Abstract
Mutational spectrum analysis has become an informative genetic tool to understand those protein functions involved in mutation avoidance pathways since specific types of mutations are often associated with particular protein defects involved in DNA replication and repair. In this study, we describe a novel, fluorescence-based procedure for direct determination of deletions and insertions with 100% accuracy. We performed two complementary directed termination PCR with near infrared dye-labeled primers, followed by visualization of termination fragments using an automated Li-cor DNA sequencer. This method is used for rapid analysis of mutational spectra generated in nuclease-defective strains of Saccharomyces cerevisiae to elucidate the role of RNase H(35) in RNA primer removal during DNA replication and in mutation avoidance. Strains deficient in RNH35 displayed a distinct spontaneous mutation spectrum of deletions characterized by a unique 4 bp deletion in a lys2-Bgl allele. This was in sharp contrast to strains deficient in rad27 that displayed duplication mutations. Further analysis of mutations in a rnh35/rad27 double mutant revealed a mixed spectrum. These results indicate that RNase H(35) may participate in a redundant pathway in Okazaki fragment processing and that mutational spectra caused by protein deficiencies may be more intermediate-specific than pathway-specific.
INTRODUCTION
Deficiencies in proteins involved in maintaining genetic stability often generate mutator phenotypes, increased genome instability and higher rates of mutations in cancer cells (1–3). Because particular protein defects are often associated with specific types of mutations, mutational spectrum analysis has led to an understanding of how particular protein functions are involved in DNA repair and mutation avoidance pathways. Deficiencies in proteins involved in different metabolic pathways give rise to different mutational spectra, while mutations in genes participating in the same pathway usually result in similar mutagenic consequences. This interpretation of the cause–effect relationship between biochemical pathway and mutagenic consequence has contributed to functional elucidation of proteins involved in DNA replication (4–10), mismatch repair (11–17) and nucleotide/base excision repair (18–21). For example, mutants deficient in nuclease Rad27, the yeast Saccharomyces cerevisiae homolog of FEN1, display a strong mutator phenotype; the mutational spectrum of their spontaneous mutations is characterized by duplications flanked by short direct repeats. This spectrum is distinct from mutations generated from cells with DNA mismatch repair defects (7). This unique mutational spectrum of duplications leads to the recognition of a novel mutation avoidance mechanism mediated by Rad27 in lagging strand DNA synthesis. Recognition of this Rad27-dependent pathway for genome maintenance prompted searches for other proteins involved in this pathway.
RNase H enzymes are ribonucleotide specific endonucleases that cleave the RNA portion of RNA–DNA/DNA or RNA/DNA duplexes (22,23). Three RNase H proteins were isolated from budding yeast. One of these, RNase H(35) (RNH35), was identified as the yeast homolog of mammalian RNase HI large subunit (24–27) that was suggested to participate in RNA primer removal during lagging-strand DNA synthesis in mammalian cells (28–30). Our recent work involving both in vitro biochemical studies and in vivo functional analyses further confirmed a role of RNase H(35) in Okazaki fragment processing (31). Although RNase H(35) deficient yeast displayed only a weak mutator phenotype, an additive effect on mutation frequency was observed in a strain with mutations in both RNH35 and RAD27 (31). This led us to propose three alternative RNA primer removal pathways involving RNase H(35) and Rad27 nucleases in yeast (31). Further genetic evidence, particularly from mutational spectra, is needed to substantiate these putative pathways. The lys2-Bgl reversion assay is informative for the analysis of frameshift mutations with only modest efforts. It is constructed by a 4 bp insertion in the lys2 gene, which is functionally equivalent to a +1 frameshift (14,32). A compensatory frameshift mutation must occur in an ∼150 bp reversion window to restore an open reading frame. Previous reports have revealed a variety of spontaneous frameshift mutations throughout this window, suggesting little functional constraint in this sequence (14,16,32). The relatively large reversion window and unbiased sampling of all possible frameshift mutations that occur within this region make this assay very useful for the study of different types of frameshift mutations caused by nuclease deficiencies.
Mutation characterization for mutational spectrum analysis remains a major technical and financial challenge, despite the convenience of mutant isolation through phenotypic selection in marker genes, and has solely relied on a two-step procedure: cloning or PCR followed by dideoxy DNA sequencing. We previously described a PCR-based method [directed termination PCR (DT–PCR)] that allows generation of nested termination fragments by integrating both selective DNA amplification and directed chain termination into a single PCR reaction (33,34). This method was successfully used for large scale screening of nucleotide sequence diversity in mitochondrial DNA by coupling it to an SSCP procedure (DT–SSCP) (35,36). However, DT–SSCP analysis does not allow characterization of the actual sequence changes because the mobility of terminating DNA fragments is conformation-dependent under non-denaturing conditions. DT–PCR, on the other hand, is a bi-directional sequencing reaction, which contains half of the sequence information of a template DNA. Two complementary DT–PCR reactions with limiting dATP and dCTP followed by size-dependent electrophoresis under denaturing conditions provide all the information needed for mutation characterization.
In this study, we describe a novel procedure based on fluorescent DT–PCR for direct characterization of frameshift mutations without DNA sequencing. This method is used for rapid analysis of reversion spectra generated by defective nucleases in the lys2-Bgl reversion assay (13,14) to elucidate the role of RNase H(35) in mutation avoidance. The rnh35 deficient mutants display characteristic 4 bp deletion mutations but have little impact on Rad27-dependent duplication mutations.
MATERIALS AND METHODS
Strain construction
The strains constructed in this study were based on those used by Qiu et al. (31). The wild-type (RKY2672) and rad27 null mutant (RKY2608) strains that contained three assay genes, CANr, HOM3 and LYS2 on the chromosome were generously provided by R. Kolodner (University of California, San Diego, CA). The single knockout mutant of rnh35 was constructed based on RKY2672 and the double knockout mutant of rnh35/rad27 was based on RKY2608. Gene knocking-out was performed using the gene-targeting cassette with KanMX4 gene flanked by the two DNA fragments homologous to the 5′ and 3′ regions of RNH35, respectively. These four strains were isogenic and used for assaying mutation rates and patterns caused by nuclease deficiencies.
Mutant isolation
Mutation rate analysis was reported previously (31). Independent lys2-Bgl revertants were isolated for mutational spectral analysis. The yeast strains of four genotypes were grown up in YPD liquid medium and were subsequently plated onto YPD agar medium. About 25 independent colonies on the YPD medium were selected from each strain. An aliquot of culture from these colonies was then plated onto SD-LYS minimal medium and only one lys2-Bgl revertant was picked from each culture. These independent revertants from each strain were grown in SD-LYS liquid minimal medium and DNA was extracted using Hoffman and Winston’s method (37). Extra lys2-Bgl revertants selected in a rapid procedure were used for method validation.
Fluorescent DT–PCR
To characterize lys2-Bgl revertants, nt 270–583 of the lys2 gene that contain a 150 bp reversion window were analyzed. The oligonucleotides 5′-CCAACGTGGTCATTTAATGAG (2628F700, forward) and 5′-GTAAATTGGTCCGCAACAATGG (2629R800, reverse) were used as PCR primers (13). These primers, synthesized by Li-cor Inc. (Lincoln, NE), were 5′-end-labeled with near infrared dyes (IRD) to fluoresce at 700 and 800 nm, respectively. For directed termination at dATP sites, DT–PCR products were amplified using 1.5 pmol each of end-labeled primers in the presence of 10 µM each of dCTP, dTTP, dGTP and 2 µM of dATP (5:1 ratio). The remaining cocktail consisted of 1× PCR buffer [50 mM KCl, 10 mM Tris–HCl, pH 8.3 and 0.001% (w/v) gelatin], 1.5 mM MgCl2, 1.5 U AmpliTaq® DNA polymerase (Perkin Elmer, Branchburg, NJ) and 10–30 ng of total yeast DNA in a 20 µl reaction. DT–PCR was performed in a PTC-100TM Programmable Thermal Controller (MJ Research Inc., Watertown, MA) under the following amplification conditions: 95°C (1 min) for the initial denaturation, followed by 35 cycles of 1 min at 94°C for denaturation, 1 min at 55°C for annealing and 1 min at 72°C for extension. The extension time for the final cycle was lengthened to 5 min. For directed termination at dCTP sites, the DT–PCR reactions were performed under the same conditions as above except that 2 µM dCTP was provided in the presence of 10 µM each of dATP, dTTP and dGTP. Fluorescent DT–PCR products could be stored for several months at –20°C without signal reduction. Yellow light illumination was used to prevent photo-degradation at the 800 nm fluorochrome.
Mutation identification using a Li-cor DNA sequencer
Fluorescent DT–PCR products were directly analyzed for mutations using a dual dye automated DNA sequencer (Li-cor Model 4200 IR2 system). Prior to electrophoresis, 8 µl from each 20 µl DT–PCR reaction was mixed with 2 µl of 5× IR2 stop/loading buffer (Li-cor) plus a drop of mineral oil. After 2 min of denaturation at 95°C, samples were chilled on ice and 2 µl of each was loaded onto a 41 cm length gel with 36 wells (6% Sequagel, DiaMed Inc., Atlanta, GA). The gel was mounted on the Model 4200 IR2 sequencing system. Electrophoresis was performed according to the manufacturer’s recommendations. Termination fragments were detected in real time and the image files generated were visually processed in Adobe Photoshop 4.0.
Dideoxy DNA sequencing
Representative lys2-Bgl reverants with unique mutations identified by fluorescent DT–PCR were re-examined by standard dideoxy DNA sequencing. A 900 bp fragment containing the 313 bp region used for mutation characterization was amplified using primers 5′-GTAACCGGTGACGATGAT (forward) and 5′-CCAATTGTCCAGCAGCTC (reverse) (14) in a 50 µl regular PCR reaction. Each PCR product was purified by electrophoresis in a 1.2% agarose gel. The segment of agarose containing the desired DNA band was removed and the DNA was recovered using the DIAEX II Gel Extraction Kit (Qiagen Inc., Valencia, CA). The purified products were sequenced using the unlabeled 2629R as an internal primer and the ABI Taq FS determinator kit. Sequencing was carried out with an ABI 377 automated DNA sequencer.
Dideoxy sequence ladders for the 313 bp fragment were generated using a Thermal Sequenase Cycle Sequencing Kit (Amersham Pharmacia Biotech, Cleveland, OH) and two IRD-labeled primers. A bi-directional dATP or dCTP ladder was performed in the presence of either 4 µl ddATP mix (2.5 mM dNTP/1.25% ddATP) or 4 µl ddCTP mix (2.5 mM dNTP/2.5% ddCTP) in a 20 µl reaction. The remaining cocktail consisted of 1× reaction buffer, 1 pmol each of IRD-labeled primers 2628F700 and 2629R800, 50–100 ng of purified DNA (the 900 bp fragment) and 1 U of Thermal Sequenase. Sequencing reactions were thermal cycled for an initial incubation of 60 s at 95°C, followed by 30 cycles at 95°C for 30 s, 56°C for 30 s and 72°C for 120 s. The extension time for the last cycle was extended for an extra 5 min and the reaction was stopped by adding 5 µl of 5× IR2 stop/loading buffer.
Calculation of expected proportion and statistic analysis
Expected proportions of two characteristic reversion mutations, the duplication mediated by short direct repeats and the 4 bp deletion, were calculated for the rnh35/rad27 double mutant under the assumptions that the mutator effects of the rnh35 and rad27 single mutants were additive and the contribution from wild-type was negligible. The expected contribution of two single knockouts to the mutation rate in the double mutant was determined by the mutation rate of each single mutant (MRrnh35 or MRrad27) divided by the sum of two rates (MRrnh35 + MRrad27). Therefore, the expected proportions for both duplication (Iexp) and 4 bp deletion (Dexp) were determined as follows: Iexp = Iobs × MRrad27 / (MRrnh35 + MRrad27), Dexp = Dobs × MRrnh35 / (MRrnh35 + MRrad27). Iobs and Dobs are duplication and deletion proportions observed in the rad27 and rnh35 single mutants, respectively. Chi-square (χ2) was used to test the null hypothesis that the expected and observed proportions in the double mutant were similar.
RESULTS
Visualization of DT–PCR products using IRD-labeled primers
Detection of DT–PCR products using two IRD-labeled primers and a Li-cor DNA sequencing system provided a sensitive and non-radioactive approach for independent visualization of two sets of termination fragments generated in the same DT–PCR reaction (Fig. 1). A digitized gel image obtained after a 4 h run on the Li-cor was as sensitive as that obtained by 1–2 day exposure of a similar experiment using radioisotope labeling (i.e. [α-35S]dATP/dCTP incorporation or [γ-33P]ATP-end-labeling) (33). Moreover, the near infrared fluorescent signal was more than 10 times stronger than the visual fluorescence of the ABI sequencing system (J.Z.Chen, personal observation). This improved sensitivity with near infrared detection and a constant separation between adjacent bands provided by the Li-cor system allowed accurate determination of any band gain and loss in the entire array of termination fragments.
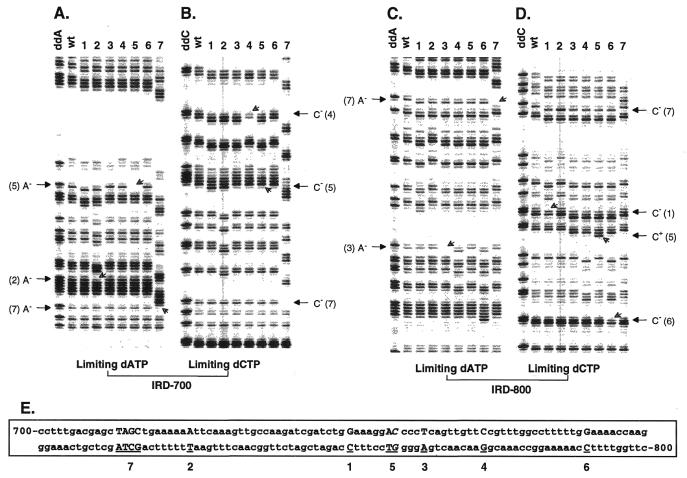
Figure 1.
Fluoroimages showing identification of frameshift mutations in a 313 bp fragment of the lys2-Bgl allele in S.cerevisiae. Two fluorescent DT–PCR reactions with limiting dATP and dCTP were carried out for eight samples including a wild-type molecule (wt) and electrophoresed side by side on a denaturing 6% polyacrylamide gel mounted on an automated Li-cor DNA sequencer. The partial fluoroimages containing all sequence changes were processed in Adobe Photoshop 4.0. The left panel including groups A and B shows terminating fragments visualized by primer 2628F end-labeled with IRD-700 and the right panel including groups C and D visualized by primer 2629R end-labeled with IRD-800. The first lane in each group is a dideoxy sequence ladder generated as described in Materials and Methods. Open arrows on the gel indicate positions of band gains and losses. Letters on each side of the fluoroimages provide nucleotide changes. The wild-type nucleotide sequence with its PCR labels is showing below the fluoroimages (E). Frameshift mutations in each sample are indicated by underlined upper case nucleotides in the sequence. Samples 1, 2, 3, 4 and 6 had a single base pair deletion as indicated by nucleotides above each number. Sample 7 had a 4 bp deletion of TAGC, while sample 5 had both an A:T base pair deletion and a C:G to G:C base pair substitution (as indicated by italic letters at an adjacent site). See text for interpretations on how frameshift mutations were actually determined in three representative samples 2, 5 and 7.
Although the banding pattern of termination fragments generated by fluorescent DT–PCR corresponded well to that of a dideoxy sequence ladder, these two banding patterns were not identical. Instead of a single band, a doublet was observed in the DT–PCR reaction at each limiting nucleotide site and a triplet was observed at two consecutive limiting sites (Fig. 1). Each doublet was composed of a relatively dark band attributed to pausing one nucleotide upstream of the limiting nucleotide and a relatively weak band attributed to a mis-incorporated 3′ nucleotide. This observation was consistent with previous reports (33,34). Even though the relative intensities of terminating bands varied among sites and within each doublet, the banding patterns of termination fragments were reproducible and the doublet phenotype assisted the recognition of band gains and losses (see below).
Characterization of frameshift mutations
The procedure of dual dye labeling coupled with automated fluorescent detection allowed inspection of any sequence change in a DNA fragment using two complementary DT–PCR reactions with limiting dATP and dCTP. A deletion or insertion in the template DNA not only resulted in band loss or gain, but also led to downward or upward shifts of bands downstream of the mutation. These two features always occurred together and form the visual basis for the accurate determination of the nature and position of frameshift mutations. The partial fluoroimages in Figure 1 demonstrate characterization of frameshift mutations in a 313 bp fragment of the lys2-Bgl allele in yeast. Three examples of the data are explained as follows. (i) Lane 2 in section A had a band missing in a string of adenines as indicated by an open arrow in the limiting dATP reaction. This was accompanied by a 1 bp downward band shift, showing that an A:T base pair was deleted at the location in sample 2. (ii) Sample 7 had a deletion involving multiple nucleotides. Lane 7 in section A had a band missing in the limiting dATP reaction and this was accompanied by a downward shift of multiple bands. This indicates that several nucleotides were deleted at the location of the shift. Based on the size of shift, the cause of the change, a 4 bp deletion could easily be determined. A band was also missing in section B of the limiting dCTP reaction, indicating a deletion of dCTP. In the right panel, lane 7 had an adenine band missing in section C and a loss of cytosine band in section D as indicated by arrows. Because all the missing bands corresponded to the same deletion in the sequence, we know exactly that nucleotides TAGC were deleted in sample 7 at the position shown in the sequence below. (iii) Lane 5 shows two mutational events. Although it involved the loss of both an adenine band in section A and a cytosine band in section B, only a 1 bp downward banding shift was observed in each reaction. In addition, since the missing bands took place at two adjacent sites and since the gain of a cytosine band was observed in section D, these observations indicate that two mutational events occurred in this sample and that they involved both a single base pair deletion and substitution. Because an A:T base pair was observed at the same position in other samples, sample 5 involved a deletion of an A:T base pair and a substitution from C:G to G:C at an adjacent site. Spontaneous reversions in other samples were characterized in the same fashion (Fig. 1 legend).
Detection efficiency of frameshift mutations
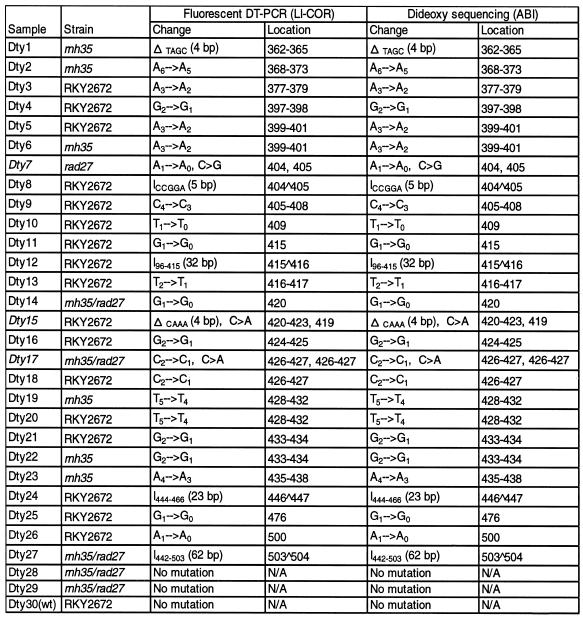
After analyzing 120 revertants in the lys2-Bgl allele, 23 unique frameshift mutations were identified. To test the detection efficiency of fluorescent DT–PCR for accurately characterizing frameshift mutations, 30 samples were selected for dideoxy DNA sequencing using the ABI automated sequencer. These samples included all 23 unique mutations, four samples with identical mutations, two samples with no detectable change and a wild-type molecule. A comparison of results from the two methods indicated that fluorescent DT–PCR correctly identified all simple frameshift mutations including several relatively large changes (Table 1). Furthermore, secondary base substitution events were also identified in three samples (Dty-7, -15 and -17) apart from their primary frameshift mutations, in which Dty-7 was the same sample shown in lane 5 of Figure 1. A novel secondary substitution was detected in subsequent analysis. The comparative study demonstrated that fluorescent DT–PCR analysis not only had 100% efficiency in determining simple frameshift mutations, but also allowed identification of secondary substitution events.
Table 1. Efficiency of fluorescent DT–PCR on the characterization of frameshift mutations in the lys2-Bgl allele of S.cerevisiae.
Symbols Δ and I stand for deletions and insertions involving more than one nucleotide, respectively. Samples in italics include both deletion and secondary substitution events.
Reversion spectra in lys2-Bgl allele
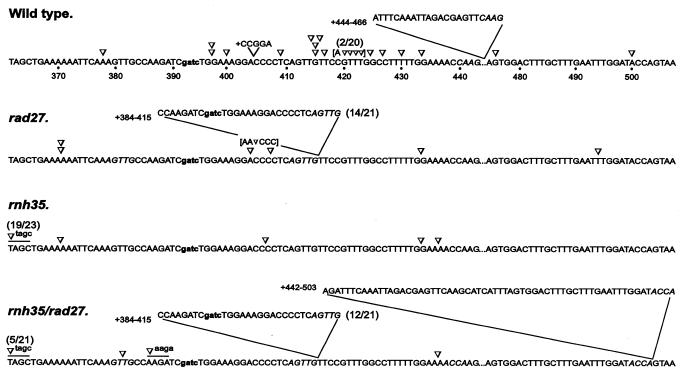
For rapid spectrum analysis of frameshift mutations in the lys2-Bgl allele, the first fluorescent DT–PCR with limiting dATP provided rapid scanning of different types of frameshift mutations in each group of revertant samples. The second DT–PCR reaction was necessary only to complete the characterization of all possible sequence changes. Figure 2 summarizes the mutation spectra of both the wild-type and three defective strains in a 150 bp reversion window in the lys2-Bgl allele. Fifteen unique frameshift events were identified in 20 revertants of the wild-type strain; they occurred throughout the entire reversion window and no hotspot mutations were found. In contrast, frameshift mutations in the rad27 mutant were characterized by a 32 bp duplication mutation flanked by a short direct repeat of AGTTG; this is consistent with a previous report of this particular duplication (7). The reversion spectrum of the rnh35 mutant was very different from that of the rad27 mutant and it was characterized by a unique 4 bp deletion. The rnh35/rad27 double mutant, on the other hand, showed a spectrum containing both the 32 bp duplication mutation and the 4 bp deletion that were characteristic of their rad27 and rnh35 single mutants. A unique 62 bp duplication mutation flanked by a short direct repeat of ACCA and a novel 4 bp deletion were also identified in the double mutant, but not in the parental single mutants.
Figure 2.
Reversion spectra of the lys2-Bgl allele in a wild-type and three nuclease-deficient strains. The reversion window comprising nt 362–443 and 475–507 is shown and the 4 bp insertion that created the lys2-Bgl allele is indicated in lower case. Each reversion is indicated at the position of changes with symbols ∇ and V for single base pair deletions and insertions. Four base pair deletions are underlined and the short direct repeats flanking inserted sequences are italicized. The frequency of mutations is indicated in parentheses when more than one event is involved and reversions involving both deletions and base substitutions are shown in brackets. The number of revertants analyzed was: 20 for wild-type, 21 for rad27, 23 for rnh35 and 21 for rnh35/rad27.
Epistatic analysis
To understand epistatic relationships between RAD27 and RNH35, both reversion frequency and spectrum of the lys2-Bgl allele were analyzed in nuclease single- and double-knockout mutants. In an earlier study, we observed a weak mutator phenotype in the rnh35 mutant, which led to a 5-fold increase in spontaneous mutation rate compared to a 48-fold increase in the rad27 mutant (Table 2). The mutation rate in the rnh35/rad27 double mutant was higher than expected from additive but lower than expected from multiplication of the frequencies of two single mutants, suggesting that RNase H(35) may participate in a redundant or different pathway in Rad27-mediated RNA primer removal. To further analyze possible interactions between these two gene products, the expected proportions of the two characteristic reversion mutations, the duplication and the 4 bp deletion mutations were calculated for the rnh35/rad27 double mutant under the assumption that the mutation rates of two single mutants are additive (Table 2). In the double mutant, the expected proportion of duplication mutations was virtually identical to the observed proportion, suggesting that rnh35 plays little role in the generation of duplication mutations. On the other hand, the observed proportion of 4 bp deletions was more than three times higher than the expected proportion, although this difference was not significant due to relatively small sampling sizes. This result suggests that the mixed spectrum observed in the double mutant was similar to the predicted sum of mutation spectra of the corresponding single mutants.
Table 2. Epistasis analysis of characteristic frameshift mutations in nuclease-defective strains.
| Strain | Mutation ratea | Proportion of characteristic mutations | |
|---|---|---|---|
| Duplication | 4 bp deletion | ||
| Wild-type |
1.3 × 10–8 |
1/20 |
0/20 |
|
rad27 |
6.1 × 10–7 |
14/21 (66.7%) |
0/21 |
|
rnh35 |
6.6 × 10–8 |
0/23 |
19/23 (82.6%) |
|
rnh35/rad27 |
1.5 × 10–6 |
13/21 (61.9%) |
6/21 (26.8%) |
| Expected proportionb | 12.6/21 (60.2%)c | 1.7/21 (8.1%)d | |
aMutation rates measured in our previous study (31).
bExpected proportions for both duplication and 4 bp deletion were calculated for the rnh35/rad27 double mutant.
cχ2 = 0.009.
dχ2 = 3.0 (0.05 < P < 0.1).
DISCUSSION
Increased frequencies of spontaneous frameshift mutations are often caused by altered DNA metabolism. The deleterious effects of frameshifts have been implicated in etiological studies of inheritable diseases and cancers (7,38–42). Since specific types of mutations are often associated with particular protein defects involved in DNA replication and repair, mutational spectrum analysis has become an informative genetic tool to understand those protein functions involved in mutation avoidance. However, reliable data require sequencing a large number of mutants to obtain saturated mutational spectra for the mutation class of interest (43). The practical use of spectral analysis is often hampered by the effort and cost involved in the multi-step sequencing analysis. In this study, we describe a fluorescence-based DT–PCR procedure for direct identification of deletions and insertions with 100% efficiency and demonstrate its usefulness in rapid spectral analysis of frameshift mutations to understand protein functions in yeast.
Characterization of frameshift mutations using fluorescent DT–PCR
DT–PCR is a one-step and bi-directional sequencing reaction. Its unique biphasic kinetics in DNA amplification leads to the generation of two sets of nested termination fragments that serve to identify all the sites requiring the incorporation of the limiting nucleotide in a template DNA. A limiting dATP reaction provides information concerning changes at A and T positions while a second reaction with limiting dCTP provides information concerning alterations at G and C sites. These two complementary DT–PCR reactions should provide all the information needed for complete mutation characterization. The procedure of dual dye labeling coupled with automated fluorescent detection described in this study provided an excellent system to retrieve the sequence information. The IRD provide satisfactory signal levels for DNA sequence analysis because of their high molar absorbtivity and satisfactory quantum efficiency (44). Meanwhile, the improved sensitivity for near infrared detection and a constant distance in band separation provided by the Li-cor automated DNA sequencer allow accurate determination of any band gain, band loss or band shift.
Characterization of frameshift mutations using fluorescent DT–PCR relies on two kinds of band pattern alteration. Band gains or losses determine the nature and exact positions of a frameshift, while band shifts identify the size of the insertions or deletions. Since band shifts are manifested by all insertions and deletions, the first fluorescent DT–PCR reaction can serve as a genotyping step for rapid scanning of different types of frameshift mutations in large scale analysis. The second and complementary DT–PCR reaction subsequently serves to unambiguously characterize each insertion or deletion. Furthermore, the two complementary DT–PCR reactions together allow identification of possible secondary substitution events that might have escaped detection after analyzing results from the first reaction alone. For example, a base substitution simultaneously results in both band gain and loss in two complementary strands without a concomitant band shift. We have demonstrated in a separate study that fluorescent DT–PCR characterized all single base substitutions in the supF gene of a shuttle vector plasmid (J.Z.Chen, L.Smith, P.G.Pfeifer and P.G.Holmquist, submitted).
Successful identification of a possible mutation is also influenced by the specificity of terminating profiles at all limiting nucleotide sites including those with mononucleotide runs. Despite the existence of several possible combinations, complementary reactions with limiting dATP and dCTP have provided the best terminating banding patterns for mutation characterization when coupled with a Taq DNA polymerase. It is important to note that the actual generation of termination fragments is complicated by the doublet nature of chain termination at each limiting nucleotide site. As a result, the terminating banding patterns are both sequence specific and reproducible, but not identical to corresponding dideoxy sequence ladders. However, this difference does not compromise DT–PCR’s ability to characterize each mutation. In fact, the doublets can assist the recognition of a nucleotide change by magnifying the signal with simultaneous gain or loss of two related terminating fragments. Other factors, such as sequence context effects, DNA polymerases and the kinetic complexity of template DNA may also impact mutation characterization and are currently under investigation.
Reversion spectra of frameshift mutations in rnh35 deficient strains
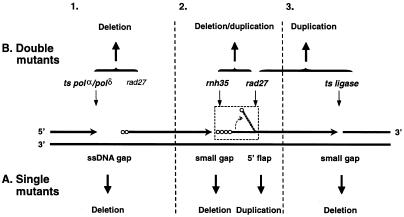
Fluorescent DT–PCR was successfully applied to the analysis of reversion spectra in a lys2-Bgl frameshift assay to elucidate the role of RNase H(35) in the avoidance of duplication mutations mediated by Rad27 in yeast. An rnh35 null mutant displayed a distinct spectrum of deletions dominated by a unique 4 bp deletion in the lys2-Bgl allele. This was in sharp contrast with that of duplication mutations detected in a rad27 mutant. Further analysis of mutations in an rnh35/rad27 double mutant revealed a mixed spectrum similar to the predicted sum of mutation spectra of the corresponding single mutants. It is possible that RNase H(35) is not involved in Okazaki fragment processing in yeast. Although this speculation is consistent with the conventional wisdom that deficiencies in proteins closely involved in the same metabolic pathway result in similar mutagenic consequences, it fails to account for the in vitro biochemical data (31). A more plausible explanation for this result is that RNase H(35) plays a minor role or participates in a redundant pathway in lagging strand maturation. It has been suggested that the 5′ flap structure generated during Okazaki fragment processing is a prerequisite for Rad27-mediated duplication mutations (8,9,45). Failure to remove ribonucleotides from Okazaki fragments would attenuate the process of the 5′ flap formation, leading to the generation of small single-stranded DNA (ssDNA) gaps. These gaps would subsequently be processed into characteristic deletion mutations through unidentified repair pathways (Fig. 3, section 2). As evidence supporting this model, a deletion-dominated spectrum was observed in a ligase deficient strain of Schizosaccharomyces pombe that supposedly resulted in accumulation of small gap intermediates (9). Although further substantiation of this model is required, this explanation seems to be consistent with all existing data.
Figure 3.
Specificity of mutational spectra caused by deficiencies in enzymes involved in lagging strand DNA synthesis in yeast. Four consecutive Okazaki fragments are shown to illustrate three joining sections between nascent lagging strands. Horizontal arrows indicate the 3′ end of an elongating Okazaki fragment and open circles show non-excised ribonucleotides at the 5′ end of an Okazaki fragment. Light vertical arrows indicate normal substrates or structures metabolized by DNA replication enzymes. Bold vertical arrows indicate the mutagenic consequences caused by accumulated intermediates (ssDNA gap versus flap structure) in either single mutant strains that correspond to enzymes as indicated by each light vertical arrow or double mutant strains as indicated by each connecting symbol. Two vertical broken lines divide the figure into three sections. Section 2 shows a proposed mechanism for the unique deletion spectra involving RNase H(35) deficiency (see text for explanation). Sections 1 and 3 depict previously suggested mechanisms (8,9). The specificity of mutational spectra in defective strains in genes involved in Okazaki fragment processing is illustrated below the template strands (A) for single gene knockouts and above the template strands (B) for double gene knockouts. (A) For single gene mutations, the mutational spectrum is specific to the metabolic intermediates accumulated in a defective cell. (B) For double gene mutations, the spectrum is determined critically by defects in upstream proteins involved in the same metabolic process.
To put this result in perspective with other published work, especially results from Kokoska et al. (8) and Liu et al. (9), a framework is emerging for the specificity of mutational spectra in Okazaki fragment processing (Fig. 3). For single knockout of genes involved in DNA replication, the specificity of their associated spontaneous mutational spectra was determined by the intermediate substrates that accumulated in a cell (i.e. ssDNA gaps versus flap structure) rather than the biochemical pathways involved. In other words, deficiencies in proteins participating in different steps of the same process could result in different mutagenic consequences (Fig. 3A). In terms of double gene knockouts, however, the mutational spectrum was determined critically by defects in upstream proteins in the same metabolic process (Fig. 3B). For example, the formation of duplications in S.pombe rad2 (homologue of RAD27) was reduced in a temperature sensitive replication mutator background, but was not impacted in a mutant ligase background (9). A similar result was observed in microsatellite instabilities stimulated by a rad27 mutant in a pol3-t mutant background in S.cerevisiae (8). The additional 4 bp deletions in the rnh35/rad27 double mutant that we observed in this study are consistent with this trend. This framework will be instrumental to guide further spectrum analysis.
In summary, fluorescent DT–PCR provides a simple alternative to dideoxy DNA sequencing for highly effective characterization of frameshift mutations. Because of its generality, this method should be applicable to other marker genes in large scale analysis of frameshift mutations. The rnh35 null mutant displays unique 4 bp deletion mutations but has little impact on Rad27-dependent duplication mutations. This may be because mutational spectra caused by defective proteins are specific to metabolic intermediates rather than metabolic pathways.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr J. Juang and F. Wu for their assistance in yeast genomic DNA extraction. We are also grateful to Dr G. P. Pfeifer and the reviewers for their useful comments on earlier versions of the manuscript. This work was supported by NIH grants CA69449 to G.P.H. and CA73764/CA85344 to B.H.S.
REFERENCES
- 1.Loeb L.A. (1991) Cancer Res., 51, 3075–3079. [PubMed] [Google Scholar]
- 2.Loeb L.A. (1994) Cancer Res., 54, 5059–5063. [PubMed] [Google Scholar]
- 3.Loeb L.A. (1998) Adv. Cancer Res., 72, 25–56. [DOI] [PubMed] [Google Scholar]
- 4.Fowler R.G., Schaaper,R.M. and Glickman,B.W. (1986) J. Bacteriol., 167, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison A., Johnson,A.L., Johnson,L.H. and Sugino,A. (1993) EMBO J., 12, 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison A. and Sugino,A. (1994) Mol. Gen. Genet., 242, 289–296. [DOI] [PubMed] [Google Scholar]
- 7.Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- 8.Kokoska R.J., Stefanovic,L., Tran,H.T., Resnick,M.A., Gordenin,D.A. and Petes,T.D. (1998) Mol. Cell. Biol., 18, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu V.F., Bhaumik,D. and Wang,T.S.-F. (1999) Mol. Cell. Biol., 19, 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Merrill,B.J., Lau,P.J., Holm,C. and Kolodner,R.D. (1999) Mol. Cell. Biol., 19, 7801–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaaper R.M. and Dunn,R.L. (1987) Proc. Natl Acad. Sci. USA, 84, 6220–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R.E., Kovvali,G.K., Prakash,L. and Prakash,S. (1995) Science, 269, 238–240. [DOI] [PubMed] [Google Scholar]
- 13.Marsischky G.T., Filosi,N., Kane,M.F. and Kolodner,R.D. (1996) Genes Dev., 10, 407–420. [DOI] [PubMed] [Google Scholar]
- 14.Greene C.N. and Jinks-Robertson,S. (1997) Mol. Cell. Biol., 17, 2844–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran H.T., Keen,D., Kricker,M., Resnick,M.A. and Gordenin,D.A. (1997) Mol. Cell. Biol., 17, 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores-Rozas H. and Kolodner,R.D. (1998) Proc. Natl Acad. Sci. USA, 95, 12404–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harfe B.D. and Jinks-Robertson,S. (1999) Mol. Cell. Biol., 19, 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunz B.A., Kohalmi,L., Kand,X.L. and Magnusson,K.A. (1990) J. Bacteriol., 172, 3009–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montelone B.A., Gilbertson,L.A., Nassar,R., Giroux,C. and Malone,R.E. (1992) Mutat. Res., 267, 55–66. [DOI] [PubMed] [Google Scholar]
- 20.Roche H., Gietz,R.D. and Kunz,B.A. (1994) Genetics, 137, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clairmont C.A., Narayanan,L., Sun,K.-W., Glazer,P.M. and Sweasy,J.B. (1999) Proc. Natl Acad. Sci. USA, 96, 9580–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wintersberger U. (1990) Pharmacol. Ther., 48, 259–280. [DOI] [PubMed] [Google Scholar]
- 23.Crouch R.J. and Toulme,J.J. (1998) Ribonucleases H. Editions INSERM, Paris, France.
- 24.Karwan R., Blutsch,H. and Wintersberger,U. (1983) Biochemistry, 22, 5500–5502. [Google Scholar]
- 25.Itaya M., Mckelvin,D., Chatterjie,S.K. and Crouch,R.J. (1991) Mol. Gen. Genet., 227, 438–445. [DOI] [PubMed] [Google Scholar]
- 26.Frank P., Braunshofer-Reiter,C. and Wintersberger,U. (1998) FEBS Lett., 421, 23–26. [DOI] [PubMed] [Google Scholar]
- 27.Frank P., Braunshofer-Reiter,C., Wintersberger,U., Grimm,R. and Busen,W. (1998) Proc. Natl Acad. Sci. USA, 95, 12872–12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turchi J.J., Huang,L., Kim,Y. and Bambara,R.A. (1994) Proc. Natl Acad. Sci. USA, 91, 9803–9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bambara R.A., Murante,R.S. and Henricksen,L.A. (1998) J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- 30.Murante R.S., Henricken,L.A. and Bambara,R.A. (1998) Proc. Natl Acad. Sci. USA, 95, 2244–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu J., Qian,Y., Frank,P., Wintersberger,U. and Shen,B. (1999) Mol. Cell. Biol., 19, 8361–8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele D.F. and Jinks-Robertson,S. (1992) Genetics, 132, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J. and Hebert,P.D.N. (1998) Nucleic Acids Res., 26, 1546–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J.Z. and Hebert,P.D.N. (1999) Genome, 42, 72–79. [PubMed] [Google Scholar]
- 35.Chen J.Z. and Hebert,P.D.N. (1999) Mutat. Res., 434, 205–217. [DOI] [PubMed] [Google Scholar]
- 36.Chen J.Z. and Hebert,P.D.N. (1999) Mutat. Res., 434, 219–231. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman C.S. and Winston,F. (1987) Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi Y., Ando,H., Nagase,H., Nishisho,I., Horii,A., Miki,Y., Utsunomiya,J., Baba,S., Petersen,G. et al. (1992) Proc. Natl Acad. Sci. USA, 89, 4452–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyoshi Y., Nagase,H., Ando,H., Horii,A., Ichii,A., Nakatsuru,S., Aoki,T., Miki,Y., Mori,T. and Nakamura,Y. (1992) Hum. Mol. Genet., 1, 229–233. [DOI] [PubMed] [Google Scholar]
- 40.Miyaki M., Konishi,M., Kikuchi-Yanoshita,R., Enomoto,M., Igari,T., Tanaka,K., Muraoka,M., Takahashi,H., Amada,Y., Fukayama,M. et al. (1994) Cancer Res., 54, 3011–3020. [PubMed] [Google Scholar]
- 41.Jego N., Thomas,G. and Hamelin,R. (1993) Oncogene, 8, 209–213. [PubMed] [Google Scholar]
- 42.Greenblatt M.S., Grollman,A.P. and Harris,C.C. (1996) Cancer Res., 56, 2130–2136. [PubMed] [Google Scholar]
- 43.Ripley L.S. (1990) Annu. Rev. Genet., 24, 189–213. [DOI] [PubMed] [Google Scholar]
- 44.Middendorf L., Amen,J., Bruce,R., Draney,D., DeGraff,D., Geweeke,J., Grone,D., Humphrey,P., Little,G., Lugade,A. et al. (1998) In Daehne,D., Resch-Genger,U. and Wolfbeis,O.S. (eds), Near-Infrared Dyes for High Technology Applications. Kluwer Academic Publishers, Holland, pp. 21–54.
- 45.Bornarth C.J., Ranalli,T.A., Henricken,L.A., Wahl,A.F. and Bambara,R.A. (1999) Biochemistry, 38, 13347–13354. [DOI] [PubMed] [Google Scholar]