Duno-Miranda et al. investigate the E525K mutation in human β-cardiac myosin, a protein crucial for heart contraction, and its role in causing dilated cardiomyopathy (DCM). They demonstrate that the length of the myosin tail influences its self-inhibition and that the E525K mutation strengthens this effect, potentially reducing heart contractility in DCM.
Abstract
Dilated cardiomyopathy (DCM) is a condition characterized by impaired cardiac function, due to myocardial hypo-contractility, and is associated with point mutations in β-cardiac myosin, the molecular motor that powers cardiac contraction. Myocardial function can be modulated through sequestration of myosin motors into an auto-inhibited “super-relaxed” state (SRX), which may be further stabilized by a structural state known as the “interacting heads motif” (IHM). Here, we sought to determine whether hypo-contractility of DCM myocardium results from reduced function of individual myosin molecules or from decreased myosin availability to interact with actin due to increased IHM/SRX stabilization. We used an established DCM myosin mutation, E525K, and characterized the biochemical and mechanical activity of wild-type and mutant human β-cardiac myosin constructs that differed in the length of their coiled-coil tail, which dictates their ability to form the IHM/SRX state. We found that short-tailed myosin constructs exhibited low IHM/SRX content, elevated actin-activated ATPase activity, and fast velocities in unloaded motility assays. Conversely, longer-tailed constructs exhibited higher IHM/SRX content and reduced actomyosin ATPase and velocity. Our modeling suggests that reduced velocities may be attributed to IHM/SRX-dependent sequestration of myosin heads. Interestingly, longer-tailed E525K mutants showed no apparent impact on velocity or actomyosin ATPase at low ionic strength but stabilized IHM/SRX state at higher ionic strength. Therefore, the hypo-contractility observed in DCM may be attributable to reduced myosin head availability caused by enhanced IHM/SRX stability in E525K mutants.
Introduction
Dilated cardiomyopathy (DCM) is characterized by left ventricular dilation and systolic dysfunction, resulting in reduced cardiac output (Rosenbaum et al., 2020) and eventually heart failure. Approximately 35% of DCM cases are monogenic, with mutations to genes encoding the heart muscle’s cytoskeletal and contractile proteins (McNally and Mestroni, 2017). One etiologic hypothesis for DCM-associated mutations of β-cardiac myosin, the molecular motor that powers the heart (Vincent, 2008), is that these mutations negatively impact myosin function, which leads to myocardial hypo-contractility. Therefore, defining the impact of DCM-associated mutations on molecular motor function is the first step in therapeutic design.
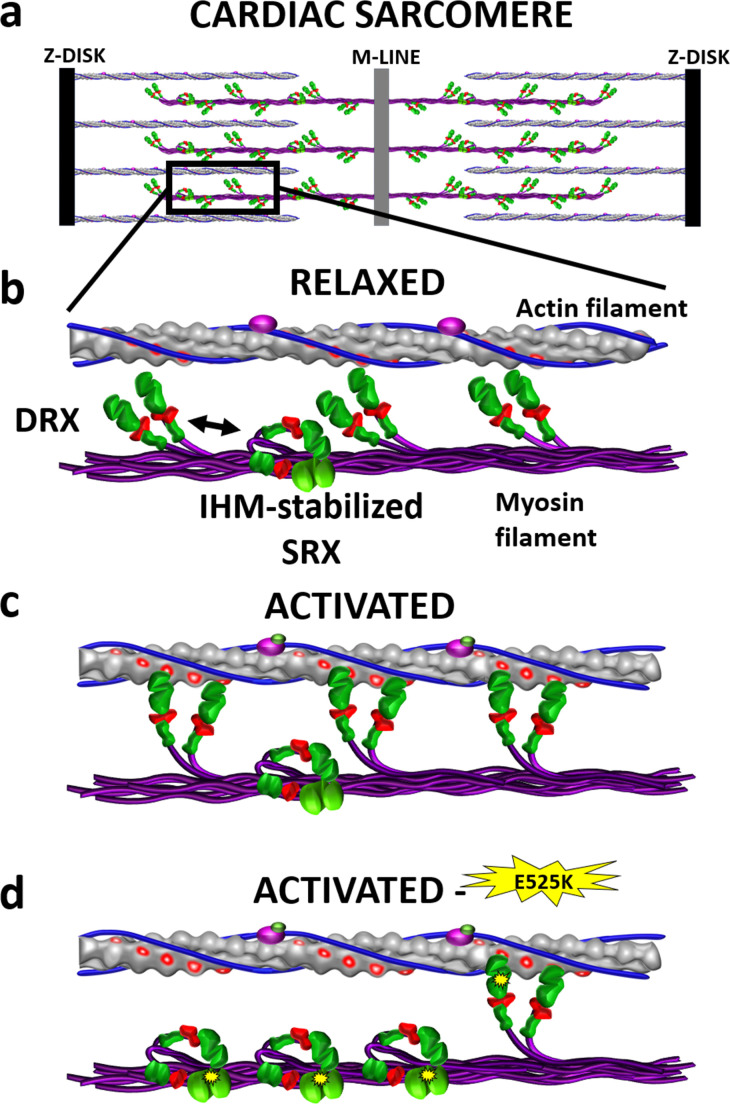
Upon cardiac muscle activation, ventricular force and motion generation originate within the sarcomere, the most basic contractile unit of cardiac muscle (Brunello and Fusi, 2023). The sarcomere is composed of myosin thick filaments from which the myosin heads emanate (Fig. 1a), capturing the chemical energy of adenosine triphosphate (ATP) hydrolysis and converting it into mechanical energy as the motor interacts cyclically with the actin thin filament (Brunello and Fusi, 2023). Thus, peak systolic force is determined by the number of attached myosin motors at any point in time, each of which generates an ∼1–2 pN intrinsic force (Palmiter et al., 1999; Finer et al., 1994). Until recently, it was assumed that upon muscle activation, all myosin heads were available to interact with the thin filament. However, in relaxed striated muscles, including cardiac muscle, myosin motors adopt one of two distinct biochemical states, an auto-inhibited, super-relaxed (SRX) state, incapable of binding to the thin filament, and a disordered-relaxed (DRX) state that can bind to the thin filament upon muscle activation to generate force (Brunello and Fusi, 2023; Hooijman et al., 2011) (Fig. 1, b and c). Therefore, the SRX/DRX ratio would determine the number of available motors that can attach to the thin filament and generate force and motion (Spudich, 2014). Although the SRX state is a biochemical state that is inherent to each of the myosin’s two heads (Anderson et al., 2018), the SRX state may be further stabilized by intramolecular electrostatic interactions, whereby the two heads fold back and interact with each other and the myosin coiled-coil tail (Anderson et al., 2018; Wendt et al., 1999) (Fig. 1 b and Fig. 2); an asymmetric structure called the “interacting heads motif” (IHM) (Woodhead et al., 2005). The IHM was first observed in electron micrographs of single myosin molecules (Wendt et al., 1999), with its presence now confirmed by cryo-electron microscopy of isolated native cardiac thick filaments (Dutta et al., 2023) and thick filaments within intact sarcomeres (Tamborrini et al., 2023). The IHM and its potential stabilization of the auto-inhibited SRX state offer a mechanism by which the number of available myosin heads competent to generate force can be regulated.
Figure 1.
Cardiac myosin conformations and interactions with actin filaments in relaxed and activated muscle. (a) Representation of a cardiac sarcomere composed of interdigitated myosin filaments (purple) connected at the M-line and actin filaments anchored at the Z-disk. (b) In relaxed muscle, myosin motors that emanate from the myosin filament backbone exist either in the enzymatically auto-inhibited, SRX state, which can be stabilized by adopting the IHM and precludes thin filament binding, or in the DRX state that is capable actin filament attachment and force generation upon cardiac activation. (c) Once activated, only DRX (i.e., active heads) myosin is recruited to generate force. (d) Proposed mechanism by which the DCM-associated E525K β-cardiac myosin mutation (yellow starburst) stabilizes the IHM/SRX state, reducing the available pool of myosin motors for force generation, leading to cardiac hypo-contractility. Since most humans are heterozygote for cardiac myosin mutations, only one myosin head is depicted with the mutation.
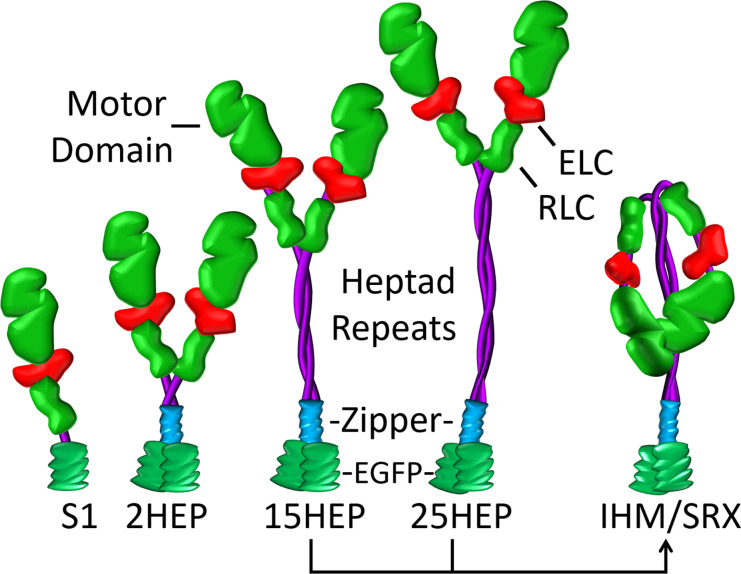
Figure 2.
Various human single- and double-headed β-cardiac myosin constructs. Expressed human β-cardiac myosin constructs (single-headed: S1; double-headed with varying heptad [HEP] tail length: 2HEP, 15HEP, and 25HEP). All constructs have a regulatory light chain (RLC), essential light chain (ELC), and a C-terminal EGFP, with the double-headed constructs also containing a GCN4 leucine zipper to ensure dimerization. Only the 15HEP and 25HEP can adopt the IHM conformation, which presumably stabilizes the SRX state.
In contrast to DCM, hypertrophic cardiomyopathy (HCM) is also associated with β-cardiac myosin mutations and may present clinically with myocardial hyper-contractility (Spudich, 2014, 2019). HCM-linked mutations have been shown to destabilize the IHM/SRX state, allowing more myosin heads to interact with the thin filament, offering a mechanistic explanation for the hyper-contractile HCM phenotype (Rohde et al., 2018; Heitner et al., 2019; Olivotto et al., 2020). In fact, Mavacamten, an FDA-approved, small-molecule therapeutic for HCM patients (Claassen et al., 2023), targets myosin and restores normal contractility by increasing the probability of the myosin IHM/SRX state. Accordingly, we hypothesize that the hypo-contractile phenotype observed with DCM-associated β-cardiac myosin mutations arises from enhanced stabilization of the IHM/SRX state, reducing the number of myosin motors available to generate the power required for normal cardiac output (Spudich, 2014) (Fig. 1 d), although DCM-associated myosin mutations may also alter the motors’ intrinsic force and motion generation (Debold et al., 2007). Therefore, in this study, we chose to define the functional impact on the enzymatic and molecular mechanics of expressed human β-cardiac myosin harboring an established DCM-associated mutation (E525K), which localizes to an intramolecular contact region believed to stabilize the IHM (Rasicci et al., 2022).
We build on our recently published biochemical and structural studies using single- and double-headed β-cardiac myosin constructs of both wild-type (WT) and the DCM-associated E525K mutant, which demonstrated that only the double-headed construct readily adopts the IHM/SRX state (Rasicci et al., 2022). The present study provides biochemical and mechanical characterization of human β-cardiac myosin heavy meromyosin (HMM) constructs with varying lengths of coiled-coil domain (2, 15, and 25 heptad repeats, Fig. 2) to determine whether: (1) the presence of the IHM/SRX state in WT myosin affects in vitro actin filament motility, a model system for unloaded muscle shortening (Huxley, 1990), and (2) the E525K DCM-associated mutation affects either the motor’s intrinsic enzymatic and mechanical performance and/or the capacity to adopt the IHM/SRX state. In fact, our new biochemical and in vitro motility data suggest that double-headed, longer-tailed (15 and 25 heptad) HMM constructs readily adopt the IHM/SRX state, leading to lower actin-activated ATPase activity and slower actin filament velocities compared to constructs incapable of adopting the IHM/SRX state. Surprisingly, in these longer-tailed HMM, the E525K DCM-associated mutation had little impact on the actin-activated ATPase activity and in vitro actin filament motility, under the low ionic strength conditions necessary to make these measurements. However, single ATP turnover studies that characterize the IHM/SRX state proportion over a range of ionic strengths indicate that the E525K mutation does stabilize the IHM/SRX state and may be the dominant factor that reduces the number of available motors upon activation in vivo, potentially explaining the hypo-contractile DCM phenotype.
Materials and methods
Protein expression and purification
The human β-cardiac myosin constructs were all derived from the MYH7 gene (GenBank accession no. AAA51837.1) (Fig. 2). As previously described (Rasicci et al., 2022), the β-cardiac myosin HMM construct (amino acids 1–946), with 15 heptads of the coiled-coil (15HEP), is followed first by a GCN4 leucine zipper (sequence: MKQLEDKVEELLSKNYHLENEVARLKKLVGER), then a short linker (GSGKL), and a C-terminal series of EGFP, Avi-tag (GLNDIFEAQKIEWHE), and FLAG-tag (DYKDDDDK). HMM constructs with either a shorter 2 heptad coiled-coil (2HEP) (amino acids 1–855) or a longer 25 heptad coiled-coil (25HEP) (amino acids 1–1016) had the same C-terminal, leucine zipper, linker, EGFP, and Avi- and FLAG-tags. The β-cardiac myosin subfragment 1 construct (S1) (amino acids 1–842) had only the C-terminal EGFP, Avi-, and FLAG-tags. The E525K mutation was introduced by Quikchange site-directed mutagenesis (200523; Agilent). The constructs were cloned into the pDual shuttle vector, and the initial recombinant adenovirus stock was produced by Vector Biolabs at a titer of 108 plaque-forming units per ml (pfu/ml). As previously described (Swenson et al, 2017), the virus was expanded by the infection of Ad293 cells (240085; Agilent) at a multiplicity of infection (MOI) of 3–5. The virus was harvested from the cells and purified by CsCl density sedimentation, giving a final virus titer of 1010–1011 pfu/ml.
The mouse skeletal muscle-derived C2C12 cell line (CRL-1772; ATCC) was used to express the various β-cardiac myosin constructs as described previously (Chow et al., 2002; Wang et al., 2003; Winkelmann et al., 2015). Briefly, C2C12 cells were grown to ∼90% confluency on tissue culture plates (145/20 mm) in growth media (DMEM with 9% FBS, 1% penicillin–streptomycin). On the day of infection, 20 plates were differentiated by changing the media to contain horse serum (DMEM with 9% horse serum, 1% FBS, 1% penicillin–streptomycin) and simultaneously infected with virus at 4 × 107 pfu/ml. The cells were harvested for myosin purification 10–12 days after infection, and β-cardiac myosin constructs were purified by FLAG affinity chromatography. The reported concentration of β-cardiac myosin is given on a per head basis. At least three biological replicates of each construct were expressed and the same replicates were used for both biochemical and motility experiments described below. Actin was purified using acetone powder from rabbit skeletal muscle (41995; Pel-Freez) (Pardee and Spudich, 1982).
Steady-state actin-activated ATPase assay
We utilized the NADH-coupled ATPase assay to examine the actin-activated ATPase activity at a range of actin concentrations (0–60 µM). Briefly, each construct (S1, 2HEP, 15HEP, and 25HEP) at 0.1 µM, was assayed in the presence of 1 mM ATP (A2383; Sigma-Aldrich), and the NADH (N6005; Sigma-Aldrich) absorbance was monitored for a 200 s period in the stopped-flow apparatus (Applied Photophysics) at 25°C in MOPS 20 buffer (10 mM MOPS, pH 7.0, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT). For S1 with 0 µM actin, 1.0 µM of construct was used to improve signal to noise. The linear fit of the NADH absorbance as function of time was converted to the ADP produced per unit time using a standard curve with known ADP (A2754; Sigma-Aldrich) concentrations. The ATPase was expressed as the amount of product produced per second per molar concentration of myosin heads. The ATPase data plotted as a function of actin concentration were fit to the Michaelis–Menten equation to determine the maximal ATPase activity (kcat) and actin concentration at which ATPase is one-half maximal (KATPase). The ATPase activity without actin (v0) was also determined.
Single turnover measurements
The fluorescence of 3′-O-(N-methyl-anthraniloyl)-2′-deoxyadenosine-5′-triphosphate (mantATP; NU-203S; Jena Biosciences) was measured with 290 nm excitation and a 395 nm long-pass emission filter in a stopped-flow apparatus (Applied Photophysics) at 25°C. Each myosin sample was incubated on ice for about 10 min in MOPS 20 buffer at varying KCl concentrations (20–150 mM) prior to each experiment. Single mantATP turnover reactions were performed by incubating 0.5 μM myosin constructs (S1, 2HEP, 15HEP, and 25HEP) with 2 μM mantATP for 30 s at room temperature. Subsequently, the complex was mixed with saturating ATP (2 mM) in the stopped flow (final concentrations: 0.25 µM myosin, 1 µM mantATP, and 1 mM ATP). The mantATP fluorescence increases upon binding myosin, stays elevated during hydrolysis and phosphate release, and decreases upon mantADP release (Woodward et al., 1991). The rate of mant fluorescence decay is limited by the slowest step in the ATPase cycle without actin present, which is thought to be phosphate release (Geeves, 2016). Fluorescence decays were examined over a 1,000 s period and were typically best fit by a two-exponential function to determine the SRX and DRX rate constants (Fig. S1). However, when the fraction of the slow or fast phase is <5% of the total fluorescence decay, it becomes difficult to differentiate between a single versus a double-exponential fit. Photobleaching was found to be ≤0.5% of the fluorescence signal. Relative amplitudes of the fast and slow phase rate constants determined the fraction of myosin in the DRX and SRX states, respectively.
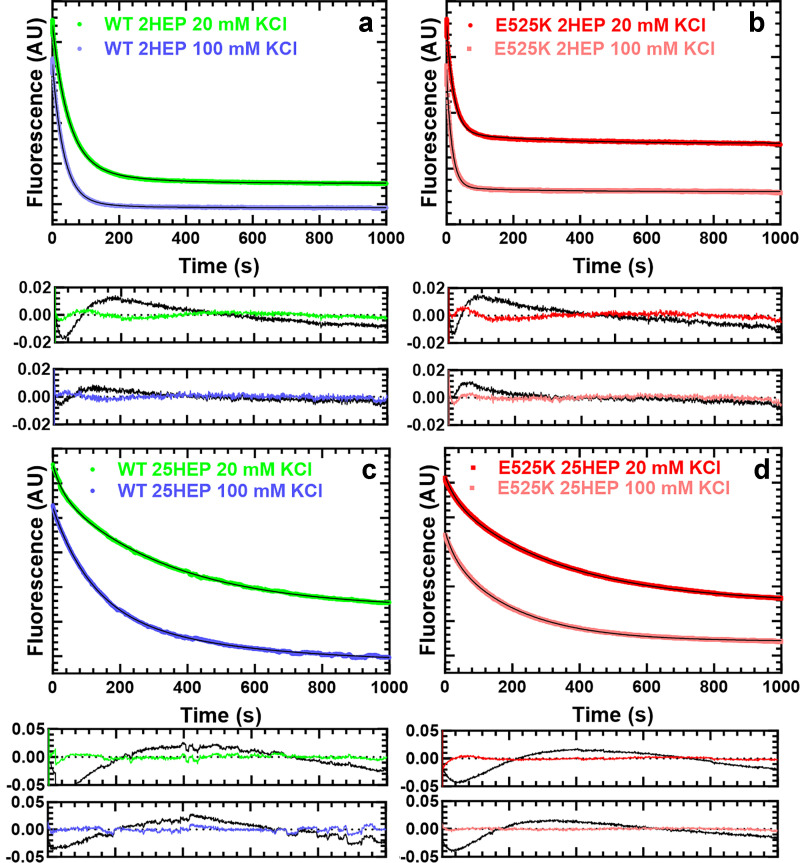
Figure S1.
Single mantATP turnover fluorescence decays. Examples of single mantATP turnover reactions for WT and E525K 2HEP (a and b), and 25HEP (c and d) constructs as a function of ionic strength (20 and 100 mM KCl). Representative fluorescence decay fits (upper graph) using two-exponentials (thin solid black line) and their respective residuals (lower pair of graphs) are presented (the residuals from the single-exponential fits are shown in black for comparison). Relative amplitudes of the fast and slow phase-exponential fit rate constants determined the fraction of myosin in the DRX and SRX states, respectively. See Table 2 for summary of values.
In vitro motility assay
The movement of fluorescently labeled actin filaments over a β-cardiac myosin-coated microscope slide surface was characterized using a modified version of previously established protocols (Rasicci et al., 2022; Kron et al., 1991; Warshaw et al., 1990). In brief, the in vitro motility assay occurs within a sealed flow-through experimental chamber (i.e., flow-cell) constructed using a nitrocellulose-coated microscope coverslip. To minimize non-specific surface interactions that may compromise the motor’s capacity to propel actin filaments, all myosin constructs were attached to the surface via an anti-GFP nanobody (Sommese et al., 2016). Preliminary in vitro motility data confirmed that the motion-generating capacity of both the S1 and 2HEP constructs was compromised when the constructs were directly adhered non-specifically to the nitrocellulose surface (Fig. S2). This was not the case for the longer-tailed 15HEP and 25HEP constructs. Regardless, all constructs were attached via the nanobody, which was diluted to 700 nM in actin buffer (25 mM KCl, 25 mM imidazole, 1 mM EGTA, and 10 mM DTT, pH 7.4) and infused into the flow-cell, incubated for 2 min, followed by a bovine serum albumin (BSA; A3059; Sigma-Aldrich) wash (0.5 mg/ml BSA in actin buffer) to block any exposed nitrocellulose surface. Prior to myosin addition to the flow-cell, all constructs were subjected to a “dead-head” spindown to reduce the amount of non-functional myosin. Specifically, myosin was centrifuged in the presence of equimolar actin and 1 mM ATP (A7699; Sigma-Aldrich) in myosin buffer (300 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, and 10 mM DTT, pH 7.4) with the supernatant recovered for motility. Myosin constructs were then infused into the flow-cell at concentrations ranging from 5 to 700 nM in myosin buffer and incubated for 2 min. To eliminate any remaining non-functional myosin heads within the assay, unlabeled actin filaments (1 μM in actin buffer) were infused into the flow-cell and incubated for 1 min, after which an actin buffer wash with 1 mM added MgATP released actin filaments from only the ATP-sensitive, functional myosin heads. This approach effectively removed damaged heads due to their ATP-insensitive and irreversible actin filament binding. Next, an actin buffer wash followed by infusion of 1–2.5 nM vortexed (∼1 s) Alexa-532-phalloidin-labeled actin filaments in actin buffer with 20% vol/vol O2 scavengers (0.1 μg/ml glucose oxidase [G2133; Sigma-Aldrich], 0.018 μg/ml catalase [C9322; Sigma-Aldrich], and 2.3 μg/ml glucose) and then incubated for 1 min. Finally, the flow-cell was washed extensively with actin buffer, with the final infusion of motility buffer (actin buffer with the addition of 1 mM ATP, 0.5% methylcellulose [M0512; Sigma-Aldrich], 2% vol/vol O2 scavengers, 1 mM creatine phosphate [P7936; Sigma-Aldrich], 1 mg creatine phosphate kinase [C3755; Sigma-Aldrich]), initiating actin filament motility. O2 scavengers were used to prevent oxidative damage, while methylcellulose was used to maintain actin filaments near the motility surface by reducing diffusional excursions.
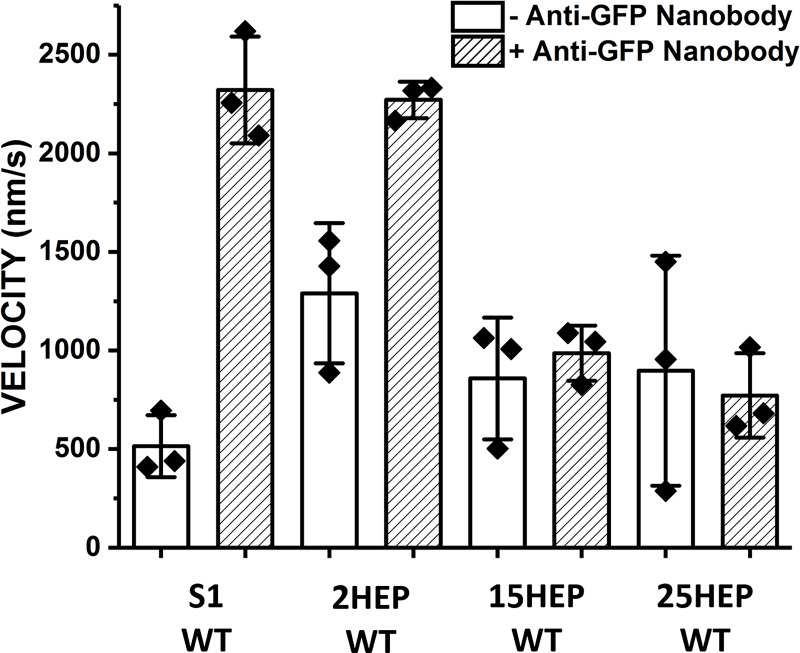
Figure S2.
β-cardiac myosin motility surface attachment strategy. The use of an anti-GFP nanobody to attach myosin constructs to the motility surface improves velocity of the S1 and 2HEP WT constructs, which are incapable of adopting the IHM/SRX state, by at least a factor of 2. This result suggests that the nanobody attachment strategy allows myosin heads to interact properly with actin without surface interference. Data represent mean values ± SD from three independent protein preparations.
For experiments where 2′-deoxyadenosine-5′-triphosphate (dATP; C01577; GenScript) was used to limit the myosin SRX state (Walklate et al., 2022), dATP was substituted for ATP in all ATP-containing buffers. For myosin mixture experiments, the same protocol described above was used, with the exception that 25 nM WT 2HEP was infused first, the flow-cell was then washed with actin buffer, and after 2 min, between 0 and 125 nM WT 15HEP was infused into the flow-cell and incubated for 2 min with the remaining steps the same. All motility experiments were imaged in total internal reflection fluorescence (TIRF) microscopy using a Nikon Eclipse TiU with Plan Apo objective (100×, 1.35 NA) and a Lumen 200W metal arc lamp (Prior Scientific) with actin filament images captured on a Turbo 620G camera (Stanford Photonics, Inc.) running Piper Controlled Imaging Desktop Software (Stanford Photonics, Inc.). Individual motility movies of 30 s in length (10 fps) were recorded at three different 45 × 60 μm regions of interest within a single flow-cell. All experiments were performed at 30°C.
β-Cardiac myosin motility surface occupancy
To assess the myosin motility surface occupancy, we took advantage of each myosin construct having an EGFP C-terminal tag (see above). EGFP fluorescence in the flow-cell was imaged on the same TIRF microscope described above for the motility experiments. Therefore, each motility experiment had an accompanying estimate of the myosin surface occupancy. Then, 3 s long, EGFP-myosin fluorescence movies (10 fps) were taken at three different 45 × 60 μm regions of interest within the same flow-cell after recording motility. For each movie, we took the first 20 frames and created an average intensity z projection (ImageJ) and recorded its integrated total intensity. Then for the given flow-cell and myosin concentration, the total intensities for the three movies were averaged and used as a readout of EGFP fluorescence. The range of EGFP fluorescence intensity in arbitrary units associated with the different myosin concentrations was normalized to that obtained for the maximum 700 nM myosin used. However, it was critical that the camera system gain be adjusted, so that the EGFP fluorescence for the 700 nM myosin concentration did not saturate the system’s fluorescence intensity readout. Once set, the same gain was used for flow-cells with lesser myosin concentrations.
In vitro motility data analysis
For each motility movie that had 10–40 filaments per movie, depending on conditions, a mean actin filament velocity and fraction motile filaments were determined using a combination of the ImageJ (Schindelin et al., 2012) plugin MTrack2 (https://imagej.net/plugins/mtrack2) and Mean Squared Displacement (MSD) analysis. MSD analysis was employed here as it allows an accurate assessment of velocity, independent of tracking error, across a very wide range of velocities. Briefly, x,y trajectories for individual actin filaments were obtained using MTrack2 and used to calculate MSD according to
where N is the total number of frames in the trajectory, n is the number of frames for the time intervals over which the MSD is calculated (such that n ≤ N/4), Δt is the time between frames (100 ms), and xi and yi are the positions of the actin filament in frame i. As the relationship between MSD and time can be generalized as MSD∼(nΔt)α, we determined the diffusive exponent (α) as the slope of the relationship between MSD and the time interval (nΔt) on log–log axes. Filament trajectories of ≥20 data points that demonstrated directed motion (α > 1; Fig. S3 d) were used to calculate a velocity from the slope of the relationship between √MSD and nΔt, while the fraction of moving filaments was calculated as the fraction of all trajectories with α > 1. As velocity is only determined for moving filaments, we define a “Velocity Index” as the product of the mean velocity and the fraction of moving filaments to better describe the overall motion-generating activity for each condition. At least three biological replicates were characterized for all constructs and the replicate data for a given construct combined into mean velocity, mean fraction moving, and mean Velocity Index versus myosin surface occupancy (see Fig. S3, a–c, for 2HEP WT example). Only the 2HEP WT Velocity Index versus myosin surface occupancy data were fit with a modified Hill dose–response equation using a Levenberg–Marquard algorithm (Origin(Pro) 2017, OriginLab Corporation), while all other construct relations were fit to predicted curves from an analytical model (see below).
Figure S3.
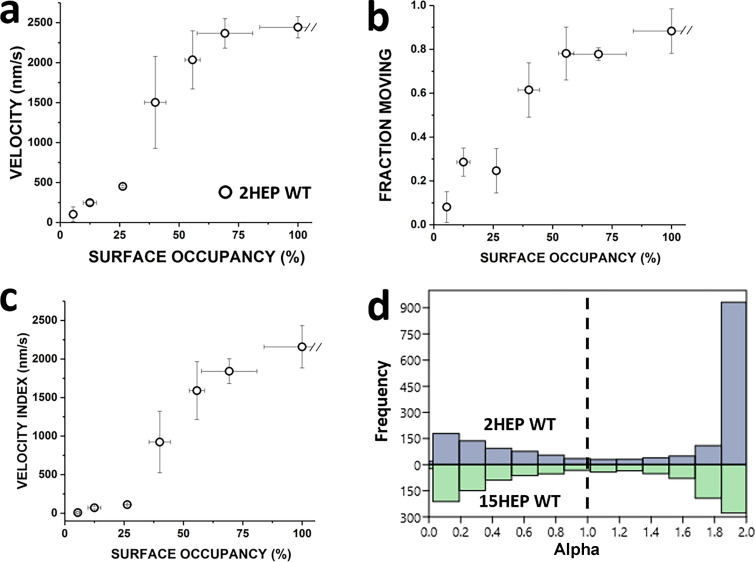
Velocity times fraction motile = Velocity Index. The Velocity Index offers a more complete characterization of the motility behavior by multiplying velocity data by the fraction of moving filaments, which can be reduced at limiting myosin surface densities. (a) 2HEP WT velocity increases in proportion to the surface occupancy. (b) 2HEP WT fraction moving is reduced at low myosin surface occupancy where the numbers of active motors becoming limiting. (c) 2HEP WT Velocity Index = (velocity) × (fraction moving) using data in a and b. Data points represent mean values ± SD of three experiments from separate protein preparations. (d) Bihistogram of pooled alpha values for 2HEP WT and 15HEP WT across all surface occupancies (n = 3, see Statistics).
Statistics
Statistical comparisons between data sets were as follows. For kinetic experiments and measurements of SRX, multiple t tests with Holm–Sidak correction were used to compare WT versus E525K with the same construct, at the same salt concentration (e.g., WT S1 at 20 mM KCl versus E525K S1 at 20 mM KCl). For comparisons within groups (e.g., WT S1 versus WT 2HP versus WT 15 HP versus WT 25 HP), a two-way ANOVA with Tukey’s post hoc was performed. Both of these statistical analyses were completed in GraphPad Prism (GraphPad Software).
For both the motility surface EGFP fluorescence intensity versus myosin surface concentration and the Velocity Index versus myosin surface occupancy within WT groups or versus the E525K groups, data were compared using the non-parametric Kruskal–Wallis one-way ANOVA. For experiments where the Velocity Index for constructs was compared at 100% myosin surface occupancy with and without the presence of dATP, the Student’s t test was used for statistical comparisons. For both the Kruskal–Wallis and Student’s t test, a P value ≤ 0.05 was considered significantly different. These statistical analyses were performed in Origin(Pro) 2017, OriginLab Corporation.
Online supplemental material
Fig. S1 shows the single mantATP turnover fluorescence decays. Fig. S2 shows the β-cardiac myosin motility surface attachment strategy. Fig. S3 shows the velocity times fraction motile = Velocity Index. Fig. S4 shows the model of the %IHM/SRX impact on β-cardiac myosin Velocity Index. Video 1 shows the motility movie of WT 2HEP myosin at 100% surface occupancy. Video 2 shows the motility movie of WT 2HEP myosin at 10% surface occupancy. Video 3 shows the motility movie of WT 15HEP myosin at 100% surface occupancy. Video 4 shows the motility movie of WT 15HEP myosin at 10% surface occupancy. Table S1 shows the P values for comparisons of steady-state actin-activated ATPase measurements. Table S2 shows the P values for comparisons between WT and E525K actin-activated ATPase. Table S3 shows the P values for comparisons of SRX fraction as a function of salt concentration from single mantATP turnover measurements. Table S4 shows the P values for comparisons of SRX fraction comparing different constructs from single mantATP turnover measurements. Supplemental text at the end of the PDF contains modeling details, additional data, and statistical P values.
Figure S4.
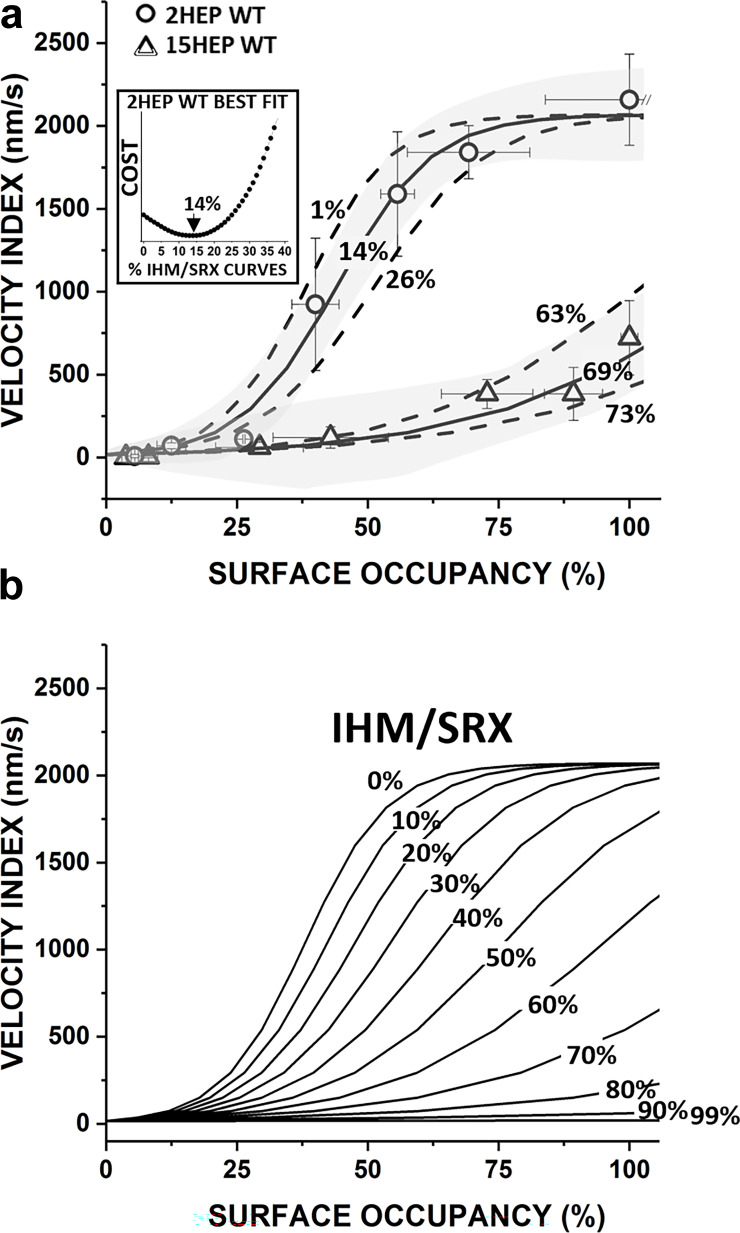
Model of the %IHM/SRX impact on β-cardiac myosin Velocity Index. (a) Velocity Index versus surface occupancy data for 2HEP WT were fit to the modified Hill equation (solid line) (Supplemental text) and used to represent an β-cardiac myosin population with 16% SRX that allowed a family of curves to be generated for hypothetical IHM/SRX percentages ranging from 0 to 100% (b). The 95% confidence limits (gray shaded area) for the 2HEP WT fit were used to estimate the error in the model predicted %IHM/SRX for a given construct. All fits were performed by calculating the minimum COST function from the least squared error analysis (example for 2HEP WT, inset). For 2HEP WT, the model best fit returns a best fit of 14% IHM/SRX with an estimate error of ±13% IHM/SRX. Velocity Index versus surface occupancy data for 15HEP WT with model predicted 69% IHM/SRX curve and 95% confidence limits (gray shaded area) error estimates of ±6% IHM/SRX. Predicted %IHM/SRX and the errors in the estimate for all constructs were as follows: WT: 2HEP (14 ± 13%), 15HEP (69 ± 6%), 25HEP (76 ± 4%); E525K: 2HEP (16 ± 12%), 15HEP (62 ± 6%), and 25HEP (71 ± 4%). (b) Family of curves for hypothetical %IHM/SRX ranging from 0 to 100% based on the original WT 2HEP modified Hill dose–response curve in a, which assumed a 16%IHM/SRX (see Supplemental text for details).
Video 1.
Motility video of WT 2HEP myosin at 100% surface occupancy. Frames were collected every 0.1 s for 30 s, for a total of 300 frames. Playback is 10× real time.
Video 2.
Motility video of WT 2HEP myosin at 10% surface occupancy. Frames were collected every 0.1 s for 30 s, for a total of 300 frames. Playback is 10× real time.
Video 3.
Motility video of WT 15HEP myosin at 100% surface occupancy. Frames were collected every 0.1 s for 30 s, for a total of 300 frames. Playback is 10× real time.
Video 4.
Motility video of WT 15HEP myosin at 10% surface occupancy. Frames were collected every 0.1 s for 30 s, for a total of 300 frames. Playback is 10× real time.
Results
WT steady-state ATPase activity
We previously studied the E525K DCM mutation and its impact on β-cardiac myosin structure and enzymatic activity (Rasicci et al., 2022). However, we only compared the single-headed S1 and double-headed HMM 15HEP constructs. Here, we have compared these same constructs to both a shorter-tailed 2HEP and longer-tailed 25HEP as part of a more comprehensive study on how both the E525K mutation and β-cardiac myosin tail length impact biochemical and mechanical function.
The NADH-coupled ATPase assay was used to examine the steady-state actin-activated ATPase activity of each of our WT and E525K β-cardiac myosin constructs (S1, 2HEP, 15HEP, 25HEP; Fig. 2) in the absence and presence of actin (0, 5, 10, 20, 40, and 60 µM) (Fig. 3). Since kcat displayed considerable uncertainty in the long-tail constructs (15HEP and 25HEP), we compared the actin-activated ATPase at 60 µM actin to describe the relative differences between constructs and the impact of the mutation.
Figure 3.
Steady-state ATPase activity of β-cardiac myosin constructs. (a–d) Steady-state ATPase kinetics of WT and E525K β-cardiac myosin constructs were determined using an NADH-coupled assay across varying actin concentrations. E525K mutations in S1 and 2HEP constructs significantly increased actin-activated ATPase activity (∼3-fold) and decreased the actin concentration for half-maximal activity (KATPase) by ∼18-fold. In contrast, 15HEP and 25HEP constructs showed reduced ATPase activities, with no significant difference between WT and E525K. Data are presented as mean ± SD of three experiments from separate protein preparations. Data in c originally published in Rasicci et al. (2022). Comparison p-values provided in Tables S1 and S2.
The actin-activated ATPase activity of the WT S1 construct containing an EGFP tag was similar to our previously published results without the EGFP tag (Rasicci et al., 2022), suggesting that the C-terminal EGFP did not alter the ATPase activity (Table 1). The WT 15HEP construct, as previously published (Rasicci et al., 2022), showed a ∼10-fold reduced actin-activated ATPase activity compared to S1 (Fig. 3, a and c; and Table 1), which we previously interpreted as evidence for the 15HEP construct adopting the IHM/SRX state (Rasicci et al., 2022). Interestingly, the short-tailed, double-headed WT 2HEP construct displayed similar actin-activated ATPase activity to WT S1 (Fig. 3, a and b), with similar KATPase and kcat (Table 1). Therefore, on a per-head basis, the S1 and 2HEP were enzymatically indistinguishable (Tables S1 and S2). As with the 15HEP construct, the longer-tailed 25HEP construct displayed a reduced actin-activated ATPase activity compared to either the S1 or short-tailed 2HEP (Fig. 3 d, Table 1, and Table S1), consistent with its ability to form the auto-inhibited, IHM/SRX state.
Table 1.
Steady-state actin-activated ATPase measurements
| Construct |
v0 (s−1) (± SD) |
ATPase @ 60 µM actin (s−1) (± SD) |
KATPase (µM) (± SE) |
kcat (s−1) (± SE) |
|---|---|---|---|---|
| WT S1, n = 3 | 0.02 ± 0.01 | 3.8 ± 0.2 | 26.5 ± 5.6 | 5.7 ± 0.5 |
| WT 2HEP, n = 4 | 0.03 ± 0.03 | 3.4 ± 1.0 | 23.4 ± 11.5 | 4.7 ± 1.0 |
| WT 15HEP, n = 5 | 0.02 ± 0.02 | 0.4 ± 0.1 | 99.9 ± 64.5 | 1.1 ± 0.5 |
| WT 25HEP, n = 4 | 0.03 ± 0.02 | 0.6 ± 0.3 | ND | ND |
| E525K S1, n = 3 | 0.04 ± 0.01 | 9.9 ± 1.4** | 1.5 ± 0.6* | 11.3 ± 0.5** |
| E525K 2HEP, n = 3 | 0.03 ± 0.04 | 9.4 ± 0.3** | 2.2 ± 0.8 | 9.5 ± 0.5* |
| E525K 15HEP, n = 3 | 0.01 ± 0.02 | 0.2 ± 0.1* | 53.3 ± 20.8 | 0.4 ± 0.1 |
| E525K 25HEP, n = 4 | 0.02 ± 0.02 | 0.7 ± 0.2 | ND | ND |
WT single mantATP turnover assays to measure the myosin SRX state population
The single mantATP turnover assay is a powerful technique to understand the kinetics of a single round of ATP hydrolysis by a given enzyme. In the case of β-cardiac myosin in the absence of actin, the single ATP turnover assay has been particularly useful in examining the fraction of myosin molecules that populate the SRX state, as it hydrolyzes ATP 5–10-fold slower than the uninhibited myosin in the DRX state. Therefore, we compared single mantATP turnover results to determine the SRX fraction in each of our WT and E525K β-cardiac myosin constructs (S1, 2HEP, 15HEP, 25HEP; Fig. 4 and Fig. S1). The fluorescence decays were best fit to a double-exponential function, and in most cases, we found a significant improvement in the residuals of the fit when comparing the double- to the single-exponential fits (Fig. S1).
Figure 4.
Assessment of IHM/SRX state fraction via single mantATP turnover. (a–d) Single turnover of mantATP by β-cardiac myosin constructs was analyzed at different KCl concentrations to deduce the fraction of myosin heads in the IHM/SRX state. WT and E525K constructs at 0.25 µM were preincubated with mantATP (1 µM) for ∼30 s before introducing saturating ATP (2 mM). The fluorescence transients were fitted to a two-exponential decay function to infer the SRX state’s fraction and kinetics. Notably, long-tailed (15 and 25HEP; c and d) constructs displayed a greater SRX fraction than S1 or 2HEP (a and b), which was sensitive to salt concentration only in the WT constructs with significant differences identified by asterisks (*) (15HEP, P < 0.005; 25HEP, P < 0.05). The mean ± SD of three experiments from separate protein preparations is reported in panels a, b, and d. Data in c originally published in Rasicci et al. (2022). Comparison P values provided in Tables S3 and S4.
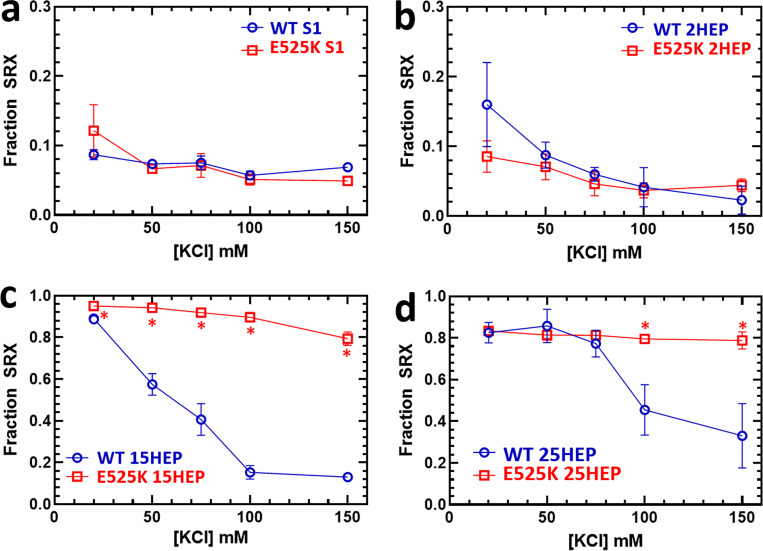
Single-headed WT S1 constructs exhibit an inherently low percentage of SRX heads regardless of the ionic strength (Fig. 4 a; and Tables 2 and 3). Conversely, our previously published results showed that the double-headed, long-tailed WT 15HEP construct populated the SRX state in an ionic strength-dependent manner, with nearly 90% SRX myosin heads at low salt (20 mM KCl) and 10% SRX at high salt (150 mM KCl) (Fig. 4 c and Tables 2 and 3; Rasicci et al., 2022). These data provide evidence that the reduced steady-state actin-activated ATPase for the 15HEP described above at low salt (Fig. 3 c and Table 1) reflects electrostatic intramolecular interactions between the heads and long coiled-coil tail stabilizing the auto-inhibited IHM/SRX state. The tail length is critical to this inhibition, as the WT 2HEP construct showed a small SRX fraction (2–16%) at all KCl concentrations similar to the WT S1 (Fig. 4 b and Tables 2 and 3). Interestingly, the longer-tailed WT 25 HEP construct demonstrated a high SRX content, but with a less pronounced ionic strength-dependence compared to the WT 15HEP, suggesting that the additional 10 heptads of coiled-coil further stabilize the auto-inhibited state (Fig. 4 d, Tables 2 and 3, and Tables S3 and S4).
Table 2.
Single mantATP turnover measurements for WT and E525K constructs (n = 3 ± SD)
| KCl (mM) | DRX rate (s−1) | SRX rate (s−1) | SRX fraction (%) |
|---|---|---|---|
| WT S1 | |||
| 20 | 0.034 ± 0.002 | 0.0020 ± 0.0002 | 8.7 ± 0.7 |
| 50 | 0.034 ± 0.001 | 0.0029 ± 0.0007 | 7.4 ± 0.3 |
| 75 | 0.030 ± 0.001 | 0.0016 ± 0.0001 | 7.5 ± 1.0 |
| 100 | 0.028 ± 0.002 | 0.0029 ± 0.0012 | 5.7 ± 0.6 |
| 150 | 0.025 ± 0.001 | 0.0028 ± 0.0008 | 6.8 ± 0.3 |
| E525K S1 | |||
| 20 | 0.072 ± 0.005 | 0.0019 ± 0.0005 | 12.1 ± 3.8 |
| 50 | 0.064 ± 0.003 | 0.0023 ± 0.0003 | 6.6 ± 0.3 |
| 75 | 0.063 ± 0.005 | 0.0017 ± 0.0004 | 7.1 ± 1.7 |
| 100 | 0.064 ± 0.006 | 0.0023 ± 0.0008 | 5.1 ± 0.8 |
| 150 | 0.061 ± 0.001 | 0.0024 ± 0.0005 | 4.9 ± 0.2** |
| KCl (mM) | DRX rate (s −1 ) | SRX rate (s −1 ) | SRX fraction (%) |
| WT 2HEP | |||
| 20 | 0.022 ± 0.001 | 0.0066 ± 0.0017 | 16.0 ± 6.0 |
| 50 | 0.025 ± 0.001 | 0.0053 ± 0.0006 | 8.7 ± 1.9 |
| 75 | 0.028 ± 0.001 | 0.0047 ± 0.0031 | 6.0 ± 1.0 |
| 100 | 0.030 ± 0.001 | 0.0047 ± 0.0027 | 4.1 ± 2.8 |
| 150 | 0.031 ± 0.002 | 0.0038 ± 0.0036 | 2.3 ± 2.0 |
| E525K 2HEP | |||
| 20 | 0.045 ± 0.002 | 0.0033 ± 0.0005 | 8.5 ± 2.3 |
| 50 | 0.059 ± 0.002 | 0.0073 ± 0.0030 | 7.1 ± 1.9 |
| 75 | 0.063 ± 0.003 | 0.0052 ± 0.0013 | 4.6 ± 1.7 |
| 100 | 0.062 ± 0.001 | 0.0040 ± 0.0018 | 3.7 ± 1.1 |
| 150 | 0.068 ± 0.001 | 0.0042 ± 0.0046 | 4.4 ± 0.9 |
| KCl (mM) | DRX rate (s −1 ) | SRX rate (s −1 ) | SRX fraction (%) |
| WT 15HEP | |||
| 20 | 0.014 ± 0.002 | 0.0033 ± 0.0001 | 89.1 ± 0.8 |
| 50 | 0.013 ± 0.001 | 0.0054 ± 0.0005 | 57.4 ± 5.2 |
| 75 | 0.015 ± 0.002 | 0.0069 ± 0.0009 | 40.6 ± 7.6 |
| 100 | 0.016 ± 0.001 | 0.0058 ± 0.0004 | 15.3 ± 3.3 |
| 150 | 0.018 ± 0.001 | 0.0071 ± 0.0008 | 13.0 ± 1.7 |
| E525K 15HEP | |||
| 20 | 0.027 ± 0.008 | 0.0022 ± 0.0002 | 95.0 ± 0.7** |
| 50 | 0.019 ± 0.002 | 0.0026 ± 0.0001 | 94.2 ± 0.7** |
| 75 | 0.016 ± 0.006 | 0.0030 ± 0.0001 | 91.8 ± 1.6** |
| 100 | 0.014 ± 0.003 | 0.0033 ± 0.0001 | 89.6 ± 2.3** |
| 150 | 0.013 ± 0.002 | 0.0042 ± 0.0001 | 79.4 ± 3.2** |
| KCl (mM) | DRX rate (s −1 ) | SRX rate (s −1 ) | SRX fraction (%) |
| WT 25HEP | |||
| 20 | 0.036 ± 0.009 | 0.0030 ± 0.0001 | 82.5 ± 5.0 |
| 50 | 0.033 ± 0.016 | 0.0039 ± 0.0004 | 85.8 ± 8.0 |
| 75 | 0.025 ± 0.012 | 0.0039 ± 0.0004 | 77.3 ± 6.4 |
| 100 | 0.011 ± 0.003 | 0.0029 ± 0.0005 | 45.4 ± 12.1 |
| 150 | 0.010 ± 0.001 | 0.0032 ± 0.0008 | 33.1 ± 15.4 |
| E525K 25HEP | |||
| 20 | 0.029 ± 0.003 | 0.0030 ± 0.0001 | 83.3 ± 0.2 |
| 50 | 0.027 ± 0.006 | 0.0034 ± 0.0002 | 81.4 ± 0.2 |
| 75 | 0.027 ± 0.004 | 0.0041 ± 0.0006 | 81.3 ± 1.4 |
| 100 | 0.034 ± 0.003 | 0.0048 ± 0.0002 | 79.5 ± 1.4* |
| 150 | 0.039 ± 0.011 | 0.0061 ± 0.0002 | 78.8 ± 4.0* |
Table 3.
Comparison of SRX percentages for all β-cardiac myosin constructs at varying salt concentrations
| KCl | WT S1 | WT 2HEP | WT 15HEP | WT 25HEP |
|---|---|---|---|---|
| 20 | 8.7 ± 0.7 | 16.0 ± 6.0 | 89.1 ± 0.8** | 82.5 ± 5.0** |
| 50 | 7.4 ± 0.3 | 8.7 ± 1.9 | 57.4 ± 5.2** | 85.8 ± 8.0**^^ |
| 75 | 7.5 ± 1.0 | 6.0 ± 1.0 | 40.6 ± 7.6** | 77.3 ± 6.4**^^ |
| 100 | 5.7 ± 0.6 | 4.1 ± 2.8 | 15.3 ± 3.3 | 45.4 ± 12.1**^^ |
| 150 | 6.8 ± 0.3 | 2.3 ± 2.0 | 13.0 ± 1.7 | 33.1 ± 15.4**^^ |
| KCl | E525K S1 | E525K 2HEP | E525K 15 HEP | E525K 25HEP |
| 20 | 12.1 ± 3.8 | 8.5 ± 2.3 | 95.0 ± 0.7** | 83.3 ± 0.2**^^ |
| 50 | 6.6 ± 0.3 | 7.1 ± 1.9 | 94.2 ± 0.7** | 81.4 ± 0.2**^^ |
| 75 | 7.1 ± 1.7 | 4.6 ± 1.7 | 91.8 ± 1.6** | 81.3 ± 1.4**^^ |
| 100 | 5.1 ± 0.8 | 3.7 ± 1.1 | 89.6 ± 2.3** | 79.5 ± 1.4**^^ |
| 150 | 4.9 ± 0.2 | 4.4 ± 0.9 | 79.4 ± 3.2** | 78.8 ± 4.0** |
Defining myosin surface occupancy in the motility assay
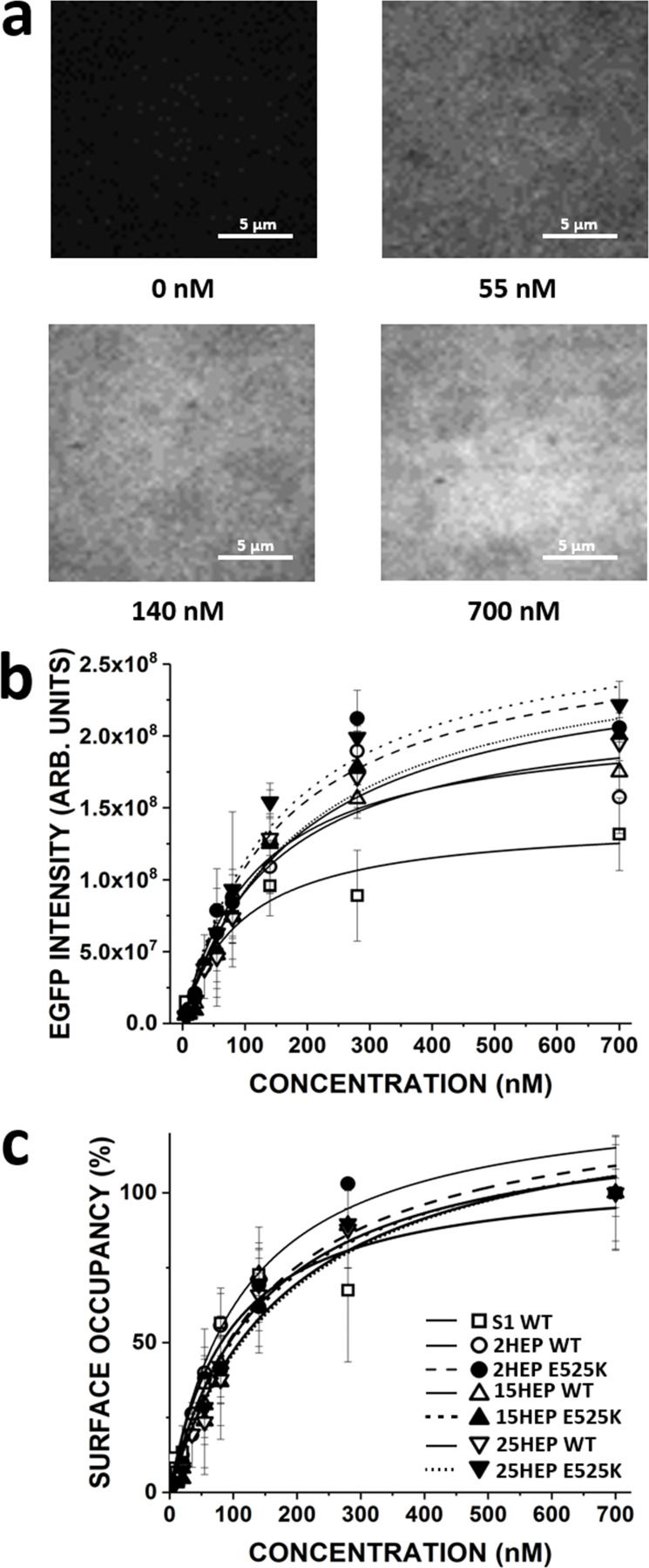
Knowing that the various myosin constructs can adopt an IHM/SRX state, do these auto-inhibited motors impact in vitro motility? If so, then it is critical that the total motor number on the motility surface be estimated, which would include both the active and auto-inhibited (IHM/SRX) motors. Therefore, the C-terminal EGFP tag on each construct served both as a fluorescence readout for motor counting, as well as an attachment strategy to the motility surface via an anti-GFP nanobody (see Materials and methods). All WT and E525K mutant constructs were introduced into the flow-cell at various concentrations (0–700 nM). With increasing myosin concentration, the overall fluorescence intensity increased (Fig. 5 a) and was well fit with a hyperbolic saturation binding model (Fig. 5 b). The apparent surface affinity as characterized by an averaged Kapp of 147 ± 33 nM for the WT double-headed constructs (2HEP, 15HEP, and 25HEP) was not different than the corresponding E525K mutants with an averaged Kapp of 165 ± 22 nM, suggesting that neither the tail length, nor E525K mutation impact the binding to the nanobody (P = 0.99). As expected, given that S1 has a single EGFP tag, the absolute fluorescence was lower at all WT S1 concentrations when compared to the double-headed WT 2HEP (Fig. 5 b). Accordingly, WT S1 surface binding showed a lower Kapp of 93 ± 27 nM.
Figure 5.
β-cardiac myosin construct motility assay surface occupancy estimation. (a) β-cardiac myosin constructs were attached to the motility assay glass surface via their C-terminal EGFP to preadhered anti-GFP nanobodies. Increasing the applied myosin concentration increased surface EGFP fluorescence intensity. (b) Surface EGFP fluorescence intensity increased as a function of β-cardiac myosin construct concentration infused into motility assay flow-cell. Fluorescence intensity followed saturable binding behavior with an apparent dissociation constant, Kapp (nM): KappS1 WT = 93 ± 27, Kapp2HEP WT = 116 ± 42, Kapp15HEP WT = 143 ± 32, Kapp25HEP WT = 181 ± 36, Kapp2HEP E525K = 152 ± 38, Kapp15HEP E525K = 191 ± 45, and Kapp25HEP E525K = 153 ± 34. No difference in the surface EGPF intensity across WT and E525K mutants (P = 0.98). (c) Data in b normalized to fluorescence at 700 nM β-cardiac myosin concentration and converted to surface occupancy. Data are mean ± SD of at least three experiments from separate protein preparations.
Once defined, each surface fluorescence curve was normalized to the fluorescence intensity at the 700 nM construct loading concentration and converted to percent surface occupancy (Fig. 5 c). This allowed in vitro motility data across all constructs to be compared as a function of myosin surface occupancy regardless of the functional capacity of the construct (i.e., active versus auto-inhibited IHM/SRX).
WT myosin in vitro motility
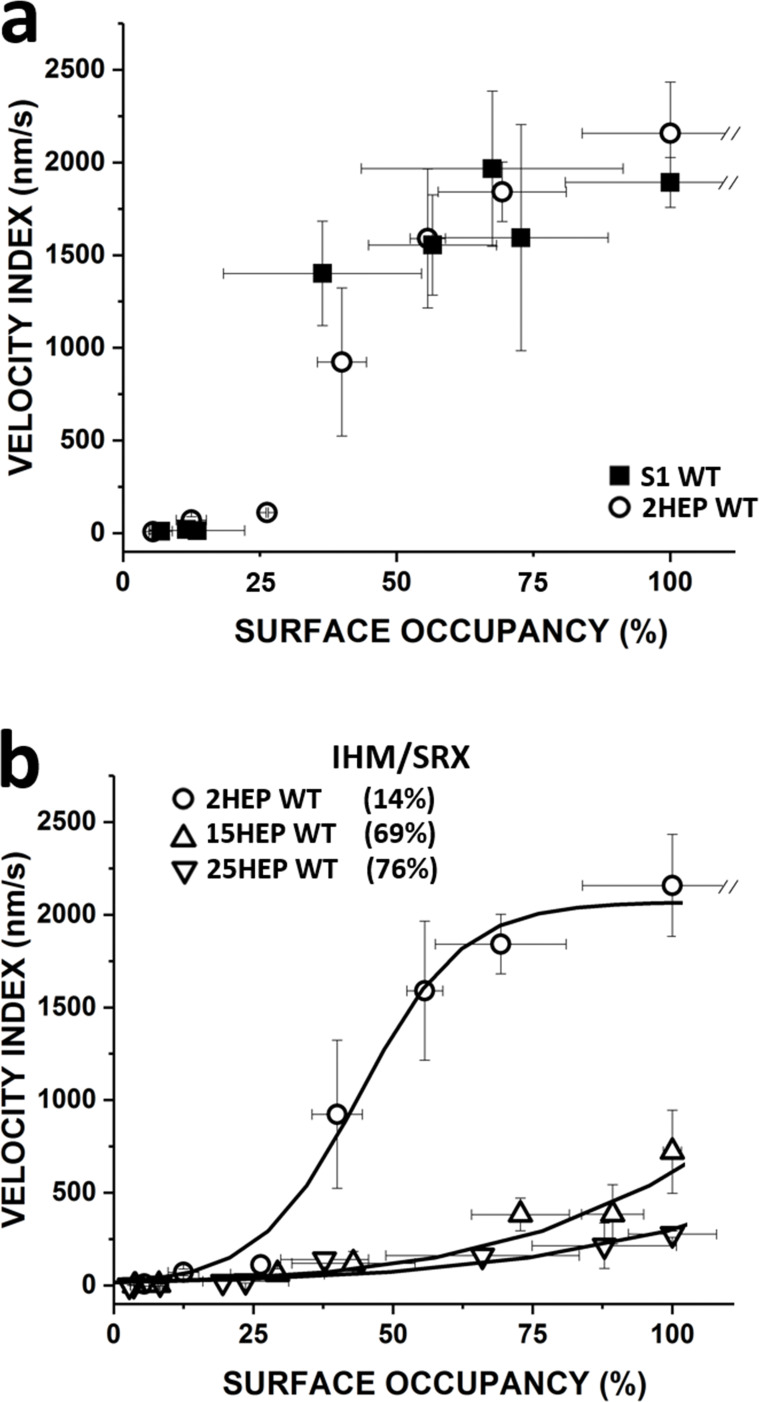
The in vitro motility for the various WT constructs was characterized by a Velocity Index (see Materials and methods, Fig. 6), which is the product of the motile fraction of filaments and their velocity, providing a better metric of activity for very low densities of myosin. For limiting densities of low duty ratio (∼5%) motors, such as muscle myosin (Harris and Warshaw, 1993), actin velocity is limited by the rate of cross-bridge attachment. Thus, actin velocity increases with the number of motors interacting with the actin filament (Fig. S3 a), saturating when at least one motor contributes to actin filament movement at any point in time. However, if additional motors simultaneously interact with the actin filament, no further increase in velocity is observed (Uyeda et al., 1990). Therefore, if changes in the IHM/SRX state occur due either to the construct tail length or E525K mutation, which could alter the effective number of available motors, then comparing maximum velocities alone would be an insensitive measure. Therefore, changes in Velocity Index across a range of myosin surface occupancies will allow effective alterations in motor number due to construct differences or mutation to be identified.
Figure 6.
Motility analysis of WT human β-cardiac myosin constructs. (a) The Velocity Index versus surface occupancy for WT S1 and 2HEP constructs showed a dose–response-like characteristic that saturated at ∼2,000 nm/s and were indistinguishable (P = 0.87). (b) The Velocity Index versus surface occupancy for WT 2HEP, 15HEP, and 25HEP constructs. The longer-tailed WT 15HEP and 25HEP constructs had indistinguishable Velocity Indices (P = 0.87), but slower at each myosin surface occupancy compared to WT 2HEP. The WT 2HEP Velocity Index versus surface occupancy data were fitted to a Hill dose–response relationship (solid curve) and then used as the basis for an analytical model (see Results) to predict the IHM/SRX content for each construct. Predicted IHM/SRX percentages (legend in figure) and associated fitted curve (solid curve) are shown. All in vitro motility experiments were performed in low salt (25 mM KCl). Data points are presented as mean ± SD of at least three experiments from separate protein preparations.
The Velocity Index relationships for the WT S1 and 2HEP constructs showed a dose–response-like characteristic that saturated at ∼2,000 nm/s (Fig. 6 a) and were indistinguishable (P = 0.87). The increasing Velocity Index with myosin surface occupancy suggests that motor number was rate limiting, which is supported by the fraction moving filaments being low at the low myosin surface occupancies (see Fig. S3 b). Non-motile filaments were stationary and did not exhibit reptation (Videos 1, 2, 3, and 4), a back and forth, diffusive-like motion commonly observed on sparsely coated myosin surfaces (Harris and Warshaw, 1993; Uyeda et al., 1990). The filaments being stationary suggest that a non-specific, motility surface-filament interaction must exist to keep the filament close to the surface and that filaments must engage active myosin heads for directed motion to occur. The similarity in the Velocity Indices for the WT S1 and 2HEP constructs may reflect the similarity in their steady-state actin-activated and single turnover ATPase measurements (Fig. 3 and Fig. 4). Although the longer-tailed WT 15HEP and 25HEP constructs had Velocity Indices which were not different from each other (P = 0.87) (Fig. 6 b), their Velocity Indices were slower at each myosin surface occupancy compared to the WT S1 and 2HEP constructs, being >60% slower at 100% myosin surface occupancy (Fig. 6, a and b).
Modeling the effect of the IHM/SRX on in vitro motility
With reductions in the actin-activated ATPase activities of the long-tailed WT 15HEP and 25HEP (Fig. 3, c and d) compared to the WT S1 and 2HEP (Fig. 3, a and b) constructs being attributed to the presence of IHM/SRX heads (Fig. 4), we developed a simple analytical model whereby the presence of IHM/SRX myosin for the long-tailed constructs accounts for a reduction in the Velocity Index as a function of myosin surface occupancy (Fig. 6). The model (see Supplemental text at the end of the PDF for model fitting details) was based on the following assumptions.
-
1
The myosin surface occupancy reflects the total number of myosin motors on the surface (Fig. 5).
-
2
An IHM/SRX motor does not generate or impede actin movement, i.e., it is mechanically silent, thus effectively reducing the number of active heads on the motility surface.
-
3
Each construct’s proportion of active and IHM/SRX is maintained at all myosin surface occupancies.
-
4
The Velocity Index versus myosin surface occupancy of the WT 2HEP construct (Fig. 6 a) reflects a population of motors having 84% active and 16% IHM/SRX heads (Fig. 4 b and Table 2).
-
5
As they share the same motor domain, the maximum theoretical Velocity Index for all constructs is that experimentally observed for saturating WT 2HEP (2,300 nm/s) (Fig. 6).
Based on these assumptions, the WT 2HEP Velocity Index data were fitted to a modified Hill dose–response relationship and then used as a baseline curve for a construct that has 16% IHM/SRX content (Fig. 4 b, Fig. 6 b, and Table 2). Using this same curve, a family of curves was generated with the only free parameter being the percent IHM/SRX, which was varied between 0 and 100% IHM/SRX (Supplemental text and Fig. S4 b). Using this family of curves, the best least squared error fits to the WT 2HEP, 15HEP, and 25HEP Velocity Index data predict IHM/SRX percentages of 14 ± 13%, 69 ± 6%, and 76 ± 4%, respectively (Fig. 6 b, solid curves; Supplemental text; and Fig. S4 for fitting and estimate error determination), in close agreement with the results from single turnover experiments (Fig. 4).
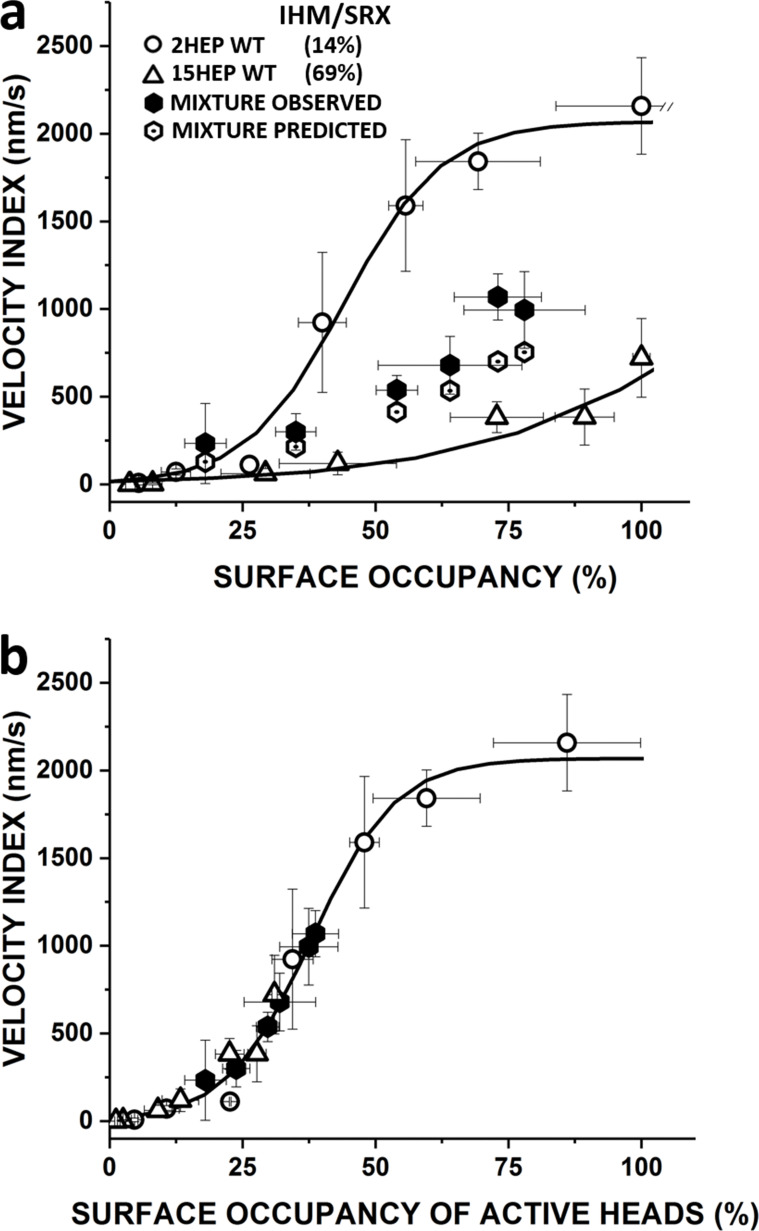
To test the predictive nature of our model and its underlying assumptions, most importantly that for each construct, the proportion of IHM/SRX content does not change with protein concentration, we performed a mixtures motility assay by adhering a constant 25 nM WT 2HEP (14% IHM/SRX) with an additional 0–125 nM WT 15HEP (69% IHM/SRX). The resultant Velocity Index versus myosin surface occupancy for these mixtures was midway between the 2HEP and 15HEP curves (Fig. 7 a). However, replotting these data as a function of the surface occupancy of active (DRX) myosin (the product of myosin surface occupancy and predicted percent DRX {i.e., 100% − [predicted %IHM/SRX content]}, Fig. 7 b) demonstrates that the same relationship determines the velocity profiles for both the 2HEP and 15HEP constructs, as well as mixtures of the two. This suggests that the Velocity Index reflects the actin filament motility generated by active (DRX) motors only, regardless of tail length and that IHM/SRX motors are mechanically silent, offering no resistance or internal load to the active motor population.
Figure 7.
Influence of IHM/SRX content on motility of WT 2HEP:15HEP mixture. (a) The Velocity Index versus surface occupancy of WT 2HEP, 15HEP, and a mixture of these two constructs. Addition of increasing amounts (0, 25, 50, 75, 100, and 125 nM) of 15HEP WT (76% IHM/SRX) to a constant amount (25 nM) of 2HEP WT (14% IHM/SRX) results in increasing Velocity Index values (filled octagons) that compare favorably to predicted values (open octagons; see Table S1), supporting the model’s predictive nature, which assumes that a given construct has a defined IHM/SRX content and that the Velocity Index reflects actin filament motility generated by only active motors. (b) Data in a were transformed to Velocity Index versus surface occupancy of active heads by replotting the Velocity Index data in a after calculating the surface occupancy of active (DRX) myosin heads from the product of observed total myosin surface occupancy and predicted percent DRX (i.e., 100% − [predicted %IHM/SRX content]). The solid curve represents the 0% IHM/SRX curve (Fig. S4). The 2HEP (open circles), 15HEP (open triangles), and observed mixture (closed hexagons) data, once transformed to represent only active motors, are well fit by the 0% IHM/SRX curve. Data points represent mean values ± SD of three experiments from separate protein preparations.
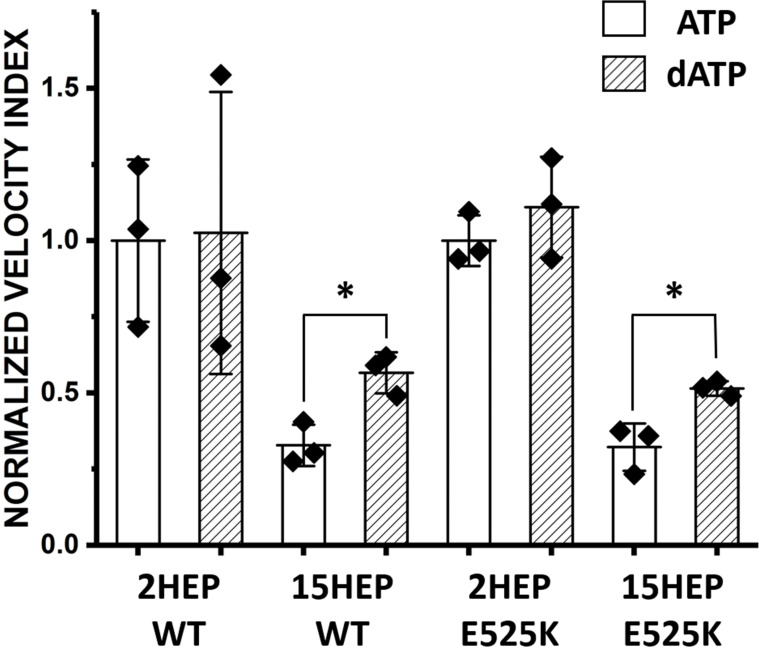
dATP reduces the IHM/SRX population
deoxy ATP (dATP) has been shown to shift myosin out of the IHM/SRX state, while still supporting myosin’s biochemical and mechanical functions (Walklate et al., 2022). Therefore, dATP should increase the Velocity Index for constructs with significant IHM/SRX content. To test this, dATP was exchanged for ATP in the motility assay with the WT 2HEP and 15HEP constructs at 100% myosin surface occupancy. Fig. 8 shows no significant change in the WT 2HEP Velocity Index in the presence of dATP compared to that with ATP, (P = 0.94). In contrast, dATP increased the WT 15HEP Velocity Index by ∼50% (P = 0.01), which our modeling would predict is due to a ∼10% reduction in IHM/SRX (Fig. 7 b). These results support the biochemical and motility data that suggest a low IHM/SRX population in the WT 2HEP construct, whereas dATP apparently reduces, but does not eliminate the high IHM/SRX population in the WT 15HEP construct.
Figure 8.
Deoxy-ATP (dATP) impact on IHM/SRX motors. The SRX inhibitor, dATP, was exchanged for ATP in the motility assay with the WT and E525K 2HEP and 15HEP constructs at 100% myosin surface occupancy. Data are normalized to the respective (i.e., WT and E525K) 2HEP mean Velocity Index data in the presence of ATP. * identifies significantly different than control (P = 0.01). Data points represent mean values ± SD of three experiments from separate protein preparations.
Impact of E525K mutation on ATPase activity and in vitro motility
Steady-state and single turnover ATPase
Introduction of the E525K mutation in both the S1 and 2HEP constructs enhanced the steady-state, actin-activated ATPase activity ∼3-fold as well as dramatically decreasing by ∼18-fold the actin concentration at which ATPase activity is one-half maximal (KATPase) (Fig. 3, a and b; and Table 1), as reported previously (Rasicci et al., 2022) for the S1. The lowest actin concentration measured (5 µM) was higher than the KATPase determined for E525K S1 and 2HEP, which may have reduced the precision of this value. However, both the E525K 15HEP and 25HEP actin-activated ATPase activity appeared insensitive to the mutation compared to WT and continued to show reduced actin-activated ATPase activity compared to the E525K S1 and 2HEP constructs (Fig. 3 and Table 1).
As for single ATP turnover measurements that were used to assess a construct’s fraction of SRX, the E525K mutation did not alter the S1 and 2HEP SRX percentage, which remained small (5–10%) across all KCl concentrations (Fig. 4, a and b; Tables 2 and 3). The SRX rate constants were unchanged in the E525K S1 and 2HEP constructs, whereas the DRX rate constant was accelerated approximately twofold by the mutation at almost all KCl concentrations (Table 2). Compared to WT, the E525K mutation in both the 15HEP and 25HEP constructs stabilized the SRX state against the changes in ionic strength (Fig. 4, c and d; Tables 2 and 3). Notably, in the E525K 25HEP construct, the SRX rate constants were universally decreased approximately twofold, whereas the DRX rate constants were primarily unchanged, except at the higher salt concentrations (100 and 150 mM KCl) where they were accelerated approximately threefold (Table 2).
In vitro motility
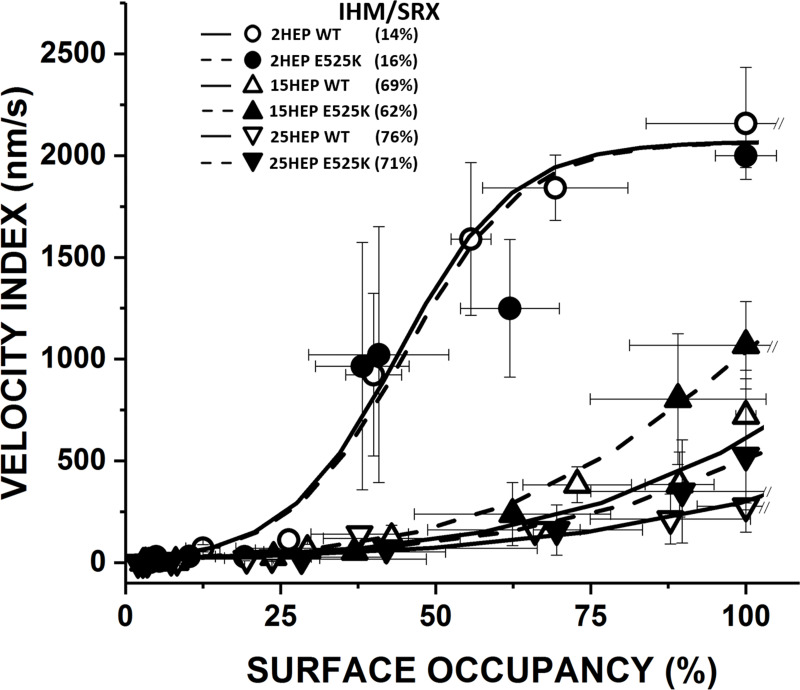
The Velocity Index versus myosin surface occupancy relationship for the E525K short- (2HEP) and long-tailed (15HEP and 25HEP) constructs was compared to WT relations (Fig. 9). All three E525K constructs were not significantly different from their WT counterparts (2HEP, P = 0.75; 15HEP, P = 0.75; 25HEP, and P = 0.87). Using our model to estimate the %IHM/SRX for the various E525K constructs (2HEP, 15HEP, 25HEP) (see above), the least squared error fits to the Velocity Index versus myosin surface occupancy relations (Fig. 9, dashed curves) suggest IHM/SRX percentages of 16 ± 12% (E525K 2HEP), 62 ± 6% (E525K 15HEP), and 71 ± 4% (E525K 25HEP). Given the error in the predicted %IHM/SRX estimate for the various model fits (Fig. S4), the predicted %IHM/SRX for all of the E525K mutants are not different than WT. Additionally, the lack of difference between WT and E525K 2HEP constructs, which do not contain a large IHM/SRX population (Fig. 4 b), suggests that the mutation does not impact the motor’s inherent motion-generating capacity under the unloaded conditions in the motility assay. In addition, the E525K mutation does not appear to further stabilize or alter the long-tailed 15HEP and 25HEP ability to adopt the IHM/SRX state at least for the low salt conditions used in the motility assay.
Figure 9.
Impact of the E525K mutation on β-cardiac myosin motility. Velocity Index versus surface occupancy for WT and E525K 2HEP, 15HEP, and 25HEP constructs. WT data and fitted curves from Fig. 6 b. All three E525K constructs were not significantly different than their WT controls (2HEP, P = 0.75; 15HEP, P = 0.75; 25HEP, P = 0.87). Dashed curves are the least squared error model fits to the Velocity Index versus surface occupancy relations in order to estimate the %IHM/SRX for the various E525K constructs (see legend in figure). The predicted %IHM/SRX for all of the E525K mutants are no different than WT. All in vitro motility experiments performed in low salt (25 mM KCl). Data points are presented as mean ± SD of at least three experiments from separate protein preparations.
As with the WT constructs, we exchanged dATP for ATP in the motility assay for the E525K constructs to determine whether in contrast to the WT 15HEP construct, the mutation may prevent the transition out of the IHM/SRX state due to the presence of dATP. As might be expected for the E525K 2HEP construct due to its low IHM/SRX percentage (Fig. 4 b; and Tables 2 and 3), dATP had no significant effect on the normalized Velocity Index (P = 0.38) (Fig. 8). In contrast, the presence of dATP produced an ∼40% increase in the normalized Velocity Index for the E525K 15HEP construct (P = 0.01) (Fig. 8), suggesting that additional motors are recruited out of the IHM/SRX state. Notably, the similar increase in Velocity Index for both the WT and the E525K 15HEP due to dATP further suggests that the E525K mutation does not enhance the stability of the IHM/SRX state in the 15HEP construct at least under the low ionic strength conditions of the motility assay.
Discussion
HCM and DCM are considered diseases of the sarcomere, as these myopathies can involve mutations in β-cardiac myosin and other sarcomeric proteins critical to muscle function (Spudich, 2014; Barefield et al., 2023; Lehman et al., 2022; Alamo et al., 2017). Although cardiac contractility in HCM and DCM patients differs dramatically, i.e., hyper-contractile (Tuohy et al., 2020) and hypo-contractile (Reichart et al., 2019), respectively, the underlying myofilament-based mechanisms that dictate these contractile differences may be common. Specifically, cardiac muscle power generation is related to the number of force-generating myosin motors recruited upon activation and each motor’s intrinsic force and motion generation while attached to the actin-thin filament (Fig. 1). Therefore, HCM- and DCM-associated mutations to myosin could impact either or both of these critical processes, with evidence in the literature for both (Spudich, 2014; Kawana et al., 2022). Garnering much attention is the SRX, auto-inhibited myosin state that can be stabilized by intramolecular interactions between myosin’s two heads and its coiled-coil tail domain (i.e., IHM) (Chu et al, 2021; Gollapudi et al, 2021b; Rasicci et al, 2022; Nelson et al, 2023). Therefore, the proportion of myosin that adopt this auto-inhibited IHM/SRX state effectively determines the number of available force-generating myosin heads upon activation (Fig. 1). In fact, HCM-associated myosin mutations appear to disrupt the IHM/SRX state, increasing myosin head availability upon cardiac activation, which may explain the observed hyper-contractility (Kawana et al., 2022). In contrast to HCM, DCM-associated β-cardiac myosin mutations may further stabilize the IHM/SRX state, thus reducing the number of functionally available myosin heads, which would lead to cardiac hypo-contractility (Fig. 1 d). In fact, we previously reported that the E525K DCM mutation, in expressed human β-cardiac myosin with a 15HEP, did stabilize the IHM structural state and thus the SRX biochemical state as well (Rasicci et al., 2022). Here, we extend these studies in both WT and E525K DCM mutant human β-cardiac myosin to better define the extent that the IHM/SRX state is dependent on the myosin tail length, and how the IHM/SRX state impacts both myosin’s ATPase activity and molecular mechanical function with and without the E525K mutation.
Here we show, as have others (Anderson et al., 2018; Grinzato et al., 2023), that myosin’s ability to adopt the IHM/SRX state, based on steady-state actomyosin ATPase and single ATP turnover experiments, requires both heads and at least 15 heptads of tail length. This tail length was originally reported as the minimum for IHM formation in smooth muscle myosin (Trybus et al., 1997), and subsequently in cardiac myosin (Rasicci et al, 2022). Therefore, as expected, a double-headed β-cardiac myosin construct with a short 2HEP tail is enzymatically indistinguishable from a single-headed myosin S1 construct, which is incapable of adopting the IHM state (Fig. 3, a and b), whereas both the WT 15HEP and longer-tailed 25HEP constructs, which can adopt the IHM/SRX state, exhibit significantly reduced steady-state actomyosin ATPase activity compared to WT S1 and 2HEP (Fig. 3 and Table 1). The IHM/SRX state in the long-tailed constructs is stabilized by intramolecular electrostatic interactions (Spudich, 2014; Anderson et al., 2018; Rasicci et al., 2022; Woodhead et al., 2005), as evidenced by a reduced fraction of myosin in the IHM/SRX state with increasing ionic strength (i.e., 20–150 mM KCl) (Fig. 4, c and d). Although the WT 25HEP construct was less sensitive to ionic strength (Fig. 4 d), suggesting that increased tail length and presumably additional intramolecular interactions lead to a more stable IHM/SRX state in solution (Gollapudi et al, 2021a).
Myosin is a mechanoenzyme, having both enzymatic and mechanical activities. Although these dual activities are generally correlated, the rate-limiting steps for the enzymatic and mechanical cycles differ (Huxley, 1990; Tyska and Warshaw, 2002). Therefore, the in vitro motility assay has become a standard for characterizing the motor’s motion and force-generating capabilities (Palmiter et al., 1999; Huxley, 1990; Kron et al., 1991; Warshaw et al., 1990), which we used here to define how the β-cardiac myosin tail impacts myosin mechanical function. Specifically, the actin filament Velocity Index versus myosin surface occupancy relation in the motility assay should reflect both myosin’s intrinsic motion generation and the number of motors available to interact and propel the actin filament, both of which might be altered by a DCM mutation. The nonlinear, Velocity Index dependence on surface occupancy (i.e., motor number) (Fig. 6) is due to β-cardiac myosin being a low duty ratio motor (Tyska and Warshaw, 2002), i.e., spending a small fraction of its enzymatic cycle attached to actin and generating motion. If the IHM/SRX state dictates the number of available motors, then the shape of the Velocity Index versus myosin surface occupancy relation should be sensitive to the IHM/SRX state stability. Therefore, we took advantage of the β-cardiac myosin construct C-terminal, EGFP tag both as a site-specific attachment strategy to the motility surface and as a fluorescence indicator of the total number of motors on the motility surface (i.e., active and inactive [IHM/SRX]) at any myosin concentration. Using an anti-GFP nanobody attachment to the surface, myosin heads are positioned to interact properly with actin without surface interference (see Fig. S2) and are free to transition between the inactive IHM/SRX and active DRX states.
Based on each construct’s EGFP fluorescence, we confirmed that the myosin surface occupancy (i.e., total number of attached motors) was the same for any double-headed construct (Fig. 5), which if not the case would have confounded the interpretation of Velocity Index differences. Interestingly, the maximum Velocity Indexes for the WT 15HEP and 25HEP constructs were threefold slower than the WT single-headed S1 and double-headed, short 2HEP construct (Fig. 6). Based on the enzymatic characterization, these same long-tailed constructs (i.e., 15HEP and 25HEP) can adopt the IHM/SRX state (see Results). Therefore, is the reduced WT 15HEP and 25HEP Velocity Index due to a reduced number of available heads? In support of this scenario, substituting dATP, an SRX inhibitor (Walklate et al., 2022), for ATP in the motility assay, the WT 15HEP Velocity Index increased by ∼50% (Fig. 8), suggesting that dATP shifted the myosin state equilibrium out of the IHM/SRX state, effectively making more active myosin available on the motility surface.
With the Velocity Index versus myosin surface occupancy sensitive to the number of active myosin on the motility surface, we developed a simple analytical model (see Results and Supplemental text) to predict the proportion of WT 15HEP and 25HEP motors in the IHM/SRX state. For this model, we assumed, based on single ATP turnover experiments (Fig. 4 b), that the WT 2HEP Velocity Index versus myosin surface occupancy relation was that of a myosin population with 16% in the IHM/SRX state. Additionally, we assumed that IHM/SRX myosins are inactive and do not contribute to actin movement, thus effectively reducing the number of active heads at any myosin surface occupancy. Based on the model, the WT 15HEP and 25HEP constructs are predicted to have 69 and 76% IHM/SRX state probability, respectively (Fig. 6). These model estimates are in relatively good agreement with the ∼85% IHM/SRX proportion observed in the single ATP turnover experiments for these same constructs. (Fig. 4). In support of the model assumption that the IHM/SRX state is mechanically silent, thus effectively reducing the number of available motors, we conducted a mixture experiment using WT 2HEP and 15HEP in various ratios. This experiment allowed us to vary the proportion of IHM/SRX myosin on the motility surface, given the predicted 14 ± 13% and 69 ± 6% IHM/SRX state probability for the two constructs, respectively. The agreement between the observed and predicted Velocity Index versus myosin surface occupancy for the various WT myosin mixtures (Fig. 7) further supports a mechanism by which the IHM/SRX state effectively limits the number of active motors that contribute to cardiac contractility. In this mixture assay, the observed Velocity Index is determined solely by the surface occupancy of DRX myosin (Fig. 7 b), regardless of the myosin construct, or even mixtures of myosin constructs. Critically, in mixtures where the longer-tailed construct (i.e., 15HEP) contributes the majority of the DRX myosin, the Velocity Index still superimposes on that of the shorter-tailed construct (i.e., 2HEP), in spite of a much greater overall myosin occupancy, and hence, a much greater presence of IHM/SRX myosin (Fig. 7 b). This provides strong evidence that the presence of IHM/SRX myosin contributed by the 15HEP myosin to the mixture does not impose a significant drag or resistive load.
With both the WT myosins construct enzymatic and motile properties characterized, we then used the same experimental approaches to determine whether the E525K DCM mutation impacts these properties and specifically whether the mutation further stabilizes the IHM/SRX state, as suggested by our earlier study using only the S1 and 15HEP constructs. The E525K (Rasicci et al., 2022; Lakdawala et al., 2012) DCM-associated mutation is located in the “activation loop,” part of the actin-binding face of myosin’s lower 50 kD domain (Grinzato et al., 2023), where it is clustered with four additional DCM-linked myosin mutations (Grinzato et al., 2023). E525 is involved in electrostatic intramolecular interactions between the “blocked” head and the S2 domain of the “free” head that stabilizes the IHM state (Alamo et al., 2017; Woodhead and Craig, 2020). The charge reversal of the E525K mutation eliminates one of the few negatively charged residues within the “mesa trail” of the myosin head, a region that is already rich in positive charges (Rasicci et al., 2022). Studies designed to examine the impact of the E525K mutation on intrinsic motor function have proposed that the E525K has a direct impact on the allosteric mechanism of actin-activated phosphate release and lever arm rotation (Bodt et al., 2023, Preprint). Additionally, in the asymmetric structure of the IHM, E525K within the blocked head may instead form a new salt bridge with D900 of the free head myosin S2 (Bodt et al., 2023, Preprint). This new salt bridge may strengthen and thus further stabilize the folded IHM state.
Interestingly, as we reported previously (Rasicci et al., 2022), the E525K S1 showed approximately threefold enhanced actomyosin ATPase activity relative to WT (Fig. 3 a and Table 1). As expected, this enhanced ATPase activity was also observed in the E525K 2HEP (Fig. 3, a and b; and Table 1). However, no such ATPase enhancement was observed for the E525K 15HEP as reported previously (Rasicci et al., 2022), and similarly for the E525K 25HEP (Fig. 3, c and d; and Table 1). With the myosin heads being identical for all of the mutant constructs, the lack of any effect of the E525K mutation on the 15HEP and 25HEP ATPase activities suggests that these long-tailed construct’s ability to adopt the IHM/SRX state may effectively mask the enhanced ATPase activities of the individual mutated heads. With respect to in vitro motility, there were no statistical differences in the Velocity Index versus myosin surface occupancy relations for any of the double-headed E525K constructs compared to their WT controls (Fig. 9). The fact that the approximately threefold greater actomyosin ATPase activity for the E525K 2HEP did not translate into a faster Velocity Index may reflect differences in the rate-limiting steps for solution-based ATP hydrolysis versus surface-attached motion generation (Huxley, 1990; Tyska and Warshaw, 2002).
If no differences in enzymatic and motile activities are apparent in the E525K 15HEP and 25HEP constructs compared to WT, then how is the hypo-contractile DCM phenotype possible? Both the actomyosin ATPase and motility assay measurements are performed at low ionic strength (25 mM KCl) to maximize actomyosin interactions in these reductionist assays, whereas in vivo, the close proximity between the actin-thin and myosin-thick filaments within the muscle sarcomere makes their effective concentrations infinite and thus overcomes the electrostatic shielding effects of the high ionic strength in muscle. Based on the single ATP turnover data, at 20 mM KCl, the IHM/SRX fraction (∼85%) for the E525K 15HEP and 25HEP is unchanged compared to WT. If the IHM/SRX state is the dominant factor dictating the actomyosin ATPase activity and actin filament motility in vitro, then it’ is not surprising that the Velocity Indices were the same at low ionic strength for both E525K and WT long-tailed constructs. However, single ATP turnover measurements can be performed over a range of ionic strengths (Fig. 4 and Table 3) and interestingly, the IHM/SRX probabilities for both E525K 15HEP and 25HEP constructs remained high and were insensitive to ionic strength up to 150 mM KCl (Fig. 4 C). These data suggest that the E525K DCM-associated mutation does stabilize the IHM/SRX state. Therefore, if both the steady-state actomyosin ATPase activity and in vitro motility assay could be performed at physiological ionic strengths, where the WT IHM/SRX state is destabilized, one might expect significantly reduced enzymatic and motile activity from the DCM mutant myosin, which might explain the cardiac hypo-contractility in DCM patients. In the future, power generation, a more relevant physiological measure of cardiac muscle contractility, can be assessed at higher physiological ionic strengths in the optical trap (Finer et al., 1994; Debold et al., 2007; Guilford et al., 1997), whereby an actin filament can be brought into contact with a mutant myosin ensemble regardless of the ionic strength. By this approach, the true impact of DCM-associated mutations on human β-cardiac myosin can be defined.
Given myosin’s enzymatic and mechanical activities, which are each multi-step processes, there is no a priori reason to assume that every HCM or DCM mutation will impact these processes identically. Whether or not alterations in the IHM/SRX stability are the governing factor that dictates the overall biochemical or motile properties for every mutant myosin, which may be the case for the E525K mutation reported here, is yet to be established. DCM remains a formidable challenge due to its heterogeneous nature and the lack of a unifying molecular mechanism for monogenetic cases. Extending these findings to clinical settings and studying a broader spectrum of DCM mutations compared to E525K will hopefully provide more comprehensive insights into DCM’s underlying mechanisms.
Supplementary Material
shows the P values for comparisons of steady-state actin-activated ATPase measurements.
shows the P values for comparisons between WT and E525K actin-activated ATPase.
shows the P values for comparisons of SRX fraction as a function of salt concentration from single mantATP turnover measurements.
shows the P values for comparisons of SRX fraction comparing different constructs from single mantATP turnover measurements.
Acknowledgments
Henk L. Granzier served as editor.
The authors thank Guy Kennedy from the Instrumentation and Model Facility at the University of Vermont for his microscopy expertise, Samantha Beck-Previs for her technical expertise, and the entire Warshaw lab for critical discussions.
This work was supported by the funds from the American Heart Association (23PRE1013839 to S. Duno-Miranda), National Institutes of Health (HL150953 and HL163585 to D.M. Warshaw, C.M. Yengo, and S. Sivaramakrishnan), National Science Foundation (GRFP2237827 to D. Vang), and a generous gift from Arnold and Mariel Goran to D.M. Warshaw.
Author Contributions: S. Duno-Miranda contributed to conceptualization, methodology, software, formal analysis, investigation, resources, data curation, writing—original draft, writing—review and editing, visualization, funding acquisition; S.R. Nelson contributed to methodology, software, formal analysis, writing—original draft, writing—review and editing; D.V. Rasicci contributed to formal analysis, investigation, resources; S.M.L. Bodt contributed to formal analysis, investigation, resources; J.A. Cirilo, Jr. contributed to formal analysis, investigation, resources; D. Vang contributed to resources; S. Sivaramakrishnan contributed to conceptualization, funding acquisition; C.M. Yengo contributed to conceptualization, writing—review and editing, supervision, project administration, funding acquisition; D.M. Warshaw contributed to conceptualization, writing—review and editing, supervision, project administration, funding acquisition.
Data availability
Data are available from the corresponding author upon reasonable request.
References
- Alamo, L., Ware J.S., Pinto A., Gillilan R.E., Seidman J.G., Seidman C.E., and Padrón R.. 2017. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. Elife. 6:e24634. 10.7554/eLife.24634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, R.L., Trivedi D.V., Sarkar S.S., Henze M., Ma W., Gong H., Rogers C.S., Gorham J.M., Wong F.L., Morck M.M., et al. 2018. Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. USA. 115:E8143–E8152. 10.1073/pnas.1809540115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barefield, D.Y., Alvarez-Arce A., and Araujo K.N.. 2023. Mechanisms of sarcomere protein mutation-induced cardiomyopathies. Curr. Cardiol. Rep. 25:473–484. 10.1007/s11886-023-01876-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodt, S.M.L., Ge J., Ma W., Rasicci D.V., Desetty R., McCammon Y.A., and Yengo C.M.. 2023. Dilated cardiomyopathy mutation in beta-cardiac myosin enhances actin activation of the power stroke and phosphate release. bioRxiv. 10.1101/2023.11.10.566646 (Preprint posted November 13, 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunello, E., and Fusi L.. 2023. Regulating striated muscle contraction: Through thick and thin. Annu. Rev. Physiol. 86:255–275. 10.1146/annurev-physiol-042222-022728 [DOI] [PubMed] [Google Scholar]
- Chow, D., Srikakulam R., Chen Y., and Winkelmann D.A.. 2002. Folding of the striated muscle myosin motor domain. J. Biol. Chem. 277:36799–36807. 10.1074/jbc.M204101200 [DOI] [PubMed] [Google Scholar]
- Chu, S., Muretta J.M., and Thomas D.D.. 2021. Direct detection of the myosin super-relaxed state and interacting-heads motif in solution. J. Biol. Chem. 297:101157. 10.1016/j.jbc.2021.101157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen, W.J., Baelde R.J., Galli R.A., de Winter J.M., and Ottenheijm C.A.C.. 2023. Small molecule drugs to improve sarcomere function in those with acquired and inherited myopathies. Am. J. Physiol. Cell Physiol. 325:C60–C68. 10.1152/ajpcell.00047.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debold, E.P., Schmitt J.P., Patlak J.B., Beck S.E., Moore J.R., Seidman J.G., Seidman C., and Warshaw D.M.. 2007. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. Am. J. Physiol. Heart Circ. Physiol. 293:H284–H291. 10.1152/ajpheart.00128.2007 [DOI] [PubMed] [Google Scholar]
- Dutta, D., Nguyen V., Campbell K.S., Padrón R., and Craig R.. 2023. Cryo-EM structure of the human cardiac myosin filament. Nature. 623:853–862. 10.1038/s41586-023-06691-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer, J.T., Simmons R.M., and Spudich J.A.. 1994. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 368:113–119. 10.1038/368113a0 [DOI] [PubMed] [Google Scholar]
- Geeves, M.A. 2016. Review: The ATPase mechanism of myosin and actomyosin. Biopolymers. 105:483–491. 10.1002/bip.22853 [DOI] [PubMed] [Google Scholar]
- Gollapudi, S.K., Yu M., Gan Q.F., and Nag S.. 2021a. Synthetic thick filaments: A new avenue for better understanding the myosin super-relaxed state in healthy, diseased, and mavacamten-treated cardiac systems. J. Biol. Chem. 296:100114. 10.1074/jbc.RA120.016506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapudi, S.K., Ma W., Chakravarthy S., Combs A.C., Sa N., Langer S., Irving T.C., and Nag S.. 2021b. Two classes of myosin inhibitors, para-nitroblebbistatin and mavacamten, stabilize β-cardiac myosin in different structural and functional states. J. Mol. Biol. 433:167295. 10.1016/j.jmb.2021.167295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinzato, A., Auguin D., Kikuti C., Nandwani N., Moussaoui D., Pathak D., Kandiah E., Ruppel K.M., Spudich J.A., Houdusse A., and Robert-Paganin J.. 2023. Cryo-EM structure of the folded-back state of human β-cardiac myosin. Nat. Commun. 14:3166. 10.1038/s41467-023-38698-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford, W.H., Dupuis D.E., Kennedy G., Wu J., Patlak J.B., and Warshaw D.M.. 1997. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys. J. 72:1006–1021. 10.1016/S0006-3495(97)78753-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D.E., and Warshaw D.M.. 1993. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 268:14764–14768. 10.1016/S0021-9258(18)82398-5 [DOI] [PubMed] [Google Scholar]
- Heitner, S.B., Jacoby D., Lester S.J., Owens A., Wang A., Zhang D., Lambing J., Lee J., Semigran M., and Sehnert A.J.. 2019. Mavacamten treatment for obstructive hypertrophic cardiomyopathy: A clinical trial. Ann. Intern. Med. 170:741–748. 10.7326/M18-3016 [DOI] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: A potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, H.E. 1990. Sliding filaments and molecular motile systems. J. Biol. Chem. 265:8347–8350. 10.1016/S0021-9258(19)38888-X [DOI] [PubMed] [Google Scholar]
- Kawana, M., Spudich J.A., and Ruppel K.M.. 2022. Hypertrophic cardiomyopathy: Mutations to mechanisms to therapies. Front. Physiol. 13:975076. 10.3389/fphys.2022.975076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron, S.J., Toyoshima Y.Y., Uyeda T.Q., and Spudich J.A.. 1991. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 196:399–416. 10.1016/0076-6879(91)96035-P [DOI] [PubMed] [Google Scholar]
- Lakdawala, N.K., Thune J.J., Colan S.D., Cirino A.L., Farrohi F., Rivero J., McDonough B., Sparks E., Orav E.J., Seidman J.G., et al. 2012. Subtle abnormalities in contractile function are an early manifestation of sarcomere mutations in dilated cardiomyopathy. Circ. Cardiovasc. Genet. 5:503–510. 10.1161/CIRCGENETICS.112.962761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, S.J., Crocini C., and Leinwand L.A.. 2022. Targeting the sarcomere in inherited cardiomyopathies. Nat. Rev. Cardiol. 19:353–363. 10.1038/s41569-022-00682-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, E.M., and Mestroni L.. 2017. Dilated cardiomyopathy: Genetic determinants and mechanisms. Circ. Res. 121:731–748. 10.1161/CIRCRESAHA.116.309396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S., Beck-Previs S., Sadayappan S., Tong C., and Warshaw D.M.. 2023. Myosin-binding protein C stabilizes, but is not the sole determinant of SRX myosin in cardiac muscle. J. Gen. Physiol. 155:e202213276. 10.1085/jgp.202213276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto, I., Oreziak A., Barriales-Villa R., Abraham T.P., Masri A., Garcia-Pavia P., Saberi S., Lakdawala N.K., Wheeler M.T., Owens A., et al. 2020. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 396:759–769. 10.1016/S0140-6736(20)31792-X [DOI] [PubMed] [Google Scholar]
- Palmiter, K.A., Tyska M.J., Dupuis D.E., Alpert N.R., and Warshaw D.M.. 1999. Kinetic differences at the single molecule level account for the functional diversity of rabbit cardiac myosin isoforms. J. Physiol. 519:669–678. 10.1111/j.1469-7793.1999.0669n.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee, J.D., and Spudich J.A.. 1982. Purification of muscle actin. Methods Enzymol. 85:164–181. 10.1016/0076-6879(82)85020-9 [DOI] [PubMed] [Google Scholar]
- Rasicci, D.V., Tiwari P., Bodt S.M.L., Desetty R., Sadler F.R., Sivaramakrishnan S., Craig R., and Yengo C.M.. 2022. Dilated cardiomyopathy mutation E525K in human beta-cardiac myosin stabilizes the interacting-heads motif and super-relaxed state of myosin. Elife. 11:e77415. 10.7554/eLife.77415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichart, D., Magnussen C., Zeller T., and Blankenberg S.. 2019. Dilated cardiomyopathy: From epidemiologic to genetic phenotypes: A translational review of current literature. J. Intern. Med. 286:362–372. 10.1111/joim.12944 [DOI] [PubMed] [Google Scholar]
- Rohde, J.A., Roopnarine O., Thomas D.D., and Muretta J.M.. 2018. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci. USA. 115:E7486–E7494. 10.1073/pnas.1720342115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, A.N., Agre K.E., and Pereira N.L.. 2020. Genetics of dilated cardiomyopathy: Practical implications for heart failure management. Nat. Rev. Cardiol. 17:286–297. 10.1038/s41569-019-0284-0 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommese, R.F., Hariadi R.F., Kim K., Liu M., Tyska M.J., and Sivaramakrishnan S.. 2016. Patterning protein complexes on DNA nanostructures using a GFP nanobody. Protein Sci. 25:2089–2094. 10.1002/pro.3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J.A. 2014. Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106:1236–1249. 10.1016/j.bpj.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J.A. 2019. Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Pflugers Arch. 471:701–717. 10.1007/s00424-019-02259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson, A.M., Tang W., Blair C.A., Fetrow C.M., Unrath W.C., Previs M.J., Campbell K.S., and Yengo C.M.. 2017. Omecamtiv mecarbil enhances the duty ratio of human β-cardiac myosin resulting in increased calcium sensitivity and slowed force development in cardiac muscle. J. Biol. Chem. 292:3768–3778. 10.1074/jbc.M116.748780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborrini, D., Wang Z., Wagner T., Tacke S., Stabrin M., Grange M., Kho A.L., Rees M., Bennett P., Gautel M., and Raunser S.. 2023. Structure of the native myosin filament in the relaxed cardiac sarcomere. Nature. 623:863–871. 10.1038/s41586-023-06690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trybus, K.M., Freyzon Y., Faust L.Z., and Sweeney H.L.. 1997. Spare the rod, spoil the regulation: Necessity for a myosin rod. Proc. Natl. Acad. Sci. USA. 94:48–52. 10.1073/pnas.94.1.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuohy, C.V., Kaul S., Song H.K., Nazer B., and Heitner S.B.. 2020. Hypertrophic cardiomyopathy: The future of treatment. Eur. J. Heart Fail. 22:228–240. 10.1002/ejhf.1715 [DOI] [PubMed] [Google Scholar]
- Tyska, M.J., and Warshaw D.M.. 2002. The myosin power stroke. Cell Motil. Cytoskeleton. 51:1–15. 10.1002/cm.10014 [DOI] [PubMed] [Google Scholar]
- Uyeda, T.Q., Kron S.J., and Spudich J.A.. 1990. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 214:699–710. 10.1016/0022-2836(90)90287-V [DOI] [PubMed] [Google Scholar]
- Vincent, J.L. 2008. Understanding cardiac output. Crit. Care. 12:174. 10.1186/cc6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walklate, J., Kao K., Regnier M., and Geeves M.A.. 2022. Exploring the super-relaxed state of myosin in myofibrils from fast-twitch, slow-twitch, and cardiac muscle. J. Biol. Chem. 298:101640. 10.1016/j.jbc.2022.101640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Moncman C.L., and Winkelmann D.A.. 2003. Mutations in the motor domain modulate myosin activity and myofibril organization. J. Cell Sci. 116:4227–4238. 10.1242/jcs.00709 [DOI] [PubMed] [Google Scholar]
- Warshaw, D.M., Desrosiers J.M., Work S.S., and Trybus K.M.. 1990. Smooth muscle myosin cross-bridge interactions modulate actin filament sliding velocity in vitro. J. Cell Biol. 111:453–463. 10.1083/jcb.111.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt, T., Taylor D., Messier T., Trybus K.M., and Taylor K.A.. 1999. Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J. Cell Biol. 147:1385–1390. 10.1083/jcb.147.7.1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann, D.A., Forgacs E., Miller M.T., and Stock A.M.. 2015. Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat. Commun. 6:7974. 10.1038/ncomms8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead, J.L., Zhao F.Q., Craig R., Egelman E.H., Alamo L., and Padrón R.. 2005. Atomic model of a myosin filament in the relaxed state. Nature. 436:1195–1199. 10.1038/nature03920 [DOI] [PubMed] [Google Scholar]
- Woodhead, J.L., and Craig R.. 2020. The mesa trail and the interacting heads motif of myosin II. Arch. Biochem. Biophys. 680:108228. 10.1016/j.abb.2019.108228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, S.K., Eccleston J.F., and Geeves M.A.. 1991. Kinetics of the interaction of 2′(3′)-O-(N-methylanthraniloyl)-ATP with myosin subfragment 1 and actomyosin subfragment 1: Characterization of two acto-S1-ADP complexes. Biochemistry. 30:422–430. 10.1021/bi00216a017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the P values for comparisons of steady-state actin-activated ATPase measurements.
shows the P values for comparisons between WT and E525K actin-activated ATPase.
shows the P values for comparisons of SRX fraction as a function of salt concentration from single mantATP turnover measurements.
shows the P values for comparisons of SRX fraction comparing different constructs from single mantATP turnover measurements.
Data Availability Statement
Data are available from the corresponding author upon reasonable request.