Abstract
The use of medical implants continues to grow as the population ages. Biofilm-related implant infection is the leading cause of medical implant failure and remains difficult to diagnose and treat. Recent technologies have enhanced our understanding of the composition and complex functions of microbiota occupying various body site niches. In this review, we leverage data from molecular sequencing technologies to explore how silent changes in microbial communities from various sites can influence the development of biofilm-related infections. Specifically, we address biofilm formation and recent insights of the organisms involved in biofilm-related implant infections; how composition of microbiomes from skin, nasopharyngeal, and nearby tissue can impact biofilm-formation, and infection; the role of the gut microbiome in implant-related biofilm formation; and therapeutic strategies to mitigate implant colonization.
Keywords: biofilm-related implant infection, gut microbiome, metagenomic sequencing, prosthetic joint infections, Staphylococcus aureus
As medical and surgical techniques have evolved over the last several decades, the use of implants has increased in parallel. Medical implants can be used to restore form and function to patients who would otherwise be crippled by their disease process including but not limited to ambulation-limiting knee osteoarthritis, debilitating ventral hernia formation with loss of domain, or profound cardiac failure requiring mechanical support device insertion. Infection remains the leading cause of medical implant failure, and requires a wide range of therapy spanning prolonged intravenous antibiotic therapy to device explantation; the patient impact may be devastating.1
Biofilm-related infections are estimated to cause 80% to 100% of infections of surgically implanted devices, and account for more than 65% of human infections of any kind.2,3 Unfortunately, the presence of biofilm makes implant infections more difficult to both diagnose and treat. Although newer molecular sequencing techniques are not yet routinely incorporated in practice, their recent use in clinical studies has significantly increased our appreciation of the complexity and variability of the host–microbe interactions governing biofilm formation. In this review, we focus on evidence from recent clinical studies exploring the role of resident microbiota from various sites in development of biofilm-related implant infections. A better understanding of these microbial communities both informs current practices and may shape the development of future therapeutic strategies for combating biofilm-related implant infection.
Biofilm Formation, Clinical Implications, and Recent Insights
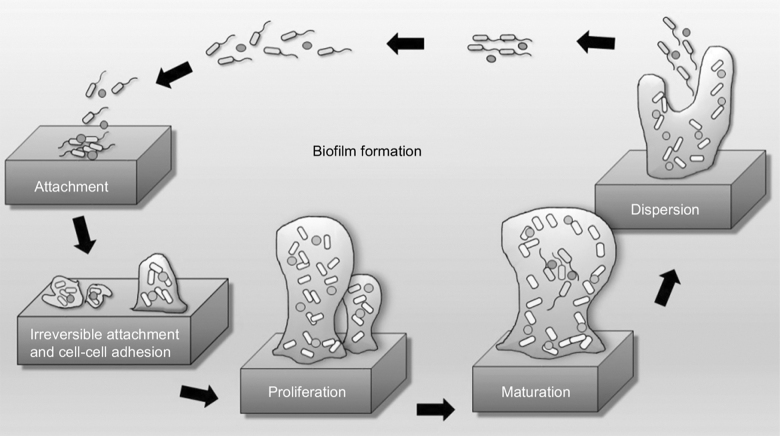
Biofilm is a heterogeneous and cooperative community of bacteria embedded in an extracellular matrix attached to a surface.4 It comprises a sessile adherent community of cells, as well as a free-floating planktonic segment, and essentially creates a protected niche to support growth and diversity (Fig. 1). The bacteria appear to transition from free-living, planktonic cells to sessile, surface-attached cells in response to a nutrient-rich medium to which they attach.5 Organisms within biofilm cooperatively maintain and strengthen the ecosystem via nutrient and metabolic regulation and production of polysaccharides. This ecosystem further provides a protective niche, guarding against innate host defense mechanisms such as complement, white blood cells, and immunoglobulins, thereby reducing the efficacy of antibiotic agents and making biofilm-based bacterial eradication exceedingly difficult.6,7
FIG. 1.
Stages of biofilm formation. This schematic depicts the five stages of the classic model of biofilm formation starting with attachment and developing through dispersion. Continued maturation forms a three-dimensional structure providing protection against host defense mechanisms and antibiotic agents. Reproduced with permission under the Creative Commons Attribution (CC BY) license from: Bakar MA, McKimm J, Haque SZ, et al. Chronic tonsillitis and biofilms: a brief overview of treatment modalities. J Inflamm Res 2018;11:329.
Organisms responsible for biofilm-related implant infections are understood to enter the wound site and colonize the implant during, or soon after implantation; hematogenous spread from infection at a distal site is also described.8 Biofilm formation is initiated by microbial adherence to a conditioning film overlying the implant surface and is formed within minutes of device insertion. As early as two to four hours after adherence, stable microcolonies of attached bacteria are observed with biofilm maturation complete as early as two to four days later.9,10 Importantly, although gram-positive aerobes are often primary colonizers, the architecture of the biofilm facilitates engagement and proliferation of gram-positive bacteria, gram-negative bacteria, anaerobes, and yeasts as well.11 Development of biofilm-related infection is a function of the structural properties of the implant material as well the interplay between the organism or organisms creating the biofilm and the environment in which the implant is placed.
In the United States, the most common medical implants aside from catheters are fracture fixation devices, joint prostheses, dental implants, vascular grafts, cardiac pacemakers, mechanical heart valves, and ventricular assist devices (VADs).12 Although substantial variability exists, clinical studies using culture-based techniques most frequently implicate Staphylococcus and Streptococcus species, followed by other common skin commensals including Corynebacterium and Cutibacterium acnes (formerly Propionibacterium acnes), in addition to Enterococci, Enterobacteriaceae, and Pseudomonas spp.11,13 However, conventional diagnostic modalities used in clinical practice rely on the presence of free-floating cells and growth in culture media and therefore often lack the sensitivity to detect organisms resident within a biofilm. As such, the diagnosis of biofilm-related implant infection remains a challenge. No standardized diagnostic methodology exists, and current guidelines are based on clinical and laboratory signs of infection coupled with recognition of antibiotic therapy failure in the setting of a recent implant.14
The increasing use of techniques such as sequencing of the 16S rRNA subunit and metagenomic analysis in clinical settings has both highlighted the insensitivity of culture-based methods and underscored the variability in biofilm microbial communities. It is likely that the incidence of implant infections is much higher than reported, and that anaerobes and atypical organisms are under-diagnosed contributors. For example, in the largest study exploring diagnostic utility of metagenomic sequencing in prosthetic joint infections (PJIs), investigators identified pathogenic organisms in synovial fluid sonicate in 43% of 190 culture-negative PJIs, with sensitivities as high as 80% in similar smaller studies.15,16 Organisms that were identified by sequencing but not culture included Staphylococcus, Streptococcus, and Enterococcus spp., as well as more unusual species such as Clostridium, Anaerococcucus, Pasteurella, and Candida spp.15 Nonetheless, these techniques are not without limitations, and time and cost remain major barriers to their use in clinical practice.
As with culture, low quantities of planktonic bacterial DNA in fluid render samples more susceptible to contamination and false-positive results. However, several studies have been able to incorporate mechanical or enzymatic digestion of the implant to facilitate recovery of biofilm. Although currently limited to investigations of indwelling catheters, they suggest that multispecies biofilms may be more prevalent in clinical practice than previously recognized. In a study of biofilm-related infection in patients with central venous catheters (CVCs), investigators identified more than 30 bacterial species in catheters from patients with and without signs of clinical infection, although biofilm architecture and dominant species were specific to the infected group.17 Metagenomic sequencing of endotracheal tubes from patients with ventilator-associated pneumonia (VAP) further demonstrated that all assessed biofilms comprised multispecies consortia. Importantly, specific biofilm community structures were strongly associated with clinical outcomes.18 Specifically, a low abundance of Pseudomonaceae and a high abundance of Staphylococcus epidermis favored lower mortality, while co-occurrence of Staphylococcus epidermis, Serratia marcescens, and Klebsiella spp. were predictive of VAP development.18
Can the Composition of the Resident Microbial Communities Predict Biofilm Formation?
Although our knowledge of key contributors to biofilm-related infections has increased, our understanding of how compositional changes in resident microbiota influence susceptibility to infection remains far from complete. Certain microbiome features, namely nasopharyngeal carriage of Staphylococcus aureus increase the risk of biofilm-related infections. Particularly in orthopedic and cardiac surgery, nasal colonization by Staphylococcus aureus has been identified as a pre-operative risk factor for post-operative infection with methicillin-sensitive and methicillin-resistant strains.19 In a study of surgical site infections, investigators demonstrated that 80% of staphylococcal strains causing infection were concomitantly detectable in the nares of affected patients.20 However, not all organisms demonstrate the same predictive potential. The previously noted investigation of CVCs did not identify any differences in the microbial communities from skin samples taken from patients with and without CVC infection.17 Similarly, comparison of 79 Cutibacterium acnes strains recovered from PJIs versus samples collected from the skin of healthy controls demonstrated that strains isolated from healthy controls were similar in distribution and just as capable of forming biofilm as the strains involved in PJIs.21
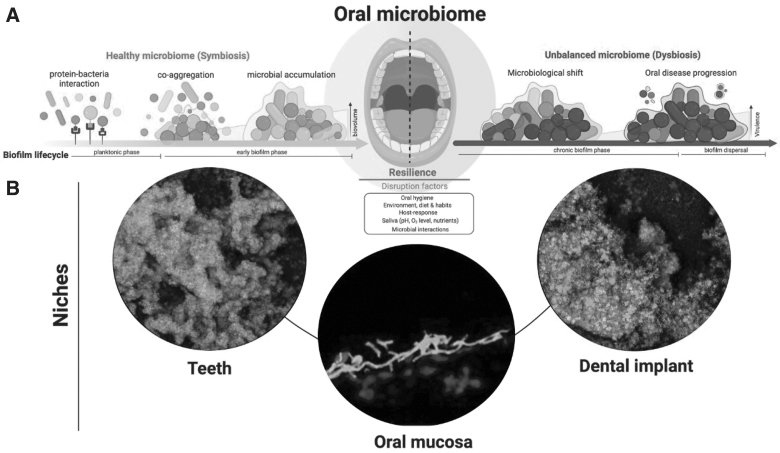
The significance of microbial composition is better established for communities within sites adjacent to a colonized or infected implant. Peri-implantitis and mucositis are biofilm-associated infections of the oral cavity affecting an estimated 20% to 40% of dental implants.22 Metagenomic sequencing of tissue affected by peri-implantitis and healthy oral tissue from the same individuals revealed that the presence of several bacteria including Fusobacterium nucleatum was specific to tissue surrounding dental implant (Fig. 2). The investigators further showed that identification of this consortia was able to predict periimplantitis as well as mucositis.23 Importantly, this finding suggests that infection arises when certain disease-associated organisms are present amid other host and implant-related factors.
FIG. 2.
Biofilm formation in the oral cavity. The relation of the oral microbiome, a dental implant, and peri-implantitis is depicted. (A) In a symbiotic relation, the oral bacteria coupled to deposited proteins from oral fluids (i.e., saliva and plasma) bind through adhesin receptor interactions. Through co-aggregation between different species biofilm accumulation is promoted on dental implants and teeth. A disruption of the symbiotic state in the mouth leads to a dysbiotic environment in which pathogenic bacteria can overgrow and trigger periimplantitis. (B) In the wake of dysbiosis, polymicrobial films can accumulate and induce oral disease in the peri-oral niches of the native dentition, the mucosa, and the dental implant. Reproduced with permission under the Creative Commons Attribution (CC BY) license from: Bertolini M, Costa RC, Barão VA, et al. Oral microorganisms and biofilms: New insights to defeat the main etiologic factor of oral diseases. Microorganisms 2022;10(12):2413.
Furthermore, this observation establishes the importance of multispecies interactions in biofilm generation and implant infection. Cross-kingdom interactions in the surrounding environment clearly influence biofilm infections. For example, Candida albicans augments Staphylococcus aureus adhesion and colonization with a resultant increase in biofilm formation. In a murine model of oral candidiasis, bacterial–fungal interactions were found to increase susceptibility to both oral and systemic bacterial infection by Staphylococcus aureus—findings that were prevented if Candida albicans was eradicated.24 Together, these studies indicate that the presence of certain cooperative relations adjacent to the implant contribute to the clinical course but also suggest potential alterations of surgical site infection prophylaxis agents.
The microbial communities residing in tissues adject to an implant have additional implications in infection recurrence. Preclinical studies demonstrate that Staphylococcus aureus and Staphylococcus epidermidis colonize and replicate in peri-implant tissue even after implant removal.25,26 In patients, bacteria responsible for infection have been identified in breast tissue surrounding implants, and fibroblasts in bone adjacent to infected hip prostheses.27 These findings further emphasize the difficulty in treating biofilm-related infection, and the importance of adequate tissue debridement upon device explant for to achieve adequate source control. Defining the limits of adequate tissue debridement remains problematic during the intra-operative segment of care and may ultimately rely on techniques that detect the presence (or absence) of bacterial or fungal DNA at the resection margin.
Composition of the Gut Microbiota Influences Biofilm-Related Infection and the Case for (Against) Dysbiosis?
Increasing evidence suggests that the composition of the gut microbiota may also influence the development of remote biofilm-related infection. The gut microbiota plays a central role in intestinal and systemic immune responses in three inter-related ways.28–30 First, the gut microbiome promotes mucin production and enhances the gut barrier integrity. Second, during periods of homeostasis, gut commensals prevent overgrowth and dissemination of enteric pathogens. Third, certain taxa utilize direct signaling pathways as well as the production of metabolites like short-chain fatty acids to govern T-cell responses, neutrophil activation, and cytokine secretion. In preclinical studies, derangements in gut microbial community structure or function—or dysbiosis—render the host more susceptible to infections including enteritis, bacteremia, pneumonia, and sepsis.31,32
Although causative associations are more difficult to establish in clinical studies, similar complications are observed in patients with dysbiosis and immunosuppression or increased gut barrier permeability, the latter event is well-characterized as a result of sepsis and shock. In patients undergoing hematopoietic stem cell transplantation (HCT), Taur et al.34 demonstrated that patients with peri-operative gut microbial excess proportion of Enterococcus had a nine-fold increased risk of developing vancomycin-resistant Enterococcus (VRE) bacteremia after transplantation, and that a predominance of Proteobacteria conferred a five-fold increased risk of gram-negative bacteremia.33 Another study in this population used a novel platform to demonstrate that in patients with blood stream infections after HCT, the majority of pathogens detected in the blood were present in stool samples collected in the weeks preceding bacteremia. In critically ill patients certain organisms recovered from intensive care unit (ICU) admission fecal swabs (Escherichia coli, Pseudomonas spp., Klebsiella spp., Clostridium difficile, and VRE) correlate with subsequent infection by the same organisms but at remote sites during the index episode of critical care.35
In the peri-operative period, numerous factors reduce microbial diversity and alter the community landscape. Antimicrobials represent the most pervasive culprit but supplemental oxygen (hyperoxia), general anesthesia, nothing by mouth status, and multiple other routinely used therapies also reduce the abundance of certain beneficial commensals.36–39 In this sense, surgical patients may be considered an at-risk group with impaired mucosal defenses allowing for enhanced entry of enteric pathogens into the systemic circulation.40 In the setting of recent device implantation, could these translocated opportunists find refuge by forming biofilms on newly implanted material, ultimately accounting for subsequent development of overt as well as subclinical infection? Although a compelling theory, it remains uncertain if this mechanism contributes to biofilm formation. Nonetheless, recent data suggest that this hypothesis is quite plausible.
In a retrospective study of more than 100,000 primary total hip arthroplasty patients, Moran et al.41 assessed the root cause underpinning the need for revision surgery between healthy controls and those with inflammatory bowel disease (IBD; n = 20,000). Patients with IBD are characterized by gut dysbiosis and decreased barrier function, conditions that could reasonably influence PJI. The investigators found that patients with IBD had a higher risk of peri-prosthetic infection requiring revision (p = 0.004), compared with disease-free controls who primarily underwent revision for aseptic causes. A separate study of PJIs revealed that a number of organisms identified from affected tissue adjacent to the infected implant were gut commensals, and that increased intestinal barrier permeability correlated with the likelihood of identifying a gut-derived organism.42 Rather than direct translocation, gut-derived Staphylococcus aureus may seed distal sites via carriage by neutrophils (i.e., the Trojan horse theory).43 Pre-clinical models of surgical site infection and PJIs have validated this mechanism, demonstrating that lysis of neutrophils and release of pathogenic Staphylococcus aureus results in development of remote site infection.44
Finally, specific alterations in the gut microbiota may influence biofilm infections in more indirect ways. Specific taxonomic alterations in the gut microbiota can modify innate and adaptive immune responses to infectious and inflammatory stimuli.39,45 A pre-clinical study evaluating host responses to various implanted materials demonstrated that antibiotic-treated and germ-free mice exhibit maladaptive responses to implant placement including reduced macrophage and myofibroblast recruitment to the fibrous capsule, and differential expression of angiogenic and inflammatory markers.46 Importantly, fecal microbial transplantation prior to implantation was able to reverse these findings. Inflammatory responses were also associated with changes in the gut microbiota in an extensive study of pre-operative characteristics and infectious complications in patients with heart failure undergoing left ventricular assist device (LVAD) implantation.47 Investigators discovered that the presence of certain gut commensals (including Veillonella and Phascolarctobacterium) was associated with distinct inflammatory phenotypes, as well as a significantly greater risk of post-LVAD infection by any organism. Moreover, identification of these microbes also demonstrated superior predictive power for patients at-risk for infection than clinical decision making alone. Thus, an abnormal microbiome may derail adaptive aspects of wound repair as well as invasive pathogen detection and management.
Therapeutic Strategies to Mitigate Biofilm-Related Infection Risk
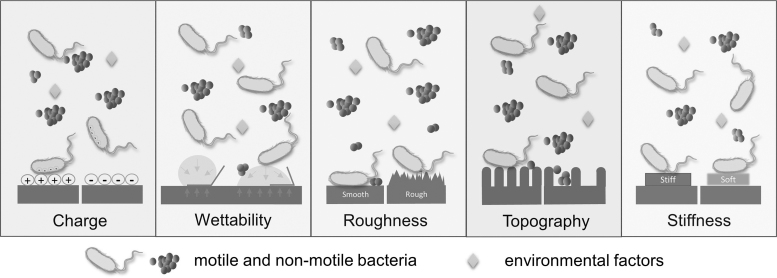
Although more precise characterizations of biofilm communities provide insights into their structure, function, and management, that knowledge also raises additional questions. Findings from recent studies underscore the importance of fundamental infection control strategies and can inform development of new therapeutic interventions. First and foremost, proper sterile technique and decontamination strategies continue to be the mainstay of prevention. In cases of implant infection requiring removal, concern for ongoing reservoirs of infection may influence intra-operative techniques, or the timing of surgery if re-implantation is necessary. In certain settings, modifications of implants that reduce bacterial adherence are commercially available. For example, antibiotic coatings for endotracheal tubes, urinary catheters, and CVCs are increasingly utilized but are not yet standard practice. Although most permanent implants have lagged in the commercial translation of specific drug–device combinations, antimicrobial-coated biologic and synthetic hernia repair meshes are readily available.48 When selecting a specific mesh, one must consider the hydrophobicity or hydrophilicity of the implant given that hydrophobic polymers such as polypropylene demonstrate less bacterial adherence compared with polytetrafluoroethylene or polyester (Fig. 3). When considering orthopedic implants, surface topography and roughness plays a role in cell or bacterial attachment as does surface charge, thereby advocating for adaptive surface design as an effective means of reducing implant infection susceptibility. In particular, deliberate nanodesign may shape how both durable implanted devices (i.e., artificial joints) as well as temporary percutaneous devices (i.e., CVC) surfaces are designed to reduce biofilm formation and infection risk.
FIG. 3.
Surface nanometric elements that promote biofilm formation. This illustration depicts the surface elements that govern bacterial adhesion to implant surfaces. Surface charge, charge density, wettability, roughness, topography, and stiffness all play substantial roles in cellular and bacterial adhesion. This schematic illustrates conditions under which material adhesion is enhanced or diminished. Reproduced with permission under the Creative Commons Attribution (CC BY) license from: Zheng S, Bawazir M, Dhall A, et al. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front Bioeng Biotechnol 2021;9:643722.
Outside of the operating room, decisions regarding antibiotic agents should be made thoughtfully. Many commonly prescribed antibiotic agents are unable to reach bacteria in protected spaces such as biofilm or an intracellular site. Moreover, if an antibiotic is able to penetrate into a space characterized by a low partition coefficient, metabolically quiescent bacteria demonstrate reduced susceptibility. Certain antibiotic agents demonstrate enhanced cellular penetrance and offer a selective advantage when used in combination with a related agent. For example, combination therapy that includes rifampicin appears to be more effective for intracellularly resident bacteria as well as multi-drug–resistant organisms such as Cutibacterium acnes.9,49 Although antibiotic agents are essential in managing implant infection, with or without device explantation, treatment should be narrowed and stopped as early as possible to prevent prolonged periods of antibiotic-driven dysbiosis.50 Although intuitively attractive, there is insufficient data to recommend the routine use of pre-, pro-, or synbiotics to modulate the gut microbiome to help abrogate biofilm related infection.
Conclusions
Implant-related infection is largely linked to biofilm formation. Biofilm formation is a rapid event that is influenced by both the local microbiota as well as remote microbiota such as those in the gut. Specific conditions increase the likelihood of gut dysbiosis and appear to influence implant infection from remote organisms unrelated to the surgical site. Specific microbiome elements also appear to shape implant as well as other infection risk, especially during an episode of critical care. These observations offer the potential for deliberate microbiome preservation or therapeutic alteration to reduce implant infection risk in particular, and infection risk in general. Device design alterations that impede bacterial attachment offer promise as a mechanism to reduce biofilm formation that may be particularly useful for patients whose underlying conditions (e.g., inflammatory bowel disease) are difficult to alter to adjust their impact on the microbiome. Such design modifications may extend to temporary devices such as transcutaneous catheters that are widely used in the operating room, the interventional radiology suite, the ICU, and other complex care environments.
Authors' Contributions
Conceptualization: Fernandez-Moure. Data curation: all authors. Formal analysis: all authors. Writing–original draft: all authors. Writing–review and editing: Fernandez-Moure. All authors approved the final version.
Funding Information
No funding was received.
Author Disclosure Statement
No competing financial interests exist.
References
- 1. Fernandez-Moure JS, Mydlowska A, Shin C, et al. Nanometric considerations in biofilm formation. Surg Infect 2019;20(3):167–173; doi: 10.1089/sur.2018.237 [DOI] [PubMed] [Google Scholar]
- 2. McLean RJ, Lam JS, Graham LL. Training the biofilm generation—A tribute to J. W. Costerton. J Bacteriol 2012;194(24):6706–6711; doi: 10.1128/JB.01252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Williams DL, Costerton JW. Using biofilms as initial inocula in animal models of biofilm-related infections. J Biomed Mater Res B Appl Biomater 2012;100(4):1163–1169; doi: 10.1002/jbm.b.31979 [DOI] [PubMed] [Google Scholar]
- 4. Beloin C, Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol 2005;13(1):16–19; doi: 10.1016/j.tim.2004.11.008 [DOI] [PubMed] [Google Scholar]
- 5. O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol 2000;54:49–79; doi: 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- 6. O'Gara JP, Humphreys H. Staphylococcus epidermidis biofilms: importance and implications. J Med Microbiol 2001;50(7):582–587; doi: 10.1099/0022-1317-50-7-582 [DOI] [PubMed] [Google Scholar]
- 7. Dasgupta MK. Biofilms and infection in dialysis patients. Semin Dial 2002;15(5):338–346; doi: 10.1046/j.1525-139x.2002.00084.x [DOI] [PubMed] [Google Scholar]
- 8. Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2004;2(2):95–108; doi: 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 9. Hall MR, McGillicuddy E, Kaplan LJ. Biofilm: Basic principles, pathophysiology, and implications for clinicians. Surg Infect 2014;15(1):1–7; doi: 10.1089/sur.2012.129 [DOI] [PubMed] [Google Scholar]
- 10. Costerton JW. The etiology and persistence of cryptic bacterial infections: A hypothesis. Rev Infect Dis 1984;6(Suppl 3):S608–S616; doi: 10.1093/clinids/6.supplement_3.s608 [DOI] [PubMed] [Google Scholar]
- 11. Percival SL. Importance of biofilm formation in surgical infection. Br J Surg 2017;104(2):e85–e94; doi: 10.1002/bjs.10433 [DOI] [PubMed] [Google Scholar]
- 12. Laverty G, Gorman SP, Gilmore BF. Biofilms and implant-associated infections. In: Biomaterial and Medical Device Associated Infection. (Barnes L, Cooper I, eds.) Woodhead Publsihing: 2014; pp. 19–45; doi: 10.1533/9780857097224.1.19 [DOI] [Google Scholar]
- 13. Brandwein M, Steinberg D, Meshner S. Microbial biofilms and the human skin microbiome. NPJ Biofilms Microbiomes 2016;2(1):1–6; doi: 10.1038/s41522-016-0004-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Høiby N, Bjarnsholt T, Moser C, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 2015;21:S1–S25; doi: 10.1016/j.cmi.2014.10.024 [DOI] [PubMed] [Google Scholar]
- 15. Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, et al. Identification of prosthetic joint infection pathogens using a shotgun metagenomics approach. Clin Infect Dis 2018;67(9):1333–1338; doi: 10.1093/cid/ciy303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cai Y, Fang X, Chen Y, et al. Metagenomic next generation sequencing improves diagnosis of prosthetic joint infection by detecting the presence of bacteria in periprosthetic tissues. Int J Infect Dise 2020;96:573–578; doi: 10.1016/j.ijid.2020.05.125 [DOI] [PubMed] [Google Scholar]
- 17. Okuda KI, Yoshii Y, Yamada S, et al. Detection of bacterial DNA from central venous catheter removed from patients by next generation sequencing: a preliminary clinical study. Ann Clin Microbiol Antimicrob 2018;17(1):44; doi: 10.1186/s12941-018-0297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hotterbeekx A, Xavier BB, Bielen K, et al. The endotracheal tube microbiome associated with Pseudomonas aeruginosa or Staphylococcus epidermidis. Sci Rep 2016;6(1):1–11 doi: 10.1038/srep36507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakr A, Bregeon F, Mege JL, et al. Staphylococcus aureus nasal colonization: An update on mechanisms, epidemiology, risk factors, and subsequent infections. Front Microbiol 2018;9:2419; doi: 10.3389/fmicb.2018.02419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perl TM, Cullen JJ, Wenzel RP, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med 2002;346(24):1871–1877; doi: 10.1056/NEJMoa003069 [DOI] [PubMed] [Google Scholar]
- 21. Salar-Vidal L, Aguilera-Correa JJ, Brüggemann H, et al. Microbiological characterization of Cutibacterium acnes strains isolated from prosthetic joint infections. Antibiotics 2022;11(9):1260; doi: 10.3390/antibiotics11091260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derks J, Tomasi C. Peri-implant health and disease. A systematic review of current epidemiology. J Clin Periodontol 2015;42(Suppl 16):S158–S171; doi: 10.1111/jcpe.12334 [DOI] [PubMed] [Google Scholar]
- 23. Ghensi P, Manghi P, Zolfo M, et al. Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics. NPJ Biofilms and Microbiomes 2020;6(1):1–12; doi: 10.1038/s41522-020-00155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kong EF, Kucharikova S, Van Dijck P, et al. Clinical implications of oral candidiasis: Host tissue damage and disseminated bacterial disease. Infect Immun 2015;83(2):604–613; doi: 10.1128/IAI.02843-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Engelsman AF, van der Mei HC, Francis KP, et al. Real time noninvasive monitoring of contaminating bacteria in a soft tissue implant infection model. J Biomed Mater Res B Appl Biomater 2009;88(1):123–129; doi: 10.1002/jbm.b.31158 [DOI] [PubMed] [Google Scholar]
- 26. Broekhuizen CA, Sta M, Vandenbroucke-Grauls CM, et al. Microscopic detection of viable Staphylococcus epidermidis in peri-implant tissue in experimental biomaterial-associated infection, identified by bromodeoxyuridine incorporation. Infect Immun 2010;78(3):954–962; doi: 10.1128/IAI.00849-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ochsner PE, Hailemariam S. Histology of osteosynthesis associated bone infection. Injury 2006;37(Suppl 2):S49–S58; doi: 10.1016/j.injury.2006.04.009 [DOI] [PubMed] [Google Scholar]
- 28. Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol 2015;33:227–256; doi: 10.1146/annurev-immunol-032713-120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 2016;167(4):1125–1136; doi: 10.1016/j.cell.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lopetuso LR, Scaldaferri F, Petito V, et al. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathog 2013;5(1):1–8; doi: 10.1186/1757-4749-5-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivera-Chavez F, Zhang LF, Faber F, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe 2016;19(4):443–454; doi: 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becattini S, Littmann ER, Carter RA, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med 2017;214(7):1973–1989; doi: 10.1084/jem.20170495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012;55(7):905–914; doi: 10.1093/cid/cis580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamburini FB, Andermann TM, Tkachenko E, et al. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med 2018;24(12):1809–1814; doi: 10.1038/s41591-018-0202-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freedberg DE, Zhou MJ, Cohen ME, et al. Pathogen colonization of the gastrointestinal microbiome at intensive care unit admission and risk for subsequent death or infection. Intensive Care Med 2018;44(8):1203–1211; doi: 10.1007/s00134-018-5268-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lange K, Buerger M, Stallmach A, et al. Effects of antibiotics on gut microbiota. Dig Dis 2016;34(3):260–268; doi: 10.1159/000443360 [DOI] [PubMed] [Google Scholar]
- 37. Serbanescu MA, Mathena RP, Xu J, et al. General anesthesia alters the diversity and composition of the intestinal microbiota in mice. Anesth Analg 2019;129(4):e126-e129; doi: 10.1213/ANE.0000000000003938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ashley SL, Sjoding MW, Popova AP, et al. Lung and gut microbiota are altered by hyperoxia and contribute to oxygen-induced lung injury in mice. Sci Transl Med 2020;12(556):eaau9959; doi: 10.1126/scitranslmed.aau9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lukovic E, Moitra VK, Freedberg DE. The microbiome: Implications for perioperative and critical care. Curr Opin Anaesthesiol 2019;32(3):412–420; doi: 10.1097/ACO.0000000000000734 [DOI] [PubMed] [Google Scholar]
- 40. Alverdy JC, Krezalek MA. Collapse of the microbiome, emergence of the pathobiome, and the immunopathology of sepsis. Crit Care Med 2017;45(2):337–347; doi: 10.1097/CCM.0000000000002172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moran MM, Wessman P, Rolfson O, et al. The risk of revision following total hip arthroplasty in patients with inflammatory bowel disease, a registry based study. PLoS One 2021;16(11):e0257310; doi: 10.1371/journal.pone.0257310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chisari E, Cho J, Wouthuyzen-Bakker M, et al. Periprosthetic joint infection and the Trojan horse theory: Examining the role of gut dysbiosis and epithelial integrity. J Arthroplasty 2022;37(7):1369–1374; doi: 10.1016/j.arth.2022.03.030 [DOI] [PubMed] [Google Scholar]
- 43. Krezalek MA, Hyoju S, Zaborin A, et al. Can methicillin-resistant Staphylococcus aureus silently travel from the gut to the wound and cause postoperative infection? Modeling the “Trojan horse hypothesis.” Ann Surg 2018;267(4):749–758; doi: 10.1097/SLA.0000000000002173 [DOI] [PubMed] [Google Scholar]
- 44. Zhu H, Jin H, Zhang C, et al. Intestinal methicillin-resistant Staphylococcus aureus causes prosthetic infection via 'Trojan Horse' mechanism: Evidence from a rat model. Bone Joint Res 2020;9(4):152–161; doi: 10.1302/2046-3758.94.BJR-2019-0205.R1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Manor O, Dai CL, Kornilov SA, et al. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun 2020;11(1):1–12; doi: 10.1038/s41467-020-18871-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen S-L, Lundy DJ, Ruan S-C, et al. The gut microbiota regulates acute foreign body reaction and tissue repair after biomaterial implantation. Biomaterials 2022;289:121807; doi: 10.1016/j.biomaterials.2022.121807 [DOI] [PubMed] [Google Scholar]
- 47. Yuzefpolskaya M, Lumish HS, Javaid A, et al. Association of preoperative infections, nasal Staphylococcus aureus colonization and gut microbiota with left ventricular assist device outcomes. Eur J Heart Failure 2021;23(8):1404–1415; doi: 10.1002/ejhf.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guillaume O, Pérez-Tanoira R, Fortelny R, et al. Infections associated with mesh repairs of abdominal wall hernias: Are antimicrobial biomaterials the longed-for solution? Biomaterials 2018;167:15–31; doi: 10.1016/j.biomaterials.2018.03.017 [DOI] [PubMed] [Google Scholar]
- 49. Coenye T, Spittaels KJ, Achermann Y. The role of biofilm formation in the pathogenesis and antimicrobial susceptibility of Cutibacterium acnes. Biofilm 2022;4:100063; doi: 10.1016/j.bioflm.2021.100063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA 2011;108(Suppl 1):4554–4561; doi: 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]