-2164-14-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salih KJ, Duncan O, Li L, O'Leary B, Fenske R, Trösch J, Millar AH. Impact of oxidative stress on the function, abundance, and turnover of the Arabidopsis 80S cytosolic ribosome. Plant J. 2020;103:128–139. doi: 10.1111/tpj.14713. [DOI] [PubMed] [Google Scholar]
- 91.Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH. TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc Natl Acad Sci USA. 2018;115:12823–12828. doi: 10.1073/pnas.1809526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bakshi A, Moin M, Madhav MS, Kirti PB. Target of rapamycin, a master regulator of multiple signalling pathways and a potential candidate gene for crop improvement. Plant Biol (Stuttg) 2019;21:190–205. doi: 10.1111/plb.12935. [DOI] [PubMed] [Google Scholar]
- 93.Pereyra CM, Aznar NR, Rodriguez MS, Salerno GL, Martínez-Noël GMA. Target of rapamycin signaling is tightly and differently regulated in the plant response under distinct abiotic stresses. Planta. 2019;251:21. doi: 10.1007/s00425-019-03305-0. [DOI] [PubMed] [Google Scholar]
- 94.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moore M, Smith A, Wesemann C, Schmidtpott S, Wegener M, Farooq MA, Seidel T, Pogson B, Dietz K-J. Retrograde control of cytosolic translation targets synthesis of plastid localized proteins and nuclear responses for efficient light acclimation. bioRxiv. 2021 doi: 10.1101/2021.02.18.431817. [DOI] [Google Scholar]
- 96.Ahn CS, Lee DH, Pai HS. Characterization of Maf1 in Arabidopsis: function under stress conditions and regulation by the TOR signaling pathway. Planta. 2019;249:527–542. doi: 10.1007/s00425-018-3024-5. [DOI] [PubMed] [Google Scholar]
- 97.Torrent M, Chalancon G, de Groot NS, Wuster A, Madan Babu M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci Signal. 2018 doi: 10.1126/scisignal.aat6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lalande S, Merret R, Salinas-Giegé T, Drouard L. Arabidopsis tRNA-derived fragments as potential modulators of translation. RNA Biol. 2020;17:1137–1148. doi: 10.1080/15476286.2020.1722514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cognat V, Morelle G, Megel C, Lalande S, Molinier J, Vincent T, Small I, Duchêne AM, Maréchal-Drouard L. The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 2020;48:8812–8813. doi: 10.1093/nar/gkaa651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lokdarshi A, Guan J, Urquidi Camacho RA, Cho SK, Morgan PW, Leonard M, Shimono M, Day B, von Arnim AG. Light activates the translational regulatory kinase GCN2 via reactive oxygen species emanating from the chloroplast. Plant Cell. 2020;32:1161–1178. doi: 10.1105/tpc.19.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu X, Afrin T, Pajerowska-Mukhtar KM. Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun Biol. 2019;2:302. doi: 10.1038/s42003-019-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell. 2017;29:445–460. doi: 10.1105/tpc.16.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giorgio M, Dellino GI, Gambino V, Roda N, Pelicci PG. On the epigenetic role of guanosine oxidation. Redox Biol. 2020;29:101398. doi: 10.1016/j.redox.2019.101398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 106.Bazin J, Langlade N, Vincourt P, Arribat S, Balzergue S, El-Maarouf-Bouteau H, Bailly C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell. 2011;23:2196–2208. doi: 10.1105/tpc.111.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhan Y, Dhaliwal JS, Adjibade P, Uniacke J, Mazroui R, Zerges W. Localized control of oxidized RNA. J Cell Sci. 2015;128:4210–4219. doi: 10.1242/jcs.175232. [DOI] [PubMed] [Google Scholar]
- 108.Chantarachot T, Bailey-Serres J. Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 2018;176:254–269. doi: 10.1104/pp.17.01468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maruri-López I, Figueroa NE, Hernández-Sánchez IE, Chodasiewicz M. Plant Stress Granules: Trends and Beyond. Front Plant Sci. 2021;12:722643. doi: 10.3389/fpls.2021.722643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emara MM, Fujimura K, Sciaranghella D, Ivanova V, Ivanov P, Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem Biophys Res Commun. 2012;423:763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lokdarshi A, Conner WC, McClintock C, Li T, Roberts DM. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol. 2016;170:1046–1059. doi: 10.1104/pp.15.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nguyen CC, Nakaminami K, Matsui A, Kobayashi S, Kurihara Y, Toyooka K, Tanaka M, Seki M. Oligouridylate binding protein 1b plays an integral role in plant heat stress tolerance. Front Plant Sci. 2016;7:853. doi: 10.3389/fpls.2016.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chodasiewicz M, Sokolowska EM, Nelson-Dittrich AC, Masiuk A, Beltran JCM, Nelson ADL, Skirycz A. Identification and characterization of the heat-induced plastidial stress granules reveal new insight into Arabidopsis stress response. Front Plant Sci. 2020;11:595792. doi: 10.3389/fpls.2020.595792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Uniacke J, Zerges W. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 2008;182:641–646. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Møller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- 116.Rodrigues O, Reshetnyak G, Grondin A, Saijo Y, Leonhardt N, Maurel C, Verdoucq L. Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc Natl Acad Sci USA. 2017;114:9200–9205. doi: 10.1073/pnas.1704754114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat Commun. 2017;8:49. doi: 10.1038/s41467-017-00074-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Caplan JL, Kumar AS, Park E, Padmanabhan MS, Hoban K, Modla S, Czymmek K, Dinesh-Kumar SP. Chloroplast stromules function during innate immunity. Dev Cell. 2015;34:45–57. doi: 10.1016/j.devcel.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R. Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco bright yellow-2 cells. Plant Physiol. 2007;143:1817–1826. doi: 10.1104/pp.106.090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roschzttardtz H, Séguéla-Arnaud M, Briat JF, Vert G, Curie C. The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell. 2011;23:2725–2737. doi: 10.1105/tpc.111.088088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.El-Esawi M, Arthaut LD, Jourdan N, d’Harlingue A, Link J, Martino CF, Ahmad M. Blue-light induced biosynthesis of ROS contributes to the signaling mechanism of Arabidopsis cryptochrome. Sci Rep. 2017;7:13875. doi: 10.1038/s41598-017-13832-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vivancos PD, Dong Y, Ziegler K, Markovic J, Pallardó FV, Pellny TK, Verrier PJ, Foyer CH. Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 2010;64:825–838. doi: 10.1111/j.1365-313X.2010.04371.x. [DOI] [PubMed] [Google Scholar]
- 123.Zechmann B, Stumpe M, Mauch F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta. 2011;233:1–12. doi: 10.1007/s00425-010-1275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Go YM, Jones DP. Redox control systems in the nucleus: mechanisms and functions. Antioxid Redox Signal. 2010;13:489–509. doi: 10.1089/ars.2009.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gaber A, Ogata T, Maruta T, Yoshimura K, Tamoi M, Shigeoka S. The Involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012;53:1596–1606. doi: 10.1093/pcp/pcs100. [DOI] [PubMed] [Google Scholar]
- 126.Delorme-Hinoux V, Bangash SA, Meyer AJ, Reichheld JP. Nuclear thiol redox systems in plants. Plant Sci. 2016;243:84–95. doi: 10.1016/j.plantsci.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 127.Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ. Characterization of two thioredoxin h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol. 2001;46:361–371. doi: 10.1023/a:1010697331184. [DOI] [PubMed] [Google Scholar]
- 128.Ying Y, Yue W, Wang S, Li S, Wang M, Zhao Y, Wang C, Mao C, Whelan J, Shou H. Two h-Type Thioredoxins Interact with the E2 Ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in Rice. Plant Physiol. 2017;173:812–824. doi: 10.1104/pp.16.01639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Serrato AJ, Cejudo FJ. Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta. 2003;217:392–399. doi: 10.1007/s00425-003-1009-4. [DOI] [PubMed] [Google Scholar]
- 130.Kim J-H. Multifaceted chromatin structure and transcription changes in plant stress response. Int J Mol Sci. 2021;22:2013. doi: 10.3390/ijms22042013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014;55:1859–1863. doi: 10.1093/pcp/pcu125. [DOI] [PubMed] [Google Scholar]
- 132.Bilichak A, Kovalchuk I. Transgenerational response to stress in plants and its application for breeding. J Exp Bot. 2016;67:2081–2092. doi: 10.1093/jxb/erw066. [DOI] [PubMed] [Google Scholar]
- 133.Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci Adv. 2016;2:e1501340. doi: 10.1126/sciadv.1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sani E, Herzyk P, Perrella G, Colot V, Amtmann A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013;14:r59. doi: 10.1186/gb-2013-14-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singh D, Chaudhary P, Taunk J, Singh CK, Sharma S, Singh VJ, Singh D, Chinnusamy V, Yadav R, Pal M. Plant epigenomics for extenuation of abiotic stresses: Challenges and future perspectives. J Exp Bot. 2021 doi: 10.1093/jxb/erab337. [DOI] [PubMed] [Google Scholar]
- 136.Sudan J, Raina M, Singh R. Plant epigenetic mechanisms: role in abiotic stress and their generational heritability. 3 Biotech. 2018;8:172. doi: 10.1007/s13205-018-1202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Matsuda M, Shimomura I. Increased oxidative stress in obesity: implications for metabolic syndrome, diabetes, hypertension, dyslipidemia, atherosclerosis, and cancer. Obes Res Clin Pract. 2013;7:330–341. doi: 10.1016/j.orcp.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 138.Dimauro I, Paronetto MP, Caporossi D. Exercise, redox homeostasis and the epigenetic landscape. Redox Biol. 2020;35:101477. doi: 10.1016/j.redox.2020.10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rm SK, Wang Y, Zhang X, et al. Redox Components: Key Regulators of Epigenetic Modifications in Plants. Int J Mol Sci. 2020 doi: 10.3390/IJMS21041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bazopoulou D, Knoefler D, Zheng Y, et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature. 2019;576:301–305. doi: 10.1038/s41586-019-1814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Buzas DM. Emerging links between iron-sulfur clusters and 5-methylcytosine base excision repair in plants. Genes Genet Syst. 2016;91:51–62. doi: 10.1266/ggs.16-00015. [DOI] [PubMed] [Google Scholar]
- 143.Lamadema N, Burr S, Brewer AC. Dynamic regulation of epigenetic demethylation by oxygen availability and cellular redox. Free Radic Biol Med. 2019;131:282–298. doi: 10.1016/j.freeradbiomed.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 144.Lindermayr C, Rudolf EE, Durner J, Groth M. Interactions between metabolism and chromatin in plant models. Mol Metab. 2020;38:100951. doi: 10.1016/j.molmet.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 146.Schader T, Löwe O, Reschke C, Malacarne P, Hahner F, Müller N, Gajos-Draus A, Backs J, Schröder K. Oxidation of HDAC4 by Nox4-derived H2O2 maintains tube formation by endothelial cells. Redox Biol. 2020;36:101669. doi: 10.1016/j.redox.2020.101669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mengel A, Ageeva A, Georgii E, Bernhardt J, Wu K, Durner J, Lindermayr C. Nitric Oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 2017;173:1434–1452. doi: 10.1104/pp.16.01734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ageeva-Kieferle A, Georgii E, Winkler B, Ghirardo A, Albert A, Hüther P, Mengel A, Becker C, Schnitzler JP, Durner J, Lindermayr C. Nitric oxide coordinates growth, development, and stress response via histone modification and gene expression. Plant Physiol. 2021;187:336–360. doi: 10.1093/plphys/kiab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 150.Nott A, Nitarska J, Veenvliet JV, Schacke S, Derijck AA, Sirko P, Muchardt C, Pasterkamp RJ, Smidt MP, Riccio A. S-nitrosylation of HDAC2 regulates the expression of the chromatin-remodeling factor Brm during radial neuron migration. Proc Natl Acad Sci USA. 2013;110:3113–3118. doi: 10.1073/pnas.1218126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kreuz S, Fischle W. Oxidative stress signaling to chromatin in health and disease. Epigenomics. 2016;8:843–862. doi: 10.2217/epi-2016-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Galligan JJ, Marnett LJ. Histone Adduction and Its Functional Impact on Epigenetics. Chem Res Toxicol. 2017;30:376–387. doi: 10.1021/acs.chemrestox.6b00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, Marnett LJ. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J Am Chem Soc. 2014;136:11864–11866. doi: 10.1021/ja503604t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Borg M, Berger F. Chromatin remodelling during male gametophyte development. Plant J. 2015;83:177–188. doi: 10.1111/tpj.12856. [DOI] [PubMed] [Google Scholar]
- 155.Zheng M, Lin J, Liu X, Chu W, Li J, Gao Y, An K, Song W, Xin M, Yao Y, Peng H, Ni Z, Sun Q, Hu Z. Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 2021;186:1951–1969. doi: 10.1093/plphys/kiab187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sanders YY, Liu H, Liu G, Thannickal VJ. Epigenetic mechanisms regulate NADPH oxidase-4 expression in cellular senescence. Free Radic Biol Med. 2015;79:197–205. doi: 10.1016/j.freeradbiomed.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 157.Brewer AC. Physiological interrelationships between NADPH oxidases and chromatin remodelling. Free Radic Biol Med. 2021;170:109–115. doi: 10.1016/j.freeradbiomed.2021.01.052. [DOI] [PubMed] [Google Scholar]
- 158.Zelko IN, Folz RJ. Regulation of Oxidative Stress in Pulmonary Artery Endothelium. Modulation of Extracellular Superoxide Dismutase and NOX4 Expression Using Histone Deacetylase Class I Inhibitors. Am J Respir Cell Mol Biol. 2015;53:513–524. doi: 10.1165/rcmb.2014-0260OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Manea SA, Constantin A, Manda G, Sasson S, Manea A. Regulation of Nox enzymes expression in vascular pathophysiology: Focusing on transcription factors and epigenetic mechanisms. Redox Biol. 2015;5:358–366. doi: 10.1016/j.redox.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.He H, Van Breusegem F, Mhamdi A. Redox-dependent control of nuclear transcription in plants. J Exp Bot. 2018;69:3359–3372. doi: 10.1093/jxb/ery130. [DOI] [PubMed] [Google Scholar]
- 161.Dietz KJ. Redox regulation of transcription factors in plant stress acclimation and development. Antioxid Redox Signal. 2014;21:1356–1372. doi: 10.1089/ars.2013.5672. [DOI] [PubMed] [Google Scholar]
- 162.Mou Z, Fan W, Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 163.Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hieno A, Naznin HA, Inaba-Hasegawa K, Yokogawa T, et al. Transcriptome analysis and identification of a transcriptional regulatory network in the response to H2O2. Plant Physiol. 2019;180:1629–1646. doi: 10.1104/pp.18.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Babbar R, Karpinska B, Grover A, Foyer CH. Heat-Induced Oxidation of the Nuclei and Cytosol. Front Plant Sci. 2021;11:617779. doi: 10.3389/fpls.2020.617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Giesguth M, Sahm A, Simon S, Dietz KJ. Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 2015;589:718–725. doi: 10.1016/j.febslet.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 167.Liu Y, Zhang C, Chen J, Guo L, Li X, Li W, Yu Z, Deng J, Zhang P, Zhang K, Zhang L. Arabidopsis heat shock factor HsfA1a directly senses heat stress, pH changes, and hydrogen peroxide via the engagement of redox state. Plant Physiol Biochem. 2013;64:92–98. doi: 10.1016/j.plaphy.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 168.Petrov V, Vermeirssen V, De Clercq I, Van Breusegem F, Minkov I, Vandepoele K, Gechev TS. Identification of cis-regulatory elements specific for different types of reactive oxygen species in Arabidopsis thaliana. Gene. 2012;499:52–60. doi: 10.1016/j.gene.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 169.De Clercq I, Van de Velde J, Luo X, Liu L, Storme V, Van Bel M, Pottie R, Vaneechoutte D, Van Breusegem F, Vandepoele K. Integrative inference of transcriptional networks in Arabidopsis yields novel ROS signalling regulators. Nat Plants. 2021;7:500–513. doi: 10.1038/s41477-021-00894-1. [DOI] [PubMed] [Google Scholar]
- 170.Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE. 2012;7:e35995. doi: 10.1371/journal.pone.0035995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013;22:327–341. doi: 10.1007/s11248-012-9645-8. [DOI] [PubMed] [Google Scholar]
- 172.Sewelam N, Kazan K, Thomas-Hall SR, Kidd BN, Manners JM, Schenk PM. Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE. 2013;8:e70289. doi: 10.1371/journal.pone.0070289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dubois M, Skirycz A, Claeys H, Maleux K, Dhondt S, De Bodt S, Vanden Bossche R, De Milde L, Yoshizumi T, Matsui M, Inzé D. Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 2013;162:319–332. doi: 10.1104/pp.113.216341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shahnejat-Bushehri S, Nobmann B, Devi Allu A, Balazadeh S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal Behav. 2016;11:e1181245. doi: 10.1080/15592324.2016.1181245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chen X, Liu J, Lin G, Wang A, Wang Z, Lu G. Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 2013;32:1589–1599. doi: 10.1007/s00299-013-1469-3. [DOI] [PubMed] [Google Scholar]
- 176.Ebrahimian-Motlagh S, Ribone PA, Thirumalaikumar VP, Allu AD, Chan RL, Mueller-Roeber B, Balazadeh S. JUNGBRUNNEN1 Confers Drought Tolerance Downstream of the HD-Zip I Transcription Factor AtHB13. Front Plant Sci. 2017;8:2118. doi: 10.3389/fpls.2017.02118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vanderauwera S, Vandenbroucke K, Inzé A, van de Cotte B, Mühlenbock P, De Rycke R, Naouar N, Van Gaever T, Van Montagu MC, Van Breusegem F. AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:20113–20118. doi: 10.1073/pnas.1217516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.