Abstract
Haemaphysalis longicornis ticks, commonly found in East Asia, can transmit various pathogenic viruses, including the severe fever with thrombocytopenia syndrome virus (SFTSV) that has caused febrile diseases among humans in Hubei Province. However, understanding of the viromes of H. longicornis was limited, and the prevalence of viruses among H. longicornis ticks in Hubei was not well clarified. This study investigates the viromes of both engorged (fed) and free (unfed) H. longicornis ticks across three mountainous regions in Hubei Province from 2019 to 2020. RNA-sequencing analysis identified viral sequences that were related to 39 reference viruses belonging to unclassified viruses and seven RNA viral families, namely Chuviridae, Nairoviridae, Orthomyxoviridae, Parvoviridae, Phenuiviridae, Rhabdoviridae, and Totiviridae. Viral abundance and diversity in these ticks were analysed, and phylogenetic characteristics of the Henan tick virus (HNTV), Dabieshan tick virus (DBSTV), Okutama tick virus (OKTV), and Jingmen tick virus (JMTV) were elucidated based on their full genomic sequences. Prevalence analysis demonstrated that DBSTV was the most common virus found in individual H. longicornis ticks (12.59%), followed by HNTV (0.35%), whereas JMTV and OKTV were not detected. These results improve our understanding of H. longicornis tick viromes in central China and highlight the role of tick feeding status and geography in shaping the viral community. The findings of new viral strains and their potential impact on public health raise the need to strengthen surveillance efforts for comprehensively assessing their spillover potentials.
Keywords: Ticks, Virome, Haemaphysalis longicornis, Phylogenetic analysis, Viral abundance and diversity
Highlights
-
•
The free and engorged Haemaphysalis longicornis ticks demonstrate different viromes and vector competence.
-
•
The feeding status and geographic distribution may have impact on shaping tick virome.
-
•
New strains of tick-borne viruses are identified in H. longicornis ticks.
-
•
Dabieshan tick virus is dominantly prevalent in H. longicornis ticks.
1. Introduction
Ticks are blood-sucking arthropods that transmit several viral pathogens (Fang et al., 2015; Randolph, 2011). Over a century ago, the first tick-borne virus causing zoonotic disease was identified (Alexander and Neitz, 1933), leading to the discovery of many tick-borne viruses (TBVs) associated with human and animal diseases worldwide (Jia et al., 2020; Kodama et al., 2021; Yu et al., 2011; Ma et al., 2021; Ni et al., 2023; Shi et al., 2018; Wang et al., 2019; Xu et al., 2011). Tick-borne encephalitis virus (TBEV) and Crimean-Congo hemorrhagic fever virus (CCHFV) are the most notorious tick-borne viral pathogens associated with febrile diseases of high fatality rates, usually accompanied by severe encephalitis and hemorrhage. TBEV, transmitted by Ixodes spp. ticks across a vast area from western Europe to the eastern coast of Japan (Lindquist and Vapalahti, 2008), is the causitive agent of tick-borne encephalitis (TBE), a severe central nervous system disease, and has caused over 10,000 human infections annually in endemic regions of the Eurasian continent, especially in Russia (Gritsun et al., 2003; Lindquist and Vapalahti, 2008). In China, the first human infection with TBEV was reported in 1943, and the virus was first isolated from patients and ticks in 1952 (Wu et al., 2013). From 2007 to 2018, 3364 TBE cases and 19 deaths were reported (Chen et al., 2019), resulting in a high mortality rate of up to 25% among the forestry workers in Northeast China (Lu et al., 2008). CCHFV infection leads to Crimean-Congo hemorrhagic fever (CCHF), a deadly tick-borne disease with a human mortality rate of up to 50% (Hoogstraal, 1979; Swanepoel et al., 1987), affecting countries in Asia, Europe, and Africa (Madison-Antenucci et al., 2020). Bites of Hyalomma spp. ticks are consistently linked to human infections, making them the crucial vectors for transmitting CCHFV (Gargili et al., 2017). In China, CCHFV was first identified in Bachu County of southwestern Xinjiang in 1965 (Gao et al., 2010; Wu et al., 2013) and has been sporadically reported subsequently (Gao et al., 2010) with the last report arriving in 2005. Nonetheless, CCHFV infection risk persists, as the virus is still detected in ticks in Xinjiang province (Gao et al., 2010; Moming et al., 2018; Sun et al., 2009), and CCHFV antibodies were detected in livestock and humans in Qinghai, Inner Mongolia, and Yunnan provinces (Gao et al., 2010; Wu et al., 2013; Xia et al., 2011).
In recent years, several novel TBVs with medical significance have been identified, including severe fever with thrombocytopenia syndrome virus (SFTSV) (Yu et al., 2011), Jingmen tick virus (Qin et al., 2014), Alongshan virus (Wang et al., 2019), Tamdy virus (Moming et al., 2018), Songling virus (SGLV) (Ma et al., 2021), and Yezo virus (Kodama et al., 2021). Owing to limited knowledge about the pathogenesis of these viruses and the lack of effective antiviral treatment and vaccines, emerging TBVs pose an increasing threat to public health by challenging disease prevention and control strategies. Rapid advances in metagenomic sequencing have led to the identification of numerous novel viruses in ticks (Ni et al., 2023), expanding our understanding of the viral sphere and raising questions about current strategies of disease prevention and control.
H. longicornis is a tick specie with a broad host range that primarily inhabits East Asia and is believed to have spread to Australia, New Zealand, and several Pacific islands since 1983 (Barker and Walker, 2014; Zhao et al., 2020). H. longicornis is widely distributed in central and eastern China and is considered a major vector of SFTSV (Zhao et al., 2020). Hubei Province, located in central China, is a primary habitat for H. longicornis and a natural focus for SFTSV research. As of 2020, SFTSV infections have been reported in 19 Chinese provinces, affecting over ten thousand people with an initial high mortality rate of 30% (Li et al., 2021). However, our knowledge of the viromes of H. longicornis remains limited. A recent study that collected H. longicornis and R. microplus ticks from two different locations in Hubei Province in 2021 suggested that ecological factors may shape the virome structure of different tick species (Xu et al., 2021). Nevertheless, in-depth knowledge of the composition of the viral spectrum of H. longicornis in Hubei is missing and deserves investigation.
This study examined the viromes of H. longicornis ticks collected from three mountainous regions in Hubei Province. Viral abundance in ticks collected from fields and domestic animals was compared. We analyzed the viral abundance and diversity associated with tick feeding status and geographic location and examined the prevalence and abundance of four identified viruses in H. longicornis ticks. These results will promote our understanding of viromes in H. longicornis ticks and reveal the epidemiological features of viruses carried by H. longicornis in central China.
2. Materials and methods
2.1. Sample collection
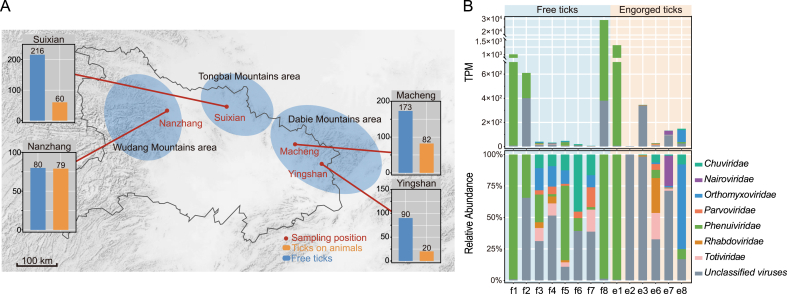
In 2019 and 2020, ticks were collected from five cities or counties located in three mountainous regions: the Wudang (Nanzhang County), Tongbai (Sui County), and Dabie (Macheng and Yingshan Counties) mountainous regions in Hubei Province, China (Supplementary Table S1). Free ticks (unfed) were collected using a 1 m2 flannel flag that was waved over vegetation and engorged ticks (fed) were collected from animals raised by herders at the same sampling locations, if possible. All ticks were transported to the laboratory and stored at −80 °C until further analysis. Tick species were morphologically identified as H. longicornis by experienced specialists under a HZ6100 microscope (Haokangbio, Guangzhou, China).
A total of 559 free H. longicornis ticks were collected from Nanzhang (n = 80), Sui (n = 216), Macheng (n = 173), and Yingshan (n = 90), and 241 engorged H. longicornis ticks were collected from animals at the same locations in Nanzhang (n = 79), Sui (n = 60), Macheng (n = 82), and Yingshan (n = 20) (Fig. 1A and Supplementary Table S1). A subset of 514 ticks was randomly selected and was prepared into 14 pools (eight pools of free ticks and six pools of engorged ticks) for RNA sequencing according to their sampling sites and feeding status. The pools were coded based on the feeding status of the ticks and numbered according to their relative relatedness at the same sampling location. For example, the pool f1 contained free ticks from the same sampling location as the pool e1 containing engorged ticks (Supplementary Table S2).
Fig. 1.
Sampling locations in Hubei Province and the viral compositions of H. longicornis ticks. A The map of Hubei Province showing the sampling locations of free and engorged H. longicornis ticks. The mountainous areas are shaded, and the cities or counties where ticks were collected are indicated by red dots. Bar charts showing the sample sizes of free and engorged ticks are provided for each location. B The abundances (upper) and proportions (bottom) of viral communities belonging to different viral families in the eight pools of free ticks and the six pools of engorged ticks.
2.2. Total RNA sequencing
The 514 ticks were divided into groups according to sampling location and feeding status (Supplementary Table S2). Each group was washed three times and homogenized in 2 mL of sterile phosphate-buffered saline (PBS) using a Tissue Cell-Destroyer (D1000, Novastar, Wuhan, China) as previously described (Zhang et al., 2018). The homogenates were centrifuged at 4 °C (6000 × g, 10 min), and supernatants were harvested. Total RNA was purified from the supernatant of each tick pool using an RNAiso Plus extraction reagent (Takara, Shiga, Japan) according to the manufacturer's instructions. The purified RNA was used for library preparation using the VAHTS Universal V8 RNA-seq Library Prep Kit (Vazyme, Nanjing, China). Subsequently, RNA sequencing was performed using a DNBSeQ-G400 sequencer (MGI Tech, Shenzhen, China).
2.3. Bioinformatic analyses
The quality of sequencing results was assessed using FastQC version 0.11.9 (Wingett and Andrews, 2018). Then, Trimmomatic version 0.39 (Bolger et al., 2014) was used to filter low-quality sequences and adapters. The resulting reads were assembled into contigs using MEGAHIT version 1.2.9 (Li D. et al., 2015). To identify virus-related sequences, the assembled contigs were compared to the NR and NT databases from GenBank, which were available until May 2021, using DIAMOND BLASTX (Buchfink et al., 2021) and BLASTN (e-value < 10−10) (Altschul et al., 1990). The abundance of mapped viral sequences was measured in transcripts per million (TPM) to avoid any bias caused by unequal gene lengths or sequencing depths between different libraries (Li and Dewey, 2011; Li P. et al., 2015).
Briefly, denotes reads mapped to transcript, is the transcript length, and corresponds to the sum of mapped reads to transcript normalized by transcript length (Zhao et al., 2021). TPM values on a log10 scale were used to show the normalized abundance of each viral contig in the heatmap. Heat maps showing the abundance of viruses in each sample bank were generated using TBtools (Chen et al., 2020).
Phylogenetic analyses were performed using MEGA 11 (Tamura et al., 2021) to construct maximum likelihood trees with 1000 bootstrap replicates of viral nucleic acid sequences. Trees were visualized using iTOL (Letunic and Bork, 2019).
Cluster analysis of the viruses in each sample pool was performed using the Vegan package in R Studio (Oksanen et al., 2008). A Bray-Curtis dissimilarity matrix was used to create a PCoA plot in ImageGP (Labbé et al., 2020) to indicate Beta diversity.
2.4. Bead-based assays and qRT-PCR
A bead-based assay was constructed to detect viral RNA in individual ticks, which enables detection of multiple viruses in one reaction according to previous studies (Höfler et al., 2014; Zhang et al., 2021), resulting in a more precise rate of virus prevalence in ticks. The homogenate from a single tick was prepared following a process similar to that used for grouped ticks as described above, and RNA was purified using the RNAiso Plus extraction reagent (Takara) according to the manufacturer's instructions. RT-PCR was performed using the One-Step PrimeScript RT-PCR Kit (Takara) with primers for viral sequences identified from RNA-seq data (Supplementary Table S3). The PCR products from each tick were hybridized with mixed beads labeled with probes (P0119-MC620, Novastar). Finally, a biochemical luminescence detector (NovaHT; Novastar) was used to detect the biotin fluorescence in the hybridized products.
Quantitative RT-PCR (qRT-PCR) was performed to determine the viral RNA copies in individual ticks. To prepare the standard curve for these viruses, a partial fragment (∼300 pb) of the nucleoprotein gene of each virus was amplified by RT-PCR from the viral RNA-positive pools, which were identified by next-generation sequencing (NGS), using the specific forward primer fused with the T7 promoter sequence (TAATACGACTCACTATAGGG) at its 5′ end and the reverse primer designed according to the virus gene sequence. The DNA products were purified and transcribed into RNA using a T7 High Yield RNA Transcription Kit (Vazyme). The RNA was purified, quantified, and serially diluted 10 times to generate standards. To measure the viral RNA copies in tick individuals, qRT-PCR was performed with 1 μL RNA from the single tick as the template and specific forward and reverse primers and probes (Supplementary Table S4) using the One Step PrimeScript RT-PCR Kit (Takara).
2.5. Statistical analysis
Various samples underwent statistical tests for comparison, with all inter-group comparisons subjected to statistical analyses using Graphpad Prism (version 8, Graphpad Software). The significance of differences between two groups was assessed through Student's t-test. In the PCoA plot of Beta diversity, the PERMANOVA test was employed to determine the statistical significance of differences between the two groups. Levels of statistical significance are indicated as follows: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001. Figures do not display non-significant differences.
3. Results
3.1. The free and engorged H. longicornis ticks in Hubei Province had different virome structures
A total of 237.90 Gb of sequencing data were generated from the pooled tick samples. The viral abundance varied in these pools, as revealed by the TPM values that ranged from 0.00016% to 2.99% of the total (Supplementary Table S2). From these pools, viral sequences related to the 39 reference viruses were identified, with nucleotide sequence similarities varying from 27.30% to 100.00% (Table 1), suggesting the existence of novel viruses and new strains. Those reference viruses comprised 12 unclassified viruses, as well as viruses from seven families: Chuviridae, Nairoviridae, Orthomyxoviridae, Parvoviridae, Phenuiviridae, Rhabdoviridae, and Totiviridae (Table 1, Fig. 1B). In free ticks, four pools (f1, f2, f5, and f8) showed a high abundance of viruses related to Phenuiviridae. In engorged ticks, the e1 pool, collected from the same sampling location as the f1 pool, had a viral composition similar to that of f1, with a high abundance of Phenuiviridae. The e8 pool had a high abundance of Orthomyxoviridae, in contrast to the high abundance of Phenuiviridae in the corresponding f8 pool (Fig. 1B).
Table 1.
Summary of viruses identified from H. longicornis tick pools by RNA-seq and their TPM values and identities compared with reference viruses.
| Event | Reference virus | Family | Coverage (%) | Identity (%) | TPM |
|---|---|---|---|---|---|
| 1 | Bole tick virus 3 | Chuviridae | 24.39–98.63 | 41.7–80.6 | 10.98 |
| 2 | Changping tick virus 2 | 18.25–91.53 | 48.7–83.1 | 5.35 | |
| 3 | Chuviridae sp. | 12.46–17.41 | 81.1–82.8 | 8.16 | |
| 4 | Hebei mivirus 2 | 35.45–95.19 | 48.9–79.5 | 0.46 | |
| 5 | Karukera tick virus | 15.56–53.85 | 54.0–85.3 | 0.63 | |
| 6 | Wuhan tick virus 2 | 57.68–100.0 | 94.4–99.4 | 13.78 | |
| 7 | Xinjiang mivirus 1 | 40.33 | 80.0 | 0.63 | |
| 8 | Huangpi tick virus 1 | Nairoviridae | 5.4–93.14 | 95.5–99.8 | 28.68 |
| 9 | Taggert virus | 14.11–86.84 | 86.7–86.7 | 2.90 | |
| 10 | Granville quaranjavirus | Orthomyxoviridae | 60.98–96.22 | 61.9–83.6 | 64.07 |
| 11 | Ohshima virus | 32.77–94.8 | 27.4–48.4 | 1.90 | |
| 12 | Zambezi tick virus 1 | 30.21–92.82 | 44.9–76.4 | 36.89 | |
| 13 | Lone star tick densovirus 1 | Parvoviridae | 24.31–99.0 | 31.6–58.5 | 8.12 |
| 14 | Dabieshan tick virus | Phenuiviridae | 22.63–100.0 | 58–100 | 30658.97 |
| 15 | Kismayo virus | 31.15–99.44 | 43.8–47.9 | 1.80 | |
| 16 | Lihan tick virus | 99.66 | 99.4 | 1.75 | |
| 17 | Lone star virus | 32.45–71.53 | 50–52.2 | 1.13 | |
| 18 | Okutama tick virus | 44.8–99.27 | 99.3–99.7 | 839.31 | |
| 19 | Rhipicephalus associated phlebovirus 1 | 97.32–98.67 | 93.8–96.7 | 2.61 | |
| 20 | Huangpi tick virus 3 | Rhabdoviridae | 11.83–98.98 | 35.0–58.9 | 2.35 |
| 21 | IRE/CTVM19 associated rhabdovirus | 14.41–89.05 | 52.6–70.2 | 1.99 | |
| 22 | Tacheng tick virus 3 | 48.14–52.91 | 58.5–58.8 | 0.78 | |
| 23 | Wuhan tick virus 1 | 62.82 | 99.0 | 3.01 | |
| 24 | Lonestar tick totivirus | Totiviridae | 15.06–99.69 | 33.3–70.7 | 15.81 |
| 25 | Xinjiang tick totivirus 2 | 33.16–99.5 | 40.5–71.2 | 2.46 | |
| 26 | Alongshan virus | Unclassified viruses | 20.18–99.7 | 65.4–100 | 18.26 |
| 27 | Jingmen tick virus | 8.25–99.96 | 61.8–100 | 735.60 | |
| 28 | Bole tick virus 4 | 13.97–95.33 | 76.2–85.7 | 0.72 | |
| 29 | Henan tick virus | 29.39–91.23 | 97.1–99.4 | 83.81 | |
| 30 | Hepelivirales sp. | 24.11–99.81 | 67.6–100 | 396.84 | |
| 31 | Hubei sobemo like virus 15 | 8.45–99.85 | 57.0–100 | 7.60 | |
| 32 | Hubei toti like virus 24 | 34.95–99.75 | 31.5–71.3 | 4.95 | |
| 33 | Ixodes scapularis associated virus 2 | 13.65–87.34 | 45.9–76.7 | 3.94 | |
| 34 | Liman tick virus | 17.08–99.66 | 36.3–68.7 | 11.63 | |
| 35 | Lone star tick associated virus-1 | 8.45 | 52.7 | 27.62 | |
| 36 | Manly virus | 37.55–99.67 | 29.9–75.0 | 6.91 | |
| 37 | Norway mononegavirus 1 | 30.43–99.64 | 27.3–79.4 | 0.76 | |
| 38 | Shanxi tick virus 1 | 55.22 | 99.2 | 1.03 | |
| 39 | Tick borne tetravirus-like virus | 88.41–95.68 | 91.2–100 | 9.89 | |
| Total TPM | 33024.07 | ||||
3.2. The free and engorged H. longicornis ticks may have different competence to carry viruses
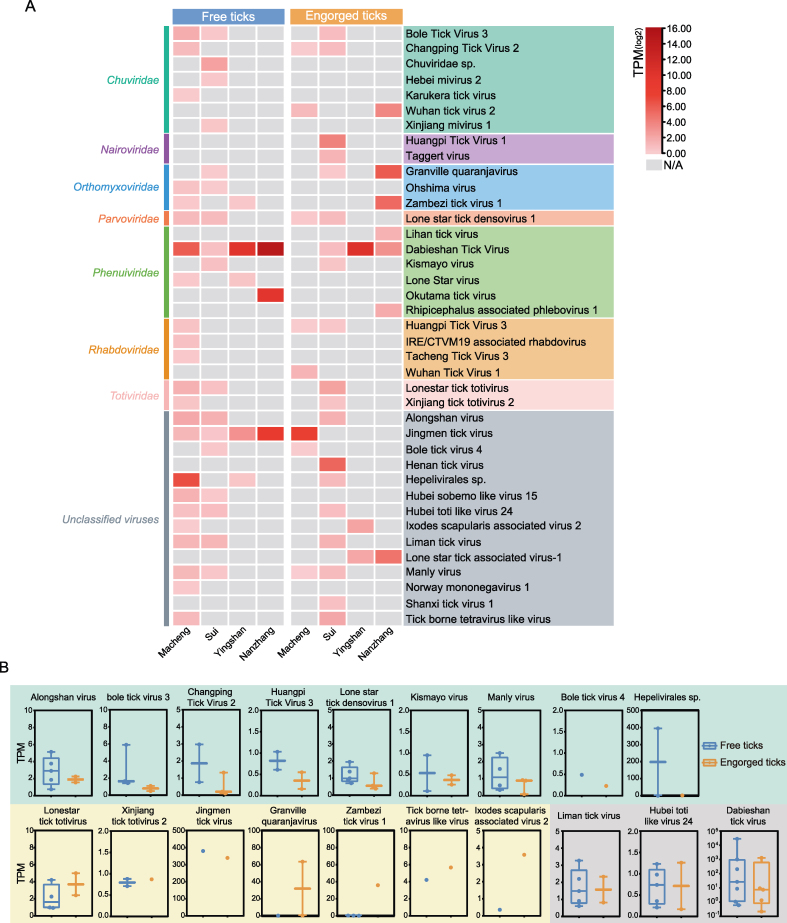
To better understand the competence of H. longicornis ticks in association with geographic distribution, the average abundances of the 39 identified viruses were calculated by merging tick pools from the same location for analysis. Subsequently, heatmaps representing the viromes of free and engorged ticks were generated for each of the four locations separately. In general, ticks from Macheng and Sui showed greater viral diversity than ticks from Yingshan and Nanzhang (Fig. 2A, Supplementary Table S5). We noted a significantly high abundance of the Dabieshan tick virus, which was commonly found in free and engorged ticks from all four locations, suggesting that this virus may have a high prevalence in H. longicornis ticks in Hubei. Of the 39 viruses, 19 were discovered in both free and engorged ticks, including Bole tick virus 3 and Changping tick virus 2 (Chuviridae), Granville quaranjavirus and Zambezi tick virus 1 (Orthomyxoviridae), Lone star tick densovirus 1 (Parvoviridae), Dabieshan tick virus and Kismayo virus (Phenuiviridae), Huangpi tick virus 3 (Rhabdoviridae), Lonestar tick totivirus and Xinjiang tick totivirus 2 (Totiviridae), Alongshan virus, Jingmen tick virus, Bole tick virus 4, Hepelivirales sp., Hubei toti-like virus 24, Ixodes scapularis associated virus 2, Liman tick virus, Manly virus, and tick-borne tetravirus-like virus (unclassified) (Supplementary Table S6). The abundances of the 19 viruses found in both free and engorged ticks were compared. We found that the free ticks primarily had higher abundances of nine viruses including Alongshan tick virus, Bole tick virus 3, Changing tick virus 2, Huangpi tick virus 3, Lone star tick densovirus 1, Kismayo virus, Manly virus, Bole tick virus 4, and Hepelivirales sp. compared to engorged ticks (Fig. 2B, shaded in green). Meanwhile, seven viruses including Lonestar tick totivirus, Xinjiang tick totivirus 2, Jingmen tick virus, Graville quaranjavirus, Zambezi tick virus 1, tick borne tetravirus-like virus, and Ixodes scapularis associated virus 2 had higher abundances in engorged ticks than free ticks (Fig. 2B, shaded in yellow). Further, three viruses (Lihan tick virus, Hubei toti-like virus 24, and Dabieshan tick virus) were abundant in free ticks, compared to engorged ticks (Fig. 2B, shaded in gray).
Fig. 2.
Analyses of viral abundance in free and engorged ticks. A A heatmap of the abundances of viruses identified in the pools of free and engorged ticks collected from different locations. TPM values were calculated by merging tick pools from the same location. B Abundances of viruses which were identified in both free and engorged ticks. Viruses having higher average abundances in free ticks than in engorged ticks are shaded in green and those with lower abundances in free ticks than in engorged ticks are shaded in yellow. Those having equal abundances in free and engorged ticks are shaded in gray.
In summary, these results revealed differences in viral abundances between free and engorged ticks. Moreover, of the 39 viruses, eleven viruses were exclusively found in free ticks, whereas nine were identified only in engorged ticks (Supplementary Table S6). These finding indicated that free and engorged H. longicornis ticks may have different vector competence in carrying viruses.
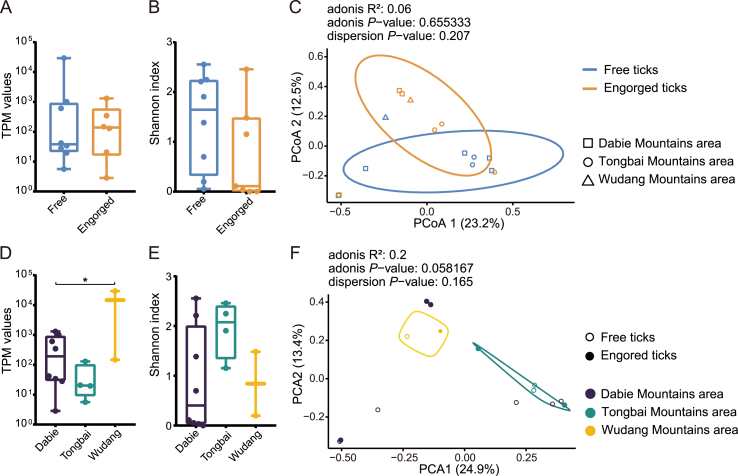
3.3. Geographic locations may affect virus abundance, richness, and diversity more than feeding status in H. longicornis ticks
Although the total TPM values of viruses did not show any significant differences between free and engorged ticks (Fig. 3A), the free ticks tended to have higher viral richness, as suggested by the higher Shannon index, than the engorged ticks. However, this difference was not significant (Fig. 3B). Based on the beta diversity analysis, free ticks from different locations formed a cluster of viral spectra, which partially overlapped with the cluster formed by engorged ticks (Fig. 3C). This finding suggested that the divergence of viral diversity between free and engorged ticks was not significant. Moreover, the viral abundance in ticks from the Wudang Mountain area was significantly higher than that in the Dabie Mountain area (Fig. 3D, P = 0.0383). Ticks from the Tongbai mountainous region had a higher viral richness than those from the Dabie and Wudang mountainous areas; however, the difference was not significant (Fig. 3E). Additionally, beta diversity analysis showed separate clusters of viral diversity in ticks from the Tongbai and Wudang mountainous regions (Fig. 3F), suggesting a differential composition of tick viral spectra in these two areas. The viral diversity of ticks from the Dabie Mountain region presented a scattered pattern (Fig. 3F), suggesting a high diversity of tick viromes in this region.
Fig. 3.
Comparison of the viral diversity and abundance between sampling sites and feeding statuses. A Total TPM of the virus in free and engorged ticks. B The Shannon index of free and engorged ticks. C PCoA analysis of viruses in free and engorged ticks. D Total TPM of the virus in the Dabie, Tongbai, and Wudang mountainous areas. E The Shannon index of ticks in the Dabie, Tongbai, and Wudang mountainous areas. F PCoA analysis of viruses based on the sampling locations of ticks.
3.4. Phylogeny of new strains of Henan tick virus (HNTV), Dabieshan tick virus (DBSTV), Okutama tick virus (OKTV), and Jingmen tick virus (JMTV)
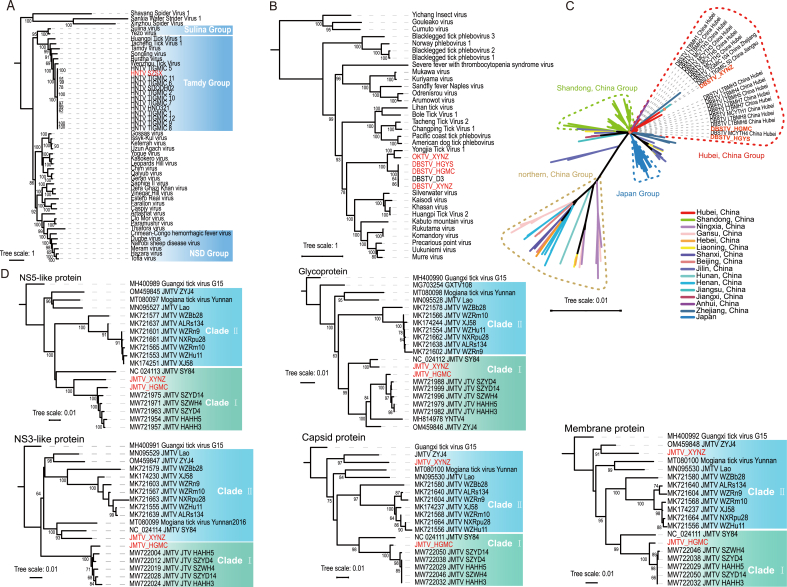
3.4.1. Nairoviridae: Henan tick virus
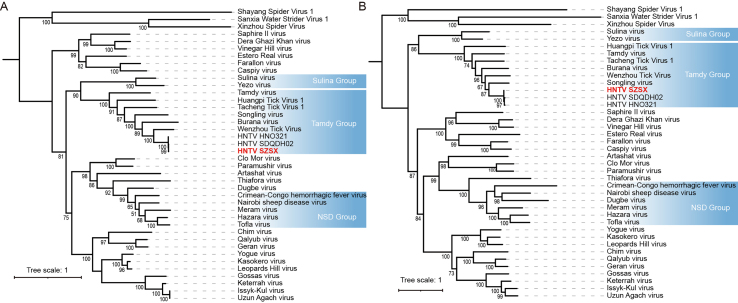
The complete genome sequence of HNTV was obtained from the engorged ticks collected in Suizhou, and contained three RNA segments, L (12022 nt), M (4820 nt), and S (1820 nt), encoding RNA-dependent RNA Polymerase (RdRp), glycoprotein (GP), and nucleoprotein (NP), respectively. Evolutionary analysis showed that in the RdRp phylogenetic tree, the HNTV strain identified in this study is most closely related to strain TIGMIC5 (ON811869.1). TIGMIC5 was identified from H. longicornis in Jiangsu, China, and shares a nucleotide similarity of 99.47% with HNTV (Fig. 4A). Due to the fewer M and S segment sequences available in GenBank compared to that for the L segment, the HNTV strain was found to be most closely related to the strain SDQDH02 (OQ513668.1) by sharing high nucleotide similarity (94.37% for NP and 94.47% for GP) with this reference strain (Supplementary Fig. S1). Therefore, the HNTV in this study is a new strain designated as SZSX, based on the sampling location. This new strain belongs to the Tamdy orthonairovirus group of the genus Orthonairovirus of family Nairoviridae and is closely related to the TS1-2 strain of Wenzhou tick virus, which was found in Haemaphysalis hystricis in Wenzhou, China, in 2013 (Li C.X. et al., 2015) (Fig. 4A, Supplementary Fig. S1).
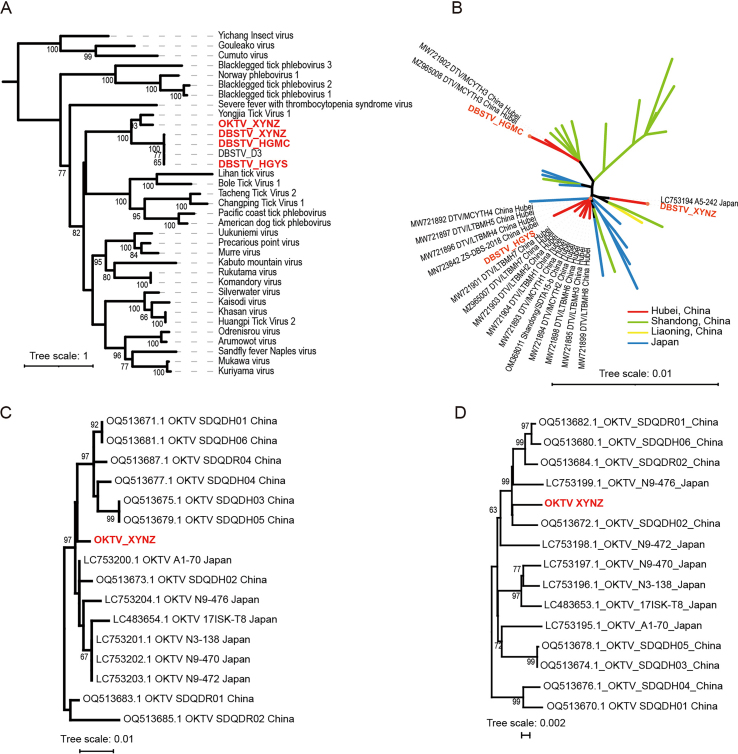
Fig. 4.
Phylogenetic trees constructed for HNTV based on the amino acid sequences of RdRp (A), for DBSTV and OKTV based on the amino acid sequences of RdRp (B), for DBSTV based on nucleotide sequences of RdRp (C), and for five proteins of JMTV (NS5-like protein, glycoprotein, NS3-like protein, capsid protein, and membrane protein) based on nucleotide sequences (D). The viruses discovered in this study are shown in red.
3.4.2. Phenuiviridae: Dabieshan tick virus and Okutama tick virus
The complete sequences of the S and L segments of the three DBSTV strains were obtained from Nanzhang, Macheng, and Yingshan, while the complete sequences of the S and L segments of the new OKTV strain (XYNZ) were obtained from engorged ticks in Nanzhang. The M segments of DBSTV and OKTV were not found in our RNA-seq data, consistent with previous studies that also reported the absence of M segments in these two viruses (Kobayashi et al., 2020; Ma et al., 2022). Evolutionary analysis of the RdRp and NP sequences showed that they are closely related to Yongjia tick virus 1 (Fig. 4B, Supplementary Fig. S2A). Previously, DBSTV variants have been discovered in H. longicornis from Japan and 14 provinces in China. Further, DBSTV variants were also found in Haemaphysalis concinna from Shandong and Jilin, Rhipicephalus turanicus from Shandong, and Rhipicephalus microplus from Hubei (Li C.X. et al., 2015; Ma et al., 2022; Mekata et al., 2023; Shao et al., 2020; Xu et al., 2021; Zhu et al., 2020). Based on the RdRp tree, we found that the DBSTV phylogeny may be associated with their geographic distribution. The DBSTV strains from Hubei, including the three strains in this study, formed a cluster (Hubei group), while the strains from Shandong and Japan clustered separately from their respective clades (Shandong and Japan groups, respectively). DBSTV strains from other provinces were much fewer than those found in Hubei, Shandong, and Japan, and those from provinces in northern China clustered in the north China group (Fig. 4C). However, we did not observe an obvious association between DBSTV evolution and geographic distribution in the tree of DBSTV NPs, probably because the NP sequences were much fewer in number than the RdRp sequences. In the DBSTV NP tree, which contained 57 sequences from the Hubei, Shandong, and Liaoning provinces of China and 10 sequences from Japan, the DBSTV strain HGYS clustered with most strains from Hubei, whereas HGMC strain clustered with two other strains (MW721902 and MZ965008) from Hubei on a different branch, and XYNZ was close to one strain from Japan (LC753194) (Supplementary Fig. S2B). This suggests a different phylogenetic relationship between the DBSTV L and S segments.
In recent years, OKTV variants have been identified in ticks from Shandong, China, and Japan (Kobayashi et al., 2020; Matsumoto et al., 2018). The OKTV XYNZ strain identified in this study was similar to a strain from Shandong (Supplementary Fig. S2C and D).
3.4.3. Unclassified: Jingmen tick virus
Complete genome sequences of two JMTV strains from engorged ticks in Macheng and free ticks in Nanzhang were obtained and designated as strains HGMC and XYNZ, respectively. Phylogenetic analysis using nucleotide sequences from five JMTV ORFs (NS5-like protein, glycoprotein, NS3-like protein, capsid protein, and membrane protein) revealed that JMTV strains generally belong to two clades. Strain HGMC was classified within clade I, including strains from Hubei across all five trees. However, the position of strain XYNZ varied among the trees. Specifically, in the trees of JMTV NS5-like protein and glycoprotein, XYNZ clustered with HGMC in clade I. In trees constructed using NS3-like protein, membrane protein, or capsid protein, it was categorized under clade II (Fig. 4D).
3.5. Prevalence and abundance of viruses in individual ticks
Regarding the high abundance detected in libraries and their evolutionary relationship to tick-borne viral pathogens, the substantial infection rates of HNTV, DBSTV, OKTV, and JMTV were investigated in the remaining 286 H. longicornis ticks using bead-based assays (Table 2). Among these ticks, one free tick from Sui was tested positive for HNTV RNA, resulting in an infection rate of 0.35%. For DBSTV, 36 positive ticks were found (12.59% of total), among which, 28 were from Yingshan (28/79, 35.44%) and 8 were from Nanzhang (8/50, 16.00%). Interestingly, free ticks in Yingshan displayed a higher DBSTV infection rate (23/50, 46.00%) compared to engorged ticks from the same location (5/29, 17.24%). No ticks tested positive for JMTV or OKTV.
Table 2.
The prevalence rates of HNTV, DBSTV, OKTV, and JMTV among 286 H. longicornis tick individuals sequenced using beads assays.
| Location | Ticks (n) | HNTV | DBSTV | OKTV | JMTV | |
|---|---|---|---|---|---|---|
| Dabie Mountain area | Yingshan | Free ticks (n = 50) | 0 | 23, 46.00% | 0 | 0 |
| Engorged ticks (n = 29) | 0 | 5, 17.24% | 0 | 0 | ||
| Subtotal (n = 79) | 0 | 28, 35.44% | 0 | 0 | ||
| Macheng | Free ticks (n = 50) | 0 | 0 | 0 | 0 | |
| Engorged ticks (n = 27) | 0 | 0 | 0 | 0 | ||
| Subtotal (n = 77) | 0 | 0 | 0 | 0 | ||
| Tongbai Mountain area | Suixian | Free ticks (n = 50) | 1, 2% | 0 | 0 | 0 |
| Engorged ticks (n = 30) | 0 | 0 | 0 | 0 | ||
| Subtotal (n = 80) | 1, 1.25% | 0 | 0 | 0 | ||
| Wudang Mountain area | Nanzhang | Free ticks (n = 50) | 0 | 8, 16.00% | 0 | 0 |
| Total (n = 286) | 286 | 1, 0.35% | 36, 12.59% | 0 | 0 | |
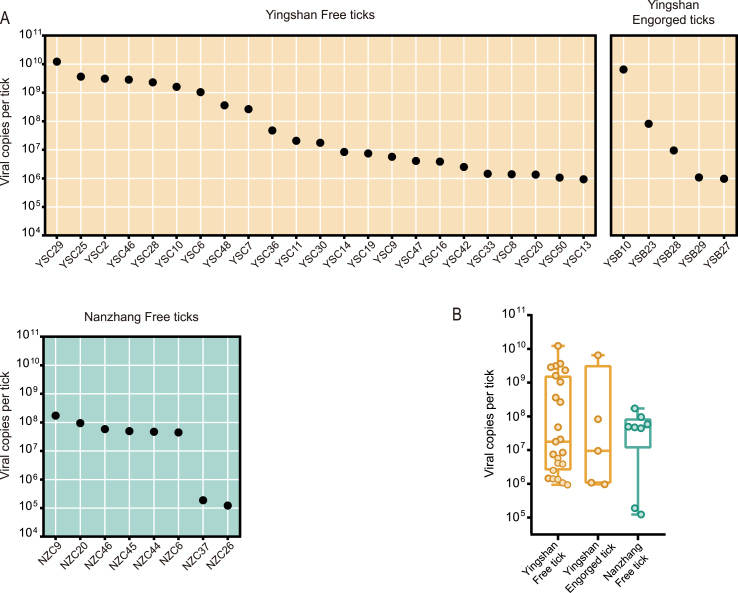
Subsequently, the viral abundances of HNTV and DBSTV in each of the positive ticks were investigated using qRT-PCR. The viral load of HNTV in the single-positive tick was 3.61 × 1010 copies. For DBSTV, the viral loads ranged from 1.23 × 105 to 1.22 × 1010 copies per tick (Fig. 5A), displaying approximately a 100,000-fold variation. Furthermore, free ticks from Yingshan frequently exhibited high DBSTV viral loads ranging from 106 to 1010 copies per tick, similar to those in engorged ticks from the same location. The lowest DBSTV RNA loads, approximately 105 copies per tick, were identified in two free ticks from Nanzhang, while other free ticks from Nanzhang had DBSTV loads ranging from 107 to 108 copies per tick, which was still less than that of Yingshan but not statistically significant (Fig. 5B).
Fig. 5.
Viral RNA loads in individual ticks were measured by qRT-PCR. A RNA copies of DBSTV in the individuals of free and engorged ticks from Yingshan and Nanzhang. B Comparison of DBSTV RNA from the free and engorged ticks of Yingshan and from the free ticks of Nanzhang.
4. Discussion
H. longicornis, the predominant tick specie in central China, plays a crucial role in transmitting tick-borne viruses such as SFTSV. The Dabie Mountain region, located between Hubei and Henan provinces, is a natural hotspot for SFTS (Liu et al., 2014). Investigations into this region have involved assessing SFTSV prevalence in hosts (i.e., ticks, animals, or humans), and tick viromes to understand the threat of SFTSV infection and prepare for its prevention and control (Liu et al., 2023; Xing et al., 2016; Xu et al., 2021; Yang et al., 2019). A few studies, which examined the viromes of H. longicornis ticks collected from the Dabie Mountain region in 2016 and 2019 (Liu et al., 2023; Xu et al., 2021; Yang et al., 2019), reported a large diversity of DNA viruses. In this study, we investigated H. longicornis tick viromes collected in 2019 and 2020 from three mountainous regions including the Dabie Mountain region locating in Hubei. RNA-seq results identified viral sequences from 39 viruses in seven RNA viral families and unclassified viruses. Although viral species varied compared to previous studies (Liu et al., 2023; Xu et al., 2021; Yang et al., 2019), Nairoviridae, Phenuiviridae, and Rhabdoviridae were the most predominant viral communities in H. longicornis ticks in Hubei, and were highly abundant in our samples. These newly discovered viral sequences suggest novel diversity and potential new viral strains. Unlike previous studies that identified SFTSV sequences in ticks (Xu et al., 2021; Yang et al., 2019), we did not find SFTSV-related sequences in our RNA-seq or bead assay results from individual ticks (data not shown). A possible explanation for this discrepancy is the differences in the site and time of sample collection. Nonetheless, consistent with a recent study (Xu et al., 2021), we observed high abundances of DBSTV and JMTV in both free and engorged H. longicornis ticks.
DBSTV exhibits a wide geographical distribution, with variants found in 14 provinces of China (Ma et al., 2022; Shao et al., 2020; Xu et al., 2021; Zhu et al., 2020) and in Japan (Mekata et al., 2023). Its evolution appears to be linked to its geographical distribution, with strains from Hubei, Shandong, and Japan clustering as dominant variants of all identified DBSTV sequences. Although other provinces had fewer strains, those from the northern provinces of China also formed clusters. Despite limited knowledge of DBSTV's pathogenicity, its wide distribution and geographic distribution-associated phylogeny indicate the need for continuous assessment of the potential transmission and disease risks. According to the phylogenetic trees built based on the ORF sequences of NS5-like proteins, glycoproteins, NS3-like proteins, membrane proteins, and capsid proteins, JMTV sequences formed two clades. Clade I mainly consisted of sequences from Hubei, whereas Clade II primarily included sequences from Wenzhou, Guizhou, Henan, and Laos (Guo et al., 2020; Temmam et al., 2019). The shifting position of strain XYNZ likely indicates genetic variation resulting from segment reassortment in this segmented RNA virus. To explore JMTV's evolutionary features regarding location, migration, and recombination or reassortment, more sequences are needed. Engorged ticks had a high abundance of HNTV, and OKTV was found in free ticks. Importantly, ours is the first study to identify HNTV in Hubei Province. HNTV belongs to the Tamdy group in the genus Orthonairovirus, which contains pathogenic viruses associated with human disease, such as Tamdy virus and Songling virus (Ma et al., 2021; Moming et al., 2021). Our study recommends to conduct further surveys of HNTV prevalence in ticks and hosts. Further, this is first time that OKTV has been identified in H. longicornis ticks in Hubei, which was previously reported from H. flava ticks from the Okutama region of Japan and later in other parts of Japan (Kobayashi et al., 2020; Matsumoto et al., 2018). OKTV was present in ticks in Shandong, China, according to the sequences deposited in GenBank recently. The phylogenetic association between OKTV identified in Hubei and Shandong suggests that this virus has a common origin and a wide spatial distribution.
H. longicornis plays a critical role in virus maintenance and transmission through taking blood meals. Therefore, feeding status affected the virome composition of these ticks in our study. We observed differences between free and engorged ticks collected from the same sampling locations. By comparing virus communities and abundances in these ticks, we observed that some viruses more abundant in free ticks, while others were more prevalent in engorged ticks. Although the depth of sequencing data may be affected by the blood substance, thus having impact on virus abundance in the engorged ticks, this observation still indicates the impact of blood feeding on virus proliferation or consumption in ticks. The host-specific factors may also contribute. The viral diversity in engorged ticks is lower compared to free ticks, possibly because ticks transmit most viruses into the host's blood through their saliva, triggering the host's immune system and neutralizing many viruses (Fogaça et al., 2021). This results in reduced tick-borne virus diversity in engorged ticks, with only a few viruses capable of successful replication in the host, and their abundance remains unaffected and still detectable in engorged ticks.
Certain viruses showed consistent abundance in both free and engorged ticks. For example, DBSTV exhibited stability in tick pools and viral RNA loads in individual ticks. Previous studies have detected DBSTV in H. longicornis, Haemaphysalis concinna, Rhipicephalus turanicus, and Rhipicephalus microplus ticks in Hubei and Shandong provinces, as well as in many other provinces in northern China (Li C.X. et al., 2015; Ma et al., 2022; Mekata et al., 2023; Shao et al., 2020; Xu et al., 2021; Zhu et al., 2020). However, serological exposure to this virus has not been reported to date. In contrast, despite a higher abundance of JMTV in free H. longicornis ticks based on RNA-seq in two different pools, individual ticks did not show JMTV RNA. JMTV has been previously found in R. microplus ticks in Hubei province of China (Qin et al., 2014). It has been increasingly detected in various regions of China, as well as in Brazil, Trinidad, Tobago, French Antilles, and Turkey, and in tick species belonging to the genera Rhipicephalus, Amblyomma, Dermacentor, Haemaphysalis, Hyalomma, and Ixodes, with a particularly high prevalence in R. microplus ticks (Colmant et al., 2022; Dinçer et al., 2019; Gondard et al., 2020; Maruyama et al., 2014; Sameroff et al., 2019; Souza et al., 2018; Temmam et al., 2019; Xu et al., 2021). This observation suggests that JMTV has a wide distribution worldwide. However, H. longicornis ticks may not be its major vector, which possibly explain why JMTV was not identified in ticks.
Free and engorged ticks exhibited varying viral richness and diversity. These results are consistent with the findings of a recent study, which suggested that tick virome structure is influenced by ecological factors such as tick feeding status (Xu et al., 2021). In addition, our study showed that H. longicornis ticks from different mountainous regions exhibited distinct viral richness and diversity, suggesting that the local ecological environment plays a key role in shaping tick viromes. Therefore, conducting an extensive epidemiological investigation of tick viromes associated with ecological factors would further promote our understanding of the composition of viral communities in H. longicornis ticks across different locations.
Although we revealed the precise viral prevalence in individual ticks, a shortcoming of our study is its limited sample size, which may have affected the complete representation of the overall virus prevalence in Hubei province. Nevertheless, we did observe a high prevalence of DBSTV in ticks from Hubei. For other viruses, a larger sample size should be included in future surveys. Together with the previous research (Xu et al., 2021), our findings of differential viral communities, richness, and abundance in free and engorged ticks suggest that blood feeding likely plays a role in shaping virome structures. However, because we lack samples from animal hosts, we could not determine whether animals were infected and/or exposed to viruses identified with differing abundances in free and engorged ticks. Nonetheless, our results provide insight into the impact of blood feeding on viral proliferation and infection in ticks and animal hosts.
5. Conclusions
This study investigated the viromes of H. longicornis ticks collected across three mountainous regions in Hubei Province during 2019–2020. Our results revealed the genetic diversity within the viral community, suggesting the potential roles of blood feeding and geographic distribution in shaping virome structures. New strains of HNTV, DBSTV, OKTV, and JMTV were identified, and novel phylogenetic insights were gained from DBSTV and JMTV. Moreover, we established a method to precisely determine virus prevalence in individual ticks and found a high prevalence of DBSTV in H. longicornis ticks. These findings significantly enhance our understanding of the H. longicornis tick virome in Hubei, emphasizing the need for increased attention on emerging prevalent viruses in ticks. To comprehensively evaluate the risk of TBV infection and spillover into hosts in China, surveys involving a larger sample size of H. longicornis ticks, along with related animal samples across more wide regions, are needed.
Data availability
The metagenomic data are available in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA1024474, and the viral genomic sequences have been deposited in GenBank nucleotide database under the accession numbers OR573899−OR573917. The metagenomic data are also available in China National GenBank Database (CNGBdb) under project: CNP0004853.
Ethics statement
This article does not contain any human or animal studies by any of the authors.
Author contributions
Jian Xiao: data curation, formal analysis, investigation, methodology, software, writing-original draft. Xuan Yao: resource, investigation, project administration. Xuhua Guan: resource, investigation. Jinfeng Xiong: resource, data curation. Yaohui Fang: data curation, investigation. Jingyuan Zhang: data curation, investigation. You Zhang: data curation, investigation. Shu Shen: conceptualization, funding acquisition, project administration, supervision, validation, writing-review & editing. Fei Deng: conceptualization, funding acquisition, project administration, supervision, validation, writing-review & editing.
Conflict of interest
Professor Fei Deng is an editorial board member for Virologica Sinica and was not involved in the editorial review or the decision to publish this article. The authors declare that they have no conflict of interest.
Acknowledgments
We would like to thank Dr. Zhixian Qiao and Xiaocui Chai at the Analysis and Testing Center of the Institute of Hydrobiology, Chinese Academy of Sciences, for their assistance with RNA-seq and data analysis.
This study was supported by the National Natural Science Foundation of China (grant number: U21A20180), the National Key R&D Program of China (2021YFC2300900, 2022YFC2305100), the Key Deployment Projects of the Chinese Academy of Sciences (KJZD-SW-L11), the Youth Project of the Wuhan Institute of Virology, Chinese Academy of Sciences (2023QNTJ-03), and the National Basic Science Data Sharing Service Platform (NBSDC-DB-13).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.02.003.
Contributor Information
Shu Shen, Email: shenshu@wh.iov.cn.
Fei Deng, Email: df@wh.iov.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- Alexander R.A., Neitz W.O. The transmission of louping-ill of sheep by ticks. Vet. J. 1933;89:320–323. [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker S.C., Walker A.R. Ticks of Australia. The species that infest domestic animals and humans. Zootaxa. 2014;3816:1–144. doi: 10.11646/zootaxa.3816.1.1. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Reuter K., Drost H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chen X., Li F., Yin Q., Liu W., Fu S., He Y., Lei W., Xu S., Liang G., Wang S., Yang G., Qi X., Wang H. Epidemiology of tick-borne encephalitis in China, 2007-2018. PLoS One. 2019;14 doi: 10.1371/journal.pone.0226712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmant A.M.G., Charrel R.N., Coutard B. Jingmenviruses: ubiquitous, understudied, segmented flavi-like viruses. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.997058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinçer E., Hacioǧlu S., Kar S., Emanet N., Brinkmann A., Nitsche A., Özkul A., Linton Y.M., Ergünay K. Survey and characterization of Jingmen tick virus variants. Viruses. 2019;11:1071. doi: 10.3390/v11111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L.Q., Liu K., Li X. Lou, Liang S., Yang Y., Yao H.W., Sun R.X., Sun Y., Chen W.J., Zuo S.Q., Ma M.J., Li H., Jiang J.F., Liu W., Yang X.F., Gray G.C., Krause P.J., Cao W.C. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect. Dis. 2015;15:1467–1479. doi: 10.1016/S1473-3099(15)00177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogaça A.C., Sousa G., Pavanelo D.B., Esteves E., Martins L.A., Urbanová V., Kopáček P., Daffre S. Tick immune system: what is known, the interconnections, the gaps, and the challenges. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.628054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Nasci R., Liang G. The neglected arboviral infections in mainland China. PLoS Negl. Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargili A., Estrada-Peña A., Spengler J.R., Lukashev A., Nuttall P.A., Bente D.A. The role of ticks in the maintenance and transmission of Crimean-Congo hemorrhagic fever virus: a review of published field and laboratory studies. Antiviral Res. 2017;144:93–119. doi: 10.1016/j.antiviral.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondard M., Temmam S., Devillers E., Pinarello V., Bigot T., Chrétien D., Aprelon R., Vayssier-Taussat M., Albina E., Eloit M., Moutailler S. RNA viruses of amblyomma variegatum and rhipicephalus microplus and cattle susceptibility in the French antilles. Viruses. 2020;12:144. doi: 10.3390/v12020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun T.S., Lashkevich V.A., Gould E.A. Tick-borne encephalitis. Antiviral Res. 2003;57:129–146. doi: 10.1016/s0166-3542(02)00206-1. [DOI] [PubMed] [Google Scholar]
- Guo J.J., Lin X.D., Chen Y.M., Hao Z.Y., Wang Z.X., Yu Z.M., Lu M., Li K., Qin X.C., Wang W., Holmes E.C., Hou W., Zhang Y.Z. Diversity and circulation of Jingmen tick virus in ticks and mammals. Virus Evol. 2020;6 doi: 10.1093/ve/veaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfler D., Nicklas W., Mauter P., Pawlita M., Schmitt M. A bead-based multiplex assay for the detection of DNA viruses infecting laboratory rodents. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraal H. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J. Med. Entomol. 1979;15:307–417. doi: 10.1093/jmedent/15.4.307. [DOI] [PubMed] [Google Scholar]
- Jia N., Wang J., Shi W., Du L., Sun Y., Zhan W., Jiang J.F., Wang Q., Zhang B., Ji P., Bell-Sakyi L., Cui X.M., Yuan T.T., Jiang B.G., Yang W.F., Lam T.T.Y., Chang Q.C., Ding S.J., Wang X.J., Zhu J.G., Ruan X.D., Zhao L., Wei J. Te, Ye R.Z., Que T.C., Du C.H., Zhou Y.H., Cheng J.X., Dai P.F., Guo W. Bin, Han X.H., Huang E.J., Li L.F., Wei W., Gao Y.C., Liu J.Z., Shao H.Z., Wang X., Wang C.C., Yang T.C., Huo Q.B., Li W., Chen H.Y., Chen S.E., Zhou L.G., Ni X.B., Tian J.H., Sheng Y., Liu T., Pan Y.S., Xia L.Y., Li J., Zhao F., Cao W.C. Large-scale comparative analyses of tick genomes elucidate their genetic diversity and vector capacities. Cell. 2020;182:1328–1340.e13. doi: 10.1016/j.cell.2020.07.023. [DOI] [PubMed] [Google Scholar]
- Kobayashi D., Murota K., Itokawa K., Ejiri H., Amoa-Bosompem M., Faizah A.N., Watanabe M., Maekawa Y., Hayashi T., Noda S., Yamauchi T., Komagata O., Sawabe K., Isawa H. RNA virome analysis of questing ticks from Hokuriku District, Japan, and the evolutionary dynamics of tick-borne phleboviruses. Ticks Tick Borne Dis. 2020;11 doi: 10.1016/j.ttbdis.2019.101364. [DOI] [PubMed] [Google Scholar]
- Kodama F., Yamaguchi H., Park E., Tatemoto K., Sashika M., Nakao R., Terauchi Y., Mizuma K., Orba Y., Kariwa H., Hagiwara K., Okazaki K., Goto A., Komagome R., Miyoshi M., Ito T., Yamano K., Yoshii K., Funaki C., Ishizuka M., Shigeno A., Itakura Y., Bell-Sakyi L., Edagawa S., Nagasaka A., Sakoda Y., Sawa H., Maeda K., Saijo M., Matsuno K. A novel nairovirus associated with acute febrile illness in Hokkaido, Japan. Nat. Commun. 2021;12:5539. doi: 10.1038/s41467-021-25857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé M., Girard C., Vincent W.F., Culley A.I. Extreme viral partitioning in a marine-derived High Arctic lake. mSphere. 2020;5 doi: 10.1128/mSphere.00334-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.X., Shi M., Tian J.H., Lin X.D., Kang Y.J., Chen L.J., Qin X.C., Xu J., Holmes E.C., Zhang Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife. 2015;4 doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li J., Li S., Yang L., Cao P., Lu J. Severe fever with thrombocytopenia syndrome virus: a highly lethal bunyavirus. Crit. Rev. Microbiol. 2021;47:112–125. doi: 10.1080/1040841X.2020.1847037. [DOI] [PubMed] [Google Scholar]
- Li P., Piao Y., Shon H.S., Ryu K.H. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data. BMC Bioinformatics. 2015;16:347. doi: 10.1186/s12859-015-0778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist L., Vapalahti O. Tick-borne encephalitis. Lancet. 2008;371:1861–1871. doi: 10.1016/S0140-6736(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Liu Q., He B., Huang S.Y., Wei F., Zhu X.Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect. Dis. 2014;14:763–772. doi: 10.1016/S1473-3099(14)70718-2. [DOI] [PubMed] [Google Scholar]
- Liu Y., Guo L., Wang G., Gao F., Tu Z., Xu D., Sun L., Yi L., Zhu G., Tu C., He B. DNA virome of ticks in the Northeast and Hubei provinces of China reveals diverse single-stranded circular DNA viruses. Parasites Vectors. 2023;16:61. doi: 10.1186/s13071-023-05684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Bröker M., Liang G. Tick-borne encephalitis in mainland China. Vector Borne Zoonotic Dis. 2008;8:713–720. doi: 10.1089/vbz.2008.0028. [DOI] [PubMed] [Google Scholar]
- Ma C., Zhang R., Zhou H., Yu G., Yu L., Li J., Cui M., Carr M.J., Zhang Z., Shi W. Prevalence and genetic diversity of Dabieshan tick virus in Shandong Province, China. J. Infect. 2022;85:90–122. doi: 10.1016/j.jinf.2022.04.002. [DOI] [PubMed] [Google Scholar]
- Ma J., Lv X.L., Zhang X., Han S.Z., Wang Z.D., Li L., Sun H.T., Ma L.X., Cheng Z.L., Shao J.W., Chen C., Zhao Y.H., Sui L., Liu L.N., Qian J., Wang W., Liu Q. Identification of a new orthonairovirus associated with human febrile illness in China. Nat. Med. 2021;27:434–439. doi: 10.1038/s41591-020-01228-y. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci S., Kramer L.D., Gebhardt L.L., Kauffman E. Emerging tick-borne diseases. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00083-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S.R., Castro-Jorge L.A., Ribeiro J.M.C., Gardinassi L.G., Garcia G.R., Brandão L.G., Rodrigues A.R., Okada M.I., Abrão E.P., Ferreira B.R., da Fonseca B.A.L., de Miranda-Santos I.K.F. Characterisation of divergent flavivirus NS3 and NS5 protein sequences detected in Rhipicephalus microplus ticks from Brazil. Mem. Inst. Oswaldo Cruz. 2014;109:38–50. doi: 10.1590/0074-0276130166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N., Masuoka H., Hirayama K., Yamada A., Hotta K. Detection and phylogenetic analysis of phlebovirus, including severe fever with thrombocytopenia syndrome virus, in ticks collected from Tokyo, Japan. J. Vet. Med. Sci. 2018;80:638–641. doi: 10.1292/jvms.17-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekata H., Kobayashi I., Okabayashi T. Detection and phylogenetic analysis of Dabieshan tick virus and Okutama tick virus in ticks collected from Cape Toi, Japan. Ticks Tick Borne Dis. 2023;14 doi: 10.1016/j.ttbdis.2023.102237. [DOI] [PubMed] [Google Scholar]
- Moming A., Shen S., Fang Y., Zhang J., Zhang Yanfang, Tang S., Li T., Hu Z., Wang H., Zhang Yujiang, Sun S., Wang L.F., Deng F. Evidence of human exposure to tamdy virus, Northwest China. Emerg. Infect. Dis. 2021;27:3166–3170. doi: 10.3201/eid2712.203532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moming A., Yue X., Shen S., Chang C., Wang C., Luo T., Zhang Yanfang, Guo R., Hu Z., Zhang Yujiang, Deng F., Sun S. Prevalence and phylogenetic analysis of Crimean-Congo hemorrhagic fever virus in ticks from different ecosystems in Xinjiang, China. Virol. Sin. 2018;33:67–73. doi: 10.1007/s12250-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X.B., Cui X.M., Liu J.Y., Ye R.Z., Wu Y.Q., Jiang J.F., Sun Y., Wang Q., Shum M.H.H., Chang Q.C., Zhao L., Han X.H., Ma K., Shen S.J., Zhang M.Z., Guo W. Bin, Zhu J.G., Zhan L., Li L.J., Ding S.J., Zhu D.Y., Zhang J., Xia L.Y., Oong X.Y., Ruan X.D., Shao H.Z., Que T.C., Liu G.Y., Du C.H., Huang E.J., Wang X., Du L.F., Wang C.C., Shi W.Q., Pan Y.S., Zhou Y.H., Qu J.L., Ma J., Gong C.W., Chen Q.Q., Qin Q., Lam T.T.Y., Jia N., Cao W.C. Metavirome of 31 tick species provides a compendium of 1,801 RNA virus genomes. Nat. Microbiol. 2023;8:162–173. doi: 10.1038/s41564-022-01275-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Kindt R., Legendre P., O'Hara B., Simpson G.L., Solymos P.M., Stevens M.H.H., Wagner H. The vegan package. Community Ecol. Packag. 2008;190 [Google Scholar]
- Qin X.C., Shi M., Tian J.H., Lin X.D., Gao D.Y., He J.R., Wang J.B., Li C.X., Kang Y.J., Yu B., Zhou D.J., Xu J., Plyusnin A., Holmes E.C., Zhang Y.Z. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6744–6749. doi: 10.1073/pnas.1324194111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph S.E. Transmission of tick-borne pathogens between co-feeding ticks: Milan Labuda’s enduring paradigm. Ticks Tick Borne Dis. 2011;2:179–182. doi: 10.1016/j.ttbdis.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Sameroff S., Tokarz R., Charles R.A., Jain K., Oleynik A., Che X., Georges K., Carrington C.V., Lipkin W.I., Oura C. Viral diversity of tick species parasitizing cattle and dogs in Trinidad and Tobago. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., Pang Z., Fu H., Chang R., Lin Z., Lv A., Wang S., Kong X., Luo M., Liu X., Yu X., Liu L., Niu G. Identification of recently identified tick-borne viruses (Dabieshan tick virus and SFTSV) by metagenomic analysis in ticks from Shandong Province, China. J. Infect. 2020;81:973–978. doi: 10.1016/j.jinf.2020.10.022. [DOI] [PubMed] [Google Scholar]
- Shi J., Hu Z., Deng F., Shen S. Tick-borne viruses. Virol. Sin. 2018;33:21–43. doi: 10.1007/s12250-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza W.M., Fumagalli M.J., Torres Carrasco A.O., Romeiro M.F., Modha S., Seki M.C., Gheller J.M., Daffre S., Nunes M.R.T., Murcia P.R., Acrani G.O., Figueiredo L.T.M. Viral diversity of Rhipicephalus microplus parasitizing cattle in southern Brazil. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Dai X., Aishan M., Wang X., Meng W., Feng C., Zhang F., Hang C., Hu Z., Zhang Y. Epidemiology and phylogenetic analysis of Crimean-Congo hemorrhagic fever viruses in Xinjiang, China. J. Clin. Microbiol. 2009;47:2536–2543. doi: 10.1128/JCM.00265-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R., Shepherd A.J., Leman P.A., McGillivray G.M., Erasmus M.J., Searle L.A., Gill D.E. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am. J. Trop. Med. Hyg. 1987;36:120–132. doi: 10.4269/ajtmh.1987.36.120. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmam S., Bigot T., Chrétien D., Gondard M., Pérot P., Pommelet V., Dufour E., Petres S., Devillers E., Hoem T., Pinarello V., Hul V., Vongphayloth K., Hertz J.C., Loiseau I., Dumarest M., Duong V., Vayssier-Taussat M., Grandadam M., Albina E., Dussart P., Moutailler S., Cappelle J., Brey P.T., Eloit M. Insights into the host range, genetic diversity, and geographical distribution of Jingmenviruses. mSphere. 2019;4 doi: 10.1128/mSphere.00645-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.D., Wang B., Wei F., Han S.-Z., Zhang L., Yang Z.T., Yan Y., Lv X.L., Li L., Wang S.C., Song M.X., Zhang H.J., Huang S.J., Chen J., Huang F.Q., Li S., Liu H.H., Hong J., Jin Y.L., Wang W., Zhou J.Y., Liu Q. A new segmented virus associated with human febrile illness in China. N. Engl. J. Med. 2019;380:2116–2125. doi: 10.1056/NEJMoa1805068. [DOI] [PubMed] [Google Scholar]
- Wingett S.W., Andrews S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 2018;7:1338. doi: 10.12688/f1000research.15931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.B., Na R.H., Wei S.S., Zhu J.S., Peng H.J. Distribution of tick-borne diseases in China. Parasites Vectors. 2013;6:119. doi: 10.1186/1756-3305-6-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Li P., Yang J., Pan L., Zhao J., Wang Z., Li Y., Zhou H., Dong Y., Guo S., Tang S., Zhang Z., Fan Z., Hu Z., Kou Z., Li T. Epidemiological survey of Crimean-Congo hemorrhagic fever virus in Yunnan, China, 2008. Int. J. Infect. Dis. 2011;15:e459–e463. doi: 10.1016/j.ijid.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Xing X., Guan X., Zhan J., Jiang H., Liu L., Li G., Xiong J., Tan L., Xu J., Jiang Y., Yao X., Zhan F., Nie S. Natural transmission model for severe fever with thrombocytopenia syndrome bunyavirus in Villages of Hubei Province, China. Medicine (United States) 2016;95 doi: 10.1097/MD.0000000000002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Liu L., Huang X., Ma H., Zhang Y., Du Y., Wang P., Tang X., Wang H., Kang K., Zhang S., Zhao G., Wu W., Yang Y., Chen H., Mu F., Chen W. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (ftls) in henan province, China: discovery of a new bunyavirus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Guo M., Hu B., Zhou H., Yang W., Hui L., Huang R., Zhan J., Shi W., Wu Y. Tick virome diversity in Hubei Province, China, and the influence of host ecology. Virus Evol. 2021;7 doi: 10.1093/ve/veab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.E., Zhao Z., Hou G., Zhang C., Liu J., Xu L., Li W., Tan Z., Tu C., He B. Genomes and seroprevalence of severe fever with thrombocytopenia syndrome virus and Nairobi sheep disease virus in Haemaphysalis longicornis ticks and goats in Hubei, China. Virology. 2019;529:234–245. doi: 10.1016/j.virol.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.J., Liang M.F., Zhang S.Y., Liu Y., Li J.D., Sun Y.L., Zhang L., Zhang Q.F., Popov V.L., Li C., Qu J., Li Q., Zhang Y.P., Hai R., Wu W., Wang Q., Zhan F.X., Wang X.J., Kan B., Wang S.W., Wan K.L., Jing H.Q., Lu J.X., Yin W.W., Zhou H., Guan X.H., Liu J.F., Bi Z.Q., Liu G.H., Ren J., Wang H., Zhao Z., Song J.D., He J.R., Wan T., Zhang J.S., Fu X.P., Sun L.N., Dong X.P., Feng Z.J., Yang W.Z., Hong T., Zhang Y., Walker D.H., Wang Y., Li D.X. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hu B., Agwanda B., Fang Y., Wang J., Kuria S., Yang J., Masika M., Tang S., Lichoti J., Fan Z., Shi Z., Ommeh S., Wang H., Deng F., Shen S. Viromes and surveys of RNA viruses in camel-derived ticks revealing transmission patterns of novel tick-borne viral pathogens in Kenya. Emerg. Microbes Infect. 2021;10:1975–1987. doi: 10.1080/22221751.2021.1986428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.F., Shen S., Fang Y., Liu J., Su Z., Liang J., Zhang Z., Wu Q., Wang C., Abudurexiti A., Hu Z., Zhang Yujiang, Deng F. Isolation, characterization, and phylogenetic analysis of two new Crimean-Congo hemorrhagic fever virus strains from the northern region of Xinjiang province, China. Virol. Sin. 2018;33:74–86. doi: 10.1007/s12250-018-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Li J., Cui X., Jia N., Wei J., Xia L., Wang H., Zhou Y., Wang Qian, Liu X., Yin C., Pan Y., Wen H., Wang Qing, Xue F., Sun Y., Jiang J., Li S., Cao W. Distribution of Haemaphysalis longicornis and associated pathogens: analysis of pooled data from a China field survey and global published data. Lancet Planet. Health. 2020;4:e320–e329. doi: 10.1016/S2542-5196(20)30145-5. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li M.C., Konaté M.M., Chen L., Das B., Karlovich C., Williams P.M., Evrard Y.A., Doroshow J.H., McShane L.M. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Transl. Med. 2021;19:269. doi: 10.1186/s12967-021-02936-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., He T., Wu T., Ai L., Hu D., Yang X., Lv R., Yang L., Lv H., Tan W. Distribution and phylogenetic analysis of Dabieshan tick virus in ticks collected from Zhoushan, China. J. Vet. Med. Sci. 2020;82:1226–1230. doi: 10.1292/jvms.20-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomic data are available in NCBI Sequence Read Archive (SRA) under Bioproject: PRJNA1024474, and the viral genomic sequences have been deposited in GenBank nucleotide database under the accession numbers OR573899−OR573917. The metagenomic data are also available in China National GenBank Database (CNGBdb) under project: CNP0004853.