Abstract
In-situ vaccination (ISV) utilizing nanoparticles (NPs) and therapeutic devices like focused ultrasound (FUS) can trigger immune-mediated killing of both treated and untreated cancer cells. However, the impact of confounding factors such as aging and gut microbiota composition on therapeutic outcomes remains poorly understood. In this study, we sequentially treated young mice (~8 weeks) and old mice (>18 months) with bilateral melanoma using FUS and calreticulin nanoparticles (CRT-NP) to enhance immunogenic cell death. The combination of CRT-NP and FUS (CFUS) demonstrated greater efficacy in inducing regression of treated and untreated tumors in young mice compared to old mice. The diminished effectiveness in older mice was associated with significant differences in gut microbiome composition, characterized by alterations in bacterial species and splenic immune cells. Specifically, young mice exposed to CFUS exhibited higher abundance of Bacteroidetes and Verrucomicrobia, which was not observed in the aged cohorts. Turicibacter, Anaerotruncus, and Ruminiclostridium demonstrated negative correlations with CD8+ T cells but positive correlations with CD4+ T cells and MDSC cells in both age groups. Taxon set enrichment analysis revealed 58 significantly enriched host gene targets in the young cluster compared to only 11 in the aged cluster. These findings highlight the relationship between ISV treatment efficacy and gut microbiome composition, suggesting that interventions such as diet modification, probiotics, or fecal microbiota transplantation may hold potential as therapeutic strategies to enhance immune responses against solid tumors.
Keywords: Microbiome, Aging, Nanoparticle, Focused ultrasound, Immunotherapy
1. Introduction
Microbiome represents a complex and dynamic ecosystem comprised of diverse microorganisms that inhabit various regions of the body (Kennedy and Chang, 2020). In particular, the gut microbiome plays a crucial role in modulating the immune system through several mechanisms, including the production of short-chain fatty acids (SCFAs) and the regulation of cytokines, chemokines, and other signaling molecules (Kim, 2021; Maldonado Galdeano et al., 2019). As individuals age, composition of the gut microbiome undergoes significant changes, often resulting in reduced microbial diversity compared to younger individuals (Badal et al., 2020). These age-related changes in microbial diversity can be attributed to shifts in the relative abundance of different bacterial species. For instance, the proportion of beneficial bacteria, such as Bifidobacterium and Lactobacillus, tends to decrease with age, while potentially harmful bacteria, such as Proteobacteria, may become more prevalent (Arboleya et al., 2016; Saraswati and Sitaraman, 2014). Alterations in the gut microbiome have been shown to significantly impact the immune system and the response to immunotherapy (Lu et al., 2022). Thus, understanding the implications of age-related changes in the gut microbiome is essential for tailoring immunotherapies and improving treatment outcomes for patients of all ages. In this study, we aimed to elucidate the therapeutic implications of combinatorial immunotherapy regimens involving the use of focused ultrasound (FUS) and immunogenic cell death (ICD)-enhancing calreticulin nanoparticles (CRT-NPs) in the gut microbiota of young and aged mice bearing melanoma tumors.
Combinatorial immunotherapy represents a cutting-edge approach in cancer treatment that incorporates multiple therapeutic agents or strategies either simultaneously or sequentially to target diverse aspects of tumor biology (Al-Lazikani et al., 2012). The primary objective of combinatorial immunotherapy is to synergize antitumor immunity while overcoming limitations associated with single-agent therapies, such as immune resistance and tumor heterogeneity (Gotwals et al., 2017). At present, >50 combinatorial protocols are being investigated in cancer clinical trials, showcasing the growing interest and potential of this approach in the field of oncology (Tan et al., 2021). FUS is a non-invasive therapeutic technique that utilizes high-intensity focused sound waves to target and treat tumors using thermal and mechanical effects (Fernando et al., 2013). Calreticulin (CRT) is a damage-associated molecular pattern (DAMP) protein that, when present on the cell surface, can promote the uptake of dying cancer cells by antigen-presenting cells (APCs) (Gorbet and Ranjan, 2020; Gorbet et al., 2020). Our CRT-NP is designed as an in-situ vaccine (ISV), and delivered directly to a tumor, rather than being injected or otherwise administered elsewhere in the body. We have previously demonstrated that locally combining focused ultrasound with CRT-NP (CRT-NP + FUS: CFUS), as well as other immunomodulators, improves anti-tumor immunity against poorly immunogenic B16F10 melanoma in a syngeneic murine model (Sethuraman et al., 2020; Singh et al., 2019).
Although age-related decline in immune function can potentially influence the response to specific immunotherapies, it remains unclear whether combinatorial ISVs such as CFUS applied directly to a tumor induce comparable or distinct changes in the gut microbiota in an age-dependent or independent manner. Understanding the relationship between age, gut microbiota composition, and the response to combinatorial therapies, can provide insights into the potential implications for immunotherapy outcomes and contribute to the development of more effective, personalized cancer treatments. This knowledge may help optimize immunotherapy regimens and improve overall treatment success for cancer patients across different age groups. Our data reveal that CFUS can modulate the gut microbiota in both young and aged mice, generating crucial therapeutic signatures that may influence the effectiveness of immunotherapy. This thereby underscores the importance of considering age-related microbiome changes when designing and evaluating combinatorial cancer therapies, as these factors could significantly impact patient responses and overall treatment success.
2. Materials and methods
2.1. Materials
DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) (890890) and cholesterol (700000) lipids were purchased from Avanti Polar Lipids, Inc., Alabaster, AL, USA. Nanoplasmid (NTC 9385R-MCS) containing the CRT gene was purchased from Aldevron (Lincoln, NE, USA). Chloroform (C2432) was purchased from Sigma-Aldrich, St. Louis, MO, USA. HEPES, sterile 1 M buffer (J848), DMEM (11965092), Fetal bovine serum, FBS (10082147), Penicillin-Streptomycin - PenStrep (15140122), PBS (10010023), Collagenase IV (17104019), were purchased from ThermoFisher/Gibco, Waltham, MA, USA. The following Fluorochrome-conjugated monoclonal antibodies (mAbs) for flow cytometry were purchased from BioLegend, San Diego, CA, USA and were listed: APC-Cy7 anti-CD45 (103115), PerCP anti-CD3 epsilon (100325), PE-Cy5 anti-CD4 (100410), PE-Cy7 anti-CD8a (100721), PE anti-Granzyme B (372208), BV510 anti-INFg (505841), BV510 anti-Ly-6G (127633), PE anti-Ly-6C (128007), BV650 anti-CD11b (101239), APC anti-CD206 (141707), BV421 anti-CD11c (117329). AF488 anti-iNOS (53-5920-82) was procured from eBiosciences/ThermoFisher, Waltham, MA, USA. Liberase (5401119001) was procured from Life Technologies, NY, USA. Transcription factor buffer set (562574) was purchased from BD Biosciences, San Jose, CA, USA. B16F10 was provided by Dr. Mary Jo Turk, Dartmouth. Aged C57BL/6 mice were obtained from NIA, Bethesda, MD, USA.
2.2. Study design and CFUS treatment protocols
To evaluate the effects of CFUS treatment across a range of human-equivalent ages, we enrolled young (6–8 weeks) and old (>18 months) C57BL/6 female mice (Quinn et al., 2018; Leins et al., 2018). In brief, B16F10 cells cultured in DMEM containing 10 % v/v fetal bovine serum (FBS) and 1 % v/v streptomycin/penicillin were harvested at 80–90 % confluency. Cells were then rinsed and diluted in sterile, cold PBS to create a dosage of 0.125 × 10^6 cells per mouse per 100 μl injection volume, which was injected into both the right and left flank regions of the mice using a 27-gauge needle (BD, Franklin Lakes, NJ, USA) to induce bilateral tumors. Mice tumor volume was measured daily by serial caliper measurements (General Tools Fraction™, New York, NY, USA) using the formula (length × width2)/2, where length was the largest dimension and width was the smallest dimension perpendicular to the length. When tumors were palpable (day 7 post-inoculation), young and aged mice were randomized into two groups (n = 8 mice/group): 1) Control and 2) CFUS. CFUS was administered by giving three CRT-NP intratumoral injections (20 μg DNA per injection) and two histotripsy treatment using an Alpinion transducer (Singh et al., 2021) with parameters: Duty cycle 1 %, Power 600 W, 5 Pulse repetition frequency, time per focal point 20 s. We aligned one of the bilateral tumors at a fixed focal depth, covering a voxel size of 1 × 1 × 10 mm, which included <10 % of the tumor volume homogenization. CRT-NP was synthesized following our previously published protocol (Sethuraman et al., 2020). In brief, we cloned the full-length DNA of human CRT into an NTC vector (Aldevron Inc. Nebraska, USA) and loaded it into cationic liposomes. We characterized the particles for size, zeta potential, and polydispersity index. On day 20 post inoculation, we sacrificed the mice for microbiome and spleen immune cell analysis (Fig. 1A schematic).
Fig. 1.
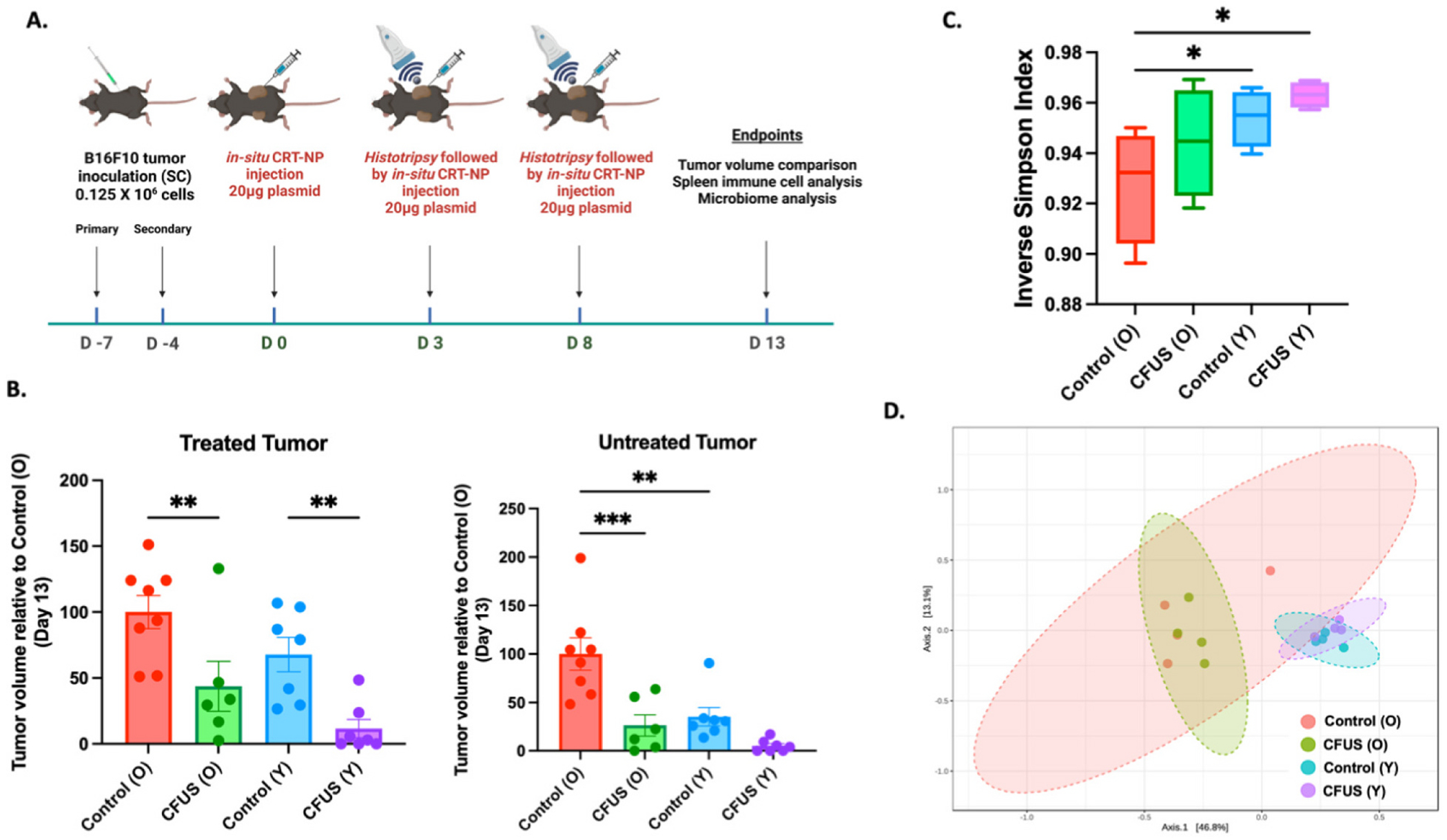
Local treatment of B16F10 melanoma tumors with CFUS reduced tumor burden and altered the gut microbiome in mice. A) Schematic of study design, B) Aged control mice showed faster tumor growth rates than young mice. When treated with CFUS, both young and aged mice exhibited a reduction in tumor volume, with young mice showing a near complete remission than old mice at treated and contralateral untreated (abscopal) locations. C) The alpha diversity of the gut microbiome was assessed using the Simpson index via 16S sequencing of fecal samples collected from young and aged CFUS treated and control groups and plotted as inverse Simpson diversity, D) 3-dimensional principal coordinate analysis (PCoA) plot created using the matrix of paired-wise distance between aged and young samples calculated by the Bray-Curtis dissimilarity using unique amplicon sequence variants (ASV). Statistical analysis was done using one-way ANOVA with Fisher’s LSD test, *p < 0.05, **p < 0.005, ***p < 0.0005.
2.3. Microbiome analysis of fecal samples
Fecal samples were processed and analyzed with the ZymoBIOMICS® Service (Zymo Research, Irvine, CA, USA). Briefly, DNA was extracted using the ZymoBIOMICS®-96 MagBead DNA Kit (Zymo Research, Irvine, CA, USA), and prepared for targeted sequencing with the Quick-16S™ NGS Library prep kit (Zymo Research, Irvine, CA, USA) from n = 4 randomly selected samples per group. The primer sets used in this project were Quick-16S™ Primer Set V3-V4 (Zymo Research, Irvine, CA, USA). The final PCR products were quantified using qPCR fluorescence readings and pooled together based on equal molarity, were cleaned with the Select-a-Size DNA Clean & Concentrator™ (Zymo Research, Irvine, CA, USA), and then quantified with TapeStation® (Agilent Technologies, Santa Clara, CA, USA) and Qubit® (Thermo Fisher Scientific, Waltham, WA, USA). The ZymoBIOMICS® Microbial community DNA standard (Zymo Research, Irvine, CA, USA) was used as a positive control for each targeted library preparation. To assess the level of bioburden introduced by the wet-lab process, negative controls, such as blank extraction and blank library preparation controls were included. The final library was sequenced on an Illumina® MiSeq™ with a v3 reagent kit (600 cycles). The sequencing was performed with 10 % PhiX spike-in. Unique amplicon sequences were inferred from raw reads using the Dada2 pipeline. Chimeric sequences were also removed with the Dada2 pipeline. Taxonomy assignment was done using Uclust from Qiime v. 1.9.1 and assigned taxonomy with the Zymo Research Database, an internally designed and curated 16S database, as a reference. Taxonomy assignment was performed using Uclust from Qiime v. 1.9.1 and assigned with the Zymo Research Database, a 16S database that was internally designed and curated, as a reference.
Marker gene data analysis was done using MicrobiomeAnalyst web tool with Qiime v. 1.9.1 for community profiling and comparative analysis. Data were filtered to remove low count (<20 %) and low variance (<10 %). Data were scaled and normalized at the total sum scale. Differences in taxonomy abundance were analyzed using Linear Discriminant Analysis Effect size (LEfSe) employing the Kruskal-Wallis rank sum test. Euclidean distance was used for community clustering with the Ward algorithm. Clustered taxa were analyzed using Taxon Set Enrichment Analysis (TSEA) available in MicrobiomeAnalyst. For Pattern search, Pearson r correlation analysis was done among different treatment groups at the genus level. Graphs and heatmaps were generated by MicrobiomeAnalyst internal scripts. For gene enrichment analysis Metascape web tool was used.
2.4. Flow cytometry analysis of spleen
Spleens were collected and minced, filtered through a 70 μm cell strainer (Corning Inc., Corning, NY, USA), and incubated in RBC lysis buffer for 10 min in dark. Cells were re-suspended in flow buffer (PBS + 2 % BSA) and stained on ice for 30 min in the dark with above mentioned fluorochrome-conjugated antibodies using a separate panel for T cells and myeloid cells. Intracellular antigen staining of IFNγ, Granzyme-B, and CD206 was performed by fixing and permeabilizing cells with a transcription factor buffer set (BD Biosciences, San Jose, C, USA) for 20 min on ice and incubating cells with specific antibodies upon washing. All acquisitions were performed on an LSR II flow cytometer (BD Biosciences) within 24 h. Compensations were performed with unstained and single-stained cells. FlowJo software v.10.2 (Treestar Inc., Ashland, OR, USA) was used for data analysis and cells were gated as follows - CD45+ (Total leukocytes), CD45+ CD3+ (Total T-cells), CD45+ CD3+ CD4+ CD8− (TH, CD4+ T helper cells), CD45+ CD3+ CD4+ CD8− (Treg), CD45+ CD3+ CD8− CD4+ INFγ (Activated CD4 T-cells), CD45+ CD3+ CD4− CD8+ (TC, CD8+ T-cells), CD45+ CD3+ CD4− CD8+ GZMB+ & INFγ (Effector cytotoxic T-cells), CD45+ CD11c+ (DC, Dendritic cells), CD45+ CD11b+ (Macrophages), CD45+ CD11b+ iNOS+ (M1 Macrophages), CD45+ CD11b+ CD206+ (M2 Macrophages), CD45+ CD11b+ Ly6CHi Ly6G− (Monocytic-Myeloid derived suppressor cells, M-MDSC) & CD45+ CD11b+ Ly6CLo Ly6G+ (Polymorphonuclear-MDSC, PMN-MDSC).
2.5. Statistical analysis
Statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA). Tumor volume data and spleen immune cell data are presented as mean ± SEM. Multiple group analyses were performed using one-way ANOVA with the Tukey test. For comparison between the two groups, two-tailed unpaired t-tests were performed. P values <0.05 were considered significant and represented as *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001. Statistical analysis of microbiome alterations was calculated by MicrobiomeAnalyst in-built tests.
3. Results
3.1. In-situ treatment of B16F10 melanoma tumor with CFUS reduces tumor burden and alters the gut microbiome
DLS characterizations of CRT-NP by DLS in physiological buffer showed a hydrodynamic diameter of 372 ± 2.63 nm, zeta-potential of +14.43 ± 3.11 mv, and a PDI <0.3. Young mice showed slower tumor growth at the primary site (about 32 % slower) and abscopal sites (about 65 % slower) compared to aged control mice. CFUS reduced tumor volume to ~50 % in aged and ~70 % in young mice, compared to their respective untreated control groups (Fig. 1B). Analysis of the alpha diversity of the microbiome using the Simpson index, based on 16S sequencing of fecal samples collected from each treatment group, revealed that the mice with the least tumor burden (CFUS (Y) group) had the highest inverse Simpson diversity score, whereas the mice with high tumor load (Control (O) group) had the least inverse Simpson score (Fig. 1C). The Principle Coordinates analysis (PCoA) 2D plot representing beta diversity shows a notable difference in microbial diversity between young and aged untreated control mice. Additionally, a slight shift in population clusters was observed in both aged and young CFUS treated mice (Fig. 1D).
3.2. Aged and young mice showed substantial differences in microbiome abundance which altered with CFUS in-situ tumor treatment
Pattern discovery analysis conducted at the phylum level showed a higher abundance of Firmicutes in aged control mice, while young control mice had a higher abundance of Bacteroidetes and Verrucomicrobia. CFUS treatment increased the abundance of Proteobacteria in both aged and young mice. Additionally, Actinobacteria increased only in young mice, whereas in aged mice, Tenericutes and Deferribacteres populations increased with treatment (Fig. 2A). At the genus level, Turicibacter was significantly more abundant in aged mice (both control and treated groups), while in the young, the population of lactobacillus was more prominent. However, with CFUS treatment in both age groups, a decrease in Turicibacter and an increase in the lactobacillus population was observed (Fig. 2B & C).
Fig. 2.
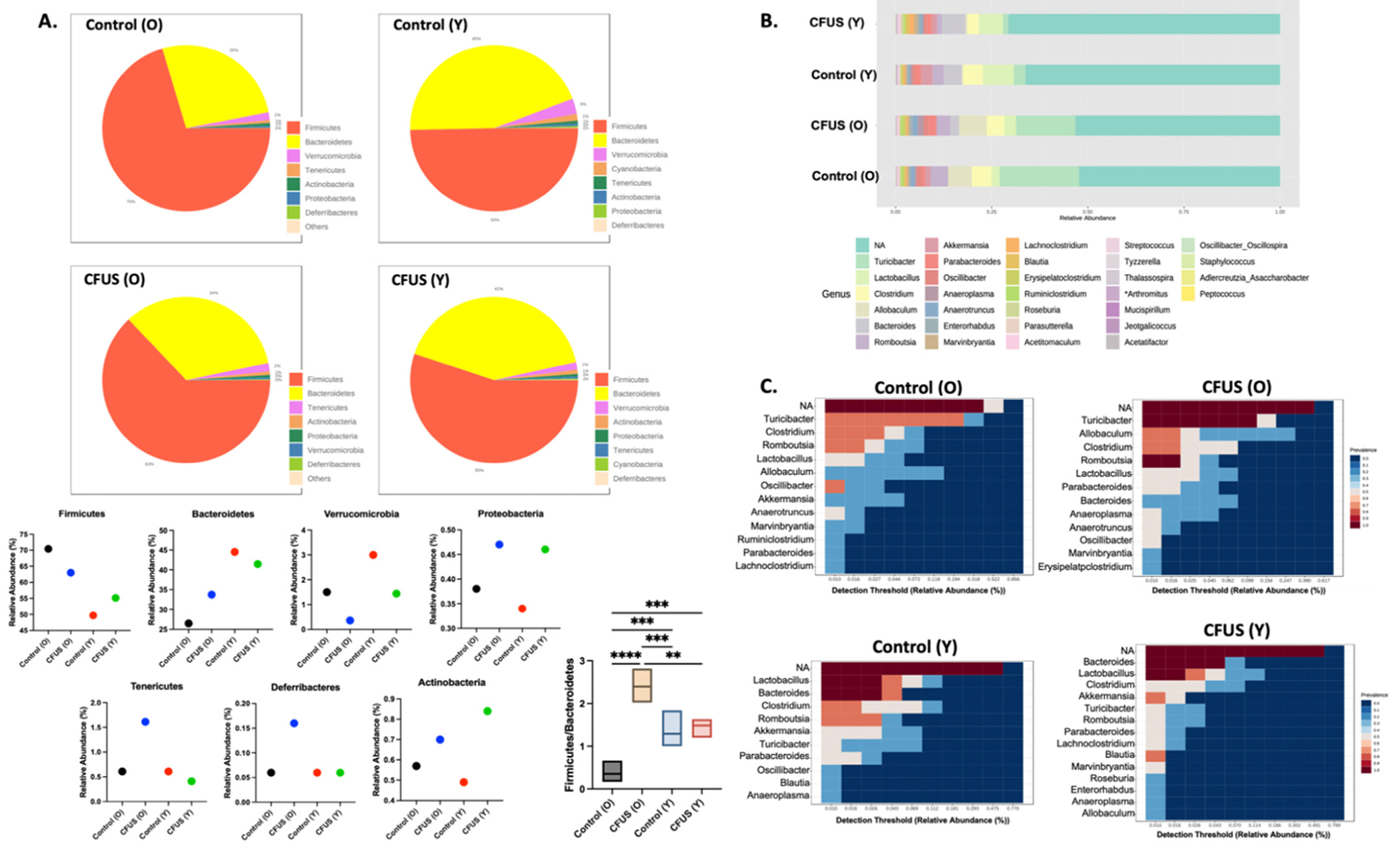
Pattern discovery analysis showed changes in microbiome abundance with age and treatments. A. Pie charts representing relative phylum abundance constituting microbial community in each treatment group. Bar graphs represent the abundance percentage of phylum present in all groups. B. Stacked bar plot of the phylogenetic composition of common bacterial taxa at genus level in different treatment groups. C. Heatmap representing the relative abundance of the most prevalent genus (>0.01 Detection threshold) in each treatment group. Statistical analysis was done using one-way ANOVA with Fisher’s LSD test, **p < 0.005, ***p < 0.0005, ****p < 0.0001.
3.3. Comparative analysis of treatment groups revealed differences in microbiome populations among the groups
Heat tree analysis, which compared the relative abundance of microbial communities using a non-parametric Wilcoxon test at different taxonomic levels, revealed a significant decrease in the abundance of the phylum Cyanobacteria (p = 0.021) and the phylum Firmicutes (p = 0.057) in aged control mice versus young. At the order level, Bacillales (which belongs to Phylum Firmicutes) showed a significant decrease in abundance in aged control mice (p = 0.026) along with Gastranaerophilales (Phylum Cyanobacteria, Class Melainabacteria) (Fig. 3A(i)). In the group-wise comparison of CFUS treated aged and young mice, a significant increase in the abundance of Firmicutes (p = 0.028) and a significant decrease in Bacteroidetes (p = 0.028) populations were observed in aged vs young (Fig. 3A(ii)). CFUS treated aged mice did not show any significant differences in microbial communities compared to untreated control, while in young mice, CFUS treatment led to a significant decrease in the abundance of Phylum Actinobacteria (p = 0.028) compared to control mice, along with unclassified Family_XIII (p = 0.029) of Phylum Bacillota (Fig. 3A(iv)). A list of the top 10 differential populations is shown in Table 1, except for the CFUS (O) vs (Y) group comparison, where 12 significantly different populations are listed.
Fig. 3.
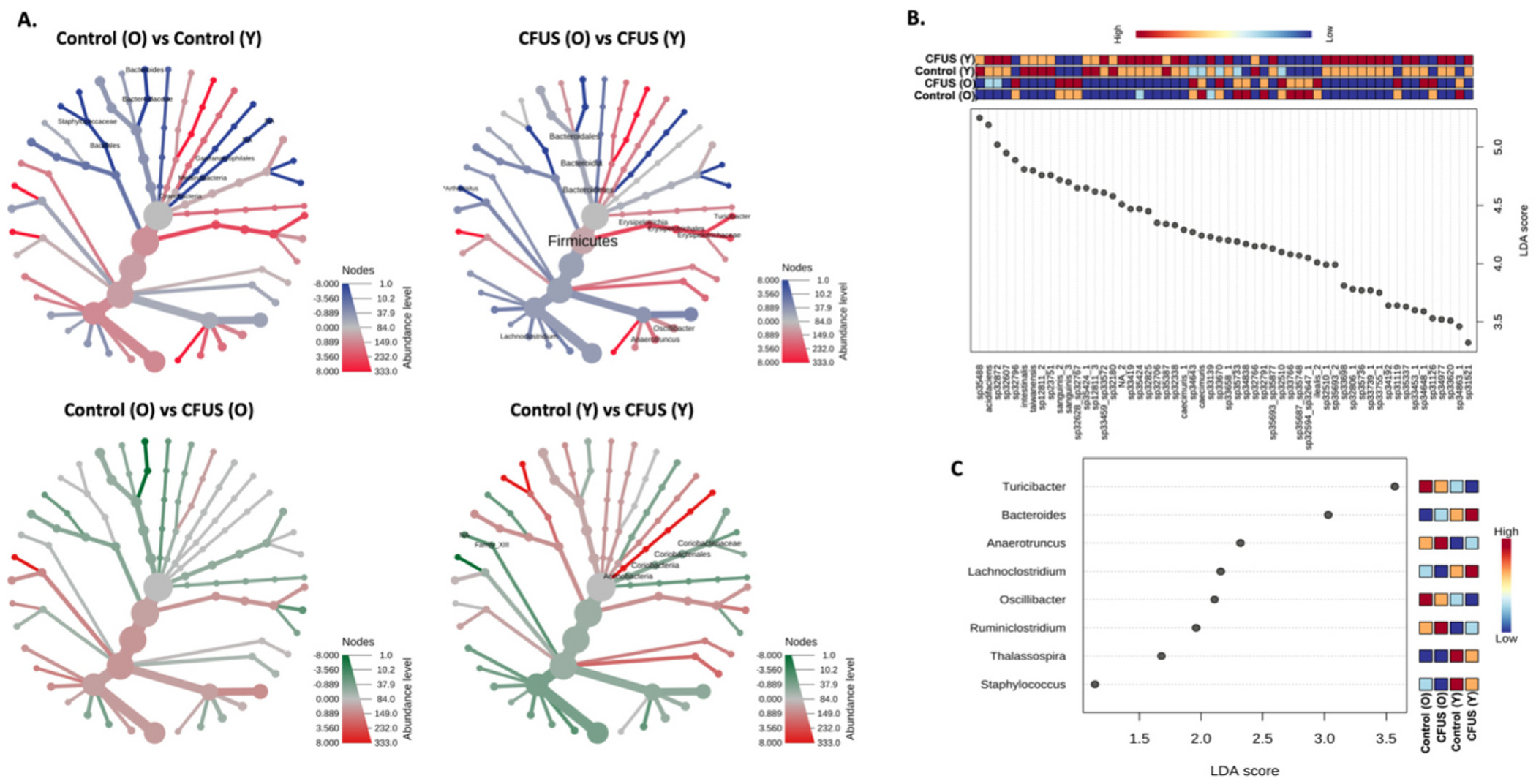
Comparative analysis among treatment groups revealed significant differences in microbial taxa with age and treatment. A. Phylogenetic tree representing comparative analysis between two treatment groups using non-parametric Wilcoxon test at different taxa levels. B. Multifactor Lefse analysis of all four treatment groups at Feature levels yielded 152 significantly different populations, mostly at species levels, out of which 56 with the highest LDA score are represented in the dot plot. Whereas, at the genus level (C.) 8 significantly different populations among treatment groups were observed.
Linear discriminant analysis effect size (LEfSe) analysis for biomarker discovery at the feature level for all treatment groups yielded 152 significantly different microbial communities with Log LDA >2, p < 0.05. The 56 communities with the highest LDA score are represented in Fig. 3B, and the LEfSe output for all significant populations is listed in Table 2. To narrow down the multi-group comparison, LEfSe analysis was performed at the genus level (Log LDA score >1, p < 0.1), which yielded eight distinguished populations among the four treatment groups. Control (O) mice had a high abundance of Turicibacter and Oscillibacter, Control (Y) had higher Thalassospira and Staphylococcus populations, CFUS (O) had higher Anaerotruncus and Ruminiclostridium, and CFUS (Y) had higher Bacteroides and Lachnoclostridium populations (Fig. 3C). The LEfSe output for all the genus with log LDA >1 is listed in Table 3.
3.4. Microbial communities identified using multi-group LEfSe analysis correlated with distinct splenic immune cell populations
Eight genera identified using LEfSe analysis of all treatment groups were correlated with different splenic immune cell populations (Figs. 4 & 5), including CD45+ overall hematopoietic cells, CD4+ TH cells, FOXP3+ Treg, CD8+ TC cells, CD11b+ macrophages, and Ly6Chi Ly6G− M-MDSCs. Pearson r analysis demonstrated that Turicibacter was negatively correlated with CD45+ hematopoietic cells, CD8+ T cells, and CD11b+ macrophages, whereas it was positively correlated with CD4+ T cells. A higher accumulation of Oscillibacter was positively correlated with M-MDSCs and negatively correlated with CD8+ T cells. Anaerotruncus and Ruminiclostridium were negatively correlated with CD45+ hematopoietic cells and CD8+ T cells, and positively correlated with M-MDSCs. Increased populations of Thalassospira negatively correlated with M-MDSCs. The Bacteroides population was found to be positively correlated with CD45+ hematopoietic cells, CD8+ T cells, Treg cells, and CD11b+ macrophages. Lachnoclostridium was positively correlated with M-MDSCs and negatively correlated with CD4+ T cells.
Fig. 4.
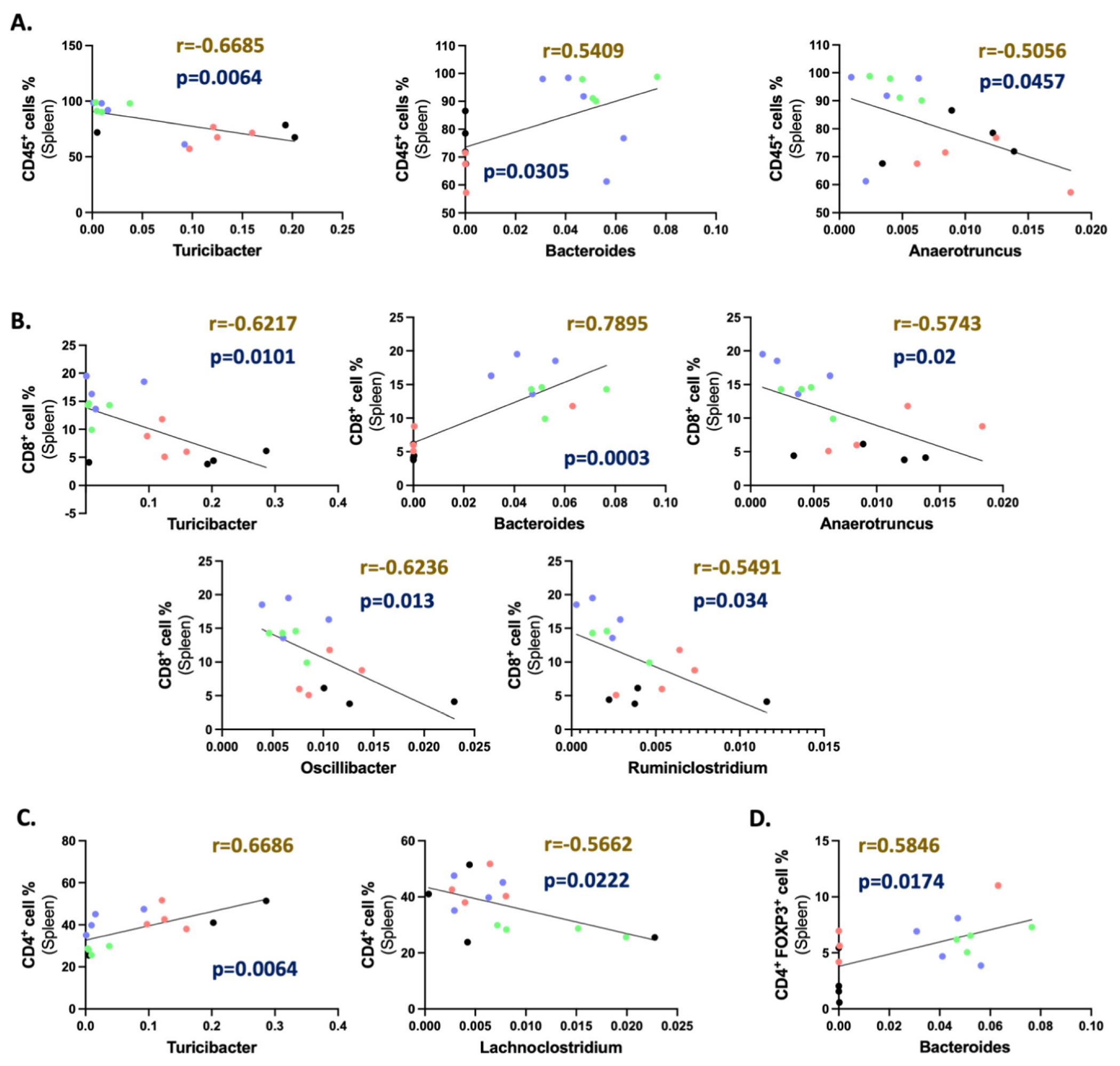
Pearson r correlation of identified genus with splenic lymphocyte population. Correlation graphs representing significant (p < 0.05) linear correlation between (A) CD45+ immune cells, (B) CD8+ T cells (Gated at CD45+ CD3+ CD4−), (C) CD4+ T cells (Gated at CD45+ CD3+ CD8−), and (D) FOXP3+ CD4 T cell populations in the spleen with the gut abundance of different genera irrespective of treatment groups. Sample identification in the correlation graphs: Control (O)- Black, CFUS (O)- Red, Control (Y)- Blue, and CFUS (Y)- Green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
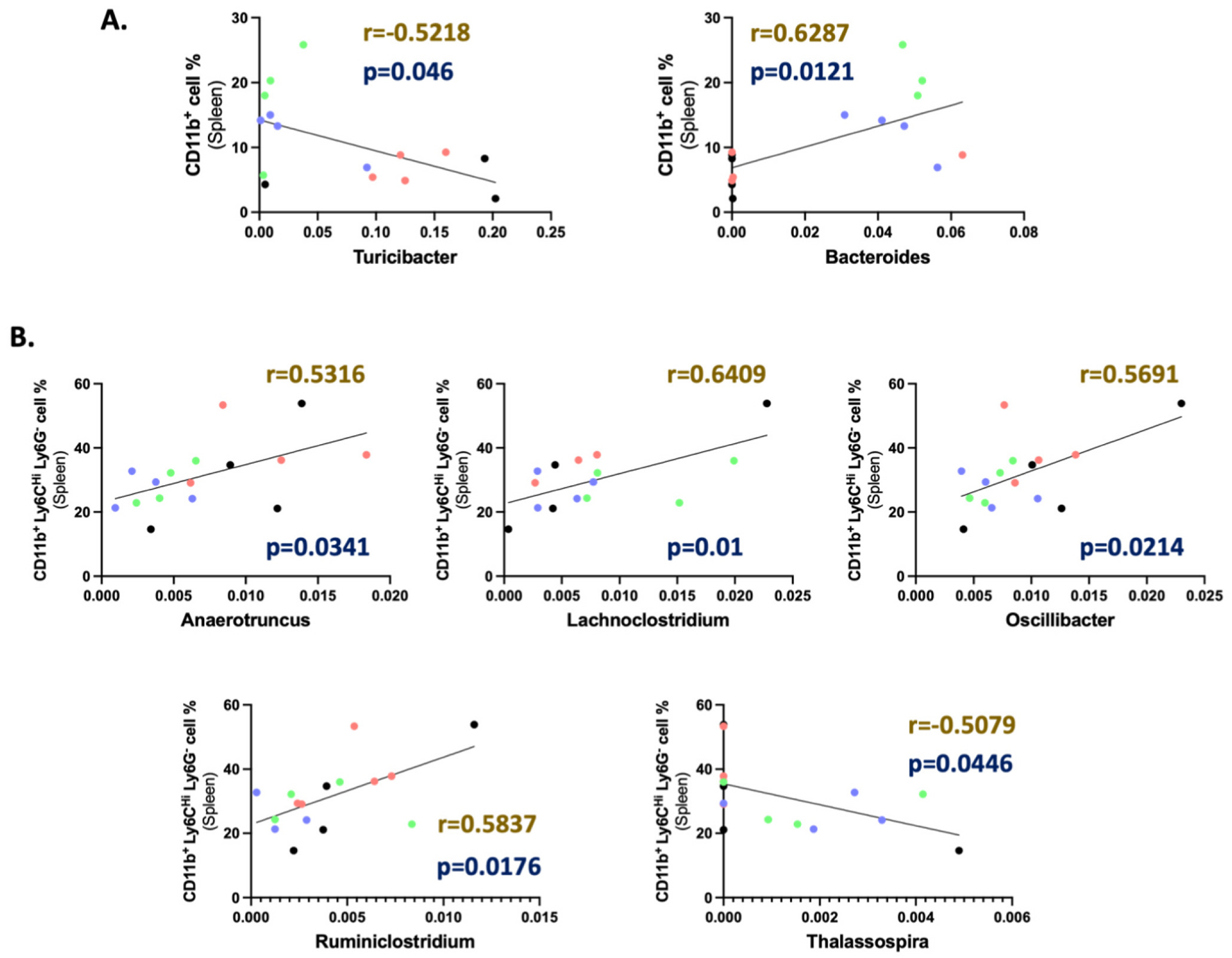
Pearson r correlation of identified genus with splenic myeloid population. Correlation graphs representing significant (p < 0.05) linear correlation between (A) CD11b+ macrophages (Gated at CD45 CD11c−), (B) Ly6CHi Ly6G− M-MDSCs (Gated at CD45+ CD11c− CD11b+) population in the spleen with the gut abundance of different genera irrespective of treatment groups. Sample identification in the correlation graphs: Control (O)- Black, CFUS (O)- Red, Control (Y)- Blue, and CFUS (Y)- Green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Community typing grouped aged and young mice genera into two hierarchical clusters
Distinct hierarchical clusters were observed among mice groups based on the age of mice, rather than treatment. Community phenotyping grouped aged and young mice genera into two separate groups using the Ward clustering algorithm (Fig. 6A). Further analysis of these clusters using Taxon set enrichment analysis (TSEA) with Host-intrinsic taxon sets database showed an association of the young mice cluster with 26 disease conditions and the old mice cluster with 20 disease conditions.
Fig. 6.
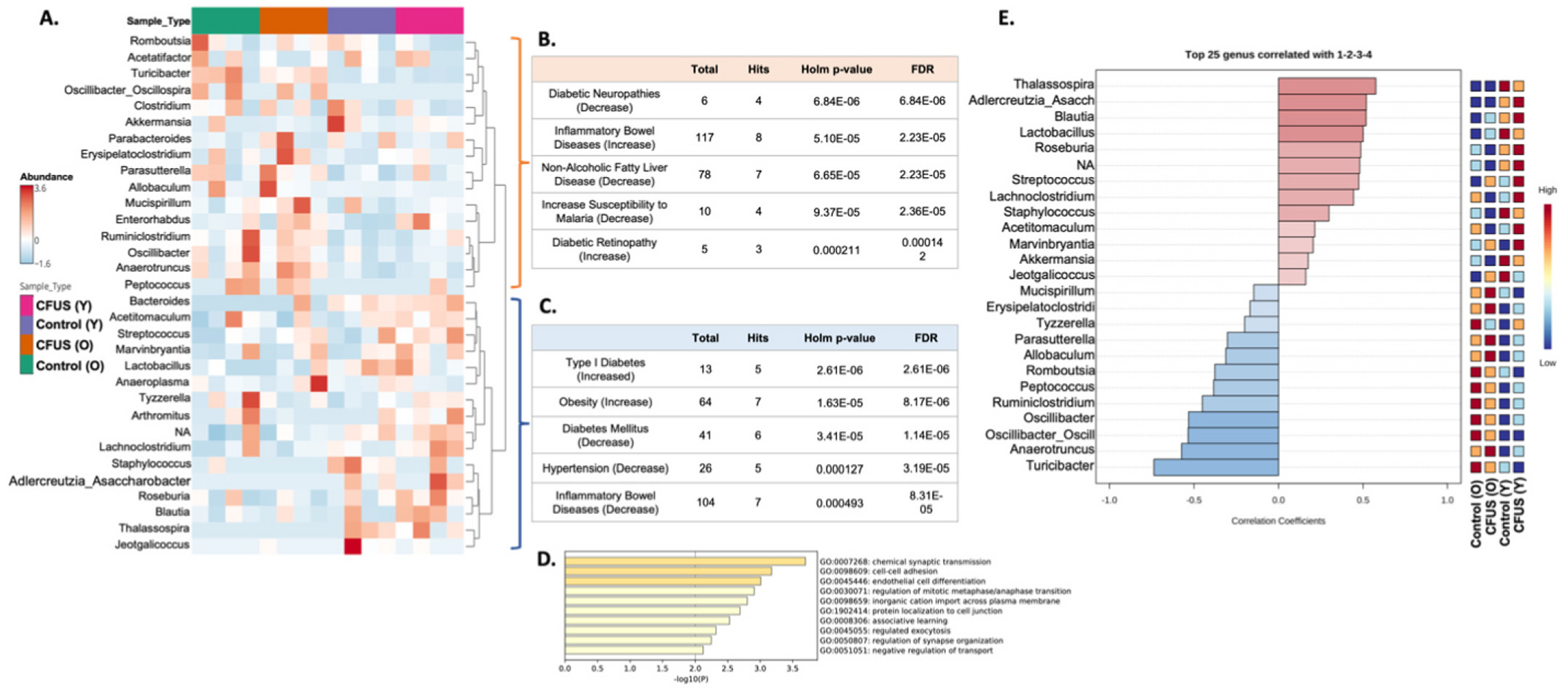
Community typing divided aged and young mice genera into two hierarchical clusters. A. Hierarchical clustering of different treatment groups divided aged and young mice populations into two clusters at the genus level. Taxon set enrichment analysis (TSEA) analysis of young mice clusters using Host-intrinsic taxon sets database hit 26 significant disease conditions (B.) and old mice cluster had 20 hits (C.) Top 5 hits for each cluster with lowest p values mentioned in tables. A complete list of significant hits is mentioned in Tables 4 and 5. TSEA using the Host genetics taxon sets database yielded 58 significantly enriched host gene targets for the young cluster whereas 11 for the aged cluster. D. Bar graph representing function groups for young cluster gene targets enriched using Metascape gene enrichment software. Genes predicted for the aged cluster did not enrich into any specific molecular or biological function group. E. Pearson r correlation graph demonstrating top 25 genera correlated to different treatment groups. Populations correlated using Pearson r analysis matches the hierarchical clustering of various genera with treatment groups.
The top 5 hits of the young cluster are presented in Fig. 6B, while the top 5 hits of the old cluster are presented in Fig. 6C. The complete list of significant associations (adjusted Holm p < 0.05) is presented in Tables 4 and 5, respectively. TSEA analysis using the Host genetics taxon sets database yielded 58 significantly enriched host gene targets for the young cluster, while only 11 for the aged cluster. Gene enrichment of these host gene targets, using Metascape software, showed the enrichment of young cluster-associated host genes into 10 GO biological and molecular function groups (Fig. 6D), whereas no significant gene enrichment was observed with aged cluster-associated host genes. Pattern search using Pearson r correlation between microbial genera and treatment groups yielded the top 25 genera that matched the mice age-based hierarchical clustering (Fig. 6E).
4. Discussion
As individuals age, typically those over the age of 65, their immune systems can weaken due to age-related changes and the presence of other medical conditions. While immunotherapy has shown promise as a potential treatment option for older patients with cancer, their limited participation in clinical trials highlights the need for additional data to fully understand the effectiveness and potential side effects in this population (Kanesvaran et al., 2018; Benguerfi et al., 2022). Age-related changes such as alterations in nutrient processing and absorption, as well as changes in the metabolism of immunotherapy drugs, can contribute to a decline in the diversity of the immune repertoire (Vétizou et al., 2015). Additionally, the dysbiosis of the gut microbiota can increase with age, which can lead to a decrease in T-cell infiltration and an increase in immunosuppressive cells (Vétizou et al., 2015). Given that cancer diagnoses are more common among older individuals, personalized treatment decisions should take into account factors such as comorbidities and functional status to achieve optimal outcomes (Franceschi and Campisi, 2014). The objective of this study was to perform ISV-based CFUS in a murine model of melanoma, and profile the gut microbiota as a functional biomarker, and correlate those with immune cells from both young and old cohorts to gain a better understanding of the therapeutic features.
We conducted three intratumoral CFUS-based exposures over the course of a week in mice with B16F10 bilateral flank tumors. Our findings indicated that in both the treated and untreated contralateral sites, tumor growth rates were relatively faster in the older mice compared to the younger cohort (Fig. 1B), and these were associated with a higher population of immunosuppressive immune cells in splenic tissues of aged mice (Supplementary Fig. 1C & D). These results are not surprising, as aging is often accompanied by inflammaging, a state of chronic, low-grade inflammation that can increase the levels of pro-inflammatory cytokines and negatively impact the immune system, reducing its response to immunotherapy (Teissier et al., 2022). It is worth noting the patterns of tumor growth in the bilateral setting in our study. Previous reports in C57BL6 and Balb/c × C57BL/6 F1 (CB6F1) models indicated slower tumor growth rates in older mice than in younger mice upon unilateral injections in the inguinal and axillary spaces, as well as via the intradermal route (Pettan-Brewer et al., 2012; Ershler et al., 1984). However, others reported similar growth rates of B16 tumors in young and old mice with unilateral inoculations (Hurez et al., 2012; Padrón et al., 2018). In our bilateral B16F10 model, we observed tumor growth rates that were similar or slightly higher in older mice than in younger mice across several independent studies. Thus, based on this premise, we propose that the bilateral B16F10 model tends to have a faster growth pattern than the unilateral model, especially in mice >18 months old. Although various factors could cause such an outcome, we specifically correlated these growth patterns with functional microbiome markers and immune cell populations. We noted that young mice with the least tumor burden with CFUS showed the highest alpha diversity, while mice with a high tumor load (Control (O) group) showed the least inverse Simpson score. Alpha diversity has been shown to be a predictor of response to immunotherapy in both humans and mice. Those who respond well to immunotherapy tend to have higher alpha diversity in their gut microbiome compared to those with a poor response (Matson et al., 2018). Mechanistically, this is linked to increased production of short-chain fatty acids (SCFAs), which have immunomodulatory effects (Blackman et al., 2022). While alpha diversity can provide a valuable tool in predicting response to immunotherapy and developing personalized treatment plans for cancer patients, our study is the first to demonstrate that ISV that is administered directly to a tumor in contrast to systemic therapy via intravenous or intraperitoneal route, can also generate a similar therapeutic response.
Numerous studies have also linked the abundance of specific bacterial taxa with an improved response to immunotherapy in cancer patients. For example, a higher abundance of Bacteroidetes and Verrucomicrobia was associated with a superior response to checkpoint blockade therapy in melanoma patients (Matson et al., 2018; Gopalakrishnan et al., 2018). Additionally, in patients with non-small cell lung cancer treated with immune checkpoint inhibitors, the abundance of Actinobacteria was found to be significantly higher in responders compared to non-responders (Routy et al., 2018). Similarly, an increased abundance of Bifidobacterium was shown to be positively associated with a response to anti-PD-1 therapy in melanoma patients (Sivan et al., 2015). Our pattern analysis of young mice showed a higher abundance of Bacteroidetes and Verrucomicrobia, however, such an abundance was missing in the aged cohorts. These findings highlight how modifications in the composition of the gut microbiota in aged mice influenced tumor growth rates and response to immunotherapy, reinforcing the development of personalized treatment strategies, especially those employing ISVs. CFUS treatment also increased the abundance of Proteobacteria in both aged and young mice, but this bacterial phylum has been shown to be an unreliable biomarker for predicting a response to immunotherapy (Frankel et al., 2017). Further, we noted a higher abundance of Actinobacteria in young CFUS mice, which was previously shown to be associated with an improved response to anti-PD-1 therapy in melanoma patients (Jin et al., 2019). In contrast, in aged mice, Tenericutes and Deferribacteres populations increased with CFUS treatment. Gopalakrishnan et al. showed higher populations of certain Firmicutes in non-responders to immune checkpoint inhibitor therapy (Gopalakrishnan et al., 2018). We similarly observed notably higher levels of Firmicutes such as Turicibacter, Oscillibacter, Anaerotruncus, and Ruminiclostridium in aged mice (Fig. 1A & 3C). These findings suggest that Firmicutes could serve as potential biomarkers for poor immunotherapy response, regardless of age or type of immunomodulation (ISV, immune checkpoint inhibitor, etc.) in both murine and human settings.
Although local ISV improves outcomes, microbiome profiling and its relationship with immune cells involved in antitumor immunity in young and aged is not fully understood. We correlated the splenic populations of various types of immune cells with microbiomes in the B16F10 model. Our findings revealed a negative correlation between Turicibacter and CD8+ T cells and macrophages, but a positive correlation with CD4+ T cells (Fig. 4B). Previous studies have also reported that the presence of Turicibacter is associated with lower populations of CD8+ T cells in the colonic mucosa of mice (Presley et al., 2010). Similarly, we found negative correlations between Anaerotruncus, Oscillibacter, and Ruminiclostridium and CD8+ T cells. Although the exact reasons of the observed correlations are currently unknown, our analysis suggests that the local tumor therapy employing the combinatorial CFUS modality led to significant alterations in immune cell functions, which we propose consequently influenced the abundance of the gut microbiome. The premise of our hypothesis stems from prior reports where gut microbiome and anti-tumor immune cells abundance in humans as well as in preclinical models of various diseases were found to be linked (Lu et al., 2022; Vijay and Valdes, 2022; Gomez et al., 2012; Wen and Duffy, 2017; Claesson et al., 2012). What is particularly intriguing is that even ISV regimens (such as CFUS) can impact age-associated dysbiosis (Supplementary Fig. 1C & D), suggesting the existence of a tumor-microbiome crosstalk in cancer patients. Indeed, Taxon set enrichment analysis (TSEA) with Host-intrinsic taxon sets database showed 58 significantly enriched host gene targets for the young cluster, compared to only 11 for the aged cluster. Although our study did not establish the basis for the apparent age-dependent influence of the gut microbiome on host gene expression, we speculate that the distinct differences in the gut microbiome composition that we observed with CFUS, including alterations in the diversity and composition of bacterial species and their metabolic activity, may have contributed to the differential host gene expression profile. Further research is currently underway in our laboratory to fully elucidate the complex interplay between the gut microbiome and host gene expression in the context of age-related diseases.
In summary, our study suggests that the gut microbiome and immune cells are related to antitumor immunity in young and aged B16F10 bearing mice following CFUS treatment. The clinical implications of our study are significant, as we hypothesize that interventions such as diet, probiotics, or fecal microbiota transplantation may be potential therapeutic strategies for improving immune responses and mitigating the risk of age-related diseases in humans. Further research is needed to fully elucidate the complex interactions between the gut microbiome and host immune system and to develop targeted interventions for improving outcomes in patients with cancer and age-related diseases.
Supplementary Material
Funding/acknowledgement
We acknowledge the National Cancer Institute of the National Institutes of Health under award numbers R37 CA239150-02 & R01CA260974, and the Kerr Endowed Chair at Oklahoma State University for supporting this research project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.exger.2023.112268.
CRediT authorship contribution statement
Ashish Ranjan conceived the project, designed the studies, helped interpret the data, and wrote the manuscript. Akansha Singh performed experiments associated with the manuscript, analyzed the microbiome and immune data, and wrote the paper along with Ashish Ranjan. Harshini Ashar assisted with the mice experiment.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Al-Lazikani B, Banerji U, Workman P, 2012. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol 30 (7), 679–692. [DOI] [PubMed] [Google Scholar]
- Arboleya S, Watkins C, Stanton C, Ross RP, 2016. Gut bifidobacteria populations in human health and aging. Front. Microbiol 7, 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badal VD, Vaccariello ED, Murray ER, Yu KE, Knight R, Jeste DV, Nguyen TT, 2020. The gut microbiome, aging, and longevity: a systematic review. Nutrients 12 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benguerfi S, Lesimple T, Houot R, Ricordel C, Legoupil D, Alleaume C, Lamy R, Deniel Lagadec D, Corre R, 2022. Immune checkpoint inhibitors in patients aged 80 or older with advanced non-small cell lung cancer or melanoma: a real-life multicentre study. Acta Oncol. 61 (11), 1339–1346. [DOI] [PubMed] [Google Scholar]
- Blackman AC, Thapa S, Venkatachalam A, Horvath TD, Runge JK, Haidacher SJ, Hoch KM, Haag AM, Luna RA, Anagnostou A, 2022. Insights into microbiome and metabolic signatures of children undergoing peanut oral immunotherapy. Children (Basel) 9 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW, 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488 (7410), 178–184. [DOI] [PubMed] [Google Scholar]
- Ershler WB, Stewart JA, Hacker MP, Moore AL, Tindle BH, 1984. B16 murine melanoma and aging: slower growth and longer survival in old mice. J. Natl. Cancer Inst 72 (1), 161–164. [DOI] [PubMed] [Google Scholar]
- Fernando R, Downs J, Maples D, Ranjan A, 2013. MRI-guided monitoring of thermal dose and targeted drug delivery for cancer therapy. Pharm. Res 30 (11), 2709–2717. [DOI] [PubMed] [Google Scholar]
- Franceschi C, Campisi J, 2014. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci 69 (Suppl. 1), S4–S9. [DOI] [PubMed] [Google Scholar]
- Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, Koh AY, 2017. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 19 (10), 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, White BA, Taneja V, 2012. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One 7 (4), e36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Sanchez E, Marcelo Riquelme, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA, 2018. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359 (6371), 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbet MJ, Ranjan A, 2020. Cancer immunotherapy with immunoadjuvants, nanoparticles, and checkpoint inhibitors: recent progress and challenges in treatment and tracking response to immunotherapy. Pharmacol. Ther 207, 107456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbet MJ, Singh A, Mao C, Fiering S, Ranjan A, 2020. Using nanoparticles for in situ vaccination against cancer: mechanisms and immunotherapy benefits. Int. J. Hyperthermia Off. J. Eur. Soc. Hyperthermic Oncol. N. Am. Hyperthermia Group 37 (3), 18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotwals P, Cameron S, Cipolletta D, Cremasco V, Crystal A, Hewes B, Mueller B, Quaratino S, Sabatos-Peyton C, Petruzzelli L, Engelman JA, Dranoff G, 2017. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 17 (5), 286–301. [DOI] [PubMed] [Google Scholar]
- Hurez V, Daniel BJ, Sun L, Liu AJ, Ludwig SM, Kious MJ, Thibodeaux SR, Pandeswara S, Murthy K, Livi CB, Wall S, Brumlik MJ, Shin T, Zhang B, Curiel TJ, 2012. Mitigating age-related immune dysfunction heightens the efficacy of tumor immunotherapy in aged mice. Cancer Res. 72 (8), 2089–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen Y, Zheng H, Yao C, Wang Y, Lu S, 2019. The diversity of gut microbiome is associated with favorable responses to anti-programmed death 1 immunotherapy in Chinese patients with NSCLC. J. Thorac. Oncol 14 (8), 1378–1389. [DOI] [PubMed] [Google Scholar]
- Kanesvaran R, Cordoba R, Maggiore R, 2018. Immunotherapy in older adults with advanced cancers: implications for clinical decision-making and future research. Am. Soc. Clin. Oncol. Educ. Book 38, 400–414. [DOI] [PubMed] [Google Scholar]
- Kennedy MS, Chang EB, 2020. The microbiome: composition and locations. Prog. Mol. Biol. Transl. Sci 176, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, 2021. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol. Immunol 18 (5), 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leins H, Mulaw M, Eiwen K, Sakk V, Liang Y, Denkinger M, Geiger H, Schirmbeck R, 2018. Aged murine hematopoietic stem cells drive aging-associated immune remodeling. Blood 132 (6), 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, Li Q, 2022. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J. Hematol. Oncol 15 (1), 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado Galdeano C, Cazorla SI, Lemme Dumit JM, Vélez E, Perdigón G, 2019. Beneficial effects of probiotic consumption on the immune system. Ann. Nutr. Metab 74 (2), 115–124. [DOI] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF, 2018. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359 (6371), 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón Á, Hurez V, Gupta HB, Clark CA, Pandeswara SL, Yuan B, Svatek RS, Turk MJ, Drerup JM, Li R, Curiel TJ, 2018. Age effects of distinct immune checkpoint blockade treatments in a mouse melanoma model. Exp. Gerontol 105, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettan-Brewer C, Morton J, Coil R, Hopkins H, Fatemie S, Ladiges W, 2012. B16 melanoma tumor growth is delayed in mice in an age-dependent manner. Pathobiol. Aging Age Relat. Dis 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley LL, Wei B, Braun J, Borneman J, 2010. Bacteria associated with immunoregulatory cells in mice. Appl. Environ. Microbiol 76 (3), 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KM, Fox A, Harland KL, Russ BE, Li J, Nguyen THO, Loh L, Olshanksy M, Naeem H, Tsyganov K, Wiede F, Webster R, Blyth C, Sng XYX, Tiganis T, Powell D, Doherty PC, Turner SJ, Kedzierska K, La Gruta NL, 2018. Age-related decline in primary CD8(+) T cell responses is associated with the development of senescence in virtual memory CD8(+) T cells. Cell Rep. 23 (12), 3512–3524. [DOI] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L, 2018. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359 (6371), 91–97. [DOI] [PubMed] [Google Scholar]
- Saraswati S, Sitaraman R, 2014. Aging and the human gut microbiota-from correlation to causality. Front. Microbiol 5, 764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman SN, Singh MP, Patil G, Li S, Fiering S, Hoopes PJ, Guha C, Malayer J, Ranjan A, 2020. Novel calreticulin-nanoparticle in combination with focused ultrasound induces immunogenic cell death in melanoma to enhance antitumor immunity. Theranostics 10 (8), 3397–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MP, Sethuraman SN, Ritchey J, Fiering S, Guha C, Malayer J, Ranjan A, 2019. In-situ vaccination using focused ultrasound heating and anti-CD-40 agonistic antibody enhances T-cell mediated local and abscopal effects in murine melanoma. Int. J. Hyperthermia 36 (sup1), 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MP, Sethuraman SN, Miller C, Malayer J, Ranjan A, 2021. Boiling histotripsy and in-situ CD40 stimulation improve the checkpoint blockade therapy of poorly immunogenic tumors. Theranostics 11 (2), 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF, 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350 (6264), 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AC, Bagley SJ, Wen PY, Lim M, Platten M, Colman H, Ashley DM, Wick W, Chang SM, Galanis E, Mansouri A, Khagi S, Mehta MP, Heimberger AB, Puduvalli VK, Reardon DA, Sahebjam S, Simes J, Antonia SJ, Berry D, Khasraw M, 2021. Systematic review of combinations of targeted or immunotherapy in advanced solid tumors. J. Immunother. Cancer 9 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissier T, Boulanger E, Cox LS, 2022. Interconnections between Inflammageing and Immunosenescence during ageing. Cells 11 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L, 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350 (6264), 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay A, Valdes AM, 2022. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr 76 (4), 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wen L, Duffy A, 2017. Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J. Nutr 147 (7), 1468S–1475S. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


