Abstract
The ability of mammals to mount adaptive immune responses culminating with the establishment of immunological memory is predicated on the ability of the mature T cell repertoire to recognize antigenic peptides presented by syngeneic MHC class I and II molecules. While it is widely believed that mature T cells are highly skewed towards the recognition of antigenic peptides originating from genetically diverse (for example, foreign or mutated) protein-coding regions, preclinical and clinical data rather demonstrate that novel antigenic determinants efficiently recognized by mature T cells can emerge from a variety of non-mutational mechanisms. In this Review, we describe various mechanisms that underlie the formation of bona fide non-mutational neoantigens, such as epitope mimicry, upregulation of cryptic epitopes, the usage of non-canonical initiation codons, alternative RNA splicing, defective ribosomal RNA processing, as well as both enzymatic and non-enzymatic post-translational protein modifications. Moreover, we discuss the implications of the immune recognition of non-mutational neoantigens for human disease.
Introduction
MHC proteins present a sampling of the cellular proteome at the cell surface for inspection by T cells as part of the immune surveillance of the body for infection or malignancy. Generally, MHC class I proteins sample peptides generated by the proteasomal degradation of cytosolic proteins, whereas MHC class II proteins sample peptides generated by the endosomal or lysosomal proteolysis of (1) cell surface proteins, (2) proteins taken up from the extracellular environment through receptor-mediated or fluid-phase endocytosis, and (3) proteins from cellular organelles engulfed by autophagosomes. Thus, the MHC system accesses all major protein degradation pathways and intracellular compartments as a means to inform T cells about the cellular state.1
Positive and negative thymic selection have evolved to ensure that the mature T cell repertoire of each individual specifically recognizes peptides that (1) are presented by autologous MHC class I and II molecules, and (2) are not generally presented by healthy tissues.2 However, central tolerance as generated by thymic selection is leaky, implying that mature T cell repertoires contain many T cell clones that are specific for unmodified self antigens.3 Importantly, a panel of mechanisms exist to prevent the activation of such autoreactive T cell clones, including (but not limited to) (1) the requirement of multiple signals beyond TCR activation for T cells to acquire effector and memory functions, and (2) the existence of antigen-specific immunosuppressive T cell populations such as CD4+CD25+FOXP3+ regulatory T (TREG) cells.4
Defects in these mechanisms of peripheral tolerance (for example, FOXP3 mutations) have been associated with various autoimmune conditions, such as IPEX syndrome.5 Moreover, modern MHC elution techniques coupled with the development of algorithms that can link peptide mass spectra with specific peptide structures have unveiled the existence of many antigenic epitopes that – because of structural modifications or overrepresentation – become able to be efficiently presented by MHC class I and II molecules, and hence elicit reactivity, but do not originate from DNA mutations.3 Such ‘non-mutational neoantigens’ can originate from a variety of post-transcriptional and post-translational mechanisms, including epitope mimicry, the usage of non-canonical initiation codons, alternative RNA splicing, defective ribosomal RNA processing, as well as both enzymatic and non-enzymatic post-translational protein modifications, such as phosphorylation, citrullination, glycation and many others.3 Importantly, preclinical and clinical data suggest that non-mutational neoantigens are involved in a number of disorders ranging from autoreactive conditions to cancer, with multiple host-related risk factors (Box 1).
Box 1. Risk factors for autoreactivity driven by non-mutational neoantigens.
Numerous factors have been associated with an increased likelihood for the development of autoreactivity against non-mutational neoantigens. These factors include mutations in important genes underlying peripheral tolerance (for example, FOXP3 mutations),5 as well as polymorphisms in gene encoding cytokines (for example, CCL21, which is linked to rheumatoid arthritis), cytokine receptors (for example., IL23R, which is associated with Crohn’s disease), death receptors and their ligands (for example, FAS and FASLG, which are linked to systemic lupus erythematosus [SLE]), Toll-like receptors (for example, TLR3, which is associated with type 1 diabetes) as well as intracellular pattern recognition receptors (for example, NOD2, which is also linked to Crohn’s disease).172, 173, 174 That said, HLA polymorphisms are by far the most common risk factors for autoimmune reactions against non-mutational neoantigens.175 To name a few examples, this applies to ankylosing spondylitis (HLA-B27), psoriasis (HLA-C06), rheumatoid arthritis (HLA-DR4), Crohn’s disease (HLA-C), SLE (HLA-DRB1, HLA-DQA1, HLA-DQB1), type 1 diabetes (HLA-DR4, HLA-DQ8, HLA-DR3/DQ2), multiple sclerosis (HLA-DR2), and celiac disease (HLA-DQ2, HLA-DQ8).175 In each of these cases except for SLE and Crohn’s disease, the disease-associated MHC allele has been shown to present an autoantigen recognized by self-reactive T cells, but the precise role of such autoreactivity in pathogenesis or disease progression is unclear. Other non-overlapping mechanisms to loosen central tolerance have been described, including increased thymocyte resistance to negative selection,176 alternate peptide docking and binding register,177, 178 low-affinity peptide binding,179, 180, 181 reduced antigen density or HLA instability,182 and abnormal regulation of HLA transcription.183, 184, 185 Conversely, defects in cytokine and PRR signaling can impair peripheral tolerance by promoting disproportioned inflammatory responses in tissues.186 Irrespective of the precise mechanism, all these risk factors predispose the host to autoreactive CD4+ and/or CD8+ T cell responses of pathological relevance.
In this Review, we summarize the molecular mechanisms leading to the generation of the autoreactive non-mutational MHC immunopeptidome and their emerging role in human disease. Conversely, we will not cover the details of physiological MHC class I and II antigen processing and presentation (reviewed elsewhere),1 nor autoreactivity to non-peptide antigens presented by non-canonical MHC proteins such as CD1 family members and major histocompatibility complex, class I-related (MR1) (reviewed elsewhere).6, 7
Epitope mimicry
Autoreactivity against non-mutational neoantigens can emerge from so-called ‘epitope mimicry’, the ability of some microbial antigens to elicit cross-reactive T or B-cell responses against the host (Figure 1).8 Mimicry can obviously result from a considerable overlap in the primary amino acid sequence between microbial and host epitopes, but also from structural similarities in the context of minimal sequence overlaps.9 Autoreactive conditions associated with epitope mimicry encompass: (1) multiple sclerosis, in which myelin basic protein (MBP) is recognized upon Epstein Barr virus (EBV) infection;10, 11 (2) rheumatic fever, which is unleashed upon infection by Streptococcus spp. and consequent cross-reactivity against cardiac glycoproteins;12 (3) Guillain-Barré syndrome, as driven by autoreactive responses against neuronal proteins elicited by EBV, cytomegalovirus (CMV) or type I herpes simplex virus (HSV-1) infection;13 (4) type 1 diabetes, in which pancreatic β cell antigens are targeted upon infection with enteroviruses or lymphocytic choriomeningitis virus (LCMV);14, 15, 16 (5) rheumatoid arthritis, which is promoted by cross-recognition of synovial tissue and cartilage antigens upon infection by type I human immunodeficiency virus (HIV-1), hepatitis B virus (HBV) or hepatitis C virus (HCV);17, 18 as well as (6) systemic lupus erythematosus (SLE), which is facilitated by DNA- and Smith (Sm)-targeting antibodies emerging after EBV, CMV, HCV or parvovirus infection.19, 20, 21 Similarly inflammatory cardiomyopathy seems to be promoted by cardiac myosin-specific TH17 cells elicited in the intestine by a commensal Bacteroides spp. epitope.22 Also, SLE, the Guillain-Barré syndrome and other autoimmune conditions such as pediatric inflammatory multisystemic syndrome or Kawasaki-like disease in children have also been linked to epitope mimicry upon SARS-CoV-2 infection.23, 24, 25
Figure 1. Mimicry and crypticity in non-mutational neo-antigenicity.
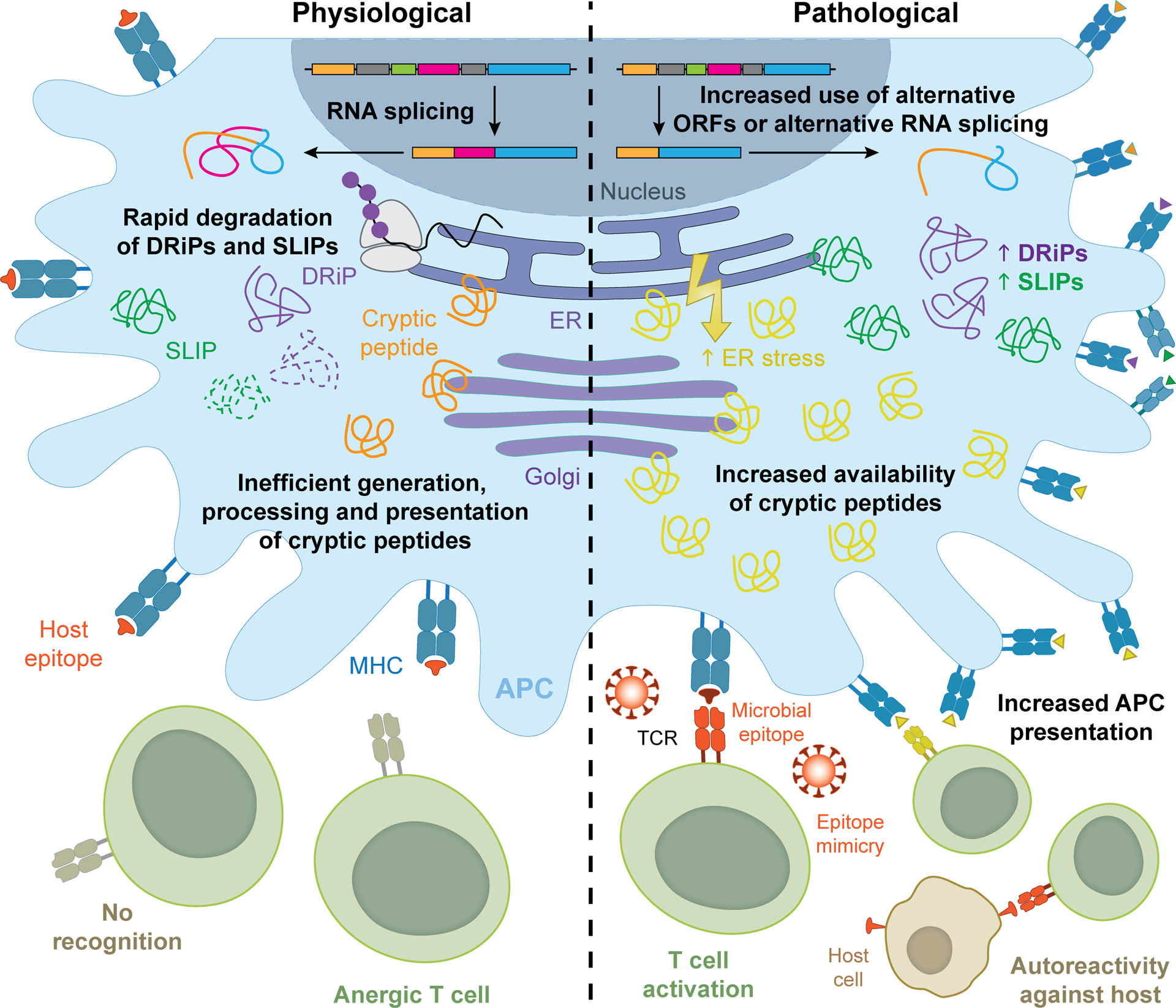
Autoreactivity against non-mutational neoantigens can emerge from a variety of mechanisms including epitope mimicry and crypticity. Such autoreactive responses are invariably driven by T cell clones escaping (leaky) thymic selection. On the one hand, pathogen-derived epitopes exhibiting considerable structural resemblance to self peptides can drive potent autoreactive responses, at least in individuals with one or more risk factors (see Box 1). On the other hand, purely self peptides (at least from a genetic standpoint), as well as defective ribosomal products (DRiPs) and short-lived proteins (SLiPs) that are normally not presented on MHC class I or II molecules, may elicit autoreactivity downstream of accrued antigen presentation as a consequence of: (1) usage of non-canonical open reading frames (ORFs), (2) alternative RNA splicing, (3) increased expression levels, (4) decreased competition for binding to MHC class I or II molecules, and/or (5) stress conditions that overall alter antigen processing and presentation. APC, antigen-presenting cell; ER, endoplasmic reticulum.
That said, the majority of patients with infectious diseases do not develop autoimmune disorders. Moreover, a comparative analysis of the human and pathogen proteome identified up to 90% overlap in 6–8 identical amino acids stretches within 9-mer epitopes.26 Thus, factors beyond epitope mimicry probably contribute to the development of autoreactive disorders after infection. Aside from polymorphisms in HLA-coding genes and other genes involved in immune regulation (Box 1),27 such factors might include the differential presentation of antigenic epitopes as a consequence of infection-related perturbations of cellular homeostasis (for example, endoplasmic reticulum stress),28 and the pathogenic relocalization of normally symbiotic microbes to ectopic locations (e.g., from the intestine to the liver).29
Of note, various microbial epitopes have also been shown to resemble tumor-associated antigens (TAAs).30 For example, similarities have been documented between Mycoplasma penetrans HF-2 epitopes and MAGE family member A6 (MAGEA6),31 CMV peptides and MAGEA10,32 HSV-1 components and melan-A (MLANA),33 as well as HBV antigens and transmembrane protein 161A (TMEM161A).34 Supporting the notion that epitope mimicry is common in neoplastic conditions, a comparison between public databases of TAA-derived epitopes (https://caped.icp.ucl.ac.be/peptide/list) and viral proteomes (Viruses tazid:10239) identified almost 100 shared sequences.35 The functional significance of epitope mimicry for patients with cancer is exemplified by studies identifying (1) cross-reactivity between microbial antigens and TAAs, (2) a positive association between microbial antigens and response to immunotherapy, and (3) the existence of T cells recognizing cross-reactive antigens.36, 37 Cross-reactivity between microbial antigens and TAAs has been detected in patients with bladder cancer treated with Bacillus Calmette-Guérin, an attenuated strain of Mycobacterium bovis,38 or with neoadjuvant immune checkpoint inhibitors (ICIs),36 as well as in individuals with renal and lung cancer receiving a programmed cell death 1 (PD-1) blocker, where CD8+ T cells recognizing an epitope from the opportunistic pathogen Enterococcus hirae cross-react with naturally-processed TAAs.37 Considerable efforts are currently being dedicated to combining bacterial or viral peptides with other immunostimulatory maneuvers to elicit tumor-targeting immune responses that may be actionable with ICIs.30 Considering that a particular TCR can recognize more than a million different peptides,39 and that different TCRs can recognize the same peptide (with different affinity),40 issues of TCR degeneracy versus specificity,41, 42 will have to be explored in the context of epitope mimicry.
In summary, although mimicry between pathogen-derived and self epitopes may elicit pathogenic T cell responses, other factors are normally required for the development of autoreactivity/autoimmunity against these antigenic determinants. Understanding these factors will enable the development of therapeutic strategies to prevent or harness epitope mimicry in patients.
Cryptic epitopes, DRiPs, SLiPs and unconventional translation products
Cryptic peptides are epitopes that are physiologically processed and presented by antigen-presenting cells (APCs) inefficiently, at low copy number, or not at all, hence failing to elicit central tolerance or peripheral T cell responsiveness.43 However, various perturbations of cellular homeostasis, including alterations in the MHC class I and II machinery, can affect the processing of native proteins, potentially resulting in an increase in the number of cryptic peptides presented by MHC molecules and hence in the initiation of an autoreactive T cell response (Figure 1).43 Conditions that may generate or upregulate cryptic peptides include: (1) reduced competition by other peptides for binding to MHC molecules; (2) enhanced protein availability (due to increased synthesis or decreased degradation); (3) protein unfolding exposing cleavage sites for alternative proteases (as in the case of endoplasmic reticulum stress); (4) changes in the cytosolic or reticular microenvironment (for example, during inflammatory conditions), resulting in activation/deactivation of specific proteases, and (5) quantitative changes in the expression of peptide processing and editing components of the MHC class I and II machinery.43, 44, 45
In the context of autoimmunity, one of the best examples of crypticity is provided by proteolipid protein 1 (PLP1), one of the main components of the myelin sheet of neurons that is targeted during autoimmune encephalomyelitis. Specifically, PLP1 exists in multiple splice variants, and the one presented in the thymus (DM20) lacks a major epitope that instead is presented in the periphery, resulting in lack of central tolerance and consequent T cell activation at neuronal terminals, and pathogenic demyelination.46 Different splice variants and cryptic peptides of MBP have also been associated with multiple sclerosis,47, 48 similar to novel PLP1 reactivities emerging after Theiler virus infection.49 Along similar lines, cartilage-derived cryptic epitopes seem to be preferentially presented in the acute, inflammatory phase of rheumatoid arthritis, likely as a consequence of extracellular matrix (ECM) protein unfolding and processing by enzymes released by immune cells.50 Finally, systemic lupus erythematosus (SLE) has been shown to involve T-cell and B-cell autoreactivity against cryptic epitopes from components of the complement cascade.51 Similar findings have been obtained in the context of autoreactive responses as elicited by viral infection. Specifically, the HIV-1 membrane glycoprotein gp120 has been shown to uncover cryptic CD4 peptides resulting in pathogenic CD4-specific autoantibodies.52 Moreover, ribosome profiling and proteomic mapping identified cryptic SARS-CoV-2 antigens associated with severe post-infection autoreactivity.53, 54, 55 These observations exemplify the broad pathogenic relevance of autoreactivity against cryptic epitopes.
Potentially autoreactive epitopes can also originate from defective ribosomal products (DRiPs), short-lived proteins (SLiPs),56 non-canonical (including non-AUG defined and novel unannotated) open-reading frames (ORFs), protein frameshift (for example, owing to the lack of a specific amino acid),57, 58, 59, 60 as well as from the translation of 5’- or 3’- untranslated regions (UTRs).61, 62 All these scenarios have been shown to be relevant for the composition of the MHC immunopeptidome.
DRiPs and SLIPs can be translated as efficiently as canonical proteins and have been shown to generate peptides 5-fold more efficiently per translation event.57 Indeed, it seems that the MHC class I and less so class II immunopeptidome contain a large proportion of DRiPs and SLIPs as compared to cryptic epitopes derived from canonical proteins.56 Importantly though, defining whether an MHC-bound epitope is a DRiP or a SLiP cannot be achieved with standard MHC elution and mass spectrometry, as these peptides share the same amino acid sequence of the corresponding full protein, calling for the use of dynamic SILAC and pulse chase quantitative mass spectrometric techniques or ribosomal profiling coupled with MHC elution.63 DRiPs/SLiPs can arise from the translation of non-canonical (including non-AUG defined) open reading frames (ORFs),59, 60 as well as from the translation of 5’- or 3’- untranslated regions (UTRs),61, 62 two scenarios that result in the generation of novel epitopes.
DRiPs/SLiPs were initially characterized in the context of viral infection, potentially offering a rapid mechanism for infected cells to enable MHC-restricted antiviral immunosurveillance.62 Albeit the vast majority of defined viral peptides derive from standard ORFs of stable proteins, several viral DRiPs and SLiPs have been shown to derive from errors in protein translation, trafficking, folding or other mostly unknown mechanisms.64, 65 For example, peptides translated from a non-AUG defined ORF have been shown to potently elicit alloreactive T cells in the context of eukaryotic translation initiation factor 2 subunit alpha (EIF2S1, best known as eIF2α) phosphorylation,66 which is one of the first cellular reactions to viral infection,67 as well as in the presence of pro-inflammatory cues including interferon gamma (IFNG).68 Of note, DRiP/SLiP-generated viral epitopes generally coupled to specific cytotoxic T cell responses69 have been documented in the context of infection by a number of viral pathogens, including influenza virus, coxsackie virus, HSV and numerous retroviruses.69, 70, 71 Moreover, at least some viral pathogens (for example, EBV, SARS-CoV-2) promote the presentation of host DRiPs/SLiPs, ultimately resulting in autoreactivity.72, 73
Non-canonical translation leading to DRiPs/SLiP accumulation has also been linked to overt autoimmunity.61 For example, Reiter’s syndrome has been associated with autoreactive T cells specific for an interleukin-10 (IL10) epitope created by a non-canonical ORF.74 Along similar lines, DRiPs/SLiPs generated from an alternative ORF in the insulin (INS)-coding mRNA have been shown to efficiently load on MHC class I and class II molecules, generating both humoral and cellular autoreactivity of pathogenic relevance in type 1 diabetes.75 In line with this notion, ribosomal profiling of human pancreatic β cells exposed to inflammatory conditions identified numerous DRiPs/SLiPs generated from non-canonical ORFs that are not present in healthy pancreatic β cells.76
Most (but not all) non-mutated cancer-associated epitopes that have been associated with natural cytotoxic T cell responses, or successfully harnessed to elicit at least some degree of tumor-specific autoreactivity, represent bona fide cryptic epitopes, as their immunogenicity largely reflects differential expression levels in malignant versus normal tissues.77 For example, melanoma-infiltrating lymphocytes seem to be highly enriched in T cells recognizing epitopes from differentiation and cancer-testis antigens,78 but also contain T cells specific for epitopes from absent in melanoma 2 (AIM2) and alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase (MGAT5) generated by non-canonical ORFs emerging from incomplete splicing.79 Importantly, such autoreactive T cells could be isolated from the peripheral blood of patients with melanoma, but not healthy volunteers or patients with neoplasms other than melanoma,79 corroborating the crypticity of these epitopes. Similar results have been obtained with atypical teratoid/rhabdoid tumors, which were shown to present several MHC class I- and II-associated cryptic epitopes (shared with glioblastomas but not extracranial tumors) associated with CD8+ and CD4+ T cell reactivity,80 as well as colorectal cancers.81
Supporting the presentation of cryptic epitopes by malignant cells, a number of clinical trials testing TAAs or epitopes thereof as therapeutic vaccines (delivered via various technologies) have documented at least some degree of tumor-targeting immune reactivity.82, 83 These studies include trials testing: (1) erb-b2 receptor tyrosine kinase 2 (ERBB2, best known as HER2) epitopes in women with HER2+ breast cancer,84 (2) glypican 3 (GPC3) epitopes in patients with hepatocellular carcinoma,85 (3) a multiepitope vaccine in individuals with melanoma,86 and (4) a premelanosome protein (PMEL, best known as gp100) epitope in patients with melanoma.87 Moreover, a cryptic NY-ESO-1 epitope (159–167) was shown to induce robust immunodominant reactivity over two other HLA-A2-binding NY-ESO-1 peptides that were associated with at least some clinical activity in HLA-A2+ patients with melanoma expressing cancer/testis antigen 1B (CTAG1B, best known as NY-ESO-1).88, 89 That said, it is now clear that therapeutic vaccination has limited clinical efficacy in patients with cancer, reflecting the potently immunosuppressive microenvironment that most neoplasms generate.77 Interestingly, diagnostic applications have also been proposed for DRiP-based and SLiP-based preparations. Specifically, it has been suggested to use naturally processed DRiPs and SLiPs contained in tumor-derived autophagosomes (which originate from an intracellular mechanism for the lysosomal degradation of cytoplasmic material)90 to interrogate tumor-targeting reactivity in cancer patients prior to immunotherapy.91
Taken together, these observations suggest that cryptic epitopes including peptides generates by DRiPs, SLiPs and non-canonical translation products are abundantly presented by cells experiencing perturbations of homeostasis. Although these epitope seem to contribute to pathogenic autoreactivity in the setting of infectious and autoimmune disorders, they might represent a therapeutically actionable mechanism for cancer therapy.
Protein modifications
Antigenic epitopes can also originate via multiple post-transcriptional mechanisms other than the usage of non-canonical ORFs, alternative splicing and the generation of non-conventional ribosomal products. These mechanisms reflect not only direct amino acid substitution at ribosomes and so-called ‘peptide splicing’, i.e., the fusion of conventional proteolytic products, but also other enzymatic and non-enzymatic post-translational modifications (PTMs) (Figure 2).
Figure 2. Protein modifications in non-mutational neo-antigenicity.
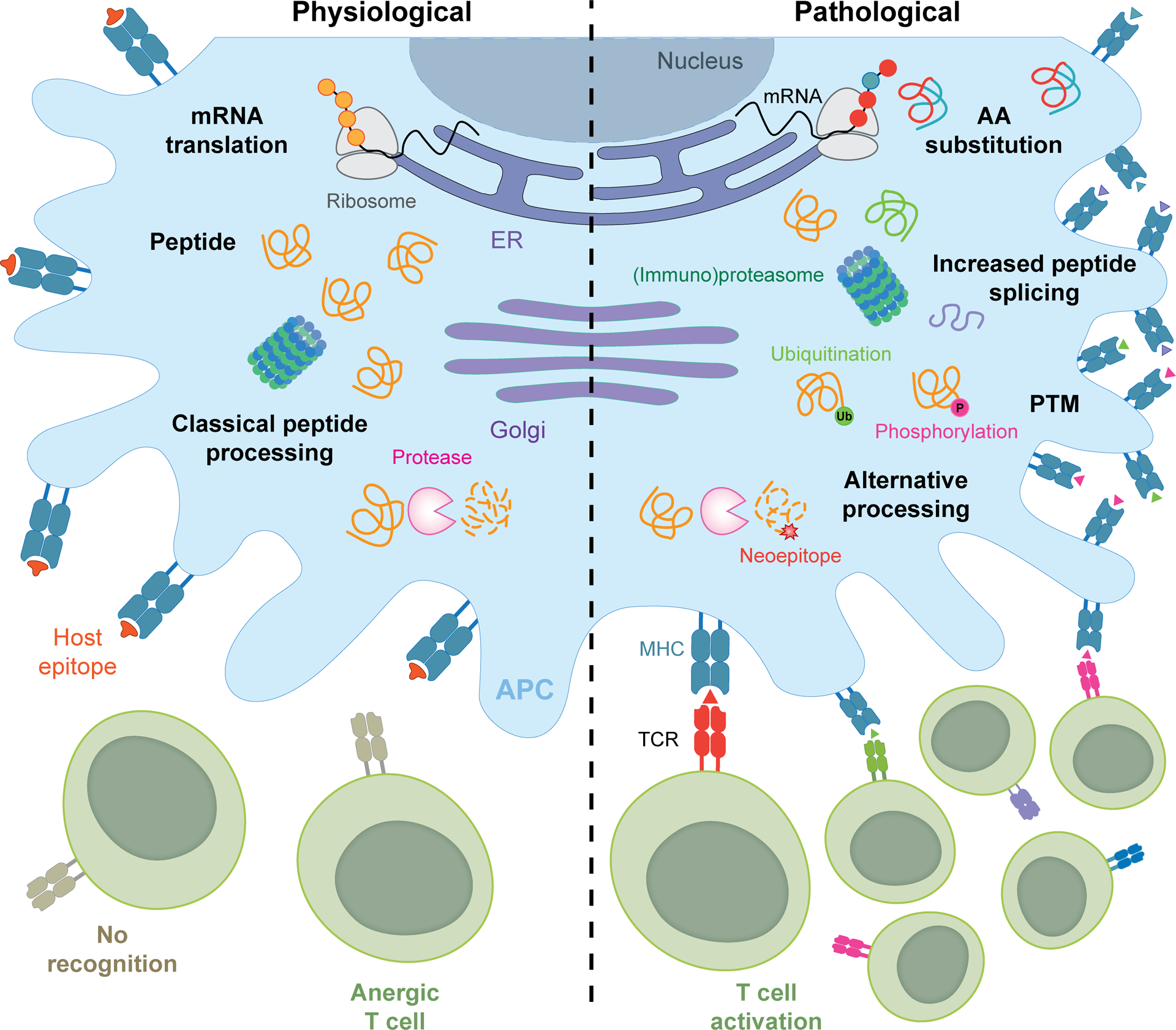
Loss of peripheral tolerance and consequent autoreactivity against non-mutated neoepitopes can emerge from a number of protein modifications associated with altered cellular homeostasis, encompassing: (1) the direct replacement of one or more amino acids during translation, (2) peptide splicing within the proteasome, endosomes/lysosomes, other protease-containing compartments, (3) a variety of enzymatic or non-enzymatic post-translational protein modifications (PTMs) such as citrullination, oxidation and glycation, and (4) altered protease activity and consequent generation of novel cleavage products. AA, amino acid; APC, antigen-presenting cell; ER, endoplasmic reticulum.
Amino acid substitution and peptide splicing.
When a specific amino acid is scarce, the correct aminoacyl tRNA can be replaced by another, resulting in the generation of mutated proteins during translation, despite the usage of purely wild-type mRNAs.92 This mechanism has been shown to promote T cell reactivity in melanoma cells depleted of tryptophan, generally resulting in phenylalanine substitutants.92 Along similar lines, approximately 1% of methionine residues in the physiological cellular proteome are inserted via non-methionyl-tRNAs, a fraction that remarkably increases in the presence of viral or chemical cues.93 Whether these findings can be extended to the depletion of other amino acids remains to be investigated. Moreover, the effect of amino acid substitution on autoimmune conditions has not been formally interrogated yet.
The term peptide splicing (or transpeptidation) refers to the generation of a polypeptide by fusion of shorter peptides from non-contiguous regions of the same (cis) or different (trans) proteins.94 Originally described for MHC class I epitopes, spliced peptides have also been reported for MHC class II epitopes, a setting in which they are often described as ‘hybrid’ peptides, reflecting an apparent preference for trans over cis splicing.94 While the cytoplasmic proteasome seems to be the major cellular site responsible for transpeptidation of MHC class I epitopes,95 resulting from the high local concentrations of peptide products in the proteasome interior that compete with water for hydrolysis of acyl-enzyme intermediates,96, 97 endosomal cathepsins and proteasomes have also been reported to efficiently catalyze this reaction.94, 98 Interestingly, viral and bacterial proteins also have been reported to undergo cis-splicing to generate novel antigenic determinants associated with cellular immunity.99, 100, 101 However, the reported abundance of spliced peptides in the MHC immunopeptidome ranges from as much as 25–30% (Refs. 102, 103) to as little as 1–3% (Refs. 56, 104, 105), with many epitopes originally attributed to splicing being alternatively explained as originating from novel unannotated ORFs.104, 106 Thus, the contribution of spliced peptides to the MHC immunopeptidome is controversial.
Supporting the relevance of spliced peptides for human autoimmune disorders, T cells from patients with type 1 diabetes have been shown to recognize an MHC class II-associated epitope resulting from the trans-splicing of pro-insulin (aa 64–71) and islet amyloid polypeptide (IAPP, aa 74–80),107 as well as MHC class I-restricted epitopes derived from the cis-splicing of IAPP108 or secretogranin V (SCG5).109 In all these settings, splicing is believed to occur in pancreatic β cell secretory granules, which contain very high concentrations of secretory hormones and processing enzymes. Data from non-obese diabetic (NOD) mice provide mechanistic support for the pathogenic relevance of these neoantigens.109, 110, 111
Immunogenic neoantigens generated by amino acid substitution and transpeptidation have also been described in the context of cancer.112 For example, an asparagine-to-aspartate substitution in tyrosinase (TYR) has been shown to generate a neoepitope resulting in the activation of melanoma-targeting T cell responses.113 Moreover, the cis-splicing of fibroblast growth factor 5 (FGF5) reportedly elicits immunoreactive epitopes in human renal cell carcinoma cells,96 as does the cis-splicing of gp100 and TYR in melanoma cells.97, 114, 115 Several other spliced neoantigens associated with tumor-targeting immune responses have been reported.112
Post-translational modifications.
Hundreds of PTMs have been documented in mammals, greatly expanding the diversity of the cellular proteome.116 These modifications encompass enzymatic alterations that are inbuilt in normal physiology and responses to stress, such as phosphorylation, ubiquitination, acetylation, and enzymatic oxidation, as well as non-enzymatic changes imposed by intracellular or extracellular conditions, such as glycation, carbonylation, and non-enzymatic oxidation.116 At least theoretically, shifts in PTMs can generate new antigenic determinants either directly, by modifying a previously existing non-immunogenic epitope, or indirectly, by influencing virtually any step of antigen presentation from proteasome processing through MHC binding and exposure. In line with this notion, an expanding literature demonstrates that both enzymatic and non-enzymatic PTMs associated with perturbations of cellular homeostasis linked to various human disorders can generate autoreactive responses of pathological significance.3
Citrullination is a PTM of arginine that can occur upon non-enzymatic oxidation, which is common in inflamed and aging tissues,117 or via the catalytic activity of protein arginine deiminase (PAD) family members.116 Citrullinated peptides have been associated with the generation of novel antigenic determinants and pathogenic humoral or cellular autoreactivity to epitopes from extracellular matrix proteins in rheumatoid arthritis,118, 119 glucokinase (GCK) in type 1 diabetes120 and MBP in multiple sclerosis.121 Whether these epitopes uniquely emerge from enzymatic or non-enzymatic citrullination, however, is unclear. Phosphorylation is a critical event in multiple signaling cascade, and phosphorylated epitopes from peripherin (PRPH) and small RNA binding exonuclease protection factor La (SSB) have been linked to the generation of pathogenic autoantibodies in type 1 diabetes122 and SLE.123, 124 Acetylation is a PTM with broad activity in cellular biology, notably in the regulation of gene expression.125 Acetylation of the amino-terminal peptide of MBP is required for induction of autoimmunity in experimental allergic encephalomyelitis, a mouse model of multiple sclerosis.126, 127 Antibodies against acetylated histones are common in patients with SLE, and their titer generally correlates with disease severity.128
Cellular stress as imposed by aging, inflammation and metabolic alterations such as hyperglycemia can result in an increased abundance of reactive species that promote non-enzymatic oxidation, carbonylation, and glycation.129 Oxidized epitopes from multiple components of lipoproteins have been shown to promote B cell and T cell autoreactivity with pathological significance for atherosclerosis and other cardiovascular conditions.130 Along similar lines, oxidative protein modifications as driven by malondialdehyde elicit autoantibodies that promote rheumatoid arthritis at least in part by stimulating osteoclast activation.131 Juvenile idiopathic arthritis has been associated not only with an increase in carbonylated albumin and immunoglobulins in the circulation,132 but also with carbonylation-related alteration in the affinity of a transthyretin (TTR)-derived epitope for HLA-DR1, culminating in pathogenic autoreactivity.133 More broadly, carbonylation has been shown to have pleiotropic effects on antigen presentation by dendritic cells (DCs), resulting in considerable qualitative and quantitative changes in their MHC class II immunopeptidome.134 Glycation is particularly common in metabolic conditions associated with hyperglycemia such as type 1 and 2 diabetes.134, 135 Aside from imposing broad qualitative and quantitative alterations to the DC MHC class II immunopeptidome,134 glycation as driven by metabolic cues has been shown to favor the emergence of a neoantigenic determinant in protein disulfide isomerase family A member 3 (PDIA3) that elicits autoreactive antibodies promoting disease progression.135 Finally, the presentation of iodinated, deaminated or nitrosylated thyroglobulin (TG) epitopes has been shown to trigger autoimmune thyroiditis.136, 137, 138 These observations exemplify the broad pathogenic effect of enzymatic and non-enzymatic PTMs in human disorders with an autoreactive or autoimmune component.
On the other hand, PTM-derived neoepitopes might be beneficial for the development of therapeutic cancer vaccines, based on their increased abundance in malignant over non-malignant cells as a result of oncogene signaling.139, 140 For example, phosphorylated epitopes from enolase 1 (ENO1), tumor protein p53 (TP53, best known as p53), insulin receptor substrate 2 (IRS2), and cell division cycle 25B (CDC25B), are overrepresented in pancreatic carcinoma (ENO1)141 and multiple other tumors (p53, IRS2, CDC25B),142, 143 whereas citrullinated epitopes from matrix metallopeptidase 21 (MMP21), ENO1 and vimentin (VIM), are overrepresented in melanoma144, 145 and metastasizing carcinomas,146 respectively. Supporting the actual immunogenicity of these PTMs, epitope-specific responses against citrullinated ENO1 or VIM peptides restricted to the MHC class II molecules HLA-DR4 or HLA-DP4 have been documented in 58% of patients with ovarian cancer.146 Aside from confirming the immunogenicity of citrullinated ENO1 and mechanistically linking it to citrullination, mouse data demonstrate that this neoantigenic epitope can be employed successfully as a therapeutic vaccine against MHC class II-positive (but not MHC class II-negative) mouse tumors established in immunocompetent, syngeneic HLA-DR4 transgenic mice, an effect that is accompanied by CD4+ T cell activation and acquisition of cytotoxic effector functions.147
Supporting the clinical relevance of these observations, an acetylated p53 epitope – but not its de-acetylated counterpart – has been shown to elicit HLA-DR-restricted CD4+ T cell responses in peripheral lymphocytes from patients with cancer, but not healthy volunteers.142 Moreover, various therapeutic vaccination approaches based on glycosylated mucin 1, cell surface associated (MUC1) have been associated with robust immunogenicity and at least some degree of efficacy in patients with a variety of tumors, although clinical efficacy remains marginal.148, 149 Despite this and other limitations, the aforementioned observations suggest that a number of PTMs generate non-mutational neoantigenic determinants that – at least in principle – might be harnessed to drive anticancer immune responses.
Alternative processing.
Proteases (the genes for which account for approx. 3% of the human coding genome) are broadly classified into: (1) cysteine proteases (for example, papain, calpains, caspases, cathepsin B, C, H, F, L, K, S, O, W), (2) serine proteases (for example, chymotrypsin, trypsin, elastase), (3) threonine proteases (for example, proteasome-associated proteases and some acyltransferases), (4) aspartic proteases (for example, renin, cathepsin D and E), and (5) metalloproteases (for example, matrix metalloproteases [MMPs], collagenase, various aminopeptidases and carboxypeptidases).150 In physiological conditions, MHC class I-restricted and class II-restricted peptides are mostly generated by (immuno)proteasomal and endolysosomal proteolysis, respectively.151 MMPs, calpains and caspases are also known to contribute to the physiological MHC immunopeptidome, reflecting their important function in physiological cellular processes including matrix degradation during immune cell trafficking (MMPs) and programmed cell death (caspases and less-so calpains).152, 153
In the presence of perturbations of cellular or microenvironmental homeostasis (for example, inflammatory stimuli, metabolic cues, oxidative stress), the overall proteolytic activity of a cell can considerably change as a consequence of the activation or inactivation of various proteases, resulting in the formation of potentially antigenic (and hence potentially pathogenic) neoepitopes.154 For example, robust effector T cell activation associated with abundant granzyme B (GZMB) release155 has been shown to promote the GZMB-dependent generation of neoepitopes from lamin B (LMNB), poly(ADP-ribose) polymerase 1 (PARP1) and RNA, U1 small nuclear 1 (RNU1–1, also known as U1) in SLE,156, 157, 158, 159 centromere protein C (CENPC) in scleroderma,160 and SSB in Sjögren’s syndrome,161 most often resulting in pathogenic B cell and/or T cell autoreactivity.158, 159, 161, 162 Along similar lines, MMPs released by immune cells responding to pro-inflammatory cytokines have been demonstrated to cleave ECM components to release neoantigenic determinants. As an example, matrix metallopeptidase 9 (MMP9) released in response to IL-1 and tumor necrosis factor (TNF) reportedly generates type II collagen-derived neoepitopes that contribute to rheumatoid arthritis163, 164 Moreover, inhibition of endoplasmic reticulum aminopeptidase 1 (ERAP1) causes profound changes in the MHC class I immunopeptidome, resulting (at least in mice) in a substantial T cell response to a H2-Qa1-restricted neoepitope.44, 165 In line with this notion, ERAP1 polymorphisms have been associated with several autoimmune disorders including ankylosing spondylitis,166 birdshot chorioretinopathy,167 psoriasis168, 169 and Beçhet disease.170
While alternative processing is likely to occur and generate non-mutational neoantigens also in malignant cells, the impact of this process on tumor-targeting immune responses is understudied. That said, neutrophil-derived proteases taken up by endosomes in lung cancer cells not only appear to improve antigen presentation by favoring the exposure of MHC class I molecules on the cell surface, but also seem to generate a number of neoantigenic determinants within these organelles.171 Importantly, CD8+ cytotoxic T cells recognizing some of these neoepitopes, including epitopes from minichromosome maintenance complex binding protein (MCMBP), ATP binding cassette subfamily A member 1 (ABCA1), and signal regulatory protein delta (SIRPD), were enriched in the tumor microenvironment of patients with lung cancer as compared to the circulation,171 suggesting at least some degree of specificity.
These observations exemplify the broad pathological effect of non-mutated neoepitopes generated by protein modifications.
Conclusion
Preclinical and clinical data suggest that non-mutational neoantigens are abundantly presented on both MHC class I and II molecules, and that humoral and cellular immune responses targeting these neoepitopes can have a major impact on disease. On the one hand, it is now clear that neoantigenic determinants generated by non-mutational sources elicit B cell and T cell responses that contribute to the etiology of various disorders with an autoimmune/autoreactive component, including (but not limited to) SLE, rheumatoid arthritis, autoimmune encephalitis, and diabetes (both type 1 and 2).3 On the other hand, non-mutational neoantigens presented by malignant cells seem to elicit at least some degree of tumor-targeting immunity. While such a natural immune response is generally unable to arrest tumor progression because of multipronged immunosuppressive mechanisms established by developing neoplasms, non-mutational neoantigens offer a valid target for the development of tumor-specific immunotherapeutics including cancer vaccines and (at least in the case of surface-exposed neoepitopes) CAR T cells.77
Importantly, the accurate identification and quantification of non-mutational neoantigens cannot be achieved from DNA/RNA sequencing data, but relies strictly on peptide elution from MHC molecules coupled with mass spectrometry. This fact has at least two important implications. On the one hand, it highlights the crucial importance of the reference libraries employed for spectral matching. As an obvious example, a mammalian library is intrinsically incompatible with the identification of pathogen-derived epitopes. On the other hand, it underscores the complexity associated with the identification of non-mutational neoantigens in clinical samples. Indeed, in most cases, the number of cells obtained from tissue biopsies may not be sufficient to achieve a sufficient peptide elution yield for high-resolution immunopeptidome analysis by mass spectrometry.
Despite such limitations, we surmise that interrogating the non-mutational neo-immunopeptidomes in increased detail will not only provide additional mechanistic insights into disorders as diverse as autoimmunity and cancer, but also may suggest new therapeutic avenues against these and other human pathologies.
Acknowledgements.
We are indebted to Vanessa Klapp (Luxembourg Institute of Health, Luxembourg City, Luxembourg) for help with figure preparation. LJS is supported by NIH (#AG067581, #AI146180, #AI143976 #AI137198, #AI127869, #AI153828, #AR080593) and the Parkinson’s Foundation. LG is/has been supported (as a PI unless otherwise indicated) by one R01 grant from the NIH/NCI (#CA271915), by two Breakthrough Level 2 grants from the US DoD BCRP (#BC180476P1, #BC210945), by a grant from the STARR Cancer Consortium (#I16-0064), by a Transformative Breast Cancer Consortium Grant from the US DoD BCRP (#W81XWH2120034, PI: Formenti), by a U54 grant from NIH/NCI (#CA274291, PI: Deasy, Formenti, Weichselbaum), by the 2019 Laura Ziskin Prize in Translational Research (#ZP-6177, PI: Formenti) from the Stand Up to Cancer (SU2C), by a Mantle Cell Lymphoma Research Initiative (MCL-RI, PI: Chen-Kiang) grant from the Leukemia and Lymphoma Society (LLS), by a Rapid Response Grant from the Functional Genomics Initiative (New York, US), by a pre-SPORE grant (PI: Demaria, Formenti) and a Clinical Trials Innovation Grant from the Sandra and Edward Meyer Cancer Center (New York, US); by startup funds from the Dept. of Radiation Oncology at Weill Cornell Medicine (New York, US), by industrial collaborations with Lytix Biopharma (Oslo, Norway), Promontory (New York, US) and Onxeo (Paris, France), as well as by donations from Promontory (New York, US), the Luke Heller TECPR2 Foundation (Boston, US), Sotio a.s. (Prague, Czech Republic), Lytix Biopharma (Oslo, Norway), Onxeo (Paris, France), Ricerchiamo (Brescia, Italy), and Noxopharm (Chatswood, Australia). LS is supported by NIH (#AI153828, #AI146180, #AI137198, #AI137198-S, #AI169723, #AI134696, #AG031782, #AT011419, #AR081493, #AI170897) and the Cure Alzheimer’s Foundation.
Footnotes
Competing Interests. LG is/has been holding research contracts with Lytix Biopharma, Promontory and Onxeo, has received consulting/advisory honoraria from Boehringer Ingelheim, AstraZeneca, OmniSEQ, Onxeo, The Longevity Labs, Inzen, Imvax, Sotio, Promontory, Noxopharm, EduCom, and the Luke Heller TECPR2 Foundation, and holds Promontory stock options. The other authors declare no competing interests.
References
- 1.Pishesha N, Harmand TJ & Ploegh HL A guide to antigen processing and presentation. Nat Rev Immunol 22, 751–764 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Klein L, Kyewski B, Allen PM & Hogquist KA Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat Rev Immunol 14, 377–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santambrogio L & Marrack P The broad spectrum of pathogenic autoreactivity. Nat Rev Immunol 23, 69–70 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ElTanbouly MA & Noelle RJ Rethinking peripheral T cell tolerance: checkpoints across a T cell’s journey. Nat Rev Immunol 21, 257–267 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Josefowicz SZ, Lu LF & Rudensky AY Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowther MD & Sewell AK The burgeoning role of MR1-restricted T-cells in infection, cancer and autoimmune disease. Curr Opin Immunol 69, 10–17 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Shahine A, Van Rhijn I, Rossjohn J & Moody DB CD1 displays its own negative regulators. Curr Opin Immunol 83, 102339 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajishengallis G & Lambris JD Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol 11, 187–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossjohn J et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol 33, 169–200 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Lang HL et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol 3, 940–943 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Schneider-Hohendorf T et al. Broader Epstein-Barr virus-specific T cell receptor repertoire in patients with multiple sclerosis. J Exp Med 219 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibofsky A & Zabriskie JB Rheumatic fever and poststreptococcal reactive arthritis. Curr Opin Rheumatol 7, 299–305 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Salmon DA et al. Association between Guillain-Barré syndrome and influenza A (H1N1) 2009 monovalent inactivated vaccines in the USA: a meta-analysis. Lancet 381, 1461–1468 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Ohashi PS et al. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell 65, 305–317 (1991). [DOI] [PubMed] [Google Scholar]
- 15.Harkonen T, Lankinen H, Davydova B, Hovi T & Roivainen M Enterovirus infection can induce immune responses that cross-react with beta-cell autoantigen tyrosine phosphatase IA-2/IAR. J Med Virol 66, 340–350 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Hiemstra HS et al. Cytomegalovirus in autoimmunity: T cell crossreactivity to viral antigen and autoantigen glutamic acid decarboxylase. Proc Natl Acad Sci U S A 98, 3988–3991 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker J & Winthrop KL Update on rheumatic manifestations of infectious diseases. Curr Opin Rheumatol 22, 72–77 (2010). [DOI] [PubMed] [Google Scholar]
- 18.de Pablo P, Dietrich T & McAlindon TE Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. J Rheumatol 35, 70–76 (2008). [PubMed] [Google Scholar]
- 19.Barzilai O, Ram M & Shoenfeld Y Viral infection can induce the production of autoantibodies. Curr Opin Rheumatol 19, 636–643 (2007). [DOI] [PubMed] [Google Scholar]
- 20.James JA et al. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest 100, 3019–3026 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanlon P, Avenell A, Aucott L & Vickers MA Systematic review and meta-analysis of the sero-epidemiological association between Epstein-Barr virus and systemic lupus erythematosus. Arthritis Res Ther 16, R3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil-Cruz C et al. Microbiota-derived peptide mimics drive lethal inflammatory cardiomyopathy. Science 366, 881–886 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Dotan A et al. The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun Rev 20, 102792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karami Fath M et al. SARS-CoV-2 Proteome Harbors Peptides Which Are Able to Trigger Autoimmunity Responses: Implications for Infection, Vaccination, and Population Coverage. Front Immunol 12, 705772 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodruff MC et al. Dysregulated naive B cells and de novo autoreactivity in severe COVID-19. Nature (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragone C et al. Molecular mimicry between tumor associated antigens and microbiota-derived epitopes. J Transl Med 20, 316 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J et al. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. Cell 183, 1264–1281.e1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Conza G, Ho PC, Cubillos-Ruiz JR & Huang SC Control of immune cell function by the unfolded protein response. Nat Rev Immunol (2023). [DOI] [PubMed] [Google Scholar]
- 29.Manfredo Vieira S et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 359, 1156–1161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zitvogel L & Kroemer G Cross-reactivity between microbial and tumor antigens. Curr Opin Immunol 75, 102171 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Vujanovic L, Shi J, Kirkwood JM, Storkus WJ & Butterfield LH Molecular mimicry of MAGE-A6 and Mycoplasma penetrans HF-2 epitopes in the induction of antitumor CD8(+) T-cell responses. Oncoimmunology 3, e954501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiaro J et al. Viral Molecular Mimicry Influences the Antitumor Immune Response in Murine and Human Melanoma. Cancer Immunol Res 9, 981–993 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loftus DJ et al. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1(27–35). J Exp Med 184, 647–657 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiou SH et al. Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery. Immunity 54, 586–602 e588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tagliamonte M et al. Molecular mimicry and cancer vaccine development. Mol Cancer 22, 75 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goubet AG et al. Escherichia coli-Specific CXCL13-Producing TFH Are Associated with Clinical Efficacy of Neoadjuvant PD-1 Blockade against Muscle-Invasive Bladder Cancer. Cancer Discov 12, 2280–2307 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Fluckiger A et al. Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage. Science 369, 936–942 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Antonelli AC, Binyamin A, Hohl TM, Glickman MS & Redelman-Sidi G Bacterial immunotherapy for cancer induces CD4-dependent tumor-specific immunity through tumor-intrinsic interferon-γ signaling. Proc Natl Acad Sci U S A 117, 18627–18637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wooldridge L et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 287, 1168–1177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song I et al. Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8(+) T cell epitope. Nat Struct Mol Biol 24, 395–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borbulevych OY et al. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 31, 885–896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishizuka J et al. Quantitating T cell cross-reactivity for unrelated peptide antigens. J Immunol 183, 4337–4345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moudgil KD & Sercarz EE Understanding crypticity is the key to revealing the pathogenesis of autoimmunity. Trends Immunol 26, 355–359 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Koumantou D et al. Editing the immunopeptidome of melanoma cells using a potent inhibitor of endoplasmic reticulum aminopeptidase 1 (ERAP1). Cancer Immunol Immunother 68, 1245–1261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olsson N, Jiang W, Adler LN, Mellins ED & Elias JE Tuning DO:DM Ratios Modulates MHC Class II Immunopeptidomes. Mol Cell Proteomics 21, 100204 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein L, Klugmann M, Nave KA, Tuohy VK & Kyewski B Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat Med 6, 56–61 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Cardamone G et al. The Characterization of GSDMB Splicing and Backsplicing Profiles Identifies Novel Isoforms and a Circular RNA That Are Dysregulated in Multiple Sclerosis. Int J Mol Sci 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderton SM, Viner NJ, Matharu P, Lowrey PA & Wraith DC Influence of a dominant cryptic epitope on autoimmune T cell tolerance. Nat Immunol 3, 175–181 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Miller SD et al. Persistent infection with Theiler’s virus leads to CNS autoimmunity via epitope spreading. Nat Med 3, 1133–1136 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Buttle DJ, Bramwell H & Hollander AP Proteolytic mechanisms of cartilage breakdown: a target for arthritis therapy? Clin Mol Pathol 48, M167–177 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu WJ, Tan Y, Liu XL, Yu F & Zhao MH C1q A08 Is a Half-Cryptic Epitope of Anti-C1q A08 Antibodies in Lupus Nephritis and Important for the Activation of Complement Classical Pathway. Front Immunol 11, 848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salemi S, Caporossi AP, Boffa L, Longobardi MG & Barnaba V HIVgp120 activates autoreactive CD4-specific T cell responses by unveiling of hidden CD4 peptides during processing. J Exp Med 181, 2253–2257 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finkel Y et al. The coding capacity of SARS-CoV-2. Nature 589, 125–130 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Schmidt N et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat Microbiol 6, 339–353 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan M et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Admon A The biogenesis of the immunopeptidome. Semin Immunol 67, 101766 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Ruiz Cuevas MV et al. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep 34, 108815 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yewdell JW Immunology. Hide and seek in the peptidome. Science 301, 1334–1335 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Yewdell JW, Anton LC & Bennink JR Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol 157, 1823–1826 (1996). [PubMed] [Google Scholar]
- 60.Ronsin C et al. A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J Immunol 163, 483–490 (1999). [PubMed] [Google Scholar]
- 61.Laumont CM et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun 7, 10238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starck SR & Shastri N Nowhere to hide: unconventional translation yields cryptic peptides for immune surveillance. Immunol Rev 272, 8–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holly J & Yewdell JW Game of Omes: ribosome profiling expands the MHC-I immunopeptidome. Curr Opin Immunol 83, 102342 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Croft NP et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc Natl Acad Sci U S A 116, 3112–3117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Purcell AW, Croft NP & Tscharke DC Immunology by numbers: quantitation of antigen presentation completes the quantitative milieu of systems immunology! Curr Opin Immunol 40, 88–95 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Schwab SR, Shugart JA, Horng T, Malarkannan S & Shastri N Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol 2, e366 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harapas CR et al. Organellar homeostasis and innate immune sensing. Nat Rev Immunol 22, 535–549 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Komov L, Melamed Kadosh D, Barnea E & Admon A The Effect of Interferons on Presentation of Defective Ribosomal Products as HLA Peptides. Mol Cell Proteomics 20, 100105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maness NJ et al. CD8+ T cell recognition of cryptic epitopes is a ubiquitous feature of AIDS virus infection. J Virol 84, 11569–11574 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang N et al. Defining Viral Defective Ribosomal Products: Standard and Alternative Translation Initiation Events Generate a Common Peptide from Influenza A Virus M2 and M1 mRNAs. J Immunol 196, 3608–3617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zanker DJ et al. Influenza A Virus Infection Induces Viral and Cellular Defective Ribosomal Products Encoded by Alternative Reading Frames. J Immunol 202, 3370–3380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smatti MK et al. Viruses and Autoimmunity: A Review on the Potential Interaction and Molecular Mechanisms. Viruses 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lodha M, Erhard F, Dolken L & Prusty BK The Hidden Enemy Within: Non-canonical Peptides in Virus-Induced Autoimmunity. Front Microbiol 13, 840911 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saulquin X et al. +1 Frameshifting as a novel mechanism to generate a cryptic cytotoxic T lymphocyte epitope derived from human interleukin 10. J Exp Med 195, 353–358 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kracht MJ et al. Autoimmunity against a defective ribosomal insulin gene product in type 1 diabetes. Nat Med 23, 501–507 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Thomaidou S et al. Long RNA Sequencing and Ribosome Profiling of Inflamed β-Cells Reveal an Extensive Translatome Landscape. Diabetes 70, 2299–2312 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Dersh D, Holly J & Yewdell JW A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat Rev Immunol 21, 116–128 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Andersen RS et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res 72, 1642–1650 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Andersen RS et al. High frequency of T cells specific for cryptic epitopes in melanoma patients. Oncoimmunology 2, e25374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcu A et al. Natural and cryptic peptides dominate the immunopeptidome of atypical teratoid rhabdoid tumors. J Immunother Cancer 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schwarz S et al. T cells of colorectal cancer patients’ stimulated by neoantigenic and cryptic peptides better recognize autologous tumor cells. J Immunother Cancer 10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bezu L et al. Trial watch: Peptide-based vaccines in anticancer therapy. Oncoimmunology 7, e1511506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vacchelli E et al. Trial watch: Dendritic cell-based interventions for cancer therapy. Oncoimmunology 2, e25771 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Disis MLN et al. Safety and Outcomes of a Plasmid DNA Vaccine Encoding the ERBB2 Intracellular Domain in Patients With Advanced-Stage ERBB2-Positive Breast Cancer: A Phase 1 Nonrandomized Clinical Trial. JAMA Oncol 9, 71–78 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sawada Y et al. Phase II study of the GPC3-derived peptide vaccine as an adjuvant therapy for hepatocellular carcinoma patients. Oncoimmunology 5, e1129483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butterfield LH et al. Immune Correlates of GM-CSF and Melanoma Peptide Vaccination in a Randomized Trial for the Adjuvant Therapy of Resected High-Risk Melanoma (E4697). Clin Cancer Res 23, 5034–5043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwartzentruber DJ et al. gp100 peptide vaccine and interleukin-2 in patients with advanced melanoma. N Engl J Med 364, 2119–2127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jäger E et al. Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc Natl Acad Sci U S A 97, 12198–12203 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gnjatic S et al. CD8(+) T cell responses against a dominant cryptic HLA-A2 epitope after NY-ESO-1 peptide immunization of cancer patients. Proc Natl Acad Sci U S A 99, 11813–11818 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klionsky DJ et al. Autophagy in major human diseases. EMBO J 40, e108863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van de Ven R et al. Autophagosome-based strategy to monitor apparent tumor-specific CD8 T cells in patients with prostate cancer. Oncoimmunology 7, e1466766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pataskar A et al. Tryptophan depletion results in tryptophan-to-phenylalanine substitutants. Nature 603, 721–727 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Netzer N et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Faridi P, Dorvash M & Purcell AW Spliced HLA-bound peptides: a Black Swan event in immunology. Clin Exp Immunol 204, 179–188 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mishto M et al. Driving forces of proteasome-catalyzed peptide splicing in yeast and humans. Mol Cell Proteomics 11, 1008–1023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hanada K, Yewdell JW & Yang JC Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 427, 252–256 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Vigneron N et al. An antigenic peptide produced by peptide splicing in the proteasome. Science 304, 587–590 (2004). [DOI] [PubMed] [Google Scholar]
- 98.Sengupta D, Graham M, Liu X & Cresswell P Proteasomal degradation within endocytic organelles mediates antigen cross-presentation. EMBO J 38, e99266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paes W et al. Contribution of proteasome-catalyzed peptide cis-splicing to viral targeting by CD8(+) T cells in HIV-1 infection. Proc Natl Acad Sci U S A 116, 24748–24759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Platteel AC et al. CD8(+) T cells of Listeria monocytogenes-infected mice recognize both linear and spliced proteasome products. Eur J Immunol 46, 1109–1118 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Platteel ACM et al. Multi-level Strategy for Identifying Proteasome-Catalyzed Spliced Epitopes Targeted by CD8(+) T Cells during Bacterial Infection. Cell Rep 20, 1242–1253 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Liepe J et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Faridi P et al. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci Immunol 3 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Mylonas R et al. Estimating the Contribution of Proteasomal Spliced Peptides to the HLA-I Ligandome. Mol Cell Proteomics 17, 2347–2357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Levy R et al. Large-Scale Immunopeptidome Analysis Reveals Recurrent Posttranslational Splicing of Cancer- and Immune-Associated Genes. Mol Cell Proteomics 22, 100519 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ouspenskaia T et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol 40, 209–217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Delong T et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science 351, 711–714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gonzalez-Duque S et al. Conventional and Neo-antigenic Peptides Presented by β Cells Are Targeted by Circulating Naïve CD8+ T Cells in Type 1 Diabetic and Healthy Donors. Cell Metab 28, 946–960.e946 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Azoury ME et al. Peptides Derived From Insulin Granule Proteins Are Targeted by CD8(+) T Cells Across MHC Class I Restrictions in Humans and NOD Mice. Diabetes 69, 2678–2690 (2020). [DOI] [PubMed] [Google Scholar]
- 110.Wiles TA et al. An insulin-IAPP hybrid peptide is an endogenous antigen for CD4 T cells in the non-obese diabetic mouse. J Autoimmun 78, 11–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jin N et al. N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes. Proc Natl Acad Sci U S A 112, 13318–13323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Faridi P et al. Spliced Peptides and Cytokine-Driven Changes in the Immunopeptidome of Melanoma. Cancer Immunol Res 8, 1322–1334 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Skipper JC et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med 183, 527–534 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ebstein F et al. Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci Rep 6, 24032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dalet A et al. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc Natl Acad Sci U S A 108, E323–331 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Conibear AC Deciphering protein post-translational modifications using chemical biology tools. Nat Rev Chem 4, 674–695 (2020). [DOI] [PubMed] [Google Scholar]
- 117.Cannizzo ES et al. Age-related oxidative stress compromises endosomal proteostasis. Cell Rep 2, 136–149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Umemoto A et al. Identification of anti-citrullinated osteopontin antibodies and increased inflammatory response by enhancement of osteopontin binding to fibroblast-like synoviocytes in rheumatoid arthritis. Arthritis Res Ther 25, 25 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Curran AM et al. Citrullination modulates antigen processing and presentation by revealing cryptic epitopes in rheumatoid arthritis. Nat Commun 14, 1061 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang ML et al. Citrullination of glucokinase is linked to autoimmune diabetes. Nat Commun 13, 1870 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deraos G et al. Citrullination of linear and cyclic altered peptide ligands from myelin basic protein (MBP(87–99)) epitope elicits a Th1 polarized response by T cells isolated from multiple sclerosis patients: implications in triggering disease. J Med Chem 51, 7834–7842 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Doran TM et al. Discovery of Phosphorylated Peripherin as a Major Humoral Autoantigen in Type 1 Diabetes Mellitus. Cell Chem Biol 23, 618–628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Terzoglou AG, Routsias JG, Avrameas S, Moutsopoulos HM & Tzioufas AG Preferential recognition of the phosphorylated major linear B-cell epitope of La/SSB 349–368 aa by anti-La/SSB autoantibodies from patients with systemic autoimmune diseases. Clin Exp Immunol 144, 432–439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Utz PJ, Hottelet M, Schur PH & Anderson P Proteins phosphorylated during stress-induced apoptosis are common targets for autoantibody production in patients with systemic lupus erythematosus. J Exp Med 185, 843–854 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F & Kroemer G Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 21, 805–821 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Sullivan BA, Kraj P, Weber DA, Ignatowicz L & Jensen PE Positive selection of a Qa-1-restricted T cell receptor with specificity for insulin. Immunity 17, 95–105 (2002). [DOI] [PubMed] [Google Scholar]
- 127.Zamvil SS et al. T-cell epitope of the autoantigen myelin basic protein that induces encephalomyelitis. Nature 324, 258–260 (1986). [DOI] [PubMed] [Google Scholar]
- 128.Liu CL et al. Specific post-translational histone modifications of neutrophil extracellular traps as immunogens and potential targets of lupus autoantibodies. Arthritis Res Ther 14, R25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cannizzo ES, Clement CC, Sahu R, Follo C & Santambrogio L Oxidative stress, inflamm-aging and immunosenescence. J Proteomics 74, 2313–2323 (2011). [DOI] [PubMed] [Google Scholar]
- 130.Taleb A, Witztum JL & Tsimikas S Oxidized phospholipids on apoB-100-containing lipoproteins: a biomarker predicting cardiovascular disease and cardiovascular events. Biomark Med 5, 673–694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sakuraba K et al. Autoantibodies targeting malondialdehyde-modifications in rheumatoid arthritis regulate osteoclasts via inducing glycolysis and lipid biosynthesis. J Autoimmun 133, 102903 (2022). [DOI] [PubMed] [Google Scholar]
- 132.Zurawa-Janicka D et al. Preferential immunoglobulin oxidation in children with juvenile idiopathic arthritis. Scand J Rheumatol 35, 193–200 (2006). [DOI] [PubMed] [Google Scholar]
- 133.Clement CC et al. Autoimmune response to transthyretin in juvenile idiopathic arthritis. JCI Insight 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Clement CC et al. Pleiotropic consequences of metabolic stress for the major histocompatibility complex class II molecule antigen processing and presentation machinery. Immunity 54, 721–736 e710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clement CC et al. PDIA3 epitope-driven immune autoreactivity contributes to hepatic damage in type 2 diabetes. Sci Immunol 7, eabl3795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Champion BR et al. Identification of a thyroxine-containing self-epitope of thyroglobulin which triggers thyroid autoreactive T cells. J Exp Med 174, 363–370 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cirrito TP, Pu Z, Deck MB & Unanue ER Deamidation of asparagine in a major histocompatibility complex-bound peptide affects T cell recognition but does not explain type B reactivity. J Exp Med 194, 1165–1170 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morris G & Maes M Oxidative and Nitrosative Stress and Immune-Inflammatory Pathways in Patients with Myalgic Encephalomyelitis (ME)/Chronic Fatigue Syndrome (CFS). Curr Neuropharmacol 12, 168–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Petroni G, Buqué A, Coussens LM & Galluzzi L Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat Rev Drug Discov 21, 440–462 (2022). [DOI] [PubMed] [Google Scholar]
- 140.Srivastava AK, Guadagnin G, Cappello P & Novelli F Post-Translational Modifications in Tumor-Associated Antigens as a Platform for Novel Immuno-Oncology Therapies. Cancers (Basel) 15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cappello P et al. An integrated humoral and cellular response is elicited in pancreatic cancer by alpha-enolase, a novel pancreatic ductal adenocarcinoma-associated antigen. Int J Cancer 125, 639–648 (2009). [DOI] [PubMed] [Google Scholar]
- 142.Kumai T et al. Induction of tumor-reactive T helper responses by a posttranslational modified epitope from tumor protein p53. Cancer Immunol Immunother 63, 469–478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zarling AL et al. MHC-restricted phosphopeptides from insulin receptor substrate-2 and CDC25b offer broad-based immunotherapeutic agents for cancer. Cancer Res 74, 6784–6795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Symonds P et al. Citrullinated Epitopes Identified on Tumour MHC Class II by Peptide Elution Stimulate Both Regulatory and Th1 Responses and Require Careful Selection for Optimal Anti-Tumour Responses. Front Immunol 12, 764462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhou W et al. Mass spectrometry analysis of the post-translational modifications of alpha-enolase from pancreatic ductal adenocarcinoma cells. J Proteome Res 9, 2929–2936 (2010). [DOI] [PubMed] [Google Scholar]
- 146.Brentville VA et al. Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res 76, 548–560 (2016). [DOI] [PubMed] [Google Scholar]
- 147.Cook K et al. Citrullinated α-enolase is an effective target for anti-cancer immunity. Oncoimmunology 7, e1390642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Scheid E et al. Tn-MUC1 DC Vaccination of Rhesus Macaques and a Phase I/II Trial in Patients with Nonmetastatic Castrate-Resistant Prostate Cancer. Cancer Immunol Res 4, 881–892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Stergiou N et al. Reduced Breast Tumor Growth after Immunization with a Tumor-Restricted MUC1 Glycopeptide Conjugated to Tetanus Toxoid. Cancer Immunol Res 7, 113–122 (2019). [DOI] [PubMed] [Google Scholar]
- 150.McShane E & Selbach M Physiological Functions of Intracellular Protein Degradation. Annu Rev Cell Dev Biol 38, 241–262 (2022). [DOI] [PubMed] [Google Scholar]
- 151.Kotsias F, Cebrian I & Alloatti A Antigen processing and presentation. Int Rev Cell Mol Biol 348, 69–121 (2019). [DOI] [PubMed] [Google Scholar]
- 152.Clement CC et al. The Dendritic Cell Major Histocompatibility Complex II (MHC II) Peptidome Derives from a Variety of Processing Pathways and Includes Peptides with a Broad Spectrum of HLA-DM Sensitivity. J Biol Chem 291, 5576–5595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vitale I et al. Apoptotic cell death in disease-Current understanding of the NCCD 2023. Cell Death Differ 30, 1097–1154 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alvarez I, Antón LC & James EA Editorial: alternative antigen processing and presentation in immune disorders. Front Immunol 13, 993393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Darrah E & Rosen A Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ 17, 624–632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang D, Beresford PJ, Greenberg AH & Lieberman J Granzymes A and B directly cleave lamins and disrupt the nuclear lamina during granule-mediated cytolysis. Proc Natl Acad Sci U S A 98, 5746–5751 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu P et al. The cytotoxic T lymphocyte protease granzyme A cleaves and inactivates poly(adenosine 5’-diphosphate-ribose) polymerase-1. Blood 114, 1205–1216 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cram DS, Fisicaro N, Coppel RL, Whittingham S & Harrison LC Mapping of multiple B cell epitopes on the 70-kilodalton autoantigen of the U1 ribonucleoprotein complex. J Immunol 145, 630–635 (1990). [PubMed] [Google Scholar]
- 159.O’Brien RM, Cram DS, Coppel RL & Harrison LC T-cell epitopes on the 70-kDa protein of the (U1)RNP complex in autoimmune rheumatologic disorders. J Autoimmun 3, 747–757 (1990). [DOI] [PubMed] [Google Scholar]
- 160.Schachna L et al. Recognition of Granzyme B-generated autoantigen fragments in scleroderma patients with ischemic digital loss. Arthritis Rheum 46, 1873–1884 (2002). [DOI] [PubMed] [Google Scholar]
- 161.Huang M et al. Detection of apoptosis-specific autoantibodies directed against granzyme B-induced cleavage fragments of the SS-B (La) autoantigen in sera from patients with primary Sjogren’s syndrome. Clin Exp Immunol 142, 148–154 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jeoung D et al. Identification of autoantibody against poly (ADP-ribose) polymerase (PARP) fragment as a serological marker in systemic lupus erythematosus. J Autoimmun 22, 87–94 (2004). [DOI] [PubMed] [Google Scholar]
- 163.Van den Steen PE et al. Cleavage of denatured natural collagen type II by neutrophil gelatinase B reveals enzyme specificity, post-translational modifications in the substrate, and the formation of remnant epitopes in rheumatoid arthritis. FASEB J 16, 379–389 (2002). [DOI] [PubMed] [Google Scholar]
- 164.Descamps FJ, Van den Steen PE, Nelissen I, Van Damme J & Opdenakker G Remnant epitopes generate autoimmunity: from rheumatoid arthritis and multiple sclerosis to diabetes. Adv Exp Med Biol 535, 69–77 (2003). [DOI] [PubMed] [Google Scholar]
- 165.Nagarajan NA, Gonzalez F & Shastri N Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat Immunol 13, 579–586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Garcia-Medel N et al. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol Cell Proteomics 11, 1416–1429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Alvarez-Navarro C, Martin-Esteban A, Barnea E, Admon A & Lopez de Castro JA Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) Polymorphism Relevant to Inflammatory Disease Shapes the Peptidome of the Birdshot Chorioretinopathy-Associated HLA-A*29:02 Antigen. Mol Cell Proteomics 14, 1770–1780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sun LD et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet 42, 1005–1009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Genetic Analysis of Psoriasis, C. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42, 985–990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen L et al. Identification of an Unconventional Subpeptidome Bound to the Behcet’s Disease-associated HLA-B*51:01 that is Regulated by Endoplasmic Reticulum Aminopeptidase 1 (ERAP1). Mol Cell Proteomics 19, 871–883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peters HL et al. Serine Proteases Enhance Immunogenic Antigen Presentation on Lung Cancer Cells. Cancer Immunol Res 5, 319–329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Goris A & Liston A The immunogenetic architecture of autoimmune disease. Cold Spring Harb Perspect Biol 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vandenbroeck K Cytokine gene polymorphisms and human autoimmune disease in the era of genome-wide association studies. J Interferon Cytokine Res 32, 139–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang Y, Liu J, Wang C, Liu J & Lu W Toll-Like Receptors Gene Polymorphisms in Autoimmune Disease. Front Immunol 12, 672346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dendrou CA, Petersen J, Rossjohn J & Fugger L HLA variation and disease. Nat Rev Immunol 18, 325–339 (2018). [DOI] [PubMed] [Google Scholar]
- 176.Castro JE et al. Fas modulation of apoptosis during negative selection of thymocytes. Immunity 5, 617–627 (1996). [DOI] [PubMed] [Google Scholar]
- 177.Madsen LS et al. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet 23, 343–347 (1999). [DOI] [PubMed] [Google Scholar]
- 178.Hahn M, Nicholson MJ, Pyrdol J & Wucherpfennig KW Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol 6, 490–496 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bulek AM et al. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat Immunol 13, 283–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cole DK et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest 126, 3626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yin Y, Li Y, Kerzic MC, Martin R & Mariuzza RA Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection. EMBO J 30, 1137–1148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miyadera H, Ohashi J, Lernmark A, Kitamura T & Tokunaga K Cell-surface MHC density profiling reveals instability of autoimmunity-associated HLA. J Clin Invest 125, 275–291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Raj P et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.O’Huigin C et al. The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles. Am J Hum Genet 89, 424–431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hu X et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet 47, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gong T, Liu L, Jiang W & Zhou R DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol 20, 95–112 (2020). [DOI] [PubMed] [Google Scholar]


