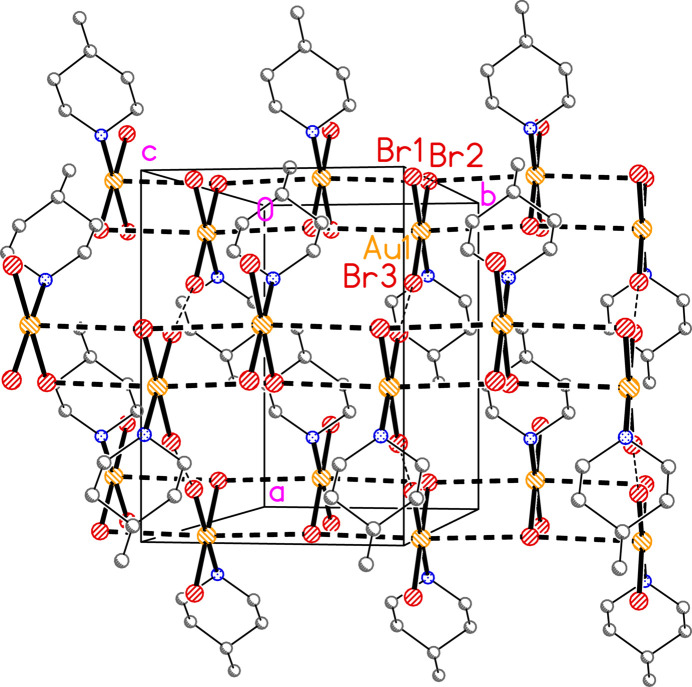
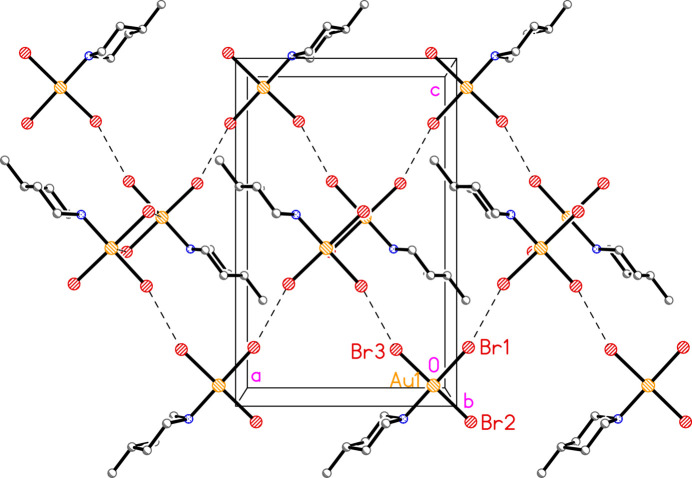
All three structures involve chains of molecules linked by Au⋯halogen contacts; these are accompanied by hydrogen bonds or halogen⋯halogen contacts.
Keywords: crystal structure, gold, methylpiperidine, hydrogen bonds, polymorph
Abstract
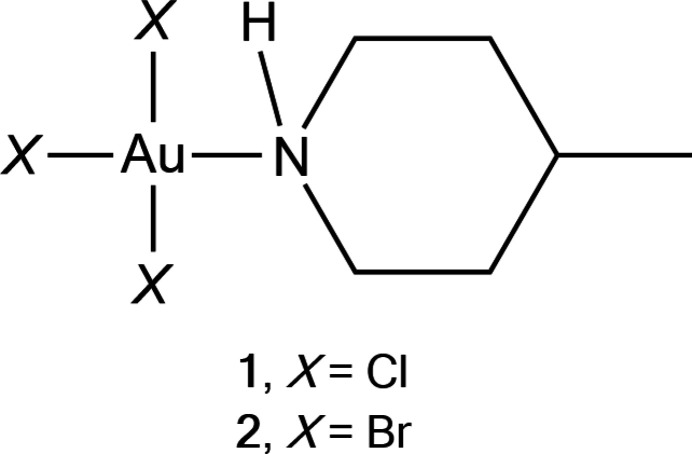
Trichlorido(4-methylpiperidine)gold(III), [AuCl3(C6H13N)], 1, crystallizes in Pbca with Z = 8. Tribromido(4-methylpiperidine)gold(III), [AuBr3(C6H13N)], 2, crystallizes as two polymorphs, 2a in Pnma with Z = 4 (imposed mirror symmetry) and 2b, which is isotypic to 1. The Au—N bonds trans to Cl are somewhat shorter than those trans to Br, and the Au—Cl bonds trans to N are longer than those cis to N, whereas the Au—Br bonds trans to N are slightly shorter than the cis bonds. The methyl and AuX 3 groups (X = halogen) occupy equatorial positions at the six-membered ring. The packing of all three structures involves chains of molecules with offset stacking of the AuX 3 moieties associated with short Au⋯X contacts; for 1 and 2b these are reinforced by N—H⋯X hydrogen bonds, whereas for 2a there are no classical hydrogen bonds and the chains are interconnected by Br⋯Br contacts.
1. Chemical context
We have published a series of articles describing the structures of amine complexes of gold. The three most recent, Parts 12–14 in the series, concerned gold(I) and gold(III) derivatives of piperidine and pyrrolidine (Döring & Jones, 2023a
▸), gold(I) complexes of morpholine (Döring & Jones, 2023b
▸) and gold(I) complexes of methylpiperidine (Döring & Jones, 2024 ▸). An extensive introduction, with details of previous results, may be found in Part 12 and will not be repeated here. Here we present the structures of the two 4-methylpiperidine complexes of gold(III) trihalides, namely trichlorido(4-methylpiperidine)gold(III) 1 and tribromido(4-methylpiperidine)gold(III) 2. The ligands piperidine and 4-methylpiperidine are henceforth abbreviated to ‘pip’ and ‘4-Me-pip’.
2. Structural commentary
The molecular structures of 1, 2a and 2b are shown in Figs. 1 ▸–3 ▸ ▸. Compound 2 crystallized as two polymorphs in the space groups Pnma (2a) and Pbca (2b); the former displays crystallographic mirror symmetry, whereby the mirror plane contains the gold and bromine atoms, the NH group, the carbon at C-4 and the methyl carbon (these atoms are numbered for 2a as C14 and C15). For all three structures, the halogen atoms are numbered such that X1 (X = halogen) is trans to the ligand nitrogen atom N11. Structures 1 and 2b are isotypic. The geometry at the gold atoms is as expected square planar. Bond lengths and angles (Tables 1 ▸–3 ▸ ▸) may be considered normal. The Au—N bonds trans to Cl are somewhat shorter than those trans to Br, and the Au—Cl bonds trans to N are longer than those cis to N, whereas the Au—Br bonds trans to N are slightly shorter than the cis bonds. Similar trends were observed for (pip)AuCl3 and (pip)AuBr3 (Döring & Jones, 2023a ▸).
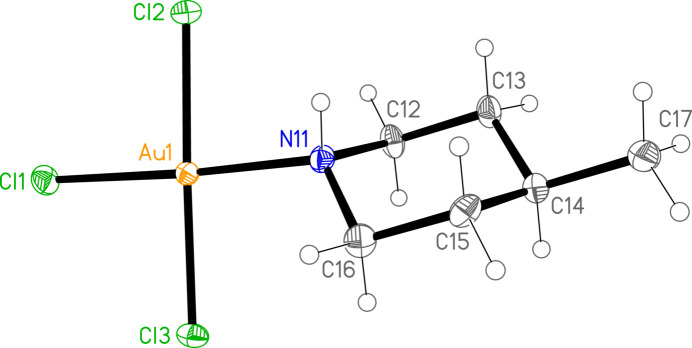
Figure 1.
The structure of compound 1 in the crystal. Ellipsoids correspond to 50% probability levels.
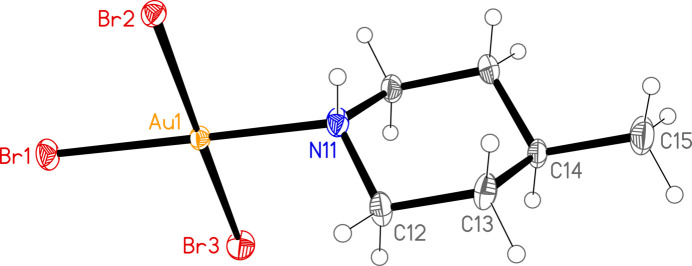
Figure 2.
The structure of compound 2a in the crystal. Ellipsoids correspond to 50% probability levels. Only the asymmetric unit is numbered.
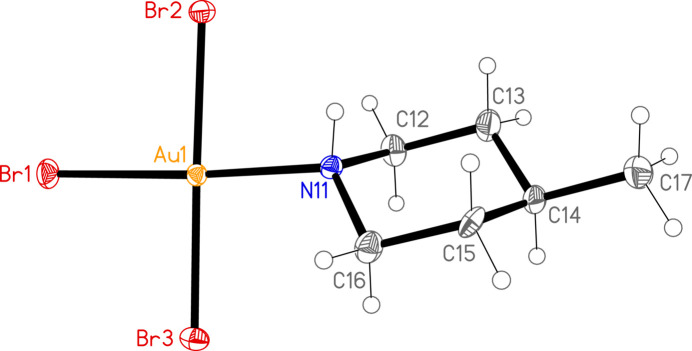
Figure 3.
The structure of compound 2b in the crystal. Ellipsoids correspond to 50% probability levels.
Table 1. Selected geometric parameters (Å, °) for 1 .
| Au1—N11 | 2.070 (3) | Au1—Cl2 | 2.2832 (10) |
| Au1—Cl3 | 2.2826 (10) | Au1—Cl1 | 2.3006 (10) |
| N11—Au1—Cl3 | 93.13 (11) | Cl2—Au1—Cl1 | 91.18 (4) |
| N11—Au1—Cl2 | 85.80 (11) | C16—N11—C12 | 110.6 (3) |
| Cl3—Au1—Cl2 | 178.07 (4) | C16—N11—Au1 | 117.8 (3) |
| N11—Au1—Cl1 | 176.94 (11) | C12—N11—Au1 | 111.0 (3) |
| Cl3—Au1—Cl1 | 89.90 (4) | ||
| Cl3—Au1—N11—C16 | −30.0 (3) | Au1—N11—C12—C13 | 169.1 (3) |
| Cl2—Au1—N11—C16 | 151.6 (3) | C12—C13—C14—C17 | 179.8 (4) |
| Cl3—Au1—N11—C12 | 98.8 (3) | C17—C14—C15—C16 | 179.4 (4) |
| Cl2—Au1—N11—C12 | −79.6 (3) | Au1—N11—C16—C15 | −172.6 (3) |
Table 2. Selected geometric parameters (Å, °) for 2a .
| Au1—N11 | 2.096 (5) | Au1—Br3 | 2.4110 (7) |
| Au1—Br1 | 2.4066 (7) | Au1—Br2 | 2.4273 (6) |
| N11—Au1—Br1 | 179.50 (15) | Br1—Au1—Br2 | 91.68 (2) |
| N11—Au1—Br3 | 91.78 (15) | Br3—Au1—Br2 | 179.60 (2) |
| Br1—Au1—Br3 | 88.72 (2) | C12i—N11—C12 | 111.2 (5) |
| N11—Au1—Br2 | 87.82 (15) | C12i—N11—Au1 | 113.4 (3) |
| Br3—Au1—N11—C12 | −64.1 (3) | Au1—N11—C12—C13 | −174.4 (3) |
| Br2—Au1—N11—C12 | 115.9 (3) | C12—C13—C14—C15 | −179.7 (4) |
Symmetry code: (i)
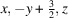 .
.
Table 3. Selected geometric parameters (Å, °) for 2b .
| Au1—N11 | 2.094 (4) | Au1—Br2 | 2.4244 (5) |
| Au1—Br1 | 2.4187 (5) | Au1—Br3 | 2.4246 (5) |
| N11—Au1—Br1 | 176.50 (11) | Br2—Au1—Br3 | 177.736 (17) |
| N11—Au1—Br2 | 86.00 (11) | C12—N11—C16 | 110.9 (4) |
| Br1—Au1—Br2 | 90.665 (18) | C12—N11—Au1 | 111.6 (3) |
| N11—Au1—Br3 | 93.62 (11) | C16—N11—Au1 | 117.8 (3) |
| Br1—Au1—Br3 | 89.751 (18) | ||
| Br2—Au1—N11—C12 | −78.0 (3) | Au1—N11—C12—C13 | 168.1 (3) |
| Br3—Au1—N11—C12 | 99.8 (3) | C12—C13—C14—C17 | −179.2 (4) |
| Br2—Au1—N11—C16 | 152.0 (3) | C17—C14—C15—C16 | 178.9 (4) |
| Br3—Au1—N11—C16 | −30.2 (3) | Au1—N11—C16—C15 | −171.6 (3) |
The relative orientation of the ligand and the AuX 3 unit is described by the torsion angles Xn—Au1—N11—H01 and Xn—Au1—N11—C, where n = 2 or 3 (torsion angles for n = 1 are meaningless because the sequence X1—Au1—N1 is linear). We observe two distinct types: either one angle Xn—Au1—N11—H01 is approximately zero, corresponding to a short H01⋯Xn contact that might be considered an intramolecular hydrogen bond, and the smallest absolute Xn—Au1—N11—C angle is around 60°, or the angle Xn—Au1—N11—H01 is approximately 30–40° and the smallest absolute Xn—Au1—N11—C angle is around 30°. The former type applies to (pip)AuCl3 and 2a [where Br2—Au1—N11—H01 is exactly zero by symmetry and H01⋯Br2 is 2.71 (6) Å], and the latter to (pip)AuBr3, 1 and 2b.
As would be expected for bulky substituents attached to cyclohexane-type rings, the methyl groups and the AuX 3 moieties occupy equatorial positions, with torsion angles C—C—N—Au and C—C—C—Cmethyl around ±180°. Our previous two papers however include several structures where a gold(I) atom occupies an axial position in similar molecules. The ‘normal’ equatorial positions observed for 1, 2a and 2b may be associated with steric effects, which should be greater for the larger AuX 3 moieties compared to the linear gold(I) centres.
3. Supramolecular features
For compound 1, the main intermolecular contacts are the hydrogen bond N1—H01⋯Cl1(
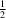 − x,
− x,
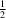 + y, z, the b glide operator) and the two Au⋯Cl contacts Au1⋯Cl3 (same operator) = 3.2980 (10) Å and Au1⋯Cl2(
+ y, z, the b glide operator) and the two Au⋯Cl contacts Au1⋯Cl3 (same operator) = 3.2980 (10) Å and Au1⋯Cl2(
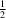 − x, −
− x, −
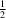 + y, z) = 3.3604 (10) Å that correspond to an offset stacking of the AuCl3 moieties. These combine to form chains of molecules parallel to the b axis (Fig. 4 ▸). In the isotypic 2b, the corresponding Au⋯Br distances are 3.4060 (5) and 3.5018 (5) Å.
+ y, z) = 3.3604 (10) Å that correspond to an offset stacking of the AuCl3 moieties. These combine to form chains of molecules parallel to the b axis (Fig. 4 ▸). In the isotypic 2b, the corresponding Au⋯Br distances are 3.4060 (5) and 3.5018 (5) Å.
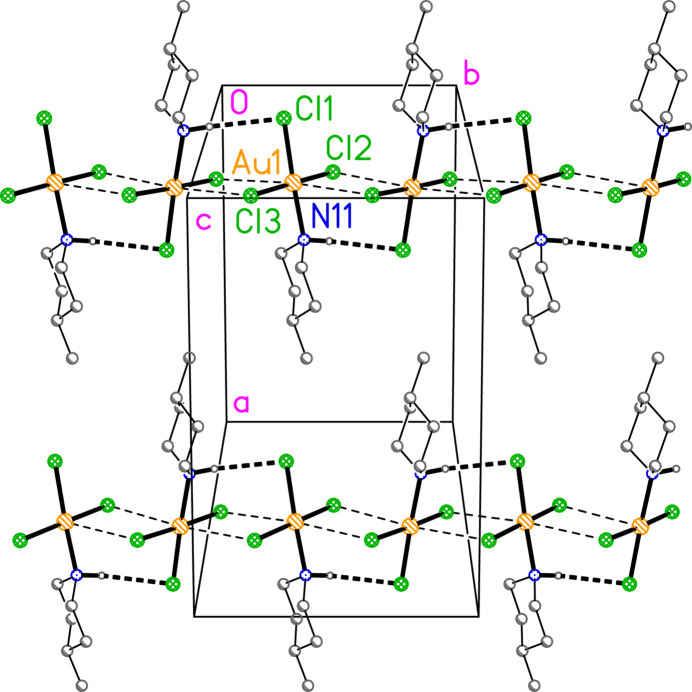
Figure 4.
Packing diagram of compound 1 viewed approximately parallel to the c axis (but rotated by ca 15° around the horizontal axis for clarity) in the region z ≃ 0.125, showing two chains of molecules parallel to the b axis. Dashed lines indicate H⋯Cl hydrogen bonds (thick) or Au⋯Cl contacts (thin). Hydrogen atoms not involved in hydrogen bonding are omitted. Atom labels indicate the asymmetric unit. Similar chains are formed in the regions z ≃ 0.375, 0.625 and 0.875.
Compound 2a forms chains analogous to those of 1, with Au1⋯Br2(−x, 1 − y, −z and −x, 2 − y, −z) = 3.5847 (2) Å; these run parallel to the b axis (Fig. 5 ▸). The chains are crosslinked by short Br⋯Br contacts involving one cis (to N) and the trans Br atom, with Br1⋯Br3(−
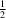 + x, y,
+ x, y,
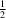 − z, the a glide operator) = 3.3686 (6) Å and angles Au1—Br1⋯Br3′ = 166.26 (3) and Au1—Br3⋯Br1′ = 162.77 (3)°. These contacts are indicated in Fig. 5 ▸ but are shown more clearly in Fig. 6 ▸; they link the molecules to form chains parallel to the b axis. The NH group is not involved in intermolecular hydrogen bonding.
− z, the a glide operator) = 3.3686 (6) Å and angles Au1—Br1⋯Br3′ = 166.26 (3) and Au1—Br3⋯Br1′ = 162.77 (3)°. These contacts are indicated in Fig. 5 ▸ but are shown more clearly in Fig. 6 ▸; they link the molecules to form chains parallel to the b axis. The NH group is not involved in intermolecular hydrogen bonding.
Figure 5.
Packing diagram of compound 2a viewed aproximately parallel to the c axis (but rotated by ca 10° about the vertical axis for clarity), showing three chains of molecules parallel to the b axis. The chains are centred on the regions (x, z) = (0, 0), (1, 0) and (1/2, 1/2). Dashed lines indicated Au⋯Br contacts (thick) or Br⋯Br contacts (thin); the latter are shown more clearly in Fig. 6 ▸. Atom labels indicate the asymmetric unit.
Figure 6.
Packing diagram of compound 2a showing two zigzag chains of molecules parallel to the b axis; the lower chain is centred in the mirror plane at y = 0.75 and the upper chain in the plane at y = 0.25. Dashed lines indicated Br⋯Br contacts (or, just visible, Au⋯Br contacts linking the two chains in the direction into the paper). Atom labels indicate the asymmetric unit.
All three structures also display C—H⋯ X contacts that might be interpreted as ‘weak’ hydrogen bonds (Tables 4 ▸–6 ▸ ▸), but none of these is strikingly short. These (and other) weak interactions might well contribute significantly to the packing energy, but it is difficult to incorporate them in easily interpretable packing diagrams.
Table 4. Hydrogen-bond geometry (Å, °) for 1 .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N11—H01⋯Cl1i | 0.93 (4) | 2.64 (4) | 3.535 (4) | 163 (4) |
| C15—H15B⋯Cl1i | 0.99 | 2.97 | 3.804 (4) | 143 |
| C13—H13B⋯Cl2ii | 0.99 | 2.82 | 3.798 (4) | 171 |
| C15—H15A⋯Cl3iii | 0.99 | 2.95 | 3.610 (4) | 125 |
| C15—H15A⋯Cl3iv | 0.99 | 2.99 | 3.728 (4) | 132 |
| C16—H16B⋯Cl3i | 0.99 | 2.91 | 3.656 (5) | 133 |
Symmetry codes: (i)
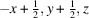 ; (ii)
; (ii)
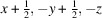 ; (iii)
; (iii)
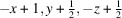 ; (iv)
; (iv)
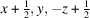 .
.
Table 5. Hydrogen-bond geometry (Å, °) for 2a .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N11—H01⋯Br2 | 0.89 (6) | 2.71 (6) | 3.146 (5) | 111 (5) |
| C12—H12B⋯Br1ii | 0.99 | 2.94 | 3.786 (5) | 145 |
| C12—H12B⋯Br2ii | 0.99 | 2.99 | 3.798 (4) | 139 |
| C15—H15A⋯Br2iii | 0.97 | 2.98 | 3.936 (7) | 169 |
| C12—H12A⋯Br3 | 0.99 | 2.96 | 3.526 (4) | 118 |
| C13—H13A⋯Br3iv | 0.99 | 3.09 | 4.002 (4) | 154 |
| C15—H15B⋯Br3iv | 0.98 | 3.05 | 3.965 (3) | 155 |
Symmetry codes: (ii)
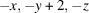 ; (iii)
; (iii)
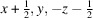 ; (iv)
; (iv)
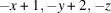 .
.
Table 6. Hydrogen-bond geometry (Å, °) for 2b .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N11—H01⋯Br1i | 0.97 (4) | 2.81 (4) | 3.759 (4) | 164 (4) |
| C12—H12A⋯Br2 | 0.99 | 2.99 | 3.542 (5) | 116 |
| C13—H13B⋯Br2ii | 0.99 | 2.93 | 3.903 (5) | 169 |
| C16—H16B⋯Br3i | 0.99 | 2.99 | 3.750 (5) | 135 |
Symmetry codes: (i)
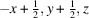 ; (ii)
; (ii)
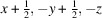 .
.
4. Database survey
The searches employed the routine ConQuest (Bruno et al., 2002 ▸), part of Version 2023.3.0 of the Cambridge Database (Groom et al., 2016 ▸). A search for short Cl⋯Cl contacts between molecules LAuCl3 (L = any atom) gave 51 hits (59 independent molecules) with contact distances from 3.086 to 3.37 Å and an average Au—Cl⋯Cl angle of 152.9°. A similar search for LAuBr3 (L = any atom) gave 28 hits (36 independent molecules) with contact distances from 3.26 to 3.67 Å and an average Au—Br⋯Br angle of 150.7°. The upper bounds for the contact distances in both cases correspond to the double van der Waals radii as stored in the CCDC. For both sets of results, the cis (to L) halogen atoms were more often involved than the trans halogen atoms (the latter corresponding to X1 in the structures presented here); for X = Cl there were 9 contacts of the form trans/trans, 5 cis/trans and 37 cis/cis, and the corresponding values for X = Br were 4, 7 and 25. In many cases, the Au—X⋯X angles were equal by symmetry, and both values were used to calculate the average values.
5. Synthesis and crystallization
The starting materials of choice would be the gold(I) complexes (4-Me-pip)AuX, but these exist in the ionic form [(4-Me-pip)2Au][AuX 2] rather than as neutral molecules (Döring & Jones, 2024 ▸).
Trichlorido(4-methylpiperidine)gold(III) (1)
A solution of bis(4-methylpiperidine)gold(I) dichloridoaurate(I) (310 mg, 0.454 mmol) in 4 mL of dichloromethane was added to a solution of PhICl2 (125 mg, 0.454 mmol) in 3 mL of dichloromethane. 2 mL of the mixed solution were divided amongst five small test-tubes and overlayered with various precipitants. The tubes were then stoppered and stored in a refrigerator at 276 K. The measured crystal was obtained using diisopropyl ether as precipitant. Elemental analysis [%]: calc. C 17.91, H 3.26, N 3.48; found C 17.64, H 3.30, N 3.65.
Tribromido(4-methylpiperidine)gold(III) (2)
Polymorph 2a: Bis(4-methylpiperidinium) bromide tetrabromidoaurate(III), {(4-Me-pip)H}2·Br·[AuBr4] (Döring, 2016 ▸) (26 mg, 0.0327 mmol) was dissolved in 1.5 mL of dichloromethane. The solution was divided amongst three small test tubes and overlayered with various precipitants. The tubes were then stoppered and stored in a refrigerator at 276 K. Using diisopropyl ether as precipitant, a mixture of crystals of the starting material (structure to be reported elsewhere) and of 2a was obtained.
Polymorph 2b: Bis(4-methylpiperidine)gold(I) dibromidoaurate(I), [(4-Me-pip)2Au][AuBr2], (90 mg, 0.239 mmol) was dissolved in 2 mL of dichloromethane and two drops of elemental bromine were added. The solution was overlayered with diisopropyl ether and stored in a refrigerator at 276 K, whereby crystals of 2b formed.
6. Refinement
Details of the measurements and refinements are given in Table 7 ▸. Structures were refined anisotropically on F 2. For all compounds, the NH hydrogen atoms were refined freely. Methylene hydrogens were included at calculated positions and refined using a riding model with C—H = 0.99 Å and H—C—H = 109.5°. Methine hydrogens were included similarly, but with C—H = 0.99 Å. Methyl groups were included as idealized rigid groups with C—H 0.98 Å and H—C—H 109.5°, and were allowed to rotate but not to tip (command ‘AFIX 137’). U values of the hydrogen atoms were fixed at 1.5 × U eq of the parent carbon atoms for methyl groups and 1.2 × U eq of the parent carbon atoms for other hydrogens. For compound 2a, an extinction correction was performed; the extinction parameter (Sheldrick, 2015 ▸) refined to 0.00051 (4).
Table 7. Experimental details.
| 1 | 2a | 2b | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | [AuCl3(C6H13N)] | [AuBr3(C6H13N)] | [AuBr3(C6H13N)] |
| M r | 402.49 | 535.87 | 535.87 |
| Crystal system, space group | Orthorhombic, P b c a | Orthorhombic, P n m a | Orthorhombic, P b c a |
| Temperature (K) | 100 | 100 | 100 |
| a, b, c (Å) | 12.5716 (6), 8.3940 (3), 20.3319 (7) | 9.9871 (5), 7.1505 (4), 15.7160 (8) | 12.6471 (5), 8.7247 (3), 21.0262 (7) |
| V (Å3) | 2145.53 (14) | 1122.32 (10) | 2320.07 (15) |
| Z | 8 | 4 | 8 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 14.40 | 23.74 | 22.96 |
| Crystal size (mm) | 0.22 × 0.03 × 0.01 | 0.27 × 0.06 × 0.03 | 0.14 × 0.04 × 0.03 |
| Data collection | |||
| Diffractometer | Oxford Diffraction Xcalibur, Eos | Oxford Diffraction Xcalibur, Eos | Oxford Diffraction Xcalibur, Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) | Multi-scan (CrysAlis PRO; Rigaku OD, 2015 ▸) |
| T min, T max | 0.702, 1.000 | 0.240, 1.000 | 0.380, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 53277, 2887, 2134 | 28605, 1864, 1581 | 38297, 3371, 2495 |
| R int | 0.080 | 0.070 | 0.074 |
| θ values (°) | θmax = 29.1, θmin = 2.6 | θmax = 31.1, θmin = 2.4 | θmax = 30.0, θmin = 2.5 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.024, 0.042, 1.05 | 0.030, 0.050, 1.11 | 0.029, 0.043, 1.04 |
| No. of reflections | 2887 | 1864 | 3371 |
| No. of parameters | 105 | 65 | 105 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.86, −0.86 | 1.56, −1.19 | 0.99, −0.99 |
Supplementary Material
Crystal structure: contains datablock(s) 1, 2a, 2b, global. DOI: 10.1107/S2056989024002822/pk2705sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989024002822/pk27051sup2.hkl
Structure factors: contains datablock(s) 2a. DOI: 10.1107/S2056989024002822/pk27052asup3.hkl
Structure factors: contains datablock(s) 2b. DOI: 10.1107/S2056989024002822/pk27052bsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
We gratefully acknowledge support by the Open Access Publication Funds of the Technical University of Braunschweig.
supplementary crystallographic information
Trichlorido(4-methylpiperidine)gold(III) (1). Crystal data
| [AuCl3(C6H13N)] | Dx = 2.492 Mg m−3 |
| Mr = 402.49 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 7041 reflections |
| a = 12.5716 (6) Å | θ = 2.6–30.3° |
| b = 8.3940 (3) Å | µ = 14.40 mm−1 |
| c = 20.3319 (7) Å | T = 100 K |
| V = 2145.53 (14) Å3 | Needle, yellow |
| Z = 8 | 0.22 × 0.03 × 0.01 mm |
| F(000) = 1488 |
Trichlorido(4-methylpiperidine)gold(III) (1). Data collection
| Oxford Diffraction Xcalibur, Eos diffractometer | 2887 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2134 reflections with I > 2σ(I) |
| Detector resolution: 16.1419 pixels mm-1 | Rint = 0.080 |
| ω scan | θmax = 29.1°, θmin = 2.6° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2015) | h = −17→17 |
| Tmin = 0.702, Tmax = 1.000 | k = −11→11 |
| 53277 measured reflections | l = −27→27 |
Trichlorido(4-methylpiperidine)gold(III) (1). Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.024 | Hydrogen site location: mixed |
| wR(F2) = 0.042 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0105P)2 + 2.0478P] where P = (Fo2 + 2Fc2)/3 |
| 2887 reflections | (Δ/σ)max = 0.001 |
| 105 parameters | Δρmax = 0.86 e Å−3 |
| 0 restraints | Δρmin = −0.86 e Å−3 |
Trichlorido(4-methylpiperidine)gold(III) (1). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Au1 | 0.24309 (2) | 0.30864 (2) | 0.12540 (2) | 0.01126 (5) | |
| Cl1 | 0.06218 (8) | 0.27039 (12) | 0.11903 (6) | 0.0183 (2) | |
| Cl2 | 0.23963 (8) | 0.47300 (12) | 0.03594 (5) | 0.0152 (2) | |
| Cl3 | 0.25068 (9) | 0.13911 (12) | 0.21305 (5) | 0.0163 (2) | |
| N11 | 0.4051 (3) | 0.3526 (4) | 0.12740 (19) | 0.0140 (7) | |
| H01 | 0.413 (3) | 0.459 (5) | 0.116 (2) | 0.016 (12)* | |
| C12 | 0.4627 (4) | 0.2518 (5) | 0.0768 (2) | 0.0164 (10) | |
| H12A | 0.461894 | 0.138753 | 0.090652 | 0.020* | |
| H12B | 0.426095 | 0.260030 | 0.033863 | 0.020* | |
| C13 | 0.5764 (3) | 0.3084 (5) | 0.0700 (2) | 0.0172 (9) | |
| H13A | 0.576749 | 0.418818 | 0.052940 | 0.021* | |
| H13B | 0.613978 | 0.240127 | 0.037781 | 0.021* | |
| C14 | 0.6356 (3) | 0.3033 (5) | 0.1356 (2) | 0.0171 (9) | |
| H14 | 0.637276 | 0.190175 | 0.150987 | 0.020* | |
| C15 | 0.5748 (4) | 0.4004 (5) | 0.1862 (2) | 0.0167 (9) | |
| H15A | 0.610893 | 0.391913 | 0.229285 | 0.020* | |
| H15B | 0.575131 | 0.513867 | 0.172913 | 0.020* | |
| C16 | 0.4608 (3) | 0.3441 (5) | 0.1930 (2) | 0.0183 (10) | |
| H16A | 0.459812 | 0.233002 | 0.209420 | 0.022* | |
| H16B | 0.422949 | 0.411771 | 0.225335 | 0.022* | |
| C17 | 0.7505 (3) | 0.3604 (5) | 0.1278 (2) | 0.0226 (9) | |
| H17A | 0.750776 | 0.472073 | 0.113702 | 0.034* | |
| H17B | 0.786546 | 0.295036 | 0.094620 | 0.034* | |
| H17C | 0.787735 | 0.350621 | 0.169907 | 0.034* |
Trichlorido(4-methylpiperidine)gold(III) (1). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Au1 | 0.01234 (7) | 0.00980 (8) | 0.01163 (7) | 0.00022 (6) | 0.00052 (7) | −0.00063 (6) |
| Cl1 | 0.0153 (5) | 0.0172 (5) | 0.0225 (6) | −0.0036 (4) | 0.0014 (5) | 0.0009 (5) |
| Cl2 | 0.0187 (5) | 0.0130 (5) | 0.0139 (5) | −0.0011 (4) | −0.0021 (4) | 0.0019 (4) |
| Cl3 | 0.0227 (5) | 0.0127 (5) | 0.0135 (4) | −0.0006 (4) | 0.0019 (5) | 0.0015 (4) |
| N11 | 0.0121 (16) | 0.0110 (17) | 0.0189 (18) | 0.0001 (13) | −0.0004 (16) | 0.0011 (16) |
| C12 | 0.014 (2) | 0.021 (3) | 0.015 (2) | 0.0004 (17) | 0.0039 (19) | −0.0070 (17) |
| C13 | 0.013 (2) | 0.023 (3) | 0.016 (2) | 0.001 (2) | 0.0022 (17) | −0.0025 (19) |
| C14 | 0.013 (2) | 0.020 (2) | 0.019 (2) | −0.0010 (18) | −0.0001 (17) | 0.0024 (19) |
| C15 | 0.018 (2) | 0.019 (2) | 0.013 (2) | 0.0014 (18) | −0.0065 (18) | −0.0034 (17) |
| C16 | 0.018 (2) | 0.025 (3) | 0.012 (2) | −0.0002 (19) | −0.0024 (17) | −0.0042 (18) |
| C17 | 0.019 (2) | 0.026 (2) | 0.022 (2) | −0.0043 (18) | −0.004 (2) | 0.007 (2) |
Trichlorido(4-methylpiperidine)gold(III) (1). Geometric parameters (Å, º)
| Au1—N11 | 2.070 (3) | C13—H13B | 0.9900 |
| Au1—Cl3 | 2.2826 (10) | C14—C15 | 1.518 (6) |
| Au1—Cl2 | 2.2832 (10) | C14—C17 | 1.531 (6) |
| Au1—Cl1 | 2.3006 (10) | C14—H14 | 1.0000 |
| N11—C16 | 1.509 (5) | C15—C16 | 1.515 (6) |
| N11—C12 | 1.516 (5) | C15—H15A | 0.9900 |
| N11—H01 | 0.93 (4) | C15—H15B | 0.9900 |
| C12—C13 | 1.512 (6) | C16—H16A | 0.9900 |
| C12—H12A | 0.9900 | C16—H16B | 0.9900 |
| C12—H12B | 0.9900 | C17—H17A | 0.9800 |
| C13—C14 | 1.528 (6) | C17—H17B | 0.9800 |
| C13—H13A | 0.9900 | C17—H17C | 0.9800 |
| N11—Au1—Cl3 | 93.13 (11) | C15—C14—C13 | 109.4 (3) |
| N11—Au1—Cl2 | 85.80 (11) | C15—C14—C17 | 112.2 (4) |
| Cl3—Au1—Cl2 | 178.07 (4) | C13—C14—C17 | 111.1 (3) |
| N11—Au1—Cl1 | 176.94 (11) | C15—C14—H14 | 108.0 |
| Cl3—Au1—Cl1 | 89.90 (4) | C13—C14—H14 | 108.0 |
| Cl2—Au1—Cl1 | 91.18 (4) | C17—C14—H14 | 108.0 |
| C16—N11—C12 | 110.6 (3) | C16—C15—C14 | 111.8 (3) |
| C16—N11—Au1 | 117.8 (3) | C16—C15—H15A | 109.3 |
| C12—N11—Au1 | 111.0 (3) | C14—C15—H15A | 109.3 |
| C16—N11—H01 | 103 (3) | C16—C15—H15B | 109.3 |
| C12—N11—H01 | 108 (3) | C14—C15—H15B | 109.3 |
| Au1—N11—H01 | 106 (3) | H15A—C15—H15B | 107.9 |
| C13—C12—N11 | 109.8 (3) | N11—C16—C15 | 110.0 (3) |
| C13—C12—H12A | 109.7 | N11—C16—H16A | 109.7 |
| N11—C12—H12A | 109.7 | C15—C16—H16A | 109.7 |
| C13—C12—H12B | 109.7 | N11—C16—H16B | 109.7 |
| N11—C12—H12B | 109.7 | C15—C16—H16B | 109.7 |
| H12A—C12—H12B | 108.2 | H16A—C16—H16B | 108.2 |
| C12—C13—C14 | 111.8 (3) | C14—C17—H17A | 109.5 |
| C12—C13—H13A | 109.3 | C14—C17—H17B | 109.5 |
| C14—C13—H13A | 109.3 | H17A—C17—H17B | 109.5 |
| C12—C13—H13B | 109.3 | C14—C17—H17C | 109.5 |
| C14—C13—H13B | 109.3 | H17A—C17—H17C | 109.5 |
| H13A—C13—H13B | 107.9 | H17B—C17—H17C | 109.5 |
| Cl3—Au1—N11—C16 | −30.0 (3) | C12—C13—C14—C15 | −55.8 (5) |
| Cl2—Au1—N11—C16 | 151.6 (3) | C12—C13—C14—C17 | 179.8 (4) |
| Cl3—Au1—N11—C12 | 98.8 (3) | C13—C14—C15—C16 | 55.7 (5) |
| Cl2—Au1—N11—C12 | −79.6 (3) | C17—C14—C15—C16 | 179.4 (4) |
| C16—N11—C12—C13 | −58.4 (4) | C12—N11—C16—C15 | 58.4 (4) |
| Au1—N11—C12—C13 | 169.1 (3) | Au1—N11—C16—C15 | −172.6 (3) |
| N11—C12—C13—C14 | 57.4 (5) | C14—C15—C16—N11 | −57.7 (5) |
Trichlorido(4-methylpiperidine)gold(III) (1). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H01···Cl1i | 0.93 (4) | 2.64 (4) | 3.535 (4) | 163 (4) |
| C15—H15B···Cl1i | 0.99 | 2.97 | 3.804 (4) | 143 |
| C13—H13B···Cl2ii | 0.99 | 2.82 | 3.798 (4) | 171 |
| C15—H15A···Cl3iii | 0.99 | 2.95 | 3.610 (4) | 125 |
| C15—H15A···Cl3iv | 0.99 | 2.99 | 3.728 (4) | 132 |
| C16—H16B···Cl3i | 0.99 | 2.91 | 3.656 (5) | 133 |
Symmetry codes: (i) −x+1/2, y+1/2, z; (ii) x+1/2, −y+1/2, −z; (iii) −x+1, y+1/2, −z+1/2; (iv) x+1/2, y, −z+1/2.
Tribromido(4-methylpiperidine)gold(III) (2a). Crystal data
| [AuBr3(C6H13N)] | Dx = 3.171 Mg m−3 |
| Mr = 535.87 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pnma | Cell parameters from 4663 reflections |
| a = 9.9871 (5) Å | θ = 3.1–30.3° |
| b = 7.1505 (4) Å | µ = 23.74 mm−1 |
| c = 15.7160 (8) Å | T = 100 K |
| V = 1122.32 (10) Å3 | Needle, orange |
| Z = 4 | 0.27 × 0.06 × 0.03 mm |
| F(000) = 960 |
Tribromido(4-methylpiperidine)gold(III) (2a). Data collection
| Oxford Diffraction Xcalibur, Eos diffractometer | 1864 independent reflections |
| Radiation source: fine-focus sealed X-ray tube | 1581 reflections with I > 2σ(I) |
| Detector resolution: 16.1419 pixels mm-1 | Rint = 0.070 |
| ω scan | θmax = 31.1°, θmin = 2.4° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2015) | h = −14→14 |
| Tmin = 0.240, Tmax = 1.000 | k = −10→10 |
| 28605 measured reflections | l = −22→22 |
Tribromido(4-methylpiperidine)gold(III) (2a). Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.030 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.050 | w = 1/[σ2(Fo2) + (0.0121P)2 + 3.4353P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.11 | (Δ/σ)max < 0.001 |
| 1864 reflections | Δρmax = 1.56 e Å−3 |
| 65 parameters | Δρmin = −1.18 e Å−3 |
| 0 restraints | Extinction correction: SHELXL2019/3 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.00051 (4) |
Tribromido(4-methylpiperidine)gold(III) (2a). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Au1 | 0.09315 (2) | 0.750000 | 0.04663 (2) | 0.01176 (7) | |
| Br1 | −0.07000 (6) | 0.750000 | 0.15933 (4) | 0.01882 (14) | |
| Br2 | −0.08081 (6) | 0.750000 | −0.06122 (4) | 0.01351 (13) | |
| Br3 | 0.26712 (6) | 0.750000 | 0.15298 (4) | 0.02039 (15) | |
| N11 | 0.2339 (5) | 0.750000 | −0.0523 (3) | 0.0161 (11) | |
| H01 | 0.184 (7) | 0.750000 | −0.099 (4) | 0.013 (17)* | |
| C12 | 0.3183 (4) | 0.9225 (6) | −0.0547 (3) | 0.0182 (9) | |
| H12A | 0.373767 | 0.928684 | −0.002574 | 0.022* | |
| H12B | 0.259617 | 1.034089 | −0.055751 | 0.022* | |
| C13 | 0.4087 (4) | 0.9241 (6) | −0.1324 (3) | 0.0193 (9) | |
| H13A | 0.465879 | 1.037237 | −0.131098 | 0.023* | |
| H13B | 0.353144 | 0.929562 | −0.184542 | 0.023* | |
| C14 | 0.4975 (6) | 0.750000 | −0.1354 (4) | 0.0152 (12) | |
| H14 | 0.554971 | 0.750002 | −0.083321 | 0.018* | |
| C15 | 0.5896 (7) | 0.750000 | −0.2129 (4) | 0.0253 (16) | |
| H15A | 0.534829 | 0.750000 | −0.264018 | 0.038* | |
| H15B | 0.647617 | 0.860443 | −0.211145 | 0.038* |
Tribromido(4-methylpiperidine)gold(III) (2a). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Au1 | 0.01034 (11) | 0.01467 (12) | 0.01027 (12) | 0.000 | 0.00112 (9) | 0.000 |
| Br1 | 0.0180 (3) | 0.0253 (3) | 0.0132 (3) | 0.000 | 0.0057 (2) | 0.000 |
| Br2 | 0.0113 (3) | 0.0163 (3) | 0.0129 (3) | 0.000 | −0.0005 (2) | 0.000 |
| Br3 | 0.0172 (3) | 0.0304 (4) | 0.0135 (3) | 0.000 | −0.0042 (2) | 0.000 |
| N11 | 0.011 (2) | 0.027 (3) | 0.010 (3) | 0.000 | 0.000 (2) | 0.000 |
| C12 | 0.018 (2) | 0.016 (2) | 0.021 (2) | 0.0031 (17) | 0.0090 (18) | 0.0020 (19) |
| C13 | 0.016 (2) | 0.022 (2) | 0.020 (2) | −0.0009 (19) | 0.0067 (19) | 0.005 (2) |
| C14 | 0.013 (3) | 0.016 (3) | 0.017 (3) | 0.000 | 0.007 (2) | 0.000 |
| C15 | 0.017 (3) | 0.033 (4) | 0.026 (4) | 0.000 | 0.009 (3) | 0.000 |
Tribromido(4-methylpiperidine)gold(III) (2a). Geometric parameters (Å, º)
| Au1—N11 | 2.096 (5) | C12—H12B | 0.9900 |
| Au1—Br1 | 2.4066 (7) | C13—C14 | 1.529 (5) |
| Au1—Br3 | 2.4110 (7) | C13—H13A | 0.9900 |
| Au1—Br2 | 2.4273 (6) | C13—H13B | 0.9900 |
| N11—C12i | 1.494 (5) | C14—C15 | 1.526 (8) |
| N11—C12 | 1.494 (5) | C14—H14 | 1.0000 |
| N11—H01 | 0.89 (6) | C15—H15A | 0.9713 |
| C12—C13 | 1.520 (6) | C15—H15B | 0.9800 |
| C12—H12A | 0.9900 | C15—H15Bi | 0.9800 |
| N11—Au1—Br1 | 179.50 (15) | C12—C13—C14 | 111.3 (4) |
| N11—Au1—Br3 | 91.78 (15) | C12—C13—H13A | 109.4 |
| Br1—Au1—Br3 | 88.72 (2) | C14—C13—H13A | 109.4 |
| N11—Au1—Br2 | 87.82 (15) | C12—C13—H13B | 109.4 |
| Br1—Au1—Br2 | 91.68 (2) | C14—C13—H13B | 109.4 |
| Br3—Au1—Br2 | 179.60 (2) | H13A—C13—H13B | 108.0 |
| C12i—N11—C12 | 111.2 (5) | C15—C14—C13i | 111.9 (3) |
| C12i—N11—Au1 | 113.4 (3) | C15—C14—C13 | 111.9 (3) |
| C12—N11—Au1 | 113.4 (3) | C13i—C14—C13 | 109.0 (5) |
| C12i—N11—H01 | 107 (2) | C15—C14—H14 | 107.9 |
| C12—N11—H01 | 107 (2) | C13i—C14—H14 | 107.9 |
| Au1—N11—H01 | 104 (4) | C13—C14—H14 | 107.9 |
| N11—C12—C13 | 111.2 (4) | C14—C15—H15A | 108.7 |
| N11—C12—H12A | 109.4 | C14—C15—H15B | 109.5 |
| C13—C12—H12A | 109.4 | H15A—C15—H15B | 110.9 |
| N11—C12—H12B | 109.4 | C14—C15—H15Bi | 109.48 (19) |
| C13—C12—H12B | 109.4 | H15A—C15—H15Bi | 110.9 |
| H12A—C12—H12B | 108.0 | H15B—C15—H15Bi | 107.4 |
| Br3—Au1—N11—C12i | 64.1 (3) | Au1—N11—C12—C13 | −174.4 (3) |
| Br2—Au1—N11—C12i | −115.9 (3) | N11—C12—C13—C14 | −56.8 (5) |
| Br3—Au1—N11—C12 | −64.1 (3) | C12—C13—C14—C15 | −179.7 (4) |
| Br2—Au1—N11—C12 | 115.9 (3) | C12—C13—C14—C13i | 55.9 (6) |
| C12i—N11—C12—C13 | 56.4 (6) |
Symmetry code: (i) x, −y+3/2, z.
Tribromido(4-methylpiperidine)gold(III) (2a). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H01···Br2 | 0.89 (6) | 2.71 (6) | 3.146 (5) | 111 (5) |
| C12—H12B···Br1ii | 0.99 | 2.94 | 3.786 (5) | 145 |
| C12—H12B···Br2ii | 0.99 | 2.99 | 3.798 (4) | 139 |
| C15—H15A···Br2iii | 0.97 | 2.98 | 3.936 (7) | 169 |
| C12—H12A···Br3 | 0.99 | 2.96 | 3.526 (4) | 118 |
| C13—H13A···Br3iv | 0.99 | 3.09 | 4.002 (4) | 154 |
| C15—H15B···Br3iv | 0.98 | 3.05 | 3.965 (3) | 155 |
Symmetry codes: (ii) −x, −y+2, −z; (iii) x+1/2, y, −z−1/2; (iv) −x+1, −y+2, −z.
Tribromido(4-methylpiperidine)gold(III) (2b). Crystal data
| [AuBr3(C6H13N)] | Dx = 3.068 Mg m−3 |
| Mr = 535.87 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbca | Cell parameters from 5682 reflections |
| a = 12.6471 (5) Å | θ = 3.2–29.7° |
| b = 8.7247 (3) Å | µ = 22.96 mm−1 |
| c = 21.0262 (7) Å | T = 100 K |
| V = 2320.07 (15) Å3 | Needle, red |
| Z = 8 | 0.14 × 0.04 × 0.03 mm |
| F(000) = 1920 |
Tribromido(4-methylpiperidine)gold(III) (2b). Data collection
| Oxford Diffraction Xcalibur, Eos diffractometer | 3371 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2495 reflections with I > 2σ(I) |
| Detector resolution: 16.1419 pixels mm-1 | Rint = 0.074 |
| ω scan | θmax = 30.0°, θmin = 2.5° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2015) | h = −17→17 |
| Tmin = 0.380, Tmax = 1.000 | k = −11→12 |
| 38297 measured reflections | l = −29→29 |
Tribromido(4-methylpiperidine)gold(III) (2b). Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.029 | Hydrogen site location: mixed |
| wR(F2) = 0.043 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0081P)2 + 2.1449P] where P = (Fo2 + 2Fc2)/3 |
| 3371 reflections | (Δ/σ)max = 0.001 |
| 105 parameters | Δρmax = 0.99 e Å−3 |
| 0 restraints | Δρmin = −0.99 e Å−3 |
Tribromido(4-methylpiperidine)gold(III) (2b). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Au1 | 0.24373 (2) | 0.30618 (2) | 0.12636 (2) | 0.01070 (5) | |
| Br1 | 0.05410 (4) | 0.27328 (5) | 0.12037 (2) | 0.01833 (11) | |
| Br2 | 0.24027 (4) | 0.47282 (5) | 0.03411 (2) | 0.01409 (10) | |
| Br3 | 0.24951 (4) | 0.13133 (5) | 0.21592 (2) | 0.01560 (10) | |
| N11 | 0.4069 (3) | 0.3465 (4) | 0.12801 (19) | 0.0115 (8) | |
| H01 | 0.416 (4) | 0.454 (5) | 0.116 (2) | 0.019 (14)* | |
| C12 | 0.4638 (4) | 0.2527 (5) | 0.0784 (2) | 0.0169 (11) | |
| H12A | 0.426683 | 0.261747 | 0.037140 | 0.020* | |
| H12B | 0.463810 | 0.143443 | 0.091066 | 0.020* | |
| C13 | 0.5769 (4) | 0.3088 (6) | 0.0714 (2) | 0.0201 (11) | |
| H13A | 0.576259 | 0.415630 | 0.055540 | 0.024* | |
| H13B | 0.613889 | 0.244725 | 0.039599 | 0.024* | |
| C14 | 0.6373 (4) | 0.3028 (5) | 0.1341 (2) | 0.0162 (10) | |
| H14 | 0.640778 | 0.193362 | 0.148091 | 0.019* | |
| C15 | 0.5762 (4) | 0.3927 (5) | 0.1845 (2) | 0.0154 (11) | |
| H15A | 0.612751 | 0.382498 | 0.225900 | 0.018* | |
| H15B | 0.575849 | 0.502588 | 0.172813 | 0.018* | |
| C16 | 0.4628 (4) | 0.3372 (5) | 0.1914 (2) | 0.0180 (11) | |
| H16A | 0.462434 | 0.229940 | 0.206773 | 0.022* | |
| H16B | 0.425261 | 0.401276 | 0.222985 | 0.022* | |
| C17 | 0.7504 (4) | 0.3616 (6) | 0.1266 (2) | 0.0253 (11) | |
| H17A | 0.748980 | 0.471144 | 0.116451 | 0.038* | |
| H17B | 0.785491 | 0.305515 | 0.092217 | 0.038* | |
| H17C | 0.789141 | 0.345693 | 0.166448 | 0.038* |
Tribromido(4-methylpiperidine)gold(III) (2b). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Au1 | 0.01025 (9) | 0.00994 (8) | 0.01190 (8) | 0.00053 (7) | 0.00084 (8) | −0.00021 (7) |
| Br1 | 0.0108 (2) | 0.0184 (2) | 0.0259 (3) | −0.00129 (18) | 0.0015 (2) | 0.0018 (2) |
| Br2 | 0.0151 (2) | 0.0137 (2) | 0.0135 (2) | −0.0008 (2) | −0.0013 (2) | 0.00201 (17) |
| Br3 | 0.0207 (3) | 0.0128 (2) | 0.0133 (2) | 0.0001 (2) | 0.0017 (2) | 0.00118 (17) |
| N11 | 0.011 (2) | 0.0125 (19) | 0.011 (2) | −0.0006 (15) | −0.0018 (17) | 0.0001 (16) |
| C12 | 0.016 (3) | 0.021 (3) | 0.013 (2) | 0.001 (2) | 0.003 (2) | −0.0075 (19) |
| C13 | 0.015 (3) | 0.025 (3) | 0.021 (3) | 0.002 (2) | 0.000 (2) | −0.001 (2) |
| C14 | 0.011 (2) | 0.018 (2) | 0.020 (3) | −0.002 (2) | −0.002 (2) | 0.002 (2) |
| C15 | 0.013 (3) | 0.016 (2) | 0.017 (3) | 0.003 (2) | −0.008 (2) | −0.002 (2) |
| C16 | 0.016 (3) | 0.021 (3) | 0.017 (3) | 0.001 (2) | −0.002 (2) | −0.003 (2) |
| C17 | 0.018 (3) | 0.034 (3) | 0.024 (3) | −0.006 (2) | −0.003 (3) | 0.007 (2) |
Tribromido(4-methylpiperidine)gold(III) (2b). Geometric parameters (Å, º)
| Au1—N11 | 2.094 (4) | C13—H13B | 0.9900 |
| Au1—Br1 | 2.4187 (5) | C14—C15 | 1.527 (6) |
| Au1—Br2 | 2.4244 (5) | C14—C17 | 1.527 (6) |
| Au1—Br3 | 2.4246 (5) | C14—H14 | 1.0000 |
| N11—C12 | 1.508 (6) | C15—C16 | 1.520 (7) |
| N11—C16 | 1.511 (6) | C15—H15A | 0.9900 |
| N11—H01 | 0.97 (4) | C15—H15B | 0.9900 |
| C12—C13 | 1.518 (7) | C16—H16A | 0.9900 |
| C12—H12A | 0.9900 | C16—H16B | 0.9900 |
| C12—H12B | 0.9900 | C17—H17A | 0.9800 |
| C13—C14 | 1.524 (7) | C17—H17B | 0.9800 |
| C13—H13A | 0.9900 | C17—H17C | 0.9800 |
| N11—Au1—Br1 | 176.50 (11) | C13—C14—C15 | 109.2 (4) |
| N11—Au1—Br2 | 86.00 (11) | C13—C14—C17 | 111.6 (4) |
| Br1—Au1—Br2 | 90.665 (18) | C15—C14—C17 | 111.9 (4) |
| N11—Au1—Br3 | 93.62 (11) | C13—C14—H14 | 108.0 |
| Br1—Au1—Br3 | 89.751 (18) | C15—C14—H14 | 108.0 |
| Br2—Au1—Br3 | 177.736 (17) | C17—C14—H14 | 108.0 |
| C12—N11—C16 | 110.9 (4) | C16—C15—C14 | 112.3 (4) |
| C12—N11—Au1 | 111.6 (3) | C16—C15—H15A | 109.1 |
| C16—N11—Au1 | 117.8 (3) | C14—C15—H15A | 109.1 |
| C12—N11—H01 | 107 (3) | C16—C15—H15B | 109.1 |
| C16—N11—H01 | 102 (3) | C14—C15—H15B | 109.1 |
| Au1—N11—H01 | 106 (3) | H15A—C15—H15B | 107.9 |
| N11—C12—C13 | 110.0 (4) | N11—C16—C15 | 109.9 (4) |
| N11—C12—H12A | 109.7 | N11—C16—H16A | 109.7 |
| C13—C12—H12A | 109.7 | C15—C16—H16A | 109.7 |
| N11—C12—H12B | 109.7 | N11—C16—H16B | 109.7 |
| C13—C12—H12B | 109.7 | C15—C16—H16B | 109.7 |
| H12A—C12—H12B | 108.2 | H16A—C16—H16B | 108.2 |
| C12—C13—C14 | 112.2 (4) | C14—C17—H17A | 109.5 |
| C12—C13—H13A | 109.2 | C14—C17—H17B | 109.5 |
| C14—C13—H13A | 109.2 | H17A—C17—H17B | 109.5 |
| C12—C13—H13B | 109.2 | C14—C17—H17C | 109.5 |
| C14—C13—H13B | 109.2 | H17A—C17—H17C | 109.5 |
| H13A—C13—H13B | 107.9 | H17B—C17—H17C | 109.5 |
| Br2—Au1—N11—C12 | −78.0 (3) | C12—C13—C14—C15 | −55.0 (5) |
| Br3—Au1—N11—C12 | 99.8 (3) | C12—C13—C14—C17 | −179.2 (4) |
| Br2—Au1—N11—C16 | 152.0 (3) | C13—C14—C15—C16 | 54.8 (5) |
| Br3—Au1—N11—C16 | −30.2 (3) | C17—C14—C15—C16 | 178.9 (4) |
| C16—N11—C12—C13 | −58.4 (5) | C12—N11—C16—C15 | 58.0 (5) |
| Au1—N11—C12—C13 | 168.1 (3) | Au1—N11—C16—C15 | −171.6 (3) |
| N11—C12—C13—C14 | 57.4 (5) | C14—C15—C16—N11 | −56.8 (5) |
Tribromido(4-methylpiperidine)gold(III) (2b). Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N11—H01···Br1i | 0.97 (4) | 2.81 (4) | 3.759 (4) | 164 (4) |
| C12—H12A···Br2 | 0.99 | 2.99 | 3.542 (5) | 116 |
| C13—H13B···Br2ii | 0.99 | 2.93 | 3.903 (5) | 169 |
| C16—H16B···Br3i | 0.99 | 2.99 | 3.750 (5) | 135 |
Symmetry codes: (i) −x+1/2, y+1/2, z; (ii) x+1/2, −y+1/2, −z.
References
- Bruker (1998). XP. Bruker Analytical X–Ray Instruments, Madison, Wisconsin, USA.
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. [DOI] [PubMed]
- Döring, C. (2016). Halogen(I)-Aminkomplexe und ihre Oxidationsprodukte. Dissertation, Technical University of Braunschweig. Germany. ISBN: 978-3-8439-2639-3.
- Döring, C. & Jones, P. G. (2023a). Acta Cryst. E79, 1017–1027. [DOI] [PMC free article] [PubMed]
- Döring, C. & Jones, P. G. (2023b). Acta Cryst. E79, 1161–1165. [DOI] [PMC free article] [PubMed]
- Döring, C. & Jones, P. G. (2024). Acta Cryst. E80, 157–165. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2015). (Formerly Oxford Diffraction and later Agilent Technologies.) CrysAlis PRO, Version 1.171.38.43 (earlier versions were also used, but are not cited separately). Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 1, 2a, 2b, global. DOI: 10.1107/S2056989024002822/pk2705sup1.cif
Structure factors: contains datablock(s) 1. DOI: 10.1107/S2056989024002822/pk27051sup2.hkl
Structure factors: contains datablock(s) 2a. DOI: 10.1107/S2056989024002822/pk27052asup3.hkl
Structure factors: contains datablock(s) 2b. DOI: 10.1107/S2056989024002822/pk27052bsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report