Abstract
In Crithidia fasciculata, the ribosomal RNA (rRNA) gene repeats range in size from ∼11 to 12 kb. This length heterogeneity is localized to a region of the intergenic spacer (IGS) that contains tandemly repeated copies of a 19mer sequence. The IGS also contains four copies of an ∼55 nt repeat that has an internal inverted repeat and is also present in the IGS of Leishmania species. We have mapped the C.fasciculata transcription initiation site as well as two other reverse transcriptase stop sites that may be analogous to the A0 and A′ pre-rRNA processing sites within the 5′ external transcribed spacer (ETS) of other eukaryotes. Features that could influence processing at these sites include two stretches of conserved primary sequence and three secondary structure elements present in the 5′ ETS. We also characterized the C.fasciculata U3 snoRNA, which has the potential for base-pairing with pre-rRNA sequences. Finally, we demonstrate that biosynthesis of large subunit rRNA in both C.fasciculata and Trypanosoma brucei involves 3′-terminal addition of three A residues that are not present in the corresponding DNA sequences.
INTRODUCTION
In many eukaryotes, ribosomal RNA (rRNA) genes are tandemly repeated, with each repeat encoding 18S, 5.8S and 28S rRNAs that are separated from each other in the primary transcript by internal transcribed spacers. Each repeat is transcribed by RNA polymerase I and the mature rRNAs are excised from the resulting long co-transcript by a complex series of processing reactions that require protein factors and small nucleolar RNAs (snoRNAs) (1,2). An IGS separates the large subunit (LSU) rRNA gene (encoding 5.8S and 28S rRNA) in one repeat from the small subunit (SSU) rRNA gene (encoding 18S rRNA) in the adjacent downstream repeat. The IGS contains the transcription initiation site (TIS), which marks the beginning of the 5′ external transcribed spacer (ETS).
In the trypanosomatid protozoon, Crithidia fasciculata, the rDNA TIS has been identified (3) and the RNA polymerase I has been characterized (4). The C.fasciculata pre-rRNA (∼10 kb) (3) is processed to yield mature SSU rRNA (5), 5.8S rRNA (6) and six discrete rRNA species that together represent the homolog of the 28S rRNA of other eukaryotes (7,8). This characteristic fragmentation of trypanosomatid 28S rRNA arises by removal of additional internal transcribed spacers from the pre-rRNA (8–10). The 5S rRNA genes are tandemly repeated in C.fasciculata, are not physically linked to genes for other RNAs and are transcribed by RNA polymerase III (4,11).
The TIS in trypanosomatid rDNA has also been mapped for several Trypanosoma (9,12–14) and Leishmania species (15–17). The rDNA promoter has been characterized by mutational analysis in Trypanosoma brucei (18–20) and Leishmania donovani (17). As well as signals that direct transcription initiation and termination (21,22), the IGS is also expected to contain pre-rRNA processing signals (1,2), DNA replication signals (23–25) and sequences involved in binding to the nucleolar matrix (26).
Repeated sequences upstream of the TIS are a common feature in trypanosomatids (12,14–17,27). Although such repeated sequences serve to enhance transcription from the rDNA promoter in some eukaryotes (21,22), the upstream ∼150 nucleotide (nt) repeats in Trypanosoma cruzi (12) do not enhance rDNA expression (28). In Leishmania chagasi, the 64 nt tandem repeats seem to have a mild enhancer effect (16); however, nearly identical repeats in L.donovani have no enhancer activity (17).
This paper reports our analysis of the IGS region of C.fasciculata rDNA, which also contains repeated sequences. The portion of the IGS responsible for rDNA length heterogeneity corresponds to the position of one of these repeats, a tandem 19mer. We also report the identification of the TIS and two putative 5′ ETS processing sites. Finally, we isolated C.fasciculata U3 snoRNA, which may be involved in 5′ ETS processing, and determined its sequence. Sequence comparisons indicate a potential for base-pairing between the U3 snoRNA and conserved sequences in the 5′ ETS.
MATERIALS AND METHODS
The strain of C.fasciculata used in these studies was obtained from Dr F. B. St C. Palmer (Dalhousie University, Halifax, Nova Scotia) and originated from Dr G. W. Kidder, Amherst College, Amherst, MA. Total C.fasciculata DNA was isolated from large-scale cultures as described (5). Clonal isolates were obtained by growing C.fasciculata on agar plates containing brain heart infusion media supplemented with hemin (10 µg/ml) (29). Liquid cultures were obtained by innoculating 2.0 ml of the same media with agar plugs containing single colonies. After growth for 3 days, half of each culture was used for nucleic acid extraction as follows. Cell pellets were washed with 25 ml of 10 mM Tris–HCl (pH 8.5), and resuspended in 200 µl of the same buffer. After addition of 0.1 vol of both 3.0 M NaOAc and 10% SDS, the solution was extracted with phenol and nucleic acids were precipitated with 600 µl of 95% ethanol. Nucleic acids were further purified by precipitation with polyethylene glycol (30).
Restriction endonuclease digests of total DNA were resolved in agarose gels, transferred to nitrocellulose, and probed with radioactive RNA as described (31). 3′-End-labeled small rRNAs (6,7) and partially hydrolyzed, 5′-end-labeled large rRNAs (5) were used as hybridization probes.
End-labeled restriction fragments of C.fasciculata rDNA clones (5,8) were subjected to chemical sequence analysis (32) using a modified protocol (D.F.Spencer, unpublished data). With a few exceptions, this approach allowed complete sequence determination for the IGS region of two PstI clones (Cf1 and Cf4) and a short stretch of a HindIII clone (CfH1) that connects the sequences of the PstI clones. Remaining sequence ambiguities were resolved using the fmol DNA Cycle Sequencing System (Promega).
In order to localize an IGS length polymorphism, clone CfH1 and total DNA were compared as templates in PCR experiments. Primers were based upon unique sequences present in the IGS; PCR reactions were performed in the presence of 5% dimethyl sulfoxide (33).
Gel-purified 5′-end-labeled primers (10 pmol) that were complementary to 5′ ETS sequences were incubated with total RNA (25–50 µg) and AMV reverse transcriptase (RT; 40 U) for 1 h at 50°C in a 50 µl reaction volume containing 60 mM NaCl, 50 mM Tris–HCl (pH 8.3), 5 mM MgCl2, 5 mM dithiothreitol, 0.05 mg/ml bovine serum albumin and 0.3 mM dNTPs. Products were extracted with phenol, precipitated with ethanol, and purified in a sequencing gel. Bands were eluted and sequenced by the chemical method, as above.
U3 snoRNA, enriched from C.fasciculata total RNA by preparative gel electrophoresis (34) or by immunoprecipitation with an antibody directed against the trimethylguanosine cap structure (35), was 3′-end labeled, further purified in a sequencing gel and subjected to chemical (36) and enzymatic (37,38) sequence analysis. The U3 snoRNA was also decapped (39), 5′-end labeled, gel purified and subjected to enzymatic sequence analysis. Terminal nucleotide analysis of end-labeled RNA was performed as described (40). Total RNA was polyadenylated and subjected to RT–PCR (35) using a 5′-end-labeled primer specific for U3 snoRNA (corresponding to U3 snoRNA positions 58–77). The RT–PCR product was gel purified and subjected to chemical sequence analysis to verify the order of residues in the 3′ region of the U3 snoRNA.
A small RNA (the 140 srRNA) (9) was gel purified from T.brucei total RNA (a gift from M. McManus and S. L. Hajduk), 3′-end labeled and re-purified in sequencing gels (a 6% followed by a 10% polyacrylamide gel). The 3′-terminal sequence of this homolog of C.fasciculata rRNA species g was determined by the chemical method.
Sequence alignments were performed manually in the Genetic Data Environment (GDE) (41). Sequences were analyzed and compared using the Basic Local Alignment Search Tool (BLAST) (42), DNASIS version 2.5 (MiraiBio Inc., Alameda, CA) and the MicroGenie Sequence Analysis Program (43). Secondary structure diagrams were generated with the program XRNA, developed by B. Weiser and H. Noller (University of California, Santa Cruz, CA).
RESULTS
Primary sequence of the C.fasciculata IGS
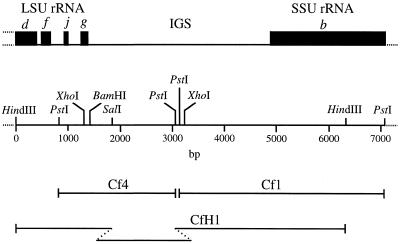
The C.fasciculata IGS, separating the genes for LSU and SSU rRNAs (Fig. 1), is 3508 nt in length (Fig. 2). This composite sequence was obtained by completing the sequences of clones that we had previously used to characterize the SSU rRNA gene (clone Cf1) (5) and the LSU rRNA gene (clone Cf4) (8). These two PstI clones are separated on the restriction map (Fig. 1) by a 55 bp PstI fragment whose sequence was obtained from a HindIII clone (clone CfH1) (8). Overlapping sequence data were obtained for both DNA strands. The data also overlap our previously published sequences (5,8), yielding a complete rDNA repeat sequence of 11 315 bp.
Figure 1.
Gene map and restriction map of clones containing the C.fasciculata rDNA IGS. Solid rectangles denote regions encoding the SSU rRNA (b) and segments (d, f, j and g) encompassing the 3′-terminal region of the discontinuous LSU rRNA. Relative locations of PstI clones (Cf1 and Cf4) and a HindIII clone (CfH1) are shown below the restriction map.
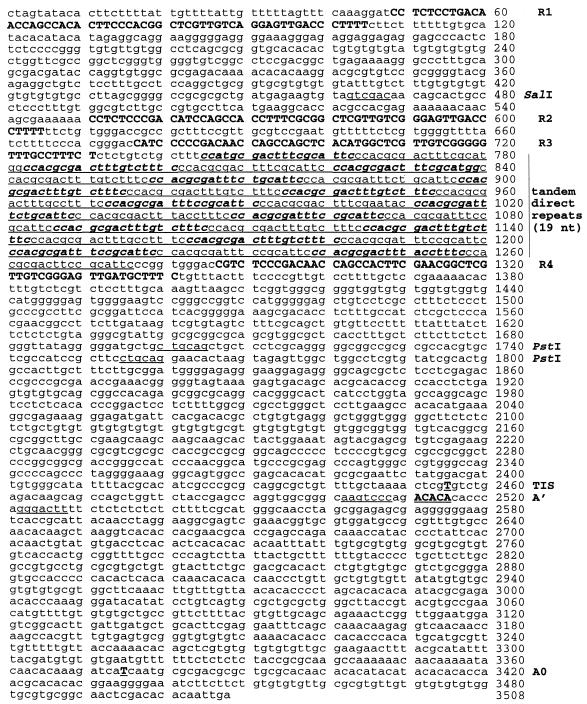
Figure 2.
Complete sequence of the C.fasciculata rDNA IGS. Restriction sites mentioned in the text are underlined (SalI and PstI). Repeated sequences R1–R4 are presented in uppercase and bold. Tandem direct repeats (19mers) are underlined with every second copy set in bold and italics. RT stop sites (TIS, A′ and A0) are uppercase, bold and underlined. Also underlined are complementary 7 nt sequences flanking a putative 13 nt loop that contains the heterogeneous A′ site.
Our sequence differs at several positions from the previously published 420 nt sequence surrounding the TIS (3). We have re-checked and verified our sequence assignment at each of these positions but we cannot rule out the possibility that at least some of these discrepancies represent real differences, either between rDNA copies or between the C.fasciculata isolates used in each study. We did detect some sequence differences between the PstI clones and the HindIII clone in regions where our partial sequence of the HindIII clone allowed comparison. It is noteworthy that the HindIII clone had four copies of a trinucleotide repeat (TTC) that is found only twice in clone Cf4 (positions 1672–1677). Furthermore, pre-rRNA transcripts derived from the 5′ ETS also contain sequence heterogeneities (see below).
Post-transcriptional oligoadenylation of trypanosomatid LSU rRNA
The IGS sequence begins immediately following position 132 of C.fasciculata LSU rRNA species g (7), which is homologous to the 3′-terminal region of the 25–28S rRNA of other eukaryotes (8,44). The three A residues present at the 3′ end of LSU rRNA species g (7) are not present in either of the two clones sequenced in this study and therefore they must be added post-transcriptionally. Three A residues are also present at the 3′ end of the homologous small RNA from T.cruzi (45) but in that case the gene sequence has not been determined. In order to establish whether this 3′-terminal addition of A residues is common among trypanosomatids, we determined the sequence at the 3′ end of the species g homolog from T.brucei, a distantly related trypanosomatid (46). This RNA also has three A residues at its 3′ end (Fig. 3) that are not present in the DNA sequence (9).
Figure 3.
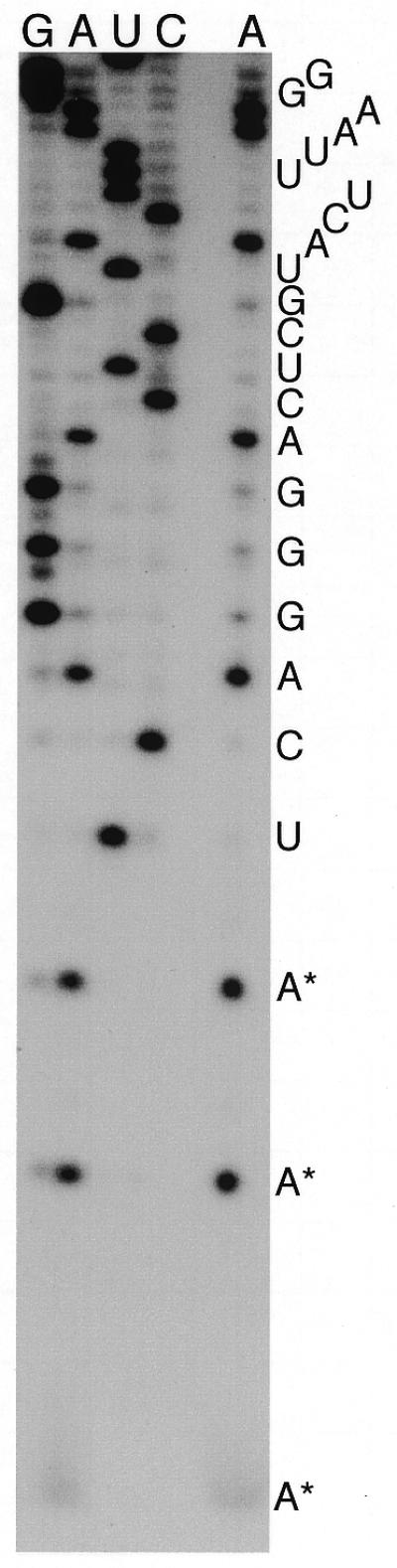
Chemical sequencing of the 3′ terminus of T.brucei LSU rRNA. The separate A lane (chemical cleavage of A>G) verifies that this homolog of C.fasciculata rRNA species g ends in three A residues (*) rather than the U residues predicted by the gene sequence (9).
Length heterogeneity within the IGS
When individual components of the fragmented C.fasciculata LSU rRNA (species c–g, i, j) were 32P-labeled and used to probe Southern blots of SalI- or BamHI-digested DNA, an rDNA repeat of ∼11–12 kb was detected (not shown). In PstI digests, each of these rRNAs, with the exception of species g and j, hybridized to a single restriction fragment: species c, d and i to a 3.1 kb PstI fragment, species d and f to a 2.0 kb PstI fragment. The SSU rRNA hybridized to a 3.9 kb PstI fragment (5). The remaining two LSU rRNA species (g and j; see Fig. 1 for the relative locations of their genes in the rDNA repeat) hybridized to a ladder of PstI fragments, the most abundant of which were in the ∼2–3 kb size range (Fig. 4). This PstI restriction fragment length heterogeneity appears to be stably inherited because it is virtually identical for two different DNA samples that had been prepared 7 years apart after many passages of a maintenance culture. DNA was also isolated from small volumes of culture that had each been innoculated with a C.fasciculata colony derived from a single cell. LSU rRNA species j hybridized to the same size and number of PstI fragments from these clonal DNA preparations as was observed with DNA from large-scale non-clonal cultures; however, the relative intensity of bands was different for each clonal DNA preparation (Fig. 4).
Figure 4.
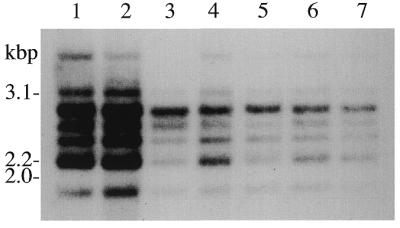
Size heterogeneity of PstI restriction fragments detected by Southern hybridization using 3′-end-labeled LSU rRNA species j as a probe (see Fig. 1). Lanes 1 and 2 contain DNA prepared 7 years apart from large-scale cultures. Lanes 3–7 contain DNA isolated from small-scale cultures of clonal isolates. Numbers listed on the left indicate the sizes of PstI inserts from clones that contain LSU rRNA coding sequences (see Results).
Comparison of restriction maps for two different clones that span this region of the rDNA repeat established that the length heterogeneity is localized to a SalI–PstI fragment within the IGS (Fig. 1). Primers that were expected to amplify within the first 641 bp of the IGS gave identically sized products in PCR experiments regardless of whether clone CfH1 or total DNA was used as template (not shown), establishing that length heterogeneity is not present in this particular region of the IGS. Similar PCR experiments also established that there is no significant length heterogeneity between position 1533 and the 3′ end of the IGS (not shown). On the other hand, PCR amplification of total DNA between IGS positions 622 and 1552 yielded multiple discrete bands, indicating that the IGS length heterogeneity lies within this region. Preliminary sequence data for sections of the CfH1 insert further localized the size difference to a region of the IGS that contains tandem direct repeats (19mers; see below).
Repeated sequences
Analysis of the IGS sequence revealed several classes of repeated sequences (Figs 2 and 5). First, the IGS has four copies of a 55–57 nt sequence (repeats R1–R4). Each copy contains an internal inverted repeat that suggests the presence of a hairpin in the putative transcript. This structure is supported by the fact that sequence changes in one strand are almost always accompanied by complementary changes in the other strand (Fig. 5 and not shown) such that base-pairing potential is maintained. Second, the IGS also contains 28 copies of a 19 nt tandem direct repeat. Minor variations within these tandem repeats allowed us to group them into 10 sequence families (Fig. 5B). In addition to these major repeats, we also detected a short G+A-rich sequence that is present twice in the IGS and also in a divergent (D3) region of the LSU rRNA (Fig. 5C). Two other IGS sequences (Fig. 5D and E) are also related to regions of rRNA that have been defined as variable (V) or divergent (D). Finally, we noted considerable dinucleotide frequency bias in regions of the IGS, with long stretches of the sequence (TG)n and its complement (CA)n being most obvious.
Figure 5.
Alignment of IGS sequences that are repeated in C.fasciculata rDNA. Positions of repeats relative to the sequence in Figure 2 are indicated. In each alignment, positions that differ from the top sequence are presented in bold. (A) The 55–57 nt repeats (R1–R4) are aligned with a similar sequence from L.infantum (Li) (58). The location of an inverted repeat (potential hairpin) is indicated by the dashed arrows above the alignment. (B) The tandemly repeated 19mers are grouped into 10 sequence families as indicated by numbers at the right of the alignment. Additional sequences in the IGS (C–E) are repeated in variable (V7 and V8) regions in the C.fasciculata SSU rRNA secondary structure as defined in Schnare et al. (5) or in divergent (D3 and D10) regions of the C.fasciculata LSU rRNA as defined by Spencer et al. (8). The LSU rRNA secondary structure (44) is available at URL http://www.rna.icmb.utexas.edu/
ETS sequence and structural conservation
Beginning 146 nt downstream of the TIS, long stretches of the C.fasciculata 5′ ETS sequence can be aligned reasonably well with ETS sequences from Leishmania species (15–17), most likely reflecting the close evolutionary relationship between these organisms. On the other hand, addition of the more distantly related T.cruzi sequence (13) to the alignment suggests functional roles for regions that can still be aligned. Two sections of the T.cruzi ETS sequence show significant similarity to the Crithidia and Leishmania sequences (Fig. 6) (15): one short stretch (Fig. 6A) at the beginning of the alignment and a longer stretch (Fig. 6B) that contains sequences complementary to U3 snoRNA (see below).
Figure 6.
Conservation of 5′ ETS primary sequences. Two regions of the C.fasciculata (Cf) 5′ ETS are aligned with homologous sequences from L.amazonensis (La) (15), L.donovani (Ld) (17), L.chagasi (Lc) (16) and T.cruzi (Tc) (13). (A) C.fasciculata IGS positions 2600–2639 [part of motif 1 of Uliana et al. (15)]. (B) Crithidia fasciculata IGS positions 3048–3187 [includes motifs 4 and 5 of Uliana et al. (15)]. Differences from the top sequence are indicated while positions that are identical are represented by dots. ETS coordinates are listed for each sequence. Sequences that are complementary to U3 snoRNA are boxed (regions I and II).
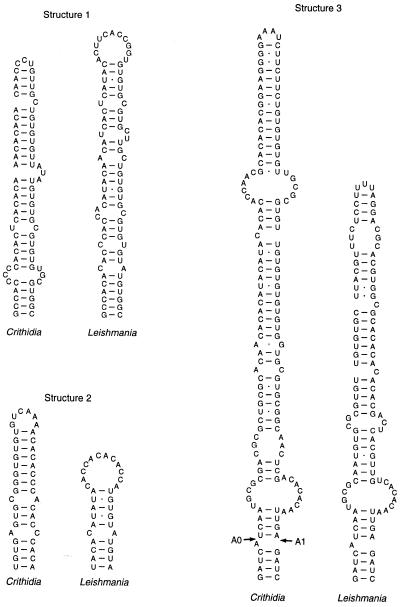
In addition to the IGS repeats described above, we also detected three imperfect inverted repeats within the 5′ ETS. We suspected that these inverted repeats might indicate the presence of higher-order structural features within the ETS (Fig. 7). This proposal gains support from the fact that Leishmania 5′ ETS sequences from the same portions of the alignment can also be folded to produce similar structures (Fig. 7), even though the Crithidia and Leishmania sequences have diverged significantly in both length and primary sequence in these three regions. The potential to form structure 3 in trypanosomatid ETS sequences has been noted previously, with similar structures also possible for T.brucei and T.cruzi (47). The base of this particular structure (including the 5′-terminal sequence of the SSU rRNA) could potentially help to define the position of processing sites (A0 and A1) in the pre-rRNA (see below).
Figure 7.
Conservation of 5′ ETS structural elements. Imperfect inverted repeats in equivalent positions of aligned 5′ ETS sequences (not shown) suggest the presence of secondary structures in C.fasciculata and Leishmania species. The L.donovani sequence (17), which is very similar to the other Leishmania sequences, was used in this figure. Structure 1 spans C.fasciculata IGS positions 2883–2953 (ETS positions 429–499); structure 2 encompasses C.fasciculata IGS positions 3191–3230 (ETS positions 737–776); structure 3 begins at C.fasciculata IGS position 3370 (ETS position 916) and ends 4 nt into the 5′ end of the SSU rRNA sequence. Arrows indicate the positions of processing sites (A0 and A1). The L.donovani structure 1 spans ETS positions 411–489, which are obviously similar to the sequence used in the C.fasciculata structure. The L.donovani structure 2 sequence (ETS positions 750–781) is not very similar to the corresponding C.fasciculata sequence; however, in both organisms this sequence follows shortly after the conserved sequence presented in Figure 6B and is immediately followed by another short stretch of sequence that is conserved between the two organisms. The L.donovani structure 3 begins at ETS position 922.
RT mapping of the pre-rRNA
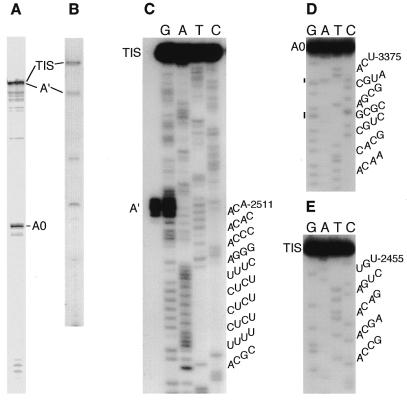
We used ETS-specific primers and RT to scan the entire 5′ ETS for the presence of pre-rRNA 5′ termini. This approach yielded an abundant RT product that maps to site A′ (Figs 2 and 8C), corresponding to IGS positions 2511–2515 (ETS positions 57–61, see below). We also detected a second abundant RT product mapping to site A0 (Figs 2 and 8D), at IGS position 3375 (ETS position 921, see below). Minor amounts of RT product corresponding in size to site A0 ±1 were also detected (Fig. 8A). Our interpretation that these RT stops represent pre-rRNA processing sites fits well with what is known about 5′ ETS processing in other eukaryotes (see Discussion). No other abundant processing intermediates were detected in this study. Notably, we saw no indication of processing around ETS position ∼90 as reported by Grondal et al. (3). These experiments also provided evidence for microsequence heterogeneity in the 5′ ETS (Fig. 8C).
Figure 8.
Chemical sequence analysis of 5′-end-labeled RT products. The A lanes represent cleavage at A>G. RT products corresponding to stops at the TIS and the A′ and A0 sites were purified by gel electrophoresis prior to sequence analysis. Sequences of the RNA transcripts are shown to the right of each gel, with the 5′-terminal position of each RT product numbered according to its location in the IGS (see Fig. 2). Representative RT reactions resolved in sequencing gels are shown in (A) (primer complementary to IGS positions 3485–3508) and (B) (primer complementary to IGS positions 2660–2680). The two lower bands in gel B represent non-target products that are not detected with other IGS primers. (C) Analysis of RT products isolated from gel (B). A sequencing ladder for the TIS product is presented. The A′ product was electrophoresed adjacent to and mixed with the G sample. Minor bands present in this gel were also detected during sequencing of the A′ product, which also gave the expected ladder (not shown). These minor bands most likely reflect microsequence heterogeneity in ETS transcripts. (D) Analysis of the A0 product isolated from gel A. Vertical lines indicate positions of band compressions in the sequencing gel. (E) Analysis of an RT product generated using an oligonucleotide complementary to IGS positions 2547–2566.
In this same set of experiments we detected an RT stop in the region of the IGS that was expected to contain the TIS (3). Our mapping approach (direct chemical sequencing of a gel-purified RT product) has several advantages over the standard method [comparison of the mobility of an RT (or S1 nuclease protection) product in one lane to the mobilities of sequencing reactions in adjacent lanes of a gel]. First, in our experiments the RT product and the sequencing reactions are in the same lanes of the gel, eliminating sample composition effects that can cause mobility artifacts. Second, direct sequencing proves that the RT product is derived from the target ETS site rather than from artifactual binding of primer to more abundant non-target (mature rRNA) transcripts. Third, non-target RT products are removed prior to sequence analysis. Finally, the actual 5′ nucleotide of the RT product can be read from chemical sequencing gels; i.e. the band just below full length (Fig. 8E, lane A) results from chemical removal of a residue from the end of the DNA molecule. Note that longer electrophoresis times and lighter film exposures verified that no additional bands were present below full length (not shown). We can therefore unequivocally state that the TIS is located 2 nt downstream of the site reported by Grondal et al. (3), at IGS position 2455 (ETS position +1).
U3 snoRNA
The sequence of C.fasciculata U3 snoRNA [identified by comparison with known U3 snoRNA sequences from other trypanosomatids (48,49)] is presented in Figure 9. This RNA has a methlyguanosine cap (mG, either 7-monomethylguanosine or N2,N2,7-trimethylguanosine) as judged by immunoprecipitation with an anti-cap antibody (35). Complete hydrolysis of end-labeled RNA followed by thin layer chromatography established that the 5′-terminal nucleoside was O2′-methyladenosine (Am) and the 3′-terminal nucleoside was C. The residue at position 2 yielded a band in the A lane of chemical sequencing gels but was not cleaved by RNases or alkali in enzymatic sequencing, suggesting the presence of another Am at this position. The presence of bands in enzymatic sequencing gels ruled out the possibility of additional O2′-methylnucleosides (Nm) at positions 3–84 or 102–142. We were unable to evaluate Nm content between positions 85 and 101 because of weak cleavage and band compressions in enzymatic sequencing gels (probably due to stable secondary structure, Fig. 9); it is within this region (position 88) that T.brucei U3 snoRNA is thought to contain a Cm residue (49). No pseudouridine residues were present in this RNA as judged by the fact that all U residues were cleaved in the hydrazine reaction during chemical sequencing.
Figure 9.
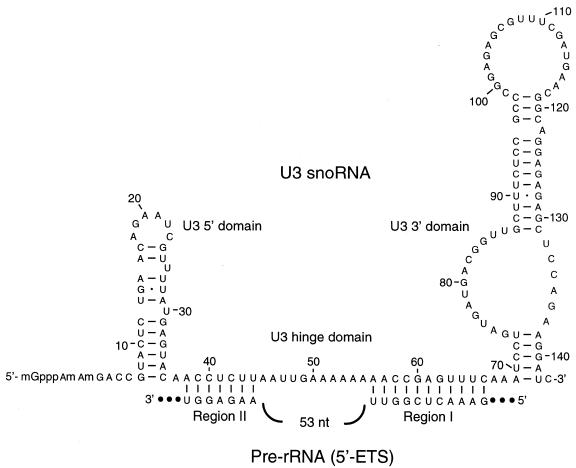
Primary sequence and potential secondary structure of C.fasciculata U3 snoRNA. Putative base-pairing interactions with sequences in the rDNA 5′ ETS (see Fig. 6) are included.
Sequences within the hinge region of U3 snoRNAs from diverse organisms are thought to interact by direct base-pairing with 5′ ETS sequences downstream of the 5′-most processing site (A′, see Discussion). We therefore searched the entire C.fasciculata ETS for sequences with the potential to pair with the U3 snoRNA hinge sequence. Interestingly, the two possible U3 snoRNA/ETS pairings that we detected (Fig. 9) involve ETS sequences (regions I and II) that are located in a stretch of the ETS that is conserved among C.fasciculata, Leishmania species and T.cruzi (Fig. 6B). With the exception of T.cruzi region I, the ETS sequences involved in the proposed interactions are identical (Fig. 6B); the T.cruzi region I interaction would require a G-C to G·U base-pair change and a 2 nt bulge. The two sequences within the U3 hinge region that are proposed to interact with ETS regions I and II are identical in U3 snoRNAs from C.fasciculata, T.cruzi (49) and Leishmania tarentolae (50).
DISCUSSION
The C.fasciculata rDNA repeat
The work reported here completes the sequence of the equivalent of an entire rDNA repeat (total length 11 315 bp) from C.fasciculata. Data linking the 3′ end of an rDNA repeat to the 5′ end of a downstream rDNA repeat could be interpreted to mean that the rDNA is either circular or tandemly repeated. Circular rDNAs, with one or a few rDNA copies per circle, have been identified in some other protozoa, notably Euglena gracilis (51), Entamoeba histolytica (52) and Naegleria gruberi (53). However, considering that the rDNA is tandemly repeated and integrated into chromosome-sized DNA molecules in several trypanosomatids (54,55), it is likely that this is also the case in C.fasciculata.
Although IGS lengths are homogeneous in some organisms, pronounced length heterogeneity has been documented in other species, both between and within individual cells (56). Because several discrete IGS size variants exist in C.fasciculata, its rDNA repeats vary in length between ∼11 and 12 kb. We have localized the rDNA length heterogeneity to a region of the IGS that contains tandem direct repeats of a 19mer sequence located upstream of the TIS. Clonal isolates of C.fasciculata all contained the same discrete IGS size variants in their rDNA population; however, the relative abundance of these variants differed. This could be explained by amplification/deletion within clustered rDNA repeats as has been proposed to account for chromosome size differences in natural isolates of Leishmania (57).
3′-Termini of trypanosomatid pre-rRNA and LSU rRNA
The ∼55 nt repeated sequences R1–R4 are likely to play an important biological role because homologous repeated sequences are also present in Leishmania species (17,58) (Fig. 5) within a region of the IGS that is otherwise poorly conserved. Requena et al. (58) recognized that these repeats (composed of hairpin structures followed by U-rich sequences) resemble bacterial rho-independent transcriptional terminators. These investigators showed that few, if any, nascent transcripts extended past the first of these repeats in Leishmania infantum. The arrangement of these repeats in the C.fasciculata IGS also resembles the arrangement of RNA polymerase I terminators in other eukaryotes (22), with three copies (R1–R3) immediately downstream of the LSU rRNA gene and a single copy (R4, a putative fail-safe terminator) located between the tandemly repeated 19mers and the TIS (Fig. 2). However, it is important to point out that in other eukaryotes, transcription by RNA polymerase I is not terminated by a bacterial type rho-independent mechanism (59); in fact, these hairpin structures could represent processing signals. In yeast, RNase III is known to be involved in pre-rRNA processing, cleaving at a hairpin downstream of the LSU rRNA sequence (60,61).
The lack of significant levels of 3′ ETS transcripts in the trypanosomatids T.brucei (9) and L.infantum (58) could be explained equally well by termination or rapid processing; at this point we cannot distinguish between these two possibilities. Assuming that transcription does continue beyond the 3′ end of trypanosomatid LSU rRNA genes [as suggested for T.brucei (9)], then production of the mature 3′-terminus would involve cleavage as well as post-transcriptional addition of three A residues. The relationship, if any, between LSU rRNA 3′ end formation in trypanosomatids and the coupled cleavage/polyadenylation at the 3′-termini of eukaryotic mRNA (62) remains to be determined.
The C.fasciculata rDNA TIS
We re-examined the C.fasciculata rDNA TIS and established that transcription starts at a T residue that is located two nucleotides downstream of the site mapped previously by Grondal et al. (3). Alignment of putative promoter sequences relative to the position of mapped initiation sites demonstrates that trypanosomatid rDNA promoters are not very highly conserved (14,17). On the other hand, some of these sequences can be aligned on the basis of primary sequence similarity (this is possible for closely related species). In these alignments (not shown), it is clear that the TIS mapped for Leishmania amazonensis (15) is located 13 bp upstream of the site identified in both L.chagasi (16) and L.donovani (17). Similarly, distinct transcription initiation sites, separated by ∼270 bp, have been mapped for two different strains of T.cruzi (12,13). This could reflect the fact that T.cruzi strains can be divided into two groups, I and II. Constructs containing group I rDNA promoters function in both groups whereas group II rDNA promoters function only in group II strains (63). Taking these studies into consideration, we cannot completely rule out the possibility that the two differently mapped C.fasciculata initiation sites [(3) versus this study] reflect real differences in rDNA promoters in the two C.fasciculata isolates.
RT stop sites (putative processing sites) within the 5′ ETS
The 5′-most RT stop site identified in the C.fasciculata 5′ ETS (site A′) is positionally analogous to the U3 snoRNA-dependent primary processing site identified in animals (64,65). Heterogeneous RT stops at site A′ in C.fasciculata are also reminiscent of what has been observed in mammals (66). Furthermore, the A′ site in mouse is located in the loop of a hairpin structure and a single-stranded conformation is necessary for processing (67); a short hairpin with the A′ site in the loop is also possible in C.fasciculata (Fig. 2). Putative homologs of the A′ processing site have also been identified in fungi (68–70) and T.brucei (9,47). It has been proposed that this particular processing site in the 5′ ETS has been universally conserved among eukaryotes and that it may play an important role in assembly of a processing complex (containing U3 and other snoRNAs) that is required for production of SSU rRNA (65,68,71). U3 snoRNA remains bound to the downstream product after cleavage at site A′ (64) and the processing complexes are represented by terminal balls in chromatin spreads of rDNA transcriptional units (72).
On the basis of secondary structure comparisons, it appears that the second C.fasciculata putative processing site detected in this study is analogous to the A0 site identified in fungi (73,74) and T.brucei (47); in each case the A0 site occurs near the base of a structural element (47,74) that places the site in close proximity to the 5′ end of the SSU rRNA sequence (site A1) (Fig. 7). Processing at sites A0 and A1 are U3 snoRNA-dependent in yeast (73,75) but there are conflicting reports concerning the possible involvement of an RNase III homolog in cleavage at site A0 (60,61). A processing site, possibly analogous to site A0, has been detected in mouse pre-rRNA 105 nt upstream of the SSU rRNA sequence (76,77); however, this site does not map to the expected region (across from site A1) in the secondary structure model proposed by Michot and Bachellerie (78).
Structure of U3 snoRNA from C.fasciculata and potential interactions with pre-rRNA
It is well documented in other systems (see below) that U3 snoRNA interacts by direct base-pairing with sequences in the pre-rRNA, including the 5′ ETS. We therefore determined the sequence of C.fasciculata U3 snoRNA in order to evaluate the potential for U3 snoRNA:pre-rRNA pairing in this organism. The C.fasciculata U3 snoRNA sequence is presented in secondary structure form (Fig. 9) (49). In this model, the U3 snoRNA molecule is divided into 5′ and 3′ structured domains separated by a single-stranded ‘hinge’ domain. Secondary structure within the U3 snoRNA 3′ domain is strongly supported by phylogenetic comparisons (49) and this domain contains determinants for RNA–protein interactions (79). Secondary structure within the 5′ domain of U3 snoRNA appears to be lineage-specific and is not conserved in broad phylogenetic comparisons (49); however, sequences within this 5′ domain and hinge region do have the potential to form phylogenetically conserved base-pairing interactions with SSU rRNA sequences in pre-rRNA (75,79,80). These proposed interactions are conserved in C.fasciculata and may be important for processing and/or pseudoknot formation at the 5′ end of the SSU rRNA. Similar interactions, involving U3-like sequences in the 5′ ETS, are possible in eubacteria and archaea (81).
We also searched for regions of complementarity between the U3 snoRNA and the 5′ ETS, identifying two stretches of ETS sequence (regions I and II in Figs 6B and 9) that could possibly be targets for U3 snoRNA pairing. The hinge region of T.brucei U3 snoRNA has been shown, by crosslinking with psoralen, to interact with pre-rRNA ETS sequences downstream of the A′ site (82). This led to the proposal of two potential region I-analogous interactions (using U3 positions ∼54–67) (47,82), and the corresponding ETS sequences were shown to be essential for SSU rRNA accumulation in T.brucei [ETS sites 1b and 3 of Hartshorne and Toyofuku (47)]. However, there is no obvious sequence similarity between the C.fasciculata ETS sequence and these T.brucei sequences; thus, it is unclear whether either of these proposed T.brucei interactions is strictly homologous to the C.fasciculata region I interaction proposed here. Similarly, the highly variable nature of rDNA ETS sequences makes it difficult to compare the putative C.fasciculata region I interaction with a similar interaction that was recently proposed for animals (using positions ∼59–66 in the C.fasciculata U3 snoRNA numbering system) (83).
A proposal has been made (68) for a yeast U3 snoRNA:ETS base-pairing interaction that involves U3 nucleotides homologous to the C.fasciculata sequence predicted to interact with ETS region II (Fig. 9). This yeast interaction is known to be of functional importance (69,84). A similar region II-analogous interaction has been proposed for T.brucei (48); this interaction also appears to be functionally important [the U3 snoRNA:site 1a interaction of Hartshorne and Toyofuku (47)]. In both of these models the U3 snoRNA pairs to ETS sequences at the 5′-most processing site; however, there is no obvious potential for base-pairing between C.fasciculata U3 snoRNA and the A′ site mapped in this study.
Acknowledgments
ACKNOWLEDGEMENTS
We thank R. Breckon and F. B. St C. Palmer for cultures of C.fasciculata, and M. McManus and S. L. Hajduk for T.brucei RNA. We also thank B. Weiser and H. Noller for the computer program XRNA, which was used to generate secondary structure figures. A grant (MT-11212) to M.W.G. from the Medical Research Council of Canada and salary support from the Canadian Institute for Advanced Research (Program in Evolutionary Biology) are gratefully acknowledged.
DDBJ/EMBL/GenBank accession nos Y00055, AF277396
REFERENCES
- 1.Eichler D.C. and Craig,N. (1994) Prog. Nucleic Acid Res. Mol. Biol., 49, 197–239. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 3.Grondal E.J.M., Evers,R. and Cornelissen,A.W.C.A. (1990) Nucleic Acids Res., 18, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köck J. and Cornelissen,A.W.C.A. (1991) Mol. Microbiol., 5, 835–842. [DOI] [PubMed] [Google Scholar]
- 5.Schnare M.N., Collings,J.C. and Gray,M.W. (1986) Curr. Genet., 10, 405–410. [DOI] [PubMed] [Google Scholar]
- 6.Schnare M.N. and Gray,M.W. (1982) Nucleic Acids Res., 10, 2085–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnare M.N., Spencer,D.F. and Gray,M.W. (1983) Can. J. Biochem., 61, 38–45. [DOI] [PubMed] [Google Scholar]
- 8.Spencer D.F., Collings,J.C., Schnare,M.N. and Gray,M.W. (1987) EMBO J., 6, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White T.C., Rudenko,G. and Borst,P. (1986) Nucleic Acids Res., 14, 9471–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell D.A., Kubo,K., Clark,C.G. and Boothroyd,J.C. (1987) J. Mol. Biol., 196, 113–124. [DOI] [PubMed] [Google Scholar]
- 11.Köck J. and Cornelissen,A.W.C.A. (1990) Mol. Biochem. Parasitol., 38, 295–298. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich P., Soares,M.B., Affonso,M.H.T. and Floeter-Winter,L.M. (1993) Gene, 125, 103–107. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Calvillo S. and Hernández,R. (1994) Gene, 142, 243–247. [DOI] [PubMed] [Google Scholar]
- 14.Downey N. and Donelson,J.E. (1999) Mol. Biochem. Parasitol., 104, 25–38. [DOI] [PubMed] [Google Scholar]
- 15.Uliana S.R.B., Fischer,W., Stempliuk,V.A. and Floeter-Winter,L.M. (1996) Mol. Biochem. Parasitol., 76, 245–255. [DOI] [PubMed] [Google Scholar]
- 16.Gay L.S., Wilson,M.E. and Donelson,J.E. (1996) Mol. Biochem. Parasitol., 77, 193–200. [DOI] [PubMed] [Google Scholar]
- 17.Yan S., Lodes,M.J., Fox,M., Myler,P.J. and Stuart,K. (1999) Mol. Biochem. Parasitol., 103, 197–210. [DOI] [PubMed] [Google Scholar]
- 18.Zomerdijk J.C.B.M., Kieft,R., Shiels,P.G. and Borst,P. (1991) Nucleic Acids Res., 19, 5153–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudenko G., Chung,H.-M.M., Pham,V.P. and Van der Ploeg,L.H.T. (1991) EMBO J., 10, 3387–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janz L. and Clayton,C. (1994) Mol. Cell. Biol., 14, 5804–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob S.T. (1995) Biochem. J., 306, 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paule M.R. and Lofquist,A.K. (1996) In Zimmermann,R.A. and Dahlberg,A.E. (eds), Ribosomal RNA. Structure, Evolution, Processing, and Function in Protein Biosynthesis. CRC Press, Boca Raton, FL, pp. 395–419.
- 23.Miller C.A. and Kowalski,D. (1993) Mol. Cell. Biol., 13, 5360–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernández P., Martin-Parras,L., Martinez-Robles,M.L. and Schvartzman,J.B. (1993) EMBO J., 12, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gencheva M., Anachkova,B. and Russev,G. (1996) J. Biol. Chem., 271, 2608–2614. [DOI] [PubMed] [Google Scholar]
- 26.Stephanova E., Stancheva,R. and Avramova,Z. (1993) Chromosoma, 102, 287–295. [DOI] [PubMed] [Google Scholar]
- 27.Novak E.M., de Mello,M.P., Gomes,H.B.M., Galindo,I., Guevara,P., Ramirez,J.L. and de Silveira,J.F. (1993) Mol. Biochem. Parasitol., 60, 273–280. [DOI] [PubMed] [Google Scholar]
- 28.Tyler-Cross R.E., Short,S.L., Floeter-Winter,L.M. and Buck,G.A. (1995) Mol. Biochem. Parasitol., 72, 23–31. [DOI] [PubMed] [Google Scholar]
- 29.Hughes D.E., Schneider,C.A. and Simpson,L. (1982) J. Parasitol., 68, 642–649. [PubMed] [Google Scholar]
- 30.Lis J.T. (1980) Methods Enzymol., 65, 347–353. [DOI] [PubMed] [Google Scholar]
- 31.Bonen L. and Gray,M.W. (1980) Nucleic Acids Res., 8, 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxam A.M. and Gilbert,W. (1980) Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 33.Shen W.-H. and Hohn,B. (1992) Trends Genet., 8, 227. [Google Scholar]
- 34.Schnare M.N. and Gray,M.W. (2000) Biochim. Biophys. Acta, 1490, 362–366. [DOI] [PubMed] [Google Scholar]
- 35.Schnare M.N. and Gray,M.W. (1999) J. Biol. Chem., 274, 23691–23694. [DOI] [PubMed] [Google Scholar]
- 36.Peattie D.A. (1979) Proc. Natl Acad. Sci. USA, 76, 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donis-Keller H., Maxam,A.M. and Gilbert,W. (1977) Nucleic Acids Res., 4, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donis-Keller H. (1980) Nucleic Acids Res., 8, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry K.L., Watkins,K.P. and Agabian,N. (1987) Proc. Natl Acad. Sci. USA, 84, 8190–8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKay R.M., Spencer,D.F., Doolittle,W.F. and Gray,M.W. (1980) Eur. J. Biochem., 112, 561–576. [DOI] [PubMed] [Google Scholar]
- 41.Smith S.W., Overbeek,R., Woese,C.R., Gilbert,W. and Gillevet,P.M. (1994) CABIOS, 10, 671–675. [DOI] [PubMed] [Google Scholar]
- 42.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 43.Queen C. and Korn,L.J. (1984) Nucleic Acids Res., 12, 581–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnare M.N., Damberger,S.H., Gray,M.W. and Gutell,R.R. (1996) J. Mol. Biol., 256, 701–719. [DOI] [PubMed] [Google Scholar]
- 45.Galvan S.C., Castro,C., Segura,E., Casas,L. and Castaneda,M. (1991) Nucleic Acids Res., 19, 2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevens J.R., Noyes,H.A., Dover,G.A. and Gibson,W.C. (1999) Parasitol., 118, 107–116. [Google Scholar]
- 47.Hartshorne T. and Toyofuku,W. (1999) Nucleic Acids Res., 27, 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartshorne T. and Agabian,N. (1993) Mol. Cell. Biol., 13, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartshorne T. and Agabian,N. (1994) Nucleic Acids Res., 22, 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi X., Chen,D.-H.T. and Suyama,Y. (1994) Mol. Biochem. Parasitol., 65, 23–37. [DOI] [PubMed] [Google Scholar]
- 51.Ravel-Chapuis P., Nicolas,P., Nigon,V., Neyret,O. and Freyssinet,G. (1985) Nucleic Acids Res., 13, 7529–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huber M., Koller,B., Gitler,C., Mirelman,D., Revel,M., Rozenblatt,S., and Garfinkel,L. (1989) Mol. Biochem. Parasitol., 32, 285–296. [DOI] [PubMed] [Google Scholar]
- 53.Clark C.G. and Cross,G.A.M. (1987) Mol. Cell. Biol., 7, 3027–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leon W., Fouts,D.L. and Manning,J. (1978) Nucleic Acids Res., 5, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van der Ploeg L.H.T., Cornelissen,A.W.C.A., Barry,J.D. and Borst,P. (1984) EMBO J., 3, 3109–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franke W.W., Scheer,U., Spring,H., Trendelenburg,M.F. and Zentgraf,H. (1979) In Busch,H. (ed.), The Cell Nucleus. Academic Press, New York, Vol. 7, pp. 49–95.
- 57.Inga R., De Doncker,S., Gomez,J., Lopez,M., Garcia,R., Le Ray,D., Arevalo,J. and Dujardin,J.-C. (1998) Mol. Biochem. Parasitol., 92, 219–228. [DOI] [PubMed] [Google Scholar]
- 58.Requena J.M., Soto,M., Quijada,L., Carrillo,G. and Alonso,C. (1997) Mol. Biochem. Parasitol., 84, 101–110. [DOI] [PubMed] [Google Scholar]
- 59.Reeder R.H. and Lang,W.H. (1997) Trends Biochem. Sci., 22, 473–477. [DOI] [PubMed] [Google Scholar]
- 60.Abou Elala S., Igel,H. and Ares,M.,Jr (1996) Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- 61.Kufel J., Dichtl,B. and Tollervey,D. (1999) RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahl E. and Rüegsegger,U. (1999) FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 63.Nunes L.R., de Carvalho,M.R.C. and Buck,G.A. (1997) Mol. Biochem. Parasitol., 86, 211–224. [DOI] [PubMed] [Google Scholar]
- 64.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 65.Mougey E.B., Pape,L.K. and Sollner-Webb,B. (1993) Mol. Cell. Biol., 13, 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kass S., Craig,N. and Sollner-Webb,B. (1987) Mol. Cell. Biol., 7, 2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Craig N., Kass,S. and Sollner-Webb,B. (1991) Mol. Cell. Biol., 11, 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beltrame M. and Tollervey,D. (1992) EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beltrame M., Henry,Y. and Tollervey,D. (1994) Nucleic Acids Res., 22, 4057–4065 (reprinted Nucleic Acids Res., 22, 5139–5147). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brulé F., Venema,J., Ségault,V., Tollervey,D. and Branlant,C. (1996) RNA, 2, 183–197. [PMC free article] [PubMed] [Google Scholar]
- 71.Enright C.A., Maxwell,E.S., Eliceiri,G.L. and Sollner-Webb,B. (1996) RNA, 2, 1094–1099 (published erratum RNA., 2, 1318). [PMC free article] [PubMed] [Google Scholar]
- 72.Mougey E.B., O’Reilly,M., Osheim,Y., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1993) Genes Dev., 7, 1609–1619. [DOI] [PubMed] [Google Scholar]
- 73.Hughes J.M.X. and Ares,M.,Jr (1991) EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Intine R.V.A., Good,L. and Nazar,R.N. (1999) J. Mol. Biol., 286, 695–708. [DOI] [PubMed] [Google Scholar]
- 75.Sharma K. and Tollervey,D. (1999) Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mishima Y., Mitsuma,T. and Ogata,K. (1985) EMBO J., 4, 3879–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mishima Y., Katayama,M. and Ogata,K. (1988) J. Biochem., 104, 515–520. [DOI] [PubMed] [Google Scholar]
- 78.Michot B. and Bachellerie,J.-P. (1991) Eur. J. Biochem., 195, 601–609. [DOI] [PubMed] [Google Scholar]
- 79.Méreau A., Fournier,R., Grégoire,A., Mougin,A., Fabrizio,P., Lührmann,R. and Branlant,C. (1997) J. Mol. Biol., 273, 552–571. [DOI] [PubMed] [Google Scholar]
- 80.Hughes J.M.X. (1996) J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 81.Dennis P.P., Russell,A.G. and Moniz de Sá,M. (1997) RNA, 3, 337–343. [PMC free article] [PubMed] [Google Scholar]
- 82.Hartshorne T. (1998) Nucleic Acids Res., 26, 2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lange T.S., Ezrokhi,M., Borovjagin,A.V., Rivera-León,R., North,M.T. and Gerbi,S.A. (1998) Mol. Biol. Cell, 9, 2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beltrame M. and Tollervey,D. (1995) EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]