Abstract
U3 small nucleolar RNA (snoRNA) is a member of the Box C/D family of snoRNAs which functions in ribosomal RNA processing. U3-55k is a protein that has been found to interact with U3 but not other members of the Box C/D snoRNA family. We have found that interaction of the U3-55k protein with U3 RNA in vivo is mediated by the conserved Box B/C motif which is unique to U3 snoRNA. Mutation of Box B and Box C, but not of other conserved sequence elements, disrupted interaction of U3-55k with U3 RNA. Furthermore, a fragment of U3 containing only these two conserved elements was bound by U3-55k in vivo. RNA binding assays performed in vitro indicate that Box C may be the primary determinant of the interaction. We have cloned the cDNA encoding the Xenopus laevis U3-55k protein and find strong homology to the human sequence, including six WD repeats. Deletion of WD repeats or sequences near the C-terminus of U3-55k resulted in loss of association with U3 RNA and also loss of localization of U3-55k to the nucleolus, suggesting that protein–protein interactions contribute to the localization and RNA binding of U3-55k in vivo.
INTRODUCTION
Eukaryotic ribosome production requires the contribution of small nucleolar RNAs (snoRNAs) in the processing and modification of ribosomal RNA (rRNA). Involvement of snoRNAs in rRNA biogenesis includes the processing of the primary rRNA transcript into the mature 18S, 5.8S and 28S rRNAs, the methylation of selected ribose moieties (2′ O-methylation) and the isomerization of specific uridine residues (pseudouridylation) (1–6). More than 150 snoRNAs have been identified and (with the exception of MRP RNA) are divided into two major classes: the Box C/D snoRNAs and the Box H/ACA snoRNAs (7,8). These two classes are distinguished by conserved sequence/secondary structure elements and by associated proteins (7,8). Both snoRNAs and their associated proteins are essential to the maturation of rRNA, indicating that the functional complexes in rRNA processing and modification are small nucleolar ribonucleoprotein particles (snoRNPs) (9).
U3 snoRNA is a Box C/D snoRNA essential for the processing of 18S rRNA from the pre-rRNA transcript (10–16). Sequence analysis of U3 snoRNA reveals six evolutionarily conserved sequence elements, termed Boxes A, A′, B, C, C′ and D (17–27). Boxes A and A′ are located in the 5′ domain of U3 snoRNA and, along with the ‘hinge’ region, base pair to sites within the pre-rRNA to direct the multiple endonucleolytic cleavages necessary for the maturation of 18S rRNA (15,16,23,25,28). Boxes B, C, C′ and D are found in the 3′ domain of U3 and have been characterized as sequences required for protein association (17,19,23,25,29). The secondary structure of the 3′ domain positions Boxes B and C, as well as Boxes C′ and D into discrete loops flanked by base paired stems to form the Box B/C and Box C′/D motifs, respectively (23,27,30). The Box C′/D motif of U3 is equivalent to the Box C/D motifs of other members of this snoRNA family in sequence/structure and function (23,27,30). The Box B/C motif is unique to U3 snoRNA. Both the Box B/C and Box C′/D motifs operate as independent units of U3 RNA nuclear retention in the Xenopus oocyte (30). The Box C′/D motif of U3 also directs the localization of the snoRNA to the nucleolus (31). The functions associated with the Box B/C and Box C′/D sequence motifs are very likely mediated by proteins that bind at these sites.
The U3 snoRNA exists as a U3 snoRNP in vivo (20,32,33). The RNA is associated with proteins common to all Box C/D snoRNAs, including fibrillarin/Nop1p (34,35), Nop56p (36) and Nop58/Nop5p (37–39) and proteins specific to the U3 snoRNP, including Sof1p (supressor of fibrillarin; 40), Mpp10 (41,42), Lcp5p (43), Imp3p (44), Imp4p (44) and U3-55k (29,45). Each of these proteins (like U3 RNA) are essential and have been shown to be specifically required for the maturation of 18S rRNA [39–41,43,44,46,47, and J.Venema and H.A.Raue (Vrije Universiteit, Amsterdam, The Netherlands) personal communication]. U3-55k, the focus of this work, was initially identified as part of a trimeric complex (p55, p50 and p15) that co-purified from CHO cells with U3 snoRNA. The co-purification of the trimeric complex with U3 snoRNA was carried out under conditions of high salt (650 mM KCl or 500 mM NaCl), suggesting that the three proteins are core components of the U3 snoRNP (29). Lubben et al. analyzed deletion mutants of human U3 RNA and determined that a central region of the RNA (nt 97–204) was important for binding of a complex that included CHO U3-55k in vitro (29). The cDNA for the U3-55k protein was subsequently isolated from a human teratocarcinoma library and the protein was characterized in HeLa cells (45). Pluk et al. found that human U3-55k (hU3-55k) co-localized with fibrillarin in nucleoli and showed the association of hU3-55k with U3 in vivo (45). In addition, characterization of several hU3-55k mutants revealed that replacement of 17 amino acids at the C-terminus disrupted localization of the protein to nucleoli and the ability of U3-55k to associate with U3 snoRNA in vivo (45).
U3-55k belongs to the WD repeat family of proteins (45), characterized by the core sequence [GH]-(X23–41)-[WD] arranged in repeats of four to eight units (48–50). β-Transducin (Gβ), a member of the same family, has seven WD repeats which fold into a propeller-like structure with each repeat motif forming a discrete blade of the propeller (51–53). Although Gβ is the only WD repeat protein whose structure has been determined by crystallography, structural analysis and homology modeling of other WD proteins indicate that the propeller structure may be common to the members of the family (54–57). The structure appears to provide a platform on which protein–protein interactions can occur, with the specificity of these interactions being directed by variable sequences within each WD repeat.
WD repeat proteins have been found to function in a variety of processes (49). Several WD repeat proteins in addition to U3-55k (e.g. PRP4, PRP17, U5-40kD and Sof1) have been found to be associated with specific RNAs (40,58–64). Like U3-55k, these proteins do not contain known RNA binding motifs and it remains to be determined whether these proteins interact directly with the associated RNAs.
We have isolated the Xenopus homolog of U3-55k (XlU3-55k) and characterized the sequences of Xenopus U3 RNA important for interaction with U3-55k both in vivo and in vitro. Our results indicate that the Box B/C motif of U3 snoRNA is both necessary and sufficient for U3-55k binding in vivo. In vitro analysis suggests that Box C is of greater importance than Box B in the interaction between U3 and U3-55k. In addition, we have examined the effects of deletion of regions of hU3-55k on nucleolar localization of the protein and U3 snoRNP assembly. While nuclear localization sequences and a glutamic acid-rich region near the N-terminus are not required, our results indicate that the WD repeats and specific sequences near the C-terminus of hU3-55k are required for localization to the nucleolus and interaction with U3 snoRNA.
MATERIALS AND METHODS
Cloning of the Xenopus laevis U3-55k
The oligonucleotides used in the cloning of XlU3-55k were: 84, GAGCTGGARGARACYGCACAGGAAAAGAA; 85, TTCTCCACCATCGGCCAAGCCTGTGCTC; 111, CAATGGCATCCTGATCCTCG; 124, CTGGGATCCTCTGACATG; 166, GGAATTCCATATGGAATTCATGTCCGGCCTTTTCATCAAAAAGAAATCAGG; 167, GAAGATCTGAATTCGTGCAACAGGGTCAACAAAATATGTCTATGTGG.
The Xenopus homolog of hU3-55k was isolated using a combination of RT–PCR and rapid amplification of cDNA ends (RACE) against mRNA isolated from stage V and VI X.laevis oocytes (mRNA Separator Kit, Clontech, CA). Degenerate oligonucleotides, 84 and 85, were designed based on sequences conserved between human (accession no. AJ001340) and potential mouse (accession no. AA170033) or yeast (accession no. U40829, ORF: YPR137W) U3-55k sequences, respectively, and used in RT–PCR (Boehringer Mannheim, Indianapolis, IN) to amplify an internal fragment (1090 bp) of the Xenopus U3-55k cDNA. Primers 111 and 124 were used in RACE to isolate the 5′ and 3′ ends of the cDNA, respectively (Marathon-Ready cDNA, Clontech, and 3′ RACE system, Gibco BRL, MD). Primers 166 (start codon in bold) and 167 (3′-UTR) were designed with incorporated EcoRI sites (underlined) immediately flanking the predicted protein coding region. Using these primers, RT–PCR was used to isolate the full-length (1451 bp) cDNA from purified Xenopus oocyte mRNA and the product was subcloned and sequenced.
Expression of recombinant Xenopus U3-55k and production of antibodies
The Xenopus U3-55k cDNA was excised by digestion at the engineered EcoRI sites flanking the encoding cDNA, and cloned in frame into the EcoRI site of the pET32a expression vector (Novagen, WI). The fusion protein was expressed in Escherichia coli BL21(DE3) and isolated by nickel chelation affinity chromatography under denaturing conditions (His-Bind Resin and Buffer Kit, Novagen) and gel purification. One New Zealand white rabbit was immunized with 250 µg of the purified recombinant protein (Animal Resources, University of Georgia, Athens, GA). Antibodies were affinity purified using purified XlU3-55k protein coupled to an N-hydroxysuccinimide-activated Sepharose column (HiTrap, Amersham Pharmacia Biotech, Piscataway, NJ).
U3 snoRNA mutant constructs
Construction of the Xenopus U3 snoRNA single box substitutions A, A′, B, C, C′ and D, the 3′ domain sub-fragment (nt 75–220), the C′/D sub-fragment (nt 75–104, tetraloop GCUU, nt 198–220) and B/C sub-fragment (nt 93–208) has been detailed (31).
Injection of RNAs into Xenopus oocytes
The procedure for microinjection of Xenopus oocyte nuclei has been described in detail (30,31). In short, a mixture of 32P-labeled RNAs was resuspended in 20 mg/ml of blue dextran (at a final concentration of 100 fmol/µl U3 snoRNA). Nuclear injections of X.laevis oocytes (stages V and VI) delivered 10 nl (1 fmol of U3) of sample and cells were incubated for 4 h at 18°C in MBSH buffer [88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 20 mM HEPES pH 7.5, 0.82 MgSO4, 0.33 mM Ca(NO3)2, 0.41 mM CaCl2, containing 10 µg/ml of penicillin and streptomycin]. After incubation, nuclear extracts were prepared by manual enucleation of injected oocytes and homogenized in Ipp100-T buffer (100 mM NaCl, 50 mM Tris–HCl, pH 7.5, and 0.05% Triton X-100).
Construction of hU3-55k deletion mutants
The oligonucleotides used to generate hU3-55k deletion mutants were: 55k140-For, GCCTCAGCTGACATTCGGATCCTACGGGGGCACCAGC; 55k140-Rev, GCTGGTGCCCCCGTAGGATCCGATGTCAGCTGAGGC; 55k193-For, GAAGGGTGCCGAGGGATCCCCCCCTGGCCACAGC; 55k193-Rev, GCTGTGGCCAGGGGGGGATCCCTCGGCACCCTTC; 55k230-For, CATTTGGGAGGCCCAGAGGATCCAGCACTTGTACACCTTC; 55k230-Rev, GAAGGTGTACAGTGCTGGATCCTCTGGGCCTCCCAAATG; 55k315-For, GGAAGATCCCCGAGGGATCCCAGCTTGTCTTCTATG; 55k315-Rev, CATAGAAGACAAGCTGGATCCCTCGGGGATCTTCC; 55k352-For, GTGGCCTTGTGGGGTGGATCCAAGAAGCGACCACTTG; 55k352-Rev, CAAGTGGTCGCTTCTTGGATCCACCCCACAAGGCCAC; 55kΔE-For, GCCTAGCTCCAAGGAAGCCTACTGCACAGGAAAAGAAGCTG; 55kΔE-Rev, CAGCTTCTTTTCCTGTGCAGTAGGCTTCCTTGGAGCTAGGC; 55kA, GCAGCCCTTCTGGATATCGTCGG; 55kB, AGAATCAAAGAGGCTCGGAATTCTTGATCTAGAGCG; and 55kC, TGCATCATCCCACTCCGCAGGTGATCTAGAGCG.
pCI-neo VSV-55k was constructed as described (45). The original hU3-55k cDNA was modified by PCR to introduce an XhoI site before the translational start codon [pGEM3Zf(+)-hU3-55k (45)].
The hU3-55k deletion mutants Δ459–475, Δ467–475 and Δ64–74 were generated via mutagenic PCR using pGEM 3Zf(+)-hU3-55k as template. Deletion mutant Δ459–475 was made using oligonucleotides 55kA and 55kB which incorporate a stop codon (indicated in bold) at amino acid position 458 followed by an in-frame XbaI restriction site (underlined). Deletion mutant Δ467–475 was made using oligonucleotides 55kA and 55kC which incorporate a stop codon (bold) at amino acid position 466, followed by an in-frame XbaI site (underlined). The Δ459–475 and Δ467–475 PCR products were digested with EcoRV and XbaI and cloned into wild-type pCI-neo VSV-55k digested with EcoRV and XbaI. Deletion mutant Δ64–74 was generated by PCR (QuickChange site-directed mutagenesis kit, Stratagene, The Netherlands) using oligonucleotides 55kΔE-For and 55kΔE-Rev and pGEM3Zf(+)-hU3-55k as template. The PCR product was digested with BglII and NarI and the 180 bp product, which lacked the E-stretch, was cloned into the wild-type pGEM3Zf(+)-hU3-55k vector digested with BglII and NarI, thereby replacing the wild-type sequence. The resulting construct, pGEM3Zf(+)-hU3-55kΔE, was subsequently digested with HindIII, made blunt with Klenow polymerase and partially digested with XhoI. The resulting hU3-55kΔE fragment was ligated into an XhoI and SmaI-digested pCI-neo vector that contains a 5′ VSV-G tag (MEIYTDIEMNRLGK). The resulting construct contained a 5′ VSV tag in-frame with the coding sequence of hU3-55k.
To construct the hU3-55k WD deletion mutants, PCR mutagenesis was performed on the pGEM3Zf(+)-hU3-55k cDNA using appropriate combinations of the oligonucleotides listed above (QuickChange site-directed mutagenesis kit). In-frame deletions of WD repeats were generated from five pGEM3Zf(+)-55k constructs that contain a BamHI site (underlined) at nucleotide positions 452 (55k452), 610 (55k610), 723 (55k723), 977 (55k977) or 1087 (55k1087), corresponding to amino acids 140, 193, 230, 315 or 352. 55kΔ140–230 was made by combining the XhoI–BamHI fragment of 55k452 with the BamHI–HindIII fragment of 55k723. 55kΔ193–315 was created by combining the XhoI–BamHI fragment of 55k610 with the BamHI–HindIII fragment of 55k977. 55kΔ193–352 was created by combining the XhoI–BamHI fragment of 55k610 with the BamHI–HindIII fragment of 55k1087.
Finally, all pGEM3Zf(+)-55k deletion mutant constructs were digested with HindIII, made blunt with Klenow polymerase and partially digested with XhoI. The resulting hU3-55k fragments were ligated into an XhoI and SmaI-digested pCI-neo vector. The resulting constructs contained a 5′ VSV tag in-frame with the coding sequence of hU3-55k. The correctness of each construct was confirmed by sequence analysis.
Expression of hU3-55k constructs in COS-1 and HEp-2 cells
hU3-55k constructs were transiently transfected into cultured cells. Monolayers of cells were grown to 80% confluence. COS-1 cells (4–5 × 106) were transfected in 400 µl phosphate-buffered saline (PBS) with 20 µg DNA. Electroporation was performed at 300 V and a capacity of 125 µFa with Gene Pulser II (Bio-Rad, Richmond, CA). HEp-2 cells (2.5 × 106) were transfected in 800 µl of Dulbecco’s modified Eagle’s medium (DMEM) with 10 µg of DNA. Electroporation was performed at 260 V and a capacity of 950 µFa with Gene Pulser II (Bio-Rad). After electroporation, cells were resuspended in 5 or 10 ml of DMEM supplemented with 10% fetal calf serum and grown for 18 h before use.
Immunoblot analysis
Analysis of Xenopus nuclear proteins with antibodies against XlU3-55k. Nuclear samples were prepared by manual dissection of nuclei from stage VI Xenopus oocytes and were electrophoresed on a 10% SDS–polyacrylamide gel with five nuclear equivalents per lane. Proteins were transferred to nitrocellulose (Bio-Rad). Immobilized proteins were blocked for 1 h with 5% non-fat milk in TBS (100 mM Tris, 0.9% NaCl, pH 7.5) and washed three times in TBS. Pre-immune serum was diluted 1:100 and affinity purified αXlU3-55k antibodies (∼2 µg/µl) were diluted 1:2000 in TTBS (0.5% Tween-20 in TBS), and incubated with membranes for 1 h at room temperature. Membranes were washed and incubated with anti-rabbit IgG (whole molecule) alkaline phosphatase conjugated antibodies (Sigma, St Louis, MO) at a dilution of 1:5000 in TTBS for 1 h at room temperature. After washing, immunoreactive proteins were visualized by incubation with NBT/BCIP (Sigma) for 10 min.
Analysis of expression of VSV-hU3-55k constructs in COS-1 cells. Extract prepared from 1.0 × 106 COS-1 cells was acetone precipitated, the proteins resolved by 10% SDS–PAGE and blotted to a nitrocellulose membrane (Protan, Schleicher & Schuell, Dassel, Germany). After 1 h incubation in blocking buffer (5% non-fat milk, 0.1% NP-40, in PBS), the blot was incubated with anti-VSV antibodies (Boehringer Mannheim, Almere, The Netherlands) diluted 1:50 in buffer B (3% non-fat milk, 0.1% NP-40, in PBS) supplemented with 1% normal rabbit serum for 1 h at room temperature. The blot was washed in buffer B, and then incubated with horseradish peroxidase-conjugated, goat anti-mouse antibodies (Dako, Denmark) for 1 h at room temperature. After washing, bound secondary antibodies were visualized via chemiluminescence.
Immunofluorescence analysis
Analysis of X1U3-55k in Xenopus cells. Indirect immunofluorescence analysis was carried out on Xenopus oocyte nuclear spreads as described (31). Briefly, Xenopus nuclear contents were centrifuged onto a slide (5140 g for 1 h), fixed with 1% paraformaldehyde in PBS for 1 h and blocked with 10% horse serum (Hyclone, Logan, UT) in PBS for 15 min at 37°C. Slides were washed and incubated with pre-immune serum (diluted 1:1000 in PBS) or affinity purified αXlU3-55k antibodies (diluted 1:1000 in PBS) for 1 h at 37°C. Slides were washed and incubated with FITC-conjugated anti-rabbit antibody (Jackson ImmunoResearch, PA) at 1:50 dilution in PBS for 1 h at 37°C. Slides were washed and mounted with 50% glycerol in PBS containing 1 mg/ml phenylenediamine, pH 9.0.
For indirect immunofluorescence of Xenopus somatic cells, XlK2 cells were grown on autoclaved glass slides to ∼80% confluence. Slides were rinsed in PBS and fixed for 10 min in 5% paraformaldehyde in PBS at 25°C. Cells were permeabilized in acetone (pre-chilled at –20°C) for 5 min, rinsed and blocked in 3% BSA in PBS for 1 h at room temperature. Slides were rinsed and incubated with pre-immune serum (diluted 1:1000) or affinity purified antibodies (diluted 1:20 000) in 3% BSA in PBS for 1 h at 37°C. Slides were washed and Texas Red-conjugated anti-rabbit antibodies (Jackson Immunoresearch) were added at 1:50 dilution in 3% BSA in PBS and incubated for 45 min at room temperature. Slides were washed and mounted as described above.
Analysis of Xenopus oocyte nuclear spreads and kidney cells was carried out on a Zeiss Axiovert S 100 inverted fluorescence microscope equipped with differential interference contrast optics (Thornwood, NY). Images were acquired at 63× magnification using a cooled charge-coupled device camera (Quantix-Photometrix, AZ) and IP Lab Spectrum software (Signal Analytics, VA).
Analysis of VSV-hU3-55k proteins in HEp-2 cells. Transiently transfected HEp-2 cells were grown for 18 h. The slides were washed with PBS, placed in methanol for 5 min at –20°C and permeabilized with acetone for 1 min at room temperature. Indirect immunofluorescence was performed using monoclonal anti-fibrillarin antibody 72B9 (65) and monoclonal anti-VSV antibody (Boehringer Mannheim, Almere, The Netherlands) diluted 1:2 and 1:50, respectively, in PBS supplemented with 1% normal rabbit serum. Fixed cells were incubated with monoclonal antibodies for 1 h, followed by goat anti-mouse antibodies coupled to fluorescein isothiocyanate (Dako; diluted 1:50 in PBS) for 1 h.
Images were aquired using an Olympus BH2 microscope (Olympus, Hamburg, Germany) in combination with an Olympus DP10 microscope digital camera system and analySIS sofware (Soft Imaging System GMBH, Münster, Germany)
Immunoprecipitations
Xenopus U3-55k. Antiserum against XlU3-55k (10 µl/immunoprecipitation) was coupled to protein A–Sepharose CL-4C beads (Sigma) in 0.5 ml of Ipp 500 buffer (500 mM NaCl, 50 mM Tris–HCl, pH 7.5, and 0.1% NP-40) at 4°C overnight. Immunoprecipitations were performed with nuclear extracts isolated from injected oocytes (three nuclei/immunoprecipitation) in Ipp100-T. Samples were incubated overnight at 4°C in the presence of 40 U RNasin (Promega, Madison, WI) and 2 mM dithiothreitol (DTT). RNA from supernatants and pellets was isolated by proteinase K digestion, phenol extraction and ethanol precipitation. Precipitated RNAs were separated by 8% urea–PAGE and visualized by autoradiography.
Human U3-55k. After 18 h of growth, transfected COS-1 cells were washed with PBS and resuspended in buffer A [25 mM Tris–HCl, pH 7.5, 100 mM KCl, 1 mM DTE (1,4-dithioerythritol), 2 mM EDTA, 0.5 mM PMSF, 0.05% NP-40] to ∼1000 cells/µl. The cell suspension was sonicated with a Branson microtip (three times for 20 s each), insoluble material was pelleted (12 000 g, 15 min) and the supernatants were used for immunoprecipitation. Rabbit anti-mouse antibodies (Dako) were coupled to 10 µl of protein A agarose beads (Biozym, Landgraaf, The Netherlands) in IPB500 (500 mM NaCl, 10 mM Tris–Cl, pH 8.0, 0.05% NP-40). The beads were washed with IPB500 and then incubated with monoclonal anti-VSV and anti-fibrillarin antibodies for 1 h at room temperature. The beads were subsequently washed twice with IPB500 and once with IPB150 (150 mM NaCl, 10 mM Tris–Cl, pH 8.0, 0.05% NP-40) and 250 µl of cell extract (∼2.5 × 105 cells) was added and incubated for 1 h at 4°C. After washing three times with IPB150, RNA was isolated from the beads by phenol–chloroform extraction and ethanol precipitation. RNA was resolved on an 8% denaturing polyacrylamide gel and blotted onto a Hybond-N+ membrane (Amersham, Buckinghamshire, UK). Northern analysis performed with anti-sense radiolabeled U3 RNA probes was done essentially as previously described (45,66).
Analysis of U3-55k association with U3 snoRNA in vitro
Biotinylated RNAs were synthesized using a 10:1 ratio of UTP to UTP-21-biotin (Clontech) and the yield estimated by ethidium bromide staining (to allow addition of comparable amounts of each RNA to reactions). In vitro translated, 35S-labeled hU3-55k was synthesized using wheat germ extract (TNT Coupled Wheat Germ Extract System, Promega) in the presence of translation grade [35S]methionine (1000 Ci/mmol; Amersham, NJ). Approximately 5% of the biotinylated RNA made in a 25 µl transcription reaction was added to 10 µl of in vitro translated hU3-55k, 40 U RNasin (Promega), 2 µg non-specific competitor yeast tRNA (Boehringer Mannheim, IN), and 2 µl of 10× binding assay buffer (100 mM Tris, pH 8.0, 1 M KCl and 0.5% NP-40). Reactions were brought to a final volume of 20 µl with H2O and allowed to incubate for 1 h at 4°C. After incubation, 10 µl of streptavidin agarose beads (Gibco BRL, Grand Island, NY) were added and the final volume was brought to 500 µl with 1× binding assay buffer. Samples were incubated for 1 h at 4°C. After incubation, pellets were washed three times with 1 ml ice-cold 1× binding assay buffer and resuspended in SDS–PAGE loading buffer. Samples were boiled and resolved by 12% SDS–PAGE. Gels were fixed in water:isopropanol:glacial acetic acid (65:25:10) for 30 min, incubated in Amplify (Amersham, NJ) for an additional 30 min, dried and exposed to film at –80°C.
RESULTS
Isolation and characterization of Xenopus U3-55k
U3-55k has been found to interact specifically with U3 snoRNA in hamster and human cells (29,45). In order to determine the U3 RNA sequences required for the association of U3-55k in vivo (using the Xenopus oocyte system), we cloned the cDNA encoding the X.laevis homolog of U3-55k. The Xenopus clone was identified by RT–PCR using degenerate oligonucleotide primers based on alignments of the human U3-55k (45) with sequences from potential mouse and yeast homologs present in the GenBank/EMBL sequence database (Materials and Methods). The predicted amino acid sequence of the Xenopus homolog, XlU3-55k, is shown with the human and yeast sequences in Figure 1. The Xenopus U3-55k protein is ∼65% identical and 73% similar to the human, but 41% identical and 54% similar to the yeast homolog. The positions of each of five WD repeats initially identified in hU3-55k (45) are conserved in XlU3-55k (Fig. 1, WD1-5). In addition, we report the presence of a sixth WD repeat in both human and Xenopus U3-55k (Fig. 1, WD6). The yeast U3-55k contains stretches of amino acids not found in the human and Xenopus proteins (e.g. between WD4 and WD5) and shares five of the six WD repeats found in human and Xenopus (WD2–6) but lacks a WD consensus sequence at WD1. A Caenorhabditis elegans homolog of U3-55k (CE21137) contains four of the six WD repeats corresponding to human/Xenopus WD3–6 (R.Terns and M.Terns, unpublished data). Putative nuclear localization signals were identified within the first 45 amino acids of U3-55k (45) and these signals appear to be conserved in Xenopus U3-55k. Xenopus U3-55k also contains a glutamic acid-rich region near the N-terminus (amino acids 60–65) that is found in the human and yeast homologs. The sequence analysis indicates that we have isolated the X.laevis homolog of U3-55k and that Xenopus and human U3-55k each contain six potential WD domains.
Figure 1.
Alignment of X.laevis U3-55k with human and yeast homologs. An alignment between the predicted amino acid sequences of X.laevis (Xl), human (h) and Saccharomyces cerevisiae (Sc) U3-55k was generated using PILEUP from the GCG Wisconsin Sequence Analysis package (Version 10). Residues that are identical in two sequences are in black boxes; similar residues are in gray boxes. WD repeat motifs were identified using the Protein Structure Analysis server at the BioMolecular Engineering Research Center (http://BMERC-www.bu.edu/ ) and are indicated with brackets. The glutamic acid-rich region defined by Pluk et al. (45) is indicated with a bar (E-rich region).
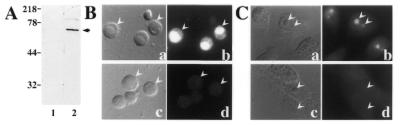
The XlU3-55k protein was expressed in E.coli and used to generate antibodies (Materials and Methods). The affinity purified antibodies recognize an ∼60 kDa protein by western blot analysis of Xenopus oocyte nuclear extract (Fig. 2A). We assessed the cellular localization of XlU3-55k in nuclear spreads prepared from Xenopus oocytes and in a Xenopus somatic cell line using the αU3-55k antibodies (Fig. 2B and C). The immunofluorescence analysis revealed that XlU3-55k is present in the nucleoli of both cell types.
Figure 2.
Antibodies against U3-55k recognize a nucleolar protein in Xenopus cells. Polyclonal antibodies were raised against recombinant Xenopus U3-55k. (A) Immunoblot analysis of extract from five Xenopus oocyte nuclei was performed using pre-immune sera (lane 1) or affinity purified αU3-55k antibodies (lane 2). The arrow indicates the primary band corresponding to an ∼60 kDa protein. Immunofluorescence analysis was carried out on Xenopus oocyte nuclear spreads (B) and Xenopus kidney cells (C). Differential interference contrast (DIC, panels a and c) and fluorescence (panels b and d) microscopy images are shown for each experiment. Representative nucleolar structures are indicated with arrowheads. Samples were analyzed using pre-immune (panel d) and affinity purified αU3-55k (panel b) antibodies.
Analysis of U3 RNA sequences required for U3-55k association in vivo
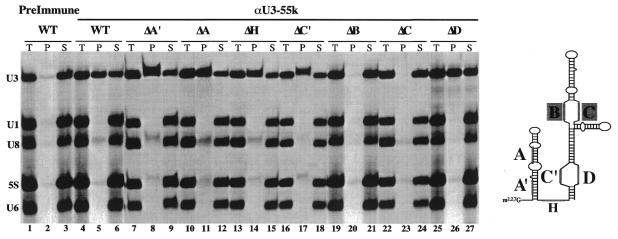
We have used the antibodies against XlU3-55k to investigate the sequences in U3 snoRNA required for interaction with U3-55k in vivo. In these experiments, radiolabeled Xenopus U3 and control RNAs (U1 and U6 snRNAs, U8 snoRNA and 5S rRNA) were co-injected into Xenopus oocytes and allowed to incorporate into RNP complexes. Four hours after injection, RNP complexes containing U3-55k were immunoprecipitated from oocyte nuclear extracts. The αU3-55k antibodies specifically immunoprecipitated a complex containing U3 RNA (Fig. 3, lane 5). In order to determine which conserved U3 RNA sequence elements are necessary for interaction with U3-55k, we substituted the sequence of each of the phylogenetically conserved U3 Box elements, A, A′, B, C, C′ and D as well as the hinge with complementary sequences (Fig. 3, Δ indicates substitution). Each of the radiolabeled U3 mutant RNAs was co-injected with the control RNAs and tested for interaction with the U3-55k protein by immunoprecipitation. We found that substitution of the Box B or C elements of U3 RNA disrupted association with the U3-55k protein (Fig. 3, U3ΔB and U3ΔC, lanes 19–24). Substitution of the other sequence elements did not prevent interaction (Fig. 3, U3ΔA′, U3ΔA, U3ΔH, U3ΔC′ and U3ΔD, lanes 7–18, 25–27). The results indicate that Boxes B and C of U3 snoRNA are necessary for interaction with U3-55k in vivo.
Figure 3.
Boxes B and C of U3 snoRNA are required for U3-55k association in vivo. In vitro transcribed, 32P-labeled U3 and U8 snoRNAs, U1 and U6 snRNAs and 5S rRNA were co-injected into the nuclei of Xenopus oocytes. After 4 h, complexes were immunoprecipitated using αU3-55k antibodies and co-immunoprecipiated RNAs were isolated from both pellet (P) and supernatant (S) fractions and analyzed by gel electrophoresis and autoradiography. For each experiment, 100% of the pellet (P) and 50% of the total (T) and supernatant (S) samples were analyzed. Immunoprecipitation using pre-immune serum is shown in lanes 1–3. Immunoprecipitation, using αU3-55k antibodies with wild-type and variant U3 snoRNAs are shown in lanes 4–27. The positions of the conserved sequence elements are indicated on the U3 RNA secondary structure model diagram.
In order to assess whether the Box B/C motif of U3 RNA is also sufficient for U3-55k binding, we tested the ability of stable fragments of U3 RNA to interact with U3-55k. Again, radiolabeled RNAs were injected and interaction was assessed by immunoprecipitation. A fragment consisting of the entire 3′ domain of U3 was bound by U3-55k in vivo (Fig. 4, 3′ Domain, lanes 1–3), but as expected U3-55k did not interact with a sub-fragment consisting of the Box C′/D motif (composed of U3 Box C′, Box D and flanking stems; Fig. 4, lanes 7–9). A sub-fragment containing Box B, Box C and the adjacent stem regions of U3 RNA was co-immunoprecipitated with αU3-55k antibodies (Fig. 4, lanes 4–6) indicating that the Box B/C motif is sufficient for interaction with U3-55k in vivo.
Figure 4.

Fragment of U3 snoRNA containing Boxes B and C is sufficient for U3-55k association in vivo. 32P-labeled U3 snoRNA fragments and U1 snRNA (negative control) were injected into the nuclei of Xenopus oocytes and tested for co-immunoprecipitation with αU3-55k antibodies as described in Figure 3. The B/C sub-fragment undergoes limited 3′-end trimming in Xenopus oocytes (30). The sub-fragments of U3 snoRNA are shown schematically above each panel.
Interaction of U3-55k with U3 RNA in vitro
We have found that Box B and Box C are each necessary for interaction of U3-55k in vivo (Fig. 3). Lubben et al. reported that interaction of a complex of proteins that included U3-55k with U3 RNA in vitro required a region of the RNA containing both Box B and Box C (29). Interestingly, we found that Box C is the primary determinant of U3-55k binding in an in vitro binding assay. In vitro translated, radiolabeled hU3-55k was incubated with biotinylated Xenopus U3 RNA, the RNA was selected using streptavidin beads and the association of U3-55k was assessed by fluorography. We found that U3-55k bound to U3 RNA (Fig. 5A). While only a small fraction (∼0.5%) of the U3-55k added to the reaction was co-precipitated, the interaction was reproducible and specific to U3. U3-55k did not interact with U8, another Box C/D snoRNA (Fig. 5A and B, lanes 7 and 4, respectively). The amount of U3-55k observed with U8 in Figure 5A (lane 7) was similar to that bound to streptavidin beads alone (data not shown).
Figure 5.
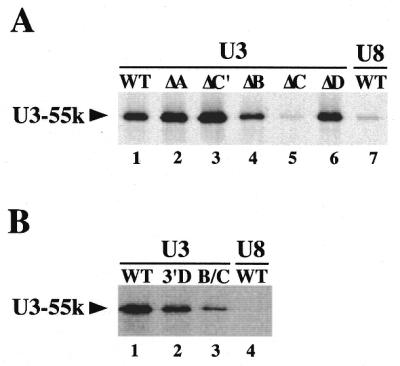
Box C of U3 snoRNA is required for U3-55k association in vitro. In vitro translated, 35S-labeled hU3-55k was added to in vitro transcribed, UTP-21-biotin-labeled RNA. The mixture was incubated for 1 h and biotinylated RNAs were selected from the mixture with streptavidin agarose beads. hU3-55k associated with the biotinylated RNA was eluted by boiling in SDS loading buffer and analyzed by SDS–PAGE and fluorography. Biotinylated RNAs used in each reaction are indicated above each lane. (A) Evaluation of U3-55k association with wild-type (lane 1) and mutant (ΔA, ΔC′, ΔB, ΔC and ΔD, lanes 2–6, respectively) U3 snoRNA as well as U8 (lane 7). (B) Evaluation of U3-55k association with U3 snoRNA 3′ domain and Box B/C sub-fragments (lanes 2 and 3). Wild-type U3 and U8 are shown in lanes 1 and 4.
Furthermore, we have found that the interaction of U3-55k with U3 RNA in vitro depends upon specific sequences in U3. Substitution of Boxes A, C′ and D of U3 did not reduce the association of U3-55k with the RNA (Fig. 5A). However, mutation of Box C eliminated the interaction of U3-55k with the U3 RNA, reducing it to the background level observed with U8 RNA (Fig. 5A). Disruption of Box B did not prevent U3-55k binding in vitro, but did appear to reduce binding relative to wild-type and other mutants (Fig. 5A). Our results indicate that the interaction of U3-55k with U3 RNA in vitro depends predominantly on Box C and to a lesser extent on Box B of U3. While it is important to note that we cannot exclude the possibility that protein components present in the wheat germ extract used to generate the U3-55k protein contributed to the U3 RNA binding, our results may indicate that U3-55k interacts directly with U3 RNA. Assessment of direct binding of purified, recombinant U3-55k expressed in E.coli to U3 RNA was not possible due to the insolubility of the protein under all tested conditions.
We tested the ability of U3-55k to interact with fragments of U3 RNA containing the Box B/C motif in vitro. Both the 3′ domain of U3 and a sub-fragment comprised of Box B, Box C and flanking stem sequences were bound by the U3-55k protein in vitro (Fig. 5B). The interactions with the fragments appeared to be reduced relative to wild-type U3 but were substantially greater than the background observed with U8 RNA. In summary, our results suggest that the Box B/C motif of U3 is necessary and sufficient for interaction of U3-55k in vivo, but that Box C may be a primary determinant of the interaction.
Analysis of U3-55k sequences required for nucleolar localization and association with U3 RNA
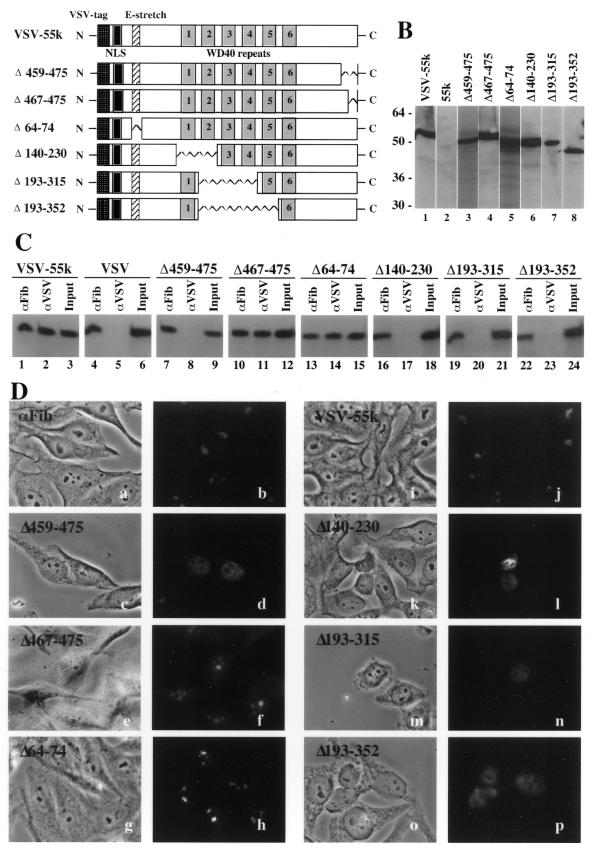
Previous work indicated that replacement of 17 amino acids at the C-terminus of hU3-55k (with 15 vector-encoded amino acids) disrupted interaction of U3-55k with U3 RNA and resulted in loss of localization of U3-55k to the nucleolus (45). Deletion of 45 amino acids from the N-terminus of the protein, including a putative nuclear localization sequence, did not prevent interaction with U3 or nucleolar localization of the protein (45). We have further investigated the role of hU3-55k sequences in these two processes in experiments using VSV-tagged hU3-55k deletion mutants (Fig. 6A) expressed in transfected cells.
Figure 6.
U3-55k sequences required for association with U3 RNA and localization of U3-55k to the nucleolus. (A) The positions of the VSV tag, nuclear localization signals, glutamic acid-rich region and WD repeats in the VSV-tagged wild-type human U3-55k protein (VSV-55k) are represented with boxes. Regions of the protein deleted in each of the mutant constructs analyzed in this work are represented with zig-zag lines. (B) Expression levels of hU3-55k deletion mutants. Western blots using anti-VSV antibodies are shown for COS-1 cells expressing untagged hU3-55k (lane 2), VSV-tagged wild-type U3-55k (lane 1) and deletion constructs Δ459–475 (lane 3), Δ467–475 (lane 4), Δ64–74 (lane 5), Δ140–230 (lane 6), Δ193–315 (lane 7) and Δ193–352 (lane 8). Lanes 3 and 5 are taken from a different blot. (C) The effect of hU3-55k deletions on U3 snoRNP association. VSV-tagged human U3-55k wild-type and deletion constructs were expressed in COS-1 cells and complexes containing the tagged proteins (or endogenous fibrillarin) were immunoprecipitated using αVSV (or αFib) antibodies. Co-immunoprecipitated U3 RNA was detected by northern blotting. Co-immunoprecipitation of U3 with fibrillarin is shown for each experiment. The input lanes contain 10% of RNA isolated from extracts prepared for immunoprecipitation. (D) Cellular localization of hU3-55k wild-type and deletion mutants. VSV-tagged U3-55k wild-type and deletion constructs were expressed in HEp-2 cells and the localization of the tagged proteins (or endogenous fibrillarin) was assessed by immunofluorescence using αVSV (panels c–p) [or αFib (panels a and b)] antibodies. Phase contrast (panels a, c, e, g, i, k, m and o) and fluorescence (panels b, d, f, h, j, l, n and p) microscopy images are shown for each experiment. Fibrillarin is found primarily in the phase-dark nucleoli of HEp-2 cells (panels a and b). VSV-tagged wild-type U3-55k (panels i and j) and deletion constructs Δ467–475 (panels e and f) and Δ64–74 (panels g and h) were localized to nucleoli. Deletion mutants Δ459–475 (panels c and d), Δ140–230 (panels k and l), Δ193–315 (panels m and n) and Δ193–352 (panels o and p) did not localize to nucleoli but accumulated in the nucleoplasm.
We first assessed the level of expression of each of the VSV-tagged proteins by western analysis of total extract of 1.0 × 106 transfected COS-1 cells using anti-VSV monoclonal antibodies. The various mutant proteins were found to be expressed at levels comparable to VSV-tagged wild-type hU3-55k (Fig. 6B).
VSV-tagged wild-type U3-55k interacted with U3 RNA and localized to nucleoli in our experimental system. We have assessed the ability of hU3-55k proteins to interact with U3 RNA by precipitation of the VSV-tagged proteins from transfected cell extracts and northern analysis of co-precipitated RNAs (Fig. 6C). While the VSV protein alone did not co-precipitate U3 RNA (Fig. 6C, lane 5), wild-type VSV-tagged hU3-55k expressed in COS-1 cells associated with U3 snoRNA as expected (Fig. 6C, lane 2). Fibrillarin is also found associated with U3 RNA in these cells (Fig. 6C) and fibrillarin immunoprecipitations were included as positive controls in each of the co-precipitation experiments that will be described. The cellular localization of expressed U3-55k proteins was evaluated by immunofluorescence using anti-VSV antibodies (Fig. 6D). As expected, wild-type VSV-tagged hU3-55k expressed in HEp-2 cells localized to nucleoli similar to fibrillarin (Fig. 6D, panels b and j).
In order to further assess the role of the C-terminal domain of U3-55k in nucleolar localization and U3 RNA association (45), two additional C-terminal mutant constructs were analyzed (Fig. 6A). We found that deletion of 17 amino acids at the C-terminus of U3-55k also disrupted U3 RNA binding and U3-55k nucleolar localization (Δ459–475, Fig. 6C and D, lanes 7–9 and panel d, respectively). Though we cannot exclude the possibility that this deletion had global effects on protein folding, the mutant protein was expressed (Fig. 6B) and appeared to accumulate in the nucleoplasm (Fig. 6D, panel d). Deletion of the last nine amino acids did not prevent RNA binding or nucleolar localization (Δ467–475, Fig. 6C and D, lanes 10–12 and panel f, respectively). These results indicate that residues located within amino acids 459–466 of hU3-55k are important for interaction with U3 RNA and localization of U3-55k to the nucleolus.
A glutamic acid-rich region was previously noted near the N-terminus of the hU3-55k protein sequence (45). This region appears to be conserved in the yeast and Xenopus proteins (Fig. 1). However, we have found that deletion of these amino acids did not prevent the interaction of the protein with U3 RNA or the localization of U3-55k to the nucleolus (Δ64–74, Fig. 6C and B, lanes 13–15 panel h, respectively).
WD domains like those found in U3-55k have been characterized to be important for dynamic protein–protein interactions (49). We were interested to assess whether the WD repeats of U3-55k are important for association of the protein with U3 snoRNA and localization of the protein to nucleoli. Three deletion mutants were tested: Δ193–352 lacking WD repeats 2–5, Δ193–315 lacking WD repeats 2–4 and Δ140–230 lacking WD repeats 1 and 2 (Fig. 6A). The effects of the deletions on the overall structure of the protein are not known but did not prevent accumulation or nuclear import of the mutant proteins. Though each of the WD deletion mutants was expressed in the transfected cells, U3 RNA was not found in association with any of the three mutants (Fig. 6B and C, lanes 6–8 and lanes 16–24, respectively). Co-precipitation of U3 was not detected even upon a 20-fold increased exposure of the northern blot (data not shown). In addition, the WD deletion mutants accumulated in the nucleus but did not localize to nucleoli (Fig. 6D, panels l, n and p). The results suggest that the WD domains of U3-55k are required for association of U3-55k with U3 snoRNA and for localization of U3-55k to nucleoli in vivo.
DISCUSSION
A detailed understanding of the composition and organization of the U3 snoRNP complex is required in order to understand U3 RNP biogenesis, transport and function. Little information is available regarding the nature of the association of either the common Box C/D-associated proteins or the U3-specific proteins with U3 RNA. In this study, we have identified sequence domains in both the U3 RNA and U3-55k protein that are required for their association.
The predominant sequence feature of U3-55k is a series of six WD repeats. WD repeats within a protein are generally thought to mediate dynamic protein–protein interactions and to provide a platform for the assembly of multicomponent complexes (49). We found that disruption of the WD repeats of U3-55k resulted in loss of association of the protein with U3 RNA in vivo (Fig. 6C) suggesting that protein–protein interactions play an important role in the protein–RNA association in vivo. At the same time, our finding that recombinant U3-55k protein interacts with U3 RNA in vitro suggests that U3-55k may interact directly with U3 RNA. The U3-55k protein sequence does not possess any recognizable RNA binding motifs, but may contain an undefined RNA-binding domain [possibly including the sequences in the C-terminus of U3-55k that we found to be required for U3 snoRNA binding (Fig. 6C)]. Conceivably, U3-55k interacts directly with U3 RNA and the interaction is strengthened by association with other proteins in vivo.
The interaction of the U3-55k protein with U3 RNA in vivo and in vitro depends upon conserved sequences in an RNA motif that is specific to U3, the Box B/C motif (Figs 3–5). Our findings are in good agreement with a previous study which demonstrated that a region of hU3 (nt 97–204) including Boxes B and C supported the assembly of a complex which included CHO U3-55k in vitro (29). We found that mutation of Box B disrupted the interaction of U3-55k with U3 RNA to a greater extent in vivo than in vitro (Figs 3 and 5). The greater importance of Box B in vivo may reflect the importance of another protein located at Box B (which is absent or limited in the in vitro assay) in the interaction of U3-55k with U3. One model consistent with our results is that U3-55k interacts primarily with Box C of U3 RNA and that the interaction is stabilized by association with other proteins (mediated by the WD repeats of U3-55k) that bind Box B.
It is noteworthy that U3 snoRNA associates with two WD repeat proteins, U3-55k and Sof1. Sof1 was identified as a high copy supressor that overcomes the temperature sensitive phenotype of yeast expressing human, rather than yeast, fibrillarin and, like U3-55k, interacts specifically with U3 RNA (40). We have cloned the Xenopus (A.A.Lukowiak, S.Mattox, R.Terns and M.P.Terns, unpublished data) and human (S.Granneman and W.J.van Venrooij, unpublished data) Sof1 homologs and also plan to characterize the manner in which this WD repeat protein is recruited into U3 snoRNP complexes.
There is evidence that fibrillarin also requires Box C for association with U3 RNA in vitro and in vivo (34,67, W.Speckmann, R.Terns and M.P.Terns, unpublished data). It is possible that fibrillarin interacts with U3 RNA through association with U3-55k (or another associated protein). On the other hand, a recent study indicates that fibrillarin interacts directly, and in the absence of other proteins, with another Box C/D snoRNA, U16 (68), raising the interesting question whether U3-55k and fibrillarin interact simultaneously or exclusively with U3 RNA. This work and another study indicate that U3-55k can associate with U3 RNA in the absence of fibrillarin (Fig. 5; 29).
U3 RNA is predominantly found within the nucleoli of various cell types where it functions in rRNA processing. It is now clear that U3-55k is also localized in the nucleoli of multiple cell types (Figs 2B and C and 6D; 45). The sequence elements involved in the localization of U3 RNA to nucleoli have been investigated (31,69,70). We have found that while the Box B/C motif of U3 is involved in the retention of the RNA in the nucleus, it is neither required nor sufficient for nucleolar localization (30,31). Thus, U3-55k may function as part of the redundant mechanisms for nuclear retention of U3, but probably does not play a role in the targeting of U3 RNA to nucleoli. Indeed, U3-55k probably relies on interaction with U3 for its own localization to the nucleolus. We observed a correlation between the ability of U3-55k variants to interact with U3 RNA and to be localized to nucleoli (Fig. 6). Mutants that were not able to interact with U3 also did not localize to nucleoli. Thus, U3-55k may be targeted to nucleoli via interaction with the U3 snoRNP.
It will be important to determine the role of U3-55k in U3 snoRNP biogenesis and function. Studies in yeast have indicated that the Box B/C motif (i.e. the U3-55k binding site) is not essential for stability of U3 RNA but is essential for viability, indicating an important role for the Box B/C motif in U3 snoRNP function (23,27). Correspondingly, it has been found that the yeast U3-55k homolog (Rrp9p) is also not required for the stability of U3 RNA, but is essential for viability and 18S rRNA production (J.Venema and H.A.Raue, personal communication). Future work will further investigate the role of U3-55k in rRNA processing by the U3 snoRNP.
Acknowledgments
ACKNOWLEDGEMENTS
We kindly thank K. M. Pollard (Scripps Research Institute, La Jolla, CA) for the Xenopus cell line (XlK2) and Antoon Huiskens (University of Nijmegen, Nijmegen, The Netherlands) for his expert technical assistance. This work was supported by the National Institutes of Health (GM54682 to M.P.T.) and The Netherlands Research Council for Chemical Sciences (NWO-CW, grant 97-033 to W.J.v.V.). A.A.L. was supported by a training grant (GM07103) from the National Institutes of Health.
REFERENCES
- 1.Bachellerie J., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier,M. (1995) Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- 2.Gerbi S. (1995) Biochem. Cell Biol., 73, 845–858. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 35, 897–933. [DOI] [PubMed] [Google Scholar]
- 4.Smith C. and Steitz,J. (1997) Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 5.Tollervey D. and Kiss,T. (1997) Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein L.B. and Steitz,J.A. (1999) Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 7.Balakin A., Smith,L. and Fournier,M. (1996) Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 8.Ganot P., Bortolin,M. and Kiss,T. (1997) Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 9.Venema J. and Tollervey,D. (1999) Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 10.Enright C., Maxwell,E. and Sollner-Webb,B. (1996) RNA, 2, 1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes J. and Ares,M.J. (1991) EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed]
- 12.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 13.Mougey E.B., Pape,L.K. and Sollner-Webb,B. (1993) Mol. Cell. Biol., 13, 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savino R. and Gerbi,S.A. (1990) EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltrame M. and Tollervey,D. (1994) Nucleic Acids Res., 22, 5139–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beltrame M. and Tollervey,D. (1995) EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartshorne T. and Agabian,N. (1994) Nucleic Acids Res., 22, 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes J.M., Konings,D.A. and Cesareni,G. (1987) EMBO J., 6, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeppesen C., Stebbins-Boaz,B. and Gerbi,S. (1988) Nucleic Acids Res., 16, 2127–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T. and Solymosy,F. (1990) Nucleic Acids Res., 18, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leader D.J., Connelly,S., Filipowicz,W. and Brown,J.W. (1994) Biochim. Biophys. Acta, 1219, 145–147. [DOI] [PubMed] [Google Scholar]
- 22.Mazan S., Gulli,M.P., Joseph,N. and Bachellerie,J.P. (1992) Eur. J. Biochem., 205, 1033–1041. [DOI] [PubMed] [Google Scholar]
- 23.Mereau A., Fournier,R., Gregoire,A., Mougin,A., Fabrizio,P., Luhrmann,R. and Branlant,C. (1997) J. Mol. Biol., 273, 552–571. [DOI] [PubMed] [Google Scholar]
- 24.Orum H., Nielsen,H. and Engberg,J. (1993) Nucleic Acids Res., 21, 2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker K. and Steitz,J. (1987) Mol. Cell. Biol., 7, 2899–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter G.L., Brennwald,P.J., Holm,K.A. and Wise,J.A. (1988) Nucleic Acids Res., 16, 10131–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samarsky D. and Fournier,M. (1998) Mol. Cell Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes J. (1996) J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 29.Lubben B., Marshallsay,C., Rottmann,N. and Luhrmann,R. (1993) Nucleic Acids Res., 21, 5377–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speckmann W., Narayanan,A., Terns,R. and Terns,M.P. (1999) Mol. Cell. Biol., 19, 8412–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanan A., Speckmann,W., Terns,R. and Terns,M. (1999) Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein P., Reddy,R. and Busch,H. (1984) Biochemistry, 23, 5421–5425. [DOI] [PubMed] [Google Scholar]
- 33.Tyc K. and Steitz,J.A. (1989) EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baserga S.J., Yang,X.D. and Steitz,J.A. (1991) EMBO J., 10, 2645–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E. (1989) EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lafontaine D.L. and Tollervey,D. (2000) Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafontaine D.L.J. and Tollervey,D. (1999) RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyman S.K., Gerace,L. and Baserga,S.J. (1999) RNA, 5, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu P., Brockenbrough,J., Metcalfe,A., Chen,S. and Aris,J. (1998) J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen R., Tollervey,D. and Hurt,E.C. (1993) EMBO J., 12, 2549–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunbar D., Wormsley,S., Agentis,T. and Baserga,S. (1997) Mol. Cell. Biol., 17, 5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westendorf J., Konstantinov,K., Wormsley,S., Shu,M., Matsumoto-Taniura,N., Pirollet,F., Klier,F., Gerace,L. and Baserga,S. (1998) Mol. Biol. Cell, 9, 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiederkehr T., Prétôt,R.F. and Minvielle-Sebastia,L. (1998) RNA, 4, 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S.J. and Baserga,S.J. (1999) Mol. Cell. Biol., 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pluk H., Soffner,J., Luhrmann,R. and van Venrooij.W.J. (1998) Mol. Cell. Biol., 18, 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tollervey D., Lehtonen,H., Carmo-Fonseca,M. and Hurt,E.C. (1991) EMBO J., 10, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Voorn L. and Ploegh,H.L. (1992) FEBS Lett., 307, 131–134. [DOI] [PubMed] [Google Scholar]
- 49.Neer E.J., Schmidt,C.J., Nambudripad,R. and Smith,T.F. (1994) Nature, 371, 297–300. [DOI] [PubMed] [Google Scholar]
- 50.Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- 51.Sondek J., Bohm,A., Lambright,D.G., Hamm,H.E. and Sigler,P.B. (1996) Nature, 379, 369–374. [DOI] [PubMed] [Google Scholar]
- 52.Wall M.A., Coleman,D.E., Lee,E., Iniguez-Lluhi,J.A., Posner,B.A., Gilman,A.G. and Sprang,S.R. (1995) Cell, 83, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 53.Lambright D.G., Sondek,J., Bohm,A., Skiba,N.P., Hamm,H.E. and Sigler,P.B. (1996) Nature, 379, 311–319. [DOI] [PubMed] [Google Scholar]
- 54.Ng J., Li,R., Morgan,K. and Simon,J. (1997) Mol. Cell. Biol., 17, 6663–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Higuera I., Fenoglio,J., Li,Y., Lewis,C., Panchenko,M.P., Reiner,O., Smith,T.F. and Neer,E.J. (1996) Biochemistry, 35, 13985–13994. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Higuera I., Gaitatzes,C., Smith,T.F. and Neer,E.J. (1998) J. Biol. Chem., 273, 9041–9049. [DOI] [PubMed] [Google Scholar]
- 57.Saxena K., Gaitatzes,C., Walsh,M.T., Eck,M., Neer,E.J. and Smith,T.F. (1996) Biochemistry, 35, 15215–15221. [DOI] [PubMed] [Google Scholar]
- 58.Bjorn S.P., Soltyk,A., Beggs,J.D. and Friesen,J.D. (1989) Mol. Cell. Biol., 9, 3698–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben Yehuda S., Dix,I., Russell,C.S., Levy,S., Beggs,J.D. and Kupiec,M. (1998) RNA, 4, 1304–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bordonne R., Banroques,J., Abelson,J. and Guthrie,C. (1990) Genes Dev., 4, 1185–1196. [DOI] [PubMed] [Google Scholar]
- 61.Ayadi L., Miller,M. and Banroques,J. (1997) RNA, 3, 197–209. [PMC free article] [PubMed] [Google Scholar]
- 62.Dalrymple M.A., Petersen-Bjorn,S., Friesen,J.D. and Beggs,J.D. (1989) Cell, 58, 811–812. [DOI] [PubMed] [Google Scholar]
- 63.Frank D., Patterson,B. and Guthrie,C. (1992) Mol. Cell. Biol., 12, 5197–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Achsel T., Ahrens,K., Brahms,H., Teigelkamp,S. and Luhrmann,R. (1998) Mol. Cell. Biol., 18, 6756–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reimer G., Pollard,K.M., Penning,C.A., Ochs,R.L., Lishwe,M.A., Busch,H. and Tan,E.M. (1987) Arthritis Rheum., 30, 793–800. [DOI] [PubMed] [Google Scholar]
- 66.Verheijen R., Wiik,A., De Jong,B.A., Hoier-Madsen,M., Ullman,S., Halberg,P. and Van Venrooij,W.J. (1994) J. Immunol. Methods, 169, 173–182. [DOI] [PubMed] [Google Scholar]
- 67.Baserga S.J., Gilmore-Hebert,M. and Yang,X.W. (1992) Genes Dev., 6, 1120–1130. [DOI] [PubMed] [Google Scholar]
- 68.Fatica A., Galardi,S., Altieri,F. and Bozzoni,I. (2000) RNA, 6, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lange T.S., Ezrokhi,M., Borovjagin,A.V., Rivera-Leon,R., North,M.T. and Gerbi,S.A. (1998) Mol. Biol. Cell, 9, 2973–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]