Abstract
Coriariaceae are a small plant family of 14–17 species and subspecies that currently have a global but disjunct distribution. All species can form root nodules in symbiosis with diazotrophic Frankia cluster-2 strains, which form the earliest divergent symbiotic clade within this bacterial genus. Studies on Frankia cluster-2 mostly have focused on strains occurring in the northern hemisphere. Except for one strain from Papua New Guinea, namely Candidatus Frankia meridionalis Cppng1, no complete genome of Frankia associated with Coriaria occurring in the southern hemisphere has been published thus far, yet the majority of the Coriariaceae species occur here. We present field sampling data of novel Frankia cluster-2 strains, representing two novel species, which are associated with Coriaria arborea and Coriaria sarmentosa in New Zealand, and with Coriaria ruscifolia in Patagonia (Argentina), in addition to identifying Ca. F. meridionalis present in New Zealand. The novel Frankia species were found to be closely related to both Ca. F. meridionalis, and a Frankia species occurring in the Philippines, Taiwan, and Japan. Our data suggest that the different Frankia cluster-2 species diverged early after becoming symbiotic circa 100 million years ago.
Keywords: actinorhizal symbiosis, Coriariaceae, Frankia, microbiome, New Zealand, Papua New Guinea, Patagonia
The distribution of the microsymbionts that induce nitrogen-fixing root nodules on Coriaria species was examined in the Southern Hemisphere; the results suggest that the microsymbionts diverged early after evolving symbiosis.
Introduction
Coriariaceae are a small plant family within the Cucurbitales, consisting of the single genus Coriaria with about 16 plant species (Yokoyama et al. 2000, Renner et al. 2020). They are found worldwide but have a very disjunct distribution (Fig. 1, Data S1). Occupying harsher environments such as riverbeds and volcanic hills, they are considered to be pioneer species (Becking 1977). The origin of the family and their biogeographic history have been discussed in several publications (Good 1930, Skog 1972, Yokoyama et al. 2000, Nouioui et al. 2014), and according to the most recent data, the family evolved around 87 million years ago (Mya), and it split into two lineages between 46 and 57 Mya (Renner et al. 2020). The majority of species belong to the lineage which dispersed in the southern hemisphere and reached America via Antarctica (Renner et al. 2020). This includes eight species native to New Zealand, Coriaria papuana in Papua New Guinea, and two subspecies of Coriaria ruscifolia in America which have dispersed as far north as Mexico. The second lineage is found in the northern hemisphere; spanning from Mediterranean Europe and northern Africa (Coriaria myrifolia), Nepal and China (Coriaria terminalis, Coriaria nepalensis, and Coriaria duthiei), Japan (Coriaria japonica), to Taiwan, and the Philippines (Coriaria intermedia; Fig. 1; Renner et al. 2020). All Coriaria species are described to form root nodules in symbiosis with Frankia spp. These specialized organs are formed when plants belonging to the nitrogen-fixing clade engage in symbiosis with soil diazotrophs; i.e. rhizobia for legumes and Parasponia spp., or Frankia spp. for actinorhizal plants, thereby allowing the host plant to access atmospheric dinitrogen in nitrogen-poor soils.
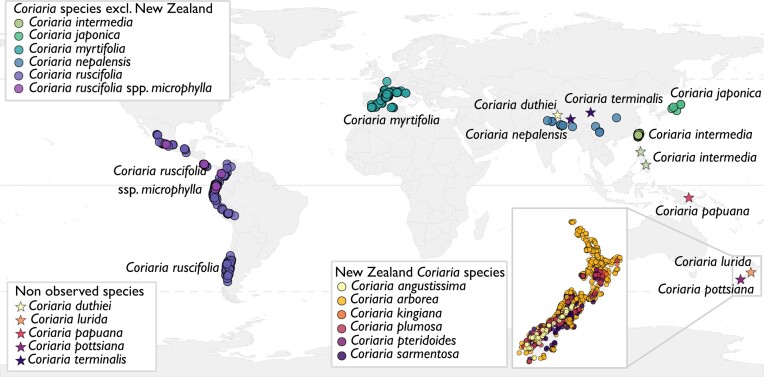
Figure 1.
Distribution of Coriariceae. Data were gathered from iNaturalist in June 2023, and only research grade level observations were used. Data from the observations are available in Table S1. The different coloured circles illustrate the different species, and stars indicate the species for which reliable observations were lacking. C. japonica is native to Japan, C. intermedia to Taiwan and the Philippines, C. myrtifolia is native to the Mediterranean, C. nepalensis is found in the foothills of the Himalayan and China, C. ruscifolia and C. ruscifolia spp. microphylla are found near the west coast of America. In New Zealand, C. arborea is the most common species, followed by C. plumosa. C. angustissima and C. sarmentosa are found only on the Southern Island, while C. kingiana and C. pteridoides are mostly observed on the Northern Island. Observations of C. duthiei (Nepal), C. terminalis (Nepal and southern China), C. papuana (Papua New Guinea), C. lurida, and C. pottsiana (New Zealand) were not recorded. Map was produced in R (RStudio Team 2022).
Frankia strains are members of the type genus of the Frankiaceae (Actinobacteria). Most of them have the capability of engaging in root nodule symbiosis with a polyphyletic group of so-called actinorhizal host plants that belong to the Fagales, Rosales, and Cucurbitales. Studies have shown that root nodule symbiosis evolved once, in the common ancestor of Fabales, Fagales, Cucurbitales and Rosales; the majority of the descending plant lineages since have lost their symbiotic capability (Griesmann et al. 2018, van Velzen et al. 2018, 2019, Libourel et al. 2023). van Velzen and colleagues et al. (2019) suggest that it is more plausible that Frankia rather than a rhizobium was the original symbiont. Frankia can be divided into four phylogenetically distinct clades, termed cluster-1 to cluster-4 (Normand et al. 1996, Sen et al. 2014, Nguyen et al. 2016). Cluster-1, -2, and -3 are symbiotic, while cluster-4 is not. The association of each cluster with its group of host plants has been discussed in several publications (Benson and Dawson 2007, Svistoonoff et al. 2014, Nguyen et al. 2016). Frankia cluster-2 is the earliest divergent symbiotic clade within the genus (Nguyen et al. 2016, 2019, Berckx et al. 2022a), and its host plants include Ceanothus (Rhamnaceae, Rosales), the Dryadoideae except for Dryas octopetala (Rosaceae, Rosales), Datiscaceae (Cucurbitales), and Coriariaceae (Cucurbitales).
Frankia cluster-2 strains are assumed to have a low saprotrophic potential and, to this date despite several unsuccessful and unpublished efforts, only two strains could be cultivated in vitro: Frankia coriariae BMG5.1 (Gtari et al. 2015, Nouioui et al. 2017) and F. coriariae BMG5.30 (Gueddou et al. 2019). Several more metagenome-assembled genomes (MAGs) of Frankia cluster-2 were sequenced from whole nodules obtained from both greenhouse cross-inoculation experiments and field samples. Here it was found that several closely related strains not only occupy a single nodule (Normand et al. 2017, Nguyen et al. 2019, Berckx et al. 2022a), but that some strains can persist as nodule-associated bacteria under unfavourable conditions and become the main symbiont in a more suitable host plant (Berckx et al. 2022a). Based on the analysis of the core genomes of Frankia cluster-2 strains from greenhouse samples, it could be concluded that the strains occurring in Eurasia, namely from France to Japan, form a single group with very low diversity (Nguyen et al. 2019). These strains were found to be the direct sister group of the Frankia cluster-2 strains occurring in North America, presumably by spreading via the Bering Strait (Nguyen et al. 2019). Further analysis of cluster-2 termed the Eurasian and North American strains the ‘continental’ lineage, to contrast with the ‘island’ lineage: those Frankia cluster-2 strains which occur on islands in the Pacific Ocean, specifically Japan, Taiwan, the Philippines, and Papua New Guinea (Nguyen et al. 2019, Berckx et al. 2022a). Our previous study highlighted the importance of sequencing field nodules: while studies using cross-inoculation studies or isolation of greenhouse nodules of D. glomerata and C. myrtifolia found continental Frankia cluster-2 strains in Japan (Gtari et al. 2015, Nguyen et al. 2019), sequencing field samples directly identified island lineage Frankia to be the main strains present in C. japonica nodules (Berckx et al. 2022a).
Initial studies of the Frankia cluster-2 phylogeny, using the marker genes glnA, dnaA and 16S rRNA, led to the conclusion that those Frankia strains associated with Coriariaceae in New Zealand form a sister group to other Frankia cluster-2 strains (Newcomb and Wood 1987, Benson et al. 1996, Nouioui et al. 2014, Nguyen et al. 2016). Interestingly, the study by Nouioui et al. (2014) found that Frankia cluster-2 strains in Mexico from C. ruscifolia subsp. microphylla were most closely related to Frankia strains from Nepal. However, it is important to point out that this study did not support a monophyletic position of C. ruscifolia in Mexico, and the Frankia phylogeny was built on only three phylogenetic marker genes. Several studies since have shown that phylogeny based on a low number of marker genes does not resolve the Frankia phylogeny as reliably as core genome or multi-locus sequence analysis does (Nguyen et al. 2016, 2019, Pozzi et al. 2018, Herrera-Belaroussi et al. 2020).
In light of the recent, whole genome analysis of Frankia cluster-2 (Berckx et al. 2022a), we were interested to see if Frankia strains occurring in New Zealand form a third lineage within cluster-2, or if they group with the strains making up the island lineage. Since Coriaria spp. in New Zealand are closely related to C. ruscifolia and C. ruscifolia subsp. microphylla in America (Yokoyama et al. 2000, Renner et al. 2020), we included nodule samples collected in Patagonia (Argentina) as well. We hypothesize that the Frankia cluster-2 strains occurring in New Zealand and Patagonia are more closely related to the island lineage than to the continental lineage, such as strains from Nepal. As New Zealand is the centre of diversity for Coriariaceae, it would be interesting to see if there is a high species diversity of Frankia cluster-2 here as well, indicating hosts and microsymbionts have co-evolved together over the last 25 million years. In addition, this would give more insights into the early evolution of root nodule symbiosis as a whole. Further, we were interested in analysing the whole bacterial community of Coriaria root nodules. Recent reports have been made of the isolation of non-nodulating bacteria isolated from actinorhizal nodules, such as Micormonospora (Trujillo et al. 2006, Carro et al. 2013, Ghodhbane-Gtari et al. 2021) or Streptomyces (Allen et al. 1966, Wollum II et al. 1966, Ghodhbane-Gtari et al. 2010, Berckx et al. 2022b). We wanted to investigate the microbiome of Frankia cluster-2 nodules and analyse whether differences could be found between sites and species.
Material and methods
Biological material
Nodules and leaves of Coriaria sp., as well as the soil around their root system, were collected along the Waimakariri riverbed near Christchurch (New Zealand), in February and March 2020. Nodules were gently washed in river water to remove excess soil. Nodules and leaves were immediately stored in pure ethanol at room temperature until arriving back in Stockholm where they were stored at −20°C until further analysis. The soil was collected in pouches and kept at 4°C. In the greenhouse, uninoculated seedlings of C. arborea, C. intermedia, C. myrtifolia, as well as Datisca glomerata were infected with the collected soil. Plants were checked for nodulation status before infection. Plants were watered with MilliQ water two times per week, and with ¼ strength Hoaglands without nitrogen one time per week (Hoagland and Arnon 1950). However, plants died six to eight weeks after infection. No nodules could be collected.
Nodules and leaves were collected from Coriaria ruscifolia, while the soil was collected from around its root system, near Lago Correntoso in Patagonia, Argentina, in March 2019. DNA isolation failed for freeze-dried nodules. The collected soil was used to infect two C. ruscifolia plants, originating from the Nymphenburg Botanical Garden (Munich, Germany), in a greenhouse at LMU Munich, in 2020. Nodules were collected sixteen weeks after infection and stored in 70% EtOH until arriving back in Stockholm where they were stored at −20°C until further analysis. In a second sampling trip, nodules and leaves of C. ruscifolia were collected from the same area in Patagonia, in October 2021. Nodules were collected in RNAlater (ThermoFisher Scientific) until arriving back in Stockholm where they were stored at −20°C until further analysis. Leaves were pressed and dried.
Vouchers of the Coriaria samples were deposited in the herbarium of the Swedish Museum of Natural History (Stockholm, Sweden), except for the field material of C. ruscifolia, which was deposited at the herbarium of Centro Regional Universitario Bariloche (San Carlos de Bariloche, Argentina). An overview of the collected specimens, geographic coordinates, and voucher numbers is given in Table S2A.
DNA isolation
Ethanol was removed from all plant parts using a speed-vac for six to eight hours, after which samples were shock-frozen and kept at −20°C until handling further. To remove RNAlater, samples were washed three times in sterile MilliQ water, patted dry to remove excess water, and immediately used for DNA isolation. Samples were ground in liquid nitrogen using a sterile mortar and pestle. DNA from leaves and nodules was extracted using the NucleoSpin Plant II kit (Macherey-Nagel, Germany). Polyclar AT (Serva, Germany) was added to the lysis buffer in all samples to remove excess polysaccharides. Ground nodules were sonicated in the lysis buffer as described by Nguyen et al. (2019). DNA of nodules was cleaned using the NucleoSpin gDNA Clean-up kit (Macherey-Nagel, Germany), and concentration was determined using Nanodrop (Thermo Scientific, Sweden) and Qubit (ThermoFisher, Sweden).
Frankia genome sequencing, assembly, phylogeny
Sequencing of the DNA from nodules was performed as previously described (Nguyen et al. 2019, Berckx et al. 2022a). The raw reads were assembled into Frankia metagenome-assembled genomes as described before (Berckx et al. 2022a). For the phylogenetic analysis and calculation of the average nucleotide identity (ANI), the EDGAR 3.0 platform was used (Blom et al. 2009, 2016, Dieckmann et al. 2021). The ANI is calculated between two genomes for consecutive 1020 nt fragments of the core genome, after which the average is taken. For the identification of nod gene operons, BLAST searches based on AA sequence were used in GenDB platform (Meyer et al. 2003). Sequences were aligned using ClustalO algoritm in SeaView 5.0 (Galtier et al. 1996, Gouy et al. 2021). Sequences were concatenated, and the phylogenetic tree was built using maximum likelihood phylogenies (PhyML) available through SeaView, with 100 iterations.
Coriaria species determination
The species determination of Coriaria was based on the trnL and ITS intergenic regions, as well as the maturase K (matK) sequence of the chloroplast as described by Renner et al. (2020), with an added mixture of trehalose/bovine serum albumin (BSA)/Tween-20 (TBT-PAR; Samarakoon et al. 2013) to enhance PCR efficiency. Primer sequences, amplified gene regions, and accession numbers of used references, are given in Table S2B. In brief, PCR conditions were as followed: 94°C for 3 min, 15 cycles of 94°C for 1 min 30 sec, 45°C for 2 min, 60°C for 3 min, 15 cycles of 94°C for 1 min 30 sec, 45°C for 2 min, 72°C for 3 min, and final elongation cycle of 72°C for 15 min. PCR products were digested using EcoRI and BamHI (Thermo Scientific, trnL and ITS) or KpnI and XhoI (Thermo Scientific, matK), and ligated pBluescript plasmids (Thermo Scientific) were used to transform One Shot™ TOP10 Chemically Competent E. coli cells (Invitrogen).
Analysis of the regions was performed using SeaView 5.0 (Galtier et al. 1996, Gouy et al. 2021), by aligning the obtained sequences to all reference sequences (Renner et al. 2020) using MUSCLE. Next, phylogenetic trees for individual regions and concatenated regions were built, using maximum likelihood phylogenies (PhyML) available through SeaView, with 100 iterations.
Microbiome analysis
The microbiomes were analysed as follows: 16S metabarcoding was performed on the same DNA extracts as presented here, as well as on previous extracts (Berckx et al. 2022a), by amplifying and sequencing the V3-V4 region. PCR amplification, sequencing and initial analysis were performed by Novogene. In brief, DNA quality was assessed by gel electrophoresis and photometrically using a NanoDrop ND-2000c UV–Vis spectrophotometer (NanoDrop Technologies). The V3-V4 region of the 16 s rRNA gene was amplified using barcoded versions of the universal primer set 341F (5′- CCT AYG GGR BGC ASC AG-3′) and 806R (5′-GGA CTA CNN GGG TAT CTA AT-3′). Amplicon quality was assessed by agarose gel electrophoresis before library preparation. Library quality was controlled using a Qubit 2.0 Fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2100 system. Libraries were sequenced on an Illumina NovaSeq 6000 platform employing 250 bp paired-end reads at Novogene Europe (Cambridge, UK). Demultiplexed reads delivered by Novogene were then trimmed using Trimmomatic version 0.38 (Bolger et al. 2014), accessed via the Galaxy portal (Jalili et al. 2020). Analysis of the reads was performed using DADA2 (Callahan et al. 2016), phyloseq (McMurdie and Holmes 2012), and metacoder packages in R. The number of initial, filtered, denoised, and non-chimaera reads are given in Table S3. The R script used is available upon request to the first author.
Results and discussion
Identification of the Coriaria species
The phylogenetic tree built on three marker genes (matK, trnL, and ITS) for the Coriaria samples are given in Fig. S1. Our samples of Coriaria ruscifolia, collected in Patagonia and obtained from the Nymphenburg Botanical Garden, are included in a clade that also comprises species of Coriaria from New Zealand. It should, however, be noted that the species delimitation of Coriaria ruscifolia appears unclear and may need further research. A previous phylogenetic study has indicated that the species is not monophyletic (Figs S3 and S4 of Renner et al. 2020). A sample of Coriaria ruscifolia included in that study was shown to be more closely related to the Mexican Coriaria ruscifolia subsp. microphylla, while two other accessions were found to be more closely related to different New Zealand (NZ) Coriaria species (Renner et al. 2020). Our analyses support this conclusion. This is interesting as due to the distance and the fact that Coriaria spp. seeds are spread by birds which do not migrate long distances (Burrows 1995), there should not have been exchange of genetic material between South American and NZ Coriaria species within the last 30 million years. Further investigation of the South American Coriaria speciation is needed, and beyond the scope of this study. As a consequence, we refer to the Patagonian sample as Coriaria ruscifolia, the Coriaria species in South America.
The New Zealand samples were unidentifiable at the growth stage, as no flowers or fruits were produced. At the time of collection, the Coriaria plants from Christchurch, New Zealand (NZ), were assumed to be Coriaria arborea or Coriaria sarmentosa, the two most likely species, based on distribution and occurrence. The phylogenetic analysis did not yield conclusive results (Fig. S1), and the species identities remain unclear.
Identification of novel Frankia cluster-2 strains
The Frankia metagenome-assembled genomes (MAGs) obtained in this study were named according to the nomenclature established before (Nguyen et al. 2019, Berckx et al. 2022a): [name of inoculum]_[initials of host plant from which DNA was isolated]_[“nod” for direct isolation of mixed plant and bacterial DNA from nodules, “vc” for isolation of DNA from vesicle clusters isolated from nodules]. The NZ samples and their associated inoculum were abbreviated to Cas, with continuous numbering. In total eight different sites were sampled, of which seven MAGs could be assembled (Tables 1a and 1b). Cas1_Cas_nod was found to be a chimeric MAG of at least two Frankia strains, which could not be separated bioinformatically, explaining the larger genome size (Table 1b). The Coriaria ruscifolia greenhouse MAG, originating from collected soil from Patagonia, was termed Cr1_Cr_nod, referring to C. ruscifolia, the only endemic Coriaria species. The MAGs Cr2_Cr_nod, Cr3_Cr_nod, Cr4_Cr_nod, and Cr5_Cr_nod originate from nodule field samples taken from root systems of C. ruscifolia growing in Patagonia.
Table 1a.
Overview of collected samples, the origin of inoculum, and accession number of bacterial MAGs. The genome names were based on the method established before (Nguyen et al. 2019, Berckx et al. 2022b). The coordinates; herbarium voucher information, and plant gene sequences for species delineation are presented in Table S2. The full accession numbers of the assembly and the contigs are presented in Table S4a. *This nodule sample was induced in a greenhouse using inoculum from Patagonian soil, whilst all other genomes came directly from field material.
| Inoculum | Host plant | Country of origin | Genome | Accession number |
|---|---|---|---|---|
| Cas1 | Coriaria sp. | New Zealand | Cas1_Cas_nod | GCA_963555895 |
| Cas2 | Coriaria sp. | New Zealand | Cas2_Cas_nod | GCA_963555855 |
| Cas3 | Coriaria sp. | New Zealand | Cas3_Cas_nod | GCA_963555955 |
| Cas4 | Coriaria sp. | New Zealand | Cas4_Cas_nod | GCA_963555915 |
| Cas5 | Coriaria sp. | New Zealand | Cas5_Cas_nod | GCA_963555945 |
| Cas7 | Coriaria sp. | New Zealand | Cas7_Cas_nod | GCA_963555905 |
| Cas8 | Coriaria sp. | New Zealand | Cas8_Cas_nod | GCA_963555845 |
| Cr1* | Coriaria ruscifolia | Argentina | Cr1_Cr_nod | GCA_963555865 |
| Cr2 | Coriaria ruscifolia | Argentina | Cr2_Cr_nod | GCA_963555885 |
| Cr3 | Coriaria ruscifolia | Argentina | Cr3_Cr_nod | GCA_963555925 |
| Cr4 | Coriaria ruscifolia | Argentina | Cr4_Cr_nod | GCA_963555875 |
| Cr5 | Coriaria ruscifolia | Argentina | Cr5_Cr_nod | GCA_963555935 |
Table 1b.
Overview of genome size and completeness, N50 values, coverage, and the number of reads from the genomes of collected samples. The complete BUSCO values for the genomes are given in Table S4B. *This nodule sample was induced in a greenhouse using inoculum from Patagonia, whilst all other genomes came directly from field material. **This MAG consists of two Frankia strains which cannot be separated.
| Inoculum | Genome | Genome size | BUSCO | N50 | Coverage | Reads |
|---|---|---|---|---|---|---|
| Cas1 | Cas1_Cas_nod | 9.9 Mbp** | 92% | 14560 | 21 | 694491 |
| Cas2 | Cas2_Cas_nod | 4.8 Mbp | 84% | 13339 | 22 | 352012 |
| Cas3 | Cas3_Cas_nod | 6.1 Mbp | 90% | 30570 | 45 | 915241 |
| Cas4 | Cas4_Cas_nod | 5.4 Mbp | 90% | 27728 | 31 | 558310 |
| Cas5 | Cas5_Cas_nod | 4.4 Mbp | 73% | 9506 | 11 | 161312 |
| Cas7 | Cas7_Cas_nod | 5.6 Mbp | 90% | 23190 | 41 | 779152 |
| Cas8 | Cas8_Cas_nod | 4.8 Mbp | 71% | 9560 | 10 | 163218 |
| Cr1* | Cr1_Cr_nod | 5.8 Mbp | 90.5% | 12065 | 18 | 348716 |
| Cr2 | Cr2_Cr_nod | 5.3 Mbp | 92.60% | 83965 | 137 | 2431428 |
| Cr3 | Cr3_Cr_nod | 5.6 Mbp | 94.60% | 80581 | 150 | 2766817 |
| Cr4 | Cr4_Cr_nod | 5.4 Mbp | 84.50% | 48191 | 62 | 1116218 |
| Cr5 | Cr5_Cr_nod | 5.2 Mbp | 71.60% | 28167 | 52 | 866514 |
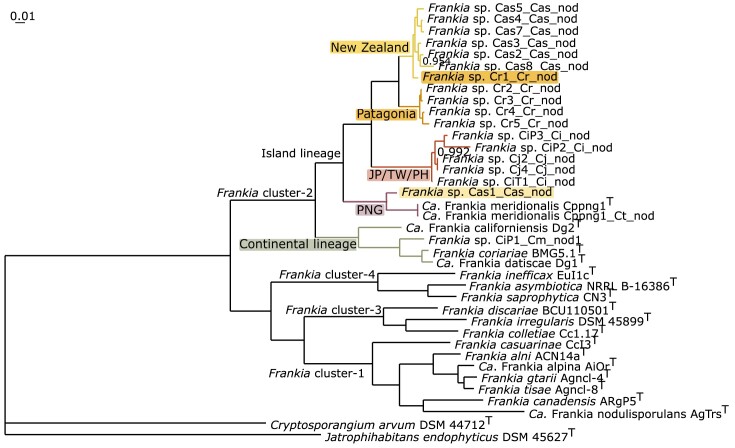
A core genome tree was constructed for the Frankia cluster-2 sequences available thus far (Fig. 2). Previous work has shown that cluster-2 can be split into two groups: the continental lineage and the island lineage (Berckx et al. 2022a). We found that the Frankia strains in this study also belonged to the island lineage, forming a direct sister group of those Frankia strains found in the Philippines, Taiwan, and Japan (Fig. 2). The novel MAGs were grouped into two sister groups based on their geographic origin, except for Cr1_Cr_nod (highlighted in Fig. 2 in orange) which grouped with the NZ MAGs, and Cas1_Cas_nod (highlighted in Fig. 2 in yellow) which grouped with Candidatus Frankia meridionalis Cppng1, which originates from Papua New Guinea (Nguyen et al. 2019).
Figure 2.
Core genome tree of Frankia cluster-2. Tree for 38 genomes, built out of a core of 359 genes per genome, 13642 in total. The core gene sets were aligned using MUSCLE (Edgar 2004) and subsequently concatenated, resuling in an alignment of 150104 AA residues per genome, 5703952 in total. The tree was calculated using FastTree 2 inference of maximum-likelihood phylogeny (Price et al. 2010), available through the EDGAR 3.0 platform (Dieckmann et al. 2021). The scale represents AA substitutions, and the branch support values were computed using the Shimodaira-Hasegawa test (Shimodaira and Hasegawa 1999). As the vast majority of the branches shows a perfect support value of 1.00, only lower values are presented in the tree. The different groups within cluster-2 are annotated in different colours and with text. Abbreviations: Ca.: Candidatus; JP/TW/PH: Japan/Taiwan/the Philippines; PNG: Papua New Guinea. The Frankia MAGs from New Zealand are depicted in yellow. Frankia sp. Cr1_Cr_nod, the greenhouse sample going back to Patagonia, is highlighted in orange and was found to group with the New Zealand MAGs. The MAGs from Patagonia are depicted in orange. The Frankia MAGs from the JP/TW/PH (Berckx et al. 2022b) are depicted in red. Candidatus Frankia meridionalis Cppng1T and Ca. F. meridionalis Cppng1_Ct_nod, from PNG, are illustrated in purple (Nguyen et al. 2019). The genome of Frankia sp. Cas1_Cas_nod is highlighted in yellow and was found to group with Ca. F. meridionalis. All these genomes comprise the island lineage. The continental lineage is given in green, represented by Ca. Frankia californiensis Dg2T (Nguyen et al. 2016), Frankia sp. CiP1_Cm_nod1 (Berckx et al. 2022b), Frankia coriariae BMG5.1T (Gtari et al. 2015), and Ca. Frankia datiscae Dg1T (Persson et al. 2015). Frankia cluster-1, cluster-3, and cluster-4 are illustrated in black. Cluster-4 is represented by Frankia inefficax Eul1cT (Nouioui et al. 2017b), Frankia asymbiotica NRRL B-16386T (Nouioui et al. 2017c), and Frankia saprophytica CN3T (Nouioui et al. 2018a). Cluster-3 is represented by Frankia discariae BCU110501T (Nouioui et al. 2017a), Frankia irregularis DSM 45899T (Nouioui et al. 2018b), and Frankia colletiae Cc1.17T (Nouioui et al. 2023a). Cluster-1 is represented by Frankia casuarinae CcI3T (Nouioui et al. 2016), Frankia alni ACN14aT (Nouioui et al. 2016), Ca. Frankia alpina AiOrT (Pozzi et al. 2020), Frankia gtarii Agncl-4T (Nouioui et al. 2023b), Frankia tisai Agncl-8T (Nouioui et al. 2023b), Frankia canadensis ArgP5T (Normand et al. 2018), and Ca. Frankia nodulisporulans AgTrsT (Herrera-Belaroussi et al. 2020). The tree is rooted by Cryptosporangium arvum DSM 44712T (Tamura et al. 1998) and Jatrophihabitans endophyticus DSM 45627T (Madhaiyan et al. 2013).
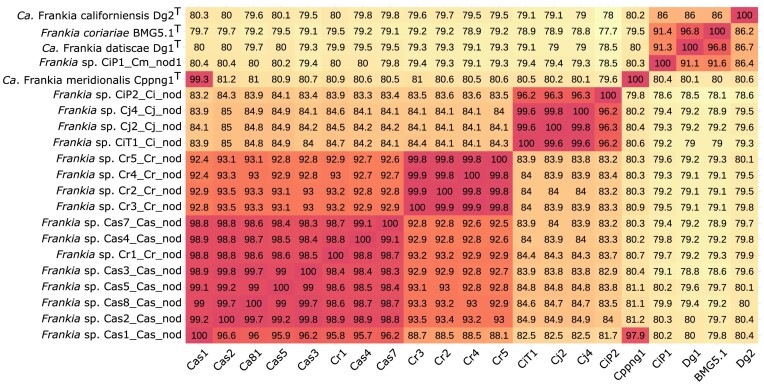
We were interested to see which species the Frankia MAGs we sequenced represented, or if they belonged to a novel species. The average nucleotide identity (ANI) was calculated for each MAG of this study compared to other cluster-2 species belonging to both the island and the continental lineage (Fig. 3 for median values; Fig. S2 for mean values). Using the accepted 95–96% threshold to separate species (Goris et al. 2007, Richter and Rosselló-Móra 2009), we found that the samples from NZ and Patagonia, respectively, represent two novel Frankia species.
Figure 3.
Median average nucleotide identity (ANI) of core genomes of Frankia cluster-2. The query-genomes are on the y-axis. Names of genomes along the x-axis have been reduced. The analysis includes the novel Frankia MAGs of this study and those representatives of all Frankia cluster-2 species published previously. Seven different groups with high ANI values (95% or more) can be distinguished. Going from left to right: the first group includes novel MAGs of New Zealand (Cas1_Cas_nod, Cas2_Cas_nod, Cas8_Cas_nod, Cas5_Cas_nod, Cas3_Cas_nod, Cas4_Cas_nod, and Cas7_Cas_nod), as well as the greenhouse nodules originating from Patagonia soil (Cr1_Cr_nod). The second group includes MAGs from all Patagonian field samples (Cr3_Cr_nod, Cr2_Cr_nod, Cr4_Cr_nod, and Cr5_Cr_nod). The third group includes MAGs from Taiwan (CiT_Ct_nod), Japan (Cj2_Cj_nod and Cj4_Cj_nod) and the Philippines (CiP2_Ci_nod) (Berckx et al. 2022b). The MAG Cas1_Cas_nod from New Zealand was found to share high ANI with MAGs of Candidatus Frankia meridionalis Cppng1 (Nguyen et al. 2019). The fifth group includes the novel species of the continental lineage, namely CiP1_Cm_nod1 (Berckx et al. 2022b). The sixth group is formed by Candidatus Frankia datiscae Dg1 (Persson et al. 2011, 2015) and Frankia coriariae BMG5.1 (Gtari et al. 2015). The seventh and last group is represented by Candidatus Frankia californiensis Dg2 (Nguyen et al. 2016). ANI calculations were made using EDGAR 3.0 platform (Dieckmann et al. 2021). See Fig. S2 for mean values.
The Frankia MAGs of all NZ samples (Cas1_Cas_nod to Cas8_Cas_nod) share between 98.3% and 99.7% ANI, indicating they belong to the same species, different from the other Frankia cluster-2 MAGs included in the analysis. Cr1_Cr_nod, the MAG of the Patagonian soil sample which had been used to inoculate C. ruscifolia in the greenhouse, was found to belong to the same species as the NZ Frankia strains (95.8%-98.9% ANI, Fig. 3). Interestingly, the MAG of Cas1_Cas_nod additionally shared high similarity to Ca. F. meridionalis Cppng1 (97.9% ANI, Fig. 3). The ANI values of Cas1_Cas_nod compared to the other NZ MAGs showed a non-reciprocal pattern: while the other genomes shared between 98.8% and 99.2% ANI with Cas1_Cas_nod, Cas1_Cas_nod only shared between 95.2% and 96.6% ANI with the other genomes. This Frankia MAG has a much larger size than the others (Table 1b). It has been reported before that more than one closely related genome cannot be separated by bioinformatics means, leading to a mixed assembly (Nguyen et al. 2016, 2019). We thus conclude that Cas1_Cas_nod contains two genomes: one of which belongs to the new species from NZ, while the other is a representative of Ca. F. meridionalis. The position of this genome in the phylogenetic tree can be attributed to the fact that it represents a mix of two species (Fig. 2).
The Frankia MAGs of field samples from Patagonia (Cr2_Cr_nod to Cr5_Cr_nod) belong to a different, novel, Frankia species based on their shared high ANI values (99.8%-99.9%). Both the novel NZ and the novel Patagonian Frankia species shared higher native to values with other Frankia MAGs from the island lineage, than with Frankia MAGs from the continental lineage.
Dispersal of Frankia cluster-2 in the southern hemisphere
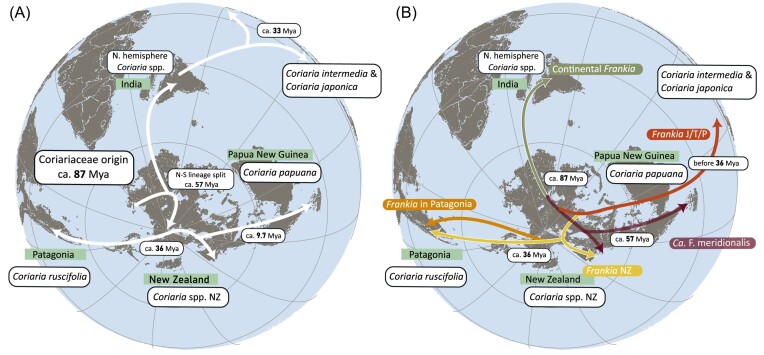
Combining the new genome data from Frankia with novel host plant biogeographic analysis, we can draw some conclusions on the dispersal of Frankia cluster-2 in the southern hemisphere (Fig. 4). Renner et al. (2020) concluded that Coriaria spp. in the southern hemisphere split between ca. 57 and 46 Mya from those in the northern hemisphere, and Coriariaceae arrived in America from Antarctica (Fig. 4A), rather than migrating over the Pacific Ocean, as proposed earlier (Yokoyama et al. 2000). The divergence of C. ruscifolia and C. ruscifolia subsp. microphylla from the other southern hemisphere Coriaria species, and the arrival to America, is estimated to have occurred before 36 Mya (Fig. 4A) (Renner et al. 2020). We found a Frankia species able to engage in symbiosis with both Patagonian C. ruscifolia (Cr1_Cr_nod) and NZ Coriaria spp. (Cas1_Cas_nod to Cas8_Cas_nod) (Fig. 2, illustrated in yellow). This species could have diverged from the other Frankia species and dispersed during the Eocene, post-Gondwana when South America and NZ were still loosely connected by Antarctica. It is currently not known how Frankia disperses, but several lines of evidence suggest through animals faeces (Burleigh and Dawson 1995, Chaia et al. 2012, Paschke and Dawson 1993). This implies the two novel Frankia species identified here, must have arrived in South America before the opening of the Drake passage, which is in agreement with the arrival of C. ruscifolia in South America at 36 Mya (Reguero et al. 2014, Renner et al. 2020).
Figure 4.
Paleogeographic reconstruction of (A) Coriariaceae and (B) Frankia cluster-2 dispersal patterns. Reconstruction of the continents is based on PALEOMAP available in GPlates 2.3 (Müller et al. 2018). (A) Coriariaceae dispersal is illustrated with white arrows. Dates are based on the most recent phylogeny (Renner et al. 2020). (B) Dispersal and speciation patterns of Frankia cluster-2. Based on the phylogeny of Frankia species and their associated host plants, we assume the continental Frankia species diverged from the island lineage already 87 Mya. The continental lineage of Frankia (green) would have dispersed together with the northern hemisphere Coriariae and Datiscaceae in India, and reached continental Eurasia via this route. In the island lineage, Ca. F. meridionalis is illustrated in purple; it is found in Papua New Guinea and New Zealand. Frankia found in Japan, Taiwan, and the Philippines (JTP) is illustrated in red. The New Zealand Frankia species is illustrated in yellow and can also be found in Patagonia. The Patagonian Frankia species is illustrated in orange.
This would imply a scenario where the host plants and Frankia species evolved together. However, this scenario does not hold up based upon closer investigation of all Frankia cluster-2 species and Coriaria spp. In NZ, we were able to identify two different Frankia species: Ca. F. meridionalis, and a novel Frankia species (Fig. 2, yellow). Ca. F. meridionalis was first identified from samples originating in Papua New Guinea (Nguyen et al. 2019). C. arborea and C. papuana, the Coriaria species endemic to Papua New Guinea, are closely related species which diverged in the late Miocene (9 to 8 Mya). Therefore, one could assume that their associated Frankia evolved together with their host plant, i.e. Ca. F. meridionalis and the novel Frankia species are direct sister groups. However, this is not the case: Ca. F. meridionalis is the earliest divergent Frankia species within the island lineage, whereas the novel NZ Frankia species diverged from the Patagonian species relatively recently.
However, as argued above, both Frankia and its host plants arrived in Patagonia ca. 36 Mya. The divergence of the two Frankia species from the other species must have occurred before this arrival. As a consequence, the Frankia species occurring in Japan/Taiwan/the Phillippines as well as Ca. F. meridionalis have to have separated more than 36 Mya. This is in agreement with our previous work, where we showed that the species in Japan/Taiwan/the Philippines dispersed from the south to the north by an unknown host plant (Fig. 4B), which has either gone extinct or lost its symbiotic capability (Berckx et al. 2022a). Upon the arrival of C. japonica and C. intermedia on these islands, this Frankia species was already present. C. japonica and C. intermedia have been dated to have separated from the other northern hemisphere Coriaria spp. at least 33 Mya.
C. terminalis has also been shown to be nodulated by Ca. F. meridionalis (Berckx et al. 2022a). C. terminalis is endemic to Nepal, and belongs to the northern hemisphere Coriaria clade, diverging from the southern clade 57–46 Mya. The most parsimonious conclusion therefore would be that Ca. F. meridionalis speciated before this time. To our knowledge, members of the island lineage are unable to nodulate Datiscaceae, whereas the members of the continental lineage can. We were unable to induce nodules on Datiscaceae by Ca. F. meridionalis (Nguyen et al. 2019), or the Frankia species from the Philippines (Berckx et al. 2022a). We were also unable to induce nodules on Datiscaceae using the soil collected in New Zealand. However, the plants died six to eight weeks post-infection, and thus we cannot exclude other factors impacting nodulation. This implies the divergence of the island lineage from the continental lineage Frankia species around the same time as the divergence of Coriariaceae from the Corynocarpaceae, ca. 86–82 Mya.
In summary, the different Frankia cluster-2 species of the island diverged between 82 and 36 Mya. Ca. F. meridionalis then split off from the other island lineage Frankia between 87 and 57 mya. We cannot determine when the next divergence of species occurred, but the most recent divergence, i.e. the Patagonian and NZ species from the Japanese/Taiwanese/Philippine species occurred more than 36 Mya. The NZ and Patagonian Frankia species are more closely related to each other, and belong to the island lineage, instead of the continental lineage. This contradicts previously published data based on the use of single phylogenetic marker genes (Nouioui et al. 2014).
Our data did not allow us to test our second hypothesis, whether NZ is a hotspot of species diversity for Frankia as well. Further investigation is needed to sequence more root nodules collected from other Coriaria spp. in other locations in New Zealand. Similarly, more sampling of soil C. ruscifolia in Patagonia and other parts of South and Central America is needed to assess the occurrence of the different Frankia species. As mentioned above, the monophyletic position of C. ruscifolia is unclear, and therefore Frankia associating with C. ruscifolia and C. ruscifolia subsp. microphylla might belong to a species different from the Frankia we identified in this study.
Frankia strains compete for host inoculation and can persist under unfavourable conditions
The genome of Cr1_Cr_nod belongs to the same species as the NZ Frankia strains (Fig. 3). This species was not identified in any of the four field nodules sequenced directly from Patagonia. It was only found in the nodules induced in the greenhouse by soil collected under C. ruscifolia in Patagonia. While the directly sequenced nodules and the soil originate from two different sampling trips, cross contamination in our greenhouse can be excluded. It can be expected that in the soil, both species are present. Our data indicates that some competition between the different Frankia species takes place, and our greenhouse conditions promoted nodulation by the NZ Frankia species on C. ruscifolia. Under field conditions, which could include many different environmental factors such as climate or pH of the soil, the Patagonian Frankia species seemed to induce nodules on C. ruscifolia. In our nodule samples, the NZ species was either present on the nodule and below the detection limit, or it was not present in or on these particular nodules altogether.
Similar phenomena have been reported before: after sequencing field nodules of C. intermedia from the Philippines, we assembled island lineage Frankia MAGs. Using the same nodules in the greenhouse on C. myrtifolia, we could only obtain continental Frankia MAGs, namely CiP1_Cm_nod1 and CiP1_Cm_nod2 (Berckx et al. 2022a). In the same study, we could assemble Frankia MAGs from the island lineage from field nodules of C. japonica (Berckx et al. 2022a). It had previously been assumed that only Frankia species of the continental lineage were present in Japan, as inoculation of Datisca glomerata (Nguyen et al. 2019) or of C. myrtifolia followed by isolation (Gtari et al. 2015) with nodules from Japan, led to nodules containing Frankia from the continental lineage. Another notable example is cluster-3 Frankia irregularis. This species was first isolated from Casuarina spp. nodules (Casuarinaceae, Fagales), which are cluster-1 host plants (Mansour et al. 2017, Nouioui et al. 2018, Pujic et al. 2015). However, upon investigation, F. irregularis was unable to re-infect Casuarina spp. but inducing nodules on Rhamnaceae (Rosales), and on Gymnostoma spp., which are cluster-3 host plants. A later study showed that F. irregularis was present on the surface of nodules of Casuarina spp. (Vemulapally et al. 2019). Lastly, non-symbiotic cluster-4 strains have been isolated from nodules of actinorhizal Cucurbitales but could not nodulate them (Hafeez 1983, Hameed et al. 1994, Mirza et al. 1994). In conclusion, Frankia strains, including those of cluster-2, can persist under non-symbiotic conditions in the soil or on the surface of actinorhizal nodules, and the symbiotic strains among them can maintain their ability to nodulate once optimal conditions occur.
Nod genes are not required for the establishment of symbiosis in Coriariaceae
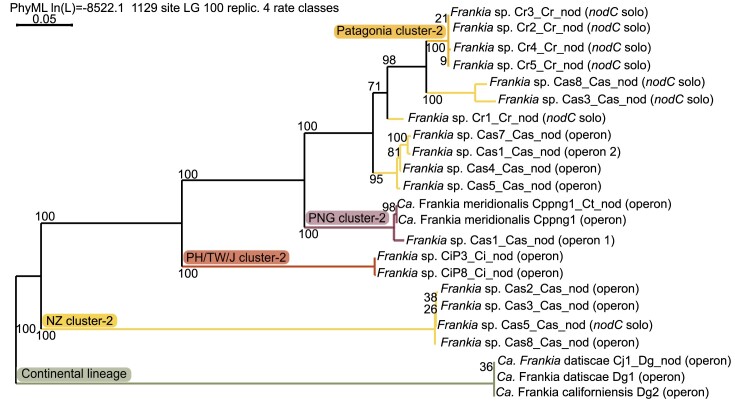
Unlike rhizobial symbiosis, actinorhizal symbiosis is not established through lipochito-oligosaccharide (LCO) Nod factor signalling as most Frankia genomes do not contain the canonical nodABC genes, encoding the proteins responsible for the Nod factor backbone production. Candidatus Frankia datiscae Dg1 was the first Frankia in which the canonical nodABC genes could be identified (Persson et al. 2011). These genes have been reported in most Frankia cluster-2 genomes of the continental lineage, and the North American strains additionally contain the sulfotransferase gene nodH (Nguyen et al. 2016, 2019). However, they have not been identified in strains infecting only Coriariaceae such as F. coriariae (Gtari et al. 2015, Gueddou et al. 2019), the new species of the continental lineage that was part of the Coriaria intermedia inoculum (CiP1_Cm_nod1, Berckx et al. 2022a), or members of the island lineage. Instead, Ca. F. meridionalis Cppng1 had been reported to contain a nodB'C operon, as well as a carbamoyl transferase gene nodU, but not a nodBA operon (Nguyen et al. 2019). In most other members of the island lineage, no nod genes could be identified (Berckx et al. 2022a). We, therefore, wanted to investigate the potential presence of nod genes in the new genomes (Table S4C). While the Frankia MAGs from NZ all contained the nodC-nltIJ-nodU operon, identical to the one found in Ca. F. meridionalis, we only able to identify nodC in the Patagonian MAGs (Fig. 5, Table S4C, Fig. S3).
Figure 5.
Phylogenetic tree of nodC-nltIJ operon. Sequences were obtained using searches for the operon and BLAST using GenDB. Concatenation of nodC, nltI, and nltJ, alignment and maximum likelihood phylogenetic tree construction based on 100 bootstrap replicates, were performed in SeaView. The branches were colored as followed: the new Frankia species from Patagonia is given in orange, the new species from NZ is given in yellow, the species from Japan/Taiwan/Philippines is given in red, Candidatus Frankia meridionalis is given in purple, and the continental lineage is given in green.
While the separation between the island and the continental lineage is clear, the nodC (Fig. S3) and the nodC-nltIJ operon phylogenies (Fig. 5) do not follow the core gene phylogeny. At some point, a gene duplication event of nodC took place in the island lineage, before the split of the Patagonian and the NZ species. With the limited data available, it is unclear whether this duplication event happened earlier and the Patagonian Frankia species contained two nodC copies at some point. It also cannot be determined if only the nodC gene was duplicated, or the entire operon, followed by sequential loss of the other genes. In the Patagonian MAGs, only one nodC copy can be found and this copy is not part of an operon including nltIJ. Both copies are present in the MAGs of Cas3_Cas_nod, Cas5_Cas_nod and Cas8_Cas_nod. However, while in Cas3_Cas_nod and in Cas8_Cas_nod the single nodC groups together with the nodC of the Patagonian species, the single nodC of Cas5_Cas_nod groups together with the nodC of other NZ MAGs in which nodC is present as part of an operon. The Cas5_Cas_nod nodC-nltIJ operon groups together with the operons of the other NZ MAGs.
As illustrated above (Fig. 2), Ca. F. meridionalis diverged first within the island lineage. The nodC-nltIJ operon of the other Frankia MAGs of the island lineage, do not follow the core phylogeny. The Frankia cluster-2 nod genes are surrounded by many transposons (Nguyen et al. 2019), and the neighbourhood of the nod genes in the MAGs of this study also contain many transposons. These transposons allow a different selection pressure, evolution, and horizontal gene transfer, compared to the rest of the genome. In addition, they enable the loss of many genes within the nod operon, i.e. nodA, nodB, nodU, nltI, nltJ, in the different species. Based on the limited data on genes which were maintained, it is unclear if the nod genes were acquired by the ancestor of symbiotic Frankia, or if multiple horizontal gene transfer events took place later.
We hypothesize that the Nod factors, i.e. the LCOs produced by the Nod proteins, play a potential role in suppressing plant immunity during nodulation, a role which has been attributed to LCOs in general (Feng et al. 2019). We previously reported that Frankia cluster-2 strains lacking in nod genes are unable to nodulate Datiscaceae (Berckx et al. 2022a), which do not occur in the southern hemisphere. If the infection of Coriariaceae does not require LCO suppression of the plant immune system, there would be no need for Frankia to retain the nod genes. Interestingly, the MAG of the continental Cm1_Cm_nod, originating from Coriaria myrtifolia (Nguyen et al. 2019), retained a nodB1A and a nodB2CnltIJ operon and was able to infect both Datiscaceae and C. myrtifolia, similar to Ca. F. datiscae Dg1 which contains both operons and was able to infect both Datiscaceae and C. nepalensis (Nguyen et al. 2019). It does not seem that the absence of nod genes is a requirement for nodulating Coriariaceae, but rather that the presence of nodA, nodB, and nodC is required for nodulating Datiscaceae. Further analysis, including cross-inoculation studies with other cluster-2 host plants such as Ceanothus spp., and with the novel Frankia species described here are needed to understand the role of nod genes, nodulation, and host-specificity in Frankia cluster-2.
Coriaria root nodules encompass more than Frankia
As outlined above, Frankia strains can be part of the inocula while not inducing nodules. Actinorhizal nodules are known to harbour a more diverse microbiome than only Frankia strains. Micromonospora spp. have been suggested to commonly occupy actinorhizal, as well as leguminous, nodules (Trujillo et al. 2006, 2010, Carro et al. 2013). We previously isolated and identified a novel Streptomyces spp. from C. intermedia, originating in the Philippines (Berckx et al. 2022b). Therefore, we set out to investigate the microbiome composition of the Coriaria field nodules sequenced in this study. In addition, field samples from Coriaria japonica were also included. This species is native to Japan, and its nodules were previously sampled and analysed (Berckx et al. 2022a). Due to the limitation in DNA extracts available, no other samples were included.
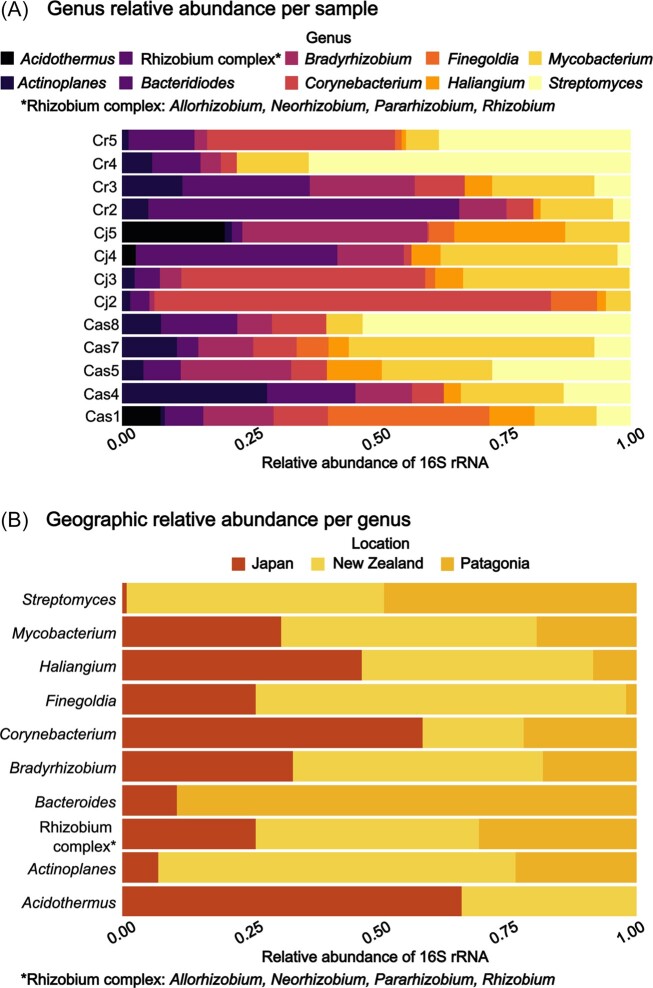
Analysing 16S rRNA gene profiles, we found that Actinobacteria, Cyanobacteria, and Proteobacteria were the three most dominant phyla in the samples (Fig. S2A). A large proportion of the Cyanobacteria reads are assumed to originate from the host plant plastids, while Frankia would account for the majority of Actinobacteria reads. Excluding Cyanobacteria, we indeed found that Frankia dominates the 16S profiles (Fig. S2B). To identify which other bacteria were present, a threshold of 2% was set to identify the nodule bacterial community, excluding Frankia and Cyanobacteria. The top 10 most abundant genera are presented in Fig. 6 (Fig. S3C for top 30).
Figure 6.
Top 10 most abundant bacterial genera identified in Coriariaceae field samples based on V3-V4 region of the 16S rRNA. The sample names refer to from which the corresponding Frankia MAG was assembled. Cr5, Cr4, Cr3, Cr2: nodules from Coriaria ruscifolia growing in Patagonia, Cj5, Cj4, Cj3, Cj2: nodules from Coriaria japonica growing in Japan, Cas8, Cas7, Cas5, Cas4, Cas1: Coriaria sp. growing in New Zealand. A) The relative abundance of each genus in each sample. Rhizobium complex refers to Allorhizobium, Neorhizobium, Pararhizobium, and Rhizobium, which were assigned to the same taxonomic group. B) The relative geographic distribution of each genus is illustrated for samples coming from Japan (red), New Zealand (yellow) and Patagonia (orange).
Our data indicate that the most abundant bacteria, overall, were rhizobia (Bradyrhizobium and Rhizobium complex, consisting of Allorhizobium, Neorhizobium, Pararhizobium, and Rhizobium i), Corynebacterium, Mycobacterium, and Streptomyces (Fig. 6A). Some differences could be seen between the samples originating from the same geographical region and plant species (Fig. 6B). Streptomyces was predominantly present in samples from New Zealand (yellow) or Patagonia (orange), but almost lacking in samples from Japan (red). Similarly, Finegoldia and Acidothermus were found to be much less abundant to lacking in Patagonian samples, compared to Japanese or New Zealand ones. Such differences could be explained by many variables, such as the host plant, soil and climate conditions, or adjacent plants. From the sequencing data alone, it also cannot be determined whether the bacteria are present inside the nodules or attached to the surface and if they are metabolically active. Further research into the roles of different nodule-occupying bacteria is needed, as well as determining which factors play an important role in determining the composition. However, our results do show that some genera, such as Mycobacterium, Haliangium, Corynebacterium, rhizobia, and Actinoplanes, are commonly present on actinorhizal nodules of Coriariaceae, regardless of the specific conditions.
Conclusion
Our study has led to the identification of two novel Frankia cluster-2 species, associated with Coriariaceae in the southern hemisphere. Phylogenetic analysis of endosymbiont and host plant, together with the geographic distribution over a geological time scale from 87 Mya, indicate that the different Frankia species diverged early in evolution and independently of host plant speciation. Although speciation of the two novel species occurred at the latest 36 Mya, little genetic changes took place. During the dispersal towards Patagonia, strains of the two species lost nodCnltIJnodU operon independently. Microbial communities associated with Coriaria spp. nodules are dominated by Mycobacterium, Haliangium, Corynebacterium, rhizobia, and Actinoplanes, regardless of specific climate or soil conditions. Our study was only able to analyse Frankia strains associated with a small number of the New Zealand Coriaria species. Further research is needed to identify if more Frankia spp. evolved together with Coriaria. in New Zealand, as well as identifying the exact factors which determine the recruitment and role of the nodule microbiome.
Supplementary Material
Acknowledgement
This work was achieved through the help of many people involved. We would like to thank Greg Stanley (Regional Council, Canterbury, New Zealand) and Eugenia Chaia (CONICET/Centro Regional Universitario Bariloche, Bariloche, Argentina) for helping with the collection of field samples. We also want to thank Ellen Fasth (Umeå University) for the initial investigation of the root nodule microbiomes, and Ayco Tack and Rachel Foster (Stockholm University) for the sharing of laboratory equipment. At LMU Munich, we would like to thank Susanne S. Renner for the donation of Coriaria ruscifolia plants, Dagmar Hann for taking care of the plants, and Xiaoyun Gong for collecting nodules from these plants.
Contributor Information
Fede Berckx, Department of Ecology, Environment and Plant Science, Stockholm University, 106 91 Stockholm, Sweden; Department of Crop Production Ecology, Swedish University of Agricultural Sciences, 756 51 Uppsala, Sweden.
Daniel Wibberg, CeBiTec, Bielefeld University, 33615 Bielefeld, Germany.
Andreas Brachmann, LMU München, Faculty of Biology, Genetics, 82152 Planegg-Martinsried, Germany.
Ciara Morrison, Department of Ecology, Environment and Plant Science, Stockholm University, 106 91 Stockholm, Sweden.
Nadia B Obaid, Department of Ecology, Environment and Plant Science, Stockholm University, 106 91 Stockholm, Sweden.
Jochen Blom, Bioinformatics and Systems Biology, Justus-Liebig University Giessen, 35392 Giessen, Germany.
Jörn Kalinowski, CeBiTec, Bielefeld University, 33615 Bielefeld, Germany.
Luis G Wall, CONICET, National Council for Scientific and Technical Research, Argentina; Department of Science and Technology, National University of Quilmes, B12876BXD Bernal, Argentina.
Katharina Pawlowski, Department of Ecology, Environment and Plant Science, Stockholm University, 106 91 Stockholm, Sweden.
Author contributions
Fede Berckx (Conceptualization, Formal analysis, Funding acquisition, Investigation, Visualization, Writing – original draft, Writing – review & editing), Daniel Wibberg (Data curation, Formal analysis, Investigation, Writing – review & editing), Andreas Brachmann (Formal analysis, Investigation, Writing – review & editing), Ciara Morrison (Investigation, Writing – review & editing), Nadia B. Obaid (Investigation, Supervision, Writing – review & editing), Jochen Blom (Resources, Software, Writing – review & editing), Jörn Kalinowski (Supervision, Writing – review & editing), Luis G. Wall (Resources, Writing – review & editing), and Katharina Pawlowski (Conceptualization, Funding acquisition, Supervision, Writing – review & editing)
Conflict of interest
None declared.
Funding
This project was funded by a grant from the Lars Hiertas Minne Foundation (FO2019-0515) to F.B. and by two grants from the Swedish Research Council Vetenskapsrådet (VR 2012-03061 and 2019-05540) to K.P. The bioinformatics support of the BMBF-funded project “Bielefeld-Gießen Center for Microbial Bioinformatics” (BiGi) and the BMBF grant FKZ 031A533 within the German Network for Bioinformatics Infrastructure (de.NBI) are gratefully acknowledged. The Galaxy server that was used for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR (de.NBI)).
References
- Allen JD, Silvester WB, Kalin M. Streptomyces associated with root nodules of Coriaria in New Zealand. N Z J Bot. 1966;4:57–65. 10.1080/0028825X.1966.10443953. [DOI] [Google Scholar]
- Becking JH. Dinitrogen-fixing associations in higher plants other than legumes. A Treatise on Dinitrogen Fixation, Sec. III: biology. New York: John Wiley, 1977, 185–275. [Google Scholar]
- Benson DR, Dawson JO. Recent advances in the biogeography and genecology of symbiotic Frankia and its host plants. Physiol Plant. 2007;130:318–30. 10.1111/j.1399-3054.2007.00934.x. [DOI] [Google Scholar]
- Benson DR, Stephens DW, Clawson ML et al. Amplification of 16S rRNA genes from Frankia strains in root nodules of Ceanothus griseus, Coriaria arborea, Coriaria plumosa, Discaria toumatou, and Purshia tridentata. Appl Environ Microbiol. 1996;62:2904–9. 10.1128/aem.62.8.2904-2909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckx F, Nguyen TV, Bandong CM et al. A tale of two lineages: how the strains of the earliest divergent symbiotic Frankia clade spread over the world. BMC Genomics. 2022a;23:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckx F, Bandong CM, Wibberg D et al. Streptomyces coriariae sp. nov., a novel streptomycete isolated from actinorhizal nodules of Coriaria intermedia. Int J Syst Evol Microbiol. 2022b;72:005603. 10.1099/ijsem.0.005603. [DOI] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D et al. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinf. 2009;10:154. 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Kreis J, Spänig S et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:W22–28. 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Dawson JO. Spores of Frankia strain HFPCcl3 nodulate Casuarina equisetifolia after passage through the digestive tracts of captive parakeets (Melopsittacus undulatus). Can J Bot. 1995;73:1527–30. 10.1139/b95-165. [DOI] [Google Scholar]
- Burrows CJ. Germination behaviour of the seeds of four New Zealand species of Coriaria (Coriariaceae). N Z J Bot. 1995;33:265–75. 10.1080/0028825X.1995.10410489. [DOI] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro L, Pujic P, Trujillo ME et al. Micromonospora is a normal occupant of actinorhizal nodules. J Biosci. 2013;38:685–93. 10.1007/s12038-013-9359-y. [DOI] [PubMed] [Google Scholar]
- Chaia EE, Sosa MC, Raffaele E. Vertebrate faeces as sources of nodulating Frankia in Patagonia. Symbiosis. 2012;56:139–45. 10.1007/s13199-012-0169-z. [DOI] [Google Scholar]
- Dieckmann MA, Beyvers S, Nkouamedjo-Fankep RC et al. EDGAR3.0: comparative genomics and phylogenomics on a scalable infrastructure. Nucleic Acids Res. 2021;49:W185–92. 10.1093/nar/gkab341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Sun J, Radhakrishnan GV et al. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun. 2019;10:5047. 10.1038/s41467-019-12999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics. 1996;12:543–8. [DOI] [PubMed] [Google Scholar]
- Ghodhbane-Gtari F, D'Angelo T, Gueddou A et al. Alone yet not alone: frankia lives under the same roof with other bacteria in actinorhizal nodules. Front Microbiol. 2021;12:749760. 10.3389/fmicb.2021.749760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodhbane-Gtari F, Essoussi I, Chattaoui M et al. Isolation and characterization of non-Frankia actinobacteria from root nodules of Alnus glutinosa, Casuarina glauca and Elaeagnus angustifolia. Symbiosis. 2010;50:51–7. 10.1007/s13199-009-0029-7. [DOI] [Google Scholar]
- Good RD. The geography of the genus Coriaria. The New Phytologist. 1930;29:170–98. [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Gouy M, Tannier E, Comte N et al. Seaview version 5: a multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. In: Katoh K (ed.), Multiple Sequence Alignment: Methods and Protocols (Book Series: Methods in Molecular Biology). New York, NY, USA: Springer Nature, 2021, 241–60. [DOI] [PubMed] [Google Scholar]
- Griesmann M, Chang Y, Liu X et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361:eaat1743. 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- Gtari M, Ghodhbane-Gtari F, Nouioui I et al. Cultivating the uncultured: growing the recalcitrant cluster-2 Frankia strains. Sci Rep. 2015;5:13112. 10.1038/srep13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueddou A, Swanson E, Hezbri K et al. Draft genome sequence of the symbiotic Frankia sp. strain BMG5.30 isolated from root nodules of Coriaria myrtifolia in Tunisia. Antonie Van Leeuwenhoek. 2019;112:67–74. 10.1007/s10482-018-1138-1. [DOI] [PubMed] [Google Scholar]
- Hafeez F. 1983. Nitrogen fixation and nodulation in Datisca cannabina L. and Alnus nitida Endl. Islamabad, Pakistan: PhD Thesis Quaid-e-Azam University. [Google Scholar]
- Hameed S, Hafeez FY, Mirza MS et al. Confirmation of an isolate from Datisca cannabina as atypical Frankia strain using PCR amplified 16S rRNA sequence analysis. Pak J Bot. 1994;26:247–51. [Google Scholar]
- Herrera-Belaroussi A, Normand P, Pawlowski K et al. Candidatus Frankia nodulisporulans sp. nov., an Alnus glutinosa-infective Frankia species unable to grow in pure culture and able to sporulate in-planta. Syst Appl Microbiol. 2020;43:126134. 10.1016/j.syapm.2020.126134. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil circular california agricultural experiment station. 1950;347. https://www.cabdirect.org/cabdirect/abstract/19500302257. [Google Scholar]
- Jalili V, Afgan E, Gu Q et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020;48:W395–402. 10.1093/nar/gkaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libourel C, Keller J, Brichet L et al. Comparative phylotranscriptomics reveals ancestral and derived root nodule symbiosis programmes. Nat Plants. 2023;9:1067–80. 10.1038/s41477-023-01441-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhaiyan M, Hu CJ, Kim SJ et al. Jatrophihabitans endophyticus gen. nov., sp. nov., an endophytic actinobacterium isolated from a surface-sterilized stem of Jatropha curcas L. Int J Syst Evol Microbiol. 2013;63(Pt 4):1241–8. 10.1099/ijs.0.039685-0. [DOI] [PubMed] [Google Scholar]
- Mansour S, Swanson E, McNutt Z et al. Permanent draft genome sequence for Frankia sp. strain CcI49, a nitrogen-fixing bacterium isolated from Casuarina cunninghamiana that infects Elaeagnaceae. J Genomics. 2017;5:119–23. 10.7150/jgen.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. 2012. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 235–46. [PMC free article] [PubMed]
- Meyer F, Goesmann A, McHardy AC et al. GenDB–an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–95. 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza MS, Hahn D, Dobritsa SV et al. Phylogenetic studies on uncultured Frankia populations in nodules of Datisca cannabina. Can J Microbiol. 1994;40:313–8. [DOI] [PubMed] [Google Scholar]
- Müller RD, Cannon J, Qin X et al. GPlates: building a virtual earth through deep time. Geochem Geophys. 2018;19:2243–61. 10.1029/2018GC007584. [DOI] [Google Scholar]
- Newcomb W, Wood SM. Morphogenesis and fine structure of Frankia (Actinomycetales): the microsymbiont of nitrogen-fixing actinorhizal root nodules. Int Rev Cytol. 1987;109:1–88. 10.1016/s0074-7696(08)61719-2. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Wibberg D, Battenberg K et al. An assemblage of Frankia Cluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. Bmc Genomics [Electronic Resource]. 2016;17:796. 10.1186/s12864-016-3140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Wibberg D, Vigil-Stenman T et al. Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: they come in teams. Genome Biol Evol. 2019;11:2273–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P, Nguyen TV, Battenberg K et al. Proposal of “Candidatus Frankia californiensis”, the uncultured symbiont in nitrogen-fixing root nodules of a phylogenetically broad group of hosts endemic to western North America. Int J Syst Evol Microbiol. 2017;67:3706–15. 10.1099/ijsem.0.002147. [DOI] [PubMed] [Google Scholar]
- Normand P, Nouioui I, Pujic P et al. Frankia canadensis sp. nov., isolated from root nodules of Alnus incana subspecies rugosa. Int J Syst Evol Microbiol. 2018;68:3001–11. 10.1099/ijsem.0.002939. [DOI] [PubMed] [Google Scholar]
- Normand P, Orso S, Cournoyer B et al. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol. 1996;46:1–9. 10.1099/00207713-46-1-1. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Fernandez MP et al. Absence of cospeciation between the uncultured Frankia microsymbionts and the disjunct actinorhizal Coriaria species. Biomed Res Int. 2014;2014:e924235. 10.1155/2014/924235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Jando M et al. Frankia colletiae sp. nov., a nitrogen-fixing actinobacterium isolated from Colletia cruciata. Int J Syst Evol Microbiol. 2023a;73:005656. 10.1099/ijsem.0.005656. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Pötter G et al. Novel species of Frankia, Frankia gtarii sp. nov. and Frankia tisai sp. nov., isolated from a root nodule of Alnus glutinosa. Syst Appl Microbiol. 2023b;46:126377. 10.1016/j.syapm.2022.126377. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Klenk HP et al. Frankia saprophytica sp. nov., an atypical, non-infective (Nod-) and non-nitrogen fixing (Fix-) actinobacterium isolated from Coriaria nepalensis root nodules. Int J Syst Evol Microbiol. 2018a;68:1090–95. 10.1099/ijsem.0.002633. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Rhode M et al. Frankia irregularis sp. nov., an actinobacterium unable to nodulate its original host, Casuarina equisetifolia, but effectively nodulates members of the actinorhizal Rhamnales. Int J Syst Evol Microbiol. 2018b;68:2883–914. 10.1099/ijsem.0.002914. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Montero-Calasanz MDC, Ghodhbane-Gtari F et al. Frankia discariae sp. nov.: an infective and effective microsymbiont isolated from the root nodule of Discaria trinervis. Arch Microbiol. 2017a;199:641–7. 10.1007/s00203-017-1337-6. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Montero-Calasanz MDC et al. Frankia inefficax sp. nov., an actinobacterial endophyte inducing ineffective, non nitrogen-fixing, root nodules on its actinorhizal host plants. Antonie Van Leeuwenhoek. 2017b;110:313–20. 10.1007/s10482-016-0801-7. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Gueddou A, Ghodhbane-Gtari F et al. Frankia asymbiotica sp. nov., a non-infective actinobacterium isolated from Morella californica root nodule. Int J Syst Evol Microbiol. 2017c;67:4897–901. 10.1099/ijsem.0.002153. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Montero-Calasanz MDC et al. Proposal of a type strain for Frankia alni (Woronin 1866) Von Tubeuf 1895, emended description of Frankia alni, and recognition of Frankia casuarinae sp. nov. and Frankia elaeagni sp. nov. Int J Syst Evol Microbiol. 2016;66:5201–10. 10.1099/ijsem.0.001496. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Rhode M et al. Frankia irregularis sp. nov., an actinobacterium unable to nodulate its original host, Casuarina equisetifolia, but effectively nodulates members of the actinorhizal Rhamnales. Int J Syst Evol Microbiol. 2018;68:2883–914. 10.1099/ijsem.0.002914. [DOI] [PubMed] [Google Scholar]
- Nouioui I, Ghodhbane-Gtari F, Rohde M et al. Frankia coriariae sp. nov., an infective and effective microsymbiont isolated from Coriaria japonica. Int J Syst Evol Microbiol. 2017;67:1266–70. 10.1099/ijsem.0.001797. [DOI] [PubMed] [Google Scholar]
- Paschke MW, Dawson JO. Avian dispersal of Frankia. Can J Bot. 1993;71:1128–31. 10.1139/b93-132. [DOI] [Google Scholar]
- Persson T, Battenberg K, Demina IV et al. Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS One. 2015;10:e0127630. https://doi.org/0.1371/journal.pone.0127630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson T, Benson DR, Normand P et al. Genome sequence of “Candidatus Frankia datiscae” Dg1, the uncultured microsymbiont from nitrogen-fixing root nodules of the dicot Datisca glomerata. J Bacteriol. 2011;193:7017–8. 10.1128/JB.06208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi AC, Bautista-Guerrero HH, Abby SS et al. Robust Frankia phylogeny, species delineation and intraspecies diversity based on Multi-Locus Sequence Analysis (MLSA) and Single-Locus Strain Typing (SLST) adapted to a large sample size. Syst Appl Microbiol. 2018;41:311–23. 10.1016/j.syapm.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Pozzi ACM, Herrera-Belaroussi A, Schwob G et al. Proposal of ‘Candidatus Frankia alpina’, the uncultured symbiont of Alnus alnobetula and A. incana that forms spore-containing nitrogen-fixing root nodules. Int J Syst Evol Microbiol. 2020;70:5453–9. 10.1099/ijsem.0.004433. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujic P, Bolotin A, Fournier P et al. Genome sequence of the atypical symbiotic Frankia R43 strain, a nitrogen-fixing and hydrogen-producing actinobacterium. Genome Announc. 2015;3:e01387–15. 10.1128/genomeA.01387-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguero MA, Gelfo JN, López GM et al. Final Gondwana breakup: the Paleogene South American native ungulates and the demise of the South America–Antarctica land connection. Global Planet Change. 2014;123:400–13. 10.1016/j.gloplacha.2014.07.016. [DOI] [Google Scholar]
- Renner SS, Barreda VD, Tellería MC et al. Early evolution of Coriariaceae (Cucurbitales) in light of a new early Campanian (ca. 82 Mya) pollen record from Antarctica. Taxon. 2020;69:87–99. 10.1002/tax.12203. [DOI] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Nat Acad Sci USA. 2009;106:19126–31. 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio Team . RStudio: Integrated Development Environment for R. RStudio. PBC, Boston, MA. 2022. http://www.rstudio.com/.
- Samarakoon T, Wang SY, Alford MH. Enhancing PCR amplification of DNA from recalcitrant plant specimens using a trehalose-based additive. Applications in Plant Sciences. 2013;1:apps.1200236. 10.3732/apps.1200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Daubin V, Abrouk D et al. Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders “Frankiales” and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. and Nakamurellales ord. nov. Int J Syst Evol Microbiol. 2014;64:3821–32. 10.1099/ijs.0.063966-0. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–6. 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Skog LE. The genus Coriaria (Coriariaceae) in the western hemisphere. Rhodora. 1972;74:242–53. [Google Scholar]
- Svistoonoff S, Hocher V, Gherbi H. Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation?. Curr Opin Plant Biol. 2014;20:11–8. 10.1016/j.pbi.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Tamura T, Hayakawa M, Hatano K. A new genus of the order Actinomycetales, Cryptosporangium gen. nov., with descriptions of Cryptosporangium arvum sp. nov. and Cryptosporangium japonicum sp. nov. Int J Syst Bacteriol. 1998;48Pt3:95–1005. 10.1099/00207713-48-3-995. [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Alonso-Vega P, Rodríguez R et al. The genus Micromonospora is widespread in legume root nodules: the example of Lupinus angustifolius. ISME J. 2010;4:10. 10.1038/ismej.2010.55. [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Kroppenstedt RM, Schumann P et al. Micromonospora coriariae sp. nov., isolated from root nodules of Coriaria myrtifolia. Int J Syst Evol Microbiol. 2006;56:2381–5. 10.1099/ijs.0.64449-0. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Doyle JJ, Geurts R. A resurrected scenario: single gain and massive loss of nitrogen-fixing nodulation. Trends Plant Sci. 2019;24:49–57. 10.1016/j.tplants.2018.10.005. [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu F et al. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc Natl Acad Sci. 2018;115:E4700–9. 10.1073/pnas.1721395115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemulapally S, Guerra T, Hahn D. Localization of typical and atypical Frankia isolates from Casuarina sp. in nodules formed on Casuarina equisetifolia. Plant Soil. 2019;435:385–93. [Google Scholar]
- Wollum AG II, Youngberg CT, Gilmour CM. Characterization of a Streptomyces sp. isolated from root nodules of Ceanothus velutinus Dougl. Soil Sci Soc Am J. 1966;30:463–7. 10.2136/sssaj1966.03615995003000040020x. [DOI] [Google Scholar]
- Yokoyama J, Suzuki M, Iwatsuki K et al. Molecular phylogeny of Coriaria, with special emphasis on the disjunct distribution. Mol Phylogenet Evol. 2000;14:11–9. 10.1006/mpev.1999.0672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.