Abstract
We present evidence of formation of an intramolecular parallel triple helix with T•A.T and G•G.C base triplets (where • represents the hydrogen bonding interaction between the third strand and the duplex while . represents the Watson–Crick interactions which stabilize the duplex). The third GT strand, containing seven GpT/TpG steps, targets the polypurine sequence 5′-AGG-AGG-GAG-GAG-3′. The triple helix is obtained by the folding back twice of a 36mer, formed by three dodecamers tethered by hydroxyalkyl linkers (-L-). Due to the design of the oligonucleotide, the third strand orientation is parallel with respect to the polypurine strand. Triple helical formation has been studied in concentration conditions in which native gel electrophoresis experiments showed the absence of intermolecular structures. Circular dichroism (CD) and UV spectroscopy have been used to evidence the triplex structure. A CD spectrum characteristic of triple helical formation as well as biphasic UV and CD melting curves have been obtained in high ionic strength NaCl solutions in the presence of Zn2+ ions. Specific interactions with Zn2+ ions in low water activity conditions are necessary to stabilize the parallel triplex.
INTRODUCTION
Triple helices are supposed to exist in eukaryotic cells involved in transcriptional regulation and in replication (1–4). Accessibility of the target sequences in cell nuclei and in vivo persistence of DNA triple helices have been proved using oligonucleotide–psoralen conjugates (5–7). Proteins have been shown to recognize specifically pyrimidine motif triplexes (8) or purine motif triplexes (9). Classically, recognition of a purine stretch in duplex DNA by an oligonucleotide residing in the major groove can be achieved either by a pyrimidine sequence oriented in a parallel direction with respect to the purine duplex strand (pyrimidine motif triplex Y•R.Y with C•G.C and T•A.T base triples, where • represents the hydrogen bonding interaction between the third strand and the duplex while . represents the Watson–Crick interactions which stabilize the duplex) or by a purine sequence (purine motif triplex R•R.Y with G•G.C and A•A.T base triples). In this case third strand orientation is usually considered to be antiparallel, although G•G.C base triplets were observed by X-ray crystal diffraction with both parallel and antiparallel orientations (10). Triple helical structures can also be obtained with a third strand containing pyrimidines and purines (GT-rich oligonucleotides) through formation of G•G.C and T•A.T base triples. Most reports concerning such GT third strand triple helices also considered an antiparallel third strand orientation (11–16). It has been proposed that the number of GpT/TpG steps in the third strand will determine the orientation of the third strand with respect to the purine duplex strand. Above three junctions the antiparallel third strand orientation should be favored (17). Molecular mechanics studies showed that in the case of a third GT strand, even if initially the parallel triplex is favored, the parallel and antiparallel orientations become energetically comparable when the number of thymines in the third strand is increased (18). A systematic experimental study of intermolecular triplex formation in the presence of Mg2+ ions has shown that, in these conditions, hybridization occurred only in an antiparallel orientation and only for high G third strand content (>65%) in sequences with less than four GpT/TpG steps. No parallel triplex has been detected in that study, and moreover, when the amount of GpT/TpG steps was higher than four no triplex at all, not even antiparallel, was formed (19). However, a recent work performed by UV and circular dichroism (CD) spectroscopy has proposed that Mg2+ ions could stabilize an intramolecular parallel triple helix with G4T4G4 as the third strand (66% G containing two GpT/TpG steps) (20).
We have shown in a previous study the formation of an antiparallel triple helix by a GT-rich third strand (66% G) containing seven GpT/TpG steps (21). This intramolecular triplex was obtained by folding back twice on itself of the 5′-CTC CTC CCT CCT -L- AGG AGG GAG GAG -L- GTG GTG GGT GGT-3′ oligonucleotide, where -L- is the hydroxyalkyl linker pO(CH2CH2O)3p. The use of such a molecule had been adopted because it: (i) avoids strand stoicheiometry problems; (ii) allows the unambiguous definition of the third strand orientation; (iii) increases the stability of the triplex when compared to a triplex formed by separate strands with identical sequences; and (iv) reduces the possibility of the formation of intermolecular structures by GT-rich oligonucleotides such as tetraplexes (22–24) or aptamers (25–28). The present investigation is aimed at studying the formation of the intramolecular triple helix involving the same strands but with a parallel third strand orientation. We have used for this study another 36mer formed by three dodecamers tethered by non-nucleotidic linkers, 5′-TGG TGG GTG GTG -L- CTC CTC CCT CCT -L- AGG AGG GAG GAG-3′. Due to the design of the oligonucleotide (respective positions of the dodecamers) the third strand orientation should be parallel. Investigation of triple helical formation has been performed by native gel electrophoresis, CD spectroscopy and thermal denaturation followed by UV and CD. Such a parallel GT triplex with a third strand containing an important number of GpT/TpG steps was expected to be difficult to form. Purine motif triple helices usually require the presence of divalent ions, but not all divalent ions have equivalent efficiencies. For instance Mg2+ ions which have been used in the above recalled studies are unable to induce R•R.Y triplexes in d(GA.TC)n sequences (29) while Zn2+ ions do (30). In many cases Zn2+ ions have proved to be particularly efficient in antiparallel triple helix stabilization (31–33). Therefore, we have investigated whether Zn2+ ions could also favor the formation of a parallel triplex. We give here evidence of the stabilization of this parallel triplex by interaction with Zn2+ ions. A new CD spectrum has been obtained for parGT in high ionic strength NaCl solutions after addition of Zn2+ ions. The presence of sodium ions is required so as to avoid interactions between the Zn2+ ions and the phosphate groups, and favor specific binding of the Zn2+ ions on the bases. Biphasic melting profiles are found by UV and CD in these ionic strength conditions. The first transition is assigned to the dissociation of the third strand, the second one, identical to that of the 24mer hairpin duplex, is assigned to the helix→coil transition of the duplex in the triple helix.
MATERIALS AND METHODS
Oligonucleotides
The following oligonucleotides were used in the UV and CD spectroscopic experiments: 36mer parGT, 5′-TGG TGG GTG GTG -L- CTC CTC CCT CCT -L- AGG AGG GAG GAG-3′; 36mer deaza-parGT, 5′-TG7G7 TG7G7 G7TG7 G7TG7 -L- CTC CTC CCT CCT -L- AGG AGG GAG GAG-3′; 24mer duplex, 5′-CTC CTC CCT CCT -L- AGG AGG GAG GAG-3′; 12mer single strand, 5′-TGG TGG GTG GTG-3′ (where G7 is 7-deaza-2′-deoxyguanine).
Concentrations of oligonucleotides were determined by UV spectroscopy. Molar concentrations in nucleotide units were computed using a mean molar extinction coefficient per residue (34). The quality of the synthesized oligonucleotides was tested by PAGE under denaturing conditions: 20% polyacrylamide gel (19:1 acrylamide:bis-acrylamide), 7 M urea in 1× TBE, pH 8.3. Their sequences were verified by the Maxam–Gilbert procedure (35). For native gel electrophoresis the oligonucleotides were 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase.
Native gel electrophoresis
Samples were prepared in an appropriate buffer by heating the oligonucleotides to 95°C followed by cooling to 3–5°C. Gel electrophoresis experiments were carried out at 4°C on a 15% polyacrylamide gel (29:1 acrylamide:bis-acrylamide), in either Tris–borate–EDTA buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3), 250 mM NaCl or Tris–borate buffer (45 mM Tris, 45 mM boric acid, pH 8.3), 100 mM NaCl, 2 mM ZnCl2. Delay after annealing is indicated in each case.
UV spectroscopy
UV absorption spectra and melting profiles have been recorded using Kontron Uvikon spectrophotometers (models 933 and 942) equipped with temperature regulated cuvette holders. Cell temperature was increased at a rate of 0.1°C per min. Absorbance measurements at 280 nm were used to follow triplex and duplex denaturation. Samples were dissolved in NaCl solutions (between 10 and 250 mM) or in high NaCl ionic strength (1 or 2 M NaCl) in the presence of ZnCl2 (concentration varied between 1 and 5 mM). DNA concentration was varied between 10–5 and 10–4 M.
Circular dichroism (CD)
CD spectra were obtained with a Jasco-715 spectropolarimeter equipped with a thermostatic cuvette holder. Optical path lengths used were 0.1 and 1 cm. Spectra were recorded between 4 and 85°C. Ionic strength conditions were the same as for UV measurements. Oligonucleotide concentrations were varied between 6 × 10–6 and 4 × 10–4 M. The experimental data were analyzed with the Microcal Origin computer program (Microcal Software Inc.).
RESULTS
Oligonucleotide structure in the presence of sodium ions
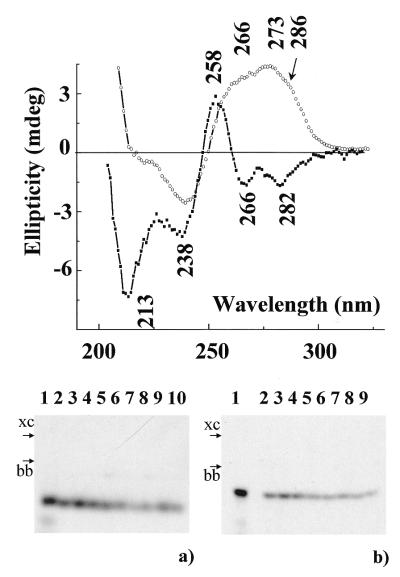
Native gel electrophoresis study. Oligonucleotide parGT was designed so that it is potentially able to fold back twice on itself to form an intramolecular triple helix containing G•G.C and T•A.T base triplets with parallel third strand orientation with respect to the purine duplex strand. In order to test the conditions of intramolecular structure formation, gel electrophoresis experiments in the presence of Na+ ions under native conditions were performed (Fig. 1a). Only one line is observed for parGT whatever the concentration used (between 4 × 10–8 and 6 × 10–5 M) and with either short (15 min, lanes 5–7) or long (90 min, lanes 8–10) annealing times. Migration of deaza-parGT annealed for 15 min is shown in lanes 2–4. Only one band is also observed. The possibility of intermolecular structure formation by deaza-parGT is low due to the absence of N7 atoms of guanines in the 5′-end segment. Thus, the band corresponding to deaza-parGT is reasonably correlated with an intramolecular structure and the similar migration of deaza-parGT and parGT allows us to propose that no intermolecular complexes are formed by parGT.
Figure 1.
(Top) CD spectra of parGT at 7°C in 50 mM NaCl (circles) and 1 M NaCl, 2.5 mM ZnCl2 (squares). (Bottom) Electrophoretic analysis of intermolecular complex formation. (a) In 250 mM NaCl at 4°C by oligonucleotides dT15 (lane 1), deaza-parGT (lanes 2–4, 15 min annealing) and parGT (lanes 5–7, 15 min annealing; lanes 8–10, 90 min annealing). Concentrations expressed in nucleotides: lanes 1, 4, 7 and 10: 4 × 10–8 M; lanes 3, 6 and 9: 2 × 10–5 M; lanes 2, 5 and 8: 6 × 10–5 M. (b) In 100 mM NaCl, 2 mM ZnCl2 at 4°C by oligonucleotides dT15 (lane 1), deaza-parGT (lanes 2–5) and parGT (lanes 6–9). Concentrations expressed in nucleotides: lanes 1, 5 and 9: 4 × 10–10 M; lanes 4 and 8: 4 × 10–6 M; lanes 3 and 7: 2 × 10–5 M; lanes 2 and 6: 6 × 10–5. xc, xylene cyanol; bb, bromophenol blue.
CD spectra. Figure 1 (top) presents the CD spectrum recorded at 7°C of parGT (circles) in 50 mM NaCl solution. The CD spectrum of parGT presents a negative band at 240 nm and a broad positive band with three contributions around 266, 273 and 286 nm. Such a spectrum looks very much like the spectrum of a B-family form duplex (20,36). Comparison of the CD spectrum of the whole oligomer supposed to form a triple helix and the sum of the CD spectra of the duplex and of the third strand recorded separately in the same ionic strength conditions allows one to evidence an interaction between the third strand and the duplex (37). In the present case very little difference is found with the simulated spectrum obtained by addition of the CD signals recorded of the duplex and the single strand (curve not shown). Upon heating the sample, a clear transition around 78°C corresponding to the melting of the duplex is observed. Experiments performed by varying the DNA concentration between 6 × 10–6 and 4 × 10–4 M yielded the same melting temperatures reflecting the intramolecular character of the studied structure.
We have checked that the 5′ extremity TGG TGG GTG GTG remains single-stranded when tethered to the duplex, and does not form, as could have been expected, a tetrameric structure. The CD spectrum of the dodecamer TGG TGG GTG GTG alone in the same conditions presents a very characteristic profile of a parallel tetraplex (spectrum not shown) with positive bands at 258 and 290 nm and a negative one around 240 nm (38) which has not been observed when the 12mer is linked to the duplex. Thus the structure adopted by the parGT oligomer should be a double helical stem (similar to the Watson–Crick duplex) with a dangling dodecamer TGG TGG GTG GTG at the 5′-end.
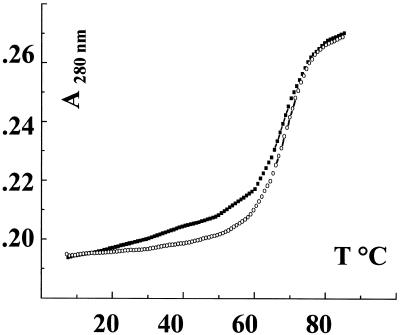
Thermal denaturation experiments. The thermal stability of parGT was investigated using UV spectroscopy. Figure 2 presents the melting curves of parGT (squares) and the 24mer duplex (circles) in 10 mM NaCl solutions. Both curves clearly present a transition around 78°C, very cooperative in the case of the duplex, with a slight slope for parGT below 60°C, too weak to be assigned to a conformational transition. This result is in agreement with the absence of triple helical structure for parGT in NaCl solutions found by CD. In these ionic strength conditions the parallel triplex is not formed.
Figure 2.
Melting curves in 10 mM NaCl of parGT (squares) and the 24mer duplex (circles).
Oligonucleotide structure in the presence of zinc ions
Native gel electrophoresis study. Figure 1b presents the native gel electrophoresis of parGT (lanes 6–9) and deaza-parGT (lanes 2–5) performed in 100 mM NaCl, 2 mM ZnCl2. Four concentrations have been used for each oligonucleotide. The higher concentrations are those used in CD experiments. Only one structure is detected in each case, which allows us to propose that in the presence of 2 mM ZnCl2 no intermolecular structure is detected for parGT.
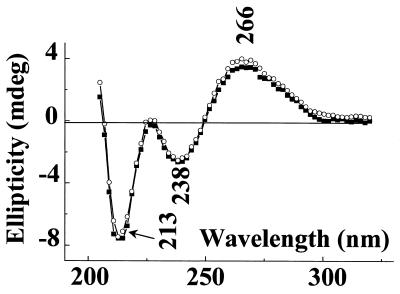
CD spectra. The effect of addition of zinc ions to the solution of parGT is presented in Figure 1 (top). The CD spectrum of parGT in 1 M NaCl, 2.5 mM ZnCl2 (squares) is obviously different from that obtained simply in the presence of sodium (circles) discussed above. Four negative bands at 213, 238, 266 and 282 nm and a positive one at 258 nm are now observed. Similar spectra were obtained in 1 M NaCl with lower ZnCl2 concentrations, 1.5 and 2 mM.
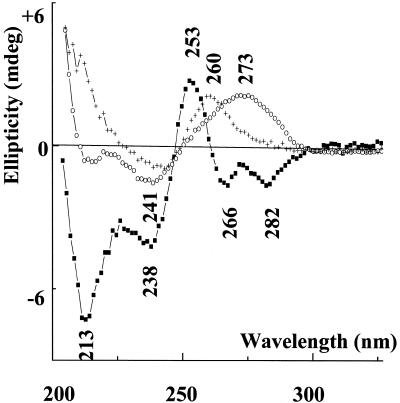
Figure 3 presents the CD spectra of parGT (squares), the 24mer duplex (circles) and the separate third strand (crosses) recorded in the same conditions (2 M NaCl, 5 mM ZnCl2). Clearly the mathematical sum of the two latter spectra cannot match the experimental spectrum of parGT. This reflects the interaction between the third strand and the duplex, and thus the formation of the parallel triplex structure.
Figure 3.
CD spectra in 2 M NaCl, 5 mM ZnCl2 of parGT (squares), 24mer duplex (circles) and 12mer third strands (crosses).
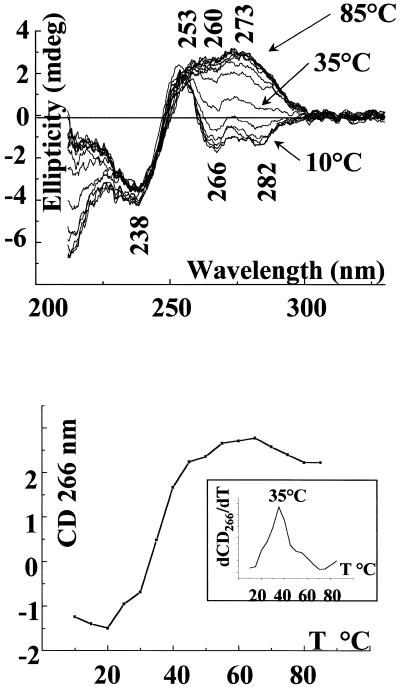
Melting of this structure is presented in Figure 4. The spectra shown have been recorded in 2 M NaCl, 5 mM ZnCl2 every 5°C between 10 and 85°C, and the experiment is totally reversible [Fig. 4 (top)]. The negative bands at 266 and 282 nm are progressively inverted by increasing the temperature above 30°C. We notice that the spectrum recorded at 40°C already looks very much like that of the duplex. Thermal denaturation can be characterized by monitoring ellipticity changes of different CD bands (39,40). Figure 4 (bottom) shows the plot of the ellipticity measured at 266 nm with increasing temperature. The inset presents the derivative of the plotted curve. We clearly observe a very cooperative transition with a midpoint located around 35°C, reflecting the separation of the third strand. The melting of the remaining duplex is not fully observed, occurring above 80°C, due to the high ionic strength of the solution.
Figure 4.
(Top) Melting of parGT in 2 M NaCl, 5 mM ZnCl2 followed by CD, spectra shown every 5°C between 10 and 85°C. (Bottom) Ellipticity at 266 nm versus temperature for parGT in 2 M NaCl, 5 mM ZnCl2 (inset: derivative of ellipticity at 266 nm versus temperature).
Figure 5 presents the CD spectrum recorded at 5°C of deaza-parGT (the same oligonucleotide as above but with deazaguanines instead of normal guanines in the third strand), in 1 M NaCl (circles) and after addition of Zn2+ ions, in 1 M NaCl, 2.5 mM ZnCl2 (squares). Both spectra are totally identical and the presence of a positive broad band at 266 nm clearly shows that the addition of Zn2+ ions does not induce in this case triplex formation.
Figure 5.
CD spectra of deaza parGT at 5°C in 1 M NaCl (circles) and 1 M NaCl, 2.5 mM ZnCl2 (squares).
UV absorbance spectroscopy. We have performed melting experiments of parGT in 2 M NaCl and increasing amounts of ZnCl2. The melting profiles presented in Figure 6 are those of parGT in 2 M NaCl, 1 mM ZnCl2 (crosses) and 2 M NaCl, 5 mM ZnCl2 (squares; the inset shows the derivative of this melting curve). The melting curve of the corresponding duplex (24mer) recorded in 2 M NaCl, 5 mM ZnCl2 is shown by circles. Denaturation has been monitored by following the absorption at 280 nm. The biphasic character of the melting profiles of parGT is obvious. The first transition is classically assigned to third strand dissociation, and as expected is zinc concentration dependent: Tm at 24°C in the presence of 1 mM, 30°C of 2.5 mM and 35°C of 5 mM ZnCl2. The second cooperative transition (Tm >80°C) can be assigned to the melting of the duplex in the triple helix by comparison with the melting profile of the duplex alone (circles). The midpoint temperatures, in identical ionic strength conditions, are in agreement with those determined earlier using CD spectroscopy.
Figure 6.
UV melting curves of parGT in 2 M NaCl, 1 mM ZnCl2 (crosses), 2 M NaCl, 5 mM ZnCl2 (squares; inset: derivative of the 280 nm absorbance versus temperature) and of the 24mer duplex in 2 M NaCl, 5 mM ZnCl2 (circles).
DISCUSSION
Formation of triple helices with third strands containing both purines and pyrimidines (GT strands) remains an intriguing problem. While third strand orientation is considered well established for pyrimidine motif triplexes (parallel to the polypurine strand) and purine motif triplexes incorporating both A and G third strand nucleosides (antiparallel to the homologous polypurine strand), the question remains unclear for mixed GT third strands with variable ratios of purine/pyrimidines. In fact if a parallel orientation has been observed for both extreme compositions (all Ts or all Gs) the simultaneous presence of both G and T nucleotides in the third strand makes the situation less clear. The non-isomorphism of the G•G.C and T•A.T base triplets induces an important destabilizing effect at each GpT/TpG step in the third strand sequence. Thus the third strand parallel orientation observed in the absence of such a GpT/TpG step becomes less favored when their amount increases (17) and it has even been proposed that above four GpT/TpG steps formation of the triplex is not possible (19). Our experiments performed on an oligonucleotide designed to form a parallel GT triple helix show that it is possible to observe such a structure even with a very large amount of such destabilizing GpT/TpG steps (in our case seven in a 12mer sequence).
Native gel electrophoresis of parGT performed in 2 mM ZnCl2, 100 mM NaCl has shown that only one structure is formed for an immediately annealed sample. Moreover the CD spectrum of parGT recorded in 4 M NaCl but without Zn2+ is similar to that recorded in low sodium ionic strength conditions, which shows that parGT is not aggregated at high concentration of Na+ ions. Thus the CD spectrum of parGT recorded in 2 M NaCl, 5 mM ZnCl2 corresponds to an intramolecular triple helical structure. We notice that the CD signature of this parallel GT triplex presents in the high wavelength part of the spectum two negative bands (266 and 282 nm) which is different from the classical CD signature of antiparallel triplexes which in this region present only one negative band around 278 nm (16,39,40).
As noticed above, the CD spectrum recorded in 4 M NaCl but without Zn2+ ions does not present negative bands at 266 and 282 nm. Similarly the spectra recorded in the presence of only 2.5 mM Zn2+ ions also do not show this inversion. Thus both neutralization of the phosphate groups and specific interactions of the Zn2+ ions on the bases seem necessary to stabilize the parGT triplex. Zinc ions play a particular role, as Mg2+ ions do not induce the inversion of the CD signal of parGT, with the obtained spectrum (not shown) reflecting an aggregation of the molecule which increases with elapsing time. This is in agreement with the reported impossibility to form triplexes in the case of d(GA.TC)n sequences using Mg2+ ions, the structure of which was, on the contrary, stabilized by Zn2+ (29). Zinc ions are known to bind to the N7 and (to a lesser extent) N3 sites of guanines (41,42). The stabilization of an antiparallel GA purine motif triplex has been obtained by Zn2+ ions. This had been explained by divalent metal coordination to the N7 atoms of the third strand purines with concomitent polarization effects on the bases resulting in the Hoogsteen type hydrogen bond enhancement (43). In our case the base pairing model proposed for parallel G•G.C triplets after crystal diffraction experiments leaves both third strand guanine N7 and N3 atoms accessible to metal interactions (10). Our experiment with deaza-parGT shows that when in the third strand guanines the N7 atoms are substituted by CH groups, thus preventing a possible interaction of the Zn2+ ions on this site, the triplex is no longer formed. It is thus possible to propose that a similar electronic displacement induced by Zn2+ ion binding on the guanines could enhance the third strand binding and stabilize the parallel GT triple helix as in the case of the antiparallel GA triple helix (43).
In summary our study has shown that a parallel triple helical structure can be formed in the presence of divalent Zn2+ counterions by a third strand containing G and T bases with many non-regularly spaced GpT/TpG steps. The parallel GT triple helical structure is observed over a broad DNA concentration range. Native gel electrophoresis experiments performed in the 10–10 to 10–5 M nucleotide concentration range have clearly shown the monomolecular character of this structure. Biphasic melting curves have been determined by UV and CD spectroscopy in the 10–5 to 10–4 M nucleotide concentration range. In comparable ionic strength conditions identical melting temperatures have been measured over the whole DNA concentration domain, again showing the intramolecular character of the formed triplexes. A new CD signature with negative bands at 266 and 282 nm, which we propose to be characteristic of the formation of parallel GT triplexes, has been obtained.
Acknowledgments
ACKNOWLEDGEMENT
E.B.K. was supported by a FEBS short term fellowship.
REFERENCES
- 1.Frank-Kamenitskii M.D. and Mirkin,S.M. (1995) Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 2.Chan P.P. and Glazer,P.M. (1997) J. Mol. Med., 75, 267–282. [DOI] [PubMed] [Google Scholar]
- 3.Lavrovsky Y., Mastyugin,V., Stoltz,R.A. and Abraham,N.G. (1996) J. Cell. Biochem., 61, 301–309. [DOI] [PubMed] [Google Scholar]
- 4.Olivas W.M. and Maher,L.J.,III (1996) Nucleic Acids Res., 24, 1758–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musso M., Wang,J.C. and Van Dyke,M.W. (1996) Nucleic Acids Res., 24, 4924–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guieysse A.L., Praseuth,D., Grigoriev,M.H., Harel Bellan,A. and Hélène,C. (1996) Nucleic Acids Res., 24, 4210–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunther E.J., Havre,P.A., Gasparro,F.P. and Glazer,P.M. (1996) Photochem. Photobiol., 63, 207–212. [DOI] [PubMed] [Google Scholar]
- 8.Guieysse A.L., Praseuth,D. and Hélène,C. (1997) J. Mol. Biol., 267, 289–298. [DOI] [PubMed] [Google Scholar]
- 9.Musso M., Nelson,L.D. and Van Dyke,M.W. (1998) Biochemistry, 37, 3086–3095. [DOI] [PubMed] [Google Scholar]
- 10.Vlieghe D., Van Meervelt,L., Dautant,A., Gallois,B., Précigoux,G. and Kennard,O. (1996) Science, 273, 1702–1705. [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan I., de los Santos,C. and Patel,D.J. (1991) J. Mol. Biol., 221, 1403–1418. [PubMed] [Google Scholar]
- 12.Beal P.A. and Dervan,P.B. (1991) Science, 251, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 13.Radhakrishnan I. and Patel,D.J. (1993) Structure, 1, 135–152. [DOI] [PubMed] [Google Scholar]
- 14.Chandler S.P. and Fox,K.R. (1996) Biochemistry, 35, 15038–15048. [DOI] [PubMed] [Google Scholar]
- 15.Li J., Hogan,M.E. and Gao,X. (1996) Curr. Biol., 4, 425–435. [Google Scholar]
- 16.Gondeau C., Maurizot,J.C. and Durand,M. (1998) J. Biomol. Struct. Dyn., 15, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 17.Sun J.S., de Bizemont,T., Duval-Valentin,G., Montenay-Garestier,T. and Hélène,C. (1991) C. R. Acad. Sci. Ser. III, 313, 585–590. [PubMed] [Google Scholar]
- 18.Kiran M.R. and Bansal,M. (1995) Indian J. Biochem. Biophys., 32, 391–403. [PubMed] [Google Scholar]
- 19.Clarenc J.P., Lebleu,B. and Léonetti,J.P. (1994) Nucl. Nucl., 13, 799–809. [Google Scholar]
- 20.Gondeau C., Maurizot,J.C. and Durand,M. (1998) Nucleic Acids Res., 26, 4996–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khomyakova E., Liquier,J., Huynh-Dinh,T., Florentiev,V., Mirzabekov,A. and Taillandier,E. (2000) J. Biomol. Struct. Dyn., 11, 227–237. [DOI] [PubMed] [Google Scholar]
- 22.Smith F.W. and Feigon,J. (1992) Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 23.Kang C., Zhang,X., Ratliff,R., Moyzis,R. and Rich,A. (1992) Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y. and Patel,D.J. (1992) Biochemistry, 31, 8112–8119. [DOI] [PubMed] [Google Scholar]
- 25.Wang K.Y., Krawczyk,S.H., Bischofberger,N., Swaminathan,S. and Bolton,P.H. (1993) Biochemistry, 32, 11285–11292. [DOI] [PubMed] [Google Scholar]
- 26.Kelly J.A., Feigon,J. and Yeates,T.O. (1996) J. Mol. Biol., 256, 417–422. [DOI] [PubMed] [Google Scholar]
- 27.Lee P.P., Ramanathan,M., Hunt,C.A. and Garovoy,M.R. (1996) Transplantation, 62, 1297–1301. [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian V., Nguyen,L.T., Balasubramanian,S.V. and Ramanathan,M. (1998) Mol. Pharmacol., 53, 926–932. [PubMed] [Google Scholar]
- 29.Bernués J., Beltran,R., Casasnovas,J.M. and Azorin,F. (1990) Nucleic Acids Res., 18, 4067–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernués J., Beltran,R., Casasnovas,J.M. and Azorin,F. (1989) EMBO J., 8, 2087–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyamichev V.I., Voloshin,O.N., Frank-Kamenetskii,M.D. and Soyfer,V.N. (1991) Nucleic Acids Res., 19, 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malkov V.A., Voloshin,O.N., Soyfer,V.N. and Frank-Kamenetskii,M.D. (1993) Nucleic Acids Res., 21, 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherny D.I., Malkov,V.A., Volodin,A.A. and Frank-Kamenetskii,M.D. (1993) J. Mol. Biol., 230, 379–383. [DOI] [PubMed] [Google Scholar]
- 34.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 35.Maxam A.M. and Gilbert,W. (1980) In Grossman,L. and Moldave,K. (eds), Methods in Enzymology, Vol. 65. Academic Press, New York, NY, pp. 499–559.
- 36.Guschlbauer W. (1988) In Encyclopedia of Polymer Science and Engineering, Vol. 12. Wiley, New York, NY, pp. 699–785.
- 37.He Y., Scaria,P.V. and Shafer,R.H. (1997) Biopolymers, 41, 431–441. [DOI] [PubMed] [Google Scholar]
- 38.Balagurumoorthy P., Brahmachari,S.K., Mohanty,D., Bansal,M. and Sasisekharan,V. (1992) Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xodo L.E. (1995) FEBS Lett., 370, 153–157. [DOI] [PubMed] [Google Scholar]
- 40.Scaria P.V., Will,S., Levenson,C. and Shafer,R.H. (1995) J. Biol. Chem., 270, 7295–7303. [DOI] [PubMed] [Google Scholar]
- 41.Saenger W. (1984) In Principles of Nucleic Acids Structure. Springer Verlag, Berlin, Germany.
- 42.Froystein N.A., Davies,J.T., Reid,B.R. and Sletten,E. (1993) Acta Chem. Scand., 47, 649–657. [DOI] [PubMed] [Google Scholar]
- 43.Potaman V.N. and Soyfer,V.N. (1994) J. Biomol. Struct. Dyn., 11, 1035–1040. [DOI] [PubMed] [Google Scholar]