Abstract
The usage of codons and nucleotide combinations varies along genes and systematic variation causes gradients in usage. We have studied such gradients of nucleotides and nucleotide combinations and their immediate context in Escherichia coli. To distinguish mutational and selectional effects, the genes were subdivided into three groups with different codon usage bias and the gradients of nucleotide usage were studied in each group. Some combinations that can be associated with a propensity for processivity errors show strong negative gradients that become weaker in genes with low codon bias, consistent with a selection on translational efficiency. One of the strongest gradients is for third position G, which shows a pervasive positive gradient in usage in most contexts of surrounding bases.
INTRODUCTION
In most organisms, synonymous codons are not used equally. Often the codon choice can be attributed to a general A+T or G+C preference that pervades the entire genome. But in many unicellular organisms, like Escherichia coli, the preferential use of some codons varies from gene to gene and the strength of the preference—the codon bias—increases in genes at high expression level (1). This suggests that there is a positive selection on codons that are translated more efficiently, either faster or more accurately. The strength of the codon bias to some extent also depends on gene length (2) and on the context of bases surrounding each codon (3–5). Average codon bias also varies within each gene, increasing in the first 100 codons (6,7), leveling off and remaining constant until it again declines in the last 20 codons (8). Some variation in average codon usage as well as in third-position G+C content along the genes of E.coli has been reported (9). A more pervasive gradient in codon usage has been found for the Phe codons (TTC and TTT) before T and C (5), where TTT is less favoured the farther from the translation start the Phe codon is. This can be interpreted as an avoidance of a nucleotide combination that is particularly prone to +1 frameshifts. The further translation has proceeded before the frameshift, the larger the cost in wasted ribosome activity and the stronger the selection to avoid it. That such frameshifts occur has been observed experimentally (10). Thus, a strong gradient in nucleotide usage could be a signal that the nucleotide combination in question is prone to errors in translational processivity (e.g. drop-off, false stops, frameshifts, etc.). For rapidly propagating organisms such as E.coli, selection against wasted ribosomal time would be important (11). The nucleotide distribution and synonymous codon choice will also influence secondary structure in the mRNA (12). Shifting secondary structure requirements along the messenger could therefore also give rise to nucleotide gradients.
In this paper we report a systematic study of the gradients in the usage of nucleotides and nucleotide combinations up to codons and their immediate context along the genes of E.coli. The main emphasis is on the variation in the internal regions of the genes beyond the first 100 codons and before the last seven codons. To set the results in perspective, some limited analyses were also performed on the genome from Bacillus subtilis. We find a number of nucleotide combinations that show significant gradients along the E.coli genes. Most depend on expression level and may be determined by translational efficiency, and a few can be directly identified as caused by an avoidance of processivity errors.
MATERIALS AND METHODS
The complete E.coli genome was acquired from GenBank FTP. The full data set was divided into three groups according to the value of their codon adaptation index (CAI) (13). The choice of CAI groups is the same as used by Berg and Silva (5). This resulted in high (H), medium (M) and low (L) bias groups (Table 1). We will use the CAI value as an estimate of the expression level of a gene. From the full data set, two further data sets were assembled: membrane proteins were identified by calculating the proportion of hydrophobic amino acids in the first 500 codon positions, using the (A-T)2 skew (14). Potential horizontally transferred (HT) genes were identified using the methods of Lawrence and Ochman (15). The HT genes were particularly numerous in the L set where they had a noticeable influence on the observed gradients. All results reported for the L set below are after removal of the putative HT genes.
Table 1. Distribution of E.coli genes by expression group and length.
| Expression group | All genes | defa | Long genes | def | CAI values |
|---|---|---|---|---|---|
| High | 776 | Hfull | 201 | H | CAI > 0.4 |
| Medium | 1284 | Mfull | 250 | M | 0.315 < CAI ≤ 0.4 |
| Lowb | 1997 | Lfull | 303 | L | CAI ≤ 0.315 |
| Total | 4057 | 754 |
adef denotes the abbreviated term used for this set of genes.
b233 presumed horizontally transferred genes have been removed from the L set.
A subset of long genes was extracted from each data set and codon frequencies were calculated only in the regions where all genes contribute, i.e. below the cutoff length. As a compromise between having a large sample number and a long common region, the long-gene cutoff was chosen to be at 1401 bp; this left ∼200 genes in each group (Table 1). This rough equality in the size of the groups provides some statistical justification for the choice of CAI values. The genes were then divided into windows of 60 bp and the frequency of codon usage calculated in each window. The gradients in the usage of all 1, 2, 3 and 4 nucleotide combinations, including codons and their immediate context, were calculated from linear regression. To avoid effects from start and stop regions, the slopes of the regression lines were calculated between windows 6 and 23, which leaves 100 codons to the start and at least 7 codons to the gene ends. Analysing only the common regions of long genes eliminates spurious gradients occurring due to differences in usage at gene ends as well as in genes of different lengths. However, for comparison, in some cases the gradients in sets including all gene lengths were also considered.
For each nucleotide combination, the intercept at window 6, p0, and the slope, S, of the regression line were calculated. To get an estimate of the strength of the position-dependent selection, we also calculated a selection coefficient, s, defined from the frequency f(j) in window j from the gene start.
f(j) = p0 + (j – 6)S ≈ p0e(j – 6)s
Thus, the positional selection coefficient is s ≈ S / p0, such that s is a measure of selection that is independent of the overall usage, p0. Both slope and selection reported below are counted per window and not per codon or bp.
The significance of the regression slopes was estimated from 10 000 binomial simulations of a position-independent codon usage and calculating the sample variance, σs2, of the random slopes that occurred. Normally, significance tests of linear regression would follow a t-distribution. However, it was decided that simulations were more informative and accurate, given the prior knowledge of the variance of the data points. As a measure for the significance in the observed gradient, slope S, for a codon we use the reference variable RV = S/σs. Values of │RV│ > 2 and │RV│ > 3 correspond to confidence levels P < 0.05 or < 0.005, respectively.
For comparison and support, a further set of genes was extracted from B.subtilis, also found in GenBank. Bacillus subtilis was chosen since it is a fully sequenced free-living bacterium that displays some codon bias and which is not too closely related to E.coli. These genes were also divided into three sets based on their CAI values and subsets of long genes were considered, analogously in both aspects to the subdivision of E.coli genes. In B.subtilis H, we found 159 long genes. For more limited comparisons, we also looked at the full sequences of yeast chromosome 4, Rickettsia prowazekii and Haemophilus influenzae. These data sets were not processed as E.coli was, but were used in the way they appear in GenBank.
Nucleotide and codon preference can have many causes. Mutational bias may have local variations, e.g. on leading and lagging strands, but these variations should not affect the gradient analysis. Bias that is caused by selection on translational efficiency will become weaker in genes at low expression level. If this efficiency is based on avoidance of missense errors, it has been suggested that the selection would be proportional to gene length, since a severe error would lead to the loss of the whole protein (2). On the other hand, for processivity errors, i.e. drop-off, false stops or frameshifts, the cost will be proportional to the length of protein produced before the error; in this case, there will be a negative gradient in the usage of error-prone sequences (5). Clearly, gradients in codon usage will also lead to an apparent dependence on gene length for the average usage.
Codon notations
In this report, we will use the following notations: the current codon will have bases denoted by 1, 2, 3, while bases in context before will have negative numbers. Subsequent bases will be numbered from 4 and up, i.e.
G │C T G│ C
-1 1 2 3 4
where vertical lines show the reading frame. G3 therefore always denotes the third position nucleotide of the current codon. Similarly, R–1 denotes a preceding third position purine and Y1 denotes a first position pyrimidine.
Correlation of gradients with gene length
Since the E.coli data sets have a highly heterogenous composition of gene lengths, it is important to investigate whether genes of different lengths have different attributes which would complicate the evaluation of gradients. For example, longer genes having higher G3 levels could be explained by either a higher general G3 content or by a positive gradient of G3 in individual genes.
To verify that there is no difference in gradients in H and the subset of long genes, the full H (Hfull) data set was compared to the subset of genes longer than 1401 bp (referred to as H). Pairwise confidence testing of gradients in both Hfull and H showed that there was no significant difference in gradients between data sets. Tests did not involve further subdivision of the Hfull data set into a short subset, since these genes would not fully contribute to as many windows as H.
However, while there was no significant difference between pairwise gradients, there may still be codon combinations with special considerations in subsets of shorter genes. For example, the C3 gradient is not significant in Hfull, but is significantly negative in H. Another example of artefactual effects from short genes is the (A-T)2 skew, discussed below. Therefore, all calculations hereafter use the long subset of genes from each data set.
RESULTS
Amino acid and codon bias gradients
Amino acid composition by position varies little in the H set. Repeats of amino acids on the protein function level have little effect on a compound of approximately 200 highly expressed genes. The non-degenerate Met and Trp have no significant gradients, therefore not affecting gradients in G3. Of 20 amino acids in H, only tyrosine has a significant gradient (RV = –2.0), indicating that the majority of gradients presented in this article are of a synonymous nature. One outlier with │RV│ > 2.0 is exactly what we would expect from a random distribution of 20 gradients with an average RV = 0 and std = 1, suggesting that there is no significant positional selection on the amino acid level in E.coli. There is no gradient in CAI by position, as previously noted (8), either in long or full sets of genes.
G3 and C3
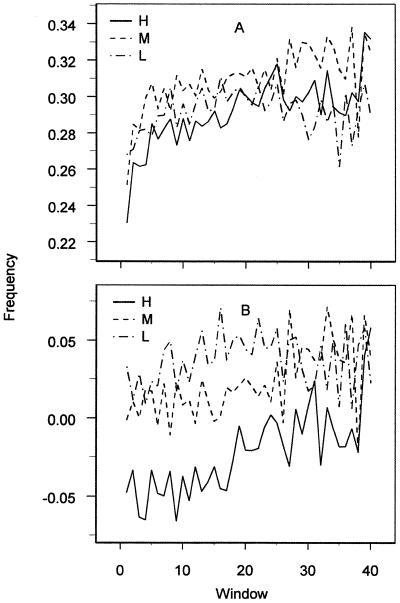
In Table 2 we have listed the results for the usage of single nucleotides at each of the three codon positions in the H set of E.coli. The most prominent gradient is third position guanine (G3). It is significant in H and M and of similar magnitude in all three E.coli data sets, though progressively weaker in genes of lower expression level (Fig. 1A). Ten out of 15 G-ending codons have positive gradients. In some cases, minor codons of amino acids have positive gradients when having a G in third position, e.g. Lys, Thr and Glu, implying that there is no correlation to the major/minor status of a codon. Although the positive G3 gradient is pervasive, there are some interesting variations depending on the immediate context: for instance, G3 followed by A shows the strongest positive gradient (significance RV = 5.1, selection s = 1.4 × 10–2), while G3 followed by C is slightly negative (except CTG│C which is strongly positive in H, RV = 3.5, s = 1.8 × 10–2). The combination NGG also has a negative slope; although not significantly different from zero (RV = –1.2, s = –5.1 × 10–3), it is significantly different from the positive slopes of the other N2G3 combinations (RV = 2.1 to 3.1, s = 4.8 × 10–3 to 8.8 × 10–3). The strongest contribution both to NGG and NNG│C comes from NGG│C. The gradient in G│NNG, i.e. for two consecutive G-ending codons, is significantly lower than expected from the single occurrences of G3 in H, but not in M. Studies of G3 in context show that G3 has a positive gradient when succeeded by any of the four nucleotides in the following first position. Moreover, G3 has a positive gradient after A2, T2 and C2, but as mentioned, not after G2. Of the 16 (N2, N4) combinations with NG│N, 11 are positive. Of these, four are significant. These observations suggest that the G3 gradient may be caused by a selection on the singlet level, i.e. G3 seems to be increasingly preferred towards the ends of genes regardless of its context and the strength of the preference increases with expression level.
Table 2. The observed singlet gradients in the E.coli H set.
| Base | p0 | s | RV | |
|---|---|---|---|---|
| N1 | A | 0.240 | 2.61 × 10–3 | 2.246 |
| C | 0.240 | –4.70 × 10–4 | –0.405 | |
| G | 0.380 | –3.71 × 10–4 | –0.487 | |
| T | 0.140 | –2.67 × 10–3 | –1.631 | |
| N2 | A | 0.318 | –9.04 × 10–4 | –0.914 |
| C | 0.222 | 6.93 × 10–4 | 0.572 | |
| G | 0.173 | 9.54 × 10–4 | 0.675 | |
| T | 0.287 | –1.12 × 10–4 | –0.105 | |
| N3 | A | 0.153 | –1.71 × 10–3 | –1.109 |
| C | 0.337 | –2.78 × 10–3 | –2.979 | |
| G | 0.269 | 5.15 × 10–3 | 4.692 | |
| T | 0.241 | –7.65 × 10–4 | –0.660 |
Columns show, in order: intercept at window 6, p0; selection parameter, s; and relative significance, RV.
Figure 1.
(A) The frequency of G3 plotted against window in E.coli data sets H, M and L. (B) The frequency of (G-C)3 plotted against window in E.coli data sets H, M and L. Windows are 60 bp long.
The C3 gradient is significant only in H (Table 2). Like NGG, NCC is positionally avoided in H. C3 is negative in 11 of all 16 (N2, N4) contexts, but only NAC│C and NCC│G are significant. For C3 in an N1, N2 context, GCC (Ala) and CGC (Arg) are two significant contributors to the C3 gradient, and both are minor codons. In contrast to G3, the gradient of C3 seems to be dominated by contributions from a few codons and nucleotide combinations. Since C3 is largely insignificant with some negative contributions and G3 is largely significantly positive with some insignificant contributions, there does not seem to be a selection acting on C3 as a singlet.
In the H set, there is a positive (G+C)3 gradient. Karlin et al. (9) found a positive correlation between (G+C)3 content and gene length; however, we find no significant difference in the (G+C)3 content in codon position 100–200 between long (>1401 bp) and short genes, indicating that (G+C)3 is more correlated to position than to gene length. Thus, the apparent difference in groups of genes with different lengths can be attributed mainly to positional gradients of G3 and C3.
(G-C)3 gradient
Since C3 is significantly negative in H and insignificant in L and M, and G3 is significantly positive in H and M, there is consequently a strong positive gradient in the (G-C)3 skew. This is displayed in Figure 1B. The (G-C)3 gradient is consistent through all three data sets, which indicates a positional selection. As noted above, the (G-C)3 gradient appears to be more an effect of a pervasive G3 gradient than of a positional preference for a high (G-C)3 on the gene level.
Genomic strand asymmetries in G3 and C3 have been studied and summarized extensively by Frank and Lobry (16), who have suggested that asymmetry is primarily caused by biases in mutational mechanisms. To test the effect of strand asymmetry on positional selection, our data set was subdivided in accordance with their genomic position and orientation (leading or lagging). When comparing leading and lagging H genes on the first 2 Mb clockwise from oriC, no significant difference in (G-C)3 skew or its gradient was observed. An analogous comparison of L genes did show a clear difference in (G-C)3 skew intercept, while both leading and lagging L genes had the same gradient. Positional selection therefore acts independently of the leading-lagging strand substitution bias. To give an idea of the size of the (G-C)3 gradient in the H data set, it can be noted that it is of the same magnitude as the difference in (G-C)3 skew between leading and lagging strands.
Rho-dependent transcription termination and (G-C)3
Transcriptional terminators serve important purposes at the end of operons and have regulatory functions when found directly after promoters or between operonic genes (17). They are also found within genes where they are activated when transcribed genes are not translated due to amino acid starvation or erroneous transcription initiation. Because rho-dependent transcription termination sites have a characteristic region of C > G (18), such sites could contribute to the (G-C)3 gradient, if they are positioned primarily in the beginning of genes. However, the exact ratio of C to G does not seem to be very stringent (19).
We propose that the (G-C)3 gradient is not an effect of rho-dependent transcription termination, due to the following observations. (i) There is no significant covariance between G3 and C3 in individual genes other than the expected multinomial covariance. (ii) G3 and C3 for the whole data set divided into windows of 20 bases show a constant covariance score for windows five and up, implying that there is no bias in position of covarying sites. (iii) The distribution of the covariance of G3 and C3 is symmetric, i.e. there are as many sites with high G3/low C3 as there were sites with low G3/high C3. A distribution biased towards C > G would be expected.
In summary, the primary factor in the (G-C)3 gradient seems to be G3 itself. There are no effects of transcription termination regions.
Potential frameshift sites
A potential frameshift-sensitive site is a codon with an out-of-frame context that codes for the same amino acid, e.g. TTT│T or A│AAG, where slippage of the ribosome could occur forwards or backwards respectively. In the case of Phe, the bias and the negative gradient of ![]() has been interpreted as an avoidance of frameshift sensitive sites in a forward direction (5). Similarly,
has been interpreted as an avoidance of frameshift sensitive sites in a forward direction (5). Similarly, ![]() may show an avoidance of backward frameshifting.
may show an avoidance of backward frameshifting.
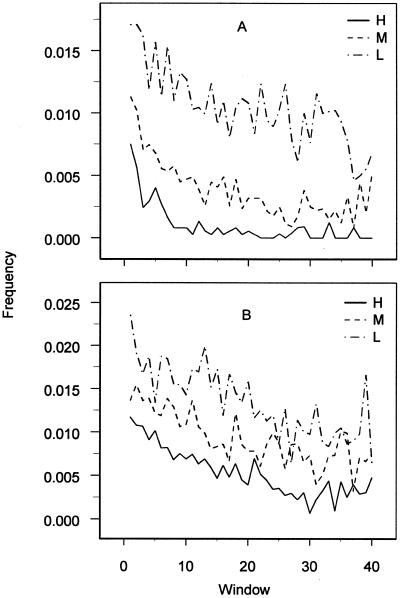
Strong negative gradients are suggestive of frameshift avoidance. This was observed for forward frameshifting of TTT in H and to a lesser degree in the M and L sets for both E.coli and B.subtilis (Fig. 2). Forward frameshifting of Lys is more complicated to assess; primarily due to the rarity of the synonymous codon AAG. There is a positional avoidance of AAA│A (RV = –2.3, s = –1.3 × 10–2), compensated for by AAG│A (RV = 3.3, s = 6.8 × 10–2). However, there is no gradient in avoidance of AAA│G, possibly due to the rarity of the synonymous AAG│G. In any case, the avoidance of Lys forward frameshifting seems weaker than for Phe forward frameshifting (TTT│Y; RV = –2.5, s = –4.4 × 10–2).
Figure 2.
(A) The frequency of the frameshift sensitive site TTT│Y plotted against window in E.coli data sets H, M and L. (B) TTT│Y in B.subtilis.
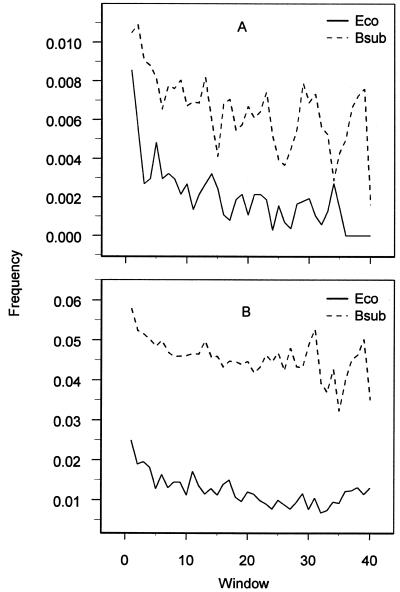
The interpretation of the potential backward frameshifting of Lys and Phe was complicated by a general expression-dependent gradient of avoidance of T│TTN and A│AAN (Table 3), where gradients in lower expression groups are less significant (data not shown). These sites are also implicated in missense errors, as discussed below. The negative gradients for T│TTR (Leu) and A│AAY (Asn), suggest sensitivity to some kind of processivity error, possibly frameshift coupled with missense (see below). Backward frameshifting of these sites seems to be of much lower probability than the corresponding forward frameshifts. For Phe backwards frameshifting, T│TTC has no gradient, which is consistent with a wobbling anticodon binding stronger to C than to T. Although the avoidance of T│TTC is weak, these frameshifts are still physically possible (20). T│TTT for E.coli H and B.subtilis H is shown in Figure 3A and A│AAN for E.coli H and B.subtilis H is shown in Figure 3B. In all data sets, the Pro codon CCC and Gly codon GGG were too rare for accurate appreciation of any avoidance of frameshifting.
Table 3. Summary of backwards frameshift- and missense-sensitive sites involving A and T in the E.coli H set.
| Codon | p0 | s | RV |
|---|---|---|---|
| T│TTN | 1.06 × 10–2 | –8.62 × 10–3 | –1.49781 |
| T│TTA | 1.22 × 10–3 | –2.18 × 10–2 | –0.8337 |
| T│TTC | 4.74 × 10–3 | 1.69 × 10–3 | 0.185098 |
| T│TTG | 1.47 × 10–3 | –7.50 × 10–4 | –0.03383 |
| T│TTT | 3.20 × 10–3 | –2.25 × 10–2 | –1.88118 |
| A│AAN | 1.69 × 10–2 | –1.73 × 10–2 | –3.70924 |
| A│AAA | 6.84 × 10–3 | –1.29 × 10–2 | –1.77375 |
| A│AAC | 7.11 × 10–3 | –1.94 × 10–2 | –2.72525 |
| A│AAG | 9.71 × 10–4 | –2.36 × 10–2 | –0.73984 |
| A│AAT | 1.95 × 10–3 | –2.18 × 10–2 | –1.24197 |
Figure 3.
(A) The frequency of T│TTT plotted against window in E.coli H and B.subtilis H. (B) The frequency of A│AAN plotted against window in E.coli H and B.subtilis H.
Potential frameshift sequences T│TTY, TTT│Y and G│GGN show negative gradients also in B.subtilis, particularly the forward frameshifting of Phe (Fig. 2B). However, the counter-selection is weaker than in E.coli, reflecting the weaker translational selection also shown by the lower codon bias of B.subtilis (21).
It has been suggested that codons of the form G-nonG-N, through their complementarity with a C-periodical section of 16S rRNA, can help the ribosome keep its correct frame during translation (22,23). We find no significant gradient in this nucleotide combination in any frame, suggesting that this sequence does not affect frame keeping.
Near-stop errors
Like frameshift errors, premature termination by release factors infer a cost of error proportional to the distance from initiation. The accuracy of RF1 and RF2 in the presence and absence of RF3 was investigated by Freistroffer et al. (24), who rate the near-stops (i.e. codons that are one base different from a real stop codon) according to the decrease in kcat/Km compared with the true stops. Since TAT is the major contributor to false termination by misreading as TAA (24), the gradients of TAT with and without context were examined. TAT in itself is significantly negative in H (RV = –3.0, s = –1.4 × 10–2) and slightly less negative in M (RV = –1.9, s = –8.4 × 10–3). In the H set, we find negative gradients for both TAT and TAC and no bias for avoidance of the error-prone TAT over TAC. As previously mentioned, tyrosine in itself has a negative gradient in H, complicating analysis of false termination errors. Given that +4 context increases the strength of termination (25,26), it is probable that release factor misreading may also be amplified by context. However, there are no particularly significant gradients in any of the +4 contexts. Nor do we find any significant gradients in other near-stop sequences.
TAT as a potential false termination signal may be supported by comparisons with the B.subtilis H data set, where TAT is avoided with a gradient similar to the E.coli M set. However, based on the weak gradients in usage observed, it seems that the avoidance of near-stop errors is slight at best.
Missense translational errors
Precup et al. (27) have shown a high level of mistranslation of TTC and TTT as Leu in a Phe-starved E.coli. In the argI gene product, there are phenylalanines at codon positions 3 and 8, TTT and TTC, respectively. Since an interchange of synonymous codons at the two positions did not change the high error rate at position 8, the context appears to be a more important factor than the actual codon used. Position 3 of the argI gene is G│TTT│T and T│TTC│C at position 8. The missense errors at position 8 would in some way be induced by the T(–1). T in context before TTN is underrepresented in the H set and not in L as is A│AAN. This suggests an avoidance based on translational selection, possibly due to potential missense errors. However, these combinations also show negative gradients and are increasingly avoided towards the gene ends, suggesting avoidance of processivity error (Table 3).
In T-rich surroundings, TGT (Cys) can be misread as Trp (28). T│TGT has an insignificantly negative gradient in M, while it is too rare in H to calculate. T in context after TGT is also too rare or suppressed in both H and M for evaluation of the gradient. Both T│TGT and TGT│T are underrepresented in H but not in L, consistent with an avoidance of missense error.
Missense avoidance is not expected to depend on position, since the ribosome will in any case complete translation of the protein. In those cases where avoidance seems to have a positional component, such as A│AAN and T│TTN, there may be risk of some additional types of errors. It has been suggested (29) that a missense error could increase the probability of processivity error by destabilizing the translation complex. Since processivity errors are costly, even a very small increase in this probability may lead to selectional avoidance and a gradient in usage.
Nucleotide combinations with significantly deviating gradients
There are a number of combinations that are significant in E.coli, which have not been covered above. In most cases, these gradients can be attributed more strongly to their individual components than to the combination itself. Prime examples of this are the NNG codons. To account for the contributions of individual nucleotides to the gradients of two or more combined nucleotides, we performed a regression test of deviation. If the gradient deviates by more than 3 standard deviations [t0.01(17) = 2.9] from that expected from the gradients of the corresponding singlets, it is considered significant. The reason for using a standard regressional test of significance, rather than simulations, is that the prior knowledge of variances is already taken into account through RV. This test simply examines the possible gradients that could arise from independent nucleotides. The results are shown in Table 4.
Table 4. Combinations with significant deviations in respect to gradient.
| Codon | RV | s | Codon | RV | s |
|---|---|---|---|---|---|
| A│AAN | –3.71 | –1.73 × 10–2 | GNG│T | 3.67 | 2.59 × 10–2 |
| A│CTG | 4.04 | 2.75 × 10–2 | T│CGT | 3.69 | 3.92 × 10–2 |
| A│CTN | 3.03 | 1.80 × 10–2 | T│GCA | 3.26 | 5.72 × 10–2 |
| A│NCG | 3.60 | 1.95 × 10–2 | T│NGT | 4.70 | 2.64 × 10–2 |
| A│NTG | 4.29 | 2.04 × 10–2 | TAC│A | 2.68 | 3.11 × 10–2 |
| A│NTT | –3.55 | –1.99 × 10–2 | TCN│A | 4.51 | 4.05 × 10–2 |
| AGG | –0.10 | –2.14 × 10–2 | TGN│G | 2.86 | 2.14 × 10–2 |
| ATN│C | 3.48 | 1.66 × 10–2 | TTT | –3.40 | –1.57 × 10–2 |
| CGT│A | 3.21 | 2.52 × 10–2 | TNC│A | 2.76 | 1.53 × 10–2 |
| CTG│C | 3.47 | 1.77 × 10–2 | NCC│G | –3.92 | –1.39 × 10–2 |
| G│CAG | 4.45 | 3.29 × 10–2 | NCG│A | 4.66 | 2.28 × 10–2 |
| G│GGT | 4.80 | 5.68 × 10–2 | NGG│C | –2.35 | –1.59 × 10–2 |
| G│TCT | 4.31 | 2.28 × 10–1 | NGG | –1.22 | –5.13 × 10–3 |
| G│TCN | 3.94 | 3.69 × 10–2 | NGT│A | 5.70 | 2.88 × 10–2 |
| G│NGG | –0.13 | –1.01 × 10–3 | NGT | 3.94 | 1.06 × 10–2 |
| GCC│G | –3.42 | –1.84 × 10–2 | NTT│A | –2.93 | –1.28 × 10–2 |
| GCG│A | 4.18 | 3.10 × 10–2 | NTT | –3.39 | –8.36 × 10–3 |
| GGT│A | 3.55 | 2.49 × 10–2 |
Effects in the first 20 and 100 codons
In all three bias groups of E.coli and B.subtilis, A3 is more preferred and G3 more avoided in the first 20 codons (smaller effects on T and C). The same effect, though a little weaker, is evident in A1 and G1, but not A2 and G2. In R.prowazekii the same trends are present, although they are stronger for A1 and G1 than for A3 and G3, possibly because A3 and G3 usage is already very extreme [synonymous (G+C)3 content = 17%] (30). This seems to be an effect of conflicting selection (7) to accommodate initiation signals or to avoid secondary structure.
In the high-bias group of E.coli, there is a gradual increase in G3 and C3 together with a decrease in A3 and T3 through the first 100–150 codons. When this variation is subdivided into the different synonymous codon choices, one finds that among the G-ending codons only the preferred ones show a significant increase, while among the A-ending codons only the minor ones show a significant decrease between positions 20 and 100. This supports the notion that the variation in codon choice in this region of the genes is caused by a reduced selection towards the beginning (6) rather than conflicting selection (7). In B.subtilis, there is no significant variation in the first 100 codons beyond the initial 20.
Distribution of membrane protein genes
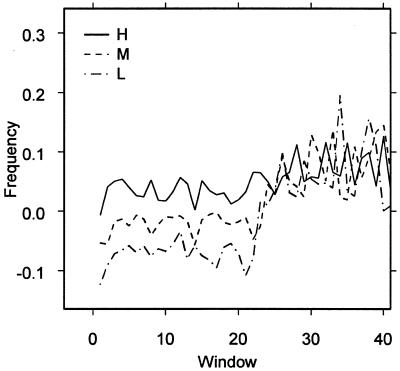
Membrane proteins have a decisively hydrophobic content, reflected in a negative (A-T)2 skew (14). This group of genes had an impact on the overall (A-T)2 skew of the full data sets. The total (A-T)2 skew was calculated for the full E.coli data set and an outlying group with a low (A-T)2 skew was extracted from the different expression groups. The distribution of membrane protein genes was found to be 5.12% in Hfull, 7.63% in Mfull and 13.90% in Lfull.
This distribution of membrane protein genes produces an (A-T)2 skew in the three full data sets with a gradient that decreases with increasing expression level. This gradient was highly deceptive; windows 6–20 had no gradient, but windows 21 and up were elevated, producing an artefactual gradient (Fig. 4). However, when the genes coding for membrane proteins were excluded from the main data sets, no (A-T)2 skew was apparent in the main sets of genes. This would imply that hydrophobic regions of genes are evenly distributed in non-membrane protein coding genes.
Figure 4.
The (A-T)2 gradient plotted against window in E.coli data sets Hfull, Mfull and Lfull.
The membrane protein coding genes are generally shorter than the other genes and, therefore, the (A-T)2 skew in these genes introduces an apparent strong gradient when the full data sets are considered. This behaviour is consistent in all five genomes we have looked at. In addition, the longest of the membrane protein coding genes have a higher hydrophilic content towards their end, further inducing a strong (A-T)2 skew gradient in the full data sets. The apparent (A-T)2 gradient disappears when the long subsets of genes are considered.
DISCUSSION
Processivity errors in translation are common and costly (31). Sequences that are prone to such errors are therefore expected to show strong negative gradients in usage. We found fewer such gradients than expected, possibly because of the limitation to four successive nucleotides. Gradients in the usage of longer sequences would be very difficult to observe due to low levels of occurrence and processivity errors that require more than four nucleotides for specification would consequently not be detected in this way. The clearest negative gradients were found in potential frameshift sensitive sites. Nucleotide combinations with positive gradients are also found, which do not seem to be an effect of compensations for other, avoided, combinations. An important point is the observation that processivity affects a positional selection which is mainly retrospective, i.e. selection rarely ‘looks downstream’. This explains differences in overall G3 levels in genes of different lengths. In other individual nucleotide combinations, large differences in gradients between full data sets and long subsets can be dismissed as chance with a high degree of confidence. Comparisons between the three E.coli data sets also show, in many cases, that the gradients change in the same direction when going from H to M and M to L. Of the 1116 nucleotide combinations studied, 175 have significant gradients in H. Of these, 115 show such consistent changes through M and L. This observation strengthens the confidence in the gradients from the individual groups. The most prominent consistent relations include G3, C3 (becomes slightly positive in L), A│AAN, TTT, AAA│A, NCC and AAG│A, while A1 does not change consistently with expression level. It can be concluded that there is a correlation between positional selection and expression level.
For triplets, changes in codon preference may occur due to speed or accuracy. We find no relationship between major/minor status of a codon and its gradient of usage. In a comparison between values for translation speeds (32) and gradients of usage, 6 out of 9 codons with positive gradients had a faster speed of translation than their synonyms. Due to the limited translation speed data, no significance tests can be made.
The choice of the single nucleotide G3 seems to be of some importance for E.coli. The gradients in (G+C)3 and the (G-C)3 skew appear primarily to be consequences of G3. The difference between the C3 and G3 gradients is that the negative C3 gradient appears to have strong contributions from a smaller number of codon combinations. Elimination of NCC from the data set renders the C3 gradient insignificant in the H set. There is a tendency for an avoidance which increases with distance from initiation of the doublets NCC, NGG and NTT.
Possible advantages of a positive G3 gradient along the genes include the following. (i) The G3 codons may simply be translated with a lower degree of processivity errors than other codons. (ii) If G3 codons are translated faster than other codons, congestion of ribosomes may be avoided by a gradual increase of the speed of translation along the gene. It is possible that ribosomal congestion may increase the frequency of translational errors, although there is little experimental data on the mechanisms of ribosome congestion. (iii) Guanine is an effective base in secondary mRNA structure formation since it binds both T and C. Secondary structure may be stabilizing or regulatory in nature. Explanations (i) and (ii) rely on translational efficiency, which correlates with a gradient in G3 that decreases with expression. The observed gradient was greatest in H and lowest in L. Explanation (iii) would suggest that a positive G3 gradient could also be a general feature in other genomes. Although B.subtilis does have a slightly positive G3, the overall average level of G3 is low in accordance with its lower GC content. The conclusion from comparisons with B.subtilis must be that a high G3 bias and gradient in E.coli is organism-specific. Furthermore, if the G3 gradient was determined by a need for secondary structure towards gene ends, one would expect that it would be coupled to an increasing avoidance of the poor structure formed by A3; this is not observed.
The strength of a negative gradient of sequence usage along genes may be used as a measure for the propensity of processivity errors in genomes with strong selection on translational efficiency. A positive gradient, like the one for G3 in E.coli, has no such obvious explanation but must also reflect some intrinsic molecular mechanism of mutation or selection.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grants from the Swedish Natural Science Research Council.
REFERENCES
- 1.Gouy M. and Gautier,C. (1982) Nucleic Acids Res., 10, 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre-Walker A. (1996) Mol. Biol. Evol., 13, 864–872. [DOI] [PubMed] [Google Scholar]
- 3.Yarus M. and Folley,L.S. (1985) J. Mol. Biol., 182, 529–540. [DOI] [PubMed] [Google Scholar]
- 4.Gouy M. (1987) Mol. Biol. Evol., 4, 426–444. [DOI] [PubMed] [Google Scholar]
- 5.Berg O.G. and Silva,P.J.N. (1997) Nucleic Acids Res., 25, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulmer M. (1988) J. Theor. Biol., 133, 67–71. [DOI] [PubMed] [Google Scholar]
- 7.Eyre-Walker A. and Bulmer,M. (1993) Nucleic Acids Res., 21, 4599–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyre-Walker A. (1996) J. Mol. Evol., 42, 73–78. [DOI] [PubMed] [Google Scholar]
- 9.Karlin S., Mrázek,J. and Campbell,A.M. (1998) Mol. Microbiol., 29, 1341–1355. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz R. and Curran,J.F. (1997) Nucleic Acids Res., 25, 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrenberg M. and Kurland,C.G. (1984) Q. Rev. Biophys., 17, 45–82. [DOI] [PubMed] [Google Scholar]
- 12.Seffens W. and Digby,D. (1999) Nucleic Acids Res., 27, 1578–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp P.M. and Li,W.-H. (1987) Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mrázek J. and Kypr,J. (1994) J. Mol. Evol., 39, 439–447. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence J.G. and Ochman,H. (1998) Proc. Natl Acad. Sci. USA, 95, 9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank A.C. and Lobry,J.R. (1999) Gene, 238, 65–77. [DOI] [PubMed] [Google Scholar]
- 17.Richardson J.P. (1991) Cell, 64, 1047–1049. [DOI] [PubMed] [Google Scholar]
- 18.Alifano P., Rivellini,F., Limauro,D., Bruni,C.B. and Carlomagno,M.S. (1991) Cell, 64, 553–563. [DOI] [PubMed] [Google Scholar]
- 19.Zalatan F. and Platt,T. (1992) J. Biol. Chem., 267, 19082–19088. [PubMed] [Google Scholar]
- 20.Barak Z., Lindsley,D. and Gallant,J. (1996) J. Mol. Biol., 256, 676–684. [DOI] [PubMed] [Google Scholar]
- 21.Shields D.C. and Sharp,P.M. (1987) Nucleic Acids Res., 15, 8023–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trifonov E.N. (1987) J. Mol. Biol., 194, 643–652. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez G., Márquez,L. and Marín,A. (1996) Nucleic Acids Res., 24, 2525–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freistroffer D.V., Kwiatkowski,M., Buckingham,R.H. and Ehrenberg,M. (2000) Proc. Natl Acad. Sci. USA, 97, 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tate W.P. and Mannering,S.A. (1996) Mol. Microbiol., 21, 213–219. [DOI] [PubMed] [Google Scholar]
- 26.Pavlov M.Y., Freistroffer,D.V., Dincbas,V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1998) J. Mol. Biol., 284, 579–590. [DOI] [PubMed] [Google Scholar]
- 27.Precup J., Ulrich,A.K., Roopnarine,O. and Parker,J. (1989) Mol. Gen. Genet., 218, 397–401. [DOI] [PubMed] [Google Scholar]
- 28.Carrier M.J. and Buckingham,R.H. (1984) J. Mol. Biol., 175, 29–38. [DOI] [PubMed] [Google Scholar]
- 29.Kurland C.G. and Ehrenberg,M. (1985) Q. Rev. Biophys., 18, 423–450. [DOI] [PubMed] [Google Scholar]
- 30.Andersson S.G.E. and Sharp,P.M. (1996) J. Mol. Evol., 42, 525–536. [DOI] [PubMed] [Google Scholar]
- 31.Jørgensen F. and Kurland,C.G. (1990) J. Mol. Biol., 215, 511–521. [DOI] [PubMed] [Google Scholar]
- 32.Curran J.F. and Yarus,M. (1989) J. Mol. Biol., 209, 65–77. [DOI] [PubMed] [Google Scholar]