Abstract
MGA (Max-gene associated) is a dual-specificity transcription factor that negatively regulates MYC-target genes to inhibit proliferation and promote differentiation. Loss-of-function mutations in MGA have been commonly identified in several hematological neoplasms, including acute myeloid leukemia (AML) with RUNX1::RUNX1T1, however, very little is known about the impact of these MGA alterations on normal hematopoiesis or disease progression. We show that representative MGA mutations identified in patient samples abolish protein-protein interactions and transcriptional activity. Using a series of human and mouse model systems, including a newly developed conditional knock-out mouse strain, we demonstrate that loss of MGA results in upregulation of MYC and E2F targets, cell cycle genes, mTOR signaling, and oxidative phosphorylation in normal hematopoietic cells, leading to enhanced proliferation. The loss of MGA induces an open chromatin state at promoters of genes involved in cell cycle and proliferation. RUNX1::RUNX1T1 expression in Mga-deficient murine hematopoietic cells leads to a more aggressive AML with a significantly shortened latency. These data show that MGA regulates multiple pro-proliferative pathways in hematopoietic cells and cooperates with the RUNX1::RUNX1T1 fusion oncoprotein to enhance leukemogenesis.
Subject terms: Acute myeloid leukaemia, Haematopoiesis
Introduction
Max-gene associated (MGA) is a transcription factor that uses an n-terminal T-box domain and a c-terminal MYC-like basic helix-loop-helix (bHLH) domain to regulate MAX-network and T-box family targets [1]. MGA interacts with MAX, L3MBTL2, E2F6, and PCGF6 as a part of the non-canonical polycomb repressive complex (ncPRC1.6) [2] and is required for PRC1.6 complex formation. In non-hematopoietic cells, the recruitment of MGA to T-box and MAX-network target genes was shown to result in the deposition of repressive histone marks such as H2AK119ub1 and H3K27me3 [2, 3]. MGA is required for embryogenesis and its depletion leads to aberrant embryonic stem cell differentiation and embryonic lethality [4–6].
Heterozygous somatic alterations of MGA, the most common of which lead to loss-of-function truncation/deletion of the MYC-like bHLH domain, occur in 5% of all cancers and are commonly seen in lung adenocarcinoma, endometrial carcinoma, and colorectal cancer [7–10]. The loss of MGA in lung adenocarcinoma cells, which occurs in ~8% of lung adenocarcinoma patients, was shown to lead to an upregulation of MYC- and E2F6-target genes, resulting in an increase in cancer cell proliferation and invasiveness [11, 12]. MGA has also been identified as a common genetic alteration in hematological neoplasms, including acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), natural killer/T-cell lymphoma, B-cell acute lymphoblastic leukemia (B-ALL), and T-cell acute lymphoblastic leukemia (T-ALL) [13–18]. In particular, we previously identified MGA mutations as recurrent alterations in AMLs with RUNX1::RUNX1T1 fusions [17]. Likewise, similar mutations have been observed in AMLs with KMT2A-PTD [16].
Despite the recurrence of these mutations, the molecular role of MGA in normal hematopoiesis, as well as in hematopoietic malignancies, has been understudied. This is partly due to the embryonic lethality of in vivo MGA deficiency models, which we circumvented in this study by developing a conditional knockout mouse model. Harnessing a multidisciplinary approach to characterize the hematopoietic function of MGA in both human and mouse cells, we establish that MGA loss leads to an increase in proliferation via the upregulation of several cell cycle pathway genes and results in a shortened latency in a mouse model of RUNX1::RUNX1T1-driven leukemia.
Results
The reported mutations of MGA in pediatric RUNX1::RUNX1T1 leukemias are heterozygous, somatic mutations at variable variant allele frequencies and result in the deletion of the MYC-like bHLH domain, potentially resulting in a protein with altered or abolished function (Fig. 1A) [17]. We first assessed the protein localization of MGA, including full-length MGA (WT-MGA) and 3 MGA truncations (MGA p.E1953*, MGA p.S812*, and MGA p.C623*) identified in patient samples, by transfecting GFP fusion expression vectors in HEK293T cells (Fig. 1A). We also included an n-terminus truncation which deletes the T-box domain; this construct expresses exons 14–23, including the bHLH domain, to serve as control along with WT-MGA. WT-MGA was localized in both the cytoplasm and nucleus, while the MGA-ex14-23 control was exclusively found in the cytoplasm, which suggests the n-terminal region is required for nuclear localization (Fig. 1B). The MGA p.S812* and MGA p.C623* mutant proteins were expressed exclusively in the nucleus, while the MGA p.E1953* truncation maintained normal localization in both the nucleus and cytoplasm. The retained nuclear localization of MGA truncations suggests these mutations could still be functional.
Fig. 1. MGA mutations lead to loss of molecular function.
A Schematic of full-length MGA with protein domains and mutations identified in RUNX1::RUNX1T1 AMLs and an experimentally-designed truncation of the T box domain [17]. Tag = GFP or Flag. Variant allele frequencies of MGA mutations identified in RUNX1::RUNX1T1 patients called by whole exome sequencing and RNA read counts, adapted from (Faber et al. [17]). B Immunofluorescent confocal microscopy of HEK293T cells expressing GFP-tagged MGA and the indicated truncations. MGA is labeled with eGFP (green), fibrillarin with Alex Fluor568 (red), tubulin with Alex Fluor647 (cyan), and nucleus with DAPI (blue). Images were collected on a Nikon C2 laser scanning confocal microscope using a 60X oil immersion optical lens. C Western blot analysis of immunoprecipitation of Flag-tagged MGA, HA-tagged-MAX, and indicated truncations in HEK293T cells probed for components of the ncPRC1.6 complex.
We then performed co-immunoprecipitation (Co-IP) assays followed by western blot to determine if MGA truncations interact with components of the ncPRC1.6 complex. We co-expressed flag-tagged WT-MGA, MGA-ex14-23, and MGA p.C623* with HA-tagged MAX in HEK293T cells and performed IP-western blots. We found that MGA p.C623* does not significantly bind L3MBTL2, PCGF6, or MAX, suggesting it loses its interaction with the ncPRC1.6 complex (Fig. 1C). Although nuclear MGA p.C623* does not interact with components of the ncPRC1.6 complex, it still could interact with the genome and hamper the genomic binding of MGA and the ncPRC1.6 complex in a dominant-negative manner at MGA binding sites. To examine this, we expressed HA-tagged MGA p.C623* in MOLM-13 cells and observed a global loss in genomic binding by Cleavage Under Targets and Release using Nuclease (CUT&RUN) assays. In addition to this genome-wide decrease in binding of the MGA mutant, binding was abolished at defined MGA target sites such as CCND2 (Supplementary Fig. 1A, B).
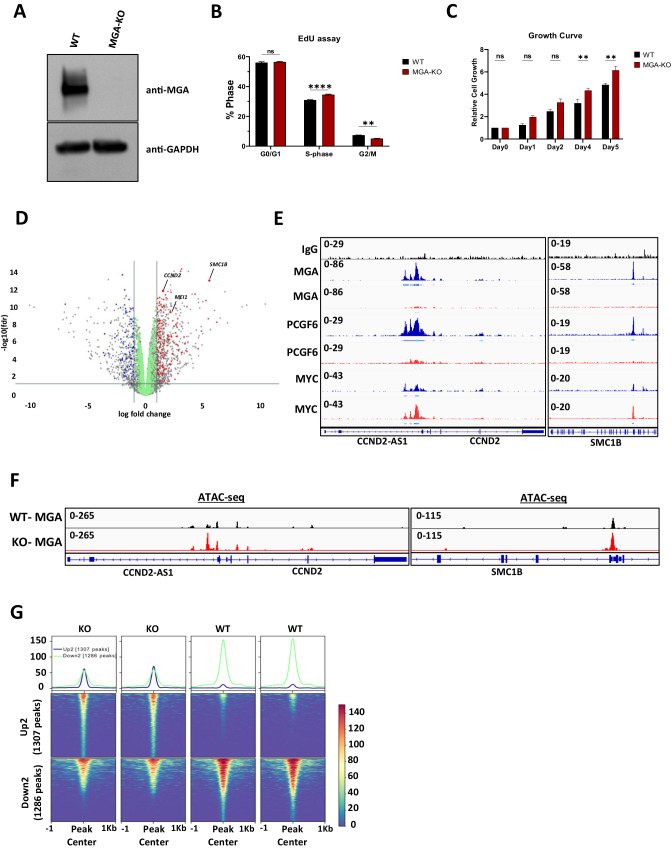
The loss of interaction with the ncPCR1.6 complex and the loss of DNA binding of MGA p.C623* supports the hypothesis that the truncating mutations of MGA identified in patients are loss-of-function mutations, leading to haploinsufficiency. To better understand the impact of MGA loss in hematopoietic cells, we designed a knock-out (KO) model of MGA using a CRISPR-based approach in MOLM-13 cells (Fig. 2A). The loss of MGA in these cells led to a slight increase in proliferation (Fig. 2B, C). RNA-sequencing (RNA-seq) analysis showed that the loss of MGA led to the upregulation of several genes that play a critical role in proliferation, genome stability, and tumorigenicity, including Cyclin D1 (CCND1), Cyclin D2 (CCND2), and Structural Maintenance of Chromosomes 1B (SMC1B) (Fig. 2D and Supplementary Fig. 1C) [19–21]. Supporting our RNA-seq data, we found that MGA binds to the CCND2 and SMC1B promoters and that MGA deletion reduces PCGF6 binding at these promoters and at the genome-wide level (Fig. 2E and Supplementary Fig. 1D). In contrast, MYC binding is minimally disrupted and potentially enhanced at CCND2 and SMC1B promoters (Fig. 2E, Supplementary Fig. 1D). We also reveal that the loss of MGA resulted in little to no change in the total number of TSS (transcription start site) occupied by H2AK119Ub1, H3K27me3, and H3K4me3. However, its deletion led to an increase in the total number of TSS with the active histone mark of H3K27Ac (Supplementary Fig. 1E, F). In addition, the intensity (log2Fold change) of active marks H3K4me3 and H3K27Ac at TSS are significantly higher, while the intensity of repressive mark H3K27me3 is lower in MGA-KO MOLM-13 cells, suggesting that MGA depletion leads to a more active epigenetic landscape (Supplementary Fig. 1G). Supporting these data, ATAC-seq analysis demonstrates that the loss of MGA leads to a more open chromatin state at the promoters of CCND2 and SMC1B with minimal global chromatin accessibility changes (Fig. 2F, G). These observations were validated in primary cord blood CD34+ cells immortalized by RUNX1::RUNX1T1, in which disruption of MGA by CRISPR leads to a reduction of PCGF6 binding to the CCND2 promoter associated with an overall increase in CCND2 expression, as well as an increase in self-renewal capacity (Supplementary Fig. 1H–J).
Fig. 2. Loss of MGA in vitro promotes cell growth.
A Western blot analysis of MGA in WT and MGA-KO cells. B Flow cytometric analysis of WT (n = 3) and MGA-KO (n = 3) MOLM-13 cells treated with EdU for 2 h and stained with DAPI. C Cell growth assay of WT and MGA-KO MOLM-13 cells over time. Statistics: Two-way ANOVA with Šídák’s multiple comparisons test; error bars indicate the standard error of the mean from three or more biological replicates. D Volcano plot of differentially expressed genes (DEGs) from MGA-KO MOLM-13 cells. Red & Blue points represent upregulated and downregulated genes, respectively, and that have wild-type MGA binding at their promoter-TSS as determined by CUT&RUN. Green points represent nonsignificant transcriptional changes with MGA binding at promoter-TSS. E CUT&RUN coverage plots of MGA, PCGF6, and MYC occupancy on CCND2 and SMC1B in WT (blue) and MGA-KO (red) MOLM-13 cells. F Chromatin occupancy at CCND2 and SMC1B promoters from ATAC-Seq in WT (black) and MGA-KO (red) MOLM-13 cells. G Replicate tornado plots of ATAC-Seq showing the signal enrichment at the most stringent threshold - Up2 and Down2 (FC > 2, FDR < 0.05).
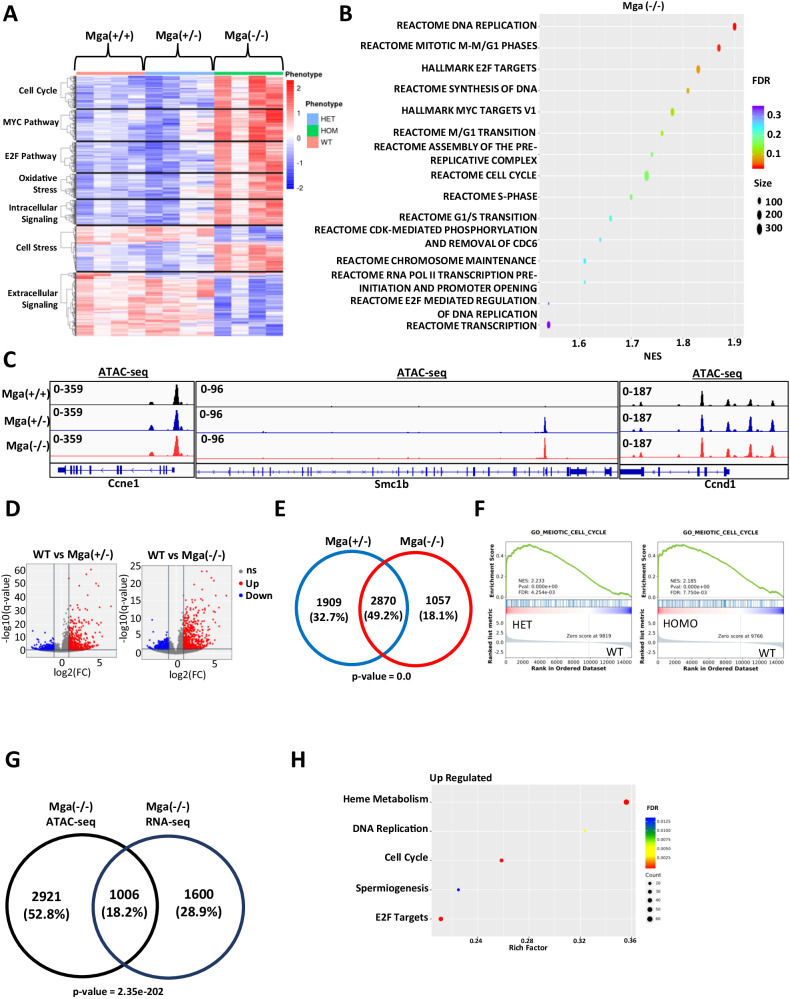
Considering that MGA loss leads to a cell growth advantage in leukemic cell lines, we wanted to assess the role of MGA in hematopoietic cell development. To this aim, we designed a conditional knock-out model in the C57BL/6 background, in which LoxP sites were introduced that flank exon 3 of Mga (Supplementary Fig. 2A). When crossed with Vav1-Cre, this leads to the deletion of exon 3 and the effective deletion of Mga in hematopoietic cells. This conditional hematopoietic cell KO model circumvented the known embryonic lethality of homozygous Mga loss [6], as offspring with both heterozygous (Vav1-cretg/+ x Mgafl/+ = Mga(±)) and homozygous (Vav1-cretg/+ x Mgafl/fl = Mga(−/−)) deletions of Mga induced by Vav1-Cre were viable with normal Mendelian ratios. Although the mutations identified in patients are heterozygous, we also established a cohort of mice with homozygous deletion of Mga to better define the functional role of Mga in hematopoiesis. RNA-seq on lineage- (lin-),cKit+ hematopoietic stem and progenitor cells (HSPCs) isolated from the bone marrow (BM) of Mga(+/+) (control), Mga(±), and Mga(−/−) mice demonstrated dysregulation of several critical pathways in Mga(−/−) but not in control or Mga(±) HSPCs, including the upregulation of Myc and E2F targets and the downregulation of extracellular signaling pathways including JAK-STAT, TGFβ, and TNFα signaling (Fig. 3A, B, Supplementary Fig. 2C). Similar to our findings in the MOLM-13 MGA knock-out model, Mga(−/−) HSPCs had significant upregulation of cell cycle and proliferation genes, including Ccne1, Ccnd1, Ccna2, Hdac2, and Smc1b (Supplementary Fig. 2D). To assess the epigenetic changes resulting from the loss of Mga, we performed CUT&RUN and ATAC-seq on cKit+ HSPCs from these mice. Unfortunately, an anti-murine MGA antibody compatible with CUT&RUN is not available. However, both heterozygous and homozygous deletions of Mga led to increases in H3K4me3 and H3K27Ac marks and a decrease in H3K27me3 at the promoters of genes such as Ccne1, Smc1b, and Cdk1 (Supplementary Fig. 3A–C). ATAC-seq analysis showed no significant changes in promoter accessibility of Ccne1 and Cdk1, however, the promoter of Smc1b, which is closed in control HSPCs, showed significantly more ATAC-seq signals in both Mga(±) and Mga(−/−) HSPCs (Fig. 3C). Globally, both heterozygous and homozygous deletions of Mga led to a significant increase in open chromatin at gene promoters compared to WT controls (5043 promoters (p < 0.05) and 3984 promoters (p < 0.05), respectively) (Fig. 3D, Supplementary Fig. 3D, E). There is a ~ 50% overlap of open promoters between Mga(±) and Mga(−/−) HSPCs leading to a nearly identical gene set enrichment with both conditions showing a strong enrichment of peaks of promoter regions of cell cycle-related genes (Fig. 3E, F). When comparing the upregulated genes from RNA-seq in Mga(−/−) HSPCs with the enriched ATAC-seq open promoter genes for Mga(−/−), we found an overlap of 1006(18.2%) genes (Fig. 3G). This overlapping gene list is enriched for cell cycle, pro-proliferation, and chromosome stability pathway activation, suggesting that these cells have improved fitness and proliferate faster than controls (Fig. 3H).
Fig. 3. Loss of Mga in vivo leads to aberrant cell cycle pathway signaling.
A Expression heatmap showing the enrichment of differentially regulated pathways in WT, Mga(±), and Mga(−/−) HSPCs isolated from BM. The z-scale is set to between 2 and −2; red = upregulation, blue = downregulation. B Rich factor plots of GSEA indicating upregulated pathways in Mga(−/−) HSPCs. The size of each dot represents gene count and the color represents FDR. C Coverage plots of ATAC-seq analysis of chromatin status at on Ccne1, Smc1b, and Ccnd1 promoters in WT (black), Mga(±) (blue), and Mga(−/−) (red) HSPCs. D ATAC-seq volcano plot for Mga(±) vs. WT and Mga(−/−) vs. WT. Statistical analysis for ATAC-seq are: Up/Down = log2(FC) > 1 and q-value < 0.05, ns = not significant. E Venn diagram of differentially expressed peaks at open promoters (p-value < 0.05) comparing Mga(±) and Mga(−/−) F GSEA plot for Mga(±) vs. WT and Mga(−/−) vs. WT. G Venn diagram of differentially expressed genes with increased accessibility (p-value < 0.05) for Mga(−/−) and upregulated genes (p-value < 0.05) in RNA-seq of Mga(−/−) HSPCs. H Rich factor plots showing the differentially regulated pathways of common up/open genes identified in G from RNA-seq/ATAC-seq analysis. The size of each dot represents gene count and the color represents FDR. Statistics: Venn diagram p-values calculated using hypergeometric distribution.
In addition to these molecular perturbations, loss of Mga in hematopoietic cells also has functional consequences. Mga(−/−) HSPCs have a significant increase in cell growth and proliferation and in self-renewal capacity when compared to controls, while Mga(±) HSPCs have only a slight advantage (Fig. 4A–D). These ex vivo analyses of Mga deficient cells propose that the loss of Mga may alter hematopoietic homeostasis. However, Mga(±) and Mga(−/−) mice at 3–6 months displayed normal CBCs and demonstrated no defect in the development of B cells (B220+), T cells (CD3e+), or myeloid cells (CD11b+) (Supplementary Fig. 3F, G). Ki67 staining of the B cell, T cell, or myeloid cell populations showed a slight increase in cycling B and T cells but not myeloid cells (Supplementary Fig. 3H). HSPC immunophenotyping by flow cytometry showed no change in the lymphoid progenitors (lin- cKit-, Sca-1low) or myeloid progenitors (lin-, cKit+) in both Mga(±) and Mga(−/−) mice, however, there was a slight decrease in MPP3/4 (Lin- cKit+ Sca-1 + CD48 + CD150-) populations for Mga(−/−) but not Mga(±) mice (Fig. 4E, F). Aged control, Mga(±), and Mga(−/−) mice (16 months) did not spontaneously develop a hematopoietic malignancy (data not shown). Despite the observed self-renewal phenotype, serial noncompetitive BM transplants of HSPCs isolated from control, Mga(±), and Mga(−/−) mice (Supplementary Fig. 4A) lead to no significant changes in WBC counts or in the ability to reestablish mature blood cell production over a 16 week period (Supplementary Fig. 4B–G). However, primary recipient mice that received Mga(−/−) HSPCs had a slight decrease in LSKs and an increase in myeloid progenitor cells (Supplementary Fig. 4B, D, E).
Fig. 4. Loss of Mga in vivo primes myeloid progenitors for a growth advantage.
A Cell growth assay of lineage-negative HSPCs from WT (n = 3), Mga(±) (n = 3), and Mga(−/−) (n = 3) mice. B Flow cytometric analysis of lineage-negative HSPCs from WT (n = 7), Mga(±) (n = 7), and Mga(−/−) (n = 7) mice treated ex vivo with EdU for 2 h and stained with DAPI. C Colony Forming Units (CFU) showing the total number of colonies from lineage-negative HSPCs from WT (n = 9), Mga(±) (n = 12), and Mga(−/−) (n = 9) mice. D Relative distribution of colony subtypes in C, E = erythroid; GEMM = granulocyte, erythrocyte, monocyte, megakaryocyte; GM = granulocyte, monocyte; M = monocyte; G = granulocyte. Percent E and absolute cell number F from the flow cytometric analysis of HSPCs from whole bone marrow harvested from WT (n = 6), Mga(±) (n = 6), and Mga(−/−) (n = 7) mice; LSK = lin-, Sca-1+, cKit+; CLP = lin-, Sca-1lo, cKit-, CD127+; Myeloid Progenitors = lin-, Sca-1-, cKit+; LT-HSC = Lin- cKit+ Sca-1+CD48- CD150 + CD34-; ST-HSC = Lin- cKit+ Sca-1 + CD48- CD150-; MPP3/4 = Lin- cKit+ Sca-1 + CD48 + CD150-; MPP2 = Lin- cKit+ Sca-1 + CD48 + CD150+. Absolute cell counts where done by dividing cell frequencies by the average number of cells isolated from both hindlimbs (hips/tibia/femur). Statistics: A One-way ANOVA with Bonferroni correction (*p < 0.05, **p < 0.01). Error bars indicate the standard error of the mean from three or more biological replicates. B–E Two-way ANOVA with Dunnett’s multiple comparisons test.
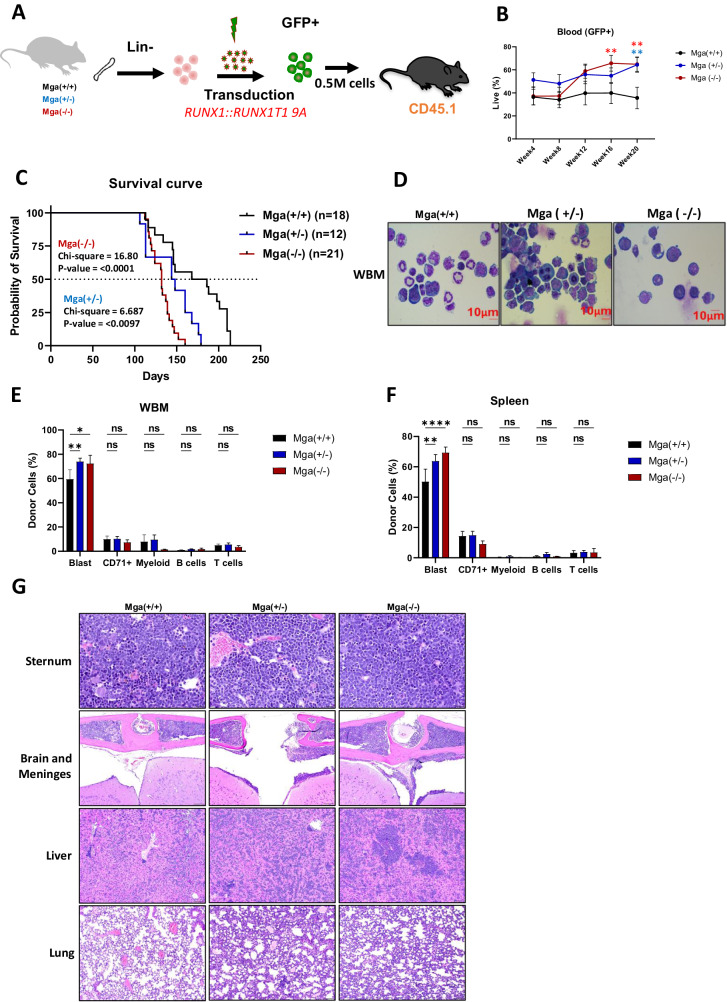
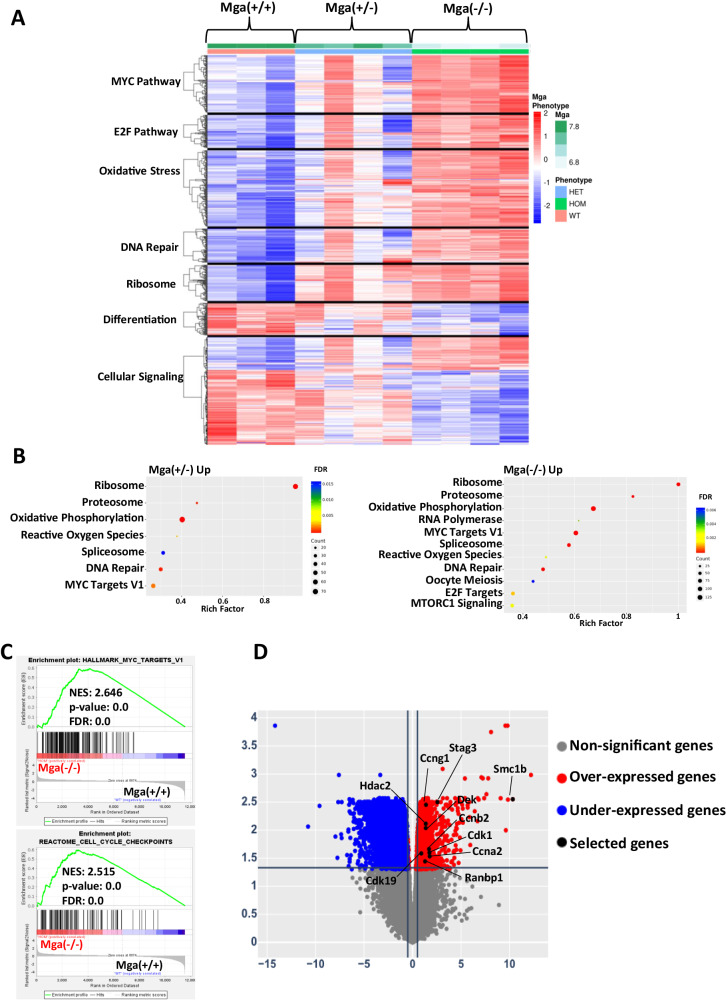
These data show that Mga deficiency is not sufficient for an overt hematopoietic malignant phenotype, but do suggest that the loss of Mga may promote a transcriptional and epigenetic landscape that could enhance the aggressive phenotypes of leukemia in the presence of an oncogenic driver. This was tested using an established murine leukemia model using retroviral expression of RUNX1::RUNX1T1 9a, which encodes a shortened form of RUNX1::RUNX1T1, that induces the rapid development of AML (Fig. 5A) [22]. Ex vivo RUNX1::RUNX1T1 9a expression in Mga(+/+), Mga(±), and Mga(−/−) HSPCs showed that Mga(−/−) HSPCs have an increase in cell growth when compared to controls, while the Mga(±) HSPCs had little to no change, however, both had a significant increase in CFU colony formation and serial replating (Supplementary Fig. 5A–C). These data suggest that Mga deficiency may enhance the development of leukemia in a RUNX1::RUNX1T1 9 A model. This was confirmed in vivo as Mga(±) and Mga(−/−) HSPCs expressing RUNX1::RUNX1T1 9 A rapidly expanded and developed into leukemias characterized by immature GFP+, cKit+ myeloid progenitor-like cells when transplanted into lethally irradiated recipient mice (Fig. 5C and Supplementary Fig. 5D, E, H). Mga deficiency in RUNX1::RUNX1T1 9 A expressing cells led to a significantly shortened leukemia latency with a median survival of 177 days for controls, 146 days for Mga(±) mice, and 132 days for Mga(−/−) mice (Fig. 5C). Immunophenotyping of the BM and spleen confirmed that RUNX1::RUNX1T1 9 A expressing tumors were cKit+ and morphologically resembled immature blast cells (Fig. 5D–F and Supplementary Fig. 5F, G). Pathological analysis of the bones (sternum), brain and meninges, liver, and lungs further show that RUNX1::RUNX1T1 9 A expression in Mga(±) and Mga(−/−) HSPCs leads to mice that are more severely infiltrated by leukemia compared to control mice (Fig. 5G). To determine if the genomic binding of HA-tagged RUNX1::RUNX1T1 is altered due to the loss of Mga, we performed CUT&RUN on GFP+ sorted tumor cells isolated from the spleen. These data show that the loss of Mga does not affect RUNX1::RUNX1T1’s target binding which is supported by our CD34 model (Supplementary Fig. 1H and Supplementary Fig. 6A, B). Transcriptional analysis by RNA-seq of sorted GFP + RUNX1::RUNX1T1 9 A expressing tumors from Mga(±) and Mga(−/−) moribund mice revealed upregulation of pro-proliferation pathways including MYC, ribosome, and oxidative phosphorylation, while Mga(−/−) tumors interestingly had downregulation of differentiation pathways including angiogenesis and hematopoietic cell lineage (Fig. 6A–C). Mga(−/−) leukemic cells demonstrated a higher expression of Smc1b as well as an increased expression of cell cycle-related genes such as Ccng1, Ccnd3, Cdk1, Cdk19, Dek, and Ranbp1 (Fig. 6D) [23, 24]. These transcriptional data support our cell growth and serial CFU replating phenotypes. These tumors were serially transplantable into sublethally irradiated mice and the secondary mice quickly developed AML 4–10 weeks post-transplantation. There was no significant difference between Mga(+/+) and Mga(±) mice, while Mga(−/−) mice had a slightly longer latency (Supplementary Fig. 7A, B).
Fig. 5. Mga deficiency enhances RUNX1::RUNX1T1 leukemia development.
A Schematic of retroviral transduction of RUNX1::RUNX1T1 9 A in lineage-negative HSPCs from WT, Mga(±), and Mga(−/−) mice (CD45.2) and transplantation into lethally irradiated recipient mice (C57BL/6, CD45.1). B GFP expression in the peripheral blood over time. C Survival curve of recipient mice. Statistics were done using Mantel-Cox test in GraphPad Prism. D Cytospins of whole bone marrow (WBM) harvested at sacrifice stained with Wright–Giemsa; magnification 60X. Flow cytometric analysis of WBM F and Spleen G harvested from terminal recipient mice stained with tumor panel from Supplemental Table 1. Blast = GFP + , cKit + , lineage-negative. G. Hematoxylin and eosin H, E -stained sections of sternum marrow, brain meninges, liver, and lungs harvested from indicated terminal recipient mice. Magnification 40X. Statistics: B One-way ANOVA with Bonferroni correction (*p < 0.05, **p < 0.01). Error bars indicate the standard error of the mean from three or more biological replicates. F, G Two-way ANOVA with Dunnett’s multiple comparisons test.
Fig. 6. Mga deficient tumors lead to hyperproliferative transcriptional profile.
A Heatmap showing the enrichment of differentially regulated pathways in RUNX1::RUNX1T1 9 A tumors (GFP + ) from WT, Mga(±), and Mga(−/−) cells. The z-scale is set to between 2 and -2; red=upregulation, blue = downregulation. B Rich factor plot of top upregulated pathways from RUNX1::RUNX1T1 9 A expression in Mga(±) and Mga(±) cells. The size of each dot represents gene count and the color represents FDR. C GSEA enrichment plots of selected upregulated pathways in RUNX1::RUNX1T1 expressing Mga(−/−) GFP+ cells vs. Mga(+/+) GFP+ cells isolated from spleens. D Volcano plot of differentially expressed genes (DEGs) from RUNX1::RUNX1T19A expression in Mga(−/−) vs. Mga(+/+)cells. Statistics: One-way ANOVA with Bonferroni correction (*p < 0.05, **p < 0.01). Error bars indicate the standard error of the mean from three or more biological replicates.
Discussion
Here we generated both human and mouse models to assess the role of MGA in normal and malignant hematopoiesis. Our data support the well-documented antiproliferative role of MGA and show that loss of MGA leads to an open and active epigenetic chromatin status, and subsequently the upregulation of several critical pathways for cell growth and proliferation (Fig. 7) [2, 3]. Previous reports in non-hematopoietic models show that components of the ncPRC1.6 complex, such as MGA and E2F6, regulate transcriptional control of critical regulators of meiosis such as STAG3, SMC1B, MEIOC, and CNTD1 and overall result in transcriptional silencing [2, 25]. While components of the ncPRC1.6 complex are known to play a critical role in embryonic stem cell maintenance, as depletion of MGA, L3MBTL2, and PCGF6 leads to defects in pluripotency, proliferation, and differentiation, the impact of disrupting this complex has not been thoroughly evaluated in hematopoietic cells [3, 6, 26]. Our hematopoietic MGA-KO models support these reports, revealing a similar transcriptional landscape, including the increased expression of genes such as Cdk1, Smc1b, and Ccne1, when Mga is disrupted. Ccne1 and Cdk1 are known regulators of cell cycle progression, while Smc1b is critical for genomic stability and is suggested to play a role in cell proliferation [20, 21, 27, 28]. We believe the upregulation of these genes and others is due to the direct loss of MGA-dependent ncPRC1.6 binding and regulation leading to the serial replating and cell growth advantage of Mga(−/−) HSPCs. Overexpression of these genes has also been observed in several cancer types including AML, hepatocellular carcinoma, and ovarian carcinoma [29–31]. The lack of changes in H2AK119Ub1 upon MGA depletion was surprising. However, the decrease in H3K27me3 and increase in H3K27Ac suggest both that there are functional redundancies with other PRC1 complexes and that the binding of MGA-dependent ncPRC1.6 to the loci is more critical for transcriptional repression [32, 33]. To further support this, the open chromatin of MGA target genes in MGA deficient cells is likely due to the loss of L3MBTL2, which can regulate chromatin compaction regardless of histone modification status [34].
Fig. 7. Model of MGA deficiency.
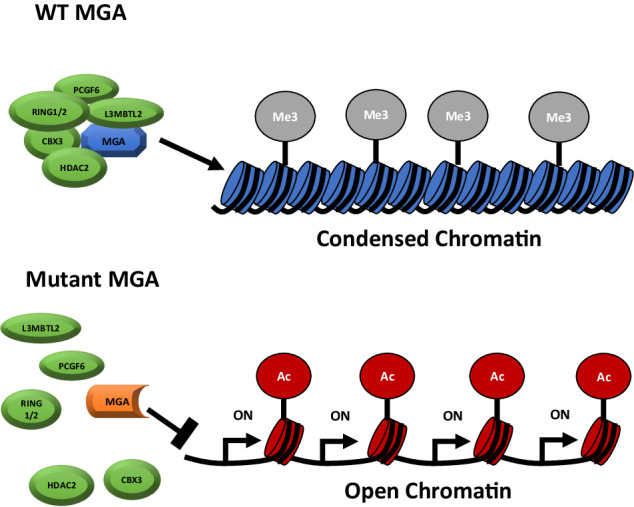
A Model of MGA deficiency in hematopoietic cells. The expression of full-length MGA leads to ncPRC1.6 complex formation and the deposition of repressive H3K27me3 of MYC target genes leading to a condensed and transcriptionally repressed chromatin status (top). Somatic mutations of MGA as seen in AML; lead to the abolishment of ncPRC1.6 complex formation leading to an open and transcriptionally active chromatin status.
Despite these clear transcriptional and epigenetic changes, the loss of MGA in the hematopoietic compartment alone is not sufficient to promote profound hematopoietic defects or the development of a hematopoietic neoplasm, even under stress conditions such as serial BM transplantation and 5-FU treatments (data not shown). These findings support the notion that the loss of function mutations in MGA observed in hematologic malignancies are cooperating mutations that provide a transcriptional state that may enhance the effects of tumor drivers. In support of this hypothesis, the latency of leukemia induced by a viral model of RUNX1::RUNX1T1 9 A was significantly shortened when expressed in both Mga(±) and Mga(−/−) hematopoietic cells, and the resulting leukemic cells displayed a more immature phenotype.
RUNX1::RUNXT1 AMLs have been shown to be dependent on the gene activation of D cyclins such as CCND2 and CCND1 [35, 36]. Overall, we propose that the shortened latency in our RUNX1::RUNXT1 9 A model is likely due to MGA-deficient cells being primed for enhanced proliferation via the upregulation of cyclin genes and MYC-targets, as shown by our transcriptional analysis of RUNX1::RUNXT1 9 A tumors. Consistent with this, our previous work identified gain-of-function mutations in CCND2 in RUNX1::RUNX1T1 AMLs, which has been supported by other studies [37]. The findings presented here suggest that MGA loss may phenocopy these CCND2 mutations. Further supporting this claim is the observation that MGA and CCND2 were mutually exclusive in our previous study [17]. Collectively, our data highlight MGA as an important regulator of pathways critical for leukemia development, such as MYC activation, cell cycle, and oxidative phosphorylation, and that loss of function mutations identified in patients can functionally cooperate with the RUNX1::RUNX1T1 oncoprotein [21, 38–41]. Importantly, we have also defined the in vivo consequences of Mga loss by using a conditional knock-out approach that circumvents the embryonic lethality of constitutional knock-out models, thus providing the scientific community with an important model.
Methods
Animals
We used Ingenious Targeting Laboratory (Stony Brook, NY, USA) to generate the conditional Mga KO model in C57BL/6 mice that bore a LoxP-flanked exon 3 of Mga using embryonic stem cell-based gene targeting. CD45.1 and C57BL/6 mice were obtained from Jackson Laboratory. Blood, WBM, and spleens were harvested as previously described [42]. HSPCs were isolated from WBM (both hips, femurs, and tibias) using EasySep™ Mouse Hematopoietic Cell Isolation kit. All Animal studies were approved by St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
RNA-Seq analysis
MOLM-13 cells or Lin-,cKit+ HSPCs were harvested and RNA was extracted using a quick-RNA Microprep kit (Zymo Research, CA). RNA-seq was done using TruSeq Stranded Total RNA library kit (Illumina, CA) as previously described [43]. The RNA-Seq paired-end reads were mapped to the mouse mm10 genome or human hg38 genome using STAR and quantified using RSEM [44, 45]. Differentially expressed gene analysis and GSEA was done as previously described [42, 46].
CUT&RUN analysis
MOLM-13 cells or Lin-, cKit+ HSPCs were harvested at 500,000 cells for each antibody probe. Genomic localization profiling was performed using Cleavage Under Targets and Release Using Nuclease kit (CUT&RUN) according to the manufacture’s protocols (Epicypher, NC) as previously described [47]. Library preparations of up to 6 ng of isolated DNA fragments were done using NEBNEXT Ultra library prep Kit with AMPure XP beads (Beckman Colter, CA) following the NEBNext® Ultra™ DNA Library Prep Protocol for Illumina® With Size Selection (E7370) V.2 (fragment size < 70 bp). CUT&RUN libraries were sequenced using Novaseq 6000 by performing 100 cycles of paired-end sequencing (200 cycles total). CUT&RUN bam files were pre-processed and peaks were called using methods previously described [48]. Peaks from replicate samples were merged and fragments covering those peaks were counted using bedtools version 2.25 [49]. Peaks differentially expressed between treatment groups were determined using a combination of the R tools limma and voom as previously described [46]. Peaks were annotated using HOMER version 4.10 [50]. The annotation was simplified by combining the HOMER annotations into 3 categories. Peaks within 3 kb of a transcription start site were called ‘TSS ± 3 kb’. Peaks annotated as intergenic were labeled as “Intergenic”. Peaks annotated as non-coding were labeled “non-coding” and all other peaks were labeled ‘Genebody’.
ATAC-seq analysis
MOLM-13 cells or Lin-,cKit+ HSPCs were harvested at 50,000 cells. Open chromatin status was assessed using the assay for transposase-accessible chromatin via sequencing (ATAC-seq) kit (Active Motif, CA) following the manufacturer’s protocols. Library prep, included in kit, was done following the manufacturer’s protocols. ATAC-seq libraries were sequenced using Novaseq 6000 by performing 100 cycles of paired-end sequencing (200 cycles total). Analysis was done by the Center for Applied Bioinformatics at SJCRH. Raw reads in fastq format were processed with Trim-Galore tool (v0.4.4, Krueger F. (2012)), in order to remove potential adapters and quality trim 3’ end of reads with cutadapt program, followed by FastQC analysis [51, 52]. A quality score cutoff of Q20 was used and the first 15 bp of each reads were also clipped to reduce the GC bias. Next, reads were mapped to the human reference genome (hg38; GRCh38.p12) with BWA mem (0.7.17-r1188), then converted to BAM format and deduplicated with fq2bam (v3.0.0.6) [53]. Subsequently, uniquely mapped properly paired reads were extracted with samtools (v1.2), and fragments were extracted with bedtools(v2.24.0) [49, 54]. Chromatin status (open/closed) was defined by a q-value of ≤ 0.05 and a log2FC of ± 2 compared to indicated WT control.
Data presentation and statistical analysis
Data graphs were created using GraphPad Prism (San Diego, CA). One-way ANOVA with Bonferroni correction, Two-way ANOVA with Dunnett’s or Šídák’s multiple comparisons test was used for statistical analyses where indicated. For survival curves, Kaplan-Meier estimates were computed in GraphPad Prism and statistical analysis was done using the Mantel-Cox test. Venn diagram p-values were calculated using hypergeometric distribution [55]. For One-way ANOVA with Bonferroni correction statistical significance was set at the following: *p < 0.05, and **p < 0.01 compared to indicated controls.
Study approval
All animal studies and experimental procedures were performed in accordance with protocols approved by St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee.
Supplementary information
Supplemental - Functional Characterization of Cooperating MGA Mutations in RUNX1-RUNX1T1 Acute Myeloid Leukemia.
Acknowledgements
The work was funded by the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital and funds from the US National Institutes of Health (NIH), including R01 HL144653 (JMK) and F32HL152484-01 (MET). The content is the responsibility of the authors and does not necessarily represent the official views of the NIH. JMK holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. Supporting resources for these studies were provided by the Shared Resources provided through the St. Jude Comprehensive Cancer Center (P30-CA21765). The authors would like to thank Juan Barajas and Masayuki Umeda for their critical review of the manuscript, and past and previous members of the Klco lab for their support and reagent generation.
Author contributions
All the authors have approved their authorship, gave input to the manuscript, and contributed as follows: Conceptualization, MET, WQ, and JMK; Methodology, MET, and JMK; Investigation, all authors; Formal Analysis, MET, MW, JM, WR, BX, and LJ; Writing Draft, MET, and JMK; Visualization, MET; Funding acquisition, MET, and JMK; Supervision, JMK.
Data availability
RNA-seq, CUT&RUN, and ATAC-seq data will be deposited into Gene Expression Omnibus (GEO) (GSE253753).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-024-02193-y.
References
- 1.Hurlin PJ, Steingrìmsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–28. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical polycomb repressive complex PRC1.6. PLoS Genet. 2018;14:e1007193. doi: 10.1371/journal.pgen.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao W, Tong H, Huang Y, Yan Y, Teng H, Xia Y, et al. Essential role for polycomb group protein Pcgf6 in embryonic stem cell maintenance and a noncanonical polycomb repressive complex 1 (PRC1) integrity. J Biol Chem. 2017;292:2773–84. doi: 10.1074/jbc.M116.763961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Wang C, Zhu Y, Su T, Dong L, Huang Y, et al. Mga safeguards embryonic stem cells from acquiring extraembryonic endoderm fates. Sci Adv. 2021;7:eabe5689. doi: 10.1126/sciadv.abe5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burn SF, Washkowitz AJ, Gavrilov S, Papaioannou VE. Postimplantation Mga expression and embryonic lethality of two gene-trap alleles. Gene Expr Patterns. 2018;27:31–5. doi: 10.1016/j.gep.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Washkowitz AJ, Schall C, Zhang K, Wurst W, Floss T, Mager J, et al. Mga is essential for the survival of pluripotent cells during peri-implantation development. Development. 2015;142:31–40. doi: 10.1242/dev.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo YS, Kim MS, Yoo NJ, Lee SH. Somatic mutation of a candidate tumour suppressor MGA gene and its mutational heterogeneity in colorectal cancers. Pathology. 2016;48:525–7. doi: 10.1016/j.pathol.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llabata P, Mitsuishi Y, Choi PS, Cai D, Francis JM, Torres-Diz M, et al. Multi-omics analysis identifies MGA as a negative regulator of the MYC pathway in lung adenocarcinoma. Mol Cancer Res. 2020;18:574–84. doi: 10.1158/1541-7786.MCR-19-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathsyaraja H, Catchpole J, Freie B, Eastwood E, Babaeva E, Geuenich M, et al. Loss of MGA repression mediated by an atypical polycomb complex promotes tumor progression and invasiveness. Elife. 2021;10:e64212. doi: 10.7554/eLife.64212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Paoli L, Cerri M, Monti S, Rasi S, Spina V, Bruscaggin A, et al. MGA, a suppressor of MYC, is recurrently inactivated in high risk chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1087–90. doi: 10.3109/10428194.2012.723706. [DOI] [PubMed] [Google Scholar]
- 14.Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47:1061–6. doi: 10.1038/ng.3358. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–8. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun QY, Ding LW, Tan KT, Chien W, Mayakonda A, Lin DC, et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD) Leukemia. 2017;31:1–10. doi: 10.1038/leu.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faber ZJ, Chen X, Gedman AL, Boggs K, Cheng J, Ma J, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–6. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratti S, Lonetti A, Follo MY, Paganelli F, Martelli AM, Chiarini F, et al. B-ALl complexity: is targeted therapy still a valuable approach for pediatric patients? Cancers. 2020;12:3498. doi: 10.3390/cancers12123498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding ZY, Li R, Zhang QJ, Wang Y, Jiang Y, Meng QY, et al. Prognostic role of cyclin D2/D3 in multiple human malignant neoplasms: a systematic review and meta-analysis. Cancer Med. 2019;8:2717–29. doi: 10.1002/cam4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannini L, Cucco F, Quarantotti V, Amato C, Tinti M, Tana L, et al. SMC1B is present in mammalian somatic cells and interacts with mitotic cohesin proteins. Sci Rep. 2015;5:18472. doi: 10.1038/srep18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi F, Wang Z, Liu J, Zhang Y, Wang Z, Xu H, et al. Structural maintenance of chromosomes protein 1: role in genome stability and tumorigenesis. Int J Biol Sci. 2017;13:1092–9. doi: 10.7150/ijbs.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–89. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei Z, Duan X, Li Q, Li Q, Wang Y. High expression of Ran binding protein 1 predicts poor outcomes in hepatocellular carcinoma patients: a Cancer Genome Atlas database analysis. J Gastrointest Oncol. 2021;12:2966–84. doi: 10.21037/jgo-21-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L, Huang X, Zhang W, Zhao H, Wu G, Lv F, et al. Critical role of DEK and its regulation in tumorigenesis and metastasis of hepatocellular carcinoma. Oncotarget. 2016;7:26844–55. doi: 10.18632/oncotarget.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storre J, Schafer A, Reichert N, Barbero JL, Hauser S, Eilers M, et al. Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J Biol Chem. 2005;280:41380–6. doi: 10.1074/jbc.M506797200. [DOI] [PubMed] [Google Scholar]
- 26.Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–32. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campaner S, Viale A, De Fazio S, Doni M, De Franco F, D’Artista L, et al. A non-redundant function of cyclin E1 in hematopoietic stem cells. Cell Cycle. 2013;12:3663–72. doi: 10.4161/cc.26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao S, Chen C, Cheng T. Cell cycle regulation of hematopoietic stem or progenitor cells. Int J Hematol. 2016;103:487–97. doi: 10.1007/s12185-016-1984-4. [DOI] [PubMed] [Google Scholar]
- 29.Hedblom A, Laursen KB, Miftakhova R, Sarwar M, Anagnostaki L, Bredberg A, et al. CDK1 interacts with RARgamma and plays an important role in treatment response of acute myeloid leukemia. Cell Cycle. 2013;12:1251–66. doi: 10.4161/cc.24313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosen DG, Yang G, Deavers MT, Malpica A, Kavanagh JJ, Mills GB, et al. Cyclin E expression is correlated with tumor progression and predicts a poor prognosis in patients with ovarian carcinoma. Cancer. 2006;106:1925–32. doi: 10.1002/cncr.21767. [DOI] [PubMed] [Google Scholar]
- 31.Nie H, Wang Y, Yang X, Liao Z, He X, Zhou J, et al. Clinical significance and integrative analysis of the SMC family in hepatocellular carcinoma. Front Med. 2021;8:727965. doi: 10.3389/fmed.2021.727965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng Z, Gao Z. Mammalian PRC1 complexes: compositional complexity and diverse molecular mechanisms. Int J Mol Sci. 2020;21:8594. doi: 10.3390/ijms21228594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scelfo A, Fernandez-Perez D, Tamburri S, Zanotti M, Lavarone E, Soldi M, et al. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol Cell. 2019;74:1037–52.e7. doi: 10.1016/j.molcel.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell. 2011;42:438–50. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Soria N, McKenzie L, Draper J, Ptasinska A, Issa H, Potluri S, et al. The oncogenic transcription factor RUNX1/ETO corrupts cell cycle regulation to drive leukemic transformation. Cancer Cell. 2018;34:626–42.e8. doi: 10.1016/j.ccell.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swart LE, Heidenreich O. The RUNX1/RUNX1T1 network: translating insights into therapeutic options. Exp Hematol. 2021;94:1–10. doi: 10.1016/j.exphem.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisfeld AK, Kohlschmidt J, Schwind S, Nicolet D, Blachly JS, Orwick S, et al. Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia. 2017;31:1278–85. doi: 10.1038/leu.2016.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C, Hao X, Lai X, Liu L, Zhu J, Shao H, et al. Oxidative phosphorylation enhances the leukemogenic capacity and resistance to chemotherapy of B cell acute lymphoblastic leukemia. Sci Adv. 2021;7:eabd6280. doi: 10.1126/sciadv.abd6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng M, Huang Y, Zhang L, Zhao X, Hou Y. Targeting mitochondrial oxidative phosphorylation eradicates acute myeloid leukemic stem cells. Front Oncol. 2022;12:899502. doi: 10.3389/fonc.2022.899502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgado MD, Leon J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–16. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boukaba A, Liu J, Ward C, Wu Q, Arnaoutov A, Liang J, et al. Ectopic expression of meiotic cohesin generates chromosome instability in cancer cell line. Proc Natl Acad Sci USA. 2022;119:e2204071119. doi: 10.1073/pnas.2204071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdelhamed S, Thomas ME, 3rd, Westover T, Umeda M, Xiong E, Rolle C, et al. Mutant Samd9l expression impairs hematopoiesis and induces bone marrow failure in mice. J Clin Invest. 2022;132:e158869. doi: 10.1172/JCI158869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz JR, Ma J, Lamprecht T, Walsh M, Wang S, Bryant V, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8:1557. doi: 10.1038/s41467-017-01590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas ME, 3rd, Abdelhamed S, Hiltenbrand R, Schwartz JR, Sakurada SM, Walsh M, et al. Pediatric MDS and bone marrow failure-associated germline mutations in SAMD9 and SAMD9L impair multiple pathways in primary hematopoietic cells. Leukemia. 2021;35:3232–44. doi: 10.1038/s41375-021-01212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson MR, Reske JJ, Holladay J, Neupane S, Ngo J, Cuthrell N, et al. ARID1A mutations promote P300-d–ependent endometrial invasion through super-enhancer hyperacetylation. Cell Rep. 2020;33:108366. doi: 10.1016/j.celrep.2020.108366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Xu B, He M, Zong X, Cunningham T, Sha C, et al. Control of Foxp3 induction and maintenance by sequential histone acetylation and DNA demethylation. Cell Rep. 2021;37:110124. doi: 10.1016/j.celrep.2021.110124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12.
- 52.Leggett RM, Ramirez-Gonzalez RH, Clavijo BJ, Waite D, Davey RP. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front Genet. 2013;4:288. doi: 10.3389/fgene.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:16438–43. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental - Functional Characterization of Cooperating MGA Mutations in RUNX1-RUNX1T1 Acute Myeloid Leukemia.
Data Availability Statement
RNA-seq, CUT&RUN, and ATAC-seq data will be deposited into Gene Expression Omnibus (GEO) (GSE253753).