ABSTRACT
The recent emergence of a SARS-CoV-2 saltation variant, BA.2.87.1, which features 65 spike mutations relative to BA.2, has attracted worldwide attention. In this study, we elucidate the antigenic characteristics and immune evasion capability of BA.2.87.1. Our findings reveal that BA.2.87.1 is more susceptible to XBB-induced humoral immunity compared to JN.1. Notably, BA.2.87.1 lacks critical escaping mutations in the receptor binding domain (RBD) thus allowing various classes of neutralizing antibodies (NAbs) that were escaped by XBB or BA.2.86 subvariants to neutralize BA.2.87.1, although the deletions in the N-terminal domain (NTD), specifically 15-23del and 136-146del, compensate for the resistance to humoral immunity. Interestingly, several neutralizing antibody drugs have been found to restore their efficacy against BA.2.87.1, including SA58, REGN-10933 and COV2-2196. Hence, our results suggest that BA.2.87.1 may not become widespread until it acquires multiple RBD mutations to achieve sufficient immune evasion comparable to that of JN.1.
KEYWORDS: SARS-CoV-2, neutralizing antibodies, humoral immunity, immune escape, saltation variant
Dear editor,
Recently, BA.2.87.1 gained wide attention for its expansion with striking number of Spike mutations [1,2]. This novel saltation variant harbours 65 mutations on the Spike glycoprotein, notably including two unique extensive NTD deletions 15-23del and 136–146del, and RBD substitutions such as K417 T, K444N, V445G, L452M, and N481 K (Figure 1A). With eight sequences originating from South Africa in the last quarter of 2023 and one travel-related sequence in USA, it was officially designated in February 2024 and has then been tracked by USA CDC (Centers for Disease Control and Prevention). Some of the prior saltation variants of SARS-CoV-2 have demonstrated profound capability of evading humoral immunity induced by vaccinations and infections due to the substantial mutations and antigenic difference, with the most notable examples Omicron BA.1, BA.2.75 and the recent JN.1 [3–5] (Figure 1B). Consequently, it is imperative to rapidly assess BA.2.87.1 potential increases in immune resistance.
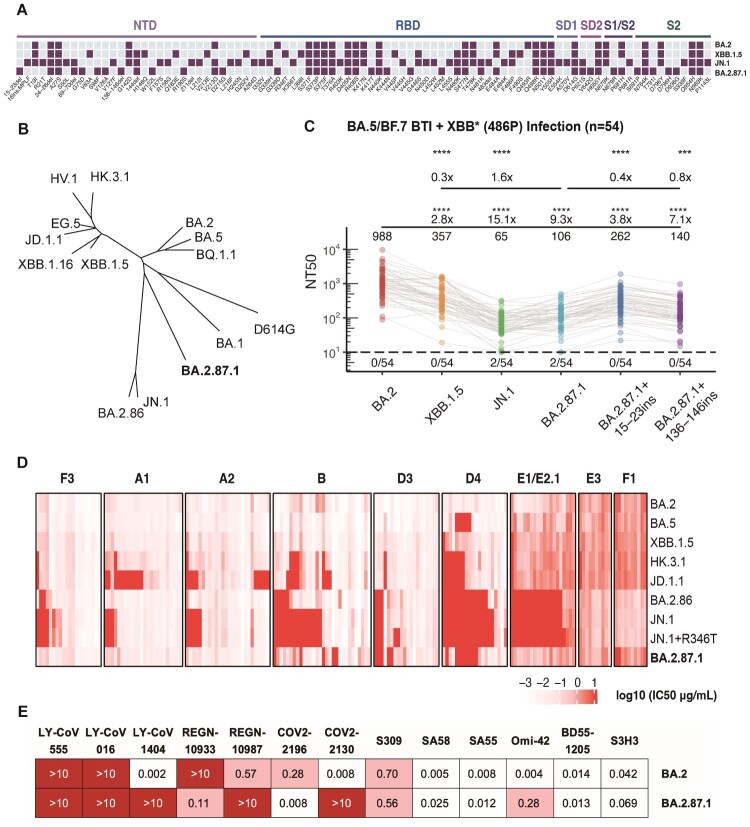
Figure 1.
Spike mutations, phylogeny and neutralization resistance of BA.2.87.1. (A) Mutations on the Spike glycoprotein of BA.2, XBB.1.5, JN.1, and BA.2.87.1. Purple square indicates the presence of the mutations in each variant, while sky blue square indicates the absence of the mutations. The domains of the mutations on the Spike protein are annotated above. (B) Unrooted phylogenetic tree of the Spike glycoprotein of prevalent SARS-CoV-2 variants. (C) The 50% neutralizing titres (NT50) of convalescent plasma from individuals who experienced breakthrough infections with BA.5 or BF.7 followed by reinfection with XBB* + 486P subvariants (n = 54) against SARS-CoV-2 variants. Geometric mean titres, relative fold changes, and statistical significance are annotated above each group. The dashed line represents the limit of detection (50% neutralizing titre = 10), and the number of negative samples is indicated below it. Paired samples were analyzed by two-tailed Wilcoxon signed-rank test. ***p < 0.001, ****p < 0.0001. (D) The 50% inhibitory concentration (IC50, μg/mL) of a panel of BA.2-effective monoclonal neutralizing antibodies targeting distinct RBD epitopes (determined by DMS) against BA.2, BA.5, XBB.1.5, HK.3.1, JD.1.1, BA.2.86, JN.1, JN.1 + R346 T, and BA.2.87.1 mutants. Epitope groups are annotated above. The intensity of red represents the magnitude of the IC50 values. (E) IC50 (μg/mL) of approved or candidate monoclonal neutralizing antibody drugs targeting the RBD or SD1 regions of the Spike protein, against BA.2 and BA.2.87.1 pseudoviruses.
First, to investigate BA.2.87.1's potential for humoral immune escape, vesicular stomatitis virus (VSV)-based pseudovirus neutralization assays were performed on plasma samples obtained from participants (n = 54) who underwent BA.5/BF.7 breakthrough infection (BTI) after three doses of ancestral-strain inactivated vaccination, and were then re-infected with XBB* + 486P (Figure 1C and Table S1). The results suggested that pseudovirus bearing the BA.2.87.1 Spike protein demonstrated superior evasion to XBB.1.5, indicated by a 70% decrease in 50% neutralizing titres (NT50). However, it exhibited lower ability to evade humoral immunity induced by BA.5/BF.7 BTI and XBB reinfection when compared to JN.1, with a 1.6-fold higher NT50. To evaluate the impact of the two long NTD deletions on plasma immune escape, two NTD segments of BA.2 (the precursor of BA.2.87.1) on the corresponding residues 15–23 (CVNLITRTQ) and 136–146 (CNDPFLDVYYH) were inserted into BA.2.87.1 pseudovirus and tested for plasma neutralization. Both the reversion of 15–23del and 136–146del significantly dampen the antibody evasion of BA.2.87.1, with 2.5-fold and 1.3-fold increase in NT50, respectively. In summary, neutralization experiments with human plasma revealed that NTD deletion profoundly contributed to the plasma escape of BA.2.87.1, allowing it to exhibit higher immune evasion capability than BA.2 and XBB.1.5. However, its overall immune evasion strength was not comparable to that of JN.1.
Subsequently, we evaluated the neutralizing potency of a collection of BA.2-effective monoclonal antibodies (mAbs) derived from repetitive Omicron infections that target various epitopes of RBD, as defined in our previous studies, to further interrogate BA.2.87.1’s immune escape capability and mechanism (Figure 1D) [6]. Most of the mAbs involved remain reactive to XBB.1.5 as well. We found that BA.2.87.1 could be easily neutralized by most of the antibodies in the A1, A2, B, and F3 epitope groups (Class 1, Class 2, and Class 1/4 antibodies), which directly compete with the binding of host angiotensin-converting enzyme 2 (ACE2) to realize potent inhibition [4]. In contrast, the previously and recently circulating evasive variants, including HK.3.1, JD.1.1, and JN.1, could efficiently escape these antibodies due to their escape mutations on the Spike receptor-binding motif (RBM). Specifically, the “FLip” mutation (L455F + F456L) of HK.3.1 improved its ability to evade A1 and A2 antibodies, and the additional A475 V mutation carried by JD.1.1 further enhanced the evasion [7]. Furthermore, the lack of F486P but the achievement of N481 K mutation of BA.2.87.1 compared to XBB.1.5 lineages demonstrated its relatively weak yet distinct escape pattern against Group B antibodies. Without the R346 T and K356 T mutations, BA.2.87.1 could not efficiently evade Class 3 antibodies (Group D3, D4, E1/E2.1) [8]. In addition, JN.1 + R346 T showed slightly enhanced resistance to antibodies from Class 3. These observations suggest that BA.2.87.1, despite being antigenically distinct, typically demonstrates less resistance to various classes of RBD-targeting antibodies compared to JN.1.
As for the therapeutic antibodies, SA55 maintained consistent effectiveness against BA.2.87.1 (Figure 1E) [9]. REGN-10933 and COV2-2196, which target the B epitope, regained their neutralizing capacity against BA.2.87.1, likely due to the absence of mutation on F486, which was usually mutated to Val, Ser, or Pro in existing BA.5, XBB.1.5, and BA.2.86 subvariants [10,11]. Similarly, the SA58 antibody also restored its potency, attributable to the lack of G339H and R346 T mutations [9]. Moreover, BA.2.87.1 was also unable to resist the SD1-targeting S3H3 without the E554 K mutation which was carried by the BA.2.86 lineages [12]. Nevertheless, due to the K444N and L452M mutations, BA.2.87.1 exhibited robust resistance to Group D1/D2 antibodies, such as REGN-10987, LY-CoV1404, and COV2-2130 [10,11,13]. As for Class 1 broad-spectrum antibodies, Omi-42 exhibited substantially compromised neutralization, while BD55-1205, which belonged to the public IGHV3-53/3-66 NAb group, maintained its neutralization potency against all tested variants [9,14].
Discussion
Consistent with the recent research, our findings highlight that the NTD deletions, particularly the 15–23del, significantly contribute to the evasion of BA.2.87.1. Such long stretches of NTD deletions are frequently observed in persistent evolution, underscoring the need to monitor the intra-host persistent evolution of SARS-CoV-2. We also observed a partial recovery of therapeutic antibodies. However, it is highlighted that BA.2.87.1 demonstrates weaker immune resistance to the currently dominant variant JN.1, which are in line with the findings of other researchers [15–18]. Our results also emphasized that BA.2.87.1 cannot counteract Group A and B antibodies without mutations such as L455S, L455F, F456L, A475V, and F486P, which are present in JN.1 and JD.1.1. Furthermore, in the absence of mutation at position R346 or K356, it exhibits weak capability of escape NAbs in the E1/E2.1 epitope group. These characteristics collectively suggest that BA.2.87.1 is not likely to outcompete current JN.1 and XBB subvariants without substantial further antigenic drift on the RBD, given the current population-level immunity established by the repeated vaccinations and infections. Although BA.2.87.1 is relatively weak in terms of antigenicity, its other virological characteristics and potential of accumulating additional RBD mutations during regional circulation should be closely evaluated and monitored.
Supplementary Material
Acknowledgments
We extend our gratitude to scientists in the community for their persistent monitoring of SARS-CoV-2 variants and valuable discussion. We thank all volunteers who contributed blood samples for this research.
Funding Statement
This work was supported by the Ministry of Science and Technology of China (2023YFC3043200), Changping Laboratory (2021A0201; 2021D0102) and the National Natural Science Foundation of China (32222030, 2023011477).
Disclosure statement
Y.C. is the inventor of the provisional patent applications for BD series antibodies, which includes BD55-5514 (SA55), BD55-5840 (SA58) and BD55-1205. Y.C. is the founder of Singlomics Biopharmaceuticals. Other authors declare no competing interests.
References
- 1.Wang X, Jiang S, Ma W, et al. Robust neutralization of SARS-CoV-2 variants including JN.1 and BA.2.87.1 by trivalent XBB vaccine-induced antibodies. bioRxiv. 2024. doi: 10.1101/2024.02.16.580615 [DOI]
- 2.Zhang L, Dopfer-Jablonka A, Nehlmeier I, et al. Virological traits of the SARS-CoV-2 BA.2.87.1 lineage. bioRxiv; 2024.
- 3.Cao Y, Song W, Wang L, et al. . Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30:1527–1539.e5. doi: 10.1016/j.chom.2022.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang S, Yu Y, Xu Y, et al. . Fast evolution of SARS-CoV-2 BA.2·86 to JN.1 under heavy immune pressure. Lancet Infect Dis. 2023;24:e70–e72. doi: 10.1016/S1473-3099(23)00744-2 [DOI] [PubMed] [Google Scholar]
- 5.Yang S, Yu Y, Jian F, et al. . Antigenicity and infectivity characterisation of SARS-CoV-2 BA.2.86. Lancet Infect Dis. 2023;23:e457–e459. doi: 10.1016/S1473-3099(23)00573-X [DOI] [PubMed] [Google Scholar]
- 6.Yisimayi A, Song W, Wang J, et al. . Repeated Omicron exposures override ancestral SARS-CoV-2 immune imprinting. Nature. 2024;625:148–156. doi: 10.1038/s41586-023-06753-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jian F, Feng L, Yang S, et al. . Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455–456 synergistically enhances antibody evasion and ACE2 binding. Wan X-F, editor. PLoS Pathog. 2023;19:e1011868. doi: 10.1371/journal.ppat.1011868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jian F, Yu Y, Song W, et al. . Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect Dis. 2022;22:1535–1537. doi: 10.1016/S1473-3099(22)00642-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y, Jian F, Zhang Z, et al. . Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. Cell Rep. 2022;41:111845. doi: 10.1016/j.celrep.2022.111845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Copin R, Baum A, Wloga E, et al. . The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184:3949–3961.e11. doi: 10.1016/j.cell.2021.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zost SJ, Gilchuk P, Case JB, et al. . Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Wang Y, Wang Y, et al. . Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein. Emerg Microbes Infect. 2022;11:351–367. doi: 10.1080/22221751.2021.2024455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westendorf K, Žentelis S, Wang L, et al. . LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39:110812. doi: 10.1016/j.celrep.2022.110812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nutalai R, Zhou D, Tuekprakhon A, et al. . Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell. 2022;185:2116–2131.e18. doi: 10.1016/j.cell.2022.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, et al. SARS-CoV-2 omicron BA.2.87.1 exhibits higher susceptibility to serum neutralization than EG.5.1 and JN.1. bioRxiv; 2024.
- 16.Li P, et al. Distinct patterns of SARS-CoV-2 BA.2.87.1 and JN.1 variants in immune evasion, antigenicity and cell-cell fusion. bioRxiv; 2024. [DOI] [PMC free article] [PubMed]
- 17.Lasrado N, Rössler A, Rowe M, et al. . Neutralization of SARS-CoV-2 omicron subvariant BA.2.87.1. Vaccine. 2024;42:2117–2121. doi: 10.1016/j.vaccine.2024.03.007 [DOI] [PubMed] [Google Scholar]
- 18.Sheward DJ, Marking U, Bladh O, et al. Neutralisation sensitivity of the SARS-CoV-2 BA. 2.87. 1 variant. bioRxiv; 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.